Abstract
2-Aminoimidazoles are an emerging class of small molecules that possess the ability to inhibit and disperse biofilms across bacterial order, class, and phylum. Herein, we report the synergistic effect between a 2-aminoimidazole/triazole conjugate and antibiotics toward dispersing preestablished biofilms, culminating with a 3-orders-of-magnitude increase of biofilm dispersion toward Staphylococcus aureus biofilms. Furthermore, we document that the 2-aminoimidazole/triazole conjugate will also resensitize multidrug-resistant strains of bacteria to the effects of conventional antibiotics, including methicillin-resistant Staphylococcus aureus (MRSA) and multidrug-resistant Acinetobacter baumannii.
Bacterial biofilms and antibiotic resistance genes represent a tremendous hurdle in human health care. First, the NIH estimates that 3 in 4 bacterial infections are biofilm based (20). Bacteria within a biofilm are upwards of 1,000-fold more resistant to antibiotics and are inherently insensitive to the host immune response (20). Thus, bacteria in a biofilm represent a significant hurdle for antibiotic treatment. Second, the dissemination of antibiotic resistance genes among diverse pathogenic bacteria, coupled with the dearth of new antibiotics that have been introduced by the biomedical community, has led to a situation in which many microbial infections are multidrug resistant and extremely difficult or impossible to treat. Examples of this include the outbreaks of methicillin-resistant Staphylococcus aureus (MRSA) infections among healthy individuals (16) and the multidrug-resistant strains of Acinetobacter baumannii (MDRAB) (22) that are infecting wounded soldiers in the Middle East. In fact, more individuals die every year in the United States from complications arising from MRSA infections than from complications related to HIV (15). The lack of new strategies to combat bacterial infections has become so dire that the Infectious Diseases Society of America has recently issued a call to action for the medical community (34).
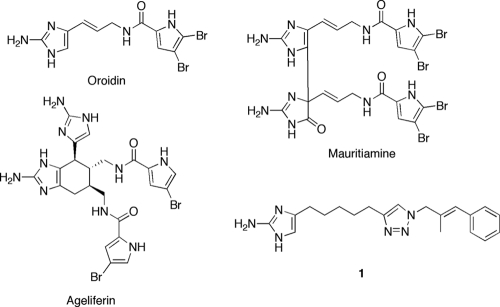
Derivatives of 2-aminoimidazoles have recently been developed as molecules that both inhibit and disperse bacterial biofilms (28). Examples of natural products in this class include oroidin, ageliferin, and mauritiamine (1) (Fig. 1), and these naturally occurring secondary metabolites have been reported to have either antibiofilm or antifouling activity (14, 35, 37). Recent efforts in our lab have provided a number of synthetically accessible 2-aminoimidazole derivatives that both inhibit and disperse bacterial biofilms (3, 4, 9, 10, 11, 18, 24-27, 29, 31-33). One class of derivatives recently reported was of 2-aminoimidazole/triazole (2-AIT) conjugates (33). The most active molecule identified in this study was compound 1, and we documented that compound 1 inhibits and disperses biofilms of Pseudomonas aeruginosa, Acinetobacter baumannii, Bordetella bronchiseptica, and Staphylococcus aureus. Growth curves and colony count assays indicated that compound 1 inhibited and dispersed bacterial biofilms without inducing cellular death.
FIG. 1.
2-Aminoimidazoles that have antibiofilm and antibiofouling properties.
As a therapeutic strategy, molecules that control bacterial biofilms through nonmicrobicidal mechanisms will most likely act synergistically with conventional antibiotics, cooperating advantageously to overcome an infectious threat that would otherwise persist if treated with either agent individually (36). The antibiofilm agent will maintain bacteria within their sensitive planktonic state, while the antibiotic will eliminate the bacterial population. Given this situation, it is important to determine how conventional antibiotics affect the ability of antibiofilm agents to control biofilm development and maintenance. To this end, we have studied the ability of 2-AIT compound 1 to disperse bacterial biofilms in the presence of approved antibiotics. As a follow-up study, we have investigated the ability of antibiofilm agents to affect the action of antibiotics on planktonic bacteria. Herein we report that antibiotics work synergistically with compound 1 to eliminate biofilm colonization, that compound 1 augments the action of conventional antibiotics and effectively suppresses multidrug resistance, and that compound 1 is not hemolytic at active concentrations.
MATERIALS AND METHODS
General experimental materials.
A. baumannii (ATCC 19606), S. aureus (ATCC 29213), Staphylococcus epidermidis (ATCC 29886), MRSA (ATCC BAA-44), MDRAB (ATCC BAA-1605), and a control strain of Escherichia coli (ATCC 35695) were obtained from the ATCC. P. aeruginosa strains PA14 and PDO300 were provided by Daniel J. Wozniak at the Department of Microbiology and Immunology, Wake Forest University School of Medicine. Chloramphenicol- and ampicillin-resistant E. coli was provided by Reza A. Ghiladi at North Carolina State University. Tetracycline-resistant E. coli K-12 ER2738 was purchased from New England Biolabs. Tetracycline hydrochloride (catalog number 0446012) and ampicillin sodium salt (catalog number BP1760) were purchased from Fisher Scientific. Penicillin G sodium salt (catalog number P3032), tobramycin (catalog number T4014), gentamicin sulfate salt (catalog number G4793), and erythromycin (catalog number 45673) were purchased from Sigma-Aldrich. Chloramphenicol (catalog number C0310) was purchased from Teknova. Mechanically defibrinated sheep blood (DSB100) was obtained from Hemostat Labs. MDRAB clinical isolates 3340, AB0043, and UH8407 were donated by Robert A. Bonomo. Mueller-Hinton medium was purchased from Fluka (catalog number 70192).
Biofilm dispersion with an antibiotic or a combination of antibiotic and antibiofilm agent.
Dispersion assays were performed by taking an overnight culture of bacterial strain and subculturing it at an optical density at 600 nm (OD600) of 0.01 into the necessary medium (LB for A. baumannii, LB without NaCl [LBNS] for PA14 and PDO300, tryptic soy broth with a 0.5% glucose supplement [TSBG] for S. aureus, and tryptic soy broth with a 0.5% glucose supplement and a 3% NaCl supplement [TGN] for S. epidermidis). The resulting bacterial suspension was aliquoted (100 μl) into the wells of a 96-well PVC microtiter plate. Plates were then wrapped in Glad Press'n Seal wrap, followed by an incubation under stationary conditions at ambient temperature to establish the biofilms. After 24 h, the media were discarded from the wells and the plates were washed thoroughly with water. Stock solutions of the chosen antibiotic (tobramycin for PA14 and PDO300, colistin for A. baumannii, and novobiocin for S. epidermidis and S. aureus) with or without compound 1 were then made in media (the antibiotic test concentration ranged initially from 0.001 μM to 100 μM to determine the highest concentration of antibiotic that would not disperse the preformed biofilm so it could be used in a combination study). These stock solutions were aliquoted (100 μl) into the wells of the 96-well PVC microtiter plate with the established biofilms. Medium alone was added to a subset of the wells to serve as a control. Plates were then incubated for 24 h at 37°C. After incubation, the media were discarded from the wells and the plates were washed thoroughly with water. Plate wells were then stained with 100 μl of a 0.1% solution of crystal violet (CV) and then incubated at ambient temperature for 30 min. Plates were washed with water again, and the remaining stain was solubilized with 200 μl of 95% ethanol. A 125-μl portion of the solubilized CV stain from each well was transferred to the corresponding wells of a polystyrene microtiter dish. Biofilm dispersion was quantitated by measuring the OD540 of each well (33).
Quantification of the resensitization effects of compound 1 toward tetracycline-resistant E. coli and MRSA.
An overnight culture of the bacterial strain was subcultured to an OD600 of 0.01 into the necessary medium (LB for E. coli and TSB for MRSA). The resulting bacterial suspension was then aliquoted (3.0 ml) into culture tubes. The sensitizing agent was then added at a nonbactericidal concentration (150.0 μM for E. coli and 45.0 μM for MRSA) to the media of the test samples. Then, the choice antibiotic (tetracycline for tetracycline-resistant E. coli and erythromycin, gentamicin, penicillin G, and tetracycline for MRSA) was added to the test samples at nonbactericidal concentrations. Control experiments were conducted by growing bacteria with either (i) medium and sensitizing agent (no antibiotic), (ii) medium only (no antibiotic/sensitizing agent), or (iii) medium and dimethyl sulfoxide (DMSO) (amount used to administer sensitizing agent). Samples were then placed in an incubator at 37°C and shaken at 200 rpm. When the OD600 of the control samples reached approximately 0.4, 100 μl was taken from each culture tube and then diluted serially into LB medium. Then, 10 μl was removed from each serial dilution and plated out on a square-gridded petri dish, followed by 16 h of incubation at 37°C to grow viable colonies, which were quantified by the track-dilution method (6, 13).
Broth microdilution method for MIC determination.
Overnight cultures of bacterial strain were subcultured to 5 × 105 CFU/ml in Mueller-Hinton medium (Fluka number 70192). The resulting bacterial suspension was aliquoted (1.0 ml) into culture tubes. Samples were prepared from these culture tubes containing either 512 μg/ml of specified antibiotic, 512 μg/ml of specified antibiotic with a nonbactericidal concentration of compound 1, 512 μg/ml of compound 1, or no test compound as a control. Samples were then aliquoted (200 μl) into the first row of wells of a 96-well microtiter plate in which subsequent wells were prefilled with 100 μl of a Mueller-Hinton medium-based 5 × 105-CFU/ml bacterial subculture (samples including compound 1 as a supplement to an antibiotic included the nonbactericidal concentration of compound 1 in this stock to keep the compound 1 concentration uniform throughout the antibiotic dilution procedure). Using the multichannel pipettor set at 100 μl, row 1 wells were mixed 8 to 10 times. Then, 100 μl was withdrawn and transferred to row 2. Row 2 wells were mixed 8 to 10 times, followed by a 100-μl transfer from row 2 to row 3. This procedure was used to serially dilute the rest of the rows of the microtiter plate. The microtiter plate sample was then covered with a microtiter plate lid and placed in a covered plastic container. The chamber was incubated under stationary conditions at 37°C. After 16 h, the lid was removed and MIC values were recorded (6).
Broth microdilution method for the determination of Ca2+, Mn2+, and Mg2+ doping on the antibiotic resensitization activity of compound 1.
Cultures were grown in the presence or absence of 25 mg/ml CaCl2, MnCl2, MgSO4, or 30.6 mg/ml CaSO4 for 10 to 12 h. The cultures were then subcultured to 5 × 105 CFU/ml in Mueller-Hinton medium (Fluka number 70192) containing their respective metal ions. The resulting bacterial suspension was aliquoted (1.0 ml) into culture tubes. Samples were prepared from these culture tubes containing either 512 μg/ml of specified antibiotic, 512 μg/ml of specified antibiotic with a nonbactericidal concentration of compound 1, 512 μg/ml of compound 1, or no test compound as a control (samples treated with compound 1 were allowed to stand for 30 min before the antibiotic was introduced). Samples were then aliquoted (200 μl) into the first row of wells of a 96-well microtiter plate in which subsequent wells were prefilled with 100 μl of a Mueller-Hinton medium-based 5 × 105-CFU/ml bacterial subculture. (Samples including compound 1 as a supplement to an antibiotic included the nonbactericidal concentration of compound 1 in this stock to keep the compound 1 concentration uniform throughout the antibiotic dilution procedure, and samples supplemented with 25 mg/ml CaCl2, MnCl2, MgSO4, or 30.6 mg/ml CaSO4 included the test concentration of these salts in the stock to keep their concentrations uniform throughout the antibiotic dilution procedure.) Using the multichannel pipettor set at 100 μl, row 1 wells were mixed 8 to 10 times. Then, 100 μl was withdrawn and transferred to row 2. Row 2 wells were mixed 8 to 10 times, followed by a 100-μl transfer from row 2 to row 3. This procedure was used to serially dilute the rest of the rows of the microtiter plate. The microtiter plate sample was then covered with a microtiter plate lid and placed in a covered plastic container. The chamber was incubated under stationary conditions at 37°C. After 16 h, the lid was removed and MIC values were recorded (6).
Microbicidal activity of compound 1 against planktonic MRSA and MDRAB.
Portions (10 μl) were taken from individual microtiter wells from untreated MRSA and MDRAB wells and compound 1 (32 μg/ml)-treated MRSA and MDRAB wells after the 16-hour incubation time from the previously described microdilution susceptibility testing assay. Serial dilutions were then made with the aliquots from the microtiter wells, plated out on tryptic soy agar plates, and incubated overnight. The colonies that formed were then counted and used to determine CFU ml−1. At 32 μg/ml (82.9 μM), compound 1 did not reduce the number of viable bacterial cells of MRSA and MDRAB in comparison to the nontreated controls.
Evolution of MRSA resistance to the effects of compound 1.
An overnight culture of the bacterial strain was subcultured to an OD600 of 0.01 in TSB medium. The resulting bacterial suspension was then aliquoted (3.0 ml) into culture tubes. A sublethal combination of antibiotic and compound 1 (25.0 μM penicillin G with 45.0 μM compound 1) was then added. Samples were then placed in an incubator at 37°C and shaken at 200 rpm for 24 h. Then, samples were subcultured at an OD600 of 0.01 into the necessary medium. Aliquots (3.0 ml) of these subcultures were then treated again with an identical amount of the antibiotic-compound 1 combination followed by an incubation of the samples at 37°C while being shaken at 200 rpm for 24 h. This antibiotic-compound 1 cycle was performed a total of seven times on the bacteria (within one week). After the final incubation, samples were subcultured at an OD600 of 0.01 into the necessary medium. The resulting bacterial suspension was then aliquoted (3.0 ml) into culture tubes. Then, an active dose (50.0 μM penicillin G with 45.0 μM compound 1) was administered, followed by an incubation of the samples at 37°C while being shaken at 200 rpm. Controls were employed in which no antibiotic was administered, as well as antibiotic by itself (50.0 μM penicillin G for MRSA). When the OD600 of the control samples reached approximately 1.2, 100 μl was taken from each culture tube and then diluted serially into the necessary medium. Then, 10 μl was removed from each serial dilution and plated out on a square-gridded petri dish, followed by 16 h of incubation at 37°C to grow viable colonies, which were quantified by the track-dilution method (13).
Red blood cell hemolysis assay.
Hemolysis assays were performed on mechanically defibrinated sheep blood (DSB100; Hemostat Labs). Blood (1.5 ml) was placed into a microcentrifuge tube and centrifuged at 10,000 rpm for 10 min. The supernatant was removed, and then the cells were resuspended with 1 ml of phosphate-buffered saline (PBS). The suspension was centrifuged, the supernatant was removed, and cells were resuspended two more times. The final cell suspension was then diluted 10-fold. Test compound solutions were made in PBS and then added to aliquots of the 10-fold suspension dilution. PBS alone was used as a negative control and as the zero hemolysis marker, whereas a 1% Triton X sample was used as a positive control and as the 100% lysis marker. Samples were then placed in an incubator at 37°C while being shaken at 200 rpm for 1 h. After 1 h, the samples were transferred to microcentrifuge tubes and then centrifuged at 10,000 rpm for 10 min. The resulting supernatant was diluted by a factor of 40 in distilled water. The absorbance of the supernatant was measured with a UV spectrometer at a 540-nm wavelength (17).
RESULTS AND DISCUSSION
The first question addressed was whether tandem addition of compound 1 with conventional antibiotics would show synergistic effects toward biofilm dispersion. It is known that sublethal doses of antibiotics will promote biofilm formation (8), and it was unclear if the effect of an antibiofilm agent would be mitigated under these conditions. To this end, the synergistic effect of compound 1 was studied with colistin against A. baumannii biofilms, with novobiocin against S. aureus and S. epidermidis, and with tobramycin against two strains of P. aeruginosa (PA14 and PDO300). Tobramycin was used because it is currently prescribed to cystic fibrosis patients to slow P. aeruginosa infections (19). Colistin was investigated because it is currently the antibiotic of last resort used for treatment of multidrug-resistant Acinetobacter baumannii (MDRAB) (7). Novobiocin was investigated because it has been used to treat S. aureus and S. epidermidis infections of indwelling medical devices (23).
To study synergistic effects, biofilms were first established in 96-well PVC plates for 24 h. Media and planktonic bacteria were then removed, and the wells were washed to remove any loosely adherent bacteria. Wells were then treated with either media alone, media containing antibiotic, media containing compound 1, or media containing a combination of compound 1 and the antibiotic. Biofilm mass was then quantified by using a crystal violet reporter assay (21). To quantify any synergistic effects between compound 1 and the antibiotic toward eliminating biofilm colonization, dose-response studies were performed to determine the EC50 of compound 1 as a function of antibiotic concentration. This value was then compared to the EC50 of compound 1 alone. Here, EC50 is defined as the concentration necessary to elicit 50% dispersion of the biofilm.
As expected, the antibiotics alone did not affect biofilm mass at all levels studied (the highest concentrations being 10 μM tobramycin for PDO300 and PA14, 1.0 μM each novobiocin and colistin for S. epidermidis and A. baumannii, respectively, and 0.1 μM novobiocin for S. aureus) compared to samples treated with medium only. For each bacterial strain, we observed synergistic activity between the antibiotic and compound 1 that is dependent on antibiotic concentration. These data are outlined in Table 1. As can be seen, dramatic effects are observed toward the clearance of Staphylococcus biofilms and, for PDO300, a mucoid variant of P. aeruginosa that is relevant for cystic fibrosis. For S. aureus, an increase in activity of 3 orders of magnitude was observed at the highest concentration of novobiocin used (EC50 of compound 1 alone = 2.6 μM, EC50 of compound 1 = 1.0 nM with 0.1. μM novobiocin), while for S. epidermidis, we observed picomolar levels of dispersion (EC50 of compound 1 alone = 395 nM, EC50 of compound 1 with 1.0 μM novobiocin = 670 pM). We observed moderate levels of synergy (ca. 5- to 6-fold) with the highest concentrations of antibiotics for PA14 and A. baumannii.
TABLE 1.
Dispersal enhancement
| Bacterial strain | EC50 | Antibiotic | Antibiotic concn | Combined EC50 |
|---|---|---|---|---|
| S. epidermidis | 325 ± 26 nM | Novobiocin | 1.0 μM | 0.67 ± 0.11 nM |
| A. baumannii | 121 ± 11 μM | Colistin | 1.0 μM | 24.8 ± 1.6 μM |
| P. aeruginosa PDO300 | 13.2 ± 1.1 μM | Tobramycin | 10.0 μM | 0.0077 ± 0.0015 μM |
| S. aureus | 2.6 ± 0.6 μM | Novobiocin | 0.1 μM | 0.001 ± 0.00011 μM |
| P. aeruginosa PA14 | 22 ± 4.5 μM | Tobramycin | 10.0 μM | 4.72 ± 0.5 μM |
After establishing that conventional antibiotics would enhance the antibiofilm properties of compound 1, we turned to investigating the effects that compound 1 in combination with antibiotics had on the growth of multidrug-resistant strains of bacteria. There are a limited number of reports documenting that molecules that inhibit bacterial communication can suppress antibiotic resistance (38). Given that (i) the formation and dispersion of bacterial biofilms are mediated by cell-to-cell communication (5) and (ii) compound 1 inhibits and disperses bacterial biofilms from both Gram-negative and Gram-positive bacteria, we hypothesized that compound 1 may be able to resensitize antibiotic-resistant bacteria (both Gram positive and Gram negative) to the effects of antibiotics. This effect was explored using two strains of antibiotic-resistant E. coli (tetracycline resistant and chloramphenicol/tetracycline resistant), a MRSA clinical isolate, and multidrug-resistant A. baumannii (MDRAB) clinical isolates. Two tests were performed to quantify this effect. The first test was measurement of viable colonies after 4 h of growth, while the second test involved determination of MIC values.
Tetracycline-resistant E. coli grew identically in the absence or presence of 10 or 50 μM tetracycline. Growth of tetracycline-resistant E. coli in the presence of 10 or 50 μM tetracycline and 150 μM compound 1 led to a >99% reduction in bacterial growth (Fig. 2A and summarized in Table 2). In comparison, growth of an E. coli strain sensitive to tetracycline was reduced by only 55% with 10 μM tetracycline. We also observed >99% reduction in bacterial growth with 50 μM tetracycline and either 50 μM or 25 μM compound 1. No reduction in bacterial growth was noted when the bacterium was grown solely in the presence of 150 μM compound 1, confirming the absence of microbicidal activity for compound 1 and synergistic activity between compound 1 and the antibiotic.
FIG. 2.
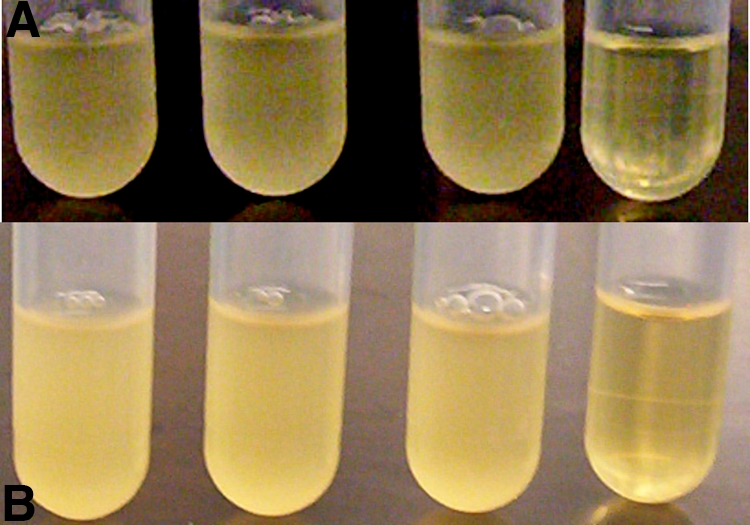
(Top) Tetracycline-resistant E. coli. From left to right: E. coli control, E. coli with 50 μM tetracycline, E. coli with 150 μM compound 1, E. coli with 50 μM tetracycline and 150 μM compound 1. (Bottom) MRSA strain BAA-44. From left to right: MRSA control, MRSA with 25 μM methicillin, MRSA with 45 μM compound 1, MRSA with 25 μM methicillin and 45 μM compound 1.
TABLE 2.
Colony count measurement of antibiotic sensitization
| Bacterial strain | Antibiotic | Concn of antibiotic (μM) | Growth reduction (%) | Concn of compound 1 (μM) added | Growth reduction (%) with antibiotic and compound 1 |
|---|---|---|---|---|---|
| MRSA | Gentamicin | 150.0 | 45.0 | 86.3 ± 0.4 | |
| MRSA | Erythromycin | 100.0 | 45.0 | 81.9 ± 2.3 | |
| MRSA | Penicillin | 25.0 | 45.0 | 94.8 ± 1.0 | |
| MRSA | Methicillin | 25.0 | 45.0 | 90.5 ± 1.7 | |
| MRSA | Tetracycline | 1.0 | 45.0 | 69.2 ± 2.3 | |
| S. aureus | Gentamicin | 150 | 99.8 ± 0.1 | ||
| S. aureus | Erythromycin | 100.0 | 89.3 ± 0.9 | ||
| S. aureus | Penicillin | 25.0 | 90.6 ± 2.7 | ||
| S. aureus | Methicillin | 25.0 | 99.9 ± 0.1 | ||
| E. coli (Tetr) | Tetracycline | 50.0 | 150.0 | 99.9 ± 0.01 | |
| E. coli (Tetr) | Tetracycline | 10.0 | 150.0 | 99.9 ± 0.1 | |
| E. coli | Tetracycline | 10.0 | 55.3 ± 3.7 |
Next, we investigated the ability of compound 1 to resensitize a multidrug-resistant strain of MRSA (BAA-44). BAA-44 grew identically in the absence or presence of the following antibiotics: (i) 150 μM gentamicin, (ii) 100 μM erythromycin, (iii) 25 μM penicillin G, (iv) 25 μM methicillin, and (v) 1 μM tetracycline. BAA-44 grew identically in the presence or absence of 45 μM compound 1 (Table 2). However, combining each antibiotic individually with bacteria preexposed to compound 1 caused a dramatic reduction in bacterial growth in each case. The combination of 150 μM gentamicin-45 μM compound 1 caused 86% reduction in BAA-44 growth, while 100 μM erythromycin-45 μM compound 1 caused 82% reduction in BAA-44 growth. In comparison, growth of antibiotic-sensitive S. aureus is reduced by >99% or 89% in the presence of 150 μM gentamicin or 100 μM erythromycin, respectively. In the presence of 45 μM compound 1 and either 25 μM penicillin G, 25 μM methicillin, or 1 μM tetracycline, BAA-44 growth was reduced 95%, 91%, and 69%, respectively (Fig. 2B). Growth of antibiotic-sensitive S. aureus was reduced by 91%, >99%, and 45% in the presence of 25 μM penicillin G, 25 μM methicillin, and 1 μM tetracycline, respectively.
After establishing that a combination of compound 1 with conventional antibiotics would affect bacterial growth, we established the MIC values of each antibiotic in the absence or pretreated presence of compound 1 against all of the aforementioned bacterial strains. The MIC measurement differs from the enumeration of viable bacteria used above in two key aspects. The first is the time frame of the assay (4 h versus 16 to 24 h), and the second is the stringency of the assay. The bacterial growth assay is a measurement of how the bacteria respond under given conditions, while the MIC assay reports the concentration at which a given treatment causes no visible bacterial growth and is the accepted standard for defining the in vitro efficacy of a potential treatment approach.
MIC values for each bacterial strain were first established for compound 1 and then for each antibiotic separately studied using the microdilution protocol (6). These results are summarized in Table 3. The MIC values determined with each antibiotic against each strain were in agreement with established MIC values (6). The MIC value of compound 1 against the MRSA strain was determined to be 64 μg/ml, while the MIC value of compound 1 against the A. baumannii strains was determined to be 64 μg/ml. MIC values of each antibiotic were then determined in the presence of compound 1. For the A. baumannii strains we tested 75 μM compound 1, while we tested 45 μM compound 1 for MRSA. Against both A. baumannii and MRSA, 82.5 μM compound 1 demonstrated no bactericidal activity. By screening at concentrations of compound 1 that did not elicit any measurable effect on bacterial growth, any reduction in MIC values could be attributed to resensitization activity. MRSA and MDRAB were chosen specifically for MIC studies because they are primary clinical isolates and thus are representative pathogens encountered in a hospital setting.
TABLE 3.
MIC measurement of antibiotic sensitization
| Organism and antibiotic | MIC μg/ml of: |
||
|---|---|---|---|
| Drug alone | Drug with 45 μMa of compound 1 | Drug with 75 μMa of compound 1 | |
| MRSA | |||
| Penicillin G | 32 | 4 | |
| Tetracycline | 16 | 16 | |
| Methicillin | 256 | 64 | |
| Ciprofloxacin | 4 | 2 | |
| Compound 1 | 64 | ||
| MDRAB | |||
| Imipenem | 16 | 2 | |
| Ciprofloxacin | 64 | 16 | |
| MDRAB 3340, imipenem | 64 | 8 | |
| MDRAB AB0043, imipenem | 16 | 4 | |
| MDRAB UH8407, imipenem | 1 | 0.125 | |
Compound 1 at 32 μg/ml (83 μM) was determined to be nontoxic to planktonic MRSA and MDRAB via colony count analysis.
The results of these experiments for MRSA are summarized in Table 3. An 8-fold (32 μg/ml to 4 μg/ml) drop and a 4-fold (256 μg/ml to 64 μg/ml) drop in MIC were noted with penicillin G and methicillin, respectively, in combination when the bacteria were pretreated with 45 μM compound 1. In comparison, the MIC values for β-lactam-sensitive S. aureus were 2 μg/ml with methicillin and 1 μg/ml with penicillin G. This effect was not noted (where we define the difference in MIC value to be 2-fold or less) for tetracycline, with which the MIC remained at 16 μg/ml, or for ciprofloxacin, with which the MIC was reduced only from 4 μg/ml to 2 μg/ml.
For the MDRAB strain obtained from the ATCC (BAA-1605), the MIC of imipenem was reduced from 16 μg/ml to 2 μg/ml after pretreatment of the bacteria with 75 μM compound 1. A similar effect was noted for ciprofloxacin, with which the MIC was reduced from 64 μg/ml to 16 μg/ml after pretreatment with 75 μM compound 1. For antibiotic-sensitive A. baumannii, the MICs were determined as 0.25 μg/ml (imipenem) and 0.5 μg/ml (ciprofloxacin).
Three additional MDRAB clinical isolate strains (AB0043, UH8407, and 3340) were then tested for resensitization effects. The most well-characterized clinical specimen in this study was AB0043, an isolate from a patient suffering from an A. baumannii infection at Walter Reed Army Medical Center. This bacterial strain was isolated in 2004 from the blood of a soldier that was deployed to Iraq/Kuwait and was found to be resistant to seven of the nine antibiotics used in the study to determine its MDR phenotype. Genetic analysis showed that this isolate demonstrated enhanced efflux pump activity via adeR expression, which typically makes isolates resistant to aminoglycosides, quinolones, tetracycline, and trimethoprim. Several aminoglycoside-modifying enzymes (AMEs) were expressed in this clinical isolate as well. Isolate AB0043 was found to express the AME gene aadB, which was often associated with tobramycin resistance of A. baumannii clinical isolates (12).
For MDRAB clinical isolate 3340 (Table 3), the MIC values of imipenem were reduced after a pretreatment with 75 μM compound 1 (64 μg/ml to 8 μg/ml), while similar effects were also noticed for strain AB0043, with which the MIC values of imipenem dropped after a pretreatment with 75 μM compound 1 (16 μg/ml to 4 μg/ml). Lastly, for clinical isolate UH8407, the MIC values of imipenem also dropped after a pretreatment with 75 μM compound 1 (1 μg/ml to 0.125 μg/ml). No significant reduction (>2-fold) in MICs of ciprofloxacin was observed.
We also wanted to investigate if the resensitization effect was due directly to maintaining bacteria in their planktonic state (i.e., biofilm inhibition or dispersion) or if it was due to a specific mechanism attributable to compound 1. To address this question, we have recently developed a 2-aminobenzimidazole (2-ABI) (Fig. 3) that inhibits and disperses Gram-positive biofilms (MRSA, S. epidermidis, and vancomycin-resistant Enterococcus faecium) at low micromolar/high nanomolar concentrations through a zinc-dependent mechanism (31). This 2-ABI conjugate was screened at 45 μM in the presence of 25 μM methicillin and was unable to elicit a resensitizing response against MRSA. Therefore, resensitization activity appears to be attributable to a specific mechanism associated with compound 1, not a generic phenomenon of simply inhibiting/dispersing biofilms.
FIG. 3.
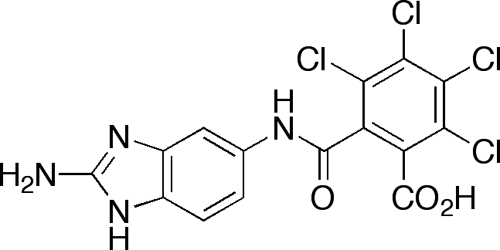
A 2-aminobenzimidazole that inhibits and disperses Gram-positive biofilms through a zinc-dependent mechanism.
We also noticed a time dependence of exposure to compound 1 to the sensitization activity efficacy before the introduction of the antibiotic. This effect was delineated by determining the MIC values of penicillin G against MRSA with both a 30-min and a 60-min exposure to 45 μM compound 1 prior to antibiotic addition. The penicillin G MIC of 45 μM compound 1 with a 30-min exposure was found to be 2 μg/ml, whereas that with a 60-min exposure was 0.25 μg/ml.
Once we had established the ability of compound 1 to resensitize MDR bacteria to the effects of conventional antibiotics, we performed preliminary mechanism of action studies. We noting that switching from standard Mueller-Hinton broth (MHB) prepared in our lab (stock solid purchased from Fluka) to preprepared cation-adjusted Mueller-Hinton broth (CAMHB) purchased from Becton Dickinson significantly suppressed the resensitization activity of compound 1. The difference between the two media is that CAMHB has been supplemented with calcium(II) and magnesium(II) ions to yield media with final concentrations of 20 to 25 mg/liter of Ca(II) and 10 to 12.5 mg/liter of Mg(II), respectively. Therefore, we initially supplemented our MHB with both CaCl2 (an additional 25 mg/liter) and MgSO4 (an additional 25 mg/liter) and evaluated the ability of compound 1 to resensitize MRSA to the effects of penicillin G. In these new media, at 45 μM compound 1, we observed only a modest 2-fold reduction in the MIC of penicillin G (32 μg/ml to 16 μg/ml), indicating that metal cations can suppress the resensitization effects. We then individually supplemented our MHB with either CaCl2 (an additional 25 mg/liter) or MgSO4 (an additional 25 mg/liter). Only the MHB supplemented with CaCl2 suppressed the resensitization activity of compound 1.
Once we had determined that calcium(II) levels modulate the resensitization activity of compound 1, we set up a number of experiments to quantify these effects (Table 4). We first set up resensitization experiments with CaSO4 and penicillin G, MRSA, and compound 1 to determine if the effect was due to Ca(II) or the chloride counterion. CaSO4 modulated the activity of compound 1 in a fashion similar to that of CaCl2, indicating that Ca(II) was responsible for suppressing the resensitization activity of compound 1. For example, in MHB supplemented with 30.7 mg/liter CaSO4 and 45 μM compound 1, the MIC of penicillin G against MRSA was 16 μg/ml. In comparison, the MIC of penicillin G against MRSA was 32 μg/ml in MHB and 4 μg/ml in MHB with 45 μM compound 1. We also reverified that the activity was due to excess calcium by determining the MIC of penicillin G against MRSA in MHB supplemented with 225 μM CaCl2 (25 mg/liter), 225 μM EGTA, and 45 μM compound 1. EGTA is a calcium(II)-specific chelator that will sequester any available Ca(II); thus, if free calcium cations are suppressing the activity of compound 1, the addition of EGTA should allow compound 1 to resensitize MRSA to the effects of antibiotics. The MIC of penicillin G in this medium (MHB, CaCl2, EGTA, compound 1) was 4 μg/ml, identical to the MIC determined in MHB supplemented with 45 μM compound 1. As a control, we determined that the MIC of penicillin G in MHB with 225 μM EGTA was 32 μg/ml, which is identical to the MIC in MHB only.
TABLE 4.
Metal doping effects on the antibiotic resensitization ability of compound 1
| Organism and antibiotic | MIC (μg/ml)a with: |
||||
|---|---|---|---|---|---|
| Mueller-Hinton | CaSO4 | CaCl2 | MgSO4 | MnCl2 | |
| MRSA | |||||
| Penicillin G | 32 | 32 | 32 | 32 | 32 |
| Penicillin G with 45 μM compound 1 | 4 | 16 | 16 | 4 | 16 |
| Penicillin G with 90 μM compound 1 | 0.125 | 2 | 2 | 0.125 | 8 |
| MDRAB | |||||
| Imipenem | 16 | 16 | 16 | 16 | |
| Imipenem with 75 μM compound 1 | 2 | 8 | 2 | 8 | |
| Imipenem with 150 μM compound 1 | 0.125 | 4 | 0.125 | 2 | |
For doping, 25 μg/ml of CaCl2, MgSO4, or MnCl2 or 30.7 μg/ml of CaSO4was used.
In terms of bioactivity, manganese(II) can typically be used as a surrogate for calcium(II) when studying calcium-dependent processes (2). Therefore, we elected to investigate if manganese(II) would also attenuate the activity of compound 1. The MIC of penicillin G against MRSA in MHB supplemented with 25 mg/liter MnCl2 and 45 μM compound 1 was calculated as 16 μg/ml, identical to the result for MHB with CaCl2 and compound 1.
After delineating this metal-dependent activity with MRSA, we explored whether these effects also applied to MDRAB. We performed parallel experiments with one of the A. baumannii isolates (BAA-1605) and observed that the resensitization activity of 75 μM compound 1 was significantly muted in MHB supplemented with an additional 25 mg/liter CaCl2 (8 μg/ml for imipenem) versus that in unsupplemented MHB (2 μg/ml for imipenem). In comparison, the MIC of imipenem against this MDRAB strain in MHB was 32 μg/ml. As with the MRSA strain, Mn(II) also suppressed the ability of compound 1 to resensitize the MDRAB strain to antibiotics. We also noted that increasing the concentration of compound 1 offset the effects of additional Ca(II) for both the MRSA and the MDRAB clone (Table 4), thus indicating that further analogue design to optimize activity can be employed if the activity of compound 1 is significantly impaired in vivo by excess calcium ions.
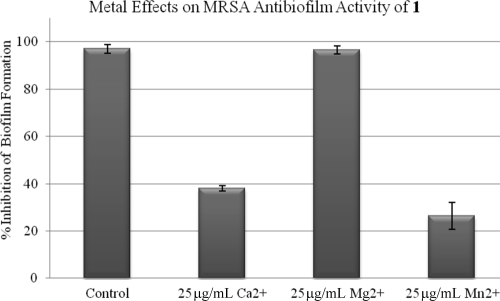
Calcium(II) is also known to have a significant effect on bacterial biofilm formation (2). Therefore, we investigated whether the addition of Ca(II) would mitigate the antibiofilm effects of compound 1 and perhaps indicate that a single bacterial target can be modulated to control both biofilm formation and certain classes of antibiotic resistance (Fig. 4). BAA-44 biofilm formation was completely inhibited by 100 μM compound 1 in tryptic soy broth with a 0.5% glucose supplement (TSBG), while in TSBG supplemented with either 25 mg/liter CaCl2 or 25 mg/liter MnCl2, 100 μM compound 1 inhibited only 40% or 27% of MRSA biofilm formation. Control experiments of biofilms grown with TSBG medium supplemented with either 25 mg/liter CaCl2 or 25 mg/liter MnCl2 showed biofilm masses identical to those of MRSA grown in TSBG only.
FIG. 4.
Metal effects on the ability of compound 1 to inhibit MRSA biofilm formation.
With any given antibacterial strategy, evolution of resistance is always a significant problem. Therefore, we evaluated the ability of MRSA clone BAA-44 to evolve resistance to compound 1. BAA-44 was inoculated into 3 ml of media containing 45 μM compound 1 and 25 μM penicillin G. Bacteria were allowed to grow for 24 h, at which time 100 μl of the bacterial solution was used to inoculate another 3 ml of medium containing 45 μM compound 1 and 25 μM penicillin G. This process was repeated for 1 week. Four-hour growth analysis of the resulting bacterial culture was then monitored in either (i) medium only, (ii) medium containing compound 1, (iii) medium containing penicillin G, (iv) medium containing methicillin, (v) medium containing compound 1 and penicillin G, or (vi) medium containing compound 1 and methicillin. Growth characteristics of the resulting bacterial culture were identical to those of growth previously determined (Table 2). Therefore, selection pressures to develop compound 1 resistance are either fully or significantly repressed.
Finally, red blood cell hemolysis analysis (17) of compound 1 was also performed using defibrinated sheep blood. The HD50 (hemolytic dose that lyses 50% of the red blood cells) was found to be 800 μM. Even more promising is the fact that hemolysis was not detected until after compound 1 reached 400 μM, a concentration that is well above the concentration of compound 1 used to elicit the sensitization response.
In conclusion, we have demonstrated that the combination of an antibiofilm agent containing a 2-aminoimidazole/triazole motif with conventional antibiotics provides an effective strategy for remediating biofilm colonization. Furthermore, we have shown that it is possible to employ the 2-aminoimidazole/triazole conjugate compound 1 as an adjuvant for resensitizing multidrug-resistant bacteria to the effects of conventional antibiotics. Currently, we are further deconvoluting the mechanism of action of compound 1 as it applied to both its antibiofilm activity and its resensitization activity to potentially provide novel therapeutic strategies/targets for the treatment of bacterial infections. Given that 2-aminoimidazoles have been shown to be nontoxic in both cell culture and model organism models of toxicity (9, 30), we are also tuning the molecular architecture of compound 1 to enhance its resensitization activity to evaluate the in vivo potential of 2-aminoimidazoles to serve as adjuvants for the treatment of multidrug-resistant bacterial infections.
Acknowledgments
We thank the UNC General Administration Competitiveness Grant and the V Foundation for funding (S.A.R. and R.W.H. as a predoctoral Jimmy V Scholar).
We also thank Robert Bonomo for providing A. baumannii clinical isolates AB0043, UH8407, and 3340.
Footnotes
Published ahead of print on 8 March 2010.
REFERENCES
- 1.Al Mourabit, A., and P. Potier. 2001. Sponge's molecular diversity through the ambivalent reactivity of 2-aminoimidazole: a universal chemical pathway to the oroidin-based pyrrole-imidazole alkaloids and their palau'amine congeners. Eur. J. Org. Chem. 2001(2):237-243. [Google Scholar]
- 2.Arrizubieta, M. J., A. T. Arana, B. Amorena, J. R. Penades, and I. Lasa. 2004. Calcium inhibits Bap-dependent multicellular behavior in Staphylococcus aureus. J. Bacteriol. 186:7490-7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballard, T. E., J. J. Richards, A. Aquino, C. S. Reed, and C. Melander. 2009. Antibiofilm activity of a diverse oroidin library generated through reductive acylation. J. Org. Chem. 74:1755-1758. [DOI] [PubMed] [Google Scholar]
- 4.Ballard, T. E., J. J. Richards, A. L. Wolfe, and C. Melander. 2008. Synthesis and antibiofilm activity of a second-generation reverse-amide oroidin library: a structure-activity relationship study. Chemistry 14:10745-10761. [DOI] [PubMed] [Google Scholar]
- 5.Camilli, A., and B. L. Bassler. 2006. Bacterial small-molecule signaling pathways. Science 311:1113-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.CLSI. 2009. Performance standards for antimicrobial susceptibility testing; 19th informational supplement, p. 152. Clinical and Laboratory Standards Institute, Wayne, PA.
- 7.Gilad, J., and Y. Carmeli. 2008. Treatment options for multidrug-resistant Acinetobacter species. Drugs 68:165-189. [DOI] [PubMed] [Google Scholar]
- 8.Hoffman, L. R., D. A. D'Argenio, M. J. MacCoss, Z. Zhang, R. A. Jones, and S. I. Miller. 2005. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436:1171-1175. [DOI] [PubMed] [Google Scholar]
- 9.Huigens, R. W., L. Ma, C. Gambino, A. Basso, P. D. R. Moeller, J. Cavanagh, D. J. Wozniak, and C. Melander. 2008. Control of bacterial biofilms with marine alkaloid derivatives. Mol. Biosyst. 4:614-621. [DOI] [PubMed] [Google Scholar]
- 10.Huigens, R. W., III, J. J. Richards, G. Parise, T. E. Ballard, W. Zeng, R. Deora, and C. Melander. 2007. Inhibition of Pseudomonas aeruginosa biofilm formation with bromoageliferin analogues. J. Am. Chem. Soc. 129:6966-6967. [DOI] [PubMed] [Google Scholar]
- 11.Huigens, R. W., III, S. A. Rogers, A. T. Steinhauer, and C. Melander. 2009. Inhibition of Acinetobacter baumannii, Staphylococcus aureus and Pseudomonas aeruginosa biofilm formation with a class of TAGE-triazole conjugates. Org. Biomol. Chem. 7:794-802. [DOI] [PubMed] [Google Scholar]
- 12.Hujer, K. M., A. M. Hujer, E. A. Hulten, S. Bajaksouzian, J. M. Adams, C. J. Donskey, D. J. Ecker, C. Massire, M. W. Eshoo, R. Sampath, J. M. Thomson, P. N. Rather, D. W. Craft, J. T. Fishbain, A. J. Ewell, M. R. Jacobs, D. L. Paterson, and R. A. Bonomo. 2006. Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrob. Agents Chemother. 50:4114-4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jett, B. D., K. L. Hatter, M. M. Huycke, and M. S. Gilmore. 1997. Simplified agar plate method for quantifying viable bacteria. Biotechniques 23:648-650. [DOI] [PubMed] [Google Scholar]
- 14.Kelly, S. R., P. R. Jensen, T. P. Henkel, W. Fenical, and J. R. Pawlik. 2003. Effects of Caribbean sponge extracts on bacterial attachment. Aquat. Microb. Ecol. 31:175-182. [Google Scholar]
- 15.Klein, E., D. L. Smith, and R. Laxminarayan. 2007. Hospitalizations and deaths caused by methicillin-resistant Staphylococcus aureus, United States, 1999-2005. Emerg. Infect. Dis. 13:1840-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuehn, B. M. 2007. MRSA infections rise. JAMA 298:1389. [Google Scholar]
- 17.Liu, Z. G., A. Brady, A. Young, B. Rasimick, K. Chen, C. H. Zhou, and N. R. Kallenbach. 2007. Length effects in antimicrobial peptides of the (RW)n series. Antimicrob. Agents Chemother. 51:597-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melander, C., P. D. R. Moeller, T. E. Ballard, J. J. Richards, R. W. Huigens, and J. Cavanagh. 2009. Evaluation of dihydrooroidin as an antifouling additive in marine paint. Int. Biodeterior. Biodegradation 63:529-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moreau-Marquis, S., B. A. Stanton, and G. A. O'Toole. 2008. Pseudomonas aeruginosa biofilm formation in the cystic fibrosis airway. Pulm. Pharmacol. Ther. 21:595-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Musk, D. J., and P. J. Hergenrother. 2006. Chemical countermeasures for the control of bacterial biofilms: effective compounds and promising targets. Curr. Med. Chem. 13:2163-2177. [DOI] [PubMed] [Google Scholar]
- 21.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 22.Perez, F., A. M. Hujer, K. M. Hujer, B. K. Decker, P. N. Rather, and R. A. Bonomo. 2007. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob. Agents Chemother. 51:3471-3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raad, I. I., R. Y. Hachem, D. Abi-Said, K. V. Rolston, E. Whimbey, A. C. Buzaid, and S. Legha. 1998. A prospective crossover randomized trial of novobiocin and rifampin prophylaxis for the prevention of intravascular catheter infections in cancer patients treated with interleukin-2. Cancer 82:403-411. [DOI] [PubMed] [Google Scholar]
- 24.Richards, J. J., T. E. Ballard, R. W. Huigens, and C. Melander. 2008. Synthesis and screening of an oroidin library against Pseudomonas aeruginosa biofilms. Chembiochem 9:1267-1279. [DOI] [PubMed] [Google Scholar]
- 25.Richards, J. J., T. E. Ballard, and C. Melander. 2008. Inhibition and dispersion of Pseudomonas aeruginosa biofilms with reverse amide 2-aminoimidazole oroidin analogues. Org. Biomol. Chem. 6:1356-1363. [DOI] [PubMed] [Google Scholar]
- 26.Richards, J. J., R. W. Huigens, T. E. Ballard, A. Basso, J. Cavanagh, and C. Melander. 2008. Inhibition and dispersion of proteobacterial biofilms. Chem. Commun. (Camb.) 2008:1698-1700. [DOI] [PubMed] [Google Scholar]
- 27.Richards, J. J., and C. Melander. 2008. Synthesis of a 2-aminoimidazole library for anti-biofilm screening utilizing the Sonogashira reaction. J. Org. Chem. 73:5191-5193. [DOI] [PubMed] [Google Scholar]
- 28.Richards, J. J., and C. Melander. 2009. Controlling bacterial biofilms. Chembiochem 10:2287-2294. [DOI] [PubMed] [Google Scholar]
- 29.Richards, J. J., C. S. Reed, and C. Melander. 2008. Effects of N-pyrrole substitution on the anti-biofilm activities of oroidin derivatives against Acinetobacter baumannii. Bioorg. Med. Chem. Lett. 18:4325-4327. [DOI] [PubMed] [Google Scholar]
- 30.Richards, J. J., S. Reyes, S. D. Stowe, A. T. Tucker, T. E. Ballard, J. Cavanagh, and C. Melander. 2009. Amide isosteres of oroidin: assessment of antibiofilm activity and C. elegans toxicity. J. Med. Chem. 52:4582-4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rogers, S. A., R. W. Huigens, and C. Melander. 2009. A 2-aminobenzimidazole that inhibits and disperses Gram-positive biofilms through a zinc-dependent mechanism. J. Am. Chem. Soc. 131:9868-9869. [DOI] [PubMed] [Google Scholar]
- 32.Rogers, S. A., M. Krayer, J. S. Lindsey, and C. Melander. 2009. Tandem dispersion and killing of bacteria from a biofilm. Org. Biomol. Chem. 7:603-606. [DOI] [PubMed] [Google Scholar]
- 33.Rogers, S. A., and C. Melander. 2008. Construction and screening of a 2-aminoimidazole library identifies a small molecule capable of inhibiting and dispersing biofilms across bacterial order, class, and phylum. Angew. Chem. Int. Ed. Engl. 47:5229-5231. [DOI] [PubMed] [Google Scholar]
- 34.Spellberg, B., R. Guidos, D. Gilbert, J. Bradley, H. W. Boucher, W. M. Scheld, J. G. Bartlett, and J. Edwards, Jr. 2008. The epidemic of antibiotic-resistant infections: a call to action for the medical community from the Infectious Diseases Society of America. Clin. Infect. Dis. 46:155-164. [DOI] [PubMed] [Google Scholar]
- 35.Tsukamoto, S., H. Kato, H. Hirota, and N. Fusetani. 1996. Mauritiamine, a new antifouling oroidin dimer from the marine sponge Agelas mauritiana. J. Nat. Prod. 59:501-503. [Google Scholar]
- 36.Wu, H., Z. Song, M. Hentzer, J. B. Andersen, S. Molin, M. Givskov, and N. Hoiby. 2004. Synthetic furanones inhibit quorum-sensing and enhance bacterial clearance in Pseudomonas aeruginosa lung infection in mice. J. Antimicrob. Chemother. 53:1054-1061. [DOI] [PubMed] [Google Scholar]
- 37.Yamada, A., H. Kitamura, K. Yamaguchi, S. Fukuzawa, C. Kamijima, K. Yazawa, M. Kuramoto, G. Y. S. Wang, Y. Fujitani, and D. Uemura. 1997. Development of chemical substances regulating biofilm formation. Bull. Chem. Soc. Jpn. 70:3061-3069. [Google Scholar]
- 38.Zeng, Z., L. Qian, L. Cao, H. Tan, Y. Huang, X. Xue, Y. Shen, and S. Zhou. 2008. Virtual screening for novel quorum sensing inhibitors to eradicate biofilm formation of Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 79:119-126. [DOI] [PubMed] [Google Scholar]