Abstract
Clonal complex 59 (CC59) community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) strains were characterized using pulsed-field gel electrophoresis, spa typing, multilocus sequence typing, diagnostic DNA microarrays, and PCRs targeting staphylococcal cassette chromosome mec (SCCmec) elements and Panton-Valentine leukocidin (PVL). Six distinct groups within CC59 were characterized. At least seven different variants of SCCmec elements were identified (IVa [2B], IVb [2B], IVd [2B], IV variant [2B], IVa [2B&5], V variant [5C2], and V [5C2&5]). (The structural type is indicated by a Roman numeral, with a lowercase letter indicating the subtype, and the ccr complex and the mec complex are indicated by an Arabic numeral and an uppercase letter, respectively. Where there is an extra ccr element, this is indicated by “&” and an Arabic numeral designating the ccr type.) The first group is similar to the American sequence type 59 (ST59) MRSA-IV CA-MRSA strain USA1000. The second group includes a PVL-negative ST87 strain with an SCCmec element of subtype IVb (2B). The third group comprises PVL-variable ST59 MRSA-IV strains harboring multiple SCCmec IV subtypes. PVL-negative ST59 MRSA strains with multiple or composite SCCmec elements (IVa [2B&5]) form the fourth group. Group 5 corresponds to the internationally known “Taiwan clone,” a PVL-positive strain with a variant SCCmec element (V [5C2&5]). This strain proved to be the most common CC59 MRSA strain isolated in Western Australia. Finally, group 6 encompasses the ST59 MRSA-V variant (5C2). The differentiation of CC59 into groups and strains indicates a rapid evolution and spread of SCCmec elements. Observed differences between groups of strains as well as intrastrain variability within a group facilitate the tracing of their spread.
Several well-characterized community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) strains predominate in different regions of the world. Many of these strains harbor the bicomponent Panton-Valentine leukocidin (PVL). Sequence type 8 (ST8) MRSA-IV (USA300), ST80 MRSA-IV, and ST93 MRSA-IV are the major CA-MRSA strains reported to occur in the United States, Europe, and Australia, respectively. In the Asia Pacific region, a distinct genotype, clonal complex 59 (CC59)/ST59, has become widespread. ST59 CA-MRSA strains are an important cause of morbidity in Taiwan (1, 13, 17, 25, 29) This so-called “Taiwan clone” has acquired a novel type V staphylococcal cassette chromosome mec (SCCmec) element (V [5C2&5], also known as VT) and ermB, a macrolide-lincosamide-streptogramin B resistance gene (28) frequently reported to be present in streptococci and other bacteria. Its properties have recently been described in detail (24). ST59 MRSA-V and other CC59 strains have now been reported to occur in several countries, including the United States (USA1000) (6), Sweden (11), Germany (20), the United Kingdom (19), Vietnam (26), and Australia (22).
In Western Australia (WA), all MRSA strains are referred to the state's central typing reference laboratory (the Gram-Positive Bacteria Typing and Research Unit) for molecular characterization (5). Multiple CC59 strains, colloquially characterized as WA MRSA-9, -15, -24, -52, -55, -56, and -73, have been identified. They differ from each other in ST designation, pulsed-field gel electrophoresis (PFGE) pattern, SCCmec element, and PVL carriage.
To better understand the molecular epidemiology of this clonal complex, all CC59 MRSA strains isolated in Western Australia were examined using PFGE, spa typing, multilocus sequence typing (MLST), diagnostic DNA microarrays, and PCRs targeting SCCmec elements and PVL.
MATERIALS AND METHODS
Isolates and patients.
From July 2003 to June 2008, 43 MRSA strains from 40 individuals living in WA were characterized as CC59 MRSA by the Gram-Positive Bacteria Typing and Research Unit. One person yielded three isolates (WA MRSA-15 04-16657, 05-17619, and 06-17484) over a 3-year period, and a second yielded two isolates (WA MRSA-9 06-17363 and 07-17830) over a 2-year period. Three isolates (WA MRSA-52 07-15076, 07-16295, and 07-16320) were obtained from three family members over a 12-month period. Isolates were recovered from 33 skin and soft tissue infections and seven nasal screening swabs.
Control strains.
Two reference strains of USA1000 ST59 MRSA-IV (NARSA483 and NARSA676) were obtained from the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) and have been included in this study for comparison.
Susceptibility testing.
An antibiogram was performed by disk diffusion on Mueller-Hinton agar according to the Clinical and Laboratory Standards Institute (CLSI) recommendations (3). A panel of eight antimicrobial drugs, i.e., erythromycin (15 μg), tetracycline (30 μg), trimethoprim (5 μg), ciprofloxacin (5 μg), gentamicin (10 μg), rifampin (5 μg), fusidic acid (10 μg), and mupirocin (5 μg), was tested. CLSI interpretive criteria (4) were used for all drugs except fusidic acid (2) and mupirocin (9).
PFGE.
Electrophoresis of chromosomal DNA was performed as previously described (23), using a contour-clamped homogeneous electric field (CHEF) DR III system (Bio-Rad Laboratories Pty Ltd). Chromosomal patterns were examined visually, scanned with a Quantity One device (Bio-Rad Laboratories Pty Ltd), and digitally analyzed using FPQuest (Bio-Rad Laboratories). CHEF patterns were grouped according to the criteria of Tenover et al. (27), and a dendrogram similarity of 80% or greater was used to assign strain relatedness. S. aureus strain NCTC 8325 was used as a reference strain.
MLST and spa typing.
Chromosomal DNA for MLST and spa typing was prepared using a DNeasy tissue kit (Qiagen Pty Ltd).
MLST was performed as specified by Enright et al. (7). The method involves bidirectional sequencing of 450- to 500-bp internal fragments of seven housekeeping genes obtained by PCR using highly conserved primer pairs. Each allele sequence is assigned a number by the curator of the MLST database (http://saureus.mlst.net), and the allelic profile determines the sequence type (ST). Allelic profiles can be compared using the BURST (based upon related sequence types) program (http://linux.mlst.net/burst.htm). Clusters of single-locus variants (SLVs) and double-locus variants are referred to as clonal clusters.
To assign an ST, sequences were compared with the sequences on the MLST website. By use of the MLST database, clones were subsequently grouped into CC59.
spa typing, a DNA sequence-based analysis of the protein A gene variable region, was performed as previously described (10) using the nomenclature as described on the Ridom website (http://spa.ridom.de/).
PVL.
PCR for the detection of PVL determinants was performed as previously described (8).
SCCmec typing.
SCCmec is a mobile genetic element that carries the mecA gene, which encodes broad-spectrum beta-lactam resistance, and unique site-specific recombinases designated cassette chromosome recombinases (ccr). SCCmec elements are classified into types and subtypes by a hierarchical system (15). Types are defined by the combination of the ccr gene complex (types 1 to 5, represented by the ccr gene allotypes [ccrA1, ccrA2, ccrA3, and ccrA4; ccrB1, ccrB2, ccrB3, and ccrB4; and ccrC1]) and the mec gene complex (classes A, B, C1, and C2). The SCCmec element also contains three nonessential components known as the “J regions.” Variations in the J regions within the same mec-ccr complex are used for defining SCCmec subtypes.
SCCmec was typed by PCR using the following strategy. The structural architecture and the mec complex were determined using primers described by Zhang et al. (30). SCCmec type IV was further subtyped using published primers (18). Cassette chromosome recombinase (ccr) typing and ccrC1 allele 2 and 8 allotyping were performed as published previously (12, 16). Nontypeable strains and the type V SCCmec were characterized using previously published primers (15). An ISSau4-like transposase (GenBank accession no. DQ680163) insertion in open reading frame (ORF) V011 (GenBank accession no. AB12129) of the type V SCCmec was detected by the production of a ca. 1,600-bp PCR product rather than the characteristic 325-bp product in the multiplex reaction performed by Zhang et al. (30) and confirmed by sequencing. SCCmec nomenclature is used as proposed by the International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC) (15). Briefly, the structural type is indicated by a Roman numeral, with a lowercase letter indicating the subtype, and the ccr complex and the mec complex are indicated by an Arabic numeral and an uppercase letter, respectively. Where there is an extra ccr element, this is indicated by “&” and an Arabic numeral designating the ccr type.
DNA microarray.
The DNA microarray used for this study covered 334 target sequences corresponding to 185 distinct genes and their allelic variants. A complete list of targets has been provided previously (20, 21). Target genes included species markers, virulence factors, resistance genes, staphylococcal superantigen-like or exotoxin-like genes (set or ssl genes), and genes encoding adhesion proteins, as well as markers for accessory gene regulator (agr) alleles and capsule types. With regard to SCCmec typing, the array included probes for mecA, ugpQ (GenBank accession no. BA000018.3, locus tag SA0036), mecI (BA000018.3, SA0040), xylR (BA000018.3, SA0041), the dcs region (BA000018.3, SA0024 [EMBL accession no. Q9XB68]), and two probes for mecR (BA000018.3, SA0039). The last two probes allowed detection and discrimination of untruncated mecR and ΔmecR, respectively. Recombinase genes ccrA1, ccrB1, ccrA2, ccrB2, ccrA3, ccrB3, ccrA4, ccrB4, and ccrC1 were also covered. A gene for a “hypothetical protein” accompanying ccrC1 was also included, and alleles from strain 85-2082 (GenBank accession no. AB037671.1, nucleotides 61250 to 62893) and strain MRSAZH47 (GenBank accession no. AM292304.1, nucleotides 5654 to 7273) were distinguished. The mercury resistance and kdp operons were also included, but they are not relevant for the CC59 strains discussed in this study.
Arrays and reagents were obtained from CLONDIAG. The principle of the assay and related procedures have previously been described in detail (20, 21). Briefly, DNA was obtained by enzymatic lysis of overnight cultures. All targets were simultaneously amplified by linear PCR, and the products were labeled by the incorporation of biotin-16-dUTP during the reaction. The labeled sample was then hybridized to the array, followed by washing steps and the addition of a blocking reagent. Horseradish peroxidase-streptavidin conjugate was added to the array, followed again by incubation and washing. Finally, Seramun Green precipitating dye (Seramun, Heidesee, Germany) was added. An image of the array was recorded and analyzed using a dedicated reader and software.
SplitsTree analysis.
To analyze similarities between DNA microarray profiles, SplitsTree software (14) was used. DNA microarray results for relevant genes (see the legend for Fig. 2) were converted into strings of information which were handled as “sequences,” using “A” for “positive” and “T” for “negative.” These “sequences” were used for tree construction using SplitsTree 4.10 (character transformation, uncorrected P; distance transformation, Neighbor-Net; and variance, ordinary least squares).
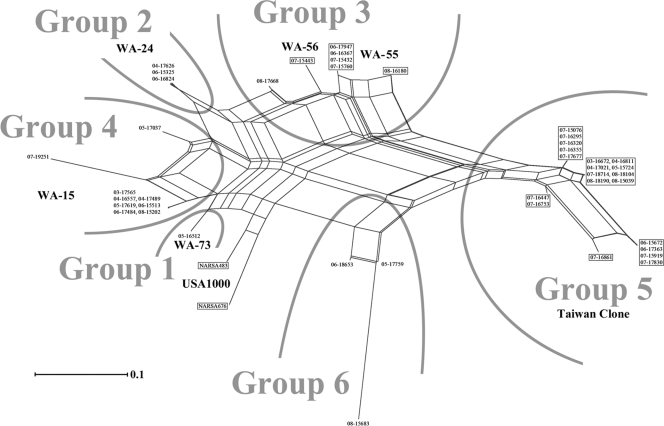
FIG. 2.
SplitsTree graph visualizing similarities of CC59 isolates and strains based on hybridization results for mecA, ΔmecR, ugpQ, the dcs region, ccrA2, ccrB2, 85-2082 ccrC1, hypothetical ORF accompanying ccrC1, blaZ, blaI, blaR, ermB, ermC, msrA, mpbBM, aphA3, sat, tetK, pC221 cat, pC223 cat, sea, seb, sek, seq, lukF-PV, lukS-PV, and sak (Table 2). Isolate designations in boxes indicate PVL-positive isolates.
RESULTS
Antibiogram analysis, PVL PCR, PFGE, spa typing, MLST, SCCmec typing, and DNA microarray analysis (data not shown) were performed on all isolates (Tables 1 and 2).
TABLE 1.
Antibiogram results, PVL PCR results, PFGE patterns, spa types, MLST results, and SCCmec types for the CC59 MRSA isolates
| Group and isolatea | Source | Antibiogram resultc | PVL PCR resultd | PFGE pattern | spa sequence | spa type | MLST sequence | ST | SCCmec type |
|---|---|---|---|---|---|---|---|---|---|
| Group 1, “WA MRSA-73” | |||||||||
| 05-16512 | Colonization | N | WA MRSA-73 | 04 | t528 | 19-23-15-2-19-20-15 | 59 | IVb (2B) | |
| Group 2, “WA MRSA-24” | |||||||||
| 04-17626 | SSTIb | Eryr | N | WA MRSA-24 | 04-20-17-20-17-31-16-34 | t216 | 19-23-15-2-41-20-15 | 87 | IVb (2B) |
| 06-15325 | SSTI | Eryr | N | WA MRSA-24 | 04-20-17-20-17-31-16-34 | t216 | 19-23-15-2-41-20-15 | 87 | IVb (2B) |
| 06-16824 | SSTI | Eryr | N | WA MRSA-24 | 04-20-17-20-17-31-16-34 | t216 | 19-23-15-2-41-20-15 | 87 | IVb (2B) |
| Group 3, “WA MRSA-55/56” | |||||||||
| 06-17947 | SSTI | Eryr Tetr | P | WA MRSA-55 | 04-20-17-20-17-25-34 | t437 | 19-23-15-2-19-20-15 | 59 | IVa (2B) |
| 06-16367 | SSTI | Eryr Tetr | P | WA MRSA-55 | 04-20-17-20-17-25-34 | t437 | 19-23-15-2-19-20-15 | 59 | IVa (2B) |
| 07-15432 | SSTI | Eryr Tetr | P | WA MRSA-55 | 04-20-17-20-17-25-34 | t437 | 19-23-15-2-19-20-15 | 59 | IVa (2B) |
| 07-15760 | SSTI | Eryr Tetr | P | WA MRSA-55 | 04-20-17-20-17-25-34 | t437 | 19-23-15-2-19-20-15 | 59 | IVa (2B) |
| 08-16180 | SSTI | Eryr Tetr | P | WA MRSA-55 | 04-20-17-20-17-25-34 | t437 | 19-23-15-2-19-20-15 | 59 | IVd (2B) |
| 08-17668 | Colonization | Eryr | N | WA MRSA-55 | 04-20-17-20-17-25-34 | t437 | 19-23-15-2-19-20-15 | 59 | IVv (2B) |
| 07-15443 | SSTI | Eryr | P | WA MRSA-56 | 04-20-17-20-17-25-34 | t437 | 19-23-15-2-19-20-15 | 59 | IVa (2B) |
| Group 4, “WA MRSA-15” | |||||||||
| 03-17565 | SSTI | N | WA MRSA-15 | 04-20-17-20-31-16-34 | t976 | 19-23-15-2-19-20-15 | 59 | IVa (2B&5) | |
| 04-16557 | SSTI | N | WA MRSA-15 | 04-20-17-20-31-16-34 | t976 | 19-23-15-2-19-20-15 | 59 | IVa (2B&5) | |
| 04-17489 | SSTI | N | WA MRSA-15 | 04-20-17-20-31-16-34 | t976 | 19-23-15-2-19-20-15 | 59 | IVa (2B&5) | |
| 05-17037 | SSTI | Eryr Tetr | N | WA MRSA-15 | 04-20-17-20-31-16-34 | t976 | 19-23-15-2-19-20-15 | 59 | IVa (2B&5) |
| 05-17619 | SSTI | N | WA MRSA-15 | 04-20-17-20-31-16-34 | t976 | 19-23-15-2-19-20-15 | 59 | IVa (2B&5) | |
| 06-15513 | SSTI | N | WA MRSA-15 | 04-20-17-20-31-16-34 | t976 | 19-23-15-2-19-20-15 | 59 | IVa (2B&5) | |
| 06-17484 | SSTI | N | WA MRSA-15 | 04-20-17-20-31-16-34 | t976 | 19-23-15-2-19-20-15 | 59 | IVa (2B&5) | |
| 07-19251 | SSTI | Eryr | N | WA MRSA-15 | 04-20-17-20-31-16-34 | t976 | 19-23-15-2-19-20-15 | 59 | IVa (2B&5) |
| 08-15202 | SSTI | N | WA MRSA-15 | 04-20-17-20-31-16-34 | t976 | 19-23-15-2-19-20-15 | 59 | IVa (2B&5) | |
| Group 5, “WA MRSA-9/52,” or “Taiwan clone” | |||||||||
| 06-15672 | SSTI | Eryr Tetr | P | WA MRSA-9 | 04-20-17-25-34 | t441 | 19-23-15-2-19-20-15 | 59 | V (5C2&5) |
| 06-17363 | SSTI | Eryr Tetr | P | WA MRSA-9 | 04-20-17-25-34 | t441 | 19-23-15-2-19-20-15 | 59 | V (5C2&5) |
| 07-16447 | SSTI | Eryr Tetr | P | WA MRSA-9 | 04-20-17-25-34 | t441 | 19-23-15-2-19-20-15 | 59 | V (5C2&5) |
| 07-17830 | SSTI | Eryr Tetr | P | WA MRSA-9 | 04-20-17-25-34 | t441 | 19-23-15-2-19-20-15 | 59 | V (5C2&5) |
| 03-16672 | SSTI | Eryr Tetr | P | WA MRSA-9 | 04-20-17-20-17-25-34 | t437 | 19-23-15-2-19-20-15 | 59 | V (5C2&5) |
| 04-16811 | SSTI | Eryr Tetr | P | WA MRSA-9 | 04-20-17-20-17-25-34 | t437 | 19-23-15-2-19-20-15 | 59 | V (5C2&5) |
| 04-17021 | SSTI | Eryr Tetr | P | WA MRSA-9 | 04-20-17-20-17-25-34 | t437 | 19-23-15-2-19-20-15 | 59 | V (5C2&5) |
| 05-15724 | SSTI | Eryr Tetr | P | WA MRSA-9 | 04-20-17-20-17-25-34 | t437 | 19-23-15-2-19-20-15 | 59 | V (5C2&5) |
| 07-15919 | SSTI | Eryr Tetr | P | WA MRSA-9 | 04-20-17-20-17-25-34 | t437 | 19-23-15-2-19-20-15 | 59 | V (5C2&5) |
| 07-16753 | SSTI | Eryr Tetr | P | WA MRSA-9 | 04-20-17-20-17-25-34 | t437 | 19-23-15-2-19-20-15 | 59 | V (5C2&5) |
| 07-16861 | SSTI | Eryr | P | WA MRSA-9 | 04-20-17-20-17-25-34 | t437 | 19-23-15-2-19-20-15 | 59 | V (5C2&5) |
| 07-18714 | SSTI | Eryr Tetr | P | WA MRSA-9 | 04-20-17-20-17-25-34 | t437 | 19-23-15-2-19-20-15 | 59 | V (5C2&5) |
| 08-15039 | SSTI | Eryr Tetr | P | WA MRSA-9 | 04-20-17-20-17-25-34 | t437 | 19-23-15-2-19-20-15 | 59 | V (5C2&5) |
| 08-18104 | SSTI | Eryr Tetr | P | WA MRSA-9 | 04-20-17-20-17-25-34 | t437 | 19-23-15-2-19-20-15 | 59 | V (5C2&5) |
| 08-18190 | Colonization | Eryr Tetr | P | WA MRSA-9 | 04-20-17-20-17-25-34 | t437 | 19-23-15-2-19-20-15 | 59 | V (5C2&5) |
| 07-17677 | Colonization | Eryr | P | WA MRSA-9 | 04-20-17-20-17-25-34 | t437 | 19-23-15-48-19-20-15 | 338 | V (5C2&5) |
| 07-15076 | Colonization | Eryr | P | WA MRSA-52 | 04-20-17-20-17-34 | t1950 | 113-23-15-2-19-20-15 | 952 | V (5C2&5) |
| 07-16295 | Colonization | Eryr | P | WA MRSA-52 | 04-20-17-20-17-34 | t1950 | 113-23-15-2-19-20-15 | 952 | V (5C2&5) |
| 07-16320 | Colonization | Eryr | P | WA MRSA-52 | 04-20-17-20-17-34 | t1950 | 113-23-15-2-19-20-15 | 952 | V (5C2&5) |
| 07-16355 | SSTI | Eryr | P | WA MRSA-9 | 04-20-16-34 | t2365 | 19-23-15-2-19-20-15 | 59 | V (5C2&5) |
| Group 6, “WA MRSA-9” | |||||||||
| 05-17759 | SSTI | N | WA MRSA-9 | 04-20-17-31-16-34 | t316 | 19-23-15-2-19-20-15 | 59 | Vv (5C2) | |
| 08-15683 | SSTI | Eryr | N | WA MRSA-9 | 04-20-17-31-16-34 | t316 | 19-23-15-2-19-20-15 | 59 | Vv (5C2) |
| 06-18653 | SSTI | Cipr | N | WA MRSA-9 | 04-20-17-25-34 | t441 | 19-23-15-2-19-20-15 | 59 | Vv (5C2) |
| NARSA control strains, “USA1000” | |||||||||
| NARSA483 | Eryr | P | USA1000 | 04-20-17-31-16-34 | t316 | 19-23-15-2-19-20-15 | 59 | IVa (2B) | |
| NARSA676 | P | USA1000 | 04-20-17-20-17-31-16-34 | t216 | 19-23-15-2-19-20-15 | 59 | IVa (2B) |
Group 1, PVL-negative ST59 MRSA-IVb (2B) (ccrA2B2 and mec complex class B); group 2, PVL-negative ST87 (59SLV) MRSA-IVb (2B) (ccrA2B2 and mec complex class B); group 3, PVL-variable ST59 MRSA-IVa, -IVd, and -IVv (2B) (ccrA2B2 and mec complex class B); group 4, PVL-negative ST59 MRSA-IVa (2B&5) (ccrA2B2, mec complex class B, and ccrC1); group 5, PVL-positive ST59/338 (59SLV)/952 (59SLV) MRSA-V (5C2&5) (mec complex class C2 and ccrC1); group 6, PVL-negative ST59 MRSA-Vv (5C2) (mec complex class C2 and ccrC1); NARSA control strains, PVL-positive ST59 MRSA-IVa (2B) (ccrA2B2 and mec complex class B).
SSTI, skin and soft tissue infection.
Eryr, erythromycin resistant; Tetr, tetracycline resistant; Cipr, ciprofloxacin resistant.
N, negative; P, positive.
TABLE 2.
Grouping of CC59 isolates based on DNA microarray hybridization results
| Group and no. of isolatesa | Presence (+) or absence (−) of: |
||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mecA | ΔmecR | ugpQ | dcs region | ccrA2 | ccrB2 | 85-2082 ccrA | MRSAZH47 ccrA | 85-2082 ccrC | blaZ | blaI | blaR | ermA | ermB | ermC | msrA | mpbBM | aphA3 | sat | tetK | pC221 cat | pC223 cat | entA | entB | entK | entQ | lukF-PV | lukS-PV | sak | |
| Group 1, “WA MRSA-73” | |||||||||||||||||||||||||||||
| 1 | + | + | + | + | + | + | − | − | − | + | + | + | − | − | − | − | − | − | − | − | − | − | − | + | + | + | − | − | + |
| Group 2, “WA MRSA-24” | |||||||||||||||||||||||||||||
| 3 | + | + | + | + | + | + | − | − | − | + | + | + | − | − | − | + | + | + | + | − | − | − | − | + | + | + | − | − | + |
| Group 3, “WA MRSA-55/56” | |||||||||||||||||||||||||||||
| 4 | + | + | + | + | + | + | − | − | − | + | + | + | − | + | − | − | − | + | + | + | − | − | − | + | + | + | + | + | + |
| 1 | + | + | + | + | + | + | − | − | − | + | + | + | − | + | − | − | − | + | + | + | − | + | − | + | + | + | + | + | − |
| 1 | + | + | + | + | + | + | − | − | − | + | + | + | − | + | − | − | − | + | + | − | − | + | − | + | + | + | − | − | + |
| 1 | + | + | + | + | + | + | − | − | − | + | + | + | − | + | − | − | − | + | + | − | − | + | + | + | + | + | + | + | + |
| Group 4, “WA MRSA-15” | |||||||||||||||||||||||||||||
| 7 | + | + | + | + | + | + | + | − | + | + | + | + | − | − | − | − | − | − | − | − | − | − | + | + | + | + | − | − | + |
| 1 | + | + | + | + | + | + | + | − | + | + | + | + | − | − | − | + | + | + | + | + | − | − | + | + | + | + | − | − | + |
| 1 | + | + | + | + | + | + | + | − | + | + | + | + | − | − | − | + | + | − | − | − | − | − | + | − | − | − | − | − | + |
| Group 5, “WA MRSA-9/52,” or “Taiwan clone” | |||||||||||||||||||||||||||||
| 4 | + | − | + | − | − | − | + | + | + | + | + | + | − | + | − | − | − | + | + | + | − | + | − | − | − | − | + | + | − |
| 2 | + | − | + | − | − | − | + | + | + | + | + | + | − | + | − | − | − | + | + | + | − | − | − | + | + | + | + | + | − |
| 8 | + | − | + | − | − | − | + | + | + | + | + | + | − | + | − | − | − | + | + | + | − | + | − | + | + | + | + | + | − |
| 5 | + | − | + | − | − | − | + | + | + | + | + | + | − | + | − | − | − | + | + | − | − | + | − | + | + | + | + | + | − |
| 1 | + | − | + | − | − | − | + | + | + | + | + | + | − | + | − | − | − | + | + | − | − | − | − | − | − | − | + | + | − |
| Group 6, “WA MRSA-9” | |||||||||||||||||||||||||||||
| 1 | + | − | + | − | − | − | + | − | + | + | + | + | − | − | − | − | − | − | − | − | − | − | − | + | + | + | − | − | − |
| 1 | + | − | + | − | − | − | + | − | + | − | − | − | − | − | + | − | − | − | − | − | + | − | − | + | + | + | − | − | − |
| 1 | + | − | + | − | − | − | + | − | + | + | + | + | − | − | − | − | − | − | − | − | − | − | − | + | + | + | − | − | + |
| NARSA control strains, “USA1000” | |||||||||||||||||||||||||||||
| 1 (NARSA483) | + | + | + | + | + | + | − | − | − | + | + | + | + | − | − | − | − | − | − | − | − | − | − | + | + | + | + | + | − |
| 1 (NARSA676) | + | + | + | + | + | + | − | − | − | + | + | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + | + | − |
Group 1, PVL-negative ST59 MRSA-IVb (2B); group 2, PVL-negative ST87 (59SLV) MRSA-IVb (2B); group 3, PVL-variable ST59 MRSA-IVa, -IVd, and -IVv (2B); group 4, PVL-negative ST59 MRSA-IVa (2B&5); group 5, PVL-positive ST59/338 (59SLV)/952 (59SLV) MRSA-V (5C2&5); group 6, PVL-negative ST59 MRSA-Vv (5C2); NARSA control strains, PVL-positive ST59 MRSA-IVa (2B).
PFGE identified seven WA CC59 CA-MRSA strains: WA MRSA-9, -15, -24, -52, -55, -56, and -73 (Fig. 1).
FIG. 1.
Representative pulsed-field gel electrophoresis patterns of the seven Western Australian CC59 MRSA strains.
Shared properties of CC59 MRSA isolates.
All isolates were agr group I and capsule type 8 and carried the gamma-hemolysin genes lukF, lukS, and hlgA, as well as the hl (GenBank accession no. CP000046.1, locus tag SACOL0921), hla, hld, and hlIII hemolysin genes. Three out of five probes for beta-hemolysin yielded signals, which could be interpreted as an indication of an as yet unsequenced allelic variant. Leukocidin genes lukD and lukE were not detected. All isolates lacked protease genes splA, splB, and splE but harbored aur, sspA, sspB, and sspP. The carriage of set and ssl genes was uniform among all isolates, including the presence of setC, ssl1 (in a Mu50/N315-like allele), ssl3, ssl4, ssl5 (an unsequenced allele giving signals with a probe derived from CC1/5/8 sequences as well as with another one based on RF122), ssl7, ssl8, ssl9, ssl10, and setB1 to setB3. The probe for ssl2 gave weaker or variable results, indicating another as yet unsequenced allelic variant.
The biofilm operon icaACD was present. The gene for a biofilm-associated protein, bap, was absent.
Genes encoding MSCRAMMs (microbial surface components recognizing adhesive matrix molecules), bbp, clfA, clfB, ebh, ebpS, eno, fib, fnbA, fnbB, map, sasG, sdrC, sdrD, and vwb were detected in all isolates. The gene for collagen-binding adhesin (cna) was absent.
Carriage of resistance genes, PVL, beta-hemolysin integrating phages, and superantigens was variable.
DNA microarray-based analysis of the CC59 isolates clustered them into six groups (Fig. 2).
Group 1, “WA MRSA-73” (PVL-negative ST59 MRSA-IVb [2B]).
“WA MRSA-73” (isolate 05-16512) is an ST59/spa type t528 strain that has acquired an SCCmec IVb (2B) (ccrA2B2 and mec complex class B) element. Although lacking PVL, this strain has a DNA microarray profile similar to that of the NARSA ST59 MRSA-IVa “USA1000” control strain (NARSA483). Both strains have acquired seb, sek, and seq enterotoxin genes and lack antimicrobial resistance genes apart from mecA and blaZ.
Group 2, “WA MRSA-24” (PVL-negative ST87 MRSA-IVb [2B]).
Three “WA MRSA-24” isolates (04-17626, 06-15325, and 06-16824), collected from unrelated patients in 2004 and 2006, were identified as ST87 (an SLV of ST59 [59SLV])/spa type t216 with a type IVb (2B) SCCmec. The beta-lactamase operon (blaZ, blaI, and blaR), the msrA, mpbBM (macrolide), aphA3 (neomycin), and sat (streptothricin) resistance genes, and the seb, sek, and seq enterotoxin genes were present in all three isolates. The isolates were also positive for staphylokinase (sak), chemotaxis-inhibiting protein (chp), and staphylococcal complement inhibitor (scn) genes, which are known to be located on beta-hemolysin integrating phages. However, the enterotoxin A (sea) gene, which is also located on beta-hemolysin integrating phages, was not detected. PVL was not detected.
Group 3, “WA MRSA-55/56” (PVL-variable ST59 MRSA-IV [2B], structural subtypes IVa, IVd, and IVv).
Group 3, a group of seven ST59/spa type t437 isolates, is comprised of four very similar strains that have acquired ermB (macrolide-lincosamide-streptogramin B), aphA3, sat, and tetK resistance genes. In this regard, they resemble the “Taiwan clone” (see below). The other three strains in this group differ by variable carriage of PVL and enterotoxin A (sea) and SCCmec type IV subtype.
The first strain, PVL-positive “WA MRSA-55,” included four isolates (06-16367, 06-17947, 07-15432, and 07-15760) collected in 2006 and 2007. All four isolates harbored an SCCmec IVa (2B) element, and their DNA microarray-based profiles were identical. The beta-lactamase operon (blaZ, blaI, and blaR), ermB, aphA3, sat, and tetK resistance genes as well as the seb, sek, and seq enterotoxin genes, PVL genes (lukF-PV and lukS-PV), and chp, scn, and sak genes were all detected in this strain.
The second strain (08-16180) was also PVL positive and, except for the absence of the sak gene and the presence of the chloramphenicol resistance gene cat, yielded the same DNA microarray hybridization pattern as the first strain in this group. However, SCCmec analysis by PCR revealed that this isolate harbored a type IVd (2B) SCCmec element.
The third strain, a PVL-negative isolate (08-17668), closely resembled the PVL-positive strain “WA MRSA-55” in terms of microarray hybridization and PFGE pattern. However, it has acquired a novel type IV (2B) SCCmec which could not be subtyped with primers specifying subtypes a to h.
The fourth strain, “WA MRSA-56,” consisted of one isolate (07-15443) collected in 2007. It was also identified as PVL-positive ST59 MRSA-IVa (2B). However, unlike the other group 3 strains, “WA MRSA-56” carried the enterotoxin gene sea.
Group 4, “WA MRSA-15” (PVL-negative ST59 MRSA-IVa [2B&5]).
Group 4 consisted of nine isolates (03-17565, 04-16557, 04-17489, 05-17037, 05-17619, 06-15513, 06-17484, 07-19251, and 08-15202) collected from seven individual patients between 2003 and 2008. One patient with various wound infections yielded three identical isolates in three subsequent years (2004 to 2006).
These isolates were identified as PVL-negative ST59 MRSA-IVa (2B&5) (ccrA2B2 mec complex class B and ccrC1)/spa type t976.
All nine isolates carried the beta-lactamase operon (blaZ, blaI, and blaR). Variable resistance genes, including msrA and mpbBM (in two isolates), aphA3 and sat (in one isolate), and tetK (in one isolate), were detected. All isolates were negative for PVL genes but positive for the enterotoxin A gene (sea). Seven out of nine isolates harbored seb, sek, and seq enterotoxin genes.
Isolate 05-17037 differed in its carriage of resistance genes (msrA, mpbBM, aphA3, sat, and tetK). DNA microarray-based markers indicated that all isolates in this group encoded an unusual SCCmec element with mec complex class B and ccr complex type 2, characteristic of SCCmec type IV, and ccrC1, which has thus far been found in SCCmec type V or SCC elements. Positive hybridization signals for SCCmec markers mecA, ΔmecR, ugpQ, the dcs region, ccrA2, ccrB2, 85-2082 ccrA, and 85-2082 ccrC1 were obtained. SCCmec PCR confirmed this observation. All isolates in the group were positive for SCCmec type IVa (2B) structural elements, and all encoded a class B mec complex, a type 2 ccr complex, and ccrC1 allele 2. This suggests either the presence of a composite IVa (2B) and V (5C2) SCCmec element or the additional presence of an SCC element encoding ccrC1 allele 2.
Group 5, “WA MRSA-9/52,” or “Taiwan clone” (PVL-positive ST59/338/952 MRSA-V [5C2&5]).
Group 5 consisted of 17 “WA MRSA-9” isolates identified as ST59/spa type t437, t441, or t2365 and three “WA MRSA-52” isolates identified as ST952 (a single-locus variant of ST59)/spa type t1950 (Table 1). The DNA microarray-based profiles for these 20 isolates were similar, and consequently the two strains were classified into one group. “WA MRSA-9” was the first SCCmec V (5C2&5) (mec complex class C2 and ccrC1) CC59 strain found in Western Australia (in 2003), and it appears to be the most common and clinically relevant strain in this clonal complex.
For SCCmec markers on the array, all isolates yielded hybridization signals with probes for mecA, ugpQ, 85-2082 ccrA, and MRSAZH47 ccrA, as well as for ccrC1. When tested by PCR, all 20 strains had mec complex C2 and ccrC1. ccrC1 allotyping revealed the presence of two ccrC1 complexes, ccrC1 allele 2 and ccrC1 allele 8, which is characteristic of the SCCmec encoded by the Taiwan clone. An ISSau4-like transposase was found inserted into the structural gene V011 of all isolates in this group.
All 20 “Taiwan clone” isolates carried the beta-lactamase operon (blaZ, blaI, and blaR) as well as ermB, aphA3, and sat. Variable resistance genes were tetK (in 14 isolates) and cat (in 17 isolates). All isolates were positive for lukF-PV and lukS-PV. Enterotoxin genes seb, sek, and seq were present in 15 of the 20 isolates.
Group 6, “WA MRSA-9” (PVL-negative ST59 MRSA-Vv [5C2]).
Although characterized as “WA MRSA-9” by PFGE, three isolates (05-17759, 08-15683, and 06-18653) were classified into group 6. These isolates carried an SCCmec type V variant (5C2) (ccrC1 and mec complex class C2) element. However, they yielded a DNA hybridization pattern different from that of the “Taiwan clone” (group 5). They were positive for mecA, ugpQ, 85-2082 ccrA, and 85-2082 ccrC1 but negative with the MRSAZH47 ccrA probe.
PCR SCCmec analysis revealed that isolates from this group did not amplify the SCCmec type V (5C2) specific structural ORF V011 (GenBank accession no. AB12129) with or without the ISSau4-like transposase insertion. However, they were positive for the type V (5C2) core genes for mec complex C2 and ccrC1. Allotyping of the ccrC1 gene revealed that it was neither allele 2 nor allele 8. On the basis of a lack of amplification of the structural gene, the SCCmec of these strains has been classified as a type V variant (Vv [5C2]). It is evident that this group of isolates harbors an SCCmec element that is significantly different from that of the Taiwan clone.
Two isolates carried the beta-lactamase operon, and one was positive for ermC (macrolide-lincosamide resistance) and cat. All three isolates harbored seb, sek, and seq enterotoxin genes as well as chp and scn. One isolate also yielded hybridization signals for the staphylokinase gene sak. All isolates were PVL negative.
DISCUSSION
In Western Australia there are at least six discernible groups of CC59 CA-MRSA strains, which can be differentiated by PFGE, MLST, determination of the presence or absence of PVL, determination of the SCCmec type, or microarray analysis. Within the study strains, at least seven different variants of SCCmec elements (IVa [2B], IVb [2B], IVd [2B], IVa [2B&5], IVv [2B], Vv [5C2], and V [5C2&5]) were distinguished. This suggests rapid evolution and/or multiple transfer events of SCCmec elements. In a recent study by Takano et al. (24), at least six SCCmec elements were described to occur in a collection of ST59 MRSA strains isolated in Taiwan, including V (5C), V (5C2&5), IVc (2B), IV (2B), and two novel elements. This diversity of SCCmec types and subtypes can be expected to cause ambiguities in nomenclature, which underscores the need for sequence information. Consequently, the novel SCCmec elements described in this study warrant further characterization, although sequencing is beyond the scope of this study. Another observation suggesting a rapid evolution within CC59 is that groups 3 and 5 (“Taiwan clone”) appear to be closely related to each other with regard to all markers but SCCmec. Both groups share aphA3, sat, ermB, and usually also cat as well as PVL. This indicates that groups 3 and 5 might represent one branch of the CC59 complex that evolved into separate groups by acquiring different SCCmec elements. Generally, CC59 displayed a high degree of variability, affecting not only SCCmec markers but also a variety of other mobile genetic elements. For instance, “USA1000” and “WA MRSA-73” differ only in the presence of PVL. It cannot yet be determined whether “USA1000” evolved from a “WA MRSA-73”-like ancestor by acquiring PVL, whether “WA MRSA-73” was a deletion variant of USA1000, or whether both represent independently evolved branches of one lineage. Similarly, isolate 08-17668 might represent either a PVL-negative ancestor or a mere deletion mutant of group 3 strains. A high degree of variability can also be detected within CC59 groups. For example, the Taiwan clone (group 5) can be subdivided based on resistance and toxin genes and spa and MLST sequences. Such variation within a supposed “clone” might be used to trace individual chains of infection, as in the case of the patients with the ST952 variant of the “Taiwan clone” (“WA MRSA-52”) who belonged to the same family.
Further studies should investigate the variability and evolution of CC59 strains in other locations where this clonal complex has been detected. Apart from data for the Taiwan clone (group 5), there are little data available on the distribution of CC59 clones outside Western Australia. It can be assumed that these strains are usually identified as “USA1000” or the “Taiwan clone” and that their true diversity remains unrecognized. This might also obscure routes of transmission of CC59 CA-MRSA strains and hinder the understanding of their international spread.
If a variety of closely related strains exist simultaneously, it can be assumed that they are in competition with each other for “ecological” resources, i.e., for susceptible, as yet uncolonized hosts. The ecological success of strains should result in wide distribution and/or relatively high prevalence. The Taiwan clone (group 5) could be regarded as the most successful strain among the CC59 CA-MRSA strains isolated in Western Australia. In our study, nearly as many isolates belonged to group 5 as to all other CC59 groups combined. Since the groups are nearly isogenic, a marker determining such success should be found among the rather limited number of genes which are variable within CC59, and it should be present in the Taiwan clone. Full genome sequencing of representative strains of CC59 may provide the answer and overcome the limitations of this study. Among the genes which were examined in this study, PVL genes could be related to the success of the Taiwan clone.
Acknowledgments
We thank Samantha Cramer, Lynne Wilson, Yi Kong Chew, Antje Ruppelt, Hanna Kanig, Susann Kolewa, Elke Müller, Ines Engelmann, and Jana Sachtschal for their excellent technical assistance. We also thank the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) for supplying reference strains and the LotteryWest State Biomedical Facility, Genomics, Department of Clinical Immunology and Immunogenetics, PathWest Laboratory Medicine WA, at Royal Perth Hospital for performing the sequencing.
We acknowledge Enno Jacobs for supporting this work and Vico Baier for developing software.
Ralf Ehricht and Peter Slickers are employees of CLONDIAG. We declare no other conflicting interests.
Footnotes
Published ahead of print on 8 March 2010.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Boyle-Vavra, S., B. Ereshefsky, C. C. Wang, and R. S. Daum. 2005. Successful multiresistant community-associated methicillin-resistant Staphylococcus aureus lineage from Taipei, Taiwan, that carries either the novel staphylococcal chromosome cassette mec (SCCmec) type VT or SCCmec type IV. J. Clin. Microbiol. 43:4719-4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.CA-SFM. 1996. Report of the Comité de l'Antibiogramme de la Société Française de Microbiologie. Clin. Microbiol. Infect. 2:S48. [DOI] [PubMed] [Google Scholar]
- 3.CLSI. 2009. Performance standards for antimicrobial disk susceptibility tests, 7th ed. Approved standard M02-A10. CLSI, Wayne, PA.
- 4.CLSI. 2009. Performance standards for antimicrobial susceptibility testing, 19th informational supplement. M100-S18. CLSI, Wayne, PA.
- 5.Coombs, G. W., H. Van Gessel, J. C. Pearson, M. R. Godsell, F. G. O'Brien, and K. J. Christiansen. 2007. Controlling a multicentre outbreak involving the New York/Japan methicillin resistant Staphylococcus aureus clone. Infect. Control Hosp. Epidemiol. 28:845-852. [DOI] [PubMed] [Google Scholar]
- 6.Diep, B. A., H. A. Carleton, R. F. Chang, G. F. Sensabaugh, and F. Perdreau-Remington. 2006. Roles of 34 virulence genes in the evolution of hospital- and community-associated strains of methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 193:1495-1503. [DOI] [PubMed] [Google Scholar]
- 7.Enright, M. C., N. P. J. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fey, P. D., B. Said-Salim, M. E. Rupp, S. H. Hinrichs, D. J. Boxrud, C. C. Davis, B. N. Kreiswirth, and P. M. Schlievert. 2003. Comparative molecular analysis of community- or hospital-acquired methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 47:196-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finlay, J. E., L. A. Miller, and J. A. Poupard. 1997. Interpretive criteria for testing susceptibility of staphylococci to mupirocin. Antimicrob. Agents Chemother. 41:1137-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harmsen, D., H. Claus, W. Witte, J. Rothganger, H. Claus, D. Turnwald, and U. Vogel. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41:5442-5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hedin, G., and H. Fang. 2007. Epidemiology of methicillin-resistant Staphylococcus aureus in southern Stockholm, 2000-2003. Microb. Drug Resist. 13:241-250. [DOI] [PubMed] [Google Scholar]
- 12.Higuchi, W., T. Takano, L.-J. Teng, and T. Yamampto. 2008. Structure and specific detection of staphylococcal cassette chromosome mec type VII. Biochem. Biophys. Res. Commun. 377:752-756. [DOI] [PubMed] [Google Scholar]
- 13.Huang, Y. C., K. P. Hwang, P. Y. Chen, C. J. Chen, and T. Y. Lin. 2007. Prevalence of methicillin-resistant Staphylococcus aureus nasal colonization among Taiwanese children in 2005 and 2006. J. Clin. Microbiol. 45:3992-3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huson, D. H., and D. Bryant. 2006. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23:254-267. [DOI] [PubMed] [Google Scholar]
- 15.International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC). 2009. Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob. Agents Chemother. 53:4961-4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kondo, Y., T. Ito, X. X. Ma, S. Watanabe, B. N. Kreiswirth, J. Etienne, and K. Hiramatsu. 2007. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 51:264-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lo, W. T., W. J. Lin, M. H. Tseng, S. R. Wang, M. L. Chu, and C. C. Wang. 2006. Methicillin-resistant Staphylococcus aureus in children, Taiwan. Emerg. Infect. Dis. 12:1267-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milheirico, C., D. C. Oliveira, and H. de Lencastre. 2007. Multiplex PCR strategy for subtyping the staphylococcal cassette chromosome mec type IV in methicillin-resistant Staphylococcus aureus: SCCmec IV multiplex. J. Antimicrob. Chemother. 60:42-48. [DOI] [PubMed] [Google Scholar]
- 19.Monecke, S., B. Berger-Bächi, C. Coombs, A. Holmes, A. Kearns, I. Kay, H. Linde, F. O'Brien, P. Slickers, and R. Ehricht. 2007. Comparative genomics and DNA-array-based genotyping of pandemic Staphylococcus aureus strains carrying Panton-Valentine leucocidin. Clin. Microbiol. Infect. 13:236-249. [DOI] [PubMed] [Google Scholar]
- 20.Monecke, S., L. Jatzwauk, S. Weber, P. Slickers, and R. Ehricht. 2008. DNA microarray based genotyping of MRSA strains from eastern Saxony. Clin. Microbiol. Infect. 14:534-545. [DOI] [PubMed] [Google Scholar]
- 21.Monecke, S., P. Slickers, and R. Ehricht. 2008. Assignment of Staphylococcus aureus isolates to clonal complexes based on microarray analysis and pattern recognition. FEMS Immunol. Med. Microbiol. 53:237-251. [DOI] [PubMed] [Google Scholar]
- 22.Nimmo, G. R., and G. W. Coombs. 2008. Community-associated methicillin-resistant Staphylococcus aureus (MRSA) in Australia. Int. J. Antimicrob. Agents 31:401-410. [DOI] [PubMed] [Google Scholar]
- 23.O'Brien, F. G., E. E. Udo, and W. B. Grubb. 2006. Contour-clamped homogeneous electric field electrophoresis of Staphylococcus aureus. Nat. Protoc. 1:3028-3033. [DOI] [PubMed] [Google Scholar]
- 24.Takano, T., W. Higuchi, T. Otsuka, T. Baranovich, S. Enany, K. Saito, H. Isobe, S. Dohmae, K. Ozaki, M. Takano, Y. Iwao, M. Shibuya, T. Okubo, S. Yabe, D. Shi, I. Reva, L. J. Teng, and T. Yamamoto. 2008. Novel characteristics of community-acquired methicillin-resistant Staphylococcus aureus strains belonging to multilocus sequence type 59 in Taiwan. Antimicrob. Agents Chemother. 52:837-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takano, T., K. Saito, L. J. Teng, and T. Yamamoto. 2007. Spread of community-acquired methicillin-resistant Staphylococcus aureus (MRSA) in hospitals in Taipei, Taiwan in 2005, and comparison of its drug resistance with previous hospital-acquired MRSA. Microbiol. Immunol. 51:627-632. [DOI] [PubMed] [Google Scholar]
- 26.Tang, C. T., D. T. Nguyen, T. H. Ngo, T. M. Nguyen, V. T. Le, S. D. To, J. Lindsay, T. D. Nguyen, V. C. Bach, Q. T. Le, T. H. Le, D. L. Le, J. Campbell, T. K. Nguyen, V. V. Nguyen, J. Cockfield, T. G. Le, V. N. Phan, H. S. Le, T. S. Huynh, V. P. Le, M. Counahan, A. Bentsi-Enchill, R. Brown, J. Simmerman, T. C. Nguyen, T. H. Tran, J. Farrar, and C. Schultsz. 2007. An outbreak of severe infections with community-acquired MRSA carrying the Panton-Valentine leukocidin following vaccination. PLoS One 2:e822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, C.-C., W.-T. Lo, M.-L. Chu, and L. K. Siu. 2004. Epidemiological typing of community-acquired methicillin-resistant Staphylococcus aureus isolates from children in Taiwan. Clin. Infect. Dis. 39:481-487. [DOI] [PubMed] [Google Scholar]
- 29.Wang, R., K. R. Braughton, D. Kretschmer, T.-H. L. Bach, S. Y. Queck, M. Li, A. D. Kennedy, D. W. Dorward, S. J. Klebanoff, A. Peschel, F. R. DeLeo, and M. Otto. 2007. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat. Med. 13:1510-1514. [DOI] [PubMed] [Google Scholar]
- 30.Zhang, K., J. A. McClure, S. Elsayed, T. Louie, and J. M. Conly. 2005. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43:5026-5033. [DOI] [PMC free article] [PubMed] [Google Scholar]