Abstract
Members of the genus Orthoreovirus replicate in cytoplasmic inclusions termed viral factories. Compelling evidence suggests that the nonstructural protein μNS forms the matrix of the factories and recruits specific viral proteins to these structures. In the first part of this study, we analyzed the properties of avian reovirus factories and μNS-derived inclusions and found that they are nonaggresome cytoplasmic globular structures not associated with the cytoskeleton which do not require an intact microtubule network for formation and maturation. We next investigated the capacity of avian reovirus μNS to form inclusions in transfected and baculovirus-infected cells. Our results showed that μNS is the main component of the inclusions formed by recombinant baculovirus expression. This, and the fact that μNS is able to self-associate inside the cell, suggests that μNS monomers contain all the interacting domains required for inclusion formation. Examination of the inclusion-forming capacities of truncated μNS versions allowed us to identify the region spanning residues 448 to 635 of μNS as the smallest that was inclusion competent, although residues within the region 140 to 380 seem to be involved in inclusion maturation. Finally, we investigated the roles that four different motifs present in μNS(448-635) play in inclusion formation, and the results suggest that the C-terminal tail domain is a key determinant in dictating the initial orientation of monomer-to-monomer contacts to form basal oligomers that control inclusion shape and inclusion-forming efficiency. Our results contribute to an understanding of the generation of structured protein aggregates that escape the cellular mechanisms of protein recycling.
Most viruses replicate within cellular compartments that are termed viroplasms, viral factories, or virus inclusion bodies. These compartments, which are held together by protein-protein interactions, are thought to concentrate those viral components required to increase the overall efficiency of the replication process (22, 23). Factories made by cytoplasmic viruses are large inclusions that usually form at pericentriolar sites close to the microtubule-organizing center (MTOC) and resemble in many ways the aggresomes formed in cells in response to protein misfolding/aggregation (14, 32, 33). Misfolded/aggregated proteins initially form small globular structures that are delivered to the MTOC by dynein-mediated retrograde transport along microtubules for subsequent degradation by proteasomes. When the degradative capacity of the proteasome is exceeded, these aggregates coalesce to form aggresomes, which require microtubules in order to form but not to be maintained. These phase-dense structures located in the perinuclear region are often ubiquitinated, contain dynein, and are surrounded by a vimentin cage to facilitate their positioning near the MTOC.
Avian reoviruses (ARVs) are members of the genus Orthoreovirus, one of the 12 genera of the family Reoviridae (1, 20). These viruses have been described as the etiological agents of several disease conditions that may cause important economic losses in the poultry industry (13, 29). They are nonenveloped viruses that replicate in the cytoplasm of infected cells and contain a genome composed of 10 double-stranded RNA (dsRNA) segments encased within two concentric protein shells (26). Avian reovirions contain at least 8 structural proteins (31). In addition, four viral proteins have been detected in infected cells, but not in viral particles, and thus were designated nonstructural proteins (2, 30, 31). The two major nonstructural proteins μNS and σNS are encoded by the M3 and S4 genes, respectively (30), whereas p10 and p17 are encoded by the first two cistrons of the tricistronic S1 gene (2).
ARVs replicate in cytoplasmic globular inclusions called viral factories, which contain structural and nonstructural viral proteins but lack membranes and cellular organelles (27). Individual expression of viral proteins in transfected cells revealed that μNS is the only avian reovirus protein capable of forming inclusions when expressed in the absence of any other viral factor (28). This, and the fact that the inclusions formed by μNS in transfected cells are very similar in appearance to the globular reovirus factories of infected cells, suggests that μNS is the minimal viral factor required for factory formation in avian-reovirus-infected cells. Examination of transfected cells coexpressing μNS and other viral proteins revealed that μNS plays an important role in the early stages of virus morphogenesis and that the recruitment of avian reovirus proteins into viral factories is a selective and temporally controlled process (27). This prompted us to extend previous studies in order to further characterize this protein. We considered as a starting reference a recent study of the domain composition of the related mammalian reovirus (MRV) μNS protein (3). Avian and mammalian (Dearing strain) reovirus μNS proteins show 28.3% deduced-sequence identity (179 positions). On the other hand, the mammalian protein is 86 amino acids longer and makes more primary contacts with other viral structural and nonstructural proteins than does the avian reovirus μNS protein (4, 5, 21, 27, 28). Although μNS proteins from all MRV strains produce globular inclusions when expressed in transfected cells, most MRV strains produce viral factories with filamentous morphology during infection (5, 25). The filamentous phenotype of MRV factories was attributed to protein μ2, because of its capacity to associate with both microtubules and MRV μNS (5). The formation of MRV factories in infected cells, whether globular or filamentous, is dependent on the microtubule network, since treatment of the infected cells with the microtubule-depolymerizing drug nocodazole induces the appearance of many smaller μNS inclusions dispersed throughout the cytoplasm (25). A similar effect was observed with the globular inclusions made by MRV μNS in transfected cells (5). These results suggest that, as with aggresomes, retrograde transport along microtubules is required for the initially small MRV-derived inclusions to coalesce into large perinuclear inclusions near the MTOC (5). Expression of truncated versions of MRV μNS in transfected cells revealed that the segment spanning residues 471 to 721 [μNS(471-721)] is the smallest MRV μNS region shown to be necessary and sufficient for forming inclusions (3). This region is predicted to contain two sequences with high probability of forming coiled-coil structures, which are linked by an intercoil region, preceded by a stretch of ∼50 residues, and followed by a C-terminal downstream tail (3, 17). MRV-μNS C-terminal residues, as well as His-570 and Cys-572 located within the intercoil region, were shown to be strictly required for forming inclusions (3). Removal of the first predicted coiled coil from MRV μNS(471-721) was accompanied by loss of the phenotype, which could be restored by replacement of this region with the green fluorescent protein (GFP), but not with an epitope of the influenza virus hemagglutinin (HA) (3). Since GFP, but not HA, is known to dimerize, the authors interpreted this to mean that the role of coiled coil 1 is in self-association, providing interaction domains that could mediate the formation of basal oligomers.
In this study, we performed experiments to further characterize the cytoplasmic globular structures made by ARV and by its μNS protein. Throughout this article, we call virus-derived globular structures “factories” and ARV μNS-derived ones “inclusions,” since, although they are related structures, the factories are thought to contain subviral particles and viral RNAs that are not present in μNS-derived inclusions. Our results demonstrate that ARV-derived factories and μNS-derived inclusions are globular structures whose formation and evolution are not dependent on the microtubule network and are not related to aggresome or autophagosome generation. Thus, they appear to be structured protein aggregates that escape the cellular mechanisms that eliminate misfolded or aberrant proteins. We also show both that ARV μNS monomers have the capacity to oligomerize inside the cell and that its C-terminal 187-residue region retains inclusion-forming activity. Finally, our results reveal that the 30-residue C-terminal tail domain is directly involved in establishing and orienting the main monomer-to-monomer contacts that dictate the inclusion-forming efficiency of ARV μNS.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
Primary cultures of chicken embryo fibroblasts (CEFs) were prepared from 9- to 10-day-old chicken embryos (18) and grown in monolayers in medium 199 (Invitrogen, Barcelona, Spain) supplemented with 10% (wt/vol) tryptose-phosphate broth and 5% (vol/vol) calf serum. Cos-7 cells were grown in monolayers in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum. The Sf9 insect cell line was grown in suspension culture in serum-free Sf-900 II medium (Invitrogen, Barcelona, Spain) at 27°C. The propagation of baculoviruses in Sf9 cells has been described previously (12). ARV S1133 was grown in monolayers of primary CEFs as previously described (9). Rabbit polyclonal antibodies against ARV S1133 μNS protein were raised in our laboratory (28). The following antibodies were purchased from Sigma-Aldrich (Madrid, Spain): anti-α-tubulin mouse monoclonal (clone DM1A), anti-γ-tubulin mouse monoclonal (clone GTU-88), anti-vimentin mouse monoclonal (clone VIM-13.2), and Alexa 594- and Alexa 488-conjugated antibodies against mouse and rabbit IgG, respectively. The mouse monoclonal antibody (MAb) FK2 against conjugated ubiquitin (7) was from Biomol International L.P. (Exeter, United Kingdom).
Transfections and immunofluorescence (IF) microscopy.
Transfections of cell monolayers were done with the Lipofectamine Plus reagent (Invitrogen, Barcelona, Spain) according to the manufacturer's instructions. Transfected cells were incubated at 37°C for 18 h unless otherwise stated.
For indirect immunofluorescence microscopy, cell monolayers grown on coverslips were infected or transfected as indicated in the figure legends. At the indicated times, the monolayers were fixed either with 4% paraformaldehyde or with 100% methanol. Paraformaldehyde-fixed cells were permeabilized with 0.5% Triton X-100 in phosphate-buffered saline (PBS) and then blocked in PBS containing 2% bovine serum albumin. For methanol-fixed cells, the permeabilization step was omitted. After incubation with primary antibodies for 1 h, the cells were incubated for 1 h with secondary antibodies and DAPI (4′,6′-diamidino-2-phenylindole). Images were obtained with an Olympus DP-71 digital camera mounted on an Olympus BX51 fluorescence microscope. Differential interference contrast (DIC) images were obtained to determine the locations of the viral inclusion bodies. The images were processed with Adobe Photoshop (Adobe Systems, CA).
Infection of CEFs with S1133 in the presence of nocodazole and determination of viral titers.
CEFs were infected with S1133 virus at a multiplicity of infection (MOI) of 5 at 4°C for 1 h. Unadsorbed virus was then removed and replaced by fresh incubation medium containing 10 μM nocodazole or without nocodazole. Infection was allowed to proceed for 18 h at 37°C, and the cells were then washed 3 times with PBS and harvested. Viral titers were determined by plaque assays of CEFs. Titers were obtained from two separate assays performed in duplicate, and no effect of the nocodazole treatment on the viral titer was observed.
Immunoblotting.
For Western blot analysis, cell extracts were resolved by SDS-PAGE, and proteins in unfixed gels were transferred to polyvinylidene difluoride (PVDF) membranes (Immobilon-P Millipore, Madrid, Spain) for 1 h at 100 mA in a semidry blotting apparatus (Bio-Rad, CA). Protein bands were detected with specific antibodies using the Immobilon Western Chemiluminiscent HRP Substrate (Millipore, Madrid, Spain).
Mammalian two-hybrid assay.
Transfections of preconfluent cell monolayers were done with the Lipofectamine Plus reagent (Invitrogen, Barcelona, Spain) according to the manufacturer's instructions. The day before transfection, Cos-7 cells were seeded in 12-well tissue culture plates at a density of approximately 5 × 104 cells per well and then transfected with the firefly luciferase plasmid pG5luc plus the pM-μNS and pVP16-μNS plasmids at a 1:1:1 ratio. The cells were also transfected with the control plasmids included in the kit (Clontech, Saint-Germain en Laye, France): pM, pVP16, pM53 (expressing GAL4 DNA-BD fused to mouse p53), and pVP16T (expressing VP16 AD fused to the simian virus 40 [SV40] large T antigen). After 48 h at 37°C, the cells were lysed, and the luciferase activity of cell extracts was determined with a Luminoskan-ascent luminometer (Thermo, Waltham, MA). The results of six replicates are expressed as the mean relative light units (RLU) per well ± the standard deviation.
Plasmid constructions.
The construction of the recombinant plasmid pCINeo-μNS, which expresses full-length μNS, has been described previously (27).To generate plasmids expressing μNS truncations, start and stop codons and restriction sites were introduced at different positions in the avian reovirus S1133 M3 gene by PCR amplification of the desired M3 region, using pGEMT-M3 as a template (27). Each PCR product was cut with EcoRI and XbaI and then ligated to the pCINeo or pCDNA3.1/Zeo vector, which had been cut with the same enzymes.
The pEGFP-C1 and pEGFP-N1 vectors (BD Biosciences, Madrid, Spain) were used to express fusions of Aequorea victoria enhanced green fluorescent protein (EGFP) to the N terminus or C terminus of the desired μNS region. EcoRI and BamHI sites were introduced at the desired positions in the M3 gene during PCR amplification. Each PCR was performed using pGEMT-M3 (27) as a template. Each PCR product was cut with EcoRI and BamHI and ligated to pEGFP-C1 or pEGFP-N1 that had been cut with the same enzymes. The construction of pEGFP-C1-M3 to express EGFP fused to the N terminus of μNS (GFP-μNS) was previously described (27).
To express the C-terminal dimerization domain of the HIV-1 capsid protein CA-C(146-231) or the mutant CA-C-M W184A(146-231) fused to the N terminus of the desired μNS region, the recombinant plasmids pET21b(+)-CA-C(146-231) and the pET21b(+)-CA-C-M W184A(146-231) (19) were subjected to PCR amplification with the following primers: the forward primer was 5′-GCGGGATTCATCATGAGCCCTACCAGCATTCTGG-3′ (the BamHI site is single underlined, and the start codon is doubled underlined), and the reverse primer was 5′-GCGGAATTCCAAAACTCTTGCCTTATGGCC-3′ (the EcoRI site is underlined). The amplification products were digested and cloned into the corresponding sites of the recombinant plasmids containing different deletion mutants of μNS that were previously generated (http://webspersoais.usc.es/export/sites/default/persoais/jose.martinez.costas/SUPPLEMENTAL_DATA.pdf, Table S3).
To generate the recombinant plasmids pM-μNS and pVP16-μNS, the recombinant plasmid pGEMT-M3 (27) was subjected to PCR amplification with the following primers. The forward primer was 5′-GCGGAATTCATCATGGCGTCAACCAAGTGG-3′ (the EcoRI site is underlined), and the reverse primer was 5′-GCGTCTAGATCACAGATCATCCACCAATTCTTC-3′ (the XbaI site is underlined, and the stop codon is double underlined). The resulting amplified product was digested and cloned into the EcoRI and XbaI sites of the pM and pVP16 vectors (Clontech, Saint-Germain en Laye, France).
The QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) was used to generate the recombinant plasmids that expressed the point mutants μNS-H487Q and μNS-C489S, using pCINeo-μNS as a template (27). The following mutagenic oligonucleotide primers were used: for the production of pCINeo-μNS(H487Q), the sense primer was 5′-GTTGGCTTATCTCAATGAACAGGTGTGTGTAAATGCTAAAGATC-3′ and the antisense primer was 5′-GATCTTTAGCATTTACACACACCTGTTCATTGAGATAAGCCAAC- 3′; for the production of pCINeo-μNS(C489S), the sense primer was 5′-GTTGGCTTATCTCAATGAACACGTGAGTGTAAATGCTAAAGATC-3′ and the antisense primer was 5′-GATCTTTAGCATTTACACTCACGTGTTCATTGAGATAAGCCAAC-3′.
The correctness of the constructs was confirmed by sequencing and by Western blot analysis of the expressed proteins. The primer sequences used for the generation of the different μNS constructions are available as supplemental data (http://webspersoais.usc.es/export/sites/default/persoais/jose.martinez.costas/SUPPLEMENTAL_DATA.pdf, Tables S1 to S3). The μNS-expressing constructs were named for the residues of μNS that the expressed proteins contained (http://webspersoais.usc.es/export/sites/default/persoais/jose.martinez.costas/SUPPLEMENTAL_DATA.pdf, Tables S1 to S3).
Construction of recombinant baculovirus.
Recombinant baculoviruses were generated using the Bac-to-Bac system (Invitrogen, Barcelona, Spain) following the supplier's protocols. For construction of the transfer vectors, sequences of the full-length μNS and μNS-Mi [μNS(448-635)] were obtained by PCR amplification using specific primers and plasmid pGEMT-M3 as a template (27). The PCR products were cloned into the EcoRI and XbaI sites of the pFastBac1 plasmid.
To express the recombinant GFP-μNS(448-605) protein, the coding sequence was obtained by PCR, using plasmid pEGFP-C1-μNS(448-605) (http://webspersoais.usc.es/export/sites/default/persoais/jose.martinez.costas/SUPPLEMENTAL_DATA.pdf, Table S2) as a template. The PCR product was cloned into the BamHI and XbaI sites of pFastBac1.
The transfer vector for the CA-C(146-231)-μNS(448-605) protein was obtained by subcloning the coding sequence obtained from plasmid pCDNA3.1/Zeo-CA-C(146-231)-μNS(448-605) into pFastBac1 using BamHI and XbaI restriction enzymes.
The pFastBac1 constructs were then used to generate the corresponding recombinant baculovirus according to the supplier's protocol. The correctness of the constructs was confirmed by sequencing them. The primer sequences used in PCRs are available upon request.
Baculovirus expression and inclusion purification.
Sf9 insect cells growing in suspension were infected with 5 PFU/cell of the different recombinant baculoviruses and incubated at 27°C for 72 h. The cells were then pelleted by centrifugation for 10 min at 1,000 × g, resuspended in hypotonic buffer (10 mM HEPES, pH 7.9, 10 mM KCl, 1.5 mM MgCl2), and placed on ice for 15 min. The extract was then centrifuged at 4°C for 10 min at 2,000 × g, and the pellet was washed twice with 10 ml of hypotonic buffer. The resulting pellet was resuspended in 10 ml of hypotonic buffer and sonicated (6 pulses at 45 cycles) to break the nuclei and to shear the genomic DNA. The sonicated extract was centrifuged at 4°C for 5 min at 250 × g, and the inclusion-containing pellet was washed and centrifuged five times under the same conditions. The final pellet was resuspended in 1 ml of hypotonic buffer and analyzed by SDS-PAGE and Western blotting.
RESULTS
ARV factories and ARV-μNS-derived inclusions are not related to aggresome or autophagosome generation, and their formation and evolution are not dependent on the microtubule network.
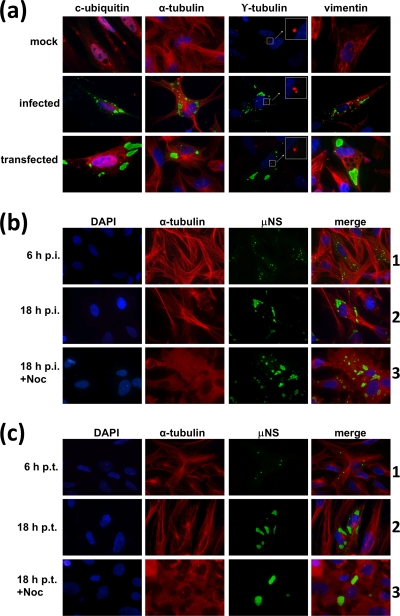
In the first part of this study, we sought to determine if the factories made by ARV in infected cells and the inclusions generated by overexpressing ARV μNS in transfected cells are related to aggresomes or autophagosomes and if their formation is dependent on an intact microtubule network. ARV-infected cells, as well as cells transfected with an ARV μNS-expressing plasmid, were immunostained with antibodies against μNS and counterstained with antibodies against (i) conjugated ubiquitin, (ii) α-tubulin (to stain microtubules), (iii) γ-tubulin (to stain the MTOC), and (iv) vimentin (to detect aggresome formation). The results revealed that ARV factories and ARV μNS-derived inclusions do not contain ubiquitinated proteins, do not colocalize with microtubules or the MTOC, and are not surrounded by a vimentin cage (Fig. 1a), indicating that these inclusions are not related to cellular aggresomes or autophagosomes.
FIG. 1.
Relationship of ARV factories and μNS-derived inclusions with the cytoskeleton and aggresome generation. (a) Semiconfluent CEF monolayers were mock infected (top row), infected with 5 PFU/cell of avian reovirus S1133 (middle row), or transfected with 1 μg of pCINeo-μNS(S1133) per well (bottom row) and then fixed at 18 h p.i. or 18 h p.t. The cells were immunostained with rabbit anti-μNS serum and then with Alexa 488-conjugated goat anti-rabbit IgG (green) and counterstained with mouse monoclonal antibodies to either conjugated ubiquitin, α-tubulin, γ-tubulin, or vimentin, as indicated above each column, followed by Alexa 594-conjugated goat anti-mouse IgG (red). Nuclei were stained with DAPI (blue). The γ-tubulin-stained MTOC is amplified for clarity. The insets are enlargements of the boxed areas. (b) Semiconfluent monolayers of CEFs were infected with 5 PFU/cell of avian reovirus S1133, and the cells were either untreated (6 h p.i. and 18 h p.i.) or incubated with 10 μM nocodazole from 0 to 18 h postinfection (18 h p.i.+Noc). All cells were fixed at the times postinfection indicated on the left and immunostained using rabbit anti-μNS serum and Alexa 488-conjugated goat anti-rabbit IgG (green). Microtubules were visualized by immunostaining them with mouse monoclonal antibodies to α-tubulin, followed by Alexa 594-conjugated goat anti-mouse IgG (red). Nuclei were counterstained with DAPI (blue). (c) Same as panel b, but the CEFs were transfected with μNS-expressing plasmid instead of infected. On the left are indicated the hours posttransfection when the cells were fixed, either untreated (6 h p.t. and 18 h p.t.) or treated with 10 μM nocodazole from 0 to 18 h posttransfection (18 h p.t.+Noc).
To determine if microtubules play a role in the formation or maturation of ARV-derived inclusions, we examined the effects of nocodazole on the morphology, evolution, and intracellular localization of these structures. In preliminary experiments using α-tubulin staining, we found that treatment of CEFs with nocodazole concentrations as low as 10 μM for 1 h completely depolymerized the cell microtubules, so we used 10 μM nocodazole for the experiments described below. CEF monolayers were infected with avian reovirus S1133 (Fig. 1b) or transfected with an ARV μNS-encoding plasmid (Fig. 1c) and subsequently treated or not with nocodazole from the beginning of the infection to 18 h postinfection (p.i.) or 18 h posttransfection (p.t.). Examination of the untreated cells with a fluorescence microscope using antibodies against ARV μNS and α-tubulin revealed that the size of the inclusions increased from 0 to 18 h postinfection or posttransfection (Fig. 1b and c, compare rows 1 and 2), suggesting that the initially formed small inclusions grew and fused with each other to form larger ones during that period. The results further showed that, in spite of the fact that microtubules were soon depolymerized after the addition of nocodazole and remained so for the next 18 h, the drug did not cause a significant change in the size and intracellular distribution of the inclusions at 18 h postinfection or posttransfection (Fig. 1b and c, compare rows 2 and 3). Furthermore, inclusion of 10 μM nocodazole from the beginning of the infection did not affect the viral titer at all (see Materials and Methods). These results demonstrate that ARV factories are not associated with cellular microtubules and that retrograde transport along microtubules is not required for the maturation of ARV-derived inclusions.
ARV-μNS homo-oligomerizes and is the main building block of μNS-derived intracellular inclusions.
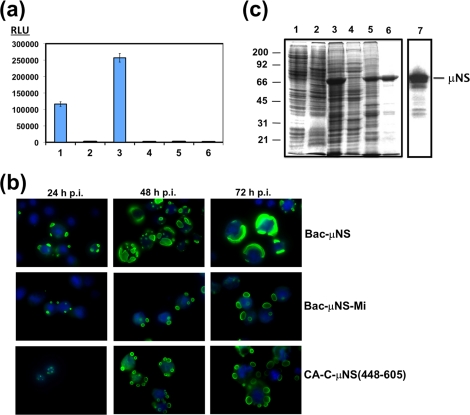
We have previously shown that transfection of CEFs with a plasmid expressing ARV μNS results in the formation of globular inclusions that resemble the viral factories made by ARV in infected cells (28), but the oligomeric status of the protein has not been demonstrated. To determine if ARV μNS is able to self-associate into oligomers within the cell, we used a mammalian two-hybrid system. For this, the ARV μNS-encoding gene was cloned into the pVP16 plasmid (to express μNS fused to the transcriptional-activation protein VP16) and into the pM plasmid (to express μNS fused to the DNA-binding domain of Gal-4). The two recombinant plasmids were transfected into Cos-7 cells, together with the luciferase reporter plasmid pG5Luc. The cells were lysed at 48 h posttransfection, and the luciferase activity of the resulting cell extracts was determined with a luminometer. The luciferase activity of the extract from cells transfected with the two ARV μNS-expressing plasmids was much higher than that of the positive-control sample (Fig. 2a, compare lanes 1 and 3), demonstrating that μNS monomers strongly self-associate to form homo-oligomers.
FIG. 2.
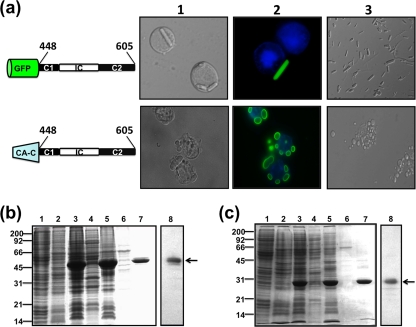
Mammalian two-hybrid analysis, and baculovirus expression of μNS-derived constructs in insect cells. (a) Mammalian Matchmaker two-hybrid analysis of μNS self-interaction. The bars represent the luciferase levels of extracts from Cos-7 cells transfected with the following plasmid combinations: 1, positive control included in the system (see Materials and Methods); 2, empty plasmids expressing activation and DNA-binding domains plus the luciferase reporter plasmid; 3, plasmids expressing μNS fused to the activation and DNA-binding domains plus luciferase reporter plasmid; 4, plasmids expressing μNS fused to the activation domain and empty plasmid expressing the DNA-binding domain plus luciferase reporter plasmid; 5, empty plasmid expressing the activation domain and plasmid expressing μNS fused to the DNA-binding domain plus luciferase reporter plasmid; 6, untransfected cells. RLU are indicated on the left. The error bars indicate standard deviations. (b) Time course analysis showing the subcellular localization of μNS, μNS-Mi, and CA-C-μNS(448-605) proteins in Sf9 cells. The last two proteins are described in Fig. 5 and 7, but they are included here for comparison. Semiconfluent monolayers of Sf9 cells were infected with a recombinant baculovirus expressing μNS (top), μNS-Mi (middle), or CA-C-μNS(448-605) (bottom). The cells were then fixed and immunostained with rabbit antibodies raised against μNS, followed by Alexa 488-conjugated goat anti-rabbit IgG (green), at the infection times indicated above the images. Nuclei were counterstained with DAPI (blue). (c) Expression, purification, and immunoblot analysis of μNS-derived inclusions. Sf9 insect cells infected with a recombinant baculovirus expressing μNS were lysed in hypotonic buffer at 72 h p.i., and the resulting cell extract (lane 3) was fractionated by centrifugation into pellet and supernatant fractions (the supernatant fraction is shown in lane 4). The pellet was then washed twice with hypotonic buffer, resuspended in the same volume of hypotonic buffer, and sonicated. The sonicated extract (lane 5) was centrifuged, and the pelleted inclusions were washed five times with hypotonic buffer (lane 6). Mock-infected and wild-type-baculovirus-infected Sf9 cell extracts, obtained by lysing the cells in hypotonic buffer at 72 h p.i., are shown in lanes 1 and 2, respectively. All samples were resolved by 10% SDS-PAGE, and the protein bands were visualized by Coomassie blue staining. The position of recombinant μNS is indicated on the right and those of the molecular weight markers on the left. The sample in lane 6 (purified μNS inclusions) was subjected to Western blot analysis with anti-μNS antibodies (lane 7).
We next purified the inclusions and analyzed their protein contents. We employed the baculovirus-insect cell system to maximize ARV μNS expression and inclusion production. The expression of recombinant ARV μNS in Sf9 insect cells was assessed by electrophoretic and immunoblotting analyses. As shown in Fig. 2c, a major protein of ∼70 kDa was detected in extracts of insect cells infected with the recombinant baculovirus (lane 3), but not in those of uninfected cells (lane 1) or of cells infected with wild-type baculovirus (lane 2). Furthermore, this protein was specifically recognized by anti-μNS polyclonal antibodies (lane 7). Visualization of the intracellular ARV μNS distribution with a fluorescence microscope revealed that the protein collected in cytoplasmic globular inclusions that were clearly visible at 24 h p.i. (Fig. 2b, top row, 24 h p.i.). At later times postinfection, the inclusions became less numerous and bigger, and they occupied most of the insect cell cytosol at 72 h p.i. At that time, some cells contained a single huge inclusion surrounding the nucleus of the spherical insect cell (Fig. 2b, top row, 48 h p.i. and 72 h p.i.). These results demonstrate that the capacity of ARV μNS to form inclusions is not dependent on cell-type-specific factors, since it forms inclusions when expressed in avian, mammalian (28), or insect cells and by different systems (ARV infection, recombinant baculovirus infection, and plasmid transfection).
To determine the protein composition of the inclusions, we purified them, avoiding the use of high salt concentrations and detergents in order to preserve their structure, as described in the legend to Fig. 2c (see also Materials and Methods). Coomassie blue staining of the SDS-PAGE gel shown in Fig. 2c revealed that recombinant ARV μNS was by far the most abundant protein present in the final pellet containing the purified inclusions (lane 6). Furthermore, some of the minor lower-molecular-mass contaminating proteins, detected as faint bands in the stained gel (lane 6), were recognized by our anti-μNS antibodies (lane 7), suggesting that they were μNS-derived proteolytic products. These results indicate that ARV μNS is the main building block in the inclusion construction in insect cells.
The last 187 amino acid residues represent the minimal inclusion-competent fraction of ARV μNS protein.
If inclusions are mainly generated by μNS-μNS interactions, each μNS monomer should contain several self-interacting domains necessary to construct the three-dimensional network required to build a large inclusion. To identify the ARV μNS domains that are necessary for inclusion formation, plasmids expressing the C-terminal and N-terminal deletion mutants shown in Fig. 3 and 4 were constructed and transfected into CEF monolayers. The identity of each construct was checked by plasmid sequencing and Western blot analysis (results not shown). The capacities of the constructs to form cytoplasmic globular inclusions were analyzed by indirect immunofluorescence, using polyclonal antibodies against ARV μNS. Anti-conjugated-ubiquitin and anti-vimentin antibodies were also used to discriminate between inclusions and structures containing aggregated/misfolded proteins (some relevant examples are shown in Fig. 4b).
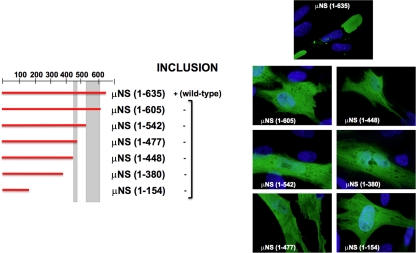
FIG. 3.
Summary of the expression of C-terminal truncations of μNS. Full-length μNS is schematically indicated by a horizontal red bar comprising residues 1 to 635 (the positions are numbered on the top and right). A similar red bar indicates each single truncation, spanning the approximate region corresponding to the construct. The positions of two previously described coiled-coil elements predicted in the sequence of μNS are indicated by two vertical gray bars. The ability of each construct to form intracellular inclusions is indicated as positive (+) or negative (−), and representative immunofluorescence images of transfected CEFs are presented on the right. The immunofluorescence analysis was performed as indicated in Fig. 2b on CEFs fixed after 18 h of transfection with plasmids expressing the indicated C-terminal μNS truncations.
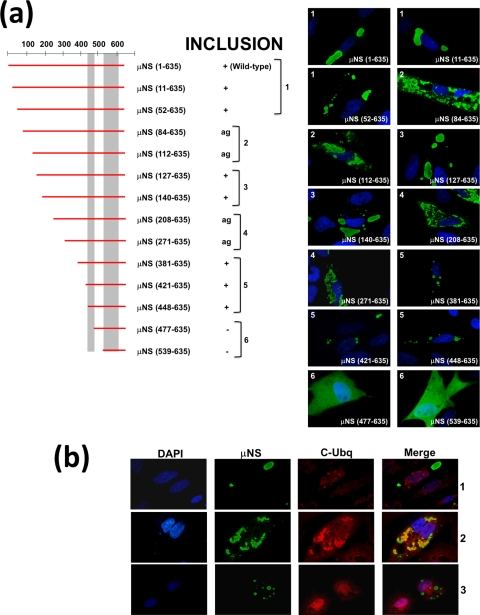
FIG. 4.
Summary of the expression of N-terminal deletions of μNS. (a) N-terminal deletions of μNS are indicated as in Fig. 3. The ability of each construct to form intracellular inclusions is indicated as positive (+), negative (−), or aggregated (ag). The immunofluorescence analysis shown on the right was performed as indicated in Fig. 2b on CEFs fixed after 18 h of transfection with plasmids expressing the indicated N-terminal μNS truncations. (b) Ubiquitination analysis of N-terminal deletions. CEFs were fixed after 18 h of transfection with plasmids expressing μNS(140-635) (row 1), μNS(112-635) (row 2), or μNS(381-635) (row 3) as examples of large, aggregated, and small inclusions, respectively. The cells were immunostained with rabbit anti-μNS antibodies (μNS) and mouse anti-conjugated ubiquitin (C-Ubq). The secondary antibodies used were Alexa 488-conjugated goat anti-rabbit IgG (green) and Alexa 594-conjugated goat anti-mouse IgG (red), respectively. In the merged image, colocalization of μNS(112-635) (row 2) and conjugated ubiquitin is indicated by yellow. Nuclei were counterstained with DAPI (blue).
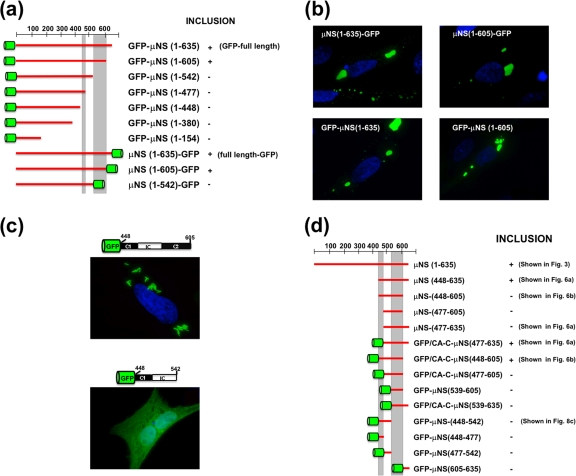
We first tested the C-terminal truncations depicted in Fig. 3. None of the truncated proteins seemed to be aggregated or misfolded, since they did not colocalize with conjugated ubiquitin or induce vimentin redistribution (results not shown). Interestingly, none of the C-terminal truncations tested was able to form inclusions but showed a diffuse intracellular distribution. These results indicate that sequences at the ARV μNS C terminus are very important for forming inclusions.
We next evaluated the importance of N-terminal sequences by analyzing the inclusion formation capacities of the N-terminal truncations shown in Fig. 4a. Deleting as many as 140 residues from the ARV μNS N terminus did not adversely affect the capacity of the viral protein to form inclusions and did not alter the inclusion morphology (Fig. 4a, compare images 1 and 3), suggesting that these residues are dispensable for inclusion formation. However, ARV μNS truncations lacking 380, 420, or 447 N-terminal residues formed inclusions that were smaller and more spherical than the ones formed by full-length μNS (Fig. 4a, compare images 1 and 3 with images 5), and the size of the small inclusions did not increase with longer times posttransfection. These results suggest that residues within the region encompassing ARV μNS residues 140 to 380 influence inclusion morphology and inclusion size. Western blot analysis of the transfected cells revealed that the smaller size of the inclusions seen in images 5 was not caused by reduced protein expression (not shown). In contrast with the above-mentioned truncations, which showed no colocalization with ubiquitin (Fig. 4b, rows 1 and 3) and did not induce vimentin reorganization (not shown), the truncations μNS(84-635), μNS(112-635), μNS(208-635), and μNS(271-635) were recognized by anti-ubiquitin antibodies in most cells (Fig. 4a, images 2 and 4, and b, row 2), suggesting that they were misfolded/aggregated. Finally, the truncations missing more than 447 N-terminal residues were no longer able to form inclusions but were uniformly distributed throughout the cell (Fig. 4a, images 6). Taken together, these results demonstrate that the segment comprising residues 448 to 635 is the minimal ARV μNS region shown to be necessary and sufficient for forming inclusions, so we designated it μNS-Mi.
As with full-length ARV μNS, μNS-Mi formed globular inclusions when expressed by recombinant baculovirus in insect cells (Fig. 2b, middle row), and these inclusions could be easily purified (not shown). The μNS-Mi-derived inclusions were smaller and more spherical than the ones made by the full-length protein (compare the top and middle rows in Fig. 2b), confirming our previous observation that residues between positions 140 and 380 influence the size and morphology of μNS-derived inclusions. Analysis of the protein composition of μNS-Mi inclusions by SDS-PAGE revealed that their main protein component is μNS-Mi (results not shown).
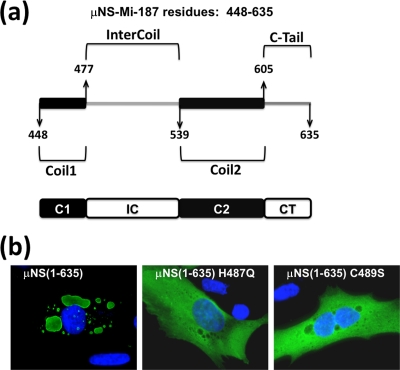
Examination of the deduced ARV μNS sequence revealed the presence of four different regions in μNS-Mi (Fig. 5a): two coiled-coil elements (one comprising residues 448 to 477, designated Coil1 or C1, and the other one comprising residues 539 to 605, designated Coil2 or C2), a 61-residue spacer region that links the two predicted coiled coils (designated Intercoil or IC), and finally, a 30-residue stretch downstream of the second predicted coiled-coil element, which encompasses residues 605 to 635 (designated C-Tail or CT). The positions of the coiled-coil elements were previously described (28) but were recalculated for this study with the Coils program (ExPASy proteomics server, Swiss Institute of Bioinformatics) (8, 15, 16), obtaining the results shown in Fig. 5A.
FIG. 5.
Domain composition of μNS-Mi and relevance of the Intercoil domain. (a) Schematic drawing of the minimal μNS region found to retain inclusion-forming ability (μNS-Mi). The four constituent domains are indicated by the amino acid residues that mark the interdomain positions: Coil1 or C1 (448 to 477), Intercoil or IC (477 to 539), Coil2 or C2 (539 to 605), and C-Tail or CT (605 to 635). (b) Immunofluorescence analysis of CEFs transfected with plasmids expressing full-length μNS(1-635) and μNS containing the single-amino-acid Intercoil mutations indicated. The Cells were immunostained as in Fig. 2b.
Although coiled coils participate in protein-protein contacts in many different proteins, such as transcription factors, myosins, and viral fibers (6, 10, 11, 24), additional interacting domains involved in inclusion formation are likely to be required for μNS-Mi to form inclusions, so we investigated the presence of these domains within the four μNS-Mi regions.
Intercoil.
It has been shown that the Intercoil region of MRV μNS contains a small consensus motif common to all of the examined μNS homologs from different reoviruses (3). This sequence contains two universally conserved residues, His570 and Cys572, and mutation of MRV μNS at each of these two residues has been demonstrated to completely eliminate its capacity to form inclusions (3). To check if the same is true for its ARV counterpart, we generated constructs encoding single mutations at equivalent residues in ARV μNS (His487 to Gln and Cys489 to Ser) and expressed the mutants in transfected cells. As shown in Fig. 5b, mutants μNS(H487Q) and μNS(C489S) were uniformly distributed throughout the cell cytoplasm, demonstrating that His487 and Cys489 are each necessary for ARV μNS to form inclusions.
Coil1 of μNS-Mi can be replaced by a foreign dimerization domain.
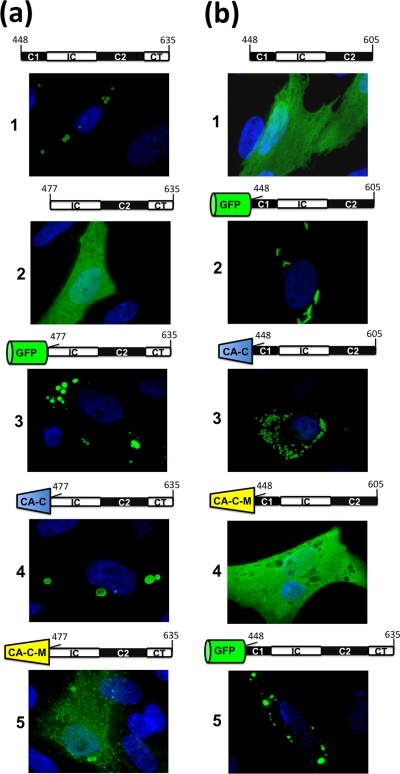
That Coil1 of ARV μNS is necessary for inclusion formation was already demonstrated by the experiments shown in Fig. 4a, since its removal from μNS-Mi to generate μNS(477-635) was accompanied by loss of the inclusion-competent phenotype. To experimentally determine the role that ARV μNS Coil1 plays in inclusion formation, we replaced Coil1 of μNS-Mi with (i) GFP, (ii) the C-terminal dimeric domain of the HIV capsid protein (CA-C) (19), and (iii) a Trp184-to-Ala point mutant of CA-C that we designated CA-C-M. We chose CA-C for two reasons. First, it is a dimer whose monomer orientation is completely different from that of GFP. Thus, while GFP monomers are oriented antiparallel in the crystal structure (34), CA-C monomers have a V-shaped parallel orientation. The second reason is that its CA-C-M point mutant has drastically reduced dimerization activity and therefore could be used as a suitable negative control (19). First, we confirmed that the different constructs shown in Fig. 6a did not colocalize with conjugated ubiquitin or cause vimentin relocalization (data not shown). As reported for MRV (3), replacement of μNS-Mi Coil1 by GFP restored the inclusion-competent phenotype (Fig. 6a, image 3), indicating that GFP is able to replace the most N-terminal predicted coiled coil of the two μNS homologs. To experimentally confirm that GFP restores the phenotype because of its dimerization activity, Coil1 was replaced by CA-C or CA-C-M. As with GFP, CA-C fused to the N terminus of μNS(477-635) displayed an inclusion-competent phenotype (Fig. 6a, image 4), indicating that Coil1 can be replaced by a dimerization domain regardless of the monomer orientation within the dimer. In contrast, the protein resulting from attaching the dimerization-deficient domain CA-C-M to the N terminus of μNS(477-635) was diffusely distributed, although some very tiny inclusions dispersed throughout the transfected cell were also observed (Fig. 6a, image 5). These results strongly suggest that Coil1 can be replaced by dimeric domains, which in turn suggests that its self-association capacity contributes to the ARV μNS inclusion-forming activity by collaborating in the generation of basal oligomers.
FIG. 6.
Immunofluorescence analysis of the intracellular expression of μNS-Mi chimeras. CEFs were transfected with plasmids expressing μNS-Mi (a1) or the different constructs indicated above each panel. The four domains present in μNS-Mi are represented following the scheme shown in Fig. 5. The green barrel represents green fluorescent protein, whereas blue and yellow truncated cones represent CA-C and CA-C-M, respectively. The transfected cells were stained as in Fig. 2b, except those containing GFP that were visualized without antibodies. (a) Substitution of Coil1. (b) Substitution of C-Tail.
The C-Tail of μNS-Mi can also be replaced by a dimerization domain, but the inclusion phenotype becomes dependent on the foreign domain.
The results shown in Fig. 3 demonstrated that the ARV μNS C-Tail is necessary for inclusion formation, since small deletions from the ARV μNS C terminus or C-Tail removal are accompanied by loss of the inclusion-competent phenotype. To better understand the role that this region plays in inclusion formation, we determined if the C-Tail of μNS-Mi could be replaced by dimeric domains.
Removal of the C-Tail from μNS-Mi to generate μNS(448-605) was accompanied by loss of the inclusion-competent phenotype (Fig. 6b, image 1), indicating that the region encompassing the two coiled coils plus the Intercoil region is not sufficient to generate inclusions. We next examined whether the C-Tail of μNS-Mi could be replaced by GFP, CA-C, and CA-C-M. We first tried to express GFP fused to the C terminus of the inclusion-deficient protein μNS(448-605), but unfortunately, the resulting construct colocalized with conjugated ubiquitin, suggesting that it was misfolded (not shown). In contrast, the proteins generated by fusing any of the three dimeric domains to the amino terminus of μNS(448-605) did not colocalize with conjugated ubiquitin or cause vimentin redistribution (results not shown), so their inclusion-forming capacities were subsequently examined. When GFP was fused to the amino terminus of μNS(448-605), the phenotype was restored, but unexpectedly, the inclusions formed by this construct (GFP-Coil1-Intercoil-Coil2) exhibited a tubular, elongated shape (Fig. 6b, image 2) instead of the typical globular appearance of the inclusions generated by all the ARV-μNS-derived mutants tested so far. For the sake of clarity, only cells containing a small number of tubular structures are shown in Fig. 6b, since they are hard to focus when taking pictures of cells containing a large number of them.
To assess whether the parallel orientation of monomers in dimeric GFP is responsible for the tubular phenotype, we next replaced GFP by CA-C or CA-C-M. Replacement by CA-C brought the inclusions back to the globular shape (Fig. 6b, image 3), while the construct generated by CA-C-M replacement was evenly distributed throughout the cell (Fig. 6b, image 4). These results indicate that (i) the orientation of monomer-monomer contacts is important for inclusion formation/shape and (ii) the C-Tail domain of μNS-Mi is probably implicated in such orientation, because its replacement by a foreign dimeric domain renders the inclusion-forming phenotype dependent on the foreign domain. To confirm the last point, and considering that the foreign domains had been fused to the N terminus of μNS-Mi whereas the C-Tail is located at the C terminus, we decided to add the C-Tail back to the construct GFP-μNS(448-605), which forms tubular inclusions. The new construct, GFP-μNS(448-635) (GFP-Coil1-Intercoil-Coil2-C-Tail) formed globular inclusions (Fig. 6b, image 5), confirming that the C-Tail is directly involved and is dominant in orienting the μNS monomer-to-monomer contacts required to form the basal oligomers that dictate the inclusion shape and inclusion-forming efficiency. This conclusion is supported by the fact that losing the C-Tail leads to an inclusion-deficient phenotype and that orientation of such contacts by a foreign dimerization domain renders the inclusion phenotype dependent on the substitute domain.
Taking into consideration the unusual shape of the rod-like inclusions and the presence of foreign epitopes, such as GFP and the CA-C domain, the possibility exists that cellular proteins are recruited to the inclusions built by these chimeras, masking or modifying the effects that GFP, CA-C, or the individual μNS-Mi domains have on the construction and orientation of the tubular and globular inclusions. To address this question, we generated recombinant baculoviruses expressing the two chimeric constructs, GFP-μNS(448-605) and CA-C-μNS(448-605), and analyzed both the shape and the protein content of the inclusions formed in infected insect cells. As in transfected CEFs, the expression of GFP-μNS(448-605) in insect cells generated fluorescent tubular inclusions (Fig. 7a, top row) that could be easily seen under light (images 1 and 3) and fluorescence (image 2) microscopy, indicating that the GFP moiety was correctly folded within the chimeric construct. Furthermore, these inclusions, whose lengths appeared to be limited only by the cell size, were thicker than the ones formed in transfected CEFs (Fig. 6b, image 2), probably because of the high expression efficiency of the baculovirus-insect cell system. This in turn suggests that the size of the tubular rods is controlled by the intracellular concentration of the inclusion-forming protein. These inclusions were purified using the protocol described in the legend to Fig. 2 for full-length μNS (Fig. 7b), and the analysis of their protein content by SDS-PAGE revealed that their main component was a 45-kDa protein (Fig. 7b, lane 7). That the 45-kDa protein was GFP-μNS(448-605) (theoretical molecular mass, 46.4 kDa) was confirmed by Western blot analysis using anti-μNS antibodies (Fig. 7b, lane 8) and anti-GFP antibodies (not shown). In contrast, CA-C-μNS(448-605) generated cytoplasmic globular inclusions in insect cells that could be immunostained with anti-μNS antibodies (Fig. 2b and 7a, bottom rows). Purified inclusions could also be seen under the light microscope (Fig. 7a, bottom row, images 1 and 3), and their main component was a 30-kDa protein (Fig. 7c, lane 7), which reacted with anti-μNS antibodies (Fig. 7c, lane 8), indicating that the protein was CA-C-μNS(448-605) (theoretical molecular mass, 28.2 kDa). The results shown in Fig. 7 also indicate that sonication of the cell extract did not dismantle the inclusions made by the μNS chimeras or change their appearance. Although the possibility still exists that such treatment might alter their protein compositions, their integrity indicates that they are mainly built using μNS monomers. The comparative analysis shown in Fig. 2b, showing a time course of the baculovirus expression of full-length μNS (top row), μNS-Mi (middle row), and CA-C-μNS(448-605) (bottom row), supports our previous suggestion that the ARV μNS region encompassing residues 140 to 380, although not required for inclusion formation, is necessary for maturation of small inclusions into bigger ones, as only the full-length protein is able to develop big, cytoplasm-wide inclusions in the insect cell.
FIG. 7.
Baculovirus expression and purification of μNS-Mi chimeras. (a) Sf9 cells were infected with baculoviruses expressing the chimeras indicated on the left. The cells were fixed 3 days after infection and then either treated with DAPI to stain the nuclei (top row) or stained with DAPI and anti-μNS antibodies and photographed under a bright-field (column 1) or fluorescence (column 2) microscope. Column 3 shows images of the tubular (top) or globular (bottom) inclusions purified from the cytoplasm of infected Sf9 cells. (b) Expression, purification, and immunoblot analysis of tubular inclusions (GFP-C1-IC-C2). Sf9 cells infected with baculovirus expressing GFP-C1-IC-C2 were lysed in hypotonic buffer at 72 h p.i., and the resulting cell extract (lane 3) was fractionated by centrifugation into pellet and supernatant fractions (the supernatant fraction is shown in lane 4). The pellet was washed twice with hypotonic buffer, resuspended in the same volume of hypotonic buffer, and sonicated. The sonicated extract (lane 5) was centrifuged and fractionated into pellet and supernatant fractions (the supernatant fraction is shown in lane 6). The pellet was then washed and centrifuged five times with hypotonic buffer (lane 7). Mock-infected or wild-type-baculovirus-infected Sf9 cell extracts lysed in hypotonic buffer at 72 h p.i. are shown in lanes 1 and 2, respectively. All samples were resolved by 12.5% SDS-PAGE, and the protein bands were visualized by Coomassie blue staining. The position of recombinant GFP-C1-IC-C2 is indicated by an arrow on the right and those of the molecular weight markers on the left. The sample in lane 7 (purified tubular inclusions) was subjected to Western blot analysis with anti-μNS antibodies (lane 8). (c) Expression, purification, and immunoblot analysis of globular inclusions (CA-C-C1-IC-C2). The expression and purification of globular inclusions was performed as for panel b. The position of recombinant CA-C-C1-IC-C2 is indicated by an arrow on the right and those of the protein markers on the left. The sample in lane 7 (purified globular inclusions) was subjected to Western blot analysis with anti-μNS antibodies (lane 8).
The C-Tail of full-length μNS can be replaced by a guide domain in the full-length μNS protein.
The results shown in Fig. 6 indicate that the C-Tail domain is directly implicated in orienting the contacts between μNS monomers, thus avoiding random interactions that would lead to reduced inclusion-forming efficiency. If this is true, ARV μNS mutants lacking the C-Tail would become inclusion competent when fused to a dimeric domain that would guide the μNS monomer-to-monomer contacts. To address this question, we constructed a series of mutants containing GFP fused to the N or C terminus of the inclusion-deficient μNS C-terminal deletions. To assess whether the GFP moiety might alter in some way the inclusion-forming capacity of μNS, we first constructed plasmids directing the expression of GFP fused to the N or the C terminus of full-length μNS. The results, summarized in Fig. 8a and b, show that (i) GFP has no effect on the inclusion-forming ability of full-length μNS (Fig. 8b, left images) and (ii) GFP is able to restore the inclusion-forming phenotype when fused to either end of C-Tail-less μNS (Fig. 8b, right images), but not when fused to larger C-terminal deletions (Fig. 8a) (http://webspersoais.usc.es/export/sites/default/persoais/jose.martinez.costas/SUPPLEMENTAL_DATA.pdf). No conjugated ubiquitin or vimentin cages were detected in the inclusions shown in Fig. 8b (not shown). These results confirm our previous conclusion that the C-Tail domain is required for μNS inclusion-forming activity because of its ability to dictate the initial monomer-to-monomer contacts that lead to the formation of correctly oriented basal oligomers.
FIG. 8.
Summary of the expression of different μNS-derived constructs. (a and d) Schematic representations of the different constructs. The presence of fused GFP or CA-C is indicated with a green barrel. +, inclusion-forming constructs; −, constructs lacking inclusion-forming activity. Constructions already shown in other figures are indicated in parentheses on the right. (b and c) Fluorescence microscope analysis of CEFs transfected with the indicated plasmids. Nuclei were stained blue with DAPI.
Coil2.
The results discussed above demonstrate that ARV μNS(448-605), which comprises the two coiled coils plus the Intercoil region, forms inclusions when its amino terminus is fused to dimeric domains, indicating that such domains can replace the absent C-Tail. To assess whether Coil2 plus the C-tail of μNS-Mi can be similarly replaced by a dimeric protein, we generated a recombinant plasmid that expressed GFP fused to the amino terminus of μNS(448-542), which lacks Coil2 plus the C-Tail. Fluorescence microscopy analysis of cells expressing GFP-μNS(448-542) revealed that the construct is evenly distributed throughout the cell cytoplasm (Fig. 8c, bottom), indicating that a dimeric protein cannot replace the absent Coil2 plus the C-Tail. On the other hand, our finding that GFP-μNS(448-605), lacking the C-Tail, forms inclusions but GFP-μNS(448-542), lacking the C-Tail plus Coil2, does not indirectly suggests that Coil2 also contains interacting domains necessary for forming inclusions (Fig. 8c, compare top and bottom images).
To identify the smallest ARV μNS region that forms inclusions when fused to dimeric domains, we generated constructs containing GFP or CA-C fused to the amino terminus of the truncated μNS-Mi versions depicted in Fig. 8d. Examination of the cells expressing these constructs with a fluorescence microscope revealed that none of them formed inclusions (Fig. 8d) (http://webspersoais.usc.es/export/sites/default/persoais/jose.martinez.costas/SUPPLEMENTAL_DATA.pdf). These results and those shown in Fig. 6 indicate that μNS(448-605) and μNS(477-635) are the smallest μNS regions that are able to form inclusions when their amino termini are fused to dimeric domains. Some of the chimeric constructs containing GFP localized to the whole cell, and some of them even appeared to accumulate preferentially at the nucleus (http://webspersoais.usc.es/export/sites/default/persoais/jose.martinez.costas/SUPPLEMENTAL_DATA.pdf). Although we do not know the exact meaning of such behavior, most of them contained very small fragments of μNS protein that were most likely carried to the nucleus by the GFP moiety.
DISCUSSION
Most viruses replicate in cellular compartments that are termed viroplasms, viral factories, or viral inclusion bodies. The replication and assembly of members of the genus Orthoreovirus also take place in phase-dense inclusions, and numerous observations suggest that their nonstructural viral protein μNS plays a key role in constructing the factories and in recruiting specific viral proteins to these structures (4, 5, 21, 25, 27, 28). The results of this study demonstrate that the construction of ARV-derived inclusions is not related to the mechanisms of aggresome and autophagosome generation, because they are not polyubiquitinated, are not surrounded by a vimentin cage, and are not positioned near the MTOC. Furthermore, in contrast to the factories formed by most MRV isolates, ARV factories are globular structures that are not associated with microtubules. The differences found in microtubule network dependence between the inclusions formed by ARV μNS and MRV μNS suggest that MRV μNS probably needs to interact with cytoskeletal proteins to travel along microtubules toward the MTOC to form large perinuclear inclusions, while its ARV counterpart is able to form large inclusions in the absence of an intact microtubule network. By using a mammalian two-hybrid system, we could demonstrate that ARV μNS monomers self-interact inside the cell. SDS-PAGE analysis of purified ARV μNS-derived inclusions formed in insect cells demonstrated that their main protein component is μNS. However, the possibility still exists that cellular proteins participate in promoting some steps of inclusion formation but either are not incorporated into mature inclusions or could be released by sonication during the purification process.
In the second part of this study, we investigated the role that ARV μNS sequences play in inclusion formation, and some of our results support the conclusions previously reached for MRV μNS: (i) sequences upstream of Coil1 (residues 1 to 448 in ARV μNS and 1 to 470 in MRV μNS) are dispensable for forming inclusions, (ii) sequences within the C-Tail are necessary, (iii) the conserved His and Cys residues within the Intercoil motif are each required for inclusion formation, and (iv) carboxy-proximal regions (residues 448 to 635 in ARV μNS and 471 to 721 in MRV μNS) are necessary and sufficient for forming inclusions. On the other hand, our finding that ARV μNS Coil1 can be replaced by GFP or CA-C, but not by CA-C-M, experimentally confirmed that this region can be replaced by a dimeric domain, supporting the previously proposed suggestion that Coil1 plays a role in self-association.
In this study, we also provided indirect evidence that ARV μNS Coil2 contains sequences required for forming inclusions, since GFP-μNS(448-605), lacking the C-Tail, forms inclusions whereas GFP-μNS(448-542), lacking both the C-Tail and Coil2, does not. However, further studies are needed to uncover the exact role of Coil2 in inclusion formation.
Although the minimal identified inclusion-forming regions of ARV and MRV μNS proteins appear to possess similar modular organizations, the MRV μNS(471-721) N terminus contains a segment of approximately 50 residues upstream of Coil1 (3) that is not present in ARV μNS-Mi. It appears that sequences within this segment are required for MRV μNS, but not for ARV μNS, to form inclusions. On the other hand, in this study, we have shown that the sequence within the ARV μNS region spanning residues 140 to 380 affects the size and morphology of the inclusions. Such an inclusion size-related domain has not been identified in MRV μNS, although residues in the region 41 to 172 of MRV μNS appear to affect inclusion morphology, since removal of this region causes the inclusions to be more elongated and “fenestrated” but not smaller. It is likely that the size of MRV μNS-derived inclusions is not influenced by a self-interacting domain but relies on their capacity to interact with cytoskeletal proteins and to travel along microtubules toward the MTOC. This hypothesis is supported by the observation that treatment with the microtubule-depolymerizing drug nocodazole prevents the formation of large inclusions by MRV μNS (5, 25), but not by ARV μNS.
Novel and interesting results were obtained when the ARV μNS C-Tail was replaced by dimeric domains. Thus, as we conclude that the C-Tail also provides self-association activity, our finding that tubular inclusions are formed when the μNS-Mi C-Tail is replaced by GFP but globular inclusions are formed when it is replaced by CA-C suggests that when the C-Tail is absent, the shape of μNS-Mi-derived inclusions becomes dependent on the foreign dimeric domain. This in turn suggests that the C-Tail dictates the initial ARV μNS monomer-to-monomer contacts required for the generation of basal oligomers with the ability to efficiently build cytoplasmic inclusions. This suggestion is further supported by our observation that adding the C-Tail back to the construct that forms tubular inclusions rescues the globular phenotype, suggesting that the C-Tail is dominant over the foreign dimeric domain. The absence of the C-Tail would probably facilitate the formation of oligomers containing disordered random interactions, which could possibly block or hide the interacting domains required for the basal oligomers to progress into inclusions.
Although we do not know the structure of GFP-μNS(448-605)-derived tubular inclusions, their observation under a scanning electron microscope suggests that they are not hollow cylinders (not shown). Furthermore, their thickness increases with the expression level, suggesting that folding intermediates are not toxic to the cell and that small rods can associate both laterally and linearly to form larger rods.
Our findings that μNS-Mi Coil1 and C-Tail can each be replaced by dimeric domains might be exploited for the identification of oligomeric proteins and oligomeric domains, since their fusion to ARV μNS(477-635) or μNS(448-605) should probably generate proteins that collect into inclusions, whereas the proteins generated by fusing monomeric domains/proteins should be diffusely distributed. We are currently investigating whether this system might have general application for the detection of dimeric domains and proteins inside living cells.
We have tried to adapt the inclusion purification protocol for the purification of viral factories from ARV-infected cells, but so far, viral factories have been resistant to purification because they seem to be less dense and more fragile than μNS inclusions. This supports the notion that ARV-μNS-derived inclusions and ARV factories are structurally and functionally different structures, since the latter should contain, in addition to μNS, other viral proteins and viral RNAs and perhaps cellular factors.
Inclusions formed by the ARV μNS protein are big protein assemblies that could be seen by the cell as protein aggregates. The wild-type protein and most inclusion-positive mutants tested produced cytoplasmic inclusions with a regular shape, nonpolyubiquitinated and not enclosed within vimentin cages, suggesting that the inclusions formed by ARV μNS are structured protein aggregates that somehow escape the cellular mechanisms of defense against protein aggregation. The availability of ubiquitination-prone mutants makes μNS a good model to study the mechanisms used by the cell to detect and fight protein aggregation.
Acknowledgments
We thank Mark van Raaij for critical reading of the manuscript and Leticia Barcia Castro for excellent technical assistance. We also thank M. Mateu for providing plasmids pET21b(+)-CA-C (146-231) and pET21b(+)-CA-C-M W184A and Laboratorios Intervet (Salamanca, Spain) for providing pathogen-free embryonated eggs.
This work was supported by grants from the European Commission under contract ERAS-CT-2003-980409 (as part of the European Science Foundation EUROCORES Programme EuroSCOPE), the Spanish Ministerio de Ciencia y Tecnología (BFU2007-61330 and BFU2005-24982-E), and the Xunta de Galicia (08CSA009203PR). A.B.-N. was the recipient of a predoctoral FPI fellowship from the Spanish Ministerio de Educación y Ciencia.
Footnotes
Published ahead of print on 24 February 2010.
REFERENCES
- 1.Attoui, H., F. Billoir, P. Biagini, P. de Micco, and X. de Lamballerie. 2000. Complete sequence determination and genetic analysis of Banna virus and Kadipiro virus: proposal for assignment to a new genus (Seadornavirus) within the family Reoviridae. J. Gen. Virol. 81:1507-1515. [DOI] [PubMed] [Google Scholar]
- 2.Bodelon, G., L. Labrada, J. Martinez-Costas, and J. Benavente. 2001. The avian reovirus genome segment S1 is a functionally tricistronic gene that expresses one structural and two nonstructural proteins in infected cells. Virology 290:181-191. [DOI] [PubMed] [Google Scholar]
- 3.Broering, T. J., M. M. Arnold, C. L. Miller, J. A. Hurt, P. L. Joyce, and M. L. Nibert. 2005. Carboxyl-proximal regions of reovirus nonstructural protein muNS necessary and sufficient for forming factory-like inclusions. J. Virol. 79:6194-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broering, T. J., J. Kim, C. L. Miller, C. D. Piggott, J. B. Dinoso, M. L. Nibert, and J. S. Parker. 2004. Reovirus nonstructural protein muNS recruits viral core surface proteins and entering core particles to factory-like inclusions. J. Virol. 78:1882-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broering, T. J., J. S. Parker, P. L. Joyce, J. Kim, and M. L. Nibert. 2002. Mammalian reovirus nonstructural protein microNS forms large inclusions and colocalizes with reovirus microtubule-associated protein micro2 in transfected cells. J. Virol. 76:8285-8297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chappell, J. D., A. E. Prota, T. S. Dermody, and T. Stehle. 2002. Crystal structure of reovirus attachment protein sigma1 reveals evolutionary relationship to adenovirus fiber. EMBO J. 21:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujimuro, M., H. Sawada, and H. Yokosawa. 1994. Production and characterisation of monoclonal antibodies specific to multi-ubiquitin chains of polyubiquitinated proteins. FEBS Lett. 349:173-180. [DOI] [PubMed] [Google Scholar]
- 8.Gasteiger, E., A. Gattiker, C. Hoogland, I. Ivanyi, R. D. Appel, and A. Bairoch. 2003. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31:3784-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grande, A., and J. Benavente. 2000. Optimal conditions for the growth, purification and storage of the avian reovirus S1133. J. Virol. Methods 85:43-54. [DOI] [PubMed] [Google Scholar]
- 10.Guardado-Calvo, P., G. C. Fox, A. L. Llamas-Saiz, and M. J. van Raaij. 2009. Crystallographic structure of the alpha-helical triple coiled-coil domain of avian reovirus S1133 fibre. J. Gen. Virol. 906:72-77. [DOI] [PubMed] [Google Scholar]
- 11.Holmes, K. C., and W. J. Lehman. 2008. Gestalt-binding of tropomyosin to actin filaments. Muscle Res. Cell Motil. 29:213-219. [DOI] [PubMed] [Google Scholar]
- 12.Hsiao, J., J. Martinez-Costas, J. Benavente, and V. N. Vakharia. 2002. Cloning, expression, and characterization of avian reovirus guanylyltransferase. Virology 296:288-299. [DOI] [PubMed] [Google Scholar]
- 13.Jones, R. C. 2000. Avian reovirus infections. Rev. Sci. Tech. 19:614-625. [DOI] [PubMed] [Google Scholar]
- 14.Kopito, R. R. 2000. Aggresomes, inclusion bodies and protein aggregation. Trends Cell. Biol. 10:524-530. [DOI] [PubMed] [Google Scholar]
- 15.Lupas, A. 1996. Prediction and analysis of coiled-coil structures. Methods Enzymol. 266:513-525. [DOI] [PubMed] [Google Scholar]
- 16.Lupas, A., M. Van Dyke, and J. Stock. 1991. Predicting coiled coils from protein sequences. Science 252:1162-1164. [DOI] [PubMed] [Google Scholar]
- 17.McCutcheon, A. M., T. J. Broering, and M. L. Nibert. 1999. Mammalian reovirus M3 gene sequences and conservation of coiled-coil motifs near the carboxyl terminus of the muNS protein. Virology 264:16-24. [DOI] [PubMed] [Google Scholar]
- 18.Martinez-Costas, J., R. Varela, and J. Benavente. 1995. Endogenous enzymatic activities of the avian reovirus S1133: identification of the viral capping enzyme. Virology 206:1017-1026. [DOI] [PubMed] [Google Scholar]
- 19.Mateu, M. G. 2002. Conformational stability of dimeric and monomeric forms of the C-terminal domain of human immunodeficiency virus-1 capsid protein. J. Mol. Biol. 318:519-531. [DOI] [PubMed] [Google Scholar]
- 20.Mertens, P. 2004. The dsRNA viruses. Virus Res. 101:3-13. [DOI] [PubMed] [Google Scholar]
- 21.Miller, C. L., T. J. Broering, J. S. Parker, M. M. Arnold, and M. L. Nibert. 2003. Reovirus sigma NS protein localizes to inclusions through an association requiring the muNS amino terminus. J. Virol. 77:4566-4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Netherton, C., K. Moffat, E. Brooks, and T. Wileman. 2007. A guide to viral inclusions, membrane rearrangements, factories, and viroplasm produced during virus replication. Adv. Virus Res. 70:101-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Novoa, R. R., G. Calderita, R. Arranz, J. Fontana, H. Granzow, and C. Risco. 2005. Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol. Cell 97:147-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Shea, E. K., J. D. Klemm, P. S. Kim, and T. Alber. 2001. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science 254:539-544. [DOI] [PubMed] [Google Scholar]
- 25.Parker, J. S., T. J. Broering, J. Kim, D. E. Higgins, and M. L. Nibert. 2002. Reovirus core protein mu2 determines the filamentous morphology of viral inclusion bodies by interacting with and stabilizing microtubules. J. Virol. 76:4483-4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spandidos, D. A., and A. F. Graham. 1976. Physical and chemical characterization of an avian reovirus. J. Virol. 19:968-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tourís-Otero, F., M. Cortez-San Martín, J. Martínez-Costas, and J. Benavente. 2004. Avian reovirus morphogenesis occurs within viral factories and begins with the selective recruitment of sigmaNS and lambdaA to microNS inclusions. J. Mol. Biol. 341:361-374. [DOI] [PubMed] [Google Scholar]
- 28.Touris-Otero, F., J. Martínez-Costas, V. N. Vakharia, and J. Benavente. 2004. Avian reovirus nonstructural protein microNS forms viroplasm-like inclusions and recruits protein sigmaNS to these structures. Virology 319:94-106. [DOI] [PubMed] [Google Scholar]
- 29.van der Heide, L. 2000. The history of avian reovirus. Avian Dis. 44:638-641. [PubMed] [Google Scholar]
- 30.Varela, R., and J. Benavente. 1994. Protein coding assignment of avian reovirus strain S1133. J. Virol. 68:6775-6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Varela, R., J. Martinez-Costas, M. Mallo, and J. Benavente. 1996. Intracellular posttranslational modifications of S1133 avian reovirus proteins. J. Virol. 70:2974-2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wileman, T. 2006. Aggresomes and autophagy generate sites for virus replication. Science 312:875-878. [DOI] [PubMed] [Google Scholar]
- 33.Wileman, T. 2007. Aggresomes and pericentriolar sites of virus assembly: cellular defense or viral design? Annu. Rev. Microbiol. 61:149-167. [DOI] [PubMed] [Google Scholar]
- 34.Yang, F., L. G. Moss, and G. N. Phillips. 1996. The molecular structure of green fluorescent protein. Nat. Biotechnol. 14:1246-1251. [DOI] [PubMed] [Google Scholar]