Abstract
Several recent studies have shown that the transcriptional induction of yeast GAL genes occurs with faster kinetics if the gene has been previously expressed. Depending on the experimental regimen, this transcriptional “memory” phenomenon can persist for 1 to 2 cell divisions in the absence of an inducer (short-term memory) or for >6 cell divisions (long-term memory). Long-term memory requires the GAL1 gene, suggesting that memory involves the cytoplasmic inheritance of high levels of Gal1 that are expressed in the initial round of expression. In contrast, short-term memory requires the SWI/SNF chromatin-remodeling enzyme, and thus, it may involve the inheritance of distinct chromatin states. Here we have reevaluated the roles of SWI/SNF, the histone variant H2A.Z, and components of the nuclear pore in both the short-term and long-term memory of GAL genes. Our results suggest that the propagation of novel chromatin structures does not contribute to the transcriptional memory of GAL genes, but rather, memory of the previous transcription state is controlled primarily by the inheritance of the Gal3p and Gal1p signaling factors.
The establishment and maintenance of transcriptional states that are heritable to progeny play a central role during the development of multicellular organisms. In many cases a transcriptional state is propagated in the absence of the original inducing signal, suggesting some type of transcriptional “memory.” Likewise, unicellular organisms must rapidly adapt to signals from their microenvironment, and this process often involves the activation of complex transcriptional networks. The ability to pass on a “memory” of altered environmental conditions and, thus, a memory of altered transcription states may provide progeny with a selective advantage. Since chromatin structure plays a key role in determining the on/off state of eukaryotic genes, the inheritance of altered chromatin structures may provide one mechanism for transcriptional memory.
The transcriptional regulation of the GAL gene cluster of budding yeast serves as a paradigm for a complex gene regulatory network that also exhibits the phenomenon of transcriptional memory. GAL genes can be separated broadly into two groups: the structural genes (GAL1, GAL5, GAL7, and GAL10) that encode enzymes to metabolize galactose and regulatory genes (GAL2, GAL3, GAL4, and GAL80) that encode products that transport galactose and control the expression of the structural genes. The transcription of many of the GAL genes is tightly controlled by the sugar present in the medium: the expressions of GAL1, GAL3, GAL4, GAL7, and GAL10 are repressed by glucose, and most GAL genes are induced 3- to 1,000-fold in the presence of galactose (GAL5 is constitutive) (3, 12, 18, 19, 28).
The galactose-dependent transcriptional activation of GAL genes involves a complex regulatory network. The Gal4 activator binds to one or more sites upstream of each inducible GAL gene, but in the absence of galactose, the activation domain of Gal4 is inactivated by an interaction with the Gal80 repressor. When galactose is added to cells, the Gal2 permease imports galactose, and the binding of galactose to the cytoplasmic Gal3 protein allows Gal3 to bind to Gal80, sequestering the repressor in the cytoplasm (3, 17, 28). Notably, the product of GAL1, galactokinase, can substitute for Gal3 when present at high concentrations (14, 15, 29). Thus, Gal3 and Gal1 function as key signal transducers that activate Gal4. The Gal2 and Gal3 proteins are expressed at low basal levels in the absence of galactose, and their expression is increased in galactose medium, creating two positive-feedback loops. The 1,000-fold increase in Gal1 expression is likely to further enhance the Gal3 feedback loop. These positive loops are antagonized by a negative-feedback loop involving the Gal80 repressor, which is induced 2- to 3-fold by galactose. The Gal3 and Gal80 loops work in concert to ensure GAL gene homeostasis (2, 30).
Several studies have demonstrated that the Gal3/Gal1 feedback loop provides cells with a “memory” of previous galactose exposure (2, 37). For instance, when naïve, glucose-grown cells are switched to galactose medium, the full induction of GAL gene transcription is rather slow, requiring 2 to 4 h (21). These slow induction kinetics presumably reflect the need to synthesize additional Gal4 and to overcome glucose repression mechanisms that occur in cis at GAL genes (19). However, if cells were previously exposed to galactose, the reinduction of GAL genes that follows a 12-h period of glucose repression occurs with much more rapid kinetics. These rapid reinduction kinetics require the Gal1 protein, and heterokaryon studies indicate that the cytoplasmic inheritance of Gal1 provides the memory of previous galactose exposure (37). Interestingly, Brickner and colleagues also reported that the histone variant H2A.Z (also known as Htz1) is also required for transcriptional memory and, furthermore, that memory correlates with the association of a GAL gene with the nuclear periphery (4).
Previously, we also described a transcriptional memory phenomenon at yeast GAL genes. In our experimental regimen, cells were grown in a neutral, nonrepressing sugar (raffinose) prior to the first round of transcriptional induction in galactose. In this case, GAL transcription reaches maximal levels by ∼1 h. GAL genes were then repressed by growth in glucose medium for 30 min to 4 h, and after transfer to galactose medium, the reinduction of GAL genes was found to reach maximal levels by ∼5 min. Recently, Laine et al. used a similar protocol to monitor GAL transcriptional memory (22). Those studies indicated that the rapid reactivation of GAL genes requires the SWI/SNF chromatin-remodeling enzyme and an intragene loop between the 5′ and 3′ ends of a GAL gene (21, 22). Interestingly, the memory of previous galactose exposure persisted through at least one cell division but was lost by 6 to 8 h of growth in glucose. This result contrasts with data from the studies described above, where memory was stable for at least 12 h (4, 37). Given the key role of the SWI/SNF remodeling enzyme in transcriptional memory, we proposed that SWI/SNF may establish an “active” chromatin structure at GAL genes that could persist through a few generations in the absence of the galactose inducer. The roles of H2A.Z, the nuclear periphery, or the Gal1 or Gal3 protein in this “short-term” memory phenomenon have not been tested.
Here we reevaluate the role of chromatin-remodeling factors, the histone variant H2A.Z, the nuclear periphery, and cytoplasmic signaling molecules in both the “short-term” and “long-term” transcriptional memory of GAL genes. We find that neither H2A.Z nor the recruitment of GAL genes to the nuclear periphery is required for the “short-term” memory of GAL genes, nor does recruitment to the nuclear periphery appear to be required for “long-term” memory. In contrast, our data suggest that Gal1 and Gal3 function redundantly to promote rapid reinduction kinetics, likely through their cytoplasmic inheritance. Furthermore, SWI/SNF does not appear to be involved in memory per se, but rather, its chromatin-remodeling activity is required in both rounds of induction to achieve rapid activation. However, the function of SWI/SNF is most apparent during reinduction, when rapid signaling by Gal3/Gal1 renders chromatin remodeling a rate-limiting step for GAL1 activation.
MATERIALS AND METHODS
Yeast strains, media, and culture conditions.
Strains used in this study are isogenic derivatives of the S288c background. Saccharomyces cerevisiae liquid cultures were grown at 30°C in YEP (1% yeast extract, 2% Bacto-peptone) medium supplemented with 2% glucose, 2% galactose, or 2% raffinose plus 0.2% sucrose depending on whether GAL1 activation or repression was required. For Gal3p-overexpressing strains, wild-type and swi2Δ strains were transformed with 2μm plasmids expressing full-length Gal3p from a constitutive ADH1 promoter or the relevant vector control. Transformants were selected and grown on synthetic media (0.67 g yeast nitrogen base without amino acids per 100 ml) containing 2% dextrose lacking uracil (URA).
RNA isolation and analysis.
Total RNA was isolated from yeast cells grown to logarithmic phase in appropriate media by the hot-phenol extraction method. The concentration of RNA was estimated by measuring the A260 after dissolving it in diethyl pyrocarbonate-treated water. Ten micrograms of total RNA from each sample was electrophoresed on 1% formaldehyde agarose gels, and Northern blotting was done. The housekeeping gene ACT1 was used as a loading control. Radioactively labeled probes for hybridization were generated by PCR amplifying the complete GAL1, GAL10, or ACT1 open reading frame (ORF) from genomic DNA.
RT-PCR.
Cells were grown to mid-log phase in YEP medium with 2% glucose, 2% galactose, or 2% raffinose plus 0.2% sucrose at 30°C. Ten milliliters of cells was harvested, and total RNA was extracted as described above. First-strand cDNA was synthesized by using 2.5 μg RNA, Superscript II RNase H− reverse transcriptase (RT) (Invitrogen), and 2 pmol each downstream primer designed for genes of interest, according to the manufacturer's instructions. Subsequently, 32P-labeled PCR was performed by using 2 μl of the first-strand cDNA reaction mixture and gene-specific primer sets to determine the relative levels of GAL1, GAL3, GAL10, and ACT1 mRNA for each strain. After 12 cycles (for GAL1, GAL10, and ACT1) or 25 cycles (for GAL3) of amplification, PCR products were electrophoresed on 10% acrylamide gels. Reactions were visualized by PhosphorImager (Molecular Dynamics) analysis and quantified by use of ImageQuant software (Amersham Biosciences).
ChIP.
Rabbit polyclonal antibody to the C terminus of histone H3 (ab1791) was obtained from Abcam, Inc. Mouse monoclonal antibody to RNA polymerase II (RNAPII) (CTD4H8) was obtained from Covance Research Products. Chromatin immunoprecipitation (IP) (ChIP) assays were performed as described previously by Li et al. (25). The immunoprecipitated DNA was amplified by using quantitative PCR performed with [α-32P]dCTP and then electrophoresed on 5% acrylamide gels. Reactions were visualized and quantified by PhosphorImager analysis.
Mononucleosome preparation.
Samples (from 100 ml of culture at an A600 of ∼0.7) were cross-linked for 30 min with 37% formaldehyde (final concentration of 2%) at 30°C. Reactions were quenched by the addition of 2.5 M glycine to a final concentration of 125 mM. Cell pellets were collected and washed with water to remove residual medium. Mononucleosomes were prepared as described previously by Dion et al. (9). An aliquot of this sample was deproteinized, and cross-links were reversed. Phenol-chloroform extraction was done, and samples were ethanol precipitated. The resulting pellet was resuspended and treated with RNase A (1 μg for 1 h at 37°C) to remove all RNA. Samples were then electrophoresed on 1.5% agarose gels to determine the best titration that yielded mononucleosomal DNA.
Nucleosome-scanning analysis.
The method for nucleosome-scanning analysis was adapted from a method described previously by Sekinger et al. (32), with modifications. Briefly, mononucleosomal chromatin was prepared as described above. This material was used for IP with histone H3 antibody. The immunoprecipitated DNA was amplified by using quantitative PCR and a set of overlapping primer pairs that were staggered 20 bp relative to each other and together covered an approximately 300-bp region of DNA. The products of all primer pairs were approximately 100 bp long. The efficiency of each primer pair was assayed by performing quantitative PCR with genomic DNA.
RESULTS
Short-term memory of GAL genes requires Gal3 and Gal1.
Transcriptional memory at GAL1 has been defined as the ability to reinduce GAL1 transcription with much faster kinetics than induction rates for the initial exposure to galactose. In our previous study, we showed that memory persisted through only a few cell divisions during glucose repression (21). In contrast, two groups described a different experimental regimen in which GAL transcriptional memory persisted for at least 12 h in glucose medium (∼4 to 6 cell divisions) (4, 37). In these cases, the initial round of GAL induction involved the transfer of repressed, glucose-grown cultures to galactose medium, whereas in our case, raffinose-grown cultures were shifted to galactose. Given the apparent differences in the durations of memory between these studies, we tested whether our strains could recapitulate long-term memory.
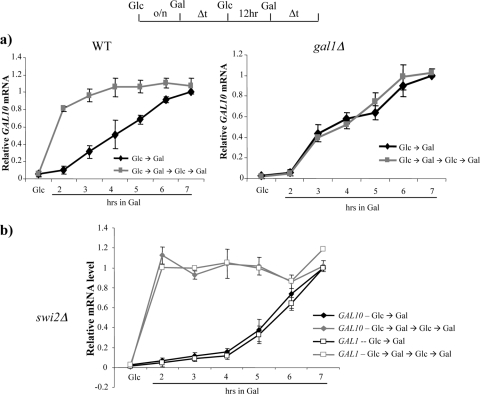
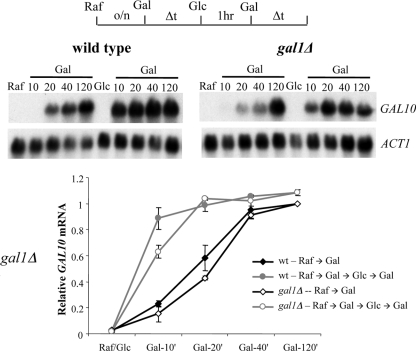
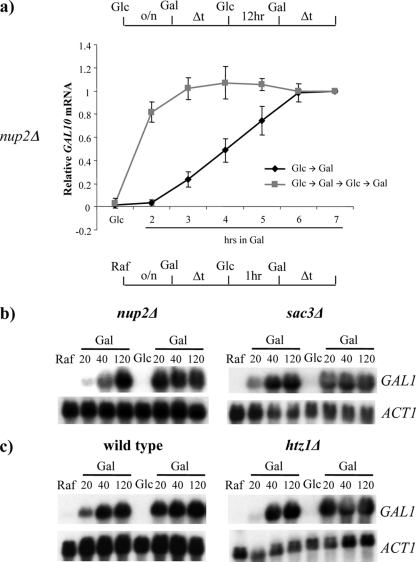
Single colonies of isogenic strains were isolated from glucose plates and grown overnight in glucose medium. GAL10 transcription was then induced by switching cultures to galactose medium (Glc→Gal), and cell aliquots were removed at various times for RNA isolation. Following 10 h in galactose medium, cells were transferred to repressive glucose medium for 12 h, followed by the reinduction of GAL genes by a switch back to galactose medium (Glc→Gal→Glc→Gal). RT-PCR analysis of GAL10 transcripts demonstrated that the initial round of expression is quite slow, with peak levels of expression in both the wild-type and gal1Δ strains requiring 6 to 7 h (Fig. 1a). In contrast, the reinduction of GAL10 in the wild-type strain was rapid, with peak levels of expression occurring within 2 to 3 h (Fig. 1a, left). Consistent with previous results, these rapid reinduction kinetics required GAL1 (Fig. 1a, right). In contrast, the inactivation of the SWI/SNF remodeling complex (swi2Δ) showed a rapid reinduction of both GAL10 and GAL1 (Fig. 1b and data not shown). Thus, in contrast to our previous study that demonstrated SWI/SNF dependence for the short-term memory of GAL gene transcription (21; see also Fig. 6), SWI/SNF is not required for long-term memory. This result is consistent with the cytoplasmic inheritance of Gal1 playing a dominant role in long-term memory (37).
FIG. 1.
Long-term memory at GAL genes requires Gal1p but not SWI/SNF. The schematic at the top depicts a regimen of growth in different carbon sources. Gal, 2% galactose; Glc, 2% glucose. (a) RT-PCR analysis of GAL10 mRNA levels following initial induction and reinduction after long-term glucose repression (12 h). The wild-type (WT) culture showed a memory of previous GAL10 induction, which was lost in the gal1Δ strain. (b) RT-PCR of GAL1 and GAL10 mRNA levels from the swi2Δ strain. The reinduction of GAL1 and GAL10 is fast compared to initial induction following long-term glucose repression (12 h). Data were averaged over three independent experiments and are represented as relative fold increases over ACT1 mRNA normalized to a maximum value of 1. Error bars represent standard deviations at each point.
FIG. 6.
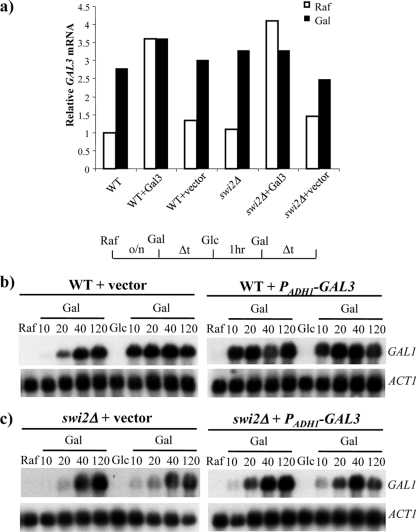
Gal3p coinducer overexpression leads to rapid GAL1 induction in the wild type. (a) GAL3 mRNA expression compared between growth in raffinose and that in galactose for the indicated strains and shown as fold induction over ACT1. (b) Northern analysis using the short-term memory regimen for wild-type cells transformed with the control plasmid (left) and a Gal3p-overexpressing plasmid (PADH1-GAL3) (right). (c) Same as panel b for the swi2Δ strain. The constitutive expression of Gal3p rapidly activates GAL1 transcription even upon initial induction in the wild type. The overexpression of Gal3 uncovers a more prominent role for SWI/SNF during the initial round of induction. All Northern blots were subsequently probed for ACT1 as a loading control.
Next, we tested whether Gal1p was also required for short-term memory. In this case, raffinose-grown cells were switched to galactose medium for the initial induction. In this case, maximal expression occurred at between 1 and 2 h. After 2 h, glucose was added to repress GAL expression for 60 min, and cells were then switched back to galactose medium (Raf→Gal→Glc→Gal). As expected, wild-type cells showed robust short-term memory at GAL10, with maximal expression occurring within 10 min (Fig. 2). Surprisingly, short-term memory was not eliminated in the gal1Δ strain, although the kinetics were reproducibly slower than those of the wild-type strain. Thus, it appears that Gal1p is not essential for the transcriptional memory of GAL10 expression after a brief period of repression.
FIG. 2.
Short-term memory at GAL10 does not require Gal1p. Shown is Northern analysis of GAL10 RNA levels in the wild-type (left) and gal1Δ (right) strains during initial induction and reinduction following short-term glucose repression (1 h). The gal1Δ strain rapidly reinduces GAL10 transcription with kinetics almost similar to those of the wild type. The graph shows data from an average of three experiments, while the top shows a representative experiment. Error bars represent standard deviations. ACT1 is the loading control for total RNA levels.
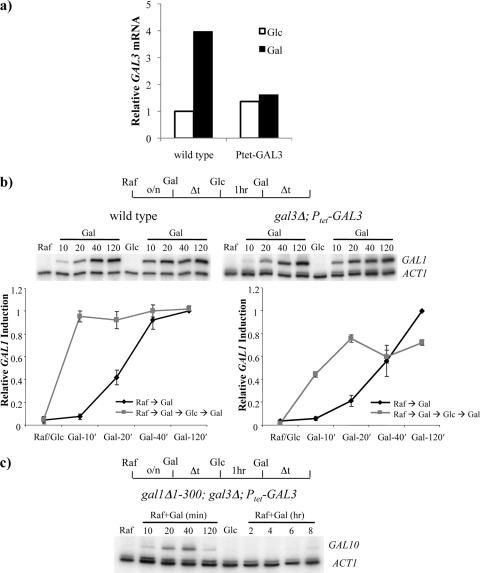
During the initial induction with galactose, the GAL3 gene was induced ∼3-fold, whereas GAL1 was induced ∼1,000-fold. Thus, we considered the possibility that memory involves the cytoplasmic inheritance of both Gal3 and Gal1; however, after many generations of growth in glucose (long-term memory), only Gal1 contributes to memory, as the considerably lower level of induced Gal3 would be depleted within a few cell divisions. Consistent with this view, GAL3 is not required for long-term memory (37). In contrast, during a short-term memory experiment, Gal3 and Gal1 might function in a redundant fashion. To test this possibility, we used strains in which Gal3 is expressed from a doxycycline-inducible promoter (Ptet-GAL3). In this strain, Gal3 was no longer inducible by galactose, but expression was maintained at a low, basal level in the absence of doxycycline (Fig. 3a) (2). In the absence of doxycycline, sufficient Gal3 is expressed to support a normal induction of GAL genes when cells are switched from raffinose to galactose medium (Fig. 3b). Likewise, when cells were then switched to glucose for 60 min, the reinduction of GAL1 was only slightly slower than that of the wild-type strain, indicating that memory was partially intact in the absence of Gal3 induction (Fig. 3b). However, the deletion of the GAL1 gene in the Ptet-GAL3 strain eliminated the rapid reinduction of GAL10, indicating that Gal1 and Gal3 play overlapping roles in transcriptional memory (Fig. 3c). In this strain, we also reproducibly observed a decrease in GAL10 transcription at later time points during the first induction and a poor accumulation of transcripts even at late time points in the second round. This is likely due to the galactose-dependent induction of the Gal80 negative-feedback loop. Since Gal3 levels were not concomitantly induced in these strains, the imbalance in the Gal80/Gal3 ratio is expected from previous studies to prevent Gal4 activation (see references 2 and 30 and references therein). These data further support the view that Gal1 and Gal3 function as redundant inducers of the GAL system and that this positive-feedback loop plays a major role in transcriptional memory.
FIG. 3.
Cytoplasmic signaling by both Gal1p and Gal3p is required for optimal GAL gene reinduction. (a) RT-PCR of wild-type and gal3Δ;Ptet-GAL3 strains showing GAL3 expression levels in glucose or galactose. (b) Minimal expression of Gal3p from a Ptet-GAL3 construct causes a partial defect in GAL1 reinduction following short-term glucose repression (1 h). Shown are GAL1 mRNA levels for the wild-type (left) and gal3Δ-Ptet-GAL3 (right) strains. At top is a representative RT-PCR, and the graphs below show data from an average of three independent experiments represented as fold increases over ACT1 mRNA normalized to a maximum value of 1. Error bars show standard deviations at each point. o/n, overnight. (c) GAL10 is induced in minimally expressing Ptet-GAL3 and an amino-terminal deletion of a GAL1 double mutant but is lost at 2 h postinduction. GAL10 reinduction is severely compromised even after 8 h following short-term glucose repression (1 h). Data in panel c are representative of five independent experiments.
Transcriptional memory of GAL genes does not require tethering at the nuclear periphery.
Several studies demonstrated that the activation of GAL1 or GAL10 transcription leads to the migration of the locus to the nuclear periphery, where it interacts with nuclear pore complexes (NPCs) (1, 4-6, 34). This relocalization event requires several components of the nuclear pore complex, such as Nup2 and Mlp1 (1, 4, 35, 36), as well as nuclear pore-associated factors that regulate mRNA export, such as Sac3 (8, 11, 16, 23). Initially, we tested whether the Nup2-dependent tethering of GAL10 to the nuclear periphery is required for long-term memory. We monitored GAL1 induction and reinduction kinetics in a nup2Δ strain (Fig. 4a) and found that the inactivation of Nup2 had no significant effect on the long-term transcriptional memory of GAL10.
FIG. 4.
GAL1 memory does not require nuclear pore localization or H2A.Z. Schematics at the top depict regimens of growth in different carbon sources. Raf, 2% raffinose; Gal, 2% galactose; Glc, 2% glucose. (a) RT-PCR analysis of GAL10 induction and reinduction in nup2Δ cells following long-term glucose repression (12 h). Long-term memory does not require Nup2p. Data represent data from three independent experiments. (b) Short-term GAL1 memory is unaffected in nup2Δ (left) and sac3Δ (right) mutants. (c) Northern analysis of wild-type (left) and htz1Δ (right) strains showing rapid GAL1 reinduction following short-term glucose repression (1 h). Data are representative of data from three independent experiments.
We then tested whether relocalization to the nuclear periphery plays a role in a short-term memory regimen. In these experiments, an isogenic set of strains that lacked either Nup2 or Sac3 was grown in galactose and then switched to glucose medium for 1 h prior to reinduction by the addition of galactose. Both of these components were previously shown to play essential roles in the relocalization of active GAL genes to the nuclear periphery (5, 20). As shown previously, high levels of GAL1 transcripts are detectable ∼10 min after the addition of galactose in the wild-type strain. Strikingly, the short-term memory of GAL transcription was intact in each of the mutants that blocked GAL localization to the periphery (Fig. 4b and data not shown). Thus, localization to the nuclear periphery is not essential for rapid GAL1 reinduction. These results are consistent with several studies demonstrating that plasmid-borne GAL genes are released from the nuclear periphery following >1 h of growth in glucose medium (1, 36; S. Kundu, C. L. Peterson, and M. Rosbash, unpublished results), and thus, peripheral localization in most strains does not appear to be maintained through more than one cell cycle.
H2A.Z does not contribute to short-term memory.
Brickner and colleagues (4) previously reported that the long-term memory of GAL1 transcription requires the histone variant H2A.Z (also known as Htz1). In our strain background (s288C), we found that an htz1Δ mutant had a significant defect in the initial induction of GAL1 or GAL10 when cells were switched from glucose to galactose medium, and this transcriptional defect made analyses of long-term memory problematic (data not shown). We found similar results for strains of the w303 background (data not shown). Our observations are consistent with data from a previous study demonstrating that Htz1 plays a key role in the recruitment of the Mediator complex during the initial activation of a glucose-repressed GAL gene (13, 24). To circumvent this issue, we asked if Htz1 was required for short-term memory. When cells were switched from raffinose to galactose medium, both the wild-type and htz1Δ strains showed identical kinetics of initial GAL1 induction (Fig. 4C). Interestingly, the htz1Δ strain also demonstrated a rapid reinduction of GAL1 transcription, indicating that Htz1 is not required for short-term memory in strains of the s288C background (Fig. 4C).
SWI/SNF promotes rapid PIC loading but does not generate alternate nucleosome positions.
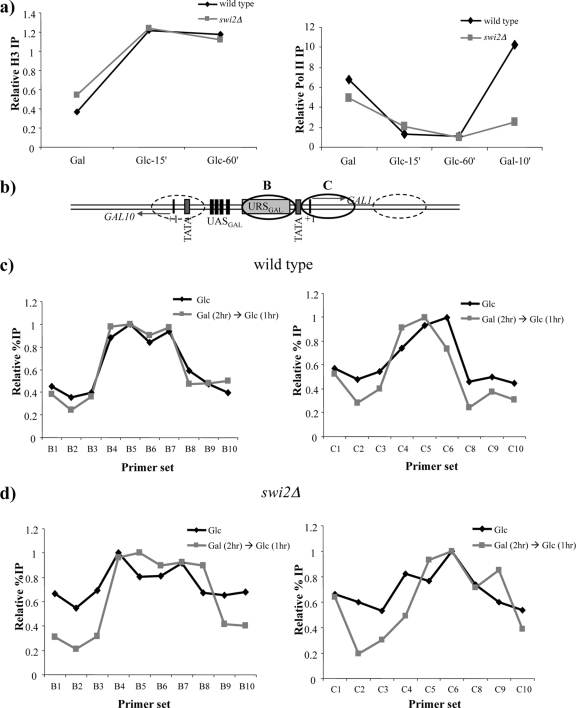
These studies indicate that SWI/SNF and Gal1/Gal3 may be the primary factors that control the transcriptional memory of GAL genes. To test if chromatin remodeling by SWI/SNF facilitates faster preinitiation complex (PIC) assembly on the GAL1 promoter during reinduction, we monitored the RNA polymerase II (RNAPII) association with the GAL1 promoter during a cycle of expression, repression, and reinduction. For both the wild-type and swi2Δ strains, RNAPII was rapidly lost from the promoter during glucose repression, as observed previously by us and others (21, 31). Glucose repression is also associated with a rapid reassembly of nucleosomes over the GAL1 promoter in both wild-type and swi2Δ strains (Fig. 5a, left). During reinduction in the wild-type strain, RNAPII was recruited to GAL1 within 10 min, paralleling the rapid appearance of GAL1 transcripts (Fig. 5a, right). Likewise, both TATA binding protein (TBP) and SWI/SNF were rapidly recruited to the GAL1 locus during reinduction, and transcriptional reinduction was associated with a rapid nucleosome loss (data not shown). In contrast, RNAPII was recruited slowly in the swi2Δ strain, reflecting the requirement for SWI/SNF to act at an early step in promoting rapid RNAPII re-recruitment during GAL gene reinduction.
FIG. 5.
SWI/SNF promotes RNA polymerase II loading but does not generate alternate nucleosome positions at the GAL1 promoter. (a, left) Histone H3 ChIP of the wild-type and swi2Δ strains to measure nucleosome occupancy at the GAL1 promoter. The loss of SWI/SNF does not inhibit the kinetics of promoter nucleosome reloading during glucose repression. (Right) RNA polymerase II ChIP of the wild-type and swi2Δ strains showing that the faster recruitment during GAL1 reinduction is dependent on SWI/SNF. RNA polymerase II and H3 levels were tested at the GAL1 promoter and normalized to a telomere sequence (Chr VI, 70 bp from the right end). (b) Schematic representation of the GAL1-10 regulatory region. UASGAL marks the Gal4p binding sites. URSGAL is the binding site for the glucose-dependent repressor Mig1p. Ovals represent previously mapped nucleosome positions. Ovals shown as solid lines represent GAL1 promoter nucleosomes that are mapped in c and d. TATA represents the TBP binding sites, and +1 represents the transcription start sites. (c) Nucleosome-scanning ChIP with histone H3 antibody in the wild-type strain for promoter nucleosomes B (left) and C (right). Black lines represent cultures grown in glucose overnight, and gray lines represent short-term (1-h) glucose-repressed cultures following a brief GAL1 induction. On the x axes of graphs, B1 to B10 represent 10 primer pairs spanning positions −302 to +3 from the translation start site. C1 to C10 represent nine primer pairs spanning positions −148 to +160 from the translation start site. On the y axis, the relative percent IP of H3 normalized to a maximum value of 1 is plotted. (d) Same as panel c but with an swi2Δ strain.
In our previous study, we proposed a model in which SWI/SNF might control transcriptional memory by influencing the positioning of nucleosomes that are reassembled onto the GAL1 promoter during glucose repression. Since SWI/SNF rapidly dissociates from the glucose-repressed GAL1 promoter (21), this model further proposes that these alternative nucleosome positions are propagated through DNA replication in the absence of SWI/SNF. Changes in nucleosome positioning might then enhance the rate of RNA polymerase II recruitment. To test this model directly, we mapped the positions of two promoter-proximal nucleosomes at the GAL1 locus (see Fig. 5b for a schematic representation) in wild-type and swi2Δ cells. Nucleosome positioning was mapped under two conditions: (i) cells that were grown continuously in glucose (long-term repression) and (ii) cells that had been grown in galactose and then repressed with glucose for 1 h (short-term repression). In each case, cells were treated with formaldehyde prior to collection and analysis of nucleosome positioning by nucleosome-scanning ChIP (32). Mononucleosomal chromatin was prepared by MNase digestion and used for ChIP with an anti-histone H3 antibody. Quantitative PCR was performed with primer pairs scanning approximately 300 bp around the predicted dyads of promoter nucleosome B (Fig. 5c, left) and promoter nucleosome C (Fig. 5c, right).
In long-term-repressed cells, nucleosomes B and C were positioned as predicted from previous studies, each protecting ∼160 bp of DNA. In contrast, no ChIP signal was detected when mononucleosomal chromatin was prepared from cells growing in galactose, consistent with a loss of GAL1 promoter nucleosomes (26, 27). However, when galactose-grown cells were exposed to glucose for 1 h, nucleosomes were reassembled, and the positioning of NucB and NucC appeared to be nearly identical between long-term-repressed and short-term-repressed cultures. Notably, this result is consistent with previous analyses of the GAL1 chromatin structure (7, 26). We repeated the same set of experiments with an swi2Δ strain and obtained similar results, confirming that SWI/SNF was not involved in nucleosome positioning at the GAL1 promoter during glucose repression (Fig. 5d). Thus, these data indicate that the role of SWI/SNF in promoting rapid GAL1 reinduction does not involve the generation of a novel pattern of nucleosome positions at the GAL1 promoter.
Gal3p coinducer overexpression leads to rapid GAL1 induction in wild-type cells.
We considered an alternate hypothesis in which SWI/SNF functions in both the first and second rounds of GAL1 induction but SWI/SNF action is only rate limiting for expression during reinduction. This model proposes that GAL1 expression is controlled by two different kinetic steps: the accumulation of signaling molecules, Gal1 and Gal3 (designated k1), which antagonize the Gal80 repressor, and chromatin remodeling (designated k2) (see model in Fig. 7). In this model, the accumulation of signaling molecules (k1) is the rate-limiting step for the initial induction of GAL genes (i.e., k2 > k1). However, in the second round of expression, signaling is very rapid due to the high levels of Gal3p and Gal1p that accumulate during initial induction, and chromatin remodeling by SWI/SNF becomes the slow step (i.e., k2 < k1). Note that in this model, SWI/SNF action must be partially redundant with other remodeling enzymes that remodel the promoter, albeit more slowly, in its absence (see Fig. 7). SWI/SNF is not required for the induced levels of Gal3 or Gal1, eliminating simple models where SWI/SNF directly regulates the Gal1/Gal3 feedback loop (18).
FIG. 7.
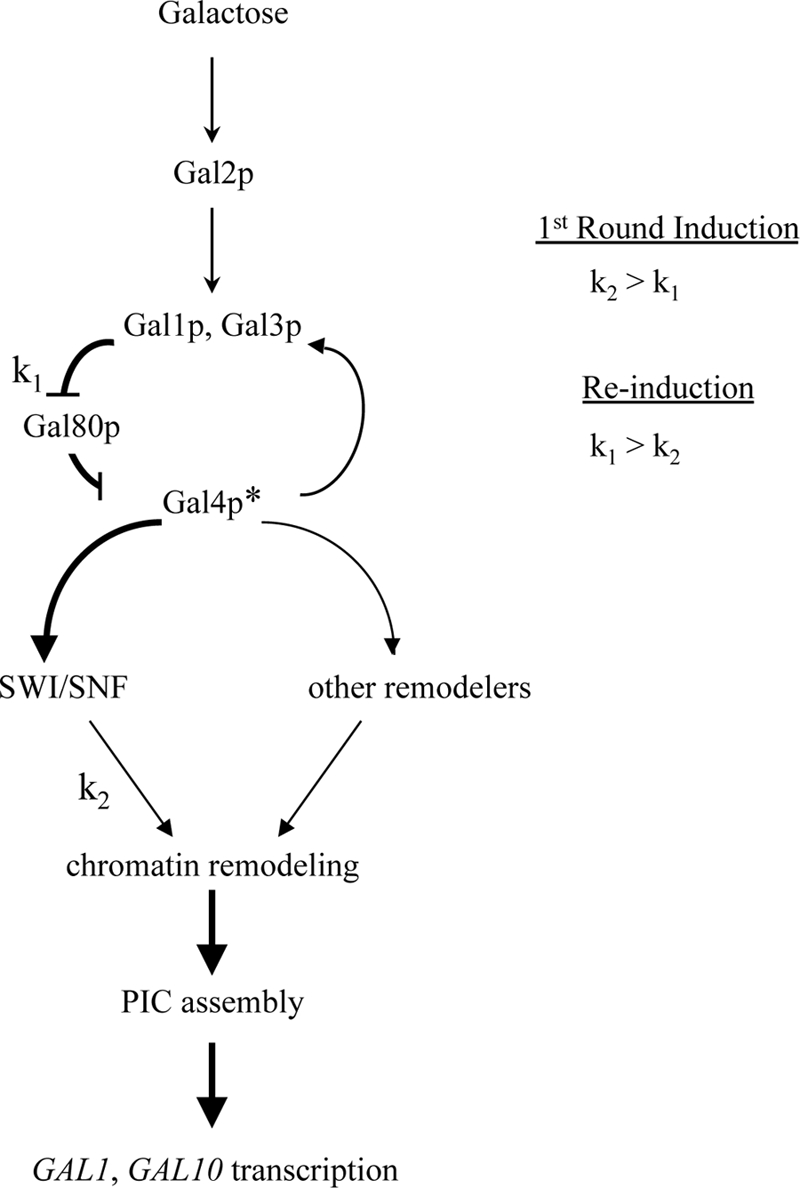
Two-step model for optimum induction kinetics of GAL1 transcription. Extracellular galactose is transported into the cytoplasm by Gal2p, and galactose-bound Gal3p or Gal1p antagonizes the Gal80 repressor, leading to the activation of the Gal4p activator and allowing it to recruit SWI/SNF. Chromatin remodeling by SWI/SNF leads to the rapid loss of promoter nucleosomes and PIC assembly, resulting in GAL1 induction. Gal4p activation (shown as Gal4p*) leads to an increased production of Gal1p and Gal3p, thereby amplifying the signal. k1 and k2 represent the rate-determining steps of this pathway, where k1 reflects the action of Gal1p and Gal3p and k2 reflects chromatin remodeling by SWI/SNF. In the absence of SWI/SNF, other nonspecific chromatin remodelers may cause promoter nucleosome loss. In the initial round of induction, Gal1p and Gal3p levels are low, making k1 the rate-determining step. After the initial round of expression and a brief period of glucose repression, Gal3p and Gal1p levels are much higher than those in the initial state. Hence, k1 is fast during GAL reinduction, making k2 the slowest and rate-determining step.
One prediction of this model is that increasing the levels of signaling molecules prior to the first round of induction should uncover a kinetic role for SWI/SNF during the initial round of expression. To test this idea, both wild-type and swi2Δ strains were transformed with a high-copy-number plasmid that harbors a constitutively expressed ADH1-GAL3 gene. The growth of these cells in raffinose medium leads to high levels of GAL3 expression in both the wild-type and swi2Δ strains (Fig. 6a). In the wild-type strain, the constitutive expression of Gal3p increased the rate of the initial induction such that the accumulation of GAL1 transcripts was nearly indistinguishable from the rate of reinduction (Fig. 6b, right). In contrast, cells transformed with the vector control showed slow kinetics of GAL1 expression in the first round and rapid kinetics during reinduction (Fig. 6b, left). Thus, constitutive, high-level expression of Gal3 appears to obviate transcriptional memory. Strikingly, the constitutive expression of GAL3 uncovered a significant role for SWI/SNF during the first round of expression (Fig. 5c, right). In fact, the GAL1 induction rates in the swi2Δ strain were identical in the first and second rounds of expression and were slow compared to wild-type rates. Thus, SWI/SNF action is generally rate limiting for GAL expression when cells contain high levels of Gal3. These data are fully consistent with the kinetic model shown in Fig. 7.
DISCUSSION
In the present study, we have carried out experiments aimed at reconciling studies that have investigated transcriptional memory at yeast GAL genes. In our previous work, we showed that the SWI/SNF remodeling enzyme was required for the rapid reinduction of GAL1 transcription following a 0.5- to 4-h period of glucose repression. In our experimental regimen, transcriptional memory was inherited through only 1 to 2 cell divisions. In contrast, two groups reported an experimental scheme in which the transcriptional memory of GAL genes was maintained for at least 12 h under glucose-repressed conditions (4 to 6 divisions). In this case, memory was shown to require a GAL1 gene product, the H2A.Z histone variant, and it correlated with peripheral nuclear localization. Here we have shown that the tethering of GAL loci to the nuclear periphery is not essential for transcriptional memory in either scheme. Our data also indicate that SWI/SNF, and likely H2A.Z, functions downstream of the transcriptional memory process, regulating the general kinetics of GAL induction. In contrast, transcriptional memory generally requires the Gal1/Gal3 feedback loop, with the function of Gal1, as a weak Gal3-like inducer, predominating in the long-term memory regimen and both factors showing overlapping functions in a short-term memory scheme. Thus, the transcriptional memory of GAL genes does not appear to involve the inheritance of chromatin states but rather involves the cytoplasmic inheritance of Gal1 and/or Gal3 signaling factors.
The kinetics of GAL gene induction are strongly influenced by the concentration of galactose in the medium, levels of the Gal3 inducer, and levels of the Gal80 repressor (2, 30). Alterations in the levels of these factors can have a strong impact on GAL transcription. Likewise, our data indicate that the levels of Gal3 can also impact the extent to which SWI/SNF is required for the optimal kinetics of GAL induction. At low, basal Gal3 concentrations, SWI/SNF action does not appear to be rate limiting for GAL expression, as the inactivation of SWI/SNF has only a minor effect on induction kinetics (Fig. 1). However, an increased expression of Gal3 leads to a more extensive GAL transcriptional defect in the absence of SWI/SNF. These results provide an explanation for why swi/snf mutations in some strain backgrounds lead to strong defects in GAL transcription even during an initial round of expression (10). Presumably, these strain backgrounds have a higher level of basal Gal3 signaling (or lower levels of the Gal80 repressor). Differences in the potencies of the Gal3 positive-feedback loop among strain backgrounds may also explain why the Htz1 histone variant plays variable roles in the initial induction of GAL genes (4, 13).
In the strain background used in our studies (s288C), Htz1 contributes significantly to the initial induction kinetics of GAL1 when cells are transferred from glucose to galactose medium, and consequently, we were unable to assess its role during the long-term memory regimen. We have also found that Htz1 is required for rapid GAL induction kinetics using a second, common strain background (w303) (our unpublished observations). Similar results were reported previously by Gligoris et al. (13). In contrast, Brickner and colleagues (4) used the JBY strain background, and thus, it remains a possibility that Htz1 may contribute to the transcriptional memory phenomenon in these strains.
Several studies demonstrated that the activation of GAL gene expression leads to the rapid localization of these loci to the nuclear periphery and interaction with NPCs. It was suggested that this NPC association may facilitate the rapid export of GAL mRNA into the cytoplasm. Brickner and colleagues (4) also reported that GAL1 maintains its association with the NPC for many cell divisions, suggesting that peripheral localization may be involved in memory. The genetic requirements for GAL1 localization to the nuclear periphery are well established, with key roles being played by the NPC component Nup2 and components of the mRNA processing and export machinery, such as Sac3. We find that the inactivation of any one of these factors has no detectable impact on GAL transcriptional memory, indicating that while GAL gene transcription tethers the loci to the nuclear periphery, tethering itself is not required for memory. We note that our results are consistent with several studies from the Rosbash group demonstrating that GAL genes are released from the periphery after 40 min to 1 h in glucose medium (see, e.g., reference 36) and thus, tethering does not generally correlate well with memory. Likewise, we have used a plasmid-based system in our strain background for the monitoring of GAL localization, and we also find that plasmid-borne GAL genes are released from the periphery after 1 h of glucose repression (36; S. Kundu, C. L. Peterson, M. Rosbash, and S. Vodala, unpublished results). Furthermore, previous studies have shown that parental and newly assembled NPCs are asymmetrically segregated during yeast cell division. Daughter cells receive only new nuclear membrane material and newly assembled NPCs, with all of the parental NPCs remaining in the mother cell (33). Thus, if a gene locus is associated with a parental NPC, a daughter cell can inherit this structure from the mother only if interactions with parental and newly assembled NPCs are dynamic. Such dynamic interactions, however, would appear to be inconsistent with a memory phenomenon that requires the faithful segregation of the parental state. Together, the data indicate that the tethering of loci to NPCs is unlikely to contribute to transcriptional memory events that are heritable to progeny.
Recently, two groups reported that the transcriptional induction of GAL genes leads to the formation of an intragene loop between the 5′ and 3′ ends of the GAL1,10 gene cluster (22, 35). Loop formation requires the general transcription factor TFIIB and components of the mRNA processing and export machinery. During a period of glucose repression, the stability of this gene loop also requires the NPC-associated protein Mlp1. Interestingly, this loop is maintained for at least 1 cell division in glucose medium, and the disruption of the intragene loop leads to the loss of the short-term transcription memory of GAL genes. SWI/SNF is not required for gene loop formation or maintenance, suggesting that the SWI/SNF action functions downstream. Those authors also showed that the intragene loop is required for rapid RNAPII recruitment during GAL1 reinduction, leading those authors to suggest that the intragene loop facilitates the reassociation of RNAPII during the second round of expression. However, since SWI/SNF is also required for RNAPII recruitment, but not loop formation, these data indicate that the loop is not sufficient for RNAPII recruitment. The intragene loop is required to maintain the Gal4 activator at the promoter during glucose repression, so one simple model that we favor is that the loop functions primarily to maintain Gal4 so that it is poised to rapidly re-recruit SWI/SNF and other key targets when galactose is reencountered in the environment.
Footnotes
Published ahead of print on 8 March 2010.
REFERENCES
- 1.Abruzzi, K. C., D. A. Belostotsky, J. A. Chekanova, K. Dower, and M. Rosbash. 2006. 3′-end formation signals modulate the association of genes with the nuclear periphery as well as mRNP dot formation. EMBO J. 25:4253-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Acar, M., A. Becskei, and A. van Oudenaarden. 2005. Enhancement of cellular memory by reducing stochastic transitions. Nature 435:228-232. [DOI] [PubMed] [Google Scholar]
- 3.Bhat, P. J., and T. V. Murthy. 2001. Transcriptional control of the GAL/MEL regulon of yeast Saccharomyces cerevisiae: mechanism of galactose-mediated signal transduction. Mol. Microbiol. 40:1059-1066. [DOI] [PubMed] [Google Scholar]
- 4.Brickner, D. G., I. Cajigas, Y. Fondufe-Mittendorf, S. Ahmed, P. C. Lee, J. Widom, and J. H. Brickner. 2007. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 5:e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cabal, G. G., A. Genovesio, S. Rodriguez-Navarro, C. Zimmer, O. Gadal, A. Lesne, H. Buc, F. Feuerbach-Fournier, J. C. Olivo-Marin, E. C. Hurt, and U. Nehrbass. 2006. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature 441:770-773. [DOI] [PubMed] [Google Scholar]
- 6.Casolari, J. M., C. R. Brown, S. Komili, J. West, H. Hieronymus, and P. A. Silver. 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117:427-439. [DOI] [PubMed] [Google Scholar]
- 7.Cavalli, G., and F. Thoma. 1993. Chromatin transitions during activation and repression of galactose-regulated genes in yeast. EMBO J. 12:4603-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chekanova, J. A., K. C. Abruzzi, M. Rosbash, and D. A. Belostotsky. 2008. Sus1, Sac3, and Thp1 mediate post-transcriptional tethering of active genes to the nuclear rim as well as to non-nascent mRNP. RNA 14:66-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dion, M. F., T. Kaplan, M. Kim, S. Buratowski, N. Friedman, and O. J. Rando. 2007. Dynamics of replication-independent histone turnover in budding yeast. Science 315:1405-1408. [DOI] [PubMed] [Google Scholar]
- 10.Ferreira, M. E., P. Prochasson, K. D. Berndt, J. L. Workman, and A. P. Wright. 2009. Activator-binding domains of the SWI/SNF chromatin remodeling complex characterized in vitro are required for its recruitment to promoters in vivo. FEBS J. 276:2557-2565. [DOI] [PubMed] [Google Scholar]
- 11.Fischer, T., K. Strasser, A. Racz, S. Rodriguez-Navarro, M. Oppizzi, P. Ihrig, J. Lechner, and E. Hurt. 2002. The mRNA export machinery requires the novel Sac3p-Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. EMBO J. 21:5843-5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frolova, E., M. Johnston, and J. Majors. 1999. Binding of the glucose-dependent Mig1p repressor to the GAL1 and GAL4 promoters in vivo: regulation by glucose and chromatin structure. Nucleic Acids Res. 27:1350-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gligoris, T., G. Thireos, and D. Tzamarias. 2007. The Tup1 corepressor directs Htz1 deposition at a specific promoter nucleosome marking the GAL1 gene for rapid activation. Mol. Cell. Biol. 27:4198-4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hawkins, K. M., and C. D. Smolke. 2006. The regulatory roles of the galactose permease and kinase in the induction response of the GAL network in Saccharomyces cerevisiae. J. Biol. Chem. 281:13485-13492. [DOI] [PubMed] [Google Scholar]
- 15.Hittinger, C. T., and S. B. Carroll. 2007. Gene duplication and the adaptive evolution of a classic genetic switch. Nature 449:677-681. [DOI] [PubMed] [Google Scholar]
- 16.Jani, D., S. Lutz, N. J. Marshall, T. Fischer, A. Kohler, A. M. Ellisdon, E. Hurt, and M. Stewart. 2009. Sus1, Cdc31, and the Sac3 CID region form a conserved interaction platform that promotes nuclear pore association and mRNA export. Mol. Cell 33:727-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang, F., B. R. Frey, M. L. Evans, J. C. Friel, and J. E. Hopper. 2009. Gene activation by dissociation of an inhibitor from a transcriptional activation domain. Mol. Cell. Biol. 29:5604-5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnston, M. 1987. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol. Rev. 51:458-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnston, M., J. S. Flick, and T. Pexton. 1994. Multiple mechanisms provide rapid and stringent glucose repression of GAL gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 14:3834-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohler, A., M. Schneider, G. G. Cabal, U. Nehrbass, and E. Hurt. 2008. Yeast ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nat. Cell Biol. 10:707-715. [DOI] [PubMed] [Google Scholar]
- 21.Kundu, S., P. J. Horn, and C. L. Peterson. 2007. SWI/SNF is required for transcriptional memory at the yeast GAL gene cluster. Genes Dev. 21:997-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laine, J. P., B. N. Singh, S. Krishnamurthy, and M. Hampsey. 2009. A physiological role for gene loops in yeast. Genes Dev. 23:2604-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lei, E. P., C. A. Stern, B. Fahrenkrog, H. Krebber, T. I. Moy, U. Aebi, and P. A. Silver. 2003. Sac3 is an mRNA export factor that localizes to cytoplasmic fibrils of nuclear pore complex. Mol. Biol. Cell 14:836-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemieux, K., M. Larochelle, and L. Gaudreau. 2008. Variant histone H2A.Z, but not the HMG proteins Nhp6a/b, is essential for the recruitment of Swi/Snf, Mediator, and SAGA to the yeast GAL1 UAS(G). Biochem. Biophys. Res. Commun. 369:1103-1107. [DOI] [PubMed] [Google Scholar]
- 25.Li, X. Y., S. R. Bhaumik, and M. R. Green. 2000. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science 288:1242-1244. [DOI] [PubMed] [Google Scholar]
- 26.Lohr, D. 1984. Organization of the GAL1-GAL10 intergenic control region chromatin. Nucleic Acids Res. 12:8457-8474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lohr, D., and J. Lopez. 1995. GAL4/GAL80-dependent nucleosome disruption/deposition on the upstream regions of the yeast GAL1-10 and GAL80 genes. J. Biol. Chem. 270:27671-27678. [DOI] [PubMed] [Google Scholar]
- 28.Lohr, D., P. Venkov, and J. Zlatanova. 1995. Transcriptional regulation in the yeast GAL gene family: a complex genetic network. FASEB J. 9:777-787. [DOI] [PubMed] [Google Scholar]
- 29.Meyer, J., A. Walker-Jonah, and C. P. Hollenberg. 1991. Galactokinase encoded by GAL1 is a bifunctional protein required for induction of the GAL genes in Kluyveromyces lactis and is able to suppress the gal3 phenotype in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:5454-5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramsey, S. A., J. J. Smith, D. Orrell, M. Marelli, T. W. Petersen, P. de Atauri, H. Bolouri, and J. D. Aitchison. 2006. Dual feedback loops in the GAL regulon suppress cellular heterogeneity in yeast. Nat. Genet. 38:1082-1087. [DOI] [PubMed] [Google Scholar]
- 31.Schwabish, M. A., and K. Struhl. 2007. The Swi/Snf complex is important for histone eviction during transcriptional activation and RNA polymerase II elongation in vivo. Mol. Cell. Biol. 27:6987-6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sekinger, E. A., Z. Moqtaderi, and K. Struhl. 2005. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol. Cell 18:735-748. [DOI] [PubMed] [Google Scholar]
- 33.Shcheprova, Z., S. Baldi, S. B. Frei, G. Gonnet, and Y. Barral. 2008. A mechanism for asymmetric segregation of age during yeast budding. Nature 454:728-734. [DOI] [PubMed] [Google Scholar]
- 34.Taddei, A., G. Van Houwe, F. Hediger, V. Kalck, F. Cubizolles, H. Schober, and S. M. Gasser. 2006. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature 441:774-778. [DOI] [PubMed] [Google Scholar]
- 35.Tan-Wong, S. M., H. D. Wijayatilake, and N. J. Proudfoot. 2009. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev. 23:2610-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vodala, S., K. C. Abruzzi, and M. Rosbash. 2008. The nuclear exosome and adenylation regulate posttranscriptional tethering of yeast GAL genes to the nuclear periphery. Mol. Cell 31:104-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zacharioudakis, I., T. Gligoris, and D. Tzamarias. 2007. A yeast catabolic enzyme controls transcriptional memory. Curr. Biol. 17:2041-2046. [DOI] [PubMed] [Google Scholar]