Abstract
Development of the cellular antiviral response requires nuclear translocation of multiple transcription factors and activation of a wide variety of cellular genes. To counteract the antiviral response, several viruses have developed an efficient means of inhibiting nucleocytoplasmic traffic. In this study, we demonstrate that the pathogenic strain of Venezuelan equine encephalitis virus (VEEV) has developed a unique mechanism of nuclear import inhibition. Its capsid protein forms a tetrameric complex with the nuclear export receptor CRM1 and the nuclear import receptor importin α/β. This unusual complex accumulates in the center channel of the nuclear pores and blocks nuclear import mediated by different karyopherins. The inhibitory function of VEEV capsid protein is determined by a short 39-amino-acid-long peptide that contains both nuclear import and supraphysiological nuclear export signals. Mutations in these signals or in the linker peptide attenuate or completely abolish capsid-specific inhibition of nuclear traffic. The less pathogenic VEEV strains contain a wide variety of mutations in this peptide that affect its inhibitory function in nuclear import. Thus, these mutations appear to be the determinants of this attenuated phenotype. This novel mechanism of inhibiting nuclear transport also shows that the nuclear pore complex is vulnerable to unusual cargo receptor complexes and sheds light on the importance of finely adjusted karyopherin-nucleoporin interactions for efficient cargo translocation.
The genus Alphavirus in the family Togaviridae contains a number of important human and animal pathogens. Alphaviruses are widely distributed on all continents and are transmitted between vertebrates, which serve as amplifying hosts, by different mosquito vectors. Several alphaviruses cause severe diseases in humans, with symptoms ranging from arthralgia to severe meningoencephalitis that can lead to a lethal outcome (24, 30). Venezuelan equine encephalitis virus (VEEV) is of particular importance because it causes major epidemics and equine epizootics (49). The alphavirus genome is packaged into an icosahedral nucleocapsid surrounded by a lipid envelope with embedded glycoprotein spikes. The genome is represented by a single-stranded RNA of positive polarity, encoding a nonstructural polyprotein translated directly from the genomic RNA, and a structural polyprotein, translated from the subgenomic RNA, which is synthesized during virus replication (44). Thus, the alphavirus genome encodes only a few proteins, which not only function in replication of the viral genome and formation of infectious viral particles, but also interfere with the host response aimed at inhibition of virus replication and viremia development.
The currently accepted hypothesis suggests that alphaviruses originated in either the New World or the Old World. Their transmission to another hemisphere most likely occurred twice between 2,000 and 3,000 years ago (38). Thus, for a long period, the New World and the Old World alphaviruses were evolving separately. They accumulated strong differences in the nonstructural and structural genes and cause different diseases in vertebrate hosts but sustained one of the important elements in their pathogenesis on a molecular level, the ability to interfere with cellular transcription and subsequent activation of the antiviral response. However, in vertebrate cells infected by geographically isolated alphaviruses, the transcriptional shutoff is mediated by different virus-specific proteins (21). The Old World alphavirus nonstructural protein 2 (nsP2) accumulates in cell nuclei and inhibits cellular RNA polymerases I and II (20), but in New World alphavirus-infected cells, the inhibition of cellular transcription is induced by capsid protein and not nsP2 (19). As do some other viral proteins, VEEV capsid protein exhibits many functions. It selectively packages the viral genome, but not cellular or viral subgenomic RNA, into viral particles. Capsid protein also possesses protease activity required for processing of the structural polyprotein. In our previous studies, we demonstrated that capsid proteins of the New World alphaviruses, VEEV and Eastern equine encephalitis virus (EEEV), are highly cytotoxic, and this effect is strongly correlated with the ability of the protein to inhibit cellular transcription (19). Further studies demonstrated that the inhibitory effect of VEEV capsid protein was independent of its protease activity and an RNA-binding domain and was determined by a short amino-terminal peptide. Importantly, a large fraction of VEEV capsid protein was detected in the cell nuclei and on the nuclear membrane, where distribution of the protein was reminiscent of the nuclear pore complex (NPC) distribution. Colocalization of VEEV capsid protein with the NPC suggested that it interferes with NPC functions. Indeed, we demonstrated that VEEV capsid protein functions as a very potent inhibitor of nucleocytoplasmic trafficking and blocks nuclear import of the proteins mediated by a variety of nuclear localization signals (NLS), if not all of them (5). These inhibitory functions suggested that capsid-induced inhibition of nucleocytoplasmic trafficking might be the key determinant of profound downregulation of cellular transcription.
Here, we present the mechanism responsible for nuclear import inhibition by VEEV capsid protein. Specifically, we demonstrate that a 39-amino-acid (aa)-long capsid-specific peptide interacts with two nuclear transport receptors, exportin and importin α/β. The resulting tetrameric complex accumulates in the central channel of the NPC and blocks nuclear import. Importantly, the same peptide derived from the nonpathogenic VEEV strain Pixuna interferes with nuclear import less efficiently and thus appears to be a contributor to an attenuated phenotype of this natural VEEV isolate.
MATERIALS AND METHODS
Cell culture.
BHK-21 cells were kindly provided by P. Olivo (Washington University, St. Louis, MO). They were maintained at 37°C in alpha minimum essential medium (αMEM) supplemented with 10% fetal bovine serum (FBS) and vitamins.
Plasmid constructs.
Plasmids encoding VEEV replicons (VEErep) with a nuclear import reporter (4×Tomato) having different NLS or a VEEV capsid-green fluorescent protein (GFP) fusion, cloned under the control of the subgenomic promoters, were described elsewhere (5). The pVEErep/H68-GFP and pVEErep/H60-GFP plasmids encoded VEEV replicons expressing the tested peptides H68 and H60 fused with GFP (Fig. 1A and D). In pVEErep/H60-GFP-3×NLS, the peptide-GFP-coding sequence was fused with 3 copies of standard simian virus 40 (SV40) T-antigen (TAg) NLS. Plasmids encoding replicons having additional mutations in the H68 peptide had the same design as pVEErep/H68-GFP. The mutations were introduced by PCR and standard cloning techniques. The H68-GFP, H60-GFP, H68AA1-GFP, and H68AA2-GFP cassettes were also cloned into the pTriEx1 plasmid with an N-terminal 6×His tag.
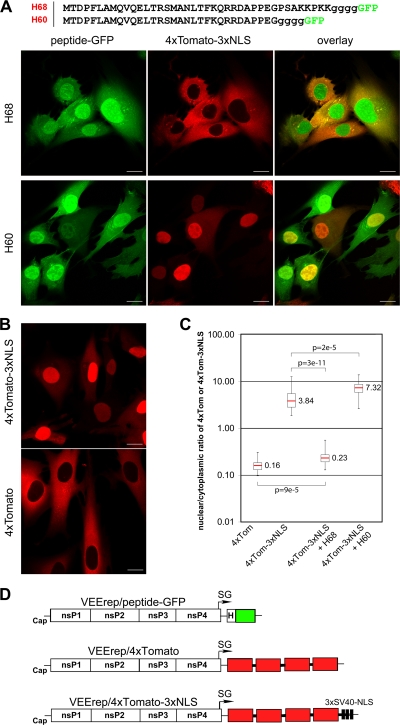
FIG. 1.
Identification of the VEEV capsid-specific peptide that inhibits nuclear import. (A) (Top) Amino acid sequences of H68 and H60 peptides derived from VEEV capsid protein. Capsid-specific sequences are indicated by uppercase letters, and the glycine linker between the peptides and GFP is indicated by lowercase letters. Methionine was introduced to promote translation initiation. (Bottom) Representative confocal images demonstrating the distribution of the nuclear import reporter 4×Tomato-3×NLS in the presence of H68-GFP or H60-GFP. BHK-21 cells were infected with the packaged replicons VEErep/H68-GFP or VEErep/H60-GFP and VEErep/4×Tomato-3×NLS and fixed at 4 h postinfection. Scale bars, 20 μm. (B) Representative confocal images of cells expressing 4×Tomato-3×NLS or 4×Tomato from corresponding VEEV replicons at 4 h postinfection. Scale bars, 20 μm. (C) Box plot showing the nuclear/cytoplasmic distribution of 4×Tomato and 4×Tomato-3×NLS (4×Tom and 4×Tom-3×NLS, respectively) alone or when 4×Tomato-3×NLS was expressed in the presence of H60-GFP or H68-GFP (H60 and H68, respectively). The nuclear/cytoplasmic ratio for distribution of 4×Tomato was used as a control demonstrating the distribution of the large protein that is incapable of translocation to the nucleus. The P values were calculated using the Mann-Whitney test (n = 30 for all experiments). Red line, median. (D) Schematic representation of the VEEV replicons used in this study. H indicates the position of a wild-type or mutant peptide derived from VEEV capsid.
Packaging of the replicons.
BHK-21 cells were coelectroporated with the in vitro-synthesized VEEV replicon RNA and two helper RNAs, HVEE/C and HVEE/GI, as described previously (46). The packaged replicons were harvested at 24 h posttransfection. Titers were determined by infecting BHK-21 cells with different dilutions of the stocks and evaluating the number of GFP-positive cells after 16 h of incubation at 30°C.
Protein production.
H68-GFP, H60-GFP, H68AA1-GFP, and H68AA2-GFP were expressed as N-terminal 6×His-tagged proteins from the pTriEx1 plasmid in Escherichia coli strain Turner(DE3)(pLysS) according to the manufacturer's instructions (Novagen). Cells were harvested by centrifugation and lysed in buffer containing 100 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 1 mg/ml of lysozyme. After the addition of Triton X-100 to 0.3% and phenylmethylsulfonyl fluoride (PMSF) to 1 mM, the cells were sonicated, and insoluble material was removed by centrifugation. The proteins were purified using Ni Sepharose High Performance according to the manufacturer's instructions (GE Healthcare). The purified proteins were dialyzed against a buffer containing 20 mM HEPES-KOH, pH 8.0, 200 mM NaCl, 1 mM dithiothreitol (DTT), and 8.75% glycerol. Recombinant CRM1, RanGTP (13), importin β (36), and importin α1 (50) were purified as previously described.
Analysis of reporter transport.
BHK-21 cells were seeded into 8-well μ-chambers (Ibidi) and infected or coinfected with packaged replicons in 150 μl of phosphate-buffered saline (PBS) supplemented with 1% FBS. The packaged replicons were used at concentrations of 5 × 106 infectious units/ml (VEErep-4×Tomato-3×NLS) and 1 × 107 infectious units/ml (all other packaged replicons). After 1 h of incubation at 37°C in 5% CO2, the medium was replaced with complete medium, and incubation continued for 4 h. The cells were then fixed with 4% paraformaldehyde in PBS. The compartmentalization of the proteins was analyzed using a Leica SP1 confocal microscope with a 60× 1.4-NA oil immersion Plan-Apochromat objective. In every experiment, quantification of nuclear/cytoplasmic ratios of 4×Tomato-3×NLS or peptide-GFP fusion proteins was done on 29 to 30 cells. The median pixel intensities were calculated from equal areas of cytoplasm and nuclei and corrected for background. P values were calculated using the Mann-Whitney test.
Immunofluorescence analysis of CRM1 and importin β distributions.
BHK-21 cells were seeded into 8-well μ-chambers (Ibidi) and infected with VEErep/H68-GFP replicon as described above. The cells were fixed at 2.5 h postinfection with 4% paraformaldehyde in PBS. At that time, the H68-GFP expression was low, and it was readily detected within the NPC in the nuclear rim. The primary antibodies against CRM1 and importin β were purchased from Calbiochem (ST1100) and Abcam (ab2811), respectively, and used at 1:1,000 dilution. The secondary Alexa Fluor 555 antibodies were purchased from Invitrogen. Images were acquired on a Zeiss LSM510 confocal microscope with a 63× 1.4-NA oil immersion Plan-Apochromat objective at identical settings for mock- and H68-GFP-expressing cells. The median pixel intensities for nuclear-envelope-specific and nuclear signals were measured for multiple cells. P values were calculated using the Mann-Whitney test (n = 8 for importin β; n = 6 for CRM1).
Leptomycin treatment.
BHK-21 cells were seeded into 8-well μ-chambers (Ibidi) and infected with VEErep/capsid-GFP replicon and replicons encoding different 4×Tomato reporters as described above. At 2 h postinfection, the medium was replaced with medium supplemented with 45 nM leptomycin B. After an additional 4 h of incubation, the cells were fixed in 4% paraformaldehyde in PBS, and images were acquired on Zeiss LSM510 confocal microscope with a 63× 1.4-NA oil immersion Plan-Apochromat objective.
Cryoimmunogold electron microscopy (EM).
HeLa cells transfected with GFP-H68 or GFP-H68AA1 were fixed, sectioned, immunolabeled with anti-GFP antibodies, and imaged as described previously (13). Images representing eight independent cells with low GFP expression were analyzed for NPC-proximal gold particles. Gold particles were considered to be NPC associated when they were present within a radius of 100 nm, which was based on the overall dimensions of the NPC, which measured 150 nm in length with a outer diameter of 125 nm (6). Within NPCs, gold was considered central when it was located between the two nuclear membranes.
Surface plasmon resonance (SPR) spectroscopy.
SPR spectroscopy was performed at 25°C on a Biacore T100 (GE Healthcare). H68-GFP or its variants were immobilized on a CM5 chip with amine coupling in 10 mM sodium acetate at pH 4.5. Recombinant proteins were streamed over the chip in 20 mM HEPES-KOH, pH 7.9, 200 mM NaCl, 0.1 mM DTT at 0.03 ml/min. Simultaneously, an empty flow cell was used as a reference. Biacore T100 evaluation software was used for analysis of the data.
RESULTS
Identification of the short peptide of VEEV capsid (H68) that is able to inhibit nuclear import.
Previously, we identified the minimal peptide of VEEV capsid protein, termed H68 (aa 30 to 68), which inhibited transcription in cells of vertebrate origin as efficiently as a full-length capsid protein (5). Further decrease in the size of this peptide to aa 30 to 60 (H60) completely abolished its inhibitory effect on cellular RNA synthesis. To further evaluate the effects of these peptides on nuclear import, both H68 and H60 peptide-coding sequences were fused with GFP (Fig. 1A). These fusion proteins mimic the size and structure of VEEV capsid protein, which has a structurally ordered carboxy-terminal domain and a disordered amino-terminal domain (aa 1 to 107) (9, 12). The 4×Tomato-3×NLS protein, a 109.5-kDa fluorescent protein whose transport into the nucleus is mediated by a triple SV40 TAg NLS located at the carboxy terminus, was used as a reporter of nuclear import, as previously described (5). All proteins were expressed from modified VEEV replicons (5) (Fig. 1D). BHK-21 cells were infected at a multiplicity of infection (MOI) sufficient for delivery of both replicons into the same cell. The cells were fixed at 4 h postinfection, and the distribution of the fluorescent proteins was analyzed by confocal microscopy. As demonstrated in Fig. 1A and C, H68-GFP efficiently blocked the accumulation of 4×Tomato-3×NLS in the nuclei, and distribution of the latter protein was very similar to that of 4×Tomato with no NLS (Fig. 1B and C). H60-GFP did not inhibit nuclear import (Fig. 1A and C), and this was correlated with its inability to inhibit cellular transcription (19). Thus, H68 is the minimum peptide sufficient for both induction of transcriptional shutoff and nuclear import inhibition, and the carboxy terminus of H68 is required for both functions.
The H68 peptide contains a functional NLS, which is essential for nuclear import inhibition.
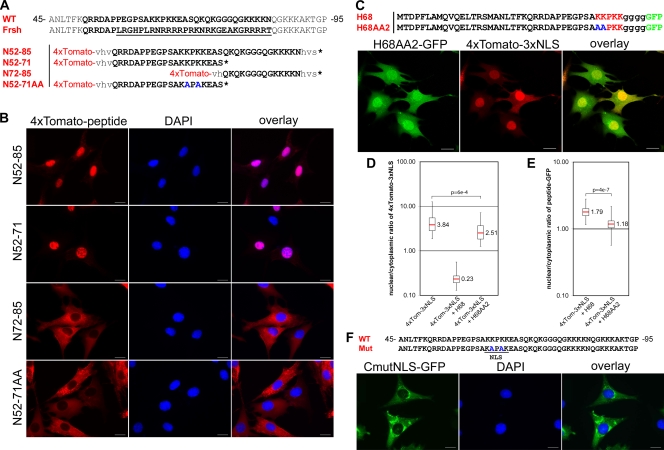
The carboxy terminus of the H68 peptide contains a short sequence of positively charged amino acids that might serve as an NLS. This sequence is a part of the large positively charged domain (aa 64 to 117 of the capsid) that is involved in encapsidation of viral genomic RNA (18, 48) and contains a number of positively charged peptides, which, based on the computer prediction, may serve as NLS. Previously, we isolated a frameshift mutant of VEEV capsid protein, CVEEfrsh (Fig. 2A), that not only lost its ability to inhibit cellular transcription and nuclear import (5, 19, 21), but also accumulated exclusively in the cytoplasm. Lack of nuclear accumulation of this mutant protein suggested that a nuclear localization signal might be present in a modified sequence between aa 57 and 85. Indeed, when the aa 57-to-85 peptide was fused with the 4×Tomato protein, which cannot translocate to the nucleus by diffusion (Fig. 1B), the resulting 4×Tomato-N52-85 protein efficiently accumulated in the nuclei (Fig. 2A and B), further suggesting the presence of an NLS sequence in the peptide. The N52-85 sequence contains two basic fragments, which are predicted to have NLS functions. We divided this sequence into two shorter peptides (Fig. 2A) and assessed their abilities to mediate nuclear import. The 4×Tomato-N52-71 fusion protein, but not the 4×Tomato-N71-81 protein, accumulated efficiently in the nuclei (Fig. 2B), suggesting that only amino acids 64-KKPKK-68 functioned as the NLS. To confirm this result, we replaced lysines at positions 65 and 67 with alanines, and the mutated peptide in 4×Tomato-N52-71AA did not mediate nuclear translocation of the protein (Fig. 2A and B). The same mutation that replaced lysines at positions 65 and 67 with alanines also abolished translocation of full-length VEEV capsid into the nucleus (Fig. 2F). Thus, the KKPKK sequence in the N52-71 peptide is the only functional capsid-specific NLS. This sequence resembles the classical monopartite NLS, which mediates nuclear import by binding with the importin α/β complex.
FIG. 2.
Identification of the NLS in VEEV capsid protein. (A) Sequence alignment of a highly positively charged VEEV capsid fragment (WT) with corresponding sequences of the previously identified frameshift noncytopathic mutant protein (Frsh) (19). The mutated fragment is underlined. The sequence used for analysis of NLS localization is indicated in boldface. N52-85, N52-71, N72-85, and N52-71AA represent peptides fused with a 4×Tomato reporter for dissecting the position of the NLS. The mutated amino acids in N52-71AA are indicated in blue. (B) Representative images of cells expressing different 4×Tomato fusions. The images were acquired on a Nikon Ti-U inverted fluorescence microscope using a 60× CFI Super Plan Fluor objective. The nuclei were stained with DAPI (4′,6′-diamidino-2-phenylindole). (C) Representative confocal images demonstrating the distribution of 4×Tomato-3×NLS in the presence of a H68AA2-GFP fusion containing mutations in the NLS. (D) Box plot demonstrating nuclear/cytoplasmic distribution of 4×Tomato-3×NLS alone or in the presence of H68-GFP or H68AA2-GFP. Note that in the presence of mutant H68AA2-GFP protein, accumulation of the reporter protein in the nucleus remained at almost the same level. (E) Box plot of the nuclear/cytoplasmic distribution of H68-GFP and H68AA2-GFP. Mutation of the NLS inhibits accumulation of the H68AA2-GFP in nuclei. The P values were calculated using the Mann-Whitney test (n = 30 for all experiments). (F) Mutations in the VEEV capsid NLS inhibit its translocation into the nuclei. BHK-21 cells were infected with a replicon expressing VEEV capsid-GFP containing mutations in the NLS. The mutations are indicated in blue. The images were acquired on a Nikon Ti-U inverted fluorescence microscope with a 60× CFI Super Plan Fluor objective. The nuclei were stained with DAPI. Mut, mutant. Scale bars, 20 μm.
To evaluate whether this NLS is essential to the ability of the H68 peptide to inhibit nuclear import, we mutated lysines 64 and 65 in the H68-GFP fusion (H68AA2). Indeed, when 4×Tomato-3×NLS and H68AA2-GFP were coexpressed in the same cell, the reporter was transported into the nucleus almost as efficiently as in the cells expressing 4×Tomato-3×NLS alone (Fig. 2C and D). As expected, the H68AA2-GFP mutant exhibited lower accumulation in nuclei than H68-GFP (Fig. 2E) but retained its localization in the NPC (data not shown). This result was a strong indication that the presence of a functional NLS is required for H68 function in nuclear import inhibition.
The H68 peptide of the VEEV capsid protein contains a functional NES that is essential for its activity in the inhibition of nuclear import.
The H68 peptide is highly conserved among the New World alphaviruses (Fig. 3A) but differs greatly from that of the Old World viruses (data not shown). The amino terminus of H68 contains a conserved helix I that was predicted to form a coiled coil and has been implicated in capsid dimerization and nucleocapsid assembly (28, 45). In our previous study, the deletions in helix I (aa 35 to 47) led to accumulation of the mutant capsid protein in nuclei, indicating that the deleted fragment might be involved in nuclear export (5). Sequence analysis of this helix (34) revealed that it is homologous to the leucine-rich nuclear export signal (NES) (Fig. 3A). The NES mediates protein export from nuclei by the nuclear export receptor CRM1 (16), and its binding to CRM1 is inhibited by leptomycin B (33). Thus, to experimentally prove that VEEV capsid protein contains NES, we treated cells expressing capsid-GFP and the nuclear import reporter 4×Tomato-3×NLS with leptomycin B (Fig. 3B). In mock-treated cells, the capsid-GFP fusion was mostly localized to the cytoplasm and strongly inhibited nuclear import of the 4×Tomato-3×NLS reporter. Upon treatment of the cells with leptomycin B, capsid-GFP was found exclusively in nuclei, and most importantly, the nuclear import of the 4×Tomato-3×NLS reporter was completely restored. This result strongly suggested that the H68 peptide is involved in CRM1-mediated nuclear export and that its binding to CRM1 is essential for H68-mediated inhibition of nuclear import. Moreover, leptomycin B treatment of cells expressing capsid-GFP and reporters having different nuclear import signals (NLS), 4×Tomato-M19 or 4×Tomato-H2B, also restored nuclear import (data not shown).
FIG. 3.
Identification of the NES in VEEV capsid protein. (A) Sequence alignment of H68 peptides derived from different New World alphaviruses with a consensus NES sequence (NES), functional NES of cyclic-AMP (c-AMP)-dependent kinase inhibitor (PKI) (51) and supraphysiological NES (S1) (13). The sequence alignment was performed using ClustalW. Conservative hydrophobic amino acids in the NES are shown in red. Amino acids that are identical between different members of the New World alphaviruses are shaded in green. Asterisks indicate identical residues; colons indicate conserved substitutions; periods indicate semiconserved substitutions. WEEV, western equine encephalitis virus. (B) BHK-21 cells were coinfected with packaged replicons expressing VEEV capsid-GFP and 4×Tomato-3×NLS and treated with leptomycin B at 2 h postinfection as described in Materials and Methods. The images were acquired after 4 h of leptomycin B treatment on a Zeiss LSM510 confocal microscope. Scale bars, 20 μm. (C) (Top) Amino acid alignments of mutated H68-GFP fusions. Conserved hydrophobic amino acids are indicated in red, and introduced point mutations are shown in blue. (Bottom) Representative confocal images of cells expressing 4×Tomato-3×NLS and mutant proteins. Scale bars, 20 μm. (D) Box plot demonstrating the nuclear/cytoplasmic distribution of 4×Tomato-3×NLS when expressed alone and the distribution of the same protein when coexpressed with mutant peptide-GFP. A small but statistically significant increase in nuclear-reporter accumulation was detected for single-amino-acid mutants. The double mutant no longer affected nuclear accumulation of 4×Tomato-3×NLS. (E) Box plot demonstrating the nuclear/cytoplasmic distributions of H68-GFP and its mutants. The increase in nuclear accumulation of the mutant-peptide-GFP fusions was correlated with their reduced efficiencies in nuclear import inhibition. The P values were calculated using the Mann-Whitney test (n = 30 for all experiments).
Next, we tested whether replacement of critical hydrophobic amino acids (Φ) in the putative H68 NES with alanines (26, 51) also abolished the inhibitory function of this peptide in nuclear trafficking. Replacement of Leu-48 (H68A1-GFP) had only a marginal effect on the ability of H68 to downregulate nuclear import (Fig. 3C and D). Replacement of Phe-50 made H68A2-GFP a poor inhibitor of nuclear import, and introducing both mutations completely abolished the inhibition of nuclear import by the H68 peptide. Introduction of both mutations also led to accumulation of H68AA1-GFP protein in the nuclei (Fig. 3C and E). Thus, these results demonstrated that the capsid protein itself and H68 contain a functional NES and that its function is a prerequisite for the peptide's ability to block receptor-mediated nuclear import.
The H68 peptide forms a tetrameric complex with CRM1 and importin α1/β.
The data presented above implied that in order to inhibit nuclear import the H68 peptide needs to bind to the nuclear export receptor CRM1 and the nuclear import receptor importin α/β at the same time. This was an intriguing finding. Many proteins that shuttle into and out of the nucleus contain both nuclear import and nuclear export signals. However, two opposing nuclear transport receptors do not normally bind a cargo at the same time because of the action of RanGTP, which assembles exportin-cargo complexes and disassembles importin-cargo complexes (7, 23).
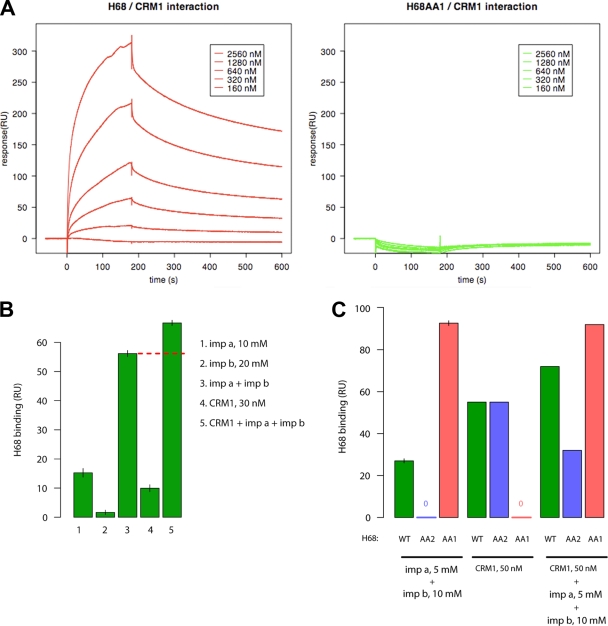
To directly analyze the binding of CRM1 and importin α/β to H68 and the possible formation of a tertrameric complex, we employed surface resonance spectroscopy (Biacore) using purified recombinant proteins. H68-GFP or its mutants were immobilized on the chip, and the surface resonance was measured upon addition of a stream of CRM1, importin α1, and/or importin β. Standard NESs measurably bind to CRM1 only in the presence of RanGTP (13). However, in our study, we found that, H68-GFP bound to CRM1 with a Kd (dissociation constant) of 2.4 μM in the absence of RanGTP (Fig. 4A). In the presence of RanGTP, the affinity of H68-GFP for CRM1 further increased about 10-fold (Kd, 230 nM). This result indicated that H68 contains a “supraNES,” an unusually strong NES that can stably interact with CRM1 without the requirement for RanGTP (13). As expected, the NES mutant H68AA1-GFP did not detectably bind to CRM1 (Fig. 4A and C), and the NLS mutant H68AA2-GFP interacted with CRM1 with similar affinity (Fig. 4C).
FIG. 4.
H68 forms a tetrameric complex with CRM1, importin α1, and importin β. (A) Surface plasmon resonance sensograms of 6×His-H68-GFP (left) and 6×His-H68AA1-GFP, an NES mutant (right), upon exposure to the indicated concentrations of CRM1. RU, relative units. (B) Surface plasmon resonance response of 6×His-H68-GFP upon exposure to importin α1 (imp a) or β (imp b) or CRM1 alone or their combinations as indicated. The red dashed line indicates the increase in response of the tetrameric complex compared to the trimeric complex. The error bars indicate standard deviations. (C) Surface plasmon resonance response of 6×His-H68-GFP or its NLS (6xHis-H68AA2-GFP) or NES (6x-His-H68AA1-GFP) mutant upon exposure to importin α1 or β or CRM1 alone or their combinations.
Next, we assessed the binding of H68-GFP to importin α1 and the importin α1/β complex. H68-GFP bound to importin α1 weakly (Fig. 4B), but the addition of importin β significantly increased the association of importin α1, as was previously reported for the standard NLS (22). The NLS mutant H68AA2-GFP did not bind importin α1 or the importin α1/β complex, while as expected, the NES mutant H68AA1-GFP bound both (Fig. 4C). Thus, the H68-GFP peptide contains a classical NLS that directly interacts with the importin α/β complex.
In order to determine whether H68-GFP can interact with CRM1 and importin α1/β simultaneously and form a tetrameric complex, both CRM1 and importin α1/β were added to the chip-bound H68-GFP. As shown in Fig. 4B, the response signal was clearly additive, and no cooperativity or competition between CRM1 and importin α1/β in binding to H68 was detected. As expected, the H68AA1 and H68AA2 mutants bound only to importin α1/β or CRM1, respectively, and did not form tetrameric complexes (Fig. 4C). We conclude that H68 contains a very strong or supraNES sequence that allows simultaneous binding of CRM1 and importin α/β in the absence of RanGTP. Although our data support the hypothesis of the formation of a tetrameric complex, future structural experiments are needed to confirm it.
The H68 peptide accumulates in the central channel of the NPC and sequesters CRM1 and importin β at the nuclear envelope (NE).
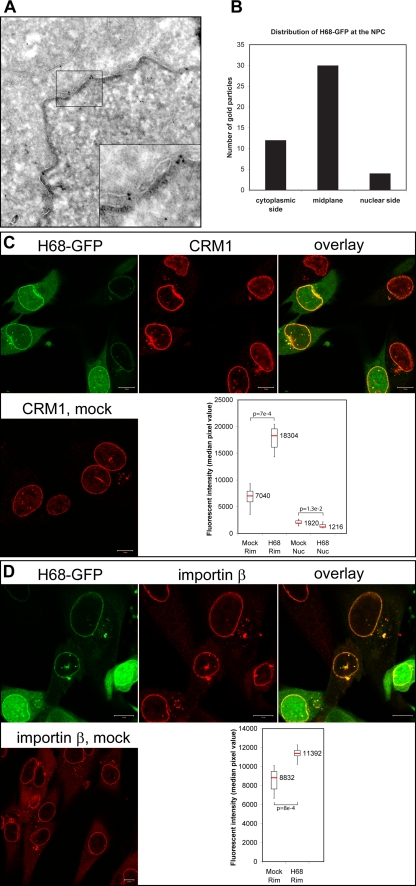
So far, the data suggested that inhibition of nuclear import by VEEV capsid protein is mediated by its simultaneous binding to CRM1 and importin α/β, resulting in a block of NPC traffic by the tetrameric complex. To test whether the localization of H68 would be consistent with such a scenario, we analyzed the presence of H68-GFP in the NPC by electron microscopy. H68-GFP was expressed from a plasmid, where it was cloned under the control of an RNA polymerase II promoter. This approach ensured a low level of H68-GFP expression due to the negative feedback on its own transcription, and thus, a large fraction of the H68-GFP was expected to accumulate in the NPC. Cells transfected with plasmids expressing H68-GFP were fixed at 24 h posttransfection, and immunogold labeling was performed on ultrathin cryosections. As demonstrated in Fig. 5A and B, H68-GFP was mainly localized in the central channel of the NPC (65%), delimited by the nuclear membranes, while the remaining NPC-associated gold particles were found on the cytoplasmic side (26%) rather than on the nuclear side (9%). The predominant accumulation of H68-GFP protein in the central channel was consistent with the detected obstruction of nuclear traffic through the NPC. The bias toward the cytoplasmic face versus the nuclear side of the NPC was correlated with the presence of a supraNES that could mediate the accumulation of H68- GFP at the cytoplasmic fibrils, as was observed for other supraNES-containing proteins (13). Very little NPC-associated gold was detected in cells expressing the GFP-H68AA1 mutant with a mutation in the CRM1 binding domain (data not shown).
FIG. 5.
H68-GFP, CRM1, and importin β are readily detectable in the NPC. (A) HeLa cells were transfected with the plasmid expressing 6×His-H68-GFP. At 24 h posttransfection, they were fixed and processed for immuno-EM using anti-GFP antibodies. The inset demonstrates 6×His-H68-GFP localization near the center of the NPC (boxed area). The cytoplasm is at the top of the image. (B) The distribution of gold particles in the midplane of the NPC and at nuclear and cytoplasmic sites was calculated using multiple images. (C) BHK-21 cells expressing H68-GFP from VEEV replicon- or mock-infected cells were stained with anti-CRM1 antibodies, followed by Alexa Fluor 555-labeled secondary antibodies. The CRM1 and H68-GFP are clearly colocalized at the nuclear rim and in small aggregates in the cytoplasm and nuclei. The box plot presents the medians of the fluorescence intensities of the CRM1-specific signals in the nuclear envelope and nucleus. (D) BHK-21 cells expressing H68-GFP from VEEV replicon- or mock-infected cells were stained with anti-importin β antibodies and Alexa Fluor 555-labeled secondary antibodies. The importin β and H68-GFP are clearly colocalized at the nuclear rim and in small aggregates in the cytoplasm, but not in the nuclei. The box plot presents the medians of the fluorescence intensities of importin β-specific signals in the nuclear envelope. The P values were calculated using the Mann-Whitney test (n = 8 for importin β; n = 6 for CRM1). Scale bars, 20 μm. Rim, nuclear rim; Nuc, nucleoplasm.
To test whether expression of H68-GFP leads to accumulation of CRM1 and importin α/β in the NPC, we quantitatively analyzed the immunostaining of two transport receptors in mock- or H68-GFP-transfected cells. In the normal cells, a strong CRM1 signal was detected at the nuclear rim and in the small granules throughout the cytoplasm, which likely represented the annulate lamellae (Fig. 5C). Weak, diffuse staining was also observed in the nucleoplasm. In the cells expressing the H68-GFP fusion, the intensity of CRM1 signal in the nuclear rim increased significantly (2.6-fold) and the nucleoplasmic signal decreased (Fig. 5C). The nuclear rim staining of CRM1 had a punctate pattern (data not shown), reminiscent of that of the NPC, and colocalized with H68-GFP. The small granules containing both H68-GFP and CRM1 were also detected in the cytoplasm and in the nucleus. The effect of the H68-GFP fusion on importin β distribution was not so dramatic, but we observed about a 30% increase in NPC-specific importin β signal (Fig. 5D). The colocalization of H68-GFP and importin β was also prominent in the small cytoplasmic and nuclear granules. These data confirmed the hypothesis that H68-GFP interacted with CRM1 and importin α/β in the cells and sequestered both receptors to the NPC. However, we have not detected accumulation of Ran in the NPC (data not shown), and this additionally indicated that the H68-GFP-CRM1-importin α/β complex does not require Ran for its formation. Taken together, these results demonstrated that H68-GFP accumulates at the center of the NPC as a tetrameric complex with CRM1 and importin α/β.
Blocking of nuclear import by the VEEV capsid protein requires specific spacing between the NLS and supraNES.
The result that the inhibitory function of H68 required binding of two nuclear receptors mediating opposing nucleocytoplasmic traffic pathways contradicted previously published data. The common approach in analyzing the efficiency of the NESs is their fusion with Rev(1.4)NLS signal (26). However, there are no reports regarding whether such constructs affect nucleocytoplasmic traffic. A distinct feature of the H68 peptide is the presence of a supraNES that, unlike a classical NES, mediates CRM1 binding in the absence of RanGTP and thus can bind CRM1, not only in the nucleus, but also in the cytoplasm. On the other hand, the nonfunctional H60 peptide also contains the supraNES but lacks the NLS. To test whether the addition of a strong NLS would restore its ability to inhibit nuclear import, we added a triple SV40 TAg NLS to the C terminus of the H60-GFP fusion protein. The resulting protein inhibited nuclear import very inefficiently (data not shown), and this correlated with its previously described inability to inhibit cellular transcription (19). These data suggested that the relative position and/or sequence between the NES and NLS are also important for the ability of the H68 peptide to block NPC function.
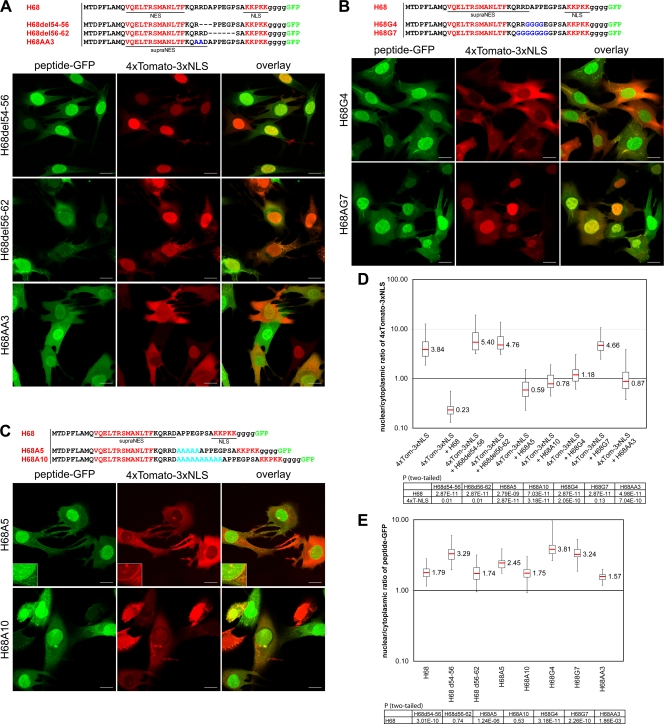
To further define the role of the linker domain between the NES and NLS, we first introduced two deletions into the sequence between the signals (Fig. 6A). Both deletion mutants completely lost the ability to inhibit nuclear import of the 4×Tomato-3×NLS reporter. Interestingly, the cellular distributions of the deletion mutants were different. H68del54-56-GFP became mostly nuclear and lost its association with the nuclear rim. On the other hand, the H68del56-62-GFP distribution was similar to that of H68-GFP, with clear accumulation at the nuclear rim. These results suggested that the sequence deleted in H68del54-56-GFP could be involved in the supraphysiological binding with CRM1. To get additional confirmation for this hypothesis, we replaced two arginines, located downstream of the NES, with alanines. This mutant, H68AA3-GFP, had reduced ability to inhibit nuclear import but did not efficiently accumulate in the nuclei (Fig. 6A, D, and E). The loss of the inhibitory function by the second mutant, H68del56-62-GFP, was more likely the result of less efficient binding of importin α/β to the dimeric complex H68-GFP-CRM1, which accumulated at the nuclear rim similarly to supraphysiological NES (13). Taken together, these results implied that the CRM1 binding site could occupy a larger area of the downstream leucine-rich domain, up to D55, and that this sequence (K49QRRD) was involved in the strong binding of the H68-GFP fusion to CRM1.
FIG. 6.
Mutations in the NES- and NLS-connecting linker affect the ability of H68 peptide to inhibit nuclear import. In all of the experiments presented, BHK-21 cells were coinfected with packaged VEEV replicons encoding mutant-peptide-GFP fusions and 4×Tomato-3×NLS. The cells were fixed at 4 h postinfection, and the distribution of fluorescent proteins was analyzed using a Leica SP1 confocal microscope. (A) (Top) Amino acid sequences of the wild-type H68 peptide and the deletion or substitution mutants. The positions of the deletions are indicated by dashed lines. (Bottom) The deletions of 3 or 5 aa in the linker peptide completely abolished nuclear import inhibition by mutant-peptide-GFP fusions. H68del54-56 accumulated more efficiently in the nuclei than H68-GFP (compare Fig. 1A and panel E). (B) (Top) Amino acid sequences of the wild-type H68 peptide and the designed mutants. Four or 7 amino acids of the linker were replaced with glycines, which are highlighted in blue. (Bottom) Substitution of 4 aa (H68G4) partially restored the nuclear import of 4×Tomato-3×NLS. Substitution of 7 amino acids (H68G7) completely abolished the ability of the mutant peptide (H68G7) to inhibit nuclear import. (C) (Top) Amino acid sequences of the wild-type H68 peptide and the insertion mutants. Five- and 10-aa-long linkers were inserted between the supra-NES and NLS. (Bottom) In the presence of both mutated GFP fusions, nuclear import of the 4×Tomato-3×NLS reporter was partially restored. The mutant peptide with the longer linker had less inhibitory effect on nuclear import. The enlarged insets demonstrate the accumulation of 4×Tomato-3×NLS and H68A5-GFP at the nuclear rim. (D) The box plot demonstrates the nuclear/cytoplasmic distribution of 4×Tomato-3×NLS expressed alone or in the presence of a mutant peptide-GFP. (E) The box plot demonstrates the nuclear/cytoplasmic distribution of H68-GFP and mutant proteins. The P values were calculated using the Mann-Whitney test (n = 30 for all experiments). Scale bars, 20 μm.
To further validate that the effect of H68 on nuclear cytoplasmic trafficking depended on the sequence of the peptide connecting the NES and NLS, we replaced 4 or 7 amino acids between the NES and NLS domains with glycines (Fig. 6B) without affecting the linker size. H68G4-GFP still partially inhibited nuclear import but efficiently accumulated in the nucleus. These data were suggestive of the less efficient interaction of mutated peptides with CRM1 and thus confirmed that the sequence downstream of the leucine-rich domain is part of the supraNES. Replacement of 7 amino acids in the linker completely restored nuclear import in the presence of mutant H68G7-GFP fusion (Fig. 6B and D). In addition, as could be predicted, this mutant protein also efficiently accumulated in the nuclei (Fig. 6E).
Next, we inserted 5 or 10 additional alanines between the NES and NLS sequences in H68. The insertion mutants (H68A5-GFP and H68A10-GFP) demonstrated strong reduction in the ability to inhibit nuclear import. The length of the insertion was negatively correlated with the efficiency of nuclear transport inhibition (Fig. 6C and D). Interestingly, in the presence of H68A5, we detected accumulation of the 4× Tomato-NLS reporter in the nuclear membrane, suggesting that it competed with the mutant H68A5-GFP for access to NPC (Fig. 6C, insets). Since the positions of the insertions were predicted to be outside the supraNES or NLS domain, we speculate that the insertions changed the spatial arrangement of CRM1 and importin α/β in the tetrameric complex and thus its overall tertiary structure.
Thus, the mutagenesis analysis of the linker peptide between the NES and NLS demonstrated that any modifications in the linker peptide attenuated the ability of the H68 peptide to inhibit nuclear import.
The nonpathogenic phenotype of VEEVs is correlated with their reduced ability to inhibit nuclear import.
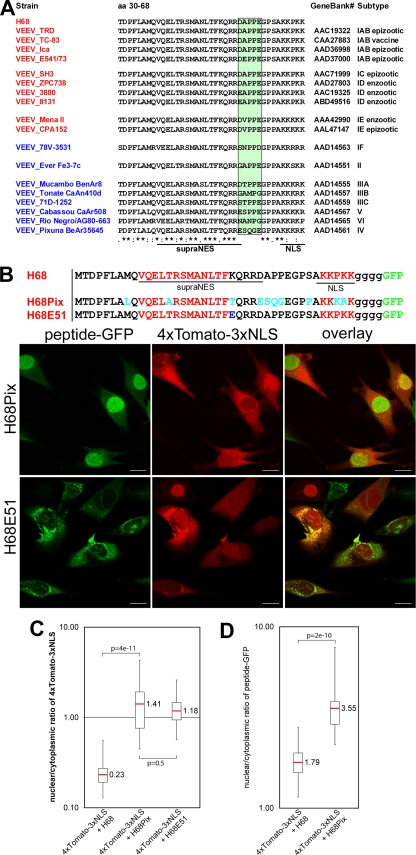
Viruses of the VEEV complex are continuously circulating in nature and cause periodic outbreaks in Latin America (2, 39, 41, 49). There are 13 subtypes of VEEV (49). Epizootic strains have been described only in subtypes IAB, IC, and IE and were shown to have emerged via mutations in viral glycoprotein E2 (3, 11). Subtypes IF and II to VI are less pathogenic and do not cause epidemics. Figure 7A presents the alignment of the H68 fragments from representative members of different subtypes. Interestingly, the most divergent part of the H68 sequence is a linker fragment between the NES and NLS domains. The mutations in subtypes IAB, IC, and IE are all conservative, suggesting that they might have no affect on the ability of H68 to inhibit nuclear import. On the other hand, the mutations in subtypes ID and II to VI are usually nonconservative. The available information about the diseases caused by these serotypes is very limited due to bias in the studies toward serotypes that cause epidemics. Pixuna virus (serotype V) was described as nonpathogenic and was not lethal even for immunosuppressed hamsters (29, 43). Therefore, to test the biological significance of our data, we examined whether the H68 peptide of the nonpathogenic Pixuna virus (subtype V) (29, 43) was capable of inhibiting nuclear import. The H68 sequence of the Pixuna virus (H68Pix) was used to replace the corresponding sequence in the above-described H68-GFP (Fig. 7B). The Pixuna-specific peptide demonstrated a very weak ability to inhibit nuclear import of the reporter (Fig. 7B and C), and H68Pix-GFP accumulated to high concentrations in the nucleus (Fig. 7B). This result was consistent with the presence of multiple mutations in the supra-NES and linker domains. To confirm that partial restoration of nuclear import can lead to noncytopathic viral infection, we analyzed H68E51-GFP, a mutant with a point mutation in the supraNES. This mutation was selected previously, and virus carrying the mutation was noncytopathic and incapable of inhibiting cellular transcription (19). Indeed, H68E51-GFP downregulated nuclear import to the same extent as did H68Pix-GFP. Thus, this result suggests that the nonpathogenic phenotype of Pixuna virus is strongly correlated with the inability of its capsid protein to efficiently inhibit nuclear import.
FIG. 7.
H68 peptide derived from nonpathogenic VEEV Pixuna is incapable of efficient inhibition of nuclear import. (A) Sequence alignment of H68 peptides derived from representative members of different VEEV subtypes. The green-shaded box indicates the most divergent sequence between different subtypes. NLS and NES are indicated by red letters. The asterisks indicate identical residues; the colons indicate conserved substitutions; the periods indicate semiconserved substitutions. (B) (Bottom) Representative confocal image of BHK-21 cells coexpressing 4×Tomato-3×NLS and H68Pix-GFP or H68E51-GFP. (Top) The mutated amino acid in the H68E51 peptide is highlighted in blue. The amino acids that differ between H68 and H68Pix are highlighted in turquoise. Scale bars, 20 μm. (C) Box plot demonstrating the nuclear/cytoplasmic distribution of 4×Tomato-3×NLS when expressed alone or in the presence of a mutant H68 peptide-GFP. The distributions of 4×Tomato-3×NLS in the presence of H68Pix and H68E51 were similar. (D) Box plot presenting the nuclear/cytoplasmic distributions of H68-GFP and H68Pix-GFP. H68Pix-GFP mostly accumulated in the nucleus. The distribution of H68E51 was not analyzed due to its high tendency to aggregate in the cytoplasm. The P values were calculated using the Mann-Whitney test (n = 30 for H68 and H68Pix; n = 29 for H68E51).
DISCUSSION
Development of the antiviral response requires de novo synthesis of a large number of cellular proteins, which either downregulate virus replication in the infected cells or are released from the cells and activate the antiviral state in yet-uninfected cells (31). New World alphaviruses have evolved a unique mechanism for interfering with the development of an antiviral response on a cellular level. Their capsid proteins efficiently inhibit nucleocytoplasmic traffic in vertebrate cells, and within 8 h postinfection, the cells exhibit profound inhibition of transcription, which is a very efficient means of interfering with the activation of the antiviral genes (1, 5, 19, 21). The same capsid protein does not affect nuclear import in mosquito cells, which demonstrate no noticeable changes in transcription and support persistent virus replication. In this study, we further defined the mechanism of capsid-specific inhibition of nuclear import and provided a plausible explanation for the less pathogenic phenotype of some natural VEEV isolates.
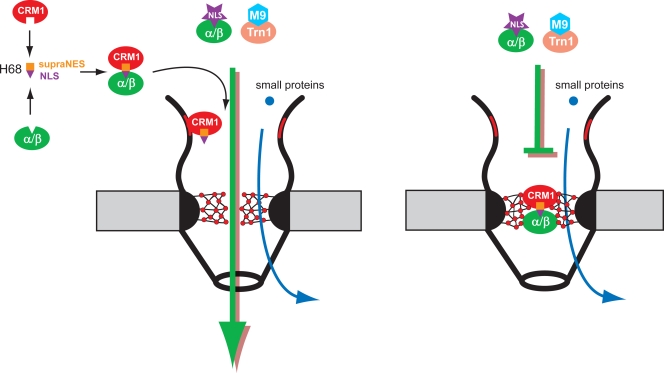
The current model of VEEV capsid protein functioning in the regulation of nuclear import is presented in Fig. 8. Our study demonstrated that its inhibitory function is mediated by a short, N-terminal, 39-aa-long peptide, H68. This peptide simultaneously binds two nuclear traffic receptors, CRM1 and importin α/β, which mediate two opposing nucleocytoplasmic traffic pathways. H68 binds strongly to CRM1 in the absence of RanGTP and thus contains a supraNES (13), which is located at the N terminus of the peptide. The core element of the supraNES is a 9-aa-long leucine-rich alpha peptide. However, an additional 5 aa, located downstream of the core, are also critical for its functioning as a supraNES. The importin α/β complex binds to the carboxy terminus of the H68 peptide through a monopartite NLS. Binding of CRM1 and importin α/β to H68 appears to be additive, and there is no detectable interaction between CRM1 and importin receptors in the final tetrameric complex. Binding of both CRM1 and importin α/β receptors is a prerequisite of nuclear import inhibition by the VEEV capsid protein, and mutations in either of the binding sites completely abolish the inhibitory function of the protein.
FIG. 8.
Model of H68-dependent inhibition of nuclear import. (Left) Binding of CRM1 and H68-GFP does not require RanGTP, and thus, the tetrameric complex H68-GFP/CRM1/importin α/β can form in the cytoplasm. Alternatively, the dimeric complex H68-GFP/CRM1 may preassemble on the cytoplasmic fibers by binding with Nup358, followed by binding with importin α/β. (Right) Next, the H68-GFP complex moves through NPC and blocks the central channel by binding to yet-unidentified nucleoporins. As a result, NPCs become inaccessible to receptor-mediated traffic but still support the diffusion of small molecules.
Since formation of the H68-specific complex does not require RanGTP, the complex may be formed either in the cytoplasm or on the cytoplasmic side of the NPC, where the H68-GFP/CRM1 complex accumulates due to its strong binding to the cytoplasmic fibrils. This phenomenon has previously been shown for another supraphysiological NES, S1 (14). The possibility of dimeric-complex formation on the cytoplasmic fibrils is supported by the detected accumulation of some GFP fusions with mutated peptides at the nuclear rim (Fig. 1 and 2, H60-GFP and H68AA2-GFP mutants). The latter mutants have intact supraNESs but lack the functional NLS domain. This hypothesis is also supported by the detection of preferential accumulation of H68-GFP on the cytoplasmic side of the NPC by EM (Fig. 4). Upon reaching the central channel of the NPC, the tetrameric complex binds strongly to the nucleoporins, which need to be further defined, and its migration either completely stops or slows drastically. We favor the hypothesis that the tetrameric complex continues to slowly migrate through the NPC, because the large capsid-GFP fusion was found in the nuclei and not only in the NPC (5). However, the direct detection of migration of the H68-GFP through the NPC is technically challenging due to its slow migration and its presence in two complexes, H68-GFP/CRM1 and H68-GFP/CRM1/importin α1/β. The very slow migration of H68-GFP-containing complexes through the NPC would also explain the incomplete inhibition of nuclear import by some mutants (such as H68A5 and H68G4) and, at the same time, accumulation of the 4×Tomato-3×NLS reporter in the nuclear rim in their presence. These mutations appear to affect the efficiency of tetrameric complex formation or its stability, and thus, the NPC would become more accessible for another import cargo, although it translocates through the NPC at lower rates.
Facilitated nucleocytoplasmic traffic is mediated by reasonably weak interactions between nuclear transport receptors, karyopherins and nucleoporins, containing FG repeats (FG nups) (40, 42). Strong evidence suggests that FG nups can form a barrier due to the binary interactions between FG repeats (17, 37). Each karyopherin contains several binding sites that bind FG repeats, thereby melting the FG meshwork to ensure rapid cargo translocation. The capsid-specific tetrameric complex contains two karyopherins and about twice the FG-binding sites of standard import or export complexes. It is tempting to conclude that this simple increase in the FG repeat-binding domain numbers slows down translocation of the tetrameric complex through the NPC. However, mutants with insertions between the supraNES and NLS in the H68 peptide, which should not affect the number of FG-binding sites in the complex but might change the relative positions of the CRM1 and importin α/β in the complex, were no longer capable of efficiently inhibiting nuclear import. Similarly, the H60-GFP carrying three NLSs on the carboxy terminus of GFP did not efficiently inhibit nuclear import. In addition, many larger cargos contain multiple karyopherins (10, 35, 36) without obstructing the NPC. Therefore, we hypothesize that the tetrameric complex tertiary structure is vital for sequestering the H68-GFP fusion and VEEV capsid protein in the NPC and that only specific orientation of the complex in the NPC allows its tight simultaneous binding with nucleoporin FG repeats. Interestingly, cargo-bound conformations of both CRM1 and importin α/β have higher affinities for several FG nups than the empty conformations (4, 8, 32). Therefore, it is likely that CRM1 and importin β in their cargo-bound states bind with high affinity to different nucleoporins and that the cooperative binding in the tetrameric complex inhibits efficient translocation through the NPC, thereby inhibiting other transport pathways. The identification of these nucleoporins will provide new insights into NPC structure and function. Importantly, VEEV capsid protein does not inhibit nuclear import in mosquito cells, and this suggests that the structures of vertebrate and invertebrate NPCs are divergent or that mosquito CRM1 does not bind as strongly to the H68 peptide.
Nucleocytoplasmic traffic is essential for the regulation of all cellular processes and cell survival. It also appears to play a critical role in the activation of cellular genes in response to infection with different viral pathogens. Thus, it is not surprising that some of the RNA viruses, which do not require nuclei for their replication, have developed efficient mechanisms for interfering with the nucleocytoplasmic traffic (25). The poliovirus and rhinovirus proteases degrade several nucleoporins and thus inhibit some of the nuclear traffic pathways. The matrix protein of vesicular stomatitis virus (VSV) is so far the only other viral protein that efficiently blocks nucleocytoplasmic traffic by binding one of the NPC nucleoporins, nup98 (27, 47). This interaction is mediated by the mRNA export factor Rae1 (15) and leads to the disruption of cellular mRNA nuclear export and, consequently, cellular transcription. Interestingly, the transcription inhibition is secondary to the disruption of NPC functioning. We have also previously demonstrated that the capsid proteins of the New World alphaviruses strongly inhibit transcription of cellular mRNAs and rRNAs at 10 to 24 h postinfection. In this study, we observed strong inhibition of nuclear import in just 4 h postinfection with a replicon expressing H68-GFP. Thus, inhibition of nuclear import appears to precede the transcriptional shutoff during alphavirus infection. Although, the resulting inhibition of transcription is a very efficient way of interfering with the antiviral response, the inhibition of nuclear import has additional benefits, such as preventing the import of transcriptional factors involved in the host response and inhibiting the export of newly synthesized mRNA. Hence, the inhibition of the antiviral response can occur more efficiently.
In conclusion, we have demonstrated that pathogenic strains of VEEV have developed a unique and efficient means of inhibiting nuclear import by forming tetrameric complexes between viral capsid protein and three karyopherins: CRM1, importin α, and importin β. Importantly, different serotypes of VEEV exhibit sequence variations in the H68 peptide, and the sequences of the H68 peptides of nonpathogenic strains are strongly divergent from those of the pathogenic strains. Our data demonstrated that the less pathogenic phenotype of Pixuna virus is strongly correlated with the inability of its capsid protein to completely inhibit nuclear import. Taken together, the experimental data presented imply that capsid protein is the major determinant of VEEV pathogenesis. These data also raise the possibility that the nonpathogenic strains of VEEV can evolve into more pathogenic phenotypes, and epidemiological studies should include structural and functional studies of capsid protein. The data also explain the high residual pathogenicity of the current experimental live vaccine against VEEV, TC-83, which contains the functional H68 peptide. Finally, this novel mechanism of nuclear transport inhibition shows that the NPC is vulnerable to unusual cargo receptor complexes and may be used by other cellular or viral proteins in a regulated manner.
Acknowledgments
We thank the NKI Protein Facility for purification of importin α1 and the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO) for financial support to the facility (grant 175.010.2007.012), Hans Jansen for cryoimmunoelectron microscopy, and Ilya Frolov for support, useful comments on the experiments, and critical reading of the manuscript. We are also thankful to Natalia Garmashova, Eugenia Volkova, and Rodion Gorchakov for excellent technical assistance.
This work was supported by Public Health Service grants AI070207 and AI073301 (E.I.F. and S.A.).
Footnotes
Published ahead of print on 10 February 2010.
REFERENCES
- 1.Aguilar, P. V., A. P. Adams, E. Wang, W. Kang, A. S. Carrara, M. Anishchenko, I. Frolov, and S. C. Weaver. 2008. Structural and nonstructural protein genome regions of eastern equine encephalitis virus are determinants of interferon sensitivity and murine virulence. J. Virol. 82:4920-4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguilar, P. V., I. P. Greene, L. L. Coffey, G. Medina, A. C. Moncayo, M. Anishchenko, G. V. Ludwig, M. J. Turell, M. L. O'Guinn, J. Lee, R. B. Tesh, D. M. Watts, K. L. Russell, C. Hice, S. Yanoviak, A. C. Morrison, T. A. Klein, D. J. Dohm, H. Guzman, A. P. Travassos da Rosa, C. Guevara, T. Kochel, J. Olson, C. Cabezas, and S. C. Weaver. 2004. Endemic Venezuelan equine encephalitis in northern Peru. Emerg. Infect. Dis. 10:880-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anishchenko, M., R. A. Bowen, S. Paessler, L. Austgen, I. P. Greene, and S. C. Weaver. 2006. Venezuelan encephalitis emergence mediated by a phylogenetically predicted viral mutation. Proc. Natl. Acad. Sci. U. S. A. 103:4994-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Askjaer, P., A. Bachi, M. Wilm, F. R. Bischoff, D. L. Weeks, V. Ogniewski, M. Ohno, C. Niehrs, J. Kjems, I. W. Mattaj, and M. Fornerod. 1999. RanGTP-regulated interactions of CRM1 with nucleoporins and a shuttling DEAD-box helicase. Mol. Cell. Biol. 19:6276-6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atasheva, S., N. Garmashova, I. Frolov, and E. Frolova. 2008. Venezuelan equine encephalitis virus capsid protein inhibits nuclear import in mammalian but not in mosquito cells. J. Virol. 82:4028-4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beck, M., F. Forster, M. Ecke, J. M. Plitzko, F. Melchior, G. Gerisch, W. Baumeister, and O. Medalia. 2004. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science 306:1387-1390. [DOI] [PubMed] [Google Scholar]
- 7.Becskei, A., and I. W. Mattaj. 2003. The strategy for coupling the RanGTP gradient to nuclear protein export. Proc. Natl. Acad. Sci. U. S. A. 100:1717-1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ben-Efraim, I., and L. Gerace. 2001. Gradient of increasing affinity of importin beta for nucleoporins along the pathway of nuclear import. J. Cell Biol. 152:411-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boege, U., M. Cygler, G. Wengler, P. Dumas, J. Tsao, M. Luo, T. J. Smith, and M. G. Rossmann. 1989. Sindbis virus core protein crystals. J. Mol. Biol. 208:79-82. [DOI] [PubMed] [Google Scholar]
- 10.Bradatsch, B., J. Katahira, E. Kowalinski, G. Bange, W. Yao, T. Sekimoto, V. Baumgartel, G. Boese, J. Bassler, K. Wild, R. Peters, Y. Yoneda, I. Sinning, and E. Hurt. 2007. Arx1 functions as an unorthodox nuclear export receptor for the 60S preribosomal subunit. Mol. Cell 27:767-779. [DOI] [PubMed] [Google Scholar]
- 11.Brault, A. C., A. M. Powers, E. C. Holmes, C. H. Woelk, and S. C. Weaver. 2002. Positively charged amino acid substitutions in the e2 envelope glycoprotein are associated with the emergence of Venezuelan equine encephalitis virus. J. Virol. 76:1718-1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi, H. K., S. Lee, Y. P. Zhang, B. R. McKinney, G. Wengler, M. G. Rossmann, and R. J. Kuhn. 1996. Structural analysis of Sindbis virus capsid mutants involving assembly and catalysis. J. Mol. Biol. 262:151-167. [DOI] [PubMed] [Google Scholar]
- 13.Engelsma, D., R. Bernad, J. Calafat, and M. Fornerod. 2004. Supraphysiological nuclear export signals bind CRM1 independently of RanGTP and arrest at Nup358. EMBO J. 23:3643-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engelsma, D., N. Valle, A. Fish, N. Salome, J. M. Almendral, and M. Fornerod. 2008. A supraphysiological nuclear export signal is required for parvovirus nuclear export. Mol. Biol. Cell 19:2544-2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faria, P. A., P. Chakraborty, A. Levay, G. N. Barber, H. J. Ezelle, J. Enninga, C. Arana, J. van Deursen, and B. M. Fontoura. 2005. VSV disrupts the Rae1/mrnp41 mRNA nuclear export pathway. Mol. Cell 17:93-102. [DOI] [PubMed] [Google Scholar]
- 16.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051-1060. [DOI] [PubMed] [Google Scholar]
- 17.Frey, S., and D. Gorlich. 2007. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell 130:512-523. [DOI] [PubMed] [Google Scholar]
- 18.Frolov, I., E. Frolova, and S. Schlesinger. 1997. Sindbis virus replicons and Sindbis virus: assembly of chimeras and of particles deficient in virus RNA. J. Virol. 71:2819-2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garmashova, N., S. Atasheva, W. Kang, S. C. Weaver, E. Frolova, and I. Frolov. 2007. Analysis of Venezuelan equine encephalitis virus capsid protein function in the inhibition of cellular transcription. J. Virol. 81:13552-13565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garmashova, N., R. Gorchakov, E. Frolova, and I. Frolov. 2006. Sindbis virus nonstructural protein nsP2 is cytotoxic and inhibits cellular transcription. J. Virol. 80:5686-5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garmashova, N., R. Gorchakov, E. Volkova, S. Paessler, E. Frolova, and I. Frolov. 2007. The Old World and New World alphaviruses use different virus-specific proteins for induction of transcriptional shutoff. J. Virol. 81:2472-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldfarb, D. S., A. H. Corbett, D. A. Mason, M. T. Harreman, and S. A. Adam. 2004. Importin alpha: a multipurpose nuclear-transport receptor. Trends Cell Biol. 14:505-514. [DOI] [PubMed] [Google Scholar]
- 23.Gorlich, D., M. J. Seewald, and K. Ribbeck. 2003. Characterization of Ran-driven cargo transport and the RanGTPase system by kinetic measurements and computer simulation. EMBO J. 22:1088-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffin, D. 1986. Alphavirus pathogenesis and immunity, p. 209-250. In S. Schlesinger and M. J. Schlesinger (ed.), The Togaviridae and Flaviviridae. Plenum Press, New York, NY.
- 25.Gustin, K. E. 2003. Inhibition of nucleo-cytoplasmic trafficking by RNA viruses: targeting the nuclear pore complex. Virus Res. 95:35-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henderson, B. R., and A. Eleftheriou. 2000. A comparison of the activity, sequence specificity, and CRM1-dependence of different nuclear export signals. Exp. Cell Res. 256:213-224. [DOI] [PubMed] [Google Scholar]
- 27.Her, L. S., E. Lund, and J. E. Dahlberg. 1997. Inhibition of Ran guanosine triphosphatase-dependent nuclear transport by the matrix protein of vesicular stomatitis virus. Science 276:1845-1848. [DOI] [PubMed] [Google Scholar]
- 28.Hong, E. M., R. Perera, and R. J. Kuhn. 2006. Alphavirus capsid protein helix I controls a checkpoint in nucleocapsid core assembly. J. Virol. 80:8848-8855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jahrling, P. B., E. Dendy, and G. A. Eddy. 1974. Correlates to increased lethality of attenuated Venezuelan encephalitis virus vaccine for immunosuppressed hamsters. Infect. Immun. 9:924-930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnston, R. E., and C. J. Peters. 1996. Alphaviruses, p. 843-898. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven, New York, NY.
- 31.Katze, M. G., J. L. Fornek, R. E. Palermo, K. A. Walters, and M. J. Korth. 2008. Innate immune modulation by RNA viruses: emerging insights from functional genomics. Nat. Rev. Immunol. 8:644-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kehlenbach, R. H., A. Dickmanns, A. Kehlenbach, T. Guan, and L. Gerace. 1999. A role for RanBP1 in the release of CRM1 from the nuclear pore complex in a terminal step of nuclear export. J. Cell Biol. 145:645-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kudo, N., B. Wolff, T. Sekimoto, E. P. Schreiner, Y. Yoneda, M. Yanagida, S. Horinouchi, and M. Yoshida. 1998. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell Res. 242:540-547. [DOI] [PubMed] [Google Scholar]
- 34.la Cour, T., L. Kiemer, A. Molgaard, R. Gupta, K. Skriver, and S. Brunak. 2004. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 17:527-536. [DOI] [PubMed] [Google Scholar]
- 35.Lo, K. Y., and A. W. Johnson. 2009. Reengineering ribosome export. Mol. Biol. Cell 20:1545-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palacios, I., M. Hetzer, S. A. Adam, and I. W. Mattaj. 1997. Nuclear import of U snRNPs requires importin beta. EMBO J. 16:6783-6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patel, S. S., B. J. Belmont, J. M. Sante, and M. F. Rexach. 2007. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell 129:83-96. [DOI] [PubMed] [Google Scholar]
- 38.Powers, A. M., A. C. Brault, Y. Shirako, E. G. Strauss, W. Kang, J. H. Strauss, and S. C. Weaver. 2001. Evolutionary relationships and systematics of the alphaviruses. J. Virol. 75:10118-10131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Powers, A. M., M. S. Oberste, A. C. Brault, R. Rico-Hesse, S. M. Schmura, J. F. Smith, W. Kang, W. P. Sweeney, and S. C. Weaver. 1997. Repeated emergence of epidemic/epizootic Venezuelan equine encephalitis from a single genotype of enzootic subtype ID virus. J. Virol. 71:6697-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ribbeck, K., and D. Gorlich. 2002. The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J. 21:2664-2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rico-Hesse, R., S. C. Weaver, J. de Siger, G. Medina, and R. A. Salas. 1995. Emergence of a new epidemic/epizootic Venezuelan equine encephalitis virus in South America. Proc. Natl. Acad. Sci. U. S. A. 92:5278-5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rout, M. P., J. D. Aitchison, A. Suprapto, K. Hjertaas, Y. Zhao, and B. T. Chait. 2000. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148:635-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shope, R. E., O. R. Causey, and A. H. De Andrade. 1964. The Venezuelan equine encephalomyelitis complex of group A arthropod-borne viruses, including Mucambo and Pixuna from the Amazon region of Brazil. Am. J. Trop. Med. Hyg. 13:723-727. [DOI] [PubMed] [Google Scholar]
- 44.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tellinghuisen, T. L., and R. J. Kuhn. 2000. Nucleic acid-dependent cross-linking of the nucleocapsid protein of Sindbis virus. J. Virol. 74:4302-4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Volkova, E., R. Gorchakov, and I. Frolov. 2006. The efficient packaging of Venezuelan equine encephalitis virus-specific RNAs into viral particles is determined by nsP1-3 synthesis. Virology 344:315-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.von Kobbe, C., J. M. van Deursen, J. P. Rodrigues, D. Sitterlin, A. Bachi, X. Wu, M. Wilm, M. Carmo-Fonseca, and E. Izaurralde. 2000. Vesicular stomatitis virus matrix protein inhibits host cell gene expression by targeting the nucleoporin Nup98. Mol. Cell 6:1243-1252. [DOI] [PubMed] [Google Scholar]
- 48.Warrier, R., B. R. Linger, B. L. Golden, and R. J. Kuhn. 2008. Role of Sindbis virus capsid protein region II in nucleocapsid core assembly and encapsidation of genomic RNA. J. Virol. 82:4461-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weaver, S. C., C. Ferro, R. Barrera, J. Boshell, and J. C. Navarro. 2004. Venezuelan equine encephalitis. Annu. Rev. Entomol. 49:141-174. [DOI] [PubMed] [Google Scholar]
- 50.Weis, K., I. W. Mattaj, and A. I. Lamond. 1995. Identification of hSRP1 alpha as a functional receptor for nuclear localization sequences. Science 268:1049-1053. [DOI] [PubMed] [Google Scholar]
- 51.Wen, W., J. L. Meinkoth, R. Y. Tsien, and S. S. Taylor. 1995. Identification of a signal for rapid export of proteins from the nucleus. Cell 82:463-473. [DOI] [PubMed] [Google Scholar]