Abstract
Runx1/AML1 is a transcription factor implicated in tissue stem cell regulation and belongs to the small Runx family of cancer genes. In the hair follicle (HF), Runx1 epithelial deletion in morphogenesis impairs normal adult hair homeostasis (cycle) and blocks adult hair follicle stem cells (HFSCs) in quiescence. Here, we show that these effects are overcome later in adulthood. By deleting Runx1 after the end of morphogenesis, we demonstrate its direct role in promoting anagen onset and HFSC proliferation. Runx1 deletion resulted in cyclin-dependent kinase inhibitor Cdkn1a (p21) upregulation. Interfering with Runx1 function in cultured HFSCs impaired their proliferation and normal G0/G1 and G1/S cell cycle progression. The proliferation defect could be rescued by Runx1 readdition or by p21 deletion. Chemically induced skin tumorigenesis in mice turned on broad Runx1 expression in regions of the skin epithelium, papillomas, and squamous cell carcinomas. In addition, it revealed reduced rates of tumor formation in the absence of Runx1 that were accompanied by decreased epithelial levels of phospho-Stat3. Runx1 protein expression was similar in normal human and mouse hair cycles. We propose that Runx1 may act as a skin oncogene by directly promoting proliferation of the epithelial cells.
Runx1 is part of the small Runt domain family of transcription factors implicated in tissue stem cell regulation (2), tissue development (14), and cancer (8). Runx1 plays developmental roles in several organs, including blood (54), muscle (65), the nervous system (26, 69), and hair follicles (HFs) (44, 47), by affecting cell survival, proliferation, and differentiation (14). Runx1 is frequently mutated in acute myeloid leukemia and myelodisplastic syndrome (8, 38, 54). Runx1 is required in mouse embryos for adult hematopoietic stem cell (HSC) emergence (11, 54), while in adult mice it affects specific hematopoietic lineages (21, 25, 46, 56). The role of Runx1 in epithelial skin and HFs, an important model system for stem cell regulation and cancer progression, has just begun to be addressed (2).
Mammalian skin is largely composed of closely interacting epithelial and mesenchymal tissues, such as the epidermis and the epithelium of skin appendages (e.g., HFs and sebaceous glands) and dermis. During mouse fetal and perinatal life, HFs bud from the overlying epidermis and then move into the skin mesenchyme (dermis and subcutis). Around postnatal day 17 (PD17), the HF initiates a unique process of cyclic organ transformation known as the hair cycle. One hair cycle lasts ∼3 weeks and has three phases: anagen, for HF growth and generation of proliferating pigmented hair shaft; catagen, for apoptosis-driven regression; and telogen, for relative quiescence. The old hair shaft (club hair) is shed from the skin in exogen (41, 45, 51).
HFs have an upper permanent (bulge) region containing infrequently dividing stem cells (15) and a temporary lower region (bulb) that dies out in catagen and is regenerated at anagen from short-lived matrix cells produced by bulge cells collapsing/migrating in telogen into the hair germ, a small epithelial structure found underneath (20, 27, 70). This is followed by bulge cell proliferation during early anagen phase (15, 70). The hair signaling center, the dermal papilla (DP), is a pocket of mesenchymal cells that lies at the hair base (41).
The hair bulge and germ cells express several proteins (K14, CD34, LGR5, and K15) used in lineage-tracing experiments to demonstrate long-term contributions of bulge and possibly germ cells to HF regeneration (7, 28, 29, 59, 70). Other cells besides bulge and germ cells might work as HFSCs (30).
Runx1 is expressed in a few HF compartments, including bulge and germ, but not in other skin epithelial structures, such as sebaceous gland and epidermis (44, 47). Constitutive epithelial deletion of Runx1 through development affects hair shaft structure, HFSC activation, and anagen onset (44, 47). However, it remained unclear if Runx1 directly and permanently affected HFSC proliferation and which factors might be implicated. Furthermore, the potential function of Runx1 in skin cancers is currently unexplored. Clarification of this role appears to be important, since HFSCs are a well-appreciated source of skin appendage tumors and of the most common malignancy of humans, i.e., basal cell carcinoma (17, 32, 35, 37).
MATERIALS AND METHODS
Mice.
The Cornell University IACUC approved all of our mouse work. To create knockout mice, we mated hemizygous K14-Cre (CD1) or β-actin-CreER (C57BL/6) mice with homozygous Runx1fl/fl (C57BL/6) mice; F1 K14-Cre (or β-actin-CreER)/Runx1fl/+ (CD1/C57BL/6) progeny were bred subsequently with homozygous Runx1fl/fl mice to generate K14-Cre (or β-actin-CreER); Runx1fl/fl mice at a 25% Mendelian frequency. The β-actin-CreER; Runx1fl/fl mice were crossed once more with the Runx1fl/fl mice to generate β-actin-Cre; Runx1fl/fl and Runx1fl/fl mice at a 1:1 ratio. Genotyping was performed as described previously (21, 23, 62). Mice from the β-actin-CreER crossing were injected with a 9-mg/40 g of body weight dose of tamoxifen dissolved in corn oil. Injections were performed once daily for 2 days. Mice were housed in cages with littermates of the same sex postweaning at PD21. Skin color of animals was assessed by visual inspection of the entire back skin without hair shaving or clipping, to avoid skin injury. Mice with any sign of black skin patches on the back were scored as in anagen. Single Cdkn1a knockout (−/−) mice (Jackson Laboratories strain B6;129S2-Cdkn1atm1Tyj/J), genotyped as instructed by the vendor (data not shown), were sequentially crossed to our transgenic mouse lines to create double knockout Cdkn1a−/−; K14-CreER; Runx1fl/fl mice. Two independent crosses were carried over in the same sequence to generate p21−/− and p21−/−; Runx1fl/fl; K14Cre littermates and p21+/+ and p21+/+; Runx1fl/fl; K14Cre littermates. Parental pairs from each cross were mated simultaneously, and newborn litters from each cross were obtained and sacrificed on the same day. Mice of all four genotypes were utilized side by side in each experiment of the cell proliferation assays.
DMBA/TPA treatments.
We used the two-step chemically induced tumorigenic protocol described in detail elsewhere (24). We employed K14-Cre/Runx1fl/fl knockout mice (n = 24) and wild-type littermate controls with no K14-Cre (n = 28) at three stages in their adulthood (as shown in Fig. 6, below). We shaved their back skin hair and applied 32 μg of 9,10-dimethyl-1,2-benzanthracene (DMBA) dissolved in 200 μl of acetone. This drug introduces point mutations in the DNA, including the locus of the H-ras proto-oncogene (24). Following that, we used the phorbol ester 12-O-tetradecanoylphorbol-13-acetate (TPA) at a dose of 12.4 μg/mouse dissolved in 200 μl of acetone. TPA propagates the mutation via protein kinase C activation (13), which induces robust proliferation of the skin epithelial cells (keratinocytes). Mice were subjected to TPA treatments twice a week for up to 15 or 20 weeks at a time. The hair was gently shaved several times during the procedure, as needed, to allow efficient delivery of the TPA solution. The appearance of tumors on the back skin and their numbers on each mouse were counted once a week during the 15 to 20 weeks of TPA treatment. Aged mice between 8 and 14 months were monitored for malignant transformation, as shown by sudden sinking of the tumor, accompanied by abrupt vascularization, attachment to the fascia, and rapid growth. Diagnoses of papillomas and squamous cell carcinomas were performed by a veterinary pathologist (Rachel Peters) and based on morphological assessments of tumor margins (infiltrative versus well demarcated), cellular atypia (well differentiated and uniform versus poorly differentiated and marked anisocytosis with dyskeratosis), and desmoplastic response. Tumors and other tissues were fixed in 10% neutral buffered formalin overnight and then transferred to 70% ethanol. They were processed overnight, embedded in paraffin, and sectioned at 4 μm with routine hematoxylin and eosin (H&E) staining. Skin tumors along with lung, kidney, and liver were then examined by light microscopy for the presence of neoplastic cells.
FIG. 6.
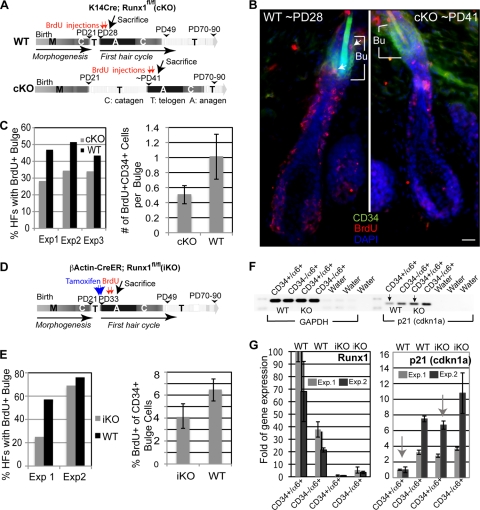
Runx1 cKO and iKO mouse cells impair bulge cell proliferation during anagen. (A) Scheme of short-term BrdU labeling in anagen in WT and K14Cre; Runx1fl/fl epithelial (cKO) mice. (B) Images of 10-μm skin sections show HFs at the same anagen stage in WT and cKO mice from panel A immunostained for markers (indicated in the corresponding color). Bu, the bulge area (contains the CD34+ cells); arrows, BrdU+/CD34+ cells. Bar, 20 μm. (C) HFs with ≥1 CD34+/BrdU+ cell are shown as the percentage of BrdU+ bulges (left). For the three experiments shown on the left (n = 185 WT and 239 cKO HFs counted), we computed the average number of BrdU+/CD34+ cells per bulge (right panel). (D) Scheme of short-term BrdU labeling in anagen in WT and β-actin-CreER; Runx1fl/fl mice injected with tamoxifen (iKO) to induce Runx1 deletion in adulthood after anagen onset. (E) HFs with ≥1 CD34+/BrdU+ cell were used to compute the fraction of BrdU+ bulges (left). For the two experiments shown on the right we computed the percentage of BrdU+ cells among all the CD34+ bulge cells (right). (F) RT-PCR of cDNA prepared from CD34+/α6+ (bulge) cells sorted from the skin cells (indicated at top) from iKO mice injected with tamoxifen after anagen onset. Note the increased expression of the Cdkn1a (p21) gene upon Runx1 knockout induction. (G) The same samples as in panel F, but results were analyzed by QRT-PCR for Runx1 and Cdkn1a expression and normalized to GAPDH levels. Note the low levels of Runx1 expression in iKO mice, as expected, and significantly increased expression of Cdkn1a in iKO CD34+/α6+ bulge cells in both experiments. Each experiment utilized a different pair of WT and iKO mice matched by sex.
BrdU labeling.
5-Bromo-3-deoxyuridine (BrdU; Sigma-Aldrich) was injected intraperitoneally in saline buffer (phosphate-buffered saline [PBS]) at 25 μg/g of body weight. To make label-retaining cells (LRCs), mice were injected on three consecutive days (PD3, -4, and -5) twice a day at 12-h intervals with a dose of 25 μg/g of body weight. BrdU label was chased for a total of 4 weeks, and mice were treated with 12.4 μg of TPA dissolved in acetone administered twice weekly for the last 2 weeks of the chase. Short-term BrdU exposure was evaluated during anagen stage as indicated by the change in skin color at a dose of 50 μg/g of body weight and at the indicated frequencies. Animals were sacrificed and skin was embedded in OCT, frozen in dry ice, sectioned using a cryostat (Richard Allan Scientific), and stained for immunofluorescence using antibodies specific to BrdU, as described previously (61).
Histology, immunofluorescence, and X-Gal staining.
Staining of skin tissue for immunofluorescence and with H&E was as described previously (61). The MOM Basic kit (Vector Laboratories) was used for mouse antibodies. Nuclei were labeled with 4′,6′-diamidino-2-phenylindole (DAPI) or Hoechst. For 5-bromo-4-chloro-3-indoxyl-beta-d-galactopyranoside (X-Gal) staining, 10-μm skin sections were fixed for 1 min in 0.1% glutaraldehyde and washed in PBS. Incubation in X-Gal solution was at 37°C for 12 to 16 h. Antibodies were from (i) rat (α6-integrin at 1:100 and CD34 at 1:150 [BD Biosciences], E-cadherin [1:500; Sigma], and BrdU [1:300; Abcam]); (ii) guinea pig (K5 and K15 [1:5,000; E. Fuchs, Rockefeller University]), (iii) rabbit (Runx1 [1:8,000; T. Jessell, Columbia University {10}], Ki67 [1:100; Novocastra], keratin 1 [1:500; Covance], Loricrin [1:200; E. Fuchs, Rockefeller University], and phosphorylated Stat3 (P-Stat3; 1:50; Cell Signaling); and (iv) mouse (myc; 1:400; Zymed).
Immunohistochemistry.
Paraffin sections of 4 μm were deparaffinized in three changes of xylene at room temperature and rehydrated through a graded ethanol-water series at ratios of 100%, 95%, and 70%, followed by rinsing in water. Antigen retrieval was performed by microwaving for 20 min in 0.01 M EDTA buffer (pH 8.0). Sections were incubated in 0.5% hydrogen peroxide to block endogenous peroxidase and washed three times in Tris-buffered saline (TBS). After blocking nonspecific antigen sites with 10% normal goat serum for 20 min, slides were incubated for 1.5 h at 37°C with antibody against phosphorylated-p44/42 mitogen-activated protein kinase (pErk1/2-Thr202/Tyr204; Cell Signaling Technology no. 4376; 1:50 dilution in TBS). Normal rabbit immunoglobulins were used as negative controls. Sections were then incubated with biotinylated goat anti-rabbit IgG (BA-1000; Vector) for 20 min. Detection was performed by a 20-min incubation with streptavidin peroxidase (50-242Z; Invitrogen), followed by a 10-min incubation with aminoethyl carbazole chromogen/substrate solution (00-2007; Invitrogen). Sections were counterstained with hematoxylin and mounted in aqueous mounting medium (0100-01; Southern Biotech).
Microscopy and image processing.
Images were acquired using the IP-Lab software (MVI) on a fluorescence light microscope (Nikon) equipped with a charge-coupled-device 12-bit digital camera (Retiga EXi; QImaging) and motorized z-stage. Single images were color combined and were assembled in montages and enhanced for brightness, contrast, and levels using Adobe Photoshop and Illustrator.
Primary cell culture, flow cytometry, and QRT-PCR.
Skin cells were cultured using low-Ca2+ keratinocyte E medium (61) by plating in triplicate 50,000, 250,000, 500,000, 1,000,000, or 1,500,000 live (not staining with trypan blue) cells on irradiated mouse embryonic fibroblasts (passage 4). Keratinocyte colonies in dishes with 50 to 200,000 cells were counted using phase-contrast microscopy or H&E staining. A similar procedure was applied to amplifying the cell line of CD34+/α6+ cells sorted from the bulge. For flow cytometry, mice were treated with tamoxifen in anagen phase and sacrificed. Skin cells extracted by trypsin treatment were stained with biotin-labeled CD34 antibody (eBioscence) followed by streptavidin-allophycocyanin (BD Biosciences) and with phycoerythrin-labeled α6-integrin (CD49f) antibody (BD Biosciences), as described previously (61). Live cells were those excluding propidium iodide (Sigma). Fluorescence-activated cell sorting (FACS) was performed using a BD Biosciences Aria apparatus at Cornell. RNA isolation from sorted cells and quantitative reverse transcription-PCR (QRT-PCR) of cDNAs were as described previously (61). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) demonstrated equal loading by RT-PCR. GAPDH was used in the QRT to normalize threshold cycle CT values and compute fold changes.
Human samples.
Fronto-temporal scalp skin specimens were obtained from female patients undergoing elective facial plastic surgery with informed patient consent according to the University of Luebeck, Faculty of Medicine Ethics Committee (Germany) approved protocol 06-109. Anonymous samples were received fresh from plastic surgery, embedded in OCT, and cut into 7-μm serial sections and kept frozen through shipping until staining, which was performed essentially as previously described for mouse skin (44).
DNA constructs.
For the K14-Runx1-myc construct, Runx1 coding sequences were amplified by PCR from mouse Aml1b cDNA previously cloned into pcDNA3.1 (a gift from Nancy Speck) as template. The 5′ forward primer was 5′-TCTAGCGGCCGCATGCGTATCCCCGTAGATGCC-3′; sequences in bold show the restriction site for NotI and underlined sequences are the first 21 bases of Runx1. The 3′ reverse primer was 5′-GCTCTAGATCACAGGTCCTCCTCTGAGATCAGCTTCTGCATTGATGCCATGGTCTCGAGGTAGGGCCGCCA CACGGC-3′; sequences in bold are restriction sites for XbaI and XhoI, respectively, sequences in italics are the myc tag followed by the stop codon, and underlined sequences are the last 18 bases of Runx1. The PCR product was purified with a gel extraction kit (Qiagen), digested with NotI (5′ end) and XbaI (3′ end), and ligated into a K14-β-globin cassette (a gift from Elaine Fuchs) at NotI and XbaI sites. The final K14-Runx1-myc construct was verified by sequencing. For the K14-enhanced green fluorescent protein (eGFP)-myc construct, Runx1 coding sequences were digested from the K14-Runx1-myc construct at SpeI and XhoI sites, and then eGFP coding sequences obtained from the pEGFP-C1 vector (Clontech) with restriction enzymes NheI and XhoI were cloned at SpeI and XhoI sites, followed by the previously cloned myc tag. For the K14-eGFP-Runt-myc construct, the Runt domain was amplified by PCR using the same Aml1b cDNA template. The 5′ forward primer was 5′-ACCGATATCTAATGGTGGAGGTACTAGCTGACCAC-3′; sequences in bold are the restriction site for EcoRV, and underlined sequences are the first 24 bases of the Runt domain. The 3′ reverse primer was 5′-TGCCTCGAGCACCTTACGCTTCTTCTTTGGCACCTTACGCTTCTTCTTTGGATCATCTAGTTTCTGCCG ATG-3′; sequences in bold are the restriction site for XhoI, sequences in italics are the nuclear localization signal (NLS) in tandem, and underlined sequences are the last 22 bases of the Runt domain (22). The PCR product was purified with a gel extraction kit (Qiagen), digested with NotI (5′ end) and XbaI (3′ end), and ligated into the K14-eGFP-myc plasmid at the NotI and XbaI sites between eGFP and the myc tag. The final K14-eGFP-Runt-myc construct was verified by sequencing.
Transfections and cell cycle analysis.
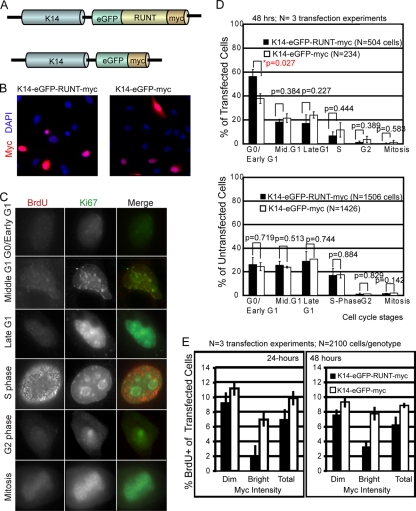
At 24 h before transfection, CD34+/α6+-sorted keratinocytes, expanded and propagated for several passages without feeders in low-Ca2+ medium (61), were plated onto six-well plates containing an 18-mm2 coverglass (Corning). The next day cells were ∼80% confluent. For transfections 1 μg of each DNA construct was mixed with 6 μl of TransFectin lipid reagent (Bio-Rad) and 125 μl of serum-free medium according to the manufacturer's instructions. The DNA/lipid complex was added to the dishes, and cells were further grown for an additional 48 h. Next, 4.4 μl of 1.7 mg/ml BrdU was added to each 35-mm plate and incubated at 37°C for 20 min. Cells were washed with PBS, fixed for 20 min with 1.6% formaldehyde, and triple stained for myc, BrdU, and Ki67 using an immunofluorescence procedure. Transfected cells were categorized positive based on a myc staining fluorescence signal intensity higher than 500 counts on a 4,095 gray-level scale with a fixed exposure. Transfected cells were further differentiated based on transgene expression levels as dim when myc staining intensity was between 500 and 1,500 gray levels and as bright when the intensity was between 1,500 and 4,095. Table 1 categorizes the cell cycle stages of transfected cells based on the presence or absence of Ki67 and BrdU signal and based on Ki67 nuclear staining patterns identified with a fluorescence microscope.
TABLE 1.
Cell cycle stages of transfected cells based on Ki67 and BrdU signals
| Growth phase | BrdU staining | Nuclear staining pattern of Ki67 foci |
|---|---|---|
| Early G1/G0 | − | Dim |
| Mid-G1 | − | Many foci |
| Late G1 | − | 2-10 foci |
| S | + | 2-5 foci |
| G2 | − | 1 large focus |
| Mitosis | − | Bright staining |
Identified using a fluorescence microscope.
Statistical analyses.
Data shown are averages and standard deviations. Student one and two-tailed t tests for tumor formation, cell cycle stages, and BrdU incorporation were performed to compare wild-type and knockout samples.
RESULTS
Runx1 cKO hair follicles spontaneously enter a new hair cycle.
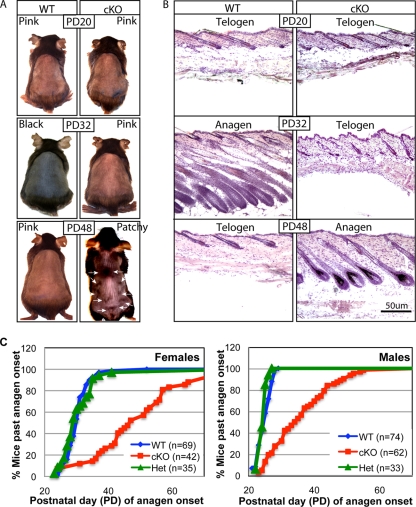
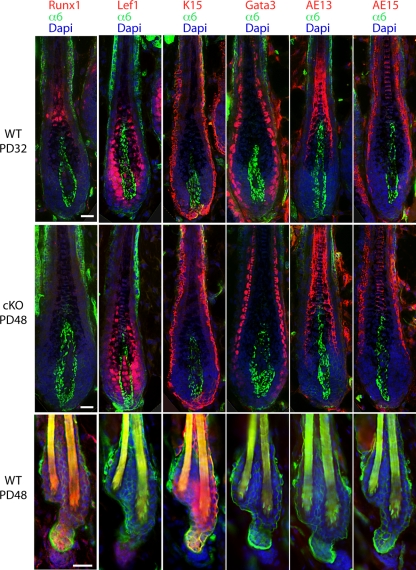
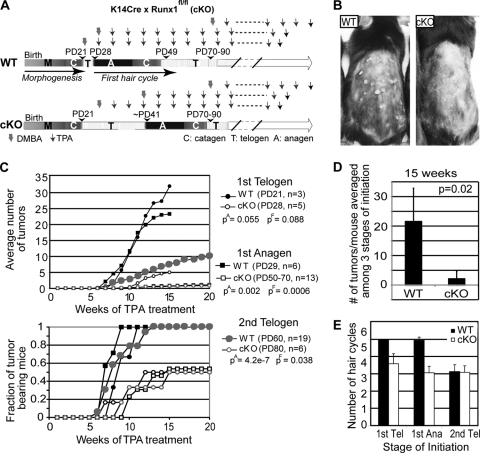
Previously we found that mice with epithelial conditional deletion of Runx1 (conditional knockout [cKO]) displayed a prolonged telogen phase, which could be overcome by skin injury (44). To see if this telogen block was overcome spontaneously later in adulthood, we crossed the K14-Cre transgenic mice (62) with mice in which loxP sites (floxed [fl]) were inserted in the Runx1 gene locus to flank the fourth exon, which encodes the Runt DNA-binding domain (21). We produced large cohorts of Runx1 cKO mice with an exon 4 deletion (Δ4), along with heterozygous (Het; Runx1Δ4/+) and wild-type (WT; RUNX1fl/fl or RUNX1fl/+) littermate controls (44) (see Materials and Methods). We visually monitored the hair cycle progression from PD20 to PD90 as evidenced by the apparent change in skin color from pink, indicative of telogen, to black, indicative of anagen, due to incorporation of melanin into the newly synthesized hair shaft (41). A few representative mice were shaved and photographed (Fig. 1A). Mice sacrificed at several stages were used to prepare frozen skin sections that were subsequently stained with hematoxylin and eosin for contrast or with fluorescently labeled antibodies for hair differentiation markers, which confirmed hair cycle stages (Fig. 1B and C and 2). All the differentiation markers were expressed correctly in cKO hair follicles upon entering anagen. WT mice changed their skin color earlier and more abruptly than cKO mice and in a head-to-tail rapid progression throughout the back skin. In contrast, the skin of epithelial Runx1 cKO mice became black not only late but also in a nonuniform or patchy manner in most animals analyzed (Fig. 1A). The patchy hair growth is reminiscent of the second telogen in WT mice (45), except the distribution of hair growth zones began from the sides and middle and was different from that observed in WT mice in the second telogen (45) (data not shown). This suggested a possible role for Runx1 in regulating the normal hair growth spreading patterns throughout the skin. Taken together these data demonstrated that the telogen arrest in the Runx1 cKO HFs is temporary, lasts ∼1 to 2 weeks, and is eventually overcome in the absence of apparent skin injury. This suggested that Runx1 activity is not an essential prerequisite for anagen induction.
FIG. 1.
Delayed first adult anagen onset in epithelial Runx1 knockout mice (cKO). (A) Typical skin color changes in WT and Runx1 cKO mice photographed at the ages indicated. Note the black skin color with new hair growth in the WT but not cKO mouse at PD32. At PD48 WT mouse skin was in the second telogen, as indicated by the pink color, while cKO mouse skin was at the first anagen onset. Note the patchy appearance at the initiation of skin color change (arrows) in the cKO mouse. (B) Frozen skin sections from WT and cKO mice at the stages indicated (stained with hematoxylin and eosin). For the PD48 cKO mouse the skin sample was collected from one of the black skin areas indicated by the arrows in panel A. (C) Fraction of Runx1 cKO, Het, and WT littermate control mice of the same sex that passed anagen onset as a function of time.
FIG. 2.
Immunostaining for hair follicle lineage markers confirmed normal expression in cKO mice after anagen onset. Frozen skin sections at the ages indicated show immunofluorescence signals from antibodies specific to markers indicated in the corresponding color. Bar, 20 μm. Normal expression in cKO skin during telogen has been previously reported (44).
Runx1 acts directly at anagen onset in adulthood.
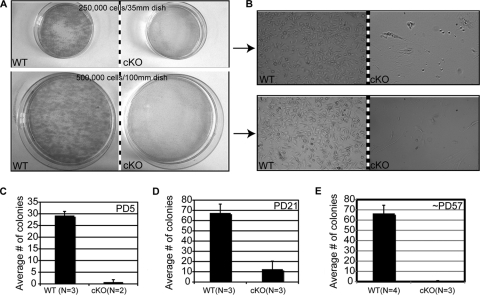
To determine if Runx1 cKO HFSCs have a delay in maturation during morphogenesis that might offset anagen onset later in adult mice, or whether HFSCs have an intrinsic defect in proliferation in cell culture irrespective of the developmental stage at which they were isolated, we performed clonogenic stem cell assays (3, 4) at different ages. We predicted that if Runx1 cKO HFSCs mature later than WT, then the clonogenic defect previously documented in early morphogenesis (44) might be overcome at the time of anagen onset. We isolated WT and Runx1 cKO skin cells from littermate mice at PD5, PD21, and PD56 to -58 (when cKO mice began the delayed anagen onset), plated equal numbers of live cells (those not stained with trypan blue), and counted keratinocyte colonies 2 weeks after plating (Fig. 3). While WT keratinocytes formed large healthy growing colonies, Runx1 cKO keratinocytes failed to proliferate under all the experimental conditions tested (Fig. 3). Plating more cells and/or a higher density of cells did not rescue the cell proliferation defect, ruling out potential lower numbers of HFSCs in the Runx1 cKO mice (Fig. 3 and data not shown). These data did not support the hypothesis that delayed maturation of HFSCs in morphogenesis might prevent colony formation by keratinocytes isolated from Runx1 cKO mice. Instead, they demonstrated that under certain stress conditions, such as those imposed by cell culture, the HFSCs, and in fact all epithelial cells isolated from skin, including those from the epidermis, fail to proliferate and survive in the absence of Runx1. This result correlates with data presented below, in which acute inhibition of Runx function in keratinocytes perturbed cell cycle regulation (see Fig. 5, below).
FIG. 3.
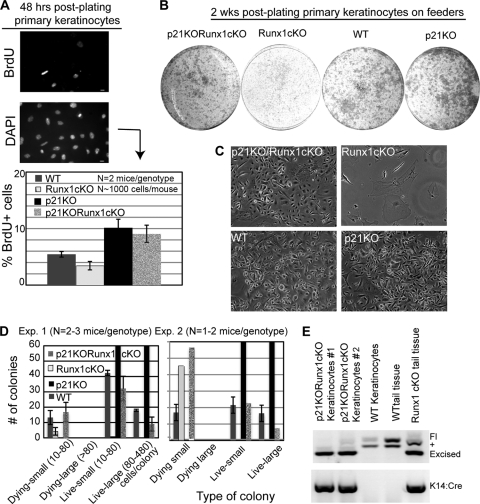
Keratinocytes isolated from young and adult Runx1 epithelial cKO skin failed to proliferate in culture. (A) Primary skin keratinocytes plated for 2 weeks on fibroblast feeders in the numbers and dish sizes indicated showed growth of large colonies from WT but not from Runx1 cKO cells by hematoxylin and eosin staining. (B) Phase-contrast images of WT and cKO keratinocytes after 2 weeks of growth. Cells in the cKO dishes are feeder fibroblasts. (C to E) Average numbers of keratinocyte colonies in WT and cKO cells isolated from mouse skin at the postnatal day indicated. N, number of mice used for each experiment.
FIG. 5.
Expression of the Runt domain in cultured CD34+/α6+-sorted bulge cells affects cell cycle progression. (A) Schematic of vector DNA containing engineered transgenes that encode the myc- and GFP-tagged versions of the dominant negative DNA binding Runt domain expressed from the keratin 14 promoter (a gift from E. Fuchs), referred to as K14-eGFP-RUNT-myc. The control was the K14-eGFP-myc construct. (B) Myc (red) and DNA DAPI (blue) staining of CD34+/α6+ cultured bulge cells after 48 h of transfection with the DNA constructs indicated at the top. (C) Examples of BrdU (red) and Ki67 (green) staining patterns, indicating distinct cell cycle stages. (D, top) Cell counts of distinct cell cycle stages are shown as the percentage of the total transfected (myc+) cells. P values shown at the top for each comparison were obtained by two-tailed t test analysis. (Bottom) Same experiment as in the top graph but for untransfected (myc-negative) cells, showing no significant differences and demonstrating comparable growth conditions among cell culture dishes. (E) Same experiment as in panel D, except cells were scored only for the percent of transfected cells BrdU+ 24 and 48 h after transfection. Cells were categorized as bright or dim based on an arbitrary cutoff value at equal exposure of ∼1,500 gray level counts for the myc immunofluorescence staining. Note the lower S-phase frequency for K14-eGFP-RUNT-myc-transfected cells.
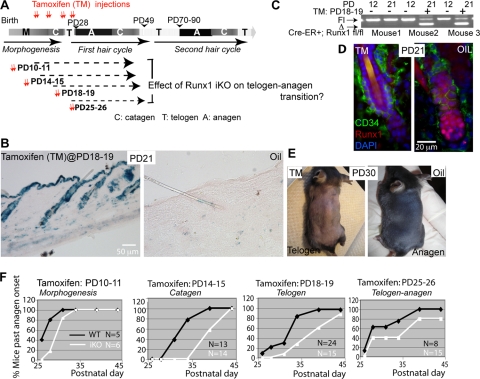
To investigate if Runx1 is needed directly at anagen onset for cell proliferation (as opposed to being required in development via HFSC maturation), we used a tamoxifen-inducible CreER transgenic system (23) to ablate Runx1 function at four stages in late postnatal morphogenesis and in adulthood (Fig. 4A). We tested several inducible Cre lines in Rosa26R reporter mice (53) and chose a β-actin-CreER mouse line (23), which showed high efficiency of tamoxifen induction and little or no leaky Cre activity in the absence of tamoxifen by X-Gal staining of frozen skin sections and by PCR (Fig. 4B and C) (see Materials and Methods). Since Runx1 is expressed in skin predominantly in the hair bulge and hair germ at anagen onset (44), we did not expect a major contribution to the phenotype by other nonepithelial skin cell types targeted by the β-actin-CreER. Runx1fl/fl; β-actin-CreER control mice injected with oil but not with tamoxifen at PD18 and PD19 and sacrificed 2 days later showed strong Runx1 protein expression in the hair germ and in rare cells at the base of the bulge by immunofluorescence analysis of skin sections in 90% of the HFs analyzed (n = 3 WT [174 HFs]. In contrast, Runx1fl/fl; tamoxifen-induced knockout (iKO) β-actin-CreER+ mice injected at the same stage with oil (n = 3 mice [126 HFs]) (Fig. 4D) lacked RUNX1 staining in 97% of HFs analyzed (Fig. 4D). Additional mice analyzed at PD29, PD32, and PD33 confirmed these results (data not shown). These data attested to the efficient and rapid depletion of the Runx1 protein in response to Runx1 KO (iKO).
FIG. 4.
Runx1 inducible knockout at several stages in adulthood delays anagen onset. (A) Scheme of tamoxifen induction to generate iKO at stages of late morphogenesis and of the first hair cycle as indicated. (B) X-Gal staining on skin sections isolated from β-actin-CreER; Rosa26 mice injected with tamoxifen (TM) dissolved in oil (left) or oil only (right) at PD18 and -19. (C) PCR with specific primers for the Runx1 floxed allele shows the lower (excision) band only in mice injected with tamoxifen. (D) Runx1 immunostaining of skin sections from mice injected with tamoxifen or oil. Note the lack of Runx1 signal in HFs from the tamoxifen-injected mouse. (E) Mice injected with tamoxifen (left) or oil (right) showed pink and black skin color, indicative of telogen and anagen, respectively. (F) Quantification of skin color changes in tamoxifen-treated β-actinCreER; Runx1fl/fl mice (iKO) and tamoxifen-treated Runx1fl/fl (no Cre) littermate mice, labeled as WT.
To induce the Runx1 KO (iKO) in adulthood, we injected the Runx1fl/fl; β-actin-CreER mice with tamoxifen during four distinct stages: PD10 to -11, PD14 to -15, PD18 to -19, and PD25 to -26, which, considering the ∼2-day lag necessary to eliminate the Runx1 protein (see above), would result in acute Runx1 loss in late morphogenesis, first catagen, first telogen, and first anagen onset, respectively (Fig. 4A). We monitored anagen onset by the change in skin color from pink to black in intact (not shaved) mouse skin. Mice in Fig. 4E were shaved for demonstrative purposes. Littermate Runx1fl/fl mice negative for β-actin-CreER were injected with tamoxifen and served as the WT control. At all time points of induction we found delayed anagen onset in Runx1 iKO mice relative to WT (Fig. 4F). These data demonstrate that the effect of Runx1 loss on adult anagen onset is due to an active requirement for Runx1 at that stage, as opposed to being a late consequence of a defect in HFSC maturation during embryonic HF morphogenesis.
Runx1 regulates G0/G1 and G1/S cell cycle phase transitions in cultured HF bulge cells.
The bulge and germ cells of Runx1 cKO mice remained quiescent for a prolonged period of time, but it was unclear if the defects were due to cell cycle effects, as seen in two blood cell lines, BaF3 and Jurkat T cells (6, 64). Thus, we inhibited Runx1 function in cell culture by using the Runt DNA-binding domain, which interferes with Runx protein function (48), and checked the immediate effect on cell cycle progression. We transfected the sorted CD34+/α6+ cultured bulge cells with a DNA construct expressing the Runt DNA-binding domain tagged with NLS (22), eGFP, and Myc, all placed in frame downstream of the keratin 14 (K14) promoter (K14-eGFP-RUNT-myc) (63). A K14-eGFP-myc transgene served as the transfection control (Fig. 5A) (see Materials and Methods for construct description). A fraction of cells stained positive for myc expression 48 h after transfection (Fig. 5B). We assessed the cell cycle phase distribution in transfected (myc-positive) and untransfected (myc-negative) cells by examining the combined BrdU and Ki67 staining patterns (Fig. 5C) (5). Transfected cells, but not untransfected cells, displayed increased numbers of cells in G0 in Runt-expressing cells relative to control cells, which suggested that interference with Runx function impaired the G0/G1 phase transition (Fig. 5D). Since S-phase cells were underrepresented in this analysis we repeated the transfections and scored solely the BrdU+ cells among transfected cells. We found a noticeable decrease in the fraction of S-phase (BrdU+ cells) in Runt-expressing cells compared with control cells at 24 and 48 h after transfection (Fig. 5E). These data suggested that inhibition of Runx function impaired G0/early G1 exit and the transition into S-phase in cultured bulge cells and might explain at least in part the prolonged quiescence of CD34+/α6+ bulge stem cells in vivo and their failure to proliferate long-term in cell culture (Fig. 3) (44).
Runx1 loss in anagen impairs proliferation of HFSCs in vivo.
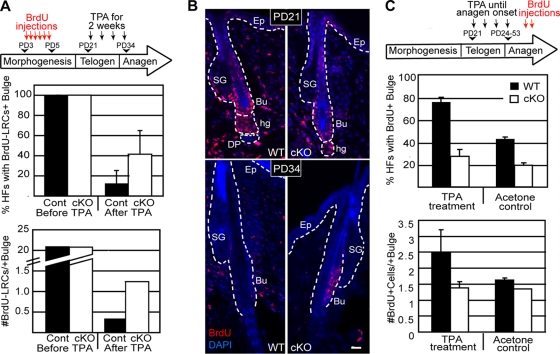
Our in vitro data thus far suggested a role of Runx1 in regulating the rates of cell cycle progression for bulge cells. This prompted us to ask whether in vivo, during the normal bulge proliferative stage, in anagen, Runx1 KO bulge cells also proliferated at a lower rate. We examined the efficiency of BrdU incorporation in bulge cells during anagen in mice with either a cKO or iKO Runx1 knockout. First, we analyzed the K14-Cre; Runx1fl/fl cKO mice and WT controls (n = 3) by injecting BrdU at anagen (see Materials and Methods), as indicated by the first sign of change in skin color from pink to black (Fig. 6A). We sacrificed the mice 12 h later and immuno-stained frozen skin sections for BrdU and for the bulge marker CD34. We then quantified the number of BrdU+/CD34+ cells per HF and the fraction of HFs with BrdU+/CD34+ (bulge) cells (Fig. 6B). HFs of WT and cKO mice were generally found in anagen 3a (20%), anagen 3b (50 to 60%), and anagen 3c (13 to 28%), as defined previously (41). HFs in both categories of mice displayed either no BrdU+/CD34+ cells or ∼1 to 2 BrdU+/CD34+ cells in thin skin sections (10 μm) (Fig. 6B and C, right). The fraction of HFs positive for BrdU was significantly higher (P = 0.04) in WT mice than in Runx1 cKO mice (Fig. 6C, left), and this was true at each subanagen stage (3a, 3b, or 3c) detected in the analysis (results not shown). Moreover, the average number of BrdU+/CD34+ cells per bulge was nearly double in WT HFs compared to cKO (Fig. 6C, right). Second, we induced the Runx1 iKO at anagen onset by injecting tamoxifen on two consecutive days in β-actin-CreER;Runx1fl/fl mice (n = 2) just prior to the pink-to-black skin color change (Fig. 6D). Mice with no β-actin-CreER and no Runx1 excision band detection by genotyping were also injected with tamoxifen and served as a WT control (n = 2). Two to 3 days after tamoxifen induction we injected BrdU four times during a 24-h time period (0, 6, 12, and 24 h), sacrificed the animals 2 h later, and assessed BrdU incorporation in bulge cells during anagen. Although more variable, these data also showed a noticeable reduction in BrdU incorporation in the CD34+ bulge cells (Fig. 6E). All these data suggested that an immediate consequence of Runx1 loss in anagen is reduced rates of bulge cell proliferation relative to WT.
Cdkn1a/p21 works downstream of Runx1 to regulate epithelial cell proliferation.
To understand the mechanisms of reduction in bulge cell proliferation rates due to Runx1 loss, we considered the cyclin-dependent kinase inhibitor Cdkn1a (p21), a key regulator of the cell cycle (31), which we previously found upregulated in the Runx1 cKO bulge cells (44). Here we checked if Cdkn1a mRNA was immediately upregulated in the freshly sorted CD34+/α6+ bulge cells following acute Runx1 deletion (iKO) during anagen (∼PD33 to -36 with tamoxifen; ∼PD36 to -39 at sacrifice) by RT-PCR (Fig. 6F) and QRT-PCR (Fig. 6G) (see Materials and Methods). In all assays acute Runx1 deletion in anagen resulted in reduced Runx1 and increased Cdkn1a mRNA levels.
To test whether Cdkn1a upregulation upon Runx1 loss might be responsible for the impaired proliferation detected in skin epithelial cells, we generated Runx1 cKO mice in which the Cdkn1a gene was also deleted (p21 KO Runx1 cKO). The generation of full Cdkn1a knockout mice has been described elsewhere (9). Newborn double KO and control pups of four different genotypes (see Materials and Methods) were sacrificed, and primary keratinocytes were isolated. Cells were plated either at high density on collagen-coated plates (BD Biosciences), labeled with BrdU, and tested for proliferation rates (Fig. 7A) or were plated at low density on irradiated mouse embryonic fibroblasts (MEF) feeder layers for colony formation assays (Fig. 7B and C). Remarkably, quantification of BrdU incorporation (Fig. 7A, bottom) and of colony formation efficiency (Fig. 7B, C, and D) indicated that Cdkn1a KO largely rescued the cKO Runx1 defect in both assays. Genotyping of cultured keratinocytes 2 weeks after plating confirmed that Runx1-deleted cells were able to grow when Cdkn1a was also deleted and that the colonies observed in these samples were not a result of incomplete Runx1 deletion due to Cre inefficiency in these mice. These data identified downregulation of CdKn1a as a major function of Runx1 in promoting skin epithelial cell proliferation.
FIG. 7.
Cdkn1a knockout rescues the proliferation impairment of Runx1 knockout keratinoyctes. (A) Primary keratinocytes freshly isolated from newborn mice plated on collagen plates and labeled and stained for BrdU (top) and DNA (bottom). (B) Same experiment as in panel A, but plating was on fibroblast feeder layers and cells were allowed to form colonies. (C) Phase-contrast images documenting morphologies of keratinocytes in each genotype. No colonies were detected in the Runx1 cKO cells. (D) Quantification of colonies shown in panel B. (E) PCR of genomic DNA isolated from cells grown from Runx1/Cdkn1a double-knockout mice after culture to detect the status of the Runx1 allele, demonstrating expansion of Cre+ cells with Runx1 excision.
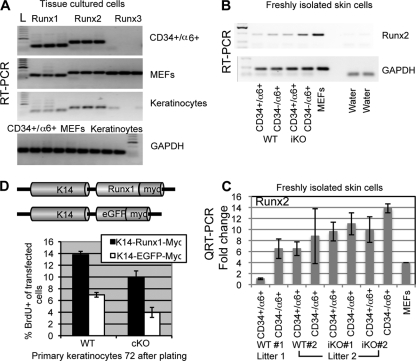
Next we tested the status of mRNA expression of the other two Runx family members, which could have redundant/compensatory effects in the Runx1 KO mice. Runx2 and Runx3 protein expression levels in the skin have been reported, but they were confined to compartments other than the proliferative hair regions (bulge, hair germ, or outer root sheath) where Runx1 is normally expressed (19, 34). Our RT-PCR analysis detected Runx1 but also Runx2 mRNAs in established cell lines grown for many passages in cultures from either mouse skin keratinocytes or sorted CD34+/α6+ bulge cells (Fig. 8A). However, in freshly isolated CD34+/α6+ bulge cells and CD34−/α6+ nonbulge basal cells, (Q)RT-PCR analysis revealed Runx2 mRNA expression levels which were low (as judged by CT values) and variable from mouse to mouse in two WT mice matched by sex from different litters, found at similar anagen stages (Fig. 8B and C). The experiments were performed three times, and MEF expression of Runx2 was used to normalize results across experiments. Since Runx2 protein has not been detected in the bulge in vivo (19, 34), it is unclear if these low and variable Runx2 mRNA levels in WT mice detected might have biological relevance. Within the same litter, Runx1 cKO mice showed a meager increase or comparable levels of Runx2 mRNA, suggesting that Runx2 mRNA is not dramatically upregulated in vivo in response to Runx1 loss (Fig. 8B and C). In light of these and previously published expression data (19, 34), it is unlikely that Runx2 plays a redundant or compensatory role with Runx1 in the hair follicle bulge, although future double KO analyses would fully address this question.
FIG. 8.
Runx1 addition to Runx1 cKO keratinocytes rescues their proliferation defect. (A) RT-PCR analysis performed in triplicate with Runx1, -2, -3 and GAPDH primer pairs as indicated. cDNA was prepared from three cell types (indicated on the right in the upper panel and at the top of the lower panel). L, DNA ladder. (B and C) RT-PCR (B) and QRT-PCR (C) analyses of mRNA levels in freshly sorted bulge (CD34+) and nonbulge (CD34−) cells. WT mice showed weak and variable expression in mice from different litters, while iKO mice showed little if any differences from WT littermates. (D) Quantification of BrdU+ cells in primary keratinocytes isolated from WT and Runx1 cKO cells transfected with the constructs schematized at the top.
Finally, to confirm that the proliferation effect detected in cultured cells is directly due to Runx1 loss, we added back Runx1 by transfecting a K14-Runx1-myc DNA construct to primary cultures of Runx1 cKO and WT keratinocytes 24 h after plating cells freshly isolated from newborn mice. Comparison with K14-eGFP-myc-transfected control cells indicated that K14-Runx1-myc restores high rates of proliferation in cKO Runx1 keratinocytes. In addition, it accelerated the rates of proliferation in WT cells, suggesting a dose effect of Runx1 on cell proliferation (Fig. 8D).
Runx1 works as an oncogene in mouse skin tumorigenesis.
Runx1 belongs to a family of cancer genes (8), and our data so far suggested that Runx1 downregulates the mRNA expression of the skin tumor suppressor gene Cdkn1a (58) in HFSCs and stimulates epithelial and HFSC proliferation. Moreover, SC overproliferation is thought to increase the risk of cancer development (12, 35). Therefore, we next asked whether the Runx1 cKO would impair papilloma and squamous cell carcinoma (SCC) formation in response to skin DMBA/TPA carcinogenic treatment (Fig. 9A). We employed a total of 28 WT and 24 Runx1 cKO mice and treated them first with DMBA to induce mutations and then with TPA to promote proliferation for 15 to 20 weeks, as previously described (24). The time as well as the hair cycle stage of DMBA initiation can have an impact on the tumor outcome (40). Because WT and cKO mice were generally not found at the same hair cycle stage at comparable ages (Fig. 1C and 9A), we initiated the DMBA mutations at three different stages during the first telogen, first anagen, and second telogen phases to account for possible stage- and/or age-dependent effects on tumor formation (Fig. 9A). Runx1 cKO mice showed delayed tumor formation, accompanied by a statistically significant reduction in the average number of tumors/mouse and even more prominently in the fraction of mice presenting tumors at each equivalent stage of DMBA initiation (Fig. 9B, C, and D). Skin initiated with DMBA in the first but not the second hair cycle and monitored for 15 weeks of TPA treatment displayed fewer number of hair cycles in Runx1 cKO mice than the WT (Fig. 9E). The second-telogen data suggested that reduced frequency of hair cycling is not the sole cause of reduced papilloma formation in Runx1 cKO mice. To verify if small papillomas formed underneath the skin but were missed in our visual inspection, we cut serial sections through half the entire back of one cKO mouse skin with no apparent tumors after DMBA/TPA treatment. This did not reveal any additional small papillomas (data not shown). These data showed that Runx1 is necessary for efficient tumor formation by chemically induced carcinogenesis in mouse skin.
FIG. 9.
Runx1 deletion impairs chemically induced skin tumorigenesis. (A) DMBA/TPA treatment of WT and K14Cre × Runx1fl/fl (cKO) littermate mice, shown on the timeline schematic of hair cycle phases. (B) Starting at 8 weeks of treatment, cKO mice showed fewer papillomas than WT mice. (C) Average numbers of tumors (top) and fractions of tumor-bearing mice (bottom) measured weekly as a function of time of TPA treatment (in weeks) for mice treated with DMBA at three different stages as indicated on the right. Note the reduction in tumor formation in cKO versus WT mice at comparable stages. P values noted as pA (top) and pF (bottom) for each stage of initiation are shown for data after 15 weeks of TPA treatment. (D) Number of tumors/mouse averaged among the three stages of initiation (n = 24 cKO and 28 WT mice). (E) Number of hair cycles, estimated by the frequency of skin color change from pink to black in mice initiated with DMBA and treated with TPA for 15 weeks.
To analyze the progression of the papillomas to malignancy in the two groups of mice treated with DMBA/TPA, we monitored their tumors weekly to ∼14 months of age and sacrificed them when tumors on the back displayed signs of malignant conversion (ulceration, vascularization, and sudden growth). Histopathological assessment of tumor morphology showed SCCs in 8 of 26 (∼30%) WT mice and 1 of 24 (∼5%) Runx1 cKO mice. The SCCs invaded deep beneath the skin and, rarely, they demonstrated pulmonary metastasis (Fig. 10B and C). Given the ∼4- to 5-fold reduction in initial papilloma formation, these data ruled out the possibility that papillomas in the Runx1 cKO mice could have in fact progressed to SCCs at a higher rate.
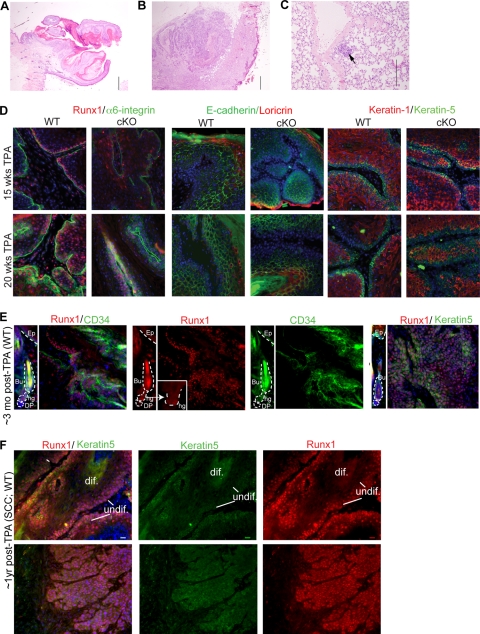
FIG. 10.
Runx1 expression in mouse skin papillomas and squamous cell carcinomas. (A) Representative squamous papilloma. Note the clearly demarcated base and outward growth of the epidermis. Hematoxylin and eosin stain was used. Magnification, ×40. (B) Representative squamous cell carcinoma. Note the infiltrative and downward growth in less differentiated cells. Hematoxylin and eosin stain was used. Magnification, ×40. (C) Lung metastatic focus from the squamous cell carcinoma in panel B. Hematoxylin and eosin stain was used. Magnification, ×200. Bar, 500 μm. (D to F) Sections of several papillomas and squamous cell carcinomas immunostained with different markers as indicated in the corresponding colors. Blue is DNA Hoescht stain. Note the Runx1 expression in zones expressing markers of undifferentiated epithelial cells (K5 and α6-integrin). (D) Note that papillomas detected in cKO mice expressed some levels of Runx1, which we attributed to incomplete Cre activity. The WT and cKO papillomas showed similar expression levels of differentiation markers (right). (E) A normal HF tissue image is shown at the same exposure next to a tumor image to compare levels. (F) Two SCCs with different levels of differentiation, showing strong and broad Runx1 expression. Bar, 20 μm.
To see if Runx1 is expressed in WT tumor cells at different stages of tumor progression, we examined in detail the expression pattern of Runx1 in papillomas and SCCs and found it abundantly present at all stages analyzed (Fig. 10D, E, and F). Runx1 protein expression, as detected by immunostaining, colocalized in many tumor cells with the epithelial undifferentiated markers K5 and α6-integrin and the bulge marker CD34+ (Fig. 10D and E), in line with a previous report of elevated Runx1 mRNA in mouse papillomas (36). Papillomas found in the Runx1 cKO mouse were likely a result of incomplete K14-Cre recombinase activity, since randomly tested cKO tumors showed detectable Runx1 expression and similar levels of differentiation markers (Fig. 10D and data not shown). The one SCC discovered in the cKO mouse did not show any Runx1 expression (data not shown), indicating that it was either formed in the complete absence of Runx1 (not as a result of incomplete K14-Cre action) or that Runx1 can be turned off at later stages in SCC formation. Of the eight SCCs found in WT mice, seven had high levels of Runx1 protein in cells largely colocalizing with K5 and low Runx1 levels in the suprabasal, more-differentiated cells (Fig. 10F, top). Runx1 remained highly expressed in invading islets of undifferentiated epithelial cells that lost the normal epithelial tissue polarity (Fig. 10F, bottom) and even in those cells that acquired spindle-like morphology, likely upon the epithelium-to-mesenchyme transition (55) (data not shown). The pattern of Runx1 expression in mouse papillomas and SCCs suggested its presence in undifferentiated proliferative cells of the tumors. Along with the apparent resistance of Runx1 cKO skin to chemically induced tumorigenesis, these data support a requirement for Runx1 in skin tumor formation and potentially in tumor cell maintenance or survival.
Characterization of early tumorigenic stages in Runx1 cKO skin.
To begin to address the mechanism by which Runx1 might affect tumor formation in mouse skin we tested (i) if in HFs Runx1 cKO bulge cells proliferated less than WT cells even upon strong promotion with TPA, and (ii) if other skin epithelial compartments might ectopically turn on Runx1 expression and respond differently at the molecular level to TPA treatment.
To check the proliferation rates in cKO and WT HFs in response to TPA, we performed pulse-chase experiments to obtain LRCs by injecting BrdU at PD3, -4, and -5, with a chase for 2 weeks, followed by TPA (no DMBA) treatment for 2 weeks prior to sacrifice to obtain information about the relative frequency of cell divisions in bulge cells during the chase period. We found that cKO mice displayed significantly more BrdU LRCs than control WT bulge cells after but not before TPA treatment (PD31) (Fig. 11A and B). Similar conclusions were obtained when we treated WT and cKO mice with TPA (not DMBA) until anagen and injected BrdU just prior to sacrifice (Fig. 11C). These data showed that Runx1 cKO bulge cells proliferated less in response to TPA stimulation, suggesting a possible link with the defective tumor formation.
FIG. 11.
TPA treatment is not sufficient to overcome the proliferation defect of Runx1 cKO bulge cells. (A) Scheme for generating bulge BrdU LRCs followed by 2 weeks of TPA treatment (top). The frequency of HFs with BrdU LRCs (middle) and average number of BrdU LRCs per positive bulge (bottom) are shown before and after 2 weeks of TPA treatment. (B) Images of HFs from skin sections before (PD21) and after (PD34) TPA treatment and immunostaining for BrdU. Higher retention of BrdU in the bulge at PD34 demonstrated fewer divisions of the LRCs upon TPA treatment. (C) Scheme of short-term BrdU labeling after TPA treatment from PD21 and up to anagen onset (top). cKO bulge cells incorporated less BrdU even upon TPA stimulation, as shown by both the percentage of BrdU+ HFs (middle) as well as the number of BrdU+ cells per positive bulge (bottom).
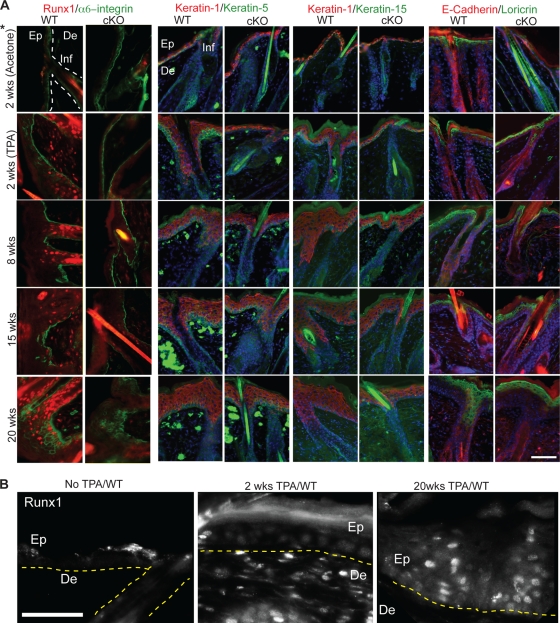
Next we asked if TPA treatment caused epithelial compartments other than HFs to turn on Runx1 expression. To that end we treated skin with DMBA at PD21 and analyzed skin after different weeks of TPA treatment. (Mice analyzed at 2 weeks of TPA treatment were not initiated with DMBA.) We checked for Runx1 expression, the progress of epidermal stratification and differentiation as shown by expression of several markers (previously described [60]), and the level of several well-known tumorigenic markers (1) (Fig. 12 and 13). Strikingly, Runx1 expression, which is normally absent or very low in the interfollicular epidermis and infundibulum (Fig. 12A, top left), became upregulated in random regions of the skin in these epithelial compartments after 2, 8, 15, and 20 weeks of TPA treatment (Fig. 12A, left panels, and B). The strength of the immunoreactivity signal seemed to correlate with the degree of stratification. Runx1 expression could be detected sporadically in skin regions of few cKO mice (due to incomplete Cre activity).
FIG. 12.
Molecular analysis of early stages of carcinogenic treatment. (A) Skin sections from mice treated with DMBA and TPA as indicated (left), immunostained with markers indicated at top. 2 wks, mice treated with TPA only. Note ectopic expression of Runx1 in the infundibulum (Inf) and epidermis (Ep) induced by TPA (left panels) not seen in normal skin (top left). Note the lower degree of epidermal expansion in cKO skin in the right panels. De, dermis. *, samples in the top panels were treated with acetone only as a control, except for the left two panels, which show untreated skin. (B) Skin sections as described for panel A, with Runx1 immunostaining shown in a single color and at a higher magnification. Bars, 75 μm.
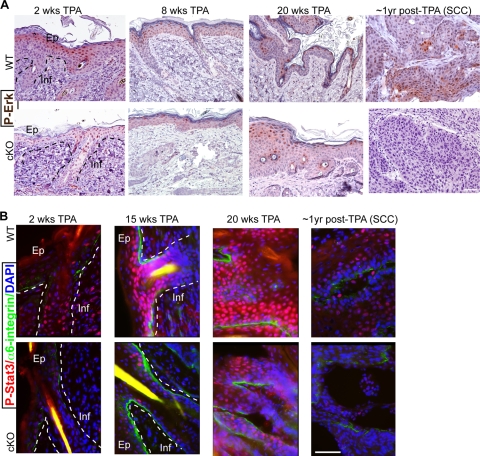
FIG. 13.
Tumorigenic marker analysis of mouse skin at early stages of carcinogenic treatment. (A) Skin sections from mice at early stages of chemically induced skin tumorigenesis sacrificed at the indicated times of treatment were immunohistochemically stained for P-Erk (brown) and counterstained with hematoxylin. Examples of the strongest-stained regions in WT and cKO skin are shown. (B) Mice treated as described for panel A, but using frozen skin sections and immunofluorescnce for P-Stat3. Note the general decrease in P-Stat3 levels in the Runx1 cKO epidermis (Ep) and infundibulum (Inf). Shown are representative pictures of skin sections of comparable epidermal thickness. Bar, 50 μm.
To check if the Runx1 cKO skin responded differently at the molecular level to TPA treatment, we checked for expression of several selected markers previously reported to be regulated during tumorigenesis (2, 60). The differentiation markers keratin 1, 5, and 15 and α6-integrin, Loricrin, and E-cadherin were expressed in both WT and cKO skin and revealed consistently lower stratification and expansion of the epidermis and the infundibulum in cKO skin (Fig. 12A). Similar to Runx1, both P-Erk and phosphorylated Stat3 (P-Stat3) immunostaining, previously shown upregulated in tumorigenesis (1), showed variable expression correlating with the degree of epidermal stratification, although the expression was broader than that of Runx1 and was present in both WT and cKO skin. P-Erk immunohistochemistry showed generally fewer stained cells in cKO samples after 2 weeks of TPA treatment. By 8 weeks of TPA treatment the differences between WT and cKO were not as apparent, and they were noticeable in only two of four cKO mice analyzed after 20 weeks of TPA treatment. P-Stat3 immunofluorescence staining at 2, 8, 15, and 20 weeks of TPA treatment showed P-Stat3 signal in both WT and cKO epidermis and infundibulum in repeat experiments with either different mice or different body regions. Despite such variability, in many samples analyzed the cKO skin showed noticeably lower P-Stat3 that WT skin. This was especially obvious at regions of hyperproliferation, which were more frequent in the WT group but were also visible in regions of similar epidermal expansion (Fig. 13B). P-Stat3 immunoreactivity was more variable, data were inconclusive after 8 weeks of TPA treatment, and the differences were less pronounced by 20 weeks of treatment in regions away from the papillomas (data not shown). The one SCC found in the cKO mouse also showed low levels of P-Stat3 and P-Erk (Fig. 13). We conclude that at early stages of tumorigenesis P-Erk and especially P-Stat3 showed lower levels in Runx1 cKO skin, which correlated with a lower degree of epidermal stratification and infundibular expansion in these mice.
Runx1 shows distinct expression patterns during all major phases of the human hair cycle.
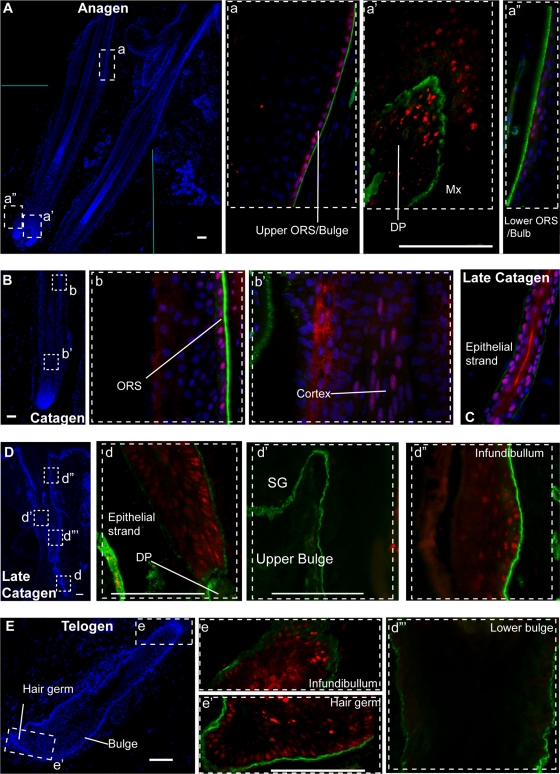
Our data thus far suggest a potential implication of Runx1 in skin cancer likely linked with its role in hair cycle and skin epithelial cell proliferation. To begin to understand if Runx1 might be a potentially interesting therapeutic target in humans, we examined the Runx1 protein expression pattern in human skin. Previously, Runx1 was reported to be expressed at anagen in the outer root sheath of human HFs, including the bulge as well as the hair shaft, and in the inner root sheath (52), similar with the pattern reported in mice in anagen (44, 47). The hair matrix keratinocytes in the lower bulb and the precortical hair matrix reportedly showed no, or very low, Runx1 expression. Here we confirm this Runx1 expression pattern in human anagen HFs using the anti-Runx1 antibody created in the Thomas Jessell laboratory (Fig. 14A) (10). In addition, we showed that Runx1 was expressed in human telogen HFs in epithelial cells at the base of the bulge and in the secondary hair germ, just as we had previously reported in the mouse (Fig. 14E) (44). Moreover, we found detectable Runx1 protein signal in the outer root sheath in catagen (Fig. 14D) and strong signal in the epithelial strand cells of late human catagen HFs (Fig. 14C and D). Runx1 expression in the bulge at late catagen is weak and diffuse. In contrast, we detected strong signal in the upper infundibular area, where Runx1 is normally weak in adult mouse skin, and also in both the outer and the inner root sheath, as previously documented for adult mouse skin (44). We conclude that the overall expression pattern of Runx1 documented here in human skin is similar to the expression found in mouse skin during the hair cycle (44, 47), suggesting conserved functions in mouse and human HFs.
FIG. 14.
Runx1 expression in human hair follicle cell cycle. HFs at anagen (A, a, a′, and a"), catagen (B, b, and b′), late catagen (C, D, d, d′, d″, and d′″) and telogen (E, e, and e′) are shown in images of frozen skin sections fixed and immunostained for Runx1 (red), α6-integrin (green), and DNA DAPI dye (blue). (A, B, D, and E) Low-magnification (×4) skin images showing HF morphology, as indicated by DAPI staining. Boxed areas are shown at a higher magnification (×40) in the corresponding lowercase letter panels as color-combined images. The DNA DAPI image was omitted in some of the panels to reveal Runx1 staining. DP, dermal papilla; Mx, matrix; ORS, outer root sheath; SG, sebaceous gland. Bar, 100 μm.
DISCUSSION
Here we provide the first evidence that Runx1 plays a direct role in adulthood in promoting anagen onset and HFSC proliferation, even though it is not absolutely essential for either process. Runx1 deletion upregulates Cdkn1a (p21), while interfering with Runx1 function impairs HFSC proliferation and cell cycle progression. Moreover, we demonstrated a role for Runx1 in mouse skin tumor formation. Since Runx1 protein expression is very similar in normal human and mouse HFs, we propose that Runx1 acts as a novel skin oncogene in murine and in human skin, which is also recruited for the control of HF cycling, by directly promoting the proliferation of epithelial cells, including HFSCs and their progeny. Manipulating Runx1 activity may, therefore, be a promising novel therapeutic target both in the management of hair growth disorders and in skin tumor prevention.
Runx1 expression in cells of the hair germ and a few cells at the base of the bulge (44, 47), where activated HFSCs begin to proliferate at the onset of anagen (18, 70), is important for HFSC activation (44). Here we have brought evidence that the HF telogen block previously reported (47) is a direct effect of Runx1 loss and is spontaneously overcome in adulthood in the absence of injury. This suggests a compensation mechanism, which removes HFs from quiescence even in the absence of Runx1. Unlike the hair cycle phenotype, the clonogenic ability of keratinocytes is strictly dependent on Runx1 (a hallmark of HFSCs in vitro [13, 43]). This underscores how differences in the environment can reveal distinct aspects of the role Runx1 plays in a context-dependent manner. Apparently, Runx1 is indispensable under certain stress conditions imposed by the culturing conditions or by rapid tumor growth (current study).
Runx1 is known to affect cell survival, proliferation, and differentiation (8, 38) and plays complex roles in cell cycle progression (6, 14, 64, 66). Cdkn1a modulates proliferation of tissue stem cells, such as blood and brain cells, during normal homeostasis and in response to stress, senescence, and aging by blocking cells in the G0/G1 and G1/S phases of the cell cycle (12). Moreover, Cdkn1a (p21) KO mouse newborn keratinocytes showed increased proliferation in vitro, accompanied by an enhanced short-term engraftment ability in vivo (58). Runx1 is known to bind to the promoter of Cdkn1a and directly repress its expression in neural cells (57). Here we show that Runx1 plays a role in setting the bulge cell proliferation in vivo and keratinocyte proliferation rates, G0/G1 exit, and S-phase transition in vitro. Moreover, we demonstrate that this role is mediated at least in part by Cdkn1a, which is as an important player downstream of Runx1 regulation of proliferation in skin epithelial cells.
What is the physiological significance of Runx1 modulation of bulge cell proliferation in epithelial skin? Runx1 modulates, but is dispensable for, HFSC proliferation, and anagen onset (reference 44 and this work). The link of bulge cell proliferation with tumorigenesis has been a recurring theme in the field, and Runx1 is a known cancer gene in blood (8, 39).
Runx1 mutations have been associated with acute myeloid leukemia (8), and recently Runx1 was found to be expressed and play a role in endometrial carcinoma in humans (16). Here we have implicated Runx1 in the repression of Cdkn1a (p21), known for its role in stem cell proliferation (see above), for tumor suppression downstream of p53 (31), and in the skin for blocking tumorigenesis in response to DMBA/TPA treatment (58). Given the dual roles Runx1 and Cdkn1a play in stem cell proliferation and tumorigenesis, it seems possible that these roles might be connected. Moreover, Runx1 interacts with other known cancer genes. It is downstream of p63 (42), another stem cell factor expressed at high levels in skin squamous cell carcinomas that is necessary for efficient tumor formation (68), and in a p53 mutant fibroblast line Runx1 was found to induce oncogenic transformation (67).
Here we found that Runx1 adopts a broad expression pattern in the epithelial skin and in papillomas and SCCs in response to carcinogenic treatment. Moreover, Runx1 was required for efficient tumor skin formation, which was associated with impairment in bulge cell proliferation and epidermal stratification even in the presence of a strong promoter such as TPA. These data correlate with the Runx1 role in control of cell cycle and proliferation in bulge cells and in keratinocytes. It follows that impairment in epithelial cell proliferation upon Runx1 loss may reduce tumorigenesis in mouse skin. Our preliminary data suggest Stat3 and Cdkn1a as potential candidates for the Runx1 mechanism in tumorigenesis, and these avenues will have to be tested more directly in the future. The Stat3 pathway has been shown essential for DMBA/TPA-induced tumorigenesis (33). The putative link of Runx1 and Stat3 in skin tumorigenesis is perhaps not surprising, given that the Runx1 mutant phenotype is strikingly similar to that shown by Stat3 knockout mice (49, 50). The relationship of these two transcription factors in skin tumorigenesis and HF regulation is intriguing and requires further exploration.
The parallel, hair cycle-dependent Runx1 expression patterns in murine and human HFs certainly encourage one to explore, next, the functional role of Runx1 not only in human hair cycle control but also in human skin appendage tumor and basal cell carcinoma development.
Acknowledgments
We thank Elaine Fuchs and Nancy Speck for mice and plasmids, Jim Smith for help with FACS, Thomas Jessell for Runx1 antibody, and Robert Weiss and John Schimenti for critical reading of the manuscript.
Funding for this work was received from NIH NIAMS AR053201 and from NYSTEM C024354.
Footnotes
Published ahead of print on 22 March 2010.
REFERENCES
- 1.Abel, E. L., J. M. Angel, K. Kiguchi, and J. DiGiovanni. 2009. Multi-stage chemical carcinogenesis in mouse skin: fundamentals and applications. Nat. Protoc. 4:1350-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appleford, P. J., and A. Woollard. 2009. RUNX genes find a niche in stem cell biology. J. Cell Biochem. 108:14-21. [DOI] [PubMed] [Google Scholar]
- 3.Barrandon, A. R. a. Y. 2004. Regeneration of epidermins from adult keratinocyte stem cells, p. 763-772. In R. Lanza (ed.), Handbook of stem cells, vol. 2. Elsevier, Inc., Boston, MA. [Google Scholar]
- 4.Barrandon, Y., and H. Green. 1987. Three clonal types of keratinocyte with different capacities for multiplication. Proc. Natl. Acad. Sci. U. S. A. 84:2302-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker, K. A., P. N. Ghule, J. A. Therrien, J. B. Lian, J. L. Stein, A. J. van Wijnen, and G. S. Stein. 2006. Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. J. Cell. Physiol. 209:883-893. [DOI] [PubMed] [Google Scholar]
- 6.Bernardin, F., and A. D. Friedman. 2002. AML1 stimulates G1 to S progression via its transactivation domain. Oncogene 21:3247-3252. [DOI] [PubMed] [Google Scholar]
- 7.Blanpain, C., W. E. Lowry, A. Geoghegan, L. Polak, and E. Fuchs. 2004. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell 118:635-648. [DOI] [PubMed] [Google Scholar]
- 8.Blyth, K., E. R. Cameron, and J. C. Neil. 2005. The RUNX genes: gain or loss of function in cancer. Nat. Rev. Cancer 5:376-387. [DOI] [PubMed] [Google Scholar]
- 9.Brugarolas, J., C. Chandrasekaran, J. I. Gordon, D. Beach, T. Jacks, and G. J. Hannon. 1995. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature 377:552-557. [DOI] [PubMed] [Google Scholar]
- 10.Chen, C. L., D. C. Broom, Y. Liu, J. C. de Nooij, Z. Li, C. Cen, O. A. Samad, T. M. Jessell, C. J. Woolf, and Q. Ma. 2006. Runx1 determines nociceptive sensory neuron phenotype and is required for thermal and neuropathic pain. Neuron 49:365-377. [DOI] [PubMed] [Google Scholar]
- 11.Chen, M. J., T. Yokomizo, B. M. Zeigler, E. Dzierzak, and N. A. Speck. 2009. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature 457:887-891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng, T. 2004. Cell cycle inhibitors in normal and tumor stem cells. Oncogene 23:7256-7266. [DOI] [PubMed] [Google Scholar]
- 13.Claudinot, S., M. Nicolas, H. Oshima, A. Rochat, and Y. Barrandon. 2005. Long-term renewal of hair follicles from clonogenic multipotent stem cells. Proc. Natl. Acad. Sci. U. S. A. 102:14677-14682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coffman, J. A. 2003. Runx transcription factors and the developmental balance between cell proliferation and differentiation. Cell Biol. Int. 27:315-324. [DOI] [PubMed] [Google Scholar]
- 15.Cotsarelis, G. 2006. Epithelial stem cells: a folliculocentric view. J. Invest. Dermatol. 126:1459-1468. [DOI] [PubMed] [Google Scholar]
- 16.Doll, A., M. Gonzalez, M. Abal, M. Llaurado, M. Rigau, E. Colas, M. Monge, J. Xercavins, G. Capella, B. Diaz, A. Gil-Moreno, F. Alameda, and J. Reventos. 2009. An orthotopic endometrial cancer mouse model demonstrates a role for RUNX1 in distant metastasis. Int. J. Cancer 125:257-263. [DOI] [PubMed] [Google Scholar]
- 17.Fuchs, E. 2007. Scratching the surface of skin development. Nature 445:834-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuchs, E. 2009. The tortoise and the hair: slow-cycling cells in the stem cell race. Cell 137:811-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glotzer, D. J., E. Zelzer, and B. R. Olsen. 2008. Impaired skin and hair follicle development in Runx2 deficient mice. Dev. Biol. 315:459-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greco, V., T. Chen, M. Rendl, M. Schober, H. A. Pasolli, N. Stokes, J. Dela Cruz-Racelis, and E. Fuchs. 2009. A two-step mechanism for stem cell activation during hair regeneration. Cell Stem Cell 4:155-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Growney, J. D., H. Shigematsu, Z. Li, B. H. Lee, J. Adelsperger, R. Rowan, D. P. Curley, J. L. Kutok, K. Akashi, I. R. Williams, N. A. Speck, and D. G. Gilliland. 2005. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood 106:494-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayashi, K., W. Natsume, T. Watanabe, N. Abe, N. Iwai, H. Okada, Y. Ito, M. Asano, Y. Iwakura, S. Habu, Y. Takahama, and M. Satake. 2000. Diminution of the AML1 transcription factor function causes differential effects on the fates of CD4 and CD8 single-positive T cells. J. Immunol. 165:6816-6824. [DOI] [PubMed] [Google Scholar]
- 23.Hayashi, S., and A. P. McMahon. 2002. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev. Biol. 244:305-318. [DOI] [PubMed] [Google Scholar]
- 24.Hennings, H., A. B. Glick, D. T. Lowry, L. S. Krsmanovic, L. M. Sly, and S. H. Yuspa. 1993. FVB/N mice: an inbred strain sensitive to the chemical induction of squamous cell carcinomas in the skin. Carcinogenesis 14:2353-2358. [DOI] [PubMed] [Google Scholar]
- 25.Ichikawa, M., T. Asai, T. Saito, S. Seo, I. Yamazaki, T. Yamagata, K. Mitani, S. Chiba, S. Ogawa, M. Kurokawa, and H. Hirai. 2004. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat. Med. 10:299-304. [DOI] [PubMed] [Google Scholar]
- 26.Inoue, K., T. Shiga, and Y. Ito. 2008. Runx transcription factors in neuronal development. Neural Dev. 3:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ito, M., K. Kizawa, K. Hamada, and G. Cotsarelis. 2004. Hair follicle stem cells in the lower bulge form the secondary germ, a biochemically distinct but functionally equivalent progenitor cell population, at the termination of catagen. Differentiation 72:548-557. [DOI] [PubMed] [Google Scholar]
- 28.Ito, M., Y. Liu, Z. Yang, J. Nguyen, F. Liang, R. J. Morris, and G. Cotsarelis. 2005. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat. Med. 11:1351-1354. [DOI] [PubMed] [Google Scholar]
- 29.Jaks, V., N. Barker, M. Kasper, J. H. van Es, H. J. Snippert, H. Clevers, and R. Toftgard. 2008. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat. Genet. 40:1291-1299. [DOI] [PubMed] [Google Scholar]
- 30.Jensen, K. B., C. A. Collins, E. Nascimento, D. W. Tan, M. Frye, S. Itami, and F. M. Watt. 2009. Lrig1 expression defines a distinct multipotent stem cell population in mammalian epidermis. Cell Stem Cell 4:427-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jung, Y. S., Y. Qian, and X. Chen. 25 January 2010, posting date. Examination of the expanding pathways for the regulation of p21 expression and activity. Cell Signal. [Epub ahead of print.] [DOI] [PMC free article] [PubMed]
- 32.Kangsamaksin, T., H. J. Park, C. S. Trempus, and R. J. Morris. 2007. A perspective on murine keratinocyte stem cells as targets of chemically induced skin cancer. Mol. Carcinog. 46:579-584. [DOI] [PubMed] [Google Scholar]
- 33.Kim, D. J., J. M. Angel, S. Sano, and J. DiGiovanni. 2009. Constitutive activation and targeted disruption of signal transducer and activator of transcription 3 (Stat3) in mouse epidermis reveal its critical role in UVB-induced skin carcinogenesis. Oncogene 28:950-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levanon, D., O. Brenner, V. Negreanu, D. Bettoun, E. Woolf, R. Eilam, J. Lotem, U. Gat, F. Otto, N. Speck, and Y. Groner. 2001. Spatial and temporal expression patterns of Runx3 (Aml2) and Runx1 (Aml1) indicate non-redundant functions during mouse embryogenesis. Mech. Dev. 109:413-417. [DOI] [PubMed] [Google Scholar]
- 35.Lorz, C., C. Segrelles, and J. M. Paramio. 2009. On the origin of epidermal cancers. Curr. Mol. Med. 9:353-364. [DOI] [PubMed] [Google Scholar]
- 36.Malanchi, I., H. Peinado, D. Kassen, T. Hussenet, D. Metzger, P. Chambon, M. Huber, D. Hohl, A. Cano, W. Birchmeier, and J. Huelsken. 2008. Cutaneous cancer stem cell maintenance is dependent on beta-catenin signalling. Nature 452:650-653. [DOI] [PubMed] [Google Scholar]
- 37.Massoumi, R., M. Podda, R. Fassler, and R. Paus. 2006. Cylindroma as tumor of hair follicle origin. J. Invest. Dermatol. 126:1182-1184. [DOI] [PubMed] [Google Scholar]
- 38.Mikhail, F. M., K. K. Sinha, Y. Saunthararajah, and G. Nucifora. 2006. Normal and transforming functions of RUNX1: a perspective. J. Cell. Physiol. 207:582-593. [DOI] [PubMed] [Google Scholar]
- 39.Miller, S. J., T. T. Sun, and R. M. Lavker. 1993. Hair follicles, stem cells, and skin cancer. J. Invest. Dermatol. 100:288S-294S. [DOI] [PubMed] [Google Scholar]
- 40.Miller, S. J., Z. G. Wei, C. Wilson, L. Dzubow, T. T. Sun, and R. M. Lavker. 1993. Mouse skin is particularly susceptible to tumor initiation during early anagen of the hair cycle: possible involvement of hair follicle stem cells. J. Invest. Dermatol. 101:591-594. [DOI] [PubMed] [Google Scholar]
- 41.Muller-Rover, S., B. Handjiski, C. van der Veen, S. Eichmuller, K. Foitzik, I. A. McKay, K. S. Stenn, and R. Paus. 2001. A comprehensive guide for the accurate classification of murine hair follicles in distinct hair cycle stages. J. Invest. Dermatol. 117:3-15. [DOI] [PubMed] [Google Scholar]
- 42.Ortt, K., E. Raveh, U. Gat, and S. Sinha. 2008. A chromatin immunoprecipitation screen in mouse keratinocytes reveals Runx1 as a direct transcriptional target of deltaNp63. J. Cell Biochem. 104:1204-1219. [DOI] [PubMed] [Google Scholar]
- 43.Oshima, H., A. Rochat, C. Kedzia, K. Kobayashi, and Y. Barrandon. 2001. Morphogenesis and renewal of hair follicles from adult multipotent stem cells. Cell 104:233-245. [DOI] [PubMed] [Google Scholar]
- 44.Osorio, K. M., S. E. Lee, D. J. McDermitt, S. K. Waghmare, Y. V. Zhang, H. N. Woo, and T. Tumbar. 2008. Runx1 modulates developmental, but not injury-driven, hair follicle stem cell activation. Development 135:1059-1068. [DOI] [PubMed] [Google Scholar]
- 45.Plikus, M. V., J. A. Mayer, D. de la Cruz, R. E. Baker, P. K. Maini, R. Maxson, and C. M. Chuong. 2008. Cyclic dermal BMP signalling regulates stem cell activation during hair regeneration. Nature 451:340-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Putz, G., A. Rosner, I. Nuesslein, N. Schmitz, and F. Buchholz. 2006. AML1 deletion in adult mice causes splenomegaly and lymphomas. Oncogene 25:929-939. [DOI] [PubMed] [Google Scholar]
- 47.Raveh, E., S. Cohen, D. Levanon, V. Negreanu, Y. Groner, and U. Gat. 2006. Dynamic expression of Runx1 in skin affects hair structure. Mech. Dev. 123:842-850. [DOI] [PubMed] [Google Scholar]
- 48.Sakakura, C., Y. Yamaguchi-Iwai, M. Satake, S. C. Bae, A. Takahashi, E. Ogawa, A. Hagiwara, T. Takahashi, A. Murakami, K. Makino, et al. 1994. Growth inhibition and induction of differentiation of t(8;21) acute myeloid leukemia cells by the DNA-binding domain of PEBP2 and the AML1/MTG8(ETO)-specific antisense oligonucleotide. Proc. Natl. Acad. Sci. U. S. A. 91:11723-11727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sano, S., S. Itami, K. Takeda, M. Tarutani, Y. Yamaguchi, H. Miura, K. Yoshikawa, S. Akira, and J. Takeda. 1999. Keratinocyte-specific ablation of Stat3 exhibits impaired skin remodeling, but does not affect skin morphogenesis. EMBO J. 18:4657-4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sano, S., M. Kira, S. Takagi, K. Yoshikawa, J. Takeda, and S. Itami. 2000. Two distinct signaling pathways in hair cycle induction: Stat3-dependent and -independent pathways. Proc. Natl. Acad. Sci. U. S. A. 97:13824-13829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schneider, M. R., R. Schmidt-Ullrich, and R. Paus. 2009. The hair follicle as a dynamic miniorgan. Curr. Biol. 19:R132-R142. [DOI] [PubMed] [Google Scholar]
- 52.Soma, T., Y. Ishimatsu-Tsuji, M. Tajima, and J. Kishimoto. 2006. Runx1 transcription factor is involved in the regulation of KAP5 gene expression in human hair follicles. J. Dermatol. Sci. 41:221-224. [DOI] [PubMed] [Google Scholar]
- 53.Soriano, P. 1999. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 21:70-71. [DOI] [PubMed] [Google Scholar]
- 54.Speck, N. A., and D. G. Gilliland. 2002. Core-binding factors in haematopoiesis and leukaemia. Nat. Rev. Cancer 2:502-513. [DOI] [PubMed] [Google Scholar]
- 55.Stoler, A. B., F. Stenback, and A. Balmain. 1993. The conversion of mouse skin squamous cell carcinomas to spindle cell carcinomas is a recessive event. J. Cell Biol. 122:1103-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taniuchi, I., M. Osato, T. Egawa, M. J. Sunshine, S. C. Bae, T. Komori, Y. Ito, and D. R. Littman. 2002. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell 111:621-633. [DOI] [PubMed] [Google Scholar]
- 57.Theriault, F. M., H. N. Nuthall, Z. Dong, R. Lo, F. Barnabe-Heider, F. D. Miller, and S. Stifani. 2005. Role for Runx1 in the proliferation and neuronal differentiation of selected progenitor cells in the mammalian nervous system. J. Neurosci. 25:2050-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Topley, G. I., R. Okuyama, J. G. Gonzales, C. Conti, and G. P. Dotto. 1999. p21WAF1/Cip1 functions as a suppressor of malignant skin tumor formation and a determinant of keratinocyte stem-cell potential. Proc. Natl. Acad. Sci. U. S. A. 96:9089-9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trempus, C. S., R. J. Morris, C. D. Bortner, G. Cotsarelis, R. S. Faircloth, J. M. Reece, and R. W. Tennant. 2003. Enrichment for living murine keratinocytes from the hair follicle bulge with the cell surface marker CD34. J. Invest. Dermatol. 120:501-511. [DOI] [PubMed] [Google Scholar]
- 60.Tsakiridis, T., J. C. Cutz, G. Singh, H. Hirte, G. Okawara, T. Corbett, R. Sur, W. Cai, T. Whelan, and J. R. Wright. 2008. Association of phosphorylated epidermal growth factor receptor with survival in patients with locally advanced non-small cell lung cancer treated with radiotherapy. J. Thorac. Oncol. 3:716-722. [DOI] [PubMed] [Google Scholar]
- 61.Tumbar, T. 2006. Epithelial skin stem cells. Methods Enzymol. 419:73-99. [DOI] [PubMed] [Google Scholar]
- 62.Vasioukhin, V., L. Degenstein, B. Wise, and E. Fuchs. 1999. The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc. Natl. Acad. Sci. U. S. A. 96:8551-8556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vassar, R., M. Rosenberg, S. Ross, A. Tyner, and E. Fuchs. 1989. Tissue-specific and differentiation-specific expression of a human K14 keratin gene in transgenic mice. Proc. Natl. Acad. Sci. U. S. A. 86:1563-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang, S., Y. Zhang, J. Soosairajah, and A. S. Kraft. 2006. Regulation of RUNX1/AML1 during the G2/M transition. Leuk. Res. 31:839-851. [DOI] [PubMed] [Google Scholar]
- 65.Wang, X., C. Blagden, J. Fan, S. J. Nowak, I. Taniuchi, D. R. Littman, and S. J. Burden. 2005. Runx1 prevents wasting, myofibrillar disorganization, and autophagy of skeletal muscle. Genes Dev. 19:1715-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wolyniec, K., S. Wotton, A. Kilbey, A. Jenkins, A. Terry, G. Peters, C. Stocking, E. Cameron, and J. C. Neil. 2009. RUNX1 and its fusion oncoprotein derivative, RUNX1-ETO, induce senescence-like growth arrest independently of replicative stress. Oncogene 28:2502-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wotton, S. F., K. Blyth, A. Kilbey, A. Jenkins, A. Terry, F. Bernardin-Fried, A. D. Friedman, E. W. Baxter, J. C. Neil, and E. R. Cameron. 2004. RUNX1 transformation of primary embryonic fibroblasts is revealed in the absence of p53. Oncogene 23:5476-5486. [DOI] [PubMed] [Google Scholar]
- 68.Wrone, D. A., S. Yoo, L. K. Chipps, and R. L. Moy. 2004. The expression of p63 in actinic keratoses, seborrheic keratosis, and cutaneous squamous cell carcinomas. Dermatol. Surg. 30:1299-1302. [DOI] [PubMed] [Google Scholar]
- 69.Zagami, C. J., M. Zusso, and S. Stifani. 2009. Runx transcription factors: lineage-specific regulators of neuronal precursor cell proliferation and mitotic neuron subtype development. J. Cell Biochem. 107:1063-1072. [DOI] [PubMed] [Google Scholar]
- 70.Zhang, Y. V., J. Cheong, N. Ciapurin, D. J. McDermitt, and T. Tumbar. 2009. Distinct self-renewal and differentiation phases in the niche of infrequently dividing hair follicle stem cells. Cell Stem Cell 5:267-278. [DOI] [PMC free article] [PubMed] [Google Scholar]