Abstract
Mediator is a multisubunit assemblage of proteins originally identified in humans as a coactivator bound to thyroid hormone receptors (TRs) and essential for thyroid hormone (T3)-dependent transcription. Cyclin-dependent kinase 8 (CDK8), cyclin C, MED12, and MED13 form a variably associated Mediator subcomplex (termed the CDK8 module) whose functional role in TR-dependent transcription remains unclear. Using in vitro and cellular approaches, we show here that Mediator complexes containing the CDK8 module are specifically recruited into preinitiation complexes at the TR target gene type I deiodinase (DioI) together with RNA polymerase II (Pol II) in a TR- and T3-dependent manner. We found that CDK8 is essential for robust T3-dependent Dio1 transcription and that CDK8 knockdown via RNA interference decreased Pol II occupancy, and also the recruitment of the Pol II kinase CDK9, at the DioI promoter. Chromatin immunoprecipitation revealed CDK8 occupancy at the DioI promoter concurrent with active transcription, thus suggesting CDK8 involvement in transcriptional reinitiation. Mutagenesis assays showed that CDK8 kinase activity is necessary for full T3-dependent DioI activation, whereas in vitro kinase studies indicated that CDK8 may contribute to Pol II phosphorylation. Collectively, our data suggest CDK8 plays an important coactivator role in TR-dependent transcription by promoting Pol II recruitment and activation at TR target gene promoters.
The physiologic action of thyroid hormone (T3) in mammals is mediated primarily through thyroid hormone receptors (TRs), members of the nuclear hormone receptor superfamily that regulate transcription from target genes bearing T3 response elements (TREs) (62, 66). There are two different yet highly homologous TR subtypes, TRα and TRβ, each encoded on a separate gene. TRs typically bind to the TREs of positively regulated T3-responsive target genes as heterodimers with retinoid X receptors (RXRs) (31). RXR/TR heterodimers activate transcription on target genes containing positive TREs by recruiting coactivator complexes in a T3-dependent manner (62, 66). Two key TR coactivators are the p160/SRC-containing complexes (17, 32) and the Mediator complex (3, 29). The p160/SRC family of coactivators contain multiple leucine-rich LXXLL motifs important for T3-dependent binding to TRs and act as platforms for the recruitment of potent histone lysine acetyltransferases (17, 32, 54) and histone arginine methyltransferases (47, 61). Importantly, acetylation of lysine residues, and methylation of arginine residues, on histones H3 and H4 near the positive TREs of target genes results in a modified chromatin structure that facilitates transcriptional activation (17, 32, 47, 54, 61).
The evolutionarily conserved Mediator complex plays an essential coregulatory role in eukaryotic transcription (23). Originally isolated from human cells as a coactivator activity bound to TR in the presence of T3 (13), the complex is thought to bridge DNA-bound nuclear hormone receptors and other signal-activated transcription factors with the basal transcriptional apparatus, thereby facilitating the assembly and activation of the RNA polymerase II (Pol II) and its associated general factors at target gene promoters (3, 25, 29, 49). The human Mediator complex is comprised of over 30 subunits (45) and arranged into four subcomplexes termed the head, middle, tail, and cyclin-dependent kinase 8 (CDK8) modules (3, 29). Twenty-two of the human subunits are homologs of proteins found within Saccharomyces cerevisiae Mediator, a large 25-subunit complex that directly associates with Pol II and is essential for yeast viability (23). Electron microscopy and biochemical studies of both yeast and human Mediator reveal that subunits in the head, middle, and tail modules directly contact Pol II, whereas subunits in all four modules have been implicated in binding to signal-activated, gene-specific transcription factors (23, 25, 29). Similar to the p160/SRC proteins, the MED1 subunit of the mammalian Mediator complex contains two LXXLL motifs and targets Mediator to TR and other nuclear receptors in a ligand-dependent manner (3, 40).
Biochemical studies show that Mediator subunits MED12, MED13, cyclin C, and CDK8 comprise a distinct separable subcomplex (termed the CDK8 module) that is variably associated with the core Mediator complex (5, 22). Several lines of evidence indicate that the CDK8 module has an intrinsic potential to negatively regulate transcription. First, studies in both human and yeast systems indicate that association of the CDK8 module with the core Mediator complex can block interactions with Pol II (21, 36, 44). Second, in humans, CDK8 can inactivate the kinase activity of TFIIH (CDK7), which normally targets the C-terminal domain (CTD) of the largest subunit of Pol II for activation (1). Third, both yeast and mammalian CDK8 are able to phosphorylate gene-specific transcriptional activators, thus targeting them for ubiquitination and proteasome-based degradation (7, 14).
On the other hand, numerous studies point to positive roles for CDK8 in transcriptional regulation. For instance, studies in yeast have revealed that the CDK8 module is associated with Mediator complexes in the genome upstream of active genes (2) and is essential for efficient transcriptional activation by both the Gal4 and Sip4 activators (19, 24, 57). Similarly, human Mediator complexes containing the CDK8 module are clearly recruited to target gene promoters in an activator-dependent manner both in vitro and in vivo and appear to promote, and in some cases are essential for, transcriptional activation (6, 9, 10, 16, 20, 28, 39, 58-60). Interestingly, CDK8 can specifically phosphorylate the CTD of Pol II (18, 26, 41, 42, 48), and CDK8 kinase activity in yeast specifically promotes Pol II transcription as well as the formation of a transcription reinitiation scaffold complex (26). For specific human genes within the serum response network, CDK8-Mediator complexes can also facilitate the recruitment of CDK9 (10), another key CTD kinase implicated in regulating Pol II transcriptional elongation (15). Taken together, these studies reveal multiple positive and negative roles for the CDK8 module in transcriptional regulation that are likely manifested in an activator- or gene-specific manner.
In this study, T3-dependent activation of the human type I deiodinase (DioI) gene was utilized as a biological paradigm in order to investigate the functional role of CDK8 in TR-regulated transcription. Chromatin immunoprecipitation (ChIP) and immobilized template assays showed that Mediator complexes containing the CDK8 module are specifically recruited to the DioI promoter along with Pol II in a T3- and TR-dependent manner. We found that CDK8 is essential for robust T3-dependent transcription of the DioI gene in T3-responsive human cells and that CDK8 knockdown via RNA interference (RNAi) decreased both Pol II occupancy and recruitment of CDK9 at the DioI promoter. Mutagenesis and in vitro assays demonstrate that intrinsic CDK8 kinase activity is necessary for full T3-dependent DioI gene activation and may contribute to Pol II phosphorylation at the DioI promoter. Interestingly, ChIP assays further reveal significant levels of CDK8 at the DioI promoter in vivo during active transcription, suggesting that the CDK8 module may be involved in transcriptional reinitiation. In sum, our findings suggest that CDK8 plays an important coactivator role in T3-dependent transcription by promoting Pol II recruitment and activation.
MATERIALS AND METHODS
Antibodies and reagents.
Affinity-purified antibodies against MED1, MED13, MED12, MED26, MED6, MED7, MED17, CDK8, CDK9, Cyclin C, TFIIF-RAP30, TFIIF-RAP74, TAFII-100, hTBP, RPB6, Brg1, PCAF, SRC1, p300, TFIIB, and tubulin were all from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies against CDK7 were from BioLegend (San Diego, CA). Mouse monoclonal antibodies against the largest subunit of Pol II were via the hybridoma 8WG16 provided by Richard Burgess (University of Wisconsin). Ser-5-phosphorylated and Ser-2-phosphorylated Pol II CTD antibodies were from Covance (Madison, WI). Anti-FLAG antibodies and anti-FLAG-agarose beads were from Sigma (St. Louis, MO). Antibodies against BAF170 and BAF155 were from Naoko Tanese (New York University), and antibodies against PRMT1 were from Harvey Herschman (UCLA). Rabbit polyclonal antibodies against MED24 and MED1 (used for immunoblotting) were described previously (65). Horseradish peroxidase-conjugated secondary anti-rabbit and anti-mouse IgG were from Cell Signaling (Beverly, MA). Immunoblotting was as described previously (37). The H7 kinase inhibitor and T3 were purchased from Sigma.
Plasmids.
To generate the pCIN4-FLAG-MED17 plasmid, the full-length MED17 cDNA was first subcloned into FLAG(AS)-pGEM7 and, subsequently, FLAG-MED17 was subcloned into pCIN4. The luciferase reporter gene 2×T3RE-tk-Luc (37) and the in vitro transcription reporter template TRE3Δ53 template were described earlier (4, 12). The pIRESneo2-CDK8 wild-type and pIRESneo2-CDK8 kinase mutant (D151A) mammalian expression vectors were from Yoshiaki Ohkuma (University of Toyama, Japan) (16). The pQE9-12CAS-GST and pQE9-12CAS-GST-CTD bacterial expression vectors have been described previously (52). The recombinant baculoviruses expressing human CDK8 and human cyclin C were provided by Krassimir Yankulov (University of Guelph, Canada) and have been described elsewhere (38).
Cell culture.
The stable FLAG-MED17-expressing cell line was generated by transfecting pCIN4-FLAG-MED17 into HeLa(s) cells using Lipofectamine 2000. Stable clones were selected with 0.6 mg/ml of G418 (Invitrogen) and then expanded for f:MED17 expression analyses via immunoblotting. One clone exhibiting substoichiometric f:MED17 expression relative to endogenous MED17 was chosen for further expansion and CDK8-Mediator purification (see below). The α-2 cell line stably expressing FLAG-TRα was described earlier (13). HeLa-f:MED17 and α-2 cells were routinely cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS; Gemini Bioproducts), 10 mM HEPES, and penicillin and streptomycin (Invitrogen, Carlsbad, CA). For experiments involving T3 stimulation, the cells were cultured in charcoal/dextran-stripped (CDS) FBS (Gemini Bioproducts) for 72 h prior to addition of hormone.
RNA interference.
Smart pool small interfering RNAs (siRNAs) specific for MED1 and MED17 were from Dharmacon Research, Inc., as previously described (37, 56). The CDK8 and cyclin C siRNAs were generated using the siRNA synthesis kit (Ambion). For each factor, combinations of two siRNAs were used: for CDK8, 5′-AAG ATG CCT GAA CAT TCA ACA-3′ and 5′-AAA TAG CAT TAC TTC GAG AGC-3′; for cyclin C, 5′-AAT GGA TTG TTG CTT GAT AGT-3′ and 5′-AAA CCA CCT CCA AAC AGT GAA-3′. A scrambled siRNA smart pool (Dharmacon) was used as a control. For transfections, 3 × 105 cells were seeded the day before transfection in cell culture medium containing no antibiotics. After 24 h, 100 nM siRNA was transfected using the Lipofectamine 2000 reagent (Invitrogen).
Luciferase assays.
Transient transfections were carried out in 12-well plates using the Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. α-2 cells (105) were transfected with siRNAs (100 nM final concentration) specific for MED1, CDK8, cyclin C, or both CDK8/cyclin C along with the 2×T3RE-tk-Luc luciferase reporter gene (250 ng) and pSV-β-gal plasmid (100 ng) as a transfection control. After culturing for 48 h in CDS-FBS, the cells were treated with 100 nM T3 for 16 h. Cells were then harvested and luciferase activity was measured using the luciferase assay system (Promega) and a luminometer. Luciferase activity was normalized for both protein concentration and β-galactosidase activity.
Expression and purification of recombinant proteins in Sf9 cells and Escherichia coli.
Recombinant human FLAG-RXRα and -TRα were baculovirally expressed in insect Sf9 cells and purified via anti-FLAG immunoaffinity chromatography as described earlier (4, 12). Recombinant baculoviruses expressing His-tagged human CDK8 and human cyclin C were baculovirally expressed in insect Sf9 cells and purified as described previously (38). The glutathione S-transferase (GST) and GST-CTD constructs (52) were expressed in BL21 E. coli and purified as described previously (40).
In vitro kinase assay.
A 100-ng aliquot of GST-CTD was incubated with 50 ng of CDK8/cyclin C in 20 μl kinase buffer (20 mM HEPES [pH 7.9], 8 mM MgCl2, 0.5% glycerol, 0.1% Triton X-100, 1 mM dithiothreitol) containing either 2 μCi [γ-32P]ATP or nonradioactive ATP (0.1 mM) for 30 min at 30°C. For experiments involving the H7 inhibitor, GST-CTD was preincubated with H7 (at the concentrations indicated below) for 30 min prior to incubation with CDK8/cyclin C. The reaction was stopped by adding 2× SDS loading buffer, and the samples were resolved via 8% SDS-PAGE. The gel was either dried and exposed by autoradiography or transferred to nitrocellulose and processed for immunoblotting.
Purification of CDK8-Mediator complex.
HeLa f:MED17 cells were adapted to spinner culture in Joklik's medium (Sigma) containing 10% FBS and expanded into 25 liters at the Biovest International/National Cell Culture Center (Minneapolis, MN). Nuclear extract was prepared as described in detail elsewhere (28). The nuclear extract was dialyzed against BC100 (20 mM Tris-HCl [pH 7.9], 20% glycerol, 0.2 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride, 5 mM β-mercaptoethanol, and 100 mM KCl) and then fractionated on a phosphocellulose (P11) ion-exchange column as described in detail elsewhere (30). The 0.5 M KCl fraction was dialyzed against BC100 for 6 h, centrifuged at 18,000 rpm for 30 min at 4°C to remove insoluble debris, and then subjected to immunoaffinity chromatography using anti-FLAG M2 monoclonal antibody-conjugated agarose resin (Sigma). The M2 column was washed three times with BC300 (same as BC100 except containing 300 mM KCl) and then once with BC100-0.1% NP-40. The CDK8-Mediator complex was eluted with 0.3 mg/ml FLAG peptide.
Immunodepletion of Mediator from nuclear extracts and in vitro transcription assay.
HeLa cell nuclear extract was immunodepleted of Mediator by passing it through two columns prepared with affinity-purified antibodies against MED1 and MED6 conjugated to protein G-agarose beads as described previously (30). Preimmune normal rabbit IgG was used to prepare a mock control column. In vitro transcription assays were carried out by incubating the TRE3Δ53 G-free cassette template (50 ng) in Mediator-depleted or mock-depleted nuclear extract (50 mg) together with 40 ng recombinant TRα/RXRα in the presence of T3 (100 nM) as described previously (4, 12, 13).
Chromatin immunoprecipitation.
α-2 cells were seeded in CDS-FBS-containing medium for 72 h prior and then treated with 100 nM T3 for 1 h. ChIP assays were performed as described earlier (46) using the antibodies indicated in the figure legends. The two sets of PCR primers for the DioI promoter and coding regions were described earlier (46, 63). The PCR primers for the FAS and ADRB2 gene promoters were also detailed previously (8).
Real-time PCR.
α-2 cells (1 × 106) were either cultured alone or transfected with siRNAs or mammalian expression vectors using Lipofectamine 2000 as outlined above. The cells were then treated with 100 nM T3 for 1 h, and total RNA was extracted using TRIzol reagent (Invitrogen). For experiments involving the H7 kinase inhibitor, the cells were preincubated with H7 (10 or 25 μM) for 30 min prior to T3 treatment. First-strand cDNA synthesis was generated by processing 1 μg of total RNA using a reverse transcription assay kit (Invitrogen). The cDNA was PCR amplified using Taq DNA polymerase (Roche) together with specific primers for DioI and β-actin. For real-time PCR, the cDNA was first normalized for β-actin expression and then a PCR was performed using a SYBR green PCR kit (Invitrogen) and an Opticon continuous fluorescence detection system (MJ Research) together with the following specific DioI primers: forward, 5′-GAAGAGGCTCTGGGTGCTCTTG-3′; reverse, 5′-ACTCCCAAATGTTGCACCTCTGT-3′. The increase in mRNA expression was calculated by the comparative Ct method (27). In some cases, PCR products were presented semiquantitatively by running the reactions on a 1% ethidium bromide-stained agarose gel.
Immobilized DNA template assay.
The 740-bp DioI template was PCR amplified from the sonicated genomic DNA using the following 5′-biotinylated primers: forward, 5′-TCG AGC CTG TAA TCC CAG CAC-3′, and reverse, 5′-GCC AGA GTA AGC TCT GAG TTC-3′. The template was fractionated on a 1% agarose gel, purified using a gel extraction kit (Qiagen), and quantitated by spectrophotometry. M-280 streptavidin Dynal beads (Invitrogen) were concentrated with a magnetic particle concentrator (MPC; Dynal) and washed twice with binding buffer (5 mM Tris-HCl [pH 7.5], 1 mM EDTA, 1 M NaCl, 0.003% NP-40) and then resuspended in binding buffer. The equilibrated Dynal beads were conjugated with 40 ng biotinylated DioI template in binding buffer for 30 min at room temperature with constant agitation. The immobilized template was then concentrated with the MPC and washed twice in binding buffer and once with transcription buffer (20 mM HEPES [pH 7.6], 4 mM MgCl2, 60 mM KCl, 0.08 mM EDTA, 8 mM dithiothreitol, 10% glycerol, 0.4 mg bovine serum albumin/ml, and 0.05% NP-40). The beads were then concentrated with the MPC, resuspended in 20 μl transcription buffer, and incubated with 50 ng of TR/RXR along with 100 nM T3 for 30 min at room temperature followed by the addition of 100 μg HeLa nuclear extract for another 50 min. The Dynal beads were then washed three times with transcription buffer and resuspended in 2× SDS loading buffer, resolved on 8% SDS-PAGE, and detected via immunoblotting. For transcription initiation experiments, the immobilized template-bound proteins were resuspended in 100 μl transcription buffer containing 100 μM ribonucleoside triphosphates (rNTPs) for 2 to 20 min at room temperature. The Dynal beads were then washed three times in 300 μl of transcription buffer, concentrated with the MPC, boiled with 2× SDS-loading buffer, and then processed for SDS-PAGE followed by immunoblotting.
RESULTS
CDK8-Mediator activates TR-dependent transcription in vitro.
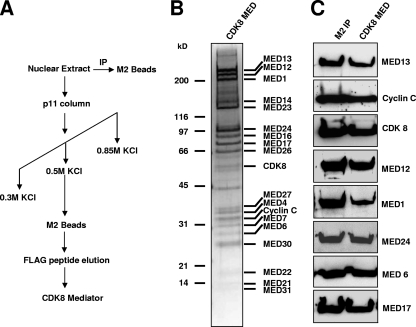
It was previously reported that Mediator complexes containing the CDK8 module are significantly less active in supporting activator-dependent transcription in vitro than complexes lacking the module (50). To investigate whether a purified Mediator complex containing the CDK8 module (hereafter termed CDK8-Mediator) can support TR-mediated transcription in vitro, we set up a cell-free TR-dependent transcription assay utilizing unfractionated HeLa nuclear extract as a source of basal transcription factors, Pol II, and other coregulatory factors (4, 12). As a source of purified CDK8-Mediator, we generated a HeLa-derived cell line stably expressing FLAG-tagged MED17 (f:MED17) and purified the complex from nuclear extracts using phosphocellulose fractionation followed by anti-FLAG immunoaffinity chromatography (Fig. 1A). MED17 was chosen for purification given its integral component status within the core Mediator complex (29, 49) and in light of the fact that it's been identified in nearly every mammalian Mediator preparation reported to date (reference 45 and references therein). Mediator complexes containing the CDK8 module can be separated from core Mediator lacking the module via phosphocellulose fractionation and elution with 0.5 M KCl (28, 35, 45, 60). Indeed, silver staining and immunoblot analyses showed that the purified CDK8-Mediator complex is highly enriched in CDK8, cyclin C, MED12, and MED13, as well as other subunits from the core Mediator complex (Fig. 1B and C).
FIG. 1.
Purification of CDK8-Mediator complex from HeLa cells. (A) Purification scheme for isolation of human CDK8-Mediator from nuclear extract prepared from HeLa cells stably expressing f:MED17. (B) Silver staining of purified human CDK8-Mediator. (C) Immunoblot analyses of purified CDK8-Mediator complexes isolated directly by using an anti-FLAG affinity column (M2 IP) or purified via P11 phosphocellulose chromatography followed by anti-FLAG affinity purification (CDK8 MED).
To test the ability of purified CDK8-Mediator to facilitate TR-mediated transcription in vitro, we utilized a naked DNA reporter plasmid containing three TREs inserted upstream of the adenovirus major late minimal promoter (−53 to +10; TRE3Δ53). In order to specifically address the functional role of the purified CDK8-Mediator complex in TR-dependent transcription, we immunodepleted Mediator from HeLa cell nuclear extracts using affinity-purified antibodies against MED1 and MED6 conjugated to protein G-agarose beads. The resulting immunodepleted nuclear extract (ΔMED) was >80% devoid of Mediator (based on the loss of the specific subunits assayed for here) (Fig. 2A), yet still replete for components of the Pol II basal transcription machinery, the SNF/SWI chromatin remodeling complex, and other nuclear receptor coactivators (Fig. 2A and B). Importantly, addition of purified human RXR/ΤR plus T3 to the mock-depleted nuclear extract efficiently activated transcription from the TRE3Δ53 template, but not when added to the ΔMED nuclear extract (Fig. 2C, compare lanes 2 and 4). Consistent with the notion that Mediator containing the CDK8 module can effectively support TR-mediated transcription in vitro, addition of the purified CDK8-Mediator complex restored RXR/TR/T3-dependent transcriptional activation in the ΔMED nuclear extract in a dose-dependent manner but had no significant effect on basal transcription (Fig. 2D).
FIG. 2.
CDK8-Mediator facilitates TR-dependent transcription in vitro. (A and B) Immunodepletion of Mediator from HeLa cell nuclear extracts, as shown by results of immunoblot analysis of HeLa cell nuclear extracts incubated with either anti-MED1/anti-MED6 antibodies or preimmune serum (mock). Specific antibodies used for immunoblotting are indicated to the right of the panels. (C) Mediator depletion abrogates TR-dependent transcription. Transcription was measured in vitro by incubating the TRE3Δ53 reporter gene in Mediator-depleted (ΔMED) or mock-depleted nuclear extract together with purified baculovirus-expressed TRα, RXRα, and T3 (10−7 M) as described previously (4). (D) Purified CDK8-Mediator restores TR-dependent transcription in a concentration-dependent manner. In vitro transcription assays were performed exactly as described for panel C, except that increasing concentrations of purified CDK8-Mediator were added to the reaction mixtures as indicated.
TR-dependent recruitment of CDK8-Mediator into a PIC at the DioI promoter coincides with the recruitment of Pol II.
Pol II and its associated general transcription factors (GTFs; TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH) assemble into a preinitiation complex (PIC) at the core promoters of all class II eukaryotic genes, and in the presence of NTPs, Pol II dissociates from the PIC and initiates transcription (43). Gene-specific activators like TR are thought to act in concert with the Mediator complex to promote the assembly, activation, and reinitiation of PICs at target gene promoters, thereby enhancing the rate of transcription (3, 20, 25, 29, 49). While the findings in Fig. 2 implicate CDK8-Mediator in facilitating TR-dependent transcriptional activation, it remained plausible that our presumptive purified CDK8-Mediator preparation additionally contained core Mediator complexes lacking the CDK8 module, or that the CDK8 module was disassociating from the core complex prior to the recruitment of Pol II into a PIC.
To more carefully examine whether CDK8-Mediator is directly recruited to promoter-bound TR and plays a functional role in promoting transcriptional initiation, we carried out immobilized template assays using a biotinylated human DioI gene promoter fragment spanning the two positive TREs in the proximal promoter as well as the transcription start site (46, 53) (Fig. 3A). The DioI promoter was immobilized on magnetic streptavidin beads and then incubated with purified human RXR/TR plus T3 in a HeLa nuclear extract allowing PIC assembly. The PICs were then washed and the bound proteins eluted, fractionated by SDS-PAGE, and analyzed by immunoblotting. In the absence of RXR/TR/T3, the GTFs TBP/TAFII100 (TFIID), TFIIB, and RAP74 (TFIIF) were all detected at the DioI promoter along with low levels of Pol II (Fig. 3C and D). Interestingly, addition of RXR/TR/T3 triggered the recruitment of subunits in the CDK8 module, the core Mediator complex, and Pol II to the immobilized DioI promoter (Fig. 3C and D). These data thus indicate that ligand-activated TR induces the recruitment of both the CDK8-Mediator complex and Pol II into a PIC at the DioI core promoter.
FIG. 3.
TR-dependent recruitment of CDK8-Mediator and Pol II into a functional PIC at the DioI promoter in vitro. (A) Schematic representation of the DioI promoter, showing the locations of TREs and biotin-conjugated PCR primers. (B) Single-round immobilized PIC assembly and transcriptional initiation assay. (C) TR-dependent recruitment of CDK8-Mediator and Pol II. Biotinylated immobilized DioI template was incubated with or without purified baculovirus-expressed TRα, RXRα, or T3 (10−7 M) in HeLa cell nuclear extract for 40 min. The PICs were then isolated by using streptavidin beads, washed, fractionated by SDS-PAGE, and then probed by immunoblotting using the specific antibodies indicated to the right of each panel. (D and E) CDK8 and Pol II dissociate from the PIC upon transcription initiation. PIC assembly on immobilized DioI promoter templates was carried out as described for panel C. The PICs were then isolated by using streptavidin beads, washed, and then resuspended in transcription buffer containing NTPs (100 μM) for the times indicated (D) or containing ATP or NTPs (100 μM) for 2 min (E). The streptavidin conjugates were then precipitated, washed, fractionated by SDS-PAGE, and then probed by immunoblotting using the specific antibodies indicated to the right of each panel.
In yeast immobilized PIC studies, a subset of GTFs (TFIIA, TFIID, TFIIE, and TFIIH) together with components of the core Mediator complex (MED6, MED15, MED17, and MED20) remain bound at the core promoter following transcription initiation, presumably forming a scaffold complex allowing for reinitiation (64). We were therefore interested in investigating whether TR-recruited CDK8 likewise remains bound at the DioI promoter upon transcriptional initiation. Toward this end, RXR/TR/T3-dependent PICs were again assembled at the DioI promoter in nuclear extract and washed as before (see above), but this time NTPs were then added to the immobilized PICs to initiate transcription. After another round of washing, the DNA-bound complexes were eluted and analyzed by immunoblotting. Interestingly and consistent with the earlier yeast studies (64), TBP/TAFII100 (TFIID) and the core Mediator subunits MED6, MED7, and MED17 all remained bound at the promoter, whereas TFIIB, TFIIF, and CDK8 all dissociated from the PIC along with Pol II upon transcription initiation (Fig. 3D). Addition of ATP alone also triggered CDK8 and Pol II disengagement from the immobilized PIC, thus indicating that ATP hydrolysis is sufficient for CDK8 dissociation (Fig. 3E).
In contrast to previous mammalian immobilized PIC studies using hybrid activators (GAL-p53 or GAL-VP16) and showing that the Mediator subunit MED1 dissociates from the PIC upon transcription initiation (28, 55), we observed here that MED1 remains firmly bound at the DioI promoter following transcription initiation, as well as in the presence of ATP (Fig. 3D and E). This observation is presumably accounted for by MED1's ability to directly bind RXR/TR (which also remains stably bound at the promoter) and underscores the importance of the promoter-specific activator in influencing the composition of the recruited Mediator complex during both PIC assembly and PIC dissociation. Significantly, these findings indicate that MED1 serves as a component of the reinitiation scaffold at TR-regulated core promoters and as such, may play an important in recycling Pol II and other distinct Mediator components back to the promoter (see Discussion). Collectively, our findings show that TR specifically recruits a Mediator complex containing the CDK8 module into a functional PIC at the DioI promoter in vitro and that this event coincides with the recruitment and subsequent activation of Pol II.
CDK8-Mediator is recruited to the DioI gene promoter in vivo in a T3-dependent manner, and its presence coincides with ongoing active transcription.
To investigate whether CDK8-Mediator is directly recruited to the T3-responsive human DioI gene in vivo, we carried out ChIP assays using human α-2 cells stably expressing human TRα (13, 46). To that end, the cells were first cultured in charcoal/dextran-stripped serum for 72 h and then treated with or without T3 for 1 h. ChIP was then carried out using antibodies specific for different Mediator subunits together with PCR primers spanning the two positive TREs in the DioI promoter region (Fig. 4A). Consistent with our immobilized template data, and in agreement with earlier studies showing activator-dependent recruitment of CDK8 to transcriptionally active gene promoters in vivo (9, 34, 58, 59), treatment of α-2 cells with T3 triggered a marked occupancy of both core Mediator subunits and the CDK8 module at the DioI promoter (Fig. 4B). Our data also revealed the T3-dependent recruitment of MED26, a subunit previously reported to be only associated with Mediator complexes lacking the CDK8 module (49).
FIG. 4.
T3-dependent recruitment of CDK8-Mediator to the DioI promoter in vivo coincides with active transcription. (A) Schematic representation of the human DioI promoter. TREs and the specific location of the PCR primers are indicated. (B) T3-dependent recruitment of CDK8-Mediator to the DioI promoter in T3-responsive cells. Chromatin was prepared from α-2 cells cultured with or without T3 (10−7 M) for 1 h and then immunoprecipitated using the specific antibodies indicated on the right. The immunoprecipitates were subjected to semiquantitative PCR using specific primers spanning the TREs in the promoter region (shown in panel A). (C and D) T3-dependent activation of DioI mRNA expression. Total RNA was extracted from α-2 cells cultured with or without T3 (10−7 M) for 1 h and then assayed by RT-PCR semiquantitatively (C) or in real time (D) using primers specific for DioI or for β-actin as a control.
Phosphorylation of specific serine residues in the CTD of the largest subunit of Pol II are important regulatory events controlling both transcriptional initiation and elongation (33). In mammals, the CTD consists of 52 repeats of the heptapeptide YSPTSPS with phosphorylation of serine 5 (Ser-5P) important for transcriptional initiation and phosphorylation of serine 2 (Ser-2P) important for elongation (15, 33). In general, phosphorylation of Ser-5 is believed to be predominantly mediated by CDK7, the kinase subunit of TFIIH (33), whereas phosphorylation of Ser-2 is thought to be catalyzed by CDK9, the kinase subunit of the positive transcription elongation factor b (P-TEFb) (15). In agreement with our immobilized PIC findings, we detected significant T3-dependent Pol II recruitment at the DioI promoter, as well as both Ser-5 and Ser-2 phosphorylation of the Pol II CTD (Fig. 4B). Consistent with the observed T3-induced recruitment and phosphorylation of Pol II at the DioI promoter, we found that stimulation of α-2 cells with T3 for 1 h significantly activated DioI mRNA expression as measured by both semiquantitative and real-time PCR (6- to 7-fold) (Fig. 4C and D). Taken together, these data are indicative of high levels of active gene transcription by Pol II due to elevated rates of transcriptional initiation and reinitiation at the DioI promoter. It is notable in this regard that significant levels of CDK8 are clearly present at the DioI promoter during this time period and are suggestive of potential positive roles for CDK8 in the transcriptional initiation and/or reinitiation process.
CDK8 promotes both Pol II occupancy and CDK9 recruitment at T3-responsive promoters and is required for T3-dependent activation of DioI mRNA expression.
Given the occupancy of CDK8-Mediator at the DioI promoter upon T3 treatment and during active transcription, we were interested in determining whether CDK8 or its heterodimeric binding partner cyclin C are functionally required for T3-dependent activation of DioI mRNA expression. Accordingly, α-2 cells were transfected with siRNA specific for either CDK8 or cyclin C and then assayed for T3-dependent DioI activation via real-time PCR (Fig. 5A and B). As positive controls for RNAi-induced Mediator impairment, we included siRNAs specific for MED1 that directly targets Mediator to TR and for MED17 that serves as an integral structural component of the core Mediator complex. Interestingly, knockdown of CDK8 or cyclin C inhibited T3-stimulated DioI mRNA expression ∼2.3- and 1.8-fold, respectively, thus revealing a functional requirement for these cofactors in T3-dependent DioI activation in vivo (Fig. 5B). Similarly, we found that RNAi silencing of either CDK8, or CDK8 together with cyclin C, abolished T3-dependent activation of a transiently transfected TRE-linked reporter gene (Fig. 5C). Together these results suggest that CDK8 serves as a positive coactivator for TR-mediated transcription.
FIG. 5.
CDK8 is required for T3-dependent activation of DioI mRNA expression. (A) RNAi knockdown of Mediator subunits in α-2 cells. Whole-cell extract was prepared from α-2 cells transfected with siRNAs specific for MED1, MED17, CDK8, cyclin C, or a nonspecific scrambled control siRNA and then probed with the specific antibodies indicated on the right. The immunoblots were then stripped and reprobed with antibodies against α-tubulin. (B and C) Loss of CDK8 or cyclin C inhibits T3-dependent gene expression. α-2 cells transfected with either control or Mediator-specific siRNAs were treated with or without T3 (10−7 M) for 1 h (B). Total RNA was then extracted processed for quantitative RT-PCR in real time using primers specific for DioI. Alternatively, α-2 cells were transfected with 2×TRE-tk-Luc along with either control or Mediator-specific siRNAs for 48 h and then treated with or without T3 (10−7 M) for 16 h (C). The cells were then harvested and assayed for luciferase activity, which was normalized against expression from a cotransfected β-galactosidase control vector. In the lower panels, equal amounts of cellular lysate from the harvested cells were probed by immunoblotting using the antibodies indicated on the right.
In light of our findings showing that T3-dependent recruitment of CDK8-Mediator at the DioI promoter coincides with Pol II recruitment, we next examined whether CDK8 silencing influences T3-dependent Pol II recruitment at the DioI gene in vivo. To perform these experiments, α-2 cells were first transfected with CDK8 siRNA or a scrambled control and then cultured with or without T3. ChIP was then carried out using PCR primers spanning the DioI proximal promoter or spanning an intragenic region ∼1 kb downstream in the DioI transcription start site (Fig. 6A). Consistent with the observed loss of activated DioI gene expression, knockdown of CDK8 in α-2 cells by RNAi resulted in a significant decrease in Pol II recruitment at the proximal DioI promoter as well as a decrease in actively transcribing Pol II within the intragenic region (Fig. 6B and C). Knockdown of CDK8 expression in α-2 cells also decreased Pol II occupancy at the promoter regions of two other T3-responsive genes, fatty acid synthase (FAS) and β-adrenergic receptor (ADRB2) (Fig. 6E and F).
FIG. 6.
Loss of CDK8 expression reduces Pol II occupancy at T3-responsive gene promoters and decreases recruitment of CDK9 at the DioI gene. (A) Schematic representation of the DioI gene, showing the locations of PCR primers in the proximal promoter and downstream intragenic regions. (B, C, and D) α-2 cells were transfected with CDK8 siRNA or a control siRNA for 72 h and then treated with or without T3 (10−7 M) for 1 h. The cells were then processed for ChIP analyses using the antibodies shown to the right of the panels and specific PCR primer sets indicated below the panels. (E and F) ChIP was carried out exactly as described above except that PCR primers specific for the T3-responsive FAS and ADRB2 gene promoters were used.
A notable reduction in Pol II CTD Ser-2 and Ser-5 phosphorylation was also observed at the DioI proximal promoter in the absence of CDK8 (Fig. 6B), possibly accounted for by a concomitant decrease in either CDK7/TFIIH and/or CDK9/PTEF-b recruitment. To explore this issue, ChIP was carried out on T3-stimulated α-2 cells transfected with or without CDK8 siRNA using antibodies specific for CDK7 and CDK9 (Fig. 6D). Interestingly, we found that CDK8 knockdown markedly decreased CDK9 recruitment at the Dio1 promoter as well as elicited a modest inhibitory effect on CDK7 recruitment. Taken together, our findings suggest that loss of CDK8 expression in α-2 cells decreases T3-dependent DioI mRNA expression primarily at the level of reduced Pol II recruitment and reduced recruitment of CTD kinases that regulate Pol II activity.
To further investigate whether the kinase activity of CDK8 itself is functionally required for T3-dependent DioI mRNA expression, α-2 cells transfected with CDK8 siRNA were additionally transfected with expression vectors for wild-type CDK8 or a kinase-deficient CDK8 mutant (D151A) (16). The transfected cells were then treated with or without T3 for 1 h, and DioI mRNA expression was measured by real-time PCR. As shown in Fig. 7, ectopic overexpression of wild-type CDK8 partially restored T3-dependent DioI activation, whereas overexpression of the kinase-deficient CDK8 mutant had no effect. We suspect that the inability of wild-type CDK8 to fully restore T3-dependent DioI expression in these assays is likely due to partial RNAi knockdown of the ectopically expressed CDK8 protein as evidenced from the immunoblotting (Fig. 7, lower panels). Nonetheless, these data indicate that CDK8 kinase activity is functionally required for full T3-dependent DioI mRNA expression in vivo.
FIG. 7.
CDK8 kinase activity is required for full T3-dependent DioI mRNA expression. α-2 cells were transfected with either scrambled or CDK8-specific siRNAs along with either a wild-type CDK8 expression vector or a mutant CDK8 expression vector (D151A) deficient in kinase activity. At 72 h posttransfection, the cells were treated with or without T3 for 1 h and then harvested. Total RNA was processed by quantitative RT-PCR in real time using primers specific for DioI, while whole-cell lysate was analyzed by immunoblotting using antibodies specific for CDK8 and α-tubulin.
H7 inhibits both CDK8 phosphorylation of the Pol II CTD and T3-dependent DioI expression.
To gain further insights into the functional significance of CDK8 kinase activity, we expressed full-length human CDK8 and cyclin C together in insect Sf9 cells via recombinant baculovirus and then copurified the proteins as a heterodimer (Fig. 8A). Consistent with previous studies reporting that CDK8 can specifically phosphorylate the CTD of the largest subunit of Pol II (16, 18, 26, 41, 42), we found that incubation of the purified CDK8/cyclin C pair with a GST-CTD fusion protein containing 27 hexapeptide repeats robustly facilitated CTD-specific phosphorylation in a dose-dependent manner (Fig. 8B). To identify specific residues within the Pol II CTD that are phosphorylated by CDK8, we incubated CDK8/cyclin C with GST-CTD and subsequently performed immunoblot analyses utilizing monoclonal antibodies specific for phosphorylated serine 5 or serine 2 of the hexapeptide repeat. In agreement with previous findings (48), the purified CDK8/cyclin C pair facilitated phosphorylation of both Ser-2 and Ser-5 at the Pol II CTD (Fig. 8D and E). These findings confirm that the Pol II CTD is a specific substrate for CDK8 and that CDK8 phosphorylates specific serine residues within the CTD hexapeptide repeats that are functionally associated with transcriptional initiation and elongation.
FIG. 8.
CDK8/cyclin C specifically phosphorylates the Pol II CTD in vitro. (A) Recombinant human CDK8 and cyclin C expressed in Sf9 cells via baculovirus were purified and stained with Coomassie blue. (B) Purified recombinant CDK8/cyclin C phosphorylates GST-Pol II-CTD in a concentration-dependent manner (see Materials and Methods for further information on the in vitro kinase assay). (C to E) H7 inhibits CDK8/cyclin C kinase activity. (C) GST-Pol II-CTD was preincubated with various concentrations of H7 prior to incubation with CDK8/cyclin C in the presence of [γ32]ATP. (D and E) GST-Pol II-CTD was incubated with CDK8/cyclin C together with unlabeled ATP in the presence or absence of H7 (10 μM). The reaction mixtures were resolved by SDS-PAGE and then probed by immunoblotting using antibodies specific for the Pol II CTD, phosphorylated Pol II CTD Ser-2P, or phosphorylated Pol II CTD Ser-5P.
The small-molecule kinase inhibitor H7 has been shown to selectively inhibit CDK8 kinase activity in vitro (41). To examine whether H7 inhibits CDK8/cyclin C phosphorylation of the Pol II CTD, we incubated CDK8/cyclin C with various concentrations of H7 and then assayed the heterodimer for phosphorylation of GST-CTD. We found that H7 treatment completely inhibited phosphorylation of the Pol II CTD at concentrations as low as 10 μM (Fig. 8C) and that the chemical inhibitor specifically blocked phosphorylation at both Ser-2 and Ser-5 of the CTD hexapeptide repeat (Fig. 8D and E). In light of our data showing that CDK8 kinase activity is functionally required for full T3-dependent DioI mRNA expression in vivo, we further examined whether treatment of cultured α-2 cells with H7 could block T3-dependent DioI activation. Interestingly, we found that H7 inhibited T3-dependent DioI mRNA activation at concentrations similar to those used to block CDK8 phosphorylation of the Pol II CTD in vitro (Fig. 9A).
FIG. 9.
H7 inhibits both T3-dependent DioI expression and Pol II phosphorylation at the DioI promoter. (A) α-2 cells were cultured with or without T3 (10−7 M) in the presence or absence of H7 (10 or 25 μM, as indicated) for 1 h and then harvested. Total RNA was processed by quantitative RT-PCR in real time using primers specific for DioI. (B) H7 inhibits Pol II CTD phosphorylation at the DioI promoter in vitro. PICs were assembled on biotinylated DioI promoter templates in the presence of RXR/TR/T3 and HeLa nuclear extract essentially as described for Fig. 3 except that H7 (10 μM) was added to the reaction mixtures as indicated (lanes 4 and 6). The PICs were then isolated by using streptavidin beads and washed, and in selected reactions, transcription was initiated upon the addition of NTPs (100 μM) for 2 min (lanes 5 and 6). The streptavidin conjugates for all reactions were then precipitated, fractionated by SDS-PAGE, and then probed by immunoblotting using antibodies specific for CDK8, Pol II, and phosphorylated Ser-5 Pol II CTD. In reaction 5 (lane 5), the transcription initiation reaction eluate was trichloroacetic acid precipitated and processed for immunoblotting as described above. (C) H7 inhibits Pol II CTD phosphorylation at the DioI promoter in vivo. α-2 cells were cultured with or without T3 in the presence or absence of H7 (10 μM) and then processed for ChIP analyses exactly as described in the Fig. 4 legend.
To investigate whether inhibition of CDK8 kinase activity via H7 negatively influences Pol II recruitment into a TR-dependent PIC, immobilized PIC assays at the DioI promoter were carried out as before (Fig. 3), but this time in the presence or absence of H7. Although H7 had no significant affect on Pol II recruitment into the PIC, addition of the inhibitor blocked NTP-induced Ser-5 phosphorylation of the Pol II CTD as well as Pol II dissociation from the PIC (Fig. 9B). Similarly, ChIP assays revealed that H7 treatment of α-2 cells dramatically decreased T3-dependent Ser-5 phosphorylation of the Pol II CTD at the DioI promoter but did not appear to significantly influence Pol II recruitment (Fig. 9C). While we cannot rule out some concomitant inhibition of CDK7/TFIIH kinase activity by H7, it is important to note that the 50% inhibitory concentration (IC50) of H7 for CDK8 is 10-fold less than that for CDK7 (41). Our findings thus suggest that the kinase activity of CDK8 may play a contributory role in phosphorylating (and thus activating) the CTD of Pol II at TR-regulated gene promoters and as such, are reminiscent of previous studies in yeast showing that CDK8 kinase activity promotes Pol II transcription and formation of reinitiation scaffold complex at distinct activator-dependent target genes (26).
DISCUSSION
Mediator was originally purified from human cells as a multimeric nuclear protein complex bound to TRs and was later shown to be an essential transcriptional coactivator for TR and other nuclear hormone receptors (3), as well as for other types of signal-activated transcription factors (25, 29). The CDK8 module is a variably associated subcomplex of Mediator implicated in both negative and positive transcriptional regulation, yet its functional role in nuclear hormone receptor signaling has remained unclear. In this study, we utilized in vitro and cellular assays to investigate the functional role of the CDK8 module during TR-regulated transcription of the T3-responsive human DioI gene. Our results indicate the following: (i) CDK8-Mediator complexes are directly recruited into PICs at the DioI promoter together with Pol II in a TR- and T3-dependent manner, (ii) the CDK8 module is present at the DioI promoter in vivo concurrent with multiple rounds of active transcription, (iii) CDK8 is essential for robust T3-dependent transcription of the DioI gene in T3-responsive human cells, (iv) loss of CDK8 expression markedly reduces Pol II occupancy and CDK9 recruitment at TR-target gene promoters, and (v) CDK8 kinase activity is required for full T3-dependent DioI gene activation in cultured cells and can phosphorylate the Pol II CTD in vitro. Collectively, our in vitro and in vivo experiments suggest that CDK8-Mediator plays positive functional roles during at least two separate steps of TR-dependent transcription: (i) recruitment of Pol II into a PIC and (ii) facilitating the phosphorylation and activation of Pol II.
The results of our study showing that TR specifically recruits CDK8-Mediator to the DioI gene promoter parallel earlier reports showing that Mediator complexes containing the CDK8 module are directly recruited to actively transcribed eukaryotic gene promoters (2, 9, 10, 58, 59). Furthermore, and consistent with the findings here, a functional requirement for CDK8 has been demonstrated for activator- and gene-specific transcriptional activation in both yeast (19, 24, 57) and humans (9, 10, 16). Nonetheless, our findings suggesting that TR recruitment of CDK8-Mediator facilitates Pol II recruitment at the DioI promoter appear to contradict earlier in vitro studies showing that core Mediator, but not CDK8-Mediator, functionally interacts with Pol II (21, 36). We propose that the T3-induced binding of TR to Mediator elicits a distinct conformational change that promotes or stabilizes the association of both Pol II and the CDK8 module and allows for the subsequent integration of this holocomplex into the PIC assembled at the core promoter of genes bearing positive TREs. In support of this supposition, electron microscopy studies show that core Mediator complexes bound to TR adopt a conformational state distinctly different from Mediator bound to other unrelated activators (e.g., VP16 and SREBP-1a) (51). Indeed, when compared to VP16-bound Mediator, TR-bound Mediator was demonstrated to expose an alternate Pol II binding pocket on the opposite face of the complex. This observation suggests that T3-induced TR binding to Mediator may expose novel Pol II binding sites that are otherwise inaccessible in the presence of the CDK8 module and/or other types of activators. Considering that the loss of CDK8 expression via RNAi significantly reduced Pol II occupancy at the T3-responsive DioI, FAS, and ADRB2 promoters (Fig. 6), we further hypothesize that when in association with TR-Mediator, the CDK8 module serves to stabilize, rather than inhibit, the interaction with Pol II.
The TR-induced shift in Mediator structure is dependent on direct TR interactions with the MED1 subunit and the subsequent rearrangement of MED1 within the complex (51). Interestingly, MED1 has been reported to exist only in a specific Mediator subpopulation (less than 20% of the total pool) that is highly enriched for specific Mediator subunits including the CDK8 module and associated with near-stoichiometric levels of Pol II (67). Thus, in accordance with the findings here, it appears that TR (via MED1) preferentially targets Mediator complexes containing the CDK8 module and Pol II. Given that MED1 contains distinct binding motifs for both TR and its heterodimeric partner RXR (40), it is conceivable that the binding of a RXR/TR heterodimer to MED1 might further promote and/or stabilize the association of the CDK8 module and Pol II with Mediator. Along these same lines, the specific DNA structure/sequence of a positive TRE within a T3-responsive promoter region may additionally influence the conformational state of the RXR/TR-Mediator complex in such a manner that promotes the association of both the CDK8 module and Pol II during PIC assembly.
Previous in vitro immobilized template studies showed that CDK8-Mediator can be directly recruited into a PIC in an activator-dependent fashion (6, 20, 28, 55) and, analogous to this study, both CDK8 and Pol II were shown to dissociate from the PIC upon one cycle of transcription (28, 55). Interestingly, we observed here that MED1, together with RXR/TR, remain firmly associated with the RXR/TR/T3-induced PIC assembled at the DioI promoter upon transcriptional initiation (Fig. 3E), whereas in the other immobilized PIC studies using unrelated activators, MED1 was observed to rapidly disengage from the PIC (28, 55). Thus, at TRE-linked genes in which TR remains firmly bound at the promoter, MED1 appears to serve as part of the reinitiation scaffold. Given the MED1 specificity for Mediator complexes containing the CDK8 module and Pol II (67), it is intriguing to speculate that a MED1-containing scaffold complex might selectively promote the recycling of CDK8 and Pol II back to the TR-bound core promoter at reinitiation. Alternatively, existing MED1-CDK8-Mediator complexes in association with Pol II might be newly recruited to promoter-bound RXR/TR heterodimers at the reinitiation step. In either case, such a model is consistent with our data showing that significant levels of CDK8 are present at the DioI promoter in vivo coincident with active ongoing transcription (Fig. 4) and that loss of CDK8 or MED1 expression via RNAi significantly reduces Pol II occupancy at the DioI promoter (Fig. 6 and data not shown).
The Pol II CTD is the target of dynamic phosphorylation events that regulate its transcriptional activity (33). Whereas a nonphosphorylated Pol II is initially recruited into a PIC, transcriptional initiation is marked by the phosphorylation on Ser-5 of the CTD heptapeptide repeats by the kinase subunit of TFIIH (CDK7), whereas productive transcriptional elongation is marked by phosphorylation of Ser-2 by the CDK9 subunit of P-TEFb (15). Strikingly, we observed here that loss of CDK8 expression in T3-stimulated α-2 cells was accompanied by a marked decrease in CDK9 recruitment at the Dio1 promoter as well as a modest reduction in CDK7 recruitment (Fig. 6D). Notably in this regard, the CDK8 submodule of the Mediator complex was recently found to interact with P-TEFb, including the CDK9 subunit (10). Thus, CDK8-Mediator may act as a transcriptional coactivator by facilitating the corecruitment of CDK9 to target gene promoters, which in turn phosphorylates Ser-2 of the Pol II CTD and promotes transcriptional elongation. Along these same lines, a positive role for the Mediator complex in facilitating the recruitment of CDK7/TFIIH into a functional PIC has also been reported in yeast (11), but whether or not the CDK8 module is specifically required for CDK7/TFIIH recruitment remains unclear.
CDK8 itself is also uniquely capable of directly phosphorylating both Ser-2 and Ser-5 of the Pol II CTD (26, 41, 42, 48). Using a chemical genetics approach, Hahn and coworkers found that inhibition of yeast CDK8 (Srb10) kinase activity resulted in loss of activator-dependent transcription in vitro, decreased Pol II occupancy at the endogenous ADH1 and PMA1 gene promoters in vivo, and decreased Pol II CTD phosphorylation and dissociation from a PIC (26). We observed here that the CDK8-selective kinase inhibitor H7 failed to significantly influence TR- or T3-dependent recruitment of Pol II at the DioI promoter either in vitro or in vivo (Fig. 9B and C). However, and consistent with the notion that CDK8 kinase activity can promote activator-specific transcription in mammals, we found that ectopic overexpression of wild-type CDK8, but not a kinase-defective CDK8 mutant, restored T3-dependent DioI activation in human cells lacking endogenous CDK8 expression (Fig. 7). Moreover, the H7 inhibitor markedly decreased Pol II phosphorylation at the DioI promoter and inhibited Pol II phosphorylation and dissociation from a PIC (Fig. 9B and C). While we cannot rule out some concomitant inhibition of CDK7 by H7 in these assays, our findings are consistent with the yeast studies indicating that CDK7 and CDK8 may have overlapping roles in promoting phosphorylation-dependent Pol II dissociation from a PIC and formation of a reinitiation scaffold (26).
In addition to the Pol II CTD, it remains plausible that CDK8 may further phosphorylate novel components of the basal transcription apparatus or possibly other accessory factors that have functional consequences for PIC assembly and activation. Interestingly, we have observed that MED1 is a specific substrate for CDK8 phosphorylation in vitro (M. Belakavadi and J. D. Fondell, unpublished data). This observation parallels earlier reports showing that MED1 is a regulatory target for mitogen-activated protein kinases (4, 37) and leaves open the possibility that CDK8 phosphorylation of MED1 (and potentially other Mediator subunits) might in turn promote PIC assembly and/or formation of a MED1-containing reinitiation scaffold. In view of the fact that MED1 is the primary binding target for DNA-bound TR, such a model is entirely consistent with our findings here demonstrating a unique requirement for the CDK8 module in TR-dependent transcriptional activation. Future studies will be required to precisely identify the MED1 phosphorylation sites and to determine how these modifications affect Pol II recruitment and activation at TR target genes.
Acknowledgments
We thank Krassimir Yankulov for providing the recombinant baculovirus expressing CDK8 and cyclin C, Yoshi Ohkuma for providing the CDK8 mutant expression vector, Pradeep Pandey for the pCIN4-MED17 vector, and Ravi Vijayvargia for generating Mediator siRNAs.
This work was funded by a grant from the National Institutes of Health (DK054030) awarded to J.D.F.
Footnotes
Published ahead of print on 15 March 2010.
REFERENCES
- 1.Akoulitchev, S., S. Chuikov, and D. Reinberg. 2000. TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature 407:102-106. [DOI] [PubMed] [Google Scholar]
- 2.Andrau, J. C., L. van de Pasch, P. Lijnzaad, T. Bijma, M. G. Koerkamp, J. van de Peppel, M. Werner, and F. C. Holstege. 2006. Genome-wide location of the coactivator mediator: binding without activation and transient Cdk8 interaction on DNA. Mol. Cell 22:179-192. [DOI] [PubMed] [Google Scholar]
- 3.Belakavadi, M., and J. D. Fondell. 2006. Role of the mediator complex in nuclear hormone receptor signaling. Rev. Physiol. Biochem. Pharmacol. 156:23-43. [DOI] [PubMed] [Google Scholar]
- 4.Belakavadi, M., P. K. Pandey, R. Vijayvargia, and J. D. Fondell. 2008. MED1 phosphorylation promotes its association with mediator: implications for nuclear receptor signaling. Mol. Cell. Biol. 28:3932-3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borggrefe, T., R. Davis, H. Erdjument-Bromage, P. Tempst, and R. D. Kornberg. 2002. A complex of the Srb8, -9, -10, and -11 transcriptional regulatory proteins from yeast. J. Biol. Chem. 277:44202-44207. [DOI] [PubMed] [Google Scholar]
- 6.Cantin, G. T., J. L. Stevens, and A. J. Berk. 2003. Activation domain-mediator interactions promote transcription preinitiation complex assembly on promoter DNA. Proc. Natl. Acad. Sci. U. S. A. 100:12003-12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chi, Y., M. J. Huddleston, X. Zhang, R. A. Young, R. S. Annan, S. A. Carr, and R. J. Deshaies. 2001. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev. 15:1078-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi, K. C., S. Y. Oh, H. B. Kang, Y. H. Lee, S. Haam, H. I. Kim, K. Kim, Y. H. Ahn, K. S. Kim, and H. G. Yoon. 2008. The functional relationship between co-repressor N-CoR and SMRT in mediating transcriptional repression by thyroid hormone receptor alpha. Biochem. J. 411:19-26. [DOI] [PubMed] [Google Scholar]
- 9.Donner, A. J., S. Szostek, J. M. Hoover, and J. M. Espinosa. 2007. CDK8 is a stimulus-specific positive coregulator of p53 target genes. Mol. Cell 27:121-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donner, A. J., C. J. Ebmeier, D. J. Taatjes, and J. M. Espinosa. 2010. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat. Struct. Mol. Biol. 17:194-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esnault, C., Y. Ghavi-Helm, S. Brun, J. Soutourina, N. Van Berkum, C. Boschiero, F. Holstege, and M. Werner. 2008. Mediator-dependent recruitment of TFIIH modules in preinitiation complex. Mol. Cell 31:337-346. [DOI] [PubMed] [Google Scholar]
- 12.Fondell, J. D. 2002. Gene activation by thyroid hormone receptor in vitro and purification of the TRAP coactivator complex. Methods Mol. Biol. 202:195-214. [DOI] [PubMed] [Google Scholar]
- 13.Fondell, J. D., H. Ge, and R. G. Roeder. 1996. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc. Natl. Acad. Sci. U. S. A. 93:8329-8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fryer, C. J., J. B. White, and K. A. Jones. 2004. Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol. Cell 16:509-520. [DOI] [PubMed] [Google Scholar]
- 15.Fuda, N. J., M. B. Ardehali, and J. T. Lis. 2009. Defining mechanisms that regulate RNA polymerase II transcription in vivo. Nature 461:186-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furumoto, T., A. Tanaka, M. Ito, S. Malik, Y. Hirose, F. Hanaoka, and Y. Ohkuma. 2007. A kinase subunit of the human mediator complex, CDK8, positively regulates transcriptional activation. Genes Cells 12:119-132. [DOI] [PubMed] [Google Scholar]
- 17.Glass, C. K., and M. G. Rosenfeld. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14:121-141. [PubMed] [Google Scholar]
- 18.Hengartner, C. J., V. E. Myer, S. M. Liao, C. J. Wilson, S. S. Koh, and R. A. Young. 1998. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol. Cell 2:43-53. [DOI] [PubMed] [Google Scholar]
- 19.Hirst, M., M. S. Kobor, N. Kuriakose, J. Greenblatt, and I. Sadowski. 1999. GAL4 is regulated by the RNA polymerase II holoenzyme-associated cyclin-dependent protein kinase SRB10/CDK8. Mol. Cell 3:673-678. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, K. M., J. Wang, A. Smallwood, C. Arayata, and M. Carey. 2002. TFIID and human mediator coactivator complexes assemble cooperatively on promoter DNA. Genes Dev. 16:1852-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knuesel, M. T., K. D. Meyer, C. Bernecky, and D. J. Taatjes. 2009. The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes Dev. 23:439-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knuesel, M. T., K. D. Meyer, A. J. Donner, J. M. Espinosa, and D. J. Taatjes. 2009. The human CDK8 subcomplex is a histone kinase that requires Med12 for activity and can function independently of mediator. Mol. Cell. Biol. 29:650-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kornberg, R. D. 2005. Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 30:235-239. [DOI] [PubMed] [Google Scholar]
- 24.Larschan, E., and F. Winston. 2005. The Saccharomyces cerevisiae Srb8-Srb11 complex functions with the SAGA complex during Gal4-activated transcription. Mol. Cell. Biol. 25:114-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis, B. A., and D. Reinberg. 2003. The mediator coactivator complex: functional and physical roles in transcriptional regulation. J. Cell Sci. 116:3667-3675. [DOI] [PubMed] [Google Scholar]
- 26.Liu, Y., C. Kung, J. Fishburn, A. Z. Ansari, K. M. Shokat, and S. Hahn. 2004. Two cyclin-dependent kinases promote RNA polymerase II transcription and formation of the scaffold complex. Mol. Cell. Biol. 24:1721-1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔCT) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 28.Malik, S., H. J. Baek, W. Wu, and R. G. Roeder. 2005. Structural and functional characterization of PC2 and RNA polymerase II-associated subpopulations of metazoan Mediator. Mol. Cell. Biol. 25:2117-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malik, S., and R. G. Roeder. 2005. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem. Sci. 30:256-263. [DOI] [PubMed] [Google Scholar]
- 30.Malik, S., and R. G. Roeder. 2003. Isolation and functional characterization of the TRAP/mediator complex. Methods Enzymol. 364:257-284. [DOI] [PubMed] [Google Scholar]
- 31.Mangelsdorf, D. J., U. Borgmeyer, R. A. Heyman, J. Y. Zhou, E. S. Ong, A. E. Oro, A. Kakizuka, and R. M. Evans. 1992. Characterization of three RXR genes that mediate the action of 9-cis retinoic acid. Genes Dev. 6:329-344. [DOI] [PubMed] [Google Scholar]
- 32.McKenna, N. J., and B. W. O'Malley. 2002. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell 108:465-474. [DOI] [PubMed] [Google Scholar]
- 33.Meinhart, A., T. Kamenski, S. Hoeppner, S. Baumli, and P. Cramer. 2005. A structural perspective of CTD function. Genes Dev. 19:1401-1415. [DOI] [PubMed] [Google Scholar]
- 34.Meyer, K. D., A. J. Donner, M. T. Knuesel, A. G. York, J. M. Espinosa, and D. J. Taatjes. 2008. Cooperative activity of cdk8 and GCN5L within Mediator directs tandem phosphoacetylation of histone H3. EMBO J. 27:1447-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mittler, G., E. Kremmer, H. T. Timmers, and M. Meisterernst. 2001. Novel critical role of a human Mediator complex for basal RNA polymerase II transcription. EMBO Rep. 2:808-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naar, A. M., D. J. Taatjes, W. Zhai, E. Nogales, and R. Tjian. 2002. Human CRSP interacts with RNA polymerase II CTD and adopts a specific CTD-bound conformation. Genes Dev. 16:1339-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pandey, P. K., T. S. Udayakumar, X. Lin, D. Sharma, P. S. Shapiro, and J. D. Fondell. 2005. Activation of TRAP/mediator subunit TRAP220/Med1 is regulated by mitogen-activated protein kinase-dependent phosphorylation. Mol. Cell. Biol. 25:10695-10710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pinhero, R., P. Liaw, K. Bertens, and K. Yankulov. 2004. Three cyclin-dependent kinases preferentially phosphorylate different parts of the C-terminal domain of the large subunit of RNA polymerase II. Eur. J. Biochem. 271:1004-1014. [DOI] [PubMed] [Google Scholar]
- 39.Rachez, C., Z. Suldan, J. Ward, C. P. Chang, D. Burakov, H. Erdjument-Bromage, P. Tempst, and L. P. Freedman. 1998. A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes Dev. 12:1787-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ren, Y., E. Behre, Z. Ren, J. Zhang, Q. Wang, and J. D. Fondell. 2000. Specific structural motifs determine TRAP220 interactions with nuclear hormone receptors. Mol. Cell. Biol. 20:5433-5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rickert, P., J. L. Corden, and E. Lees. 1999. Cyclin C/CDK8 and cyclin H/CDK7/p36 are biochemically distinct CTD kinases. Oncogene 18:1093-1102. [DOI] [PubMed] [Google Scholar]
- 42.Rickert, P., W. Seghezzi, F. Shanahan, H. Cho, and E. Lees. 1996. Cyclin C/CDK8 is a novel CTD kinase associated with RNA polymerase II. Oncogene 12:2631-2640. [PubMed] [Google Scholar]
- 43.Roeder, R. G. 1996. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 21:327-335. [PubMed] [Google Scholar]
- 44.Samuelsen, C. O., V. Baraznenok, O. Khorosjutina, H. Spahr, T. Kieselbach, S. Holmberg, and C. M. Gustafsson. 2003. TRAP230/ARC240 and TRAP240/ARC250 Mediator subunits are functionally conserved through evolution. Proc. Natl. Acad. Sci. U. S. A. 100:6422-6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sato, S., C. Tomomori-Sato, T. J. Parmely, L. Florens, B. Zybailov, S. K. Swanson, C. A. Banks, J. Jin, Y. Cai, M. P. Washburn, J. W. Conaway, and R. C. Conaway. 2004. A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol. Cell 14:685-691. [DOI] [PubMed] [Google Scholar]
- 46.Sharma, D., and J. D. Fondell. 2002. Ordered recruitment of histone acetyltransferases and the TRAP/Mediator complex to thyroid hormone-responsive promoters in vivo. Proc. Natl. Acad. Sci. U. S. A. 99:7934-7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stallcup, M. R. 2001. Role of protein methylation in chromatin remodeling and transcriptional regulation. Oncogene 20:3014-3020. [DOI] [PubMed] [Google Scholar]
- 48.Sun, X., Y. Zhang, H. Cho, P. Rickert, E. Lees, W. Lane, and D. Reinberg. 1998. NAT, a human complex containing Srb polypeptides that functions as a negative regulator of activated transcription. Mol. Cell 2:213-222. [DOI] [PubMed] [Google Scholar]
- 49.Taatjes, D. J., M. T. Marr, and R. Tjian. 2004. Regulatory diversity among metazoan co-activator complexes. Nat. Rev. Mol. Cell Biol. 5:403-410. [DOI] [PubMed] [Google Scholar]
- 50.Taatjes, D. J., A. M. Naar, F. Andel III, E. Nogales, and R. Tjian. 2002. Structure, function, and activator-induced conformations of the CRSP coactivator. Science 295:1058-1062. [DOI] [PubMed] [Google Scholar]
- 51.Taatjes, D. J., T. Schneider-Poetsch, and R. Tjian. 2004. Distinct conformational states of nuclear receptor-bound CRSP-Med. complexes. Nat. Struct. Mol. Biol. 11:664-671. [DOI] [PubMed] [Google Scholar]
- 52.Thompson, C. M., A. J. Koleske, D. M. Chao, and R. A. Young. 1993. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell 73:1361-1375. [DOI] [PubMed] [Google Scholar]
- 53.Toyoda, N., A. M. Zavacki, A. L. Maia, J. W. Harney, and P. R. Larsen. 1995. A novel retinoid X receptor-independent thyroid hormone response element is present in the human type 1 deiodinase gene. Mol. Cell. Biol. 15:5100-5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsai, C. C., and J. D. Fondell. 2004. Nuclear receptor recruitment of histone-modifying enzymes to target gene promoters. Vitam Horm. 68:93-122. [DOI] [PubMed] [Google Scholar]
- 55.Uhlmann, T., S. Boeing, M. Lehmbacher, and M. Meisterernst. 2007. The VP16 activation domain establishes an active mediator lacking CDK8 in vivo. J. Biol. Chem. 282:2163-2173. [DOI] [PubMed] [Google Scholar]
- 56.Vijayvargia, R., M. S. May, and J. D. Fondell. 2007. A coregulatory role for the mediator complex in prostate cancer cell proliferation and gene expression. Cancer Res. 67:4034-4041. [DOI] [PubMed] [Google Scholar]
- 57.Vincent, O., S. Kuchin, S. P. Hong, R. Townley, V. K. Vyas, and M. Carlson. 2001. Interaction of the Srb10 kinase with Sip4, a transcriptional activator of gluconeogenic genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:5790-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang, G., M. A. Balamotis, J. L. Stevens, Y. Yamaguchi, H. Handa, and A. J. Berk. 2005. Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Mol. Cell 17:683-694. [DOI] [PubMed] [Google Scholar]
- 59.Wang, S., K. Ge, R. G. Roeder, and O. Hankinson. 2004. Role of mediator in transcriptional activation by the aryl hydrocarbon receptor. J. Biol. Chem. 279:13593-13600. [DOI] [PubMed] [Google Scholar]
- 60.Wu, S. Y., T. Zhou, and C. M. Chiang. 2003. Human mediator enhances activator-facilitated recruitment of RNA polymerase II and promoter recognition by TATA-binding protein (TBP) independently of TBP-associated factors. Mol. Cell. Biol. 23:6229-6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wysocka, J., C. D. Allis, and S. Coonrod. 2006. Histone arginine methylation and its dynamic regulation. Front. Biosci. 11:344-355. [DOI] [PubMed] [Google Scholar]
- 62.Yen, P. M. 2001. Physiological and molecular basis of thyroid hormone action. Physiol. Rev. 81:1097-1142. [DOI] [PubMed] [Google Scholar]
- 63.Yoon, H. G., Y. Choi, P. A. Cole, and J. Wong. 2005. Reading and function of a histone code involved in targeting corepressor complexes for repression. Mol. Cell. Biol. 25:324-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yudkovsky, N., J. A. Ranish, and S. Hahn. 2000. A transcription reinitiation intermediate that is stabilized by activator. Nature 408:225-229. [DOI] [PubMed] [Google Scholar]
- 65.Zhang, J., and J. D. Fondell. 1999. Identification of mouse TRAP100: a transcriptional coregulatory factor for thyroid hormone and vitamin D receptors. Mol. Endocrinol. 13:1130-1140. [DOI] [PubMed] [Google Scholar]
- 66.Zhang, J., and M. A. Lazar. 2000. The mechanism of action of thyroid hormones. Annu. Rev. Physiol. 62:439-466. [DOI] [PubMed] [Google Scholar]
- 67.Zhang, X., A. Krutchinsky, A. Fukuda, W. Chen, S. Yamamura, B. T. Chait, and R. G. Roeder. 2005. MED1/TRAP220 exists predominantly in a TRAP/ Mediator subpopulation enriched in RNA polymerase II and is required for ER-mediated transcription. Mol. Cell 19:89-100. [DOI] [PubMed] [Google Scholar]