Abstract
A high-resolution melt (HRM) assay using a Rotor-Gene 6000 instrument was developed to characterize the codon for glycine 54 in the cyp51A genes from 13 reference isolates and 12 clinical isolates of Aspergillus fumigatus. Mutations in this codon confer reduced susceptibility to itraconazole and posaconazole. The assay is simple to perform, and a result of “wild type” or “mutant” is available after approximately 1 h following DNA extraction using commercially available reagents and conventional primers.
Aspergillus fumigatus infections cause high rates of mortality and morbidity in immunocompromised hosts (15, 16). Strains are intrinsically resistant to fluconazole, but itraconazole, voriconazole, posaconazole, and ravuconazole are typically active (10, 18, 23, 26). Recent reports suggest that resistance rates are rising (13, 26, 29). Since azoles can be both fungicidal and fungistatic, infected patients treated with azole drugs may develop resistant strains (2, 5, 6, 7, 10, 12). Resistant strains, possibly environmental strains exposed to agricultural azole compounds, have infected patients who have not had previous exposure to azole drugs (28, 29, 33). Azole drugs block the biosynthesis of ergosterol, a bulk sterol component of the cell walls of fungi. Although not found in human cells, it is nearly identical to cholesterol. Triazole drugs bind to the active site of the fungal cytochrome P450 enzyme 14-alpha lanosterol demethylase, which catalyzes the demethylation of ergosterol precursors (1, 19) and is encoded by the cyp51A and cyp51B genes in A. fumigatus (1, 21). Amino acid substitutions encoded at several codon sites in the cyp51A gene, including glycine 54, methionine 220, glycine 138, and leucine 98, have been recently shown to result in reduced susceptibility to itraconazole and posaconazole or azole cross-resistance (1, 2, 7, 10, 12, 20, 22, 23, 24, 34). Molecular assays have been used to rapidly identify mutations in these codons from clinical isolates and/or specimens using probes or sequencing (1, 10, 32). We have developed a novel high-resolution melt (HRM) assay to characterize the codon for glycine 54 in the cyp51A gene for isolates of A. fumigatus. Mutations in this codon confer reduced susceptibility to itraconazole and posaconazole.
HRM is a recent enhancement of traditional melting analysis of a PCR product. Using specialized instrumentation, highly controlled temperature transitions, and data acquisition during the melt phase allows the detection of single nucleotide polymorphisms with the use of low-cost fluorescent intercalating dyes (4, 8). Reagent costs are similar to those of traditional PCR. HRM assays have been recently used for microbiological applications using various platforms (3, 8, 9, 11, 14, 17, 25, 27, 31). We evaluated the Rotor-Gene 6000 instrument, which can perform both real-time PCR and HRM, to characterize the codon for glycine 54 in the cyp51A gene of A. fumigatus isolates. Pyrosequencing (Qiagen, Hilden, Germany) was used for confirmation.
Thirteen well-characterized clinical and laboratory mutant strains of A. fumigatus and chromosomal DNA of strain R7-1 were used to develop our assay. These strains represented 7 different genotypes, including those corresponding to itraconazole-susceptible wild-type Gly54 (encoded by GGG) and itraconazole-resistant mutant alleles of the cyp51A gene, including Glu54 (encoded by GAG and GAA), Lys54 (AAG), Arg54 (AGG), Val54 (GTG), and Trp54 (TGG) (1). Twelve clinical isolates were tested as unknowns. A 10-μl loopful from 48-h growth on potato dextrose agar was suspended in 500 μl lysis buffer (1% Triton X-100, 0.5% Tween 20, 10 mM Tris-HCl [pH 8.0], 1 mM EDTA) containing approximately 100 mg of 710- to 1,180-μm glass beads (Sigma, Deissenhogen, Germany). After vortexing for 15 min and heating for 10 min at 100°C, 200 μl was extracted on a MagNA Pure LC system using the MagNA Pure total nucleic acid kit (Roche Diagnostics, Mannheim, Germany). PCR primers were designed to amplify a 107-bp fragment of the cyp51A gene (GenBank accession no. AF338659) covering codon 54. Amplicon sequences representing the different genotypes were analyzed by the Poland melting software program to predict melting behavior (http://www.biophys.uni-duesseldorf.de/local/POLAND//poland.html) (30). The reverse primer was biotinylated for pyrosequencing. SensiMix HRM master mix (Quantace, London, England) containing EvaGreen fluorescent intercalating dye was used according to the manufacturer's instructions. The forward primer (5′ GAACCGAACAGAACCGCCAAT 3′) and reverse primer (5′ biotin CCTTTTCTCTGCACGCAAAGAAGA 3′) were used at a concentration of 300 nM in a volume of 25 μl with 1× Sensimix HRM, 1× EvaGreen dye, and 2 μl of DNA target (approximately 10 to 100 ng). Instrument parameters were as follows: 95°C for 10 min, 35 cycles (95°C 10 s, 60°C 20 s), acquiring on the green channel, HRM (65 to 85°C), rising by 0.1° each step, 2-s hold at each increment, and hold at 40°C for 1 min. The template was normalized so that samples would have similar cycle thresholds. The PCR was optimized so that samples had similar amplification efficiencies (4). Our method was not able to discriminate all 7 genotypes. However, an isolate identification as non-wild type irrespective of specific genotype is enough to suspect reduced susceptibility to itraconazole and posaconazole. A control of wild-type GGG was selected, and the HRM melts of all other samples were compared to this control using a difference plot (Fig. 1) generated by the instrument software. A confidence threshold of 90% was selected for software analysis. A score of 90% or above identified the sample as genotype GGG, while a score below 90% identified the sample as a variation (mutant). The amplified products were directly tested with pyrosequencing. Fifteen microliters of PCR amplicon was used with a previously described sequencing primer (5′ TCTGGGTAGTACCATCAGT 3′) (31). Ten base pairs were analyzed using pyrosequencing software. Pyrograms (Fig. 2) were verified by manual reads.
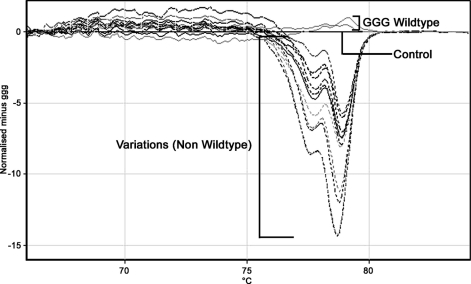
FIG. 1.
HRM difference plot comparing the Gly54 wild-type GGG reference control strain to all other strains. The control is displayed as a straight line. Strains are identified as having wild-type GGG or as variations (mutants) based on melt differences as determined by the software. Two samples were correctly identified as wild-type GGG. Eleven mutant strains, including those with Glu54 (GAG and GAA), Lys54 (AAG), Arg54 (AGG), Val54 (GTG), and Trp54 (TGG), were correctly identified as variations.
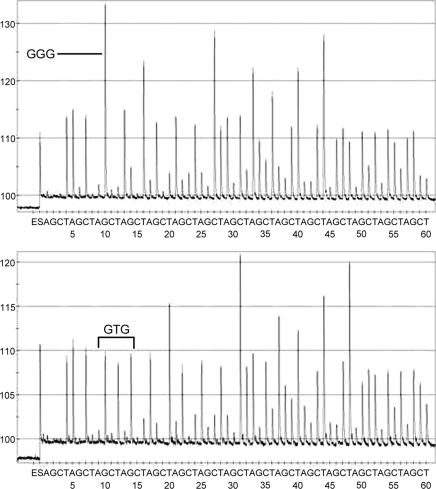
FIG. 2.
Representative pyrosequencing pyrograms of a wild-type isolate with GGG and a GTG mutant isolate. HRM-amplified products were directly tested with pyrosequencing using a previously described sequencing primer (32).
Using one wild-type reference strain, H11-20, with GGG, as the control, 12 reference strains were correctly identified as having wild-type GGG (≥90% confidence) or a cyp51a mutant (variation) at a ≤90% confidence level. All were confirmed by pyrosequencing (Table 1). Twelve clinical isolates were tested as unknowns. Controls were reference strain H11-20 (wild-type GGG) and mutant (variation) strains RIT12 (GAG) and SO/3829 (GTG). The GTG control was chosen specifically because it had the melt pattern closest to the wild-type pattern during the assay optimization. All 12 clinical isolates were identified as having wild-type GGG, with a confidence range of 93.51% to 99.43%, confirmed by pyrosequencing. The two mutant controls were identified as variations at 43.09% and 82.32% confidence ranges, respectively.
TABLE 1.
Comparison of results of HRM assay with pyrosequencing and itraconazole susceptibilities of reference A. fumigatus strainsa
| Strain | 54th codon of cyp51A | Amino acid | HRM genotype | HRM confidence (%) | Pyrosequence | Itraconazolec MIC (μg/ml) |
|---|---|---|---|---|---|---|
| H11-20b | GGG | Gly | GGG | 100 | GGG | 0.5 |
| Af41 | GGG | Gly | GGG | 98.27 | GGG | 0.25 |
| RIT12 | GAG | Glu | Variation | 51.58 | GAG | >16 |
| Af72 | GAG | Glu | Variation | 69.66 | GAG | >16 |
| RIT18 | GAA | Glu | Variation | 24.08 | GAA | >16 |
| RIT32 | GAA | Glu | Variation | 19.82 | GAA | >16 |
| RIT15 | AAG | Lys | Variation | 8.24 | AAG | >16 |
| RIT38 | AAG | Lys | Variation | 8.11 | AAG | >16 |
| RIT41 | AGG | Arg | Variation | 53.22 | AGG | >16 |
| RIT51 | AGG | Arg | Variation | 50.59 | AGG | >16 |
| SO/3827A | GTG | Val | Variation | 65.0 | GTG | >16 |
| SO/3829 | GTG | Val | Variation | 75.98 | GTG | >16 |
| R7-1 | TGG | Trp | Variation | 41.11 | TGG | >16 |
Characterized strains including itraconazole MIC data were obtained from Balashov et al. (1).
Used as GGG (wild-type) control for HRM assay. The HRM data of all other strains were compared to those of this wild-type control for identification as GGG (wild type) at ≥90% confidence or as a variation (mutant) if <90% confidence.
Genetic mutations in codon 54 of the cyp51A gene of A. fumigatus can be detected using our novel HRM assay. Although we were unable to specifically genotype the 6 mutant alleles, the characterization of an isolate as mutant is sufficient to suspect reduced susceptibility to itraconazole and posaconazole. This assay could be used to screen for wild-type genotypes of codon 54 of the cyp51A gene, and variations could be pyrosequenced directly to identify specific mutant cyp51A codon sequences if necessary.
Acknowledgments
We have no conflicts of interest to disclose.
Footnotes
Published ahead of print on 1 March 2010.
REFERENCES
- 1.Balashov, S. V., R. Gardiner, S. Park, and D. S. Perlin. 2005. Rapid, high-throughput, multiplex, real-time PCR for identification of mutations in the cyp51A gene of Aspergillus fumigatus that confer resistance to itraconazole. J. Clin. Microbiol. 43:214-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, J., H. Li, R. Li, D. Bu, and Z. Wan. 2005. Mutations in the cyp51A gene and susceptibility to itraconazole in Aspergillus fumigatus serially isolated from a patient with lung aspergilloma. J. Antimicrob. Chemother. 55:31-37. [DOI] [PubMed] [Google Scholar]
- 3.Cheng, J. C., C. L. Huang, C. C. Lin, C. C. Chen, Y. C. Chang, S. S. Chang, and C. P. Tseng. 2006. Rapid detection and identification of clinically important bacteria by high-resolution melting analysis after broad-range ribosomal RNA real-time PCR. Clin. Chem. 52:1997-2004. [DOI] [PubMed] [Google Scholar]
- 4.Corbett Research. 2006. High resolution melt assay design and analysis: instruction manual. Corbett Research, Mortlake, Australia.
- 5.Dannaoui, E., E. Borel, M. F. Monier, M. A. Piens, S. Picot, and F. Persat. 2001. Acquired itraconazole resistance in Aspergillus fumigatus. J. Antimicrob. Chemother. 47:333-340. [DOI] [PubMed] [Google Scholar]
- 6.Denning, D. W., S. A. Radford, K. L. Oakley, L. Hall, E. M. Johnson, and D. W. Warnock. 1997. Correlation between in-vitro susceptibility testing to itraconazole and in-vivo outcome of Aspergillus fumigatus infection. J. Antimicrob. Chemother. 40:401-414. [DOI] [PubMed] [Google Scholar]
- 7.Diaz-Guerra, T. M., E. Mellado, M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2003. A point mutation in the 14α-sterol demethylase gene cyp51A contributes to itraconazole resistance in Aspergillus fumigatus. Antimicrob. Agents. Chemother. 47:1120-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erali, M., K. V. Voelkerding, and C. T. Wittwer. 2008. High resolution melting applications for clinical laboratory medicine. Exp. Mol. Pathol. 85:50-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fortini, D., A. Ciammaruconi, R. De Santis, A. Fasanella, A. Battisti, R. D'Amelio, F. Lista, A. Cassone, and A. Carattoli. 2007. Optimization of high-resolution melting analysis for low-cost and rapid screening of allelic variants of Bacillus anthracis by multiple locus variable-number tandem repeat analysis. Clin. Chem. 53:1377-1380. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Effron, G., A. Dilger, L. Alcázar-Fuoli, S. Park. E. Mellado, and D. S. Perlin. 2008. Rapid detection of triazole antifungal resistance in Aspergillus fumigatus. J. Clin. Microbiol. 46:1200-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoek, K. G. P., N. C. Gey van Pittius, H. Moolman-Smook, K. Carelse-Tofa, A. Jordaan, G. D. van der Spuy, E. Streicher, T. C. Victor, P. D. van Helden, and R. M. Warren. 2008. Fluorometric assay for testing rifampin susceptibility of Mycobacterium tuberculosis complex. J. Clin. Microbiol. 46:1369-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howard, S. J., I. Webster, C. B. Moore, R. E. Gardiner, S. Park, D. S. Perlin, and D. W. Denning. 2006. Multi-azole resistance in Aspergillus fumigatus. Int. J. Antimicrob. Agents 28:450-453. [DOI] [PubMed] [Google Scholar]
- 13.Howard, S. J., D. Cerar, M. J. Anderson, A. Albarrag, M. C. Fisher, A. C. Pasqualotto, M. Laverdiere, M. C. Arendrup, D. S. Perlin, and D. W. Denning. 2009. Frequency and evolution of azole resistance in Aspergillus fumigatus associated with treatment failure. Emerg. Infect. Dis. 15:1068-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeffery, N., R. B. Gasser, P. A. Steer, and A. H. Noormohammadi. 2007. Classification of Mycoplasma synoviae strains using single-strand conformation polymorphism and high-resolution melting-curve analysis of the vlhA gene single-copy region. Microbiology 153:2679-2688. [DOI] [PubMed] [Google Scholar]
- 15.Kontoyiannis, D. P., and G. P. Bodey. 2002. Invasive aspergillosis in 2002: an update. Eur. J. Clin. Microbiol. Infect. Dis. 21:161-172. [DOI] [PubMed] [Google Scholar]
- 16.Latgé, J.-P. 1999. Aspergillus fumigatus and aspergillosis. Clin. Microbiol. Rev. 12:310-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin, J.-H., C. P. Tseng, Y. J. Chen, C. Y. Lin, S. S. Chang, H. S. Wu, and J. C. Cheng. 2008. Rapid differentiation of influenza A virus subtypes and genetic screening for virus variants by high-resolution melting analysis. J. Clin. Microbiol. 46:1090-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin, S. J., J. Schranz, and S. M. Teutsch. 2001. Aspergillosis case-fatality rate: systematic review of the literature. Clin. Infect. Dis. 32:358-366. [DOI] [PubMed] [Google Scholar]
- 19.Lupetti, A., R. Danesi, M. Campa, M. Del Tacce, and S. Kelly. 2002. Molecular basis of resistance to azole anti-fungals. Trends Mol. Med. 8:76-81. [DOI] [PubMed] [Google Scholar]
- 20.Mann, P., R. M. Parmegiani, S. Wei, C. A. Mendrick, X. Li, D. Loebenberg, B. DiDomenico, R. S. Hare, S. S. Walker, and P. M. McNicholas. 2003. Mutations in Aspergillus fumigatus resulting in reduced susceptibility to posaconazole appear to be restricted to a single amino acid in the cytochrome P450 14α-demethylase. Antimicrob. Agents Chemother. 47:577-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mellado, E., T. M. Diaz-Guerra, M. Cuenca-Estrella, and J. L. Rodríguez-Tudela. 2001. Identification of two different 14-α sterol demethylase-related genes (cyp51A and cyp51B) in Aspergillus fumigatus and other Aspergillus species. J. Clin. Microbiol. 39:2431-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mellado, E., G. Garcia-Effron, L. Alcázar-Fuoli, M. Cuenca-Estrella, and J. L. Rodríguez-Tudela. 2004. Substitutions at methionine 220 in the 14-α sterol demethylase (Cyp51A) of Aspergillus fumigatus are responsible for resistance in vitro to azole antifungal drugs. Antimicrob. Agents Chemother. 48:2747-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mellado, E., G. Garcia-Effron, L. Alcázar-Fuoli, W. J. G. Melchers, P. E. Verweij, M. Cuenca-Estrella, and J. L. Rodriguez-Tudela. 2007. A new Aspergillus fumigatus resistance mechanism conferring in vitro cross-resistance to azole antifungals involves a combination of cyp51A alterations. Antimicrob. Agents Chemother. 51:1897-1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nascimento, A. M., G. H. Goldman, S. Park, S. A. E. Marras, G. Delmas, U. Oza, K. Lolans, M. N. Dudley, P. A. Mann, and D. S. Perlin. 2003. Multiple resistance mechanisms among Aspergillus fumigatus mutants with high-level resistance to intraconazole. Antimicrob. Agents Chemother. 47:1719-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Odell, I. D., J. L. Cloud, M. Seipp, and C. T. Wittwer. 2005. Rapid species identification within the Mycobacterium chelonae-abscessus group by high-resolution melting analysis of hsp65 PCR products. Am. J. Clin. Pathol. 123:96-101. [DOI] [PubMed] [Google Scholar]
- 26.Pfaller, M. A., S. A. Messer, L. Boyken, C. Rice, S. Tendolkar, R. J. Hollis, and D. J. Diekema. 2008. In vitro survey of triazole cross-resistance among more than 700 clinical isolates of Aspergillus species. J. Clin. Microbiol. 46:2568-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slinger, R., D. Bellfov, M. Desjardins, and F. Chan. 2007. High-resolution melting assay for the detection of gyrA mutations causing quinolone resistance in Salmonella enterica serovars Typhi and Paratyphi. Diagn. Microbiol. Infect. Dis. 57:455-458. [DOI] [PubMed] [Google Scholar]
- 28.Snelders, E., R. A. G. Huis in't Veld, A. J. M. M. Rijs, H. A. L. van der Lee, J. Kuijpers, W. J. G. Melchers, and P. E. Verweij. 2008. Environmental Aspergillus fumigatus isolates resistant to triazoles are genetically related to resistant clinical isolates, abstr. O372. 18th European Congress of Clinical Microbiology and Infectious Diseases, Barcelona, Spain, 19 to 22 April 2008.
- 29.Snelders, E., H. A. L. van der Lee, J. Kuijpers, A. J. M. M. Rijs, J. Varga, R. A. Samson, E. Mellado, A. R. T. Donders, W. J. G. Melchers, and P. E. Verweij. 2008. Emergence of azole resistance in Aspergillus fumigatus and spread of a single resistance mechanism. PLoS Med. 5:1629-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steger G. 1994. Thermal denaturation of double-stranded nucleic acids: prediction of temperatures critical for gradient gel electrophoresis and polymerase chain reaction. Nucleic Acids Res. 22:2760-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stephens, A. J., J. Inman-Bamber, P. M. Giffard, and F. Huygens. 2008. High-resolution melting analysis of the spa repeat region of Staphylococcus aureus. Clin. Chem. 54:432-436. [DOI] [PubMed] [Google Scholar]
- 32.Trama, J. P., E. Mordechai, and M. E. Adelson. 2005. Detection of Aspergillus fumigatus and a mutation that confers reduced susceptibility to itraconazole and posaconazole by real-time PCR and pyrosequencing. J. Clin. Microbiol. 43:906-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verweij, P. E., M. Mellado, and W. J. Melchers. 2007. Multiple-triazole-resistant aspergillosis. N Engl. J Med. 356:1481-1483. [DOI] [PubMed] [Google Scholar]
- 34.Xiao, L., V. Madison, A. S. Chau, D. Loebenberg, R. E. Palermo, and P. M. McNicholas. 2004. Three-dimensional models of wild-type and mutated forms of cytochrome P450 14α-sterol demethylases from Aspergillus fumigatus and Candida albicans provide insights into posaconazole binding. Antimicrob. Agents Chemother. 48:568-574. [DOI] [PMC free article] [PubMed] [Google Scholar]