Abstract
The pandemic H1N1 virus of 2009 (2009 H1N1) continues to cause illness worldwide, primarily in younger age groups. To better understand the pathogenesis of these viruses in mammals, we used a mouse model to evaluate the relative virulence of selected 2009 H1N1 viruses and compared them to a representative human triple-reassortant swine influenza virus that has circulated in pigs in the United States for over a decade preceding the current pandemic. Additional comparisons were made with the reconstructed 1918 virus, a 1976 H1N1 swine influenza virus, and a highly pathogenic H5N1 virus. Mice were inoculated intranasally with each virus and monitored for morbidity, mortality, viral replication, hemostatic parameters, cytokine production, and lung histology. All 2009 H1N1 viruses replicated efficiently in the lungs of mice and possessed a high degree of infectivity but did not cause lethal disease or exhibit extrapulmonary virus spread. Transient weight loss, lymphopenia, and proinflammatory cytokine and chemokine production were present following 2009 H1N1 virus infection, but these levels were generally muted compared with a triple-reassortant swine virus and the 1918 virus. 2009 H1N1 viruses isolated from fatal cases did not demonstrate enhanced virulence in this model compared with isolates from mild human cases. Histologically, infection with the 2009 viruses resulted in lesions in the lung varying from mild to moderate bronchiolitis with occasional necrosis of bronchiolar epithelium and mild to moderate peribronchiolar alveolitis. Taken together, these studies demonstrate that the 2009 H1N1 viruses exhibited mild to moderate virulence in mice compared with highly pathogenic viruses.
The 2009 (H1N1) influenza pandemic has resulted in laboratory-confirmed cases in over 200 countries with greater than 15,000 deaths worldwide (5). The majority of infected individuals have experienced uncomplicated, upper respiratory tract illness; cases have been distinguished by symptoms which include gastrointestinal distress and vomiting in approximately 40% of patients (7, 36). While the 2009 pandemic represents the greatest incidence of human infection with influenza viruses of swine origin to date, antigenically related swine lineage viruses have previously been associated with sporadic cases of human disease and death (11, 37). Prior to 2009, the largest cluster of H1N1 swine influenza cases occurred during an outbreak in 1976 which resulted in the infection of up to 230 soldiers at Fort Dix, NJ, with 13 severe cases and one fatality (12). The outbreak was limited to Fort Dix, possibly due to the poor transmissibility of this virus (21). Triple-reassortant swine H1N1 influenza viruses (containing avian, human, and swine genes) have additionally been associated with human infection since 2005 (11, 37). While gastrointestinal symptoms following seasonal influenza virus are uncommon, diarrhea was reported in 40% of patients infected with triple-reassortant swine H1N1 viruses, similar to cases early in the 2009 pandemic (37).
The hemagglutinin (HA) gene of 2009 H1N1 belongs to the classical swine lineage, which was first introduced into swine populations circa 1918 and shares antigenic similarity with the 1918 pandemic virus as well as the 1976 H1N1 virus and the more contemporary triple-reassortant swine influenza viruses (11, 16, 44). The 1918 HA gene has been previously shown to be essential for severe pulmonary lesion development and optimal virulence (19, 40). Thus, it is important to compare the relative virulence of the 2009 H1N1 viruses to that of other H1N1 viruses that have circulated over a span of more than 90 years. Mammalian models serve an invaluable role for the study of disease severity and outcome following influenza virus infection. Previous research evaluating classical swine influenza viruses has revealed that these viruses do not consistently exhibit high virulence in the mouse model. A/NJ/8/76 virus, isolated from the outbreak in Fort Dix, was lethal only for specific strains of mice or following mouse adaptation (9, 13). Two H1N1 viruses antigenically similar to the reconstructed 1918 virus, A/Swine/IA/15/30 and A/Swine/IA/31, replicated to high titers in the lungs of mice and caused substantial weight loss at the height of infection; however, only the 1931 virus isolate exhibited lethality in this model (26, 28). However, these studies largely occurred in the context of evaluating vaccine and antiviral efficacy and did not extensively study virus pathogenesis or the host response following infection.
Due to the rapid emergence of 2009 H1N1 viruses in humans, collaborative research has been undertaken to characterize viruses isolated from this pandemic in mammalian models, including the mouse, ferret, nonhuman primate, and pig (18, 23, 30). However, much of this work has been limited by a small number of novel isolates tested and a paucity of extensive side-by-side comparison with pertinent viruses outside seasonal H1N1 isolates. To better understand the capacity of viruses isolated from the 2009 pandemic to cause disease in the context of related viruses of swine origin or highly pathogenic viruses with pandemic potential, we expanded upon a mouse model for the study of 2009 H1N1 influenza viruses associated with human infection (23). Assessment of pathogenicity in the mouse model included histopathology analysis, hemostatic measurements, and cytokine production in the lung. This work revealed that a panel of swine origin H1N1 viruses exhibited lower pathogenicity relative to an H5N1 virus and the pandemic 1918 virus. Furthermore, a triple-reassortant H1N1 virus of swine origin isolated in 2007 possessed enhanced virulence in this model compared with swine origin 2009 pandemic H1N1 viruses or the swine origin virus that sickened humans in 1976.
MATERIALS AND METHODS
Viruses.
Influenza A viruses of the H1N1 and H5N1 subtypes used in this study are shown in Table 1. Virus stocks were cultured consistent with the original isolate passage either in the allantoic cavity of 10-day-old embryonated hens' eggs (Mex/4108, Mex/4487, NJ/8, and VN/1203) or in Madin-Darby canine kidney (MDCK) cells (CA/4, TX/15, Mex/4482, OH/2, and 1918) at 37°C for 24 to 48 h as previously described (24, 40). Pooled allantoic fluid or cell supernatant was clarified by centrifugation and frozen in aliquots at −70°C. With the exception of NJ/8 (egg passage 14 [E14]), virus stocks were low-passage-number viruses (E1-E2 or cell passage 1 or 2 [C1-C2]), and the identities of virus genes were confirmed by sequence analysis to verify that no inadvertent mutations were present during the generation of virus stocks. The 50% egg infectious dose (EID50) titer for egg-grown stocks was calculated by the method of Reed and Muench (35), following serial titration in eggs. Cell-grown stocks were titrated by standard plaque assay in MDCK cells as previously described for determination of PFU titer (46). All animal experiments with 2009 H1N1 viruses were conducted under biosafety level 3 enhanced (BSL3+) containment in accordance with guidelines of the World Health Organization (https://www.who.int/csr/resources/publications/swineflu/Laboratorybioriskmanagement.pdf).
TABLE 1.
H1N1 and H5N1 viruses used in this study
| Virus | Name in study | Subtype | Patient data |
|---|---|---|---|
| A/California/4/09 | CA/4 | H1N1 | Pediatric uncomplicated, upper respiratory tract illness |
| A/Texas/15/09 | TX/15 | H1N1 | Fatal pediatric respiratory illness |
| A/Mexico/4108/09 | Mex/4108 | H1N1 | Hospitalized respiratory illness |
| A/Mexico/4482/09 | Mex/4482 | H1N1 | Severe respiratory illness |
| A/Mexico/InDRE4487/09 | Mex/4487 | H1N1 | Severe respiratory illness |
| A/New Jersey/8/76 | NJ/8 | H1N1 | Severe respiratory illness |
| A/Ohio/2/07a | OH/2 | H1N1 | Pediatric uncomplicated, upper respiratory tract illness |
| rgA/South Carolina/1/18a | 1918 | H1N1 | NAb |
| A/Vietnam/1203/04a | VN/1203 | H5N1 | Fatal pediatric respiratory illness |
Mouse in vivo experiments.
Female BALB/c mice (Charles River Laboratories, Wilmington, MA), 6 to 8 weeks of age, were deeply anesthetized with 2,2,2-tribromoethanol in tert-amyl alcohol (Avertin; Sigma-Aldrich, St. Louis, MO) and inoculated intranasally (i.n.) with 50 μl of infectious virus diluted in sterile phosphate-buffered saline (PBS). The 50% mouse infectious dose (MID50) and 50% lethal dose (LD50) were determined as previously described (22). Briefly, mice were inoculated with 10-fold dilutions (from 106 to 100 EID50 or PFU) of each virus. Three mice per group were euthanatized on day 3 postinoculation (p.i.), and homogenized lungs were serially titrated in MDCK cells by standard plaque assay to determine the MID50 calculated by the method of Reed and Muench (35). Five mice per virus were monitored daily for 14 days p.i. for morbidity, as measured by weight loss, and mortality to determine the LD50. Any mouse that lost >25% of its preinoculation body weight was euthanatized. On days 3 and 6 p.i., three mice inoculated with 105 EID50 or PFU of each virus were euthanatized for the collection of lungs, nose, spleen, intestine, thymus, and brain to determine replication and systemic spread of virus. Tissues were homogenized in 1 ml of cold PBS, and clarified homogenates were titrated in MDCK cells to determine virus infectivity, starting with undiluted sample (limit of detection, 10 PFU). Statistical significance for all experiments was determined using Student's t test.
Ocular inoculation of mice with H1N1 viruses was performed using 106 EID50 or PFU of indicated viruses in a volume of 5 μl following corneal scarification as previously described (2).
Pathology and immunohistochemistry.
Lung sections were fixed by submersion in 10% neutral buffered formalin, routinely processed, and embedded in paraffin. Sections were made at 5 μm and were stained with hematoxylin and eosin (HE). A duplicate 5-μm section was immunohistochemically stained to demonstrate influenza A virus nucleoprotein by first microwaving the sections in Antigen Retrieval Citra solution (Biogenex, San Ramon, CA) for antigen exposure. A 1:2,000 dilution of a mouse-derived monoclonal antibody (P13C11) specific for a type A influenza virus nucleoprotein was applied and allowed to incubate for 12 h at 4°C. The primary antibody was then detected by the application of biotinylated goat anti-mouse IgG secondary antibody using the Mouse on Mouse system (M.O.M. kit; Vector Laboratories, Inc., Burlingame, CA) per the manufacturer's instructions. The 3-amino-9-ethylcarbazole (AEC) peroxidase substrate kit (Vector Laboratories Inc.) was used as the substrate chromogen, and hematoxylin was used as a counterstain. Histopathological lesions and immunohistochemical viral antigen detection were scored. For lesions, scoring was as follows: 1, no lesion; 2, mild inflammation and no or rare necrosis; 3, moderate inflammation with frequent necrotic cells; 4, severe inflammation with common necrosis and edema. For immunohistochemical viral antigen scoring, the following was used: 0, no antigen; 1, rare positive cells; 2, infrequent positive cells; 3, common positive cells; 4, widespread positive cells. For histopathological changes, morphological descriptions were provided.
Peripheral cell counts.
On days 0, 3, and 6 p.i., blood was collected from the brachial artery of three to six mice inoculated with 105 EID50 or PFU of each virus. Blood was immediately placed in EDTA Vacutainer tubes (BD, Franklin Lakes, NJ), and complete blood counts were quantified using a Hemavet HV950FS instrument per the manufacturer's instructions (Drew Scientific, Inc., Oxford, CT). Absolute thymocyte counts were performed with single-cell suspensions of homogenized thymus diluted 1:10 with Turk's solution (2% acetic acid, 0.01% [vol/vol] crystal violet, double-distilled water), representative of three to six mice per group. Statistical significance of thymocyte counts was determined by analysis of variance (ANOVA) and Student's t test.
Cytokine quantification.
Clarified day 3 and 6 p.i. lung homogenates from mice inoculated with 105 EID50 or PFU of each virus indicated (n = 3) were analyzed by enzyme-linked immunosorbent assay (ELISA) according to the manufacturer's protocol (BD Biosciences, San Diego, CA). Cytokines or chemokines analyzed were interleukin-6 (IL-6), IL-10, IL-12 (p40), and monocyte chemotactic protein 1 (MCP-1) (assay sensitivity, 15.6 pg/ml).
Sequence analysis.
Genetic analyses of protein sequences were performed using the programs BioEdit and MUSCLE (8, 15).
RESULTS
Characterization of H1N1 viruses in mice.
Human influenza A viruses of the H1 and H3 subtypes which cause seasonal epidemics do not typically replicate efficiently in the mouse model, unlike avian influenza A viruses of the H5 and H7 subtypes, which replicate to high titers in the lungs of mice without prior adaptation and are capable of causing severe disease and death in this model (2, 10, 14, 22, 24). BALB/c mice were inoculated intranasally (i.n.) with multiple H1N1 viruses associated with human infection (Table 1). 2009 H1N1 viruses chosen for this study were isolated from either pediatric or adult patients with uncomplicated, severe, or fatal cases of respiratory illness.
All 2009 H1N1 viruses tested possessed low MID50 values (100.25 to 102.75 EID50 or PFU), indicating a high degree of infectivity in this model without prior adaptation, similar to the 1918 and H5N1 viruses (Table 2). However, in contrast to the virulent 1918 and H5N1 viruses, the 2009 H1N1 viruses did not mount lethal infections in mice (LD50, >106 EID50 or PFU), and with the exception of Mex/4482 virus, which caused 19% weight loss at the height of infection, morbidity was mild with <10% transient loss of initial body weight. All 2009 H1N1 viruses tested replicated to high titers (104.7 to 106.9 EID50 or PFU) in the lungs of mice on days 3 and 6 postinoculation (p.i.). Virus was detected infrequently and at low titers in the nose at these times p.i. (<102 EID50 or PFU) (Table 2). Replication of all 2009 H1N1 viruses was restricted to respiratory tract tissues, indicating a lack of systemic spread of these viruses in this model (data not shown).
TABLE 2.
Mouse infectivity and tissue viral titers of H1N1 and H5N1 viruses
| Virus | % wt loss (day p.i.)a | MID50b | LD50b | Viral titerc |
|||
|---|---|---|---|---|---|---|---|
| Day 3 p.i. |
Day 6 p.i. |
||||||
| Lung | Nose | Lung | Nose | ||||
| CA/4 | 5.3 (8) | 101.5 | >106 | 5.9 ± 0.9 | 1.5 ± 0.2 | 6.2 ± 0.1 | NDd |
| TX/15 | 1.5 (1) | 100.5 | >106 | 5.4 ± 1.0 | ND | 4.7 ± 0.8 | ND |
| Mex/4108 | 3.5 (3) | 101.5 | >106 | 6.9 ± 0.3 | 1.6 ± 0.5 | 5.6 ± 0.3 | 1.3 ± 0.3 |
| Mex/4482 | 19.0 (7) | 100.25 | >106 | 6.4 ± 0.2 | 1.2 ± 0.3 | 6.3 ± 0.4 | ND |
| Mex/4487 | 9.7 (8) | 102.75 | >106 | 6.9 ± 0.4 | 1.7 ± 0.1 | 5.9 ± 0.2 | 2.6 ± 1.0 |
| NJ/8 | 2.7 (1) | 102.75 | >106 | 6.0 ± 0.2 | ND | 5.9 ± 0.9 | ND |
| OH/2 | 14.5 (7) | 101.75 | 105.8 | 7.0 ± 0.3 | 1.4 ± 0.5 | 4.5 ± 0.1 | ND |
| 1918 | 22.4 (6) | 100.75 | 103.5 | 6.7 ± 0.1 | 1.5 ± 0.4 | 6.2 ± 0.1 | ND |
| VN/1203 | 23.4 (5) | 101.5 | 101.3 | 7.3 ± 0.1 | 2.0 ± 0.5 | 6.1 ± 0.4 | 3.5 ± 1.5 |
Mean maximum percent weight loss (5 mice per group) following inoculation with 105 PFU or EID50.
Fifty percent mouse infectious dose (MID50) and 50% lethal dose (LD50) are expressed as the PFU or EID50 required to give 1 MID50 or 1 LD50, respectively.
Virus endpoint titers are expressed as the mean log10 PFU/ml plus standard deviation of 3 to 6 mice per tissue.
ND, not detected. The limit of virus detection was 10 PFU/ml.
We next compared the virulence of 2009 H1N1 viruses to that of previous swine lineage influenza A viruses which have been associated with human disease. NJ/8 virus was isolated from throat swab material of a young military recruit during an outbreak at Fort Dix, NJ, in 1976. OH/2 virus, a triple-reassortant H1N1 virus containing avian, human, and swine genes, was isolated in 2007 from a 10-year-old female with uncomplicated, upper respiratory illness (37). Both viruses replicated in the respiratory tract of mice to titers comparable to those of the 2009 H1N1 isolates (Table 2). However, while NJ/8 virus caused insignificant morbidity, the triple-reassortant OH/2 virus resulted in greater weight loss, reached peak mean lung titers at day 3 p.i. of 107 PFU, and was lethal to mice at the highest dose of inoculum administered (LD50, 105.8 PFU) (Table 2).
In contrast to all 2009 H1N1 viruses tested, which were highly infectious but not lethal in the mouse, the reconstructed 1918 H1N1 virus and the highly pathogenic avian influenza (HPAI) H5N1 virus VN/1203 demonstrated enhanced morbidity and mortality in this model (Table 2) (24, 40). Mice inoculated with 1918 and VN/1203 viruses lost >20% of their initial body weight by day 5 p.i. and possessed LD50 values of 103.5 PFU and 101.3 EID50, respectively. In contrast to all H1N1 subtype viruses tested, VN/1203 virus exhibited extrapulmonary spread following infection, with virus detected in the spleen, intestine, thymus, and brain (data not shown). Viral titers in the gastrointestinal tract were observed only with the H5N1 virus (log10 2.7 ± 0.9 and 3.4 ± 1.4 PFU at days 3 and 6 p.i., respectively).
Reports of conjunctivitis following exposure to seasonal or avian influenza viruses indicate that influenza A viruses of multiple subtypes are capable of using the eye as a portal of entry (20). A recent study from our laboratory demonstrated the ability of selected H1N1 viruses to replicate in the lungs of mice following ocular inoculation (3). To assess the ability of 2009 H1N1 viruses to cause disease by this route, mice were inoculated by the ocular route following corneal scarification with Mex/4108, Mex/4482, NJ/8, or OH/2 virus. Mice inoculated by the ocular route did not exhibit substantial morbidity, and virus was not detected in the eye, nose, or lung of any mouse on day 3 or day 6 p.i. (data not shown). These results suggest that, unlike the case with some avian influenza viruses, the eye is not an efficient route of entry for swine origin H1N1 viruses in this model.
Histological pathology observed in H1N1 virus-infected mice.
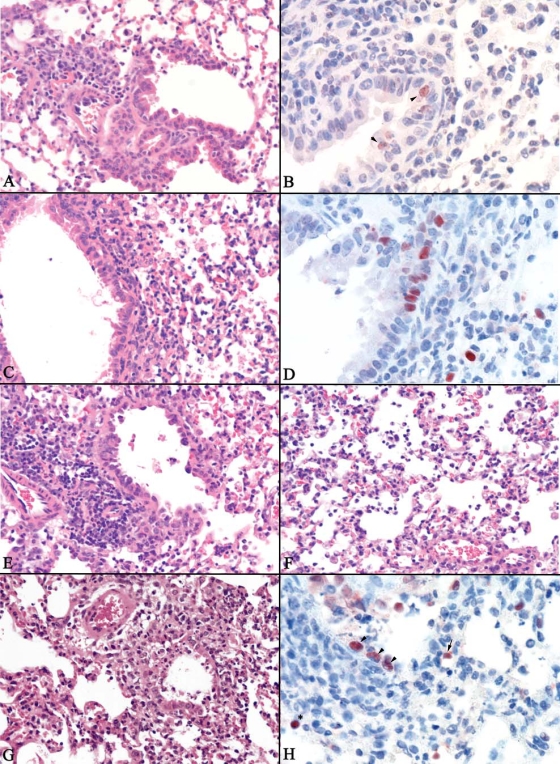
Histologically, all H1N1 virus infections produced lesions typical of influenza A virus infections: bronchiolitis and bronchitis with accompanying necrosis of respiratory epithelium and associated neutrophilic to histiocytic alveolitis (Table 3). However, the severity and character of necrosis and inflammation varied with individual virus strains. The mildest lesions were seen with TX/15, Mex/4108 and NJ/8 viruses (mean histological lesion score, 1.6 to 2.0), which produced mild bronchiolitis with minimal to no respiratory epithelial necrosis and only mild histiocytic alveolitis associated with terminal bronchioles (Fig. 1A and Table 3). Avian influenza viral antigen was rare to infrequent in alveolar epithelium and respiratory epithelium in bronchioles (Fig. 1B). Infection with Mex/4487, Mex/4482, OH/2, and CA/4 viruses resulted in lesions (mean histological lesion score, 2.3 to 3.0) in the lung varying from mild to moderate bronchiolitis with occasional necrosis of bronchiolar epithelium and mild to moderate peribronchiolar alveolitis, mainly histiocytic but with some neutrophils (Fig. 1C). Avian influenza viral antigen was rare to common in alveolar epithelium and macrophages and rare to common in respiratory epithelium in bronchioles depending on individual animals and area of the lung (Fig. 1D). The most severe lesions observed (mean histological lesion score, 3.2 to 3.3) were with VN/1203 and 1918 viruses, which produced mild to severe necrosis of bronchiolar epithelium with neutrophilic inflammation and peribronchiolar to diffuse neutrophilic alveolitis with necrosis of alveolar epithelium and prominent alveolar to interlobular edema at the hilum (Fig. 1E to G). Avian influenza virus antigen was common to widespread in alveolar epithelium and macrophages and infrequent to common in respiratory epithelium of the bronchioles (Fig. 1H). In summary, 2009 H1N1 viruses were capable of causing histological lung pathology following infection in the mouse; however, the severity of necrosis and inflammation observed in alveoli was less pronounced compared with the highly pathogenic viruses VN/1203 and 1918. These highly pathogenic viruses had necrotic and neutrophilic inflammatory lesions focused on alveoli and with greater detection of viral antigen within the alveoli compared to 2009 H1N1 or seasonal H1N1 viruses (Table 3).
TABLE 3.
Histopathological lesion and immunohistochemical viral antigen scores for mice intranasally inoculated with H1N1 influenza A viruses and sampled at 6 days p.i.
| Virus strain | Alveolar |
Bronchiolar and bronchial |
Additional lesion features | ||
|---|---|---|---|---|---|
| Histopathology scorea | Viral antigenb | Histopathology scorea | Viral antigenb | ||
| CA/4 | 2.3 | 1.3 | 2 | 1.7 | Histiocytic peribronchiolar alveolitis, some neutrophils; bronchiolitis with some necrosis |
| TX/15 | 1.6 | 0.3 | 1.3 | 0 | Histiocytic alveolitis; bronchiolitis without necrosis |
| Mex/4108 | 1.6 | 1.7 | 2 | 2 | Histiocytic alveolitis with uncommon neutrophils; bronchiolitis without necrosis |
| Mex/4482 | 2.6 | 2 | 2.6 | 1.5 | Histiocytic peribronchiolar alveolitis, common neutrophils; bronchiolitis with some necrosis |
| Mex/4487 | 3 | 1.5 | 3 | 3 | Histiocytic peribronchiolar alveolitis, common neutrophils; histiocytic to neutrophilic bronchiolitis with necrosis |
| NJ/8 | 2 | 1.5 | 2 | 2 | Histiocytic alveolitis; lymphocytic bronchiolitis without necrosis |
| OH/2 | 2.6 | 1.6 | 2.6 | 1.7 | Histiocytic peribronchiolar alveolitis; bronchiolitis with some necrosis |
| 1918 | 3.3 | 3 | 2.5 | 0 | Neutrophilic alveolitis with alveolar epithelial necrosis and edema and interlobular edema; neutrophilic bronchiolitis with severe necrosis |
| VN/1203 | 3.2 | 2 | 2.6 | 2.5 | Neutrophilic peribronchiolar to diffuse alveolitis with edema and histiocytes; neutrophilic bronchiolitis with severe necrosis |
| Mock | 1 | 0 | 1 | 0 | No lesions |
Mean histopathological lesion scores for 2 or 3 mice. Histopathological lesion scores per mouse were determined as follows: 1, no lesion; 2, mild lesion; 3, moderate lesion; 4, severe lesion.
Mean immunohistochemical viral antigen scores for 2 or 3 mice. Immunohistochemical viral antigen scores per mouse were determined as follows: 0, no antigen; 1, rare positive cells; 2, infrequent positive cells; 3, common positive cells; 4, widespread positive cells.
FIG. 1.
Histopathologic changes and immunostaining of tissues from H1N1 virus-infected mice. Photomicrographs of hematoxylin-and-eosin-strained and immunohistochemically stained lung sections from mice at 6 days p.i. are shown. (A) Mild bronchiolitis with peribronchiolar lymphocytes and adjacent mild histiocytic alveolitis in an NJ/8 virus-infected mouse. (B) Weak influenza viral antigen staining in a few respiratory epithelial cells (arrowheads) in a bronchiole of an NJ/8 virus-infected mouse. (C) Mild epithelial necrosis in bronchiole and moderate histiocytic alveolitis with some neutrophils in a CA/4 virus-infected mouse. (D) Influenza viral antigen staining in respiratory epithelium bronchioles and in alveolar epithelium and histiocytes in alveoli of a CA/4 virus-infected mouse. (E) Moderate necrosis of bronchiole with alveoli containing edema and inflammatory cells in a 1918 virus-infected mouse. (F) Moderate neutrophilic alveolitis with edema, some histiocytes, and necrosis of alveolar epithelium in a 1918 virus-infected mouse. (G) Severe necrosis of bronchiole and neutrophilic alveolitis with edema and alveolar epithelial necrosis in a VN/1203 virus-infected mouse. (H) Influenza viral antigen commonly staining positive in respiratory epithelium of bronchioles (arrowheads) and in alveolar epithelium (arrow) and histiocytes (*) in alveoli in a 1918 virus-infected mouse.
Analysis of lymphocyte populations in whole blood and thymus from 2009 H1N1 virus-infected mice.
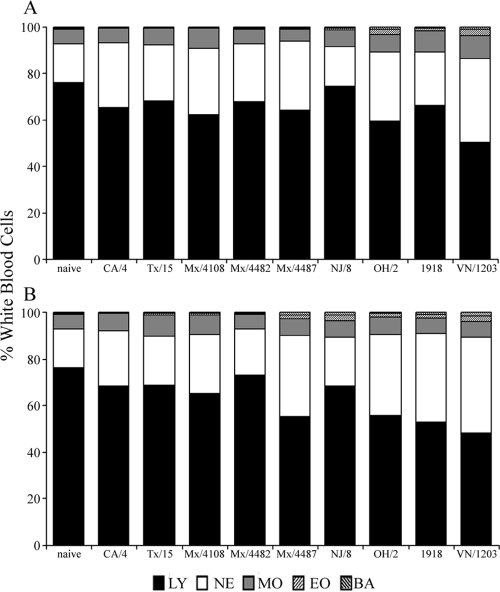
Infection with influenza viruses can lead to changes within the lymphohemopoietic system, and leukopenia and lymphopenia have been associated with both avian H5N1 human infection and human cases of triple-reassortant swine influenza (1, 37). This impairment is recapitulated in the mouse model, as previous work has noted severe depletion of lymphocytes in blood, lungs, and lymphoid tissue of mice infected with HPAI viruses (29, 41). To determine the extent of leukopenia resulting from infection with 2009 H1N1 viruses in mice, complete blood counts from mice inoculated with each virus were performed with a Hemavet multispecies hematology analyzer. Leukopenia was detected among all H1N1 viruses tested by day 6 p.i., with a statistically significant reduction in circulating white blood cells (WBCs) among TX/15- and Mex/4108-infected mice at this time p.i. (Table 4; Fig. 2). Differential blood counts of 2009 H1N1-infected mice revealed, on average, a 10% decrease of peripheral lymphocytes compared with uninfected mice on days 3 and 6 p.i. (Table 4; Fig. 2). The lethal OH/2 virus caused twice the reduction in the percentage of lymphocytes (approximately a 20% reduction from baseline levels) by day 6 p.i., which was significantly lower (P < 0.01) than that for all 2009 H1N1 viruses tested. Comparable levels of significant lymphopenia (>20% reduction from baseline levels) were also observed in mice infected with the highly virulent VN/1203 (H5N1) or 1918 virus. These levels were significantly lower than the lymphopenia induced by all 2009 H1N1 viruses tested (P < 0.005), demonstrating the enhanced virulence of these viruses in the mouse model. Because the pathogenesis of lethal influenza virus infections has been associated with depletion of thymocytes (41), we also determined the absolute thymocyte counts on days 3 and 6 p.i. (Table 4). All viruses which exhibited pronounced morbidity of >14% weight loss (Mex/4482, OH/2, VN/1203, and 1918) possessed significantly reduced (2.5- to 5-fold) total thymocyte counts by day 6 p.i. (P < 0.01). The greatest effect on the thymus followed 1918 virus infection: the number of leukocytes harvested from 1918-infected thymus tissue was 8-fold lower than the number of thymocytes from naïve mice. NJ/8 virus infection resulted in no depletion of leukocytes from the thymus, consistent with the insignificant morbidity and mortality observed.
TABLE 4.
Impact of viral infection on the mouse lymphohemopoietic systemd
| Virus | Day 3 p.i. |
Day 6 p.i. |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| WBCa | % LYb | % NEb | % MOb | Leukocytes/thymusc | WBCa | % LYb | % NEb | % MOb | Leukocytes/thymusc | |
| None (naïve mice) | 2.0 | 76.2 | 16.7 | 6.2 | 7.7 ± 0.1 | 2.0 | 76.2 | 16.7 | 6.2 | 7.7 ± 0.1 |
| CA/4 | 1.3 | 65.4* | 27.9* | 6.4 | 7.8 ± 0.2 | 1.5 | 68.2** | 23.9* | 7.4 | 7.7 ± 0.2 |
| TX/15 | 2.2 | 68.3* | 24.3 | 7.0 | 7.7 ± 0.1 | 1.0** | 68.9* | 21.0 | 8.9* | 7.6 ± 0.2 |
| Mex/4108 | 0.8** | 62.2** | 28.7** | 8.8 | 7.7 ± 0.1 | 0.8** | 65.3** | 25.2** | 8.5* | 7.5 ± 0.2 |
| Mex/4482 | 1.0** | 67.9** | 24.8** | 6.4 | 7.3 ± 0.1** | 1.5 | 73.2 | 19.5 | 6.6 | 7.3 ± 0.1** |
| Mex/4487 | 1.8 | 64.4** | 29.4** | 5.2 | 7.2 ± 0.1** | 1.7 | 55.5** | 34.9** | 6.9 | 7.0 ± 0.1** |
| NJ/8 | 1.2** | 74.5 | 17.3 | 6.9 | 7.7 ± 0.1 | 1.3* | 68.3* | 21.1 | 7.0 | 7.6 ± 0.1 |
| OH/2 | 1.0** | 59.7** | 29.4** | 7.8 | 7.5 ± 0.2 | 1.8 | 55.8** | 34.7** | 7.7 | 7.3 ± 0.2** |
| 1918 | 1.2* | 66.3* | 23.1** | 8.9 | 7.1 ± 0.1** | 1.6 | 52.9** | 38.1** | 6.7 | 6.8 ± 0.2** |
| VN/1203 | 1.3* | 50.3** | 36.3** | 9.9* | 7.0 ± 0.1** | 1.4* | 48.2** | 41.0** | 6.8 | 7.1 ± 0.1** |
Number of white blood cells (WBC) in whole blood, expressed as thousands of WBCs per microliter of whole blood.
Mean percentage of leukocytes that are lymphocytes (LY), neutrophils (NE), or monocytes (MO) from 3 to 7 mice per group.
Numbers of leukocytes are expressed as the mean log10 ± standard deviation of 3 to 6 thymi.
Statistical significance: *, P < 0.05, and **, P < 0.005, compared with naïve controls.
FIG. 2.
Kinetic analysis of circulating lymphocytes following influenza virus infection. BALB/c mice (3 or 4 mice/group) were inoculated i.n. with 105 EID50 or PFU of each virus shown. Blood was collected on day 3 (A) or 6 (B) p.i. in EDTA Vacutainer tubes and analyzed with a hematology scanner. Blood collected immediately prior to inoculation was included as a baseline control (naïve). The average percentages of lymphocytes (LY), neutrophils (NE), monocytes (MO), eosinophils (EO), and basophils (BA) in whole blood are shown.
Thrombocytopenia was documented following a human infection with triple-reassortant swine influenza virus in 2008 and is also a common observation among H5N1-infected patients with severe illness (37, 39, 43). However, platelet counts did not drop below baseline levels during the height of infection with any virus tested in mice (data not shown). While reduced levels of hemoglobin have been detected among confirmed H5N1-infected patients, erythrocyte counts did not consistently fall below baseline levels in these experiments (data not shown) (39, 42). In summary, we found that lymphopenia in peripheral blood and a reduction in thymus cellularity were associated with swine, avian, and previous pandemic influenza viruses possessing increased pathogenicity in the mouse model.
Cytokine production following primary infection with 2009 H1N1 viruses in mice.
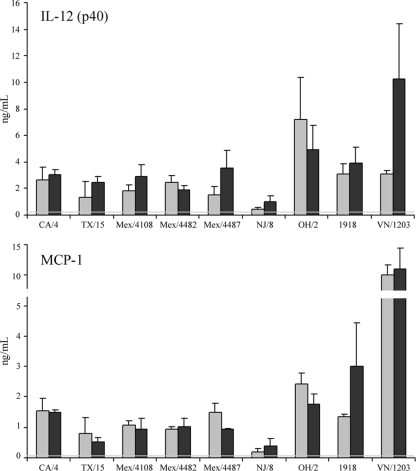
Despite efficient replication in mice of all pandemic 2009 H1N1 isolates tested, morbidity following infection varied between strains. To determine if there was a correlation between severe disease and proinflammatory cytokine production in the mouse lung with 2009 H1N1 viruses, tissue homogenates from mice inoculated with 105 EID50 or PFU were tested by ELISA for the presence of key cytokines previously shown to be differentially induced following seasonal and 2009 pandemic H1N1 virus infection (18). Interleukin 12 [IL-12 (p40)] is involved with T-cell proliferation and contributes toward the stimulation of gamma interferon (IFN-γ) and other cytokines (34). Production of IL-12 did not differ significantly between 2009 H1N1 viruses tested (Fig. 3). Compared with 2009 isolates, OH/2 virus-infected lungs possessed 3-fold-higher levels of IL-12 on day 3 p.i. (P < 0.001) and VN/1203 virus-infected lungs possessed 3-fold-higher levels on day 6 p.i. (P < 0.001). Influenza virus infection results in the production of several chemokines, including monocyte chemotactic protein 1 (MCP-1), which plays a role in the recruitment of leukocytes to sites of infection. Excessive production of this and other chemokines has been associated with the severe disease observed with H5N1 virus infection (6). MCP-1 production did not differ significantly between 2009 H1N1 viruses (Fig. 3). However, production of MCP-1 was on average 2-fold higher following infection with OH/2 virus on days 3 and 6 p.i. (P < 0.001), with 1918-infected lungs producing similarly elevated levels of MCP-1 by day 6 p.i. (P < 0.001) compared with 2009 H1N1 viruses. VN/1203-infected lungs possessed significantly higher levels of MCP-1, 10- to 14-fold above those of 2009 H1N1 viruses (P < 0.001, days 3 and 6 p.i.). Levels of both IL-12 and MCP-1 in NJ/8 virus-infected lungs were significantly lower (2- to 5-fold) than those in 2009 H1N1 viruses tested (P < 0.005, days 3 and 6 p.i.). Production of the cytokines IL-6 and IL-10 did not vary substantially between viruses tested (data not shown). In summary, while differences between 2009 H1N1 virus isolates were not observed, we found that OH/2 virus infection consistently produced elevated levels of cytokines in the lungs compared with 2009 isolates, while NJ/8 virus infection consistently produced reduced levels.
FIG. 3.
Determination of IL-12 and MCP-1 levels in H1N1- and H5N1-infected mouse lungs. BALB/c mice (3 or 4 mice/group) were inoculated i.n. with 105 EID50 or PFU of each virus shown. Lungs were removed on day 3 (gray bars) or 6 (black bars) p.i. and frozen at −70°C until processed. Clarified cell lysates from lungs homogenized in 1 ml PBS were assayed by ELISA and are presented as the mean levels plus standard deviations. Constitutive levels of IL-12 (0.38 ng/ml) or MCP-1 (0.07 ng/ml) present in the lungs were determined by harvesting normal, uninfected BALB/c lungs and are denoted in the figure by a gray line.
Molecular analysis of 2009 H1N1 viruses.
The molecular determinants of pathogenicity in mice infected with HPAI H5N1 and 1918 viruses have been characterized (32, 38). The absence of known molecular determinants that confer pathogenicity in 2009 H1N1 viruses was previously discussed in the work of Garten et al. (11). To identify a possible correlation between molecular factors and virus pathogenicity in mammalian species, we searched for molecular differences among 2009 H1N1 viruses and between these and other viruses used in this study. As shown in Table 5, the 2009 H1N1 viruses in this study possess amino acid differences in 8 out of 11 viral proteins. Overall, these amino acid changes do not map to known functional sites in viral proteins that readily explain variations in virus function, replication, or pathogenicity in mice. Two 2009 H1N1 viruses, Mex/4482 and Mex/4487, possessed amino acid changes in NA and HA proteins, respectively. Mex/4482 was one of the first viruses isolated from a fatal case that contained two amino acid changes in the NA (V106I and N247D). Mex/4487 virus contained an HA amino acid change located in the receptor binding site (Q226R).
TABLE 5.
Amino acid differences among 2009 H1N1 viruses used in this study
| Virus | Amino acidc |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PB2; 82 | PA |
HAa |
NP |
NAb |
M2; 82 | NS1; 45 | NS2; 67 | |||||||||||||
| 224 | 275 | 581 | 83 (91) | 108 (115) | 197 (200) | 210 (213) | 223 (226) | 321 (323) | 444 | 16 | 100 | 101 | 373 | 106 | 248 (247) | |||||
| CA/4 | N | P | L | M | P | V | T | F | Q | I | V | G | V | D | T | V | N | S | G | E |
| TX/15 | N | S | L | M | S | M | A | L | Q | V | V | D | V | D | T | V | N | S | R | K |
| Mex/4108 | N | S | L | L | S | V | A | F | Q | V | I | G | V | G | I | V | N | N | G | E |
| Mex/4482 | N | S | L | M | S | V | A | F | Q | V | V | G | I | D | T | I | D | S | G | E |
| Mex/4487 | S | S | I | M | S | V | A | F | R | V | V | G | V | D | T | V | N | S | G | E |
H3 numbering is in parentheses.
N2 numbering is in parentheses.
Nonconserved amino acids are highlighted by boldface.
The enhanced virulence of 1918 and VN/1203 viruses in this study prompted us to compare molecular features of these viruses to those of the H1N1 viruses examined here. The presence of a lysine at position 627 in PB2 is a known correlate of pathogenicity in the murine model (17, 38). In this study, only VN/1203, 1918, and NJ/8 viruses were found to possess a lysine at this position. A full-length PB1-F2 protein (90 amino acids [aa]) was present in VN/1203, 1918, and OH/2 viruses but not in NJ/8 virus or any of the 2009 H1N1 isolates. A full-length NS1 protein (230 aa) was present in the 1918 virus only; all 2009 H1N1 viruses tested possessed a 219-aa-long NS1 protein. In summary, molecular features which are frequently found among viruses of high pathogenicity in mammalian models were not detected in 2009 H1N1 viruses used in this study.
DISCUSSION
The rapid introduction of swine origin H1N1 viruses into human populations in 2009 necessitated the detailed study of these viruses to better understand their capacity to cause disease in mammalian species. Because of the similarity in the HA proteins of the 2009 H1N1 viruses and classical swine H1N1, including the 1918 pandemic virus, a side-by-side comparison was made among these viruses representing a span of approximately 91 years. This animal model has proven a useful tool for the study of influenza viruses with pandemic potential, due to its utility in measuring infectivity, pathogenesis, and subsequent application for the evaluation of vaccine and antiviral candidates. In this study, we used this model to assess the relative virulence of 2009 H1N1 viruses isolated from human patients compared with both swine lineage influenza viruses previously associated with human infection and highly pathogenic H1N1 and H5N1 viruses. Importantly, these studies revealed that 2009 H1N1 viruses exhibited mild to moderate virulence in mice compared with highly pathogenic avian H5N1 or 1918 viruses, as defined by morbidity, mortality, hemostatic assessments, lung histopathology, and lung cytokine production. Viruses isolated from severe or fatal human cases did not possess heightened virulence in this model. As the 2009 H1N1 pandemic continues to cause human infection and disease, this information will contribute toward a more comprehensive understanding of these viruses in the context of previous human infections with swine lineage influenza viruses and provide a well-characterized model for the study of antiviral and other therapeutic strategies against this emergent pandemic strain.
Viruses isolated from 2009 pandemic H1N1 patients replicated efficiently in the lungs of mice and sporadically and to lower titers in the noses and were restricted to the murine respiratory tract. This pattern of virus replication is similar to that observed with the 1918 reconstructed virus in mice (40). A recent study by Itoh et al., which assessed 2009 H1N1 virus replication in the mouse, confirmed this lack of systemic spread but detected virus at high titers in nasal turbinates (18). This study also reported that select 2009 H1N1 viruses were lethal to mice at high inoculum doses, a finding not recapitulated here. Differences between laboratories in anesthesia agents used, in addition to laboratory-specific conditions, may have contributed to the discordance in severity of disease observed between studies. In agreement with previous work, the 2009 H1N1 viruses tested here produced pulmonary pathology similar to those of other influenza A viruses and included mild to moderate bronchiolitis and alveolitis (18). Necrosis was not a common feature, and neutrophilic responses were typically mild. In contrast, the 1918 and VN/1203 viruses produced moderate to severe inflammation, and necrosis of bronchiolar and alveolar epithelium was frequent, as was alveolar and hilar interstitial edema.
Gastrointestinal symptoms were reported in approximately 40% of 2009 H1N1 virus-infected patients, and virus was recovered from the intestinal tract of ferrets following inoculation with 2009 H1N1 viruses; however, the 2009 H1N1 viruses tested here were not recovered from intestinal mouse tissue (7, 23). We also assessed the ability of 2009 H1N1 viruses to cause disease following ocular inoculation, as some influenza virus subtypes can use this portal of entry to initiate virus infection (3). The inability of 2009 H1N1 viruses to mount a productive infection in mice following ocular inoculation recapitulates the absence of conjunctivitis reported following 2009 H1N1 human infection and further suggests that the primary mode of 2009 H1N1 virus transmission is via the respiratory tract (7, 23, 30).
Collection of blood from mice during the acute phase of infection was performed to assess the influence of 2009 H1N1 virus infection on lymphocyte populations and other hemostatic parameters. Severe influenza virus infection in humans with H5N1 and 2009 H1N1 viruses has been associated with leukopenia, lymphopenia, and thrombocytopenia; these clinical characteristics have not been consistently observed among mild cases of 2009 H1N1 virus infection in humans (4, 37, 39). Highly virulent viruses, including the reconstructed 1918 virus and the H5N1 virus used in this study, have previously been shown to result in peripheral blood lymphopenia in the mouse model (24, 33). While transient lymphopenia was observed in mice following 2009 H1N1 virus infection, there was significantly less overall lymphocyte depletion compared with highly virulent viruses or the triple-reassortant OH/2 virus (Table 4; Fig. 2). Further clinical data from 2009 H1N1 virus-infected patients will contribute toward a comprehensive understanding of the change in hemostatic parameters following infection with this pandemic virus. There is currently a paucity of information pertaining to the consequence of influenza virus infection on complete blood counts in the mouse model as presented in this study in Table 4. By comparing viruses of different subtypes and degrees of virulence, we are better able to place in context the results obtained from 2009 H1N1 virus-infected mice.
The dysregulation of cytokines and chemokines during infection with H5N1 avian influenza viruses has been proposed to contribute to the severity of disease in human patients (25). Currently, there is little information on the proinflammatory response induced by 2009 H1N1 viruses and its relevance to disease severity in humans. A recent study described the increased production of several cytokines in mice infected with the 2009 H1N1 virus CA/4 compared with a seasonal H1N1 virus (18). As these experiments were performed with only one 2009 H1N1 isolate and did not offer a side-by-side comparison with viruses known to elicit hypercytokinemia following infection, selected analytes were chosen for further study here to more broadly assess the induction of cytokines and chemokines following infection with multiple human 2009 H1N1 isolates. We did not observe substantial differences in mouse cytokine production among 2009 H1N1 viruses isolated from patients in the early stages of the pandemic. Levels of IL-12 and MCP-1 following 2009 H1N1 virus infection were significantly higher compared with NJ/8 virus, similar to the results in the study by Itoh et al., which demonstrated elevated levels of cytokines from CA/4 virus compared with a less virulent seasonal H1N1 virus (18). Conversely, cytokine production following OH/2 virus infection was elevated compared with 2009 H1N1 viruses, which correlates with the heightened pathogenicity observed with the triple-reassortant virus in mice. Furthermore, production of IL-12 and MCP-1 by H1N1 viruses in this study was significantly reduced compared with the H5N1 virus VN/1203, similarly to a previous report demonstrating excessive release of cytokines by H5N1 viruses compared with H1N1 viruses, including the 1918 reconstructed virus (33). Our results agree with a recent study which found weak cytokine production following 2009 H1N1 virus infection in human macrophages and dendritic cells (31). These findings suggest that hypercytokinemia is not a general feature of 2009 H1N1 viruses, including those viruses isolated from fatal cases.
Similarly to previously published studies, we did not observe molecular features among 2009 H1N1 viruses which are known to correlate with virus pathogenicity in the mouse model. The E627K substitution in PB2 has been associated with enhanced virulence of HPAI viruses and is also present in the reconstructed 1918 virus (17, 40). The presence of this amino acid in NJ/8 virus indicates that not all H1N1 viruses which bear this substitution possess high virulence in the mouse model. The accessory protein PB1-F2 has known roles in virus pathogenicity and inflammation following influenza virus infection (27, 45). All viruses in this study which possessed a full-length PB1-F2 protein (VN/1203, 1918, and OH/2), and only these viruses, were capable of mounting a lethal infection in the mouse model. Future study addressing the potential for enhanced pathogenicity of 2009 H1N1 viruses following the introduction of a full-length PB1-F2 is warranted. The triple-reassortant H1N1 virus OH/2 demonstrated enhanced morbidity and mortality in mice compared with 2009 H1N1 viruses, along with elevated levels of proinflammatory cytokines and more pronounced dysregulation of hemostatic and lymphostatic parameters. The genetic composition of OH/2 virus, a member of the swH1-gamma lineage, generally aligns with 2009 H1N1 viruses, with the exception of Eurasian and not classical swine lineage NA and M genes present in 2009 isolates (11, 37). Triple-reassortant H1N1 viruses, including OH/2 virus, remain antigenically similar to the 2009 H1N1 isolates (11). Detailed study of the contribution of individual genes to the heightened pathogenicity of OH/2 virus found in this study compared with 2009 isolates will allow for a greater understanding of the potential for these viruses to cause severe disease.
Well-characterized mammalian models have afforded the opportunity for a more comprehensive understanding of previous pandemic viruses and those viruses considered to hold pandemic potential (24, 40). Establishing similar models for 2009 H1N1 viruses, such as the mouse model presented here, will accelerate the identification of those molecular correlates which contribute to the unique features of this newest pandemic strain and support efforts to limit the spread and severity of disease via the use of vaccines and antivirals. Direct comparison in vivo between 2009 H1N1 viruses and previous viruses of swine origin associated with human infection additionally presents the opportunity to more closely examine viruses within this lineage as they evolved from causing sporadic human infections to acquiring properties which resulted in the first pandemic of the 21st century.
Acknowledgments
We thank P. Blair (Naval Health Research Center, San Diego, CA), G. J. Demmler (Texas Children's Hospital, Houston, TX), and C. Alpuche-Aranda (Instituto de Diagnóstico y Referencia Epidemiológicos, Mexico) for facilitating access to viruses. We also give thanks to Frank Plummer (National Microbiology Laboratory, Winnipeg, Manitoba, Canada) for kindly providing Mex/4487 virus. We give special thanks to the Influenza Division sequencing facility for sequencing assistance.
The findings and conclusions in this report are those of the authors and do not necessarily reflect the views of the funding agency.
Footnotes
Published ahead of print on 24 February 2010.
REFERENCES
- 1.Abdel-Ghafar, A. N., T. Chotpitayasunondh, Z. Gao, F. G. Hayden, D. H. Nguyen, M. D. de Jong, A. Naghdaliyev, J. S. Peiris, N. Shindo, S. Soeroso, and T. M. Uyeki. 2008. Update on avian influenza A (H5N1) virus infection in humans. N. Engl. J. Med. 358:261-273. [DOI] [PubMed] [Google Scholar]
- 2.Belser, J. A., X. Lu, T. R. Maines, C. Smith, Y. Li, R. O. Donis, J. M. Katz, and T. M. Tumpey. 2007. Pathogenesis of avian influenza (H7) virus infection in mice and ferrets: enhanced virulence of Eurasian H7N7 viruses isolated from humans. J. Virol. 81:11139-11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belser, J. A., D. A. Wadford, J. Xu, J. M. Katz, and T. M. Tumpey. 2009. Ocular infection of mice with influenza A (H7) viruses: a site of primary replication and spread to the respiratory tract. J. Virol. 83:7075-7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bin, C., L. Xingwang, S. Yuelong, J. Nan, C. Shijun, and X. Xiayuan. 2009. Clinical and epidemiologic characteristics of 3 early cases of influenza A pandemic (H1N1) 2009 virus, People's Republic of China. Emerg. Infect. Dis. 15:1418-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2010. 26 February 2010, posting date. 2009 H1N1 flu: international situation update. Centers for Disease Control and Prevention, Atlanta, GA. http://www.cdc.gov/h1n1flu/updates/international/.
- 6.Chan, M. C., C. Y. Cheung, W. H. Chui, S. W. Tsao, J. M. Nicholls, Y. O. Chan, R. W. Chan, H. T. Long, L. L. Poon, Y. Guan, and J. S. Peiris. 2005. Proinflammatory cytokine responses induced by influenza A (H5N1) viruses in primary human alveolar and bronchial epithelial cells. Respir. Res. 6:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dawood, F. S., S. Jain, L. Finelli, M. W. Shaw, S. Lindstrom, R. J. Garten, L. V. Gubareva, X. Xu, C. B. Bridges, and T. M. Uyeki. 2009. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl. J. Med. 360:2605-2615. [DOI] [PubMed] [Google Scholar]
- 8.Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ennis, F. A., T. G. Wise, C. McLaren, and M. W. Verbonitz. 1977. Serological responses to whole and split A/New Jersey vaccines in humans and mice following priming infection with influenza A viruses. Dev. Biol. Stand. 39:261-266. [PubMed] [Google Scholar]
- 10.Gao, P., S. Watanabe, T. Ito, H. Goto, K. Wells, M. McGregor, A. J. Cooley, and Y. Kawaoka. 1999. Biological heterogeneity, including systemic replication in mice, of H5N1 influenza A virus isolates from humans in Hong Kong. J. Virol. 73:3184-3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garten, R. J., C. T. Davis, C. A. Russell, B. Shu, S. Lindstrom, A. Balish, W. M. Sessions, X. Xu, E. Skepner, V. Deyde, M. Okomo-Adhiambo, L. Gubareva, J. Barnes, C. B. Smith, S. L. Emery, M. J. Hillman, P. Rivailler, J. Smagala, M. de Graaf, D. F. Burke, R. A. Fouchier, C. Pappas, C. M. Alpuche-Aranda, H. Lopez-Gatell, H. Olivera, I. Lopez, C. A. Myers, D. Faix, P. J. Blair, C. Yu, K. M. Keene, P. D. Dotson, Jr., D. Boxrud, A. R. Sambol, S. H. Abid, K. St. George, T. Bannerman, A. L. Moore, D. J. Stringer, P. Blevins, G. J. Demmler-Harrison, M. Ginsberg, P. Kriner, S. Waterman, S. Smole, H. F. Guevara, E. A. Belongia, P. A. Clark, S. T. Beatrice, R. Donis, J. Katz, L. Finelli, C. B. Bridges, M. Shaw, D. B. Jernigan, T. M. Uyeki, D. J. Smith, A. I. Klimov, and N. J. Cox. 2009. Antigenic and genetic characteristics of swine-origin 2009 A (H1N1) influenza viruses circulating in humans. Science 325:197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaydos, J. C., F. H. Top, Jr., R. A. Hodder, and P. K. Russell. 2006. Swine influenza a outbreak, Fort Dix, New Jersey, 1976. Emerg. Infect. Dis. 12:23-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grunert, R. R., and C. E. Hoffmann. 1977. Sensitivity of influenza A/New Jersey/8/76 (Hsw1N1) virus to amantadine-HCl. J. Infect. Dis. 136:297-300. [DOI] [PubMed] [Google Scholar]
- 14.Guo, Y. J., S. Krauss, D. A. Senne, I. P. Mo, K. S. Lo, X. P. Xiong, M. Norwood, K. F. Shortridge, R. G. Webster, and Y. Guan. 2000. Characterization of the pathogenicity of members of the newly established H9N2 influenza virus lineages in Asia. Virology 267:279-288. [DOI] [PubMed] [Google Scholar]
- 15.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 16.Hancock, K., V. Veguilla, X. Lu, W. Zhong, E. N. Butler, H. Sun, F. Liu, L. Dong, J. R. Devos, P. M. Gargiullo, T. L. Brammer, N. J. Cox, T. M. Tumpey, and J. M. Katz. 2009. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N. Engl. J. Med. 361:1945-1952. [DOI] [PubMed] [Google Scholar]
- 17.Hatta, M., P. Gao, P. Halfmann, and Y. Kawaoka. 2001. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293:1840-1842. [DOI] [PubMed] [Google Scholar]
- 18.Itoh, Y., K. Shinya, M. Kiso, T. Watanabe, Y. Sakoda, M. Hatta, Y. Muramoto, D. Tamura, Y. Sakai-Tagawa, T. Noda, S. Sakabe, M. Imai, Y. Hatta, S. Watanabe, C. Li, S. Yamada, K. Fujii, S. Murakami, H. Imai, S. Kakugawa, M. Ito, R. Takano, K. Iwatsuki-Horimoto, M. Shimojima, T. Horimoto, H. Goto, K. Takahashi, A. Makino, H. Ishigaki, M. Nakayama, M. Okamatsu, D. Warshauer, P. A. Shult, R. Saito, H. Suzuki, Y. Furuta, M. Yamashita, K. Mitamura, K. Nakano, M. Nakamura, R. Brockman-Schneider, H. Mitamura, M. Yamazaki, N. Sugaya, M. Suresh, M. Ozawa, G. Neumann, J. Gern, H. Kida, K. Ogasawara, and Y. Kawaoka. 2009. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature 460:1021-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobasa, D., A. Takada, K. Shinya, M. Hatta, P. Halfmann, S. Theriault, H. Suzuki, H. Nishimura, K. Mitamura, N. Sugaya, T. Usui, T. Murata, Y. Maeda, S. Watanabe, M. Suresh, T. Suzuki, Y. Suzuki, H. Feldmann, and Y. Kawaoka. 2004. Enhanced virulence of influenza A viruses with the haemagglutinin of the 1918 pandemic virus. Nature 431:703-707. [DOI] [PubMed] [Google Scholar]
- 20.Kumlin, U., S. Olofsson, K. Dimock, and N. Arnberg. 2008. Sialic acid tissue distribution and influenza virus tropism. Influenza Other Respir. Viruses 2:147-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lessler, J., D. A. Cummings, S. Fishman, A. Vora, and D. S. Burke. 2007. Transmissibility of swine flu at Fort Dix, 1976. J. R. Soc. Interface 4:755-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu, X., T. M. Tumpey, T. Morken, S. R. Zaki, N. J. Cox, and J. M. Katz. 1999. A mouse model for the evaluation of pathogenesis and immunity to influenza A (H5N1) viruses isolated from humans. J. Virol. 73:5903-5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maines, T. R., A. Jayaraman, J. A. Belser, D. A. Wadford, C. Pappas, H. Zeng, K. M. Gustin, M. B. Pearce, K. Viswanathan, Z. H. Shriver, R. Raman, N. J. Cox, R. Sasisekharan, J. M. Katz, and T. M. Tumpey. 2009. Transmission and pathogenesis of swine-origin 2009 A (H1N1) influenza viruses in ferrets and mice. Science 325:484-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maines, T. R., X. H. Lu, S. M. Erb, L. Edwards, J. Guarner, P. W. Greer, D. C. Nguyen, K. J. Szretter, L. M. Chen, P. Thawatsupha, M. Chittaganpitch, S. Waicharoen, D. T. Nguyen, T. Nguyen, H. H. Nguyen, J. H. Kim, L. T. Hoang, C. Kang, L. S. Phuong, W. Lim, S. Zaki, R. O. Donis, N. J. Cox, J. M. Katz, and T. M. Tumpey. 2005. Avian influenza (H5N1) viruses isolated from humans in Asia in 2004 exhibit increased virulence in mammals. J. Virol. 79:11788-11800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maines, T. R., K. J. Szretter, L. Perrone, J. A. Belser, R. A. Bright, H. Zeng, T. M. Tumpey, and J. M. Katz. 2008. Pathogenesis of emerging avian influenza viruses in mammals and the host innate immune response. Immunol. Rev. 225:68-84. [DOI] [PubMed] [Google Scholar]
- 26.Matassov, D., A. Cupo, and J. M. Galarza. 2007. A novel intranasal virus-like particle (VLP) vaccine designed to protect against the pandemic 1918 influenza A virus (H1N1). Viral Immunol. 20:441-452. [DOI] [PubMed] [Google Scholar]
- 27.McAuley, J. L., F. Hornung, K. L. Boyd, A. M. Smith, R. McKeon, J. Bennink, J. W. Yewdell, and J. A. McCullers. 2007. Expression of the 1918 influenza A virus PB1-F2 enhances the pathogenesis of viral and secondary bacterial pneumonia. Cell Host Microbe 2:240-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Memoli, M. J., T. M. Tumpey, B. W. Jagger, V. G. Dugan, Z. M. Sheng, L. Qi, J. C. Kash, and J. K. Taubenberger. 2009. An early ‘classical’ swine H1N1 influenza virus shows similar pathogenicity to the 1918 pandemic virus in ferrets and mice. Virology 393:338-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mori, I., T. Komatsu, K. Takeuchi, K. Nakakuki, M. Sudo, and Y. Kimura. 1995. In vivo induction of apoptosis by influenza virus. J. Gen. Virol. 76:2869-2873. [DOI] [PubMed] [Google Scholar]
- 30.Munster, V. J., E. de Wit, J. M. van den Brand, S. Herfst, E. J. Schrauwen, T. M. Bestebroer, D. van de Vijver, C. A. Boucher, M. Koopmans, G. F. Rimmelzwaan, T. Kuiken, A. D. Osterhaus, and R. A. Fouchier. 2009. Pathogenesis and transmission of swine-origin 2009 A(H1N1) influenza virus in ferrets. Science 325:481-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osterlund, P., J. Pirhonen, N. Ikonen, E. Ronkko, M. Strengell, S. M. Makela, M. Broman, O. J. Hamming, R. Hartmann, T. Ziegler, and I. Julkunen. 2010. Pandemic H1N1 2009 influenza A virus induces weak cytokine response in human macrophages and dendritic cells and is highly sensitive to antiviral actions of interferons. J. Virol. 84:1414-1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pappas, C., P. V. Aguilar, C. F. Basler, A. Solorzano, H. Zeng, L. A. Perrone, P. Palese, A. Garcia-Sastre, J. M. Katz, and T. M. Tumpey. 2008. Single gene reassortants identify a critical role for PB1, HA, and NA in the high virulence of the 1918 pandemic influenza virus. Proc. Natl. Acad. Sci. U. S. A. 105:3064-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perrone, L. A., J. K. Plowden, A. Garcia-Sastre, J. M. Katz, and T. M. Tumpey. 2008. H5N1 and 1918 pandemic influenza virus infection results in early and excessive infiltration of macrophages and neutrophils in the lungs of mice. PLoS Pathog. 4:e1000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perussia, B., S. H. Chan, A. D'Andrea, K. Tsuji, D. Santoli, M. Pospisil, D. Young, S. F. Wolf, and G. Trinchieri. 1992. Natural killer (NK) cell stimulatory factor or IL-12 has differential effects on the proliferation of TCR-alpha beta+, TCR-gamma delta+ T lymphocytes, and NK cells. J. Immunol. 149:3495-3502. [PubMed] [Google Scholar]
- 35.Reed, L. J., and H. A. Muench. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 36.Riquelme, A., M. Alvarez-Lobos, C. Pavez, P. Hasbun, J. Dabanch, C. Cofre, J. Jimenez, and M. Calvo. 2009. Gastrointestinal manifestations among Chilean patients infected with novel influenza A (H1N1) 2009 virus. Gut 58:1567-1568. [DOI] [PubMed] [Google Scholar]
- 37.Shinde, V., C. B. Bridges, T. M. Uyeki, B. Shu, A. Balish, X. Xu, S. Lindstrom, L. V. Gubareva, V. Deyde, R. J. Garten, M. Harris, S. Gerber, S. Vagasky, F. Smith, N. Pascoe, K. Martin, D. Dufficy, K. Ritger, C. Conover, P. Quinlisk, A. Klimov, J. S. Bresee, and L. Finelli. 2009. Triple-reassortant swine influenza A (H1) in humans in the United States, 2005-2009. N. Engl. J. Med. 360:2616-2625. [DOI] [PubMed] [Google Scholar]
- 38.Subbarao, E. K., W. London, and B. R. Murphy. 1993. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J. Virol. 67:1761-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tran, T. H., T. L. Nguyen, T. D. Nguyen, T. S. Luong, P. M. Pham, V. C. Nguyen, T. S. Pham, C. D. Vo, T. Q. Le, T. T. Ngo, B. K. Dao, P. P. Le, T. T. Nguyen, T. L. Hoang, V. T. Cao, T. G. Le, D. T. Nguyen, H. N. Le, K. T. Nguyen, H. S. Le, V. T. Le, C. Dolecek, T. T. Tran, M. de Jong, C. Schultsz, P. Cheng, W. Lim, P. Horby, and J. Farrar. 2004. Avian influenza A (H5N1) in 10 patients in Vietnam. N. Engl. J. Med. 350:1179-1188. [DOI] [PubMed] [Google Scholar]
- 40.Tumpey, T. M., C. F. Basler, P. V. Aguilar, H. Zeng, A. Solorzano, D. E. Swayne, N. J. Cox, J. M. Katz, J. K. Taubenberger, P. Palese, and A. Garcia-Sastre. 2005. Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science 310:77-80. [DOI] [PubMed] [Google Scholar]
- 41.Tumpey, T. M., X. Lu, T. Morken, S. R. Zaki, and J. M. Katz. 2000. Depletion of lymphocytes and diminished cytokine production in mice infected with a highly virulent influenza A (H5N1) virus isolated from humans. J. Virol. 74:6105-6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wiwanitkit, V. 2005. Anemia in the recent reported cases of bird flu infection in Thailand and Vietnam. J. Infect. 51:259. [DOI] [PubMed] [Google Scholar]
- 43.Wiwanitkit, V. 2008. Hemostatic disorders in bird flu infection. Blood Coagul. Fibrinolysis 19:5-6. [DOI] [PubMed] [Google Scholar]
- 44.Yu, X., T. Tsibane, P. A. McGraw, F. S. House, C. J. Keefer, M. D. Hicar, T. M. Tumpey, C. Pappas, L. A. Perrone, O. Martinez, J. Stevens, I. A. Wilson, P. V. Aguilar, E. L. Altschuler, C. F. Basler, and J. E. Crowe, Jr. 2008. Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature 455:532-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zamarin, D., M. B. Ortigoza, and P. Palese. 2006. Influenza A virus PB1-F2 protein contributes to viral pathogenesis in mice. J. Virol. 80:7976-7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeng, H., C. Goldsmith, P. Thawatsupha, M. Chittaganpitch, S. Waicharoen, S. Zaki, T. M. Tumpey, and J. M. Katz. 2007. Highly pathogenic avian influenza H5N1 viruses elicit an attenuated type I interferon response in polarized human bronchial epithelial cells. J. Virol. 81:12439-12449. [DOI] [PMC free article] [PubMed] [Google Scholar]