Abstract
The Capnocytophaga sputigena isolate NOR, responsible for septicemia, was resistant to amoxicillin and narrow-spectrum cephalosporins. In a cloning experiment, a new gene, blaCSP-1, was identified; this gene encodes a novel extended-spectrum β-lactamase (ESBL) that shares only 52% and 49% identities with the CME-1 and VEB-1 β-lactamases, respectively. The G+C content of this gene, its genetic environment, the absence of conjugation transfer, and its detection in two reference strains suggested that it was an intrinsic resistance gene located on the chromosome.
Capnocytophaga sputigena is a Gram-negative rod of the oral flora of healthy humans (8) that is responsible for systemic infections in immunocompromised patients with granulocytopenia and oral ulcerations, especially in children (5, 6, 8, 11, 13). Most of the clinical isolates of C. sputigena express β-lactamases (7). Almost 50% of these enzymes were previously identified as the transposon-encoded CfxA2 or CfxA3 enzyme (7, 10, 20). The other β-lactamases, which confer a drug resistance phenotype consistent with that of extended-spectrum β-lactamases (ESBLs), remained unidentified (7). The aims of this study were to perform the molecular characterization of a novel ESBL produced by a C. sputigena clinical isolate and to investigate its distribution among reference strains.
The C. sputigena isolate NOR was recovered at the teaching hospital in Amiens, France, in November 2008 from a 6-year-old child suffering from lymphoblastic leukemia. Ten days after the initiation of vincristine, the patient developed severe mucositis accompanied by a high fever. Despite ceftazidime and amikacin treatment, the patient's temperature remained ≥38°C. Two blood cultures and a superficial sample of the necrotic oral lesions allowed isolation of C. sputigena NOR, which was identified at the genus level by the API 32ANA system (bioMérieux, Marcy l'Etoile, France) and at the species level by 16S rRNA sequencing. Once the bacteria was determined, the treatment was changed to imipenem (50 mg/kg of body weight daily) for 15 days. The course of the infection was favorable.
The NOR isolate was susceptible to fluoroquinolones (4). Moreover, its pattern of resistance was consistent with ESBL production (Table 1). PCR screening of cfxA-type genes (7) and conjugation experiments using the azide-resistant Escherichia coli J53 as the recipient strain failed (18). Cloning was attempted as previously described (3), giving rise to the recombinant clone E. coli TOP10(pNOR1). The recombinant plasmid was purified and sequenced on both strands. Nucleotide sequence analysis failed to detect a genetic mobile element, such as an insertion sequence (IS) or class 1 integron. A 918-bp open reading frame (ORF), which encoded a 305-amino-acid protein that showed, in a BLASTx search, the highest similarity scores with other class A β-lactamases, was identified. This novel β-lactamase, which was named CSP-1 (for Capnocytophaga sputigena 1), had the following identities with the indicated β-lactamases: 52% identity with CME-1 (17), 49% with VEB-1 (15), 48% with TLA-1 (19), 42% with PER-1 (12), 41% with CblA (20), 40% with PER-2 (2), 38% with CepA (16), and 32% with CfxA (14).
TABLE 1.
MICs of β-lactams for C. sputigena NOR and other C. sputigena and E. coli strainsa
| β-Lactam | MIC (μg/ml) of β-lactam for: |
||||
|---|---|---|---|---|---|
| C. sputigena NOR | E. coli TOP10 (pNOR-1) | C. sputigena CRBIP 17.39 | C. sputigena CIP 104301T | E. coli TOP10 | |
| Amoxicillin | 256 | >512 | 0.25 | 0.25 | 2 |
| Amoxicillin-CLAb | 0.25 | 4 | 0.25 | 0.25 | 2 |
| Amoxicillin-SULb | 0.25 | 8 | 0.25 | 0.25 | 2 |
| Ticarcillin | 4 | >512 | 0.25 | 0.25 | 1 |
| Ticarcillin-CLAb | 0.25 | 4 | 0.25 | 0.25 | 1 |
| Piperacillin | 0.5 | 16 | 0.25 | 0.25 | 1 |
| Piperacillin-TZBb | 0.25 | 2 | 0.25 | 0.25 | 1 |
| Cephalothin | 64 | >512 | 0.125 | 0.25 | 4 |
| Cefoxitin | 0.5 | 2 | 0.5 | 0.5 | 2 |
| Cefuroxime | 64 | 256 | 0.5 | 0.5 | 0.5 |
| Cefotaxime | 0.25 | 8 | <0.06 | <0.06 | <0.06 |
| Ceftazidime | 4 | 512 | <0.06 | <0.06 | <0.06 |
| Ceftazidime-CLAb | <0.06 | 0.5 | <0.06 | <0.06 | <0.06 |
| Aztreonam | 0.125 | 16 | 0.125 | 0.125 | 0.06 |
| Cefepime | 0.125 | 4 | <0.06 | <0.06 | 0.06 |
| Imipenem | 0.06 | 0.06 | 0.06 | 0.06 | 0.06 |
The MICs of β-lactams for C. sputigena clinical isolate NOR, two C. sputigena reference strains (CRBIP 17.39 and CIP 104301T), and the recombinant clone E. coli TOP10(pNOR-1) producing the CSP-1 enzyme are given.
CLA, clavulanic acid at 2 μg/ml; SUL, sulbactam at 4 μg/ml; TZB, tazobactam at 4 μg/ml.
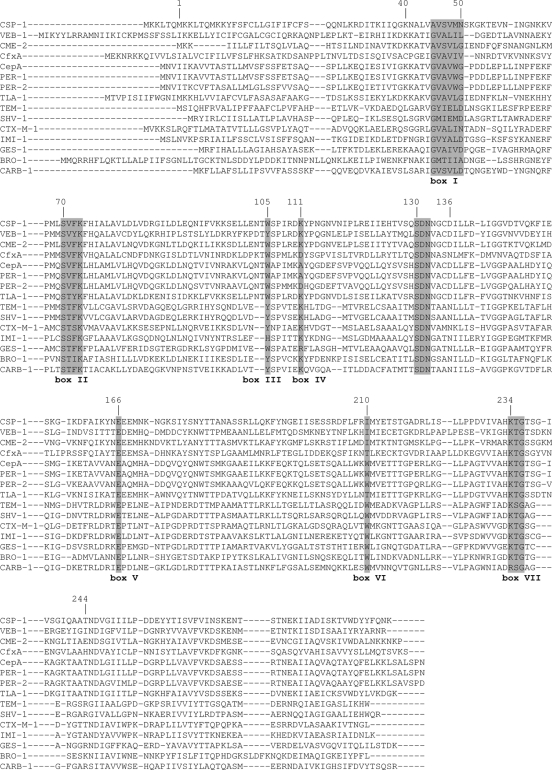
Within the mature protein sequence of CSP-1, seven boxes characteristic of β-lactamases possessing a serine active site were found (Fig. 1) (9). CSP-1 had the four conserved elements for class A β-lactamases according to Ambler et al. and Joris et al. (1, 9): the Ser-X-X-Lys consensus active site serine residue at position 70, the SDN loop at position 130, the conserved Glu166, and the KTG sequence at position 234. Moreover, the CSP-1 enzyme shares the aspartic acid residue at position 136 and the insertion of four amino acids in the region from positions 238 to 242 that are the landmarks of the PER-1 lineage, which includes the PER and VEB β-lactamases (the genes encoding these enzymes are located on an integron). The peculiar Ω loop fold, which resulted from the Asp136 together with the inserted four residues, causes a significant increase in the size of the substrate binding pocket, leading to an extended hydrolysis spectrum as revealed by the crystallographic study of PER-1 (21).
FIG. 1.
Multiple alignment of the amino acid sequences of the closely related class A β-lactamases with that of CSP-1. Boxes I through VI correspond to those described by Joris et al. (9). TLA-1, E. coli (GenBank accession no. AF148067); CME-1, Chryseobacterium meningosepticum (EMBL accession no. AJ006275); VEB-1, E. coli (EMBL accession no. O87489); PER-1, Pseudomonas aeruginosa (SwissProt accession no. P37321); PER-2, Salmonella enterica serovar Typhimurium (EMBL accession no. X93314); CblA, Bacteroides uniformis (SwissProt accession no. P30898); CepA, Bacteroides fragilis (GenBank accession no. L13472); CfxA, Bacteroides vulgatis (SwissProt accession no. P30899); TEM-1 (GenBank accession no. AAR25033); SHV-1 (GenBank accession no. AAK69828); CTX-M-1 (GenBank accession no. CAJ01342); IMI-1 (GenBank accession no. AAA93461); GES-1 (GenBank accession no. AAO32356); BRO-1 (GenBank accession no. AAA92126); CARB-1 (GenBank accession no. P16897).
DNA sequence analysis of the regions flanking the blaCSP-1 gene revealed an ORF located upstream encoding a putative peptidase E sharing 92% identity with that of Capnocytophaga gingivalis, and two ORFs located downstream, which encoded a putative HopPmaJ type III effector protein (virulent factor) sharing 60% identity with that of Chryseobacterium gleum, and a TonB-dependent receptor plug domain protein (porin family), respectively.
The G+C content of the blaCSP-1 gene is 34%, which is similar to the values for other sequenced C. sputigena genes already deposited in the EMBL sequence database (overall G+C content, 38%; GenBank accession no. NZ_A BZV00000000).
To investigate the distribution of the blaCSP-1 gene in the C. sputigena species, a PCR screening was performed using whole-cell DNA from C. sputigena reference strains CIP 104301T and CRBIP 17.39 (Pasteur Institute, Paris, France), primers CSP-1-A (5′-CAGAGAATAGATTTGCGACC-3′) and CSP-1-B (5′-TGTAAGCAATCACCTCATCG-3′), and a standard PCR protocol (denaturation for 10 min at 94°C; 35 cycles of 1 min at 94°C, 1 min at 55°C, and 2 min at 72°C; and a final extension step of 10 min at 72°C), which gave rise to two specific amplicons.
Isoelectric focusing (IEF) analysis of the β-lactamase content (3) from C. sputigena NOR gave a single band with a pI value of 6.4 that comigrated with the β-lactamase extract from E. coli TOP10(pNOR-1). The MICs of several β-lactams, which were determined as previously described (3, 6), are reported in Table 1. E. coli TOP10(pNOR-1) showed a resistance pattern that mirrored that of the parental strain, thus indicating that β-lactamase production constituted the main mechanism of resistance. E. coli TOP10(pNOR-1) was resistant to amoxicillin, ticarcillin, narrow-spectrum cephalosporins, ceftazidime, cefotaxime, and aztreonam (4). The addition of clavulanic acid, sulbactam, and tazobactam restored the susceptibility to penicillins. The clinical isolate and its recombinant clone were both susceptible to piperacillin, cefoxitin, and imipenem (Table 1). The two reference strains were susceptible to β-lactams, thus indicating that the level of expression of the blaCSP-1-like genes may vary, which might be attributable to the promoter strength. Further experiments are required to investigate this discrepancy.
The detection of a β-lactamase activity in a clinical C. sputigena isolate that did not harbor the cfxA-type gene yielded the characterization of a novel ESBL. The blaCSP-1 gene is very likely an intrinsic resistance marker resident in the chromosome of C. sputigena as suggested by its genetic environment, the G+C content relatedness with the overall G+C content in C. sputigena (38%), the absence of conjugation transfer, and the detection of blaCSP-1-like genes in the two reference strains.
Nucleotide sequence accession number.
The nucleotide sequence of the complete blaCSP-1 gene from C. sputigena isolate NOR has been deposited in the GenBank nucleotide database under accession no. GQ217533.
Footnotes
Published ahead of print on 22 March 2010.
REFERENCES
- 1.Ambler, R. P., A. F. W. Coulson, J. Frère, J. M. Ghuysen, B. Joris, M. Forsman, R. C. Levesque, G. Tiraby, and S. G. Waley. 1991. A standard numbering scheme for class A β-lactamases. Biochem. J. 276:269-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauernfeind, A., I. Stemplinger, R. Jungwirth, P. Mangold, S. Amann, E. Akalin, O. Ang, C. Bal, and J. M. Casellas. 1996. Characterization of β-lactamase gene blaPER-2, which encodes an extended-spectrum class A β-lactamase. Antimicrob. Agents Chemother. 40:616-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bellais, S., L. Poirel, T. Nass, D. Girlich, and P. Nordmann. 2000. Genetic-biochemical analysis and distribution of the Ambler class A β-lactamase CME-2, responsible for extended-spectrum cephalosporin resistance in Chryseobacterium (Flavobacterium) meningosepticum. Antimicrob. Agents Chemother. 44:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clinical and Laboratory Standards Institute. 2010. Performance standards for antimicrobial susceptibility testing: twentieth informational supplement. M100-S20. Clinical and Laboratory Standards Institute, Wayne, PA.
- 5.Forlenza, S. W., M. G. Newman, A. I. Lipsey, S. E. Siegel, and U. Blachman. 1980. Capnocytophaga sepsis: a newly recognised clinical entity in granulocytopenic patients. Lancet i:567-568. [DOI] [PubMed] [Google Scholar]
- 6.Gomez-Garces, J.-L., J.-I. Alos, J. Sanchez, and R. Cogollos. 1994. Bacteremia by multidrug-resistant Capnocytophaga sputigena. J. Clin. Microbiol. 32:1067-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Handal, T., C. Giraud-Morin, D. A. Caugant, I. Madinier, I. Olsen, and T. Fosse. 2005. Chromosome- and plasmid-encoded β-lactamases in Capnocytophaga spp. Antimicrob. Agents Chemother. 49:3940-3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jolivet-Gougeon, A., J. L. Sixou, Z. Tamanai-Shacoori, and M. Bonnaure-Mallet. 2007. Antimicrobial treatment of Capnocytophaga infections. Int. J. Antimicrob. Agents 29:367-373. [DOI] [PubMed] [Google Scholar]
- 9.Joris, B., P. Ledent, O. Dideberg, E. Fonze, J. Lamotte-Brasser, J. A. Kelly, J. M. Ghuysen, and J. M. Frère. 1991. Comparison of the sequences of the class A β-lactamases and the secondary structure elements of penicillin-recognizing proteins. Antimicrob. Agents Chemother. 35:2294-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Madinier, I., T. Fosse, J. Giudicelli, and R. Labia. 2001. Cloning and biochemical characterization of a class A β-lactamase from Prevotella intermedia. Antimicrob. Agents Chemother. 45:2386-2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsumoto, T., M. Matsubara, K. Oana, E. Kasuga, T. Suzuki, E. Hidaka, T. Shigemura, K. Yamauchi, T. Honda, H. Ota, and Y. Kawakami. 2008. First case of bacteriemia due to chromosome-encoded CfxA3-β-lactamase-producing Capnocytophaga sputigena in a pediatric patient with acute erythroblastic leukemia. Eur. J. Med. Res. 13:133-135. [PubMed] [Google Scholar]
- 12.Nordmann, P., and T. Naas. 1994. Sequence analysis of PER-1 extended-spectrum β-lactamase from Pseudomonas aeruginosa and comparison with class A β-lactamases. Antimicrob. Agents Chemother. 38:104-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parenti, D. M., and D. R. Snydman. 1985. Capnocytophaga species: infections in nonimmunocompromised and immunocompromised hosts. J. Infect. Dis. 151:140-147. [DOI] [PubMed] [Google Scholar]
- 14.Parker, A. C., and C. J. Smith. 1993. Genetic and biochemical analysis of a novel Ambler class A β-lactamase responsible for cefoxitin resistance in Bacteroides species. Antimicrob. Agents Chemother. 37:1028-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poirel, L., T. Naas, M. Guibert, E. Chaibi, R. Labia, and P. Nordmann. 1999. Molecular and biochemical characterization of VEB-1, a novel class A extended-spectrum β-lactamase encoded by an Escherichia coli integron gene. Antimicrob. Agents Chemother. 43:573-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rogers, M. B., A. C. Parker, and C. J. Smith. 1993. Cloning and characterization of the endogenous cephalosporinase gene, cepA, from Bacteroides fragilis reveals a new subgroup of Ambler class A β-lactamases. Antimicrob. Agents Chemother. 37:2391-2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rossolini, G. M., N. Franceschini, L. Lauretti, B. Caravelli, M. L. Riccio, M. Galleni, J. M. Frère, and G. Amicosante. 1999. Cloning of a Chryseobacterium (Flavobacterium) meningosepticum chromosomal gene (blaACME) encoding an extended-spectrum class A β-lactamase related to the Bacteroides cephalosporinases and the VEB-1 and PER β-lactamases. Antimicrob. Agents Chemother. 43:2193-2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 19.Silva, J., C. Aguilar, G. Ayala, M. A. Estrada, U. Garza-Ramos, R. Lara-Lemus, and L. Ledezma. 2000. TLA-1: a new plasmid-mediated extended-spectrum β-lactamase from Escherichia coli. Antimicrob. Agents Chemother. 44:997-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith, C. J., T. K. Bennett, and A. C. Parker. 1994. Molecular and genetic analysis of the Bacteroides uniformis cephalosporinase gene, cblA, encoding the species-specific β-lactamase. Antimicrob. Agents Chemother. 38:1711-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tranier, S., A.-T. Bouthors, L. Maveyraud, V. Guillet, W. Sougakoff, and J.-P. Samama. 2000. The high resolution crystal structure for class A β-lactamase PER-1 reveals the bases for its increase in breadth of activity. J. Biol. Chem. 275:28075-28082. [DOI] [PubMed] [Google Scholar]