Abstract
We compared the affinities of ceftaroline for all penicillin-binding proteins (PBPs) with those of ceftriaxone and cefotaxime in 6 Staphylococcus aureus and 7 Streptococcus pneumoniae isolates with various resistance phenotypes. Ceftaroline MICs were ≤1 μg/ml against all S. aureus isolates and were ≤0.25 μg/ml for 4 of 7 isolates of S. pneumoniae. Ceftaroline affinities for penicillin-susceptible S. pneumoniae strains were in the order PBP2X and -3 > PBP1A, -1B, and -2A > PBP2B, and ceftaroline had ≥4-fold higher 50% inhibitory concentrations (IC50s) (0.1 to 4 μg/ml) for PBP2X, -2A, -2B, and -3 than those for the other cephalosporins tested. Among 3 penicillin-resistant S. pneumoniae strains, ceftaroline had a high affinity for PBP2X (IC50, 0.1 to 1 μg/ml), a primary target for cephalosporin PBP binding activity, and high affinities for PBP2B (IC50, 0.5 to 4 μg/ml) and PBP1A (IC50, 0.125 to 0.25 μg/ml) as well, both of which are also known as major targets for PBP binding activity of cephalosporins. Ceftaroline PBP affinities in methicillin-susceptible S. aureus strains were greater than or equal to those of the 3 other β-lactams tested. Ceftaroline bound to PBP2a in methicillin-resistant S. aureus (IC50, 0.01 to 1 μg/ml) with up to 256-fold-higher affinity than those of other agents. Ceftaroline demonstrated very good PBP affinity against all S. aureus and S. pneumoniae strains tested, including resistant isolates.
β-Lactam antibiotics exert their antibacterial effect through covalent interactions with penicillin-binding proteins (PBPs), thus blocking the terminal step in cell wall biosynthesis. β-Lactam resistance in Streptococcus pneumoniae is usually caused by amino acid substitutions in the penicillin-binding domains of 1 or more of its 6 PBPs, resulting from point mutations or mosaic genes following recombination (21-23, 35). Altered PBP1A, PBP2X, and PBP2B are the most important PBPs for β-lactam resistance among clinical pneumococcal isolates (2, 3, 31, 46, 57).
In staphylococci, PBPs 1, 2, and 3, which have high affinities for most β-lactam antibiotics, are essential for cell growth and survival of methicillin-susceptible strains. Binding of β-lactams by these PBPs is lethal (6). Low-molecular-weight PBP4, although it may be important in normal cell wall synthesis and may participate to a limited extent in resistance, is not considered a critical target and may be dispensable (17, 41). Methicillin resistance in methicillin-resistant staphylococci (MRSA) is due to expression of a special PBP, PBP2a, which is not present in methicillin-susceptible staphylococci (6, 58, 59).
Ceftaroline fosamil is the developmental intravenous prodrug form of the broad-spectrum cephalosporin ceftaroline (formerly called PPI-0903 M or TAK-91825), which is active against MRSA and has a high affinity for PBP2a (PBP2′). It is also active against streptococci, including S. pneumoniae (3, 15, 16, 43, 45).
In the current study, we determined the affinities of ceftaroline, ceftriaxone, cefotaxime, and penicillin G for PBPs from 6 Staphylococcus aureus and 4 S. pneumoniae isolates with various resistance phenotypes.
MATERIALS AND METHODS
Bacterial strains and MIC determination.
MICs of ceftaroline, ceftriaxone, cefotaxime, and penicillin G were determined by CLSI macrodilution (8) for 6 clinical S. aureus and 7 S. pneumoniae strains. S. aureus strains used in the study involved 5 MRSA strains, including 1 heteroresistant vancomycin-intermediate S. aureus (hVISA) strain, 3 vancomycin-intermediate S. aureus (VISA) strains (including 1 daptomycin-resistant [29] and 1 linezolid-resistant [28] strain), and 1 vancomycin-resistant S. aureus (VRSA) strain, and 1 methicillin-susceptible S. aureus (MSSA) strain (Table 1).
TABLE 1.
MICs for all strains testeda
| Strain | Species | Country of origin | Yr isolated | Phenotype | MIC(μg/ml) |
Reference | |||
|---|---|---|---|---|---|---|---|---|---|
| PEN | CRO | CTX | CPT | ||||||
| 1564b | S. pneumoniae | Romania | 1996 | PEN resistantc | 16 | 32 | 32 | 2 | 32, 34 |
| 2688b | S. pneumoniae | Poland | 2000 | PEN resistantc | 8 | 8 | 16 | 0.5 | 32, 34 |
| 1394b | S. pneumoniae | Slovakia | 1999 | PEN resistantc | 4 | 2 | 4 | 0.25 | 32, 34 |
| 24 | S. pneumoniae | South Africa | Before 1998 | PEN resistantc | 4 | 2 | 1 | 0.25 | 32, 34 |
| 3413 | S. pneumoniae | Slovakia | 2000 | PEN resistantc | 4 | 2 | 2 | 0.125 | 34 |
| 2527 | S. pneumoniae | Croatia | 2000 | PEN resistantc | 2 | 0.03 | 0.015 | 0.015 | 34 |
| 1076 | S. pneumoniae | Austria | 1996 | PEN susceptiblec | 0.03 | 0.03 | 0.03 | 0.015 | 34 |
| ATCC 29213 | S. aureus | Reference strain | 1981 | MSSA, VSSA | 1 | 2 | 2 | 0.5 | www.atcc.org |
| 873 | S. aureus | USA | 2006 | MRSA, hVISA, β-lactamase negative | 8 | >64 | >64 | 0.5 | 33 |
| 510 (VRS2) | S. aureus | USA | 2002 | MRSA, VRSA, β-lactamase positive | 32 | >64 | >64 | 1 | 4, 5 |
| 2149A | S. aureus | USA | 2006 | MRSA, VISA, linezolid resistant, β-lactamase positive | 64 | 64 | 32 | 0.5 | 28 |
| 1287 | S. aureus | USA | 2007 | MRSA, VISA, β-lactamase positive | 32 | 8 | 4 | 0.5 | 33 |
| 25 | S. aureus | USA | 2005 | MRSA, VISA, daptomycin resistant, β-lactamase positive | 64 | >64 | >64 | 0.5 | 29 |
Abbreviations: PEN, penicillin G; CRO, ceftriaxone; CTX, cefotaxime; CPT, ceftaroline; MRSA, methicillin-resistant S. aureus; MSSA, methicillin-susceptible S. aureus; VRSA, vancomycin-resistant S. aureus; VISA, vancomycin-intermediate S. aureus; hVISA, heteroresistant vancomycin-intermediate S. aureus; VSSA, vancomycin-susceptible S. aureus.
No demonstrable PBP binding affinity observed, except for PBP3.
Classified according to 2006 CLSI M100-S17 interpretive criteria (9).
Among 7 clonally unrelated S. pneumoniae isolates from different countries chosen for this study, 6 were penicillin G resistant (MIC ≥ 2 μg/ml) and 1 was penicillin G susceptible (MIC ≤ 0.06 μg/ml) according to 2006 CLSI M100-S17 interpretive criteria (9) (Table 1).
Detection of β-lactamase enzymes in MRSA strains.
A cefinase disk test (BD Diagnostics, Sparks, MD) was used as a screening test for the presence of β-lactamase enzymes in MRSA strains. The test was performed according to the manufacturer's directions, with a positive control included. A color change from yellow to red within 1 h on the area where the culture was applied was considered positive.
Screening for optimal PBP expression among S. aureus strains. (i) Isolation of whole cells.
For each S. aureus strain, the growth curve was determined and whole cells were isolated in selected intervals within a range of optical densities at 600 nm (OD600) of 0.2 to 2, as described previously (52). Cultures were grown in 1.5 liters of brain heart infusion broth (BD Diagnostics) and harvested at OD600 values of 0.2, 0.3, 0.4, 0.5, 0.6, and 2. Cell pellets were resuspended in 50 mM NaPO4 buffer, pH 7.0, containing 100 mM NaCl and were treated with 1 mg/ml lysozyme (Sigma Inc., St. Louis, MO) and 0.01 mg/ml lysostaphin (Sigma Inc.) for 45 min at 37°C. Afterwards, 50 mM NaPO4 buffer, pH 7.0, containing 10 mM MgSO4 and 0.1 mg/ml DNase I (Promega, Madison, WI) was added and incubated for 15 min at 37°C. The 10 μl of the whole-cell suspension was subjected to the competition assay described below. Based upon PBP affinity screening at different points of bacterial growth, the optimal OD600 was selected and used for membrane isolation and PBP labeling.
(ii) Isolation of crude membranes.
Membranes containing PBPs from S. aureus were isolated using previously described methods (11, 25, 36, 48, 55, 60). Optimal OD600 values of 0.3 to 0.4 were determined for all strains except strain 25, which was harvested in the stationary growth phase (OD600 > 2.00). Whole cells were resuspended in 50 mM NaPO4 buffer, pH 7.0, containing 100 mM NaCl and were treated with lysozyme, lysostaphin, and DNase I (Promega, Madison, WI) as described above. Furthermore, cells were additionally lysed by three successive freeze-thaw cycles, using a dry ice-ethanol bath and a 37°C water bath, and then sonicated (Sonicator 3000; Misonix, Newtown, CT) in an ice bath four times at 30 s (50% duty, maximal output, pulsed), with a 60-s cooling period between rounds of sonication (48, 60). Unlysed cells were removed by centrifugation for 15 min at 5,000 × g. Fragments of S. aureus membranes containing PBPs were collected by ultracentrifugation at 100,000 × g for 90 min at 4°C and resuspended in 1 ml 50 mM NaPO4 buffer, pH 7.0 (11, 36, 48, 55, 60). The total protein concentrations in the membrane preparations were determined by a protein assay (Bio-Rad Laboratories), with bovine serum albumin as the standard.
S. pneumoniae whole-cell isolation.
Cultures of each strain were grown to an OD600 of 0.3 in Todd-Hewitt broth supplemented with 0.5% yeast extract (BD Diagnostics). Cells were collected by centrifugation for 15 min at 5,000 × g and then resuspended in 1 ml of 50 mM NaPO4 buffer, pH 7.0 (56, 65).
PBP labeling and competition assays.
PBPs were labeled using whole cells (S. aureus or S. pneumoniae) or membrane preparations (S. aureus), using methods based on the work of Sifaoui et al. (56). Membrane mixtures (100 μg of total protein per sample) or whole cells (108 to 5 × 108 CFU/ml; 20 μl for S. pneumoniae and 10 μl for S. aureus) were incubated with each of the tested antibiotics in turn (ceftaroline, ceftriaxone, and cefotaxime) for 10 min at 37°C, and a reference sample for each experiment was prepared for each mixture without incubation with antibiotic. Additionally, membranes or whole cells of all MRSA strains were preincubated with 1 mg/ml clavulanic acid (Glaxo SmithKline Laboratories, Collegeville, PA) to saturate all PBPs except for the low-affinity PBP2a (7, 11, 38, 39).
Visualization of S. aureus and S. pneumoniae PBPs and 50% inhibitory concentration (IC50) determination.
S. aureus and S. pneumoniae PBPs were labeled at 37°C for 15 min with 40 μg/ml and 10 μg/ml Bocillin FL (Invitrogen, Carlsbad, CA), respectively, as described previously (52, 65). Proteins were separated by SDS-PAGE, using 4 to 12% Bis-Tris Novex gels (Invitrogen) in morpholinepropanesulfonic acid (MOPS)-SDS running buffer (Invitrogen) for 2.5 h at 175 V (S. aureus) or for 110 min at 160 V (S. pneumoniae).
Labeled PBPs were visualized using a ChemiDoc XRS imager system at the 520-nm setting (Bio-Rad Laboratories). Affinities of the β-lactams for the PBPs were calculated as IC50s, which represent the β-lactam concentration needed to cause 50% inhibition of Bocillin FL binding, measured using Quantity One software (Bio-Rad Laboratories). The total number of membrane isolations for S. aureus was 3, and for S. pneumoniae isolates, the number of experiments was 2. The provisional IC50 in each experiment for each of the PBPs was determined and was within 1 double dilution of the antibiotic concentration used for competition assays.
Nucleotide sequence accession numbers.
All sequences obtained for pbp1A (nucleotides 870 to 1950), encoding 350 amino acids, pbp2B (nucleotides 655 to 2028), encoding 458 amino acids, pbp2X (nucleotides 301 to 2034), encoding 578 amino acids, and pbp3 (nucleotides 1 to 1242), encoding 413 amino acids, have been described previously (32, 34). Their respective GenBank accession numbers are listed in Table 3.
TABLE 3.
Binding affinities of ceftaroline, cefotaxime, and ceftriaxone for pneumococcal PBPs
| PBP | Strain (PEN susceptibility)a | GenBank accession no.b | IC50 (μg/ml) |
||
|---|---|---|---|---|---|
| Ceftaroline | Cefotaxime | Ceftriaxone | |||
| PBP1A | 1076 (S) | FJ439534 | 0.25 | 0.1 | 0.1 |
| 24 (R) | EU863721 | 0.125 | 0.125 | 0.25 | |
| 3413 (R) | FJ439539 | 0.25 | 0.25 | 0.25 | |
| 2527 (R) | FJ439536 | 0.25 | 0.25 | 0.25 | |
| PBP1B | 1076 (S) | NS | 0.25 | 0.25 | 0.1 |
| 24 (R) | NS | 0.1 | 8 | 1 | |
| 3413 (R) | NS | 0.25 | 4 | 4 | |
| 2527 (R) | NS | 0.25 | 0.25 | 0.125 | |
| PBP2X | 1076 (S) | FJ439542 | 0.1 | 0.25 | 0.1 |
| 24 (R) | EU863690 | 1 | 1 | 1 | |
| 3413 (R) | FJ439547 | 0.25 | 4 | 4 | |
| 2527 (R) | FJ439544 | 0.1 | 0.1 | 0.1 | |
| PBP2A | 1076 (S) | NS | 0.25 | 0.25 | 0.25 |
| 24 (R) | NS | 0.5 | 0.5 | 1 | |
| 3413 (R) | NS | 0.25 | 0.25 | 1 | |
| 2527 (R) | NS | 0.25 | 0.25 | 0.25 | |
| PBP2B | 1076 (S) | FJ439550 | 4 | >32 | >32 |
| 24 (R) | EU863659 | 0.5 | 16 | 16 | |
| 3413 (R) | FJ439555 | 2 | 16 | 16 | |
| 2527 (R) | FJ439552 | 4 | 4 | 4 | |
| PBP3 | 1076 (S) | FJ441591 | 0.1 | 0.1 | 0.1 |
| 24 (R) | FJ441590 | 0.1 | 0.1 | 0.25 | |
| 3413 (R) | FJ441599 | 0.1 | 0.1 | 0.25 | |
| 2527 (R) | FJ441595 | 0.25 | 0.1 | 0.25 | |
RESULTS AND DISCUSSION
MICs (μg/ml) for ceftaroline and comparators for strains used in this study are shown in Table 1. Affinities of ceftaroline and comparators for the individual PBPs in the studied isolates are indicated as IC50s and are reported in Table 2 for the isolates of S. aureus and in Table 3 for S. pneumoniae. Ceftaroline demonstrated the lowest MICs for all 7 pneumococcal strains tested, with a range of 0.015 to 2 μg/ml. Three S. pneumoniae isolates (1564, 2688, and 1394), with penicillin G MICs of 16, 8, and 4 μg/ml, respectively, did not show demonstrable PBP profiles due to affected growth (low cells count and early cell lysis occurred) resulting in poor Bocillin FL binding, which may be explained by lytic cell death, as reported by Regev-Yochay and coworkers (53). PBP binding studies of penicillin G-resistant S. pneumoniae isolates defined according to current CLSI nonmeningeal penicillin G breakpoints (susceptible, ≤2 μg/ml; intermediate, 4 μg/ml, and resistant, ≥8 μg/ml) (10) were difficult because of low bacterial cell counts (OD600 < 0.1) and the fact that cell lysis complicated preparation of sufficient amounts of cell material with all active penicillin-binding proteins (only minimal PBP3 affinity was present) for binding studies with Bocillin FL (K. Kosowska-Shick, unpublished information).
TABLE 2.
Binding affinities of β-lactams for S. aureus PBPs
| PBP | Strain | IC50 (μg/ml)a |
|||
|---|---|---|---|---|---|
| Ceftaroline | Cefotaxime | Ceftriaxone | Penicillin G | ||
| PBP1 | ATCC 29213 | 0.5 | 0.5 | 0.25 | 4 |
| 873 | 8 | 4 | 2 | 0.5 | |
| 510 | 0.5 | 2 | 0.5 | 128 | |
| 2149A | 1 | 0.5 | 4 | 2 | |
| 1287 | 0.125 | 4 | 1 | 4 | |
| 25 | 0.5 | 1 | 16 | 0.5 | |
| PBP2 | ATCC 29213 | 0.25 | 1 | 0.25 | 8 |
| 873 | 0.5 | 0.5 | 0.5 | 0.5 | |
| 510 | 0.125 | 0.5 | 0.25 | 64 | |
| 2149A | 1 | 2 | 1 | >128 | |
| 1287 | 4 | 1 | 1 | 4 | |
| 25 | 0.25 | 0.5 | 2 | 1 | |
| PBP2a | ATCC 29213 | NP | NP | NP | NP |
| 873 | 0.5 | >128 | >128 | 64 | |
| 510 | 0.25 | >128 | 1 | 64 | |
| 2149A | 1 | >128 | >128 | 2 | |
| 1287 | 1 | 4 | 2 | 4 | |
| 25 | 0.01 | 0.5 | 0.25 | 4 | |
| PBP3 | ATCC 29213 | 0.125 | 1 | 1 | 1 |
| 873 | 0.125 | 0.125 | 0.25 | 0.03 | |
| 510 | 0.125 | 0.25 | 0.25 | 4 | |
| 2149A | 0.5 | 4 | 2 | 2 | |
| 1287 | 0.1 | 1 | 1 | 0.25 | |
| 25 | 0.25 | 0.25 | 1 | 0.5 | |
| PBP4 | ATCC 29213 | >8 | >8 | >8 | >8 |
| 873 | >128 | >128 | >128 | >128 | |
| 510 | >128 | >128 | >128 | 64 | |
| 2149A | >128 | >128 | >128 | >128 | |
| 1287 | 64 | >128 | >128 | 4 | |
| 25 | ND | ND | ND | ND | |
NP, ATCC 29213, as a MSSA strain, lacks PBP2a. ND, no demonstrable binding of Bocillin FL to PBP4 was observed (17).
Ceftaroline had the lowest MICs against the 6 S. aureus isolates tested among those for cefotaxime, ceftriaxone, and penicillin G. The ceftaroline MIC was 0.5 μg/ml for all isolates except for 1 vancomycin-resistant MRSA isolate (VRSA 510), for which the MIC was 1 μg/ml. In all strains tested, ceftaroline bound to all PBPs with the same or a higher affinity (IC50 of ≤0.5 μg/ml for PBP1, -2, -2a, and -3 in MSSA strain and ≤1 μg/ml for PBP1, -2, -2a, and -3 in MRSA strains, with 4 exceptions: PBP1 in strains ATCC 29213, 873, and 2149A and PBP2 in strain 1287) than those of all β-lactam comparators (Table 2). All antibiotics tested had a weak affinity for PBP4 (IC50 > 8 μg/ml). Moisan et al. observed similar weak affinities of ceftaroline and ceftriaxone for PBP4 in strain ATCC 29213 (42). A fluorogram of PBP resolutions in methicillin-susceptible S. aureus ATCC 29213 is presented in Fig. 1, and Fig. 2 shows PBP2a profiles for all MRSA strains tested.
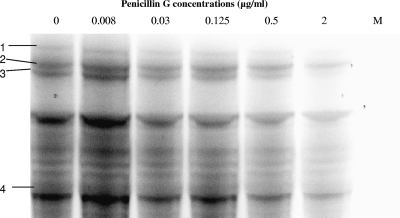
FIG. 1.
Competition assays for penicillin G binding to PBPs in MSSA (ATCC 29213).
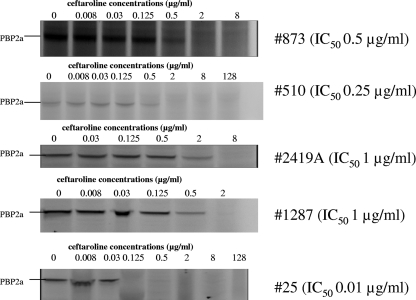
FIG. 2.
Competition assays for ceftaroline binding to PBP2a in MRSA strains.
Methicillin resistance in S. aureus is caused by production of the mecA-encoded protein PBP2a. The affinity of ceftaroline for PBP2a in all 5 MRSA strains was up to >128 times higher than those of other agents. We also observed that the PBP2a affinity levels in MRSA strains differed, depending on the growth phase during which cells were harvested for membrane isolation. PBP2a affinities for cephalosporins were higher for four strains from which membranes were isolated in the exponential growth phase (OD600 = 0.3) than for one strain (strain 25) isolated in the stationary growth phase (OD600 > 2.00). In strain 25, despite five attempts, there was no measurable PBP affinity observed during the exponential growth phase. The ceftaroline IC50 of 0.01 μg/ml for this strain was 50, 25, and 400 times lower than those of cefotaxime, ceftriaxone, and penicillin G, respectively, but the ceftaroline MIC was ≤128 times lower than those of the latter antibiotics. Hence, the higher PBP2a affinity resulted in lower MICs. We did not see a strictly proportional correlation between IC50 and the MIC for the β-lactams used in the study, which is a known phenomenon and reflects many other factors involved in the resistance level (7, 19, 26, 30, 49). We observed such differences between PBP2a affinity and the MIC for strain 25 for all antibiotics tested. This lack of correlation was also seen for strain 510 (the vanA-containing Hershey VRSA strain [4, 5]) and ceftriaxone (MIC > 64 μg/ml; IC50 = 1 μg/ml), for penicillin G and linezolid-resistant strain 2419A (28) (MIC = 64 μg/ml; IC50 = 2 μg/ml) or for ceftriaxone and this strain (MIC = 8 μg/ml; IC50 = 2 μg/ml), and for penicillin G (MIC 32 μg/ml; IC50 = 4 μg/ml) and strain 1287 (a VISA strain isolated at Hershey Medical Center [33]). Many factors may help to explain these differences. One factor could be that β-lactams, especially penicillin G, may have a relatively good affinity for PBP2a but that their susceptibility to hydrolysis by penicillinase results in high MICs (19). This may explain lower affinities of penicillin G for PBP2a in strains 2149A and 1287, which are β-lactamase positive (Table 2).
There is no reported direct relationship between the amount of expressed PBP2a protein and the β-lactam MIC (49). Additionally, not only PBP2a production but also a sufficient supply of peptidoglycan precursors would be required to mediate β-lactam resistance in MRSA. Also, altered peptidoglycan composition indicates the presence of factors other than PBP2a (12) which are involved in methicillin resistance, such as mutations in femA or genes involved in staphylococcal cell wall synthesis (12, 13, 49). In our previous study, we analyzed the cell wall composition of strain 25 and demonstrated reduced muropeptide cross-linking and a reduction in muramic acid O-acetylation (29). This strain was isolated in our hospital from a patient whose blood MRSA isolate developed vancomycin and then daptomycin resistance while the patient was on therapy with both agents (29). These facts may contribute to the observed high MICs for cefotaxime, ceftriaxone, and penicillin G, with relatively low PBP2a affinities. Many changes in the peptidoglycan composition and thickness have been noted in other VISA and VRSA strains (1), and in our collection, four strains (873, 25, 1287, and 510) had an hVISA, VISA, or VRSA phenotype, which may indirectly suggest the presence of β-lactam resistance mechanisms other than PBP2a. In strain 510, the PBP2a affinities for ceftriaxone (1 μg/ml) and cefotaxime (>128 μg/ml) varied compared to the MICs (>64 μg/ml), and this observation cannot be explained without additional experiments. The above-mentioned facts (a sufficient supply of peptidoglycan precursors and differences in peptidoglycan composition caused by altered gene expression involved in cell wall synthesis) may partially explain the differences as well as the possible mutations in the mecA gene encoding the PBP2a protein (30). It is important that affinity studies of this nature have not, to our knowledge, been published for hVISA and VISA strains, so there is no current basis for comparison of the results. These strains may well represent heterogeneous groups, each with its own specific resistance mechanisms. Villegas-Estrada et al. (61), using different determination and calculation methodology, tested the same (Hershey) VRSA strain examined in the current study as well as one different linezolid-resistant MRSA strain. Ceftaroline was found to be very active, with MICs of 0.25 to 2 μg/ml. Comparison between the IC50s found in the latter study and those found in the current study are not valid because of differing methodologies.
The range of IC50s for ceftaroline in our study with PBP2a was 0.5 to 1 μg/ml for membranes isolated in exponential growth phase and 0.01 μg/ml for strain 25, whose PBPs could be harvested only in stationary growth phase. Previous papers using comparable techniques reported average IC50s of 0.16 to 0.9 μg/ml for ceftaroline in MRSA strains (27, 42, 64). Differences between our results and those reported previously (27) may be explained at least partially by differences in strains and culture conditions. The higher affinity of ceftaroline for PBP2a than those of other β-lactams may be explained by the presence of longer side chains in the chemical structure, which increase interactions with the active-site groove of PBP2a (6, 37) and/or facilitate allosteric interactions that promote access to the active site (61).
In MRSA strains, it is known that PBP2a may replace the transpeptidase function of PBP2, although the transglycosylase function of the latter becomes critical for MRSA growth in the presence of β-lactams (51). In our study, ceftaroline showed very good affinity for PBP2, and in only 1 VISA strain (1287) was its affinity 2-fold lower than those of other cephalosporins (Table 2). The mechanisms of low affinity for PBP2 of penicillin G in strains 510 (VRSA) and 2149A (linezolid-resistant MRSA) compared to those of cephalosporins are unknown and may reflect the sequence mutations present in the gene encoding PBP2, which may affect binding (20).
PBP4 has been reported to be responsible for production of more highly cross-linked oligomers (36). The impaired function of PBP4 may be compensated by PBP2 activity (36). Also, PBP4 is linked to low-level methicillin resistance in strains lacking PBP2a (36, 41). PBP4 significance in β-lactam resistance has been reported to be more important in community-acquired MRSA (CA-MRSA) strains (41), which were not tested in the current study. In our study, PBP4 binding was weak (IC50s > 8 to 128 μg/ml) for all tested β-lactams, with the exception of VISA isolate 1287 with penicillin G (IC50 of 4 μg/ml) (Table 2). For VISA strain 25 (resistant to daptomycin and isolated from the blood of a patient who failed vancomycin and daptomycin therapy [29]), we could not obtain visible binding of Bocillin FL to PBP4; this phenomenon cannot be explained without additional investigations.
PBP1 may play a role in cell division of S. aureus (47) and is essential for growth in both MSSA and MRSA, and its function cannot be replaced by PBP2a (50). Ceftaroline showed good affinity for PBP1, with IC50s of ≤1 μg/ml for all isolates whose PBPs were harvested in exponential growth phase, with the exception of hVISA 873, for which the IC50 was 8 μg/ml. The IC50 of PBP1 in strain 25, whose PBPs were harvested in the stationary growth phase, was 0.5 μg/ml.
PBP3 is an essential PBP, and β-lactam binding to PBP3 results in cell enlargement and the cessation of septation (17). Ceftaroline had high PBP3 binding affinity relative to those of comparator β-lactams for 4 of 5 MRSA isolates tested, and in the remaining MRSA strain, hVISA strain 873, the affinity was only 2-fold lower than those of other cephalosporins.
Cephalosporin resistance in pneumococci is due to specific alterations and undefined changes in PBPs and may be influenced by mutations in other genes (14, 18, 21-23, 40, 54, 57, 62). As expected, the penicillin G-susceptible strain 1076 had no changes in the penicillin-binding domains of PBP1A (99% homology to strain R6), -2X (99% homology to strain R6), or -2B (96% homology to strain R6), which are known to be associated with β-lactam resistance or effects on PBP binding (2, 3, 31, 34, 46, 57).
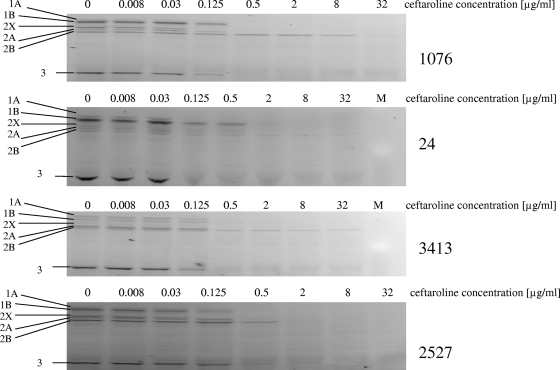
SDS-PAGE gels of competitive PBP binding experiments for ceftaroline are shown in Fig. 3 for all S. pneumoniae strains. In our studies on S. pneumoniae, the order of ceftaroline binding affinities for PBPs in penicillin-susceptible strain 1076 was PBP2X > PBP1A, -1B, and -2A > PBP3 > PBP2B, and ceftaroline had ≥2-fold higher IC50s for PBP1A and -1B than those of ceftriaxone and cefotaxime and lower IC50s for the other PBPs in this isolate (Table 3), against which all 3 cephalosporins had low MICs (≤0.03 μg/ml). PBP1A and -2X are known to be primary targets for cephalosporins in pneumococci, and all 3 cephalosporins tested showed high affinities for these PBPs, with IC50s of <0.25 μg/ml (18, 44, 63). Ceftaroline showed a higher affinity for PBP2B than those of ceftriaxone and cefotaxime in the penicillin G-susceptible pneumococcal strain 1076. Low affinities of cefotaxime and ceftriaxone for PBP2B have been described previously (18, 24, 44).
FIG. 3.
Competition assays for ceftaroline binding to PBPs from Streptococcus pneumoniae strains.
Among 3 penicillin G-resistant strains, ceftaroline had the lowest MICs (≤0.25 μg/ml) and higher or equal affinities for PBP1A, -2B, -2A, and -1B relative to those of the comparators (Table 3). The affinity of ceftaroline for PBP2X was higher than or equal to that for the most active comparator, cephalosporin, for any given isolate. The IC50s of ceftaroline for PBP3 ranged from 0.1 to 0.25 μg/ml, similar to those of comparators, although S. pneumoniae PBP3 has not been implicated in the killing action of β-lactam antibiotics (59). Ceftaroline showed the same or improved affinity for PBP2B relative to those of cefotaxime and ceftriaxone (Table 3), despite the presence of a T446A substitution in all penicillin-resistant strains (34). In penicillin-resistant strain 24, the ceftaroline MIC was 0.25 μg/ml, 2- to 4-fold lower than those of cefotaxime and ceftriaxone, with the same affinity for PBP2X (IC50 = 1 μg/ml). The lower MICs in this strain may result from improved PBP2B ceftaroline affinity, which was 16-fold higher than those of ceftriaxone and cefotaxime. In strain 3413, the ceftaroline MIC was 8-fold higher than those of cefotaxime and ceftriaxone, which may reflect improved ceftaroline affinity (4- to 8-fold lower IC50) for PBP2X and PBP2B. In penicillin-resistant strain 2527, for which all cephalosporins had MICs of ≤0.03 μg/ml, cephalosporin affinities for PBP2X and PBP2B were identical (IC50, 0.1 and 4 μg/ml, respectively). It should be noted that the role of individual PBP IC50s in contributing to the pneumococcal MIC is potentially complex and may be influenced by the constellation of mutations present in PBP genes and other genes, as well as the accessibility of the PBPs in whole bacteria (22, 23).
In summary, ceftaroline demonstrated potent binding to multiple PBPs in S. aureus, including PBP2a, responsible for methicillin resistance of MRSA, and to PBPs in S. pneumoniae, including PBP2B, PBP2X, and PBP1A, which are important in penicillin resistance. Our results help to explain the improved in vitro activity of this cephalosporin against S. aureus, including MRSA with impaired vancomycin susceptibility, and multidrug-resistant S. pneumoniae. These findings have important potential implications and support the use of ceftaroline for treatment of infections caused by resistant staphylococci and pneumococci.
Acknowledgments
This study was supported by a grant from Forest Laboratories, Inc., New York, NY. Funding for editorial assistance was provided by Forest Laboratories, Inc.
We thank Ronald Jones (JMI Laboratories, Liberty, IA) for providing the linezolid-resistant MRSA strain. Scientific Therapeutics Information, Inc., Springfield, NJ, provided editorial assistance on the manuscript.
Footnotes
Published ahead of print on 1 March 2010.
REFERENCES
- 1.Appelbaum, P. C. 2006. The emergence of vancomycin-intermediate and vancomycin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 12(Suppl. 1):16-23. [DOI] [PubMed] [Google Scholar]
- 2.Asahi, Y., Y. Takeuchi, and K. Ubukata. 1999. Diversity of substitutions within or adjacent to conserved amino acid motifs of penicillin-binding protein 2X in cephalosporin-resistant Streptococcus pneumoniae isolates. Antimicrob. Agents Chemother. 43:1252-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asahi, Y., and K. Ubukata. 1998. Association of a Thr-371 substitution in a conserved amino acid motif of penicillin-binding protein 1A with penicillin resistance of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 42:2267-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bozdogan, B., D. Esel, C. Whitener, F. A. Browne, and P. C. Appelbaum. 2003. Antibacterial susceptibility of a vancomycin-resistant Staphylococcus aureus strain isolated at the Hershey Medical Center. J. Antimicrob. Chemother. 52:864-868. [DOI] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2002. Vancomycin-resistant Staphylococcus aureus—Pennsylvania, 2002. MMWR Morb. Mortal. Wkly. Rep. 51:902. http://www.cdc.gov/mmwr//preview/mmwrhtml/mm5126a1.htm. [PubMed] [Google Scholar]
- 6.Chambers, H. F. 1999. Penicillin-binding protein-mediated resistance in pneumococci and staphylococci. J. Infect. Dis. 179(Suppl. 2):S353-S359. [DOI] [PubMed] [Google Scholar]
- 7.Chambers, H. F., and M. Sachdeva. 1990. Binding of β-lactam antibiotics to penicillin-binding proteins in methicillin-resistant Staphylococcus aureus. J. Infect. Dis. 161:1170-1176. [DOI] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M07-A8. Eighth edition. Clinical Laboratory Standards Institute, Wayne, PA.
- 9.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing. Approved standard M100-S17. Seventeenth informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA.
- 10.Clinical and Laboratory Standards Institute. 2009. Performance standards for antimicrobial susceptibility testing. Approved standard M100-S19. Nineteenth informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA.
- 11.Davies, T. A., M. G. Page, W. Shang, T. Andrew, M. Kania, and K. Bush. 2007. Binding of ceftobiprole and comparators to the penicillin-binding proteins of Escherichia coli, Pseudomonas aeruginosa, Staphylococcus aureus, and Streptococcus pneumoniae. Antimicrob. Agents Chemother. 51:2621-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Jonge, B. L., Y. S. Chang, D. Gage, and A. Tomasz. 1992. Peptidoglycan composition of a highly methicillin-resistant Staphylococcus aureus strain. The role of penicillin binding protein 2A. J. Biol. Chem. 267:11248-11254. [PubMed] [Google Scholar]
- 13.de Jonge, B. L., T. Sidow, Y. S. Chang, H. Labischinski, B. Berger-Bachi, D. A. Gage, and A. Tomasz. 1993. Altered muropeptide composition in Staphylococcus aureus strains with an inactivated femA locus. J. Bacteriol. 175:2779-2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.del Campo, R., F. Cafini, M. I. Morosini, A. Fenoll, J. Linares, L. Alou, D. Sevillano, R. Canton, J. Prieto, and F. Baquero. 2006. Combinations of PBPs and MurM protein variants in early and contemporary high-level penicillin-resistant Streptococcus pneumoniae isolates in Spain. J. Antimicrob. Chemother. 57:983-986. [DOI] [PubMed] [Google Scholar]
- 15.Fenoll, A., L. Aguilar, O. Robledo, M. J. Gimenez, J. J. Granizo, D. Biek, and D. Tarrago. 2008. In vitro activity of ceftaroline against Streptococcus pneumoniae isolates exhibiting resistance to penicillin, amoxicillin, and cefotaxime. Antimicrob. Agents Chemother. 52:4209-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ge, Y., D. Biek, G. H. Talbot, and D. F. Sahm. 2008. In vitro profiling of ceftaroline against a collection of recent bacterial clinical isolates from across the United States. Antimicrob. Agents Chemother. 52:3398-3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Georgopapadakou, N. H., B. A. Dix, and Y. R. Mauriz. 1986. Possible physiological functions of penicillin-binding proteins in Staphylococcus aureus. Antimicrob. Agents Chemother. 29:333-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grebe, T., and R. Hakenbeck. 1996. Penicillin-binding proteins 2b and 2x of Streptococcus pneumoniae are primary resistance determinants for different classes of β-lactam antibiotics. Antimicrob. Agents Chemother. 40:829-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guignard, B., J. M. Entenza, and P. Moreillon. 2005. Beta-lactams against methicillin-resistant Staphylococcus aureus. Curr. Opin. Pharmacol. 5:479-489. [DOI] [PubMed] [Google Scholar]
- 20.Hackbarth, C. J., T. Kocagoz, S. Kocagoz, and H. F. Chambers. 1995. Point mutations in Staphylococcus aureus PBP 2 gene affect penicillin-binding kinetics and are associated with resistance. Antimicrob. Agents Chemother. 39:103-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hakenbeck, R. 1999. Beta-lactam-resistant Streptococcus pneumoniae: epidemiology and evolutionary mechanism. Chemotherapy 45:83-94. [DOI] [PubMed] [Google Scholar]
- 22.Hakenbeck, R. 1998. Mosaic genes and their role in penicillin-resistant Streptococcus pneumoniae. Electrophoresis 19:597-601. [DOI] [PubMed] [Google Scholar]
- 23.Hakenbeck, R., A. Konig, I. Kern, M. van der Linden, W. Keck, D. Billot-Klein, R. Legrand, B. Schoot, and L. Gutmann. 1998. Acquisition of five high-Mr penicillin-binding protein variants during transfer of high-level beta-lactam resistance from Streptococcus mitis to Streptococcus pneumoniae. J. Bacteriol. 180:1831-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hakenbeck, R., S. Tornette, and N. F. Adkinson. 1987. Interaction of non-lytic β-lactams with penicillin-binding proteins in Streptococcus pneumoniae. J. Gen. Microbiol. 133:755-760. [DOI] [PubMed] [Google Scholar]
- 25.Hebeisen, P., I. Heinze-Krauss, P. Angehrn, P. Hohl, M. G. Page, and R. L. Then. 2001. In vitro and in vivo properties of Ro 63-9141, a novel broad-spectrum cephalosporin with activity against methicillin-resistant staphylococci. Antimicrob. Agents Chemother. 45:825-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Higashi, Y., A. Wakabayashi, Y. Matsumoto, Y. Watanabe, and A. Ohno. 1999. Role of inhibition of penicillin binding proteins and cell wall cross-linking by β-lactam antibiotics in low- and high-level methicillin resistance of Staphylococcus aureus. Chemotherapy 45:37-47. [DOI] [PubMed] [Google Scholar]
- 27.Ishikawa, T., N. Matsunaga, H. Tawada, N. Kuroda, Y. Nakayama, Y. Ishibashi, M. Tomimoto, Y. Ikeda, Y. Tagawa, Y. Iizawa, K. Okonogi, S. Hashiguchi, and A. Miyake. 2003. TAK-599, a novel N-phosphono type prodrug of anti-MRSA cephalosporin T-91825: synthesis, physicochemical and pharmacological properties. Bioorg. Med. Chem. 11:2427-2437. [DOI] [PubMed] [Google Scholar]
- 28.Jones, R. N., T. R. Fritsche, H. S. Sader, and J. E. Ross. 2007. LEADER surveillance program results for 2006: an activity and spectrum analysis of linezolid using clinical isolates from the United States (50 medical centers). Diagn. Microbiol. Infect. Dis. 59:309-317. [DOI] [PubMed] [Google Scholar]
- 29.Julian, K., K. Kosowska-Shick, C. Whitener, M. Roos, H. Labischinski, A. Rubio, L. Parent, L. Ednie, L. Koeth, T. Bogdanovich, and P. C. Appelbaum. 2007. Characterization of a daptomycin-nonsusceptible vancomycin-intermediate Staphylococcus aureus strain in a patient with endocarditis. Antimicrob. Agents Chemother. 51:3445-3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katayama, Y., H. Z. Zhang, and H. F. Chambers. 2004. PBP 2a mutations producing very-high-level resistance to β-lactams. Antimicrob. Agents Chemother. 48:453-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kosowska, K., M. R. Jacobs, S. Bajaksouzian, L. Koeth, and P. C. Appelbaum. 2004. Alterations of penicillin-binding proteins 1A, 2X, and 2B in Streptococcus pneumoniae isolates for which amoxicillin MICs are higher than penicillin MICs. Antimicrob. Agents Chemother. 48:4020-4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kosowska-Shick, K., L. M. Ednie, P. McGhee, and P. C. Appelbaum. 2009. Comparative antipneumococcal activities of sulopenem and other drugs. Antimicrob. Agents Chemother. 53:2239-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kosowska-Shick, K., L. M. Ednie, P. McGhee, K. Smith, C. D. Todd, A. Wehler, and P. C. Appelbaum. 2008. Incidence and characteristics of vancomycin nonsusceptible strains of methicillin-resistant Staphylococcus aureus at Hershey Medical Center. Antimicrob. Agents Chemother. 52:4510-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kosowska-Shick, K., P. McGhee, and P. C. Appelbaum. 2009. Binding of faropenem and other β-lactam agents to penicillin-binding proteins of pneumococci with various β-lactam susceptibilities. Antimicrob. Agents Chemother. 53:2176-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laible, G., B. G. Spratt, and R. Hakenbeck. 1991. Interspecies recombinational events during the evolution of altered PBP 2x genes in penicillin-resistant clinical isolates of Streptococcus pneumoniae. Mol. Microbiol. 5:1993-2002. [DOI] [PubMed] [Google Scholar]
- 36.Leski, T. A., and A. Tomasz. 2005. Role of penicillin-binding protein 2 (PBP2) in the antibiotic susceptibility and cell wall cross-linking of Staphylococcus aureus: evidence for the cooperative functioning of PBP2, PBP4, and PBP2A. J. Bacteriol. 187:1815-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lim, D., and N. C. Strynadka. 2002. Structural basis for the β-lactam resistance of PBP2a from methicillin-resistant Staphylococcus aureus. Nat. Struct. Biol. 9:870-876. [DOI] [PubMed] [Google Scholar]
- 38.Mainardi, J. L., D. M. Shlaes, R. V. Goering, J. H. Shlaes, J. F. Acar, and F. W. Goldstein. 1995. Decreased teicoplanin susceptibility of methicillin-resistant strains of Staphylococcus aureus. J. Infect. Dis. 171:1646-1650. [DOI] [PubMed] [Google Scholar]
- 39.Malouin, F., J. Blais, S. Chamberland, M. Hoang, C. Park, C. Chan, K. Mathias, S. Hakem, K. Dupree, E. Liu, T. Nguyen, and M. N. Dudley. 2003. RWJ-54428 (MC-02,479), a new cephalosporin with high affinity for penicillin-binding proteins, including PBP 2a, and stability to staphylococcal β-lactamases. Antimicrob. Agents Chemother. 47:658-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McGee, L., D. Biek, Y. Ge, M. Klugman, M. du Plessis, A. M. Smith, B. Beall, C. G. Whitney, and K. P. Klugman. 2009. In vitro evaluation of the antimicrobial activity of ceftaroline against cephalosporin-resistant isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 53:552-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Memmi, G., S. R. Filipe, M. G. Pinho, Z. Fu, and A. Cheung. 2008. Staphylococcus aureus PBP4 is essential for β-lactam resistance in community-acquired methicillin-resistant strains. Antimicrob. Agents Chemother. 52:3955-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moisan, H., M. Pruneau, and F. Malouin. 22 January 2010, posting date. Binding of ceftaroline to penicillin-binding proteins of Staphylococcus aureus and Streptococcus pneumoniae. J. Antimicrob. Chemother. [Epub ahead of print.] doi: 10.1093/jac/dkp503. [DOI] [PubMed]
- 43.Morrissey, I., Y. Ge, and R. Janes. 2009. Activity of the new cephalosporin ceftaroline against bacteraemia isolates from patients with community-acquired pneumonia. Int. J. Antimicrob. Agents 33:515-519. [DOI] [PubMed] [Google Scholar]
- 44.Munoz, R., C. G. Dowson, M. Daniels, T. J. Coffey, C. Martin, R. Hakenbeck, and B. G. Spratt. 1992. Genetics of resistance to third-generation cephalosporins in clinical isolates of Streptococcus pneumoniae. Mol. Microbiol. 6:2461-2465. [DOI] [PubMed] [Google Scholar]
- 45.Mushtaq, S., M. Warner, Y. Ge, K. Kaniga, and D. M. Livermore. 2007. In vitro activity of ceftaroline (PPI-0903M, T-91825) against bacteria with defined resistance mechanisms and phenotypes. J. Antimicrob. Chemother. 60:300-311. [DOI] [PubMed] [Google Scholar]
- 46.Nagai, K., T. A. Davies, M. R. Jacobs, and P. C. Appelbaum. 2002. Effects of amino acid alterations in penicillin-binding proteins (PBPs) 1a, 2b, and 2x on PBP affinities of penicillin, ampicillin, amoxicillin, cefditoren, cefuroxime, cefprozil, and cefaclor in 18 clinical isolates of penicillin-susceptible, -intermediate, and -resistant pneumococci. Antimicrob. Agents Chemother. 46:1273-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okonogi, K., Y. Noji, M. Nakao, and A. Imada. 1995. The possible physiological roles of penicillin-binding proteins of methicillin-susceptible and methicillin-resistant Staphylococcus aureus. J. Infect. Chemother. 1:50-58. [Google Scholar]
- 48.Okonogi, K., Y. Noji, M. Kondo, A. Imada, and T. Yokota. 1989. Emergence of methicillin-resistant clones from cephamycin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 24:637-645. [DOI] [PubMed] [Google Scholar]
- 49.Parvez, M. A., H. Shibata, T. Nakano, S. Niimi, N. Fujii, N. Arakaki, and T. Higuti. 2008. No relationship exists between PBP 2a amounts expressed in different MRSA strains obtained clinically and their β-lactam MIC values. J. Med. Invest. 55:246-253. [DOI] [PubMed] [Google Scholar]
- 50.Pereira, S. F. F., A. O. Henriques, M. G. Pinho, H. de Lencastre, and A. Tomasz. 2007. Role of PBP1 in cell division of Staphylococcus aureus. J. Bacteriol. 189:3525-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pinho, M. G., H. de Lencastre, and A. Tomasz. 2001. An acquired and a native penicillin-binding protein cooperate in building the cell wall of drug-resistant staphylococci. Proc. Natl. Acad. Sci. U. S. A. 98:10886-10891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pucci, M. J., and T. J. Dougherty. 2008. A method to assay penicillin-binding proteins. Methods Mol. Med. 142:131-141. [DOI] [PubMed] [Google Scholar]
- 53.Regev-Yochay, G., K. Trzcinski, C. M. Thompson, M. Lipsitch, and R. Malley. 2007. SpxB is a suicide gene of Streptococcus pneumoniae and confers a selective advantage in an in vivo competitive colonization model. J. Bacteriol. 189:6532-6539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanbongi, Y., T. Ida, M. Ishikawa, Y. Osaki, H. Kataoka, T. Suzuki, K. Kondo, F. Ohsawa, and M. Yonezawa. 2004. Complete sequences of six penicillin-binding protein genes from 40 Streptococcus pneumoniae clinical isolates collected in Japan. Antimicrob. Agents Chemother. 48:2244-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sieradzki, K., M. G. Pinho, and A. Tomasz. 1999. Inactivated pbp4 in highly glycopeptide-resistant laboratory mutants of Staphylococcus aureus. J. Biol. Chem. 274:18942-18946. [DOI] [PubMed] [Google Scholar]
- 56.Sifaoui, F., M. D. Kitzis, and L. Gutmann. 1996. In vitro selection of one-step mutants of Streptococcus pneumoniae resistant to different oral β-lactam antibiotics is associated with alterations of PBP2x. Antimicrob. Agents Chemother. 40:152-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith, A. M., and K. P. Klugman. 1995. Alterations in penicillin-binding protein 2B from penicillin-resistant wild-type strains of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 39:859-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Song, M. D., M. Wachi, M. Doi, F. Ishino, and M. Matsuhashi. 1987. Evolution of an inducible penicillin-target protein in methicillin-resistant Staphylococcus aureus by gene fusion. FEBS Lett. 221:167-171. [DOI] [PubMed] [Google Scholar]
- 59.Spratt, B. G. 1994. Resistance to antibiotics mediated by target alterations. Science 264:388-393. [DOI] [PubMed] [Google Scholar]
- 60.Utsui, Y., and T. Yokota. 1985. Role of an altered penicillin-binding protein in methicillin- and cephem-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 28:397-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Villegas-Estrada, A., M. Lee, D. Hesek, S. B. Vakulenko, and S. Mobashery. 2008. Co-opting the cell wall in fighting methicillin-resistant Staphylococcus aureus: potent inhibition of PBP 2a by two anti-MRSA β-lactam antibiotics. J. Am. Chem. Soc. 130:9212-9213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Williamson, R., R. Hakenbeck, and A. Tomasz. 1980. In vivo interaction of β-lactam antibiotics with the penicillin-binding proteins of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 18:629-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zerfass, I., R. Hakenbeck, and D. Denapaite. 2009. An important site in PBP2x of penicillin-resistant clinical isolates of Streptococcus pneumoniae: mutational analysis of Thr338. Antimicrob. Agents Chemother. 53:1107-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhanel, G. G., G. Sniezek, F. Schweizer, S. Zelenitsky, P. R. Lagace-Wiens, E. Rubinstein, A. S. Gin, D. J. Hoban, and J. A. Karlowsky. 2009. Ceftaroline: a novel broad-spectrum cephalosporin with activity against methicillin-resistant Staphylococcus aureus. Drugs 69:809-831. [DOI] [PubMed] [Google Scholar]
- 65.Zhao, G., T. I. Meier, S. D. Kahl, K. R. Gee, and L. C. Blaszczak. 1999. BOCILLIN FL, a sensitive and commercially available reagent for detection of penicillin-binding proteins. Antimicrob. Agents Chemother. 43:1124-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]