Abstract
Proteus mirabilis is known to be highly resistant to the action of polymyxin B (PB). However, the mechanism underlying PB resistance is not clear. In this study, we used Tn5 transposon mutagenesis to identify genes that may affect PB resistance in P. mirabilis. Two genes, ugd and galU, which may encode UDP-glucose dehydrogenase (Ugd) and UDP-glucose pyrophosphorylase (GalU), respectively, were identified. Knockout mutants of ugd and galU were found to be extremely sensitive to PB, presumably because of alterations in lipopolysaccharide (LPS) structure and cell surface architecture in these mutants. These mutants were defective in swarming, expressed lower levels of virulence factor hemolysin, and had lower cell invasion ability. Complementation of the ugd or galU mutant with the full-length ugd or galU gene, respectively, led to the restoration of wild-type phenotypic traits. Interestingly, we found that the expression of Ugd and GalU was induced by PB through RppA, a putative response regulator of the bacterial two-component system that we identified previously. Mutation in either ugd or galU led to activation of RpoE, an extracytoplasmic function sigma factor that has been shown to be activated by protein misfolding and alterations in cell surface structure in other bacteria. Activation of RpoE or RpoE overexpression was found to cause inhibition of FlhDC and hemolysin expression. To our knowledge, this is the first report describing the roles and regulation of Ugd and GalU in P. mirabilis.
Cationic antimicrobial polypeptides (CAP) play an important role in host defense against microbial infection and are key effectors of the host innate immune response (24). Microbial pathogens have evolved distinct mechanisms to resist killing by CAP, including expelling CAP through pumps and cleaving CAP with proteases (47). One of the important mechanisms of resistance to CAP in Gram-negative bacteria involves modification of lipopolysaccharide (LPS) with positively charged substituents, which leads to the repulsion of CAP (47). In a large number of bacterial species, the genes conferring resistance to CAP, including polymyxin B (PB), are regulated by bacterial two-component systems (31, 37, 39, 41, 42, 46). In Salmonella enterica serovar Typhimurium, evasion of CAP killing is regulated in part by the PmrA-PmrB two-component regulatory system which upregulates genes involved in covalent modifications of LPS (21, 22). The LPS modifications reduce the negative charge of LPS and consequently decrease attraction and binding of CAP to the outer membrane. The PhoP-PhoQ two-component system, a master regulator of S. enterica serovar Typhimurium virulence functions, also has been shown to be involved in regulating resistance to CAP (18). The activation of PhoP-PhoQ increases the expression of PmrD (31), which in turn leads to the activation of PmrA, resulting in modification of LPS. The PhoP-PhoQ system is activated by micromolar concentration of magnesium (18, 19), and transcription of PhoP-activated genes is upregulated by sublethal concentration of CAP (4, 8). Modulation of resistance to CAP by the PhoP-PhoQ and PmrA-PmrB two-component systems has also been observed with Pseudomonas aeruginosa (37, 41).
Proteus mirabilis exhibits a form of multicellular behavior known as swarming migration (35, 36). It is believed that the ability of P. mirabilis to colonize the urinary tract is associated with its swarming motility. The swarming behavior of P. mirabilis is under the control of a complex regulatory network that may include bacterial two-component systems (34, 36, 49, 58, 59). In this respect, we have identified a gene, rsbA, which may encode a histidine-containing phosphotransmitter of the bacterial two-component system and act as a repressor of swarming and virulence factor expression in P. mirabilis (7, 34, 36). That swarming and virulence factor expression can be coregulated has been reported previously (2, 3, 35). It has been demonstrated that swarming and CAP resistance may be coregulated (1, 30, 40). For example, activation of the PhoP-PhoQ two-component system, which is known to enhance CAP resistance, can lead to inhibition of swarming through repressing the expression of flagellin in S. enterica serovar Typhimurium (1). Moreover, in P. mirabilis, LPS has been shown to play a critical role in swarming (6, 40), and LPS modification can affect both swarming and PB resistance (40).
UDP-glucose pyrophosphorylase (GalU) is the enzyme for the biosynthesis of UDP-glucose from UTP and glucose-1-phosphate (45). UDP-glucose is the precursor for synthesis of different surface structures, LPS, and extracellular polysaccharides (EPS). In many Gram-negative pathogens, mutation in galU leads to attenuated virulence, mainly because of changes in LPS or capsular structures (16, 45, 57). UDP-glucose dehydrogenase (Ugd) is an enzyme that converts UDP-glucose into UDP-glucuronic acid (10). UDP-glucuronic acid is also necessary for the synthesis of EPS and LPS in many pathogenic bacteria (10, 21, 43, 53). Formation of these polysaccharides is critical to bacterial virulence (10, 28) because it enables the bacteria to evade attacks by host immune systems. Recent studies demonstrate that ugd mutation in Cryptococcus neoformans alters cell integrity and the mutant cells also become temperature sensitive and fail to grow in an animal model (17). Transcription of Salmonella ugd is controlled by three regulatory systems that respond to different signals (43, 44). The participation of multiple regulatory systems in the control of ugd expression suggests a role for the ugd gene product in a broad spectrum of environments. Till now, nothing has been known about the roles of galU and ugd in P. mirabilis.
P. mirabilis is known to be highly resistant to the action of CAP, such as PB (40, 52). Although the detailed mechanisms underlying P. mirabilis resistance to PB are not clear, studies have shown that modification of LPS plays an important role in modulating CAP resistance in P. mirabilis (40, 52). Previously, we reported that RppA, a putative response regulator of the two-component system, can regulate PB susceptibility through modulating LPS modification in P. mirabilis (58). How RppA regulates LPS modification is not known. In this study, we used a Tn5 transposon mutagenesis approach to identify genes that may affect PB susceptibility in P. mirabilis. Two genes, ugd and galU, whose products may be involved in LPS synthesis and modification were identified. Knockout mutants of these genes were found to be extremely sensitive to PB, presumably because of changes in LPS. These mutants also had lower ability to swarm and express virulence factors. More importantly, we found that the expression of these genes was under the control of RppA. To our knowledge, this is the first report describing the roles and regulation of Ugd and GalU in P. mirabilis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth condition.
The bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were routinely cultured at 37°C in Luria-Bertani (LB) medium. A medium, referred to as LSW− agar, was prepared to prevent the phenotypic expression of swarming motility and used for selecting Tn5-mutagenized clones (5).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or relevant phenotype | Source or reference |
|---|---|---|
| Proteus mirabilis strains | ||
| N2 | Wild type; Tcr | Clinical isolate |
| ns2 | N2 derivative; ugd Tn5-mutagenized mutant; PBs Kmr | This study |
| ns5 | N2 derivative; galU Tn5-mutagenized mutant; PBs Kmr | This study |
| dG1 | N2 derivative; galU knockout mutant; PBs Kmr | This study |
| dG1c | dG1 containing pACYC184-galU; GalU-complemented strain; Cmr | This study |
| dU2 | N2 derivative; ugd knockout mutant; PBs Smr | This study |
| dU2c | dU2 containing pACYC184-ugd; Ugd-complemented strain; Cmr | This study |
| dA10 | N2 derivative; rppA knockout mutant; PBs Kmr | 58 |
| dA10c | dA10 containing pGEM-T Easy-rppA; RppA-complemented strain; Ampr | This study |
| rpoE-over | N2 containing pGEM-T Easy-rpoE; RpoE-overexpressed strain; Apr | Our unpublished data |
| ΔrseA mutant | N2 derivative; rseA knockout mutant; Kmr | Our unpublished data |
| E. coli strains | ||
| TOP10 | F′ mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG | Invitrogen |
| S17-1 λ pir | λ pir lysogen of S17-1 (thi pro ΔhsdR hsdM+recA RP4 2-Tc::Mu-Km::Tn7 [Tpr Smr]); permissive host able to transfer suicide plasmids requiring the Pir protein by conjugation to recipient cells | 14 |
| Plasmids | ||
| pGEM-T Easy | High-copy TA cloning vector; Ampr | Promega |
| pGEM-4Z | High-copy cloning vector; Ampr | Promega |
| yT&A-xylE | High-copy TA cloning vector containing xylE coding region; Ampr | Yang Tsuey-Ching (27) |
| pUT/mini-Tn5(Km) | Suicide plasmid requiring the Pir protein for replication and containing a mini-Tn5 cassette containing Kmr gene | 14 |
| pACYC184 | Low-copy cloning vector, P15A replicon; Cmr Tetr | 11 |
| pACYC184-ugd | pACYC184 containing intact ugd sequence including its promoter; Cmr | This study |
| pACYC184-galU | pACYC184 containing intact galU sequence including its promoter; Cmr | This study |
Transposon mutagenesis and identification of the mutated gene.
The mini-Tn5 transposon mutagenesis was performed as described previously (58). The Tn5-mutagenized cells were plated on the kanamycin (Km)-containing LSW− plates and then replica plated onto the LSW− plates containing 400 μg/ml PB. The matching colonies that grew on Km-containing plates but not on PB-containing plates were isolated. Chromosomal DNA was extracted from these PB-susceptible mutants and partially digested with AluI, and fragments over 4 kb were cloned into SmaI-digested pGEM-4Z (Promega). Following the transformation of E. coli TOP10, Km-resistant Tn5 Km-containing clones were selected. The nucleotide sequences of the cloned DNA fragments were determined and subjected to BLAST analysis at http://www.ncbi.nlm.nih.gov/. We then searched the sequence in the released genome sequence of P. mirabilis (http://www.sanger.ac.uk/) and cloned the ugd and galU genes, including their promoter, by PCR TA cloning with primer pairs ugd-353F/ugd-compR and galUF1/galUR1, respectively (Table 2). The nucleotide sequence was determined step by step by using a 373A DNA sequencer (Applied Biosystems). P. mirabilis Ugd and GalU alignment with other Ugd and GalU proteins was performed using the DNAMAN software (version 4.15).
TABLE 2.
Primers used in this study
| Primer | Sequence (5′ to 3′) | Gene amplified | Size of product (bp) | Description |
|---|---|---|---|---|
| Km1-I out | GAGCTCGAATTCGGCCTAG | —a | — | For identification of transposon-inserted genes |
| Km1-O out | CCTGCAGGCATGCAAGCTTC | — | — | For identification of transposon-inserted genes |
| ugd-compR | CCAGCAGCACCTGTGACTAAA | ugd promoter and coding region | 1,562 | For ugd cloning and complementation. Paired with “ugd-353F” |
| ugd-353F | GAATTAGCCCAAGCAGATT | For ugd cloning, complementation and reporter assay | ||
| ugd-353R | GGTGATTTTCATTTTTTATCCT | ugd promoter | 353 | For ugd reporter assay. Paired with “ugd-353F” |
| rpoE promoterF | CCAGTTCCAATCTTGGGTCA | rpoE promoter | 323 | For rpoE reporter assay. Paired with “rpoE promoterR” |
| rpoE promoterR | TAGAGATGGACTCTCCCGAAG | For rpoE reporter assay | ||
| ugd realtimeF | GAACTCGACTTTACAGCAAC | ugd | 133 | For ugd real-time PCR. Paired with “ugd realtimeR” |
| ugd realtimeR | GAATAACCGACTCCACAGAA | For ugd real-time PCR | ||
| flhDC realtimeF | CGCACATCAGCCTGCAAGT | flhDC | 90 | For flhDC real-time PCR. Paired with “flhDC realtimeR” |
| flhDC realtimeR | GCAGGATTGGCGGAAAGTT | For flhDC real-time PCR | ||
| galU realtimeF | GGTGATAAGCTTCTACAGGC | galU | 131 | For galU real-time PCR. Paired with “galU realtimeR” |
| galU realtimeR | TGGTCGTTATGTGTTGTCTG | For galU real-time PCR | ||
| galUF1 | CTGCGCTAAACGCTATCATG | galU promoter and coding region | 1,377 | For galU cloning and complementation. Paired with “galUR1” |
| galUR1 | GTGTGTGGGAATAGATTGGC | For galU cloning and complementation | ||
| galU-promoter F | TACTTTACGCACCACTGATG | galU promoter | 331 | For galU reporter assay. Paired with “galU-promoter R” |
| galU-promoter R | CCTTTAGGAACACTACACTC | For galU reporter assay and galU knockout | ||
| XbaI-galU-up F | TCTAGATACTTTACGCACCACTGATG | galU upstream | 331 | For galU knockout. Paired with “galU-promoter R” |
| galU-down F | CAATTTGGGTTGGGTTGAGG | galU downstream | 1,028 | For galU knockout. Paired with “XbaI-galU-down R” |
| XbaI-galU-down R | TCTAGAGAGTTTGGCGAAGAGTTCAC | For galU knockout |
—, not applicable.
Gene knockout by homologous recombination.
Full-length ugd, including its promoter region, was amplified by PCR using primers ugd-353F and ugd-compR (Table 2) and cloned into pGEM-T Easy (Promega) to generate pGugd. A streptomycin-resistant (Smr) cassette (14) was inserted into HindIII-digested pGugd to generate pGugd-Sm, which contains the Smr cassette-inserted ugd gene. The DNA fragment containing the Smr cassette-inserted ugd gene was cleaved from pGugd-Sm and ligated into SalI/SphI-digested pUT/mini-Tn5(Km) to generate pUTugd-Sm. Gene inactivation mutagenesis by homologous recombination and confirmation of mutants with double-crossover events were performed as described previously (58).
Sequences flanking the galU gene were amplified by PCR using primer pairs XbaI-galU-up F/galU-promoter R and galU-down F/XbaI-galU-down R (Table 2), respectively, and cloned into pGEM-T Easy (Promega) to generate pGgalU-up and pGgalU-dn. pGgalU-up was digested with SalI/XbaI, and the galU upstream sequence-containing fragment was ligated to SalI/XbaI-digested pGgalU-dn to produce the pGgalU-updn plasmid, which contains the combined upstream and downstream sequences of galU. A Kmr cassette was inserted into the XbaI-digested pGgalU-updn plasmid to generate pGgalU-updn-Km. The DNA fragment containing the combined upstream and downstream sequence of the galU gene inserted with the Kmr gene was cleaved from pGgalU-updn-Km and ligated into SalI/SphI-digested pUT/mini-Tn5(Km) (14) to generate pUTgalU-Km. Gene inactivation mutagenesis by homologous recombination and confirmation of mutants with double-crossover events were performed as described previously (58).
Construction of the Ugd- and the GalU-complemented strains.
Full-length ugd and galU, including their promoter regions, were amplified by PCR using primer pairs ugd-353F/ugd-compR and galUF1/galUR1, respectively (Table 2), and cloned into pGEM-T Easy (Promega) to generate pGugd and pGgalU. DNA fragments containing full-length ugd and galU with their promoter regions were excised from pGugd and pGgalU, respectively, with SalI and SphI. The DNA fragments were ligated into a SalI/SphI-digested low-copy plasmid, pACYC184, to generate the ugd and the galU complementation plasmids, pACYC184-ugd and pACYC184-galU. The pACYC184-ugd and pACYC184-galU plasmids were then transformed into the ugd knockout mutant and the galU knockout mutant, respectively, to generate the Ugd-complemented strain and the GalU-complemented strain.
MIC assay.
In vitro determination of the MIC for PB was performed by the broth microdilution method according to the guidelines proposed by the Clinical and Laboratory Standard Institute (13). Stock solution of PB (40,960 μg/ml) prepared in sterile water was added to 96-well microtiter plates in 2-fold serial dilutions. Aliquots of bacterial culture (5 × 104 CFU) were then dispensed into each well and incubated for 16 to 18 h. The MIC was defined as the lowest PB concentration at which no visible growth occurred. Determination of sodium dodecyl sulfate (SDS) susceptibility was performed as the MIC assay using SDS concentrations ranging from 62.5 to 4,000 μg/ml.
Preparation and analysis of LPS.
LPS extraction and analysis were performed as described previously (58).
AFM.
The overnight bacterial culture was diluted 100-fold and incubated at 37°C with vigorous shaking (225 rpm) for 2 h. Prior to imaging analysis, bacteria were gently washed with distilled water and were resuspended in water (250 μl). A final bacterial concentration of about 107 cells/ml was used for atomic force microscopy (AFM) analysis. Precleaned slides were treated with 0.1% (wt/vol) poly-l-lysine and left to dry. A drop of bacterial suspension in distilled water (20 μl) was applied onto treated slides. After adsorption for 30 min, distilled water was added to remove the unabsorbed cells. AFM imaging was performed as described by Hsieh et al. (26). An SPI 3800 atomic force microscope (Seiko, Japan) was used.
DNA uptake ability.
The overnight bacterial culture was diluted 100-fold in 20 ml LB medium and grown at 37°C to an optical density at 600 nm (OD600) of 0.4. The solution was centrifuged for 10 min at 4,000 rpm at 4°C. The pellet was then resuspended in 10 ml cold 0.1 M CaCl2 solution and put on ice for 40 min. After centrifugation, the pellet was resuspended in residual buffer to yield the final competent cell suspension. A total of 50 μl competent cells was transformed with 100 ng pUC19 circular DNA by heat shock at 42°C for 1 min and then recovered in LB medium for 1 h. Finally, 100 μl solution was spread on ampicillin-containing LB agar plates to determine the colony number. The ratio of the colony count obtained with mutant and complemented strains to that of the wild type was represented as relative DNA uptake ability.
Swarming migration assay.
The swarming migration assay was performed as described previously (23, 34). Briefly, an overnight bacterial culture (5 μl) was inoculated centrally onto the surface of dry LB swarming agar (2% [wt/vol]) plates, which were then incubated at 37°C. The swarming migration distance was measured by following swarm fronts of the bacterial cells and recording progress at 60-min intervals.
Measurement of cell differentiation, flagellin level, and hemolysin activity.
Overnight LB cultures of the wild type and related strains were inoculated onto the surface of dry LB swarming agar plates, which were then incubated at 37°C. Preparation of cells for cell differentiation, hemolysin, and flagellin assays was performed as described previously (34, 36). Cell morphology was observed after Gram-staining and examined by light microscopy at a magnification of 1,000-fold under oil immersion with an Olympus BH2 microscope equipped with a graticule. Flagellin level and cell membrane-associated hemolysin activity were assayed as described previously (34, 36).
Cell invasion assay.
The overnight culture was diluted 100-fold and incubated for 3 h before the cell invasion assay, which was performed according to the protocol of Liaw et al. (34), with some modifications. Briefly, human urothelial NTUB1 cells were grown and then infected at 37°C with 1 ml of a bacterial suspension containing 5 × 107 cells for 1.5 h. Urothelial cells were then washed twice with PBS and incubated at 37°C in 1 ml of RPMI 1640 medium containing streptomycin (250 μg/ml) for another 1.5 h. Cells were washed twice again and then lysed by incubation with 1 ml of lysis solution at 37°C for 30 min. Cell lysates were diluted serially in saline, and viable bacteria were counted by plating on LSW− agar plates. Cell invasion ability was expressed as the percentage of viable bacteria that survived the streptomycin treatment versus the total inoculum.
Real-time RT-PCR.
To study the effect of rppA mutation and PB on the expression of ugd and galU mRNA, overnight LB cultures of the wild-type, the rppA knockout mutant, and the RppA-complemented strains were washed with N minimal medium [5 mM KCl, 7.5 mM (NH4)2SO4, 0.5 mM K2SO4, 1 mM KH2PO4, 0.2% glucose, 0.01% Casamino Acids, 0.1 mM Tris-HCl, pH 7.4], diluted to an optical density at 600 nm of 0.3 to 0.4 in N minimal medium without or with 1 μg/ml PB, and grown for 4 h at 37°C. Total RNA was extracted, and real-time reverse transcription (RT)-PCR was performed as described previously (58) to monitor the expression of ugd and galU mRNA. The levels of ugd and galU mRNA were normalized against 16S rRNA. For determination of flhDC mRNA, cells were plated on the LB agar plates and incubated for 4 h before collecting all the cells on the plate with LB broth. The level of flhDC mRNA in the cells was then determined by real-time RT-PCR to investigate the effect of ugd and galU mutations or the effect of RpoE on the expression of flhDC mRNA.
Reporter assay.
The 0.353-kb promoter region of ugd amplified by primers ugd-353F and ugd-353R was cloned into pGEM-T Easy to create pGEMugd. The xylE-containing yT&A plasmid (27) was cut by SacI/PstI, and the xylE-containing fragment was ligated to the SacI/PstI-cleaved pGEMugd plasmid to create pGEMugd-xylE. The pGEMugd-xylE plasmid was cut with SmaI/SphI, and the ugd promoter-xylE fusion DNA fragment was ligated to EcoRV/SphI-cleaved pACYC184 to construct the ugd-xylE reporter plasmid. The galU-xylE reporter plasmid was constructed in the same way using galU-promoter F and galU-promoter R as primers. The ugd-xylE or galU-xylE reporter plasmid containing the wild-type strain and the rppA knockout mutant were grown overnight in LB broth with 20 μg/ml chloramphenicol, diluted 100-fold in the same medium, and grown again with or without PB, and then XylE activity was monitored as described previously (27) for 3 to 5 h at 1-h intervals. Briefly, a 1-ml cell suspension at each time point was centrifuged, resuspended in assay buffer, and subjected to kinetic assay of enzyme activities after adding the substrate (catechol). Meanwhile, the OD600 value of the cell suspension was recorded. One unit of enzyme activity was defined as the amount of enzyme that converts 1 nmol substrate per minute. The specific activity of the enzyme was defined in terms of units per OD600 unit. Each experiment was repeated three times. The specific activity of xylE (nmole/min/liter/OD600) was calculated as ΔOD375 × Δmin−1 × ɛ−1 × OD600−1 × dilute factor (ɛ = 22,000 M−1cm−1). The rpoE-xylE reporter plasmid (containing the 0.343-kb promoter fragment) was constructed in the same way. rpoE-xylE reporter activity in the wild type, the ugd knockout mutant, and the galU knockout mutant was monitored after cells were plated on the LB agar plates for 2 to 5 h to investigate the effect of ugd and galU mutation on the expression of rpoE mRNA.
Nucleotide sequence accession numbers.
The nucleotide sequences of P. mirabilis N2 ugd and galU have been deposited in the DDBJ/EMBL/GenBank databases under accession no. FJ865582 and GQ358626, respectively.
RESULTS
Isolation of PB-sensitive P. mirabilis mutants.
To isolate PB-sensitive mutants of P. mirabilis, a mini-Tn5 transposon mutagenesis approach was performed as described previously (58), and the PB-containing LSW− plates were used. Two mutants, ns2 and ns5, which were over 20,480 and 10,240-fold, respectively, more sensitive to PB than the wild-type P. mirabilis N2, were isolated (MIC of 2 and 4 versus >40,960 μg/ml).
The nucleotide sequence was obtained from the cloned DNA fragment flanking the mini-Tn5 in the mutants ns2 and ns5. Through searching the released P. mirabilis genome sequence (http://www.sanger.ac.uk/) using the sequence we had obtained, we found that Tn5 was inserted into a gene which we named ugd (UDP glucose dehydrogenase) and galU (UDP-glucose pyrophosphorylase). The ugd gene is between cpsF (PMI3190, a capsule synthesis gene) and wbnF (PMI3188, a nucleotide sugar epimerase). The galU gene is between a putative wbnF gene (PMI1489) and a gene locus, PMI1491. The ugd and galU genes, with their respective upstream regions, were cloned and sequenced. Analysis of the upstream sequences of ugd and galU genes revealed putative binding sites for Salmonella RcsB and PhoP in the upstream region of ugd and only the putative RcsB site in that of galU gene, according to the report of Mouslim et al. (44).
The nucleotide sequence of ugd was found to be 97% identical to the corresponding sequence of the sequenced P. mirabilis strain HI4320. Ugd is homologous to S. enterica serovar Typhimurium Ugd (77% identity, 88% similarity), Escherichia coli Ugd (75% identity, 90% similarity), and Pseudomonas aeruginosa Ugd (27% identity, 48% similarity). The nucleotide sequence of galU was found to be 99% identical to the corresponding sequence of the sequenced P. mirabilis strain HI4320. GalU is homologous to S. enterica serovar Typhimurium GalU (76% identity, 87% similarity), E. coli GalU (75% identity, 86% similarity), and P. aeruginosa GalU (46% identity, 64% similarity).
The P. mirabilis ugd knockout and galU knockout mutants exhibit increased susceptibility to PB.
To demonstrate the role of Ugd and GalU in regulating PB susceptibility, we sought to construct ugd and galU mutants through allelic exchange mutagenesis. The ugd (dU2) and galU (dG1) knockout mutants were constructed by homologous recombination using plasmid pUT (see Materials and Methods). Southern blot analysis indicated that the mutants contained a single disrupted ugd or galU gene and no wild-type allele (data not shown). The MICs of PB for wild-type P. mirabilis N2 and the knockout mutants dU2 and dG1 were determined. While the MIC of PB for the wild-type strain was >40,960 μg/ml, those for the dU2 and dG1 mutants were 2 and 4 μg/ml, respectively. To further confirm that Ugd and GalU can affect PB susceptibility, we constructed the Ugd- and GalU-complemented strains dU2c and dG1c by transforming pACYC184-ugd and pACYC184-galU into the dU2 and dG1 mutants, respectively. We found that the MICs of PB for the dU2c and dG1c strains were >40,960 μg/ml. Together, these data suggest that both Ugd and GalU are determinants of PB resistance in P. mirabilis.
The P. mirabilis ugd knockout and galU knockout mutants have altered LPS profiles.
LPS modification plays an important role in PB susceptibility in many Gram-negative bacteria, including Salmonella, Yersinia, Pseudomonas, E. coli, and P. mirabilis (40, 41, 50, 56). To investigate the underlying cause of PB sensitivity in the ugd knockout and the galU knockout mutants, we compared the LPS profile of the two knockout mutants (dU2 and dG1) with those of the wild-type strain (N2) and the complemented strains (dU2c and dG1c). The LPS was extracted from equal amounts of these bacterial cells and was subjected to SDS-PAGE analysis. As shown in Fig. 1, while the LPS profiles of the wild-type and the complemented strains were similar, those of the ugd knockout and the galU knockout mutants were markedly different. In both knockout mutants, the O-antigen LPS ladder was lost, and there was a deficiency in the lower part of LPS bands (Fig. 1, compare lane 1 with lanes 2 and 4). These data indicate that the ugd and galU mutants have a defect in LPS in its outer membrane. To determine whether the two knockout mutants synthesized amounts of LPS similar to those of the wild-type and complemented strains, the LPS was extracted from equal amounts of these bacterial cells, and the concentration of LPS was determined (see Materials and Methods). As shown in Table 3, while the wild-type and the Ugd-complemented and the GalU-complemented strains synthesized similar amounts of LPS, the two knockout mutants synthesized much less LPS than the wild-type and complemented strains. Together, the above data indicate that the ugd knockout and the galU knockout mutants have undergone qualitative and quantitative changes in LPS and these changes may cause the ugd and galU mutants to become more sensitive to PB.
FIG. 1.
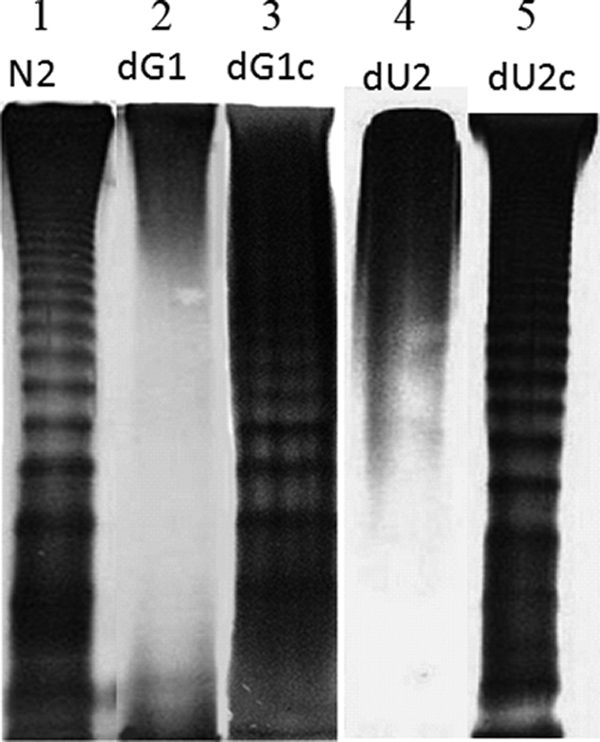
LPS profiles of wild-type P. mirabilis (N2), the galU knockout mutant (dG1), the GalU-complemented strain (dG1c), the ugd knockout mutant (dU2), and the Ugd-complemented strain (dU2c). Six microliters of LPS purified from the same number of cells (OD600 × volume [in ml] = 100) of the respective strains was subjected to SDS-PAGE analysis.
TABLE 3.
Phenotypic traits of the wild type (N2), the ugd knockout mutant (dU2), the Ugd-complemented strain (dU2c), the galU knockout mutant (dG1), and the GalU-complemented strain (dG1c)
| Trait | N2 | dU2 | dU2c | dG1 | dG1c |
|---|---|---|---|---|---|
| SDS MIC (μg/ml) | 2,000 | 500 | 2,000 | 500 | 2,000 |
| Relative DNA uptake abilitya | 1 | 3 ± 0.3 | 1 ± 0.2 | 4.9 ± 0.8 | 1.2 ± 0.2 |
| Total LPS (mg/ml) | 13.2 ± 1.2 | 5.0 ± 0.5 | 14.0 ± 0.9 | 4.8 ± 0.4 | 13.7 ± 0.9 |
| Relative invasion abilityb | 1 | 0.2 ± 0.04 | 0.9 ± 0.09 | 0.3 ± 0.05 | 0.8 ± 0.04 |
| AFMc | |||||
| Valley to peak (nm) | 186.70 ± 51.17 | 362.17 ± 127.74 | ND | 262.70 ± 48.94 | ND |
| Roughness (nm) | 182.82 ± 55.13 | 343.23 ± 120.99 | ND | 262.65 ± 49.84 | ND |
Relative ratio of DNA uptake ability. The DNA uptake ability of the wild type was set at 1.
Relative ratio of cell invasion ability. The cell invasion ability of the wild type was set at 1.
At least five cells in three 1-μm2 areas from three independent experiments; each result is expressed as the mean and the standard deviation.
Cellular surface architecture and cell permeability of the P. mirabilis ugd knockout and galU knockout mutants.
Since bacterial Ugd and GalU enzymes have been shown to be involved in the synthesis of different surface structures, we next studied the detailed cellular surface topography of the log-phase cells using AFM. A clear trend emerged, showing that the cell surface roughness and valley-to-peak distance of the ugd knockout mutant and the galU knockout mutant were increased compared to those of the wild-type cells (Table 3). These data suggest that Ugd and GalU are determinants of P. mirabilis cell surface topology and that defects in these two enzymes can lead to an aberrant cell surface topology. Consistent with this, we also found that the ugd knockout mutant and the galU knockout mutant were more sensitive to SDS and had an increased DNA uptake ability compared to the wild-type strain (Table 3). Together, these data indicate that Ugd and GalU proteins are involved in the maintenance of cell surface topology and cell permeability in P. mirabilis.
The swarming behavior of the P. mirabilis ugd knockout and galU knockout mutants.
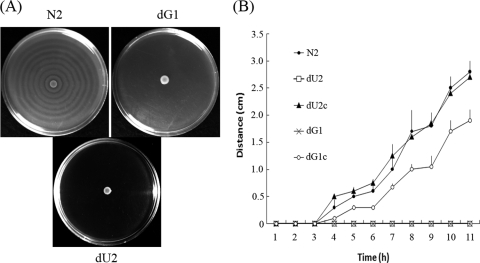
As described in the introduction, swarming and CAP susceptibility could be coregulated. We thus tested the swarming behavior of the ugd and galU mutants. We found that the ns2 and ns5 mutant strains, which we isolated originally from the Tn5 transposon mutagenesis, could not swarm on the LB swarming agar plate after 18-h incubation (data not shown). To further confirm the effect of ugd and galU mutations on swarming, we compared the swarming behavior of the wild-type strain (N2), the ugd knockout mutant (dU2), the galU knockout mutants (dG1), the Ugd-complemented strain (dU2c), and the GalU-complemented strain (dG1c). Figure 2A shows that the ugd knockout mutant and the galU knockout mutant could not swarm at all on LB swarming agar plates after 18-h incubation. Figure 2B shows that while the ugd knockout and the galU knockout mutants could not migrate after 11-h incubation, the Ugd-complemented strain exhibited migration ability similar to that of the wild-type strain. The GalU-complemented strain restored partial swarming ability of the wild-type strain. These data indicate that the integrity of the Ugd and GalU proteins is essential for maintenance of swarming ability and that changes in Ugd and GalU enzymatic activities may affect the swarming behavior of P. mirabilis.
FIG. 2.
(A) Swarming migration of wild-type P. mirabilis (N2), the ugd knockout mutant (dU2), and the galU knockout mutant (dG1) on LB swarming plates. Aliquots (5 μl) of overnight culture were inoculated centrally onto the plates. The plates were incubated at 37°C, and the representative pictures were taken at 18 h after incubation. (B) The swarming migration of wild-type P. mirabilis (N2), the ugd knockout mutant (dU2), the Ugd-complemented strain (dU2c), the galU knockout mutant (dG1), and the GalU-complemented strain (dG1c). Aliquots (5 μl) of overnight culture were inoculated centrally onto the LB swarming plates. The plates were incubated at 37°C, and the migration distance was measured hourly after inoculation. The data represent the averages of three results of independent experiments with standard deviations.
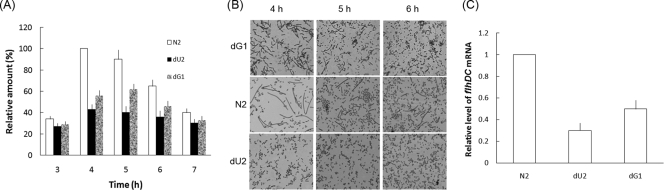
Swarming migration in P. mirabilis involves the coordinated differentiation of short vegetative cells bearing a few peritrichous flagella into long multinucleate swarm cells with a much greater surface density of flagella (2, 34, 35). To further confirm that changes in Ugd and GalU enzymatic activities may affect the swarming behavior of P. mirabilis, we measured the amount of flagellin synthesized in the ugd knockout mutant (dU2), the galU knockout mutant (dG1), and the wild-type strain (N2) during one differentiation-dedifferentiation cycle of the bacteria. As shown in Fig. 3A, the ugd and galU mutants synthesized lower levels of flagellin than did the wild-type strain at each time point. Consistent with this, we also found that the ugd and galU mutants synthesized a smaller amount of mRNA of flhDC, which is a master regulator controlling the expression of flagellum genes (49), than did the wild-type strain (Fig. 3C). We also tested whether the differentiation of P. mirabilis was affected by ugd and galU mutations. To this end, we compared the cell lengths of the ugd knockout mutant (dU2) and the galU knockout mutant (dG1) with that of the wild-type strain (N2) during one differentiation-dedifferentiation cycle of the bacteria. The ugd and the galU mutants formed shorter cells than did the wild type during the differentiation cycle (Fig. 3B), indicating that ugd and galU mutations can affect the differentiation of P. mirabilis. Together, these data further confirm that the enzymatic activities of Ugd and GalU are required for the maintenance of the swarming ability of P. mirabilis.
FIG. 3.
(A) Flagellin level of wild-type P. mirabilis (N2), the ugd knockout mutant (dU2), and the galU knockout mutant (dG1). The flagellin level was determined at different time points after seeding the strains on LB agar plates. The value obtained with the wild-type cells at 4 h postseeding was set at 100%, and all other values were expressed relative to this value. The data represent the averages of results of three independent experiments with standard deviations. (B) Microscopic observation of cell differentiation of wild-type P. mirabilis (N2), the ugd knockout mutant (dU2), and the galU knockout mutant (dG1). Cells were Gram-stained and viewed under oil (magnification, ×1,000). Three independent experiments were performed, and the representative pictures showing cell differentiation at 4, 5, and 6 h postseeding on LB agar plates are shown. The increase in cell length was taken as a sign of cell differentiation. (C) Expression of flhDC mRNA in wild-type P. mirabilis (N2), the ugd knockout mutant (dU2), and the galU knockout mutant (dG1). Total RNA was isolated from respective cells at 4 h postseeding on LB agar plates and was then subjected to real-time RT-PCR for the measurement of flhDC mRNA. The value obtained with the wild-type cells was set at 1. The data represent the averages of results of three independent experiments with standard deviations.
Hemolysin expression and cell invasion ability of the P. mirabilis ugd knockout and galU knockout mutants.
Both swarming and CAP susceptibility are correlated with the expression of virulence factors in the bacterium (3, 18, 19, 35). We thus examined the expression of hemolysin during one differentiation-dedifferentiation cycle of the bacteria by measuring the cell membrane-associated hemolysin activity. As shown in Fig. 4, while the ugd knockout mutant and the galU knockout mutant expressed lower levels of hemolysin activity than did the wild-type strain, the complemented strains restored the wild-type hemolysin activity during the incubation period. The ability of the bacteria to invade human urothelial NTUB1 cells was also measured. As shown in Table 3, the ugd knockout mutant and the galU knockout mutant had a much lower cell invasion ability than the wild-type strain.
FIG. 4.

Hemolysin activities of wild-type P. mirabilis (N2), the ugd knockout mutant (dU2), the Ugd-complemented strain (dU2c), the galU knockout mutant (dG1), and the GalU-complemented strain (dG1c). Hemolysin activity was determined at different time points after seeding the strains on LB agar plates. The value obtained with the wild-type cells at 4 h postseeding was set at 100%, and all other values were expressed relative to this value. The data represent the averages of results of three independent experiments with standard deviations.
Expression of Ugd and GalU is induced by PB through an RppA-dependent pathway.
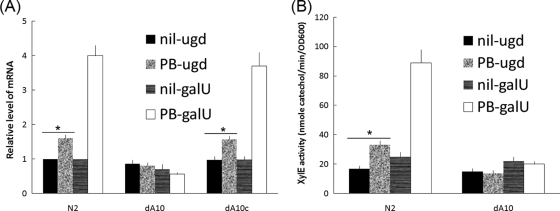
It has been shown that the expression of Ugd is regulated by two-component signal transduction systems in Salmonella (43, 44). We figured that the expression of Ugd and GalU might be regulated by RppA, a putative response regulator of the two-component system, because the latter has been shown by us to be able to regulate PB susceptibility by modulating LPS modification in P. mirabilis (58). The expression of Ugd and GalU in wild-type P. mirabilis (N2), the rppA knockout mutant (dA10), and the RppA-complemented strain (dA10c) in the presence or absence of PB (1 μg/ml) was measured by real-time RT-PCR. We found that in the absence of PB, the expression levels of Ugd and GalU in all three strains were similar. Interestingly, when PB was added to the culture media, the expression of Ugd and GalU was induced in the wild-type and RppA-complemented strains but not in the rppA knockout mutant (Fig. 5A). These data suggest that PB can induce Ugd and GalU expression through the RppA-dependent pathway. To confirm this, the reporter assays using the ugd-xylE or galU-xylE transcriptional fusion constructs were performed. As shown in Fig. 5B, in the absence of PB, xylE expression (activity) levels driven by the ugd or galU promoter were about the same in the wild-type and the rppA mutant strains. However, in the presence of PB (1 μg/ml), xylE expression (activity) from both the ugd and galU promoters was induced in the wild type but not in the rppA mutant strain. Therefore, PB seems to be able to serve as a signal that can induce Ugd and GalU expression through the RppA-dependent pathway.
FIG. 5.
(A) Effect of rppA mutation on the expression of ugd and galU mRNA in the presence or absence of PB (1 μg/ml). The mRNA amounts of ugd and galU in the wild type, the rppA knockout mutant, and the RppA-complemented strain were quantified by real-time RT-PCR at 4 h postseeding on LB agar plates as described in Materials and Methods. The value obtained with the wild-type cells in the absence of PB was set at 1. The data represent the averages of results of four independent experiments with standard deviations. (B) XylE activities of ugd-xylE and galU-xylE reporters in the wild type and the rppA knockout mutant in the presence or absence of 1 μg/ml PB. Representative data of activities for cells grown on the LB agar plates for 4 h are shown. The data represent the averages of results of three independent experiments with standard deviations. *, P < 0.01, for comparison of the value in the absence of PB with that in the presence of PB; N2, wild type; dA10, rppA knockout mutant; dA10c, RppA-complemented strain.
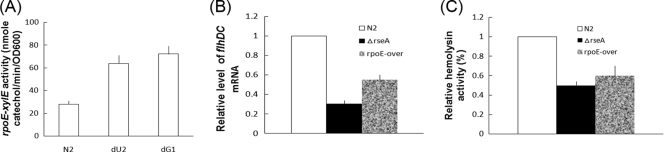
ugd and galU mutations cause the activation of RpoE, which in turn inhibits the expression of FlhDC and hemolysin.
It has been shown that membrane stress/disruption causes the activation of RpoE, an extracytoplasmic function sigma factor which controls the expression of an array of genes that are involved in pathogenesis, the folding of outer membrane proteins, and the synthesis of cell envelope proteins, phospholipid, and LPS (25, 51). Since ugd and galU mutations can cause the disturbance of the cell membrane, we figured that ugd and galU mutations may lead to the activation of RpoE. As described above, activation of RpoE causes the induction of an array of genes, including rpoE itself (51). We thus tested whether the rpoE promoter was activated in the ugd and galU mutants by using the reporter plasmid containing the rpoE-xylE transcriptional fusion sequences. As shown in Fig. 6A, the XylE activity was much higher in the ugd and galU mutants than in the wild-type strain, suggesting that ugd and galU mutations cause the activation of RpoE. To test whether RpoE could control the expression of virulence factors, such as FlhDC and hemolysin, the expression of flhDC and hemolysin was measured for the wild-type strain, the RpoE constitutively active mutant (Fig. 6, ΔrseA; RseA is the inhibitor of RpoE) (25, 51), and the RpoE-overexpressed strain (Fig. 6, rpoE-over). The expression of flhDC mRNA (Fig. 6B) and the hemolysin activity (Fig. 6C) were lower in the RpoE constitutively active mutant and the RpoE-overexpressed strain than in the wild-type strain, suggesting that RpoE can inhibit the expression of FlhDC and hemolysin in P. mirabilis.
FIG. 6.
(A) Effect of ugd and galU mutation on the promoter activity of rpoE. XylE activities of rpoE-xylE reporter were determined for the wild type, the ugd knockout mutant (dU2), and the galU knockout mutant (dG1). Representative data of activities for cells grown on the LB agar plates for 4 h are shown. (B) Effect of RpoE on the expression of flhDC mRNA. The flhDC mRNA amounts in the wild type (N2), the rseA mutant (ΔrseA), and the RpoE-overexpressed strain (rpoE-over) were quantified by real-time RT-PCR at 4 h postseeding on LB agar plates, as described in Materials and Methods. The value obtained with the wild-type cells was set at 1. The data represent the averages of results of three independent experiments with standard deviations. (C) Effect of RpoE on the hemolysin activities of the wild type (N2), the rseA mutant (ΔrseA), and the RpoE-overexpressed strain (rpoE-over). Hemolysin activity was determined at 4 h after seeding the strains on LB agar plates. The value obtained with the wild-type cells was set at 1, and all other values were expressed relative to this value. The data represent the averages of results of three independent experiments with standard deviation.
DISCUSSION
In this study, we identified two genes, ugd and galU, that are involved in controlling PB resistance in P. mirabilis. In Salmonella enterica and E. coli, both ugd and galU have been shown to be involved in the production of 4-aminoarabinose (16, 20, 21, 48), which is necessary for modifying LPS to become more positively charged. Mutation in either ugd or galU can lead to an alteration in LPS structure in Salmonella, E. coli, Burkholderia, Aeromonas, and Vibrio (9, 21, 22, 45, 57). Here we also found that the ugd and galU mutants of P. mirabilis had altered LPS profiles and cell surface topologies (Fig. 1 and Table 3). Since alterations in LPS are known to affect resistance to CAP, including PB (15, 21, 22), we believe that LPS alterations caused by ugd and galU mutations contribute to the PB-sensitive phenotype of these two mutants of P. mirabilis. That the ugd or galU mutation can cause increased sensitivity to PB has been reported previously for Salmonella, E. coli, and Vibrio (21, 22, 45).
Previously we reported that RppA, a response regulator of the two-component system, can regulate PB sensitivity in P. mirabilis (58) and that the rppA-defective mutant exhibits an increased sensitivity to PB compared to the wild type. However, how RppA modulates PB sensitivity is not known. In this study, we found that the expression of Ugd and GalU could be stimulated in the presence of PB in the wild type but not in the rppA knockout mutant (Fig. 5). Since PB has been shown to be able to serve as a signal to activate RppA (58), we believe that, in the presence of PB, Ugd and GalU are induced by RppA, leading to modification of LPS and repulsion of PB. Moreover, our preliminary data indicate that RppA could bind to the galU promoter (data not shown). RppA can also induce the expression of pmrIp, a gene involved in an LPS modification, leading to PB resistance (29).
We found that both the ugd knockout and galU knockout mutants could not swarm at all after 11-h incubation on LB swarming agar plates (Fig. 2). Several lines of evidence highlight the importance of P. mirabilis Ugd and GalU in swarming motility. First, in comparison to the wild type, both ugd and galU mutants produced reduced amounts of flagellin and flhDC mRNA and exhibited impaired cell differentiation (Fig. 3). Second, both the P. mirabilis ugd and galU mutants had defective LPS, which has been shown to be essential for swarming migration (55). Third, we have found that both the P. mirabilis ugd and galU mutants expressed lower levels of fliA and flaA mRNA (data not shown). The products of the fliA and flaA genes are known to be involved in modulating the swarming behavior of P. mirabilis (12). That the galU mutation can affect swarming in P. mirabilis has also been reported previously (6).
Our data indicate that ugd and galU mutations not only affect PB sensitivity but also lead to lower expression of virulence factors, lower cell invasion ability, and defective swarming phenotype (Fig. 2 and 4 and Table 3). How could mutation in the genes that are involved in synthesis of cell surface structure affect such a broad spectrum of phenotypic traits? Two possible mechanisms may explain this. First, cell surface alterations caused by ugd or galU mutation may activate two-component systems that in turn regulate swarming and virulence factor expression. The observation that bacterial two-component systems can sense and be regulated by alterations in the membrane has been reported previously. For instance, Serratia marcescens RssA, a sensor kinase, has been suggested to be able to sense and be activated by alteration in membrane fluidity, leading to a change in swarming motility (32). Moreover, the RcsC-RcsD-RcsB two-component system is known to be activated by cationic amphipathic molecules that can insert into the lipid bilayer and perturb the bacterial membrane (38). It is possible that RppB, which is the membrane sensor kinase of the RppA-RppB two-component system, can sense and be activated by membrane perturbation caused by ugd or galU mutation. As we reported previously, activation of the RppA-RppB two-component system can lead to inhibition of swarming and virulence factor expression (58). Therefore, activation of the RppA-RppB two-component system can explain why ugd or galU mutation causes such a broad spectrum of effects. Our preliminary data indicated that the ugd or galU mutation led to an increase in RppA expression (data not shown). Since RppA is auto-induced by itself (58), these data support the hypothesis that the ugd or galU mutation can cause activation of the RppA-RppB two-component system and subsequent broad effects.
The alternative mechanism that may explain the broad effects of ugd and galU mutations is that cell surface alterations caused by these mutations lead to the activation of RpoE, an extracytoplasmic function sigma factor. RpoE regulates the expression of a large number of genes involved in stress response, the synthesis of surface structure, and pathogenesis (51). The activity of RpoE is negatively controlled by the anti-sigma factor RseA, and deletion of rseA leads to constitutively active RpoE (25, 51). Previous studies indicate that RpoE can be activated by protein misfolding, membrane disturbance, and changes in LPS (51, 54). In this study, we found that the promoter activity of RpoE was activated by mutation in ugd or galU (Fig. 6A), presumably because these two mutations caused alterations in LPS and the cell surface (Fig. 1 and Table 3). Moreover, we found that constitutively active RpoE or RpoE overexpression could decrease swarming (data not shown) and cause inhibition of FlhDC and hemolysin expression (Fig. 6). Therefore, activation of RpoE caused by ugd or galU mutation could explain why the ugd or galU mutant is defective in swarming and virulence factor expression. The observation that RpoE can negatively regulate FlhDC expression has been reported previously for Azotobacter (33).
In conclusion, we have identified two genes, ugd and galU, that are involved in the maintenance of cell surface architecture in P. mirabilis. Mutation in either ugd or galU leads to increased PB sensitivity, defective swarming, and lower virulence factor expression. These data suggest that inhibition of Ugd and GalU enzymatic activities can make P. mirabilis, which is inherently resistant to PB, become more vulnerable to PB treatment and less virulent. It is tempting to suggest that these two enzymes are potential targets for antibacterial drug development.
Acknowledgments
This work was supported by grants from the National Science Council and National Taiwan University Hospital, Taipei, Taiwan.
We thank Yeong-Shiau Pu (National Taiwan University Hospital) for providing the NTUB1 cell line, Yang Tsuey-Ching for giving us the yT&A-xylE plasmid, and Shiming Lin for technical assistance and data interpretation of AFM.
Footnotes
Published ahead of print on 16 February 2010.
REFERENCES
- 1.Adams, P., R. Fowler, N. Kinsella, G. Howell, M. Farris, P. Coote, and C. D. O'Connor. 2001. Proteomic detection of PhoPQ- and acid-mediated repression of Salmonella motility. Proteomics 1:597-607. [DOI] [PubMed] [Google Scholar]
- 2.Allison, C., N. Coleman, P. L. Jones, and C. Hughes. 1992. Ability of Proteus mirabilis to invade human urothelial cells is coupled to motility and swarming differentiation. Infect. Immun. 60:4740-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allison, C., H. C. Lai, and C. Hughes. 1992. Co-ordinate expression of virulence genes during swarm-cell differentiation and population migration of Proteus mirabilis. Mol. Microbiol. 6:1583-1591. [DOI] [PubMed] [Google Scholar]
- 4.Bader, M. W., W. W. Navarre, W. Shiau, H. Nikaido, J. G. Frye, M. McClelland, F. C. Fang, and S. I. Miller. 2003. Regulation of Salmonella typhimurium virulence gene expression by cationic antimicrobial peptides. Mol. Microbiol. 50:219-230. [DOI] [PubMed] [Google Scholar]
- 5.Belas, R., D. Erskine, and D. Flaherty. 1991. Transposon mutagenesis in Proteus mirabilis. J. Bacteriol. 173:6289-6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belas, R., M. Goldman, and K. Ashliman. 1995. Genetic analysis of Proteus mirabilis mutants defective in swarmer cell elongation. J. Bacteriol. 177:823-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belas, R., R. Schneider, and M. Melch. 1998. Characterization of Proteus mirabilis precocious swarming mutants: identification of rsbA, encoding a regulator of swarming behavior. J. Bacteriol. 180:6126-6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brodsky, I. E., R. K. Ernst, S. I. Miller, and S. Falkow. 2002. mig-14 is a Salmonella gene that plays a role in bacterial resistance to antimicrobial peptides. J. Bacteriol. 184:3203-3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burtnick, M. N., and D. E. Woods. 1999. Isolation of polymyxin B-susceptible mutants of Burkholderia pseudomallei and molecular characterization of genetic loci involved in polymyxin B resistance. Antimicrob. Agents Chemother. 43:2648-2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campbell, R. E., R. F. Sala, I. van de Rijn, and M. E. Tanner. 1997. Properties and kinetic analysis of UDP-glucose dehydrogenase from group A streptococci. Irreversible inhibition by UDP-chloroacetol. J. Biol. Chem. 272:3416-3422. [DOI] [PubMed] [Google Scholar]
- 11.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clemmer, K. M., and P. N. Rather. 2008. The Lon protease regulates swarming motility and virulence gene expression in Proteus mirabilis. J. Med. Microbiol. 57:931-937. [DOI] [PubMed] [Google Scholar]
- 13.Clinical and Laboratory Standards Institute. 2006. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 7th ed. Approved standard M7-A7. Clinical and Laboratory Standards Institute, Wayne, PA.
- 14.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ernst, R. K., T. Guina, and S. I. Miller. 2001. Salmonella typhimurium outer membrane remodeling: role in resistance to host innate immunity. Microbes Infect. 3:1327-1334. [DOI] [PubMed] [Google Scholar]
- 16.Genevaux, P., P. Bauda, M. S. DuBow, and B. Oudega. 1999. Identification of Tn10 insertions in the rfaG, rfaP, and galU genes involved in lipopolysaccharide core biosynthesis that affect Escherichia coli adhesion. Arch. Microbiol. 172:1-8. [DOI] [PubMed] [Google Scholar]
- 17.Griffith, C. L., J. S. Klutts, L. Zhang, S. B. Levery, and T. L. Doering. 2004. UDP-glucose dehydrogenase plays multiple roles in the biology of the pathogenic fungus Cryptococcus neoformans. J. Biol. Chem. 279:51669-51676. [DOI] [PubMed] [Google Scholar]
- 18.Groisman, E. A. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Groisman, E. A., and C. Mouslim. 2006. Sensing by bacterial regulatory systems in host and non-host environments. Nat. Rev. Microbiol. 4:705-709. [DOI] [PubMed] [Google Scholar]
- 20.Gunn, J. S. 2008. The Salmonella PmrAB regulon: lipopolysaccharide modifications, antimicrobial peptide resistance and more. Trends Microbiol. 16:284-290. [DOI] [PubMed] [Google Scholar]
- 21.Gunn, J. S., K. B. Lim, J. Krueger, K. Kim, L. Guo, M. Hackett, and S. I. Miller. 1998. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol. 27:1171-1182. [DOI] [PubMed] [Google Scholar]
- 22.Gunn, J. S., S. S. Ryan, J. C. Van Velkinburgh, R. K. Ernst, and S. I. Miller. 2000. Genetic and functional analysis of a PmrA-PmrB-regulated locus necessary for lipopolysaccharide modification, antimicrobial peptide resistance, and oral virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 68:6139-6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gygi, D., M. J. Bailey, C. Allison, and C. Hughes. 1995. Requirement for FlhA in flagella assembly and swarm-cell differentiation by Proteus mirabilis. Mol. Microbiol. 15:761-769. [DOI] [PubMed] [Google Scholar]
- 24.Hancock, R. E., and M. G. Scott. 2000. The role of antimicrobial peptides in animal defenses. Proc. Natl. Acad. Sci. U. S. A. 97:8856-8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayden, J. D., and S. E. Ades. 2008. The extracytoplasmic stress factor, sigmaE, is required to maintain cell envelope integrity in Escherichia coli. PLoS One 3:e1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsieh, C. H., Y. H. Lin, S. Lin, J. J. Tsai-Wu, C. H. Herbert Wu, and C. C. Jiang. 2008. Surface ultrastructure and mechanical property of human chondrocyte revealed by atomic force microscopy. Osteoarthritis Cartilage 16:480-488. [DOI] [PubMed] [Google Scholar]
- 27.Hu, R. M., K. J. Huang, L. T. Wu, Y. J. Hsiao, and T. C. Yang. 2008. Induction of L1 and L2 beta-lactamases of Stenotrophomonas maltophilia. Antimicrob. Agents Chemother. 52:1198-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hung, R. J., H. S. Chien, R. Z. Lin, C. T. Lin, J. Vatsyayan, H. L. Peng, and H. Y. Chang. 2007. Comparative analysis of two UDP-glucose dehydrogenases in Pseudomonas aeruginosa PAO1. J. Biol. Chem. 282:17738-17748. [DOI] [PubMed] [Google Scholar]
- 29.Jiang, S.-S., M.-C. Liu, L.-J. Teng, W.-B. Wang, P.-R. Hsueh, and S.-J. Liaw. 1 February 2010. Proteus mirabilis pmrI orthologue, an RppA-regulated gene necessary for polymyxin B resistance, biofilm formation and urothelial cell invasion. Antimicrob. Agents Chemother. doi: 10.1128/AAC.01219-09. [DOI] [PMC free article] [PubMed]
- 30.Kim, W., T. Killam, V. Sood, and M. G. Surette. 2003. Swarm-cell differentiation in Salmonella enterica serovar Typhimurium results in elevated resistance to multiple antibiotics. J. Bacteriol. 185:3111-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kox, L. F., M. M. Wosten, and E. A. Groisman. 2000. A small protein that mediates the activation of a two-component system by another two-component system. EMBO J. 19:1861-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lai, H. C., P. C. Soo, J. R. Wei, W. C. Yi, S. J. Liaw, Y. T. Horng, S. M. Lin, S. W. Ho, S. Swift, and P. Williams. 2005. The RssAB two-component signal transduction system in Serratia marcescens regulates swarming motility and cell envelope architecture in response to exogenous saturated fatty acids. J. Bacteriol. 187:3407-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leon, R., and G. Espin. 2008. flhDC, but not fleQ, regulates flagella biogenesis in Azotobacter vinelandii, and is under AlgU and CydR negative control. Microbiology 154:1719-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liaw, S. J., H. C. Lai, S. W. Ho, K. T. Luh, and W. B. Wang. 2001. Characterisation of p-nitrophenylglycerol-resistant Proteus mirabilis super-swarming mutants. J. Med. Microbiol. 50:1039-1048. [DOI] [PubMed] [Google Scholar]
- 35.Liaw, S. J., H. C. Lai, S. W. Ho, K. T. Luh, and W. B. Wang. 2003. Role of RsmA in the regulation of swarming motility and virulence factor expression in Proteus mirabilis. J. Med. Microbiol. 52:19-28. [DOI] [PubMed] [Google Scholar]
- 36.Liaw, S. J., H. C. Lai, and W. B. Wang. 2004. Modulation of swarming and virulence by fatty acids through the RsbA protein in Proteus mirabilis. Infect. Immun. 72:6836-6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Macfarlane, E. L., A. Kwasnicka, M. M. Ochs, and R. E. Hancock. 1999. PhoP-PhoQ homologues in Pseudomonas aeruginosa regulate expression of the outer-membrane protein OprH and polymyxin B resistance. Mol. Microbiol. 34:305-316. [DOI] [PubMed] [Google Scholar]
- 38.Majdalani, N., and S. Gottesman. 2005. The Rcs phosphorelay: a complex signal transduction system. Annu. Rev. Microbiol. 59:379-405. [DOI] [PubMed] [Google Scholar]
- 39.Marceau, M., F. Sebbane, F. Ewann, F. Collyn, B. Lindner, M. A. Campos, J. A. Bengoechea, and M. Simonet. 2004. The pmrF polymyxin-resistance operon of Yersinia pseudotuberculosis is upregulated by the PhoP-PhoQ two-component system but not by PmrA-PmrB, and is not required for virulence. Microbiology 150:3947-3957. [DOI] [PubMed] [Google Scholar]
- 40.McCoy, A. J., H. Liu, T. J. Falla, and J. S. Gunn. 2001. Identification of Proteus mirabilis mutants with increased sensitivity to antimicrobial peptides. Antimicrob. Agents Chemother. 45:2030-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moskowitz, S. M., R. K. Ernst, and S. I. Miller. 2004. PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. J. Bacteriol. 186:575-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moss, J. E., P. E. Fisher, B. Vick, E. A. Groisman, and A. Zychlinsky. 2000. The regulatory protein PhoP controls susceptibility to the host inflammatory response in Shigella flexneri. Cell Microbiol. 2:443-452. [DOI] [PubMed] [Google Scholar]
- 43.Mouslim, C., and E. A. Groisman. 2003. Control of the Salmonella ugd gene by three two-component regulatory systems. Mol. Microbiol. 47:335-344. [DOI] [PubMed] [Google Scholar]
- 44.Mouslim, C., T. Latifi, and E. A. Groisman. 2003. Signal-dependent requirement for the co-activator protein RcsA in transcription of the RcsB-regulated ugd gene. J. Biol. Chem. 278:50588-50595. [DOI] [PubMed] [Google Scholar]
- 45.Nesper, J., C. M. Lauriano, K. E. Klose, D. Kapfhammer, A. Kraiss, and J. Reidl. 2001. Characterization of Vibrio cholerae O1 El Tor galU and galE mutants: influence on lipopolysaccharide structure, colonization, and biofilm formation. Infect. Immun. 69:435-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newcombe, J., J. C. Jeynes, E. Mendoza, J. Hinds, G. L. Marsden, R. A. Stabler, M. Marti, and J. J. McFadden. 2005. Phenotypic and transcriptional characterization of the meningococcal PhoPQ system, a magnesium-sensing two-component regulatory system that controls genes involved in remodeling the meningococcal cell surface. J. Bacteriol. 187:4967-4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peschel, A. 2002. How do bacteria resist human antimicrobial peptides? Trends Microbiol. 10:179-186. [DOI] [PubMed] [Google Scholar]
- 48.Raetz, C. R., C. M. Reynolds, M. S. Trent, and R. E. Bishop. 2007. Lipid A modification systems in gram-negative bacteria. Annu. Rev. Biochem. 76:295-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rather, P. N. 2005. Swarmer cell differentiation in Proteus mirabilis. Environ. Microbiol. 7:1065-1073. [DOI] [PubMed] [Google Scholar]
- 50.Rebeil, R., R. K. Ernst, B. B. Gowen, S. I. Miller, and B. J. Hinnebusch. 2004. Variation in lipid A structure in the pathogenic yersiniae. Mol. Microbiol. 52:1363-1373. [DOI] [PubMed] [Google Scholar]
- 51.Rowley, G., M. Spector, J. Kormanec, and M. Roberts. 2006. Pushing the envelope: extracytoplasmic stress responses in bacterial pathogens. Nat. Rev. Microbiol. 4:383-394. [DOI] [PubMed] [Google Scholar]
- 52.Rozalski, A., Z. Sidorczyk, and K. Kotelko. 1997. Potential virulence factors of Proteus bacilli. Microbiol. Mol. Biol. Rev. 61:65-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sieberth, V., G. P. Rigg, I. S. Roberts, and K. Jann. 1995. Expression and characterization of UDPGlc dehydrogenase (KfiD), which is encoded in the type-specific region 2 of the Escherichia coli K5 capsule genes. J. Bacteriol. 177:4562-4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tam, C., and D. Missiakas. 2005. Changes in lipopolysaccharide structure induce the sigma(E)-dependent response of Escherichia coli. Mol. Microbiol. 55:1403-1412. [DOI] [PubMed] [Google Scholar]
- 55.Toguchi, A., M. Siano, M. Burkart, and R. M. Harshey. 2000. Genetics of swarming motility in Salmonella enterica serovar Typhimurium: critical role for lipopolysaccharide. J. Bacteriol. 182:6308-6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tran, A. X., M. E. Lester, C. M. Stead, C. R. Raetz, D. J. Maskell, S. C. McGrath, R. J. Cotter, and M. S. Trent. 2005. Resistance to the antimicrobial peptide polymyxin requires myristoylation of Escherichia coli and Salmonella typhimurium lipid A. J. Biol. Chem. 280:28186-28194. [DOI] [PubMed] [Google Scholar]
- 57.Vilches, S., R. Canals, M. Wilhelms, M. T. Salo, Y. A. Knirel, E. Vinogradov, S. Merino, and J. M. Tomas. 2007. Mesophilic Aeromonas UDP-glucose pyrophosphorylase (GalU) mutants show two types of lipopolysaccharide structures and reduced virulence. Microbiology 153:2393-2404. [DOI] [PubMed] [Google Scholar]
- 58.Wang, W. B., I. C. Chen, S. S. Jiang, H. R. Chen, C. Y. Hsu, P. R. Hsueh, W. B. Hsu, and S. J. Liaw. 2008. Role of RppA in the regulation of polymyxin B susceptibility, swarming, and virulence factor expression in Proteus mirabilis. Infect. Immun. 76:2051-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, W. B., H. C. Lai, P. R. Hsueh, R. Y. Chiou, S. B. Lin, and S. J. Liaw. 2006. Inhibition of swarming and virulence factor expression in Proteus mirabilis by resveratrol. J. Med. Microbiol. 55:1313-1321. [DOI] [PubMed] [Google Scholar]