Abstract
The yeast Candida albicans is an opportunistic human fungal pathogen and the cause of superficial and systemic infections in immunocompromised patients. The classes of antifungal agents most commonly used to treat Candida infections are the azoles, polyenes, and echinocandins. In the present study, we identified changes in C. albicans protein abundance using two-dimensional polyacrylamide gel electrophoresis and matrix-assisted laser desorption ionization-time of flight mass spectroscopy following exposure to representatives of the azole (ketoconazole), polyene (amphotericin B), and echinocandin (caspofungin) antifungals in an effort to elucidate the adaptive responses to these classes of antifungal agents. We identified 39 proteins whose abundance changed in response to ketoconazole exposure. Some of these proteins are involved in ergosterol biosynthesis and are associated with azole resistance. Exposure to amphotericin B altered the abundance of 43 proteins, including those associated with oxidative stress and osmotic tolerance. We identified 50 proteins whose abundance changed after exposure to caspofungin, including enzymes involved in cell wall biosynthesis and integrity, as well as the regulator of β-1,3-glucan synthase activity, Rho1p. Exposure to caspofungin also increased the abundance of the proteins involved in oxidative and osmotic stress. The common adaptive responses shared by all three antifungal agents included proteins involved in carbohydrate metabolism. Some of these antifungal-responsive proteins may represent potential targets for the development of novel therapeutics that could enhance the antifungal activities of these drugs.
Candida albicans is an opportunistic yeast and a harmless commensal organism within the endogenous microflora of many healthy individuals; however, in immunocompromised patients, this fungus is a major cause of infections ranging from superficial mycoses to invasive, deep-seated systemic candidiasis. C. albicans is considered the most common human fungal pathogen, and overall, Candida species are the fourth leading cause of bloodstream infections in the United States (10, 33). The most effective classes of antifungal agents used to treat Candida infections are the azoles, polyenes, and echinocandins. The azole antifungals, such as ketoconazole, fluconazole, and itraconazole, inhibit the biosynthesis of ergosterol, which is the major sterol within the fungal membrane. The azoles exert their activity by specifically binding to the heme group in the active site of the cytochrome P450 enzyme 14α-lanosterol demethylase (Erg11p). The inhibition of Erg11p results in insufficient amounts of ergosterol within the fungal membrane and the accumulation of the toxic sterol 14α-methylergosta-8, 24-(28)-dien-3β,6α-diol, which also inhibits fungal growth (1, 18). The polyenes, including amphotericin B, natamycin, and nystatin, are amphipathic antimycotics that target ergosterol within the fungal cell membrane. After the binding of ergosterol, these agents expand the lipid bilayer, forming channels through the membrane, which lead to the leakage of intracellular potassium and other molecules, resulting in fungal death. The echinocandins, such as caspofungin, anidulafungin, and micafungin, are acylated cyclic hexapeptides that exert their fungicidal activity by noncompetitively inhibiting β-1,3-glucan synthase, an enzyme required for cell wall biosynthesis. Inhibition of this protein results in the absence of normally required β-glucans in the fungal cell wall, causing cell death.
These classes of antifungal agents represent a limited arsenal of drugs used to combat Candida infections; moreover, these agents also have significant limitations that restrict their routine clinical use. The azole antifungals are fungistatic, are associated with drug-drug interactions, and are hindered by the emergence of azole-resistant species of C. albicans (46, 51). The polyene agents have adverse side effects, such as nephrotoxicity, renal failure, and infusion-related toxicity (2, 3, 47). The echinocandin antifungals are somewhat costly and have been associated with fever, thrombophlebitis, and hepatic toxicity (17, 34, 36). Although the echinocandins are effective against fluconazole-resistant strains of Candida, there have been cases of resistance to this class of drugs in C. albicans (26, 28). In the face of these limitations, the identification of novel antifungal targets and the development of new antifungal agents are warranted for the effective treatment of Candida infections.
In a previous study, we used genome-wide gene expression profiling by microarray analysis to identify the changes in C. albicans gene expression in response to clinically available representatives of the polyene, azole, echinocandin, and pyrimidine classes of antifungal agents (24). That study revealed that 82, 256, and 480 genes were differentially expressed following exposure to ketoconazole, amphotericin B, and caspofungin, respectively. Briefly, ketoconazole exposure induced the expression of genes associated with ergosterol biosynthesis and azole resistance, while amphotericin B caused alterations in the expression of genes involved in small-molecule transport and oxidative stress response. Exposure to caspofungin was observed to induce the expression of genes associated with cell wall biosynthesis and maintenance, including the gene encoding the (1,3)-β-glucan synthase, GSL1.
Proteomic analysis has been applied to the study of many aspects of C. albicans, including its adaptive responses to salt, cadmium, peroxide, oxidative stress, and macrophage exposure (11, 21, 50). Proteomic analysis has also been used to study the adaptive response of C. albicans to fluconazole, itraconazole, and mulundocandin (5, 49) and to identify changes in protein abundance in matched sets of azole-susceptible and -resistant clinical isolates of C. albicans (15, 20). In the present study, we used the combination of two-dimensional polyacrylamide gel electrophoresis (2-D PAGE) and matrix-assisted laser desorption ionization-time of flight mass spectroscopy (MALDI-TOF MS) to identify the changes in protein abundance in a wild-type strain of C. albicans after exposure to ketoconazole, amphotericin B, and caspofungin. We used the same conditions that we used in our previous microarray study, which allow the direct comparison of the transcriptome and the proteome representing these adaptive responses (22). Our study also allows a comparative analysis of these three commonly used classes of antifungal agents. We report here the changes in protein abundance associated with both specific and nonspecific adaptive responses to these antifungal agents in this pathogenic fungus.
MATERIALS AND METHODS
Antifungal agents and chemicals.
Ketoconazole was purchased from Sigma Aldrich Chemical Company (St. Louis, MO), and amphotericin B was obtained from MP Biomedicals (Aurora, OH). A preparation of caspofungin acetate for injection (Cancidas; Merck) was obtained from the pharmacy at Le Bonheur Children's Medical Center. Stock solutions (10 mg/ml) of ketoconazole and amphotericin B were prepared in dimethyl sulfoxide (DMSO; Sigma), while stocks of caspofungin were made in sterile water. All antifungal stock solutions were prepared fresh before all experiments, and all other chemicals and reagents were obtained from Bio-Rad Laboratories (Hercules, CA), unless otherwise specified.
Organism and culture conditions.
C. albicans wild-type strain SC5314 was cultured overnight in 10 ml of synthetic dextrose (SD) minimal medium containing 165 mM morpholinepropanesulfonic acid, 0.67% (wt/vol) yeast nitrogen base without amino acids, and 2% (wt/vol) dextrose at pH 7.0 in a temperature-controlled incubator at 30°C. For each sample, cells were diluted into 500 ml of fresh SD medium to an optical density (OD) of 0.005 and were subsequently grown at 30°C until an OD of 1.0 was reached. The cultures were then diluted with fresh SD medium to an OD of 0.1. After growth to an OD of 0.2, the antifungal agents were added to the cultures at concentrations equivalent to their 50% inhibitory concentrations (ketoconazole, 19.13 μg/ml; amphotericin B, 0.029 μg/ml; caspofungin, 0.0075 μg/ml) and the cultures were incubated at 30°C with shaking at 250 rpm in a temperature-controlled incubator for 6 h. Due to the photosensitivity of amphotericin B, all cultures with amphotericin B were incubated in the dark. After the 6-h incubation, the ODs were measured to ensure that the antifungals used in the drug-treated cultures inhibited at least half of the fungal growth compared to the growth achieved by the DMSO-treated controls. All drug exposure experiments were conducted in triplicate.
Preparation of protein extracts.
C. albicans cells were collected by centrifugation. The pelleted cells were washed three times with ultrapure water and were broken in cold 50 mM Tris buffer (pH 7.4) containing 1 mM EDTA with 0.5-mm-diameter glass beads with a Mini-Bead Beater 8 apparatus (BioSpec Products, Inc., Bartlesville, OK). Briefly, an equal volume of glass beads was added to the cell pellet in 2-ml conical bottom tubes and the samples were beaten for six 20-s bursts. Between bursts, the samples were incubated on ice for 1 min. After disruption, the samples were centrifuged at 20,000 × g for 15 min at 4°C to remove the glass beads, unbroken cells, and particulate debris from the homogenate. After centrifugation, the soluble protein fraction was collected, the protein content was measured by the bicinchoninic acid (BCA) method (41), and the proteins were stored at −80.0°C for further analysis.
2-D PAGE.
For isoelectric focusing (IEF), samples containing 300 μg of protein were dissolved in 300 μl rehydration buffer consisting of 7 M urea, 2 M thiourea, 4% (wt/vol) 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS), 50 mM dithiothreitol (DTT), 0.2% (vol/vol) Biolyte 3-10 ampholytes, and 0.001% (wt/vol) bromophenol blue. The samples were then added to 17-cm ReadyStrip immobilized pH gradient (IPG) strips (Bio-Rad) and run on a Protean IEF cell (Bio-Rad) at 20.0°C. The protein samples were subjected to passive rehydration for 12 h and then run at 250 V for the first 15 min, followed by a linear increase in voltage to a maximum of 10,000 V, at which point the strips were run for an additional 5 h. After IEF, the IPG strips were removed and reduced in equilibration solution consisting of 1.5 M Tris-HCl (pH 8.8), 7 M urea, 2% (wt/vol) sodium dodecyl sulfate (SDS), 20% (vol/vol) glycerol, and 1% (wt/vol) DTT for 10 min at 25°C. The IPG strips were then alkylated in equilibration buffer containing 1.5 M Tris-HCl (pH 8.8), 7 M urea, 2% (wt/vol) SDS, 20% (vol/vol) glycerol, and 1.5% (wt/vol) iodoacetamide for 10 min at 25°C. The proteins were subsequently separated in the second dimension on the basis of their molecular masses by using 12% polyacrylamide gels containing 2.6% N,N′-methylenebisacrylamide and run at a constant 200 V for 6 to 7 h at 15°C in a Protean Plus Dodeca cell (Bio-Rad). After electrophoresis, the gels were fixed and stained with SYPRO ruby.
Gel imaging and analysis software.
The 2-D gels were imaged on an FX imager (Bio-Rad) at the medium SYPRO ruby intensity setting at a resolution of 300 dots per inch. Spot comparison of 2-D gel images was performed with PDQuest software (version 7.1; Bio-Rad) to measure the differences in spot intensity. Protein spots between gel images were compared by using an automated matching program, and any mismatched spots were corrected by use of a manual matching feature in the software. After removal of the background, the individual spot volume for each resolved protein spot was normalized against the total spot volume and area in the gel. Protein spots that were consistently differentially expressed in the mean normalized spot volume by at least 1.5-fold in three independent experiments were selected for excision and subsequent identification. Statistical analysis was performed by Student's t test.
Protein trypsinization and extraction.
Protein spots were excised from the gels and transferred to 1.5-ml microcentrifuge tubes prewashed three times with 50% methanol-10% glacial acetic acid. The proteins were then destained twice in 200 μl of 50% acetonitrile at 23°C and vortexed for 30 min. The samples were placed in a 96-well Millipore polytetrafluoroethylene 0.45-μm-pore-size filter plate; the wells had been pretreated three times with 20% methanol-0.1% trifluoroacetic acid (TFA). The protein samples were washed with acetonitrile; subsequently digested in a 100-μl aliquot of fresh digestion buffer containing 12.5 ng/μl ultrapure sequencing-grade trypsin (Promega, Madison, WI), 25 mM ammonium bicarbonate, and 10% acetonitrile; and incubated at 37°C overnight. The tryptic polypeptides were extracted in a 25-μl aliquot of fresh extraction solution containing 50% acetonitrile and 5% TFA.
Protein identification and database search.
Protein identification was carried out by the methods described by Cummings et al. (7). Briefly, MS spectra were recorded on an Ultraflex matrix-assisted laser desorption ionization reflecting time of flight (MALDI-ToF/ToF) mass spectrometer (Bruker Daltonics, Bremen, Germany). The autolytic trypsin fragment peaks of m/z 842.509 and m/z 2,211.104 were used for internal calibration of the peptide mass spectra with mass resolutions of between m/z 2,500 and 8,000. External calibration of this instrument was carried out with a 2:1 mixture of des-R-bradykinin and adrenocorticotropin (Sigma). Mass spectra parameter settings were 20 kV for the extraction voltage and 11.8 kV for the reflector voltage. Both FlexControl software (version 2.0) and FlexAnalysis software (version 2.0) were used to record the mass spectra data and sample analysis, respectively. For each sample, the signal-to-noise ratio was calculated by using the SNAP algorithm and the default settings within the FlexAnalysis software. For each protein identified, MALDI-TOF mass fingerprint data were used to perform a BLAST search of the sequences against those in a custom database (http://genolist.pasteur.fr/CandidaDB/) containing known C. albicans open reading frame (ORF) DNA sequences by using PROWL software. For each protein identified, a Z-score value was calculated from the software that measured the significance of the identification. Z-score values of 1.20 and 1.65 ranked the mass data search to 90 and 95% levels of confidence of nonrandom matches to the specific ORF, respectively.
RESULTS
Changes in protein abundance in response to ketoconazole exposure.
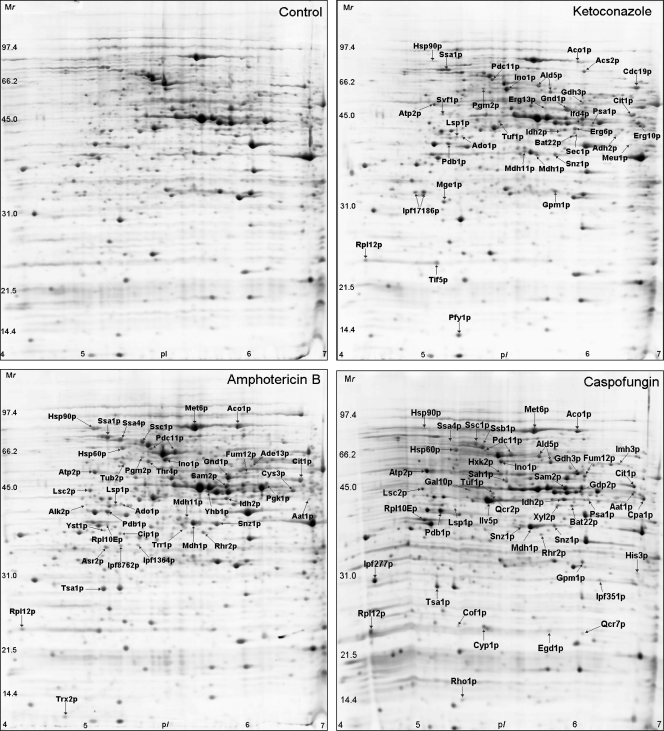
A total of 398 proteins were resolved by 2-D PAGE after exposure to ketoconazole. Among them, 116 proteins were found to be changed in abundance and 39 were identified. Of those 39 proteins, the abundance of 32 proteins was increased, while the abundance of 7 was decreased compared to the abundance for the DMSO-treated controls. The ketoconazole-responsive proteins are shown in Fig. 1. Descriptions of these proteins, including their putative biological function, molecular properties, and changes in relative levels of expressions, are displayed in Table 1. The proteins in the largest category that responded to ketoconazole exposure are involved in carbohydrate metabolism and included enzymes that function in the tricarboxylic acid (TCA) cycle (Aco1p, Cit1p, Idh2p, Mdh1p, and Mdh11p), glycolysis and gluconeogenesis (Cdc19p, Gpm1p, Pgm2p), the pentose phosphate pathway (Gnd1p), and pyruvate transformation (Pdb1p and Pdc11p). Other proteins shown to be responsive to this drug are involved in amino acid biosynthesis (Bat22p, Gdh3p, and His1p), cell stress and heat shock (Snz1p, Ssa1p, and Hsp90p), and protein biosynthesis (Tif5p and Tuf1p). Of particular interest are the enzymes involved in ergosterol biosynthesis, including Erg6p, Erg10p, and Erg13p, as well as proteins that have previously been identified as being associated with azole resistance, such as Csh1p and Orf19.251.
FIG. 1.
2-D expression profiles of the soluble proteins from C. albicans wild-type strain SC5314: untreated control, ketoconazole-treated, amphotericin B-treated, and caspofungin-treated cells after staining of the gels with SYPRO ruby.
TABLE 1.
Description of the ketoconazole-responsive proteins identified by MALDI-TOF mass spectroscopy
| Protein | CGDb systematic name | Description | pI | Mr (103) | Z score | Protein coverage (%) | No. of matched peptides | Fold change in expressionc | t test |
|---|---|---|---|---|---|---|---|---|---|
| Aco1p | Orf19.6385 | Aconitate hydrotase | 6.0 | 84.2 | 1.7 | 21 | 33 | 2.5 ± 1.4 | 0.02 |
| Acs2p | Orf19.1064 | Acetyl-CoA synthetase | 5.7 | 73.8 | 1.6 | 11 | 6 | 1.6 ± 0.1 | 0.04 |
| Adh2pa | Orf19.5113 | Aldehyde dehydrogenase I | 6.3 | 36.8 | 2.3 | 26 | 8 | 3.8 ± 0.1 | 0.00 |
| Ado1pa | Orf19.5591 | Adenosine kinase | 5 | 38.2 | 2.4 | 47 | 14 | 2.7 ± 1.1 | 0.04 |
| Ald5p | Orf19.5806 | Aldehyde dehydrogenase | 5.5 | 54.0 | 2.3 | 49 | 16 | 1.9 ± 0.8 | 0.02 |
| Ald5p | Orf19.5806 | Aldehyde dehydrogenase | 5.5 | 54.0 | 2.4 | 30 | 11 | 1.6 ± 0.2 | 0.02 |
| Atp2p | Orf19.5653 | F1F0 ATPase complex, β subunit | 4.9 | 55.7 | 2.4 | 48 | 14 | 1.5 ± 0 | 0.00 |
| Cdc19p | Orf19.3575 | Pyruvate kinase | 6.5 | 55.4 | 2.3 | 26 | 10 | 1.9 ± 0.5 | 0.02 |
| Cit1p | Orf19.4393 | Citrate synthase | 6.3 | 48.7 | 2.4 | 30 | 12 | 2 ± 0.3 | 0.00 |
| Erg6pa | Orf19.1631 | Sterol transmethylase | 5.7 | 43.1 | 2.3 | 24 | 8 | 4.3 ± 0.4 | 0.00 |
| Erg10pa | Orf19.1591 | Acetyl-CoA acetyltransferase | 6.5 | 41.9 | 1.7 | 14 | 5 | 2.4 ± 0.7 | 0.00 |
| Erg13p | Orf19.7312 | 3-Hydroxy-3-methylglutaryl-CoA synthase | 5.7 | 49.7 | 2.4 | 42 | 16 | 1.6 ± 0.2 | 0.03 |
| Gnd1p | Orf19.5024 | 6-Phosphogluconate dehydrogenase | 6.1 | 56.9 | 2.3 | 27 | 11 | 1.6 ± 0.5 | 0.10 |
| Idh2p | Orf19.5791 | Isocitrate dehydrogenase | 6.4 | 39.3 | 2.3 | 45 | 12 | 1.5 ± 0 | 0.00 |
| Ino1p | Orf19.7585 | Myoinositol-1-phosphate synthase | 5.3 | 57.8 | 2.3 | 44 | 15 | 2.6 ± 0.4 | 0.00 |
| Csh1pa | Orf19.4477 | Putative aryl-alcohol dehydrogenase | 6.0 | 38.3 | 2.0 | 31 | 10 | 2.4 ± 0.9 | 0.08 |
| Svf1p | Orf19.6068 | Unknown function | 4.9 | 42.9 | 2.3 | 20 | 7 | 1.6 ± 0.2 | 0.02 |
| Ipf17186p | Orf19.251 | Heat shock protein 31 of DJ-1/Pfpl family | 4.7 | 25.8 | 2.3 | 30 | 7 | 1.9 ± 0.1 | 0.00 |
| Ipf17186p | Orf19.251 | Heat shock protein 31 of DJ-1/Pfpl family | 4.7 | 25.8 | 2.2 | 21 | 7 | 3.1 ± 0.3 | 0.00 |
| Lsp1p | Orf19.3149 | Unknown function | 4.9 | 35.6 | 1.4 | 33 | 7 | 2.2 ± 0.2 | 0.02 |
| Meu1p | Orf19.6938 | Regulator of ADH2 expression | 7.2 | 37.6 | 1.9 | 15 | 5 | 2.9 ± 0.7 | 0.02 |
| Mdh11p | Orf19.7481 | Malate dehydrogenase | 5.4 | 36.0 | 1.5 | 24 | 5 | 3.2 ± 1.0 | 0.01 |
| Mdh1pa | Orf19.7481 | Mitochondrial malate dehydrogenase precursor | 5.7 | 34.7 | 2.3 | 37 | 8 | 1.6 ± 0.3 | 0.03 |
| Mge1p | Orf19.2524 | Heat shock protein | 5.6 | 27.2 | 2.4 | 31 | 9 | 3.8 ± 0.1 | 0.00 |
| Pdb1p | Orf19.5294 | Pyruvate dehydrogenase | 5.4 | 41.3 | 2.3 | 30 | 8 | 1.7 ± 0.1 | 0.01 |
| Pfy1p | Orf19.5076 | Actin-binding protein | 5.3 | 13.8 | 2.4 | 51 | 6 | 1.8 ± 0.2 | 0.02 |
| Pgm2p | Orf19.2841 | Phosphoglucomutase | 5.3 | 61.8 | 2.4 | 29 | 13 | 1.9 ± 0.5 | 0.06 |
| Rpl12p | Orf19.1635 | Ribosomal protein | 9.5 | 17.7 | 2.0 | 30 | 6 | 1.7 ± 0.1 | 0.03 |
| Sec1p | Orf19.13833 | Transport protein | 8.1 | 89.6 | 1.3 | 17 | 8 | 2.6 ± 0.5 | 0.03 |
| Snz1p | Orf19.2947 | Stationary-phase protein | 5.8 | 31.8 | 1.7 | 15 | 6 | 1.5 ± 0 | 0.00 |
| Tif5p | Orf19.4261 | Translation initiation factor (eIF-5A) | 5.0 | 16.4 | 1.9 | 33 | 5 | 1.6 ± 0.4 | 0.08 |
| Tuf1pa | Orf19.6047 | Translation elongation factor TU | 5.7 | 46.7 | 2.0 | 24 | 7 | 1.5 ± 0.2 | 0.03 |
| Bat22p | Orf19.6994 | Branched amino acid aminotransferase | 5.9 | 40.8 | 2.1 | 27 | 10 | 0.4 ± 0 | 0.01 |
| Gdh3p | Orf19.4716 | NADP-glutamate dehydrogenase | 5.7 | 49.6 | 2.4 | 23 | 9 | 0.3 ± 0 | 0.01 |
| Gpm1p | Orf19.903 | Phosphoglycerate mutase | 5.8 | 27.4 | 2.3 | 40 | 7 | 0.4 ± 0 | 0.00 |
| Hsp90p | Orf19.6515 | Heat shock protein | 4.8 | 80.8 | 1.8 | 21 | 14 | 0.3 ± 0.1 | 0.05 |
| Pdc11p | Orf19.2877 | Pyruvate decarboxylase | 5.4 | 62.4 | 2.3 | 30 | 14 | 0.5 ± 0.1 | 0.00 |
| Psa1p | Orf19.4943 | GDP-mannose pyrophosphorylase | 5.9 | 40.0 | 2.3 | 25 | 8 | 0.2 ± 0 | 0.00 |
| Ssa1p | Orf19.1065 | Heat shock protein of HSP family | 4.9 | 70.1 | 2.4 | 21 | 12 | 0.5 ± 0.2 | 0.08 |
Proteins whose genes were previously shown to be differentially expressed by microarray analysis.
CGD, Candida Genome Database.
Change in expression between treated cells and control cells.
Changes in protein abundance in response to amphotericin B exposure.
A total of 391 proteins were resolved by 2-D PAGE after exposure to amphotericin B. Among them, 60 were found to be changed in abundance and 43 were identified. Of those 43 proteins, the abundance of 39 proteins was increased, while the expression of 4 was decreased. These amphotericin B-responsive proteins are displayed in Fig. 1; and descriptions of these proteins, including their putative biological function, molecular properties, and changes in relative levels of expression, are shown in Table 2. The proteins in the largest class responsive to amphotericin B are involved in cell stress and heat shock (Snz1p, Ssa1p, Ssa4p, Ssb1p, Ssc1p, Hsp60p, and Hsp90p). Other proteins responsive to this drug are involved in the TCA cycle (Aco1p, Idh2p, Fum12p, Lsc2p, Mdh1p, and Mdh11p), amino acid biosynthesis (Aat1p, Met6p, Sam2p, Thr4p, and Cys3p), and glycolysis and gluconeogenesis (Pdb1p, Pdc11p, and Gnd1p); and some of these proteins had unknown functions (Ipf1364p, Ipf8762p, and Ipf19154p). Also identified were enzymes involved in oxidative stress (Cip1p, Trr1p, Trx2p, Tsa1p, and Yhb1p) and other proteins associated with osmotic tolerance (Asr2p and Rhr2p).
TABLE 2.
Description of the amphotericin B-responsive proteins identified by MALDI-TOF mass spectroscopy
| Protein | CGDb systematic name | Description of gene product | pI | Mr (103) | Z score | Protein coverage (%) | No. of matched peptides | Fold change in expressionc | t test |
|---|---|---|---|---|---|---|---|---|---|
| Aco1pa | Orf19.6385 | Aconitate hydratase | 6.3 | 84.2 | 2.4 | 19 | 10 | 2.9 ± 0.7 | 0.04 |
| Ado1p | Orf19.5591 | Adenosine kinase | 5.0 | 38.2 | 2.3 | 35 | 8 | 4.5 ± 3.8 | 0.06 |
| Alk2p | Orf19.7513 | N-Alkane inducible cytochrome P450 | 6.4 | 59.8 | 1.3 | 18 | 7 | 2.0 ± 0.2 | 0.01 |
| Asr2p | Orf19.7284 | Unknown function | 5.4 | 27.8 | 2.2 | 29 | 8 | 7.6 ± 4.1 | 0.00 |
| Atp2p | Orf19.5653 | F1F0 ATPase complex, β subunit | 4.9 | 55.7 | 2.3 | 45 | 16 | 2.7 ± 1.2 | 0.07 |
| Cip1p | Orf19.113 | Cadmium-induced protein | 5.1 | 33.0 | 1.9 | 20 | 5 | 4.7 ± 2.0 | 0.00 |
| Cit1p | Orf19.4393 | Citrate synthase | 6.3 | 48.7 | 1.4 | 17 | 6 | 2.1 ± 0.5 | 0.01 |
| Cys3pa | Orf19.6402 | Cystathionine gamma-lyase | 6.2 | 42.9 | 1.7 | 17 | 4 | 5.4 ± 4.7 | 0.00 |
| Fum12p | Orf19.6724 | Fumarate hydratase, 5′ end | 7.8 | 29.6 | 2.2 | 35 | 8 | 5.9 ± 4.9 | 0.01 |
| Gnd1p | Orf19.5024 | 6-Phosphogluconate dehydrogenase | 6.1 | 56.9 | 2.3 | 27 | 13 | 1.9 ± 0.2 | 0.01 |
| Hsp60pa | Orf19.717 | Heat shock protein 60 | 5.2 | 60.1 | 2.3 | 25 | 14 | 1.6 ± 0.2 | 0.00 |
| Hsp90p | Orf19.6515 | Heat shock protein | 4.8 | 80.8 | 1.8 | 21 | 14 | 1.8 ± 0.2 | 0.10 |
| Idh2p | Orf19.5791 | Isocitrate dehydrogenase | 6.4 | 39.3 | 2.2 | 38 | 8 | 1.8 ± 0.5 | 0.01 |
| Ino1p | Orf19.7585 | Myoinositol-1-phosphate synthase | 5.3 | 57.8 | 2.4 | 44 | 15 | 2.8 ± 1.7 | 0.00 |
| Ipf1364p | Orf19.6435 | Unknown function | 10.1 | 14.7 | 1.5 | 45 | 5 | 2.3 ± 0.2 | 0.01 |
| Ipf8762p | Orf19.822 | Unknown function | 5.2 | 21.5 | 1.4 | 19 | 5 | 2.7 ± 0.9 | 0.00 |
| Lsc2p | Orf19.1860 | Succinate-CoA ligase, β subunit 3′ end | 4.8 | 22.1 | 1.3 | 27 | 5 | 2.1 ± 0.2 | 0.01 |
| Lsp1p | Orf19.3149 | Unknown function | 4.9 | 35.6 | 1.4 | 33 | 7 | 2.1 ± 0.5 | 0.00 |
| Mdh1p | Orf19.7481 | Mitochondrial malate dehydrogenase precursor | 5.7 | 34.7 | 2.2 | 33 | 8 | 1.8 ± 0.3 | 0.00 |
| Mdh11p | Orf19.7481 | Malate dehydrogenase | 5.4 | 36.0 | 1.5 | 24 | 5 | 1.8 ± 0.8 | 0.00 |
| Pdb1p | Orf19.5294 | Pyruvate dehydrogenase | 5.4 | 41.3 | 2.3 | 30 | 8 | 1.7 ± 0.1 | 0.05 |
| Pdc11p | Orf19.2877 | Pyruvate decarboxylase | 5.4 | 62.4 | 2.3 | 28 | 10 | 1.6 ± 0.9 | 0.18 |
| Pgk1p | Orf19.3651 | Phosphoglycerate kinase | 6.1 | 45.2 | 2.4 | 51 | 20 | 3.9 ± 3.4 | 0.00 |
| Pgm2p | Orf19.2841 | Phosphoglucomutase | 5.3 | 61.8 | 2.3 | 28 | 13 | 1.7 ± 0.3 | 0.03 |
| Rhr2p | Orf19.5437 | dl-Glycerol phosphatase | 5.5 | 28.1 | 1.8 | 25 | 5 | 3.6 ± 1.1 | 0.03 |
| Rpl10e1p | Orf19.7015 | Ribosomal protein L10 | 4.7 | 33.3 | 2.4 | 36 | 12 | 1.7 ± 0.2 | 0.02 |
| Rpl12p | Orf19.1635 | Ribosomal protein | 9.5 | 17.7 | 2.0 | 30 | 6 | 1.6 ± 0.5 | 0.03 |
| Sam2p | Orf19.657 | S-Adenosylmethionine synthetase | 5.6 | 42.2 | 1.7 | 23 | 8 | 1.7 ± 0.8 | 0.17 |
| Ssa1p | Orf19.1065 | Heat shock protein of HSP family | 4.9 | 70.1 | 2.4 | 18 | 10 | 1.6 ± 0.5 | 0.05 |
| Ssa4pa | Orf19.4980 | HSP70 mRNA for heat shock protein | 5.1 | 70.3 | 2.4 | 27 | 16 | 1.5 ± 0.3 | 0.25 |
| Ssc1p | Orf19.1896 | Mitochondrial heat shock protein 70 | 5.5 | 69.7 | 2.4 | 29 | 19 | 1.8 ± 0.1 | 0.03 |
| Snz1pa | Orf19.2947 | Stationary-phase protein | 5.8 | 31.8 | 2.0 | 15 | 5 | 2.3 ± 0.5 | 0.00 |
| Thr4p | Orf19.4233 | Threonine synthase | 5.2 | 57.7 | 2.4 | 28 | 10 | 1.6 ± 0.2 | 0.02 |
| Trr1p | Orf19.4290 | Thioredoxin reductase | 5.4 | 34.7 | 2.1 | 31 | 9 | 1.6 ± 0.1 | 0.00 |
| Trx2p | Orf19.1976 | Thioredoxin | 4.9 | 13.71 | 1.2 | 33 | 3 | 1.7 ± 0.2 | 0.00 |
| Tsa1p | Orf19.7417 | Thioredoxin peroxidase | 5.0 | 21.8 | 2.4 | 33 | 6 | 2.5 ± 0.1 | 0.00 |
| Tub2p | Orf19.6034 | β-Tubulin | 4.6 | 48.77 | 2.3 | 26 | 12 | 4.0 ± 2.8 | 0.01 |
| Yhb1pa | Orf19.3707 | Flavohemoglobin | 5.6 | 45.93 | 1.5 | 31 | 10 | 2.8 ± 1.4 | 0.10 |
| Yst1p | Orf19.6975 | Ribosomal protein, exon 2 | 4.8 | 25.6 | 2.1 | 25 | 6 | 5.2 ± 2.8 | 0.01 |
| Aat1p | Orf19.3554 | Aspartate aminotransferase | 8.7 | 48.9 | 2.0 | 22 | 7 | 0.4 ± 0.4 | 0.00 |
| Ade13p | Orf19.3870 | Adenylosuccinate lyase | 5.9 | 54.4 | 2.0 | 21 | 8 | 0.5 ± 0.1 | 0.00 |
| Met6pa | Orf19.2551 | 5-Methyltetrahydropteroyltriglutamate-homocysteine methyltransferase | 5.4 | 85.7 | 2.4 | 29 | 13 | 0.5 ± 0.2 | 0.02 |
| Pdc11p | Orf19.2877 | Pyruvate decarboxylase | 5.4 | 62.4 | 2.4 | 43 | 21 | 0.1 ± 0.0 | 0.01 |
Proteins whose genes were previously shown to be differentially expressed by microarray analysis.
CGD, Candida Genome Database.
Change in expression between treated cells and control cells.
Changes in protein abundance in response to caspofungin exposure.
A total of 264 proteins were resolved by 2-D PAGE after exposure to caspofungin. Among them, 78 were found to be changed in abundance and 50 were identified. Of those 50 proteins, the abundance of 32 proteins was increased, while the abundance of 18 was downregulated. The caspofungin-responsive proteins are labeled in Fig. 1; and descriptions of these proteins, including their putative biological functions, molecular properties, and changes in relative levels of expression are provided in Table 3. The proteins in the largest class responding to caspofungin exposure are involved in glycolysis and gluconeogenesis (Cdc19p, Gnd1p, Pdb1p, Pgk1p, Pgm2p, Pdc11p, Fba1p, and Hxk2p), while other proteins shown to be responsive to this drug are involved in cell stress and heat shock (Snz1p, Ssa1p, Ssa4p, Ssb1p, Ssc1p, and Hsp90p), the TCA cycle (Aco1p, Cit1p, Idh2p, Mdh1p and Mdh11p) and amino acid biosynthesis (Bat22p, Hom2p, Gdh3p, and Sah1p). Also identified were the enzymes involved in cell wall biosynthesis, such as Gal10p, Psa1p, and the regulator of β-1,3-glucan synthase activity, Rho1p.
TABLE 3.
Description of the caspofungin-responsive proteins identified by MALDI-TOF mass spectroscopy
| Protein | CGDb Systematic name | Description | pI | Mr (103) | Z score | Protein coverage (%) | No. of matched peptides | Fold change in expressionc | t test |
|---|---|---|---|---|---|---|---|---|---|
| Aco1pa | Orf19.6385 | Aconitate hydratase | 6.0 | 84.2 | 1.7 | 19 | 10 | 3.5 ± 2.4 | 0.04 |
| Ald5p | Orf19.5806 | Aldehyde dehydrogenase | 5.5 | 54.0 | 2.3 | 38 | 14 | 1.5 ± 0.3 | 0.04 |
| Ald5p | Orf19.5806 | Aldehyde dehydrogenase | 5.5 | 54.0 | 2.3 | 29 | 10 | 1.6 ± 0.2 | 0.04 |
| Atp2p | Orf19.5653 | F1F0 ATPase complex, β subunit | 4.9 | 55.7 | 2.4 | 32 | 10 | 4.0 ± 0.2 | 0.02 |
| Cit1p | Orf19.4393 | Citrate synthase | 6.3 | 48.7 | 2.2 | 17 | 7 | 1.8 ± 0.5 | 0.05 |
| Cpa1p | Orf19.4630 | Arginine-specific carbamoylphosphate synthase, small chain | 6.5 | 47.3 | 1.3 | 12 | 4 | 2.5 ± 0.4 | 0.01 |
| Fum12p | Orf19.6724 | Fumarate hydratase, 5′ end | 7.8 | 29.6 | 2.2 | 35 | 8 | 3.2 ± 1.5 | 0.01 |
| Gpd2p | Orf19.691 | Glycerol 3-phosphate dehydrogenase | 5.2 | 40.8 | 1.3 | 19 | 5 | 1.7 ± 0.3 | 0.02 |
| Gdh3p | Orf19.4716 | NADP-glutamate dehydrogenase | 5.7 | 49.6 | 2.4 | 23 | 9 | 1.7 ± 0.4 | 0.01 |
| Hsp60p | Orf19.717 | Heat shock protein 60 | 5.2 | 60.1 | 2.0 | 24 | 11 | 2.4 ± 0.2 | 0.03 |
| Idh2p | Orf19.5791 | Isocitrate dehydrogenase | 6.4 | 39.3 | 2.4 | 57 | 16 | 1.9 ± 0.7 | 0.09 |
| Ilv5p | Orf19.88 | Keto acid reducto-isomerase | 6.2 | 44.8 | 2.4 | 45 | 16 | 1.5 ± 0.8 | 0.19 |
| Ino1p | Orf19.7585 | Myoinositol-1-phosphate synthase | 5.3 | 57.8 | 2.3 | 44 | 15 | 1.6 ± 0.2 | 0.02 |
| Ipf277p | Orf19.3268 | Human IgE-dependent histamine-releasing factor homolog | 4.3 | 18.5 | 2.1 | 33 | 5 | 1.6 ± 0.3 | 0.01 |
| Ipf351p | Orf19.7531 | Unknown function | 7.9 | 25.2 | 2.4 | 39 | 8 | 2.5 ± 1.3 | 0.10 |
| Lsc2p | Orf19.1860 | Succinate-CoA ligase, β subunit 3′ end | 4.8 | 22.1 | 1.5 | 29 | 4 | 1.5 ± 0.1 | 0.01 |
| Mdh1p | Orf19.7481 | Mitochondrial malate dehydrogenase precursor | 5.7 | 34.7 | 1.8 | 20 | 5 | 1.7 ± 0.6 | 0.11 |
| Pdb1p | Orf19.5294 | Pyruvate dehydrogenase | 5.4 | 41.3 | 2.3 | 32 | 9 | 2.7 ± 0.3 | 0.01 |
| Pdc11pa | Orf19.2877 | Pyruvate decarboxylase | 5.4 | 62.4 | 2.3 | 28 | 10 | 2.1 ± 0.3 | 0.00 |
| Qcr2p | Orf19.2644 | Ubiquinol cytochrome c reductase, chain II | 5.5 | 39.5 | 2.1 | 29 | 6 | 2.3 ± 0.4 | 0.03 |
| Qcr7p | Orf19.5629 | Ubiquinol cytochrome c reductase, subunit 7 | 5.7 | 14.4 | 2.3 | 38 | 7 | 28.3 ± 6.7 | 0.01 |
| Rho1p | Orf19.2843 | GTP-binding protein of the RHO subfamily | 5.3 | 9.3 | 2.4 | 24 | 4 | 1.5 ± 0.6 | 0.22 |
| Rhr2p | Orf19.5437 | dl-Glycerol phosphatase | 5.5 | 28.1 | 1.3 | 26 | 4 | 1.9 ± 0.1 | 0.00 |
| Rpl10ep | Orf19.7015 | Ribosomal protein L10 | 4.7 | 33.3 | 2.3 | 19 | 5 | 1.8 ± 0.4 | 0.01 |
| Rpl12p | Orf19.1635 | Ribosomal protein | 9.5 | 17.7 | 2.0 | 30 | 6 | 5.4 ± 0.7 | 0.02 |
| Sam2p | Orf19.657 | S-Adenosylmethionine synthetase | 5.6 | 42.2 | 1.5 | 29 | 8 | 1.6 ± 0.6 | 0.12 |
| Snz1p | Orf19.2947 | Stationary-phase protein | 5.8 | 31.8 | 1.7 | 15 | 6 | 2.9 ± 0.7 | 0.05 |
| Snz1p | Orf19.2947 | Stationary-phase protein | 5.8 | 31.8 | 2.3 | 24 | 7 | 1.7 ± 0.5 | 0.02 |
| Ssc1p | Orf19.1896 | Mitochondrial heat shock protein 70 | 5.5 | 69.7 | 2.4 | 41 | 28 | 2.7 ± 0.8 | 0.04 |
| Tsa1p | Orf19.7417 | Thioredoxin peroxidase | 5.0 | 21.8 | 2.3 | 40 | 7 | 2.6 ± 0.9 | 0.05 |
| Tuf1p | Orf19.6047 | Translation elongation factor TU | 5.7 | 46.7 | 2.0 | 29 | 9 | 2.3 ± 0.4 | 0.03 |
| Xyl2p | Orf19.7676 | d-Xylulose reductase | 5.6 | 38.8 | 1.8 | 17 | 5 | 1.6 ± 0.1 | 0.00 |
| Aat1pa | Orf19.3554 | Aspartate aminotransferase | 8.7 | 48.87 | 2.4 | 24 | 11 | 0.3 ± 0.1 | 0.00 |
| Bat22pa | Orf19.6994 | Branched amino acid aminotransferase | 5.9 | 40.8 | 1.8 | 20 | 6 | 0.5 ± 0.1 | 0.00 |
| Cof1p | Orf19.953.1 | Cofilin | 4.9 | 16.3 | 2.2 | 65 | 8 | 0.4 ± 0 | 0.00 |
| Cyp1pa | Orf19.6472 | Cyclophilin peptidylprolyl isomerase | 7.9 | 17.7 | 1.8 | 28 | 4 | 0.4 ± 0 | 0.00 |
| Egd1pa | Orf19.1154 | GAL4 DNA-binding enhancer protein | 5.5 | 17.0 | 2.4 | 66 | 7 | 0.4 ± 0.3 | 0.03 |
| Gpm1pa | Orf19.903 | Phosphoglycerate mutase | 5.8 | 27.4 | 1.7 | 28 | 5 | 0.3 ± 0.1 | 0.00 |
| His3p | Orf19.183 | Imidazole glycerol phosphate | 6.3 | 24.1 | 2.0 | 15 | 5 | 0.2 ± 0.1 | 0.00 |
| Hsp90pa | Orf19.6515 | Heat shock protein | 4.8 | 80.8 | 1.8 | 21 | 14 | 0.1 ± 0.1 | 0.01 |
| Hxk2p | Orf19.542 | Hexokinase II, 3′ end | 5.1 | 47.5 | 2.3 | 18 | 10 | 0.3 ± 0.1 | 0.06 |
Proteins whose genes were previously shown to be differentially expressed by microarray analysis.
CGD, Candida Genome Database.
Change in expression between treated cells and control cells.
DISCUSSION
In the present study, we used a combination of 2-D PAGE and MALDI-TOF mass spectroscopy to elucidate the specific and nonspecific adaptive responses to ketoconazole, amphotericin B, and caspofungin in C. albicans. Using this approach, we identified a total of 132 proteins corresponding to 126 genes whose abundance changed upon exposure to these antifungal agents. This work expands on our previous study in which we used microarray analysis to identify the genes in C. albicans that respond to ketoconazole, amphotericin B, and caspofungin exposures (24).
We identified 39 proteins that were differentially expressed following exposure to ketoconazole. Not surprisingly, several proteins that increased in abundance (Erg6p, Erg10p, and Erg13p) are directly involved in ergosterol biosynthesis. Sterol transmethylase (Erg6p) catalyzes the formation of zymosterol to fecosterol by using (S)-adenosyl-l-methionine as a cosubstrate. Acetyl-coenzyme A (acetyl-CoA) acetyltransferase (Erg10p) catalyzes the thiolytic cleavage of acetoacetyl-CoA to acetyl-CoA, which is the first step in the formation of mevalonate, a precursor required for the biosynthesis of ergosterol. The enzyme 3-hydroxymethylglutaryl-CoA synthase (Erg13p) catalyzes the production of hydroxymethylglutaryl-CoA from acetyl-CoA and acetoacetyl-CoA and functions in the second step in mevalonate biosynthesis. Our previous microarray analysis of C. albicans under these conditions found ERG6 to be upregulated. Moreover, in response to a longer exposure to itraconazole, De Backer et al. found ERG6, ERG10, and ERG13 to be upregulated (7a). Our data demonstrate that increased protein abundance accompanies the increased transcription of these genes in response to azole treatment. Interestingly, the increased abundance of both Erg6p and Erg10p has also been observed in azole-resistant clinical isolates of C. albicans (14, 15) as well as a laboratory-derived fluconazole-resistant strain of C. albicans produced after exposure to fluconazole (49). In one isolate, the increased abundance of these proteins was due to a G648D amino acid substitution in the transcription factor Upc2p. The upregulation of Erg6p, Erg10p, and Erg13p upon azole exposure is likely in response to the activation of Upc2p upon the depletion of ergosterol, which is consistent with the mechanism of action of this class of antifungals.
We also observed that ketoconazole exposure induced the expression of Csh1p and Orf19.251, which are proteins whose genes were previously shown to be associated with azole resistance (30). Csh1p is a putative aldo-keto reductase and is homologous to aryl-alcohol dehydrogenases in Saccharomyces cerevisiae (38). This protein has been shown to function in cell surface hydrophobicity (39). Another protein associated with azole resistance that was found to be responsive to ketoconazole was Orf19.251, a heat shock protein of the ThiJ/PfpI protein family (4). In S. cerevisiae, the ortholog of Orf19.251 is Hsp31p, and the expression of HSP31 is regulated by the transcription factor Yap1p (40). Mutants lacking orf19.251 are hypersensitive to reactive species of oxygen, which suggests that this gene functions to protect the cell against oxidative stress (40). In C. albicans, both Csh1p and Orf19.251 are known targets of the multidrug resistance regulator Mrr1p, which is a transcription factor that regulates the expression of the major facilitator superfamily drug efflux pump gene MDR1 (30). Indeed, the abundance of both proteins was increased in an azole-resistant clinical isolates and was due to gain-of-function mutations in Mrr1p. Moreover, Csh1p was among the genes that we previously found to be differentially expressed in response to ketoconazole (24). These data suggest the possibility that Mrr1p is also activated upon azole treatment. The physiological roles of Csh1p and Ipf17186 both in azole resistance and in response to ketoconazole exposure have not yet been determined.
In response to amphotericin B exposure we found several proteins to be increased in abundance, including Yhb1p, Cip1p, Trx2p, Trr1p, and Tsa1p, all of which function in response to oxidative stress. Yhb1p is a putative flavohemoglobin that functions as a dioxygenase in nitric oxide detoxification (44). In S. cerevisiae, mutants lacking YHB1 had levels of protein nitrosylation 10-fold higher than those observed in wild-type cells after exposure to donors of nitrous oxide, and the growth of the mutants was inhibited by nitrosative challenge (23). The cadmium-induced protein (Cip1p) is an enzyme with significant similarity to plant isoflavone reductases. The expression of this protein is regulated by the transcription factor Cap1p, a known regulator of oxidative stress (45, 52). This protein was also induced in the yeast-to-hypha transition and upon macrophage interaction (25, 31). Two additional amphotericin B-responsive enzymes were thioredoxin (Trx2p) and thioredoxin reductase (Trr1p), both of which are involved in the thioredoxin system. The thioredoxin system functions to maintain the cellular redox state and defend the cell against oxidative stress. Trx2p is an enzyme that reduces proteins by catalyzing thiol-disulfide reactions with two cysteine residues that shift from the reduced form [(SH)2] to the oxidized form (S2). The oxidized form returns to its reduced form in a reaction catalyzed by Trr1p by using NADPH as an electron source. Trr1p functions as a stress-induced reductase and is the key regulatory enzyme that determines the redox state of this system. In S. cerevisiae, the transcriptional regulators Yap1p and Skn7p mediate the expression of both TRX2 and TRR1 in response to hydrogen peroxide (19, 29). Another amphotericin B-responsive protein involved in oxidative stress was thioredoxin peroxidase (Tsa1p), which belongs to a family of antioxidant enzymes capable of reducing reactive forms of oxygen, nitrogen, and sulfur using Trx2p as a reducing source (16). In C. albicans, Tsa1p was found to be increased in abundance in hyphae compared to its abundance in the yeast form and has been implicated in the function of hyphal cell wall biosynthesis (6). In S. cerevisiae, the transcriptional regulators Yap1p and Skn1p are required for the H2O2-mediated induction of both TSA1 and TSA2 (22), and the disruption of TSA1 renders cells more sensitive to hydrogen peroxide (9). In the present study, it is not surprising that in response to amphotericin B we observed increases in the abundance of multiple proteins involved in oxidative stress and in the thioredoxin system, as this antifungal agent is known to cause significant oxidative damage in C. albicans (42, 43).
C. albicans exposure to amphotericin B also resulted in the increased abundance of Rhr2p and Asr2p, which are proteins involved in osmotic stress. In yeast, the response to osmotic stress is through the increased production of glycerol, which is regulated, in part, by a signaling system called the high-osmolarity glycerol (HOG) mitogen-activated protein kinase (MAPK) pathway. One amphotericin B-responsive protein regulated by this pathway is Rhr2p, a dl-glycerol-3-phosphatase that functions in glycerol biosynthesis. Previous studies with S. cerevisiae have demonstrated that the expression of RHR2 is significantly induced under conditions of hyperosmotic stress (32). Mutants lacking this gene have a low intracellular glycerol content, do not have glycerol-3-phosphatase activity, and are hypersensitive to osmotic stress. Another protein found to be responsive to amphotericin B and involved in osmotic stress is Asr2p. A previous study showed that ASR2 had significantly increased expression under conditions of osmotic stress and is regulated by cyclic AMP, and thus, the gene was designated ASR2, for adenylyl cyclase stress responsive (12). As amphotericin B is believed to compromise the structural integrity of the fungal membrane by forming pores through the membrane, it is reasonable to assume that C. albicans cells treated with amphotericin B undergo significant changes in intracellular solute concentrations, resulting in the increased abundance of proteins that function in response to cellular osmolarity.
The fungal cell wall is a dynamic structure composed of interconnecting mannoproteins, chitin, and β-glucans and is responsible for maintaining osmotic stability. Among these components, the β-1,3-glucan is the primary cell wall constituent (19, 20). The echinocandins, such as caspofungin, inhibit the activity of β-1,3-glucan synthase, which is an important enzyme required for the synthesis of β-1,3-glucans. Among the proteins that increased in abundance in response to caspofungin treatment was Rho1p, which is a low-molecular-weight GTPase that regulates the activity of β-1,3-glucan synthase in both C. albicans and S. cerevisiae. Rho1p in C. albicans is essential for yeast cell viability and functions in actin cytoskeleton reorganization and cell wall biosynthesis (48). As caspofungin noncompetitively inhibits the production of β-1,3-glucans, it is not surprising that we observed an increase in the abundance of its regulator, Rho1p, in response to this antifungal drug. Other caspofungin-responsive proteins associated with cell wall biosynthesis included GDP-mannose pyrophosphorylase (Psa1p) and UDP-glucose-4-epimerase (Gal10p), both of which were decreased in abundance in response to caspofungin exposure, and xylitol dehydrogenase (Xyl2p), which was increased in abundance in response to caspofungin exposure. Briefly, Psa1p catalyzes the formation of GDP-mannose from mannose-1-phosphate and GTP for cell wall biosynthesis. This protein is required for the incorporation of mannose into more complex N-linked and O-linked glycoproteins. In S. cerevisiae, mutants lacking PSA1 are unable to survive, and hypomorphic mutants display defects in cell wall biosynthesis and are sensitive to hyperosmolarity (13). Additionally, the Gal10p enzyme functions in galactose metabolism and catalyzes the epimerization of UDP-galactose to UDP-glucose. The absence of GAL10 in C. albicans affects the cell wall organization, the oxidative stress response, filamentation, and the formation of biofilms (37). Therefore, in addition to galactose metabolism, Gal10p appears to be required for cell wall integrity, normal hyphal growth, and resistance to oxidative stress. Furthermore, Xyl2p is a zinc-containing protein and a member of the medium-chain dehydrogenase superfamily that functions in xylulose metabolism. In S. cerevisiae, the expression of XYL2 is induced by xylose, and mutants lacking this gene have defects in their cell walls (8, 35). Although Xyl2p has been implicated in the function of cell wall biosynthesis, its specific function is unknown. The alterations in Rho1p, Psa1p, Gal10p, and Xly2p expression are characteristic of compromised cell wall integrity and are consistent with the mechanism of action of this antifungal agent.
A previous study used 2-D PAGE and MALDI-TOF MS to identify the alterations in C. albicans protein expression following exposure to the echinocandin antifungals mulundocandin and the mulundocandin derivative HMR3270 (5). That study revealed that 41 proteins were identified as being increased in abundance and 38 proteins were identified as being decreased in abundance upon exposures to mulundocandin and HMR3270. The caspofungin-influenced proteins identified in the present study that were also found to be similarly increased in abundance in response to mulundocandin and HMR3270 exposures were Cit1p, Gdh3p, Idh2p, Ino1p, Qcr2p, Sam2p, and Tsa1p. The increased abundance of these proteins in response to these two echinocandins appears to represent a common stress response to this class of antifungal agents.
The abundance of nine proteins increased in response to all three antifungals: Atp2p, Aco1p, Cit1p, Idh2p, Mdh1p, Ino1p, Pdb1p, Rpl12p, and Snz1p. Among them, Aco1p, Cit1p, Idh2p, and Mdh1p function in the TCA cycle. The TCA cycle is a central metabolic process in the mitochondria that converts acetyl-CoA to carbon dioxide and water to produce chemical energy in the form of ATP and NADH (after oxidative phosphorylation). It is also important to note that the intermediates of this process are used as precursors in the biosynthesis of amino acids, lipids, and heme. Three of these proteins have also been observed to increase in abundance in response to cadmium or upon interaction with macrophages, suggesting that they may be part of a general adaptation to environmental stress (27, 50). The increased abundance of these proteins in response to all three antifungal agents suggests an increase in energy demand during the adaptive response to these stresses.
As this proteomic analysis used conditions identical to those that we used in our previous microarray study of adaptive responses to antifungal agents, it affords a unique opportunity to compare the similarities and differences in these two types of data. Changes in protein abundance were in agreement with changes in gene expression for 6 of 39 proteins (Erg6p, Adh2p, Ado1p, Csh1p, Mdh1p, and Tuf1p) in response to ketoconazole, 7 of 43 proteins (Cys3p, Aco1p, Yhb1p, Snz1p, Hsp60p, Ssa4p, and Met6p) in response to amphotericin B, and 11 of 50 proteins (Aco1p, Sah1p, Bat22p, Met6p, Ssb1p, Egd1p, Cyp1p, Aat1p, Gpm1p, Pdc11p, and Hsp90p) in response to caspofungin. Given the limitations inherent to the methods used in our proteomic analysis, the resolution of each protein representing every gene found to be differentially expressed in our microarray analysis would not be expected. Our proteomic analysis is limited to the most abundant soluble proteins that can be resolved and visualized by 2-D PAGE that could also be successfully identified by MALDI-TOF MS. Likewise, limitations of the early-generation arrays used in our microarray analysis may have resulted in some differentially expressed genes not being detected. Moreover, changes in gene expression would be expected to precede changes in protein abundance temporally. The measurement of both gene expression and protein abundance at the same time point in these two studies may account for some of the discordance between these data. Finally, it is likely that some of the changes in protein abundance observed in response to antifungal exposure are not transcriptionally regulated.
In conclusion, our findings reveal specific and nonspecific antifungal-induced changes in protein abundance in C. albicans. This proteomic analysis expands on our previous microarray analysis, as this study provides insight into the adaptive responses to these antifungal agents at the translational level. It is possible that some of these antifungal-responsive proteins may represent potential targets for the development of novel therapeutic agents and could enhance the activities of these drugs against this pathogenic fungus.
Acknowledgments
This research was funded by NIH NIAID grant R01AI058145 and a small grant obtained through the Children's Foundation Research Center at Le Bonheur Children's Medical Center in Memphis, TN.
We thank Qing Zhang for her assistance in the laboratory.
We have no financial or commercial conflicts of interest to declare.
Footnotes
Published ahead of print on 9 February 2010.
REFERENCES
- 1.Akins, R. A. 2005. An update on antifungal targets and mechanisms of resistance in Candida albicans. Med. Mycol. 43:285-318. [DOI] [PubMed] [Google Scholar]
- 2.Arning, M., K. O. Kliche, A. H. Heer-Sonderhoff, and A. Wehmeier. 1995. Infusion-related toxicity of three different amphotericin B formulations and its relation to cytokine plasma levels. Mycoses 38:459-465. [DOI] [PubMed] [Google Scholar]
- 3.Bates, D. W., L. Su, D. T. Yu, G. M. Chertow, D. L. Seger, D. R. Gomes, and R. Platt. 2001. Correlates of acute renal failure in patients receiving parenteral amphotericin B. Kidney Int. 60:1452-1459. [DOI] [PubMed] [Google Scholar]
- 4.Braun, B. R., M. van Het Hoog, C. d'Enfert, M. Martchenko, J. Dungan, A. Kuo, D. O. Inglis, M. A. Uhl, H. Hogues, M. Berriman, M. Lorenz, A. Levitin, U. Oberholzer, C. Bachewich, D. Harcus, A. Marcil, D. Dignard, T. Iouk, R. Zito, L. Frangeul, F. Tekaia, K. Rutherford, E. Wang, C. A. Munro, S. Bates, N. A. Gow, L. L. Hoyer, G. Kohler, J. Morschhauser, G. Newport, S. Znaidi, M. Raymond, B. Turcotte, G. Sherlock, M. Costanzo, J. Ihmels, J. Berman, D. Sanglard, N. Agabian, A. P. Mitchell, A. D. Johnson, M. Whiteway, and A. Nantel. 2005. A human-curated annotation of the Candida albicans genome. PLoS Genet. 1:36-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruneau, J. M., I. Maillet, E. Tagat, R. Legrand, F. Supatto, C. Fudali, J. P. Caer, V. Labas, D. Lecaque, and J. Hodgson. 2003. Drug induced proteome changes in Candida albicans: comparison of the effect of beta (1,3) glucan synthase inhibitors and two triazoles, fluconazole and itraconazole. Proteomics 3:325-336. [DOI] [PubMed] [Google Scholar]
- 6.Choi, W., Y. J. Yoo, M. Kim, D. Shin, and H. B. Jeon. 2003. Identification of proteins highly expressed in the hyphae of Candida albicans by two-dimensional electrophoresis. Yeast 20:1053-1060. [DOI] [PubMed] [Google Scholar]
- 7.Cummings, E. D., J. M. Brown, S. T. Sarva, R. H. Waldo, and G. M. Hilliard. 2007. High-throughput proteomics processing of proteins in polyacrylamide in a multiwell format. J. Proteome Res. 6:1603-1608. [DOI] [PubMed] [Google Scholar]
- 7a.De Backer, M. D., T. Ilyind, X.-J. Ma, S. Vandoninck, W. H. M. L. Luyten, and H. Vanden Bossche. 2001. Genetic profiling of the response of Candida albicans to itraconazole treatment using a DNA microarray. Antimicrob. Agents Chemother. 45:1660-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Groot, P. W., C. Ruiz, C. R. Vazquez de Aldana, E. Duenas, V. J. Cid, F. Del Rey, J. M. Rodriquez-Pena, P. Perez, A. Andel, J. Caubin, J. Arroyo, J. C. Garcia, C. Gil, M. Molina, L. J. Garcia, C. Nombela, and F. M. Klis. 2001. A genomic approach for the identification and classification of genes involved in cell wall formation and its regulation in Saccharomyces cerevisiae. Comp. Funct. Genomics 2:124-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demasi, A. P., G. A. Pereira, and L. E. Netto. 2001. Cytosolic thioredoxin peroxidase I is essential for the antioxidant defense of yeast with dysfunctional mitochondria. FEBS Lett. 509:430-434. [DOI] [PubMed] [Google Scholar]
- 10.Edmond, M. B., S. E. Wallace, D. K. McClish, M. A. Pfaller, R. N. Jones, and R. P. Wenzel. 1999. Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin. Infect. Dis. 29:239-244. [DOI] [PubMed] [Google Scholar]
- 11.Fernandez-Arenas, E., V. Cabezon, C. Bermejo, J. Arroyo, C. Nombela, R. Diez-Orejas, and C. Gil. 2007. Integrated proteomics and genomics strategies bring new insight into Candida albicans response upon macrophage interaction. Mol. Cell. Proteomics 6:460-478. [DOI] [PubMed] [Google Scholar]
- 12.Harcus, D., A. Nantel, A. Marcil, T. Rigby, and M. Whiteway. 2004. Transcription profiling of cyclic AMP signaling in Candida albicans. Mol. Biol. Cell 15:4490-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hashimoto, H., A. Sakakibara, M. Yamasaki, and K. Yoda. 1997. Saccharomyces cerevisiae VIG9 encodes GDP-mannose pyrophosphorylase, which is essential for protein glycosylation. J. Biol. Chem. 272:16308-16314. [DOI] [PubMed] [Google Scholar]
- 14.Hoehamer, C. F., E. D. Cummings, G. M. Hilliard, J. Morschhauser, and P. D. Rogers. 2009. Upc2p-associated differential protein expression in Candida albicans. Proteomics 9:4726-4730. [DOI] [PubMed] [Google Scholar]
- 15.Hooshdaran, M. Z., K. S. Barker, G. M. Hilliard, H. Kusch, J. Morschhauser, and P. D. Rogers. 2004. Proteomic analysis of azole resistance in Candida albicans clinical isolates. Antimicrob. Agents Chemother. 48:2733-2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jang, H. H., K. O. Lee, Y. H. Chi, B. G. Jung, S. K. Park, J. H. Park, J. R. Lee, S. S. Lee, J. C. Moon, J. W. Yun, Y. O. Choi, W. Y. Kim, J. S. Kang, G. W. Cheong, D. J. Yun, S. G. Rhee, M. J. Cho, and S. Y. Lee. 2004. Two enzymes in one; two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell 117:625-635. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, M. D., and J. R. Perfect. 2003. Caspofungin: first approved agent in a new class of antifungals. Expert Opin. Pharmacother. 4:807-823. [DOI] [PubMed] [Google Scholar]
- 18.Kelly, S. L., D. C. Lamb, D. E. Kelly, N. J. Manning, J. Loeffler, H. Hebart, U. Schumacher, and H. Einsele. 1997. Resistance to fluconazole and cross-resistance to amphotericin B in Candida albicans from AIDS patients caused by defective sterol delta 5,6-desaturation. FEBS Lett. 400:80-82. [DOI] [PubMed] [Google Scholar]
- 19.Kuge, S., and N. Jones. 1994. YAP1 dependent activation of TRX2 is essential for the response of Saccharomyces cerevisiae to oxidative stress by hydroperoxides. EMBO J. 13:655-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kusch, H., K. Biswas, S. Schwanfelder, S. Engelmann, P. D. Rogers, M. Hecker, and J. Morschhauser. 2004. A proteomic approach to understanding the development of multidrug-resistant Candida albicans strains. Mol. Genet. Genomics 271:554-565. [DOI] [PubMed] [Google Scholar]
- 21.Kusch, H., S. Engelmann, D. Albrecht, J. Morschhauser, and M. Hecker. 2007. Proteomic analysis of the oxidative stress response in Candida albicans. Proteomics 7:686-697. [DOI] [PubMed] [Google Scholar]
- 22.Lee, J., C. Godon, G. Lagniel, D. Spector, J. Garin, J. Labarre, and M. B. Toledano. 1999. Yap1 and Skn7 control two specialized oxidative stress response regulons in yeast. J. Biol. Chem. 274:16040-16046. [DOI] [PubMed] [Google Scholar]
- 23.Liu, L., M. Zeng, A. Hausladen, J. Heitman, and J. S. Stamler. 2000. Protection from nitrosative stress by yeast flavohemoglobin. Proc. Natl. Acad. Sci. U. S. A. 97:4672-4676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, T. T., R. E. Lee, K. S. Barker, L. Wei, R. Homayouni, and P. D. Rogers. 2005. Genome-wide expression profiling of the response to azole, polyene, echinocandin, and pyrimidine antifungal agents in Candida albicans. Antimicrob. Agents Chemother. 49:2226-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lorenz, M. C., J. A. Bender, and G. R. Fink. 2004. Transcriptional response of Candida albicans upon internalization by macrophages. Eukaryot. Cell 3:1076-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maesaki, S., M. A. Hossain, Y. Miyazaki, K. Tomono, T. Tashiro, and S. Kohno. 2000. Efficacy of FK463, a (1,3)-beta-d-glucan synthase inhibitor, in disseminated azole-resistant Candida albicans infection in mice. Antimicrob. Agents Chemother. 44:1728-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez-Solano, L., C. Nombela, G. Molero, and C. Gil. 2006. Differential protein expression of murine macrophages upon interaction with Candida albicans. Proteomics 6(Suppl. 1):S133-S144. [DOI] [PubMed] [Google Scholar]
- 28.Moore, C. B., K. L. Oakley, and D. W. Denning. 2001. In vitro activity of a new echinocandin, LY303366, and comparison with fluconazole, flucytosine and amphotericin B against Candida species. Clin. Microbiol. Infect. 7:11-16. [DOI] [PubMed] [Google Scholar]
- 29.Morgan, B. A., G. R. Banks, W. M. Toone, D. Raitt, S. Kuge, and L. H. Johnston. 1997. The Skn7 response regulator controls gene expression in the oxidative stress response of the budding yeast Saccharomyces cerevisiae. EMBO J. 16:1035-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morschhauser, J., K. S. Barker, T. T. Liu, B. W. J. Bla, R. Homayouni, and P. D. Rogers. 2007. The transcription factor Mrr1p controls expression of the MDR1 efflux pump and mediates multidrug resistance in Candida albicans. PLoS Pathog. 3:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nantel, A., D. Dignard, C. Bachewich, D. Harcus, A. Marcil, A. P. Bouin, C. W. Sensen, H. Hogues, M. van het Hoog, P. Gordon, T. Rigby, F. Benoit, D. C. Tessier, D. Y. Thomas, and M. Whiteway. 2002. Transcription profiling of Candida albicans cells undergoing the yeast-to-hyphal transition. Mol. Biol. Cell 13:3452-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pahlman, A. K., K. Granath, R. Ansell, S. Hohmann, and L. Adler. 2001. The yeast glycerol 3-phosphatases Gpp1p and Gpp2p are required for glycerol biosynthesis and differentially involved in the cellular responses to osmotic, anaerobic, and oxidative stress. J. Biol. Chem. 276:3555-3563. [DOI] [PubMed] [Google Scholar]
- 33.Pfaller, M. A., R. N. Jones, S. A. Messer, M. B. Edmond, and R. P. Wenzel. 1998. National surveillance of nosocomial blood stream infection due to Candida albicans: frequency of occurrence and antifungal susceptibility in the SCOPE Program. Diagn. Microbiol. Infect. Dis. 31:327-332. [DOI] [PubMed] [Google Scholar]
- 34.Rex, J. H., T. J. Walsh, M. Nettleman, E. J. Anaissie, J. E. Bennett, E. J. Bow, A. J. Carillo-Munoz, P. Chavanet, G. A. Cloud, D. W. Denning, B. E. de Pauw, J. E. Edwards, Jr., J. W. Hiemenz, C. A. Kauffman, G. Lopez-Berestein, P. Martino, J. D. Sobel, D. A. Stevens, R. Sylvester, J. Tollemar, C. Viscoli, M. A. Viviani, and T. Wu. 2001. Need for alternative trial designs and evaluation strategies for therapeutic studies of invasive mycoses. Clin. Infect. Dis. 33:95-106. [DOI] [PubMed] [Google Scholar]
- 35.Richard, P., M. H. Toivari, and M. Penttila. 1999. Evidence that the gene YLR070c of Saccharomyces cerevisiae encodes a xylitol dehydrogenase. FEBS Lett. 457:135-138. [DOI] [PubMed] [Google Scholar]
- 36.Rybowicz, J., and C. Gurk-Turner. 2002. Caspofungin: the first agent available in the echinocandin class of antifungals. Proc. (Bayl. Univ. Med. Cent.) 15:97-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh, V., S. V. Satheesh, M. L. Raghavendra, and P. P. Sadhale. 2007. The key enzyme in galactose metabolism, UDP-galactose-4-epimerase, affects cell-wall integrity and morphology in Candida albicans even in the absence of galactose. Fungal Genet. Biol. 44:563-574. [DOI] [PubMed] [Google Scholar]
- 38.Singleton, D. R., and K. C. Hazen. 2004. Differential surface localization and temperature-dependent expression of the Candida albicans CSH1 protein. Microbiology 150:285-292. [DOI] [PubMed] [Google Scholar]
- 39.Singleton, D. R., J. Masuoka, and K. C. Hazen. 2001. Cloning and analysis of a Candida albicans gene that affects cell surface hydrophobicity. J. Bacteriol. 183:3582-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Skoneczna, A., A. Micialkiewicz, and M. Skoneczny. 2007. Saccharomyces cerevisiae Hsp31p, a stress response protein conferring protection against reactive oxygen species. Free Radic. Biol. Med. 42:1409-1420. [DOI] [PubMed] [Google Scholar]
- 41.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 42.Sokol-Anderson, M., J. E. Sligh, Jr., S. Elberg, J. Brajtburg, G. S. Kobayashi, and G. Medoff. 1988. Role of cell defense against oxidative damage in the resistance of Candida albicans to the killing effect of amphotericin B. Antimicrob. Agents Chemother. 32:702-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sokol-Anderson, M. L., J. Brajtburg, and G. Medoff. 1986. Amphotericin B-induced oxidative damage and killing of Candida albicans. J. Infect. Dis. 154:76-83. [DOI] [PubMed] [Google Scholar]
- 44.Ullmann, B. D., H. Myers, W. Chiranand, A. L. Lazzell, Q. Zhao, L. A. Vega, J. L. Lopez-Ribot, P. R. Gardner, and M. C. Gustin. 2004. Inducible defense mechanism against nitric oxide in Candida albicans. Eukaryot. Cell 3:715-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, Y., Y. Y. Cao, X. M. Jia, Y. B. Cao, P. H. Gao, X. P. Fu, K. Ying, W. S. Chen, and Y. Y. Jiang. 2006. Cap1p is involved in multiple pathways of oxidative stress response in Candida albicans. Free Radic. Biol. Med. 40:1201-1209. [DOI] [PubMed] [Google Scholar]
- 46.White, T. C., K. A. Marr, and R. A. Bowden. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wingard, J. R., P. Kubilis, L. Lee, G. Yee, M. White, L. Walshe, R. Bowden, E. Anaissie, J. Hiemenz, and J. Lister. 1999. Clinical significance of nephrotoxicity in patients treated with amphotericin B for suspected or proven aspergillosis. Clin. Infect. Dis. 29:1402-1407. [DOI] [PubMed] [Google Scholar]
- 48.Yamochi, W., K. Tanaka, H. Nonaka, A. Maeda, T. Musha, and Y. Takai. 1994. Growth site localization of Rho1 small GTP-binding protein and its involvement in bud formation in Saccharomyces cerevisiae. J. Cell Biol. 125:1077-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan, L., J. D. Zhang, Y. B. Cao, P. H. Gao, and Y. Y. Jiang. 2007. Proteomic analysis reveals a metabolism shift in a laboratory fluconazole-resistant Candida albicans strain. J. Proteome Res. 6:2248-2256. [DOI] [PubMed] [Google Scholar]
- 50.Yin, Z., D. Stead, J. Walker, L. Selway, D. A. Smith, A. J. Brown, and J. Quinn. 2009. A proteomic analysis of the salt, cadmium and peroxide stress responses in Candida albicans and the role of the Hog1 stress-activated MAPK in regulating the stress-induced proteome. Proteomics 9:4686-4703. [DOI] [PubMed] [Google Scholar]
- 51.Yu, D. T., J. F. Peterson, D. L. Seger, W. C. Gerth, and D. W. Bates. 2005. Frequency of potential azole drug-drug interactions and consequences of potential fluconazole drug interactions. Pharmacoepidemiol. Drug Saf. 14:755-767. [DOI] [PubMed] [Google Scholar]
- 52.Znaidi, S., K. S. Barker, S. Weber, A. M. Alarco, T. T. Liu, G. Boucher, P. D. Rogers, and M. Raymond. 2009. Identification of the Candida albicans Cap1p regulon. Eukaryot. Cell 8:806-820. [DOI] [PMC free article] [PubMed] [Google Scholar]