Abstract
tRNA nucleosides are extensively modified to ensure their proper function in translation. However, many of the enzymes responsible for tRNA modifications in mammals await identification. Here, we show that human AlkB homolog 8 (ABH8) catalyzes tRNA methylation to generate 5-methylcarboxymethyl uridine (mcm5U) at the wobble position of certain tRNAs, a critical anticodon loop modification linked to DNA damage survival. We find that ABH8 interacts specifically with tRNAs containing mcm5U and that purified ABH8 complexes methylate RNA in vitro. Significantly, ABH8 depletion in human cells reduces endogenous levels of mcm5U in RNA and increases cellular sensitivity to DNA-damaging agents. Moreover, DNA-damaging agents induce ABH8 expression in an ATM-dependent manner. These results expand the role of mammalian AlkB proteins beyond that of direct DNA repair and support a regulatory mechanism in the DNA damage response pathway involving modulation of tRNA modification.
The posttranscriptional modification of rRNA, tRNA, and other RNAs is critical for proper RNA folding, stability, and function (22). These modifications range from simple methylation of the sugar or base to complex modifications involving multiple enzymatic steps (10, 21). In tRNA alone, more than 100 different nucleoside modifications that are conserved in all kingdoms of life have been identified. A wide variety of modified nucleosides are located in the tRNA anticodon loop, which plays a fundamental role in decoding the genetic information contained within mRNAs. Many of these modifications ensure precise translation by maintaining the correct reading frame or by directing codon specificity through stabilization of codon-anticodon pairing (2). Recent studies uncovered a biological role for modified wobble uridines in the regulated translation of specific codons that are enriched in mRNAs encoding stress and DNA damage response proteins (7). In the yeast Saccharomyces cerevisiae, the last step in the formation of this particular 5-methylcarboxymethyl uridine (mcm5U) modification is catalyzed by the Trm9p methyltransferase; deletion of TRM9 diminishes the translation of specific stress response proteins, thus conferring increased sensitivity to DNA-damaging agents (6, 7, 26). tRNA modifications can thus serve critical regulatory functions by modulating the translation of a subset of mRNAs (31).
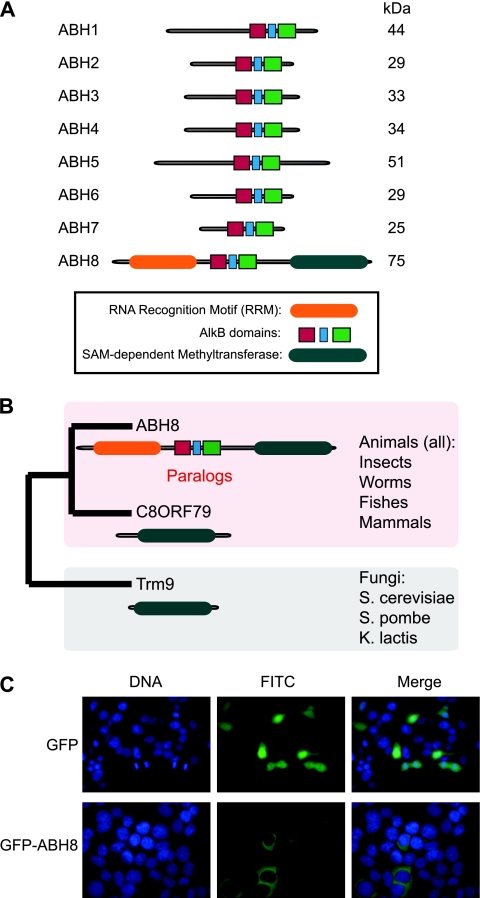
Two potential human orthologs of the yeast Trm9p tRNA methyltransferase have been identified by sequence homology, namely, the C8ORF79 gene product and the human AlkB homolog protein, ABH8, encoded by the ABH8 (or ALKBH8) gene (7, 42). Notably, in addition to the methyltransferase domain, human ABH8 contains a motif homologous to the bacterial AlkB DNA/RNA repair enzyme. Based upon phylogenetic analysis, AlkB belongs to a conserved family of nonheme, iron-dependent dioxygenases (3). Similar to other AlkB homologs, ABH8 contains a dioxygenase catalytic core domain encompassing cofactor-binding sites for iron and 2-oxoglutarate. However, ABH8 differs significantly from all other mammalian AlkB homologs due to the fusion of an RNA recognition motif (RRM) and an S-adenosyl-l-methionine (SAM)-dependent methyltransferase (MT) motif to the amino and carboxy termini of the AlkB oxygenase motif, respectively (see Fig. 1A).
FIG. 1.
Human ABH8 contains a tRNA methyltransferase motif and localizes to the cytoplasm. (A) Schematic of the human AlkB homolog (ABH) family of nonheme iron/2-oxoglutarate-dependent dioxygenase proteins. (B) Annotated gene tree of ABH8. The maximum likelihood phylogenetic tree of ABH8 and homologous proteins was generated via TreeBeST (tree building guided by species tree) using the Ensembl genome database. The complete tree with bootstrap values and distances is available on request. S. pombe, Schizosaccharomyces pombe; K. lactis, Kluyveryomyces lactis. (C) Subcellular localization of green fluorescent protein-tagged ABH8. Human 293T embryonic kidney cells were transfected with constructs expressing the indicated proteins and visualized by fluorescence microscopy. The nucleus was visualized by DAPI (4′,6-diamidino-2-phenylindole) staining. As a comparison, GFP alone was analyzed alongside.
Bacterial AlkB directly reverses the alkylated bases 1-methyladenine and 3-methylcytosine and the exocyclic base lesions ethenoadenine and ethenocytosine to normal adenine and cytosine in DNA or RNA through a distinct oxidative demethylase reaction (12, 14, 45). Bacteria deficient in AlkB are extremely sensitive to alkylating agent toxicity, underscoring the essential role of AlkB in repairing alkylation damage in vivo (27). Based upon protein fold similarity, the human genome encodes several proteins with AlkB dioxygenase motifs, including eight AlkB homologs (ABH1 through ABH8) and the recently discovered FTO (fat mass- and obesity-associated) gene product (see Fig. 1A) (3, 19, 28, 41). Mouse gene knockout models of Abh2 and Abh3 showing that ABH2 is the major repair enzyme for the in vivo reversal of 1-methyladenine (1-meA) and 3-methylcytosine (3-meC) in genomic DNA have been generated (39). While these studies define a biological role for ABH2, the cellular functions and substrates for the other mammalian AlkB homologs remain elusive.
Here, we find that human ABH8 catalyzes methylation of tRNA to form mcm5U, a key posttranscriptional modification at the wobble position of the anticodon loop. Moreover, ABH8 is required for maintaining steady-state levels of this methyl modification in vivo, and importantly, loss of ABH8 along with this modification increases the sensitivity of cells exposed to DNA-damaging agents. These studies reveal a crucial role for ABH8 in the DNA damage survival pathway through a distinct mechanism involving the regulation of tRNA modification.
MATERIALS AND METHODS
Cell culture and stable RNAi.
293T human embryonic kidney, HeLa S3 cervical carcinoma, and derivative stable cell lines were cultured in Dulbecco's minimal essential media (Invitrogen) containing 10% fetal bovine serum (FBS) and 2 mM l-glutamine. For stable RNA interference (RNAi), lentiviral RNAi constructs (Open Biosystems) expressing a nonsilencing short hairpin RNA (shRNA) (RHS4346), one of two shRNAs targeting the ABH8 transcript (RHS4430-99614583 and RHS4430-99616735), or a shRNA targeting the human URM1 transcript were cotransfected with packaging plasmids (psPAX2 and pMD2.G) into 293T cells for lentivirus production. The 293T cell line was subsequently infected with lentivirus in the presence of Polybrene, followed by stable clone selection using puromycin.
For colony-forming analysis, cells were seeded into 6-well plates in duplicate, allowed to attach for 24 h, and exposed to serum-free media alone or media containing methyl methanesulfonate (MMS) for 1 h, followed by replacement with complete media containing serum. Surviving colonies were scored 6 to 10 days later by methylene blue staining. For short-term viability assays, cells were exposed to either MMS (Sigma) for 1-hour in serum-free media or bleomycin (EMD Biosciences) for 24 h in serum-containing media before analysis. Viability in the presence of MMS was determined using a Coulter Counter coupled with trypan blue staining (total number of viable treated cells/total number of viable untreated cells), and viability in the presence of bleomycin was determined with the WST-1 cell proliferation reagent (Roche) according to the manufacturer's instructions.
Mammalian expression constructs.
The coding region for the full-length isoform of human ABH8 was amplified by reverse transcription-PCR (RT-PCR) (Easy-A; Stratagene) from human testis RNA (100179-748; VWR) and cloned into pcDNA3.1 (Invitrogen) for expression as N-terminal green fluorescent protein (GFP) or triple FLAG tag fusion proteins. The splice isoforms for ABH8 correspond to the following accession numbers: RRM, NM_138775.1; RRM-AlkB, EAW67088; MT domain alone, BX649085. The AlkB-MT variant was PCR amplified from the full-length ABH8 construct to create a cDNA encoding amino acids residues 126 to 664 of ABH8.
For stable expression of epitope-tagged ABH8, the FLAG-tagged-ABH8 coding region was cloned into the pBABEpuro or pBABEhygro retroviral vector for viral packaging using Phoenix 293T cells or stable transfection (33). HeLa S3 cells were infected with retrovirus and selected in bulk with puromycin for 2 days. The 293T ABH8-shRNA1 cell line was transfected with the pBABEhygro FLAG-ABH8 construct and selected with hygromycin. Stable clones were expanded and analyzed for expression.
Affinity purification of ABH8 from human cells.
Transient transfection and cellular extract production were performed as previously described (17). Whole-cell extract from transiently transfected cells or stable cell lines (1 mg of total protein) was rotated with 10 μl of FLAG M2 antibody resin (Sigma) for 2 h at 4°C in lysis buffer (20 mM HEPES at pH 7.9, 2 mM MgCl2, 0.2 mM EGTA, 10% glycerol, 1 mM dithiothreitol [DTT], 0.1 mM phenylmethylsulfonyl fluoride [PMSF], 0.1% NP-40) with 150 mM NaCl. Resin was washed extensively using the same buffer. Bound samples were eluted with two sequential volumes of wash buffer containing 100 μg/ml of 3×FLAG peptide (Sigma).
Protein and RNA analyses.
Cellular extracts and purified protein samples were fractionated on 4 to 12% Bis-Tris polyacrylamide gels (Invitrogen), followed by silver staining (SilverQuest kit; Invitrogen) or transfer to a nitrocellulose membrane for immunoblotting. Antibodies were against the following proteins: FLAG (F1804; Sigma), ABH8 (AV41106; Sigma), T-complex protein 1 subunit alpha (TCP-α) (sc-13869; Santa Cruz), TCP-β (sc-58864; Santa Cruz), TCP-θ (sc-13891; Santa Cruz), human Trm112 (also known as HSPC152; ab57156; Abcam), Hsp60 (sc-1052; Santa Cruz), URM1 (15285-1-AP; Protein Tech Group), GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (AV00191; GenScript). Primary antibodies were detected using IRDye-conjugated secondary antibodies (Rockland). Immunoblots were scanned using direct infrared fluorescence via the Odyssey system (Li-Cor Biosciences).
RNA from input and purified samples was extracted using TRIzol (Invitrogen) and fractionated on 7 M urea-0.5× Tris-borate-EDTA (TBE)-15 to 20% denaturing polyacrylamide gels, followed by Sybr gold nucleic acid staining (Invitrogen). RNA was visualized by 300 nM UV transillumination or a 450-nm light-emitting diode (LED) on a Storm 865 gel imaging system (GE Healthcare) and subsequently transferred to a Hybond N+ membrane (GE Healthsciences) for probe hybridization with 5′-radiolabeled oligonucleotides (Table 1).
TABLE 1.
Oligonucleotide sequences
| Probe target or substrate | Sequencea (5′-3′) |
|---|---|
| Probe targets | |
| tRNAArg(UCU) | CGACTCTGCYGGGACTCGAACCCGWAACCTTTGAATTAGAAGTCCAATGCGCTATCCATTGCGCCACAGAGCC |
| tRNAArg(ICG) | CGAGCCAGCCAGGAGTCGAACCTRGAATCTTCTGATCCGTAGTCAGACGCGTTATCCATTGCGCCACTGGCCC |
| tRNAAsn(GTT) | CGTCCCTGGGTGGGCTCGAACCACCAACCTTTCGGTTAACAGCCGAACGCGCTAACCGATTGCGCCACAGAGAC |
| tRNAAsp(GTC) | CTCCCCGTCGGGGAATCGAACCCCGGTCTCCCGCGTGACAGGCGGGGATACTCACCACTATACTAACGAGGA |
| tRNAGlu(UUC) | TTCCCAYACCGGGAGTCGAACCCGGGCCRCCTGGGTGAAAACCAGGAATCCTAACCGCTAGACCATRTGGGA |
| tRNALeu(CAA) | TGTCAGAAGTGGGATTCGAACCCACGCCTCCATTGGAGACCAGAACTTGAGTCTGGCGCCTTAGACCACTCGGCCATCC |
| tRNALys(CTT) | CGCCCAACGTGGGGCTCGAACCCACGACCCTGAGATTAAGAGTCTCATGCTCTACCGACTGAGCTAGCCGGGC |
| 5S rRNA | GGTCTCCCATCCAAGTACTAACCAGGCCCGACCCTGCTTAG |
| U6 snRNA | CGTTCCAATTTTAGTATATGTGCTGCCGAAGCGA |
| Substrates | |
| 1-meA | TAGACATTGCCATTCTCGATAGG-1-meA-TCCGGTCAAACCTAGACGAATTCCG |
| 3-meC | TAGACATTGCCATTCTCGATAGGAT-3-meC-CGGTCAAACCTAGACGAATTCCG |
| Control | TAGACATTGCCATTCTCGATAGGATCCGGTCAAACCTAGACGAATTCCG |
Boldface indicates degenerate bases.
Protein mass spectrometry.
Protein identification was performed by the MIT Center for Cancer Research Biopolymers Laboratory (http://web.mit.edu/ki/facilities/biopolymers/). Briefly, protein samples were reduced, alkylated, and digested in solution with trypsin, followed by purification and desalting on an analytical C18 column tip. Peptide samples were analyzed by chromatography on an Agilent model 1100 Nanoflow high-pressure liquid chromatography (HPLC) system coupled with electrospray ionization on a Thermo Electron model LTQ ion trap mass spectrometer. Protein identification through tandem mass spectrum correlation was performed using SEQUEST (30). Spectra had to match full tryptic peptides of at least 7 amino acids, have a normalized difference in cross-correlation scores (ΔCn) of at least 0.1, and have minimum cross-correlation scores (Xcorr) of 1.8 for singly charged, 2.5 for doubly charged, and 3.5 for triply charged spectra with at least 50% ion coverage.
Methyltransferase and demethylase activity assays.
The methyltransferase assay was carried out essentially as described previously (16). Purified samples were incubated in a total volume of 30 μl containing 1× HpaII methyltransferase buffer (New England Biolabs), 0.4 mM S-adenosyl-l-methionine 14C labeled at 50 mCi/mmol (Perkin Elmer), 10 μl of purified enzyme, and 400 to 500 ng of either small RNAs purified from the 293T control shRNA or an ABH8-shRNA1 stable cell line using the PureLink microRNA (miRNA) isolation kit (Invitrogen), single- or double-stranded DNA consisting of a 49-mer oligonucleotide (Table 1), or bovine serum albumin (New England Biolabs). The reaction mixtures were incubated at 25°C for 2 h, followed by the addition of diethyl pyrocarbonate (DEPC)-H2O to 20 μl and passage through a Quick Spin RNA column (Roche) for RNA, a Quick Spin oligonucleotide column for DNA, or a Microcon YM-30 centrifugal filter unit for protein. The eluates were added to 10 ml of scintillation fluid for scintillation counting.
Restriction enzyme-mediated oligonucleotide demethylase assays were performed essentially as described previously (39). Radiolabeled DNA oligonucleotides (150 fmol) containing 1-meA, 3-meC, or ethenoadenine lesions (Eurogentec; sequences listed in Table 1) were incubated at 25°C for 1 h in a total volume of 50 μl containing 50 mM Tris, pH 8.0, 2 mM ascorbic acid, 0.5 mM α-ketoglutarate, 40 μM FeSO4, and 10 μl of purified enzyme (∼50 ng protein) or buffer, followed by heat inactivation at 70°C for 15 min. An aliquot of each reaction mixture was digested with DpnII (New England Biolabs) for 1 h at 37°C, followed by electrophoresis on 7 M urea-18% denaturing polyacrylamide gels for 1 h at 500 V. Gels were visualized using a Cyclone phosphorimager (Perkin Elmer).
ABH8 expression analysis.
For ABH8 gene expression analysis, NHF1-hTert-lenti-LACZ (control line) and NHF1-hTert-lenti-ATM5 (ATM knockdown line) cells were constructed by methods similar to those in reference 4. Cells (4.5 × 104 cells/well) were plated in 6-well culture dishes. Twenty-four hours after plating cells were treated with 100 μM bleomycin (Research Products International, Mt. Prospect, IL) for 90 min, and the drug was removed and replaced with fresh growth medium. RNA was isolated at several time points using TRIzol reagent (Invitrogen) and subsequently purified by ethanol precipitation. Quantitative TaqMan PCR analysis was carried out with the ABI Prism 7900HT sequence detection system (Applied Biosystems, Foster City, CA) using the TaqMan one-step RT-PCR master mix reagent kit (Applied Biosystems). Assays were performed in a reaction volume of 20 μl, and reaction mixtures contained 500 ng purified RNA, 1× master mix without uracil-N-glycosylase (UNG), 1× MultiScribe and RNase inhibitor mix, and 1× probes and primer sets specific to Hs01098105_m1 (hABH8) or 1× human GAPDH. Thermal cycler parameters were as follows: incubation at 48°C for 30 min (reverse transcription step), denaturation at 95°C for 10 min, and then 40 cycles of the amplification step (denaturation at 95°C for 15 s and annealing/extension at 60°C for 1 min). All amplification reactions were performed in triplicate, and the relative quantification of ABH8 mRNA expression was determined after normalization with the endogenous control, GAPDH. Data processing and statistical analysis were performed using the ABI Prism SDS software, version 2.1 (Applied Biosystems).
Quantification of 5-carboxymethyl uridine (cm5U) and mcm5U.
RNA samples were isolated from 5 × 106 human cells using the PureLink miRNA isolation kit (Invitrogen), with the deaminase inhibitors tetrahydrouridine (50 μg/ml) and coformycin (5 μg/ml) and the antioxidants desferroxamine (0.1 mM) and butylated hydroxytoluene (0.1 mM) added to each sample to prevent postprocessing modification or degradation. Following quantification of tRNA (Agilent; 2100 Bioanalyzer), 1 pmol of [15N]5dA was added per μg of tRNA as an internal standard. RNA samples were lyophilized and redissolved at a final concentration of 120 μg of tRNA/ml in digestion buffer (30 mM sodium acetate, 2 mM zinc chloride, pH 6.8) with coformycin, tetrahydrouridine, desferroxamine, and butylated hydroxytoluene as noted earlier. RNA was hydrolyzed to nucleoside form by addition of nuclease P1 (1 unit) and RNase A (5 units) and incubation for 3 h at 37°C, followed by addition of 50 μl sodium acetate buffer (30 mM, pH 7.8), alkaline phosphatase (10 units), and phosphodiesterase I (0.5 unit) and overnight incubation at 37°C. Proteins were removed from the mixtures by ultrafiltration (Microcon; YM-10).
The chromatographic and mass spectrometric behavior of cm5U and mcm5U was initially established with tRNA from S. cerevisiae using HPLC-coupled, high-resolution quadrupole time-of-flight mass spectrometry (LC-QTOF; Agilent; 6510) operated in positive ion mode, on the basis of calculated m/z values of 303.0823 and 317.0979, respectively. Following hydrolysis and deproteination, the mixture of nucleosides was resolved by reversed-phase HPLC (Thermo Scientific; Hypersil Gold aQ, 150 by 2.1 mm, 3-μm particle size) and eluted at 0.3 ml/min and 36°C with solvent A (0.2% acetic acid in acetonitrile) and a gradient of solvent B (0.2% acetic acid in water) as follows: 0 to 18 min, 1 to 2%; 18 to 23 min, 2%; 23 to 28 min, 2 to 7%; 28 to 30 min, 7%; 30 to 31 min, 7 to 100%; 31 to 41 min, 100%. The eluting nucleosides, cm5U and mcm5U, were identified by scanning the HPLC eluate from m/z 100 to 1,000 for deglycosylation products derived from collision-induced dissociation (CID). Species with m/z values of 303.0823 and 317.0979 were observed with elution times of 4.6 and 12.8 min, respectively. The pattern of fragments produced by CID of each species revealed deglycosylation products consistent with the structures of cm5U and mcm5U (see Fig. 5B) and consistent with published observations (26). Analysis of yeast deficient in trm9 revealed a nearly complete loss of mcm5U.
FIG. 5.
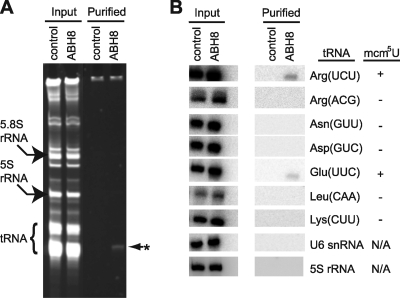
ABH8 interacts with a subset of tRNAs containing known modified wobble uridine residues. (A) Denaturing gel electrophoresis and Sybr gold stain of ABH8-associated RNAs. Arrows denote the migration pattern of the indicated RNAs, and the asterisk represents an ABH8-associated RNA species. (B) RNA blot hybridization analysis of ABH8-associated RNAs with the indicated oligonucleotide probes. The presence of a modified uridine (mcm5U) in the equivalent yeast tRNA is indicated on the right.
The relative quantities of cm5U and mcm5U in tRNA from the various cell preparations were determined by HPLC-coupled tandem quadrupole mass spectrometry (LC-MS/MS; Agilent; 6410) using the following HPLC retention times and molecular transitions: cm5U, 4.6 min, m/z 303 to 171; mcm5U, 12.8 min, m/z 317 to 185, and [15N]5dA, 10.1 min, m/z 257 to 141. The mass spectrometer was operated with an electrospray ionization source operated in positive ion mode and the following parameters: gas temperature, 350°C; gas flow, 10 liter/min; nebulizer pressure, 20 lb/in2; capillary voltage, 3,500 V; unit resolution. The signal intensity for each species was normalized by the signal for the [15N]5dA internal standard.
RESULTS
Affinity purification and proteomic analysis of ABH8 complexes.
Human ABH8 protein is the largest identified member of the mammalian AlkB homologs due to the fusion of an RNA recognition motif (RRM) and an S-adenosyl-l-methionine (SAM)-dependent methyltransferase (MT) motif to the amino and carboxy termini of the AlkB oxygenase motif, respectively (Fig. 1A). Among the identified AlkB homologs, ABH8 is the only member containing these additional domains. While the RRM motif represents a putative nucleic acid binding fold found in many RNA-associated proteins, the ABH8 MT domain is most closely related to the Trm9p tRNA methyltransferase found in yeast (Fig. 1B; data not shown). Phylogenetic analysis reveals ABH8 orthologs in higher eukaryotes, including all sequenced animal species, as well as a paralogous protein encoded by the C8ORF79 gene (Fig. 1B). Intriguingly, database searches have identified several ABH8 transcripts encoding different ABH8 variants encompassing various combinations of the individual domains (Fig. 2A). The modular domain format of the ABH8 coding exons suggests that alternative splicing could produce functional proteins with different activities.
FIG. 2.
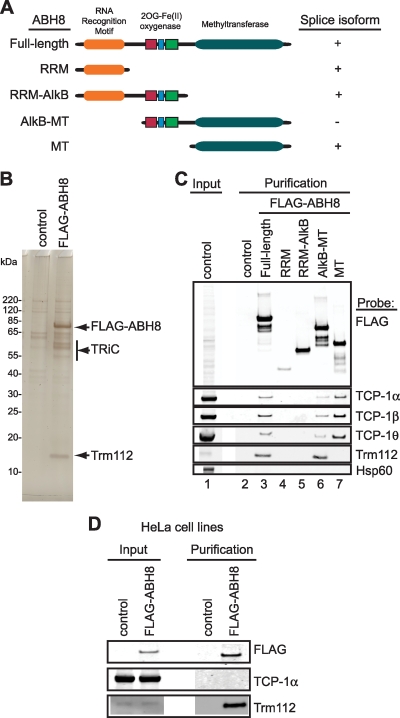
Purification and analysis of human ABH8 complexes. (A) Schematic of ABH8 variants and domains. RRM, RNA recognition motif; 2OG, 2-oxoglutarate; Fe(II), iron; SAM, S-adenosyl-l-methionine; AlkB, dioxygenase domain; MT, methyltransferase. (B) Protein profile of purified ABH8 complexes by silver stain analysis. Arrows indicate proteins specifically identified in purified ABH8 samples. (C) Confirmation and characterization of interactions between ABH8 with TRiC subunits and Trm112. (D) Purification of ABH8 complexes from HeLa human cells.
Green fluorescent protein tagging and expression revealed that the largest ABH8 variant displays a primarily cytoplasmic localization, with considerably less in the nucleus in human cells (Fig. 1C). Analysis of the remaining ABH8 variants reveals a similar pattern of cytoplasmic localization (D. Fu and L. Samson, unpublished observations). We note that these fluorescence results reveal only the steady-state localization since ABH8 could shuttle through or translocate to the nucleus under certain conditions.
To identify ABH8-interacting factors that would link ABH8 to a particular cellular process, we used affinity purification to isolate ABH8 complexes from human cells. Mammalian expression constructs encoding human ABH8 fused to the FLAG epitope tag were transfected into 293T human embryonic kidney cells, followed by whole-cell extract preparation and affinity purification on anti-FLAG antibody resin. Polyacrylamide gel fractionation and silver staining of the purified samples revealed several polypeptides that were detected specifically in the purified ABH8 sample (Fig. 2B). Several ABH8-interacting proteins were found in the 55- to 65-kDa range along with a prominent low-molecular-mass protein of ∼15 kDa.
After processing the eluates from parallel FLAG-ABH8 and mock purifications as complex mixtures for peptide identification by liquid chromatography-mass spectrometry, we compiled an inventory of proteins recovered specifically with ABH8 (Table 2). As expected, numerous peptide sequences confirmed the recovery of ABH8 itself. In addition, mass spectrometric analysis of the purified ABH8 samples revealed all eight subunits of the TCP ring complex (TRiC) (Fig. 2B; Table 2). TRiC forms a large, cylindrical chaperone assembly that is required for the folding of numerous cytosolic substrates (9). However, none of the AlkB homologs or SAM-dependent MTs was previously identified as a TRiC-associated protein.
TABLE 2.
Proteins identified by mass spectrometry specifically in purified human ABH8 samplesa
| Purification | Accession no. | Protein | Probabilityb | No. of unique peptides | MWc |
|---|---|---|---|---|---|
| 1 | Q96BT7 | Alkylated DNA repair protein AlkB homolog 8 | 7.16E−13 | 7 | 75 |
| Q9UI30 | TRM112-like protein | 1.27E−11 | 8 | 14 | |
| Q99832 | T-complex protein 1 subunit eta | 2.48E−11 | 3 | 59 | |
| P48643 | T-complex protein 1 subunit epsilon | 1.12E−09 | 4 | 60 | |
| P78371 | T-complex protein 1 subunit beta | 1.59E−09 | 5 | 57 | |
| P50991 | T-complex protein 1 subunit delta | 4.04E−09 | 2 | 57 | |
| P07900 | Heat shock protein HSP90 alpha | 5.47E−09 | 3 | 85 | |
| P50990 | T-complex protein 1 subunit theta | 7.80E−09 | 5 | 60 | |
| P17987 | T-complex protein 1 subunit alpha | 2.11E−08 | 4 | 60 | |
| P40227 | T-complex protein 1 subunit zeta | 4.62E−07 | 4 | 58 | |
| Q59H77 | Chaperonin containing TCP1, subunit 3 | 8.31E−07 | 6 | 64 | |
| 2 | P78371 | T-complex protein 1 subunit beta | 5.66E−14 | 13 | 57 |
| P40227 | T-complex protein 1 subunit zeta | 6.98E−13 | 10 | 58 | |
| Q99832 | T-complex protein 1 subunit eta | 3.51E−12 | 9 | 59 | |
| Q96BT7 | Alkylated DNA repair protein AlkB homolog 8 | 2.78E−11 | 11 | 75 | |
| P17987 | T-complex protein 1 subunit alpha | 6.24E−11 | 6 | 60 | |
| Q9UI30 | Trm112-like protein | 2.66E−10 | 9 | 14 | |
| P50991 | T-complex protein 1 subunit delta | 5.46E−10 | 9 | 58 | |
| P50990 | T-complex protein 1 subunit theta | 7.78E−10 | 16 | 60 | |
| P48643 | T-complex protein 1 subunit epsilon | 8.88E−10 | 6 | 60 | |
| Q59H77 | Chaperonin containing TCP1, subunit 3 | 1.61E−08 | 8 | 64 |
Inventories of proteins were compiled from independent FLAG-ABH8 purifications compared to a mock control purification for subtraction of nonspecific protein contaminants.
Probability of the best peptide matched to a peptide spectrum of a particular protein in the human protein database.
MW, molecular weight (in thousands).
In addition to the TRiC chaperonin, we identified a low-molecular-mass protein of 14 kDa termed human “Trm112-like” in the purified ABH8 samples, henceforth referred to as Trm112 (Fig. 2B; Table 2). In the yeast Saccharomyces cerevisiae, the orthologous Trm112p is a zinc finger protein subunit of three different methyltransferases, including the protein methylase Mtq2p and the tRNA methylases Trm9p and Trm11p (18). While Trm112p itself lacks a methyltransferase domain and does not catalyze methylation, it is an essential protein that is required for tRNA methylation in vivo (24, 38, 44). Like yeast Trm112, human Trm112 has been shown to interact with the human Mtq2 ortholog (15). However, an association between Trm112 and a human AlkB protein was not previously described.
Interaction analysis of ABH8-associated proteins.
The TRiC chaperonin and Trm112 represent the first putative proteins associated with the human ABH8 protein. To confirm these proteins as bona fide components of ABH8 complexes, we used coimmunoprecipitation followed by immunoblot analysis. To further characterize the significance of these interactions, we also tested the association of putative ABH8 variants with components of TRiC and with Trm112. The known ABH8 splice variants were expressed in human cells as FLAG-tagged fusion proteins; these were RRM alone, RRM with the dioxygenase motif, and the MT domain alone. In addition, we expressed the ABH8 dioxygenase motif fused to the MT domain (Fig. 2A). Whole-cell extracts from cells expressing tagged ABH8 proteins were subjected to affinity purification and immunoblot analysis. Using antibodies against TRiC components, we detected the specific copurification of three TRiC subunits with full-length ABH8 (Fig. 2C). Importantly, we did not detect the abundant Hsp60 chaperone in the purified ABH8 sample, indicating a specific interaction between ABH8 and TRiC (Fig. 2C). From the ABH8 domain purifications, we found that TRiC could associate only with ABH8 and its variants that contain the MT domain, including the MT domain alone (Fig. 2C). We infer that the ABH8 MT domain is necessary and sufficient for association with TRiC.
Next, we probed the purified ABH8 samples for the presence of Trm112. As was found for TRiC, we detected specific copurification of Trm112 with full-length ABH8 but not with the control, RRM, or RRM-AlkB (Fig. 2C). However, unlike the TRiC-ABH8 association, the ABH8 MT domain was not sufficient for Trm112 interaction; rather, human Trm112 requires both the AlkB motif and the MT domain to interact with ABH8 since we could readily detect an association between Trm112 and the AlkB-MT variant (Fig. 2C). Because an AlkB homolog has not been identified in S. cerevisiae, the AlkB motif-dependent interaction between human Trm112 and ABH8 is distinct from the yeast Trm112p interaction with the Trm9p tRNA methylase. Together, these results reveal TRiC and Trm112 as potential components within an ABH8 complex.
To confirm the interactions described above, we generated HeLa human cell lines that stably express lower levels of FLAG-tagged full-length ABH8. Using these cell lines, we detect robust copurification of Trm112 with ABH8 in the absence of the TRiC chaperone (Fig. 2D). These results verify a stable ABH8-Trm112 complex, which associates with TRiC upon ABH8 overexpression. Given the stable interaction between ABH8 and Trm112 along with the presence of an MT domain in human ABH8, we focused on the potential role of ABH8 in tRNA modification.
ABH8 catalyzes RNA methylation through its methyltransferase domain.
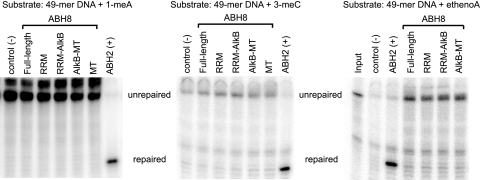
The initial identification of ABH8 as a putative AlkB dioxygenase that also contains a SAM-dependent MT domain suggests that ABH8 possesses multiple enzymatic activities. However, we were unable to detect oxidative demethylation activity for any of the ABH8 variants on canonical AlkB substrates, namely, 1-meA, 3-meC, or ethenoadenine bases present in DNA substrates (Fig. 3). While ABH8 does not display AlkB-like activity on these substrates, its potential targets could encompass bases in DNA or RNA that remain to be identified.
FIG. 3.
ABH8 does not demethylate canonical AlkB DNA substrates. The full-length ABH8 and domain variants were tested for oxidative demethylase activity on DNA substrates using a restriction enzyme-mediated oligonucleotide demethylase assay (see Materials and Methods). Purified ABH8 variants were incubated with a 49-mer DNA oligonucleotide containing 1-methyladenine (1-meA), 3-methylcytosine (3-meC), or ethenoadenine (ethenoA) (Table 1), followed by digestion with the methylation-sensitive restriction enzyme DpnII and gel electrophoresis. Unrepaired substrate and repaired product are indicated. The ABH2 protein, a known DNA/RNA demethylase, was included as a control.
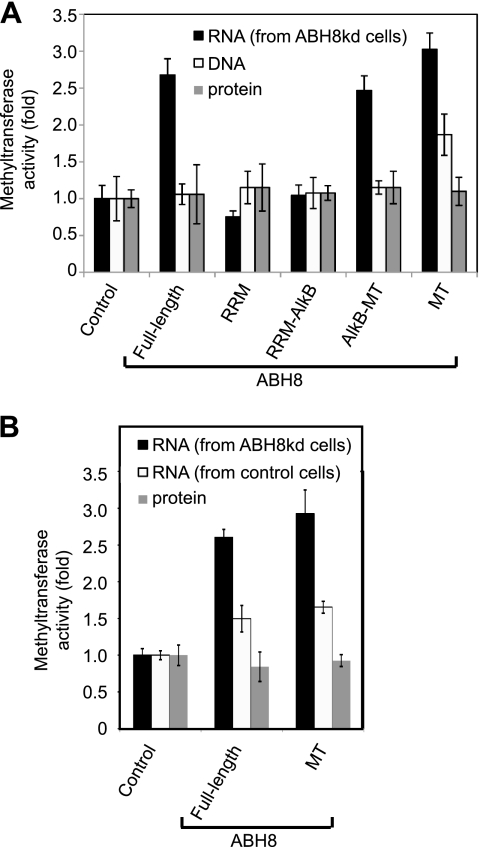
Based upon the copurification of the Trm112 subunit with ABH8, we next tested the purified ABH8 samples for methyltransferase activity using a previously described methylase assay (16). The purified ABH8 proteins were incubated with DNA, RNA, or protein substrates in the presence of SAM radiolabeled in the transferable methyl, followed by removal of unincorporated SAM and scintillation counting of labeled product. First, we tested RNA highly enriched for tRNAs purified from human cells depleted of ABH8 (described below) to increase the levels of potential ABH8 methyl acceptor substrates. Control reactions with a mock purification produced very low levels of methyl transfer to RNA, DNA, or protein. In contrast, purified full-length human ABH8 catalyzed robust RNA methylase activity, with a 2.5-fold increase in methyl transfer above background (Fig. 4; Table 3). Furthermore, the ABH8 AlkB-MT variant and the MT domain alone catalyzed RNA methylation to levels similar to that for the full-length ABH8 isoform. Importantly, we found that ABH8 variants lacking the MT domain yielded only background activity levels. Using the same reaction conditions as for RNA, we did not detect any methylase activity above background for either single- or double-stranded DNA substrates with full-length ABH8 (Fig. 4; Table 3). However, the ABH8 MT domain alone exhibited weak methylase activity on DNA, suggesting that the RRM and/or AlkB motif confers substrate specificity to the ABH8 MT domain. As a final specificity control, we found that none of the ABH8 variants could methylate bovine serum albumin protein. Significantly, the full-length and MT domain variants of ABH8 display much greater methyltransferase activity on RNA that is enriched for cm5U (Fig. 4B). Combined with enzymatic assays performed with recombinant human ABH8-Trm112 complexes purified from bacteria (L. Songe-Møller et al., personal communications), these results uncover a previously unidentified RNA methylase activity for human ABH8 catalyzed by its MT domain.
FIG. 4.
ABH8 catalyzes RNA methylation. Shown is in vitro methyltransferase activity of purified ABH8 variants on the indicated substrates. A mock purification sample (control) or purified ABH8 variants were incubated with purified human RNA enriched for tRNAs (from control or ABH8-depleted cells), DNA oligonucleotides, or bovine serum albumin protein followed by methyl donor removal and measurement of the eluted samples via scintillation counting. Methyltransferase activity is expressed as increase relative to the control. The average 14C incorporation for each sample is listed in Table 3.
TABLE 3.
Absolute 14C incorporation values for RNA, DNA, and protein substrates with ABH8
| Substrate | Form of ABH8 | cpma | cpm above control | Fold above control |
|---|---|---|---|---|
| RNA | Control (buffer) | 1,314 | 0 | 1.0 |
| Full length | 3,519 | 2,205 | 2.7 | |
| RRM | 990 | −324 | 0.8 | |
| RRM-AlkB | 1,371 | 57 | 1.0 | |
| AlkB-MT | 3,241 | 1,927 | 2.5 | |
| MT | 3,976 | 2,662 | 3.0 | |
| DNA (single stranded) | Control (buffer) | 455 | 0 | 1.0 |
| Full length | 482 | 27 | 1.1 | |
| RRM | 524 | 69 | 1.2 | |
| RRM-AlkB | 490 | 35 | 1.1 | |
| AlkB-MT | 524 | 69 | 1.2 | |
| MT | 850 | 395 | 1.9 | |
| Protein (bovine serum | Control (buffer) | 455 | 0 | 1.0 |
| albumin) | Full length | 482 | 27 | 1.1 |
| RRM | 524 | 69 | 1.2 | |
| RRM-AlkB | 490 | 35 | 1.1 | |
| AlkB-MT | 524 | 69 | 1.2 | |
| MT | 500 | 45 | 1.1 | |
| DNA (double stranded) | Control | 650 | 0.00 | 1.0 |
| Full length | 590 | 135 | 0.9 | |
| RRM | 675 | 220 | 1.0 | |
| RRM-AlkB | 610 | 155 | 0.9 | |
| AlkB-MT | 684 | 229 | 1.1 | |
| MT | 940 | 485 | 1.4 |
Average counts per minute of the radiolabeled products from the indicated reaction samples.
ABH8 interacts with tRNAs containing known modifications on their wobble uridines.
The presence of an active RNA methyltransferase domain in ABH8 that is homologous to tRNA MTs, along with the copurification of Trm112, suggests that ABH8 plays a role in the posttranscriptional modification of tRNA. We therefore analyzed the purified ABH8 complexes for the presence of copurifying RNAs. As expected, RNA purified from the input cell extracts displayed a complex pattern of small RNAs (Fig. 5A). In contrast, RNA purified from the ABH8 complex ran as a single, prominent RNA species migrating at position analogous to that for tRNAs in the total RNA input (Fig. 5A). This RNA band is specific for ABH8 since we did not detect any RNAs in the control mock purification sample.
To determine the identity of the ABH8-associated RNA molecules, they were hybridized with a panel of oligonucleotides complementary to specific tRNAs. We classified tRNAs into four groups based upon their canonical wobble nucleosides (U, G, C, or inosine) and selected at least one tRNA from each group for probing. Conspicuously, we detected two wobble uridine-containing tRNAs associated with the ABH8 complex, namely, tRNAArg(UCU) and tRNAGlu(UUC) (Fig. 5B). This interaction was specific to these particular wobble uridine tRNAs since two other tRNAs with wobble uridines, tRNALys(UUU) and tRNASec(UGA), did not copurify with ABH8 (Fu and Samson, unpublished). In contrast, none of the other tested tRNAs containing G, C, or inosine at the wobble position copurified with ABH8. As additional controls, we did not detect 5S rRNA or U6 snRNA in the purified ABH8 samples. It is important to note that yeast counterparts of tRNAGlu(UUC) and tRNAArg(UCU) are distinguished by the presence of a modified wobble uridine in the anticodon loop; the wobble uridine is initially converted to cm5U, with subsequent methylation by the Trm9p tRNA methylase to form mcm5U (26). Moreover, mcm5U and its thiolated version, mcm5s2U, has been found in the wobble position of mammalian tRNAs as well, including tRNAGlu(UUC), tRNAArg(UCU), tRNALys(UUU), and tRNASec(UGA) (23, 43). In humans, the putative methylase catalyzing this methylation event was unknown. That purified ABH8 displays RNA methylase activity and interacts with tRNAs subject to wobble uridine methylation suggests that ABH8 plays a role in mcm5U formation in tRNA.
ABH8 is required for maintaining mcm5U modification.
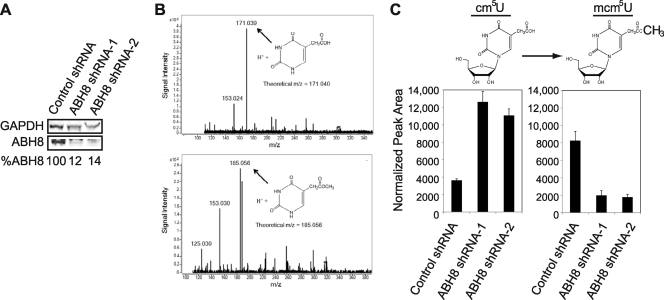
To determine whether ABH8 is involved in tRNA modification in vivo, we used small hairpin RNA (shRNA) to specifically deplete ABH8 from human cells. Two different shRNA constructs targeting distinct regions of the ABH8 mRNA and a nonsilencing shRNA construct with no known human targets were used. The retroviral shRNA constructs were integrated into 293T human embryonic kidney cells to generate stable cell lines expressing each shRNA. We successfully depleted ABH8 with both ABH8 shRNA constructs, as judged by immunoblot analysis (Fig. 6A). The ABH8-depleted cell lines exhibited no overt changes in morphology or growth rate.
FIG. 6.
ABH8 is required for maintaining mcm5U levels in vivo. (A) Confirmation of ABH8 depletion in human cells by immunoblot analysis. The depletion of ABH8 is expressed as a percentage relative to the control shRNA cell line normalized to the GAPDH loading control. (B) Identification of cm5U and mcm5U by HPLC-coupled tandem quadrupole mass spectrometry. Shown are extracted ion chromatograms of molecular species consistent with cm5U and mcm5U nucleobases based upon theoretical m/z. (C) Detection and comparison of cm5U and mcm5U levels in small RNAs purified from control and ABH8-depleted human cells. The levels of cm5U and mcm5U nucleosides were detected by mass spectrometry and quantified by integration of the normalized peak intensity for each nucleoside signal.
To determine whether modulation of ABH8 levels had any effect on mcm5U levels in cells, small RNAs highly enriched for tRNAs were purified from the control and ABH8-depleted cell lines and analyzed for mcm5U by mass spectrometry (Fig. 6B). Consistent with previous studies in wild-type yeast (26), the levels of mcm5U were much greater than that of cm5U in RNA purified from the control human cell line (Fig. 6C). In contrast, RNA from ABH8-depleted cells displayed a dramatic decrease in mcm5U, with a concomitant rise in the cm5U precursor (Fig. 6C). Importantly, the increase in cm5U levels found in the RNA of ABH8-depleted cell lines indicates that ABH8 is required for the final methyl modification to generate mcm5U rather than being involved in an upstream step in modified wobble uridine formation. Taken together, our results demonstrate that human ABH8 catalyzes the last step of mcm5U formation in vivo.
ABH8 is required for DNA damage survival.
In yeast, Trm9p-mediated wobble uridine methylation plays a critical role in the regulated translation of stress and DNA damage response pathway proteins (7, 26). Consequently, trm9 deletion mutants are extremely sensitive to gamma irradiation and the alkylating agent methyl methanesulfonate (MMS) (5-8). Based upon our finding that ABH8 is required for wobble uridine modification in human cells, we hypothesized that mammalian ABH8 could also play an essential role in DNA damage survival.
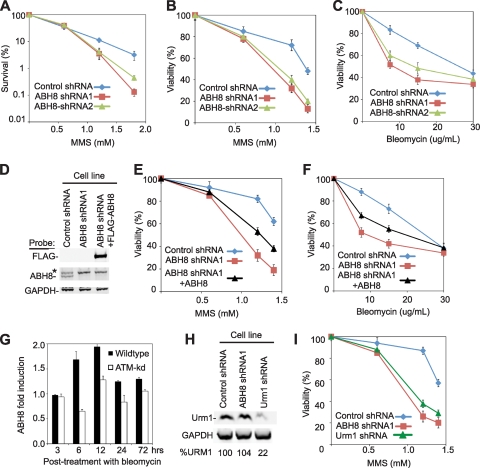
We determined whether ABH8 influences MMS sensitivity using colony formation and cellular viability assays. Human cell lines depleted for ABH8 displayed sensitivity to MMS compared to the control shRNA cell line, as measured by both assays (Fig. 7A and B). To determine whether the sensitivity phenotype exhibited by ABH8-depleted cells is specific to alkylation damage, we also tested sensitivity to bleomycin, which damages DNA through a mechanism different from that of MMS. Bleomycin intercalates between DNA base pairs and catalyzes free radical production to induce double-strand breaks (13). We found that ABH8-depleted cell lines displayed significant sensitivity to bleomycin, similar to their sensitivity to MMS (Fig. 7C). Thus, ABH8 increases the survival of human cells upon exposure to at least two damaging agents, namely, MMS and bleomycin. This sensitivity of ABH8-depleted human cells is comparable to that observed in cells depleted of characterized DNA damage response proteins, including the central DNA damage kinase, ATM, and the base excision repair protein, alkyladenine glycosylase (4, 36). Moreover, ABH8-depleted cells display significantly more sensitivity to MMS than cells completely devoid of ABH2, which is known to repair DNA in vivo (39).
FIG. 7.
Depletion of ABH8 diminishes cellular survival after exposure to DNA-damaging agents. (A) Colony formation assay of human cell lines after treatment with the indicated doses of MMS. (B and C) Viability of control and ABH8-depleted human cell lines after treatment with the indicated doses of MMS or bleomycin as measured by trypan blue dye exclusion or by the WST-1 cell proliferation and viability assay. (D) Verification of full-length ABH8 expression in human cell lines. Whole-cell extracts prepared from the indicated human cell lines were immunoblotted with antibody probes against the respective proteins. The asterisk denotes a protein that cross-reacts with the anti-ABH8 antibody. (E and F) Viability of human cell lines after treatment with MMS or bleomycin. (G) Real-time RT-PCR analysis of ABH8 transcripts from the indicated human cells. (H) Confirmation of URM1 depletion in human cells. (I) Viability of control and URM1-depleted human cell lines after treatment with MMS.
To confirm the role of ABH8 in DNA damage survival, we generated ABH8-depleted cell lines containing either an integrated empty vector or a construct expressing a siRNA-resistant form of the full-length ABH8 variant. Since the ABH8 shRNA1 cell line was generated using a shRNA construct targeting the 3′ untranslated region (UTR) of endogenous ABH8 transcripts, we utilized the ABH8 expression construct described in Fig. 2 to express a transcript containing the full-length FLAG-tagged-ABH8 open reading frame (ORF) lacking any ABH8 3′ UTR sequences. Unfortunately, we could not detect the FLAG-ABH8 protein with the anti-ABH8 antibody used in this study, most likely due to steric hindrance by the amino-terminal FLAG tag against the antibody, which recognizes only the first 50 amino acids of ABH8. Thus, we confirmed stable expression of the recombinant ABH8 in the ABH8 shRNA1 cell line using anti-FLAG antibodies (Fig. 7D). We find that expression of full-length ABH8 can partially complement the phenotype of sensitivity of ABH8-depleted cells to either MMS or bleomycin, as demonstrated by the increased damage resistance of ABH8-depleted cells expressing ABH8 compared to that of cells expressing the empty vector alone (Fig. 7E and F). The partial rescue of DNA damage sensitivity by full-length ABH8 could be due to the contributions of the remaining ABH8 splice isoforms to DNA damage survival. These results indicate that expression of at least full-length ABH8 contributes to viability after DNA damage. However, full rescue of damage sensitivity in ABH8-depleted cells could require expression of multiple ABH8 variants.
The requirement for ABH8 in DNA damage survival suggests that ABH8-catalyzed tRNA modification could be linked to the DNA damage response; therefore, we tested whether ABH8 gene expression is modulated upon exposure to bleomycin. Indeed, bleomycin exposure induced ABH8 expression, with peak induction occurring at 6 to 12 h postdamage (Fig. 7D). Notably, we find that cells depleted of the central upstream DNA damage response kinase, ATM, lacked this dynamic induction of ABH8 after bleomycin treatment, suggesting that ABH8 induction requires an intact DNA damage response pathway. The requirement for ABH8 in DNA damage survival and the ATM-dependent modulation of ABH8 expression suggest a link between ABH8, tRNA modification, and the DNA damage response in human cells.
The correlation between ABH8 depletion and DNA damage sensitivity suggests that maintenance of proper tRNA modifications is necessary for DNA damage survival. Thus, to provide additional evidence that modification of the tRNA wobble position is required for DNA damage survival, we analyzed the effect of depleting URM1 (ubiquitin-related modifier 1), a small sulfur carrier protein that is required for the thiol modification of tRNAs at the wobble positions, including those of tRNAGlu(UUC) and tRNAArg(UCU) (29, 40). Strikingly, we find that URM1 depletion leads to an MMS sensitivity phenotype comparable to that of ABH8-depleted cells (Fig. 7H and I). These results provide independent evidence that proper modification of wobble uridines in specific tRNAs is required for DNA damage survival.
DISCUSSION
The formation of diverse tRNA modifications requires many cellular pathways involving numerous enzymes. In mammals, several putative tRNA methyltransferases have been identified by sequence homology with bacterial or yeast proteins (11), but the majority of these enzymes remain uncharacterized, and their requirement in tRNA methylation awaits verification. Interestingly, the DNMT2 enzyme of the DNA cytosine methyltransferase family is one of the few known mammalian proteins with a confirmed role in tRNA methylation (20). DNMT2 methylates tRNAAsp using a DNA methyltransferase-like catalytic mechanism, but the function of this modification is unknown. Here, we show that human ABH8 protein contains an active RNA methyltransferase domain that is essential for maintaining the levels of a critical tRNA modification in human cells.
In addition to tRNA methylation, the DNA damage sensitivity phenotype of ABH8-depleted cells suggests a role for ABH8 in DNA damage survival through the regulation of tRNA modification, potentially in concert with any oxidative demethylase activity conferred by the AlkB dioxygenase domain of ABH8. Support for this notion comes first from our confirmation that ABH8 localizes primarily to the cytoplasm and second from our finding that ABH8 protects against bleomycin, a damaging agent that does not methylate nucleic acids. Future studies will be devoted to understanding the relative contributions of the ABH8 dioxygenase and methyltransferase domains to survival after DNA damage.
In yeast, mRNA transcripts encoding stress response proteins are significantly enriched for codons decoded by tRNAs containing the Trm9p-dependent mcm5U wobble modification (6). The mcm5U wobble base generated by Trm9 has been shown to modulate tRNA-mRNA pairing and enhance binding with the cognate codon (2). Thus, tRNA modification enzymes can directly regulate the translation of specific proteins by modulating a subset of codon-anticodon interactions. Our results indicate that a conserved mechanism in which ABH8-catalyzed tRNA modification regulates the translation of specific proteins that are essential for surviving genotoxic stress could be operating in human cells. Notably, previous reports have demonstrated the preferential translation of mRNAs encoding DNA damage response and repair proteins in human cells after cellular stress (37). Due to the presence of mcm5U in the wobble position of tRNASec(UGA), ABH8 could also modulate the specific translation of the entire repertoire of selenocysteine proteins. It will be of great interest to identify the specific proteins whose translation is regulated by tRNA modification catalyzed by ABH8.
The fusion of a putative dioxygenase motif to an active methyltransferase domain in ABH8 suggests the intriguing possibility of reversible RNA modification. While the substrates of the ABH8 dioxygenase motif remain to be discovered, ABH8 could demethylate wobble uridines or other modified nucleosides in tRNA under particular conditions. Alternatively, given the RNA repair capacity demonstrated by certain AlkB proteins (1, 34), ABH8 could reverse aberrant methylation of tRNA, either caused by spurious methylase activity of the methyltransferase domain or induced by endogenous or exogenous alkylating agents. Alkylating agents have been shown to alter or inactivate mRNA, tRNA, or rRNA function, leading to ribosome stalling, miscoding, or translational blocks, with the production of truncated or mutant polypeptides (32, 34). While TRiC could function as a chaperone for the correct folding of a multidomain protein such as ABH8, it could also represent a mechanism by which incompletely synthesized translation products caused by a stalled ribosome are stabilized by TRiC while ABH8 rescues the damaged RNAs.
The potential substrates of ABH8 could also encompass protein targets since the AlkB enzymes belong to a superfamily of iron-dependent dioxygenases that include the JmjC domain histone demethylases, which have an enzymatic mechanism identical to that of AlkB to demethylate lysine residues in histone proteins (35). In addition, oxidative cleavage of amino acids by iron-dependent dioxygenases such as cysteine dioxygenase plays important roles in maintaining proper levels of particular amino acids (25). In the case of ABH8, oxidative cleavage of specific amino acids or proteins could occur if their levels are upregulated under particular conditions of stress or damage.
Of significance, ABH8 is highly expressed in many urothelial cancers, with a positive correlation between ABH8 expression and high-grade, invasive carcinomas (46). Consistent with these observations, silencing of ABH8 significantly suppresses the angiogenesis and growth of bladder cancers in vivo (42). Thus, tRNA modifications catalyzed by methyltransferases such as ABH8 could be an important factor in the growth and survival of both normal and transformed cells.
Acknowledgments
We thank Peter Svensson and Emma Wang for the URM1 knockdown cell line and the Klungland and Falnes labs for sharing unpublished results.
This work was supported by NIH grants CA055042 and ES002109 to L.D.S. and ES01701 to T.J.B., by the Intramural Research Program of the NIH (R.S.P.), and by the MIT Westaway Fund and NCRR grant S10-RR023783 to P.C.D. D.F. is supported by an American Cancer Society TJX Postdoctoral Fellowship, and L.D.S. is an American Cancer Society Research Professor.
Footnotes
Published ahead of print on 22 March 2010.
REFERENCES
- 1.Aas, P. A., M. Otterlei, P. O. Falnes, C. B. Vagbo, F. Skorpen, M. Akbari, O. Sundheim, M. Bjoras, G. Slupphaug, E. Seeberg, and H. E. Krokan. 2003. Human and bacterial oxidative demethylases repair alkylation damage in both RNA and DNA. Nature 421:859-863. [DOI] [PubMed] [Google Scholar]
- 2.Agris, P. F., F. A. Vendeix, and W. D. Graham. 2007. tRNA's wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 366:1-13. [DOI] [PubMed] [Google Scholar]
- 3.Aravind, L., and E. V. Koonin. 2001. The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol. 2:RESEARCH0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arlander, S. J., B. T. Greene, C. L. Innes, and R. S. Paules. 2008. DNA protein kinase-dependent G2 checkpoint revealed following knockdown of ataxia-telangiectasia mutated in human mammary epithelial cells. Cancer Res. 68:89-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Begley, T. J., A. S. Rosenbach, T. Ideker, and L. D. Samson. 2002. Damage recovery pathways in Saccharomyces cerevisiae revealed by genomic phenotyping and interactome mapping. Mol. Cancer Res. 1:103-112. [PubMed] [Google Scholar]
- 6.Begley, T. J., A. S. Rosenbach, T. Ideker, and L. D. Samson. 2004. Hot spots for modulating toxicity identified by genomic phenotyping and localization mapping. Mol. Cell 16:117-125. [DOI] [PubMed] [Google Scholar]
- 7.Begley, U., M. Dyavaiah, A. Patil, J. P. Rooney, D. DiRenzo, C. M. Young, D. S. Conklin, R. S. Zitomer, and T. J. Begley. 2007. Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol. Cell 28:860-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bennett, C. B., L. K. Lewis, G. Karthikeyan, K. S. Lobachev, Y. H. Jin, J. F. Sterling, J. R. Snipe, and M. A. Resnick. 2001. Genes required for ionizing radiation resistance in yeast. Nat. Genet. 29:426-434. [DOI] [PubMed] [Google Scholar]
- 9.Brackley, K. I., and J. Grantham. 2009. Activities of the chaperonin containing TCP-1 (CCT): implications for cell cycle progression and cytoskeletal organisation. Cell Stress Chaperones 14:23-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Czerwoniec, A., S. Dunin-Horkawicz, E. Purta, K. H. Kaminska, J. M. Kasprzak, J. M. Bujnicki, H. Grosjean, and K. Rother. 2009. MODOMICS: a database of RNA modification pathways. 2008 update. Nucleic Acids Res. 37:D118-D121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Crecy-Lagard, V. 2007. Identification of genes encoding tRNA modification enzymes by comparative genomics. Methods Enzymol. 425:153-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delaney, J. C., L. Smeester, C. Wong, L. E. Frick, K. Taghizadeh, J. S. Wishnok, C. L. Drennan, L. D. Samson, and J. M. Essigmann. 2005. AlkB reverses etheno DNA lesions caused by lipid oxidation in vitro and in vivo. Nat. Struct. Mol. Biol. 12:855-860. [DOI] [PubMed] [Google Scholar]
- 13.Ehrenfeld, G. M., J. B. Shipley, D. C. Heimbrook, H. Sugiyama, E. C. Long, J. H. van Boom, G. A. van der Marel, N. J. Oppenheimer, and S. M. Hecht. 1987. Copper-dependent cleavage of DNA by bleomycin. Biochemistry 26:931-942. [DOI] [PubMed] [Google Scholar]
- 14.Falnes, P. O., R. F. Johansen, and E. Seeberg. 2002. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature 419:178-182. [DOI] [PubMed] [Google Scholar]
- 15.Figaro, S., N. Scrima, R. H. Buckingham, and V. Heurgue-Hamard. 2008. HemK2 protein, encoded on human chromosome 21, methylates translation termination factor eRF1. FEBS Lett. 582:2352-2356. [DOI] [PubMed] [Google Scholar]
- 16.Frye, M., and F. M. Watt. 2006. The RNA methyltransferase Misu (NSun2) mediates Myc-induced proliferation and is upregulated in tumors. Curr. Biol. 16:971-981. [DOI] [PubMed] [Google Scholar]
- 17.Fu, D., and K. Collins. 2006. Human telomerase and Cajal body ribonucleoproteins share a unique specificity of Sm protein association. Genes Dev. 20:531-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gavin, A. C., P. Aloy, P. Grandi, R. Krause, M. Boesche, M. Marzioch, C. Rau, L. J. Jensen, S. Bastuck, B. Dumpelfeld, A. Edelmann, M. A. Heurtier, V. Hoffman, C. Hoefert, K. Klein, M. Hudak, A. M. Michon, M. Schelder, M. Schirle, M. Remor, T. Rudi, S. Hooper, A. Bauer, T. Bouwmeester, G. Casari, G. Drewes, G. Neubauer, J. M. Rick, B. Kuster, P. Bork, R. B. Russell, and G. Superti-Furga. 2006. Proteome survey reveals modularity of the yeast cell machinery. Nature 440:631-636. [DOI] [PubMed] [Google Scholar]
- 19.Gerken, T., C. A. Girard, Y. C. Tung, C. J. Webby, V. Saudek, K. S. Hewitson, G. S. Yeo, M. A. McDonough, S. Cunliffe, L. A. McNeill, J. Galvanovskis, P. Rorsman, P. Robins, X. Prieur, A. P. Coll, M. Ma, Z. Jovanovic, I. S. Farooqi, B. Sedgwick, I. Barroso, T. Lindahl, C. P. Ponting, F. M. Ashcroft, S. O'Rahilly, and C. J. Schofield. 2007. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science 318:1469-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goll, M. G., F. Kirpekar, K. A. Maggert, J. A. Yoder, C. L. Hsieh, X. Zhang, K. G. Golic, S. E. Jacobsen, and T. H. Bestor. 2006. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 311:395-398. [DOI] [PubMed] [Google Scholar]
- 21.Grosjean, H. 2009. DNA and RNA modification enzymes: structure, mechanism, function, and evolution. Landes Bioscience, Austin, TX.
- 22.Grosjean, H. 2005. Fine-tuning of RNA functions by modification and editing. Springer, Berlin, Germany.
- 23.Hatfield, D. L., and V. N. Gladyshev. 2002. How selenium has altered our understanding of the genetic code. Mol. Cell. Biol. 22:3565-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heurgue-Hamard, V., M. Graille, N. Scrima, N. Ulryck, S. Champ, H. van Tilbeurgh, and R. H. Buckingham. 2006. The zinc finger protein Ynr046w is plurifunctional and a component of the eRF1 methyltransferase in yeast. J. Biol. Chem. 281:36140-36148. [DOI] [PubMed] [Google Scholar]
- 25.Joseph, C. A., and M. J. Maroney. 2007. Cysteine dioxygenase: structure and mechanism. Chem. Commun. (Cambridge) 2007:3338-3349. [DOI] [PubMed] [Google Scholar]
- 26.Kalhor, H. R., and S. Clarke. 2003. Novel methyltransferase for modified uridine residues at the wobble position of tRNA. Mol. Cell. Biol. 23:9283-9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kataoka, H., Y. Yamamoto, and M. Sekiguchi. 1983. A new gene (alkB) of Escherichia coli that controls sensitivity to methyl methane sulfonate. J. Bacteriol. 153:1301-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurowski, M. A., A. S. Bhagwat, G. Papaj, and J. M. Bujnicki. 2003. Phylogenomic identification of five new human homologs of the DNA repair enzyme AlkB. BMC Genomics 4:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leidel, S., P. G. Pedrioli, T. Bucher, R. Brost, M. Costanzo, A. Schmidt, R. Aebersold, C. Boone, K. Hofmann, and M. Peter. 2009. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature 458:228-232. [DOI] [PubMed] [Google Scholar]
- 30.MacCoss, M. J., C. C. Wu, and J. R. Yates III. 2002. Probability-based validation of protein identifications using a modified SEQUEST algorithm. Anal. Chem. 74:5593-5599. [DOI] [PubMed] [Google Scholar]
- 31.Maraia, R. J., N. H. Blewett, and M. A. Bayfield. 2008. It's a mod mod tRNA world. Nat. Chem. Biol. 4:162-164. [DOI] [PubMed] [Google Scholar]
- 32.Masta, A., P. J. Gray, and D. R. Phillips. 1995. Nitrogen mustard inhibits transcription and translation in a cell free system. Nucleic Acids Res. 23:3508-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morgenstern, J. P., and H. Land. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18:3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ougland, R., C. M. Zhang, A. Liiv, R. F. Johansen, E. Seeberg, Y. M. Hou, J. Remme, and P. O. Falnes. 2004. AlkB restores the biological function of mRNA and tRNA inactivated by chemical methylation. Mol. Cell 16:107-116. [DOI] [PubMed] [Google Scholar]
- 35.Ozer, A., and R. K. Bruick. 2007. Non-heme dioxygenases: cellular sensors and regulators jelly rolled into one? Nat. Chem. Biol. 3:144-153. [DOI] [PubMed] [Google Scholar]
- 36.Paik, J., T. Duncan, T. Lindahl, and B. Sedgwick. 2005. Sensitization of human carcinoma cells to alkylating agents by small interfering RNA suppression of 3-alkyladenine-DNA glycosylase. Cancer Res. 65:10472-10477. [DOI] [PubMed] [Google Scholar]
- 37.Powley, I. R., A. Kondrashov, L. A. Young, H. C. Dobbyn, K. Hill, I. G. Cannell, M. Stoneley, Y. W. Kong, J. A. Cotes, G. C. Smith, R. Wek, C. Hayes, T. W. Gant, K. A. Spriggs, M. Bushell, and A. E. Willis. 2009. Translational reprogramming following UVB irradiation is mediated by DNA-PKcs and allows selective recruitment to the polysomes of mRNAs encoding DNA repair enzymes. Genes Dev. 23:1207-1220. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Purushothaman, S. K., J. M. Bujnicki, H. Grosjean, and B. Lapeyre. 2005. Trm11p and Trm112p are both required for the formation of 2-methylguanosine at position 10 in yeast tRNA. Mol. Cell. Biol. 25:4359-4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ringvoll, J., L. M. Nordstrand, C. B. Vagbo, V. Talstad, K. Reite, P. A. Aas, K. H. Lauritzen, N. B. Liabakk, A. Bjork, R. W. Doughty, P. O. Falnes, H. E. Krokan, and A. Klungland. 2006. Repair deficient mice reveal mABH2 as the primary oxidative demethylase for repairing 1meA and 3meC lesions in DNA. EMBO J. 25:2189-2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schlieker, C. D., A. G. Van der Veen, J. R. Damon, E. Spooner, and H. L. Ploegh. 2008. A functional proteomics approach links the ubiquitin-related modifier Urm1 to a tRNA modification pathway. Proc. Natl. Acad. Sci. U. S. A. 105:18255-18260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sedgwick, B., P. A. Bates, J. Paik, S. C. Jacobs, and T. Lindahl. 2007. Repair of alkylated DNA: recent advances. DNA Repair (Amsterdam) 6:429-442. [DOI] [PubMed] [Google Scholar]
- 42.Shimada, K., M. Nakamura, S. Anai, M. De Velasco, M. Tanaka, K. Tsujikawa, Y. Ouji, and N. Konishi. 2009. A novel human AlkB homologue, ALKBH8, contributes to human bladder cancer progression. Cancer Res. 69:3157-3164. [DOI] [PubMed] [Google Scholar]
- 43.Sprinzl, M., and K. S. Vassilenko. 2005. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 33:D139-D140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Studte, P., S. Zink, D. Jablonowski, C. Bar, T. von der Haar, M. F. Tuite, and R. Schaffrath. 2008. tRNA and protein methylase complexes mediate zymocin toxicity in yeast. Mol. Microbiol. 69:1266-1277. [DOI] [PubMed] [Google Scholar]
- 45.Trewick, S. C., T. F. Henshaw, R. P. Hausinger, T. Lindahl, and B. Sedgwick. 2002. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature 419:174-178. [DOI] [PubMed] [Google Scholar]
- 46.Tsujikawa, K., K. Koike, K. Kitae, A. Shinkawa, H. Arima, T. Suzuki, M. Tsuchiya, Y. Makino, T. Furukawa, N. Konishi, and H. Yamamoto. 2007. Expression and subcellular localization of human ABH family molecules. J. Cell. Mol. Med. 11:1105-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]