Abstract
Trimethylated lysine 27 of histone H3 (H3K27me3) is an epigenetic mark for gene silencing and can be demethylated by the JmjC domain of UTX. Excessive H3K27me3 levels can cause tumorigenesis, but little is known about the mechanisms leading to those cancers. Mutants of the Drosophila H3K27me3 demethylase dUTX display some characteristics of Trithorax group mutants and have increased H3K27me3 levels in vivo. Surprisingly, dUTX mutations also affect H3K4me1 levels in a JmjC-independent manner. We show that a disruption of the JmjC domain of dUTX results in a growth advantage for mutant cells over adjacent wild-type tissue due to increased proliferation. The growth advantage of dUTX mutant tissue is caused, at least in part, by increased Notch activity, demonstrating that dUTX is a Notch antagonist. Furthermore, the inactivation of Retinoblastoma (Rbf in Drosophila) contributes to the growth advantage of dUTX mutant tissue. The excessive activation of Notch in dUTX mutant cells leads to tumor-like growth in an Rbf-dependent manner. In summary, these data suggest that dUTX is a suppressor of Notch- and Rbf-dependent tumors in Drosophila melanogaster and may provide a model for UTX-dependent tumorigenesis in humans.
Mammalian UTX, UTY, and JmjD3 and Drosophila UTX (dUTX) are histone demethylases that specifically demethylate di- and trimethylated lysine 27 on histone H3 (H3K27me2 and H3K27me3, respectively) (1, 20, 32, 42, 43, 69). The catalytic domain of this activity is the Jumonji C (JmjC) domain, located at the C terminus of these proteins (Fig. 1H). The N-terminal domains of UTX, UTY, and dUTX contain several tetratricopeptide repeats (TPRs) thought to be required for protein-protein interactions (4).
FIG. 1.
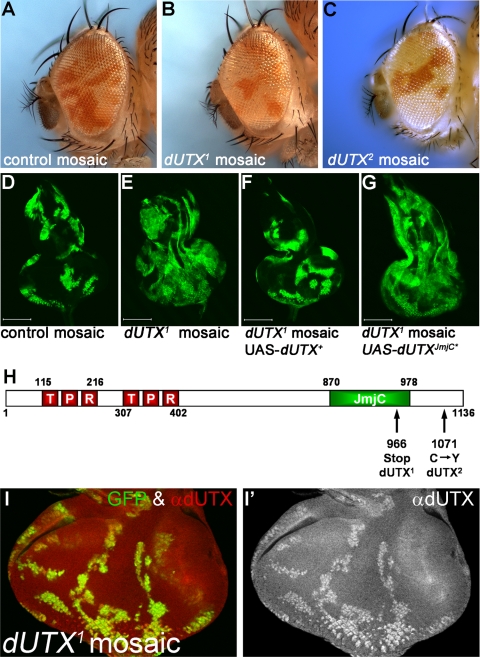
Identification of dUTX alleles as overrepresentation mutants in mosaic eyes. (A to C) Representative examples of mosaic eyes of wild-type controls (A), dUTX1 mosaics (B), and dUTX2 mosaics (C). Note the overrepresentation of the dUTX mutant tissue, marked in white, compared to the twin spots, marked in red (B and C). (D to G) Representative examples of mosaic eye-antennal imaginal discs of wild-type control (D), dUTX1 mosaics (E), dUTX1 mosaics expressing a UAS-dUTX+ rescue construct (F), and dUTX1 mosaics expressing a UAS-dUTXJmjC* catalytic mutant construct (G) using the MARCM system (44). Clones are positively labeled by GFP (green). Scale bars represent 100 μm. (H) Domain structure of dUTX and location of mutations. (I and I′) Mosaic eye imaginal discs of dUTX1 were labeled with anti-dUTX antibody. (I) Merged GFP and antibody channels. (I′) Antibody-only channels. Clones are marked by the absence of GFP. The dUTX1 allele produces no or strongly reduced levels of the dUTX protein. Note the strong overrepresentation phenotype of dUTX1 clones in this disc. Genotypes were as follows: ey-FLP; y+ FRT40A/P[w+] FRT40A (A), ey-FLP; dUTX1 FRT40A/P[w+] FRT40A (B), ey-FLP; dUTX2 FRT40A/P[w+] FRT40A (C), hs-FLP UAS-CD8:GFP; P[y+] FRT40A/P[tub-GAL80] FRT40A/P[tub-GAL4] (D), hs-FLP UAS-CD8:GFP; dUTX1 FRT40A/P[tub-GAL80] FRT40A/P[tub-GAL4] (E), hs-FLP UAS-CD8:GFP; dUTX1 FRT40A UAS-dUTX+/P[tub-GAL80] FRT40A/P[tub-GAL4] (F), hs-FLP UAS-CD8:GFP; dUTX1 FRT40A UAS-dUTXJmjC*/P[tub-GAL80] FRT40A/P[tub-GAL4](G), and ey-FLP; dUTX1 FRT40A/P[ubi-GFP] FRT40A (I and I′).
H3K27me3 is a histone mark for Polycomb (Pc)-mediated genomic silencing and transcriptional repression and is associated with animal body patterning, X-chromosome inactivation, genomic imprinting, and stem cell maintenance (51, 59, 71). H3K27 methylation is catalyzed by Polycomb repressive complex 2 (PRC2), which in Drosophila is composed of the catalytic subunit enhancer of zeste [E(z)] (EZH2 in mammals), extra sex combs (Esc), suppressor of zeste 12 [Su(z)12], and nucleosome remodeling factor 55 (Nurf55) (11, 16, 36, 41, 50, 52). H3K27me3 is recognized by the chromodomain of Pc, which is a component of a different silencing complex, called PRC1, which, in addition to Pc, contains Polyhomeotic (Ph), posterior sex combs (Psc), and dRING (27, 49, 66). The wild-type function of UTX is to demethylate H3K27me3 and, thus, to antagonize Polycomb-mediated silencing.
UTX is also a component of mixed-lineage leukemia complex 3 (MLL3) and MLL4 (15, 34, 56). MLL complexes are histone methyltransferases for H3K4. The function of UTX in MLL3 and MLL4 is unknown. However, it appears that UTX is not required for the H3K4 methyltransferase activity of MLL3 and MLL4 (43).
The best-characterized targets of H3K27me3/Pc-mediated silencing are homeotic genes, which are critical regulators of animal patterning (33, 57). However, many other genes are also enriched for H3K27 methylation and Pc binding (5, 6, 45, 53, 65, 72, 76). Furthermore, elevated H3K27me3 levels due to an increased activity of the methyltransferase EZH2 could be a leading cause of certain human cancers (7, 37, 39, 64, 78). Recently, mutations that inactivate UTX, and which are thus expected to cause increased H3K27me3 levels, have been linked to the development and progression of human cancer (77). However, the precise mechanisms by which this occurs are largely unknown.
Notch is the receptor of a highly conserved signaling pathway involved in many biological processes, including lateral inhibition, stem cell maintenance, and proliferation control (reviewed in reference 8). The binding of Delta or Serrate, the two ligands in Drosophila melanogaster, triggers the proteolytic processing of Notch, resulting in the release and translocation of the Notch intracellular domain (NICD) into the nucleus, where it regulates gene expression (reviewed in reference 8). Aberrant, oncogenic Notch signaling has been linked to tumor development in humans, including T-cell acute lymphoblastic leukemias (T-ALLs), pancreatic cancer, medulloblastoma, and mucoepidermoid carcinoma (68, 81). Thus, an improved understanding of Notch signaling will have significant implications for human health.
In Drosophila, the Notch signaling pathway also controls the growth of the eye primordium and wing margin formation during development (3, 10, 17, 18, 21, 55, 61, 73). Although the mechanistic details are unclear, one way by which Notch signaling controls proliferation during Drosophila eye development is through the negative regulation of the Retinoblastoma (Rb) family member Rbf (3). Rbf inactivation has also been implicated in Notch-induced eye tumors in Drosophila (26). Rb is a tumor suppressor that negatively regulates cell cycle progression through the inhibition of the transcription factor E2F (2, 13, 14, 23). Rb binds directly to E2F and represses its transcriptional activity. The release of Rb activates E2F to induce the transcription of cell cycle regulators such as cyclin E and PCNA (24, 30, 46, 48). Therefore, the inactivation of Rbf by increased Notch signaling can trigger increased proliferation, which may lead to cancerous growth.
Here, we genetically characterize loss-of-function mutations of dUTX. dUTX mutants display some of the characteristics of Trithorax group mutants and have increased H3K27me3 levels in vivo. Surprisingly, dUTX mutations also affect H3K4me1 levels in a JmjC-independent manner. We show that dUTX mutant tissue has an H3K27me3-dependent growth advantage over wild-type tissue due to increased proliferation in the developing eye. The growth advantage of dUTX mutant tissue is caused by increased Notch activity, demonstrating that dUTX is a Notch antagonist. The inactivation of Rbf contributes to the growth advantage of dUTX mutant tissue. Moreover, an excessive activation of Notch in dUTX mutant cells leads to tumor-like growth in an Rbf-dependent manner. In summary, these data suggest that dUTX is a suppressor of Notch- and Rbf-dependent tumors in Drosophila and may provide a model for UTX-dependent tumorigenesis in humans.
MATERIALS AND METHODS
Drosophila genetics.
The dUTX1 and dUTX2 alleles were obtained in a mutagenesis screen for chromosome arm 2L as described previously (58). Clones in larval imaginal discs and in the adult eye were induced with the ey-FLP/FRT system (80) by using an FLP that was expressed under the control of the eyeless (ey) promoter (54). Clones in Fig. 1D to G and 6F to I were induced by use of the mosaic analysis with a repressible cell marker (MARCM) system (44). MARCM clones were induced by a 60-min heat shock at 37°C in late-first-instar larvae.
FIG. 6.
dUTX interacts genetically with Notch. (A) Change of mRNA levels in dUTX mutant larvae. qRT-PCR analysis of transcript levels of the indicated genes from homozygous mutant dUTX1 and dUTXPB third-instar larvae. The levels are normalized to 100% for wild-type larvae (red line). Note that dUTX mRNA levels in dUTX1 and dUTXPB mutant larvae are also reduced. (B and B′) Anti-Notch antibody labeling of dUTX1 mosaic eye imaginal discs. The posterior is to the right, and the dorsal half is up. (B) Merged GFP and antibody channels. (B′) Antibody-only channel. dUTX clones are marked by the absence of GFP. Clones in the ventral half of the eye disc, especially in the morphogenetic furrow, contain elevated levels of the Notch protein. The anti-Notch antibody was raised against the intracellular domain. (C) Heterozygosity of Notch suppresses the overrepresentation phenotype of dUTX mosaic eyes (compare to Fig. 1B and 4C). (D and E) Wing-notching phenotype caused by the loss of one gene dose of Notch (N) (D) and Serrate (Ser) (E). (D′, D″, E′, E″, and E‴) Heterozygosity of dUTX dominantly suppresses the wing-notching phenotype of N and Ser. The suppression varies from mild (D′ and E′) to complete (D″ and E‴), which is quantified in F and G for the three dUTX alleles. Genotypes are as follows: ey-Flp; dUTX1 FRT40A/P[ubi-GFP] FRT40A (B and B′), ey-FLP/N264-39; dUTX1 FRT40A/P[w+] FRT40A (C), N8/+; P[y+] FRT40A/+ (D), N8/+; dUTX1 FRT40A/+ (D′ and D″), Ser1/+ (E), and dUTX1 FRT40A/+; Ser1/+ (E′ to E‴).
In dUTXJmjC*, His883 was changed to Ala (CAC→GCC) and Glu885 was changed to Ala (GAG→GCG) by site-directed mutagenesis. These changes disrupt the catalytic activity as demethylase (38). To obtain UAS-dUTX+ and UAS-dUTXJmjC* transgenes, the corresponding cDNA sequences were cloned into the KpnI site of pUAST-attB and inserted at 53B7 on the right arm of chromosome 2. The expression levels of both transgenes are similar (data not shown).
The following mutants and fly stocks were used: UTX1, dUTX2, dUTXPB (also known as PBac{WH}UTXf01321), UAS-dUTX+, and UAS-dUTXJmjC* (this study); RbfΔ14 and UAS-Rbf (a gift from Nick Dyson); UAS-rasV12S35 (35); N8, UAS-Delta (line 170), and hs-FLP UAS-CD8:GFP; P[tub-GAL80]; P[tub-Gal4] (a gift from Hugo Bellen) (single colons separate the different chromosomes on which the genes are located); and eyeful (ey-Gal4 UAS-Dl GS88A8) (26) (a gift of Maria Dominguez and Wouter Bossyut). All other stocks [dUTXPB, E(z)731, Pc1, N264-39, Ser1, en-Gal4, and UAS-GFP] were obtained from the Bloomington Stock Center. The eyeful crosses with dUTX alleles were performed at 18°C, because at higher temperatures, the eyeful phenotype was too strong to be visibly modified by dUTX heterozygosity.
Immunohistochemistry and antibodies.
Eye-antennal imaginal discs were dissected, fixed, and immunolabeled according to standard procedures. dUTX mutant clones were identified by either the absence or the presence (MARCM) of green fluorescent protein (GFP). The following antibodies were used to characterize the specificity of dUTX (Fig. 2 and Table 1): H3 (Abcam), H3K4me1 (Abcam), H3K4me2 (Abcam), H3K4me3 (Abcam), H3K9me1 (Upstate), H3K9me3 (Lake Placid), H3K27me2 (Abcam), H3K27me3 (Lake Placid), H3K36me1 (Abcam), H3K36me3 (Abcam), H3K79me1 (Lake Placid), and H4K20me1 (Lake Placid). dUTX antibodies were raised against the N-terminal part of dUTX (amino acids [aa] 1 to 153). The corresponding dUTX cDNA was cloned with NheI/BamHI into pET-28b(+). The recombinant dUTX protein was purified via Ni-chelate affinity chromatography and injected into rabbits. Monoclonal anti-Notch antibody (clone C17.9C6) was raised against the intracellular domain of Notch (NICD) and obtained from the Developmental Studies Hybridoma Bank (DSHB) in Iowa.
FIG. 2.
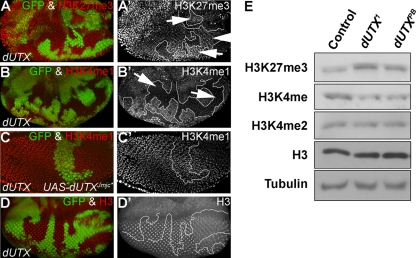
dUTX controls H3K27me3 demethylation and H3K4 monomethylation. (A to D) Eye imaginal discs were labeled with antibodies specific for the indicated histone methyl modification. GFP fluorescence was used to identify dUTX clones. Left panels are the merged GFP and antibody channels. Right panels are the antibody-only channels. To ease the identification of dUTX clones, clonal boundaries are indicated with white lines. (A and A′) Global levels of H3K27me3 are increased in dUTX mutant clones (arrows) in eye imaginal discs. dUTX mutant clones are negatively marked by the absence of GFP. (B and B′) Global levels of H3K4me1 are reduced in dUTX mutant clones (arrows) in eye imaginal discs. dUTX mutant clones are negatively marked by the absence of GFP. (C and C′) Expression of the JmjC catalytic mutant dUTXJmjC* in dUTX mutant clones by the MARCM system rescues the H3K4me1 methylation defect. Clones are positively marked by GFP. (D and D′) Global levels of histone H3 are not altered in dUTX mutant clones in eye imaginal discs. dUTX mutant clones are negatively marked by the absence of GFP. (E) Western blot analysis of larval extracts of the indicated genotype probed with the antibodies listed at the left. Genotypes are as follows: ey-FLP; dUTX1 FRT40A/P[ubi-GFP] FRT40A (A, B, and D) and hs-FLP UAS-CD8:GFP; dUTX1 FRT40A UAS-dUTXJmjC*/P[tub-GAL80] FRT40A/P[tub-GAL4] (C).
TABLE 1.
Analysis of global histone methyl modifications in dUTX mutantsa
| Histone modification | Change of methylation pattern |
|---|---|
| H3K4me1 | Downregulated in dUTX clones |
| H3K4me2 | − |
| H3K4me3 | − |
| H3K9me1 | − |
| H3K9me3 | − |
| H3K27me1 | − |
| H3K27me2 | − |
| H3K27me3 | Upregulated in dUTX clones |
| H3K36me1 | − |
| H3K36me3 | − |
| H3K79me1 | − |
| H3K20me1 | − |
Antibodies specific for the indicated histone methyl modifications were tested in dUTX mosaic eye imaginal discs. Only global levels of H3K4me1 and H3K27me3 were altered in dUTX mutant clones (Fig. 2). − indicates no change in the histone methylation pattern compared to that of the wild type.
Western blots.
For the Western blots shown in Fig. 2E, extracts of wild-type control (w; FRT40) and homozygous dUTX1 and dUTXPB larvae were prepared in 4× SDS loading buffer and separated by 18% SDS-PAGE. For alpha-tubulin labeling, extracts were separated by 12% gels. After Western transfer, the blots were probed with H3K27me3-, H3K4me1-, and H3K4me2-specific antibodies and H3 and alpha-tubulin antibodies as controls. Anti-H3, -H3K4me1, and -H3K4me2 antibodies were obtained from Abcam, anti-H3K27me3 antibody was obtained from Active Motif, and anti-alpha-tubulin antibody was obtained from Sigma.
RT-PCR and dUTX RNA interference (RNAi).
All quantitative reverse transcription (RT) (qRT)-PCRs were performed in triplicate with the QuantiTect SYBR green RT-PCR kit from Qiagen (catalog number 204243) with 100 ng of RNA and 0.2 μM each primer per reaction.
Primers for RT-PCR were dUTX forward primer AATGTTGGACCCTTGACTGC, dUTX reverse primer TCCTTGCAAGATTCCAGCTT, rp49 forward primer CCAGTCGGATCGATATGCTAA, rp49 reverse primer GTTCGATCCGTAACCGATGT, numb forward primer TGAGCCAACTGTGTCAGGAG, numb reverse primer AATGCTGCGATTGTTGTTGA, α-adaptin forward primer TTAGCAATCGGGAGACCAAC, α-adaptin reverse primer CAAAATGACCACCTCCTGGT, four-jointed forward primer CTGGTGCAGCGACTAATTGA, four-jointed reverse primer GGCGTTCCATTGAAAGTTGT, big brain forward primer ACAGCAACAGCAACAAGTGG, big brain reverse primer GCTGCATCTGATTCTGGACA, Rbf forward primer TCACTGCATAACTCGCCAAG, Rbf reverse primer GGTATTTCGCCAGCAAAGC, nitric oxide synthase forward primer ATCTTCTGGCCCGATTCTTT, and nitric oxide synthase reverse primer AATCGTTGACCAGCAACTCC.
The following primers did not show any changes (data not shown) and served as controls in addition to rp49: CBP forward primer CAAATGCAACACCAGCAACT, CBP reverse primer TGGGCATTGAGTTGACCATA, Notch forward primer CTACAAGGGCGTGGATTGTT, and Notch reverse primer TGGGAGCAATTGCATCTGTA.
The following primer pairs were used for PCR to create double-stranded RNA (dsRNA): primer pair dUTX 1 (forward primer TAATACGACTCACTATAGGGAGGCGCATTTGTACGAAGTTCA and reverse primer TAATACGACTCACTATAGGGAGAGCAAGCATAGGCATCTCGT), primer pair dUTX 2 (forward primer TAATACGACTCACTATAGGGCATATCCTTCGACCTTTTCGGAACT and reverse primer TAATACGACTCACTATAGGGTCTCGTAATTGACCACAGCTCTCAT), primer pair dUTX 3 (forward primer TAATACGACTCACTATAGGGCTGGATAAAGACCATAAAGCCG and reverse primer TAATACGACTCACTATAGGGTACTGGACTTTGCTTTGTGACC), and primer pair puc19 control (forward primer TAATACGACTCACTATAGGGAGGAATGCTTAATCAGTGAGGCACC and reverse primer TAATACGACTCACTATAGGGAGGAAAGCCATACCAAACGACGAGC).
The MEGAscript RNAi kit from Ambion was used to amplify dsRNA.
BrdU incorporation of S2 cells.
Bromodeoxyuridine (BrdU) incorporation was performed with the APC BrdU Flow kit from BD Biosciences. S2 cells were pulsed with BrdU for 30 min.
ChIP-seq.
Chromatin immunoprecipitation-sequencing (ChIP-seq) experiments against H3K27me3 were conducted with third-instar larval tissue from dUTX1 and dUTXPB mutants and two FRT40 controls, respectively. The H3K27me3-specific antibody was purchased from Active Motif (catalog number 39155). Data from the ChIP-seq analysis will be published elsewhere (our unpublished data).
RESULTS
dUTX mutant clones are overrepresented in eye imaginal discs.
We conducted a genetic screen of Drosophila for mutations that allow mutant tissue to outgrow wild-type tissue in mosaic eyes (74). In the screen of the left arm of chromosome 2, the relative representations of mutant tissue (marked in white) and wild-type tissue (marked in red) were compared to the reference ratio obtained with the wild-type parental chromosome (Fig. 1A). Mutations that inactivate a negative growth regulator are expected to provide the mutant tissue with a growth advantage and result in an increased relative representation of mutant tissue in mosaic eyes. We refer to this phenotype as an “overrepresentation” phenotype. In our screen, we isolated two alleles of dUTX based on their overrepresentation phenotype (Fig. 1B and C). The overall size of dUTX mosaic eyes is normal; however, dUTX mutant tissue represents a larger fraction of the eye (Fig. 1B and C). The overrepresentation phenotype is already apparent in dUTX mosaic eye-antennal imaginal discs. dUTX clones (marked by GFP using the MARCM technique [44] in Fig. 1E) occupy a much larger fraction of the eye disc than do control clones (Fig. 1D). A dUTX+ rescue construct reverts the growth advantage of dUTX1 (Fig. 1F), suggesting that it is indeed the result of the loss of dUTX. The overrepresentation phenotype is caused by a loss of the catalytic activity of dUTX, because a transgenic dUTX construct that contains a mutant JmjC domain (JmjC*) lacking two residues (His883 and Glu885) critical for its catalytic function fails to rescue the overrepresentation phenotype (Fig. 1G). As a control, the expression levels of dUTX+ and dUTXJmjC* were found to be similar in this experiment (data not shown).
Sequencing of the dUTX alleles revealed a premature termination codon in the JmjC domain in dUTX1 and a missense mutation changing Cys1071 to Tyr in the C terminus of dUTX2 (Fig. 1H). Cys1071 is a conserved residue in UTX proteins ranging from flies to humans and may be involved in zinc binding (42). A third allele, dUTXPB, carries a Piggyback transposon insertion in the fourth intron. A dUTX-specific antibody raised against the N-terminal 153 residues failed to detect the dUTX protein in dUTX1 mutant clones (Fig. 1I and I′). Because dUTX1 carries a premature termination codon at position 966 (Fig. 1H), the lack of the dUTX protein is likely caused by nonsense-mediated mRNA decay (NMD). Therefore, although dUTX1 is not a molecularly defined null allele, the lack of protein suggests that it encodes a strong hypomorphic, if not a null, allele. In dUTX2 and dUTXPB mutant clones, reduced levels of mutant dUTX proteins were detected (data not shown), implying that these alleles either encode an unstable protein (dUTX2) or produce less protein due to the Piggyback transposon insertion (dUTXPB). Because of the putative null character of dUTX1, most experiments reported in this paper were performed with this allele.
dUTX is required for H3K27me3 demethylation and for H3K4me1 methylation in vivo.
By use of previously applied biochemical approaches in vitro, dUTX and its mammalian homologs were shown to be H3K27me3 demethylases (1, 20, 32, 42, 43, 69). However, this has not been confirmed in vivo. Therefore, using antibodies specific for H3K27me3, we found increased immunofluorescence in dUTX clones, indicating a global increase in H3K27me3 levels in vivo (Fig. 2A and A′). The increase of H3K27me3 levels in dUTX mutant tissue is very subtle by immunohistochemistry. Nevertheless, Western blot analysis of dUTX mutant larvae further confirms the global increase of H3K27me3 levels (Fig. 2E, top), suggesting that dUTX indeed encodes an H3K27me3 demethylase. As a control, the total levels of histone H3 were found to be unaltered in dUTX mutant tissue by both immunohistochemistry and Western blotting (Fig. 2D, D′, and E). Levels of H3K27me2 and H3K27me1 are unaltered in dUTX clones in vivo (Fig. 3A, A ′, B, and B′), suggesting that dUTX is specific for the global demethylation of H3K27me3.
FIG. 3.
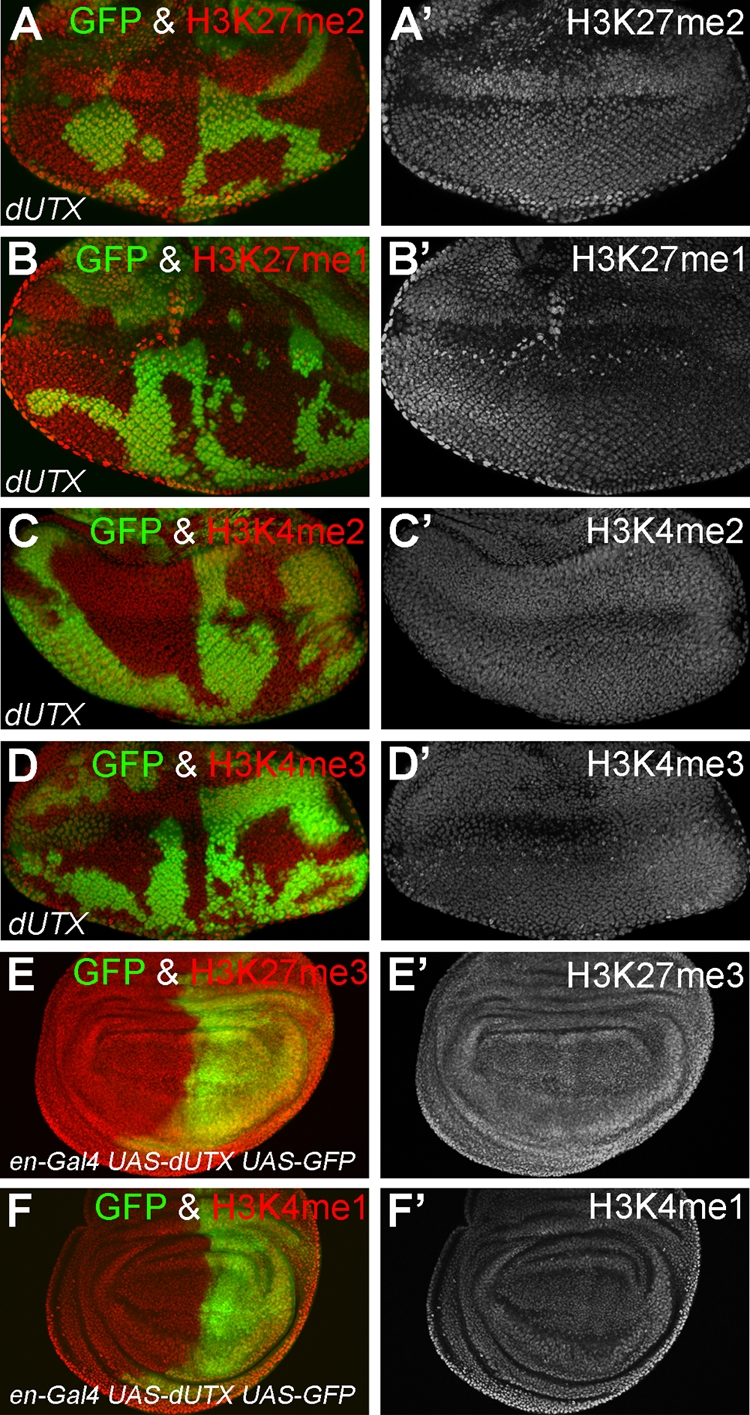
Analysis of the effect of the loss and overexpression of dUTX on global levels of H3K27 and H3K4 methylation. (A to D) Eye imaginal discs were labeled with antibodies specific for the indicated histone methyl modifications. GFP fluorescence was used to identify dUTX clones. Global levels of H3K27me2 (A), H3K27me1 (B), H3K4me2 (C), and H3K4me3 (D) in dUTX mutant clones were unchanged. Left panels are the merged GFP and antibody channels. The right panels are the antibody-only channels. (E and F) Wing imaginal discs that overexpress dUTX in the posterior compartment marked by GFP (green) were labeled with antibodies specific for H3K27me3 (E) and H3K4me1 (F). Left panels are the merged GFP and antibody channels. The right panels are the antibody-only channels. Global levels of H3K27me3 and H3K4me1 are unchanged. Similar data were obtained for the catalytic mutant dUTXJmjc* (data not shown). Genotypes are as follows: ey-FLP; dUTX1 FRT40A/P[ubi-GFP] FRT40A (A to D) and en-Gal4 UAS-dUTX UAS-GFP (E and F).
In addition to H3K27 methylation, we also tested a number of other histone modifications in dUTX mutants (Table 1). Surprisingly, we found a significant reduction of H3K4me1 levels in dUTX clones by immunohistochemistry and Western blotting (Fig. 2B, B′, and E). This is an interesting finding because mammalian UTX is a component of the mixed-lineage leukemia complexes (MLL3 and MLL4), which have methyltransferase activity toward H3K4 (15, 34, 56). The function of UTX in the MLL complexes is unknown. Our finding that Drosophila dUTX affects H3K4me1 methylation suggests that the incorporation of dUTX into MLL3 and MLL4 complexes is conserved and that in these complexes, dUTX is required for H3K4me1 methylation. H3K4me2 and H3K4me3 levels are normal in dUTX clones (Fig. 2E and 3C, C′, D, and D′), suggesting that dUTX has a specific and critical function for the establishment or maintenance of global H3K4me1 levels.
The requirement of dUTX for H3K4me1 methylation is independent of its JmjC domain. The dUTXJmjC* catalytic mutant is able to rescue the H3K4me1 methylation defect (Fig. 2C and C′), suggesting that the structural integration of dUTX into MLL complexes is necessary and sufficient for H3K4me1 methylation. This result also implies that the increased H3K27me3 levels in dUTX mutants are not a direct cause of the H3K4me1 methylation defect.
The overexpression of dUTX+ or dUTXJmjc* (data not shown) in wing imaginal discs did not alter the global methylation status of H3K4me1 and H3K27me3 (Fig. 3E, E′, F, and F′ and data not shown).
dUTX is essential for animal survival, and dUTX mutants display a Trithorax-like phenotype.
To determine the consequences of a loss of dUTX function at the organismal level, we investigated the phenotypes of dUTX mutants. Homozygous dUTX animals are semilethal. While most animals die during pupal stages, a small percentage of homozygotes (<5%) develop as viable adults. Nevertheless, these homozygous escapers die immediately after eclosion, suggesting that dUTX is an essential gene for viability.
Most interestingly, the sex combs of homozygous dUTX males contain fewer teeth (6 to 8 teeth) than those of wild-type males (∼10 teeth) (Fig. 4A and B). This is interesting because it may indicate a partial homeotic transformation of first-leg (prothoracic) to second-leg (mesothoracic) identity, which was observed previously for several Trithorax group genes (67). Thus, this genetic behavior may classify dUTX as a Trithorax group gene and further supports the notion that dUTX counteracts Polycomb repression. Other dUTX phenotypes include rough eyes (Fig. 4C and D) and wings that are wrinkled, have wing vein defects, and sometimes contain fluid-trapped blisters (Fig. 4E to G). Homozygous wings are down-curved, and the wing margin is irregular (Fig. 4F′).
FIG. 4.
Phenotypes of homozygous dUTX mutants. (A and B) Sex combs of wild-type (A) and dUTX1/dUTXPB (B) males. (C and D) Eyes of homozygous dUTX1 flies (D) have a rough appearance compared to that of wild-type (w−) eyes (C). (E to G) Wings of dUTX1/dUTXPB flies have wing vein defects and display bristle patterning defects on the wing margin (arrow in F′). Mutant wings are curved downwards and sometimes form blisters (arrow in G).
Increased proliferation in dUTX mutant cells correlates with increased H3K27me3 levels.
After this general analysis of the dUTX mutant phenotypes, we determined the cause of the overrepresentation of dUTX tissue over wild-type tissue in mosaic eyes. This phenotype could result from an increased proliferation of mutant cells, an inhibition of proliferation or apoptosis in neighboring wild-type cells due to cell competition, or a combination of both processes. By use of terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assays and cleaved caspase-3 antibody labeling (25), no significant change in apoptosis was observed for dUTX mosaics during eye imaginal disc development (data not shown), excluding apoptosis as cause of the overrepresentation phenotype. To address the effects of reduced dUTX function on cell proliferation, we used RNAi in cultured S2 cells. In independent experiments using three different double-stranded RNAs, the rate of BrdU incorporation is approximately 20% higher than that for controls (Fig. 5A and B). Thus, the overrepresentation phenotype of dUTX mutant clones in mosaic eyes is likely caused by an increased proliferation of mutant cells.
FIG. 5.
dUTX regulates the cell cycle. (A) Quantification of BrdU incorporation of S2 cells without RNAi treatment (S2 control) and with pUC19 RNAi (negative control) and three different double-stranded RNAs (dsRNAs) against dUTX. (B) Assessment of dUTX mRNA knockdown of the three different dUTX dsRNAs used in the BrdU incorporation assay (A) relative to pUC19 RNAi controls. mRNA levels of rp49 (negative control) are unaffected. (C to F) Heterozygosity for E(z) (D) and Pc (E) suppresses the overrepresentation phenotype of dUTX1 mosaics (C). Genotypes are as follows: ey-FLP; dUTX1 FRT40A/P[w+] FRT40A (C), ey-FLP; dUTX1 FRT40A/P[w+] FRT40A; E(z)731/+ (D), and ey-FLP; dUTX1 FRT40A/P[w+] FRT40A; Pc1/+ (E).
A loss of the JmjC domain of dUTX causes increased H3K27me3 levels and is sufficient to cause the overrepresentation phenotype of dUTX clones (Fig. 1G). Therefore, we further investigated the relationship between the increased proliferation of dUTX mutant tissue and H3K27me3 levels. First, we lowered H3K27me3 levels in dUTX mosaics by reducing the activity of the H3K27 methyltransferase E(z) in PRC2. Heterozygosity (i.e., a 50% reduction) of E(z) suppressed the overrepresentation phenotype of dUTX mosaics (Fig. 5C and D). Thus, the loss of the H3K27me3 demethylase dUTX can be compensated for by the simultaneous reduction of H3K27 methyltransferase activity. Second, we tested whether the reader of H3K27me3, Polycomb (Pc) in PRC1, is required for the overrepresentation phenotype. The heterozygosity of Pc suppresses the overrepresentation phenotype of dUTX (Fig. 5E). Together, these data further confirm that the increased levels of H3K27me3 due to the inactivation of dUTX participate in the overrepresentation phenotype and, thus, in the increased proliferation of dUTX mutant clones.
dUTX interacts genetically with Notch and restricts Notch signaling.
It is commonly accepted that H3K27me3 is a histone mark for gene silencing (51). Therefore, we wondered whether the increased levels of H3K27me3 may affect genes involved in an important cell cycle checkpoint in dUTX mutants. To address this question, we performed ChIP-seq experiments using H3K27me3-specific antibodies (62) to identify genes with increased H3K27me3 levels in dUTX mutants. Consistent with previous reports of mammalian UTX, several homeotic genes (Deformed, Ubx, Abd-B, Scr, and Antp) were found to contain increased levels of H3K27me3 (data not shown). In addition to this positive control, another group of genes containing increased H3K27me3 levels is involved in Notch signaling in Drosophila. This includes targets of the Notch pathway, such as genes of the E(spl) complex, α-adaptin, atonal, four-jointed (fj), and big brain (bib), as well as genes that are known to negatively regulate Notch, such as numb and roughened eye (roe) (data not shown) (9, 19, 28, 29, 47, 70). Because H3K27me3 is a histone mark for transcriptional silencing, we confirmed for some of these genes by qRT-PCR that the mRNA levels are indeed reduced in two dUTX mutants (dUTX1 and dUTXPB) compared to levels of wild-type controls (Fig. 6A). Levels of Rbf are unchanged compared to those of controls (Fig. 6A). mRNA levels of other genes tested (CBP and Notch) were also not affected in dUTX mutants (data not shown). Interestingly, not all genes associated with elevated H3K27me3 levels are transcriptionally downregulated. For instance, mRNA levels of the Drosophila nitric oxide synthase gene (dNOS) were found to be increased by 2.5-fold (Fig. 6A).
Because of this link to the Notch pathway and because eye growth during the larval stages is dependent on the Notch pathway (21, 55, 61, 73), we focused our analysis of the overrepresentation phenotype of dUTX clones on Notch. Indeed, Notch protein levels are increased in dUTX mutant clones (Fig. 6B and B′), which may be caused by a downregulation of negative regulators of Notch. The increase in the protein level of Notch is very prominent in the morphogenetic furrow (MF) (Fig. 6B′) and is also detectable posterior to the MF (Fig. 6B′). Furthermore, the increase in the protein level is confined largely to the ventral half of the eye disc, likely because the dorsal half is refractory to Notch signaling (12, 22).
Because of these links to Notch, we determined whether Notch signaling accounts for the overrepresentation phenotype by testing for genetic interactions between dUTX and Notch. Indeed, the heterozygosity of Notch suppresses the overrepresentation phenotype of dUTX clones (Fig. 6C). We also performed the reciprocal experiment and tested whether dUTX mutants can modify a Notch phenotype. Notch is required for wing margin formation, and heterozygous Notch mutations cause a dominant wing-notching phenotype (17, 18) (Fig. 6D). A similar dominant wing-notching phenotype is caused by the heterozygosity of Serrate (Ser), encoding one of the ligands of Notch (Fig. 6E) (60, 75). The Notch and Ser wing-notching phenotypes are dominantly suppressed by the heterozygosity of dUTX (Fig. 6D′, D″, E′, E″, and E‴), suggesting that the loss of dUTX increases Notch activity, thereby restoring the wing margin defect caused by the heterozygosity of either Notch or Ser. The degree of rescue varies from mild to complete but is consistent for all three dUTX alleles (Fig. 6D′, D″, E′, E″, E‴, F, and G). In summary, these genetic interaction studies between dUTX and Notch indicate that they function antagonistically and are consistent with the possibility that the loss of dUTX causes elevated Notch activity, which, in turn, may contribute to the overrepresentation phenotype of mutant tissue. Thus, the wild-type function of dUTX is to restrict Notch signaling.
To further characterize the antagonistic role of dUTX for Notch signaling, we tested dUTX mutants under a condition where excessive Notch activity induces overgrowth and tumors. The activation of Notch signaling by the expression of the Notch ligand Delta with the simultaneous expression of the Polycomb genes lola and pipsqueak can induce metastatic eye tumors in flies, which was previously referred to as the eyeful phenotype (26). We tested whether dUTX mutants can modify the eyeful phenotype. This analysis was performed at 18°C because at higher temperatures, the eyeful phenotype was too strong to be visibly modified by dUTX heterozygosity. At 18°C, the eyeful mutant causes only mild overgrowth compared to the wild type (Fig. 7A and B). However, the heterozygosity of dUTX can dominantly enhance the eyeful phenotype at 18°C. Approximately 80% of eyeful flies (n = 200) in the heterozygous dUTX background show enlarged ventral halves of the eyes (Fig. 7C). The remaining 20% of flies enhance the eyeful phenotype more dramatically, with ventrally located outgrowth and ectopic eye tissue (Fig. 7D and E). Therefore, these enhanced phenotypes further confirm that dUTX is a Notch antagonist.
FIG. 7.
Genetic interaction between Notch, Rbf, and dUTX. (A) Wild-type eye from Canton S. (B) The eyeful phenotype at 18°C. The eyeful phenotype is caused by the overexpression of UAS-Delta and GS88A8 (expressing lola and pipsqueak) driven by ey-Gal4 (26). (C to E) The eyeful phenotype at 18°C is dominantly enhanced by heterozygous dUTX alleles. The enhancement ranges from mild (C) to extreme (E). Arrows indicate areas of overgrowth or ectopic outgrowth. (F) Overexpression of Delta in otherwise wild-type eyes using the MARCM system (44) causes only a mild overgrowth phenotype. (G) Overexpression of Delta in dUTX clones using the MARCM system causes massive overgrowth of the eye. (H) Overexpression of rasV12S35 in dUTX clones using the MARCM system does not cause severe overgrowth. (I) Simultaneous overexpression of Rbf suppresses the overgrowth observed for B. (J) Heterozygosity of Rbf strongly enhances the overrepresentation phenotype of dUTX mosaics. Genotypes are as follows: Canton S (A), eyeful (ey-Gal4 UAS-Delta GS88A8) (26) (B), eyeful; dUTX1/+ (C to E), hs-FLP UAS-CD8:GFP; P[y+] FRT40A/P[tub-GAL80] FRT40A; P[tub-GAL4]/UAS-Delta (F), hs-FLP UAS-CD8:GFP; dUTX1 FRT40A/P[tub-GAL80] FRT40A; P[tub-GAL4]/UAS-Delta (G), hs-FLP UAS-CD8:GFP; dUTX1 FRT40A/P[tub-GAL80] FRT40A; P[tub-GAL4]/UAS-UAS-rasV12S35 (H), hs-FLP UAS-CD8:GFP;dUTX1 FRT40A/P[tub-GAL80] FRT40A; P[tub-GAL4]/UAS-Delta UAS-Rbf (I), and ey-FLP/Rbf14; dUTX1 FRT40A/P[ubi-GFP] FRT40A (J).
However, the eyeful phenotype is a very synthetic phenotype, which is caused not only by increased Notch signaling but also by the overexpression of the Polycomb epigenetic silencers Lola and Pipsqueak (26). Therefore, to specifically test the role of dUTX in the control of Notch signaling, we expressed only the Notch ligand Delta in dUTX mutant clones. The overexpression of Delta in otherwise wild-type clones produces a mild overgrowth phenotype in the adult eye (Fig. 7F). However, the overexpression of Delta in dUTX clones causes a massive overgrowth of eyes and heads (Fig. 7G). This overgrowth phenotype is so strong that only a few mosaic flies are recovered as adult animals; most die during pupal stages. As a control, the expression of another potent oncogene, rasV12, in dUTX clones did not cause such a striking overgrowth phenotype (Fig. 7H). Thus, dUTX mutant clones are specifically prone to Notch/Delta-induced overgrowth, suggesting that under normal conditions, dUTX is a suppressor of Notch-induced tumors.
Loss of Rbf promotes overrepresentation of dUTX clones.
We also addressed the question of how Notch stimulates proliferation in dUTX mutants. Although the mechanistic details are unclear, Notch can induce proliferation through the inhibition of Rbf, the Retinoblastoma (Rb) family member in Drosophila (3). Rbf inactivation was also implicated in Notch-induced eyeful tumors during eye development in Drosophila (26).
Therefore, we characterized the potential link between Notch signaling and Rbf further and related it to the dUTX phenotype. First, the strong-overgrowth phenotype caused by the overexpression of Delta in dUTX clones (Fig. 7G) can be suppressed by the simultaneous expression of Rbf (Fig. 7I), demonstrating that increased Rbf expression levels can normalize tissue growth. However, the overexpression of Rbf inhibits the cell cycle, and thus, the suppression of the overgrowth phenotype caused by Delta expression in dUTX clones may be indirect. Therefore, to more specifically address a link between Rbf and dUTX, we tested whether Rbf mutants can modify the dUTX overrepresentation phenotype. Significantly, a reduction of the Rbf gene dosage of 50% strongly enhances the dUTX overrepresentation phenotype. Eyes of all dUTX mosaics heterozygous for Rbf are composed of almost 100% dUTX tissue (phenotypically white in Fig. 7J). In dUTX mosaics, such a strong overrepresentation phenotype was observed for only a small fraction of the animals (<5%). The removal of one copy of Rbf transforms this phenotype to 100% penetrance. Most dUTX mosaics in a heterozygous Rbf background are trapped inside the pupal case and fail to eclose. Thus, a further reduction in levels of Rbf enhances the overrepresentation phenotype of dUTX mosaics, providing genetic evidence that the inactivation of Rbf contributes to the overrepresentation phenotype. This may occur in a Notch-dependent manner, because the expression of Rbf can suppress the strong overgrowth phenotype obtained by Delta expression in dUTX clones (Fig. 7G and I). Because we did not identify Rbf as a gene with increased H3K27me3 levels in dUTX mutants by ChIP-seq and qRT-PCR analysis (Fig. 6A), Rbf expression is likely not epigenetically silenced by H3K27me3 in dUTX clones. It is more likely that increased Notch activity reduces Rbf activity directly or indirectly, causing the overrepresentation phenotype.
DISCUSSION
dUTX is a Trithorax group gene affecting global levels of H3K27me3 and H3K4me1.
Based on the enzymatic activity of the JmjC catalytic domain as H3K27me3 demethylases, UTX proteins are predicted to counteract Polycomb function. Consistently, we found that dUTX mutants display genetic characteristics of Trithorax group genes. In vitro studies have shown that dUTX and UTX demethylate H3K27me2 and H3K27me3. However, dUTX mutants affect the global levels of only H3K27me3 (Fig. 2A) but not of H3K27me2 (Fig. 3A). Nevertheless, this observation does not mean that dUTX does not demethylate H3K27me2 in vivo. There may be fewer genes regulated by dUTX at the H3K27me2 level such that the global levels are not detectably altered in dUTX mutants.
Interestingly, dUTX mutants also affect global levels of H3K4me1, which are significantly reduced in mutant tissue (Fig. 2B and E). Mammalian UTX is a component of the MLL3 and MLL4 methyltransferase complexes (15, 34, 56), and based on the reduction of H3K4me1 levels, we predict that dUTX is also a component of the Drosophila equivalent of the MLL3/MLL4 methyltransferase complex, which contains Trithorax-related (Trr) as a histone methyltransferase. The function of UTX in MLL3 and MLL4 complexes is currently unknown (43). It was suggested previously that UTX is not required for H3K4 methylation, but in these studies, only H3K4me2 and H3K4me3 were investigated (43). Consistently, the global levels of H3K4me2 and H3K4me3 are not affected in dUTX mutant clones. Our data demonstrate that dUTX is required for the monomethylation of H3K4. Interestingly, the JmjC demethylase domain of dUTX is not required for H3K4me1 methylation, suggesting that other domains of dUTX, such as the TPR domains, may be necessary for mediating this function. The finding that the global levels of H3K4me2 and H3K4me3 are not affected in dUTX mutants is also quite interesting, as it implies that the monomethylation of H3K4 is not required for the di- or trimethylation of H3K4.
dUTX is a Notch antagonist for cell cycle control.
The epigenetic control of gene expression has been best studied for the control of homeotic gene expression (33, 57), which is established during embryogenesis and maintained throughout animal life. However, not only homeotic genes are regulated through epigenetic modifications. Other genes in different developmental processes are also subject to epigenetic control. Here, by analyzing the dUTX mutant phenotype, we establish a role of H3K27me3 levels in cell cycle control. Our data suggest that increased H3K27me3 levels in dUTX clones cause the epigenetic silencing of several genes involved in Notch signaling. This includes both positive and negative regulators of Notch signaling activity as well as target genes that are either positively or negatively regulated by the Notch pathway. Such an incoherent control of gene expression by the Notch pathway was recently reported (40), suggesting that the final outcome of Notch activity may be determined by the relative expression levels of positive or negative regulators. Because we determined that the overrepresentation phenotype of dUTX clones is caused by elevated levels of Notch signaling, it appears that the silencing of Notch inhibitors is dominant over the silencing of Notch activators, resulting in a net increase of Notch activity. However, this increased Notch activity may be specific for the cell cycle phenotype of dUTX mutants, as we did not find increased Notch activity for other Notch-dependent paradigms, such as E(spl)m8-lacZ (data not shown). This is also consistent with the finding that E(spl) genes contain increased H3K27me3 levels in dUTX mutants. Thus, the wild-type function of dUTX is to restrict the cell cycle through the negative control of Notch. Therefore, our data link H3K27me3-dependent Notch activity with enhanced tissue growth, implying that dUTX is a Notch antagonist regarding the cell cycle and explaining the overrepresentation phenotype of dUTX mutant clones.
However, this phenotype is subtle compared to that of mutants in growth control pathways such as the Hippo pathway (63). Nevertheless, the overgrowth of dUTX clones is strongly potentiated by the additional activation of Notch. The expression of Delta in dUTX clones causes a strong tumor-like growth phenotype. Thus, dUTX functions as a suppressor of Notch-induced tumors under normal conditions. This synergistic interaction between the loss of dUTX and increased Notch activity is a clear example that tumor development requires several hits for progression (31).
The overrepresentation phenotype of dUTX clones can be dominantly enhanced by the genetic loss of Rbf, suggesting that the reduction of Rbf contributes to the overrepresentation phenotype. However, the reduction of Rbf activity in dUTX clones is not caused by direct epigenetic silencing at the Rbf locus. We did not find increased H3K27me3 levels at the Rbf locus in dUTX mutants, and mRNA levels of Rbf were unchanged (Fig. 6A). Instead, Rbf is negatively regulated by the Notch pathway during eye growth (3). Thus, the increased activity of Notch in dUTX clones leads to a partial inactivation of Rbf and increased proliferation, causing the overrepresentation phenotype. Currently, it is unknown how Notch regulates Rbf.
The control of cell cycle progression by UTX proteins is likely conserved in mammals. A parallel study performed by Wang and coworkers showed that the loss of mammalian UTX also results in elevated levels of proliferation (79). Consistent with our work, those authors also implicated the inactivation of Rb function in increased proliferation in response to UTX knockdown. Similar to our study, Rb itself is not subject to increased H3K27m3 silencing, but the promoters of several genes in the Rb network were found to be occupied and likely controlled by UTX (79). Thus, although the mechanisms of Rb control by UTX proteins (Notch in this study and the Rb network in the study reported previously by Wang et al. [79]) are distinct, both studies established the control of the Rb pathway as a common element of cell cycle control by UTX proteins. Wang et al. (79) also demonstrated a link between UTX and Rb during vulval development in Caenorhabditis elegans. Thus, these studies combined suggest a well-conserved function of UTX proteins for Rb control.
Although these studies establish a link between UTX genes and Rb for cell cycle control, it should be noted that the loss of dUTX (and likely mammalian UTX) affects many genes. While the deregulation of individual genes may not cause a significant phenotype on its own, the combined deregulation may disrupt gene regulatory networks, which accounts for the growth phenotype of dUTX mutants. Thus, while we identify aberrant Notch signaling as an important element of the overrepresentation phenotype of dUTX mutants, other genes and signal transduction pathways may also contribute to this phenotype. For example, we also identified genes involved in growth control by the Hippo pathway (four-jointed [fj] and warts) associated with increased H3K27me3 levels in dUTX mutants and showed reduced transcript levels for fj (Fig. 6A). Thus, it is possible that the Hippo pathway and other genes contribute to the overrepresentation phenotype of dUTX mutants.
Implications for tumorigenesis in humans.
These observations have important implications for the initiation and development of human tumors. Increased levels of H3K27me3 due to the elevated activity of the H3K27me3 methyltransferase EZH2 have been associated with human cancer (7, 37, 78). Furthermore, mutations that inactivate UTX have been linked to human cancer (77), and low UTX activity correlates with poor patient prognosis (79). Our study establishes that increased levels of H3K27me3 affect Notch activity, which in turn affects Rbf activity. Rb is a well-known tumor suppressor, the loss of which causes human tumors. Therefore, tumors associated with the loss of UTX and, thus, increased H3K27me3 levels may be caused by decreased Rb activity. It should also be noted that aberrant Notch signaling is the cause of several human cancers, including T-cell acute lymphoblastic leukemias (T-ALLs), pancreatic cancer, medulloblastoma, and mucoepidermoid carcinoma (68). In summary, these data demonstrate that the appropriate control of H3K27 methylation is critical for normal tissue homeostasis, and increased H3K27me3 levels may contribute to cancer through the inactivation of Rb.
Acknowledgments
We thank Howard Chang for communicating results prior to publication. Nick Dyson, Hugo Bellen, Maria Dominguez, Wouter Bossuyt, the Developmental Studies Hybridoma Bank (DSHB), and the Bloomington Stock Center provided antibodies and fly stocks; Ruihong Zhu and Jeff Haug, from the cytometry facility at the Stowers Institute, provided advice and help with the BrdU assays. A.B. also thanks an anonymous donor for a generous gift.
H.-M.H. is a recipient of a fellowship by the Jane Coffins Child Memorial Fund. This work was supported by the National Institutes of Health (grants GM068016, GM081543, and GM074977 to A.B.; GM61672 to I.K.H.; and CA089455 and GM069905 to A.S.) and the Welch Foundation (grant G-1496 to A.B.).
Footnotes
Published ahead of print on 8 March 2010.
REFERENCES
- 1.Agger, K., P. A. Cloos, J. Christensen, D. Pasini, S. Rose, J. Rappsilber, I. Issaeva, E. Canaani, A. E. Salcini, and K. Helin. 2007. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449:731-734. [DOI] [PubMed] [Google Scholar]
- 2.Bagchi, S., R. Weinmann, and P. Raychaudhuri. 1991. The retinoblastoma protein copurifies with E2F-I, an E1A-regulated inhibitor of the transcription factor E2F. Cell 65:1063-1072. [DOI] [PubMed] [Google Scholar]
- 3.Baonza, A., and M. Freeman. 2005. Control of cell proliferation in the Drosophila eye by Notch signaling. Dev. Cell 8:529-539. [DOI] [PubMed] [Google Scholar]
- 4.Blatch, G. L., and M. Lassle. 1999. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays 21:932-939. [DOI] [PubMed] [Google Scholar]
- 5.Boyer, L. A., K. Plath, J. Zeitlinger, T. Brambrink, L. A. Medeiros, T. I. Lee, S. S. Levine, M. Wernig, A. Tajonar, M. K. Ray, G. W. Bell, A. P. Otte, M. Vidal, D. K. Gifford, R. A. Young, and R. Jaenisch. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441:349-353. [DOI] [PubMed] [Google Scholar]
- 6.Bracken, A. P., N. Dietrich, D. Pasini, K. H. Hansen, and K. Helin. 2006. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 20:1123-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bracken, A. P., D. Pasini, M. Capra, E. Prosperini, E. Colli, and K. Helin. 2003. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 22:5323-5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bray, S. J. 2006. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7:678-689. [DOI] [PubMed] [Google Scholar]
- 9.Buckles, G. R., C. Rauskolb, J. L. Villano, and F. N. Katz. 2001. Four-jointed interacts with dachs, abelson and enabled and feeds back onto the Notch pathway to affect growth and segmentation in the Drosophila leg. Development 128:3533-3542. [DOI] [PubMed] [Google Scholar]
- 10.Cagan, R. L., and D. F. Ready. 1989. Notch is required for successive cell decisions in the developing Drosophila retina. Genes Dev. 3:1099-1112. [DOI] [PubMed] [Google Scholar]
- 11.Cao, R., L. Wang, H. Wang, L. Xia, H. Erdjument-Bromage, P. Tempst, R. S. Jones, and Y. Zhang. 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298:1039-1043. [DOI] [PubMed] [Google Scholar]
- 12.Chao, J. L., Y. C. Tsai, S. J. Chiu, and Y. H. Sun. 2004. Localized Notch signal acts through eyg and upd to promote global growth in Drosophila eye. Development 131:3839-3847. [DOI] [PubMed] [Google Scholar]
- 13.Chellappan, S. P., S. Hiebert, M. Mudryj, J. M. Horowitz, and J. R. Nevins. 1991. The E2F transcription factor is a cellular target for the RB protein. Cell 65:1053-1061. [DOI] [PubMed] [Google Scholar]
- 14.Chittenden, T., D. M. Livingston, and W. G. Kaelin, Jr. 1991. The T/E1A-binding domain of the retinoblastoma product can interact selectively with a sequence-specific DNA-binding protein. Cell 65:1073-1082. [DOI] [PubMed] [Google Scholar]
- 15.Cho, Y. W., T. Hong, S. Hong, H. Guo, H. Yu, D. Kim, T. Guszczynski, G. R. Dressler, T. D. Copeland, M. Kalkum, and K. Ge. 2007. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J. Biol. Chem. 282:20395-20406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Czermin, B., R. Melfi, D. McCabe, V. Seitz, A. Imhof, and V. Pirrotta. 2002. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111:185-196. [DOI] [PubMed] [Google Scholar]
- 17.de Celis, J. F., and A. Garcia-Bellido. 1994. Roles of the Notch gene in Drosophila wing morphogenesis. Mech. Dev. 46:109-122. [DOI] [PubMed] [Google Scholar]
- 18.de Celis, J. F., A. Garcia-Bellido, and S. J. Bray. 1996. Activation and function of Notch at the dorsal-ventral boundary of the wing imaginal disc. Development 122:359-369. [DOI] [PubMed] [Google Scholar]
- 19.del Alamo, D., and M. Mlodzik. 2008. Self-modulation of Notch signaling during ommatidial development via the Roughened eye transcriptional repressor. Development 135:2895-2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Santa, F., M. G. Totaro, E. Prosperini, S. Notarbartolo, G. Testa, and G. Natoli. 2007. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 130:1083-1094. [DOI] [PubMed] [Google Scholar]
- 21.Dominguez, M., and J. F. de Celis. 1998. A dorsal/ventral boundary established by Notch controls growth and polarity in the Drosophila eye. Nature 396:276-278. [DOI] [PubMed] [Google Scholar]
- 22.Dominguez, M., D. Ferres-Marco, F. J. Gutierrez-Avino, S. A. Speicher, and M. Beneyto. 2004. Growth and specification of the eye are controlled independently by Eyegone and Eyeless in Drosophila melanogaster. Nat. Genet. 36:31-39. [DOI] [PubMed] [Google Scholar]
- 23.Du, W., M. Vidal, J. E. Xie, and N. Dyson. 1996. RBF, a novel RB-related gene that regulates E2F activity and interacts with cyclin E in Drosophila. Genes Dev. 10:1206-1218. [DOI] [PubMed] [Google Scholar]
- 24.Duronio, R. J., and P. H. O'Farrell. 1995. Developmental control of the G1 to S transition in Drosophila: cyclin E is a limiting downstream target of E2F. Genes Dev. 9:1456-1468. [DOI] [PubMed] [Google Scholar]
- 25.Fan, Y., and A. Bergmann. 4 December 2009, posting date. The cleaved-caspase-3 antibody is a marker of caspase-9-like DRONC activity in Drosophila. Cell Death Differ. doi: 10.1038/cdd.2009.185. [DOI] [PMC free article] [PubMed]
- 26.Ferres-Marco, D., I. Gutierrez-Garcia, D. M. Vallejo, J. Bolivar, F. J. Gutierrez-Avino, and M. Dominguez. 2006. Epigenetic silencers and Notch collaborate to promote malignant tumours by Rb silencing. Nature 439:430-436. [DOI] [PubMed] [Google Scholar]
- 27.Fischle, W., Y. Wang, S. A. Jacobs, Y. Kim, C. D. Allis, and S. Khorasanizadeh. 2003. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 17:1870-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frise, E., J. A. Knoblich, S. Younger-Shepherd, L. Y. Jan, and Y. N. Jan. 1996. The Drosophila Numb protein inhibits signaling of the Notch receptor during cell-cell interaction in sensory organ lineage. Proc. Natl. Acad. Sci. U. S. A. 93:11925-11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo, M., L. Y. Jan, and Y. N. Jan. 1996. Control of daughter cell fates during asymmetric division: interaction of Numb and Notch. Neuron 17:27-41. [DOI] [PubMed] [Google Scholar]
- 30.Hamel, P. A., R. M. Gill, R. A. Phillips, and B. L. Gallie. 1992. Transcriptional repression of the E2-containing promoters EIIaE, c-myc, and RB1 by the product of the RB1 gene. Mol. Cell. Biol. 12:3431-3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanahan, D., and R. A. Weinberg. 2000. The hallmarks of cancer. Cell 100:57-70. [DOI] [PubMed] [Google Scholar]
- 32.Hong, S., Y. W. Cho, L. R. Yu, H. Yu, T. D. Veenstra, and K. Ge. 2007. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc. Natl. Acad. Sci. U. S. A. 104:18439-18444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hueber, S. D., and I. Lohmann. 2008. Shaping segments: Hox gene function in the genomic age. Bioessays 30:965-979. [DOI] [PubMed] [Google Scholar]
- 34.Issaeva, I., Y. Zonis, T. Rozovskaia, K. Orlovsky, C. M. Croce, T. Nakamura, A. Mazo, L. Eisenbach, and E. Canaani. 2007. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol. Cell. Biol. 27:1889-1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karim, F. D., and G. M. Rubin. 1998. Ectopic expression of activated Ras1 induces hyperplastic growth and increased cell death in Drosophila imaginal tissues. Development 125:1-9. [DOI] [PubMed] [Google Scholar]
- 36.Ketel, C. S., E. F. Andersen, M. L. Vargas, J. Suh, S. Strome, and J. A. Simon. 2005. Subunit contributions to histone methyltransferase activities of fly and worm Polycomb group complexes. Mol. Cell. Biol. 25:6857-6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kleer, C. G., Q. Cao, S. Varambally, R. Shen, I. Ota, S. A. Tomlins, D. Ghosh, R. G. Sewalt, A. P. Otte, D. F. Hayes, M. S. Sabel, D. Livant, S. J. Weiss, M. A. Rubin, and A. M. Chinnaiyan. 2003. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc. Natl. Acad. Sci. U. S. A. 100:11606-11611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klose, R. J., E. M. Kallin, and Y. Zhang. 2006. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 7:715-727. [DOI] [PubMed] [Google Scholar]
- 39.Kondo, Y., L. Shen, A. S. Cheng, S. Ahmed, Y. Boumber, C. Charo, T. Yamochi, T. Urano, K. Furukawa, B. Kwabi-Addo, D. L. Gold, Y. Sekido, T. H. Huang, and J. P. Issa. 2008. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat. Genet. 40:741-750. [DOI] [PubMed] [Google Scholar]
- 40.Krejci, A., F. Bernard, B. E. Housden, S. Collins, and S. J. Bray. 2009. Direct response to Notch activation: signaling crosstalk and incoherent logic. Sci. Signal. 2:ra1. [DOI] [PubMed] [Google Scholar]
- 41.Kuzmichev, A., K. Nishioka, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 2002. Histone methyltransferase activity associated with a human multiprotein complex containing the enhancer of Zeste protein. Genes Dev. 16:2893-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lan, F., P. E. Bayliss, J. L. Rinn, J. R. Whetstine, J. K. Wang, S. Chen, S. Iwase, R. Alpatov, I. Issaeva, E. Canaani, T. M. Roberts, H. Y. Chang, and Y. Shi. 2007. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature 449:689-694. [DOI] [PubMed] [Google Scholar]
- 43.Lee, M. G., R. Villa, P. Trojer, J. Norman, K. P. Yan, D. Reinberg, L. Di Croce, and R. Shiekhattar. 2007. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science 318:447-450. [DOI] [PubMed] [Google Scholar]
- 44.Lee, T., and L. Luo. 2001. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 24:251-254. [DOI] [PubMed] [Google Scholar]
- 45.Lee, T. I., R. G. Jenner, L. A. Boyer, M. G. Guenther, S. S. Levine, R. M. Kumar, B. Chevalier, S. E. Johnstone, M. F. Cole, K. Isono, H. Koseki, T. Fuchikami, K. Abe, H. L. Murray, J. P. Zucker, B. Yuan, G. W. Bell, E. Herbolsheimer, N. M. Hannett, K. Sun, D. T. Odom, A. P. Otte, T. L. Volkert, D. P. Bartel, D. A. Melton, D. K. Gifford, R. Jaenisch, and R. A. Young. 2006. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125:301-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lees, J. A., K. J. Buchkovich, D. R. Marshak, C. W. Anderson, and E. Harlow. 1991. The retinoblastoma protein is phosphorylated on multiple sites by human cdc2. EMBO J. 10:4279-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ligoxygakis, P., S. Y. Yu, C. Delidakis, and N. E. Baker. 1998. A subset of notch functions during Drosophila eye development require Su(H) and the E(spl) gene complex. Development 125:2893-2900. [DOI] [PubMed] [Google Scholar]
- 48.Lin, B. T., S. Gruenwald, A. O. Morla, W. H. Lee, and J. Y. Wang. 1991. Retinoblastoma cancer suppressor gene product is a substrate of the cell cycle regulator cdc2 kinase. EMBO J. 10:857-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Min, J., Y. Zhang, and R. M. Xu. 2003. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 17:1823-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Montgomery, N. D., D. Yee, A. Chen, S. Kalantry, S. J. Chamberlain, A. P. Otte, and T. Magnuson. 2005. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr. Biol. 15:942-947. [DOI] [PubMed] [Google Scholar]
- 51.Moss, T. J., and L. L. Wallrath. 2007. Connections between epigenetic gene silencing and human disease. Mutat. Res. 618:163-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muller, J., C. M. Hart, N. J. Francis, M. L. Vargas, A. Sengupta, B. Wild, E. L. Miller, M. B. O'Connor, R. E. Kingston, and J. A. Simon. 2002. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111:197-208. [DOI] [PubMed] [Google Scholar]
- 53.Negre, N., J. Hennetin, L. V. Sun, S. Lavrov, M. Bellis, K. P. White, and G. Cavalli. 2006. Chromosomal distribution of PcG proteins during Drosophila development. PLoS Biol. 4:e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Newsome, T. P., B. Asling, and B. J. Dickson. 2000. Analysis of Drosophila photoreceptor axon guidance in eye-specific mosaics. Development 127:851-860. [DOI] [PubMed] [Google Scholar]
- 55.Papayannopoulos, V., A. Tomlinson, V. M. Panin, C. Rauskolb, and K. D. Irvine. 1998. Dorsal-ventral signaling in the Drosophila eye. Science 281:2031-2034. [DOI] [PubMed] [Google Scholar]
- 56.Patel, S. R., D. Kim, I. Levitan, and G. R. Dressler. 2007. The BRCT-domain containing protein PTIP links PAX2 to a histone H3, lysine 4 methyltransferase complex. Dev. Cell 13:580-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pearson, J. C., D. Lemons, and W. McGinnis. 2005. Modulating Hox gene functions during animal body patterning. Nat. Rev. Genet. 6:893-904. [DOI] [PubMed] [Google Scholar]
- 58.Pellock, B. J., E. Buff, K. White, and I. K. Hariharan. 2007. The Drosophila tumor suppressors Expanded and Merlin differentially regulate cell cycle exit, apoptosis, and Wingless signaling. Dev. Biol. 304:102-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Plath, K., J. Fang, S. K. Mlynarczyk-Evans, R. Cao, K. A. Worringer, H. Wang, C. C. de la Cruz, A. P. Otte, B. Panning, and Y. Zhang. 2003. Role of histone H3 lysine 27 methylation in X inactivation. Science 300:131-135. [DOI] [PubMed] [Google Scholar]
- 60.Rebay, I., R. J. Fleming, R. G. Fehon, L. Cherbas, P. Cherbas, and S. Artavanis-Tsakonas. 1991. Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell 67:687-699. [DOI] [PubMed] [Google Scholar]
- 61.Reynolds-Kenneally, J., and M. Mlodzik. 2005. Notch signaling controls proliferation through cell-autonomous and non-autonomous mechanisms in the Drosophila eye. Dev. Biol. 285:38-48. [DOI] [PubMed] [Google Scholar]
- 62.Robertson, G., M. Hirst, M. Bainbridge, M. Bilenky, Y. Zhao, T. Zeng, G. Euskirchen, B. Bernier, R. Varhol, A. Delaney, N. Thiessen, O. L. Griffith, A. He, M. Marra, M. Snyder, and S. Jones. 2007. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods 4:651-657. [DOI] [PubMed] [Google Scholar]
- 63.Saucedo, L. J., and B. A. Edgar. 2007. Filling out the Hippo pathway. Nat. Rev. Mol. Cell Biol. 8:613-621. [DOI] [PubMed] [Google Scholar]
- 64.Schlesinger, Y., R. Straussman, I. Keshet, S. Farkash, M. Hecht, J. Zimmerman, E. Eden, Z. Yakhini, E. Ben-Shushan, B. E. Reubinoff, Y. Bergman, I. Simon, and H. Cedar. 2007. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat. Genet. 39:232-236. [DOI] [PubMed] [Google Scholar]
- 65.Schwartz, Y. B., T. G. Kahn, D. A. Nix, X. Y. Li, R. Bourgon, M. Biggin, and V. Pirrotta. 2006. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat. Genet. 38:700-705. [DOI] [PubMed] [Google Scholar]
- 66.Schwartz, Y. B., and V. Pirrotta. 2007. Polycomb silencing mechanisms and the management of genomic programmes. Nat. Rev. Genet. 8:9-22. [DOI] [PubMed] [Google Scholar]
- 67.Shearn, A. 1989. The ash-1, ash-2 and trithorax genes of Drosophila melanogaster are functionally related. Genetics 121:517-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sjolund, J., C. Manetopoulos, M. T. Stockhausen, and H. Axelson. 2005. The Notch pathway in cancer: differentiation gone awry. Eur. J. Cancer 41:2620-2629. [DOI] [PubMed] [Google Scholar]
- 69.Smith, E. R., M. G. Lee, B. Winter, N. M. Droz, J. C. Eissenberg, R. Shiekhattar, and A. Shilatifard. 2008. Drosophila UTX is a histone H3 Lys27 demethylase that colocalizes with the elongating form of RNA polymerase II. Mol. Cell. Biol. 28:1041-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Spana, E. P., and C. Q. Doe. 1996. Numb antagonizes Notch signaling to specify sibling neuron cell fates. Neuron 17:21-26. [DOI] [PubMed] [Google Scholar]
- 71.Sparmann, A., and M. van Lohuizen. 2006. Polycomb silencers control cell fate, development and cancer. Nat. Rev. Cancer 6:846-856. [DOI] [PubMed] [Google Scholar]
- 72.Squazzo, S. L., H. O'Geen, V. M. Komashko, S. R. Krig, V. X. Jin, S. W. Jang, R. Margueron, D. Reinberg, R. Green, and P. J. Farnham. 2006. Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res. 16:890-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sun, X., and S. Artavanis-Tsakonas. 1997. Secreted forms of DELTA and SERRATE define antagonists of Notch signaling in Drosophila. Development 124:3439-3448. [DOI] [PubMed] [Google Scholar]
- 74.Tapon, N., N. Ito, B. J. Dickson, J. E. Treisman, and I. K. Hariharan. 2001. The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell 105:345-355. [DOI] [PubMed] [Google Scholar]
- 75.Thomas, U., S. A. Speicher, and E. Knust. 1991. The Drosophila gene Serrate encodes an EGF-like transmembrane protein with a complex expression pattern in embryos and wing discs. Development 111:749-761. [DOI] [PubMed] [Google Scholar]
- 76.Tolhuis, B., E. de Wit, I. Muijrers, H. Teunissen, W. Talhout, B. van Steensel, and M. van Lohuizen. 2006. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat. Genet. 38:694-699. [DOI] [PubMed] [Google Scholar]
- 77.van Haaften, G., G. L. Dalgliesh, H. Davies, L. Chen, G. Bignell, C. Greenman, S. Edkins, C. Hardy, S. O'Meara, J. Teague, A. Butler, J. Hinton, C. Latimer, J. Andrews, S. Barthorpe, D. Beare, G. Buck, P. J. Campbell, J. Cole, S. Forbes, M. Jia, D. Jones, C. Y. Kok, C. Leroy, M. L. Lin, D. J. McBride, M. Maddison, S. Maquire, K. McLay, A. Menzies, T. Mironenko, L. Mulderrig, L. Mudie, E. Pleasance, R. Shepherd, R. Smith, L. Stebbings, P. Stephens, G. Tang, P. S. Tarpey, R. Turner, K. Turrell, J. Varian, S. West, S. Widaa, P. Wray, V. P. Collins, K. Ichimura, S. Law, J. Wong, S. T. Yuen, S. Y. Leung, G. Tonon, R. A. DePinho, Y. T. Tai, K. C. Anderson, R. J. Kahnoski, A. Massie, S. K. Khoo, B. T. Teh, M. R. Stratton, and P. A. Futreal. 2009. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat. Genet. 41:521-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Varambally, S., S. M. Dhanasekaran, M. Zhou, T. R. Barrette, C. Kumar-Sinha, M. G. Sanda, D. Ghosh, K. J. Pienta, R. G. Sewalt, A. P. Otte, M. A. Rubin, and A. M. Chinnaiyan. 2002. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419:624-629. [DOI] [PubMed] [Google Scholar]
- 79.Wang, J. K., M. C. Tsai, G. Poulin, A. S. Adler, S. Chen, H. Liu, Y. Shi, and H. Y. Chang. 2010. The histone demethylase UTX enables RB-dependent cell fate control. Genes Dev. 24:327-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu, T., and G. M. Rubin. 1993. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development 117:1223-1237. [DOI] [PubMed] [Google Scholar]
- 81.Zweidler-McKay, P. A., and W. S. Pear. 2004. Notch and T cell malignancy. Semin. Cancer Biol. 14:329-340. [DOI] [PubMed] [Google Scholar]