Abstract
We have sequenced the conjugative plasmid pPR9, which carries the ileS2 gene, which had contributed to the dissemination of high-level mupirocin resistance at our institution. The plasmid backbone shows extensive genetic conservation with plasmids belonging to the pSK41/pGO1 family, but comparative analyses have revealed key differences that provide important insights into the evolution of these medically important plasmids and high-level mupirocin resistance in staphylococci and highlight the role of insertion sequence IS257 in these processes.
Plasmids carrying the ileS2 gene play an important role in high-level mupirocin resistance (Hi-Mupr) spread and consequent clinical and epidemiological problems (8, 17, 23, 26). To date, complete nucleotide sequences of two ileS2-carrying plasmids have been determined: pUSA03 (37,136 bp; GenBank entry NC_007792) was carried by an epidemic USA300 community-acquired methicillin-resistant Staphylococcus aureus (CA-MRSA) strain (10), and a partially annotated sequence is available for pV030-8 (39,041 bp; NC_010279), which was isolated from a Hi-Mupr MRSA strain from South Korea. Additionally, the ileS2-containing 34-kb plasmid pGO400 has been partially characterized (22). pUSA03, pV030-8, and pGO400 all belong to the pSK41/pGO1 family of multiresistance plasmids capable of promoting the conjugative transfer of resistance genes among bacterial strains (4, 14). In spite of their common genetic backbone, the antibiotic resistance gene contents of these plasmids are extremely diverse (14). The resistance genes are usually flanked by copies of the insertion sequence IS257, which has played a key role in the evolution of pSK41/pGO1-like plasmids (4, 13, 14, 32).
Recently we reported intrahospital Hi-Mupr dissemination among pandemic MRSA strains due to the dispersion of structurally distinct ileS2-carrying conjugative plasmids (23). In this study, we have characterized the complete nucleotide sequence of one such plasmid, pPR9, previously named pMUP9 (23). We have undertaken a comprehensive comparative analysis of the DNA sequences of pPR9 and other pSK41 family plasmids, particularly other ileS2-carrying plasmids, to ascertain their diversity and to gain insights into the evolution of Hi-Mupr in staphylococci.
DNA sequence and general overview of pPR9.
HUNSC491, the MRSA strain harboring pPR9, was isolated in 2002 at the Hospital Universitario Nuestra Señora de Candelaria (HUNSC), Tenerife, Spain. The strain belongs to ST36-SCCmecII, a pandemic genotype that has widely disseminated throughout the HUNSC (24). ST36-SCCmecII isolates had acquired different ileS2-carrying plasmids (23). DNA of one, pPR9, was obtained by using the Hi-Speed maxi-plasmid purification kit (Qiagen, Valencia, CA), and a shotgun library was constructed at Integrated Genomics Inc. (IG) (11, 12, 28). The pPR9 sequence data were submitted to the IG database and software suite, ERGO, for sequence annotation (2, 9). The predicted proteins were searched using the BLAST algorithm (1), against a nonredundant database at the National Center for Biotechnology Information (NCBI). Protein functional domains were analyzed by searching against the NCBI conserved-domain database (21) and the Pfam database (3).
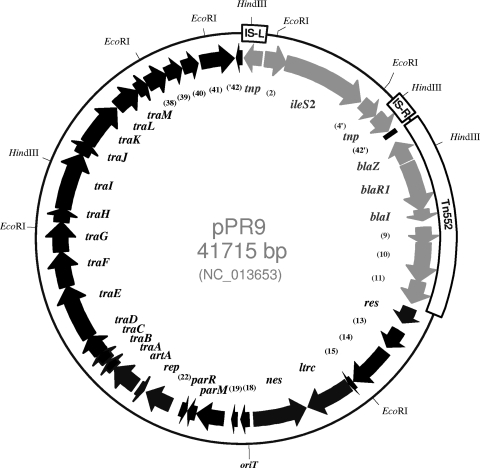
The complete nucleotide sequence of plasmid pPR9 was determined with 9.4-fold DNA coverage. The overall G+C content was 29.6%, comparable to the observed average G+C value for staphylococci (14). The sequence was found to have a 92% coding ratio with an average open reading frame (ORF) length of 915 bp; the ileS2 gene, which encodes Hi-Mupr, is the largest ORF at 3,075 bp. pPR9 possesses a mosaic structure with two clearly delineated regions: first, the backbone-encoding genes putatively involved in the replication, maintenance, and transfer of the plasmid; second, an accessory region containing antibiotic resistance genes and insertion sequences. Two copies of IS257 are evident (i.e., IS257-L and IS257-R); orf4 and orf42 are gene remnants truncated by IS257 (Fig. 1). The majority of genes in the pPR9 backbone share more than 90% amino acid identity with homologous regions of pSK41 (4) and other members of the family such as pGO1 (6), pLW1043 (32), pUSA03 (10), and pV030-8. pPR9 also contains a complete copy of a Tn552-like β-lactamase transposon (25) that shares 94% nucleotide identity with Tn552 (GenBank entry X52734). As in other pSK41/pGO1-like plasmids (20), the Tn552-like element in pPR9 is located within the resolution site of the plasmid's multimer resolution system and is flanked by the 6-bp target duplication sequence 5′-ATAGCG-3′ (Fig. 1 and 2).
FIG. 1.
Physical and genetic map of Staphylococcus aureus multiresistance plasmid pPR9 (GenBank entry NC_013653). The approximate positions of the putative origin of transfer (oriT), EcoRI and HindIII restriction sites, IS257 elements (IS257-L and IS257-R), and Tn552 are shown on the rim of the outer circle. The first G of the IS257-L terminal inverted repeat left (TIRL) was designated the first nucleotide of the plasmid, and all 42 open reading frames (ORFs; tentatively named orf1 to orf42) likely to represent translated genes are numbered in relation to this site. ORFs are represented by arrows indicating the direction of transcription; tnp marks the gene for IS257 transposase. The accessory region genes are shown in gray, and the plasmid backbone genes are shown in black. All but six ORFs were found to be transcribed in the same direction (clockwise). The nucleotide sequence was verified by comparison of in silico-generated restriction maps with experimental data. Also, the numbers and sizes of HindIII and EcoRI restriction fragments that hybridized with specific probes for the ileS2, blaZ, traK, and tnp genes corresponded to those predicted from the pPR9 sequence (data not shown). orf42, interrupted by insertion of the IS257-flanked ileS2 segment, is indicated by 42′ and ′42. The deletion of the 3′ portion of orf4 is indicated as 4′.
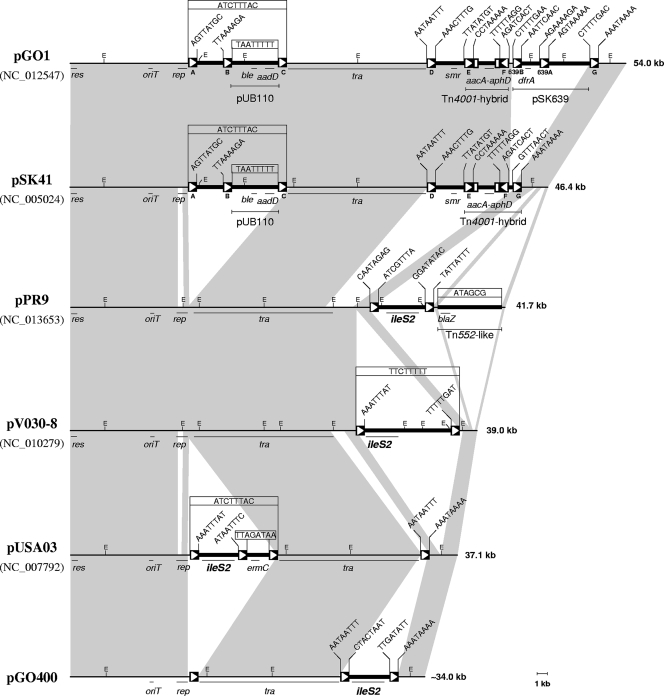
FIG. 2.
Relationships between representative pSK41/pGO1 family plasmids. Linear physical and genetic maps of staphylococcal conjugative plasmids pGO1 (6), pSK41 (4), pPR9, pV030-8, pUSA03 (10), and pGO400 (22) are presented, with accession numbers given in parentheses; plasmid sizes are shown on the right. Shaded connections between plasmid maps illustrate that they all share a conserved basic backbone. The plasmid accessory regions are shown as a thick line in each map. The genetic loci shown are aacA-aphD (gentamicin-tobramycin-kanamycin resistance), aadD (kanamycin-neomycin-paromomycin-tobramycin resistance), ble (bleomycin resistance), dfrA (trimethoprim resistance), smr (antiseptic and disinfectant resistance), ileS2 (high-level mupirocin resistance), blaZ (penicillin resistance), ermC (macrolide, lincosamide, and streptogramin B resistance), tra (conjugative transfer functions), rep (replication initiation), res (resolvase), and oriT (origin of conjugative DNA transfer). The positions and extents of the cointegrated copy of the plasmids pUB110 and pSK639, the Tn4001-IS257 hybrid structure, and the Tn552-like transposon are indicated. Inverted and truncated copies of IS256 associated with the Tn4001 hybrid structures are represented by open boxes, whereas IS257 elements are represented as solid boxes containing an arrowhead indicating the direction of the transposase transcription and hence the element's orientation. Where known, the 8-bp sequences adjacent to each IS257 element are indicated. IS257 element designations for pGO1 and pSK41 are taken from the work of Caryl and O'Neill (6) and Berg et al. (4), respectively. The location of oriT is based on homology to pGO1, where it has been mapped precisely (7). Recognition sites for the restriction endonuclease EcoRI (E) are shown. A distance scale in kb is given below the figure.
The conjugal proficiency of pPR9 (5 × 10−6 to 1 × 10−7 transconjugants per donor cell) was demonstrated using filter mating experiments (4) and is in the range of those reported for other plasmids of the same family (22, 29). Transconjugants carrying pPR9 showed mupirocin MICs of ≥1,024 μg ml−1, confirming the Hi-Mupr phenotype conferred by pPR9. In curing experiments (30), loss of pPR9 was observed at low frequencies, indicating that pPR9 is normally inherited stably.
pPR9 encodes a novel chimeric rep gene.
To date, all plasmids belonging to the pSK41/pGO1 family have utilized highly conserved RepA_N-type replication initiation genes, which are associated with plasmids or phage found in low-G+C content Gram-positive bacteria (31). pPR9 contains a putative rep gene (orf23) which encodes a protein of 341 amino acids. Surprisingly, sequence alignment between the deduced pPR9 and pSK41 Rep proteins revealed that amino acid similarity was restricted to their C-terminal ∼155 amino acids (see Fig. S1A in the supplemental material), a region most conserved between RepA_N proteins from the same genus and therefore thought to possess a host-specific function (31). The lack of similarity evident at the N-terminal end of the proteins is noteworthy because it corresponds to the RepA_N domain of pSK41 Rep, which is believed to mediate binding to four repeated sequences in the origin of replication termed Rep boxes (18, 31). Intriguingly, the N-terminal 174 amino acids of pPR9 Rep share significant similarity to the N-terminal ends of numerous hypothetical phage proteins that are annotated as replication proteins (Fig. S1A).
With the exception of the unrelated DNA segments that encode the N-terminal ends of the respective Rep proteins, nucleotide sequence comparison of the entire pPR9 and pSK41 replication regions revealed a high degree of identity (greater than 97% over 727 nucleotides [nt]), including in the upstream intergenic region that contains the pSK41 rep promoter and encodes the antisense regulator RNAI (see Fig. S1B in the supplemental material) (19). The DNA segment that has been “replaced” in pPR9 encompasses the rep start codon through to just after the four pSK41 Rep boxes. Notably, the unique segment in pPR9 contains an array of five 15-bp direct repeats, the last four in tandem organization, at a position equivalent to the pSK41 Rep boxes. It would therefore seem possible that in the pPR9 lineage of the pSK41 plasmid family, which also includes pV030-8, there has been a recombination event that has swapped the segment encoding the probable DNA binding domain of its Rep protein for that of a phage-like protein, together with its likely DNA target sequences.
To confirm the functionality of the unusual pPR9 replication region (nt 23236 to 24606; GenBank entry NC_013653) (see Fig. S1B in the supplemental material), it was cloned into the Escherichia coli vector pSK5299 (15). The resulting plasmid, pSK6846, could indeed replicate in S. aureus RN4220, and copy number analysis (15) indicated that it was maintained at approximately 5 copies per cell (data not shown), which is lower than that previously determined for the pSK41 minireplicon pSK5413 (approximately 7 copies per cell) (5). Notably, pSK6846 and pSK5413 were found to be compatible (data not shown).
Comparative analysis of ileS2-carrying plasmids.
Strikingly, although ileS2 is ubiquitously flanked by IS257 in plasmids conferring Hi-Mupr, the segment occupies a unique position within each plasmid backbone (Fig. 2). In pPR9, the ileS2 segment is inserted into orf42 (Fig. 1), which corresponds to orf138 of pSK41. In pUSA03 and pGO400 the ileS2 segments are associated with IS257s present at either end of their respective tra regions (10, 22), whereas in pV030-8, the segment is inserted into the gene corresponding to pPR9 orf41 (equivalent to pSK41 orf423, which is located immediately upstream of orf138). An important implication of these findings is that the DNA segment responsible for Hi-Mupr in staphylococci has been incorporated into pSK41/pGO1-like plasmids on at least four independent occasions, once again underlining the capacity of IS257 to mediate the capture of resistance determinants. The pathways by which the ileS2 segments have been incorporated into these plasmids cannot be inferred with any certainty; nonetheless, flanking 8-bp target duplications present in pV030-8 and pUSA03 (Fig. 2) imply that transposition has contributed to the process, and this is supported by the identity or near-identity of the flanking IS257s (data not shown).
Compared to pSK41 and pGO1, ileS2-carrying plasmids have a simpler structure with fewer copies of IS257 (Fig. 2). Notably, unlike all previously described family plasmids, pPR9 and pV030-8 lack IS257s flanking their tra genes. In the other plasmids, the distal gene in the tra region, named orf55 in pSK41, is truncated by the adjacent IS257. This gene is intact in pPR9 (orf38) and encodes a putative 243-amino-acid protein. The organization of pPR9 and pV030-8 suggests that these plasmids belong to a distinct lineage that diverged prior to the acquisition of IS257 elements at the tra termini in an evolutionary branch that has given rise to other known family members. This notion is consistent with the previous suggestion that region 1 and the tra region were probably contiguous in the pSK41 progenitor (4).
Heterogeneity of the regions surrounding the ileS2 gene.
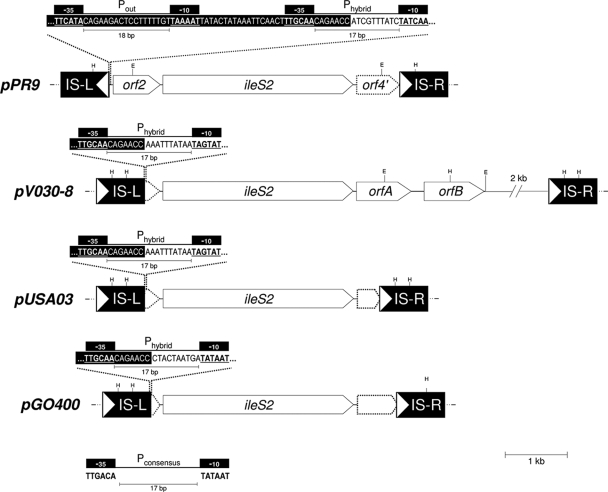
pPR9 is unique in that the IS257s flanking ileS2 are in inverted orientation with respect to each other (Fig. 3). pPR9 has the most intact form of the IS257-ileS2 upstream region. The upstream (L) copies of IS257 in pV030-8, pUSA03, and pGO400 truncate orf2. pV030-8 has the largest ileS2 downstream region, and orfA is truncated to various degrees in pPR9 (orf4′), pUSA03, and pGO400 by IS257-R. Even with the heterogeneity in extents, the flanking sequences that are present, up to the IS257 boundaries, are 100% identical between the plasmids, implying acquisition from a common source but various truncations. The organization suggests that ileS2 was part of a multigene operon.
FIG. 3.
Structural organization of the regions encompassing the ileS2 genes of plasmids pPR9, pV030-8, pUSA03, and pGO400. The respective plasmid names are shown on the left. Restriction endonuclease cleavage sites are abbreviated as follows: E, EcoRI; H, HindIII. IS257 elements (IS-L and IS-R) flanking the ileS2 gene are represented by solid boxes; the white arrows indicate the direction of IS257 transposase transcription. The ileS2 gene and the predicted ORFs upstream and downstream are represented as arrows with the arrowhead indicating their orientation. A segment of approximately 2 kb on pV030-8 has been omitted for clarity. Truncated ORFs are shown using a dotted outline. The positions of the −35 and −10 sequences for the putative promoters Phybrid and Pout are shown above each plasmid. The spacing between the −35 and −10 promoter sequences is indicated below each putative promoter. A distance scale in kb is given below the figure.
Interestingly, the various arrangements of IS257-L raise the possibility that different IS257-derived hybrid promoters (Phybrids) could drive transcription of ileS2 in the various plasmids, which might have consequences in terms of resistance expression (Fig. 3). It has been proposed that a potential Phybrid, which is a better match to the canonical consensus than the promoter predicted initially (16), drives ileS2 transcription in pGO400 (27). Similarly, pUSA03 has a good candidate for a different Phybrid, which is likewise present in pV030-8. pPR9 also has a potential Phybrid, although it is a suboptimal match to the consensus. However, the inverted orientation of IS257-L in pPR9 raises the possibility that a previously identified complete promoter within IS257, Pout (27), might be responsible for the ileS2 transcription in this plasmid (Fig. 3). We are investigating these possibilities.
Nucleotide sequence accession number.
The annotated sequence of pPR9 has been deposited in the GenBank database under accession number NC_013653.
Supplementary Material
Acknowledgments
We are grateful to Warren B. Grubb for providing the S. aureus strain WBG541.
This study was supported by grants 2006/0002 from Fondo de Investigación Sanitaria (FIS), Government of Spain, and FUNCIS50/05, Canary Islands Autonomous Government, to S.M.-A. E.P.-R. was supported by a grant from FUNCIS. S.M.-A. was supported by a FIS-SCS (FUNCIS) contract. Work in the laboratory of S.M.K. and N.F. was supported by National Health and Medical Research Council of Australia project grant 4571029.
Footnotes
Published ahead of print on 8 March 2010.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Badger, J. H., and G. J. Olsen. 1999. CRITICA: coding region identification tool invoking comparative analysis. Mol. Biol. Evol. 16:512-524. [DOI] [PubMed] [Google Scholar]
- 3.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khana, M. Marshall, S. Moxon, E. L. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berg, T., N. Firth, S. Apisiridej, A. Hettiaratchi, A. Leelaporn, and R. A. Skurray. 1998. Complete nucleotide sequence of pSK41: evolution of staphylococcal conjugative multiresistance plasmids. J. Bacteriol. 180:4350-4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berg, T. 2000. Conjugative and mobilisable antimicrobial resistance plasmids from staphylococci. Ph.D. thesis. University of Sydney, Sydney, NSW, Australia.
- 6.Caryl, J. A., and A. J. O'Neill. 2009. Complete nucleotide sequence of pGO1, the prototype conjugative plasmid from the staphylococci. Plasmid 62:35-38. [DOI] [PubMed] [Google Scholar]
- 7.Climo, M. W., V. K. Sharma, and G. L. Archer. 1996. Identification and characterization of the origin of transfer (oriT) and a gene (nes) encoding a single-stranded endonuclease on the staphylococcal plasmid pGO1. J. Bacteriol. 178:4975-4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cookson, B. D. 1998. The emergence of mupirocin resistance: a challenge to infection control and antibiotic prescribing practice. J. Antimicrob. Chemother. 41:11-18. [DOI] [PubMed] [Google Scholar]
- 9.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27:4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diep, B. A., S. R. Gill, R. F. Chang, T. H. Phan, J. H. Chen, M. G. Davidson, F. Lin, J. Lin, H. A. Carleton, E. F. Mongodin, G. F. Sensabaugh, and F. Perdreau-Remington. 2006. Complete genome sequence of USA300, an epidemic clone of community-acquired methicillin-resistant Staphylococcus aureus. Lancet 367:731-739. [DOI] [PubMed] [Google Scholar]
- 11.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 12.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 13.Firth, N., and R. A. Skurray. 1998. Mobile elements in the evolution and spread of multiple-drug resistance in staphylococci. Drug Resist. Updat. 1:49-58. [DOI] [PubMed] [Google Scholar]
- 14.Firth, N., and R. A. Skurray. 2006. Genetics: accessory elements and genetic exchange, p. 413-426. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. ASM Press, Washington, DC.
- 15.Grkovic, S., M. H. Brown, K. M. Hardie, N. Firth, and R. A. Skurray. 2003. Stable low-copy-number Staphylococcus aureus shuttle vectors. Microbiology 149:785-794. [DOI] [PubMed] [Google Scholar]
- 16.Hodgson, J. E., S. P. Curnock, K. G. H. Dyke, R. Morris, D. R. Sylvester, and M. S. Gross. 1994. Molecular characterization of the gene encoding high-level mupirocin resistance in Staphylococcus aureus J2870. Antimicrob. Agents Chemother. 38:1205-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hurdle, J. G., A. J. O'Neill, L. Mody, I. Chopra, and S. F. Bradley. 2005. In vivo transfer of high-level mupirocin resistance from Staphylococcus epidermidis to methicillin-resistant Staphylococcus aureus associated with failure of mupirocin prophylaxis. J. Antimicrob. Chemother. 56:1166-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwong, S. M., R. A. Skurray, and N. Firth. 2004. Staphylococcus aureus multiresistance plasmid pSK41: analysis of the replication region, initiator protein binding and antisense RNA regulation. Mol. Microbiol. 51:497-509. [DOI] [PubMed] [Google Scholar]
- 19.Kwong, S. M., R. A. Skurray, and N. Firth. 2006. Replication control of staphylococcal multiresistance plasmid pSK41: an antisense RNA mediates dual-level regulation of Rep expression. J. Bacteriol. 188:4404-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LeBard, R. J., S. O. Jensen. I. A. Arnaiz, R. A. Skurray, and N. Firth. 2008. A multimer resolution system contributes to segregational stability of the prototypical staphylococcal conjugative multiresistance plasmid pSK41. FEMS Microbiol. Lett. 284:58-67. [DOI] [PubMed] [Google Scholar]
- 21.Marchler-Bauer, A., J. B. Anderson, C. DeWeese-Scott, N. D. Fedorova, L. Y. Geer, S. He, D. I. Hurwitz, J. D. Jackson, A. R. Jacobs, C. J. Lanczycki, C. A. Liebert, C. Liu, T. Madej, G. H. Marchler, R. Mazumder, A. N. Nikolskaya, A. R. Panchenko, B. S. Rao, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, S. Vasudevan, Y. Wang, R. A. Yanashita, J. J. Yin, and S. H. Bryant. 2003. CDD: a curated Entrez database of conserved domain alignments. Nucleic Acids Res. 31:383-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morton, T. M., J. L. Johnston, J. Patterson, and G. L. Archer. 1995. Characterization of a conjugative staphylococcal mupirocin resistance plasmid. Antimicrob. Agents Chemother. 39:1272-1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pérez-Roth, E., C. López-Aguilar, J. Alcoba-Florez, and S. Méndez-Álvarez. 2006. High-level mupirocin resistance within methicillin-resistant Staphylococcus aureus pandemic lineages. Antimicrob. Agents Chemother. 50:3207-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pérez-Roth, E., F. Lorenzo-Díaz, N. Batista, A. Moreno, and S. Méndez-Álvarez. 2004. Tracking methicillin-resistant Staphylococcus aureus clones during a 5-year period (1998 to 2002) in a Spanish hospital. J. Clin. Microbiol. 42:4649-4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rowland, S. J., and K. G. Dyke. 1990. Tn552, a novel transposable element from Staphylococcus aureus. Mol. Microbiol. 4:961-975. [DOI] [PubMed] [Google Scholar]
- 26.Simor, A. E., T. L. Stuart, L. Louie, C. Watt, M. Ofner-Agostini, D. Gravel, M. Mulvey, M. Loeb, A. McGeer, E. Bryce, A. Matlow, and the Canadian Nosocomial Infection Surveillance Program. 2007. Mupirocin-resistant, methicillin-resistant Staphylococcus aureus strains in Canadian hospitals. Antimicrob. Agents Chemother. 51:3880-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simpson, A. E., R. A. Skurray, and N. Firth. 2000. An IS257-derived hybrid promoter directs transcription of a tetA(K) tetracycline resistance gene in the Staphylococcus aureus chromosomal mec region. J. Bacteriol. 182:3345-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tettelin, H., D. Radune, S. Kasif, H. Khouri, and S. L. Salzberg. 1999. Optimized multiplex PCR: efficiently closing a whole-genome shotgun sequencing project. Genomics 62:500-507. [DOI] [PubMed] [Google Scholar]
- 29.Thomas, W. D., Jr., and G. L. Archer. 1989. Identification and cloning of the conjugative transfer region of Staphylococcus aureus plasmid pGO1. J. Bacteriol. 171:684-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Udo, E. E., and L. E. Jacob. 1998. Conjugative transfer of high-level mupirocin resistance and the mobilization of non-conjugative plasmids in Staphylococcus aureus. Microb. Drug Resist. 4:185-193. [DOI] [PubMed] [Google Scholar]
- 31.Weaver, K. E., S. M. Kwong, N. Firth, and M. V. Francia. 2009. The RepA_N replicons of Gram-positive bacteria: a family of broadly distributed but narrow host range plasmids. Plasmid 61:94-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weigel, L. M., D. B. Clewell, S. R. Gill, N. C. Clark, L. K. McDougal, S. E. Flannagan, J. F. Kolonay, J. Shetty, G. E. Killgore, and F. C. Tenover. 2003. Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 302:1569-1571. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.