Abstract
Virus infection induces host antiviral responses, including induction of type I interferons. Transcription factor interferon regulatory factor 3 (IRF3) plays a pivotal role and is tightly regulated in this process. Here, we identify HERC5 (HECT domain and RLD 5) as a specific binding protein of IRF3 by immunoprecipitation. Ectopic expression or knockdown of HERC5 could, respectively, enhance or impair IRF3-mediated gene expression. Mechanistically, HERC5 catalyzes the conjugation of ubiquitin-like protein ISG15 onto IRF3 (Lys193, -360, and -366), thus attenuating the interaction between Pin1 and IRF3, resulting in sustained IRF3 activation. In contrast to results for wild-type IRF3, the mutant IRF3(K193,360,366R) interacts tightly with Pin1, is highly polyubiquitinated, and becomes less stable upon Sendai virus (SeV) infection. Consistently, host antiviral responses are obviously boosted or crippled in the presence or absence of HERC5, respectively. Collectively, this study characterizes HERC5 as a positive regulator of innate antiviral responses. It sustains IRF3 activation via a novel posttranslational modification, ISGylation.
The mammalian immune system consists of innate and adaptive branches, which cooperate to protect the host against microbial invasion (45). In order to contain virus spread, potent responses are elicited in host cells before intervention by the immune system is staged. Central to this process is the recognition of conserved and invariant molecular patterns of virus to induce the production of type I interferons (IFNs) and other cytokines (17). Retinoic acid-inducible gene 1 (RIG-I) and melanoma differentiation-associated gene 5 (MDA5) have recently been characterized as ubiquitous sensors for detecting cytosolic viral RNA in host cells (16, 36, 46, 51, 52). Once RIG-I/MDA5 sense viral RNAs, they initiate the formation of a protein complex on the outside membrane of mitochondria, which includes mitochondrial antiviral signaling protein (MAVS) (also known as IPS1/VISA/CARDIF) (18, 29, 43, 49), tumor necrosis factor (TNF) receptor-associated factor 3 (TRAF3) (11, 34), tumor necrosis factor receptor 1-associated death domain protein (TRADD) (30), and NAK-associated protein1 (NAP1) (41). This process is regulated by NLRX-1, STING, and DUBA (15, 19, 32). Consequently, TANK-binding kinase 1 (TBK1) and IκB kinase ɛ (IKKɛ) are activated, which further phosphorylates and activates interferon regulatory factor 3 (IRF3) on a series of Ser/Thr residues at its C terminus (8, 44). The phosphorylated IRF3 forms a homodimer and translocates into the nucleus, recruiting coactivators such as p300/CBP (24, 26, 42). This ultimately induces the early production of antiviral proteins (e.g., IFN-β) and is critical for establishing an antiviral state in host cells. Hopefully, these insights will be helpful to develop vaccines and control infectious diseases.
The intricacy of IRF3 activation and modulation is currently under intensive study and is fundamental to understanding the general mechanism of primary antiviral responses. Several proteins have been implicated in these regulations, including TRIM21, Pin1, Cull-1, GRX-1, and JNK (Jun N-terminal protein kinase) (3, 37, 40, 50, 54). A striking feature is the posttranslational modification of IRF3. Besides phosphorylation, IRF3 is subjected to ubiquitination, sumoylation, S glutathionylation, and ISGylation, which help to shape the strength and duration of IRF3 action (3, 23, 27, 37). Although the ubiquitin (Ub) E3 ligase for IRF3 remains to be identified, phosphorylation-dependent ubiquitination is well established to terminate IRF3 function by proteasome-dependent degradation (3, 26). Pin1, a peptidyl-prolyl isomerase, interacts with phosphorylated IRF3 and promotes its ubiquitination via an unknown mechanism (40). Similarly to NF-κB signal transduction (4), ubiquitination displays both positive and negative regulatory functions in triggering IRF3 activation (1, 9). Interestingly, virus proteins could influence the modifications of IRF3 (2, 39). It is a great challenge to dissect the biochemical processes of all of these modifications and understand the dynamic relationship among them in the context of virus infection.
Interferon-stimulated gene 15 (ISG15) consists of two similar domains and displays structural homology to ubiquitin (33). It is robustly induced by type I interferons, lipopolysaccharide (LPS), or viruses (20, 21, 38). ISG15 is conjugated onto host proteins via its conserved C-terminal motif (152-LRLRGG-157). Like ubiquitination, the process of ISGylation is catalyzed by E1 (UBE1L), E2 (UbcH8), and E3 ligases. This conjugation cascade is also inducible by interferons (6, 53, 55, 58). ISGylation is apparently linked to regulating some biological processes (28, 35). Taking a proteomics approach, more than 100 proteins are identified as potential targets of ISG15 modification (47, 48, 56). These proteins cover a wide spectrum of biological processes, including transcriptional regulation, signal transduction, inflammation, and control of cell growth. A caveat is that ISG15 with or without its conjugating system (E1, E2, and E3) was overexpressed in these experiments, which made the observations possibly artificial. Indeed, only a dozen of the proposed candidates have been validated as authentic substrates in vivo, and the functional implications remain largely unknown.
HERC5, initially described as cyclin E-binding protein 1 (Ceb1), belongs to the HERC protein family, which is characterized by the presence of a HECT domain and one or more RCC1-like domains (RLD) (12, 31). HERC proteins are suggested to have E3 ligase activity. In particular, HERC3 has been shown to be a ubiquitin binding protein (5). The expression of HERC5 is especially high in testis and fetal brain but low in most tissues (22). Interestingly, HERC5 is induced by the proinflammatory cytokines TNF-α and interleukin-1β (IL-1β) (22). HERC5 is also markedly induced by type I IFNs and functions as an ISG15 E3 ligase to promote ISGylation (6, 48). However, the authentic substrates and biological function of Herc5-mediated ISGylation remain largely unknown. It is open to address whether HERC5, as an ISG15 E3 ligase, plays a role in the antiviral response.
By immunoprecipitation, we have identified HERC5 as a novel regulator of IRF3 function. We report that HERC5 is significantly induced upon RNA virus infection and interacts with IRF3. Ectopic expression or knockdown of HERC5 could, respectively, enhance or impair IRF3-mediated gene expression, with a consequent reduction or promotion of virus replication. Cys994 of the HECT domain is indispensable for potentiating IRF3 activity upon virus infection. Interestingly, HERC5 catalyzes the conjugation of ISG15 onto IRF3 (Lys193, -360, and -366). This modification attenuates the interaction between Pin1 and IRF3, thus antagonizing IRF3 ubiquitination and degradation. Consistently, host antiviral responses are boosted or crippled in the presence or absence of HERC5, respectively. Collectively, our study characterizes HERC5 as a positive regulator of antiviral innate immune responses, which maintains IRF3 stability via catalyzing ISGylation of IRF3.
MATERIALS AND METHODS
Cell culture.
HEK293T and HEK293 cells were cultured using Dulbecco's modified Eagle's medium (DMEM) plus 10% fetal bovine serum (FBS), supplemented with 1% penicillin-streptomycin (Invitrogen). Transient transfection was performed with Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. IRF3−/− murine embryonic fibroblasts (MEF) were kindly provided by Genhong Cheng (University of California Los Angeles). Vero cells were a gift from Ke Lan (Shanghai Pasteur Institute, CAS).
Plasmids.
Plasmid pcDNA3-HERC5, a gift from Jon M. Huibregtse (University of Texas at Austin), was subcloned into relevant expression vectors. Site-directed mutagenesis was performed by using a QuikChange XL kit (Stratagene). All constructs were confirmed by sequencing.
Real-time PCR.
Eight hours after Sendai virus (SeV) infection, total cellular RNA was isolated with TRIzol (Invitrogen) according to the manufacturer's instructions. Reverse transcription of purified RNA was performed using an oligo(dT) primer. The quantification of gene transcripts was determined by real-time PCR using SYBR green I dye (Invitrogen). All values were normalized to the level of β-actin mRNA. The primers used were as follows: β-actin, sense (5-AAAGACCTGTACGCCAACAC-3) and antisense (5-GTCATACTCCTGCTTGCTGAT-3); IL-8, sense (5-AGGTGCAGTTTTGCCAAGGA-3) and antisense (5-TTTCTGTGTGGCGCAGTGT-3); IFN-β, sense (5-ATTGCCTCAAGGACAGGATG-3) and antisense (5-GGCCTTCAGGTAATGCAGAA-3); ISG54, sense (5-TGCAACCTACTGGCCTATCTA-3) and antisense (5-CAGGTGACCAGACTTCTGATT-3); and RANTES, sense (5-TACACCAGTGGCAAGTGCTC-3) and antisense (5-ACACACTTGGCGGTTCTTTC-3).
Luciferase reporter assays.
Luciferase reporter assays were performed as described previously (50).
Immunoblot analysis and immunoprecipitation assay.
For immunoblotting, immunoprecipitates or whole-cell lysates were resolved by SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane (Bio-Rad). The immunoblots were probed with the following antibodies (purchased from Santa Cruz Biotechnology unless indicated otherwise): anti-Flag (1/5,000; Sigma-Aldrich), antihemagglutinin (anti-HA) (1/2,000), anti-His (1/1,000), anti-β-actin (1/10,000; Sigma-Aldrich), anti-IRF3 (1/500, monoclonal; 1/1,000, polyclonal), anti-glutathione S-transferase (anti-GST) (1/1,000), anti-myc (1/1,000), antiubiquitin (1/1,000), anti-HERC5 (1/1,000; AbMART), and anti-ISG15 (1/1,000; AbMART). The proteins were visualized by using a nitroblue tetrazolium (NBT)-5-bromo-4-chloro-3-indolylphosphate (BCIP) Western blotting system (Promega) or a SuperSignal West Pico chemiluminescence ECL kit (Pierce). For immunoprecipitation, cells were collected and then lysed in Nonidet P-40 buffer or sonicated in Tris-buffered saline (TBS) buffer supplemented with a complete protease inhibitor cocktail (Roche). After cell lysates were precleared with normal mouse IgG and protein A/G agarose beads for 1 h at 4°C, whole-cell lysates were used for immunoprecipitation with various antibodies. Generally, 0.5 to 1 μg of commercial antibody was added to 0.5 ml of cell lysate and then incubated for 2 to 4 h at 4°C. After addition of protein A/G agarose beads, the incubation was continued for 2 to 10 h. Immunoprecipitates were washed extensively with lysis buffer and eluted with sodium dodecyl sulfate (SDS) loading buffer by being boiled for 5 min.
Ni-NTA-agarose pulldown assays.
For the Ni-nitrilotriacetic acid (NTA)-agarose pulldown assays, cells were lysed in 6 M urea. Equal amounts of cell extracts (1 mg) and 20 μl of Ni-NTA-agarose beads (Qiagen) were incubated overnight at 4°C. Precipitates were washed three times with the same buffer and subjected to SDS-PAGE followed by immunoblotting.
GST pulldown assays.
HEK293 cells were lysed completely with Nonidet P-40 buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 1 mM phenylmethylsulfonyl fluoride [PMSF], 0.5% [vol/vol] Nonidet P-40) supplemented with a complete protease inhibitor cocktail (Roche). The lysates were incubated with 20 μl of glutathione Sepharose 4B beads (GE Healthcare Bio) at 4°C for 2 h. Precipitates were washed extensively with lysis buffer and subjected to SDS-PAGE followed by immunoblotting.
Protein purification and MS.
Twenty-four hours after transfection with a vector expressing Flag-IRF3, HEK293T cells were mock infected or infected with SeV. Sixteen hours later, the cells were collected and lysed with Nonidet P-40 buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 1 mM PMSF, 0.5% [vol/vol] Nonidet P-40) supplemented with a complete protease inhibitor cocktail (Roche). Postcentrifuged supernatants were precleared with protein A/G beads at 4°C for 0.5 h. Precleared lysates were mixed with 1 μg of mouse anti-Flag antibody at 4°C for 1 h. Then, protein A/G beads were added, and the binding reaction mixture was incubated for 2 h at 4°C. Precipitates were washed extensively with lysis buffer. Proteins bound to protein A/G beads were separated on 8% SDS-polyacrylamide gels. After silver staining (Sigma-Aldrich), specific protein bands were excised and analyzed by ion trap mass spectrometry (MS).
siRNA knockdown.
Small interfering RNAs (siRNAs) for the nonspecific control (NC), HERC5, and ISG15 were purchased from GenePharma and transfected using Lipofectamine 2000 (Invitrogen). The sequences of the siRNA oligonucleotides used in this study were as follows: for NC siRNA, 5-UUCUCCGAAGGUGUCACGU-3; for HERC5 siRNA, 5-GGACUAGACAAUCAGAAAGTT-3; for mutant HERC5 (mtHERC5) siRNA, 5-GGACUAGACGCUCAGAGCCTT-3; and for ISG15 siRNA, 5-UGAGCACCGUGUUCAUGAATT-3. HEK293 cells (8 × 104) were plated in 12-well plates in antibiotic-free DMEM. At 50% confluence, 40 pmol of siRNA was transfected into cells. To determine efficiency of protein knockdown, 48 h posttransfection, cells were lysed in Nonidet P-40 buffer and immunoblotted with various antibodies.
Rescue experiments.
HEK293 cells were transfected with control or HERC5-specific siRNA for 24 h, and then the cells were transfected with siRNA-resistant HERC5 or control plasmids, followed by SeV infection the next day. The HERC5 siRNA-resistant forms were generated by introducing silent mutations into the HERC5 siRNA target sequence (715-GGTTTAGACAATCAAAAAGTT-735).
Measurement of IFN-β production.
HEK293 cells were transfected with various plasmids or siRNA, and then cell culture supernatants were collected 6 h after virus infection and analyzed for IFN-β production by an enzyme-linked immunosorbent assay (ELISA) (PBL Biomedical Laboratories) according to the manufacturer's instructions.
Virus manipulation.
Vesicular stomatitis virus (VSV) and Newcastle disease virus-green fluorescent protein (NDV-GFP) were kindly provided by Hongbing Shu (Wuhan University) and Zhigao Bu (Chinese Academy of Agricultural Sciences), respectively. Viral infection was performed when 80% cell confluence was reached. Then, the culture medium was replaced by serum-free DMEM, and SeV, VSV, or NDV-GFP was added into the medium at various multiplicities of infection (MOI) according to the specific experiments. After 1 h, the medium was removed and the cells were fed with DMEM containing 10% FBS. For detection of IRF3 ubiquitination, 1 μM MG132 was added to the culture medium 3 h after virus infection, and cells were incubated for another 6 to 8 h.
Statistics.
Student's t test was used for the comparison of two independent treatments. For all tests, a P value of less than 0.05 was considered statistically significant.
RESULTS
HERC5 is a new IRF3-binding protein.
Because degradation of IRF3 is normally delayed for several hours after its phosphorylation, we reasoned that there might be an unknown protein(s) that could interact with IRF3 to regulate its activation and function. To explore this possibility, Flag-IRF3 was transfected into HEK293T cells, followed by Sendai virus (SeV) infection or no infection. The IRF3 immunoprecipitates were analyzed by silver staining. A band (approximately 110 kDa) was preferentially coimmunoprecipitated with Flag-IRF3 upon SeV infection (Fig. 1A). Mass spectrometry analysis revealed it as exclusively HERC5. The identity was further confirmed by using a specific antibody against HERC5 (Fig. 1B). Recently, HERC5 was reported to be inducible by viruses and beta interferon. We have also confirmed these observations (data not shown).
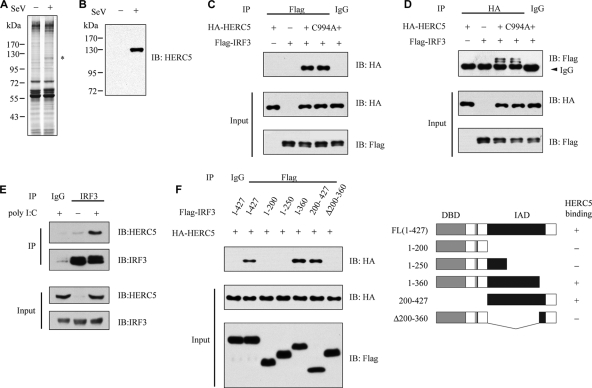
FIG. 1.
Identification of HERC5 in the IRF3 complex. (A) HEK293T cells transfected with Flag-IRF3 were mock infected or infected with SeV (MOI of 0.2) for 16 h, and then the cell lysates were subjected to immunoprecipitation with anti-Flag monoclonal antibody. The immunoprecipitates were resolved by SDS-PAGE followed by silver staining. The specific band indicated by the asterisk was excised for MS identification. (B) The same samples were immunoblotted (IB) with anti-HERC5 antibody. (C) HEK293T cells were cotransfected with the indicated constructs. Then, equal amounts of cell lysates were immunoprecipitated (IP) with anti-IgG or anti-Flag antibody. The immunoprecipitates were immunoblotted with the indicated antibodies. (D) HEK293T cells were cotransfected with the indicated constructs. Then, equal amounts of cell lysates were immunoprecipitated with anti-IgG or anti-HA antibody. The immunoprecipitates were immunoblotted with the indicated antibodies. (E) After mock or poly(I-C) (transfected, 2 μg/ml) stimulation, lysates from HEK293 cells were immunoprecipitated with anti-IRF3 antibody or anti-IgG and then immunoblotted with anti-HERC5 antibody. (F) Schematic diagram of IRF3 and its truncation mutants (right) (DBD, DNA-binding domain; IAD, IRF association domain; FL, full length). Flag-IRF3 mutants were individually transfected into HEK293T cells along with HA-HERC5. The cell lysates were immunoprecipitated with anti-IgG or anti-Flag antibody and immunoblotted with the indicated antibodies (left).
To confirm the interaction between IRF3 and HERC5, HA-HERC5 and Flag-IRF3 were cotransfected into HEK293T cells. The cell lysates were immunoprecipitated with either control IgG or anti-Flag antibody. As shown in Fig. 1C, HERC5 was coimmunoprecipitated with Flag-IRF3 but not with control IgG. Consistently, IRF3 was also detected in the immunoprecipitates of HA-HERC5 but not in those of control IgG (Fig. 1D). Bioinformatic analysis reveals that a cysteine in the HECT domain is critical for the action of the family members (14). Therefore, we generated HERC5 C994A, with the critical cysteine in the HECT domain mutated. The mutant HERC5 C994A bound to IRF3 as well (Fig. 1C and D). We next confirmed the interaction between HERC5 and IRF3 at the endogenous level (Fig. 1E). To map the critical domain, a series of Flag-IRF3 deletion mutants (Fig. 1F) were generated and individually transfected into HEK293T cells along with HA-HERC5. The middle region of IRF3 (amino acids [aa] 200 to 360) was responsible for this interaction (Fig. 1F). Collectively, these results indicate that HERC5 is a new IRF3-binding protein in vivo.
HERC5 synergizes IRF3 activation.
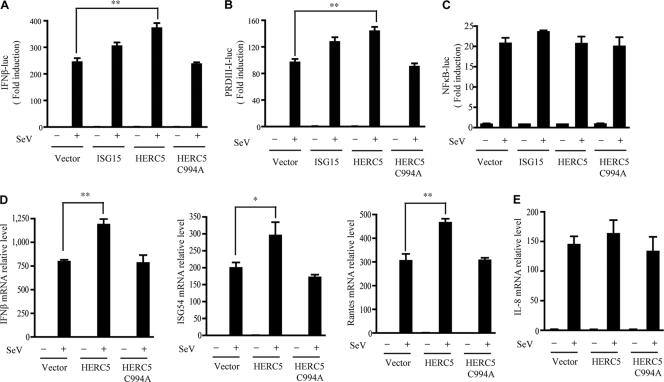
To explore the functional relevance of these findings, we investigated whether HERC5 could modulate IRF3-mediated gene expression through IFN-β and positive regulatory domain III-I (PRDIII-I) luciferase reporter assays. Introduction of wild-type (WT) HERC5 into HEK293 cells potentiated IRF3-dependent transcriptional activation upon SeV infection (Fig. 2A and B). However, the expression of an NF-κB luciferase reporter was unaffected by HERC5 under the same conditions (Fig. 2C). To make this more physiologically relevant, we investigated whether HERC5 affected the induction of IRF3-responsive genes (IFN-β, ISG54, and RANTES) by SeV infection, using a quantitative PCR (Q-PCR) approach. As shown in Fig. 2D, HERC5 displayed synergic effects on induction of the IRF3-responsive genes. However, this did not apply to induction of IL-8 mRNA, which is regulated by NF-κB (Fig. 2E). Collectively, our data suggested that HERC5 could positively regulate IRF3 transcriptional activity.
FIG. 2.
HERC5 synergizes IRF3 activation. (A to C) Equal amounts of the indicated plasmids (50 ng) were transfected into HEK293 cells along with the IFN-β (A), PRDIII-I (B), or NF-κB (C) reporter plasmid. Twenty-four hours after transfection, cells were infected with or without SeV (MOI of 0.2). The luciferase (luc) assay was performed 12 h postinfection. A pTK-Renilla reporter was used to normalize data. (D and E) Induction of IFN-β, ISG54, RANTES (D), and IL-8 (E) mRNA by SeV infection (MOI of 0.2) in the presence of a control and the indicated plasmids (50 ng) was measured by Q-PCR. Data are presented as means ± standard deviations (SD) (n = 3 replicates). *, P < 0.05; **, P < 0.01.
Knockdown of HERC5 attenuates IRF3 activation.
Alternatively, we took the knockdown approach to probe HERC5 function. The effective siRNA oligonucleotides were screened out and could effectively reduce endogenous and exogenous expression of HERC5 protein (Fig. 3A). Initially, we measured the effect of HERC5 knockdown on IRF3-responsive luciferase reporters. Knockdown of endogenous HERC5 inhibited the activation of both IFN-β and PRDIII-I luciferase reporters upon SeV infection (Fig. 3C). In contrast, mtHERC5 siRNA, a mutant form of HERC5 siRNA without silencing activity, failed to do so (Fig. 3C). As a control, NF-κB luciferase reporter activity was not affected by HERC5 knockdown (Fig. 3E). Interestingly, knockdown of HERC5 also impaired the induction of both the IFN-β and the PRDIII-I reporter by the constitutively active IRF3 5D (phosphorylation mimic mutant) (Fig. 3D).
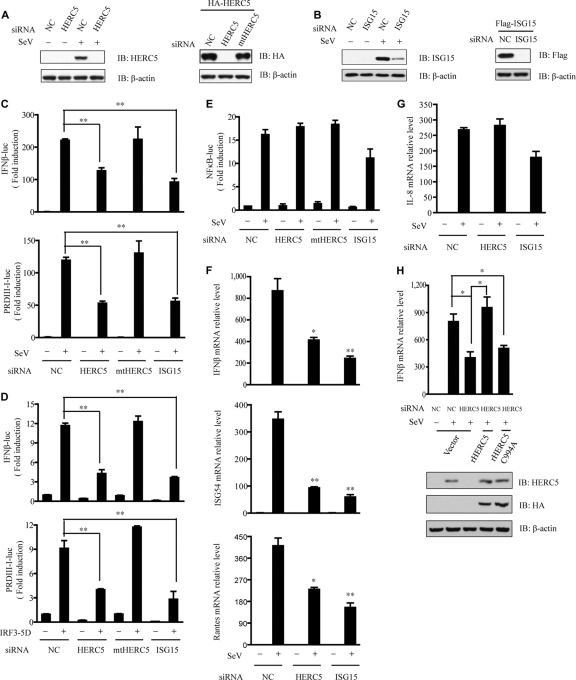
FIG. 3.
Knockdown of HERC5 attenuates IRF3 activation. (A) HEK293 cells were transfected with NC or HERC5 siRNA and then mock infected or infected with SeV. Cell lysates were immunoblotted with anti-HERC5 antibody (left). HEK293 cells were transfected with HA-HERC5 and then treated with NC, HERC5, or mtHERC5 siRNA. Cell lysates were immunoblotted with anti-HA antibody (right). (B) HEK293 cells were transfected with NC or ISG15 siRNA and then mock infected or infected with SeV (MOI of 0.2). Cell lysates were immunoblotted with anti-ISG15 antibody (left). HEK293 cells were transfected with Flag-ISG15 and then treated with NC or ISG15 siRNA. Cell lysates were immunoblotted with anti-Flag antibody (right). (C) The indicated siRNAs were transfected into HEK293 cells together with the IFN-β or PRDIII-I reporter plasmid. Forty-eight hours after transfection, cells were infected with SeV (MOI of 0.2) or not infected. The luciferase assay was performed 12 h postinfection. A pTK-Renilla reporter was used to normalize the data. (D) The indicated siRNAs were transfected into HEK293 cells together with the IFN-β or PRDIII-I reporter plasmid, and 24 h later, cells were transfected with IRF3 5D (S396, S398, S402, T404, and S405 all mutated to D). (E) The indicated siRNAs were transfected into HEK293 cells together with the NF-κB luciferase or pTK-Renilla reporter. Forty-eight hours later, cells were infected with SeV (MOI of 0.2). (F and G) Induction of IFN-β, ISG54, RANTES (F), and IL-8 (G) mRNA by SeV infection in the presence of a control and the indicated siRNAs was measured by Q-PCR. (H) HEK293 cells were transfected with the indicated siRNAs for 24 h. Then, siRNA-resistant HA-rHERC5 (50 ng) or HA-rHERC5 C994A (50 ng) was transfected into the knockdown cells. After SeV infection, induction of IFN-β mRNA was measured by Q-PCR. Data are presented as means ± SD (n = 3 replicates). NC, nonspecific control; mtHERC5 siRNA, a mutant form of HERC5 siRNA without silencing activity. *, P < 0.05; **, P < 0.01.
We further analyzed the effect of HERC5 knockdown on the induction of endogenous IRF3-responsive genes by SeV infection. As expected, HERC5 knockdown attenuated the induction of IRF3-responsive genes, such as IFN-β, ISG54, and RANTES (Fig. 3F), but not that of IL-8 (Fig. 3G). To rule out potential off-target effects of the HERC5 siRNA, we generated an RNA interference (RNAi)-resistant wild-type HERC5 construct (rHERC5), in which silent mutations were introduced into the sequence targeted by the siRNA without changing the amino acid sequence of the protein. HEK293 cells were first transfected with control or HERC5 siRNA followed by transfection of control or rHERC5 plasmids, respectively. Then, the induction of IFN-β mRNA was measured by real-time PCR after SeV infection. As shown in Fig. 3H, the induction of IFN-β mRNA by SeV infection was restored by rHERC5.
In addition, exogenous expression of ISG15 synergized IRF3-dependent transcriptional activation upon SeV infection (Fig. 2A and B). Knockdown of ISG15 attenuated IRF3 activation (Fig. 3C, D, and F). Collectively, these results strongly suggest that HERC5 is a positive regulator of IRF3. It plays an important role during the primary induction of type I interferons.
Cysteine 994 of the HECT domain is essential for HERC5 to modulate IRF3.
Since HERC5 has a HECT domain that potentially has ISG15 E3 ligase activity (6, 48), we went on to address whether this activity was critical for regulating IRF3 activation. We used HERC5 C994A, a HECT domain mutant deprived of the potential ISG15 E3 ligase activity, to investigate how HERC5 regulated IRF3 activation. As shown in Fig. 2A and B, exogenous expression of HERC5 C994A failed to synergize the induction of IRF3-reponsive reporters upon SeV infection, compared with results for wild-type HERC5. Consistently, the induction of IRF3-responsive genes (IFN-β, ISG54, and RANTES) was not potentiated in the presence of HERC5 C994A after infection with SeV (Fig. 2D). Furthermore, the expression of IFN-β induced by SeV infection was not rescued by rHERC5 C994A (RNAi-resistant HECT domain mutant) in HERC5 knockdown cells compared with the results for wild-type rHERC5 (Fig. 3H). Collectively, these data strongly suggest that HERC5-mediated ISGylation may play a positive role in regulating IRF3 activation.
HERC5 is an ISG15 E3 ligase for IRF3.
Recently, more than 100 proteins have been identified as potential targets of ISGylation through a proteomics approach. However, only a dozen of them have been validated as authentic substrates in vivo (47, 48, 56). The function of HERC5-mediated ISGylation remains largely unknown. Given the importance of HERC5 cysteine 994 in regulating IRF3 activity, we hypothesized that HERC5 could catalyze ISGylation of IRF3.
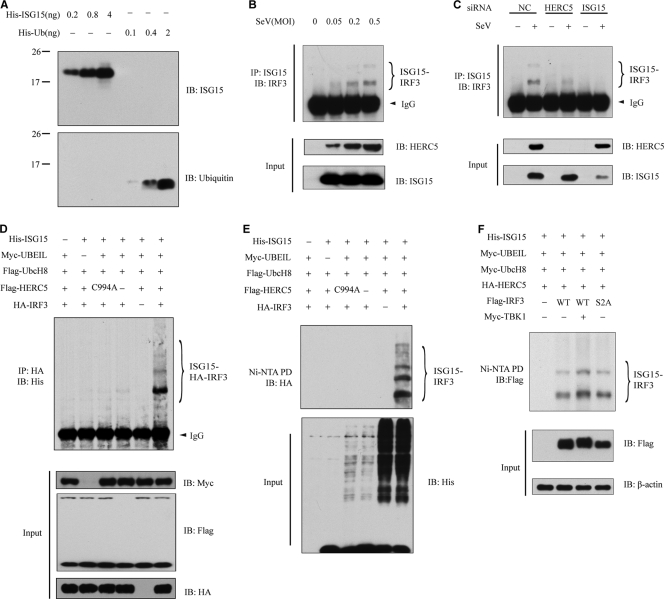
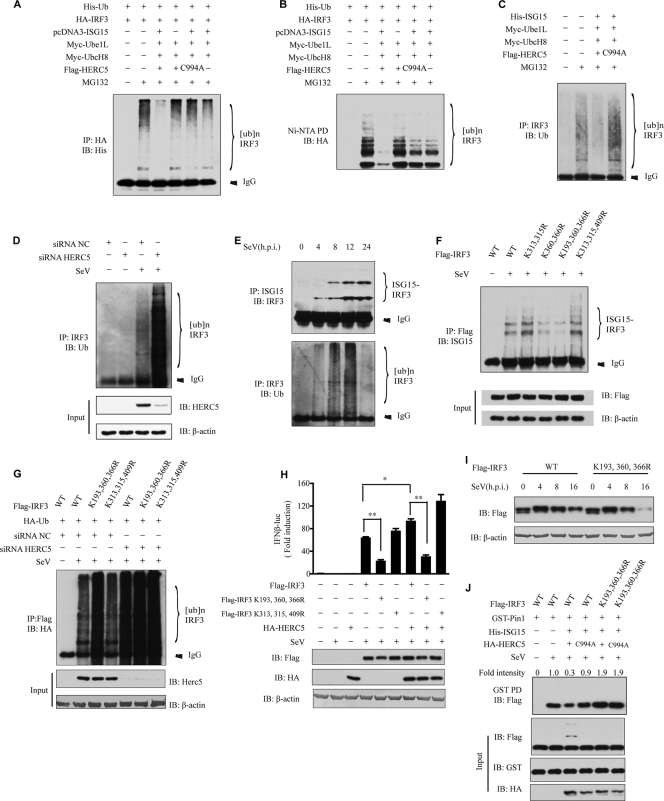
We first determined the specificities of monoclonal antibodies against ubiquitin (Ub) and ISG15. Both antibodies recognized their corresponding antigens without cross-reaction with the other proteins (Fig. 4A). To address the above-described hypothesis, HEK293 cells were infected with SeV. Cell lysates were immunoprecipitated with anti-ISG15 antibody and then probed with anti-IRF3 antibody. Interestingly, endogenous IRF3 was apparently modified by ISG15. The ISGylation of IRF3 was markedly enhanced with the increase in SeV (MOI), which correlated well with the induction of HERC5 (Fig. 4B). Consistently, endogenous knockdown of either HERC5 or ISG15 sharply attenuated the ISGylation of IRF3 (Fig. 4C).
FIG. 4.
HERC5 catalyzes the conjugation of ISG15 onto IRF3. (A) Recombinant ISG15 and ubiquitin were resolved by SDS-PAGE and probed with anti-ISG15 (top) or anti-Ub (bottom). (B) HEK293 cells were mock infected or infected with increasing doses of SeV. Cell lysates were subjected to immunoprecipitation and then immunoblotted with the indicated antibodies. The positions of ISGylated IRF3 are shown by braces at right. (C) HEK293 cells were transfected with the indicated siRNAs. After SeV infection, cell lysates were subjected to immunoprecipitation and immunoblotted with the indicated antibodies. (D and E) HEK293T cells were transfected with the indicated plasmids. Twenty-four hours after transfection, cell lysates were subjected to immunoprecipitation (D) or Ni-NTA pulldown (PD) (E) and then immunoblotted with the indicated antibodies. (F) HEK293T cells expressing the indicated plasmids were subjected to Ni-NTA pulldown and then immunoblotted with the indicated antibodies. S2A, IRF3(S385,386A).
Alternatively, we carried out an ISGylation assay to further substantiate the observation. HEK293T cells were transfected with different combinations of Ube1L, UbcH8, His-ISG15, HERC5, and HA-IRF3. The cell lysates were subjected to immunoprecipitation of HA-IRF3 (Fig. 4D) or Ni-NTA pulldown of His-ISG15 (Fig. 4E). Then, the precipitates were probed with various antibodies. As expected, ISGylation of IRF3 was readily detectable when all of the components were present. In the absence of HERC5 or any of other components, no significant ISGylation was observed (Fig. 4D and E). In addition, HERC5 C994A could not catalyze ISGylation of IRF3. Since TBK1 phosphorylates IRF3 during virus infection, we wondered whether phosphorylation of IRF3 had any impact on ISGylation. Various combinations of proteins were expressed in HEK293T cells, and then the cell lysates were subjected to Ni-NTA pulldown. As shown in Fig. 4F, levels of HERC5-catalyzed ISGylation of IRF3 in the presence and absence of TBK1 were comparable. Likewise, IRF3(S385,386A) (phosphorylation dead mutant) apparently could be modified by ISG15 in the presence of HERC5, suggesting that the IRF3 phosphorylation status did not affect ISGylation. Taken together, our results strongly indicated that IRF3 was modified by ISGylation upon virus infection. HERC5 is a novel ISG15 E3 ligase for IRF3.
HERC5 prevents proteasome-mediated degradation of IRF3.
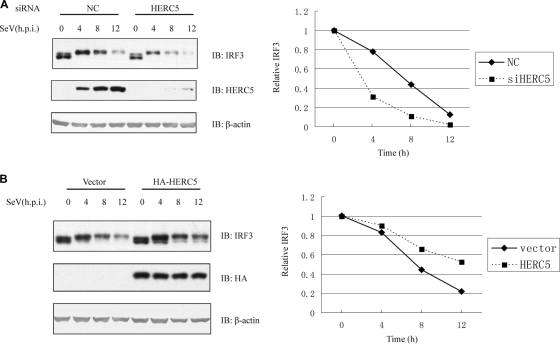
Activation of IRF3 would result in its being modified by Ub and then degraded by the 26S proteasome, which thus terminates its transcriptional activation (3, 26). Since HERC5 catalyzed IRF3 ISGylation, we investigated whether HERC5 regulates IRF3 protein stability. Notably, a decrease in endogenous HERC5 exacerbated IRF3 degradation in a time-dependent manner upon SeV infection, compared with the results for control siRNA (Fig. 5A). Moreover, exogenous expression of HERC5 apparently delayed IRF3 degradation compared with the result for the controls (Fig. 5B). Collectively, this indicates that HERC5 enhances the stability of IRF3, which suggests that HERC5 could antagonize IRF3 ubiquitination.
FIG. 5.
HERC5 enhances IRF3 stability. (A) HEK293 cells were transfected with the indicated siRNAs and then infected with SeV (MOI of 1.0) for the indicated times (h.p.i., hours postinfection). Cell lysates were immunoblotted with anti-IRF3 antibody. (B) HEK293 cells were transfected with HA-HERC5 (200 ng) or control. After infection with SeV (MOI of 1.0) for the indicated times, cell lysates were immunoblotted with anti-IRF3 antibody. Representative results are shown, and the amounts of IRF3 were measured densitometrically (right). Each datum point represents single-well samples.
HERC5-mediated ISGylation of IRF3 inhibits its ubiquitination.
Modifications by ubiquitin-like proteins (e.g., SUMO) have been shown to interfere with the ubiquitination of target proteins (7, 10, 13). To examine whether HERC5-mediated ISGylation could influence IRF3 ubiquitination, HEK293T cells were transfected with different combinations of Ube1L, UbcH8, ISG15, and HERC5, together with HA-IRF3 and His-Ub. After infection with SeV, cell lysates were subjected to immunoprecipitation with HA-IRF3 (Fig. 6A) or Ni-NTA pulldown with His-Ub (Fig. 6B) and then probed with various antibodies. As expected, IRF3 was polyubiquitinated upon SeV infection. Intriguingly, IRF3 polyubiquitination was reduced in the presence of the complete ISG15 conjugation system (with HERC5 as E3). However, IRF3 ubiquitination was not affected when the component of the ISGylation system was left out (Fig. 6A and B). In particular, HERC5 C994A failed to prevent IRF3 ubiquitination. Furthermore, polyubiquitination of endogenous IRF3 was consistently attenuated upon SeV infection, when HERC5-mediated ISGylation of IRF3 was enhanced (Fig. 6C). In contrast, knockdown of HERC5 significantly increased the polyubiquitination of endogenous IRF3 (Fig. 6D). Interestingly, we observed that the ISGylation of IRF3 was gradually increased whereas the ubiquitination of IRF3 was decreased 12 h postinfection (Fig. 6E).
FIG. 6.
HERC5 inhibits IRF3 ubiquitination. (A and B) HEK293T cells were transfected with the indicated plasmids and infected with SeV (MOI of 0.5). Cell lysates were subjected to immunoprecipitation (A) or Ni-NTA pulldown (B) and then immunoblotted with the indicated antibodies. (C) HEK293T cells were transfected with the indicated plasmids and infected with SeV (MOI of 0.5). Cell lysates were immunoprecipitated with rabbit anti-IRF3 antibody and then immunoblotted with mouse anti-Ub antibody. (D) HEK293 cells were treated with the indicated siRNAs. After mock infection or SeV infection, cell lysates were immunoprecipitated with rabbit anti-IRF3 antibody and then immunoblotted with mouse anti-Ub antibody. (E) HEK293 cells were infected with SeV (MOI of 0.5) for the indicated times. Cell lysates were immunoprecipitated with anti-ISG15 antibody and then immunoblotted with anti-IRF3 antibody (top) or immunoprecipitated with anti-IRF3 antibody and then immunoblotted with anti-Ub antibody (bottom). (F) HEK293 cells were transfected with Flag-IRF3 lysine mutants and then infected with SeV (MOI of 0.5). Cell lysates were subjected to immunoprecipitation with anti-Flag antibody and immunoblotted with anti-ISG15 antibody. (G) HEK293 cells were transfected with NC or siHERC5 for 24 h and then transfected with the indicated plasmids before infection with SeV (MOI of 0.5). Cell lysates were subjected to immunoprecipitation and then immunoblotted with the indicated antibodies. (H) The indicated plasmids (Flag-IRF3 lysine mutants at 500 ng per well and HA-HERC5 at 400 ng per well) were transfected into IRF3−/− MEF along with the IFN-β reporter plasmid. Twenty-four hours after transfection, cells were mock infected or infected with SeV (MOI of 0.8). The luciferase assay was performed 18 h postinfection. A pTK-Renilla reporter was used to normalize the data. The data are presented as means ± SD (n = 3). *, P < 0.05; **, P < 0.01. (I) IRF3−/− MEF were transfected with 200 ng of Flag-tagged WT IRF3 or Flag-IRF3(K193,360,366R) and then infected with SeV (MOI of 2) for the indicated times. Cell lysates were immunoblotted with anti-Flag antibody. (J) HEK293 cells were transfected with the indicated plasmids and then infected with SeV for 4 h. Cell lysates were subjected to GST pulldown and then immunoblotted with the indicated antibodies. The relative amounts of IRF3 were quantified by densitometry and normalized with respect to that shown in lane 2.
We carried out lysine scanning (K-to-R point mutation) to map the potential ISGylation site(s) on IRF3. The Flag-tagged mutants were transfected into HEK293 cells, followed by SeV infection. The cell lysates were immunoprecipitated with anti-Flag antibody and then probed with anti-ISG15 antibody. Although the ISGylation of IRF3(K313,315,409R) remained the same as that of WT IRF3, the ISGylation of IRF3(K360,366R) markedly decreased. Apparently, much less ISGylation was observed for IRF3(K193,360,366R) (Fig. 6F). We also tested other combinations of lysine point mutations and found that they could be modified by ISGylation to the same extent as the wild-type IRF3 (data not shown). These data suggested that Lys193, -360, and -366 are the predominant sites of ISGylation on IRF3.
We went on to explore the impact of IRF3 ISGylation on ubiquitination by introducing the lysine mutants of IRF3 into HEK293 cells. Upon SeV infection, the ubiquitination of IRF3(K313,315,409R) remained the same as that of WT IRF3, but the ubiquitination of IRF3(K193,360,366R) markedly increased. In addition, knockdown of HERC5 significantly increased the ubiquitination of WT IRF3 and IRF3(K313,315,409R) but did not influence that of IRF3(K193,360,366R) (Fig. 6G). As expected, SeV infection failed to activate the IFN-β luciferase reporter in IRF3−/− MEF. This activation could be rescued when WT IRF3 was transfected. Notably, the IFN-β reporter was marginally activated by SeV when IRF3(K190,360,366R) was transfected. Consistently, HERC5 potentiated the activation of WT IRF3 and IRF3(K313,315,409R) upon SeV infection but failed to do so with IRF3(K190,360,366R) (Fig. 6H).
Furthermore, degradation of IRF3(K193,360,366R) was accelerated upon SeV infection in IRF3−/− MEF compared with that of WT IRF3 (Fig. 6I). Pin1 was recently demonstrated to interact with phosphorylated IRF3 and plays a critical role in initiating IRF3 ubiquitination (40). An attractive model for HERC5 action is that HERC5-mediated ISGylation of IRF3 prevents Pin1 from binding to IRF3. To explore this, we conducted a competition assay by expressing GST-Pin1 and IRF3 constructs in HEK293 cells, along with the ISG15 conjugation system (HERC5 as E3). Consistently, Pin1 interacted with IRF3 only after its phosphorylation. This interaction was disrupted when HERC5-mediated ISGylation of IRF3 occurred. In contrast, HERC5 C994A failed to prevent Pin1 binding to IRF3, although HERC5 C994A could bind to IRF3 as well (Fig. 6J and 1C and D). In addition, Pin1 bound more tightly to IRF3(K193,360,366R) than to wild-type IRF3, even in the presence of HERC5-mediated ISGylation (Fig. 6J). These observations strongly suggested that HERC5 enhanced IRF3 activation by disrupting Pin1 binding and thus impairing IRF3 ubiquitination.
HERC5 enhances IRF3-mediated antiviral responses.
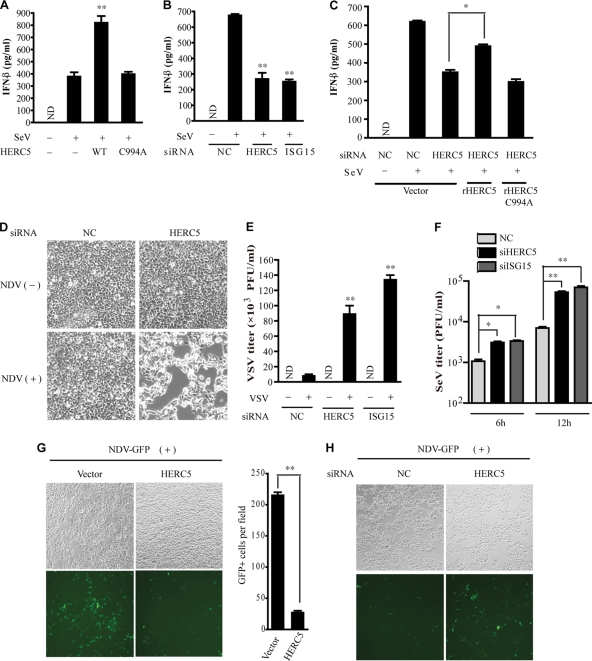
Since HERC5 positively regulates IRF3 activation, we investigated whether HERC5 functions as a new antiviral protein. The robust induction of IFN-β is the hallmark of the immediate host responses to virus infection. HERC5 or its mutants were transfected into HEK293 cells, followed by SeV infection. By ELISA, we observed that HERC5 could significantly promote IFN-β protein production whereas HERC5 C994A was unable to do so (Fig. 7A). In contrast, knockdown of endogenous HERC5 impaired IFN-β protein production upon SeV infection (Fig. 7B). Likewise, ISG15 knockdown cells produced much less IFN-β protein (Fig. 7B). Moreover, rHERC5 could rescue the decreased production of IFN-β in cells that were treated with HERC5 siRNA, but rHERC5 C994A could not do so (Fig. 7C). These results prompted us to explore the cytopathic effect of HERC5 knockdown in response to virus invasion. Therefore, HEK293 cells were transfected with siRNA, followed by Newcastle disease virus (NDV) infection. It was found that HERC5 knockdown cells were more sensitive to NDV infection than were wild-type HEK293 cells (Fig. 7D). We next investigated whether HERC5 modulated virus replication by infecting cells with VSV, SeV, or NDV-GFP. HEK293 cells were transfected with siRNAs and then infected with SeV for 10 h. The culture supernatants were applied to fresh HEK293 cells, followed by vesicular stomatitis virus (VSV) infection. The titers of VSV were analyzed by a standard plaque assay. As shown in Fig. 7E, HERC5 knockdown resulted in an apparently 10-fold increase in virus titer compared to that for controls. To determine whether Herc5-mediated ISG15 conjugation directly influenced SeV replication, we carried out a plaque assay with SeV, after treating cells with siRNAs. As shown in Fig. 7F, knockdown of HERC5 or ISG15 significantly enhanced SeV replication. For NDV-GFP infection, exogenous expression of HERC5 significantly suppressed NDV-GFP virus replication in HEK293 cells (Fig. 7G). Furthermore, HEK293 cells with HERC5 knockdown showed remarkably increased levels of NDV-GFP-positive cells (Fig. 7H). Taken together, our results convincingly demonstrate that HERC5 functions as the E3 ligase for IRF3 ISGylation and positively regulates antiviral responses in host cells.
FIG. 7.
HERC5 modulates IRF3-mediated antiviral responses. (A and B) HEK293 cells were transfected with the indicated plasmids (50 ng) (A) or siRNAs (B). Six hours after SeV infection, IFN-β production was determined by ELISA. (C) The indicated siRNAs were transfected into HEK293 cells in the presence or absence of siRNA-resistant HERC5. Six hours after SeV infection, IFN-β production was determined by ELISA. (D) HEK293 cells were transfected with the indicated siRNAs. Twenty-four hours posttransfection, the cells were infected with NDV for an additional 20 h. The cytopathic effects (CPE) were observed by differential interference contrast DIC microscopy. Original magnification, ×100. (E) HEK293 cells were transfected with the indicated siRNAs. Twenty-four hours posttransfection, the cells were infected with SeV for an additional 10 h. Equal amounts of the culture supernatants (approximately 100 μl) were applied to fresh HEK293 cells for 6 h, followed by VSV infection. The titers of VSV were determined by a standard plaque assay. (F) HEK293 cells were transfected with the indicated siRNAs. Twenty-four hours posttransfection, the cells were infected with SeV for different times. Equal amounts of the conditioned medium with anti-IFN-α/β (20 μg/ml) were applied to Vero cells. A plaque assay for SeV was carried out using Vero cells as described previously. (G and H) NDV-GFP replication in HEK293 cells transfected with exogenous HERC5 (G) or HERC5 siRNA (H) was visualized by fluorescence microscopy. Original magnification, ×100. GFP-positive cells were quantified (G, right). Data are presented as means ± SD (n = 3 replicates). ND, not detected. *, P < 0.05; **, P < 0.01.
DISCUSSION
Mammalian hosts have evolved sophisticated means for detecting and eradicating viruses. Meanwhile, the antiviral responses are under stringent modulations to avoid damage of host tissues. Recently, TLR3 and RIG-I/MDA5 have, respectively, been characterized as membrane and cytosolic sensors of viral RNA, which ultimately activate IRF3 and induce production of type I interferons (17, 46). The corresponding signal transduction pathways serve as the first line of host mobilization against virus invasion. IRF3, as a critical hub for signal integration, is subjected to dynamic and precise regulations, including protein posttranslational modifications. It is important to dissect the biochemical processes of relevant modifications and understand the functional implications of these regulatory processes. A good strategy to address this challenge is to search for an additional IRF3-interacting protein(s) in the context of virus infection. Here, we identify HERC5 as a novel IRF3-binding protein and demonstrate that it positively regulates IRF3 activation. Notably, our study establishes that HERC5 functions as an ISG15 E3 ligase and catalyzes a new type of IRF3 posttranslational modification, ISGylation. To our knowledge, this is the first report in which HERC5-mediated ISGylation is functionally characterized in terms of an authentic substrate, especially during microbial invasion.
Several lines of evidence highlight the important function of HERC5 in regulating IRF3. First, exogenous expression of HERC5 specifically potentiates the induction of IRF3 target genes upon SeV infection but does not affect the induction of the NF-κB target gene. Second, knockdown of HERC5 unequivocally results in a significant reduction in IRF3-responsive gene expression but not that of the NF-κB target gene. Additionally, this attenuation could be rescued by exogenously expressing a siRNA-resistant rHERC5. Third, loss or gain of HERC5 could, respectively, attenuate or enhance IFN-β protein production upon SeV infection. Fourth, reduction of endogenous HERC5 expression significantly sensitizes cells to virus infection and results in a much higher level of production of VSV, SeV, or NDV-GFP, whereas exogenous expression of HERC5 significantly represses NDV-GFP virus replication. Fifth, HERC5 and IRF3, when ectopically expressed, display strong binding affinity to each other. Endogenous HERC5 binds to IRF3 marginally in resting cells, but this association is apparently enhanced upon viral infection. Since HERC5 is robustly induced upon virus challenge, this enhancement of interaction is probably due to an increased abundance of HERC5. The action of HERC5 represents a potential positive feedback for the antiviral response.
As an effective mechanism of regulation, ubiquitylation is also reported to regulate the RIG-I/MDA5 signaling pathway. Several RING domain proteins (RNF125, TRIM25, RNF5, and TRIM21) have recently been demonstrated to be positive or negative modulators of this pathway (1, 9, 50, 57). RNF125, TRIM25, and RNF5 are found to catalyze ubiquitination of proteins upstream of IRF3. IRF3 per se is ubiquitinated and degraded after virus-induced activation. However, the identity of a ubiquitin E3 ligase for IRF3 remains elusive. The HECT domain is a signature of a subfamily of ubiquitin E3 ligases, which harbors a conserved cysteine for temporarily conjugating ubiquitin. We have tested whether HERC5 could catalyze IRF3 ubiquitination and found that this does not hold true. Instead, we found that knockdown of HERC5 apparently enhances IRF3 ubiquitylation and promotes its degradation upon SeV infection. To our surprise, the critical cysteine 994 in the HECT domain of HERC5 is indispensable for potentiating IRF3 activation, which suggests that a ubiquitin-like modification may be involved.
The robust induction of ubiquitin-like protein ISG15 is one of the immediate cellular responses to virus infection. It is believed that modification of host proteins by ISG15 might play a critical role in the antiviral response. Recently, it has been found that mice deficient in ISG15 have increased susceptibility to infection with several viruses, including Sindbis virus, herpes simplex virus type 1 (HSV-1), and murine gammaherpesvirus (25). Interestingly, HERC5 was implicated as a potential E3 ligase for ISGylation. However, it remained to identify the authentic substrate(s) and address the biological function of HERC5-mediated ISGylation, especially upon virus infection. Several lines of evidence in this study indicate that HERC5 serves as an ISG15 E3 ligase for IRF3 and that this ISGylation plays an essential role in regulating IRF3 activation. First, knockdown of either HERC5 or ISG15 attenuates IRF3 activation upon virus infection. Second, virus infection induces ISGylation of IRF3 endogenously, which is notably attenuated when HERC5 or ISG15 is knocked down. Third, ISGylation of IRF3 could be observed when all components of the ISGylation system, including ISG15, UBE1L, UbcH8, and HERC5, are ectopically expressed. However, this ISGylation of IRF3 is abolished when HERC5 is not transfected. In addition, HERC5 C994 could not catalyze ISGylation of IRF3. Fourth, the sites of ISGylation are mapped predominantly to lysines 193, 360, and 366 of IRF3. Mutation of these sites apparently attenuated IRF3 activation during SeV infection. Fifth, HERC5-catalyzed IRF3 ISGylation inhibits IRF3 ubiquitylation, resulting in stabilization of IRF3. It is therefore conceivable that induction of HERC5 contributes to sustaining the strength and duration of IRF3 transcriptional action, augmenting the innate immune response. This is confirmed by evaluating virus proliferation in the presence or absence of HERC5.
The destruction of IRF3 by Ub-mediated proteolysis has been proposed as an effective mechanism to terminate its transcriptional activity. However, premature degradation of IRF3 will be sure to attenuate induction of critical antiviral proteins and adversely lead to insufficient host defense against viral infection. An emerging theme is that the ubiquitin-like protein could antagonize ubiquitination by modifying the target protein. For example, sumoylation is found to inhibit ubiquitination of IκBα, PCNA, and CREB via preventing the binding of ubiquitin E3 ligases (10). Until now, it has largely been unknown how ISG15 modification affects relevant regulatory processes. In this study, we observed that ISGylation of IRF3 disrupted the binding of Pin1 to IRF3, an established prerequisite for IRF3 ubiquitination. It will be interesting to explore whether ISGylation of IRF3 could disintegrate the ubiquitin ligase complex once it is identified.
Recently, we have reported that TRIM21 positively regulates type I IFN induction by inhibiting IRF3 ubiquitination. TRIM21 also interacts specifically with IRF3 and thus prevents Pin1 from binding to IRF3 (50). We have noticed that ISGylation takes place at the N-terminal part of IRF3, whereas TRIM21 targets IRF3 mainly at its C terminus. In addition, knockdown of TRIM21 did not affect the ISGylation of IRF3 (our unpublished data), which suggests that Herc5 and TRIM21 are functionally redundant in regulating IRF3 activation. However, we could not rule out the possibility that Herc5 and TRIM21 may function differently in response to infections with diverse viruses. Given that IRF3 plays a key role in innate antiviral responses, we believe that more regulatory proteins of IRF3 will be identified in future studies, as was the case for p53 and NF-κB.
In conclusion, our study characterizes IRF3 as the first authentic substrate for HERC5. HERC5 catalyzes ISGylation of IRF3 at Lys193, -360, and -366. This modification inhibits ubiquitylation and degradation of IRF3, thus augmenting innate immunity. Our work uncovers a novel positive feedback of IRF3 regulation and sheds light on the critical role of ISGylation in antiviral responses.
Acknowledgments
We thank Genhong Cheng (University of California), Jon M. Huibregtse (University of Texas), Hongbing Shu (Wuhan University, China), Hong Tang (Institute of Biophysics, CAS), and Zhigao Bu (Chinese Academy of Agricultural Sciences, China) for providing reagents used in this study.
This work was supported by grants from the Ministry of Science and Technology of Shanghai (09XD1404800), the Ministry of Science and Technology of China (2006CB504301, 2007CB914504, and 2009ZX10004-105), and the Chinese Academy of Sciences (KSCX1-YW-R-06).
Footnotes
Published ahead of print on 22 March 2010.
REFERENCES
- 1.Arimoto, K., H. Takahashi, T. Hishiki, H. Konishi, T. Fujita, and K. Shimotohno. 2007. Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc. Natl. Acad. Sci. U. S. A. 104:7500-7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barro, M., and J. T. Patton. 2005. Rotavirus nonstructural protein 1 subverts innate immune response by inducing degradation of IFN regulatory factor 3. Proc. Natl. Acad. Sci. U. S. A. 102:4114-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bibeau-Poirier, A., S. P. Gravel, J. F. Clement, S. Rolland, G. Rodier, P. Coulombe, J. Hiscott, N. Grandvaux, S. Meloche, and M. J. Servant. 2006. Involvement of the IkappaB kinase (IKK)-related kinases tank-binding kinase 1/IKKi and cullin-based ubiquitin ligases in IFN regulatory factor-3 degradation. J. Immunol. 177:5059-5067. [DOI] [PubMed] [Google Scholar]
- 4.Chen, Z. J. 2005. Ubiquitin signalling in the NF-kappaB pathway. Nat. Cell Biol. 7:758-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cruz, C., F. Ventura, R. Bartrons, and J. L. Rosa. 2001. HERC3 binding to and regulation by ubiquitin. FEBS Lett. 488:74-80. [DOI] [PubMed] [Google Scholar]
- 6.Dastur, A., S. Beaudenon, M. Kelley, R. M. Krug, and J. M. Huibregtse. 2006. Herc5, an interferon-induced HECT E3 enzyme, is required for conjugation of ISG15 in human cells. J. Biol. Chem. 281:4334-4338. [DOI] [PubMed] [Google Scholar]
- 7.Desterro, J. M., M. S. Rodriguez, and R. T. Hay. 1998. SUMO-1 modification of IkappaBalpha inhibits NF-kappaB activation. Mol. Cell 2:233-239. [DOI] [PubMed] [Google Scholar]
- 8.Fitzgerald, K. A., S. M. McWhirter, K. L. Faia, D. C. Rowe, E. Latz, D. T. Golenbock, A. J. Coyle, S. M. Liao, and T. Maniatis. 2003. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 4:491-496. [DOI] [PubMed] [Google Scholar]
- 9.Gack, M. U., Y. C. Shin, C. H. Joo, T. Urano, C. Liang, L. Sun, O. Takeuchi, S. Akira, Z. Chen, S. Inoue, and J. U. Jung. 2007. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446:916-920. [DOI] [PubMed] [Google Scholar]
- 10.Geiss-Friedlander, R., and F. Melchior. 2007. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8:947-956. [DOI] [PubMed] [Google Scholar]
- 11.Hacker, H., V. Redecke, B. Blagoev, I. Kratchmarova, L. C. Hsu, G. G. Wang, M. P. Kamps, E. Raz, H. Wagner, G. Hacker, M. Mann, and M. Karin. 2006. Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature 439:204-207. [DOI] [PubMed] [Google Scholar]
- 12.Hochrainer, K., H. Mayer, U. Baranyi, B. Binder, J. Lipp, and R. Kroismayr. 2005. The human HERC family of ubiquitin ligases: novel members, genomic organization, expression profiling, and evolutionary aspects. Genomics 85:153-164. [DOI] [PubMed] [Google Scholar]
- 13.Hoege, C., B. Pfander, G. L. Moldovan, G. Pyrowolakis, and S. Jentsch. 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419:135-141. [DOI] [PubMed] [Google Scholar]
- 14.Huibregtse, J. M., M. Scheffner, S. Beaudenon, and P. M. Howley. 1995. A family of proteins structurally and functionally related to the E6-AP ubiquitin-protein ligase. Proc. Natl. Acad. Sci. U. S. A. 92:2563-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishikawa, H., and G. N. Barber. 2008. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455:674-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kato, H., O. Takeuchi, S. Sato, M. Yoneyama, M. Yamamoto, K. Matsui, S. Uematsu, A. Jung, T. Kawai, K. J. Ishii, O. Yamaguchi, K. Otsu, T. Tsujimura, C. S. Koh, C. Reis e Sousa, Y. Matsuura, T. Fujita, and S. Akira. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101-105. [DOI] [PubMed] [Google Scholar]
- 17.Kawai, T., and S. Akira. 2008. Toll-like receptor and RIG-I-like receptor signaling. Ann. N. Y. Acad. Sci. 1143:1-20. [DOI] [PubMed] [Google Scholar]
- 18.Kawai, T., K. Takahashi, S. Sato, C. Coban, H. Kumar, H. Kato, K. J. Ishii, O. Takeuchi, and S. Akira. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981-988. [DOI] [PubMed] [Google Scholar]
- 19.Kayagaki, N., Q. Phung, S. Chan, R. Chaudhari, C. Quan, K. M. O'Rourke, M. Eby, E. Pietras, G. Cheng, J. F. Bazan, Z. Zhang, D. Arnott, and V. M. Dixit. 2007. DUBA: a deubiquitinase that regulates type I interferon production. Science 318:1628-1632. [DOI] [PubMed] [Google Scholar]
- 20.Kim, M. J., A. G. Latham, and R. M. Krug. 2002. Human influenza viruses activate an interferon-independent transcription of cellular antiviral genes: outcome with influenza A virus is unique. Proc. Natl. Acad. Sci. U. S. A. 99:10096-10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korant, B. D., D. C. Blomstrom, G. J. Jonak, and E. Knight, Jr. 1984. Interferon-induced proteins. Purification and characterization of a 15,000-dalton protein from human and bovine cells induced by interferon. J. Biol. Chem. 259:14835-14839. [PubMed] [Google Scholar]
- 22.Kroismayr, R., U. Baranyi, C. Stehlik, A. Dorfleutner, B. R. Binder, and J. Lipp. 2004. HERC5, a HECT E3 ubiquitin ligase tightly regulated in LPS activated endothelial cells. J. Cell Sci. 117:4749-4756. [DOI] [PubMed] [Google Scholar]
- 23.Kubota, T., M. Matsuoka, T. H. Chang, P. Tailor, T. Sasaki, M. Tashiro, A. Kato, and K. Ozato. 2008. Virus infection triggers SUMOylation of IRF3 and IRF7, leading to the negative regulation of type I interferon gene expression. J. Biol. Chem. 283:25660-25670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumar, K. P., K. M. McBride, B. K. Weaver, C. Dingwall, and N. C. Reich. 2000. Regulated nuclear-cytoplasmic localization of interferon regulatory factor 3, a subunit of double-stranded RNA-activated factor 1. Mol. Cell. Biol. 20:4159-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lenschow, D. J., C. Lai, N. Frias-Staheli, N. V. Giannakopoulos, A. Lutz, T. Wolff, A. Osiak, B. Levine, R. E. Schmidt, A. Garcia-Sastre, D. A. Leib, A. Pekosz, K. P. Knobeloch, I. Horak, and H. W. T. Virgin. 2007. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc. Natl. Acad. Sci. U. S. A. 104:1371-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin, R., C. Heylbroeck, P. M. Pitha, and J. Hiscott. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18:2986-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu, G., J. T. Reinert, I. Pitha-Rowe, A. Okumura, M. Kellum, K. P. Knobeloch, B. Hassel, and P. M. Pitha. 2006. ISG15 enhances the innate antiviral response by inhibition of IRF-3 degradation. Cell. Mol. Biol. (Noisy-le-grand) 52:29-41. [PubMed] [Google Scholar]
- 28.Malakhova, O. A., and D. E. Zhang. 2008. ISG15 inhibits Nedd4 ubiquitin E3 activity and enhances the innate antiviral response. J. Biol. Chem. 283:8783-8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meylan, E., J. Curran, K. Hofmann, D. Moradpour, M. Binder, R. Bartenschlager, and J. Tschopp. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167-1172. [DOI] [PubMed] [Google Scholar]
- 30.Michallet, M. C., E. Meylan, M. A. Ermolaeva, J. Vazquez, M. Rebsamen, J. Curran, H. Poeck, M. Bscheider, G. Hartmann, M. Konig, U. Kalinke, M. Pasparakis, and J. Tschopp. 2008. TRADD protein is an essential component of the RIG-like helicase antiviral pathway. Immunity 28:651-661. [DOI] [PubMed] [Google Scholar]
- 31.Mitsui, K., M. Nakanishi, S. Ohtsuka, T. H. Norwood, K. Okabayashi, C. Miyamoto, K. Tanaka, A. Yoshimura, and M. Ohtsubo. 1999. A novel human gene encoding HECT domain and RCC1-like repeats interacts with cyclins and is potentially regulated by the tumor suppressor proteins. Biochem. Biophys. Res. Commun. 266:115-122. [DOI] [PubMed] [Google Scholar]
- 32.Moore, C. B., D. T. Bergstralh, J. A. Duncan, Y. Lei, T. E. Morrison, A. G. Zimmermann, M. A. Accavitti-Loper, V. J. Madden, L. Sun, Z. Ye, J. D. Lich, M. T. Heise, Z. Chen, and J. P. Ting. 2008. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature 451:573-577. [DOI] [PubMed] [Google Scholar]
- 33.Narasimhan, J., M. Wang, Z. Fu, J. M. Klein, A. L. Haas, and J. J. Kim. 2005. Crystal structure of the interferon-induced ubiquitin-like protein ISG15. J. Biol. Chem. 280:27356-27365. [DOI] [PubMed] [Google Scholar]
- 34.Oganesyan, G., S. K. Saha, B. Guo, J. Q. He, A. Shahangian, B. Zarnegar, A. Perry, and G. Cheng. 2006. Critical role of TRAF3 in the Toll-like receptor-dependent and -independent antiviral response. Nature 439:208-211. [DOI] [PubMed] [Google Scholar]
- 35.Okumura, F., W. Zou, and D. E. Zhang. 2007. ISG15 modification of the eIF4E cognate 4EHP enhances cap structure-binding activity of 4EHP. Genes Dev. 21:255-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pichlmair, A., O. Schulz, C. P. Tan, T. I. Naslund, P. Liljestrom, F. Weber, and C. Reis e Sousa. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science 314:997-1001. [DOI] [PubMed] [Google Scholar]
- 37.Prinarakis, E., E. Chantzoura, D. Thanos, and G. Spyrou. 2008. S-glutathionylation of IRF3 regulates IRF3-CBP interaction and activation of the IFNbeta pathway. EMBO J. 27:865-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ritchie, K. J., M. P. Malakhov, C. J. Hetherington, L. Zhou, M. T. Little, O. A. Malakhova, J. C. Sipe, S. H. Orkin, and D. E. Zhang. 2002. Dysregulation of protein modification by ISG15 results in brain cell injury. Genes Dev. 16:2207-2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saira, K., Y. Zhou, and C. Jones. 2007. The infected cell protein 0 encoded by bovine herpesvirus 1 (bICP0) induces degradation of interferon response factor 3 and, consequently, inhibits beta interferon promoter activity. J. Virol. 81:3077-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saitoh, T., A. Tun-Kyi, A. Ryo, M. Yamamoto, G. Finn, T. Fujita, S. Akira, N. Yamamoto, K. P. Lu, and S. Yamaoka. 2006. Negative regulation of interferon-regulatory factor 3-dependent innate antiviral response by the prolyl isomerase Pin1. Nat. Immunol. 7:598-605. [DOI] [PubMed] [Google Scholar]
- 41.Sasai, M., M. Shingai, K. Funami, M. Yoneyama, T. Fujita, M. Matsumoto, and T. Seya. 2006. NAK-associated protein 1 participates in both the TLR3 and the cytoplasmic pathways in type I IFN induction. J. Immunol. 177:8676-8683. [DOI] [PubMed] [Google Scholar]
- 42.Sato, M., H. Suemori, N. Hata, M. Asagiri, K. Ogasawara, K. Nakao, T. Nakaya, M. Katsuki, S. Noguchi, N. Tanaka, and T. Taniguchi. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13:539-548. [DOI] [PubMed] [Google Scholar]
- 43.Seth, R. B., L. Sun, C. K. Ea, and Z. J. Chen. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122:669-682. [DOI] [PubMed] [Google Scholar]
- 44.Sharma, S., B. R. tenOever, N. Grandvaux, G. P. Zhou, R. Lin, and J. Hiscott. 2003. Triggering the interferon antiviral response through an IKK-related pathway. Science 300:1148-1151. [DOI] [PubMed] [Google Scholar]
- 45.Stetson, D. B., and R. Medzhitov. 2006. Type I interferons in host defense. Immunity 25:373-381. [DOI] [PubMed] [Google Scholar]
- 46.Takeuchi, O., and S. Akira. 2008. MDA5/RIG-I and virus recognition. Curr. Opin. Immunol. 20:17-22. [DOI] [PubMed] [Google Scholar]
- 47.Takeuchi, T., S. Inoue, and H. Yokosawa. 2006. Identification and Herc5-mediated ISGylation of novel target proteins. Biochem. Biophys. Res. Commun. 348:473-477. [DOI] [PubMed] [Google Scholar]
- 48.Wong, J. J., Y. F. Pung, N. S. Sze, and K. C. Chin. 2006. HERC5 is an IFN-induced HECT-type E3 protein ligase that mediates type I IFN-induced ISGylation of protein targets. Proc. Natl. Acad. Sci. U. S. A. 103:10735-10740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu, L. G., Y. Y. Wang, K. J. Han, L. Y. Li, Z. Zhai, and H. B. Shu. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19:727-740. [DOI] [PubMed] [Google Scholar]
- 50.Yang, K., H. X. Shi, X. Y. Liu, Y. F. Shan, B. Wei, S. Chen, and C. Wang. 2009. TRIM21 is essential to sustain IFN regulatory factor 3 activation during antiviral response. J. Immunol. 182:3782-3792. [DOI] [PubMed] [Google Scholar]
- 51.Yoneyama, M., M. Kikuchi, K. Matsumoto, T. Imaizumi, M. Miyagishi, K. Taira, E. Foy, Y. M. Loo, M. Gale, Jr., S. Akira, S. Yonehara, A. Kato, and T. Fujita. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 175:2851-2858. [DOI] [PubMed] [Google Scholar]
- 52.Yoneyama, M., M. Kikuchi, T. Natsukawa, N. Shinobu, T. Imaizumi, M. Miyagishi, K. Taira, S. Akira, and T. Fujita. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730-737. [DOI] [PubMed] [Google Scholar]
- 53.Yuan, W., and R. M. Krug. 2001. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. EMBO J. 20:362-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang, B., M. Li, L. Chen, K. Yang, Y. Shan, L. Zhu, S. Sun, L. Li, and C. Wang. 2009. The TAK1-JNK cascade is required for IRF3 function in the innate immune response. Cell Res. 19:412-428. [DOI] [PubMed] [Google Scholar]
- 55.Zhao, C., S. L. Beaudenon, M. L. Kelley, M. B. Waddell, W. Yuan, B. A. Schulman, J. M. Huibregtse, and R. M. Krug. 2004. The UbcH8 ubiquitin E2 enzyme is also the E2 enzyme for ISG15, an IFN-alpha/beta-induced ubiquitin-like protein. Proc. Natl. Acad. Sci. U. S. A. 101:7578-7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao, C., C. Denison, J. M. Huibregtse, S. Gygi, and R. M. Krug. 2005. Human ISG15 conjugation targets both IFN-induced and constitutively expressed proteins functioning in diverse cellular pathways. Proc. Natl. Acad. Sci. U. S. A. 102:10200-10205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhong, B., L. Zhang, C. Lei, Y. Li, A. P. Mao, Y. Yang, Y. Y. Wang, X. L. Zhang, and H. B. Shu. 2009. The ubiquitin ligase RNF5 regulates antiviral responses by mediating degradation of the adaptor protein MITA. Immunity 30:397-407. [DOI] [PubMed] [Google Scholar]
- 58.Zou, W., and D. E. Zhang. 2006. The interferon-inducible ubiquitin-protein isopeptide ligase (E3) EFP also functions as an ISG15 E3 ligase. J. Biol. Chem. 281:3989-3994. [DOI] [PubMed] [Google Scholar]