Abstract
Combined deficiencies of poly(ADP)ribosyl polymerase 1 (PARP1) and ataxia telangiectasia mutated (ATM) result in synthetic lethality and, in the mouse, early embryonic death. Here, we investigated the genetic requirements for this lethality via analysis of mice deficient for PARP1 and either of two ATM-regulated DNA damage response (DDR) factors: histone H2AX and 53BP1. We found that, like ATM, H2AX is essential for viability in a PARP1-deficient background. In contrast, deficiency for 53BP1 modestly exacerbates phenotypes of growth retardation, genomic instability, and organismal radiosensitivity observed in PARP1-deficient mice. To gain mechanistic insights into these different phenotypes, we examined roles for 53BP1 in the repair of replication-associated double-strand breaks (DSBs) in several cellular contexts. We show that 53BP1 is required for DNA-PKcs-dependent repair of hydroxyurea (HU)-induced DSBs but dispensable for RPA/RAD51-dependent DSB repair in the same setting. Moreover, repair of mitomycin C (MMC)-induced DSBs and sister chromatid exchanges (SCEs), two RAD51-dependent processes, are 53BP1 independent. Overall, our findings define 53BP1 as a main facilitator of nonhomologous end joining (NHEJ) during the S phase of the cell cycle, beyond highly specialized lymphocyte rearrangements. These findings have important implications for our understanding of the mechanisms whereby ATM-regulated DDR prevents human aging and cancer.
DNA double-strand breaks (DSBs) arise constantly in mammalian cells from endogenous and exogenous sources (35). Defective DSB repair leading to cellular senescence or apoptosis, or aberrant repair to form chromosomal rearrangements, has been linked to aging and cancer in humans (22, 31). To prevent these deleterious outcomes, mammalian cells have evolved the DNA damage response (DDR), a network of factors that sense and signal DSBs to promote their repair (29). ataxia telangiectasia mutated (ATM) is a phophoinositide 3-kinase (PI3K)-like kinase that regulates the functions of hundreds of substrates during DDR, including histone H2AX and 53BP1 (41). Ultimately, the DDR restores DNA strand continuity via either of two main DSB repair pathways: homologous recombination (HR), an error-free pathway that operates only during the S/G2 phases (70), or nonhomologous end joining (NHEJ), a versatile but error-prone pathway that operates throughout the cell cycle (37, 44).
Poly(ADP)ribosyl polymerase 1 (PARP1) regulates, among other processes, transcription, cell death, and DNA repair (57). In the last context, PARP1 senses single-strand breaks (SSBs) and promotes their repair via base excision repair (BER) (15). Although PARP1 is not a component of the BER pathway per se, PARP1-deficient cells accumulate SSBs, which become substrates for HR-mediated repair upon replication (10, 21). ATM and other DDR factors may play important roles in DSB recognition and recruitment of the HR machinery in this setting (42). In addition, PARP1 also catalyzes PAR formation at de novo-generated DNA DSBs (28), where it presumably promotes repair via multiple mechanisms, including direct poly(ADP)ribosylation of DSB repair factors (2, 51) and ATM activation (1, 27). Given the complexity of the interactions between PARP1 and the ATM-regulated DDR outlined above, studies that define the hierarchy and physiological relevance of specific interactions are needed. In this regard, mice deficient for PARP1 and ATM die at gastrulation (43), precluding a mechanistic analysis of their interaction.
Mice deficient for H2AX and 53BP1 share overlapping phenotypes with ATM-deficient mice, including growth retardation, radiation sensitivity, and genomic instability (3, 4, 12, 46, 68), suggesting that H2AX and 53BP1 may similarly play nonoverlapping roles with PARP1 during organismal development. In this context, γ-H2AX is known to colocalize with RAD51 at DSBs (55) and to support HR-mediated repair (13, 71), implying that combined H2AX and PARP1 deficiencies might lead to synthetic lethality. Similarly, a role for 53BP1 in the regulation of HR is supported by observations of defective DSB repair during replication (16, 26, 59, 73) and altered frequency of sister chromatid exchanges (SCEs) (63) in 53BP1-deficient cells.
Here, we made use of mouse genetics to further investigate differential functions for H2AX and 53BP1 in DSB repair at PARP1-deficient chromatin. We found that, like ATM, H2AX is essential for viability in a PARP1-deficient background. In contrast, analysis of mice and cells deficient for 53BP1 and/or PARP1 indicated mostly distinct nonsynthetic roles for these two factors in NHEJ- and HR-mediated repair, respectively.
MATERIALS AND METHODS
Mice.
Mice deficient for PARP1 (67), 53BP1 (68), H2AX (4), ATM (6), and DNA-PKcs (25) were previously described. To generate B lymphocytes in a DNA-PKcs-deficient background, DNA-PKcs-deficient mice were bred to mice harboring a preassembled IgH (B1-8-HC) inserted (knocked in) into the endogenous JH locus and an IgL (3-83k-LC) knocked in to the J region of the Ig(κ) locus, as described previously (50, 60). Compound mice were generated by breeding of single mutants; double heterozygous (either PARP1+/−/H2AX+/− or PARP1+/−/53BP1+/−) mice appeared grossly normal in size, development, and fertility and were interbred to generate the corresponding double mutants and controls. Mice 2 to 4 months of age were used for all experiments. All mouse experiments were conducted in accordance with IACUC-approved protocols.
Drugs.
5-Bromo-2′-deoxyuridine (BrdU), mitomycin C (MMC), and hydroxyurea (HU) were all obtained from Sigma. BrdU was stored at −80°C in single-use aliquots; HU and MMC were stored at 4°C, following the manufacturer's recommendations.
Immunophenotyping.
Thymi and spleens of 8- to 12-week-old mice were disaggregated, and the total number of cells was determined using a Cellometer Auto T4 (Nexcelom Bioscience). One million cells were stained with rat anti-mouse antibodies to T-cell (CD4, CD8a, and CD3) or B-cell [IgM, B220, and Ig(κ)] markers directly conjugated with either fluorescein isothiocyanate (FITC) or phycoerythrin (PE), as described previously (23). The stained cells were analyzed using a BD LSR flow cytometer and CellQuest software.
B-cell activation and quantification of CSR.
B-lineage splenocytes were purified by negative selection with CD43 beads (Miltenyi) and activated by incubation with the B-cell mitogens α-CD40 antibody (1 μg/ml; BD Pharmingen) and interleukin 4 (IL-4) (20 ng/ml; R&D Systems) for 4 days. Class switch recombination (CSR) was assessed by flow cytometry after the cells were stained with FITC-anti-mouse IgG1 and PE-anti-mouse B220 antibodies, as described previously (23).
Irradiation studies.
Cohorts of 2-month-old mice (n = 5 males and 5 females per genotype) were irradiated using a GammaCell40 irradiator equipped with a cesium-137 source at a rate of 52 cG/min. Mouse survival and fitness were monitored for 4 weeks after irradiation. For histological analysis of irradiated organs, mice were euthanized 5 days after exposure to 5 Gy, and organs were fixed in 10% buffered formalin and embedded in paraffin following standard procedures. Five-micrometer sections were made of major organs, mounted on glass slides, and stained with hematoxylin and eosin (HE). The HE-stained sections were used for evaluation by a veterinary pathologist (D.L.H.) and for imaging of representative sections from each group.
Telomere fluorescence in situ hybridization (FISH).
Activated B cells were incubated in colcemid (KaryoMAX; Gibco), swollen in 30 mM sodium citrate, fixed in methanol-acetic acid (3/1), and hybridized with a telomere peptide nucleic acid (PNA) probe as described previously (23). Briefly, slides were fixed in 4% formaldehyde and digested in pepsin prior to denaturation at 80°C for 3 min. The slides were then hybridized with a Cy3-labeled telomeric (TTAGGG)3 probe (Applied Biosystems) in 70% formamide for 2 h, washed, dehydrated, and mounted in Vectashield with DAPI (4′,6′-diamidino-2-phenylindole) (Vector Laboratories, Burlingame, CA). Images were obtained using a Zeiss Axioplan Imager Z.1 microscope equipped with a Zeiss AxioCam and an HXP120 mercury lamp (Jena GmbH) and dedicated software (Zeiss Axiovision Rel 4.6). At least 30 metaphases per sample were scored for chromosomal aberrations.
IgH FISH.
We detected the 3′ end of the IgH locus using BAC199 and the 5′ end using BAC207, as described previously (23). Briefly, bacterial artificial chromosomes (BACs) were labeled with either biotin (Biotin Nick Translation Mix; Roche) or digoxigenin (Dig-Nick Translation Mix; Roche) following the manufacturer's instructions. Two hundred nanograms of each BAC was precipitated with mouse Cot1 DNA (Invitrogen), resuspended in hybridization buffer (50% formamide, 2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 10% dextran sulfate, 0.15% SDS), and codenatured on slides at 76°C for 5 min. After incubation at 37°C for 16 h, the slides were washed, incubated with secondary antibodies (avidin-Cy3 and anti-digoxigenin-FITC, both from Roche), and mounted in Vectashield with DAPI. Images were captured as described for telomere FISH, and at least 50 metaphases per slide were scored for IgH locus breaks.
SCE assay.
B cells activated with α-CD40 plus IL-4 for 24 h were incubated in 10 μM BrdU for 24 h, which was found to represent two cell divisions for most cells in the culture. Cells were also incubated with 0.05 μM colcemid during the last 2 h and fixed in methanol-acetic acid after swelling in hypotonic buffer. For BrdU detection, slides were incubated in 2× SSC with 0.5 μg/ml of Hoetstch 33258 for 15 min, cross-linked for 15 min in 2× SSC, and dehydrated through serial ethanols. After air drying, the slides were blocked in 1% bovine serum albumin (BSA)-phosphate-buffered saline (PBS), denatured in 0.07 N NaOH for 2 min, neutralized in PBS (pH 8.5) for 5 min, permeabilized in 0.5% Tween 20-1% BSA-PBS for 5 min twice, and incubated with a FITC-conjugated anti-BrdU antibody (347583; BD Biosciences) diluted 1:1 in 0.5% Tween 20-3% BSA-PBS. The slides were then washed in 0.1% Tween 20-PBS for 5 min three times, dehydrated, air dried, and mounted in Vectashield with DAPI (Vector Laboratories). For experiments with HU, cells were first incubated in 10 μM BrdU for 12 h (one cell division), followed by 0.25 mM HU in the presence of 10 μM BrdU for 4 h. The HU was then removed, and the cells were incubated in 10 μM BrdU for 8 additional hours before metaphase harvesting. For experiments with MMC, B cells were incubated with 10 μM BrdU and 10 ng/ml MMC for 24 h prior to metaphase harvesting. Metaphases were obtained as described above for telomere FISH.
Indirect immunofluorescence (IF).
Activated B cells were centrifuged onto slides at 800 rpm for 3 min using a Sandon Cytospin, air dried, fixed in 4% paraformaldehyde (PFA) for 15 min at room temperature (RT), permeabilized in 0.2% Triton X-PBS for 5 min, blocked in 2% BSA-PBS for 30 min, and incubated with primary and secondary antibodies prior to being mounted in Vectashield with DAPI. The primary antibodies were anti-Rad51 rabbit polyclonal (Ab-1; PC130T; Calbiochem; 1:500), anti-53BP1 rabbit polyclonal (NB100-34; Novus Biologicals; 1:500), anti-RPA rat monoclonal (clone 4E4; Cell Signaling; 1:500), and anti-γ-H2AX mouse monoclonal (clone JBW301; Millipore; 1:800). The secondary antibodies were Cy3-goat anti-mouse IgG (115-166-071; Jackson ImunoResearch), Cy3-goat anti-rabbit IgG (111-166-003; Jackson ImmunoResearch), AlexaFluor 488-goat anti-rabbit IgG (A-11034; Invitrogen), AlexaFluor 488-goat anti-mouse IgG (A-11029; Invitrogen), and AlexaFluor 488-anti-rat IgG (4416; Cell Signaling). Slides were mounted in Vectashield with DAPI (Vector Laboratories). Images were captured using the Zeiss imaging system described above for telomere FISH. For studies of colocalization via confocal microscopy, we used a Nikon EZ CI confocal microscope to obtain 0.2-μm sections.
Statistical analysis.
We performed Student's t test to establish statistical significance between 3 to 5 data points per genotype, obtained from 3 to 5 independent experiments.
RESULTS
H2AX, but not 53BP1, is required for organismal viability in a PARP1-deficient background.
To investigate a genetic requirement for specific components of the ATM-regulated network in the embryonic lethality observed in PARP1/ATM double-knockout (DKO) mice (43), we attempted generation of mice deficient for PARP1 and either of two ATM-regulated substrates in the DDR, H2AX or 53BP1. PARP1+/−/H2AX+/− mice were apparently normal in size and development and were bred to generate PARP1−/−/H2AX−/− and their corresponding controls. However, no PARP1/H2AX DKO mice were observed among 101 live-born mice genotyped, while PARP1−/−/H2AX+/− and PARP1+/−/H2AX−/− mice were obtained at approximately Mendelian ratios (Table 1). Moreover, intercrosses of these genotypes also failed to generate DKO newborns (Table 1) and resulted in markedly reduced litter size (not shown). In contrast, PARP1/53BP1 DKO mice were born from PARP1+/−/53BP1+/− intercrosses at Mendelian ratios (Table 1); both DKO males and females appeared to have normal fertility (not shown). We conclude that, like ATM, H2AX is required for viability in a PARP1-deficient background, while 53BP1 is dispensable in this setting.
TABLE 1.
Mendelian ratios in live-born PARP1/H2AX and PARP1/53BP1 mice
| Intercross (no. of litters) | Resulting phenotype |
No. observed (%) | No. expected (%) | Ratio | |
|---|---|---|---|---|---|
| PARP1 | H2AX or 53BP1 | ||||
| PARP1+/−/H2AX+/−(14) | +/+ | +/+ | 5 (6.8) | 6.3 (6.25) | 1/16 |
| +/+ | +/− | 9 (12.2) | 12.6 (12.5) | 1/8 | |
| +/+ | −/− | 1 (1.4) | 6.3 (6.2) | 1/16 | |
| +/− | +/+ | 23 (28.4) | 12.6 (12.5) | 1/8 | |
| +/− | +/− | 32 (41.9) | 25.3 (25) | 1/4 | |
| +/− | −/− | 11 (13.5) | 12.6 (12.5) | 1/8 | |
| −/− | +/+ | 4 (5.4) | 6.3 (6.25) | 1/16 | |
| −/− | +/− | 16 (18.9) | 12.6 (12.5) | 1/8 | |
| −/− | −/− | 0 (0) | 6.3 (6.25) | 1/16 | |
| Total | 101 | ||||
| PARP1−/−/H2AX+/− × PARP1+/−/H2AX+/− (4) | +/− | +/+ | 2 (11.1) | 2.25 (12.5) | 1/8 |
| +/− | +/− | 7 (38.9) | 4.5 (25) | 1/4 | |
| +/− | −/− | 4 (22.2) | 2.22 (12.5) | 1/8 | |
| −/− | +/+ | 1 (5.6) | 2.25 (12.5) | 1/8 | |
| −/− | +/− | 4 (22.2) | 4.5 (25) | 1/4 | |
| −/− | −/− | 0 (0) | 2.25 (12.5) | 1/8 | |
| Total | 18 | ||||
| PARP1−/−/H2AX+/− × PARP1−/−/H2AX+/− (2) | −/− | +/+ | 3 (50.0) | 1.5 (25) | 1/4 |
| −/− | +/− | 3 (50.0) | 3 (50) | 1/2 | |
| −/− | −/− | 0 (0) | 1.5 (25) | 1/4 | |
| Total | 6 | ||||
| PARP1+/−/53BP1+/− (12) | +/+ | +/+ | 7.0 (8.4) | 5.2 (6.25) | 1/16 |
| +/+ | +/− | 8.0 (9.6) | 10.3 (12.5) | 1/8 | |
| +/+ | −/− | 9.0 (10.8) | 5.2 (6.25) | 1/16 | |
| +/− | +/+ | 9.0 (10.8) | 10.3 (12.5) | 1/8 | |
| +/− | +/− | 22.0 (26.5) | 20.7 (25) | 1/4 | |
| +/− | −/− | 10.0 (12.0) | 10.3 (12.5) | 1/8 | |
| −/− | +/+ | 5.0 (6.0) | 5.2 (6.25) | 1/16 | |
| −/− | +/− | 9.0 (10.8) | 10.3 (12.5) | 1/8 | |
| −/− | −/− | 4.0 (4.8) | 5.2 (6.25) | 1/16 | |
| Total | 83 | ||||
53BP1 deficiency accentuates growth retardation in a PARP1-deficient background.
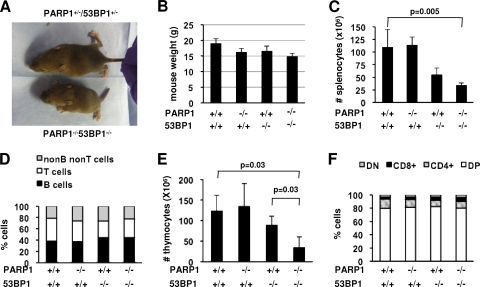
Although PARP1/53BP1 DKO mice were born at Mendelian ratios, they appeared smaller than their single-KO littermates from birth on (Fig. 1A). At 8 weeks of age, the weights of PARP1−/−, 53BP1−/−, and DKO females were, on average, 85.7, 87.3, and 78.3% of those of wild-type littermate mice (Fig. 1B); a similar decrease was observed for males (not shown).
FIG. 1.
PARP1 deficiency aggravates growth retardation in 53BP1-deficient mice. (A) Photograph of a 2.5-week-old PARP1/53BP1 DKO mouse and a heterozygous littermate. (B) Weights of 8 week-old mice of the indicated genotypes. The bars represent averages and standard deviations of 5 to 10 mice per genotype. (C) Spleens were disaggregated into single-cell suspensions, and the splenocytes were counted after red-cell lysis. The bars represent averages and standard deviations of 3 to 5 mice per genotype. (D) Splenocytes were stained with antibodies to B-cell markers (B220 and IgM) and T-cell markers (CD4 and CD8) and analyzed via flow cytometry to determine the relative proportions of B, T, and non-B/non-T cells in the spleen for the indicated genotypes. (E) Thymi were disaggregated, and total cells were counted. The bars represent averages and standard deviations of 3 to 5 mice per genotype. (F) Thymocytes were stained with antibodies to CD4 and CD8, and the relative proportions of CD4− CD8− (DN), CD4+ CD8+ (DP), CD4+ CD8−, and CD4− CD8+ lymphocytes were calculated to determine any developmental arrest.
To investigate the effects of combined PARP1/53BP1 deficiencies on specific lineages, we next quantified spleen and thymus cellularity in adult mice. As expected (68), the size of 53BP1−/− spleens was markedly reduced relative to those of wild-type littermates (Fig. 1C). Spleen cellularity was further decreased in DKO mice, with an average of 30 million splenocytes per adult (8-week-old) mouse (Fig. 1C). Splenocyte immunophenotyping with antibodies to B-cell [B220, IgM, and Ig(κ)] and T-cell (CD3, CD4, and CD8) markers indicated a marked reduction in both B- and T-cell compartments (Fig. 1D; see Fig. S1 in the supplemental material). Similarly, thymic cellularity was markedly decreased in DKO relative to 53BP1−/− mice, with averages of 123.3 ± 38.7, 134.3 ± 56.0, 88.7 ± 22.0, and 34.0 ± 26.3 thymocytes for wild-type, PARP1−/−, 53BP1−/−, and DKO mice, respectively (Fig. 1E). However, the percentages of double-negative (DN) (CD4− CD8−), double-positive (DP) (CD4+ CD8+), and single-positive (SP) (either CD4+ CD8− or CD4− CD8+) thymocytes were similar for all genotypes (Fig. 1F; see Fig. S1 in the supplemental material). We conclude that PARP1 and 53BP1 function in noncomplementary pathways in organismal development.
53BP1 and PARP1 function to suppress chromosomal breaks and translocations in mostly nonoverlapping pathways.
Cultured mouse cells deficient for 53BP1 or PARP1 harbor more frequent chromosomal aberrations than their wild-type counterparts (17, 23, 46). To test the hypothesis that growth retardation and immune cell depletion in PARP1/53BP1-deficient mice reflects on their functional interaction in the repair of DNA DSBs, we next quantified chromosomal aberrations via telomere FISH in metaphase spreads of activated DKO and control B cells (Fig. 2), following our established protocols (23, 24). These studies revealed a higher frequency of metaphases harboring at least one aberration in PARP1/53BP1 DKO cells relative to wild-type cells (Fig. 2A and Table 2; see Table S1 in the supplemental material for individual data) (n = 4 independent experiments). Similarly, the total number of aberrations was significantly increased in double mutants (Fig. 2B and Table 2; see Table S1 in the supplemental material). These differences were statistically significant regardless of the cytokines used for activation (P = 0.0004 for lipopolysaccharide [LPS] and P = 0.002 for α-CD40 plus IL-4, for aberrations in DKO cells versus wild-type cells). Moreover, DKO cultures had significantly more metaphases containing chromosomal aberrations (P = 0.04) and total aberrations (P = 0.004) than 53BP1 mutants activated in parallel. Of note, 59/70 (84.3%) aberrations observed in PARP1/53BP1 DKO cells (n = 5 mice) were of “chromosome type,” while “chromatid-type” aberrations were rare (Fig. 2C and D). This observation suggests that, as previously described for 53BP1-deficient cells (23), most unrepaired DSBs in this experimental setting originated prior to replication.
FIG. 2.
PARP1 and 53BP1 functionally interact to suppress genomic instability. (A and B) Wild-type, PARP1−/−, 53BP1−/−, and PARP1−/−/53BP1−/− B lymphocytes were activated with α-CD40 plus IL-4 for 4 days, and metaphase spreads were hybridized with a Cy3-labeled telomere-specific PNA probe and scored for chromosomal aberrations. The percentage of metaphases containing at least one aberration (A) and the total number of aberrations per metaphase (B) are shown. The bars represent the averages and standard deviations of 4 or 5 mice in 4 independent experiments. (C) Chromosomal aberrations observed in cells deficient for PARP1, 53BP1, or both were further classified as “chromosome type” or “chromatid type”; n indicates the total number of aberrations for each genotype. (D) The yellow arrows point to chromosome breaks in a partial metaphase spread of a PARP1−/−/53BP1−/− B cell after telomere FISH. Red, (TTAGGG)3 probe; blue, DAPI. (E) Wild-type, PARP1−/−, 53BP1−/−, and PARP1−/−/53BP1−/− B cells were activated with α-CD40 plus IL-4 for 4 days, and the expression of surface IgG1 (sIgG1) was measured by flow cytometry. The bars represent the averages and standard deviations of 3 independent experiments. (F) Metaphase spreads of wild-type, PARP1−/−, 53BP1−/−, and PARP1−/−/53BP1−/− B cells were used to quantify chromosomal breaks specifically at the IgH locus. FISH was performed using BAC199 and BAC207, which recognize sequences 3′ and 5′ to the locus, respectively. The total number of breaks in the same samples, quantified via telomere FISH, is shown for comparison. The bars represent the averages and standard deviations of 3 independent experiments.
TABLE 2.
Analysis of general genomic stability in B cells deficient for PARP1 and/or 53BP1 via telomere FISH
| Phenotype | Cytokine | No. of mice | No. of metaphases | No. of metaphases with aberrations (%) | No. of aberrations (%) | Types of aberrationsa |
|---|---|---|---|---|---|---|
| WTb | LPS | 4 | 120 | 3 (2.5) | 4 (3.3) | 4 cb |
| PARP1−/− | LPS | 4 | 120 | 6 (5.0) | 8 (6.7) | 2 CB, 4 cb |
| 53BP1−/− | LPS | 4 | 116 | 12 (10.4) | 14 (12.1) | 10 CB, 4 cb |
| PARP1−/−/53BP1−/− | LPS | 5 | 142 | 20 (14.1) | 28 (19.1) | 23 CB, 5 cb |
| WT | α-CD40 + IL-4 | 4 | 120 | 5 (4.2) | 5 (4.2) | 3 CB, 2 cb |
| PARP1−/− | α-CD40 + IL-4 | 4 | 96 | 11 (12.5) | 13 (14.2) | 6 CB, 6 cb, 1 dic |
| 53BP1−/− | α-CD40 + IL-4 | 4 | 95 | 9 (11.7) | 9 (11.7) | 4 CB, 4 cb, 1 dic |
| PARP1−/−/53BP1−/− | α-CD40 + IL-4 | 5 | 150 | 34 (22.7) | 41 (27.3) | 34 CB, 6 cb, 1 |
CB, chromosome break; cb, chromatid break; dic, dicentrics.
WT, wild type.
A fraction of B cells activated under our conditions underwent CSR, a process whereby activation-induced cytidine deaminase (AID)-dependent DSBs are introduced at the immunoglobulin heavy chain (IgH) locus (14). We and others previously demonstrated that, in the absence of 53BP1, AID-dependent DSBs are either repaired via internal deletion or left unrepaired, resulting in chromosomal breaks at the IgH locus and a severe CSR defect (23, 52, 53). In contrast, PARP1-deficient B cells underwent CSR with efficiency similar to that of wild-type cells (54, 67). However, PARP1 modulates at least a subset of repair events in this setting (54) and could therefore potentially interact with 53BP1 to augment IgH locus-specific genomic instability. To determine whether increased genomic instability in DKO-activated B cells is CSR related, we next quantified the contribution of IgH locus breaks via a locus-specific FISH assay (23). Despite no defect in proliferation (not shown), DKO B cells failed to undergo appreciable CSR to IgG1, similar to 53BP1-deficient B cells (39, 69) (Fig. 2E; see Table S2 in the supplemental material). Direct comparison of IgH locus-specific breaks (via two-color IgH FISH with BAC probes) and “general” chromosomal breaks (via telomere FISH) on the same population indicated that most observed genomic instability in DKO cells originated outside the IgH locus (Fig. 2F).
In summary, the additive rather than synergistic nature of observed phenotypes in the double mutants suggests that PARP1 and 53BP1 may function in distinct nonsynthetic pathways in the repair of DSBs that arise spontaneously in culture. These observations suggest a more restricted role for 53BP1 in NHEJ-mediated repair.
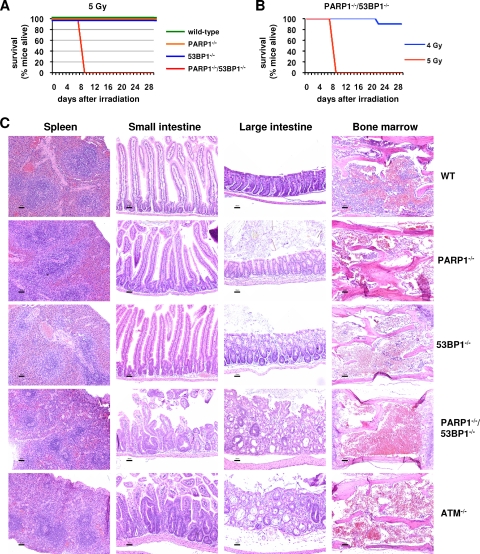
PARP1 and 53BP1 cooperate to prevent IR-induced lethality.
We next used our in vivo models to examine a functional interaction between PARP1 and 53BP1 in radioprotection (17, 40, 68). Previous reports indicated that the 50% lethal dose (LD50) for PARP1-deficient mice is approximately 6 Gy (40). In line with that report, we found that wild-type, PARP1−/−, and 53BP1−/− mice all survived an acute exposure to 5 Gy of ionizing radiation (IR) (Fig. 3A). In contrast, all irradiated DKO mice (n = 10 mice, 5 males and 5 females) became acutely ill and died between days 9 and 10 postirradiation (Fig. 3A, red line). Additional experiments indicated that the LD50 for PARP1/53BP1 DKO mice is approximately 4.5 Gy (Fig. 3B), similar to that reported for ATM-deficient mice (3).
FIG. 3.
PARP1 and 53BP1 synergize in the repair of IR-induced DSBs. (A) Mice (n = 10; 5 males and 5 females) of the indicated genotypes were exposed to 5 Gy of IR, and survival was monitored for 4 weeks after irradiation. (B) PARP1/53BP1 DKO mice (n = 10 mice, 5 males and 5 females) were exposed to 5 Gy or 4 Gy of IR, and survival was monitored for 4 weeks after irradiation. (C) Mice of the indicated genotypes were exposed to 5 Gy of IR, and their organs were fixed 5 days after exposure. Paraffin-embedded sections were stained with HE. Representative histologic images are shown of spleen, small intestine, colon, and bone marrow for each genotype. WT, wild type. Scale bars = 50 μm.
To determine whether increased radiation sensitivity in DKO mice was due to a global defect or to sensitivity of specific organs, we next performed histopathological analysis on HE-stained sections of mouse organs 5 days after exposure to 5 Gy (Fig. 3C). PARP1/53BP1 DKO spleens showed severe lymphoid depletion, comparable to spleens of ATM−/− mice irradiated in parallel. In contrast, splenic cellularity and architecture were restored in wild-type and single-mutant mice. Similarly, marked villous shortening and loss, crypt necrosis. and crypt regeneration were observed in the small and large intestines of either DKO or ATM−/− mice, suggesting that early death was likely related to loss of gastrointestinal barrier function and fulminant sepsis. In contrast, crypts of wild-type or single-mutant mice showed near-complete recovery, as previously described by others (32). Finally, although some degree of bone marrow hypoplasia was observed in all mutants, it was clearly most severe in DKO and ATM−/− mice (Fig. 3C). Thus, although combined deficiency for PARP1 and 53BP1 appeared to have an additive rather than synergistic effect on development and steady-state genomic stability (Fig. 1 and 2), clastogen challenge uncovered significant functional complementarity for the two factors in the prevention of lethal radiation injury.
53BP1 deficiency does not alter the frequency of SCEs in a PARP1-proficient or -deficient background.
The modest phenotypes observed in PARP1/53BP1 DKO mice suggested that 53BP1 may be generally dispensable for HR, instead promoting NHEJ. However, a previous report suggested a role for 53BP1 in the suppression of HU-induced SCE (63), an HR-mediated process. This observation prompted us to examine the effect of 53BP1 deficiency on the frequency of SCEs in PARP1−/− cells, where an increased number of HR substrates correlates with increased frequency of spontaneous SCEs (17). To this end, we measured the frequency of SCE in metaphase spreads of wild-type, PARP1−/−, 53BP1−/−, and PARP1/53BP1 DKO B cells activated in parallel (Fig. 4 and Table 3; see Table S3 in the supplemental material for data from individual experiments). As previously described (7), the frequency of spontaneous SCE was markedly increased in PARP1−/− B cells (0.38 and 0.99 SCEs/cell in wild-type and PARP1−/− B cells, respectively; P = 0.009; n = 4 independent experiments) (Fig. 4 and Table 3; see Table S3 in the supplemental material). In contrast, the frequency of spontaneous SCE in 53BP1−/− or in DKO B cells was similar to that in wild-type or PARP1-deficient controls, respectively (P = 0.62 and P = 0.47, respectively). Moreover, exposure to hydroxyurea (0.25 mM for 8 h) or mitomycin C (10 ng/ml for 24 h) resulted in similar increases in the frequency of SCE in wild-type and 53BP1−/− cells (Table 3; see Table S3 in the supplemental material). Finally, we observed no change in the distribution of SCEs/cell between wild-type and 53BP1−/− cells or between PARP1−/− and DKO cells, either spontaneously or after exposure to HU or MMC (Fig. 4). Of note, the same dose and treatment schedule employed for our SCE assays induced frequent chromatid breaks in 53BP1−/− cells, indicating that 53BP1 regulates the repair of a subset of HU-induced lesions other than SCEs (see below).
FIG. 4.
Frequencies of SCEs in lymphocytes deficient for PARP1 and/or 53BP1. (A) B cells of the indicated genotypes were activated with α-CD40 plus IL-4 for 24 h and incubated in 10 μM BrdU for an additional 24 h (two cell divisions), and metaphase spreads were hybridized with a FITC-labeled anti-BrdU antibody. For studies of drug-induced SCEs, metaphases were obtained after exposure to 10 ng/ml MMC for 24 h or 0.25 mM HU for 4 h, followed by a washout period of 8 h. For all experiments, colcemid was used at a concentration of 0.05 μg/ml for 2 h prior to cell harvesting. The histograms represent the number of SCEs per chromosome; n is the number of chromosomes analyzed for each condition. (B) Examples of HU-treated metaphases of the indicated genotypes, with representative chromosomes magnified in the insets.
TABLE 3.
Frequency of spontaneous or drug-induced SCEs in cells deficient for PARP1 and 53BP1
| Phenotype | Treatment | No. of chromosomes | No. of SCEs | No. of SCEs/ chromosome | Pa |
|---|---|---|---|---|---|
| Wild type | 2,331 | 892 | 0.38 | ||
| PARP1−/− | 2,373 | 2,343 | 0.99 | 0.009 | |
| 53BP1−/− | 1,473 | 593 | 0.40 | 0.62 | |
| PARP1−/−/53BP1−/− | 1,490 | 1,456 | 0.98 | 0.031 | |
| Wild type | MMC | 1,900 | 1,300 | 0.70 | |
| PARP1−/− | MMC | 1,915 | 2,532 | 1.32 | 0.002 |
| 53BP1−/− | MMC | 1,229 | 947 | 0.77 | 0.14 |
| PARP1−/−/53BP1−/− | MMC | 1,521 | 2,022 | 1.33 | 0.03 |
| Wild type | HU | 1,322 | 1,563 | 1.18 | |
| PARP1−/− | HU | 1,401 | 2,603 | 1.86 | 0.07 |
| 53BP1−/− | HU | 1,213 | 1,338 | 1.1 | 0.92 |
| PARP1−/−/53BP1−/− | HU | 997 | 1,942 | 1.95 | 0.05 |
Relative to wild type.
PARP1 and 53BP1 suppress genomic instability induced by HU, but not MMC.
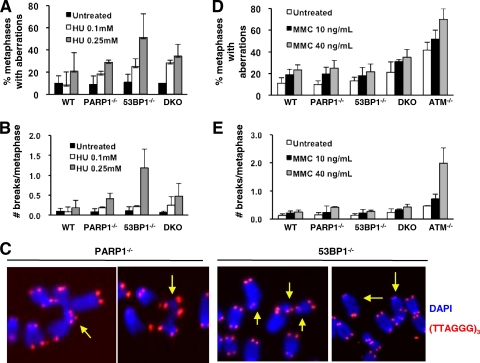
Most chromosomal breaks observed in 53BP1-deficient cells are “chromosome type” (23) (Fig. 2), suggesting that 53BP1 normally facilitates NHEJ in prereplicative stages of the cell cycle. This notion is supported by our observation of efficient HR-mediated repair in PARP1/53BP1 DKO mice mentioned above. However, several studies have demonstrated a requirement for 53BP1 in the context of replicative stress (63, 73). To further clarify functions for 53BP1 in NHEJ versus HR outside G1, we next quantified genomic instability in PARP1/53BP1 single and double mutants exposed to the replication inhibitor HU or the interstrand cross-linker MMC, both of which induce DSBs upon replication. Moreover, we took advantage of previous observations that both HR and NHEJ contribute significantly to the repair of HU-dependent lesions (38, 56) to establish the relative contribution of 53BP1 to each pathway during S phase (see below).
Exposure to HU induced a dose-dependent increase in the frequency of metaphases containing chromosomal aberrations and the number of chromosomal aberrations in cells deficient for 53BP1 or, to a lesser extent, PARP1 (Fig. 5A and B; see Table S4 in the supplemental material for data on individual samples). On average, we observed 1.19 and 0.19 chromosome breaks/metaphase in 53BP1−/− and wild-type cells, respectively, exposed to 0.25 mM HU (P = 0.04) (Fig. 5B). The frequency of aberrations was also increased in PARP1−/− and DKO cells (0.41 and 0.48 aberrations per metaphase, respectively, although these numbers did not reach statistical significance) (Fig. 5B; see Table S4 in the supplemental material). As expected from their origin at replication, most aberrations consisted of chromatid breaks, which often appeared to be engaged in complex rearrangements (see Fig. 5C for examples). In contrast, MMC induced only a modest increase in the frequency of metaphases with aberrations or the number of aberrations in cells deficient for PARP1 and/or 53BP1, comparable to that observed in wild-type cells (Fig. 5D and E; see Table S5 in the supplemental material). As a positive control, ATM−/− B cells treated in parallel harbored frequent chromosomal aberrations (0.25 and 1.98 aberrations per metaphase for wild-type and ATM−/− cells exposed to 40 ng/ml of MMC, respectively; P = 0.00027), in agreement with previous findings in a mouse model (36). We conclude that 53BP1 functions in the repair of a subset of DSBs associated with replication inhibition. In contrast, efficient repair of interstrand cross-links, which preferentially employs HR over NHEJ, necessitates ATM, but not 53BP1, at least under our experimental conditions.
FIG. 5.
PARP1 and 53BP1 function in the repair of HU-induced DSBs but are dispensable for the repair of MMC-induced DSBs. (A to C) B cells of the indicated genotypes were treated with 0.25 mM HU for 15 h, and metaphases were obtained after an 8-h washout period. Chromosomes were hybridized with a telomere-specific (TTAGG)3 PNA probe, and chromosomal aberrations were scored in 30 metaphases per sample. The percentage of metaphases containing at least one chromosomal aberration (A) and the number of aberrations per metaphase (B) are shown. (C) Representative examples of partial metaphases; the yellow arrows point to chromatid breaks and fusions. Red, (TTAGGG)3 probe; blue, DAPI. (D and E) Activated B cells of the indicated genotypes were treated with MMC at the indicated doses for 36 h, and metaphases were scored for aberrations via telomere FISH. The percentage of metaphases containing at least one chromosomal aberration (D) and the number of aberrations per metaphase (E) are shown. For all experiments, the bars represent the averages and standard deviations of 3 to 5 mice per genotype in 3 independent experiments.
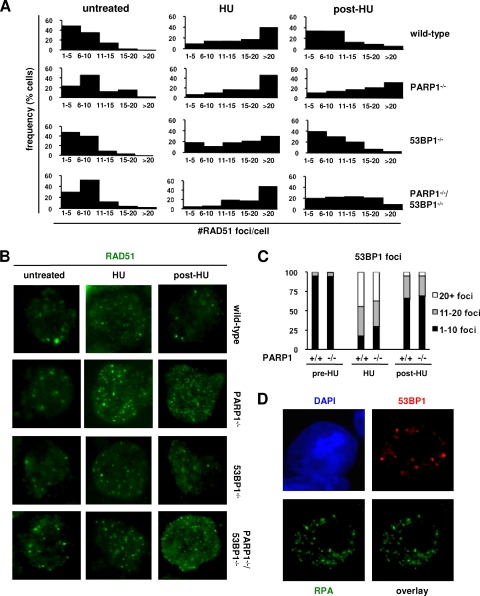
53BP1 is dispensable for RAD51-dependent repair of HU-induced lesions.
To determine whether NHEJ and/or HR is compromised during S phase in the absence of 53BP1, we first compared the dynamics of assembly and resolution of RAD51 foci, a marker of HR, in wild-type and 53BP1−/− cells exposed to HU. As expected, replication inhibition markedly increased the frequency of RAD51 foci in wild-type cells, with most cells accumulating >10 foci at the end of a 15-h exposure period and most foci resolving 8 h after drug withdrawal (Fig. 6A and B). In contrast, most PARP1-deficient cells still harbored >10 RAD51 foci at the same time point (Fig. 6A and B), supporting the notion that PARP1 is required for efficient HR-mediated repair of HU-induced lesions (9). Of note, the dynamics of RAD51 foci in 53BP1−/− cells was indistinguishable from that in wild-type cells (n = 5 independent experiments) (Fig. 6A and B). Moreover, the dynamics of PARP1/53BP1 cells closely resembled those of PARP1-deficient cells (Fig. 6A and B). Altogether, these findings suggest that 53BP1 is dispensable for the normal assembly of the recombination machinery at HU-induced lesions. In further support of this notion, parallel studies on the dynamics of foci containing RPA, a second marker of HR upstream of RAD51, were also similar in wild-type and 53BP1−/− cells (see Fig. S2 in the supplemental material).
FIG. 6.
53BP1 is dispensable for HR-mediated repair of HU-induced lesions. (A and B) Activated B cells of the indicated genotypes were treated with HU, and the RAD51 foci were quantified via immunofluorescence using a rabbit polyclonal anti-RAD51 antibody and counterstaining with DAPI. The histograms in panel A show the number of RAD51 foci per nucleus in control untreated cells, cells treated with 0.25 mM HU for 15 h (HU), and cells harvested 8 h after HU withdrawal to allow cell cycle progression and repair (post-HU). Each histogram represents a pool of 2 to 4 mice per genotype; n = 100 cells per mouse. Representative examples are shown in panel B. (C) Distribution of 53BP1 foci in HU-treated wild-type and PARP1−/− cells under the same experimental conditions as for panels A and B. The data represent an average of 7 independent B-cell activations in 6 independent experiments. (D) Combined immunofluorescence for 53BP1 (red) and RPA (green) in wild-type cells after HU exposure shows localization to distinct foci. Nuclei were counterstained with DAPI (blue).
In contrast to the observed delay in RAD51 focus clearance, the dynamics of 53BP1 foci in cells deficient for PARP1 were similar to those in wild-type cells (Fig. 6C; see Fig. S3 in the supplemental material), suggesting that PARP1 is dispensable for the recruitment of 53BP1 to HU-induced lesions. Finally, combined IF for 53BP1 and RPA in HU-treated cells indicated that the two factors localize to distinct foci (Fig. 6D). Collectively, these findings strongly suggest that, in response to HU-induced DNA damage, 53BP1 functions in a pathway that is distinct from the PARP1/RPA/RAD51 axis (9), likely NHEJ.
53BP1 promotes repair of HU-induced DSBs by DNA-PKcs-dependent NHEJ.
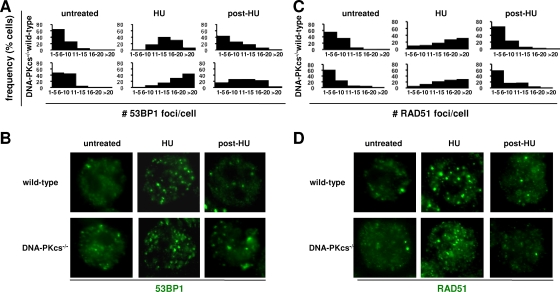
To more directly establish the link between 53BP1 and NHEJ, we next investigated the dynamics of HU-dependent 53BP1, RAD51, and RPA focus formation in cells deficient for DNA-PKcs, an NHEJ factor required for the repair of a subset of HU-induced DSBs (38). Relative to wild-type controls, the number of 53BP1 foci/cell was markedly increased in DNA-PKcs-deficient B cells after 15 h of exposure to 0.25 mM HU (Fig. 7A and B). Moreover, compared to wild-type counterparts, persistent 53BP1 foci were observed in DNA-PKcs-deficient cells 8 h after HU withdrawal (Fig. 7A and B). In contrast, the frequencies of RAD51 foci at the same time points were similar for wild-type and DNA-PKcs-deficient cells (Fig. 7C and D), consistent with efficient repair by HR in the absence of DNA-PKcs. Similar observations were obtained by counting RPA foci (see Fig. S4 in the supplemental material). We conclude that, in the context of HU-dependent replication inhibition, 53BP1 is required for the repair of a subset of replication-associated DSBs that employ NHEJ.
FIG. 7.
53BP1 promotes DSB repair via DNA-PKcs-dependent NHEJ. (A and B) DNA-PKcs−/− and control wild-type cells were activated for 24 h and exposed to HU. The number of 53BP1 foci per nucleus was determined by IF in untreated cells, in cells treated with 0.25 mM HU for 15 h (HU), or in cells obtained 8 h after the drug was washed off (post-HU). Each histogram represents the number of 53BP1 foci in two mice per genotype analyzed in two independent experiments; n = 100 cells/sample. Representative examples are shown in panel B. (C and D) To measure the dynamics of HR in HU-treated DNA-PKcs-deficient cells, IF for RAD51 was performed on the same cells and time points as in panel A; representative examples are shown in panel D.
DISCUSSION
Here, we have taken a genetic approach to investigate functional interactions between the DNA damage sensor PARP1 and the ATM-regulated network. We found that PARP1 plays nonoverlapping functions with both histone H2AX and 53BP1 in organismal development and DSB repair. Importantly, we demonstrated that H2AX is essential for viability in a PARP1-deficient background, while mice deficient for PARP1 and 53BP1 show apparently additive phenotypes. Together with our genetic dissection of the repair pathways activated in response to HU-induced DNA damage, these differential phenotypes provide strong in vivo evidence for a restricted role for 53BP1 in NHEJ-mediated repair throughout the cell cycle, beyond specialized rearrangements in developing lymphocytes.
We found that, like PARP1/ATM-deficient mice (43), PARP1/H2AX mice are embryonic lethal. As ATM generates γ-H2AX at DSBs (11), this finding may suggest a common mechanism for embryonic lethality in PARP1/ATM and PARP1/H2AX mice. In this regard, both ATM and H2AX function to facilitate repair via HR, pointing to synthetic lethality due to combined impairment of BER and HR as a potential mechanism operating in both models (8, 10, 21, 42, 66). In this context, ATM-dependent formation of γ-H2AX promotes HR via the establishment of a platform that facilitates cohesin-mediated assembly of sister chromatids prior to recombination (64), kinase cross talk (13), and possibly other mechanisms (71). However, ATM may also facilitate HR via H2AX-independent mechanisms, including activation of cell cycle checkpoints and end-processing activities (5, 33, 48). Moreover, combined deficiencies for ATM and H2AX lead to embryonic lethality (74), pointing to noncomplementary functions in vivo. In the future, analysis of PARP1/H2AX embryos will allow us to determine whether deficiency for H2AX allows survival beyond deficiency for ATM (i.e., beyond gastrulation) and, if so, to determine the contributions of specific DSB repair pathways to embryo demise.
In contrast to PARP1/ATM and PARP1/H2AX DKO mice, PARP1/53BP1 DKO mice are viable and show only modest exacerbation of phenotypes observed in single mutants. Although 53BP1 is a direct target of ATM (11) and binds to the γ-H2AX/MDC1 complex (20), it has become increasingly clear that histone modifications other than γ-H2AX are key for its recruitment to chromatin surrounding DSBs (49, 61, 65). Moreover, 53BP1 and γ-H2AX promote DSB repair via clearly distinct mechanisms in multiple settings, including lymphocyte V(D)J recombination (18), class switch recombination (53), and end-to-end chromosomal fusions (19). However, 53BP1 functions at these highly specialized settings may differ from its role in a more general setting. Moreover, several previous studies indicated that 53BP1-deficient cells are hypersensitive to drugs that induce DSBs in the S phase of the cell cycle, when HR is active. However, as NHEJ remains operative during the S and G2 phases of the cell cycle (5, 45), this previous work did not clearly establish a role for 53BP1 in HR.
To evaluate pathway specificity, we have characterized in detail the dynamics of DSB repair after exposure to HU. This experimental system is particularly useful to dissect roles for 53BP1 in HR versus NHEJ because both pathways contribute significantly to repair in this setting (38, 56). Using this approach, we provide here several lines of evidence that support a role for 53BP1 specifically in NHEJ-mediated DSB repair during S phase. First, the frequency of spontaneous RPA and RAD51 foci in 53BP1-deficient cells, as well as their dynamics after exposure to HU, is indistinguishable from that in wild-type cells. In contrast, increased numbers of spontaneous RPA/RAD51 foci and delayed resolution after HU exposure were observed in PARP1-deficient cells, consistent with previous studies (9, 58, 72). Second, the dynamics of 53BP1 focus formation were comparable in HU-treated wild-type and PARP1−/− cells. As the accumulation of MRN and HR factors is invariably impaired in the absence of PARP1 (9), this observation suggests that 53BP1 functions in a pathway other than the PARP1/MRN/RPA/RAD51 axis. Third, 53BP1 and RPA localize to distinct foci, as previously reported in the context of camptothecin-treated cells (73). Lastly, 53BP1 deficiency did not alter the frequency of HU-induced SCEs, even in PARP1-deficient cells with an increased load of HR substrates. The apparent discrepancy between these findings and a previous report (63) may reflect differences in our experimental systems (mouse primary 53BP1 knockout cells in our study versus human immortalized knockdown cell lines in the other study). Altogether, these experiments strongly suggest that HR-mediated repair of HU-dependent lesions proceeds efficiently in the absence of 53BP1.
Evidence for a specific role in NHEJ-mediated repair of such lesions comes from our studies on cells lacking the NHEJ factor DNA-PKcs. In this context, we observed a significant delay in the resolution of 53BP1 foci, but not RAD51 or RPA foci, in DNA-PKcs-deficient cells relative to their wild-type counterparts. We note that DNA-PKcs deficiency did not result in a compensatory increase in HR (measured here by the number of RPA or RAD51 foci). This observation may suggest that DNA end-bound Ku precludes HR activity in a subset of DSBs (34) or, alternatively, reflect differential kinetics for the two pathways over time (56). Finally, although our studies suggest that PARP1 functions mostly in HR in this setting, we cannot exclude a contribution to the regulation of NHEJ via functional interactions with DNA-PK (30, 47) or others.
Despite the modest aggravation of phenotypes observed in PARP1/53BP1 DKO mice, combined deficiency for the two factors may have dramatic effects in recovery from clastogenic stress, as illustrated by our studies of organismal radiation sensitivity. In this regard, the LD50 of PARP1/53BP1 DKO mice is approximately 4.5 Gy, significantly lower than for single mutants and similar to the previously reported LD50 for ATM-deficient mice (3). Moreover, histological analysis of irradiated tissues deficient for ATM or PARP1 plus 53BP1 showed severe pathology in all highly proliferative organs (spleen, bone marrow, and intestinal crypts) under conditions where single mutants have achieved near-complete recovery. If extrapolated to human cells, our observations suggest that a combination of PARP inhibitors currently in clinical use and 53BP1 inhibition may prove a useful strategy to radiosensitize tumors with acceptable toxicity for nonirradiated cells. Finally, our findings may also have implications for the selective use of PARP inhibitors for the treatment of tumors with decreased expression of 53BP1, such as some lymphomas (62).
Supplementary Material
Acknowledgments
We are grateful to Fred Alt for mice deficient for H2AX, ATM, and DNA-PKcs; Klaus Rajewsky for mice harboring immunoglobulin transgenes; Junjie Chen for mice deficient for 53BP1; and Ted Dawson for mice deficient for PARP1. We also thank Shaida Andrabi for advice on PARP1 genotyping and immunofluorescence, Juan Fu for technical assistance with B-cell purification, and Lillian Dasko-Vincent and Leslie Meslzer (in memoriam) at the Sidney Kimmel Comprehensive Cancer Center (SKCCC) Cell Imaging Core Facility for assistance with confocal microscopy and flow cytometry analysis.
We declare no conflict of interest.
Footnotes
Published ahead of print on 15 March 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aguilar-Quesada, R., J. A. Munoz-Gamez, D. Martin-Oliva, A. Peralta, M. T. Valenzuela, R. Matinez-Romero, R. Quiles-Perez, J. Menissier-de Murcia, G. de Murcia, M. Ruiz de Almodovar, and F. J. Oliver. 2007. Interaction between ATM and PARP-1 in response to DNA damage and sensitization of ATM deficient cells through PARP inhibition. BMC Mol. Biol. 8:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahel, D., Z. Horejsi, N. Wiechens, S. E. Polo, E. Garcia-Wilson, I. Ahel, H. Flynn, M. Skehel, S. C. West, S. P. Jackson, T. Owen-Hughes, and S. J. Boulton. 2009. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science 325:1240-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barlow, C., S. Hirotsune, R. Paylor, M. Liyanage, M. Eckhaus, F. Collins, Y. Shiloh, J. N. Crawley, T. Ried, D. Tagle, and A. Wynshaw-Boris. 1996. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell 86:159-171. [DOI] [PubMed] [Google Scholar]
- 4.Bassing, C. H., H. Suh, D. O. Ferguson, K. F. Chua, J. Manis, M. Eckersdorff, M. Gleason, R. Bronson, C. Lee, and F. W. Alt. 2003. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell 114:359-370. [DOI] [PubMed] [Google Scholar]
- 5.Beucher, A., J. Birraux, L. Tchouandong, O. Barton, A. Shibata, S. Conrad, A. A. Goodarzi, A. Krempler, P. A. Jeggo, and M. Lobrich. 2009. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J. [DOI] [PMC free article] [PubMed]
- 6.Borghesani, P. R., F. W. Alt, A. Bottaro, L. Davidson, S. Aksoy, G. A. Rathbun, T. M. Roberts, W. Swat, R. A. Segal, and Y. Gu. 2000. Abnormal development of Purkinje cells and lymphocytes in Atm mutant mice. Proc. Natl. Acad. Sci. U. S. A. 97:3336-3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouffler, S. D., P. Finnon, M. A. Blasco, and E. Ainsbury. 2008. A possible role for telomerase RNA and telomere length in global mitotic recombination. Cytogenet. Genome Res. 122:292-296. [DOI] [PubMed] [Google Scholar]
- 8.Bryant, H. E., and T. Helleday. 2006. Inhibition of poly(ADP-ribose) polymerase activates ATM which is required for subsequent homologous recombination repair. Nucleic Acids Res. 34:1685-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bryant, H. E., E. Petermann, N. Schultz, A. S. Jemth, O. Loseva, N. Issaeva, F. Johansson, S. Fernandez, P. McGlynn, and T. Helleday. 2009. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 28:2601-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bryant, H. E., N. Schultz, H. D. Thomas, K. M. Parker, D. Flower, E. Lopez, S. Kyle, M. Meuth, N. J. Curtin, and T. Helleday. 2005. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434:913-917. [DOI] [PubMed] [Google Scholar]
- 11.Burma, S., B. P. Chen, M. Murphy, A. Kurimasa, and D. J. Chen. 2001. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 276:42462-42467. [DOI] [PubMed] [Google Scholar]
- 12.Celeste, A., S. Petersen, P. J. Romanienko, O. Fernandez-Capetillo, H. T. Chen, O. A. Sedelnikova, B. Reina-San-Martin, V. Coppola, E. Meffre, M. J. Difilippantonio, C. Redon, D. R. Pilch, A. Olaru, M. Eckhaus, R. D. Camerini-Otero, L. Tessarollo, F. Livak, K. Manova, W. M. Bonner, M. C. Nussenzweig, and A. Nussenzweig. 2002. Genomic instability in mice lacking histone H2AX. Science 296:922-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chanoux, R. A., B. Yin, K. A. Urtishak, A. Asare, C. H. Bassing, and E. J. Brown. 2009. ATR and H2AX cooperate in maintaining genome stability under replication stress. J. Biol. Chem. 284:5994-6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaudhuri, J., U. Basu, A. Zarrin, C. Yan, S. Franco, T. Perlot, B. Vuong, J. Wang, R. T. Phan, A. Datta, J. Manis, and F. W. Alt. 2007. Evolution of the immunoglobulin heavy chain class switch recombination mechanism. Adv. Immunol. 94:157-214. [DOI] [PubMed] [Google Scholar]
- 15.Dantzer, F., V. Schreiber, C. Niedergang, C. Trucco, E. Flatter, G. De La Rubia, J. Oliver, V. Rolli, J. Menissier-de Murcia, and G. de Murcia. 1999. Involvement of poly(ADP-ribose) polymerase in base excision repair. Biochimie 81:69-75. [DOI] [PubMed] [Google Scholar]
- 16.Davalos, A. R., P. Kaminker, R. K. Hansen, and J. Campisi. 2004. ATR and ATM-dependent movement of BLM helicase during replication stress ensures optimal ATM activation and 53BP1 focus formation. Cell Cycle 3:1579-1586. [DOI] [PubMed] [Google Scholar]
- 17.de Murcia, J. M., C. Niedergang, C. Trucco, M. Ricoul, B. Dutrillaux, M. Mark, F. J. Oliver, M. Masson, A. Dierich, M. LeMeur, C. Walztinger, P. Chambon, and G. de Murcia. 1997. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc. Natl. Acad. Sci. U. S. A. 94:7303-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Difilippantonio, S., E. Gapud, N. Wong, C. Y. Huang, G. Mahowald, H. T. Chen, M. J. Kruhlak, E. Callen, F. Livak, M. C. Nussenzweig, B. P. Sleckman, and A. Nussenzweig. 2008. 53BP1 facilitates long-range DNA end-joining during V(D)J. recombination. Nature 456:529-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dimitrova, N., Y. C. Chen, D. L. Spector, and T. de Lange. 2008. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature 456:524-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eliezer, Y., L. Argaman, A. Rhie, A. J. Doherty, and M. Goldberg. 2009. The direct interaction between 53BP1 and MDC1 is required for the recruitment of 53BP1 to sites of damage. J. Biol. Chem. 284:426-435. [DOI] [PubMed] [Google Scholar]
- 21.Farmer, H., N. McCabe, C. J. Lord, A. N. Tutt, D. A. Johnson, T. B. Richardson, M. Santarosa, K. J. Dillon, I. Hickson, C. Knights, N. M. Martin, S. P. Jackson, G. C. Smith, and A. Ashworth. 2005. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434:917-921. [DOI] [PubMed] [Google Scholar]
- 22.Finkel, T., M. Serrano, and M. A. Blasco. 2007. The common biology of cancer and ageing. Nature 448:767-774. [DOI] [PubMed] [Google Scholar]
- 23.Franco, S., M. Gostissa, S. Zha, D. B. Lombard, M. M. Murphy, A. A. Zarrin, C. Yan, S. Tepsuporn, J. C. Morales, M. M. Adams, Z. Lou, C. H. Bassing, J. P. Manis, J. Chen, P. B. Carpenter, and F. W. Alt. 2006. H2AX prevents DNA breaks from progressing to chromosome breaks and translocations. Mol. Cell 21:201-214. [DOI] [PubMed] [Google Scholar]
- 24.Franco, S., H. J. van de Vrugt, P. Fernandez, M. Aracil, F. Arwert, and M. A. Blasco. 2004. Telomere dynamics in Fancg-deficient mouse and human cells. Blood 104:3927-3935. [DOI] [PubMed] [Google Scholar]
- 25.Gao, Y., J. Chaudhuri, C. Zhu, L. Davidson, D. T. Weaver, and F. W. Alt. 1998. A targeted DNA-PKcs-null mutation reveals DNA-PK-independent functions for KU in V(D)J. recombination. Immunity 9:367-376. [DOI] [PubMed] [Google Scholar]
- 26.Gorgoulis, V. G., L. V. Vassiliou, P. Karakaidos, P. Zacharatos, A. Kotsinas, T. Liloglou, M. Venere, R. A. Ditullio, Jr., N. G. Kastrinakis, B. Levy, D. Kletsas, A. Yoneta, M. Herlyn, C. Kittas, and T. D. Halazonetis. 2005. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434:907-913. [DOI] [PubMed] [Google Scholar]
- 27.Haince, J. F., S. Kozlov, V. L. Dawson, T. M. Dawson, M. J. Hendzel, M. F. Lavin, and G. G. Poirier. 2007. Ataxia telangiectasia mutated (ATM) signaling network is modulated by a novel poly(ADP-ribose)-dependent pathway in the early response to DNA-damaging agents. J. Biol. Chem. 282:16441-16453. [DOI] [PubMed] [Google Scholar]
- 28.Haince, J. F., D. McDonald, A. Rodrigue, U. Dery, J. Y. Masson, M. J. Hendzel, and G. G. Poirier. 2008. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J. Biol. Chem. 283:1197-1208. [DOI] [PubMed] [Google Scholar]
- 29.Harper, J. W., and S. J. Elledge. 2007. The DNA damage response: ten years after. Mol. Cell 28:739-745. [DOI] [PubMed] [Google Scholar]
- 30.Henrie, M. S., A. Kurimasa, S. Burma, J. Menissier-de Murcia, G. de Murcia, G. C. Li, and D. J. Chen. 2003. Lethality in PARP-1/Ku80 double mutant mice reveals physiological synergy during early embryogenesis. DNA Repair 2:151-158. [DOI] [PubMed] [Google Scholar]
- 31.Hoeijmakers, J. H. 2009. DNA damage, aging, and cancer. N. Engl. J. Med. 361:1475-1485. [DOI] [PubMed] [Google Scholar]
- 32.Ishizuka, S., K. Martin, C. Booth, C. S. Potten, G. de Murcia, A. Burkle, and T. B. Kirkwood. 2003. Poly(ADP-ribose) polymerase-1 is a survival factor for radiation-exposed intestinal epithelial stem cells in vivo. Nucleic Acids Res. 31:6198-6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jazayeri, A., J. Falck, C. Lukas, J. Bartek, G. C. Smith, J. Lukas, and S. P. Jackson. 2006. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 8:37-45. [DOI] [PubMed] [Google Scholar]
- 34.Karanjawala, Z. E., N. Adachi, R. A. Irvine, E. K. Oh, D. Shibata, K. Schwarz, C. L. Hsieh, and M. R. Lieber. 2002. The embryonic lethality in DNA ligase IV-deficient mice is rescued by deletion of Ku: implications for unifying the heterogeneous phenotypes of NHEJ mutants. DNA Repair 1:1017-1026. [DOI] [PubMed] [Google Scholar]
- 35.Khanna, K. K., and S. P. Jackson. 2001. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 27:247-254. [DOI] [PubMed] [Google Scholar]
- 36.Kirshner, M., M. Rathavs, A. Nizan, J. Essers, R. Kanaar, Y. Shiloh, and A. Barzilai. 2009. Analysis of the relationships between ATM and the Rad54 paralogs involved in homologous recombination repair. DNA Repair 8:253-261. [DOI] [PubMed] [Google Scholar]
- 37.Lieber, M. R. 2008. The mechanism of human nonhomologous DNA end joining. J. Biol. Chem. 283:1-5. [DOI] [PubMed] [Google Scholar]
- 38.Lundin, C., K. Erixon, C. Arnaudeau, N. Schultz, D. Jenssen, M. Meuth, and T. Helleday. 2002. Different roles for nonhomologous end joining and homologous recombination following replication arrest in mammalian cells. Mol. Cell. Biol. 22:5869-5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manis, J. P., J. C. Morales, Z. Xia, J. L. Kutok, F. W. Alt, and P. B. Carpenter. 2004. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat. Immunol. 5:481-487. [DOI] [PubMed] [Google Scholar]
- 40.Masutani, M., T. Nozaki, K. Nakamoto, H. Nakagama, H. Suzuki, O. Kusuoka, M. Tsutsumi, and T. Sugimura. 2000. The response of Parp knockout mice against DNA damaging agents. Mutat. Res. 462:159-166. [DOI] [PubMed] [Google Scholar]
- 41.Matsuoka, S., B. A. Ballif, A. Smogorzewska, E. R. McDonald III, K. E. Hurov, J. Luo, C. E. Bakalarski, Z. Zhao, N. Solimini, Y. Lerenthal, Y. Shiloh, S. P. Gygi, and S. J. Elledge. 2007. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316:1160-1166. [DOI] [PubMed] [Google Scholar]
- 42.McCabe, N., N. C. Turner, C. J. Lord, K. Kluzek, A. Bialkowska, S. Swift, S. Giavara, M. J. O'Connor, A. N. Tutt, M. Z. Zdzienicka, G. C. Smith, and A. Ashworth. 2006. Deficiency in the repair of DNA damage by homologous recombination and sensitivity to poly(ADP-ribose) polymerase inhibition. Cancer Res. 66:8109-8115. [DOI] [PubMed] [Google Scholar]
- 43.Menisser-de Murcia, J., M. Mark, O. Wendling, A. Wynshaw-Boris, and G. de Murcia. 2001. Early embryonic lethality in PARP-1 Atm double-mutant mice suggests a functional synergy in cell proliferation during development. Mol. Cell. Biol. 21:1828-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mills, K. D., D. O. Ferguson, and F. W. Alt. 2003. The role of DNA breaks in genomic instability and tumorigenesis. Immunol. Rev. 194:77-95. [DOI] [PubMed] [Google Scholar]
- 45.Mills, K. D., D. O. Ferguson, J. Essers, M. Eckersdorff, R. Kanaar, and F. W. Alt. 2004. Rad54 and DNA ligase IV cooperate to maintain mammalian chromatid stability. Genes Dev. 18:1283-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morales, J. C., Z. Xia, T. Lu, M. B. Aldrich, B. Wang, C. Rosales, R. E. Kellems, W. N. Hittelman, S. J. Elledge, and P. B. Carpenter. 2003. Role for the BRCA1 C-terminal repeats (BRCT) protein 53BP1 in maintaining genomic stability. J. Biol. Chem. 278:14971-14977. [DOI] [PubMed] [Google Scholar]
- 47.Morrison, C., G. C. Smith, L. Stingl, S. P. Jackson, E. F. Wagner, and Z. Q. Wang. 1997. Genetic interaction between PARP and DNA-PK in V(D)J recombination and tumorigenesis. Nat. Genet. 17:479-482. [DOI] [PubMed] [Google Scholar]
- 48.Morrison, C., E. Sonoda, N. Takao, A. Shinohara, K. Yamamoto, and S. Takeda. 2000. The controlling role of ATM in homologous recombinational repair of DNA damage. EMBO J. 19:463-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Panier, S., and D. Durocher. 2009. Regulatory ubiquitylation in response to DNA double-strand breaks. DNA Repair 8:436-443. [DOI] [PubMed] [Google Scholar]
- 50.Pelanda, R., S. Schwers, E. Sonoda, R. M. Torres, D. Nemazee, and K. Rajewsky. 1997. Receptor editing in a transgenic mouse model: site, efficiency, and role in B cell tolerance and antibody diversification. Immunity 7:765-775. [DOI] [PubMed] [Google Scholar]
- 51.Pleschke, J. M., H. E. Kleczkowska, M. Strohm, and F. R. Althaus. 2000. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 275:40974-40980. [DOI] [PubMed] [Google Scholar]
- 52.Ramiro, A. R., M. Jankovic, E. Callen, S. Difilippantonio, H. T. Chen, K. M. McBride, T. R. Eisenreich, J. Chen, R. A. Dickins, S. W. Lowe, A. Nussenzweig, and M. C. Nussenzweig. 2006. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature 440:105-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reina-San-Martin, B., J. Chen, A. Nussenzweig, and M. C. Nussenzweig. 2007. Enhanced intra-switch region recombination during immunoglobulin class switch recombination in 53BP1-/- B cells. Eur. J. Immunol. 37:235-239. [DOI] [PubMed] [Google Scholar]
- 54.Robert, I., F. Dantzer, and B. Reina-San-Martin. 2009. Parp1 facilitates alternative NHEJ, whereas Parp2 suppresses IgH/c-myc translocations during immunoglobulin class switch recombination. J. Exp. Med. 206:1047-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rodrigue, A., M. Lafrance, M. C. Gauthier, D. McDonald, M. Hendzel, S. C. West, M. Jasin, and J. Y. Masson. 2006. Interplay between human DNA repair proteins at a unique double-strand break in vivo. EMBO J. 25:222-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saintigny, Y., F. Delacote, G. Vares, F. Petitot, S. Lambert, D. Averbeck, and B. S. Lopez. 2001. Characterization of homologous recombination induced by replication inhibition in mammalian cells. EMBO J. 20:3861-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schreiber, V., F. Dantzer, J. C. Ame, and G. de Murcia. 2006. Poly(ADP-ribose): novel functions for an old molecule. Nat. Rev. Mol. Cell Biol. 7:517-528. [DOI] [PubMed] [Google Scholar]
- 58.Schultz, N., E. Lopez, N. Saleh-Gohari, and T. Helleday. 2003. Poly(ADP-ribose) polymerase (PARP-1) has a controlling role in homologous recombination. Nucleic Acids Res. 31:4959-4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sengupta, S., A. I. Robles, S. P. Linke, N. I. Sinogeeva, R. Zhang, R. Pedeux, I. M. Ward, A. Celeste, A. Nussenzweig, J. Chen, T. D. Halazonetis, and C. C. Harris. 2004. Functional interaction between BLM helicase and 53BP1 in a Chk1-mediated pathway during S-phase arrest. J. Cell Biol. 166:801-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sonoda, E., Y. Pewzner-Jung, S. Schwers, S. Taki, S. Jung, D. Eilat, and K. Rajewsky. 1997. B cell development under the condition of allelic inclusion. Immunity 6:225-233. [DOI] [PubMed] [Google Scholar]
- 61.Stewart, G. S. 2009. Solving the RIDDLE of 53BP1 recruitment to sites of damage. Cell Cycle 8:1532-1538. [DOI] [PubMed] [Google Scholar]
- 62.Takeyama, K., S. Monti, J. P. Manis, P. Dal Cin, G. Getz, R. Beroukhim, S. Dutt, J. C. Aster, F. W. Alt, T. R. Golub, and M. A. Shipp. 2008. Integrative analysis reveals 53BP1 copy loss and decreased expression in a subset of human diffuse large B-cell lymphomas. Oncogene 27:318-322. [DOI] [PubMed] [Google Scholar]
- 63.Tripathi, V., T. Nagarjuna, and S. Sengupta. 2007. BLM helicase-dependent and -independent roles of 53BP1 during replication stress-mediated homologous recombination. J. Cell Biol. 178:9-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Unal, E., A. Arbel-Eden, U. Sattler, R. Shroff, M. Lichten, J. E. Haber, and D. Koshland. 2004. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol. Cell 16:991-1002. [DOI] [PubMed] [Google Scholar]
- 65.van Attikum, H., and S. M. Gasser. 2009. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 19:207-217. [DOI] [PubMed] [Google Scholar]
- 66.Wang, X., L. Liu, C. Montagna, T. Ried, and C. X. Deng. 2007. Haploinsufficiency of Parp1 accelerates Brca1-associated centrosome amplification, telomere shortening, genetic instability, apoptosis, and embryonic lethality. Cell Death Differ. 14:924-931. [DOI] [PubMed] [Google Scholar]
- 67.Wang, Z. Q., B. Auer, L. Stingl, H. Berghammer, D. Haidacher, M. Schweiger, and E. F. Wagner. 1995. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev. 9:509-520. [DOI] [PubMed] [Google Scholar]
- 68.Ward, I. M., K. Minn, J. van Deursen, and J. Chen. 2003. p53 binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol. Cell. Biol. 23:2556-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ward, I. M., B. Reina-San-Martin, A. Olaru, K. Minn, K. Tamada, J. S. Lau, M. Cascalho, L. Chen, A. Nussenzweig, F. Livak, M. C. Nussenzweig, and J. Chen. 2004. 53BP1 is required for class switch recombination. J. Cell Biol. 165:459-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.West, S. C. 2003. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 4:435-445. [DOI] [PubMed] [Google Scholar]
- 71.Xie, A., N. Puget, I. Shim, S. Odate, I. Jarzyna, C. H. Bassing, F. W. Alt, and R. Scully. 2004. Control of sister chromatid recombination by histone H2AX. Mol. Cell 16:1017-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang, Y. G., U. Cortes, S. Patnaik, M. Jasin, and Z. Q. Wang. 2004. Ablation of PARP-1 does not interfere with the repair of DNA double-strand breaks, but compromises the reactivation of stalled replication forks. Oncogene 23:3872-3882. [DOI] [PubMed] [Google Scholar]
- 73.Yoo, E., B. U. Kim, S. Y. Lee, C. H. Cho, J. H. Chung, and C. H. Lee. 2005. 53BP1 is associated with replication protein A and is required for RPA2 hyperphosphorylation following DNA damage. Oncogene 24:5423-5430. [DOI] [PubMed] [Google Scholar]
- 74.Zha, S., J. Sekiguchi, J. W. Brush, C. H. Bassing, and F. W. Alt. 2008. Complementary functions of ATM and H2AX in development and suppression of genomic instability. Proc. Natl. Acad. Sci. U. S. A. 105:9302-9306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.