Abstract
The essential osteoblast-related transcription factor Runx2 and the female steroid hormone estrogen are known to play pivotal roles in bone homeostasis; however, the functional interaction between Runx2- and estrogen-mediated signaling in skeletal tissues is minimally understood. Here we provide evidence that aromatase (CYP19), a rate-limiting enzyme responsible for estrogen biosynthesis in mammals, is transcriptionally regulated by Runx2. Consistent with the presence of multiple Runx2 binding sites, the binding of Runx2 to the aromatase promoter was demonstrated in vitro and confirmed in vivo by chromatin immunoprecipitation assays. The bone-specific aromatase promoter is activated by Runx2, and endogenous aromatase gene expression is upregulated by Runx2 overexpression, establishing the aromatase gene as a target of Runx2. The biological significance of the Runx2 transcriptional control of the aromatase gene is reflected by the enhanced estrogen biosynthesis in response to Runx2 in cultured cells. Reduced in vivo expression of skeletal aromatase gene and low bone mineral density are evident in Runx2 mutant mice. Collectively, these findings uncover a novel link between Runx2-mediated osteoblastogenic processes and the osteoblast-mediated biosynthesis of estrogen as an osteoprotective steroid hormone.
Runx2 is an essential transcription factor in bone formation during embryogenesis (40). The ablation or C-terminal truncation of Runx2 prevents bone formation (3, 16), and a heterozygous mutation of Runx2 results in cleidocranial dysplasia in both mice and humans (15, 17). Mechanistically, Runx2 positively regulates osteoblast proliferation through epigenetic control (30) and differentiation through the upregulation of bone-related target gene expression (1, 5). Moreover, Runx2 accelerates chondrocyte differentiation in response to the upregulation of Runx2 target genes, which include the Indian hedgehog (Ihh) and type X collagen (Col10A1) genes (32, 55). The control of chondrogenesis by Runx2 is further confirmed by the chondrocyte-specific overexpression of a dominant negative form of Runx2 in transgenic mice (42, 50). Runx2 also plays an important role in estrogen deficiency-mediated bone resorption in adult mice (20). Together, these studies establish Runx2 as a master regulator of bone formation through the modulation of the activity of a cohort of target genes.
Estrogen is essential for the maintenance of bone health in both men and women (41). Estrogen deficiency is associated with increased bone turnover, resulting in reduced bone mineral density (BMD) and increased fracture risk. One of the key roles of estrogen is to modulate osteoclast survival, revealed by a loss of osteoclast apoptosis in osteoclast-specific estrogen receptor alpha (ER-α) conditional knockout mice (24). In addition, estrogen inhibits osteoclast formation through the upregulation of osteoprotegerin production (53). Therefore, the mutation or ablation of genes in the pathway related to estrogen synthesis causes a disruption of bone homeostasis. For example, a mutation of the ER-α gene in males can lead to osteopenia and even osteoporosis (11). Mutations of the aromatase gene result in unfused epiphyses and osteoporosis in males, and aromatase-deficient mice exhibit osteoporosis (11).
The human aromatase gene (CYP19; P450arom) is comprised of a 30-kb coding region containing nine exons and a 93-kb regulatory region containing 10 untranslated exons I (2, 38). The gene is expressed in many tissues, including the ovaries (8), muscle, skin, adipose tissue (18, 19, 54), placenta (28), and bone (25, 26, 33, 34, 43). The unusually large regulatory region contains 10 tissue-specific promoters that are alternatively used in various cell types (2, 38). Each promoter is regulated by a distinct set of regulatory sequences in the DNA and transcription factors that bind to these specific sequences. The promoters specific for the ovary tissues (promoter II), bone tissues (I.4 and I.6), brain (I.f), endothelial cells (I.7), fetal tissues (I.5), and placenta (2a; I.1) are localized in tandem at ∼0, 73, 1, 33, 36, 43, 78, and 93 kb upstream of the ATG translational start site in exon II. Previous findings suggested that the local expression of aromatase may regulate estrogen production and perhaps affect local bone homeostasis. The majority of aromatase transcripts in bone tissues contain exon 1.4 and exon 1.6 in the 5′ untranslated region (5′-UTR) (35, 52); however, there is little information on the regulation of the aromatase gene in bone.
Recent studies revealed that Runx2 is involved in the estrogen pathway through interactions with ER-α (21) or as a downstream target gene of estrogen or SERM (45, 48). Furthermore, Runx2 was recently shown to control the expression of GPR30/GPER, which represents a nongenomic cell surface receptor for estrogen that is essential for osteoblast proliferation in cell culture (47). Although accumulating evidence implicates a functional association of Runx2 with estrogen signaling (13, 21, 45, 48), Runx2 has not been directly shown to control estrogen biosynthesis. Aromatase is a rate-limiting enzyme in the conversion of testosterone into estrogen and plays a pivotal role in estrogen synthesis. Hence, in the present study, we addressed a possible functional link between aromatase and Runx2. We found that Runx2 increases aromatase gene expression, establishing the functional involvement of Runx2 in the estrogen biosynthesis pathway.
MATERIALS AND METHODS
Cell culture.
Human mesenchymal stem cells (hMSC) and media (mesenchymal stem cell growth medium [MSCGM]) were purchased from Lonza (Walkersville, MD). HOS and MG63 cells (human osteosarcoma cell lines), HCS-2/8 cells (human chondrocyte cell line), primary cultured human chondrocytes, and HeLa cells were cultured and maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS; Gibco), 100 U/ml penicillin, and 100 μg/ml streptomycin at 37°C in a humidified atmosphere containing 5% CO2. CHO cells were cultured in α-modified minimum essential medium supplemented with 10% FBS.
Total RNA, mRNA isolation, and RT-PCR analysis.
Total RNA was isolated from cells by using TRIzol reagent (Invitrogen, Carlsbad, CA) and mRNA was extracted by using an Oligotex mRNA Midi kit (Qiagen, Hilden, Germany) according to the manufacturers' instructions. Reverse transcriptase PCR (RT-PCR) was performed by using Superscript II RT (Invitrogen) according to the manufacturer's instructions. The PCR conditions were 94°C for 10 min, followed by 35 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 30 s, with a final extension step at 72°C for 10 min. Primers used for aromatase transcripts I.4 and I.6 in bone cells were as follows: exon I.4-F (5′-GAC CAA CTG GAG CCT GAC AG-3′), exon I.6-F (5′-CAC AGC AGA ACC AGC ACA TCA-3′), and exon 2-R (5′-GTG CCC TCA TAA TTC CAC AC-3′).
Real-time RT-PCR analysis.
Real-time PCR was performed for the relative quantification of aromatase mRNA levels by using SYBR green PCR master mix (Applied Biosystems, Foster City, CA) and primers designed by using PrimerExpress software (Applied Biosystems). Total RNA (3 μg) was reverse transcribed, and 200 ng of cDNA was used as a template for each PCR. SYBR green PCR master mix (25 μl) and 1 μl of specific primers (10 pmol) were combined with template and water in a total volume of 50 μl. A negative control with no template was also included in each assay. PCR was performed at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Primers for the human aromatase gene were 5′-TCA CTG GCC TTT TTC TCT TGG T-3′ and 5′-GGG TCC AAT TCC CAT GCA-3′, primers for the mouse aromatase gene were 5′-AAA TGC TGA ACC CCA TGC AG-3′ and 5′-AAT CAG GAG AAG GAG GCC CAT-3′, and primers for the GAPDH (glyceraldehyde-3-phosphate dehydrogenase) gene were 5′-TGG GCT ACA CTG AGC ACC AG-3′ and 5′-GGG TGT CGC TGT TGA AGT CA-3′.
Construction of Luc reporter genes.
To clone the human aromatase gene I.4 and I.6 promoters, PCR products (0.74 kb/0.14 kb and 1.0 kb, respectively) were amplified from human blood genomic DNA by using the following primers. Forward primers for the 0.74-kb and 0.14-kb I.4 promoters were KpnI-P1 (5′-ACC GGT ACC CTC TGG TCA GAT ATT TTG ATC ATG-3′) and KpnI-P2 (5′-ACC GGT ACC TTA TTG GAA AGA TGC ACA TTG AAT-3′), respectively, used with common reverse primer XhoI-P2 (5′-ATT GCT CGA GTT TCC CAA TCT AGT TTC TAG ATT T-3′). Primers for the I.6 promoter were KpnI-P1.6F (5′-AGG TAC CAA CAG GTT TTT TAA AAG TGC TTG C-3′) and XhoI-P1.6R (5′-ATT GCT CGA GTA GGT AGT CTG TAG CTG AAG-3′). The PCR products were subcloned into the TA cloning vector (Invitrogen) and subsequently inserted into the pGL3 Luc reporter vector (Promega, Madison, WI). Luciferase (Luc) reporter vectors containing promoters I.4 and I.6 were named pI.4-741-Luc/pI.4-137-Luc and pI.6-1023-Luc, respectively.
We performed site-directed mutagenesis of the Runx2 binding site (RBS) in the pI.4-741-Luc and pI.6-1023-Luc vectors by using the QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). Primers for the site-directed mutagenesis of pI.4-741-Luc were 5′-AAA TAT GTa cTT CAT GCT GGA ATG CTG GAC ATA gtA CAT TAT T-3′ and 5′-AAT AAT GTa cTA TGT CCA GCA TTC CAG CAT GAA gtA CAT ATT T-3′ (lowercase indicates mutation sites in the Runx2 binding site). Primers used for the site-directed mutagenesis of pI.6-1023-Luc were 5′-TTA AAA AAA AAA CAA gtA CAA AAC TGC TCA ACA T-3′ and 5′-ATG TTG AGC AGT TTT GTa cTT GTT TTT TTT TTA A-3′. The resulting plasmids, pI.4-741-Rm-Luc and pI.6-1023-Rm-Luc, contained pI.4-741-Luc with two RBS mutations and pI.6-1023-Luc with one RBS mutation, respectively. The DNA sequences of all cloned constructs were confirmed by sequencing.
Cell transfection and Luc reporter gene assay.
HeLa and CHO cells in six-well plates were plated at a density of 4 × 105 cells/well for transfection experiments. Cells were transfected with a total of 2 μg DNA, including 0.8 μg of the aromatase promoter I.4 gene-Luc reporter construct, 0.2 μg pSV-β-galactosidase, and various amounts of pcDNA3.1 and pcDNA3.1-Runx2 plasmids by using Lipofectamine 2000 (Invitrogen), and were grown for 16 h. The ER-α expression vector (kindly provided by K. O. Han, Kwandong University, Seoul, South Korea) was cotransfected with the reporter vector containing six copies of the RBS into CHO cells. pSV-β-galactosidase plasmids were included as an internal control for transfection efficiency. All transfections were performed in triplicate. At 16 h or 24 h after transfection, cells were washed twice with phosphate-buffered saline (PBS) (pH 7.4) and harvested with lysis buffer (Promega). Luc activity was determined with the luciferase assay system (Promega) and measured with a luminometer (AutoLumat LB953; EG & Gberthold, Germany). The ratio of Luc activity relative to β-galactosidase activity and the total protein content in the lysates were used to normalize for transfection efficiency. Each transfection was conducted in triplicate, and the results are presented as means ± standard deviations (SD).
EMSA.
The human osteosarcoma cell line HOS was maintained in DMEM (Gibco) containing 10% FBS. The methods used for nuclear extraction preparation and electrophoretic mobility shift analysis (EMSA) were described previously (14). Sequences of the wild-type and mutant oligonucleotides used are as follows: 5′-GGA AAT ATG TGG TTC ATG-3′ and 5′-CAG CAT GAA CCA CAT ATT-3′, respectively, for 1.4-Runx2-1; 5′-GGA CAT ACC ACA TTA TTG GAA AGA-3′ and 5′-GCA TCT TTC CAA TAA TGT GGT ATG-3′, respectively, for 1.4-Runx2-2; 5′-GGA AAT ATG Tac TT C ATG-3′ and 5′-CAG CAT GAA gtA CAT ATT-3′, respectively, for 1.4-Runx2-1-mut; and 5′-GGA CAT Agt ACA T TA TTG GAA AGA-3′ and 5′-GCA TCT TTC CAA TAA TGT acT ATG-3′, respectively, for 1.4-Runx2-2-mut. Probes were end labeled with [α-32P]dCTP by use of the Megaprime DNA labeling system (GE Healthcare UK Ltd., Buckinghamshire, United Kingdom). Nuclear extract (15 μg) was incubated with 1× gel shift binding buffer containing 15 mM Tris (pH 7.5), 1 mM MgCl2, 0.5 mM EDTA, 0.5 mM dithiothreitol, 50 mM NaCl, 10% glycerol, and 1 μg poly(dI-dC) for 30 min at 4°C. Competition was performed with a 100-fold molar excess of unlabeled DNA probe. Reaction mixtures were separated on 6% nondenaturing polyacrylamide gels in 0.5× Tris-borate-EDTA (TBE) buffer (50 mM Tris-HCl [pH 8.5], 50 mM borate, and 1 mM EDTA). For supershift assays, the nuclear extract was pretreated with mouse monoclonal Runx2 antiserum (30) at room temperature for 30 min before the addition of the labeled probe.
ChIP analysis.
Chromatin immunoprecipitation (ChIP) assays were performed with a human osteosarcoma cell line, HOS. HOS cells (2 × 106 cells) were exposed to 1% formaldehyde in DMEM for 10 min at 37°C. Glycine was added to 0.125 M, and the cultures were incubated for 2 min. The cells were collected in cold PBS, pelleted by centrifugation, and incubated for 10 min on ice in 0.3 ml SDS lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl [pH 8.1]). Lysed samples were sonicated at 30% power with a Sonic Dismembrator model 500 sonicator (Fisher Scientific Inc., Pittsburgh, PA) for 12 10-s pulses at 4°C. The reaction mixtures were centrifuged for 10 min at 4°C to remove debris and stored at −80°C. A 200-μl aliquot was diluted with 1.8 ml of buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl [pH 8.1], 167 mM NaCl) and precleared with 80 μl salmon sperm DNA-protein A-Sepharose for 2 h at 4°C. After the removal of the Sepharose beads by centrifugation, 2 μl anti-Runx2, 2 μl anti-hemagglutinin (HA) (Covance Inc., CA) (unrelated antibody), 5 μg normal mouse IgG (Santa Cruz Biotechnology, Santa Cruz, CA), or no antibody was added to the supernatant, and the reaction mixture was incubated overnight at 4°C. Antibody-protein-DNA complexes were isolated by incubation with 45 μl of blocked protein A-Sepharose for 2 h at 4°C. After extensive washing, bound DNA fragments were eluted and analyzed by PCR. Cycling parameters were 1 cycle of 95°C for 10 min and 28 cycles of 95°C for 30 s, 56°C for 30 s, and 72°C for 30 s. The primers used for the amplification of the aromatase promoter were 5′-CAA AGT GCT TGT CCC CTC A-3′ and 5′-TGT CGG TCA TTC AAT GTG CA-3′. The negative-control primers used for the amplification of a ∼3.7-kb region of aromatase promoter I.4 were 5′-TAA CAA CAG CAG AAG CAG CA-3′ and 5′-GCT GAG CAG AGA ACA AGC A-3. Amplified products were electrophoresed through a 2% agarose gel and visualized by ethidium bromide staining.
Adenovirus infection.
Adenovirus particles expressing Runx2 were generated by using the adEasy Adenovector system (Qbiogene, Carlsbad, CA) (9, 31). HOS and MG63 cells in six-well plates were plated at a density of 1.2 × 105 to 2 × 105 cells/well. One day after seeding, cells were infected with Runx2-expressing adenovirus in DMEM with 1% FBS for 4 h and then cultured for 24, 36, and 60 h in DMEM with 10% FBS.
Hormone assays.
The concentration of estrogen and progesterone in the culture medium was measured by a combined method between an enzyme immunoassay sandwich and a final fluorescent detection called an enzyme-linked fluorescent assay (ELFA). Concentrations of 17-β-estradiol (β-E2) and progesterone in the culture medium (0.2 ml) were measured with a Vidas instrument (bioMérieux Inc., Durham, NC) using Vidas estradiol II and a Vidas progesterone kit (bioMérieux Inc.) according to the manufacturer's protocols.
Western blot analysis.
Whole cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris-HCl [pH 7.4], 0.15 M NaCl, 0.5% SDS, 1% NP-40, 1% Na-deoxycholate, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 μg/ml pepstatin, and 1 μg/ml leupeptin), and the protein concentration was determined by using the Bradford protein assay kit (Bio-Rad, CA). Proteins were separated on 10% SDS-polyacrylamide gels and transferred onto polyvinylidene difluoride (PVDF) membranes. Blots were blocked with 5% skim-milk powder in Tris-buffered saline (TBS) (20 mM Tris-HCl, 137 mM NaCl [pH 7.6]) containing 0.1% Tween 20 (TBS-T buffer) for 1 h at room temperature. Western blot analyses were performed with anti-Runx2 (30), antiaromatase (Serotec, Oxford, United Kingdom, or Acris Antibodies GmbH, Hiddenhausen, Germany), anti-MMP13 (Calbiochem, La Jolla, CA), and anti-α-tubulin (Santa Cruz Biotechnology) antibodies. Primary antibodies were added in TBS-T buffer at a 1:1,000 dilution and incubated for 90 min at room temperature prior to incubation with horseradish peroxidase (HRP)-conjugated secondary antibodies (1:5,000 dilution; Santa Cruz Biotechnology) for 1 h at room temperature. Proteins were detected by using an enhanced chemiluminescence detection system (GE Healthcare).
Animals and genotyping.
Animal care and experiments were carried out in accordance with the Institutional Animal Care and Use Committees of Kyungpook National University. Animals were maintained on a 12-h light-12-h darkness cycle at 22°C to 25°C under specific-pathogen-free (SPF) conditions and fed with standard rodent chow and water ad libitum. Genotyping of Runx2 null and Runx2ΔC mice was performed as previously described (10). For sex determinations, genomic DNA from pups was obtained and analyzed for the presence of Sry by PCR as previously described (10).
Immunohistochemical analysis.
A Vectastain Elite-ABC kit (Vector Laboratories, Inc., CA) was used for immunohistochemistry. The lower limbs of embryonic day 17.5 (E17.5) embryos were fixed in 4% paraformaldehyde in 0.1 M PB (sodium phosphate buffer) for 16 to 42 h at 4°C. Specimens were dehydrated, embedded in paraffin, and cut into sections of a 5-μm thickness. Sections were deparaffinized, quenched in 1% H2O2 in methanol, washed in PBS, and incubated for 2 h at room temperature with diluted normal goat blocking serum prior to overnight incubation at 4°C with anti-aromatase primary antibody (diluted 1:200 in PBS). The slides were washed three times for 10 min with PBS, incubated for 1 h with Vectastain Elite ABC reagent, and washed three times with PBS. Slides were developed by using a Dako (Glostrup, Denmark) Liquid DAB+ substrate-chromogen system, with counterstaining with 1% methyl green in double-distilled water (ddH2O). In control experiments, the antibody was replaced with normal rabbit IgG. The signal was captured by using a Nikon light microscope.
Isolation of bone marrow stromal cells.
Bone marrow stromal cells (BMSC) were collected from 8-week-old female wild-type and Runx2+/− mice (10). The mice were sacrificed, and femora and tibiae were isolated. After the dissection of attached muscle and connective tissue from the bones, metaphyses of femurs and tibias were cut aseptically, and the diaphysis cavities were flushed with DMEM (Gibco) supplemented with 20% FBS (Gibco), 100 U/ml penicillin, and 100 μg/ml streptomycin using a 26-gauge needle. After 24 h, nonadherent cells and debris were removed, and the adherent cells were cultured continuously. BMSC at passage 3 were used for experiments.
BMSC were plated in six-well plates at a density of 1 × 105 cells in DMEM supplemented with 20% FBS. One day after seeding, the culture medium was used for measurements of β-E2 and progesterone as described above.
Micro-computed tomography (micro-CT) analysis.
To determine three-dimensional bone structure in vivo, we performed histomorphometric analysis with the eXplore Locus SP scanner (GE Healthcare) at an 8-μm resolution. All morphometric parameters were determined by using eXplore MicroView, version 2.2 (GE Healthcare). In the femora, scanning regions were confined to the distal metaphysis extending proximally 1.7 mm from the proximal tip of the primary spongiosa.
Establishment of a stable Runx2-expressing cell line.
HeLa cells were transfected with the pcDNA3.1 or pcDNA3.1-Runx2 expression vector by using LipofectAMINE 2000 reagent (Invitrogen), and transfected cells were selected and maintained in culture medium containing G418 (0.4 mg/ml). Selected cells were plated in a six-well plate at a density of 1 × 105 cells in DMEM with 10% FBS. The culture medium was used for measurements of β-E2 and progesterone at 16 h or 4 days after seeding.
Statistical analysis.
Data were expressed as the means ± SD from three or more independent experiments. Differences between two groups were evaluated by using a Student's t test. Results were considered significant at a P value of <0.05.
RESULTS
Runx2 increases aromatase gene expression.
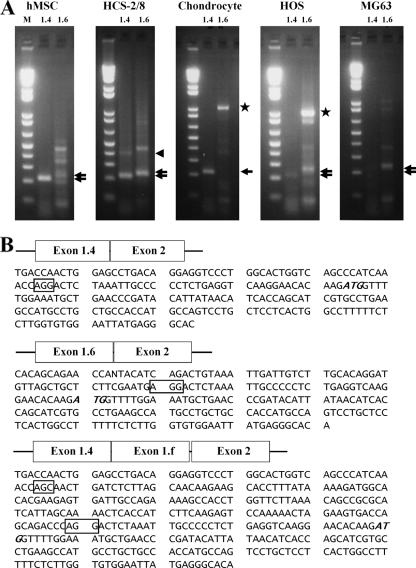
Tissue-specific aromatase gene expression is determined by alternative promoter usage. Among the multiple aromatase gene promoters, I.4 and I.6 have been shown to be bone specific (2, 7, 35, 36, 52). To confirm aromatase promoter utilization in bone cells, we performed RT-PCR analysis of RNA from various bone cell lines. The amplified PCR products were portions of the 5′-UTR derived from promoter I.4 or promoter I.6, and both transcripts were successfully detected based on their expected size (Fig. 1A). Identities of the PCR products were confirmed by sequencing (Fig. 1B). Both promoters I.4 and I.6 were active in human primary mesenchymal stem cells, human chondrocytic HCS-2/8 cells, and the human osteoblast cell lines HOS and MG63 (Fig. 1A). Interestingly, we also identified novel 5′-UTR sequences of aromatase mRNA containing exon 1.4 and exon 1.f joined to the exon 2 open reading frame in chondrocytes (Fig. 1B and 2A). The function of these novel 5′-UTR variants in chondrocytes remains to be established. Our results confirm that aromatase is expressed in bone cells, including chondrocytes, osteoblasts, and mesenchymal stem cells, as previously reported (2, 7, 33, 35, 36, 52).
FIG. 1.
Aromatase gene expression and promoter activity in bone cells. (A) Expression of exons I.4 and I.6 in primary cultured human mesenchymal stem cells (hMSCs), the human chondrocyte HCS-2/8 cell line, primary cultured human chondrocytes (chondrocyte), and human osteosarcoma HOS cells and MG63 cells. RT-PCR was performed by using specific primers derived from the sequence of the coding region from exon I.4 or exon I.6 to exon II of the human aromatase gene. The expected PCR products (220 bp and 240 bp) were identified by 2% agarose gel electrophoresis. The top and bottom arrows indicate exon I.6 and exon I.4, respectively. The 1-kb bands seen in chondrocytes and HOS cells are contaminating genomic DNA (stars). (B) Sequences of transcripts for exons I.4 and 1.f linked to exon II. A novel transcript of the aromatase gene 5′-UTR corresponding to exons I.4 and I.f linked to exon II was observed for the human chondrocyte HCS-2/8 cell line (arrowhead in panel A). Boxes indicate exon-intron junctions, and the ATG (italic type) in each transcript indicates the translation start site. M, 1-kb molecular size ladder.
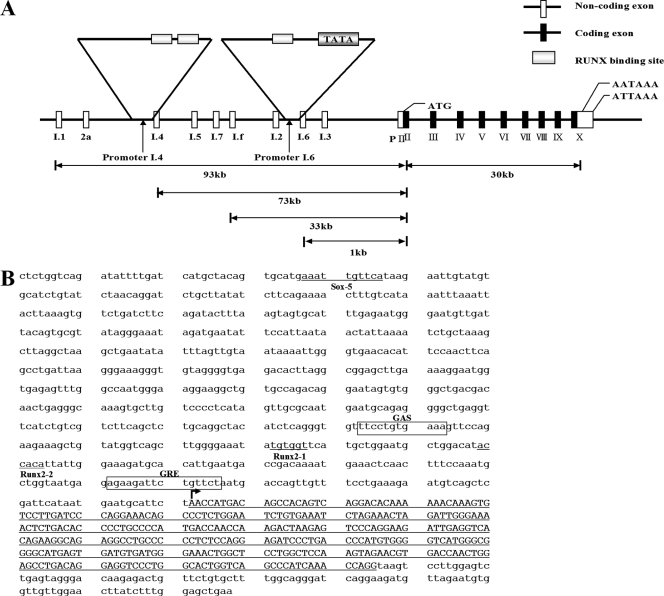
FIG. 2.
Runx2 binding sites in the aromatase promoter. (A) Promoter and coding exons of the aromatase gene. Promoter I.4 and promoter I.6 are indicated by arrows. (B) Sequences of promoter I.4 and Runx2 binding sites. Runx2 binding sites are indicated, from distal (site 1, Runx2-1) to proximal (site 2, Runx2-2). Exon sequences of the promoter I.4-derived transcript are indicated by capital letters.
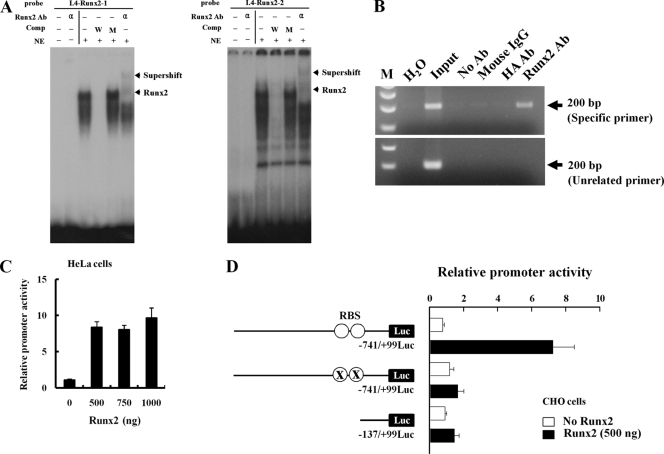
The promoter sequences for the bone-specific I.4 and I.6 aromatase gene transcripts were examined by using the TF search program (http://www.cbrc.jp/research/db/TFSEARCH.html). Putative Runx2 binding sites were identified in both the I.4 and I.6 promoters (Fig. 2A and B). To investigate the aromatase gene as a downstream target gene of Runx2, we focused on the I.4 promoter because this promoter showed strong activity in mesenchymal stem cells, chondrocytes, and osteoblasts (Fig. 1A). First, we determined whether the putative Runx2 binding sites in the I.4 aromatase promoter exhibited Runx2 binding activity. Electrophoretic mobility shift analysis (EMSA) showed Runx2 DNA binding activity for site 1 (TGTGGT) and site 2 (ACCACA) (Fig. 3A). The specificity of Runx2 binding was revealed by supershifting mediated by anti-Runx2 antibody. Furthermore, Runx2 binding activity on the endogenous aromatase gene promoter was confirmed by ChIP analysis (Fig. 3B). There were no positive signals in various controls, including the PCR control (H2O), immunoprecipitation controls, and a 3.7-kb upstream unrelated primer control (Fig. 3B). In transient transfection assays, the forced expression of Runx2 increased pI.4-741-Luc activity in HeLa cells (Fig. 3C). The enhancing effect of Runx2 on pI.4-741-Luc activity was also observed for CHO cells. To determine the contribution of Runx2 binding sites to pI.4-741-Luc activity, vectors pI.4-137-Luc and pI.4-741-Rm-Luc, containing deleted or mutated Runx2 binding sites, respectively, were constructed. As expected, Runx2 increased pI.4-741-Luc activity, but the mutant Luc reporters with deleted or mutated Runx2 binding sites exhibited basal levels of activity (Fig. 3D). We also determined the effect of Runx2 on the activity of promoter I.6. Runx2 increased pI.6-1023-Luc activity, and site-directed mutagenesis in the single Runx2 binding site of the pI.6-1023-Luc reporter abrogated this effect (data not shown). Taken together, these results indicate that Runx2 is a positive regulator of bone-specific aromatase gene expression.
FIG. 3.
Runx2 binds directly to aromatase promoter I.4. (A) EMSA was conducted with probes containing the two wild-type Runx2 motifs. 32P-labeled wild-type oligonucleotides were incubated with nuclear extracts from HOS cells in the presence of a 100-fold molar excess of unlabeled specific oligonucleotides or unlabeled mutant oligonucleotides or in the presence of anti-Runx2 antibody (α). The bottom arrows indicate the positions of the major nuclear protein-DNA complexes, and the top arrows indicate Runx2-supershifted complexes. (B) ChIP analysis was performed with formaldehyde-cross-linked chromatin isolated from HOS cells and antibody against Runx2. PCR amplification was carried out as described in Materials and Methods for a total of 28 cycles, and amplified fragments were analyzed by 2% agarose gel electrophoresis. (C) HeLa cells were cotransfected with the promoter I.4 reporter construct and Runx2 expression vector together with a β-galactosidase (β-Gal) internal control vector. The results were normalized to the protein concentration and β-Gal activity to account for transfection efficiency. (D) Runx2 did not stimulate Luc activity from a promoter with a mutant Runx2 binding site (RBS) or 0.14-kb aromatase promoter I.4 in CHO cells. Data represent the means ± SD from three independent experiments. Ab, antibody; Comp, competitor; NE, nuclear extract; W, wild-type oligonucleotide; M, mutant oligonucleotide; ○, RBS (TGTGGT and ACCACA); X, RBS mutation (TGTacT and AgtACA).
Runx2-mediated upregulation of aromatase gene expression increases estrogen production.
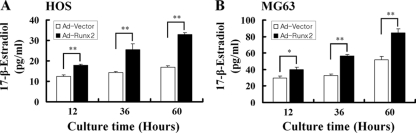
Our previous findings indicated that Runx2 is a positive regulator of aromatase gene expression (Fig. 3). We therefore investigated whether the Runx2-mediated upregulation of aromatase results in increased estrogen production. To test this hypothesis, the bone cell lines HOS and MG63 were infected with adenovirus-producing Runx2 (Fig. 4). The forced expression of Runx2 increased estrogen production in HOS cells (Fig. 4A) and MG63 cells (Fig. 4B); however, the progesterone concentration in the adenovirus-transduced culture medium was not changed (data not shown). These results were recapitulated in Runx2-negative HeLa cells only after making stably expressing Runx2. HeLa cells expressing Runx2 showed a strong correlation between aromatase mRNA and protein and estrogen production (data not shown). Interestingly, bone-type aromatase promoter I.4 and I.6 transcripts were not detected in HeLa cells (data not shown), suggesting that another promoter(s) may be involved in Runx2-mediated aromatase gene expression and the upregulation of estrogen production in Runx2-expressing HeLa cells. Runx2-expressing HeLa cells showed no morphological change (data not shown). These results suggest that the Runx2-mediated upregulation of aromatase gene expression increases estrogen production.
FIG. 4.
Forced expression of Runx2 increases estrogen production in bone cells. The concentration of estrogen in culture medium was measured after infection with adenovirus expressing Runx2 in two bone cell lines, HOS and MG63. The forced expression of Runx2 increased estrogen production in both HOS (A) and MG63 (B) cell lines. Ad, adenovirus. The values represent the means ± SD from three experiments. *, P < 0.05; **, P < 0.005.
Aromatase gene expression is downregulated in Runx2 mutant mice.
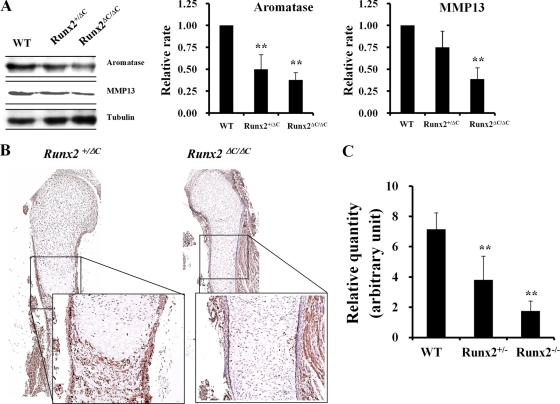
Increased estrogen production in response to Runx2-mediated aromatase gene expression in vitro is consistent with the notion that Runx2 controls steroidogenesis. We further investigated the relationship between Runx2, aromatase, and estrogen biosynthesis in vivo. Our strategy was to determine whether a downregulation of Runx2 decreases aromatase gene expression and estrogen production in intact animals by measuring aromatase expression and estrogen concentrations in Runx2 C-terminally truncated Runx2ΔC/ΔC mice and bone marrow-derived stromal cells (BMSC) of Runx2 heterozygous mice, respectively. Western blot analysis showed decreased levels of expression of aromatase in total proteins isolated from vertebrae and ribs of embryonic day 17.5 (E17.5) embryos of Runx2ΔC/ΔC mice (Fig. 5A). This decrease was comparable to the reduction in expression of MMP13, a well-known target of Runx2 (Fig. 5A) (9). A decreased level of expression of aromatase in the periosteum of Runx2ΔC/ΔC mice by immunohistochemical staining (Fig. 5B) and in the vertebra of E17.5 embryos of Runx2 knockout mice by Western blot analysis (data not shown) was also observed. To confirm aromatase gene expression in skeletal tissues, we isolated skeletal tissues separately from 19 E17.5 embryos. Consistent with the immunohistochemical results, total aromatase mRNA expression was significantly decreased in the limbs of E17.5 Runx2-null embryos (Fig. 5C). Aromatase gene expression levels were similar for male and female embryos from each group, based on sex identification by PCR analysis of the Sry gene (data not shown).
FIG. 5.
Downregulation of aromatase gene expression in the long bone of Runx2ΔC/ΔC mice. (A) Expression of aromatase in vertebrae and ribs from E17.5 wild-type (WT), Runx2+/ΔC, and Runx2ΔC/ΔC mice. Total proteins were isolated from vertebrae and ribs of E17.5 embryos for Western blotting. A Runx2 downstream target gene, the MMP13 gene, was included as a positive control, and tubulin was used as an internal control. The values represent means ± SD from three mice per group. **, P < 0.005. (B) Immunohistochemical staining of tibial sections with antiaromatase antibody in Runx2+/ΔC and Runx2ΔC/ΔC mice at embryonic day 17. 5. (C) Real-time RT-PCR showed a reduced aromatase mRNA level in limbs from E17.5 Runx2 heterozygous or null mice (**, P < 0.005). The numbers of embryos in each group were 5 wild-type, 11 heterozygote, and 3 homozygote embryos. Data are expressed as means ± SD.
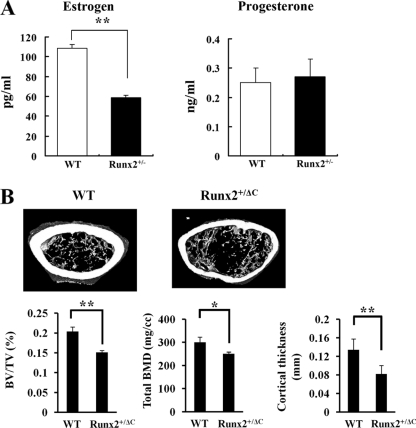
Runx2 is also expressed in BMSC. To explore estrogen production in BMSC from Runx2 heterozygotes, the amount of estrogen and progesterone in culture medium was determined after 1 day of culture. The level of estrogen production was decreased in Runx2 heterozygous BMSC without a change in progesterone production (Fig. 6A), consistent with the results for the forced expression of Runx2 in human osteosarcoma cell lines (Fig. 4). Taken together, the expression of aromatase shows a strong correlation with the production of estrogen, indicating that Runx2 functions as a positive regulator of aromatase in Runx2-expressing tissues.
FIG. 6.
Estrogen production in BMSC and micro-CT analysis of Runx2 heterozygotes. (A) Primary cultured BMSC were obtained from 8-week-old wild-type (WT) and Runx2+/− mice. Estrogen and progesterone concentrations were measured by ELFA in 1-day-accumulated medium as described in Materials and Methods. The values represent means ± SD from three experiments. **, P < 0.005. (B) Five-month-old Runx2 C-terminally truncated heterozygotes showed decreased trabecular bone volume, cortical bone thickness, and total BMD. BV/TV, bone volume per tissue volume. Data are means ± SD from eight mice per group. *, P < 0.05; **, P < 0.005.
Low bone mass in Runx2 heterozygous mice.
Recently, Sjörgren et al. reported that osteoblast-specific aromatase gene expression resulted in high bone mass (39), and those authors attributed this finding to the upregulation of osteoprotegerin production in aromatase-transgenic mice. Based on our in vitro and in vivo results, it seems likely that Runx2 heterozygotes with reduced aromatase gene expression levels in bone tissue would have low bone mass. To prove this hypothesis, we determined various bone parameters by micro-CT analysis. Compared with wild-type mice, Runx2 C-terminally truncated heterozygotes showed lower values with regard to trabecular bone volume, cortical bone thickness, and total bone mineral density (BMD) (Fig. 6B).
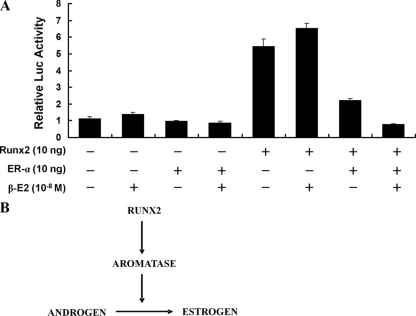
We next determined the role of ER-α in the Runx2-mediated transactivation function on downstream target genes in the presence or absence of estrogen. As shown in Fig. 7A, ER-α inhibited the Runx2 transactivation function, and this inhibition was enhanced in the presence of estrogen. This result suggests a negative regulatory loop in the Runx2-aromatase-estrogen axis. Our data indicate that the local activity of aromatase in bone tissue is important for the maintenance of bone homeostasis and that Runx2 is an upstream positive regulator for aromatase gene expression in bone tissues (Fig. 7B).
FIG. 7.
ER-α plus estrogen inhibit Runx2 transactivation function. (A) Six-Runx2-binding-site Luc reporter and Runx2 expression vectors were cotransfected with an ER-α expression vector into CHO cells, and relative Luc activity was measured at 24 h after transfection. The expression of ER-α decreased Runx2 transactivation function, and treatment with 17-β-estradiol (β-E2) enhanced this effect. Data represent the means ± SD from three independent experiments. (B) Relationship between Runx2 and the estrogen pathway through aromatase gene expression.
DISCUSSION
In this study, we demonstrate that Runx2 stimulates aromatase gene expression in skeletal tissues. The aromatase gene was identified as a Runx2 target gene by four independent criteria. First, Runx2 interacts directly with the aromatase gene promoter, as confirmed by EMSA and ChIP assays. Second, the transient transfection of Runx2 increases aromatase promoter activity. Third, the estrogen production level is elevated in bone cell lines after the forced expression of Runx2 but is decreased in BMSC of Runx2 heterozygotes. Fourth, Runx2-deficient mice exhibited decreased aromatase expression levels in bone tissues. Together, these results indicate that the aromatase gene is a downstream target gene of Runx2 and that Runx2 functions as a positive regulator for aromatase. We also demonstrated that mice heterozygous for a defective Runx2 protein lacking its C terminus exhibit a low bone mineral density by micro-CT analysis compared with wild-type mice. These results suggest that the local production of estrogen may contribute to the maintenance of bone homeostasis.
Aromatase catalyzes the conversion of C19 steroids to estrogen and is encoded by a single gene (CYP19) in the human and mouse genomes (4, 37, 38). Mechanisms controlling aromatase gene expression are complicated by the existence of multiple tissue-specific promoters (2). Aromatase exons I.4 and I.6 were previously reported to be expressed in bone cells, and strong aromatase promoter activity in several bone cells was observed in this study. Dexamethasone (25), oncostatin M, forskolin (51, 52), interleukin-1β (IL-1β) (23, 34), IL-11, tumor necrosis factor alpha (TNF-α), transforming growth factor β (TGF-β) (6, 36), and 1,25-dihydroxyvitamin D3 (44) are known upstream regulators of aromatase gene expression. Runx2 is a master regulator of osteoblast differentiation. Consistent with its role in the physiological control of osteoblastogenesis, Runx2 anabolically modulates bone formation and controls transcriptional signaling pathways that are linked to glucocorticoids and vitamin D3 (27, 29). In this study, we demonstrate that Runx2 is also a strong positive regulator of aromatase gene expression in osteoblastic cells. The Runx2-mediated increase in cellular estrogen production through the upregulation of bone-specific aromatase gene expression may contribute to the maintenance of bone mineral density in conjunction with endocrine effects. Furthermore, the possibility arises that the exogenous expression of Runx2 may perhaps contribute to the control of steroidogenesis during breast cancer tumorigenesis and metastasis (31).
Two reports support the linkage of Runx2 with steroid hormone-mediated bone homeostasis; in one report, high doses of estrogen were administered (12), and in the other report, bone marrow ablation was performed for animals with reduced levels of Runx2 (49). Those two studies showed that Runx2 heterozygotes are less responsive to estrogen and exhibit delayed healing. The observed results may be due to a compromised relationship between Runx2 and an estrogen biosynthesis pathway or a modified regulatory circuit involving Runx2. The precise relationship between estrogen levels and bone phenotypic properties in Runx2 heterozygotes requires further confirmation. Interestingly, Runx2 dominant negative transgenic mice show low bone turnover and low bone mass (20). In our study, Runx2ΔC/+ C-terminally truncated mice also showed a low bone mass. High bone turnover mediated by an estrogen deficiency requires intact Runx2 expression in bone tissues (20). Moreover, osteoblast-specific aromatase gene expression in transgenic mice results in high bone mass (39). Together, these findings suggest that the fidelity of Runx2 function is important for aromatase-estrogen-mediated bone homeostasis.
While these studies were in progress, it was revealed that osteoblasts produce endogenous estrogen receptor agonists (22). Moreover, Teplyuk and colleagues showed that Runx2 controls enzymes (e.g., Cyp11a1) that support the production of pregnenolone (46) as well as the expression of the nongenomic receptor for estrogen (Gpr30/Gper), which is necessary for osteoblast proliferation in cell culture (47). Our study shows that Runx2 increases the expression of aromatase, the key enzyme for the conversion of androgen to estrogen. Together, these findings strongly suggest that Runx2 expression in osteoblasts renders these cells competent to produce functional anabolic steroids that stimulate osteoblast function. More interestingly, the local production of estrogen in bone may be related to the Runx2-mediated transactivation function because ER-α interacts directly with Runx2 and modulates its transcriptional function in the presence of estrogen (13). Results from this and previous studies indicate a Runx2-aromatase-estrogen axis that may be controlled by negative- and positive-feedback mechanisms.
In summary, we conclude from our findings that the gene for aromatase, the key enzyme for estrogen synthesis from androgen, is a downstream target gene of Runx2 and a component of a physiological regulatory network between Runx2 and the estrogen pathway that may contribute to skeletal development and bone homeostasis.
Acknowledgments
This study was supported by grants from the Korea Health 21 R&D Project (Ministry of Health, Welfare, and Family Affairs, Republic of Korea, grants A010252 and A030003); the Ministry of Education, Science and Technology (the Regional Core Research Program); and the Brain Korea 21 Project in 2010.
Footnotes
Published ahead of print on 15 March 2010.
REFERENCES
- 1.Banerjee, C., L. R. McCabe, J. Y. Choi, S. W. Hiebert, J. L. Stein, G. S. Stein, and J. B. Lian. 1997. Runt homology domain proteins in osteoblast differentiation: AML3/CBFA1 is a major component of a bone-specific complex. J. Cell. Biochem. 66:1-8. [DOI] [PubMed] [Google Scholar]
- 2.Bulun, S. E., S. Sebastian, K. Takayama, T. Suzuki, H. Sasano, and M. Shozu. 2003. The human CYP19 (aromatase P450) gene: update on physiologic roles and genomic organization of promoters. J. Steroid Biochem. Mol. Biol. 86:219-224. [DOI] [PubMed] [Google Scholar]
- 3.Choi, J. Y., J. Pratap, A. Javed, S. K. Zaidi, L. Xing, E. Balint, S. Dalamangas, B. Boyce, A. J. van Wijnen, J. B. Lian, J. L. Stein, S. N. Jones, and G. S. Stein. 2001. Subnuclear targeting of Runx/Cbfa/AML factors is essential for tissue-specific differentiation during embryonic development. Proc. Natl. Acad. Sci. U. S. A. 98:8650-8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conley, A., and M. Hinshelwood. 2001. Mammalian aromatases. Reproduction 121:685-695. [DOI] [PubMed] [Google Scholar]
- 5.Ducy, P., R. Zhang, V. Geoffroy, A. L. Ridall, and G. Karsenty. 1997. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89:747-754. [DOI] [PubMed] [Google Scholar]
- 6.Enjuanes, A., N. Garcia-Giralt, A. Supervia, X. Nogues, L. Mellibovsky, J. Carbonell, D. Grinberg, S. Balcells, and A. Diez-Perez. 2003. Regulation of CYP19 gene expression in primary human osteoblasts: effects of vitamin D and other treatments. Eur. J. Endocrinol. 148:519-526. [DOI] [PubMed] [Google Scholar]
- 7.Enjuanes, A., N. Garcia-Giralt, A. Supervia, X. Nogues, S. Ruiz-Gaspa, M. Bustamante, L. Mellibovsky, D. Grinberg, S. Balcells, and A. Diez-Perez. 2005. Functional analysis of the I.3, I.6, pII and I.4 promoters of CYP19 (aromatase) gene in human osteoblasts and their role in vitamin D and dexamethasone stimulation. Eur. J. Endocrinol. 153:981-988. [DOI] [PubMed] [Google Scholar]
- 8.Hinshelwood, M. M., M. E. Smith, B. A. Murry, and C. R. Mendelson. 2000. A 278 bp region just upstream of the human CYP19 (aromatase) gene mediates ovary-specific expression in transgenic mice. Endocrinology 141:2050-2053. [DOI] [PubMed] [Google Scholar]
- 9.Javed, A., G. L. Barnes, J. Pratap, T. Antkowiak, L. C. Gerstenfeld, A. J. van Wijnen, J. L. Stein, J. B. Lian, and G. S. Stein. 2005. Impaired intranuclear trafficking of Runx2 (AML3/CBFA1) transcription factors in breast cancer cells inhibits osteolysis in vivo. Proc. Natl. Acad. Sci. U. S. A. 102:1454-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeong, J. H., J. S. Jin, H. N. Kim, S. M. Kang, J. C. Liu, C. J. Lengner, F. Otto, S. Mundlos, J. L. Stein, A. J. van Wijnen, J. B. Lian, G. S. Stein, and J. Y. Choi. 2008. Expression of Runx2 transcription factor in non-skeletal tissues, sperm and brain. J. Cell. Physiol. 217:511-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones, M. E., W. C. Boon, J. Proietto, and E. R. Simpson. 2006. Of mice and men: the evolving phenotype of aromatase deficiency. Trends Endocrinol. Metab. 17:55-64. [DOI] [PubMed] [Google Scholar]
- 12.Juttner, K. V., and M. J. Perry. 2007. High-dose estrogen-induced osteogenesis is decreased in aged RUNX2(+/−) mice. Bone 41:25-32. [DOI] [PubMed] [Google Scholar]
- 13.Khalid, O., S. K. Baniwal, D. J. Purcell, N. Leclerc, Y. Gabet, M. R. Stallcup, G. A. Coetzee, and B. Frenkel. 2008. Modulation of Runx2 activity by estrogen receptor-alpha: implications for osteoporosis and breast cancer. Endocrinology 149:5984-5995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, H. J., J. H. Kim, S. C. Bae, J. Y. Choi, H. J. Kim, and H. M. Ryoo. 2003. The protein kinase C pathway plays a central role in the fibroblast growth factor-stimulated expression and transactivation activity of Runx2. J. Biol. Chem. 278:319-326. [DOI] [PubMed] [Google Scholar]
- 15.Kim, H. J., S. H. Nam, H. J. Kim, H. S. Park, H. M. Ryoo, S. Y. Kim, T. J. Cho, S. G. Kim, S. C. Bae, I. S. Kim, J. L. Stein, A. J. van Wijnen, G. S. Stein, J. B. Lian, and J. Y. Choi. 2006. Four novel RUNX2 mutations including a splice donor site result in the cleidocranial dysplasia phenotype. J. Cell. Physiol. 207:114-122. [DOI] [PubMed] [Google Scholar]
- 16.Komori, T., H. Yagi, S. Nomura, A. Yamaguchi, K. Sasaki, K. Deguchi, Y. Shimizu, R. T. Bronson, Y. H. Gao, M. Inada, M. Sato, R. Okamoto, Y. Kitamura, S. Yoshiki, and T. Kishimoto. 1997. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89:755-764. [DOI] [PubMed] [Google Scholar]
- 17.Lee, B., K. Thirunavukkarasu, L. Zhou, L. Pastore, A. Baldini, J. Hecht, V. Geoffroy, P. Ducy, and G. Karsenty. 1997. Missense mutations abolishing DNA binding of the osteoblast-specific transcription factor OSF2/CBFA1 in cleidocranial dysplasia. Nat. Genet. 16:307-310. [DOI] [PubMed] [Google Scholar]
- 18.Mahendroo, M. S., G. D. Means, C. R. Mendelson, and E. R. Simpson. 1991. Tissue-specific expression of human P-450AROM. The promoter responsible for expression in adipose tissue is different from that utilized in placenta. J. Biol. Chem. 266:11276-11281. [PubMed] [Google Scholar]
- 19.Mahendroo, M. S., C. R. Mendelson, and E. R. Simpson. 1993. Tissue-specific and hormonally controlled alternative promoters regulate aromatase cytochrome P450 gene expression in human adipose tissue. J. Biol. Chem. 268:19463-19470. [PubMed] [Google Scholar]
- 20.Maruyama, Z., C. A. Yoshida, T. Furuichi, N. Amizuka, M. Ito, R. Fukuyama, T. Miyazaki, H. Kitaura, K. Nakamura, T. Fujita, N. Kanatani, T. Moriishi, K. Yamana, W. Liu, H. Kawaguchi, K. Nakamura, and T. Komori. 2007. Runx2 determines bone maturity and turnover rate in postnatal bone development and is involved in bone loss in estrogen deficiency. Dev. Dyn. 236:1876-1890. [DOI] [PubMed] [Google Scholar]
- 21.McCarthy, T. L., W. Z. Chang, Y. Liu, and M. Centrella. 2003. Runx2 integrates estrogen activity in osteoblasts. J. Biol. Chem. 278:43121-43129. [DOI] [PubMed] [Google Scholar]
- 22.McCarthy, T. L., M. E. Clough, C. M. Gundberg, and M. Centrella. 2008. Expression of an estrogen receptor agonist in differentiating osteoblast cultures. Proc. Natl. Acad. Sci. U. S. A. 105:7022-7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morioka, M., K. Shimodaira, Y. Kuwano, H. Fujikawa, H. Saito, and T. Yanaihara. 2000. Effect of interleukin-1beta on aromatase activity and cell proliferation in human osteoblast-like cells (HOS). Biochem. Biophys. Res. Commun. 268:60-64. [DOI] [PubMed] [Google Scholar]
- 24.Nakamura, T., Y. Imai, T. Matsumoto, S. Sato, K. Takeuchi, K. Igarashi, Y. Harada, Y. Azuma, A. Krust, Y. Yamamoto, H. Nishina, S. Takeda, H. Takayanagi, D. Metzger, J. Kanno, K. Takaoka, T. J. Martin, P. Chambon, and S. Kato. 2007. Estrogen prevents bone loss via estrogen receptor alpha and induction of Fas ligand in osteoclasts. Cell 130:811-823. [DOI] [PubMed] [Google Scholar]
- 25.Nawata, H., S. Tanaka, S. Tanaka, R. Takayanagi, Y. Sakai, T. Yanase, S. Ikuyama, and M. Haji. 1995. Aromatase in bone cell: association with osteoporosis in postmenopausal women. J. Steroid Biochem. Mol. Biol. 53:165-174. [DOI] [PubMed] [Google Scholar]
- 26.Oz, O. K., R. Millsaps, R. Welch, J. Birch, and J. E. Zerwekh. 2001. Expression of aromatase in the human growth plate. J. Mol. Endocrinol. 27:249-253. [DOI] [PubMed] [Google Scholar]
- 27.Paredes, R., G. Arriagada, F. Cruzat, A. Villagra, J. Olate, K. Zaidi, A. van Wijnen, J. B. Lian, G. S. Stein, J. L. Stein, and M. Montecino. 2004. Bone-specific transcription factor Runx2 interacts with the 1alpha,25-dihydroxyvitamin D3 receptor to up-regulate rat osteocalcin gene expression in osteoblastic cells. Mol. Cell. Biol. 24:8847-8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pezzi, V., J. M. Mathis, W. E. Rainey, and B. R. Carr. 2003. Profiling transcript levels for steroidogenic enzymes in fetal tissues. J. Steroid Biochem. Mol. Biol. 87:181-189. [DOI] [PubMed] [Google Scholar]
- 29.Phillips, J. E., C. A. Gersbach, A. M. Wojtowicz, and A. J. Garcia. 2006. Glucocorticoid-induced osteogenesis is negatively regulated by Runx2/Cbfa1 serine phosphorylation. J. Cell Sci. 119:581-591. [DOI] [PubMed] [Google Scholar]
- 30.Pratap, J., M. Galindo, S. K. Zaidi, D. Vradii, B. M. Bhat, J. A. Robinson, J. Y. Choi, T. Komori, J. L. Stein, J. B. Lian, G. S. Stein, and A. J. van Wijnen. 2003. Cell growth regulatory role of Runx2 during proliferative expansion of preosteoblasts. Cancer Res. 63:5357-5362. [PubMed] [Google Scholar]
- 31.Pratap, J., J. B. Lian, A. Javed, G. L. Barnes, A. J. van Wijnen, J. L. Stein, and G. S. Stein. 2006. Regulatory roles of Runx2 in metastatic tumor and cancer cell interactions with bone. Cancer Metastasis Rev. 25:589-600. [DOI] [PubMed] [Google Scholar]
- 32.Pratap, J., J. J. Wixted, T. Gaur, S. K. Zaidi, J. Dobson, K. D. Gokul, S. Hussain, A. J. van Wijnen, J. L. Stein, G. S. Stein, and J. B. Lian. 2008. Runx2 transcriptional activation of Indian Hedgehog and a downstream bone metastatic pathway in breast cancer cells. Cancer Res. 68:7795-7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sasano, H., M. Uzuki, T. Sawai, H. Nagura, G. Matsunaga, O. Kashimoto, and N. Harada. 1997. Aromatase in human bone tissue. J. Bone Miner. Res. 12:1416-1423. [DOI] [PubMed] [Google Scholar]
- 34.Shozu, M., and E. R. Simpson. 1998. Aromatase expression of human osteoblast-like cells. Mol. Cell. Endocrinol. 139:117-129. [DOI] [PubMed] [Google Scholar]
- 35.Shozu, M., Y. Zhao, S. E. Bulun, and E. R. Simpson. 1998. Multiple splicing events involved in regulation of human aromatase expression by a novel promoter, I.6. Endocrinology 139:1610-1617. [DOI] [PubMed] [Google Scholar]
- 36.Shozu, M., Y. Zhao, and E. R. Simpson. 2000. TGF-beta1 stimulates expression of the aromatase (CYP19) gene in human osteoblast-like cells and THP-1 cells. Mol. Cell. Endocrinol. 160:123-133. [DOI] [PubMed] [Google Scholar]
- 37.Simpson, E., G. Rubin, C. Clyne, K. Robertson, L. O'Donnell, S. Davis, and M. Jones. 1999. Local estrogen biosynthesis in males and females. Endocr. Relat. Cancer 6:131-137. [DOI] [PubMed] [Google Scholar]
- 38.Simpson, E. R., C. Clyne, G. Rubin, W. C. Boon, K. Robertson, K. Britt, C. Speed, and M. Jones. 2002. Aromatase—a brief overview. Annu. Rev. Physiol. 64:93-127. [DOI] [PubMed] [Google Scholar]
- 39.Sjörgren, K., M. Lagerquist, S. Moverare-Skrtic, N. Andersson, S. H. Windahl, C. Swanson, S. Mohan, M. Poutanen, and C. Ohlsson. 2009. Elevated aromatase expression in osteoblasts leads to increased bone mass without systemic adverse effects. J. Bone Miner. Res. 24:1263-1270. [DOI] [PubMed] [Google Scholar]
- 40.Stein, G. S., J. B. Lian, A. J. van Wijnen, J. L. Stein, M. Montecino, A. Javed, S. K. Zaidi, D. W. Young, J. Y. Choi, and S. M. Pockwinse. 2004. Runx2 control of organization, assembly and activity of the regulatory machinery for skeletal gene expression. Oncogene 23:4315-4329. [DOI] [PubMed] [Google Scholar]
- 41.Syed, F., and S. Khosla. 2005. Mechanisms of sex steroid effects on bone. Biochem. Biophys. Res. Commun. 328:688-696. [DOI] [PubMed] [Google Scholar]
- 42.Takeda, S., J. P. Bonnamy, M. J. Owen, P. Ducy, and G. Karsenty. 2001. Continuous expression of Cbfa1 in nonhypertrophic chondrocytes uncovers its ability to induce hypertrophic chondrocyte differentiation and partially rescues Cbfa1-deficient mice. Genes Dev. 15:467-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanaka, S., M. Haji, Y. Nishi, T. Yanase, R. Takayanagi, and H. Nawata. 1993. Aromatase activity in human osteoblast-like osteosarcoma cell. Calcif. Tissue Int. 52:107-109. [DOI] [PubMed] [Google Scholar]
- 44.Tanaka, S., M. Haji, R. Takayanagi, S. Tanaka, Y. Sugioka, and H. Nawata. 1996. 1,25-Dihydroxyvitamin D3 enhances the enzymatic activity and expression of the messenger ribonucleic acid for aromatase cytochrome P450 synergistically with dexamethasone depending on the vitamin D receptor level in cultured human osteoblasts. Endocrinology 137:1860-1869. [DOI] [PubMed] [Google Scholar]
- 45.Taranta, A., M. Brama, A. Teti, V. De luca, R. Scandurra, G. Spera, D. Agnusdei, J. D. Termine, and S. Migliaccio. 2002. The selective estrogen receptor modulator raloxifene regulates osteoclast and osteoblast activity in vitro. Bone 30:368-376. [DOI] [PubMed] [Google Scholar]
- 46.Teplyuk, N. M., Y. Zhang, Y. Lou, J. R. Hawse, M. Q. Hassan, V. I. Teplyuk, J. Pratap, M. Galindo, J. L. Stein, G. S. Stein, J. B. Lian, and A. J. van Wijnen. 2009. The osteogenic transcription factor runx2 controls genes involved in sterol/steroid metabolism, including CYP11A1 in osteoblasts. Mol. Endocrinol. 23:849-861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teplyuk, N. M., M. Galindo, V. I. Teplyuk, J. Pratap, D. W. Young, D. Lapointe, A. Javed, J. L. Stein, J. B. Lian, G. S. Stein, and A. J. van Wijnen. 2008. Runx2 regulates G protein-coupled signaling pathways to control growth of osteoblast progenitors. J. Biol. Chem. 283:27585-27597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tou, L., N. Quibria, and J. M. Alexander. 2001. Regulation of human cbfa1 gene transcription in osteoblasts by selective estrogen receptor modulators (SERMs). Mol. Cell. Endocrinol. 183:71-79. [DOI] [PubMed] [Google Scholar]
- 49.Tsuji, K., T. Komori, and M. Noda. 2004. Aged mice require full transcription factor, Runx2/Cbfa1, gene dosage for cancellous bone regeneration after bone marrow ablation. J. Bone Miner. Res. 19:1481-1489. [DOI] [PubMed] [Google Scholar]
- 50.Ueta, C., M. Iwamoto, N. Kanatani, C. Yoshida, Y. Liu, M. Enomoto-Iwamoto, T. Ohmori, H. Enomoto, K. Nakata, K. Takada, K. Kurisu, and T. Komori. 2001. Skeletal malformations caused by overexpression of Cbfa1 or its dominant negative form in chondrocytes. J. Cell Biol. 153:87-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watanabe, M., S. Ohno, and S. Nakajin. 2005. Forskolin and dexamethasone synergistically induce aromatase (CYP19) expression in the human osteoblastic cell line SV-HFO. Eur. J. Endocrinol. 152:619-624. [DOI] [PubMed] [Google Scholar]
- 52.Watanabe, M., E. R. Simpson, N. Pathirage, S. Nakajin, and C. D. Clyne. 2004. Aromatase expression in the human fetal osteoblastic cell line SV-HFO. J. Mol. Endocrinol. 32:533-545. [DOI] [PubMed] [Google Scholar]
- 53.Zallone, A. 2006. Direct and indirect estrogen actions on osteoblasts and osteoclasts. Ann. N. Y. Acad. Sci. 1068:173-179. [DOI] [PubMed] [Google Scholar]
- 54.Zhao, Y., C. R. Mendelson, and E. R. Simpson. 1995. Characterization of the sequences of the human CYP19 (aromatase) gene that mediate regulation by glucocorticoids in adipose stromal cells and fetal hepatocytes. Mol. Endocrinol. 9:340-349. [DOI] [PubMed] [Google Scholar]
- 55.Zheng, Q., G. Zhou, R. Morello, Y. Chen, X. Garcia-Rojas, and B. Lee. 2003. Type X collagen gene regulation by Runx2 contributes directly to its hypertrophic chondrocyte-specific expression in vivo. J. Cell Biol. 162:833-842. [DOI] [PMC free article] [PubMed] [Google Scholar]