Abstract
JNJ-Q2, a novel fluorinated 4-quinolone, was evaluated for its antibacterial potency by broth and agar microdilution MIC methods in studies focused on skin and respiratory tract pathogens, including strains exhibiting contemporary fluoroquinolone resistance phenotypes. Against a set of 118 recent clinical isolates of Streptococcus pneumoniae, including fluoroquinolone-resistant variants bearing multiple DNA topoisomerase target mutations, an MIC90 value for JNJ-Q2 of 0.12 μg/ml was determined, indicating that it was 32-fold more potent than moxifloxacin. Against a collection of 345 recently collected methicillin-resistant Staphylococcus aureus (MRSA) isolates, including 256 ciprofloxacin-resistant strains, the JNJ-Q2 MIC90 value was 0.25 μg/ml, similarly indicating that it was 32-fold more potent than moxifloxacin. The activities of JNJ-Q2 against Gram-negative pathogens were generally comparable to those of moxifloxacin. In further studies, JNJ-Q2 exhibited bactericidal activities at 2× and 4× MIC levels against clinical isolates of S. pneumoniae and MRSA with various fluoroquinolone susceptibilities, and its activities were enhanced over those of moxifloxacin. In these studies, the activity exhibited against strains bearing gyrA, parC, or gyrA plus parC mutations was indicative of the relatively balanced (equipotent) activity of JNJ-Q2 against the DNA topoisomerase target enzymes. Finally, determination of the relative rates or frequencies of the spontaneous development of resistance to JNJ-Q2 at 2× and 4× MICs in S. pneumoniae, MRSA, and Escherichia coli were indicative of a lower potential for resistance development than that for current fluoroquinolones. In conclusion, JNJ-Q2 exhibits a range of antibacterial activities in vitro that is supportive of its further evaluation as a potential new agent for the treatment of skin and respiratory tract infections.
Since the introduction of nalidixic acid into clinical use in 1962, successive enhancements of the antimicrobial potency and spectrum of the 4-quinolone class of agents have facilitated their expanded clinical utility in the treatment of bacterial infections in both community and health care settings. The potent activities of levofloxacin and moxifloxacin against both Gram-negative and Gram-positive respiratory tract pathogens, combined with their established safety and tolerability profiles, have led to their adoption as key agents in the treatment of community-acquired respiratory tract infections.
4-Quinolones exhibit their antibacterial activity through perturbation of the normal function of the type II DNA topoisomerases, DNA gyrase and/or DNA topoisomerase IV, each of which has distinct and essential roles in nucleic acid metabolism. The improved balance in the potencies of more recently developed 4-quinolone agents as inhibitors of both DNA topoisomerase targets has been proposed to underlie their improved resistance development properties through effective narrowing of the so-called mutant selection window (21). However, the development of resistance to 4-quinolone agents can also arise through mutations that elevate the basal levels of expression of efflux pumps. In Gram-negative organisms, elevated levels of expression of the resistance-nodulation-cell division (RND) class of efflux pumps are commonly associated with reduced fluoroquinolone susceptibility, while increased levels of expression of the major facilitator superfamily (MFS) or the multidrug and toxin extrusion (MATE) class of pumps are more frequently involved in mediated fluoroquinolone efflux in Gram-positive organisms (40). The improved profile of activity of some contemporary agents against Gram-positive clinical isolates that exhibit elevated levels of efflux of ciprofloxacin and norfloxacin can be attributed to the introduction of structural features that reduce their apparent affinity for specific efflux pumps (34, 44).
Despite the widespread use of fluoroquinolones for the treatment of respiratory tract infections in the community setting over the last 40 years or so, resistance in pneumococci has been relatively slow to become established compared to the rates of development of resistance to some other classes of antibiotics. For instance, the rate of resistance to levofloxacin among contemporary U.S. isolates of Streptococcus pneumoniae has remained 1% or less (17); and the rate of resistance to other fluoroquinolones is reported to be less than 4% among U.S. isolates (19), European isolates (18), and global isolates (32, 33), although a higher incidence of 11.6% was recently reported among isolates from elderly patients in Canada (1). The introduction of the seven-valent pneumococcal conjugate vaccine (PCV7) has been associated with the increased prevalence of non-PCV7 serotypes (16, 17), including several predominant multidrug-resistant (MDR) clones disseminated worldwide (4, 13). However, current reports of elevated rates of fluoroquinolone resistance among localized populations of S. pneumoniae are best attributed to sporadic, independent mutational events as opposed to vaccine-driven clonal expansion (15, 18, 43), and the subsequent decline in the numbers of such strains in surveillance populations has been ascribed to their apparent decreased fitness (16). Nevertheless, the potential for the clonal expansion and dissemination of MDR strains exhibiting fluoroquinolone resistance, combined with reports of treatment failures associated with the use of fluoroquinolones for the treatment of fluoroquinolone-resistant pneumococcal pneumonia (26, 35), provides the impetus to identify new compounds in the fluoroquinolone class that retain their activities against strains resistant to contemporary agents.
In contrast to S. pneumoniae, the relatively high incidence of fluoroquinolone resistance in contemporary isolates of Staphylococcus aureus and some Gram-negative species significantly limits the utility of currently approved fluoroquinolone agents. In the case of S. aureus, the introduction of ciprofloxacin into clinical use was associated with the rapid emergence of resistance in methicillin-resistant S. aureus (MRSA) strains in the nosocomial setting (8). This phenomenon can be attributed to the relative DNA topoisomerase target imbalance of ciprofloxacin in this species (20, 23), the propensity for ciprofloxacin to select for efflux-based resistance (39, 41), and the apparent increased fitness of ciprofloxacin-resistant MRSA strains in some nosocomial settings (7). In contemporary surveillance studies of health care-associated MRSA isolates, the rate of ciprofloxacin resistance is routinely reported to be ≥50%, and currently approved fluoroquinolones are indicated for use only for the treatment of staphylococcal infections mediated by methicillin-susceptible strains. However, a series of investigational fluoroquinolones have been reported to exhibit good in vitro activities against MRSA, including ciprofloxacin-resistant strains (2, 5, 25, 29, 45), and several of these agents are reported to be undergoing clinical evaluation for the treatment of complicated skin and skin structure infections (cSSSIs), including infections caused by MRSA strains.
Herein we describe the key attributes of the in vitro profile of antibacterial activity of a new fluoroquinolone agent, JNJ-Q2, with a focus on its activity against key pathogens associated with community-acquired bacterial pneumonia and cSSSIs.
(Portions of this work were previously presented at the combined 48th Annual Interscience Conference on Antimicrobial Agents and Chemotherapy [ICAAC] and the Infectious Diseases Society of America [IDSA] 46th Annual Meeting, 2008 [24, 27, 38].)
MATERIALS AND METHODS
Antimicrobial agents.
JNJ-Q2, 7-[(3E)-3-(2-amino-1-fluoroethylidene)-1-piperidinyl]-1-cyclopropyl-6-fluoro-1,4-dihydro-8-methoxy-4-oxo-3-quinolinecarboxylic acid, was synthesized at Johnson & Johnson Pharmaceutical Research and Development (J&JPRD). Moxifloxacin, gemifloxacin, and linezolid were purchased from their respective commercial manufacturers. Penicillin, erythromycin, ciprofloxacin, vancomycin, ethidium bromide, benzalkonium chloride, and reserpine were obtained from Sigma-Aldrich Chemical Company (St. Louis, MO).
Bacterial isolates.
The bacterial isolates included in this study were from the J&JPRD culture collection. S. pneumoniae strains bearing defined mutations in the quinolone resistance-determining region (QRDR) were described previously (36, 37).
In vitro susceptibility.
MICs were determined in accordance with the Clinical and Laboratory Standards Institute (CLSI) methodology (CLSI M07-A7) by either the broth microdilution or the agar dilution method with cation-adjusted Mueller-Hinton (MH) medium or MH medium supplemented with 5% lysed horse blood for S. pneumoniae (10). The susceptibility data presented in Tables 1 to 4 were determined by broth microdilution, and the MIC values used for the resistance studies (Tables 5 and 6) were determined by the agar dilution method. The following CLSI quality control strains were included, as appropriate, in the susceptibility tests: S. pneumoniae ATCC 49619, S. aureus ATCC 29213, Enterococcus faecalis ATCC 29212, Pseudomonas aeruginosa ATCC 27853, and Escherichia coli ATCC 25922.
TABLE 1.
In vitro activities of JNJ-Q2 and comparators against isolates of Gram-positive pathogens
| Organism | No. of isolates | Compound | MIC (μg/ml) |
||
|---|---|---|---|---|---|
| Range | 50% | 90% | |||
| S. pneumoniae, all isolates | 118 | JNJ-Q2 | ≤0.004-0.5 | 0.015 | 0.12 |
| 118 | Moxifloxacin | 0.06-16 | 0.25 | 4 | |
| 118 | Gemifloxacin | ≤0.004-4 | 0.03 | 0.5 | |
| 98 | Penicillin | ≤0.008-4 | 0.06 | 2 | |
| 98 | Erythromycin | ≤0.015->16 | 0.06 | >16 | |
| S. pneumoniae, ciprofloxacin resistant (characterized | 19 | JNJ-Q2 | 0.06-0.5 | 0.12 | 0.25 |
| QRDR mutationsa) | 19 | Ciprofloxacin | 8-64 | 32 | 64 |
| 19 | Levofloxacin | 8-64 | 16 | 64 | |
| 19 | Moxifloxacin | 1-16 | 4 | 16 | |
| 19 | Gemifloxacin | 0.12-4 | 0.5 | 2 | |
| S. aureus, all isolates | 406 | JNJ-Q2 | 0.002-2 | 0.12 | 0.25 |
| 406 | Ciprofloxacin | 0.12->256 | 16 | >32 | |
| 406 | Moxifloxacin | 0.06->16 | 2 | 8 | |
| 384 | Vancomycin | 0.5-2 | 1 | 1 | |
| MSSA | 26 | JNJ-Q2 | 0.008-0.015 | 0.015 | 0.015 |
| 26 | Ciprofloxacin | 0.5-1 | 0.5 | 0.5 | |
| 26 | Moxifloxacin | 0.06-0.12 | 0.06 | 0.12 | |
| 26 | Vancomycin | 1-2 | 1 | 2 | |
| MRSA isolates collected from 2004 to 2006 | 345 | JNJ-Q2 | 0.002-2 | 0.12 | 0.25 |
| 345 | Ciprofloxacin | 0.12->32 | 16 | >32 | |
| 345 | Moxifloxacin | 0.06->32 | 2 | 8 | |
| 333 | Vancomycin | 0.5-2 | 1 | 1 | |
| Ciprofloxacin-resistant MRSAb isolates collected | 25 | JNJ-Q2 | 0.12-1 | 0.5 | 0.5 |
| from 1991 to 1997 | 25 | Ciprofloxacin | 8->32 | >32 | >32 |
| 25 | Moxifloxacin | 1-16 | 4 | 8 | |
| 25 | Vancomycin | 1-2 | 1 | 2 | |
| Ciprofloxacin-resistant MRSA isolates collected from | 256 | JNJ-Q2 | 0.015-2 | 0.25 | 0.25 |
| 2004 to 2006c | 256 | Ciprofloxacin | 4->256 | 32 | 64 |
| 256 | Moxifloxacin | 0.25->16 | 4 | 8 | |
| 248 | Vancomycin | 0.5-2 | 1 | 1 | |
| MSSE | 30 | JNJ-Q2 | 0.008-0.03 | 0.015 | 0.015 |
| 30 | Ciprofloxacin | 0.12-0.5 | 0.25 | 0.5 | |
| 18 | Levofloxacin | 0.12-0.25 | 0.12 | 0.25 | |
| 30 | Moxifloxacin | 0.03-0.12 | 0.06 | 0.12 | |
| 30 | Vancomycin | 1-2 | 2 | 2 | |
| MRSE, all isolates | 34 | JNJ-Q2 | 0.008-1 | 0.12 | 0.25 |
| 34 | Ciprofloxacin | 0.12-128 | 16 | 64 | |
| 34 | Levofloxacin | 0.12-128 | 8 | 16 | |
| 34 | Moxifloxacin | 0.03-32 | 2 | 4 | |
| 34 | Vancomycin | 1-2 | 2 | 2 | |
| Ciprofloxacin-resistant MRSE | 21 | JNJ-Q2 | 0.06-1 | 0.12 | 0.25 |
| 21 | Ciprofloxacin | 4-128 | 64 | 128 | |
| 21 | Levofloxacin | 4-128 | 16 | 16 | |
| 21 | Moxifloxacin | 0.5-32 | 2 | 4 | |
| 21 | Vancomycin | 1-2 | 2 | 2 | |
| Streptococcus pyogenes | 21 | JNJ-Q2 | 0.008-0.06 | 0.008 | 0.015 |
| 21 | Ciprofloxacin | 0.25-2 | 0.5 | 1 | |
| 21 | Moxifloxacin | 0.12-0.25 | 0.12 | 0.25 | |
| Streptococcus agalactiae | 19 | JNJ-Q2 | 0.008-0.03 | 0.015 | 0.015 |
| 19 | Ciprofloxacin | 0.5-2 | 1 | 1 | |
| 19 | Moxifloxacin | 0.12-0.25 | 0.25 | 0.25 | |
| Streptococcus spp., group C | 10 | JNJ-Q2 | 0.004-0.015 | 0.008 | 0.015 |
| 10 | Ciprofloxacin | 0.12-2 | 0.25 | 1 | |
| 10 | Moxifloxacin | 0.06-0.25 | 0.12 | 0.25 | |
| Enterococcus faecalis | 12 | JNJ-Q2 | 0.03-1 | 0.06 | 0.5 |
| 12 | Ciprofloxacin | 0.5->16 | 1 | >16 | |
| 12 | Moxifloxacin | 0.25-16 | 0.25 | 16 | |
| 12 | Vancomycin | 1->16 | 2 | >16 | |
| Enterococcus faecium | 13 | JNJ-Q2 | 0.25-4 | 0.5 | 4 |
| 13 | Ciprofloxacin | 1->16 | 4 | >16 | |
| 13 | Moxifloxacin | 1->16 | 4 | >16 | |
| 13 | Vancomycin | 1->32 | >16 | >16 | |
For S. pneumoniae, ciprofloxacin resistance was defined as an MIC of ≥8 μg/ml. Among the isolates in this set, each isolate contains at least two QRDR mutations in DNA gyrase and/or DNA topoisomerase IV.
For S. aureus, ciprofloxacin resistance was defined as an MIC of ≥4 μg/ml.
Ciprofloxacin-resistant isolates are a subset of the MRSA isolates collected between 2004 and 2006.
TABLE 4.
Characterization of in vitro bactericidal activities of JNJ-Q2 and moxifloxacin against methicillin-resistant S. aureus clinical isolates by time-kill methods
| MRSA isolate | QRDR genotypeb | MIC (μg/ml) |
Log10 decrease (Δlog10 CFU/ml) in viable cells recovereda at: |
||||
|---|---|---|---|---|---|---|---|
| 2× MIC |
4× MIC |
||||||
| MXFg | JNJ-Q2 | MXF | JNJ-Q2 | MXF | JNJ-Q2 | ||
| OC 11223 | NDf | 0.06 | 0.008 | 1.0 | 2.5 | 1.5 | 3.9 |
| OC 8525d | Wild type | 0.12 | 0.008 | 0.0 | 5.1 | 3.2 | 5.0 |
| OC 11696d | ND | 1 | 0.06 | 3.1 | 6.5e | 3.2 | 6.5e |
| OC 12501d | ND | 1 | 0.06 | 3.1 | 6.2e | 3.2 | 6.2e |
| OC 8530d | gyrAS84L | 2 | 0.25 | >−2.0c | 6.3e | 2.3 | 6.3e |
| parCI143V | |||||||
| OC 4222d | gyrAS84L | 2 | 0.5 | 2.4 | 5.9 | 2.7 | 4.2 |
| parCS80F | |||||||
| OC 2838 | ND | 4 | 0.25 | 2.6 | 4.1 | 3.2 | 4.5 |
| OC 17042d | gyrAS84L, G106D | 8 | 0.5 | 0.9 | 3.8 | 1.7 | 3.5 |
| parCS80FparED432N | |||||||
| OC 11521d | gyrAS84L, S85P | 32 | 1 | 2.0 | 2.3 | 3.4 | 2.3 |
| parCE84G | |||||||
Log10 decrease in the numbers of CFU/ml recovered at 24 h in comparison to the numbers recovered at 0 h.
parC and parE were used in place of the gene names grlA and grlB, respectively.
Moxifloxacin was not effective at suppressing the elaboration of the starting inoculum.
Isolates display phenotypic evidence of activated efflux, as the MICs for norfloxacin, benzalkonium chloride, and ethidium bromide were ≥4-fold lower in the presence of 20 μg/ml reserpine.
No colonies were cultured from these samples, indicating sterilization.
ND, not determined.
MXF, moxifloxacin.
TABLE 5.
Frequency of selection of S. pneumoniae resistant mutants by JNJ-Q2 or moxifloxacin on agar at 2× and 4× MICs
| Strain | QRDR genotype | Agent | MIC (μg/ml) | Mutation frequency at: |
|
|---|---|---|---|---|---|
| 2× MIC | 4× MIC | ||||
| R6 | Wild type | JNJ-Q2 | 0.015 | 1.8 × 10−10 | <1.8 × 10−10 |
| MXFd | 0.12 | 2.8 × 10−10 | <1.8 × 10−10 | ||
| OC 7452a | parCS79Y | JNJ-Q2 | 0.015 | 1.3 × 10−9 | ≤2.9 × 10−10c |
| MXF | 0.25 | 1.8 × 10−9 | 1.6 × 10−9 | ||
| OC 7453a | gyrAS81F | JNJ-Q2 | 0.03 | ≤1.3 × 10−11 | <1.5 × 10−11 |
| MXF | 0.5 | 2.2 × 10−10 | 3.9 × 10−11 | ||
| OC 6755b | gyrAS81F | JNJ-Q2 | 0.12 | 1.6 × 10−11 | ≤3.1 × 10−11 |
| parCS79Y | MXF | 8 | 2.3 × 10−11 | ≤3.1 × 10−11 | |
Strains were derived from strain R6 through transformation with alleles with single mutations (36).
TRUST surveillance isolate collected from 2000 to 2002.
Values with the ≤ symbol indicate that a total of one colony was observed on duplicate plates.
MXF, moxifloxacin.
TABLE 6.
Rates of resistance to JNJ-Q2 and ciprofloxacin in methicillin-resistant S. aureus determined by a modified Luria-Delbruck fluctuation assay
| MRSA isolate | QRDR genotype | MIC (μg/ml)a |
Resistance rateb |
||
|---|---|---|---|---|---|
| CIPd | JNJ-Q2 | CIP | JNJ-Q2 | ||
| OC 8532c | Wild type | 0.6 | 0.008 | 3.7 × 10−12 | 2.9 × 10−12 |
| OC 8525c | Wild type | 3 | 0.012 | 1.0 × 10−12 | 1.4 × 10−12 |
| OC 11150 | gyrAS84LparCS80Y | 12 | 0.12 | 6.0 × 10−10 | 6.2 × 10−12 |
| OC 15425c | gyrAS84LparCS80F | 12 | 0.12 | 1.8 × 10−9 | 3.2 × 10−12 |
Agar MICs were determined through the use of arithmetic dilutions.
The number of mutations isolated per cell per division.
Isolates display phenotypic evidence of activated efflux, as the MICs for norfloxacin, benzalkonium chloride, and ethidium bromide were ≥4-fold lower in the presence of 20 μg/ml reserpine.
CIP, ciprofloxacin.
Time-kill studies.
Time-kill assays were used to assess the relative bactericidal activity of JNJ-Q2 and the comparators. For the S. aureus isolates, time-kill assays were performed in cation-adjusted Mueller-Hinton broth (CAMHB; BBL), and for the S. pneumoniae isolates, CAMHB supplemented with 5% lysed horse blood (Hardy Diagnostics, CA) was used. Test strains cultured overnight in the appropriate medium were diluted into fresh medium to yield starting initial cell densities of approximately 5 × 106 CFU/ml. Antimicrobial agents were diluted from 50×-concentrated stocks prepared in water for moxifloxacin or in acidified water (25 μl glacial acetic acid per 10 ml water) for JNJ-Q2. Increases or decreases in viable cell titers in the freshly inoculated cultures were determined by sampling at 0, 1, 2, 4, 6, 8, and 24 h postinoculation or at 6 and 24 h postinoculation, followed by plating of serial dilutions of the culture sample on Trypticase soy agar (TSA) plates and enumeration of the numbers of viable CFU following 20 to 24 h incubation at 37°C. TSA supplemented with 5% sheep blood was employed in studies of S. pneumoniae, and agar plates were incubated in a 5% CO2 environment.
Determination of resistance frequencies.
Liquid cultures of S. pneumoniae in the mid-logarithmic phase of growth (optical density at 600 nm, 0.8 to 0.95) were concentrated 25-fold by centrifugation and resuspension in phosphate-buffered saline, followed by plating of approximately 1 × 1010 to 2 × 1011 CFU onto TSA plates supplemented with 5% sheep blood and containing 2× or 4× MIC of drug. The plates used for resistance selection were incubated at 37°C in 5% CO2. The colonies on the plates were enumerated following 24 h of incubation, and the final CFU counts were determined following reassessment at 48 h. The inoculum titers were determined by plating serial dilutions onto drug-free TSA-blood plates. The frequency of selection of resistant mutants was calculated as the ratio of the average number of resistant colonies arising on duplicate plates at 24 h to the number of viable cells plated.
Determination of resistance rates.
The rates of resistance for methicillin-resistant S. aureus were determined through the use of fluctuation assays performed by a modification of the method described by Crane et al. (12). Briefly, a total of 15 1-ml sister cultures were inoculated in parallel with 50 to 100 CFU of each isolate (prepared by serial dilution of the same overnight culture and incubation for 24 h at 37°C with shaking at 200 rpm). Each sister culture was then serially diluted and aliquots were plated onto Mueller-Hinton agar (MHA) plates without selection for CFU enumeration. One hundred microliters of each sister culture was inoculated onto MHA plates containing 2× MIC of drug, with the MIC values being determined in this case by using an arithmetic agar dilution scheme to more precisely determine the MIC. Selective plates were incubated at 37°C for 24 h, followed by an additional 16 to 18 h of incubation at room temperature. The resistant colonies were counted, and the mutation rates were estimated by using the median estimator method defined by Jones et al. (30).
PCR amplification and DNA sequence analysis.
The QRDRs of the gyrA, gyrB, parC (grlA), and parE (grlB) genes and the norA promoter DNA sequences were determined from PCR products amplified from selected MRSA isolates. For PCR templates, the cultures were treated with 0.5 mg/ml lysostaphin (Sigma) and 15 mg/ml lysozyme (Sigma) at room temperature for 20 min (28). Amplification of the QRDRs of the gyrA, gyrB, parC, and parE genes was performed with the following primer pairs: gyrA Forward (5′-GCGATGAGTGTTATCGTTGCT-3′) plus gyrA Reverse (5′-CAGGACCTTCAATATCCTCC-3′), parC Forward (5′-GATGAGGAGGAAATCTAG-3′ plus parC Reverse (5′-GTTGGAAAATCAGGACCTT-3′), parE Forward (5′-GACAATTGTCTAAATCACTTGTA-3′) plus parE Reverse (5′-CATCAGTCATAATAATTACAC-3′), and gyrB Forward (5′-CAGCGTTAGATGTAGCAAGC-3′) plus gyrB Reverse (5′-CCGATTCCTGTACCAAATGC-3′) (42). Amplification was performed in a Techne TC-512 instrument (Bibby Scientific, Burlington, NJ) with Platinum PCR SuperMix (Invitrogen) and 10 μM each primer. For QRDR product amplification, an initial cycle of 5 min of denaturation at 95°C was followed by 30 cycles of 30 s at 95°C, 30 s at 50°C, and 30 s at 72°C, with a final extension step of 5 min at 72°C and slow cooling to 4°C. The 5′ end of the norA gene and the upstream regulatory region were amplified by the use of similar cycling parameters and the primers norA Forward (5′-TGTTAAGTCTTGGTCATCTGCA-3′) plus norA Reverse (5′-AGCAGCAACAAGTAACCCTAAA-3′). The PCR products were purified with QIAquick spin columns (Qiagen, Chatsworth, CA). DNA sequencing of the PCR products was performed by ACGT, Inc. (Wheeling, IL), by use of the amplification primers.
RESULTS
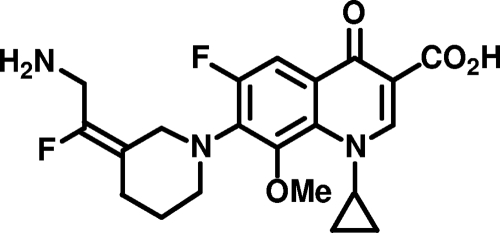
Figure 1 shows the chemical structure of JNJ-Q2, one of the most potent antistaphylococcal agents in a series of aminoethylidenylpiperidine fluoroquinolones with drug-like properties, including a low molecular weight (419.4) and acceptable solubility and lipophilicity. Specifically, the experimentally determined values of pKa (6.13), pKb (8.59), and the logarithm of the distribution coefficient (D) at pH 7.4 (log D7.4 = 0.37) for JNJ-Q2 are suggestive of absorption and permeability characteristics equivalent to those of currently approved fluoroquinolones, while the aqueous solubility of 4.52 mg/ml for the hydrochloride salt of JNJ-Q2 is consistent with the development of oral and parenteral dosage forms. The level of plasma protein binding of JNJ-Q2 for species relevant to the rodent models of infection used for preclinical studies ranged from 56% to 76%, and there was some evidence of concentration dependence.
FIG. 1.
Chemical structure of JNJ-Q2.
As shown in Table 1, JNJ-Q2 and key comparators were evaluated for their in vitro activities against recent clinical isolates of S. pneumoniae, including susceptible and multidrug-resistant isolates. JNJ-Q2 demonstrated the most potent in vitro activity of all agents tested, with an MIC90 value of 0.12 μg/ml being determined against all isolates combined (n = 118), and therefore, it was 4-fold and 32-fold more potent than the comparator fluoroquinolones gemifloxacin and moxifloxacin, respectively. Similarly, JNJ-Q2 exhibited activity superior to the activities of the comparator fluoroquinolones against a subset of S. pneumoniae isolates displaying resistance to penicillin or erythromycin, including isolates exhibiting an MDR phenotype (data not shown). The activity of JNJ-Q2 against ciprofloxacin- and levofloxacin-resistant clinical isolates of S. pneumoniae was also tested by using a set of 19 nonoverlapping strains that possess between two and five defined mutations in DNA gyrase and/or DNA topoisomerase IV. These isolates were provided from the 2000-2003 Tracking Resistance in the United States Today (TRUST) surveillance program (TRUST 4 through 7) and represent a range of serotypes. The QRDR mutations present in these strains include the prevalent mutant variants of DNA gyrase (GyrA Ser81 or Glu85 substitutions in 15 of 19 isolates) or DNA topoisomerase IV (ParC Ser79 substitutions in 13 of 19 isolates) or in both target enzymes (10 isolates) (14, 36). The MIC90 value determined for JNJ-Q2 against this set of isolates was 0.25 μg/ml, which was 8- to 256-fold lower than the MIC90 values of 64, 64, 16, and 2 μg/ml determined for ciprofloxacin, levofloxacin, moxifloxacin, and gemifloxacin, respectively.
JNJ-Q2 also displayed greater in vitro potency than the comparator fluoroquinolones and vancomycin against a collection of S. aureus strains, including methicillin-susceptible S. aureus (MSSA) and MRSA isolates and all subsets thereof (Table 1). The activity of JNJ-Q2 was tested against a set of 345 MRSA clinical isolates cultured from 2004 to 2006 from patients with skin and respiratory tract infections and from diverse geographical areas. The set included isolates with various staphylococcal cassette chromosome mec (SCCmec) genotypes and pulsed-field gel patterns indicative of both community and nosocomial lineages. Against those isolates, JNJ-Q2 displayed an MIC90 of 0.25 μg/ml, a value 32-fold lower than that for moxifloxacin determined in parallel. Seventy-four percent of the isolates (n = 256) were ciprofloxacin resistant (ciprofloxacin MIC ≥ 4 μg/ml), and JNJ-Q2 retained its activity against this subset of isolates, with the MIC90 value being 0.25 μg/ml, which is again 32-fold lower than that of moxifloxacin. Against an older set of ciprofloxacin-resistant MRSA isolates obtained between 1991 and 1997, JNJ-Q2 exhibited an MIC90 of 0.5 μg/ml, 16-fold lower than that of moxifloxacin. Finally, JNJ-Q2 was the most potent agent tested against methicillin-susceptible and -resistant Staphylococcus epidermidis (MSSE and MRSE, respectively) isolates, with the MIC90 values for contemporary isolates, including those exhibiting ciprofloxacin resistance, being 0.015 and 0.25 μg/ml, respectively.
In addition to S. pneumoniae and S. aureus, a broad range of Gram-positive and Gram-negative pathogens relevant to clinical indications for use of the currently approved fluoroquinolones were also tested for their in vitro susceptibilities to JNJ-Q2 and the comparator agents. JNJ-Q2 was more potent against the beta-hemolytic streptococci than the comparator fluoroquinolones moxifloxacin and ciprofloxacin (Table 1). For Streptococcus pyogenes, Streptococcus agalactiae, and Streptococcus sp. group C, the MIC90 values for JNJ-Q2 were all 0.015 μg/ml, whereas they were 0.25 and 1 μg/ml for moxifloxacin and ciprofloxacin, respectively. JNJ-Q2 displayed an MIC90 value against Enterococcus faecalis that was 32-fold lower than that of moxifloxacin, and the activities of JNJ-Q2 against the E. faecium isolates were greater than those of the comparator fluoroquinolones and vancomycin (Table 1).
Against Gram-negative pathogens, JNJ-Q2 generally displayed MICs comparable to or lower than those of moxifloxacin (Table 2), although against several species, the MIC values for JNJ-Q2 and moxifloxacin were higher than those determined for ciprofloxacin. Against Haemophilus influenzae and Moraxella catarrhalis, JNJ-Q2 displayed potent in vitro activity (MIC90 values ≤ 0.015 μg/ml). Against collections of Klebsiella pneumoniae, ciprofloxacin-susceptible and -intermediate Escherichia coli, Enterobacter cloacae, Enterobacter aerogenes, and Proteus mirabilis isolates, JNJ-Q2 exhibited MIC90 values of ≤0.5 μg/ml, comparable to those of moxifloxacin. The activities of JNJ-Q2 against collections of Serratia marcescens and Citrobacter freundii isolates were also comparable to those of moxifloxacin. In contrast, JNJ-Q2 exhibited MICs 8-fold and 16-fold lower than those of moxifloxacin and ciprofloxacin, respectively, against a collection of ciprofloxacin-resistant E. coli isolates. All ciprofloxacin-resistant E. coli isolates were determined to bear substitutions at the active-site serine residues of both DNA gyrase (GyrA Ser83) and DNA topoisomerase IV (ParC Ser80). The MIC90 value for JNJ-Q2 against a collection of Providencia stuartii isolates was 16-fold lower than the MIC90 values of the comparator fluoroquinolones. Finally, the in vitro activity of JNJ-Q2 against Pseudomonas aeruginosa was found to be comparable to that of levofloxacin.
TABLE 2.
In vitro activities of JNJ-Q2 and comparators against Gram-negative clinical isolates
| Organism | No. of isolates | Compound | MIC (μg/ml) |
||
|---|---|---|---|---|---|
| Range | 50% | 90% | |||
| Haemophilus influenzae | 24 | JNJ-Q2 | 0.008-0.015 | 0.008 | 0.015 |
| 24 | Ciprofloxacin | 0.008-0.015 | 0.008 | 0.015 | |
| 24 | Moxifloxacin | 0.008-0.06 | 0.015 | 0.03 | |
| 24 | Gemifloxacin | 0.002-0.015 | ≤0.004 | 0.008 | |
| Moraxella catarrhalis | 11 | JNJ-Q2 | ≤0.015 | ≤0.015 | ≤0.015 |
| 11 | Ciprofloxacin | ≤0.015-0.25 | 0.06 | 0.06 | |
| 11 | Moxifloxacin | 0.06 | 0.06 | 0.06 | |
| 11 | Gemifloxacin | ≤0.015-0.03 | ≤0.015 | ≤0.015 | |
| Escherichia coli, ciprofloxacin susceptible and intermediate | 30 | JNJ-Q2 | 0.015-0.5 | 0.06 | 0.25 |
| 30 | Ciprofloxacin | 0.008-2 | 0.015 | 0.25 | |
| 30 | Levofloxacin | 0.015-2 | 0.03 | 0.5 | |
| 30 | Moxifloxacin | 0.03-2 | 0.06 | 0.5 | |
| E. coli, ciprofloxacin resistant | 20 | JNJ-Q2 | 1-16 | 4 | 16 |
| 20 | Ciprofloxacin | 16->256 | 64 | 256 | |
| 16 | Levofloxacin | 4-128 | 16 | 64 | |
| 20 | Moxifloxacin | 8->256 | 32 | 128 | |
| 20 | Gemifloxacin | 16->128 | 32 | 64 | |
| Enterobacter cloacae | 26 | JNJ-Q2 | 0.03-0.25 | 0.12 | 0.12 |
| 26 | Ciprofloxacin | ≤0.004-0.25 | 0.015 | 0.06 | |
| 26 | Levofloxacin | 0.008-0.12 | 0.06 | 0.12 | |
| 26 | Moxifloxacin | 0.06-0.25 | 0.06 | 0.12 | |
| Enterobacter aerogenes | 26 | JNJ-Q2 | 0.06-1 | 0.12 | 0.25 |
| 26 | Ciprofloxacin | 0.008-0.25 | 0.03 | 0.03 | |
| 26 | Levofloxacin | 0.03-0.5 | 0.06 | 0.06 | |
| 26 | Moxifloxacin | 0.03-1 | 0.12 | 0.25 | |
| Klebsiella pneumoniae | 27 | JNJ-Q2 | ≤0.015-1 | 0.12 | 0.25 |
| 27 | Ciprofloxacin | ≤0.004-1 | 0.03 | 0.25 | |
| 27 | Levofloxacin | 0.015-1 | 0.06 | 0.12 | |
| 27 | Moxifloxacin | 0.015-1 | 0.12 | 0.25 | |
| Serratia marcescens | 13 | JNJ-Q2 | ≤0.015-1 | 0.5 | 1 |
| 13 | Ciprofloxacin | 0.06-0.5 | 0.12 | 0.25 | |
| 13 | Moxifloxacin | 0.06-1 | 0.5 | 1 | |
| 13 | Gemifloxacin | 0.015-1 | 0.25 | 0.5 | |
| Proteus mirabilis | 11 | JNJ-Q2 | 0.12-0.5 | 0.25 | 0.5 |
| 11 | Ciprofloxacin | 0.015-0.06 | 0.03 | 0.06 | |
| 11 | Moxifloxacin | 0.25-0.5 | 0.5 | 0.5 | |
| 11 | Gemifloxacin | 0.06-0.25 | 0.12 | 0.25 | |
| Providencia stuartii | 10 | JNJ-Q2 | 0.5-4 | 2 | 4 |
| 10 | Ciprofloxacin | 1-128 | 32 | 64 | |
| 10 | Levofloxacin | 4-64 | 16 | 64 | |
| 10 | Moxifloxacin | 8-64 | 32 | 64 | |
| Citrobacter freundii | 13 | JNJ-Q2 | 0.06-8 | 2 | 8 |
| 13 | Ciprofloxacin | 0.015->4 | 2 | >4 | |
| 13 | Moxifloxacin | 0.06->8 | 2 | >8 | |
| 13 | Gemifloxacin | 0.015->4 | 1 | >4 | |
| Pseudomonas aeruginosa | 28 | JNJ-Q2 | 0.5-4 | 1 | 2 |
| 28 | Ciprofloxacin | 0.12-1 | 0.25 | 0.5 | |
| 14 | Levofloxacin | 0.5-2 | 0.5 | 2 | |
| 28 | Moxifloxacin | 0.5-8 | 2 | 4 | |
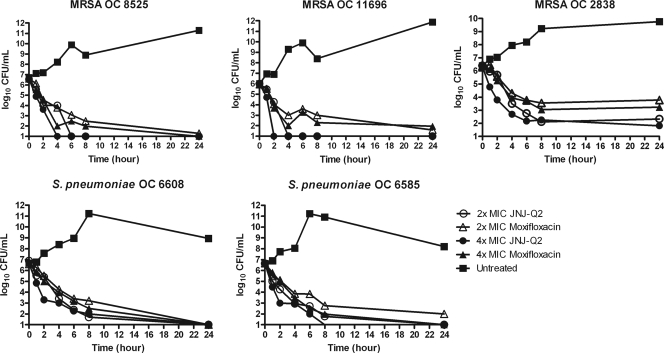
Time-kill curve analysis methods were employed to characterize the comparative bactericidal properties of JNJ-Q2 and moxifloxacin against a series of clinical isolates of S. pneumoniae categorized as susceptible, intermediate, or resistant to moxifloxacin on the basis of current CLSI breakpoints (11). The decreases in viable cell titers determined 6 and 24 h following exposure to either test agent are presented (in log10 units) in Table 3 for all S. pneumoniae isolates tested. The results of time-kill curve analyses performed at six time points are presented in Fig. 2 for a moxifloxacin-intermediate isolate, isolate OC 6608 (gyrAS81F, parCS79F), and a moxifloxacin-resistant isolate, isolate OC 6585 (gyrAE85K, parCS79F, K137N). JNJ-Q2 demonstrated bactericidal activity in vitro against all S. pneumoniae isolates tested, in that 6 h of exposure of each culture at 2× and 4× MICs resulted in ≥3 log10 decreases in viable cell titers. At a concentration of 2× MIC, JNJ-Q2 and moxifloxacin demonstrated comparable bactericidal activities against the moxifloxacin-susceptible isolate (isolate OC 7189), whereas at 6 h, JNJ-Q2 reduced the titers of moxifloxacin-intermediate isolates (strains OC 6608, OC 5462, and OC 7376) to a greater degree than moxifloxacin. These results were significantly different (t test, P < 0.05) for OC 6608 and OC 7376. In general, JNJ-Q2 demonstrated greater bactericidal activity against the moxifloxacin-resistant isolates at 2× and 4× MIC, yielding decreases in the bacterial titers of 0.6 to 1.4 log10 units above those for moxifloxacin following 6 h of exposure. Following 24 h of exposure, 2× and 4× MICs of JNJ-Q2 and moxifloxacin resulted in reductions in the bacterial titers of more than 3 log10 units for all isolates tested with the exception of isolate OC 5462, against which moxifloxacin at 2× MIC failed to achieve reductions that met this criterion for bactericidal activity. Differences in bactericidal activity between JNJ-Q2 and moxifloxacin were less pronounced at 24 h than at 6 h, although the reduction in the numbers of CFU in the untreated control cultures at 24 h (Fig. 2) indicates that the data obtained at the 6-h time point may better reflect antibacterial activity.
TABLE 3.
Characterization of in vitro bactericidal activities of JNJ-Q2 and moxifloxacin against S. pneumoniae clinical isolates by time-kill methods
| Isolate | Agent | MIC (μg/ml) | Log10 decrease (Δlog10 CFU/ml) in viable cells recovered ata: |
|||
|---|---|---|---|---|---|---|
| 6 h |
24 h |
|||||
| 2× MIC | 4× MIC | 2× MIC | 4× MIC | |||
| OC 7189 | JNJ-Q2 | 0.015 | 4.0 | 4.2 | 7.5 | 7.2 |
| MXFb | 0.25 | 3.3 | 4.1 | 7.1 | 7.4 | |
| OC 6608 | JNJ-Q2 | 0.06 | 3.8 | 4.7 | 5.8 | 7.1 |
| MXF | 2 | 3.3 | 3.7 | 5.5 | 6.6 | |
| OC 5462 | JNJ-Q2 | 0.06 | 3.2 | 3.4 | 3.3 | 6.8 |
| MXF | 1 | 2.3 | 2.7 | 2.7 | 6.8 | |
| OC 7376 | JNJ-Q2 | 0.12 | 4.4 | 5.3 | 6.8 | 6.3 |
| MXF | 2 | 3.0 | 3.6 | 6.8 | 6.8 | |
| OC 6790 | JNJ-Q2 | 0.12 | 3.6 | 3.6 | 7.2 | 6.8 |
| MXF | 4 | 2.7 | 2.7 | 6.7 | 6.0 | |
| OC 7368 | JNJ-Q2 | 0.12 | 4.0 | 3.7 | 7.2 | 6.9 |
| MXF | 4 | 2.6 | 2.3 | 5.8 | 6.3 | |
| OC 7348 | JNJ-Q2 | 0.12 | 3.8 | 3.6 | 7.0 | 6.5 |
| MXF | 8 | 2.8 | 3.0 | 6.7 | 6.6 | |
| OC 6585 | JNJ-Q2 | 0.25 | 3.9 | 4.3 | 6.6 | 6.6 |
| MXF | 8 | 2.8 | 3.4 | 4.7 | 6.6 | |
| OC 6610 | JNJ-Q2 | 0.5 | 3.4 | 3.5 | 5.0 | 6.9 |
| MXF | 16 | 2.5 | 2.5 | 5.2 | 4.4 | |
Log10 decrease in the numbers of CFU/ml recovered at 6 or 24 h in comparison to the numbers recovered at 0 h.
MXF, moxifloxacin.
FIG. 2.
Results of in vitro time-kill studies comparing the bactericidal activities of JNJ-Q2 and moxifloxacin against three MRSA isolates and two S. pneumoniae isolates.
Similarly, the relative bactericidal activity of JNJ-Q2 and moxifloxacin against nine MRSA isolates categorized as susceptible, intermediate, or resistant to moxifloxacin by use of the current CLSI breakpoints (11) was assessed by time-kill curve analysis. Among the moxifloxacin-resistant strains were strains bearing multiple mutations in the DNA topoisomerase target genes, in combination with activated fluoroquinolone efflux mediated by the upregulated expression of a reserpine-sensitive efflux pump(s). The log10 decreases in bacterial titers at 24 h from the starting inoculum level are presented in Table 4 for each of the isolates tested. Time-kill curves are presented in Fig. 2 for three MRSA isolates, including a moxifloxacin-susceptible isolate (isolate OC 8525), a moxifloxacin-intermediate isolate (isolate OC 11696), and a moxifloxacin-resistant isolate (isolate OC 2838). JNJ-Q2 demonstrated bactericidal activity in vitro against moxifloxacin-susceptible and -resistant S. aureus MRSA isolates, in that the exposure of each culture to 4× MIC of JNJ-Q2 resulted in >3-log10 decreases in bacterial titers within 24 h of exposure for all isolates except highly moxifloxacin-resistant isolate OC 11521, against which 2.3-log10 killing was observed. In contrast, at 4× MIC, moxifloxacin failed to achieve 3-log10 decreases in bacterial titers for four of the nine isolates tested. Overall, JNJ-Q2 reduced the titers of MRSA in the cultures of all nine isolates by an average of 2.0 log10 more than moxifloxacin did at 4× MIC. Of particular note, JNJ-Q2 reduced the bacterial titers below the limit of detection (100 CFU) in studies with three non-moxifloxacin-susceptible isolates (isolates OC 8530, OC 11696, and OC 12501) at both 2× and 4× MICs, effectively sterilizing the cultures.
To ascertain the relative potential of JNJ-Q2 and moxifloxacin to elicit spontaneous resistance in S. pneumoniae, four strains were used to determine the frequencies at which resistant mutants were selected by either JNJ-Q2 or moxifloxacin in agar medium. The S. pneumoniae strains included wild-type strain R6, two derived mutants bearing a single mutation (gyrAS81F or parCS79Y), and a clinical isolate (isolate OC 6755) bearing both the gyrAS81F and the parCS79Y mutations (36, 37). The resistance frequencies for wild-type strain R6 and mutant OC 6755 with double mutations were comparable for JNJ-Q2 and moxifloxacin, ranging from 1.8 × 10−10 to ≤3.1 × 10−11 (Table 5). JNJ-Q2 and moxifloxacin each selected resistant mutants of parCS79Y mutant strain OC 7452 at a frequency of approximately 1 × 10−9 at 2× MIC, but at 4× MIC, JNJ-Q2 selected resistant mutants at a frequency nearly 50 times lower than that of moxifloxacin (Table 5). At 2× MIC, moxifloxacin selected first-step mutants of strain OC 7453 (gyrAS81F) at a frequency 14 times higher than that for JNJ-Q2 (Table 5). At 4× MIC, mutants of OC 7453 were selected by moxifloxacin, while no mutants were selected with JNJ-Q2 at that concentration.
Finally, to ascertain the relative potential of JNJ-Q2 and ciprofloxacin to elicit spontaneous resistance in S. aureus, a modified Luria-Delbruck fluctuation assay (12) was employed to determine the relative rates of spontaneous mutation. These studies encompassed two ciprofloxacin-susceptible and two ciprofloxacin-resistant MRSA isolates. As shown in Table 6, the mutation rates achieved with ciprofloxacin and JNJ-Q2 were comparable for the ciprofloxacin-susceptible MRSA isolates, at less than 10−11 mutations per cell per division. Against the ciprofloxacin-resistant MRSA isolates, the rate at which mutants were selected with JNJ-Q2 was 100- to 560-fold lower than the rate observed for ciprofloxacin.
DISCUSSION
Despite the generally low prevalence of resistance to the currently approved fluoroquinolone agents in contemporary surveillance isolates of S. pneumoniae (15, 18, 19, 32), incidences of resistance as high as 11.6% have been observed among isolates from certain patient populations in Canada (1), and the judicious use of fluoroquinolones is recommended to preserve their clinical utility (22). The long-term potential for the emergence, clonal expansion, and subsequent dissemination of strains resistant to current fluoroquinolones has driven the search for new compounds in the class that retain activity against strains resistant to contemporary agents. Such agents may provide a new option to clinicians for the empirical treatment of pneumococcal infections in locations and patient settings in which elevated rates of quinolone resistance are known or suspected. Similarly, new orally active compounds in the class that retain activity against staphylococci that exhibit resistance to current fluoroquinolone agents could potentially expand the range of options for the management of MRSA and MRSE infections in the community setting. To date, medicinal chemistry efforts have yielded a series of novel 4-quinolone class agents that exhibit in vitro activities against ciprofloxacin-resistant pneumococci and staphylococci, including delafloxacin (ABT-492) (2), garenoxacin (3, 25, 32), DC-159a (9, 29, 31), and WCK-771 (5, 6). However, no members of the 4-quinolone class are currently approved for use for the treatment of infections mediated by MRSA or fluoroquinolone-resistant pathogens.
The in vitro profile of antibacterial activity described herein for JNJ-Q2 includes the retention of potency and bactericidal activity against fluoroquinolone-resistant pneumococci and staphylococci. Indeed, while direct head-to-head comparative studies were not undertaken with the aforementioned experimental agents, the MIC90 value of 0.25 μg/ml determined for JNJ-Q2 against collections of ciprofloxacin-resistant MRSA, MRSE, and S. pneumoniae isolates appears to be favorable with regard to its relative potency against such pathogens. More importantly, these studies included isolates that bear multiple, prevalent DNA topoisomerase target mutations in combination with activated efflux pathways. Data from in vitro studies performed to determine the rate or frequency of spontaneous resistance to JNJ-Q2 at relatively modest concentrations (2× and 4× MICs) are indicative of a low potential for the selection of resistance in S. pneumoniae and S. aureus compared to the potentials of current agents. Notably, the resistance selection studies whose results are described in Table 6 utilized two ciprofloxacin-resistant MRSA strains with JNJ-Q2 MICs of 0.12 μg/ml. Those studies demonstrated that JNJ-Q2 is less likely than ciprofloxacin to select for resistance by at least 2 orders of magnitude, even in strains that bear two topoisomerase mutations, and suggest that JNJ-Q2 has activity against MRSA more robust than the activities exhibited by previous 4-quinolone class agents. In combination, these data support the notion that JNJ-Q2 has target-directed activity against strains expressing prevalent variants of DNA gyrase and DNA topoisomerase IV that confer ciprofloxacin resistance. In contrast, the relatively poorer retention of activity of JNJ-Q2 against ciprofloxacin-resistant isolates of E. coli (MIC90, 16 μg/ml) could reflect a lack of activity against prevalent variants of the DNA topoisomerase targets that confer ciprofloxacin resistance in this species and/or could indicate that JNJ-Q2 is a substrate for efflux mediated by the RND class of pumps that typically exhibit a broader substrate specificity than the MFS and MATE class of pumps of Gram-positive organisms. The potential impact of additional, non-target-based fluoroquinolone resistance mechanisms on susceptibility to JNJ-Q2, such as qnrA, qnrB, qnrS, and/or aac(6′)-I-cr in key Gram-negative pathogens is currently unknown and warrants investigation. Genetic and biochemical studies are ongoing to further define the antibacterial mode of action of JNJ-Q2 in key Gram-positive and Gram-negative pathogens and to determine target- and non-target-based mechanisms of resistance. Overall, the in vitro profile of JNJ-Q2 supports its consideration for development as a potential therapeutic agent for the treatment of respiratory and skin infections caused by susceptible pathogenic bacteria.
Acknowledgments
We thank Ellyn Wira and Ashok Vasant for supporting experimental contributions and Todd Davies for helpful comments on the manuscript.
This work was supported by J&JPRD.
All authors are or were (Karen Bush) employees of J&JPRD and own stock or stock equivalents in the company.
Footnotes
Published ahead of print on 22 February 2010.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Adam, H. J., D. J. Hoban, A. S. Gin, and G. G. Zhanel. 2009. Association between fluoroquinolone usage and a dramatic rise in ciprofloxacin-resistant Streptococcus pneumoniae in Canada, 1997-2006. Int. J. Antimicrob. Agents 34:82-85. [DOI] [PubMed] [Google Scholar]
- 2.Almer, L. S., J. B. Hoffrage, E. L. Keller, R. K. Flamm, and V. D. Shortridge. 2004. In vitro and bactericidal activities of ABT-492, a novel fluoroquinolone, against gram-positive and gram-negative organisms. Antimicrob. Agents Chemother. 48:2771-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderegg, T. R., and R. N. Jones. 2004. Bactericidal activity of garenoxacin tested by kill-curve methodology against wild type and QRDR mutant strains of Streptococcus pneumoniae. Diagn. Microbiol. Infect. Dis. 50:213-217. [DOI] [PubMed] [Google Scholar]
- 4.Ardanuy, C., D. Rolo, A. Fenoll, D. Tarrago, L. Calatayud, and J. Liñares. 2009. Emergence of a multidrug-resistant clone (ST320) among invasive serotype 19A pneumococci in Spain. J. Antimicrob. Chemother. 64:507-510. [DOI] [PubMed] [Google Scholar]
- 5.Bhagwat, S. S., P. McGhee, K. Kosowska-Shick, M. V. Patel, and P. C. Appelbaum. 2009. In vitro activity of the quinolone WCK 771 against recent U.S. hospital and community-acquired Staphylococcus aureus pathogens with various resistance types. Antimicrob. Agents Chemother. 53:811-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhagwat, S. S., L. A. Mundkur, S. V. Gupte, M. V. Patel, and H. F. Khorakiwala. 2006. The anti-methicillin-resistant Staphylococcus aureus quinolone WCK 771 has potent activity against sequentially selected mutants, has a narrow mutant selection window against quinolone-resistant Staphylococcus aureus, and preferentially targets DNA gyrase. Antimicrob. Agents Chemother. 50:3568-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bisognano, C., P. Vaudaux, P. Rohner, D. P. Lew, and D. C. Hooper. 2000. Induction of fibronectin-binding proteins and increased adhesion of quinolone-resistant Staphylococcus aureus by subinhibitory levels of ciprofloxacin. Antimicrob. Agents Chemother. 44:1428-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blumberg, H. M., D. Rimland, D. J. Carroll, P. Terry, and I. K. Wachsmuth. 1991. Rapid development of ciprofloxacin resistance in methicillin-susceptible and -resistant Staphylococcus aureus. J. Infect. Dis. 163:1279-1285. [DOI] [PubMed] [Google Scholar]
- 9.Clark, C., K. Smith, L. Ednie, T. Bogdanovich, B. Dewasse, P. McGhee, and P. C. Appelbaum. 2008. In vitro activity of DC-159a, a new broad-spectrum fluoroquinolone, compared with that of other agents against drug-susceptible and -resistant pneumococci. Antimicrob. Agents Chemother. 52:77-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.CLSI. 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 8th ed. CLSI document M07-A8, vol. 29, no. 2. CLSI, Wayne, PA.
- 11.CLSI. 2009. Performance standards for antimicrobial susceptibility testing; 19th informational supplement. CLSI document M100-S19, vol. 29, no. 3. CLSI, Wayne, PA.
- 12.Crane, G. J., S. M. Thomas, and M. E. Jones. 1996. A modified Luria-Delbruck fluctuation assay for estimating and comparing mutation rates. Mutat. Res. 354:171-182. [DOI] [PubMed] [Google Scholar]
- 13.Dagan, R., N. Givon-Lavi, E. Leibovitz, D. Greenberg, and N. Porat. 2009. Introduction and proliferation of multidrug-resistant Streptococcus pneumoniae serotype 19A clones that cause acute otitis media in an unvaccinated population. J. Infect. Dis. 199:776-785. [DOI] [PubMed] [Google Scholar]
- 14.Davies, T. A., R. Goldschmidt, S. Pfleger, M. Loeloff, K. Bush, D. F. Sahm, and A. Evangelista. 2003. Cross-resistance, relatedness and allele analysis of fluoroquinolone-resistant US clinical isolates of Streptococcus pneumoniae (1998-2000). J. Antimicrob. Chemother. 52:168-175. [DOI] [PubMed] [Google Scholar]
- 15.Davies, T. A., Y. C. Yee, K. Amsler, R. Goldschmidt, D. F. Sahm, and A. T. Evangelista. 2008. Sporadic occurrences of fluoroquinolone-resistant Streptococcus pneumoniae in the United States: longitudinal analysis of institutions from the New England and West South Central regions during the TRUST 4-9 (2000-2005) surveillance studies. Postgrad. Med. 120:25-31. [DOI] [PubMed] [Google Scholar]
- 16.Davies, T. A., Y. C. Yee, K. Bush, D. Sahm, A. Evangelista, and R. Goldschmidt. 2008. Effects of the 7-valent pneumococcal conjugate vaccine on U.S. levofloxacin-resistant Streptococcus pneumoniae. Microb. Drug Resist. 14:187-196. [DOI] [PubMed] [Google Scholar]
- 17.Davies, T. A., Y. C. Yee, R. Goldschmidt, D. F. Sahm, and A. T. Evangelista. 2008. Decline in the prevalence of pandemic clones Spain23F-1 and Spain9V-3 among US fluoroquinolone-resistant Streptococcus pneumoniae TRUST surveillance isolates since 2001. Postgrad. Med. 120:39-45. [DOI] [PubMed] [Google Scholar]
- 18.de la Campa, A. G., C. Ardanuy, L. Balsalobre, E. Perez-Trallero, J. M. Marimon, A. Fenoll, and J. Linares. 2009. Changes in fluoroquinolone-resistant Streptococcus pneumoniae after 7-valent conjugate vaccination, Spain. Emerg. Infect. Dis. 15:905-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Draghi, D. C., M. E. Jones, D. F. Sahm, and G. S. Tillotson. 2006. Geographically-based evaluation of multidrug resistance trends among Streptococcus pneumoniae in the USA: findings of the FAST surveillance initiative (2003-2004). Int. J. Antimicrob. Agents 28:525-531. [DOI] [PubMed] [Google Scholar]
- 20.Drlica, K., and M. Malik. 2003. Fluoroquinolones: action and resistance. Curr. Top. Med. Chem. 3:249-282. [DOI] [PubMed] [Google Scholar]
- 21.Drlica, K., and X. Zhao. 2007. Mutant selection window hypothesis updated. Clin. Infect. Dis. 44:681-688. [DOI] [PubMed] [Google Scholar]
- 22.File, T. 2009. The science of selecting antimicrobials for community-acquired pneumonia (CAP). J. Manag. Care Pharm. 2(Suppl.):S5-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Firsov, A. A., S. N. Vostrov, I. Y. Lubenko, K. Drlica, Y. A. Portnoy, and S. H. Zinner. 2003. In vitro pharmacodynamic evaluation of the mutant selection window hypothesis using four fluoroquinolones against Staphylococcus aureus. Antimicrob. Agents Chemother. 47:1604-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foleno, B., B. J. Morrow, E. Wira, M. Macielag, and K. Bush. 2008. Broad spectrum in vitro activity of JNJ-Q2, a new fluoroquinolone, abstr. F1-2033. Abstr. 48th Annu. Intersci. Conf. Antimicrob. Agents Chemother. (ICAAC)-Infect. Dis. Soc. Am. (IDSA) 46th Annu. Meet. American Society for Microbiology and Infectious Diseases Society of America, Washington, DC.
- 25.Fritsche, T. R., H. S. Sader, and R. N. Jones. 2007. Potency and spectrum of garenoxacin tested against an international collection of skin and soft tissue infection pathogens: report from the SENTRY antimicrobial surveillance program (1999-2004). Diagn. Microbiol. Infect. Dis. 58:19-26. [DOI] [PubMed] [Google Scholar]
- 26.Fuller, J. D., and D. E. Low. 2005. A review of Streptococcus pneumoniae infection treatment failures associated with fluoroquinolone resistance. Clin. Infect. Dis. 41:118-121. [DOI] [PubMed] [Google Scholar]
- 27.He, W., K. Amsler, K. Bush, and B. J. Morrow. 2008. In vitro Staphylococcus aureus activities and resistance selection in MRSA with the new fluoroquinolone JNJ-Q2, p. F1-2035. Abstr. 48th Annu. Intersci. Conf. Antimicrob. Agents Chemother. (ICAAC)-Infect. Dis. Soc. Am. (IDSA) 46th Annu. Meet. American Society for Microbiology and Infectious Diseases Society of America, Washington, DC.
- 28.Hookey, J. V., V. Edwards, B. D. Cookson, and J. F. Richardson. 1999. PCR-RFLP analysis of the coagulase gene of Staphylococcus aureus: application to the differentiation of epidemic and sporadic methicillin-resistant strains. J. Hosp. Infect. 42:205-212. [DOI] [PubMed] [Google Scholar]
- 29.Hoshino, K., K. Inoue, Y. Murakami, Y. Kurosaka, K. Namba, Y. Kashimoto, S. Uoyama, R. Okumura, S. Higuchi, and T. Otani. 2008. In vitro and in vivo antibacterial activities of DC-159a, a new fluoroquinolone. Antimicrob. Agents Chemother. 52:65-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones, M. E., S. M. Thomas, and A. Rogers. 1994. Luria-Delbruck fluctuation experiments: design and analysis. Genetics 136:1209-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones, R. N., T. R. Fritsche, and H. S. Sader. 2008. Antimicrobial activity of DC-159a, a new fluoroquinolone, against 1,149 recently collected clinical isolates. Antimicrob. Agents Chemother. 52:3763-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones, R. N., T. R. Fritsche, H. S. Sader, and M. G. Stilwell. 2007. Activity of garenoxacin, an investigational des-F(6)-quinolone, tested against pathogens from community-acquired respiratory tract infections, including those with elevated or resistant-level fluoroquinolone MIC values. Diagn. Microbiol. Infect. Dis. 58:9-17. [DOI] [PubMed] [Google Scholar]
- 33.Jones, R. N., H. S. Sader, M. G. Stilwell, and T. R. Fritsche. 2007. Garenoxacin activity against isolates form patients hospitalized with community-acquired pneumonia and multidrug-resistant Streptococcus pneumoniae. Diagn. Microbiol. Infect. Dis. 58:1-7. [DOI] [PubMed] [Google Scholar]
- 34.Kaatz, G. W., V. V. Moudgal, and S. M. Seo. 2002. Identification and characterization of a novel efflux-related multidrug resistance phenotype in Staphylococcus aureus. J. Antimicrob. Chemother. 50:833-838. [DOI] [PubMed] [Google Scholar]
- 35.Kays, M. B., G. G. Zhanel, M. A. Reimann, J. Jacobi, G. A. Denys, D. W. Smith, and M. F. Wack. 2007. Selection of a gyrA mutation and treatment failure with gatifloxacin in a patient with Streptococcus pneumoniae with a preexisting parC mutation. Pharmacotherapy 27:221-226. [DOI] [PubMed] [Google Scholar]
- 36.Korzheva, N., T. A. Davies, and R. Goldschmidt. 2005. Novel Ser79Leu and Ser81Ile substitutions in the quinolone resistance-determining regions of ParC topoisomerase IV and GyrA DNA gyrase subunits from recent fluoroquinolone-resistant Streptococcus pneumoniae clinical isolates. Antimicrob. Agents Chemother. 49:2479-2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korzheva, N., T. A. Davies, and R. Goldschmidt. 2005. Susceptibilities to fluoroquinolones and incidences of various Ser79 ParC and Ser81 GyrA amino acid substitutions among Streptococcus pneumoniae, abstr. C1-1041. Abstr. 45th Annu. Intersci. Conf. Antimicrob. Agents Chemother. (ICAAC), Washington, DC.
- 38.Morrow, B. J., B. Foleno, R. Goldschmidt, W. He, K. Amsler, M. Macielag, and K. Bush. 2008. In vitro activity of JNJ-Q2, a new fluoroquinolone, against susceptible and multi-drug resistant Streptococcus pneumoniae, F1-2052. Abstr. 48th Anni. Intersci. Conf. Antimicrob. Agents Chemother. (ICAAC)-Infect. Dis. Soc. Am. (IDSA) 46th Annu. Meet. American Society for Microbiology and Infectious Diseases Society of America, Washington, DC.
- 39.Nakanishi, N., S. Yoshida, H. Wakebe, M. Inoue, T. Yamaguchi, and S. Mitsuhashi. 1991. Mechanisms of clinical resistance to fluoroquinolones in Staphylococcus aureus. Antimicrob. Agents Chemother. 35:2562-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nikaido, H. 2009. Multidrug resistance in bacteria. Annu. Rev. Biochem. 78:119-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmitz, F. J., A. C. Fluit, M. Luckefahr, B. Engler, B. Hofmann, J. Verhoef, H. P. Heinz, U. Hadding, and M. E. Jones. 1998. The effect of reserpine, an inhibitor of multidrug efflux pumps, on the in-vitro activities of ciprofloxacin, sparfloxacin and moxifloxacin against clinical isolates of Staphylococcus aureus. J. Antimicrob. Chemother. 42:807-810. [DOI] [PubMed] [Google Scholar]
- 42.Schmitz, F. J., M. E. Jones, B. Hofmann, B. Hansen, S. Scheuring, M. Luckefahr, A. Fluit, J. Verhoef, U. Hadding, H. P. Heinz, and K. Kohrer. 1998. Characterization of grlA, grlB, gyrA, and gyrB mutations in 116 unrelated isolates of Staphylococcus aureus and effects of mutations on ciprofloxacin MIC. Antimicrob. Agents Chemother. 42:1249-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song, J.-H., S.-I. Jung, K. S. Ko, N. Y. Kim, J. S. Son, H.-H. Chang, H. K. Ki, W. S. Oh, J. Y. Suh, K. R. Peck, N. Y. Lee, Y. Yang, Q. Lu, A. Chongthaleong, C.-H. Chiu, M. K. Lalitha, J. Perera, T. T. Yee, G. Kumarasinghe, F. Jamal, A. Kamarulzaman, N. Parasakthi, P. H. Van, C. Carlos, T. So, T. K. Ng, and A. Shibl. 2004. High prevalence of antimicrobial resistance among clinical Streptococcus pneumoniae isolates in Asia (an ANSORP study). Antimicrob. Agents Chemother. 48:2101-2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takenouchi, T., F. Tabata, Y. Iwata, H. Hanzawa, M. Sugawara, and S. Ohya. 1996. Hydrophilicity of quinolones is not an exclusive factor for decreased activity in efflux-mediated resistant mutants of Staphylococcus aureus. Antimicrob. Agents Chemother. 40:1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watanabe, S., T. Ito, and K. Hiramatsu. 2007. Susceptibilities of healthcare- and community-associated methicillin-resistant staphylococci to the novel des-F(6)-quinolone DX-619. J. Antimicrob. Chemother. 60:1384-1387. [DOI] [PubMed] [Google Scholar]