Abstract
The type III secretion system (T3SS) is a clinically important virulence mechanism in Pseudomonas aeruginosa that secretes and translocates up to four protein toxin effectors into human cells, facilitating the establishment and dissemination of infections. To discover inhibitors of this important virulence mechanism, we developed two cellular reporter assays and applied them to a library of 80,000 compounds. The primary screen was based on the dependence of the transcription of T3SS operons on the T3SS-mediated secretion of a negative regulator and consisted of a transcriptional fusion of the Photorhabdus luminescens luxCDABE operon to the P. aeruginosa exoT effector gene. Secondary assays included direct measurements of the T3SS-mediated secretion of a P. aeruginosa ExoS effector-β-lactamase fusion protein as well as the detection of the secretion of native ExoS by the sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of culture supernatants. Five inhibitors in three chemical classes were demonstrated to inhibit type III secretion selectively with minimal cytotoxicity and with no effects on bacterial growth or on the type II-mediated secretion of elastase. These inhibitors also block the T3SS-mediated secretion of a YopE effector-β-lactamase fusion protein from an attenuated Yersinia pestis strain. The most promising of the inhibitors is a phenoxyacetamide that also blocks the T3SS-mediated translocation of effectors into mammalian cells in culture. Preliminary studies of structure-activity relationships in this phenoxyacetamide series demonstrated a strict requirement for the R-enantiomer at its stereocenter and indicated tolerance for a variety of substituents on one of its two aromatic rings.
The type-three secretion system (T3SS) is a complex multiprotein apparatus that facilitates the secretion and translocation of effector proteins from the bacterial cytoplasm directly into the mammalian cytosol. This complex protein delivery device is shared by more than 15 species of gram-negative human pathogens, including Salmonella spp., Shigella flexneri, Pseudomonas aeruginosa, Yersinia spp., enteropathogenic and enteroinvasive Escherichia coli, and Chlamydia spp. (23, 25, 43). In the opportunistic pathogen P. aeruginosa, the T3SS is the major virulence factor contributing to the establishment and dissemination of acute infections (19). Four T3SS effectors have been identified in P. aeruginosa strains: ExoS, ExoT, ExoY, and ExoU. ExoS and ExoT are bifunctional proteins consisting of an N-terminal small G-protein-activating protein (GAP) domain and a C-terminal ADP ribosylation domain, ExoY is an adenylate cyclase, and ExoU is a phospholipase (reviewed in reference 11). In studies with strains producing each effector separately, ExoU and ExoS contributed significantly to persistence, dissemination, and mortality, while ExoT produced minor effects on virulence in a mouse lung infection model, and ExoY did not appear to play a major role in the pathogenesis of P. aeruginosa (51). While not a prototypical effector toxin, flagellin (FliC) also may be injected into the cytoplasm of host cells from P. aeruginosa via the T3SS machinery, where it triggers the activation of the innate immune system through the nod-like receptor NLRC4 inflammasome (13, 33).
The presence of a functional T3SS is significantly associated with poor clinical outcomes and death in patients with lower-respiratory and systemic infections caused by P. aeruginosa (48). In addition, T3SS reduces survival in P. aeruginosa animal infection models (49) and is required for the systemic dissemination of P. aeruginosa in a murine acute pneumonia infection model (56). T3SS appears to contribute to the development of severe pneumonia by inhibiting the ability of the host to contain and clear the bacterial infection of the lung. The secretion of T3SS toxins, particularly ExoU, blocks phagocyte-mediated clearance at the site of infection and facilitates the establishment of an infection (9). The result is a local disruption of an essential component of the innate immune response, which creates an environment of immunosuppression in the lung. This not only allows P. aeruginosa to persist in the lung but also facilitates superinfection with other species of bacteria.
While several antibacterial agents are effective against P. aeruginosa, the high rates of mortality and relapse associated with serious P. aeruginosa infections even in patients with hospital-acquired pneumonia (HAP) receiving antibiotics active against the causative strain reflect the increasing incidence of drug-resistant strains and highlight the need for new therapeutic agents (10, 46, 52). Conventional bacteriostatic and bactericidal antibiotics appear insufficient to adequately combat these infections, and new treatment approaches such as inhibitors of P. aeruginosa virulence determinants may prove useful as adjunctive therapies (58).
The potential for T3SS as a therapeutic target has prompted several groups to screen for inhibitors of T3SS in various bacterial species, including Salmonella enterica serovar Typhimurium, Yersinia pestis, Y. pseudotuberculosis, and E. coli (reviewed in references 5 and 25). However, only a single screen for inhibitors of P. aeruginosa T3SS inhibitors has been reported, and it yielded specific inhibitors of one of the T3SS effectors, ExoU (27), rather than inhibitors of the T3SS machinery. High levels of sequence conservation among various proteins comprising the T3SS apparatus suggest that inhibitors of T3SS in one species also are active in related species. The broad-spectrum activity of T3SS inhibitors identified in a screen against Yersinia has been demonstrated in Salmonella, Shigella, and Chlamydia (22, 57, 59). However, the need for new, potent anti-pseudomonal agents argues for additional direct screening for P. aeruginosa T3SS inhibitors. To address this unmet need, we developed and applied a cell-based bioluminescent reporter assay for the identification of inhibitors of the P. aeruginosa T3SS and qualified the hits through a series of secondary assays. In this report, we describe the features of the most potent and selective inhibitors from the screen, including a new phenoxyacetamide inhibitor that blocks T3SS-mediated secretion and the translocation of toxin effectors from P. aeruginosa and exhibits minimal cytotoxicity. This inhibitor also is active against Yersinia and Chlamydia T3SS. Preliminary structure-activity relationships (SARs) indicate that the stereocenter is crucial for activity and suggest regions of the molecule that could be altered to optimize potency.
MATERIALS AND METHODS
Strains, plasmids, and growth media.
Bacterial strains and plasmids used for assays are described in Table 1. All P. aeruginosa strains were derivatives of PAO1 (21), PAK (1), or PA14 (45). E. coli TOP10 (Invitrogen), E. coli DB3.1 (Gateway host, Invitrogen), E. coli SM10 (7), and E. coli S17-1 (ATCC 47055) were used as hosts for molecular cloning. Luria-Bertani (LB) medium (liquid and agar) was purchased from Difco. LB was supplemented with 30 μg/ml gentamicin (LBG) with or without 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and 5 mM EGTA (LBGI and LBGIE, respectively).
TABLE 1.
Strains and plasmids
| Strain | Genotype or features | Reference or source |
|---|---|---|
| P. aeruginosa | ||
| MDM852 | PA01::pGSV3-′exoT′-luxCDABE | This study |
| MDM1355 | PA01 ΔpscC::pGSV3-′exoT′-luxCDABE | This study |
| MDM973 | PAK/pUCP24GW-lacIQ-lacPO-exoS::blaM | This study |
| MDM974 | PAK ΔpscC/pUCP24GW-lacIQ-lacPO-exoS::blaM | This study |
| MDM1156 | PAO-LAC/pUCP24GW-lacPO-luxCDABE | This study |
| PAKΔC | PAK ΔpscC; T3SS defective | 28 |
| PAKΔS | PAK ΔexoS; secretes ExoT as its only cytotoxic T3SS effector | 28 |
| PAKΔSTYexoU | PAK ΔexoS::miniCTX-exoU-spcU; secretes ExoU as its only cytotoxic T3SS effector | 28 |
| PAKΔTY | PAK ΔexoT ΔexoY; secretes ExoS as its only T3SS effector | 28 |
| MDM1387 | PA14 xcpQ::MrT7; PAMr_nr_mas_02_2:H7; defective in type II secretion | 29 |
| Y. pestis | ||
| JG153/pMM85 | KIM Δpgm pPCP1− pCD1+/pHSG576 yopE::blaM | 31, 44 |
PCR and primers.
Synthetic oligonucleotide primers (from Operon, Inc.) were designed using the published genome sequence for P. aeruginosa (53) and web-based PRIMER3 (Whitehead Institute) (Table 2). Primers were used at 10 μM in PCR amplifications with Failsafe polymerase (Epicentre), buffer G (Epicentre), and 4% dimethylsulfoxide (DMSO) for P. aeruginosa chromosomal DNA templates.
TABLE 2.
Primers used
| No. | Name | Sequence |
|---|---|---|
| 1 | exoT-F+EcoRI | TACTACGAATTCCCAGGAAGCACCGAAGG |
| 2 | exoT-R+EcoRI | CATTACGAATTCCTGGTACTCGCCGTTGGTAT |
| 3 | exoT-out-F | TAGGGAAAGTCCGCTGTTTT |
| 4 | luxC-R | CCTGAGGTAGCCATTCATCC |
| 5 | exoS-F+GWL | TACAAAAAAGCAGGCTAGGAAACAGACATGCATATTCAATCGCTTCAG |
| 6 | exoS(234)-R | ATCTTTTACTTTCACCAGCGTTTCTGGGTGACCGTCGGCCGATACTCTGCT |
| 7 | BLA-F | CACCCAGAAACGCTGGTGAA |
| 8 | BLA-R+GWR | TACAAGAAAGCTGGGTTTGGTCTGACAGTTACCAATGC |
| 9 | GW-attB1 | GGGGACAAGTTTGTACAAAAAAGCAGGCT |
| 10 | GW-attB2 | GGGGACCACTTTGTACAAGAAAGCTGGGT |
| 11 | lux-F+GWL | TACAAAAAAGCAGGCTAGGAAACAGCTATGACGAAGAAGATCAGTTTTATAATTAACGGCCAGGTTGAAATC |
| 12 | lux-R+GWR | TACAAGAAAGCTGGGTGTTTTCCCAGTCACGACGTT |
Screening compounds.
Compounds screened in this study were purchased from Chembridge (San Diego, CA) and Timtec (Newark, DE), diluted in 96-well master plates at 2.5 mM in DMSO, and stored at −20°C.
Luciferase transcriptional reporter screen.
A transcriptional fusion of the Photorhabdus luminescens lux operon (luxCDABE) to effector gene exoT (PA0044) was constructed by inserting an internal fragment of the exoT gene (712 bp generated by PCR with primers exoT-F+EcoRI and exoT-R+EcoRI) (Table 2) into EcoRI-cut reporter plasmid pGSV3-lux-Gm (37) as described previously (35). The resulting plasmid was introduced into E. coli SM10 cells and transferred into P. aeruginosa PAO1 and PA01 ΔpscC cells by conjugation (35) to generate recombinant reporter strains MDM852 and MDM1355, respectively. Insertion at the exoT chromosomal locus was confirmed by PCR with a primer outside the cloned locus (exoT-out-F) and a primer within the luxC gene (luxC-R) (Table 2).
For inhibitor screening, compound master plates were thawed at room temperature on the day of the screen, and 1 μl of compound (final content, 45 μM compound and 1.8% DMSO) was added to the 384-well opaque black screening plates using a Sciclone ALH 3000 liquid-handling robot (Caliper, Inc.) and a Twister II Microplate Handler (Caliper, Inc.). Reporter strain MDM852 was grown at 37°C in LBGI to an optical density at 600 nm (OD600) of ∼0.025 to 0.05 and transferred into microplates (50 μl/well) containing test compounds and EGTA (5 μl of 0.1 M stock solution), which were covered with a translucent gas-permeable seal (catalog no. AB-0718; Abgene, Inc.). Control wells contained cells with fully induced T3SS (EGTA and DMSO, microplate columns 1 and 2) and uninduced T3SS (DMSO only, microplate columns 23 and 24). Plates were incubated at room temperature for 300 min. Luminescence was measured in an Envision Multilabel microplate reader (PerkinElmer) (Fig. 1A and B). The screening window coefficient, Z′ factor (60), defined as the ratio of the positive- and negative-control separation band to the signal dynamic range of the assay, averaged 0.7 for the screen. All screening data, including the Z score, and confirmation and validation data were stored in one central database (CambridgeSoft's ChemOffice 11.0). Validated hits were reordered from the vendor and confirmed to be >95% pure and to be of the expected mass by liquid chromatography-mass spectrometry (LC-MS) analysis. Compounds for SAR analysis were ordered from Chembridge, Inc.
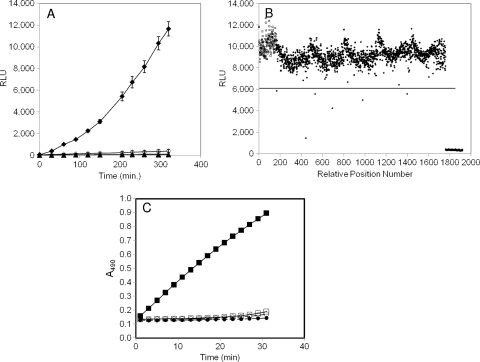
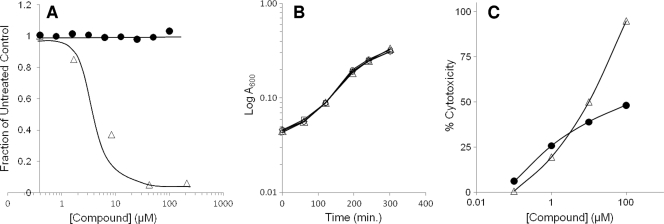
FIG. 1.
Characterization of bioluminescent and chromogenic reporter strains for identification of T3SS inhibitors. (A) Luminescence (in relative light units, RLU) from a chromosomal transcriptional fusion of exoT to the P. luminescens luxCDABE operon in wild-type (strain MDM852) or ΔpscC (strain MDM1355) P. aeruginosa PAO1 cells. Overnight cultures were diluted at time zero to A600 ∼ 0.025 and induced with 5 mM EGTA or left uninduced. RLU values were measured in 96-well opaque microplates throughout a 320-min time course. ⧫, MDM852 with EGTA; ⋄, MDM852 without EGTA; ▴, MDM1355 with EGTA; ▵, MDM1355 without EGTA. (B) Luminescence (RLU) from five 384-well microplates containing reporter strain MDM852 in a high-throughput screen for T3SS inhibitors. RLU values are shown at 300 min for 160 negative controls (□; fully induced by EGTA) in positions 1 to 160, for 160 positive controls (▴; no induction by EGTA) in positions 1,761 to 1,920, and for 1,600 samples (•) in positions 161 to 1,760. Six samples were designated hits because their RLU values displayed Z scores of >4 (i.e., >4 standard deviations below the average sample value, denoted as a horizontal line at 6,084 RLU). Compound 1 at position 443 was the most potent hit (Z score = 10) (Table 3). (C) Detection of ExoS′-βLA secretion from P. aeruginosa strains MDM973 (PAK) and MDM974 (PAK ΔpscC) carrying pUCP24GW-lacIQ-lacPO-exoS′-blaM, as measured by the hydrolysis of nitrocefin. A490 values are plotted versus time for MDM973 in the presence (▪) and absence (□) of 5 mM EGTA and for strain MDM974 in the presence (•) and absence (○) of 5 mM EGTA.
Effector-β-lactamase secretion assays. (i) P. aeruginosa.
A gene encoding an ExoS′-β-lactamase (βLA) fusion protein (comprised of 234 codons of P. aeruginosa effector ExoS fused to the TEM1 β-lactamase gene lacking secretion signal codons) was constructed by splicing by overlap extension PCR (SOE-PCR) (4) using primers 5 to 10 (Table 2), sequence confirmed, cloned into lacIQ-containing Gateway vector pUCP24GW (36) behind the lac promoter, and introduced into P. aeruginosa by electroporation (3). The secretion of fusion proteins was detected by measuring the hydrolysis of the chromogenic β-lactamase substrate nitrocefin in clear 96-well microplates in a modification of a previously described assay (27). Cells of strain MDM973 (PAK/pUCP24GW-exoS::blaM) were subcultured in the morning from overnight growths in LBG into 0.1 ml of LBGIE with or without test compounds and grown for 150 min. Nitrocefin (100 μg/ml final) was added, and A490 measurements were taken every minute for 15 min in a Victor3V 1420 Multilabel HTS Counter (PerkinElmer). Slopes were calculated as a relative measure of the quantity of the effector-βLA fusion protein secreted and were absolutely dependent on induction with IPTG and EGTA and the presence of a functional pscC gene in the P. aeruginosa cells (Fig. 1C). Typical signal:background ratios were 6-10.
(ii) Yersinia pestis.
Attenuated Y. pestis strain JG 153 (gift of Jon Goguen, University of Massachusetts Medical School, Worcester, MA) carrying plasmid pMM85 (yopE::blaM) was grown in LB plus 20 μg/ml chloramphenicol at 30°C to prevent T3SS induction and the loss of the pCD1 plasmid encoding T3SS. To induce T3SS, cells were shifted from 30 to 37°C, and EGTA was added to a 1 mM final concentration. Cell culture (0.1 ml) was added to clear 96-well microplates containing test compound and incubated for 3 h at 37°C. Nitrocefin was added (100 μg/ml final), and A490 measurements were taken every minute for 10 min in an Envision Multilabel microplate reader (PerkinElmer). Slopes were plotted versus the inhibitor concentration to determine the 50% inhibitory concentrations (IC50s).
Counterscreen for inhibition of bioluminescence of lac-promoted luxCDABE.
The complete Photorhabdus luminescens luxCDABE locus was amplified from pGSV3-lux (37) by PCR with Phusion polymerase (NEB, Beverly, MA) and primers lux-F+GWL and lux-R+GWR, followed by a second PCR with primers GW-attB1 and GW-attB2 to provide the full Gateway recognition sequence (Table 2). The ∼5.8-kb product was gel purified and inserted into pDONR221 with BPClonase (Invitrogen) and then into pUCP24GW (36) with LRClonase (Invitrogen). The resulting pUCP24GW-lacPO-luxCDABE plasmid was introduced into the P. aeruginosa PAO-LAC strain carrying one chromosomal copy of the lac repressor, lacIQ, at the phiCTX locus (20) by electroporation, selecting for gentamicin resistance (3). To measure the effects of T3SS inhibitors on lac-promoted luciferase production, the resulting strain MDM1156 was subcultured from overnight LBG growths into LBGI at an A600 of ∼0.05 and grown for 3 h in the presence or absence of inhibitors at 50 μM. The percent inhibition by compounds of the luminescence (in relative light units [RLU]) produced by lac-promoted versus exoT-promoted luciferase was calculated and used as an indication of the T3SS selectivity of the screening hits.
Detection of inhibition of T3SS-mediated ExoS secretion into culture broths.
P. aeruginosa strain PAKΔTY, which produces the ExoS, but not the ExoT or ExoY, T3SS effectors, was grown overnight in LB and treated essentially as described previously (28). Bacteria were subcultured 1:1,000 in LB supplemented with 5 mM EGTA and grown for 3 h at 37°C with aeration in the presence or absence of inhibitors at the indicated concentrations. Bacteria were sedimented by centrifugation at 3,220 × g for 15 min at 4°C. Culture supernatant was collected, and proteins were concentrated by precipitation with 12.5% trichloroacetic acid followed by a wash with acetone or by ultrafiltration. Proteins were resuspended according to original culture density (A600), separated by sodium dodecyl sulfate-12.5% polyacrylamide gel electrophoresis (SDS-12.5% PAGE), and stained with Coomassie blue. Stained gel image files were processed with ImageJ software (version 1.42q; NIH) by subtracting the background, inverting the image, and integrating the density of each band.
Inhibition of P. aeruginosa ExoU-dependent CHO cell killing.
The rescue of CHO cells from the T3SS-mediated cytotoxicity of translocated effector protein ExoU was measured using a lactate dehydrogenase (LDH) release assay as previously reported (28), except that infection with P. aeruginosa was carried out for 2 h in the absence of gentamicin. Percent cytotoxicity (percent LDH release) was calculated relative to that of the uninfected control, which was set at 0% LDH release, and that of cells infected with P. aeruginosa unprotected by test compound (100% LDH release). LDH released from unprotected, infected cells reached at least 80% of the value obtained from complete lysis with 1% Triton X-100 in the 2-h timeframe of this experiment. Pseudolipasin, which acts by the direct inhibition of the ExoU phospholipase, was used as the control inhibitor (27).
Gentamicin protection assays of bacterial internalization.
Experiments were carried out using a modification of a previously published method (18). A total of 2 × 105 HeLa cells were seeded into each well of a 12-well plate containing 2 ml per well of minimal essential medium (MEM) supplemented with 10% fetal calf serum (FCS) and incubated at 37°C in 5% CO2 for 24 h. After two washes with PBS, 1 ml of MEM containing 1% FCS was added to the HeLa cells. MBX 1641 was added to half the wells at a 50 μM final concentration (DMSO at 0.2% final concentration). P. aeruginosa strains PAKΔC (negative control) and PAKΔS (positive control) were grown overnight in LB medium at 37°C with shaking, diluted 1:1,000 in the morning, and grown to an OD600 of 0.3 (∼108 cells/ml). Bacteria were washed in phosphate-buffered saline (PBS), resuspended in 1 ml of MEM, and added to the HeLa cells at a multiplicity of infection (MOI) of 10 in the presence or absence of MBX 1641. Infected HeLa cells were incubated at 37°C in 5% CO2 for 2 h. After two washes with PBS, 1 ml of MEM containing 50 μg/ml gentamicin was added, and cells were incubated for an additional 2 h. After three washes with PBS, the cells were lysed in PBS containing 0.25% Triton X-100, and dilutions were plated on LB agar plates to count the number of bacteria internalized within HeLa cells.
Elastase secretion assay.
The effect of test compounds on the type II-mediated secretion of elastase from P. aeruginosa was determined by a modification of a previously described method (42). P. aeruginosa PA14 cells were cultured from a starting density of A600 ∼ 0.05 for 16 h to saturation in LB in the presence or absence of test compound at 50 μM. Cells were removed by centrifugation in a microcentrifuge, and 0.2 ml of cleared supernatant was added to 0.4 ml of a suspension of elastin-Congo Red (5 mg/ml; Sigma) in buffer consisting of 0.1 M Tris-HCl, pH 7.4, and 1 mM CaCl2 in capped microcentrifuge tubes. Tubes were incubated at 37°C with shaking for 6 h. A volume of 0.4 ml of buffer consisting of 0.7 M sodium phosphate (pH 6.0) was added, tubes were centrifuged in a microcentrifuge to remove undigested elastin-Congo Red, and the A495 of the cleared supernatants was measured. Readings were normalized to the original cell density (OD600), and the percent inhibition of elastase secretion was determined relative to that of untreated PA14 (no-inhibition control) and untreated type II secretion-defective PA14 xcpQ::MrT7 (29) (strain MDM1387) (Table 1) (complete inhibition control).
Chlamydia trachomatis growth inhibition assay.
The inhibition of the growth of Chlamydia trachomatis strain L2 by compounds was measured in 24-well plates essentially according to the method of Wolf et al. (59). Confluent monolayer Hep-2 cells were infected with L2 at an MOI of 0.5 and treated with compounds at the indicated concentrations for 48 h. The cultures then were collected and sonicated. Entire lysates were used for counting inclusion-forming units (IFUs) as a measurement of the production of chlamydia progeny elementary bodies (EBs) by replating onto fresh HeLa monolayers. An uninhibited control (DMSO only) and a complete inhibition control (200 μg/ml chloramphenicol) were included. Experiments were done in triplicate.
MIC.
MIC determination was done by the broth microdilution method described in the CLSI (formerly NCCLS) guidelines (39) and expressed in μM to facilitate comparisons with IC50 and cytotoxic concentration (CC50) values.
Determination of mammalian cytotoxicity.
The CC50 of a compound for cultured mammalian cells (HeLa; ATCC CCL-2; American Type Culture Collection, Manassas, VA) was determined as the concentration of compound that inhibits 50% of the conversion of MTS to formazan (32). Briefly, 96-well plates were seeded with HeLa cells at a density of 4 × 103 per well in VP-SFM medium without serum (14) in the presence or absence of serial dilutions of a compound dissolved in DMSO. Following incubation for 3 days at 37°C in VP-SFM, cell viability was measured with the vital tetrazolium salt stain 3-(4,5-dimethylthiazol-2-yl)-2,5 diphenyltetrazolium bromide according to the manufacturer's instructions (Promega, Madison, WI). Values were determined in duplicate using dilutions of inhibitory compound from 100 to 0.2 μM.
Chemistry.
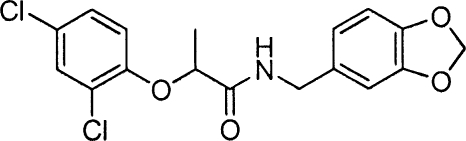
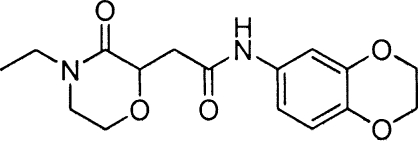
Phenoxyacetamides MBX 1685, MBX 1684, and MBX 1686, related to screening hit MBX 1641, all were prepared from 2,4-dichlorophenol. The alkylation of 2,4-dichlorophenol with ethyl 2-bromo-2-methylpropanoate (K2CO3, CH3CN) provided ethyl 2-(2,4-dichlorophenoxy)-2-methylpropanoate, which was hydrolyzed (KOH, ethyl alcohol) and coupled (HOAT, EDCI, DMF, DIPEA) (2) with 3,4-methylenedioxybenzylamine to provide MBX 1685. The Mitsunobu coupling (34) of 2,4-dichlorophenol with (S)-ethyl 2-hydroxypropanoate (PPh3, DIAD, THF) provided ethyl (R)-2-(2,4-dichlorophenoxy)propanoate, which was hydrolized (LiOH·H2O, CH3CN, H2O) and then coupled as described above with 3,4-methylenedioxybenzylamine to give MBX 1684. The corresponding S-enantiomer (MBX 1686) was prepared in precisely the same fashion but using methyl (R)-2-hydroxypropanoate with 2,4-dichlorophenol in the Mitsunobu coupling protocol. Hit compound MBX 1641 and desmethyl compound MBX 1668 were prepared directly from commercially available 2-(2,4-dichlorophenoxy)propanoic acid and 2,4-dichlorophenoxyacetic acid, respectively, by being coupled with 3,4-methylenedioxybenzylamine as described above.
RESULTS
Identification of inhibitors of P. aeruginosa T3SS.
A P. aeruginosa cell-based bioluminescent reporter screen for the identification of T3SS inhibitors was constructed in a fashion analogous to that described previously for Yersinia (24). Due to the tight coupling of T3SS gene regulation in P. aeruginosa with the type III secretion of the negative regulator ExsE, a reduced type III secretion capability results in the decreased expression of all T3SS operons (47, 54). We constructed P. aeruginosa strains carrying a transcriptional fusion of the T3SS effector gene exoT to the luxCDABE operon of Photorhabdus luminescens and evaluated their luminescence production under T3SS-inducing and -repressing conditions. When Ca2+ levels remain high (no EGTA addition [55]) or a key component of the T3SS assembly is deleted (the pscC gene encoding the secreton component of T3SS [27]), T3SS is not functional and luminescence is significantly reduced compared to that of wild-type cells grown in low levels of free Ca2+ (addition of 5 mM EGTA) (Fig. 1A). The application of the wild-type transcriptional fusion strain was optimized for screening in 384-well microplates, and about 80,000 discrete chemical compounds were screened at 50 μM to identify inhibitors of T3SS. Screening results are shown graphically for five representative 384-well assay plates in Fig. 1B. The substantial signal-to-background ratio (>20) and the very modest coefficients of variation (standard deviation/average signal) for samples as well as positive and negative controls (all <10%) are representative of those observed in the entire screen. A total of 331 compounds (0.4% of the library) were detected as primary hits due to the inhibition of RLU values by at least 4 standard deviations below the sample average (Z score ≥ 4; solid line in Fig. 1B), and more than 60% of them (208 compounds) were confirmed as inhibitors when retested in the same assay in triplicate. However, more than 80% of these putative inhibitors were eliminated by requiring that they inhibit luminescence from the exoT-lux screening strain >2-fold more potently than that from a non-T3SS-regulated lux strain (lac-regulated luxCDABE in strain MDM1156). The absence of T3SS specificity observed for most screening hits likely is the result of the many non-T3SS-related mechanisms capable of reducing luminescence (e.g., inhibition of growth, energy metabolism, transcription, or translation).
Validation of inhibitors of P. aeruginosa T3SS-mediated secretion.
The remaining T3SS-selective hits were evaluated directly for the inhibition of T3SS-mediated secretion. Measurements were carried out using a cellular assay consisting of an effector-reporter fusion protein. Codons for the type III secretion signals (8) and the GAP domain of P. aeruginosa ExoS (17) were fused to the TEM1 β-lactamase gene lacking its secretion signal. The construct was cloned into the exogenously replicating plasmid pUCP24GW, resulting in the production of ExoS′-βLA fusion protein under lac regulation in P. aeruginosa cells. In this assay, secreted β-lactamase activity is detected by the hydrolysis of the β-lactamase chromogenic substrate nitrocefin, resulting in an increased A490. Signal generation is dependent on the presence of EGTA and IPTG and is eliminated in T3SS-defective ΔpscC mutant cells (Fig. 1C). Almost all (41 of 43) of the T3SS-selective inhibitors identified in the transcriptional fusion reporter assays also inhibited the secretion of the effector-reporter fusion protein by at least 50% when added at a concentration of 50 μM during the induction of T3SS and the effector fusion. No inhibition was observed when compounds were added after induction at the time of chromogenic substrate addition, indicating that the compounds inhibit the appearance of extracellular β-lactamase rather than β-lactamase catalysis itself.
Finally, the inhibitors were evaluated for the potency of ExoS′-βLA fusion protein secretion inhibition (IC50) and counterscreened for cytotoxicity (CC50), yielding five validated T3SS inhibitors with IC50s of ≤25 μM and CC50s of ≥100 μM (Table 3). These five inhibitors exhibited no detectable MIC (MIC > 100 μM) against P. aeruginosa and did not inhibit the growth rate of P. aeruginosa cells (data not shown), confirming that they are not reducing luminescence or β-lactamase secretion by inhibiting bacterial cell growth or viability. These five validated T3SS inhibitors can be categorized into three structural classes, indicated in Table 3 as series A (phenoxyacetamides), B (malic diamides), and C (N-phenyl maleimide adducts).
TABLE 3.
Summary of selective inhibitors of type III secretion
Percent inhibition of exoT-lux RLU/percent inhibition of lac-lux RLU, both at 50 μM compound.
Percent inhibition of exoT-lux RLU in the absence of serum/percent inhibition of exoT-lux RLU in the presence of 10% fetal calf serum, both at 50 μM compound.
Compound concentration at which secretion of ExoS-βLA fusion protein from P. aeruginosa strain MDM973 is reduced by 50%. All IC50 and CC50 values are presented in μM.
Compound concentration at which the viability of HeLa cells cultured in serum-free medium is reduced by 50%.
Selectivity of T3SS inhibition as measured by the ratio of potency of the compound in the HeLa cell viability assay vs the T3SS inhibition assay.
Compound concentration at which secretion of YopE-βLA fusion protein from Y. pestis strain JG153/pMM85 is reduced by 50%.
Inhibition of T3SS-mediated secretion of native effectors.
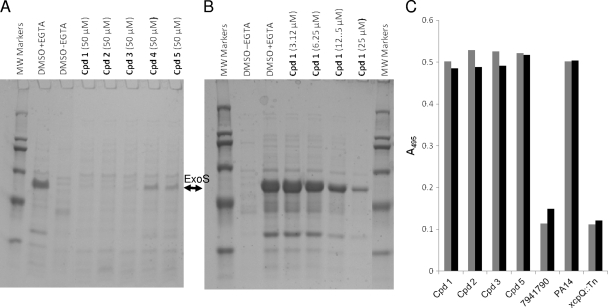
To confirm that inhibitors identified by the cell-based reporter assays inhibit the T3SS-mediated secretion of natural effectors, we concentrated conditioned culture medium from a P. aeruginosa ExoS-secreting strain exposed to each of the five T3SS inhibitors at 50 μM during growth for 3 h under T3SS-inducing conditions and visualized the secreted effectors on SDS-PAGE (Fig. 2A). All five compounds inhibited the secretion of ExoS from P. aeruginosa cells by at least 75%. The concentration dependence of the inhibition of native ExoS secretion was examined in detail for compound 1 and was found to be very similar to that observed in the ExoS′-βLA inhibition assay (IC50 of ∼12.5 μM) (Fig. 2B). The inhibitory effect appeared specific for type III secretion, since members of all three structural classes failed to inhibit type II-mediated elastase secretion when added to type II secretion-competent P. aeruginosa PA14 cells at 50 μM (Fig. 2C). Control inhibitor 7941790 (Chembridge, Inc.) reduced elastase secretion to the level observed in a type II-deficient PA14 strain carrying a transposon insertion in the secreton gene xcpQ, while the three series of T3SS inhibitors had no detectable effect.
FIG. 2.
Evaluation of inhibition of type III and type II secretion in P. aeruginosa. P. aeruginosa ExoS-secreting strain PAKΔTY was grown under T3SS-inducing conditions (LB plus 5 mM EGTA) for 3 h in the presence of the indicated concentrations of compounds, and culture medium (1 ml) was concentrated in SDS-PAGE sample buffer, separated by SDS-12.5% PAGE, and stained with Coomassie blue. The positive control, DMSO plus EGTA, was treated with 5 mM EGTA but not inhibitors, and the negative control, DMSO without EGTA, was treated with neither EGTA nor inhibitors. The identity and molecular weights of protein markers are as follows: porcine myosin (200K), E. coli β-galactosidase (116K), rabbit muscle phosphorylase B (97K), bovine albumin (66K), ovalbumin (45K), and bovine carbonic anydrase (29K). (A) Secreted proteins from cells treated with EGTA and the five validated T3SS inhibitors (Table 3). The band corresponding to 49K ExoS is marked. (B) Secreted proteins from cells treated with EGTA and serial dilutions of T3SS inhibitor compound 1. (C) Effects of T3SS inhibitors on type II secretion of elastase. P. aeruginosa PA14 cells were grown in LB medium for 16 h in the presence of 50 μM of the indicated compounds. As controls, PA14 and PA14 xcpQ::Tn cells were grown in LB in the presence of the equivalent concentration of DMSO, and PA14 was grown in the presence of a type II secretion inhibitor (compound 7941790; Chembridge, Inc.). Culture medium corresponding to equivalent numbers of cells was harvested by centrifugation and incubated with shaking for 6 h with Congo Red-elastin. Digested soluble Congo Red was measured by the A495 in two independent assays and plotted (gray and black bars).
Inhibition of T3SS-mediated effects on mammalian cells.
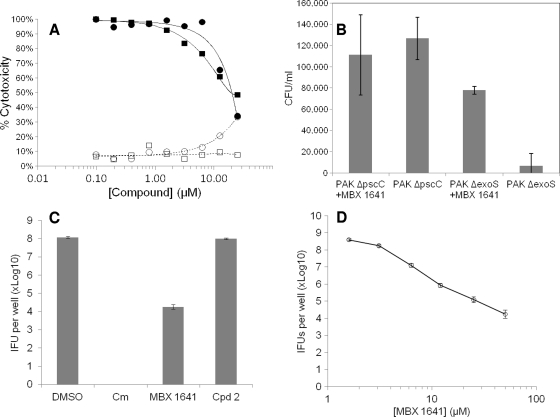
To assess their effects on the T3SS-mediated translocation of effectors, the five specific inhibitors of type III secretion were tested in a cellular activity assay for T3SS effector translocation into mammalian cells (27). The compounds were added to CHO cells simultaneously with the addition of P. aeruginosa ExoU-producing cells to determine whether the inhibitors were capable of blocking CHO cell death due to the cytotoxic activity of translocated ExoU. Only compound 1 was capable of reproducibly rescuing CHO cells from the ExoU-secreting P. aeruginosa cells (Fig. 3A), and its potency in this assay (IC50 of ∼15 μM) was similar to its potencies in the ExoS′-βLA assay (Table 3) and in the inhibition of the secretion of native ExoS (Fig. 2B). These results demonstrate that the phenoxyacetamide compound 1 not only blocks the T3SS-mediated secretion of effectors from P. aeruginosa into culture medium but also blocks the translocation of effectors into mammalian cells. Rescue from ExoU cytotoxicity by compound 1 was limited somewhat due to the cytotoxicity of the compound itself in the absence of P. aeruginosa cells, which reaches about 30% at 25 μM (Fig. 3A, open circles) and 50% at 75 μM (not shown). This CC50 is somewhat lower than the values obtained with HeLa cells (102 μM) (Table 3; also see Fig. 5C) and 293T cells (110 μM) (data not shown) in the absence of serum. The difference probably reflects the facts that three different cell types were employed and that the CHO cells were under stress due to the sudden reduction in serum levels from 10 to 1% just prior to infection with P. aeruginosa cells. In any case, there is a clear margin of efficacy for compound 1 in this CHO rescue experiment. A known ExoU inhibitor, pseudolipasin (27), also rescued CHO cells from ExoU toxicity with a similar potency. We resynthesized compound 1 and verified that the resulting compound, MBX 1641, exhibits the same T3SS inhibition potency and selectivity as the original compound 1.
FIG. 3.
Inhibition of T3SS-mediated effects on mammalian cells in culture. (A) Concentration-dependent rescue of CHO cells from ExoU cytotoxicity by T3SS inhibitor MBX 1641. ExoU-secreting P. aeruginosa strain PAKΔSTYexoU was mixed with CHO cells at an MOI of 5 in the presence of MBX 1641 (•) or the known ExoU inhibitor pseudolipasin (▪) (27) at various concentrations as indicated. The percent cytotoxicity is calculated as the percentage of LDH released from cells intoxicated with P. aeruginosa with or without inhibitor compared to LDH released from intoxicated cells that were not treated with inhibitor. The effects of pseudolipasin (□) and MBX1641 (○) also are shown in the absence of P. aeruginosa cells to evaluate the inherent cytotoxicity of the compounds themselves. (B) T3SS inhibitor MBX 1641 relieves the ExoT block of the HeLa cell internalization of P. aeruginosa. HeLa cells were infected with P. aeruginosa PAK strains secreting ExoT (PAKΔS) or deficient in T3SS (PAKΔC) at an MOI of 10. MBX 1641 was added at 50 μM to half the wells containing each strain. After 2 h, cultures were treated with gentamicin (50 μg/ml) for an additional 2 h. HeLa cells were lysed with Triton, and serial dilutions were plated to determine the number of P. aeruginosa cells that had been protected from gentamicin by internalization. The CFU/ml of P. aeruginosa cells from lysed HeLa cells were determined in triplicate and plotted as the averages ± standard deviations. (C) MBX 1641 but not compound 2 inhibits the growth of C. trachomatis L2 cells in Hep-2 cells in culture. Confluent monolayer Hep-2 cells were infected with L2 at an MOI of 0.5 and treated with compounds at the indicated concentrations, followed by sonication and the measurement of IFUs on HeLa monolayers. Experiments were done in triplicate, and averages ± standard deviations are shown. Chloramphenicol (Cm) was used at 200 μg/ml as a positive control, and compound diluent (DMSO) was used as a negative control. (D) Concentration dependence of the inhibition of C. trachomatis L2 growth in Hep-2 cells by MBX 1641.
FIG. 5.
Evaluation of the effects of MBX 1641 on bacterial and mammalian cell growth. (A) Determination of the MIC of MBX 1641 for P. aeruginosa. P. aeruginosa PAO1 cells were grown in the presence of the indicated concentrations of MBX 1641 (•) or tetracycline (▵) for 16 h in clear 96-well microplates, and the OD600 was determined. The OD600 as a fraction of that of DMSO-treated control cells is plotted. (B) Growth rate of P. aeruginosa cells treated with MBX 1641. P. aeruginosa PAO1 cells were grown in the presence of three different concentrations of MBX 1641 for 5 h in clear 96-well microplates, and the OD600 was measured periodically as indicated as a measure of cell density. MBX 1641 was present at 100 (□), 50 (•), or 25 μM (○), or cells were treated with an equivalent concentration (2%) of DMSO only (▵). (C) HeLa cell cytotoxicity of MBX 1641. HeLa cells were cultured in VP-SFM medium without serum in the presence of the indicated concentrations of MBX 1641 (•) or novobiocin (▵) for 3 days, and cytotoxicity was determined by the ability of remaining live cells to reduce a vital tetrazolium salt stain. Results are plotted as the percentage of cytotoxicity relative to levels for DMSO-treated and Triton X-100-lysed control cells.
ExoS and ExoT appear to block the uptake of P. aeruginosa cells by both epithelial and phagocytic cells in culture, suggesting that the T3SS functions as a virulence factor by preventing the phagocytic cell clearance of P. aeruginosa cells during infection (6, 15). The inhibition of the T3SS-mediated secretion and translocation of ExoS or ExoT by mutation results in the increased internalization of bacteria (6, 15, 18, 50). MBX 1641 was tested to determine if its T3SS inhibition facilitated the internalization of P. aeruginosa cells by HeLa cells in culture. The addition of the compound at 50 μM to HeLa cells simultaneously with the addition of ExoT-producing P. aeruginosa cells at a multiplicity of infection of 10 resulted in a stimulation of the internalization of bacterial cells by more than 11-fold, as measured by the protection of bacteria from gentamicin (Fig. 3B, columns 3 and 4). In the presence of MBX 1641, the number of internalized P. aeruginosa ExoT-secreting cells increased to nearly the number of T3SS-deficient ΔpscC cells taken up by HeLa (Fig. 3B, columns 2 and 3). As expected, MBX 1641 had no significant effect on the already-high levels of the uptake of a T3SS-deficient ΔpscC mutant strain (Fig. 3B, columns 1 and 2).
Bacterial spectrum of activity.
The intracellular pathogen Chlamydia trachomatis expresses a T3SS thought to be responsible for injecting effectors into the host cytosol (23). Recently, Yersinia T3SS inhibitors INP0007 and INP0400, both members of an acylated hydrazone series (40), were demonstrated to arrest the growth of C. trachomatis in mammalian cell hosts (38, 59), suggesting that the T3SS plays an essential role in the Chlamydia development cycle. MBX 1641 and compound 2 (Table 3) were tested for the ability to block the growth of C. trachomatis L2 in Hep-2 cells. The results reveal that MBX 1641, but not compound 2, significantly reduced the growth of C. trachomatis when added at 50 μM (Fig. 3C). In addition, MBX 1641 exhibited a concentration-dependent effect on C. trachomatis growth in Hep-2 cells (Fig. 3D). These results suggest that MBX 1641 is capable of inhibiting T3SS in Chlamydia.
We also examined the ability of MBX 1641 to inhibit the T3SS of Yersinia pestis. As shown in Fig. 4A, MBX 1641 inhibits the T3SS-dependent secretion of a YopE-βLA effector fusion protein from attenuated Y. pestis strain JG153 with a potency about 3-fold poorer (IC50 of ∼38 μM) than that observed for its inhibition of ExoS′-βLA secretion from P. aeruginosa. It is interesting that the other four validated T3SS inhibitors of P. aeruginosa type III secretion also inhibit Y. pestis T3SS-mediated secretion (Table 3), which is consistent with the fact that the structural components of these two T3S systems share considerable sequence homology (23).
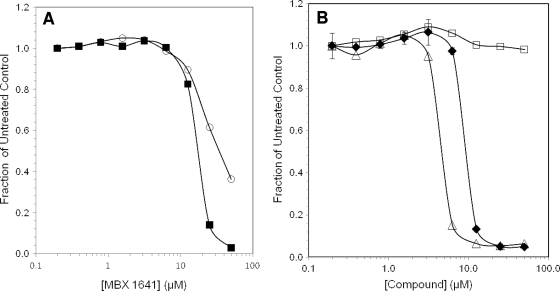
FIG. 4.
Inhibition of T3SS-mediated secretion of effector-β-lactamase fusion proteins. (A) Cells growing under T3SS-inducing conditions were treated for 3 h with MBX 1641, and secreted β-lactamase activity was measured by the cleavage of nitrocefin as the ΔA490/min. The rate of nitrocefin cleavage as a fraction of that of the untreated control is plotted versus the compound concentration. Bacterial species and effector βLA fusions were P. aeruginosa ExoS′-βLA (▪) and Y. pestis YopE-βLA (○). (B) Effects of MBX 1641 and its R- and S-enantiomers on ExoS′-βLA secretion from P. aeruginosa. Concentration dependence for MBX 1641 and its two stereo isomers, MBX 1684 and MBX 1686, were determined by the rate of nitrocefin cleavage by secreted ExoS′-βLA and calculated as the fraction of cleavage in the absence of inhibitor. Racemic mixture MBX 1641 (⧫), R-enantiomer MBX 1684 (Δ), and S-entantiomer MBX 1686 (□) are indicated.
Preliminary SAR for phenoxyacetamide T3SS inhibitors.
Results described above demonstrate that MBX 1641 inhibits both T3SS-mediated secretion and translocation. In addition, it does so with minimal effects on the extent (Fig. 5A) and rate (Fig. 5B) of the growth of P. aeruginosa cells and on the viability of HeLa cells (Fig. 5C), yielding a favorable selectivity index (CC50/IC50) of approximately 8. To explore the structure-activity relationships (SAR) of the phenoxyacetamide series represented by MBX 1641, a total of 114 analogs were purchased (Chembridge, Inc.) and assayed for T3SS inhibition at a single concentration (50 μM) (see Table S1 in the supplemental material). IC50s were determined for several key analogs by using the ExoS′-βLA assay (Table 4). The results indicate that very few alterations are acceptable on ring A, but there is considerable flexibility in the substituents tolerated on ring B. Results also suggest that the linker region cannot be lengthened by one methylene unit, but a tertiary amine is tolerated with some loss of activity. The discovery of inhibitory analogs in series A supports the validity of this chemotype as a T3SS inhibitor and provides a basis for the further optimization of the potency of this class of inhibitors.
TABLE 4.
Preliminary structure-activity relationships
See Table S1 in the supplemental material for structures.
Further SAR studies focused on the single stereocenter of MBX 1641, which is a racemic mixture. Since pure enantiomers were not available for purchase, we synthesized the two stereoisomers, MBX 1684 (R-isomer) and MBX 1686 (S-isomer). Also, to evaluate the effect of eliminating the stereocenter, we synthesized analogs of MBX 1641 lacking the methyl group at the stereocenter in the linker region (MBX 1668) and containing two methyl groups at the stereocenter (MBX 1685). The concentration-dependent inhibition of T3SS by these compounds was measured in the ExoS′-βLA reporter assay, and the results unambiguously establish the importance of the stereocenter for T3SS inhibitory activity. Only the R-isomer was active, and it was almost twice as potent as the racemic mixture (IC50 of ∼6 μM for MBX 1684 and ∼10 μM for MBX 1641) (Fig. 4B, Table 4). Both analogs lacking the stereocenter, the desmethyl and dimethyl compounds, were inactive (IC50s of >100 μM) (Table 4), as was the S-isomer (Fig. 4B).
DISCUSSION
In this study, a bioluminescent cellular reporter screen and multiple secondary assays were employed to identify and validate five new selective inhibitors of P. aeruginosa T3SS-mediated secretion and one inhibitor of T3SS-mediated translocation. These inhibitors display minimal cytotoxicity (CC50 ≥ 100 μM) and moderate potency (IC50s ≤ 15 μM) and exhibit no significant effects on the extent or rate of the growth of P. aeruginosa cells, nor do they inhibit the type II secretion system as determined by measurements of secreted elastase. The compounds represent three different chemotypes (series A, B, and C) (Table 3), but series A and B appear to be structurally related and contain a stereocenter, which was demonstrated to be critical for activity for series A. Compound 1 (MBX 1641) in series A reproducibly inhibits both T3SS-mediated secretion and translocation and was an effective antagonist in three mammalian cell assays that depend on T3SS intoxication of CHO cells by ExoU-producing P. aeruginosa, blockage by P. aeruginosa of HeLa cell internalization, and the growth of C. trachomatis in Hep-2 cells. The potency and selectivity of inhibitors in series A suggest that this class of T3SS inhibitors is suitable for further chemical optimization to produce a clinically useful inhibitor.
It is unclear why the other four validated inhibitors of T3SS-mediated secretion failed to inhibit T3SS-mediated translocation as measured by the rescue of CHO cells from ExoU intoxication. Most secretion inhibitors would be expected to inhibit translocation, since many aspects of T3SS-mediated secretion also are required for translocation. At least four possible explanations could account for this discrepancy. First, the inhibitors may interact with the T3SS apparatus at a site that is inaccessible when the P. aeruginosa needle is docked to the mammalian cell membrane. Second, the inherent cytotoxicity of the inhibitors may preclude our ability to detect the rescue of CHO cells from ExoU-mediated cytotoxicity. Some cytotoxicity was evident even in the successful inhibition by MBX 1641, and it limited our ability to achieve the complete rescue of CHO cells. While the four secretion inhibitors do not appear to be more cytotoxic than MBX 1641, even subtle increases in cytotoxicity may be sufficient to mask CHO cell rescue in this assay. Third, the secretion inhibitors may bind extensively to serum proteins and be unavailable for activity in the mammalian cell-based translocation assay. In fact, compounds 2, 3, and 5 do display greater loss of activity in the presence of serum than does compound 1 (MBX 1641) (Table 3, serum effect). A fourth formal possibility is that the inhibitors block T3SS induced by low levels of Ca2+ but not by mammalian cell contact. However, the speeds with which the inhibitors function seem to preclude action at the level of transcription regulation (see below).
The phenoxyacetamide MBX 1641 does not appear to be related structurally to any of the T3SS inhibitors reported previously. Results have been described for T3SS inhibitor screens of Yersinia pseudotuberculosis (24, 41), Y. pestis (44), enteropathogenic Escherichia coli (EPEC) (16), Salmonella enterica serovar Typhimurium (12), and P. aeruginosa (27). All have utilized cell-based assays, both for the direct identification of compounds active against whole cells and because the complexity of the molecular machine renders biochemical screens of component parts of T3SS particularly challenging. The only previously described screen for P. aeruginosa T3SS inhibitors was based on the reducing potential of remaining live CHO cells and consequently could detect inhibitors of any step in the secretion, translocation, and toxin activity leading to mammalian cell death (27). The validated inhibitors identified in the screen were shown to inhibit the ExoU toxin directly rather than the T3SS process itself. However, one series of hits described in that study displays structural similarity to MBX 1641. Two compounds in that series, 5929052 and 5925831 (27 and Table S2 in the supplemental material), failed to exhibit detectable inhibition in the ExoS′-βLA assay described here (IC50s > 100 μM; unpublished results). The absence of detectable inhibition is not surprising, since those compounds were identified as ExoU inhibitors and since they lack the stereocenter demonstrated to be crucial for the T3SS-inhibitory activity of MBX 1641 (e.g., see the desmethyl analog in Table 4). It is particularly interesting to compare the previously reported inhibitors of Y. pseudotuberculosis and Y. pestis T3SS to the inhibitors identified in this study, because the Pseudomonas T3SS proteins exhibit more sequence similarity to those of Yersinia than to those of any other genus (23). Two Y. pseudotuberculosis T3SS inhibitors, compounds 8 and 11 (41), were present in our screening collection. While they do inhibit P. aeruginosa T3SS moderately, they failed to inhibit the exoT-lux primary reporter screen with sufficient potency to be selected as primary hits (unpublished observations). One Y. pestis T3SS inhibitor, compound 2 (44), also was present in our screening collection, and it proved to be a potent inhibitor of P. aeruginosa T3SS in the primary and secondary screens applied here (IC50 of ≤10 μM in the ExoS′-βLA assay) but was not pursued due to high serum protein binding. The ability of three different Yersinia T3SS inhibitors to block P. aeruginosa T3SS is consistent with the high sequence homology observed for T3SS components in the two genera and with the ability of the five P. aeruginosa T3SS inhibitors described in this study to inhibit Y. pestis T3SS-mediated secretion.
The molecular target(s) of these P. aeruginosa T3SS inhibitors is not known; however, the results described here provide some evidence that these compounds specifically inhibit the activity of the T3SS apparatus. First, we have shown that the compounds are not simply inhibiting one of the effector toxins, because they specifically affected the secretion or the translocation of three different effectors: ExoS (SDS-PAGE), ExoT (HeLa cell internalization), and ExoU (rescue of CHO cells). Second, the inhibitors do not affect the extent (MIC) or rate of growth of P. aeruginosa cells. Third, the compounds do not appear to be general inhibitors of gene expression or virulence gene expression, because they demonstrate differential effects on the generation of luminescence by strains carrying exoT-lux and lac-lux transcriptional fusions, and they do not inhibit the production or secretion of another virulence factor, elastase, which utilizes the type II secretion mechanism. Fourth, the inhibition of ExoS′-βLA secretion by MBX 1641 is equally potent when measured in a multiple-efflux-pump knockout strain, P. aeruginosa strain PAO397 (26 and unpublished observations) (provided by Herbert Schweizer, Colorado State University). This suggests that T3SS inhibitors are not effluxed and/or do not need to enter P. aeruginosa cells to act, and the latter possibility is more likely, since few small molecules enter and are retained in P. aeruginosa cells (30). Fifth, MBX 1641 acts equally potently to block ExoS′-βLA secretion whether administered during or after the 2.5-h EGTA induction of T3SS, suggesting that the compound is not blocking T3SS gene expression or the assembly of the type III apparatus (unpublished observations). Finally, the strict requirement for the R-isomer configuration at the stereocenter of the phenoxyacetamide series indicates that the inhibitor is interacting with a specific target or targets and is not acting by a promiscuous nonspecific mechanism. The observed spectrum of activity against T3SS in three bacterial species points to a conserved target, but the sequence conservation is high across species among many of the T3SS gene products.
In addition to establishing the importance of the stereocenter in the linker region of the phenoxyacetamide series (series A), the initial SAR described here provides some clear directions for improving the potency of the inhibitor. The low tolerance for alterations to ring A (Table 4) suggests that this region of the molecule, together with the stereocenter, is involved in important contacts with the target. The further chemical optimization of these regions may provide improved potency. In contrast, the considerable tolerance demonstrated for various substituents on ring B (Table 4) suggests that few target contacts are made on that side of the compounds, perhaps providing a location for a tethered photoreactive group for target identification or for other modifications to provide pharmacologic or toxicologic benefits.
The results of this study suggest that MBX 1641 is capable of inhibiting the T3SS of three different bacterial species, P. aeruginosa, Y. pestis, and C. trachomatis. Multiple different assays demonstrate the inhibition of P. aeruginosa T3SS, while the inhibition of T3SS in the other two species is based on a single assay in each case. Nevertheless, effector-β-lactamase fusion proteins appear to be reliable reporters of T3SS function. In the absence of a manipulable genetic system in Chlamydia, it has not been possible to firmly establish the essentiality of the T3SS for intracellular growth. We cannot rule out the possibility that MBX 1641 is arresting C. trachomatis growth by mechanisms other than T3SS inhibition, but the compound has not demonstrated promiscuous behavior in a variety of assays and does not appear to be overtly cytotoxic or to block gene expression.
In summary, the potency and selectivity of the phenoxyacetamide compound MBX 1641 to block both the T3SS-mediated secretion and translocation of P. aeruginosa effectors justifies further study of this class of T3SS inhibitors. The absolute requirement for the R-stereoisomer indicates that the phenoxyacetamides target a specific component required for type III secretion. The structure-activity relationships demonstrated here suggest approaches to optimize this compound series to achieve higher potency and reduced cytotoxicity. Such optimized compounds could be evaluated in animal models either alone or in combination with antibiotics to determine their benefit in potential therapeutic applications.
Supplementary Material
Acknowledgments
We thank Jon Goguen (University of Massachusetts Medical School) for advice and for the Y. pestis reporter strain and Donald Woods (University of Calgary) for plasmid pGSV3-Lux. We thank Timothy Opperman for advice and for a critical reading of the manuscript. We thank Alicia Allaire for assistance in strain construction and Ming Di for performing elastase secretion assays as well as for strain construction.
This work was supported in part by DHHS/NIH grant R43 AI-068185 from the National Institute of Allergy and Infectious Diseases (NIAID).
Footnotes
Published ahead of print on 22 February 2010.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Bradley, D. E. 1974. The adsorption of Pseudomonas aeruginosa pilus-dependent bacteriophages to a host mutant with nonretractile pili. Virology 58:149-163. [DOI] [PubMed] [Google Scholar]
- 2.Carpino, L. A. 1993. 1-Hydroxy-7-azabenzotriazole. An efficient peptide coupling additive. J. Am. Chem. Soc. 115:4397-4398. [Google Scholar]
- 3.Choi, K. H., A. Kumar, and H. P. Schweizer. 2006. A 10-min method for preparation of highly electrocompetent Pseudomonas aeruginosa cells: application for DNA fragment transfer between chromosomes and plasmid transformation. J. Microbiol. Methods 64:391-397. [DOI] [PubMed] [Google Scholar]
- 4.Choi, K. H., and H. P. Schweizer. 2005. An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol. 5:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clatworthy, A. E., E. Pierson, and D. T. Hung. 2007. Targeting virulence: a new paradigm for antimicrobial therapy. Nat. Chem. Biol. 3:541-548. [DOI] [PubMed] [Google Scholar]
- 6.Cowell, B. A., D. Y. Chen, D. W. Frank, A. J. Vallis, and S. M. Fleiszig. 2000. ExoT of cytotoxic Pseudomonas aeruginosa prevents uptake by corneal epithelial cells. Infect. Immun. 68:403-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Lorenzo, V., and K. N. Timmis. 1994. Analysis and construction of stable phenotypes in gram-negative bacteria with Tn5- and Tn10-derived minitransposons. Methods Enzymol. 235:386-405. [DOI] [PubMed] [Google Scholar]
- 8.Derouazi, M., B. Toussaint, L. Quenee, O. Epaulard, M. Guillaume, R. Marlu, and B. Polack. 2008. High-yield production of secreted active proteins by the Pseudomonas aeruginosa type III secretion system. Appl. Environ. Microbiol. 74:3601-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diaz, M. H., C. M. Shaver, J. D. King, S. Musunuri, J. A. Kazzaz, and A. R. Hauser. 2008. Pseudomonas aeruginosa induces localized immunosuppression during pneumonia. Infect. Immun. 76:4414-4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.El Solh, A. A., G. Choi, M. J. Schultz, L. A. Pineda, and C. Mankowski. 2007. Clinical and hemostatic responses to treatment in ventilator-associated pneumonia: role of bacterial pathogens. Crit. Care Med. 35:490-496. [DOI] [PubMed] [Google Scholar]
- 11.Engel, J., and P. Balachandran. 2009. Role of Pseudomonas aeruginosa type III effectors in disease. Curr. Opin. Microbiol. 12:61-66. [DOI] [PubMed] [Google Scholar]
- 12.Felise, H. B., H. V. Nguyen, R. A. Pfuetzner, K. C. Barry, S. R. Jackson, M. P. Blanc, P. A. Bronstein, T. Kline, and S. I. Miller. 2008. An inhibitor of gram-negative bacterial virulence protein secretion. Cell Host Microbe 4:325-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Franchi, L., T. Eigenbrod, R. Munoz-Planillo, and G. Nunez. 2009. The inflammasome: a caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat. Immunol. 10:241-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frazzati-Gallina, N. M., R. L. Paoli, R. M. Mourao-Fuches, S. A. Jorge, and C. A. Pereira. 2001. Higher production of rabies virus in serum-free medium cell cultures on microcarriers. J. Biotechnol. 92:67-72. [DOI] [PubMed] [Google Scholar]
- 15.Garrity-Ryan, L., B. Kazmierczak, R. Kowal, J. Comolli, A. Hauser, and J. N. Engel. 2000. The arginine finger domain of ExoT contributes to actin cytoskeleton disruption and inhibition of internalization of Pseudomonas aeruginosa by epithelial cells and macrophages. Infect. Immun. 68:7100-7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gauthier, A., M. L. Robertson, M. Lowden, J. A. Ibarra, J. L. Puente, and B. B. Finlay. 2005. Transcriptional inhibitor of virulence factors in enteropathogenic Escherichia coli. Antimicrob. Agents Chemother. 49:4101-4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goehring, U. M., G. Schmidt, K. J. Pederson, K. Aktories, and J. T. Barbieri. 1999. The N-terminal domain of Pseudomonas aeruginosa exoenzyme S is a GTPase-activating protein for Rho GTPases. J. Biol. Chem. 274:36369-36372. [DOI] [PubMed] [Google Scholar]
- 18.Ha, U., and S. Jin. 2001. Growth phase-dependent invasion of Pseudomonas aeruginosa and its survival within HeLa cells. Infect. Immun. 69:4398-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hauser, A. R. 2009. The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat. Rev. Microbiol. 7:654-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoang, T. T., A. J. Kutchma, A. Becher, and H. P. Schweizer. 2000. Integration-proficient plasmids for Pseudomonas aeruginosa: site-specific integration and use for engineering of reporter and expression strains. Plasmid 43:59-72. [DOI] [PubMed] [Google Scholar]
- 21.Holloway, B. W., V. Krishnapillai, and A. F. Morgan. 1979. Chromosomal genetics of Pseudomonas. Microbiol. Rev. 43:73-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hudson, D. L., A. N. Layton, T. R. Field, A. J. Bowen, H. Wolf-Watz, M. Elofsson, M. P. Stevens, and E. E. Galyov. 2007. Inhibition of type III secretion in Salmonella enterica serovar Typhimurium by small-molecule inhibitors. Antimicrob. Agents Chemother. 51:2631-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kauppi, A. M., R. Nordfelth, H. Uvell, H. Wolf-Watz, and M. Elofsson. 2003. Targeting bacterial virulence: inhibitors of type III secretion in Yersinia. Chem. Biol. 10:241-249. [DOI] [PubMed] [Google Scholar]
- 25.Keyser, P., M. Elofsson, S. Rosell, and H. Wolf-Watz. 2008. Virulence blockers as alternatives to antibiotics: type III secretion inhibitors against Gram-negative bacteria. J. Intern. Med. 264:17-29. [DOI] [PubMed] [Google Scholar]
- 26.Kumar, A., K. L. Chua, and H. P. Schweizer. 2006. Method for regulated expression of single-copy efflux pump genes in a surrogate Pseudomonas aeruginosa strain: identification of the BpeEF-OprC chloramphenicol and trimethoprim efflux pump of Burkholderia pseudomallei 1026b. Antimicrob. Agents Chemother. 50:3460-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, V. T., S. Pukatzki, H. Sato, E. Kikawada, A. A. Kazimirova, J. Huang, X. Li, J. P. Arm, D. W. Frank, and S. Lory. 2007. Pseudolipasin A is a specific inhibitor for phospholipase A2 activity of Pseudomonas aeruginosa cytotoxin ExoU. Infect. Immun. 75:1089-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee, V. T., R. S. Smith, B. Tummler, and S. Lory. 2005. Activities of Pseudomonas aeruginosa effectors secreted by the type III secretion system in vitro and during infection. Infect. Immun. 73:1695-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liberati, N. T., J. M. Urbach, S. Miyata, D. G. Lee, E. Drenkard, G. Wu, J. Villanueva, T. Wei, and F. M. Ausubel. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. USA 103:2833-2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lomovskaya, O., M. S. Warren, A. Lee, J. Galazzo, R. Fronko, M. Lee, J. Blais, D. Cho, S. Chamberland, T. Renau, R. Leger, S. Hecker, W. Watkins, K. Hoshino, H. Ishida, and V. J. Lee. 2001. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob. Agents Chemother. 45:105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marketon, M. M., R. W. DePaolo, K. L. DeBord, B. Jabri, and O. Schneewind. 2005. Plague bacteria target immune cells during infection. Science 309:1739-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marshall, N. J., C. J. Goodwin, and S. J. Holt. 1995. A critical assessment of the use of microculture tetrazolium assays to measure cell growth and function. Growth Regul. 5:69-84. [PubMed] [Google Scholar]
- 33.Miao, E. A., R. K. Ernst, M. Dors, D. P. Mao, and A. Aderem. 2008. Pseudomonas aeruginosa activates caspase 1 through Ipaf. Proc. Natl. Acad. Sci. USA 105:2562-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mitsunobu, O. 1981. The use of diethyl azodicarboxylate and triphenylphosphine in synthesis and transformation of natural products. Synthesis 1981:1-28. [Google Scholar]
- 35.Moir, D. T., M. Di, R. A. Moore, H. P. Schweizer, and D. E. Woods. 2008. Cellular reporter screens for inhibitors of Burkholderia pseudomallei targets in Pseudomonas aeruginosa. Trans. R. Soc. Trop. Med. Hyg. 102(Suppl. 1):S152-S162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moir, D. T., M. Di, T. Opperman, H. P. Schweizer, and T. L. Bowlin. 2007. A high-throughput, homogeneous, bioluminescent assay for Pseudomonas aeruginosa gyrase inhibitors and other DNA-damaging agents. J. Biomol. Screen. 12:855-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moore, R. A., S. Reckseidler-Zenteno, H. Kim, W. Nierman, Y. Yu, A. Tuanyok, J. Warawa, D. DeShazer, and D. E. Woods. 2004. Contribution of gene loss to the pathogenic evolution of Burkholderia pseudomallei and Burkholderia mallei. Infect. Immun. 72:4172-4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muschiol, S., L. Bailey, A. Gylfe, C. Sundin, K. Hultenby, S. Bergstrom, M. Elofsson, H. Wolf-Watz, S. Normark, and B. Henriques-Normark. 2006. A small-molecule inhibitor of type III secretion inhibits different stages of the infectious cycle of Chlamydia trachomatis. Proc. Natl. Acad. Sci. USA 103:14566-14571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.NCCLS. 1997. Approved standard M7-A4. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 4th ed. National Committee for Clinical Laboratory Standards, Wayne, PA.
- 40.Negrea, A., E. Bjur, S. E. Ygberg, M. Elofsson, H. Wolf-Watz, and M. Rhen. 2007. Salicylidene acylhydrazides that affect type III protein secretion in Salmonella enterica serovar typhimurium. Antimicrob. Agents Chemother. 51:2867-2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nordfelth, R., A. M. Kauppi, H. A. Norberg, H. Wolf-Watz, and M. Elofsson. 2005. Small-molecule inhibitors specifically targeting type III secretion. Infect. Immun. 73:3104-3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohman, D. E., S. J. Cryz, and B. H. Iglewski. 1980. Isolation and characterization of Pseudomonas aeruginosa PAO mutant that produces altered elastase. J. Bacteriol. 142:836-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pallen, M. J., S. A. Beatson, and C. M. Bailey. 2005. Bioinformatics, genomics and evolution of non-flagellar type-III secretion systems: a Darwinian perspective. FEMS Microbiol. Rev. 29:201-229. [DOI] [PubMed] [Google Scholar]
- 44.Pan, N. J., M. J. Brady, J. M. Leong, and J. D. Goguen. 2009. Targeting type III secretion in Yersinia pestis. Antimicrob. Agents Chemother. 53:385-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rahme, L. G., E. J. Stevens, S. F. Wolfort, J. Shao, R. G. Tompkins, and F. M. Ausubel. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899-1902. [DOI] [PubMed] [Google Scholar]
- 46.Rello, J., D. Mariscal, F. March, P. Jubert, F. Sanchez, J. Valles, and P. Coll. 1998. Recurrent Pseudomonas aeruginosa pneumonia in ventilated patients: relapse or reinfection? Am. J. Respir. Crit. Care Med. 157:912-916. [DOI] [PubMed] [Google Scholar]
- 47.Rietsch, A., I. Vallet-Gely, S. L. Dove, and J. J. Mekalanos. 2005. ExsE, a secreted regulator of type III secretion genes in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 102:8006-8011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roy-Burman, A., R. H. Savel, S. Racine, B. L. Swanson, N. S. Revadigar, J. Fujimoto, T. Sawa, D. W. Frank, and J. P. Wiener-Kronish. 2001. Type III protein secretion is associated with death in lower respiratory and systemic Pseudomonas aeruginosa infections. J. Infect. Dis. 183:1767-1774. [DOI] [PubMed] [Google Scholar]
- 49.Schulert, G. S., H. Feltman, S. D. Rabin, C. G. Martin, S. E. Battle, J. Rello, and A. R. Hauser. 2003. Secretion of the toxin ExoU is a marker for highly virulent Pseudomonas aeruginosa isolates obtained from patients with hospital-acquired pneumonia. J. Infect. Dis. 188:1695-1706. [DOI] [PubMed] [Google Scholar]
- 50.Shaver, C. M., and A. R. Hauser. 2006. Interactions between effector proteins of the Pseudomonas aeruginosa type III secretion system do not significantly affect several measures of disease severity in mammals. Microbiology 152:143-152. [DOI] [PubMed] [Google Scholar]
- 51.Shaver, C. M., and A. R. Hauser. 2004. Relative contributions of Pseudomonas aeruginosa ExoU, ExoS, and ExoT to virulence in the lung. Infect. Immun. 72:6969-6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Silver, D. R., I. L. Cohen, and P. F. Weinberg. 1992. Recurrent Pseudomonas aeruginosa pneumonia in an intensive care unit. Chest 101:194-198. [DOI] [PubMed] [Google Scholar]
- 53.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 54.Urbanowski, M. L., G. L. Lykken, and T. L. Yahr. 2005. A secreted regulatory protein couples transcription to the secretory activity of the Pseudomonas aeruginosa type III secretion system. Proc. Natl. Acad. Sci. USA 102:9930-9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vallis, A. J., T. L. Yahr, J. T. Barbieri, and D. W. Frank. 1999. Regulation of ExoS production and secretion by Pseudomonas aeruginosa in response to tissue culture conditions. Infect. Immun. 67:914-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vance, R. E., A. Rietsch, and J. J. Mekalanos. 2005. Role of the type III secreted exoenzymes S, T, and Y in systemic spread of Pseudomonas aeruginosa PAO1 in vivo. Infect. Immun. 73:1706-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Veenendaal, A. K., C. Sundin, and A. J. Blocker. 2009. Small-molecule type III secretion system inhibitors block assembly of the Shigella type III secreton. J. Bacteriol. 191:563-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Veesenmeyer, J. L., A. R. Hauser, T. Lisboa, and J. Rello. 2009. Pseudomonas aeruginosa virulence and therapy: evolving translational strategies. Crit. Care Med. 37:1777-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wolf, K., H. J. Betts, B. Chellas-Gery, S. Hower, C. N. Linton, and K. A. Fields. 2006. Treatment of Chlamydia trachomatis with a small molecule inhibitor of the Yersinia type III secretion system disrupts progression of the chlamydial developmental cycle. Mol. Microbiol. 61:1543-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang, J. H., T. D. Chung, and K. R. Oldenburg. 1999. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 4:67-73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.