Abstract
Antibiotic kinases, which include aminoglycoside and macrolide phosphotransferases (APHs and MPHs), pose a serious threat to currently used antimicrobial therapies. These enzymes show structural and functional homology with Ser/Thr/Tyr kinases, which is suggestive of a common ancestor. Surprisingly, recent in vitro studies using purified antibiotic kinase enzymes have revealed that a number are able to utilize GTP as the antibiotic phospho donor, either preferentially or exclusively compared to ATP, the canonical phosphate donor in most biochemical reactions. To further explore this phenomenon, we examined three enzymes, APH(3′)-IIIa, APH(2″)-Ib, and MPH(2′)-I, using a competitive assay that mimics in vivo nucleotide triphosphate (NTP) concentrations and usage by each enzyme. Downstream analysis of reaction products by high-performance liquid chromatography enabled the determination of partitioning of phosphate flux from NTP donors to antibiotics. Using this ratio along with support from kinetic analysis and inhibitor studies, we find that under physiologic concentrations of NTPs, APH(3′)-IIIa exclusively uses ATP, MPH(2′)-I exclusively uses GTP, and APH(2″)-Ib is able to use both species with a preference for GTP. These differences reveal likely different pathways in antibiotic resistance enzyme evolution and can be exploited in selective inhibitor design to counteract resistance.
Antibiotic modification is a major mechanism of resistance that impacts the efficacy of numerous antimicrobial drug classes. The enzymes that catalyze these group transfer mechanisms have evolved from precursor genes that encode proteins used to accomplish numerous metabolic tasks no doubt unrelated to drug resistance. Borrowing nomenclature from the oncogene field, we term such elements protoresistance genes. Biochemical and structural evidence has shown that protoresistance genes can be found in a number of key metabolic pathways, including cell wall biosynthesis and signal transduction (23). With respect to group transfer antibiotic resistance enzymes, these protoresistance genes encode protein and other small-molecule kinases, acetyltransferases, adenylyltransferases, ADP-ribosyltransferases, and glycosyltransferases. The addition of phosphoryl, acyl, adenyl, ribosyl, and glycosyl groups to the antibiotic scaffold alters the interaction with the cellular target to such a degree that the resistance phenotype results (6).
The unifying biochemical logic of group transfer in antibiotic resistance is the co-opting of the normal cellular function of the protoresistance element by natural selection to include the modification of antibiotic molecules. Because group transfer enzymes require a second substrate (e.g., ATP, acetyl coenzyme A, or thymidine diphosphate glucose), these should retain the original specificity of the protoresistance enzyme since natural selection would not normally be expected to act on this site during resistance gene evolution.
Antibiotic kinases represent a large superfamily of enzymes that covalently modify antibiotics by phosphorylation, resulting in the addition of both steric bulk and anionic character to the target molecule. Kinases that modify aminoglycoside, macrolide, and fenicol antibiotics have been well characterized (6). The latter is a member of the shikimate group of kinases (10), whereas the former (aminoglycoside and macrolide phosphotransferases [APHs and MPHs, respectively]) share primary sequence, three-dimensional (3D) structure, and enzyme mechanism properties with the Ser/Thr/Tyr/lipid family of kinases that are chief components of eukaryotic signal transduction (24). These antibiotic kinases have largely been seen to use the phosphoryl donor ATP as the second substrate for antibiotic modification, consistent with the primacy of this nucleotide triphosphate (NTP) in primary metabolism and the intracellular concentration of ∼3 mM in bacterial cells (1).
During our efforts to characterize the structure and mechanism of macrolide antibiotic kinases, we were struck by the poor activity with ATP compared to GTP as a phosphoryl donor substrate in in vitro assays, exhibiting a selectivity that had been noted previously but was unexplored (13). This preference was surprising to us, given the reported intracellular concentrations of ATP versus GTP, which in bacterial and mammalian cells are reported to be on the order of 3- to 4-fold in excess of ATP (3; reviewed in reference 20). In order to explore this further, we have established a competitive high-performance liquid chromatography (HPLC) method to establish the NTP selectivity of antibiotic kinases under conditions that mimic intracellular concentrations of NTPs. We report a new partition coefficient (PC) parameter that functionally discriminates antibiotic kinases based on their NTP preference. This has significant implications for our understanding of the evolution of these kinases from protoresistance elements and practical application in the development of selective inhibitors of kinases that can reverse resistance.
MATERIALS AND METHODS
Antibiotics and reagents.
Erythromycin A, troleandomycin, clarithromycin, azithromycin, tylosin, spiramycin, NADH, phosphoenolpyruvate (PEP), pyruvate kinase/lactate dehydrogenase (PK/LDH), ATP, GTP, AMPPNP, and GMPPNP were purchased from Sigma-Aldrich (Oakville, Ontario, Canada). Neomycin and kanamycin were purchased from Bioshop Canada (Mississauga, Ontario, Canada), and telithromycin was from Sanofi Aventis.
Expression and purification of antibiotic kinases.
The mphA gene was amplified by PCR from pTZ3509 (15), a gift from N. Noguchi, and aph(2″)-Ib was amplified from pSCH075 (4), generously provided by Joe Chow, by using the oligonucleotide primers listed in Table 1. The mphA gene was ligated into pET28a using NdeI and HindIII sites, and aph(2″)-Ib was introduced into pDEST17 between the attR1 and attR2 sites using the Gateway system (Invitrogen).
TABLE 1.
Oligonucleotide primers for gene amplification
| Primer | Sequence |
|---|---|
| mphA-FWD | 5′-CGCGAATTCCATATGACCGTAGTCACGACCGCCGA-3′ |
| mphA-REV | 5′-CCCAAGCTTGGATCCTTGCCGATCCGGAAGAGAAAGGTA-3′ |
| aph(2″)-Ib-FWD | 5′-GGGGACAAGTTTGTACAAAAAAGCAGCCTTCATGGTTAACTTGGACGCTGAG-3′ |
| aph(2″)-Ib-REV | 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCCTAAAATATAAACATCTCTGCTTGTG-3′ |
APH(3′)-IIIa was purified using the protocol outlined by McKay et al. (14). Both MPH(2′)-I and APH(2″)-Ib were expressed in Escherichia coli BL21(DE3) (Novagen), which was grown overnight in the presence of the respective antibiotic [50 μg/ml kanamycin A (Bioshop) for pETMPHa and 100 μg/ml ampicillin for pDESTAPH(2″)-Ib] in Luria-Bertani (LB) broth. A 10-ml overnight culture was used to inoculate 1 liter of fresh LB broth with antibiotic selection and was grown to an optical density at 600 nm of about 0.6 at 37°C. It was then induced for 16 h at 16°C with 1 mM isopropyl-β-d-thiogalactopyranoside (Bioshop). Cells were harvested by centrifugation at 6,000 × g for 10 min. The cell pellet was resuspended in 15 ml of 50 mM HEPES (pH 7.5)-300 mM NaCl-10 mM imidazole-1 mM phenylmethylsulfonyl fluoride-1 μg/ml pancreatic bovine DNase (Sigma). The resuspended pellet was passed through a 20,000-lb/in2 French press cell (Thermo Fisher) four times to create a lysate. The insoluble and soluble fractions were separated by centrifugation at 48,000 × g for 30 min. The supernatant was then applied to a 5-ml Ni-nitrilotriacetic acid column (Qiagen) equilibrated with 50 mM HEPES (pH 7.5)-300 mM NaCl-10 mM imidazole (buffer A). A stepwise gradient over 20 column volumes was used to elute each enzyme with 50 mM HEPES (pH 7.5)-300 mM NaCl-250 mM imidazole (buffer B). The kinase-containing fractions were identified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, subsequently pooled, dialyzed against 50 mM HEPES (pH 7.5), concentrated to approximately 2 mg/ml, and stored at −20°C.
PK/LDH-coupled assay.
The phosphorylation of antibiotics was monitored by coupling the release of ADP/GDP with PK/LDH (19). The oxidization of NADH (ɛ = 6,220 M−1 cm−1) was monitored at 340 nm using a SpectraMax plate reader in a 96-well format. A typical reaction mixture contained 230 μl of reaction buffer (50 mM HEPES [pH 7.5], 40 mM KCl, 10 mM MgCl2, 0.3 mM NADH, 3.5 mM PEP, 0.00125 U PK/LDH, antibiotic kinase). A 10-μl solution of the appropriate antibiotic was added to the reaction mixture, which was allowed to incubate for 5 min at 37°C. The reaction was initiated with a 10-μl solution of nucleotide (ATP or GTP) and monitored for 5 min at 340 nm.
Initial rates were determined by utilizing the linear portion of the progress curve and analyzed by nonlinear least-squares fitting of equation 1:
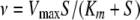 |
(1) |
or equation 2 for substrate-inhibited reactions:
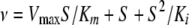 |
(2) |
To further validate interaction of the enzymes with ATP and GTP, their nonhydrolyzable analogues AMPPNP and GMPPNP were tested against the panel of enzymes using a 50% inhibitory concentration (IC50) analysis. Each analogue was diluted 2-fold and assayed against each enzyme using the PK/LDH-coupled assay. Antibiotic concentrations were kept at saturating levels, and nucleotide concentrations were held at Km concentrations. IC50s were determined using Grafit 4.0 and equation 3:
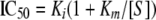 |
(3) |
Paired-ion chromatography to determine nucleotide selectivity.
To further explore the nucleotide preference of antibiotic kinases using physiological concentrations of ATP and GTP, we established an NTP competition assay with HPLC resolution of products. The concentrations of ATP and GTP reported for Salmonella enterica were used to set up the in vitro competition assay, and they were 3 and 0.9 mM, respectively (1). Enzymes were then incubated at 25°C in the presence of both nucleotides with 50 mM HEPES (pH 7.5)-40 mM KCl-10 mM MgCl2-1 mM appropriate antibiotic in a 50-μl volume. The reaction was stopped by the addition of 150 μl of 8 M urea. The reaction mixtures were then filtered through a 10-kDa Pall microcentrifugation filter at 12,100 × g for 20 min to remove the protein. The filtrate was collected and diluted two times, and 25 μl was injected onto a Waters Novapak C18 reverse-phase column (3.9 by 150 mm) that had been treated with tetrabutylammonium hydrogen sulfate (TBAHS). A linear gradient was used to elute the various forms of the purine nucleotides using a two-solvent system. Solvent A consisted of 15 mM H2KPO4/HK2PO4 and 10 mM TBAHS, and solvent B consisted of 35 mM H2KPO4/HK2PO4, 10 mM TBAHS, and 30% (vol/vol) acetonitrile. The elution was monitored using A259. Once peaks had been identified, they were integrated using the Chromeleon 6.8 HPLC software package to determine the quantity of each of the nucleotide species. The product-to-substrate ratios were determined for ADP:ATP and GDP:GTP for each time point and plotted on an x-y scattergraph. A line of best fit was applied to each set of data, and a PC was determined for each enzyme using equation 4:
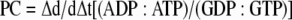 |
(4) |
RESULTS
Steady-state kinetic analysis of antibiotic kinases.
Steady-state kinetic constants for APH(3′)-IIIa (14) and APH(2″)-Ib (22), as well as antibiotic substrate specificity, have been previously reported. Steady-state constants for macrolide substrates of MPH(2′)-I were determined in this work and are reported in Table 2. MPH(2″)-I showed a higher selectivity for 14- and 15-membered macrolide antibiotics (e.g., erythromycin, clarithromycin, and azithromycin) over the 16-membered ring macrolides spiramycin and tylosin. Of the seven macrolides tested, the ketolide telithromycin was the only one to show substrate inhibition.
TABLE 2.
Kinetic constants for macrolide substrates of MPH(2′)-Ia
| Substrate | Macrocycle | Km (μM) | kcat (s−1) | Ki (mM) | kcat/Km (s−1 M−1) |
|---|---|---|---|---|---|
| Erythromycin | 14 | 37.2 ± 5.6 | 0.279 | 7.52 × 103 | |
| Clarithromycin | 14 | 45.9 ± 7.4 | 0.311 | 6.77 × 103 | |
| Telithromycinb | 14 | 7.4 ± 1.5 | 0.152 | 0.622 ± 0.196 | 2.07 × 104 |
| Azithromycin | 15 | 27.7 ± 3.4 | 0.292 | 1.05 × 104 | |
| Tylosin | 16 | 131 ± 45 | 0.174 | N/A | 1.33 × 103 |
| Spiramycin | 16 | 478 ± 55 | 0.200 | N/A | 4.19 × 102 |
All data were determined in the presence of 0.2 mM GTP.
Telithromycin was the only macrolide to show substrate inhibition.
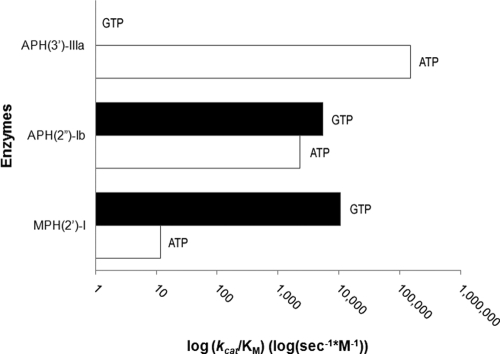
Kinetic constants for the nucleotides were determined for all antibiotic kinases (Table 3; Fig. 1). These data demonstrate that APH(3′)-IIIa is strictly ATP dependent and MPH(2′)-I has a very high selectivity for GTP. In our experiments, the N-terminally His6-tagged variant of APH(2″)-Ib shows a slight selectivity for GTP over ATP but is still able to utilize both NTPs. This contrasts with results reported for untagged APH(2″)-Ib that showed a preference for ATP but that GTP was nevertheless a good substrate (4-fold difference in Km and 4.5-fold difference in kcat) (21), and these results could be the result of minor differences in enzyme and assay conditions.
TABLE 3.
Kinetic constants of nucleotide substrates
| Substrate | Saturating antibiotica | Km (μM) | kcat (s−1) | kcat/Km (−1 M−1) |
|---|---|---|---|---|
| APH(3′)-IIIa | ||||
| ATP | Neomycin | 10.6 ± 1.4 | 1.59 | 1.50 × 105 |
| GTP | Neomycin | NCb | NC | NC |
| APH(2″)-Ib | ||||
| ATP | Kanamycin | 151 ± 12 | 0.32 | 2.30 × 103 |
| GTP | Kanamycin | 18.0 ± 1.9 | 0.10 | 5.40 × 103 |
| MPH(2′)-I | ||||
| ATP | Erythromycin | 1,890 ± 510 | 0.022 | 1.16 × 101 |
| GTP | Erythromycin | 59.2 ± 4.7 | 0.634 | 1.07 × 104 |
The concentrations of the antibiotics were held as follows: neomycin, 200 μM; kanamycin, 200 μM; erythromycin, 400 μM.
NC, no curve.
FIG. 1.
Nucleotide specificity of antibiotic kinases. The semilog bar graph shown demonstrates the nucleotide specificity of the three antibiotic kinases studied for ATP (white) versus GTP (black) based upon the Michaelis-Menten constant kcat/Km (s−1 M−1).
Inhibition profiles of antibiotic kinases as determinants of nucleotide preference.
Inhibitory profiles were determined for each enzyme by observing their dose response against the two nonhydrolyzable nucleotide analogs AMPPNP and GMPPNP. IC50s were determined for each of the enzymes and converted to Ki values using equation 3 (Table 4). Consistent with our steady-state kinetic data, APH(3′)-IIIa is only inhibited by AMPPNP, APH(2″)-Ib is inhibited by both AMPPNP and GMPPNP, and MPH(2′)-I is only inhibited by GMPPNP.
TABLE 4.
Ki values of inhibitors as probes of nucleotide selectivity
| Enzyme |
Ki value (μM) for: |
|
|---|---|---|
| AMPPNP | GMPPNP | |
| APH(3′)-IIIa | 14.2 ± 1.0 | No inhibition |
| APH(2″)-Ib | 263.0 ± 55.0 | 19.3 ± 4.0 |
| MPHa | No inhibition | 19.4 ± 1.4 |
Antibiotic kinases discriminate nucleotide usage in competition experiments.
Although in vitro kinetic data demonstrate the selectivity of these enzymes, the conditions within the bacterial cell are very different. Typically, ATP and GTP exist in a ratio of about 3:1 in most gram-negative organisms (1, 3). Therefore, the ability to use GTP as a phospho donor in steady-state kinetic experiments may not translate to a preference in the cell, where ATP dominates. In order to observe how these enzymes would behave in vivo, we established an in vitro NTP competition assay system with ATP and GTP present under physiological concentrations. NTP utilization was analyzed by paired ion chromatography (17) (Fig. 2), which uses TBAHS to interact with the phosphates, masking the hydrophilic negatively charged groups and improving HPLC resolution.
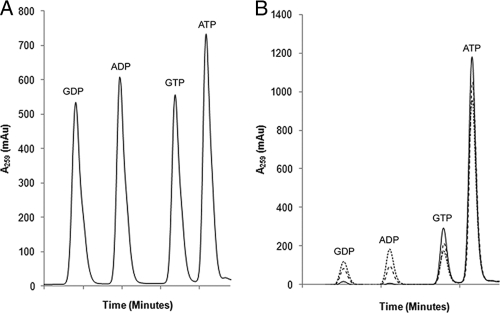
FIG. 2.
Separation of nucleotides by HPLC. (A) Standard curve demonstrating the separation of a mixture of 1 mM GDP, ADP, GTP, and ATP. (B) Time course comparison of the elution of the various nucleotide species for APH(2″)-Ib at 0 min (solid line), 60 min (coarse dashed line), and 120 min (fine dashed line).
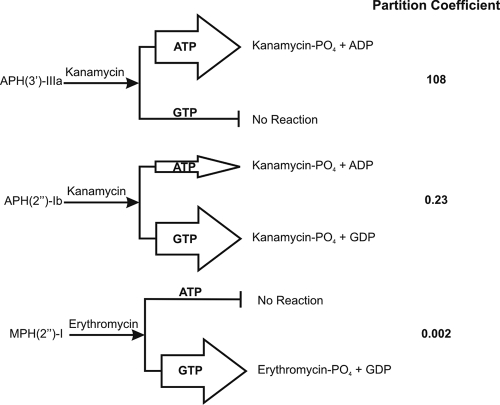
The conversion of NTPs to nucleotide diphosphates (NDPs) over time under saturating antibiotic conditions was quantitatively assessed by this method. NTP turnover was calculated from the slopes of integrated HPLC data. The slopes for ATP and GTP turnover were used to calculate the PC for each enzyme (Fig. 3). The PC is a ratio that represents the nucleotide selectivity of a given enzyme. If the PC is equal to 1, there is no bias for either ATP or GTP; if it is greater than 1, there is a preference for ATP; and if it is less than 1, the enzyme prefers GTP. MPH(2″)-I, although able to utilize ATP in steady-state kinetic experiments, did not use ATP while there was GTP present under physiological conditions and consequently shows a very low PC. On the other hand, APH(2″)-Ib utilized both nucleotides and APH(3′)-IIIa only used ATP. This experiment demonstrates that, under physiological NTP conditions, antibiotic kinases demonstrate distinct selectivity for nucleotide substrates.
FIG. 3.
NTP selectivity of antibiotic kinases. At physiological concentrations of NTP, antibiotic kinases show either exclusive or preferential usage of ATP or GTP. The width of the arrows approximates the ratio of this selectivity. The PCs listed at the right were calculated with equation 4.
DISCUSSION
Antibiotic resistance enzymes have evolved from precursor proteins, protoresistance elements, of different origins (23). Elucidation of the structure and function of resistance enzymes has been essential in linking current proteins with ancestral protoresistance enzymes. For example, the 3D structures and biochemical function analysis of aminoglycoside kinases (APHs) revealed an otherwise unexpected link with Ser/Thr/Tyr protein kinases (24). Furthermore, in silico analysis of macrolide antibiotic kinase (MPH) amino acid sequences places them in the same family as aminoglycoside and protein kinases. It is therefore highly likely that all of these proteins share an ancestor and that the protoresistance elements that gave rise to MPHs and APHs fall within their evolutionary tree.
The nucleotide selectivity of protein kinases is almost exclusively in favor of ATP. There are a few exceptions where GTP is either a preferred or an equivalent substrate (20). The best-studied example is Ser/Thr casein kinase II, CK-2, which shows a 3- to 5-fold preference for GTP over ATP (11). Another example is protein kinase C delta, which undergoes 6-fold-improved autophosphorylation with GTP (8). The human variant of the structurally unrelated small-molecule kinase PEP carboxykinase, which catalyzes the formation of PEP from oxaloacetate, will catalyze this reaction with GTP, while the E. coli homolog utilizes ATP (7). Nevertheless, GTP utilization among kinases remains rare.
There is no thermodynamic imperative for the preference of kinases for ATP over other NTPs such as GTP as a phosphodonor. The free energy associated with gamma-phosphate transfer is equivalent. Nature has nevertheless restricted most kinases to ATP utilization, and the number of such reactions is reflected in the 3- to 4-fold stoichiometric excess of cellular ATP versus GTP. GTP is utilized in cell signaling processes, e.g., tightly bound in GTP-protein-coupled receptors, and is a major source of energy during translation where each amino acid elongation requires hydrolysis of two GTP molecules. Therefore, even though they are thermodynamically equivalent, ATP versus GTP utilization is biochemically segregated. It was therefore surprising that our research, as well as several literature reports using purified APH and MPH enzymes, revealed that many of these enzymes can use both ATP and GTP (2, 13, 16). Indeed, this preference was recently used as the basis for a proposed new nomenclature for APH(2″) enzymes (21).
However, even though these in vitro studies showed that GTP was a substrate for some antibiotic kinases, this did not mean that these observations were biologically relevant where the presence of multiple NTPs at various concentrations presents a different environment than saturating concentrations of NTP in a typical enzyme assay. We therefore established a competition assay using NTPs at physiological concentrations to directly assess this phenomenon. Using this assay, we were able to demonstrate a range of GTP versus ATP utilization by aminoglycoside kinases. APH(3′)-IIIa is a “typical” APH(3′), a class that includes the popular Kanr (also known as NEO) cassette used in molecular and cellular biology, and only uses ATP. APH(2″)-Ib, which is important for clinical gentamicin resistance in gram-positive cocci and E. coli (4, 12), shows a preference, but not exclusive dependence, on GTP in our experiments. On the other hand, the partition experiment shows that the macrolide kinase MPH(2′)-I will exclusively use GTP in the cell.
The NTP preference of antibiotic kinases is fascinating and (i) could reflect the preference of the protoresistance enzyme ancestor or (ii) a structural requirement based on the nature of the second substrate, e.g., a macrolide or aminoglycoside, or (iii) it may offer an evolutionary advantage in the face of antibiotic exposure. The correlation between NTP utilization and sensitivity to NMPPNP inhibitors suggests that the second option is unlikely. Macrolides interact with the ribosome and prevent the emergence of the nascent peptide from the P site (9); furthermore, GTP is essential for protein synthesis. It could be that halting protein synthesis temporarily increases GTP stores within the cell and that this provides an advantage for the GTP preference of MPHs, consistent with the third option. On the other hand, aminoglycosides, which cause mistranslation rather than block translation, would not be expected to alter GTP levels. We explored changes in NTP and NDP levels in cells exposed to subtherapeutic antibiotics but could not detect any significant changes in NTP levels (not shown). While the biological bias of ATP or GTP selectively in antibiotic kinases therefore remains an open question, the first option, where this preference is a reflection of the electivity of the protoresistance element, appears to be the most parsimonious explanation at present.
Whatever the origin, the preference of NTP does provide a useful handle for the development of agents that selectively block antibiotic resistance, as we have demonstrated with NMPPNP inhibitors. Targeting the ATP binding site of antibiotic kinases with small-molecule inhibitors is possible with known protein kinase inhibitors (5). However, given the nonspecific nature of many of these compounds, therapeutic use to circumvent resistance has the potential to also block other, off-target, cellular kinases, with possible deleterious effects. The GTP utilization of APH(2″), an important gentamicin resistance element, and MPH, a mechanism of growing importance (18), reveals an important differentiating property that could be exploited for downstream drug discovery research into agents that block resistance and restore antibiotic function. Obtaining the 3D structures of GTP-utilizing enzymes will greatly enhance this approach.
Acknowledgments
We thank M. Morar for helpful discussions.
This research was supported by the Canadian Institutes of Health Research (MT-13536) and the Canada Research Chairs program.
Footnotes
Published ahead of print on 15 March 2010.
REFERENCES
- 1.Bochner, B. R., and B. N. Ames. 1982. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J. Biol. Chem. 257:9759-9769. [PubMed] [Google Scholar]
- 2.Boehr, D. D., D. M. Daigle, and G. D. Wright. 2004. Domain-domain interactions in the aminoglycoside antibiotic resistance enzyme AAC(6′)-APH(2″). Biochemistry 43:9846-9855. [DOI] [PubMed] [Google Scholar]
- 3.Buckstein, M. H., J. He, and H. Rubin. 2008. Characterization of nucleotide pools as a function of physiological state in Escherichia coli. J. Bacteriol. 190:718-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chow, J. W., V. Kak, I. You, S. J. Kao, J. Petrin, D. B. Clewell, S. A. Lerner, G. H. Miller, and K. J. Shaw. 2001. Aminoglycoside resistance genes aph(2″)-Ib and aac(6′)-Im detected together in strains of both Escherichia coli and Enterococcus faecium. Antimicrob. Agents Chemother. 45:2691-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daigle, D. M., G. A. McKay, and G. D. Wright. 1997. Inhibition of aminoglycoside antibiotic resistance enzymes by protein kinase inhibitors. J. Biol. Chem. 272:24755-24758. [DOI] [PubMed] [Google Scholar]
- 6.D'Costa, V., and G. D. Wright. 2009. Biochemical logic of antibiotic inactivation and modification, p. 81-96. In D. Mayers, S. Lerner, J. Sobel, and M. Ouellette (ed.), Antimicrobial drug resistance: mechanisms of drug resistance, vol. 1. Humana Press, New York, NY. [Google Scholar]
- 7.Dunten, P., C. Belunis, R. Crowther, K. Hollfelder, U. Kammlott, W. Levin, H. Michel, G. B. Ramsey, A. Swain, D. Weber, and S. J. Wertheimer. 2002. Crystal structure of human cytosolic phosphoenolpyruvate carboxykinase reveals a new GTP-binding site. J. Mol. Biol. 316:257-264. [DOI] [PubMed] [Google Scholar]
- 8.Gschwendt, M., W. Kittstein, K. Kielbassa, and F. Marks. 1995. Protein kinase C delta accepts GTP for autophosphorylation. Biochem. Biophys. Res. Commun. 206:614-620. [DOI] [PubMed] [Google Scholar]
- 9.Hermann, T. 2005. Drugs targeting the ribosome. Curr. Opin. Struct. Biol. 15:355-366. [DOI] [PubMed] [Google Scholar]
- 10.Izard, T., and J. Ellis. 2000. The crystal structures of chloramphenicol phosphotransferase reveal a novel inactivation mechanism. EMBO J. 19:2690-2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jakobi, R., and J. A. Traugh. 1992. Characterization of the phosphotransferase domain of casein kinase II by site-directed mutagenesis and expression in Escherichia coli. J. Biol. Chem. 267:23894-23902. [PubMed] [Google Scholar]
- 12.Kao, S. J., I. You, D. B. Clewell, S. M. Donabedian, M. J. Zervos, J. Petrin, K. J. Shaw, and J. W. Chow. 2000. Detection of the high-level aminoglycoside resistance gene aph(2″)-Ib in Enterococcus faecium. Antimicrob. Agents Chemother. 44:2876-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kono, M., K. O'Hara, and T. Ebisu. 1992. Purification and characterization of macrolide 2′-phosphotransferase type II from a strain of Escherichia coli highly resistant to macrolide antibiotics. FEMS Microbiol. Lett. 76:89-94. [DOI] [PubMed] [Google Scholar]
- 14.McKay, G. A., P. R. Thompson, and G. D. Wright. 1994. Broad spectrum aminoglycoside phosphotransferase type III from Enterococcus: overexpression, purification, and substrate specificity. Biochemistry 33:6936-6944. [DOI] [PubMed] [Google Scholar]
- 15.Noguchi, N., A. Emura, H. Matsuyama, K. O'Hara, M. Sasatsu, and M. Kono. 1995. Nucleotide sequence and characterization of erythromycin resistance determinant that encodes macrolide 2′-phosphotransferase I in Escherichia coli. Antimicrob. Agents Chemother. 39:2359-2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Hara, K., T. Kanda, K. Ohmiya, T. Ebisu, and M. Kono. 1989. Purification and characterization of macrolide 2′-phosphotransferase from a strain of Escherichia coli that is highly resistant to erythromycin. Antimicrob. Agents Chemother. 33:1354-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patthy, M., T. Balla, and P. Aranyi. 1990. High-performance reversed-phase ion-pair chromatographic study of myo-inositol phosphates. Separation of myo-inositol phosphates, some common nucleotides and sugar phosphates. J. Chromatogr. 523:201-216. [DOI] [PubMed] [Google Scholar]
- 18.Phuc Nguyen, M. C., P. L. Woerther, M. Bouvet, A. Andremont, R. Leclercq, and A. Canu. 2009. Escherichia coli as reservoir for macrolide resistance genes. Emerg. Infect. Dis. 15:1648-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pon, N. G., and R. J. Bondar. 1967. A direct spectrophotometric assay for pyruvate kinase. Anal. Biochem. 19:272-279. [DOI] [PubMed] [Google Scholar]
- 20.Shugar, D. 1996. The NTP phosphate donor in kinase reactions: is ATP a monopolist? Acta Biochim. Pol. 43:9-23. [PubMed] [Google Scholar]
- 21.Toth, M., J. W. Chow, S. Mobashery, and S. B. Vakulenko. 2009. Source of phosphate in the enzymic reaction as a point of distinction among aminoglycoside 2″-phosphotransferases. J. Biol. Chem. 284:6690-6696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toth, M., J. Zajicek, C. Kim, J. W. Chow, C. Smith, S. Mobashery, and S. Vakulenko. 2007. Kinetic mechanism of enterococcal aminoglycoside phosphotransferase 2″-Ib. Biochemistry 46:5570-5578. [DOI] [PubMed] [Google Scholar]
- 23.Wright, G. D. 2007. The antibiotic resistome: the nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 5:175-186. [DOI] [PubMed] [Google Scholar]
- 24.Wright, G. D., and P. R. Thompson. 1999. Aminoglycoside phosphotransferases: proteins, structure, and mechanism. Front. Biosci. 4:D9-D21. [DOI] [PubMed] [Google Scholar]