Abstract
TMC435 is a small-molecule inhibitor of the NS3/4A serine protease of hepatitis C virus (HCV) currently in phase 2 development. The in vitro resistance profile of TMC435 was characterized by selection experiments with HCV genotype 1 replicon cells and the genotype 2a JFH-1 system. In 80% (86/109) of the sequences from genotype 1 replicon cells analyzed, a mutation at NS3 residue D168 was observed, with changes to V or A being the most frequent. Mutations at NS3 positions 43, 80, 155, and 156, alone or in combination, were also identified. A transient replicon assay confirmed the relevance of these positions for TMC435 inhibitory activity. The change in the 50% effective concentrations (EC50s) observed for replicons with mutations at position 168 ranged from <10-fold for those with the D168G or D168N mutation to ∼2,000-fold for those with the D168V or D168I mutation, compared to the EC50 for the wild type. Of the positions identified, mutations at residue Q80 had the least impact on the activity of TMC435 (<10-fold change in EC50s), while greater effects were observed for some replicons with mutations at positions 43, 155, and 156. TMC435 remained active against replicons with the specific mutations observed after in vitro or in vivo exposure to telaprevir or boceprevir, including most replicons with changes at positions 36, 54, and 170 (<3-fold change in EC50s). Replicons carrying mutations affecting the activity of TMC435 remained fully susceptible to alpha interferon and NS5A and NS5B inhibitors. Finally, combinations of TMC435 with alpha interferon and NS5B polymerase inhibitors prevented the formation of drug-resistant replicon colonies.
Hepatitis C is a blood-borne infection that can ultimately result in severe liver diseases, including fibrosis, cirrhosis, and hepatocellular carcinoma (7). The chronic nature of the disease and the significant possibility of long-term liver damage have led to the current global health burden, with an estimated 180 million people being infected, of whom 130 million are chronic hepatitis C virus (HCV) carriers (54).
The current standard-of-care therapy for HCV-infected patients consists of a combination of weekly injected pegylated alpha interferon (Peg-IFN-α) and twice-daily oral ribavirin. Treatment of HCV genotype 1-infected patients with this regimen for 48 weeks has a limited success rate (a 40 to 50% sustained virological response [SVR]) and is associated with a wide range of side effects, including flu-like symptoms, anemia, and depression, leading to treatment discontinuation in a significant proportion of patients (31, 48). Therefore, specifically targeted antiviral therapies for hepatitis C (STAT-C) have been a major focus of drug discovery efforts. Treatments with several NS3/4A protease inhibitors and NS5A and NS5B polymerase inhibitors, alone or in combination with Peg-IFN-α-ribavirin, have recently shown encouraging results in clinical trials (17, 36).
HCV NS3 is an essential, bifunctional, multidomain protein that possesses protease and RNA helicase activities. NS3/4A, the viral enzyme target of TMC435, is a serine protease with a trypsin-like fold that comprises the 181-residue N-terminal protease domain of NS3 and the 54-residue NS4A cofactor. The association of the NS4A cofactor with the NS3 protease domain is required for enzymatic function, stability, and anchoring to the endoplasmic reticulum. The NS3/4A protease is responsible for cleavage of the HCV polyprotein at the junctions between NS3/NS4A, NS4A/NS4B, NS4B/NS5A, and NS5A/NS5B (reviewed by Penin et al. [37]).
Several peptidomimetic inhibitors of the NS3/4A protease are currently undergoing clinical evaluation. Two of these, telaprevir (VX-950) and boceprevir (SCH503034), possess a ketoamide moiety that reacts with the catalytic serine nucleophile to form a reversible covalent enzyme-inhibitor adduct (20, 28, 38, 52). In contrast, BILN2061, ITMN-191 (R7227), MK7009, and TMC435 are reversible noncovalent inhibitors of NS3/4A, and they all share the feature of a peptidomimetic macrocycle comprised of both backbone and side chain atoms (18, 23, 24, 31, 41, 46). The structures of various NS3/4A inhibitor complexes show that these inhibitors bind in a similar region of the enzyme active site.
The results from phase 2b clinical studies with the HCV NS3/4A inhibitors telaprevir and boceprevir have demonstrated significant improvements in cure rates (SVRs) in both treatment-naïve and treatment-experienced genotype 1-infected patients, showing that use of these inhibitors has the potential to shorten the treatment duration to 24 weeks in treatment-naïve patients (11, 16, 29, 34).
TMC435 is a competitive macrocyclic inhibitor of the HCV NS3/4A protease currently in clinical development by Tibotec (41). It has Ki values of 0.4 nM and 0.5 nM against genotype 1a and 1b enzymes, respectively, and a half-maximal (50%) effective concentration (EC50) of 8 nM in a genotype 1b replicon cell line with a luciferase readout (21). TMC435 also displayed potent inhibition of most NS3/4A proteases derived from genotypes 2 to 6, with the half-maximal (50%) inhibitory concentration (IC50) values being below 13 nM for all HCV NS3/4A enzymes tested, with the exception of a genotype 3 protease (37 nM). In vitro replicon studies have shown that the use of TMC435 with IFN-α and an HCV NS5B polymerase inhibitor results in synergistic activity and that the use of TMC435 with ribavirin results in additive activity (21). In clinical studies, a once-a-day dosing schedule of TMC435 has shown potent antiviral activity in genotype 1-infected treatment-naïve and treatment-experienced patients when it is used alone and in combination with Peg-IFN-α/ribavirin (30, 32, 42).
The importance of viral resistance on the outcome of HCV therapy in the era of direct-acting antivirals remains to be elucidated. Extensive work has identified a number of mutations associated with in vitro and in vivo exposure to HCV NS3/4A inhibitors (13). The identified positions cluster around the active site of the protease, consistent with the idea that changes at inhibitor binding sites will affect the inhibitory activity.
Here we describe the first characterization of the in vitro resistance profile of our HCV NS3/4A protease inhibitor, TMC435.
MATERIALS AND METHODS
Compounds.
The investigational HCV NS5B nucleoside inhibitor (NI) NM-107 was purchased from Toronto Research Chemicals (North York, Ontario, Canada). The HCV NS5B nonnucleoside inhibitors (NNIs) HCV-796 (3), Merck-indole (compound 55 described by Harper et al. [9]), thiophene-2 (compound 16 described by Chan et al. [4]), and GSK-625433 (8) were synthesized in-house, as described below.
The HCV NS3 protease inhibitor (PI) TMC435 (referred to as TMC435350 by Raboisson et al. [41]) was synthesized as described previously (41). The PIs boceprevir, ITMN-191, and BILN2061 were obtained from Spectrum Info (Kiev, Ukraine), Medivir (Huddinge, Sweden), and Acme Bioscience (Palo Alto, CA), respectively.
Human recombinant IFN-α was purchased from Calbiochem (La Jolla, CA). Ribavirin was purchased from Sigma Aldrich (St. Louis, MO).
Cells.
Huh7-Luc cells contain the genotype 1b (con1-based) bicistronic subgenomic replicon (clone ET), which encodes a firefly luciferase reporter with cell culture-adaptive mutations E1202G, T1280I, and K1846T in NS3 and NS4B. These cells were kindly provided by R. Bartenschlager (adapted from Lohmann et al. [25, 26]). Huh7-con1b cells (i.e., a genotype 1b [con1-based] subgenomic replicon clone with cell culture-adaptive mutation S2204I in NS5A) and Huh7-SG1a H77 cells (i.e., a genotype 1a [H77-based] subgenomic replicon clone with cell culture-adaptive mutations P1496L and S2204I in NS3 and NS5A) were obtained from Apath LLC (Brooklyn, NY) (1, 2).
Replicon cells were maintained in Dulbecco modified Eagle medium (DMEM; Sigma D-5546 medium supplemented with 10% fetal calf serum, 1% l-glutamine, 0.04% gentamicin [50 mg/ml]) containing 500 to 750 μg/ml G418. Human hepatoma Huh7-lunet cells (obtained from R. Bartenschlager) and Huh7.5 cells (Apath LLC) were maintained in DMEM.
In vitro selection experiments.
Replicon cells (Huh7-Luc, Huh7-con1b, or Huh7-SG1a cells) were seeded in 10-cm dishes (3 × 105 to 4 × 105 cells) containing DMEM supplemented with 750 μg/ml G418. Next, TMC435 was added at the appropriate concentration and the plates were incubated at 37°C in a humidified 5% CO2 atmosphere until the cells reached 70% confluence. The cells were then split at a ratio of 1:2 to 1:5 and seeded in fresh medium containing constant or increasing concentrations (steps of +10× EC50) of TMC435. After 2 to 3 weeks, significant cell death occurred (typically at a TMC435 concentration of ∼40× EC50). The surviving cell colonies were individually picked, and the replicon RNA was sequenced. For some experiments, the remaining cell colonies were removed from the cell culture dish and pooled and the replicon RNA was sequenced. The RNA of the cells was extracted with an RNeasy minikit (Qiagen, Hilden, Germany). The NS3/4A region of the HCV genome was amplified by reverse transcription-PCR (RT-PCR), and the population sequence was determined to identify the acquired mutations. Cells incubated in the absence of compound or incubated in the presence of NS5B inhibitors were analyzed in parallel and were used to determine the level of variation in NS3 in the absence of an NS3/4A protease inhibitor.
In vitro selection in HCV genotype 2 JFH-1 model.
The DNA of the JHF-1 wild type (JFH-1wt), which does not contain the reporter gene, and Jc-1-Luc, which contains the Renilla luciferase reporter gene (22, 39, 53), both of which encompassed the complete or part of the HCV genotype 2a JFH-1 genome, were prepared by using the GeneWriter synthetic gene assembly technology (Centocor, San Diego, CA). Two fragments were synthesized separately and cloned together into the pUC19 vector for further use.
Ten micrograms of pUC19 plasmid DNA containing the JFH-1wt- or Jc-1-Luc-coding sequence was linearized, and the DNA was transcribed into RNA in vitro by using a T7 MEGAscript high-yield transcription kit (Ambion, Foster City, CA), according to the manufacturer's protocol. The RNA was purified by phenol-chloroform extraction and isopropanol precipitation and was quantified spectrophotometrically with a Nanodrop instrument (Thermo Scientific, Waltham, MA).
For in vitro selection, Huh7.5 cells were washed with phosphate-buffered saline and resuspended at 107 cells per ml in Cytomix (120 mM KCl, 0.15 mM CaCl2, 10 mM K2HPO4/KH2PO4 [pH 7.6], 25 mM HEPES, 2 mM EGTA, 5 mM MgCl2) containing 2 mM ATP and 5 mM glutathione. Of this cell suspension, 400 μl was transferred to an electroporation cuvette (gap width, 0.4 cm; Bio-Rad, Hercules, CA) containing 10 μg JFH-1wt or Jc-1-Luc RNA. The cuvette was placed in the electroporation chamber (Gene-pulser; Bio-Rad) and pulsed once (settings, 960 μF; 270 V; expected time constant, ∼20 ms). After transfection, the cells were transferred to DMEM supplemented with 10% fetal calf serum (FCS), 1% l-glutamine, and 0.04% gentamicin; and 500,000 cells were seeded in a 10-cm2 dish. After 3 days, the cells were washed and cultured in the presence of TMC435 or vehicle for multiple passages. When a cytopathic effect (CPE) occurred, the cells were not split and the cells were refreshed medium with compound or vehicle until complete recovery of the cells. The RNA was extracted and analyzed by a TaqMan real-time RT-PCR at every cell passage. Relative JFH-1 RNA levels were calculated by normalization to the RPL13A RNA levels by the comparative threshold cycle (CT) method (45).
Transient replicon assay.
A plasmid encoding a genotype 1 subgenomic bicistronic replicon with the poliovirus internal ribosome entry site-driven luciferase reporter and cell culture-adaptive mutations E1202G, T1280I, and K1846T in NS3 and NS4B was kindly provided by R. Bartenschlager (adapted from Lohmann et al. [25, 26]). Site-directed mutations were inserted in the NS3 region of that construct at Eurofins Medigenomix by the use of standard site-directed mutagenesis (Ebersberg, Germany). Circular plasmid DNA was used for mutant replicon RNA preparation, as described above (plasmid linearization, however, was done with ScaI instead of XbaI).
Huh7-lunet cells were centrifuged at 1,500 rpm for 5 min, washed with phosphate-buffered saline, and electroporated with 10 μg of wild-type or mutant replicon RNA, essentially as described previously (25). After transfection, the cells were transferred into DMEM supplemented with 10% FCS, 1% l-glutamine, and 0.04% gentamicin and seeded at 4,000 cells per well in 384-well plates containing TMC435 in a nine-point dilution series. After 48 h of incubation at 37°C in a humidified 5% CO2 atmosphere, Steady-Lite Plus reagent (Perkin-Elmer, Waltham, MA) was added at a 1:1 ratio and the plates were further incubated for 10 to 20 min (in the dark), after which the luminescence was measured with a Viewlux apparatus (Perkin-Elmer). The TMC435 EC50s and changes in the EC50s compared to the parental HCV replicon EC50 were determined. Replication capacity was determined in the absence of inhibitors, essentially as described previously (27, 35), by measuring the luciferase levels at 48 h postelectroporation and normalization of those levels with the luciferase levels obtained at 4 h postelectroporation.
Colony formation assay.
Huh7-Luc replicon cells (20,000 cells) were seeded in a 10-cm dish containing DMEM and were treated with different concentrations of a single compound or a combination of two different compounds in the presence of 1,000 μg/ml G418. The cells were incubated at 37°C in a humidified 5% CO2 atmosphere. The medium plus compound was refreshed twice weekly. After 2 to 3 weeks, when significant cell death had occurred, the remaining colonies were stained with neutral red and counted.
RESULTS
In vitro selection with TMC435 in genotype 1 replicon cells.
As a first step in the determination of the in vitro profile of resistance to TMC435, human hepatoma Huh7 cells containing a replicon derived from HCV genotype 1a (Huh7-SG1a H77 cells) or genotype 1b (Huh7-Luc and Huh7-con1b cells) were incubated with increasing concentrations of TMC435 in the presence of G418. Replicon RNA was extracted and the sequence of the NS3 protease region was determined by population-based Sanger sequencing to assess the changes associated with exposure to TMC435. In total, 14 independent selection experiments were performed, and replicon RNA from 109 replicon-containing cell colonies or cell pools was analyzed (63 genotype 1b and 46 genotype 1a colonies). As a control, the sequence of the NS3 region from replicon cells incubated in the absence of an NS3/4A protease inhibitor was used to determine the background variability in this region.
Mutations representing inhibitor-associated changes as well as the background variability in replicon cells were observed in the TMC435 selection experiments (Fig. 1A). Interestingly, most replicon-containing cell colonies or cell pools remaining after TMC435 exposure contained one or multiple mutations at NS3 positions 80, 155, 156, and/or 168 of the replicon; the exceptions were four genotype 1a replicon-containing colonies that harbored the P89R plus S174K, Q41R plus E176K, Q41Q/R, and Q41Q/R plus S174K/N mutations in the NS3 protease domain of the replicon, respectively. In contrast, none of the sequences analyzed from the control experiments contained mutations at position 80, 155, 156, or 168, while one Q41R mutation was detected (Fig. 1A).
FIG. 1.
Summary of results from in vitro selection experiments. (A) Frequency of changes in the protease domain of NS3 (amino acids 1 to 181) observed in experiments conducted with different replicon-containing cell lines in the presence or the absence of TMC435. NS3 sequences from 120 control replicon colonies or cell pools (21 of genotype 1a and 99 of genotype 1b) and 109 TMC435-treated replicon colonies or cell pools (46 of genotype 1a and 63 of genotype 1b) were assessed. Positions at which mutations were present in more than three sequences are shown. Dark gray bars, TMC435-treated cells; white bars, control cells. (B) Frequency of mutations at one or more of NS3 positions 43, 80, 155, 156, and 168 in cells selected with TMC435. Experiments were performed with genotype 1a (Huh7-SG1a H77) or genotype 1b (Huh7-Luc and Huh7-con1b) replicon-containing cells. Mutations containing a mixture of the wild-type residue with a mutated residue are counted as containing the mutated residue only. The frequency was calculated for each subtype. Black bars, genotype 1a; light gray bars, genotype 1b.
The mutations that were observed most frequently in NS3 from replicons across all cell lines cultured in the presence of TMC435 were located at amino acid D168 (86 of 109 sequences, i.e., 80%), with the majority harboring a D168V or a D168A mutation (Fig. 1B). The D168A mutation was the most frequent mutation in genotype 1a replicon cells, while in genotype 1b replicon cells, the D168V mutation was the most abundant. Amino acid Y, E, H, I, T, or N was also detected at position 168, indicating that a variety of viable changes are capable of reducing susceptibility to TMC435. The nucleotide sequence encoding these residues at position 168 deviated from the wild-type sequence by one nucleotide (e.g., for amino acids V, A, and E) or two nucleotides (for amino acids I and T).
Although other amino acid changes were less frequent, an amino acid change of A156 to V, T, or G, as well as an amino acid change of Q80 to K, R, or H, was also found in these experiments. The Q80K mutation was observed only in replicons from genotype 1a cells, whereas the A156V mutation was present only in genotype 1b replicons, although both of these changes required only one nucleotide substitution in either subtype.
Seven genotype 1a replicon-containing cell colonies harboring an R155K mutation were observed after selection, whereas this mutation was not found during selection with genotype 1b replicon-containing cells. In the genotype 1a replicons, only one nucleotide substitution from the wild-type sequence was required to encode a R155K mutation, whereas at least two nucleotide changes were required in genotype 1b replicons, which could explain this genotype-specific resistance pathway. Of note, four of the seven sequences with an R155K mutation also harbored a mutation at position 80 and/or 168.
Mutations at other positions of the NS3 protease domain either were detected less frequently; were present in control replicon cells at similar frequencies (e.g., a mutation at NS3 position 71 or 109); or were detected only in combination with a mutation at position 80, 155, 156, or 168 (Fig. 1A). F43S was present only in the replicons of treated cells and was always found in combination with a mutation of residue Q80 or D168. The Q41R mutation was observed more frequently in cells selected with TMC435 than in control cells (Fig. 1A). Q41R was mainly observed in replicons from genotype 1a cells, and with the exception of three sequences, this mutation was always present in combination with mutations at position 80 and/or 168 (17 of 109 sequences in total). Mutations at residue E176 were detected in some TMC435-treated genotype 1a replicon-containing cell lines. Since mutations at this position are known cell culture-adaptive mutations, which increase the levels of replication of HCV replicons, and a E176G mutation is already present in the genotype 1b replicons used for this study, mutations at this position were not assessed further (14, 25).
A number of sequences (13 of 109) with combinations of mutations at positions 43, 80, 156 and 168 were found: F43S plus Q80R or D168E; Q80K or Q80R plus R155K; Q80R plus D168E or D168A; Q80H plus D168E; A156V plus D168V or D168A; and Q80K plus R155K plus D168A. Since these samples were analyzed by standard population sequencing, the possibility that these mutations were present on different viral genomes cannot be excluded.
In vitro selection with TMC435 in genotype 2a JFH-1 cells.
The mutation patterns for the infectious genotype 2a JFH-1 cell culture model were then determined by using human hepatoma Huh7.5 cells transfected with either JFH-1wt or Jc-1-Luc (22, 39, 53) and cultured in the presence of TMC435.
The culture of JFH-1-transfected cells in the absence of compound resulted in a clear CPE, typically after five to eight passages, followed by recovery and normal cell growth.
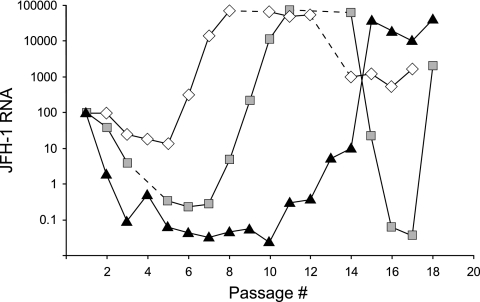
In an experiment with JFH-1wt-transfected cells, incubation with TMC435 reduced the cellular JFH-1 RNA levels and delayed the occurrence of the CPE at 500 nM TMC435, while no CPE was observed at 4,000 nM TMC435. After the initial decline, JFH-1 HCV RNA replication recovered in the presence of 500 nM TMC435 and recovered at a lower rate in the presence of 4,000 nM TMC435, and the cellular JFH-1 RNA levels increased to levels similar to those observed in the absence of compounds (Fig. 2). Interestingly, the JFH-1 RNA levels dropped at about passage 14 in the control experiment and in cells treated with 500 nM TMC435. The reasons or the relevance of this finding are currently not understood. Sequencing of NS3/4A derived from cells harvested at multiple time points between passages 12 and 19 resulted in the continuous detection of mutations D168N and F43V in cells treated with 500 nM and 4,000 nM TMC435, respectively.
FIG. 2.
Selection experiments with JFH-1. Huh7.5 cells were transfected with RNA-encoding JFH-1wt, and the cells were passaged in the presence of 500 nM or 4,000 nM TMC435 or in the absence of inhibitor (control). Cells were harvested at each passage, and the JFH-1 RNA was quantified by using quantitative RT-PCR and normalized against the quantity of the host RPL13A gene. The JFH-1 RPL13A levels 3 days after transfection (after the first passage and at the start of TMC435 treatment) were set equal to 100%, and the changes over time are shown. White diamonds, cells cultured in the absence of inhibitor; gray squares, cells cultured in the presence of 500 nM TMC435; black triangles, cells cultured in the presence of 4,000 nM TMC435. Time points at which a CPE was observed and no cells could be harvested are indicated with a dashed line.
Analysis of the NS3/4A region from two additional selection experiments in which Jc-1-Luc-transfected cells were cultured in the presence of TMC435 revealed NS3 mutations F43F/I plus A156G, D168Y, A156A/V, and D168D/V.
Interestingly, the mutations observed in the genotype 2a JFH-1 genome were located at the same positions as the mutations identified in the genotype 1 replicon systems, suggesting that the two genotypes have similar routes of escape from TMC435 drug pressure.
Impact of NS3 mutations on TMC435 susceptibility and replication capacity.
The effect of TMC435 inhibitory activity on replicons encoding the NS3 sequence mutations identified in the in vitro selection experiments with TMC435 and previously described for other investigational protease inhibitors (13) was assessed. Mutants with single or double mutations were engineered into a luciferase-containing con1b replicon construct, and the inhibitory activity was determined in a transient replicon assay. The antiviral activities of two other macrocyclic protease inhibitors (BILN2061 and ITMN-191) and two linear ketoamide inhibitors (boceprevir and telaprevir) were also assessed.
Of the mutations identified in the in vitro selection experiments with TMC435, those at positions 43, 80, 155, 156, and 168 resulted in various degrees of reduced susceptibility to TMC435, confirming the importance of these residues for TMC435 activity. In contrast, two other mutations also observed during the selection experiments, Q41R and R109K, had no significant impact on the activity of TMC435 (Table 1; see the supplemental material for a table that includes information on the interquartile ranges and the numbers of measurements).
TABLE 1.
Effects of NS3 mutations on antiviral activities of several NS3/4A protease inhibitorsa
| Mutant | RC (% of that for WT) | TMC435 |
BILN-2061 |
ITMN-191 |
Boceprevir |
Telaprevir |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| EC50 (nM) | FC | EC50 (nM) | FC | EC50 (nM) | FC | EC50 (nM) | FC | EC50 (nM) | FC | ||
| WT | 100 | 11 | NA | 0.8 | NA | 0.5 | NA | 148 | NA | 150 | NA |
| V36L | 56 | 18 | 1.7 | 1.4 | 1.0 | ND | ND | 110* | 1.6* | 300* | 3.1* |
| V36 M | 43 | 20 | 2.0 | 2.4 | 1.2 | ND | ND | 217 | 1.8 | 886 | 10 |
| V36A | 22 | 34 | 2.8 | 1.0 | 1.1 | 1.3* | 3.1* | 1,454 | 6.8 | ND | ND |
| Q41R | ND | 18 | 1.7 | 2.3 | 2.8 | ND | ND | ND | ND | ND | ND |
| F43S | 63 | 83 | 12 | 1.0 | 0.9 | 5.1* | 15* | 333 | 5.2 | 749 | 6.1 |
| F43I | ND | 562* | 89* | 0.4* | 0.5* | ND | ND | 242* | 1.1* | ND | ND |
| F43V | ND | 626* | 99* | 0.4* | 0.5* | ND | ND | 527* | 2.4* | ND | ND |
| T54A | 61 | 5.9 | 0.6 | 0.7 | 0.6 | 0.6* | 1.3* | 268 | 2.1 | 1,565 | 7.5 |
| T54S | ND | 5.4 | 1.2 | 0.9 | 1.9 | ND | ND | 1,106 | 8.5 | ND | ND |
| Q80R | 40 | 49 | 6.9 | 1.4 | 1.6 | 4.5 | 3.5 | 85 | 0.5 | 216 | 0.6 |
| Q80H | ND | 41 | 3.6 | 1.2 | 1.1 | 1.0* | 1.4* | 78 | 0.5 | ND | ND |
| Q80K | 98 | 62 | 7.7 | 1.0 | 0.9 | 0.9 | 2.3 | 94 | 0.8 | 147 | 0.5 |
| Q80G | 111 | 22 | 1.8 | 3.0 | 4.9 | 3.5 | 9.8 | 127 | 1.2 | 202 | 1.2 |
| Q80L | 169* | 12 | 2.1 | 0.8 | 1.2 | ND | ND | 133 | 0.9 | ND | ND |
| R109K | 311 | 11 | 0.7 | 1.4 | 0.7 | ND | ND | 33* | 1.0* | 66* | 0.8* |
| S138T | 19* | 77 | 4.4 | 0.5 | 3.0 | 3.0 | 3.4 | 16 | 0.1 | ND | ND |
| R155 M | 33 | 3.5 | 0.4 | 84 | 68 | 3.8* | 21* | 792 | 5.7 | 4,106 | 14 |
| R155I | ND | 6.6* | 0.8* | 29* | 39* | 7.0* | 20* | 5,343* | 28* | ND | ND |
| R155Q | 10 | 21 | 1.6 | 248 | 242 | 13* | 68.* | 174 | 1.4 | 570 | 2.5 |
| R155T | 7 | 314 | 24 | 685 | 562 | 48* | 261* | 5,463 | 51 | 22,663 | 74 |
| R155K | 40 | 260 | 30 | 489 | 496 | 135 | 447 | 743 | 4.7 | 1,470 | 10 |
| R155G | 4 | 350 | 20 | 1,002 | 811 | 79* | 324* | 4,467 | 20 | 9,631 | 20 |
| A156S | 71 | 3.0 | 0.3 | 1.7 | 2.8 | ND | ND | 525 | 3.5 | 3,107 | 20 |
| A156G | 66 | 206 | 16 | 22* | 12* | ND | ND | 65 | 2.1 | 58 | 0.6 |
| A156T | 30 | 377 | 44 | 1,066 | 1,344 | 4.8 | 41* | 7,227 | 65 | 20,326 | 105 |
| A156V | 18 | 2,149 | 177 | 1,518 | 2,085 | 12 | 63 | 9,673 | 75 | 15,470 | 112 |
| D168G | ND | 42 | 4.4 | 35 | 38 | 2.2 | 8.1* | 63 | 0.4 | ND | ND |
| D168N | 67* | 79 | 6.6 | 18 | 12 | ND | ND | 101 | 1.9 | 147 | 1.2 |
| D168E | 34 | 302 | 40 | 40 | 65 | 6.1 | 75* | 69 | 0.8 | 113 | 0.6 |
| D168T | 192 | 4,089 | 308 | 520 | 844 | 42 | 227 | 299 | 1.1 | 181 | 1.0 |
| D168Y | 30 | 6,238 | 666 | 2,099 | 2,488 | 78 | 391 | 193 | 2.4 | 187 | 1.2 |
| D168H | 104 | 5,655 | 368 | 215 | 263 | 26 | 133 | 73 | 0.7 | 102 | 0.7 |
| D168A | 90 | 6,356 | 594 | 304 | 339 | 31 | 153 | 100 | 0.7 | 46 | 0.4 |
| D168V | 94 | 17,917 | 2,591 | 1,813 | 3,213 | ND | ND | 83 | 0.5 | ND | ND |
| D168I | 36 | 23,203* | 1,807* | 20,715* | 26,822* | 104* | 195* | 279* | 1.1 | ND | ND |
| V170T | ND | 52 | 5.4 | 4.8 | 4.6 | 0.6* | 1.1* | 210* | 1.6* | ND | ND |
| V170A | 30 | 22* | 1.8* | 0.8 | 1.4 | 0.7* | 1.1* | 1,546 | 5.8 | ND | ND |
| F43S + Q80R | ND | 1,815* | 286* | 0.4* | 0.6* | ND | ND | 1,554* | 7.4* | ND | ND |
| F43S + D168E | ND | 3,607 | 694 | 5.8 | 12 | ND | ND | 417 | 3.2 | ND | ND |
| Q80K + R155K | 66 | 4,647 | 364 | 705 | 736 | 309 | 850 | 1,077 | 5.1 | 2,957 | 4.6 |
| Q80R + R155K | 157 | 2,853 | 270 | 707 | 772 | ND | ND | 680* | 2.7* | ND | ND |
| Q80R + D168E | 52* | 5,902 | 412 | 87 | 103 | 106 | 154 | 62 | 0.6 | ND | ND |
| Q80H + D168E | ND | 1,362 | 163 | 52 | 49 | 26 | 40* | 20* | 0.1* | ND | ND |
| Q80R + D168A | ND | 16,867* | 2,655* | 1,039* | 1,827* | ND | ND | 113* | 0.5* | ND | ND |
EC50s were determined in a transient replicon assay with the wild-type or mutant replicon with single- or multiple-site-directed mutations in a genotype 1b replicon backbone (clone ET) with a luciferase readout. The EC50s presented here were calculated as the medians of the EC50s derived from individual experiments. The replication capacity levels of the mutants are expressed as a percentage relative to that for the wild type. Median values are shown, and values derived from less than three independent experiments are indicated with asterisks. The mutations observed during the selection experiments with TMC435 and the data for replicons with those mutations are labeled in boldface. RC, replication capacity; WT, wild type; FC, median of the fold change in the EC50 for the mutant replicon compared to that for the wild-type replicon determined in each experiment; ND, not determined; NA, not applicable.
The mutations introduced at position 168, in particular D168V and D168I, led to the most significant decrease in the activity of TMC435 and the other two noncovalent macrocyclic inhibitors, BILN2061 and ITMN-191. The changes in the EC50s of TMC435 for replicons with a mutation at position 168 ranged from less than 10-fold for D168G and D168N to about 2,000-fold for D168V and D168I. The covalent inhibitors telaprevir and boceprevir remained fully active against the tested mutants with a mutation at residue D168, consistent with the results previously described for these inhibitors (19, 49, 50).
Residue-dependent changes in TMC435 activity were observed for several of the mutations at amino acid residues F43, Q80, R155, and A156. TMC435 generally showed a smaller change in EC50s than the changes for the other two macrocyclic protease inhibitors tested against replicons with mutations at positions 155 and 156. In contrast, mutations at position 43 led to reduced susceptibilities for TMC435 and ITMN-191, ranging from a 12-fold change in the EC50 of TMC435 for the F43S replicon to an ∼90-fold change for the F43I and F43V replicons, while BILN2061 remained fully active against these mutants. Although changes at residue Q80 had less of an impact on TMC435 activity, with the EC50 change being less than 10-fold for all mutants, this impact was slightly greater than that for the other macrocyclic inhibitors tested.
In addition, S138T, a mutation previously described as conferring reduced susceptibility to ITMN-191 (47), was also found to affect TMC435 activity, with the change in EC50s being 4-fold (Table 1), although this mutation was not identified in selection experiments with TMC435.
Mutations at amino acids V36, T54, and V170, which were observed during clinical trials with telaprevir and boceprevir, had no effect or only small effects on TMC435 activity. The change in the EC50s of TMC435 against all tested mutants with mutations at positions 36, 54, and 170 was less than 6-fold, suggesting partial complementarity of the resistance profiles of TMC435 and NS3/4A inhibitors of the ketoamide inhibitor class.
Single mutations that conferred only low or moderate changes in susceptibility to TMC435 (3- to 40-fold) led to 160- to 700-fold reductions in TMC435 activity when they were combined (a mutation at position 43 with a mutation at position 80 or 168 or a mutation at position 80 with a mutation at position 155 or 168). These mutations presumably act in concert in reducing susceptibility to TMC435. Although nucleotide changes at two or three distant positions are required for mutants with such double mutations, it is likely that 13 of the 109 selected replicon colonies harbored multiple mutations on a single genome, as inferred from the population sequencing results.
The relative replication capacity of the mutated replicons in the absence of inhibitor as a percentage of that for the parental replicon was determined to obtain a surrogate measure of the effect of the mutations on viral fitness (Table 1). Of the mutations identified in the selection experiments, the A156V mutation displayed the lowest replication capacity, which was 18% of that of the wild type, whereas some mutations at amino acid positions 80 and 168 had only a limited or no effect on the replication capacity of the mutated replicon. Interestingly, mutation R109K, which was observed in combination with other mutations, showed an ∼3-fold increased replication capacity, suggesting that it could function as a compensatory mutation by increasing the replication capacity of mutant replicons.
Colony formation assay.
To assess the effect of the use of TMC435 in combination with other HCV inhibitors, the survival frequencies of replicon colonies under culture conditions containing one or more anti-HCV compounds were determined. Huh7-Luc cells were incubated in the presence of G418 with TMC435 alone or in combination with IFN-α or HCV NS5B polymerase inhibitors. At the end of the treatment period, the surviving replicon cell colonies were counted (Fig. 3 and Table 2).
FIG. 3.
Influence of anti-HCV inhibitors used alone or in combination on the formation of replicon colonies. The images are of cells treated with TMC435 or thiophene-2 alone and in combination. Concentrations are expressed as multiples of the respective EC50s determined in a genotype 1b replicon assay with a luciferase readout.
TABLE 2.
Influence of anti-HCV inhibitors, used alone or in combination, on the formation of replicon coloniesa
| Compound | Class | Concn | No. of colonies after treatment with TMC435 at: |
|||
|---|---|---|---|---|---|---|
| 0 | 2.5 × 8 nM | 10 × 8 nM | 25 × 8 nM | |||
| / | / | / | ≫100 | NA | 90 | 11 |
| Merck-indole | NS5B NNI-1 | 10 × 200 nM | 47 | NA | 0 | 0 |
| 25 × 200 nM | 1 | NA | 0 | 0 | ||
| Thiophene-2 | NS5B NNI-2 | 10 × 600 nM | ±100 | NA | 0 | 0 |
| 25 × 600 nM | 11 | NA | 0 | 0 | ||
| / | / | / | ≫100 | NA | ±100 | 36 |
| NM-107 | NS5B NI | 1 × 4 μM | ≫100 | NA | 78 | 16 |
| 10 × 4 μM | 80 | NA | 0 | 0 | ||
| GSK-625433 | NS5B NNI-3 | 10 × 15 nM | >100 | NA | 1 | 0 |
| 25 × 15 nM | ±100 | NA | 1 | 0 | ||
| HCV-796 | NS5B NNI-4 | 5 × 14 nM | >100 | NA | 0 | 0 |
| 10 × 14 nM | 45 | NA | 0 | 0 | ||
| 25 × 14 nM | 5 | NA | 0 | 0 | ||
| / | / | / | ND | 25 | NA | 10 |
| IFN-α | Interferon | 2.5 × 0.8 IU/ml | 120 | 1 | NA | 0 |
| 5 × 0.8 IU/ml | 31 | 0 | NA | 0 | ||
The results of three independent representative experiments are shown. Concentrations are expressed as multiples of the respective EC50s determined in a genotype 1b replicon assay with a luciferase readout. EC50 values are as follows: TMC435, 8 nM; Merck-indole, 200 nM; thiophene-2, 600 nM; NM-107, 4 μM; GSK-625433, 15 nM; HCV-796, 14 nM; and IFN-α, 0.8 Iu/ml. Slashes, absence of inhibitor; ≫100, confluence of cells; NA, not applicable.
Increasing concentrations of TMC435 (up to 25× EC50) correlated with lower numbers of surviving replicon cell colonies. However, TMC435 alone did not completely inhibit the formation of these colonies at any of the concentrations tested.
The effects of HCV NS5B NNIs, which bind at each of the four best-described NS5B allosteric sites, and an NS5B NI, which binds at the active site of NS5B, were determined alone and in combination with TMC435 (6). For all NS5B inhibitors tested, a concentration-dependent reduction in the number of colonies was observed, but the complete clearance of replicon cells was not achieved at the concentrations tested when polymerase inhibitors were used alone. Dual drug combinations, consisting of TMC435 and four of the five NS5B inhibitors tested, were able to completely block the appearance of resistant colonies, in most cases, at the lowest concentrations tested (Table 2). The only exception was a combination of the NNI 3-site inhibitor GSK-625433 and TMC435, which was insufficient for the complete inhibition of colony formation. This is consistent with the appearance of more than 200 colonies when GSK-625433 was tested alone at 25× EC50 and suggests a lower genetic barrier for NNI 3-site inhibitors. Use of the combination of TMC435 and this inhibitor significantly reduced the formation of colonies but did not fully clear the replicon from the cells.
In addition, combinations of TMC435 with IFN-α completely inhibited the formation of replicon cell colonies (Table 2). In control experiments, use of a combination of these HCV inhibitors at the highest concentration tested did not affect cell growth over the 2-week time period.
These data indicate that the use of TMC435 in combination with other specifically targeted antivirals with complementary mechanisms of action could reduce or prevent the accumulation of drug resistance-conferring mutations in vitro and could result in the complete clearance of HCV replicon RNA.
DISCUSSION
The discovery of STAT-Cs for the treatment of patients infected with hepatitis C virus holds the promise of improving cure rates and shortening treatment durations. The results of recent phase 2b studies in which HCV genotype 1-infected patients were treated with an HCV NS3/4A inhibitor in combination with Peg-IFN-α-ribavirin suggest that significant improvements in HCV therapy could become possible (11, 16, 29, 34).
Given the high rate of mutation of HCV, the emergence of resistant variants can be expected in the era of treatment with STAT-Cs, which may lead to treatment failures and which could ultimately restrict future treatment options. Viral variants harboring mutations that confer reduced susceptibility to the antiviral agent have been reported for essentially all specifically targeted HCV inhibitors in in vitro studies and/or during clinical trials. However, the clinical impact of resistant variants on the treatment of HCV-infected patients remains to be fully understood. It is encouraging that the viral breakthrough and relapse rates observed in treatment-naïve genotype 1-infected patients when HCV NS3/4A protease inhibitors were combined with the standard of care (Peg-IFN-α-ribavirin) have been low, suggesting that such combinations may effectively inhibit the emergence of resistant viral variants in many patients (11, 16, 30).
As for other viral diseases, in vitro studies have been helpful in determining which mutations may occur during the clinical use of HCV inhibitors. Most mutations associated with viral resistance to HCV protease inhibitors in clinical trials either had been found in replicon selection experiments or were previously shown to confer reduced susceptibility in replicon or biochemical assays. In some cases, the specific mutations found during clinical trials had not been observed in preceding in vitro replicon selection experiments. One example is the R155K mutation observed during clinical trials for protease inhibitors, such as ITMN-191 and telaprevir, which had not been reported from in vitro studies (19, 43, 47). This may be related to the use of only genotype 1b replicon cells to characterize the in vitro resistance profiles of these compounds. In a recent study, R155K mutations were observed in vitro with telaprevir when genotype 1a replicon cells were used in the selection experiments (33). Conversely, some mutations have been identified in specifically designed in vitro selection studies but have not yet been described during clinical trials. This may be related to an insufficient effect of the mutation on the in vivo potency of the drug, the limited in vivo fitness of the mutant, or the mutations may represent in vitro artifacts.
In vitro selection experiments and transient replicon assays consistently demonstrated that changes at position 168 of NS3 are the most frequently observed mutations under selection with TMC435, with D168V and D168A being the most frequent. Also, D168V and D168A conferred the greatest shifts in the EC50s for TMC435. The D168I mutation, which reduced the TMC435 activity to a similar degree as the D168V mutation, was observed only twice in our selection experiments. This lower incidence may be related to the fact that at least two nucleotide changes are required for an amino acid change from D to I, while only one nucleotide change is needed for a V substitution. Mutations at NS3 positions 43, 80, 155, and 156 were also observed during selection experiments and conferred different levels of reduced susceptibility to TMC435, ranging from less than 10-fold (e.g., for Q80R and Q80K) to greater than 100-fold for A156V. Some genotype 1a-specific versus genotype 1b-specific differences were noted, with the A156V mutation being observed only in genotype 1b replicons and the Q80K and R155K mutations being detected only in genotype 1a replicons; the latter observation is consistent with results recently reported for telaprevir (33).
Most mutations which resulted in reduced susceptibility to TMC435 in the transient replicon assay also affected the replication capacity of the mutant replicon to various degrees. Of note, the replication capacity of the replicon carrying a D168V or a D168A mutation, as determined by this assay, was comparable to that of the parental replicon and, thus, was higher than the replication capacities previously reported for replicons with these mutations (10, 35). Further studies assessing different methods of determining in vitro replication capacity and fitness will be needed to define the optimal approach to the assessment of in vitro fitness and to obtain an understanding of the relevance of these in vitro findings for the clinical setting.
The mutations observed in association with TMC435 exposure in the genotype 2a JFH-1 system occurred at the same NS3 positions as those identified in the genotype 1 replicon cells. In contrast to genotype 1, in which an F43S mutation was observed, F43I and F43V were detected in the JFH-1 system. Analysis of the effects of these three changes on TMC435 activity in a transient genotype 1b replicon assay showed that the F43I or F43V mutation reduced the susceptibility to TMC435 by ∼90-fold, while the F43S mutation had a more modest effect on TMC435 activity (12-fold).
The TMC435-associated mutations described above were mapped onto a recently determined 2.4-Å-resolution crystal structure of the TMC435-NS3/4A protease complex (Fig. 4) (5). Consistent with the results described for other NS3/4A inhibitors, the predominant mutations arising in response to TMC435 exposure in vitro clustered around the inhibitor binding site (Fig. 4A). Residues Q80, R155, A156, and D168 contributed to the binding surface in an extended S2 subsite induced by the binding of TMC435 (Fig. 4A), consistent with the importance of these positions to the antiviral activity of TMC435 observed in vitro. The specific interactions between R155, D168, and Q80 contribute to stabilization of the conformation of the extended S2 subsite required for the binding of TMC435. Additionally, R155 forms a significant part of the surface that is buried by the quinoline of bound TMC435. A156 is also in the S2 subsite region, forming a “barrier” between the S2 and S4 regions. F43 forms the base of the S1′ subsite occupied by the cyclopropyl ring of bound TMC435, and mutation F43S confers low-level resistance to TMC435 in vitro, while F43I and F43V have more pronounced effects. The S138T mutation directly adjacent to the catalytic residue S139 likely disturbs the binding contacts with the acylsulfonamide group of TMC435.
FIG. 4.
Structural perspective on selected mutations. (A) Overall view of the NS3/4A-TMC435 complex with the key in vitro resistance-conferring mutation positions highlighted (5). (B) Close-up of the extended S2 binding region occupied by the substituted quinoline ring of bound TMC435. The figure was prepared with the PyMol program (Delano Scientific, LLC).
Some cross-resistance has been observed in vitro between linear ketoamide inhibitors and TMC435 for a few mutations at positions A156 and R155. In clinical studies with telaprevir, the R155K mutation has frequently been observed, while mutations at A156 were less frequent, suggesting the limited fitness of the latter variants in vivo (11, 44). Thus, R155K may be a key mutation that differentially reduces susceptibility to most of the macrocyclic and linear ketoamide inhibitors described today (13, 15, 43). The effect of R155K or other mutations at position 155 on TMC435 activity in vitro was limited, with a 30-fold change in EC50s being observed for replicons with the R155K mutation. Given the extensive contact between R155 and bound TMC435 (Fig. 4), the relatively small impact of mutations of residue R155 on TMC435 was unexpected (Table 1). Although the large quinoline-thiazole system of TMC435 occupies an extended S2 subsite (Fig. 4), similar to what has been shown for a close analog of BILN2061 (51) and consistent with our expectation for both BILN2061 and ITMN-191, the impact of mutations on position 155 on TMC435 activity is closer to what was observed with inhibitors that position small groups in S2 (boceprevir and telaprevir) (40). This feature appears to distinguish TMC435 from other macrocyclic inhibitors of NS3/4A.
For HIV-1, many years of genotypic and phenotypic testing and the clinical use of antiretrovirals have resulted in a well-established correlation between changes in antiviral activity in vitro and the response to treatment (12). With the introduction of antivirals for the direct treatment of HCV infections, understanding of the relevance of changes in in vitro susceptibility to a given drug on its antiviral effect in the clinic will become increasingly important. Given the exposures achieved with TMC435 in vivo, it is possible that a number of mutations with a limited effect on drug susceptibility in vitro could be still suppressed when they are present in an HCV-infected individual (42). In support of this notion, in six genotype 1-infected patients who received 200 mg TMC435 once daily for 5 days during a phase 1 monotherapy trial, the HCV RNA level remained suppressed for at least 3 days after the last dose was given, although viral variants with reduced in vitro susceptibility to TMC435 were present (42). This finding suggests that the balance between drug exposure levels and different degrees of susceptibility of specific viral variants could be an important determinant for the occurrence of viral breakthrough and treatment failure in a clinical setting.
The use of combinations of directly acting antivirals will likely be crucial to reduce the impact of resistant variants and, ultimately, to achieve high cure rates in HCV genotype-1-infected patients. We have recently reported that in vitro, the activity of TMC435 is additive with that of ribavirin and synergistic with that of IFN-α and of an NS5B inhibitor in a 3-day replicon assay (21). Here we show that the use of combinations of TMC435 with IFN-α or different NS5B inhibitors significantly reduced the survival frequency of replicon colonies or even completely eliminated the replicon colonies. In addition, mutants with the NS3 mutations tested were fully susceptible to IFN-α and NS5A and NS5B inhibitors, while TMC435 was active against mutants with mutations that confer reduced susceptibility to NS5B or NS5A inhibitors (data not shown).
In summary, TMC435 is a potent inhibitor of HCV in vivo and has an in vitro resistance profile that is partially complementary to the profiles of the ketoamide protease inhibitors. In addition, as recently described (21), the combination of TMC435 with other anti-HCV agents has an additive to synergistic effect on antiviral activity in vitro and, as shown here, can inhibit the formation of drug-resistant replicon cells, consistent with a role for TMC435 in future combination therapies. The clinical relevance of the mutations identified in vitro on the antiviral activity of TMC435 in vivo will be assessed in ongoing and future clinical trials.
Supplementary Material
Acknowledgments
Special thanks go to the colleagues from Medivir, Jinq-May Chen, Sara Felländer, and Susanne Nyström, for protein purification and crystallization. Thanks also go to Luc Geeraert for his help with the preparation of the manuscript.
All of us are employees of Tibotec or Medivir, the companies that discovered and that are developing TMC435.
Footnotes
Published ahead of print on 22 February 2010.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Blight, K. J., J. A. McKeating, J. Marcotrigiano, and C. M. Rice. 2003. Efficient replication of hepatitis C virus genotype 1a RNAs in cell culture. J. Virol. 77:3181-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burns, C. J., A. M. Del Vecchio, T. R. Bailey, B. A. Kulkarni, T. H. Faitg, S. R. Sherk, C. W. Blackledge, D. J. Rys, T. A. Lessen, J. Swestock, Y. Deng, T. J. Nitz, J. A. Reinhardt, H. Feng, and A. K. Saha. May 2004. Benzofuran compounds, compositions and methods for treatment and prophylaxis of hepatitis C viral infections and associated diseases. PCT patent WO2004041201.
- 4.Chan, L., O. Pereira, T. J. Reddy, S. K. Das, C. Poisson, M. Courchesne, M. Proulx, A. Siddiqui, C. G. Yannopoulos, N. Nguyen-Ba, C. Roy, D. Nasturica, C. Moinet, R. Bethell, M. Hamel, L. L'Heureux, M. David, O. Nicolas, P. Courtemanche-Asselin, S. Brunette, D. Bilimoria, and J. Bédard. 2004. Discovery of thiophene-2-carboxylic acids as potent inhibitors of HCV NS5B polymerase and HCV subgenomic RNA replication. Part 2. Tertiary amides. Bioorg. Med. Chem. Lett. 14:797-800. [DOI] [PubMed] [Google Scholar]
- 5.Cummings, M. D., J. Lindberg, T.-I. Lin, H. de Kock, O. Lenz, E. Lilja, S. Felländer, V. Baraznenok, S. Nyström, M. Nilsson, L. Vrang, M. Edlund, Å. Rosenquist, B. Samuelsson, P. Raboisson, and K. Simmen. 2010. Induced fit binding of the noncovalent macrocyclic inhibitor TMC435 to its HCV NS3/NS4A protease target. Angew. Chem. Int. Ed. Engl. 49:1652-1655. [DOI] [PubMed] [Google Scholar]
- 6.De Francesco, R., and A. Carfi. 2007. Advances in the development of new therapeutic agents targeting the NS3-4A serine protease or the NS5B RNA-dependent RNA polymerase of the hepatitis C virus. Adv. Drug Deliv. Rev. 59:1242-1262. [DOI] [PubMed] [Google Scholar]
- 7.Ghobrial, R. M., R. Steadman, J. Gornbein, C. Lassman, C. D. Holt, P. Chen, D. G. Farmer, H. Yersiz, N. Danino, E. Collisson, A. Baquarizo, S. S. Han, S. Saab, L. I. Goldstein, J. A. Donovan, K. Esrason, and R. W. Busuttil. 2001. A 10-year experience of liver transplantation for hepatitis C: analysis of factors determining outcome in over 500 patients. Ann. Surg. 234:384-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guidetti, R., D. Haigh, C. D. Hartley, P. D. Howes, F. Nerozzi, and S. A. Smith. May 2006. Preparation of 4-(methoxycarbonyl)pyrrolidine-2-carboxylic acid derivatives as hepatitis C virus inhibitors. PCT patent WO2006045613.
- 9.Harper, S., S. Avolio, B. Pacini, M. Di Filippo, S. Altamura, L. Tomei, G. Paonessa, S. Di Marco, A. Carfi, C. Giuliano, J. Padron, F. Bonelli, G. Migliaccio, R. De Francesco, R. Laufer, M. Rowley, and F. Narjes. 2005. Potent inhibitors of subgenomic hepatitis C virus RNA replication through optimization of indole-N-acetamide allosteric inhibitors of the viral NS5B polymerase. J. Med. Chem. 48:4547-4557. [DOI] [PubMed] [Google Scholar]
- 10.He, Y., M. S. King, D. J. Kempf, L. Lu, H. B. Lim, P. Krishnan, W. Kati, T. Middleton, and A. Molla. 2008. Relative replication capacity and selective advantage profiles of protease inhibitor-resistant hepatitis C virus (HCV) NS3 protease mutants in the HCV genotype 1b replicon system. Antimicrob. Agents Chemother. 52:1101-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hézode, C., N. Forestier, G. Dusheiko, P. Ferenci, S. Pol, T. Goeser, J.-P. Bronowicki, M. Bourlière, S. Gharakhanian, L. Bengtsson, L. McNair, S. George, T. Kieffer, A. Kwong, R. S. Kauffman, J. Alam, J.-M. Pawlotsky, S. Zeuzem, and PROVE2 Study Team. 2009. Telaprevir and peginterferon with or without ribavirin for chronic HCV infection. N. Engl. J. Med. 360:1839-1850. [DOI] [PubMed] [Google Scholar]
- 12.Johnson, V. A., F. Brun-Vézinet, B. Clotet, H. F. Gunthard, D. R. Kuritzkes, D. Pillay, J. M. Schapiro, and D. D. Richman. 2008. Update of the drug resistance mutations in HIV-1. Top. HIV Med. 16:138-145. [PubMed] [Google Scholar]
- 13.Koev, G., and W. Kati. 2008. The emerging field of HCV drug resistance. Expert Opin. Invest. Drugs 17:303-319. [DOI] [PubMed] [Google Scholar]
- 14.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kukolj, G., Y. Benhamou, M. P. Manns, M. Bourlière, S. Pol, M. Schuchmann, M. Cartier, D. Huang, L. Lagacé, G. Steinmann, and J. O. Stern. 2009. A potent HCV NS3 protease inhibitor, in treatment-naive and -experienced chronic HCV genotype-1 infection: genotypic and phenotypic analysis of the NS3 protease domain. J. Hepatol. 50:S347. [Google Scholar]
- 16.Kwo, P., E. Lawitz, J. McCone, E. Schiff, J. Vierling, D. Pound, M. Davis, J. Galati, S. Gordon, N. Ravendhran, L. Rossaro, F. Anderson, I. Jacobson, R. Rubin, K. Koury, C. Brass, E. Chaudhri, and J. Albrecht. 2009. HCV Sprint-1 final results: SVR 24 from a phase 2 study of boceprevir plus pegintron™ (peginterferon alfa-2b)/ribavirin in treatment-naive subjects with genotype-1 chronic hepatitis C. J. Hepatol. 50:S4. [Google Scholar]
- 17.Kwong, A. D., L. McNair, I. Jacobson, and S. George. 2008. Recent progress in the development of selected hepatitis C virus NS3.4A protease and NS5B polymerase inhibitors. Curr. Opin. Pharmacol. 8:522-531. [DOI] [PubMed] [Google Scholar]
- 18.Lamarre, D., P. C. Anderson, M. Bailey, P. Beaulieu, G. Bolger, P. Bonneau, M. Bos, D. R. Cameron, M. Cartier, M. G. Cordingley, A.-M. Faucher, N. Goudreau, S. H. Kawai, G. Kukolj, L. Lagace, S. R. LaPlante, H. Narjes, M.-A. Poupart, J. Rancourt, R. E. Sentjens, R. St. George, B. Simoneau, G. Steinmann, D. Thibeault, Y. S. Tsantrizos, S. M. Weldon, C.-L. Yong, and M. Llinas-Brunet. 2003. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 426:186-189. [DOI] [PubMed] [Google Scholar]
- 19.Lin, C., C. A. Gates, B. G. Rao, D. L. Brennan, J. R. Fulghum, Y.-P. Luong, J. D. Frantz, K. Lin, S. Ma, Y.-Y. Wei, R. B. Perni, and A. D. Kwong. 2005. In vitro studies of cross-resistance mutations against two hepatitis C virus serine protease inhibitors, VX-950 and BILN 2061. J. Biol. Chem. 280:36784-36791. [DOI] [PubMed] [Google Scholar]
- 20.Lin, C., A. D. Kwong, and R. B. Perni. 2006. Discovery and development of VX-950, a novel, covalent, and reversible inhibitor of hepatitis C virus NS3.4A serine protease. Infect. Disord.-Drug Targets 6:3-16. [DOI] [PubMed] [Google Scholar]
- 21.Lin, T.-I., O. Lenz, G. Fanning, T. Verbinnen, F. Delouvroy, A. Scholliers, K. Vermeiren, Å. Rosenquist, M. Edlund, B. Samuelsson, L. Vrang, H. de Kock, P. Wigerinck, P. Raboisson, and K. Simmen. 2009. In vitro activity and preclinical profile of TMC435350, a potent hepatitis C virus protease inhibitor. Antimicrob. Agents Chemother. 53:1377-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 23.Liverton, N. J., S. S. Carroll, J. DiMuzio, C. Fandozzi, D. J. Graham, D. Hazuda, M. K. Holloway, S. W. Ludmerer, J. A. McCauley, C. J. McIntyre, D. B. Olsen, M. T. Rudd, M. Stahlhut, and J. P. Vacca. 2010. MK-7009, a potent and selective inhibitor of hepatitis C virus NS3/4A protease. Antimicrob. Agents Chemother. 54:305-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Llinas-Brunet, M., M. D. Bailey, G. Bolger, C. Brochu, A.-M. Faucher, J. M. Ferland, M. Garneau, E. Ghiro, V. Gorys, C. Grand-Maitre, T. Halmos, N. Lapeyre-Paquette, F. Liard, M. Poirier, M. Rheaume, Y. S. Tsantrizos, and D. Lamarre. 2004. Structure-activity study on a novel series of macrocyclic inhibitors of the hepatitis C virus NS3 protease leading to the discovery of BILN 2061. J. Med. Chem. 47:1605-1608. [DOI] [PubMed] [Google Scholar]
- 25.Lohmann, V., S. Hoffmann, U. Herian, F. Penin, and R. Bartenschlager. 2003. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J. Virol. 77:3007-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lohmann, V., F. Körner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 27.Lu, L., T. J. Pilot-Matias, K. D. Stewart, J. T. Randolph, R. Pithawalla, W. He, P. P. Huang, L. L. Klein, H. Mo, and A. Molla. 2004. Mutations conferring resistance to a potent hepatitis C virus serine protease inhibitor in vitro. Antimicrob. Agents Chemother. 48:2260-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malcolm, B. A., R. Liu, F. Lahser, S. Agrawal, B. Belanger, N. Butkiewicz, R. Chase, F. Gheyas, A. Hart, D. Hesk, P. Ingravallo, C. Jiang, R. Kong, J. Lu, J. Pichardo, A. Prongay, A. Skelton, X. Tong, S. Venkatraman, E. Xia, V. Girijavallabhan, and F. G. Njoroge. 2006. SCH 503034, a mechanism-based inhibitor of hepatitis C virus NS3 protease, suppresses polyprotein maturation and enhances the antiviral activity of alpha interferon in replicon cells. Antimicrob. Agents Chemother. 50:1013-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manns, M., A. Muir, N. Adda, I. Jacobson, N. Afdhal, J. Heathcote, S. Zeuzem, H. Reesink, N. Terrault, M. Bsharat, S. George, J. McHutchison, and A. Di Bisceglie. 2009. Telaprevir in hepatitis C genotype-1-infected patients with prior non-response, viral breakthrough or relapse to peginterferon-alfa-2A/B and ribavirin therapy: SVR results of the Prove3 study. J. Hepatol. 50:S379. [Google Scholar]
- 30.Manns, M., H. Reesink, C. Moreno, T. Berg, Y. Benhamou, Y. Horsmans, G. Dusheiko, R. Flisiak, P. Meyvisch, O. Lenz, V. Sekar, G. van't Klooster, K. Simmen, and R. Verloes. 2009. OPERA-1 trial: interim analysis of safety and antiviral activity of TMC435 in treatment-naive genotype 1 HCV patients. J. Hepatol. 50:S7. [Google Scholar]
- 31.Manns, M. P., G. R. Foster, J. K. Rockstroh, S. Zeuzem, F. Zoulim, and M. Houghton. 2007. The way forward in HCV treatment—finding the right path. Nat. Rev. Drug Discov. 6:991-1000. [DOI] [PubMed] [Google Scholar]
- 32.Marcellin, P., H. Reesink, T. Berg, M. Cramp, R. Flisiak, H. Van Vlierberghe, R. Verloes, O. Lenz, M. Peeters, V. Sekar, and G. De Smedt. 2009. Antiviral activity and safety of TMC435 combined with peginterferon alpha-2a and ribavirin in patients with genotype 1 hepatitis C infection who failed previous IFN-based therapy. J. Hepatol. 50:S385. [Google Scholar]
- 33.McCown, M. F., S. Rajyaguru, S. Kular, N. Cammack, and I. Nájera. 2009. GT-1a or GT-1b subtype-specific resistance profiles for hepatitis C virus inhibitors telaprevir and HCV-796. Antimicrob. Agents Chemother. 53:2129-2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McHutchison, J. G., G. T. Everson, S. C. Gordon, I. M. Jacobson, M. Sulkowski, R. Kauffman, L. McNair, J. Alam, A. J. Muir, and PROVE1 Study Team. 2009. Telaprevir with peginterferon and ribavirin for chronic HCV genotype 1 infection. N. Engl. J. Med. 360:1827-1838. [DOI] [PubMed] [Google Scholar]
- 35.Mo, H., L. Lu, T. Pilot-Matias, R. Pithawalla, R. Mondal, S. Masse, T. Dekhtyar, T. Ng, G. Koev, V. Stoll, K. D. Stewart, J. Pratt, P. Donner, T. Rockway, C. Maring, and A. Molla. 2005. Mutations conferring resistance to a hepatitis C virus (HCV) RNA-dependent RNA polymerase inhibitor alone or in combination with an HCV serine protease inhibitor in vitro. Antimicrob. Agents Chemother. 49:4305-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nettles, R., C. Chien, E. Chung, A. Persson, M. Gao, M. Belema, N. A. Meanwell, M. P. DeMicco, T. C. Marbury, R. Goldwater, P. Northup, J. Coumbis, W. K. Kraft, M. R. Charlton, J. C. Lopez-Talavera, and G. Denise. 2008. BMS-790052 is a first-in-class potent hepatitis C virus (HCV) NS5A inhibitor for patients with chronic HCV infection: results from a proof-of-concept study. Hepatology 48:1025A. [Google Scholar]
- 37.Penin, F., J. Dubuisson, F. A. Rey, D. Moradpour, and J.-M. Pawlotsky. 2004. Structural biology of hepatitis C virus. Hepatology 39:5-19. [DOI] [PubMed] [Google Scholar]
- 38.Perni, R. B., S. J. Almquist, R. A. Byrn, G. Chandorkar, P. R. Chaturvedi, L. F. Courtney, C. J. Decker, K. Dinehart, C. A. Gates, S. L. Harbeson, A. Heiser, G. Kalkeri, E. Kolaczkowski, K. Lin, Y.-P. Luong, B. G. Rao, W. P. Taylor, J. A. Thomson, R. D. Tung, Y. Wei, A. D. Kwong, and C. Lin. 2006. Preclinical profile of VX-950, a potent, selective, and orally bioavailable inhibitor of hepatitis C virus NS3-4A serine protease. Antimicrob. Agents Chemother. 50:899-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pietschmann, T., A. Kaul, G. Koutsoudakis, A. Shavinskaya, S. Kallis, E. Steinmann, K. Abid, F. Negro, M. Dreux, F.-L. Cosset, and R. Bartenschlager. 2006. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. U. S. A. 103:7408-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prongay, A. J., Z. Guo, N. Yao, J. Pichardo, T. Fischmann, C. Strickland, J. J. Myers, P. C. Weber, B. M. Beyer, R. Ingram, Z. Hong, W. W. Prosise, L. Ramanathan, S. S. Taremi, T. Yarosh-Tomaine, R. Zhang, M. Senior, R.-S. Yang, B. Malcolm, A. Arasappan, F. Bennett, S. L. Bogen, K. Chen, E. Jao, Y.-T. Liu, R. G. Lovey, A. K. Saksena, S. Venkatraman, V. Girijavallabhan, F. G. Njoroge, and V. Madison. 2007. Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3-[2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3- dimethyl-1-oxobutyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034). II. Key steps in structure-based optimization. J. Med. Chem. 50:2310-2318. [DOI] [PubMed] [Google Scholar]
- 41.Raboisson, P., H. de Kock, Å. Rosenquist, M. Nilsson, L. Salvador-Oden, T.-I. Lin, N. Roue, V. Ivanov, H. Wähling, K. Wickström, E. Hamelink, M. Edlund, L. Vrang, S. Vendeville, W. Van de Vreken, D. McGowan, A. Tahri, L. Hu, C. Boutton, O. Lenz, F. Delouvroy, G. Pille, D. Surleraux, P. Wigerinck, B. Samuelsson, and K. Simmen. 2008. Structure-activity relationship study on a novel series of cyclopentane-containing macrocyclic inhibitors of the hepatitis C virus NS3 protease leading to the discovery of TMC435350. Bioorg. Med. Chem. Lett. 18:4853-4858. [DOI] [PubMed] [Google Scholar]
- 42.Reesink, H. W., G. C. Fanning, K. Abou Farha, C. Weegink, A. Van Vliet, G. Van't Klooster, O. Lenz, F. Aharchi, K. Mariën, P. Van Remoortere, H. de Kock, F. Broeckaert, P. Meyvisch, E. Van Beirendonck, K. Simmen, and R. Verloes. 2010. Rapid HCV-RNA decline with once daily TMC435: a phase I study in healthy volunteers and hepatitis C patients. Gastroenterology 138:913-921. [DOI] [PubMed] [Google Scholar]
- 43.Sarrazin, C., J. Hong, S. Lim, X. Qin, S. Susser, B. Bradford, S. Porter, S. Zeuzem, and S. Seiwert. 2009. Incidence of virologic escape observed during ITMN-191 (R7227) monotherapy is genotype dependent, associated with a specific NS3 substitution, and suppressed upon combination with peginterferon alfa-2a/ribavirin. J. Hepatol. 50:S350. [Google Scholar]
- 44.Sarrazin, C., T. L. Kieffer, D. Bartels, B. Hanzelka, U. Müh, M. Welker, D. Wincheringer, Y. Zhou, H.-M. Chu, C. Lin, C. Weegink, H. Reesink, S. Zeuzem, and A. D. Kwong. 2007. Dynamic hepatitis C virus genotypic and phenotypic changes in patients treated with the protease inhibitor telaprevir. Gastroenterology 132:1767-1777. [DOI] [PubMed] [Google Scholar]
- 45.Schmittgen, T. D., and K. J. Livak. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3:1101-1108. [DOI] [PubMed] [Google Scholar]
- 46.Seiwert, S. D., S. W. Andrews, Y. Jiang, V. Serebryany, H. Tan, K. Kossen, P. T. R. Rajagopalan, S. Misialek, S. K. Stevens, A. Stoycheva, J. Hong, S. R. Lim, X. Qin, R. Rieger, K. R. Condroski, H. Zhang, M. G. Do, C. Lemieux, G. P. Hingorani, D. P. Hartley, J. A. Josey, L. Pan, L. Beigelman, and L. M. Blatt. 2008. Preclinical characteristics of the hepatitis C virus NS3/4A protease inhibitor ITMN-191 (R7227). Antimicrob. Agents Chemother. 52:4432-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Seiwert, S. D., J. Hon, S. R. Lim, T. Wang, H. Tan, and L. M. Blatt. 2007. Sequence variation of NS3/4A in HCV replicons exposed to ITMN-191 concentrations encompassing those likely to be achieved following clinical dosing. J. Hepatol. 46:S244-S245. [Google Scholar]
- 48.Strader, D. B., T. Wright, D. L. Thomas, L. B. Seeff, and American Association for the Study of Liver Diseases. 2004. Diagnosis, management, and treatment of hepatitis C. Hepatology 39:1147-1171. [DOI] [PubMed] [Google Scholar]
- 49.Tong, X., S. Bogen, R. Chase, V. Girijavallabhan, Z. Guo, F. G. Njoroge, A. Prongay, A. Saksena, A. Skelton, E. Xia, and R. Ralston. 2008. Characterization of resistance mutations against HCV ketoamide protease inhibitors. Antiviral Res. 77:177-185. [DOI] [PubMed] [Google Scholar]
- 50.Tong, X., R. Chase, A. Skelton, T. Chen, J. Wright-Minogue, and B. A. Malcolm. 2006. Identification and analysis of fitness of resistance mutations against the HCV protease inhibitor SCH 503034. Antiviral Res. 70:28-38. [DOI] [PubMed] [Google Scholar]
- 51.Tsantrizos, Y. S., G. Bolger, P. Bonneau, D. R. Cameron, N. Goudreau, G. Kukolj, S. R. LaPlante, M. Llinas-Brunet, H. Nar, and D. Lamarre. 2003. Macrocyclic inhibitors of the NS3 protease as potential therapeutic agents of hepatitis C virus infection. Angew. Chem. Int. Ed. Engl. 42:1356-1360. [DOI] [PubMed] [Google Scholar]
- 52.Venkatraman, S., S. L. Bogen, A. Arasappan, F. Bennett, K. Chen, E. Jao, Y.-T. Liu, R. Lovey, S. Hendrata, Y. Huang, W. Pan, T. Parekh, P. Pinto, V. Popov, R. Pike, S. Ruan, B. Santhanam, B. Vibulbhan, W. Wu, W. Yang, J. Kong, X. Liang, J. Wong, R. Liu, N. Butkiewicz, R. Chase, A. Hart, S. Agrawal, P. Ingravallo, J. Pichardo, R. Kong, B. Baroudy, B. Malcolm, Z. Guo, A. Prongay, V. Madison, L. Broske, X. Cui, K.-C. Cheng, Y. Hsieh, J.-M. Brisson, D. Prelusky, W. Korfmacher, R. White, S. Bogdanowich-Knipp, A. Pavlovsky, P. Bradley, A. K. Saksena, A. Ganguly, J. Piwinski, V. Girijavallabhan, and F. G. Njoroge. 2006. Discovery of (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3- dioxopropyl]-3-[2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl- 1-oxobutyl]-6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (SCH 503034), a selective, potent, orally bioavailable hepatitis C virus NS3 protease inhibitor: a potential therapeutic agent for the treatment of hepatitis C infection. J. Med. Chem. 49:6074-6086. [DOI] [PubMed] [Google Scholar]
- 53.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. Zhao, K. Murthy, A. Habermann, H.-G. Kräusslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.World Health Organization. 2010. Hepatitis C: disease3 burden. World Health Organization, Geneva, Switzerland. http://www.who.int/vaccine_research/diseases/viral_cancers/en/index2.html.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.