Abstract
The morphogenesis of many enveloped viruses, in which viral nucleocapsid complex interacts with envelope (E) protein, is known to take place at various sites along the secretory pathway. How viral E protein retains in a particular intracellular organelle for assembly remains incompletely understood. In this study, we investigated determinants in the E protein of dengue virus (DENV) for its retention and assembly in the endoplasmic reticulum (ER). A chimeric experiment between CD4 and DENV precursor membrane/E constructs suggested that the transmembrane domain (TMD) of E protein contains an ER retention signal. Substitutions of three nonhydrophobic residues at the N terminus of the first helix (T1) and at either the N or C terminus of the second helix of the TMD with three hydrophobic residues, as well as an increase in the length of T1, led to the release of chimeric CD4 and E protein from the ER, suggesting that short length and certain nonhydrophobic residues of the TMD are critical for ER retention. The analysis of enveloped viruses assembled at the plasma membrane and of those assembled in the Golgi complex and ER revealed a trend of decreasing length and increasing nonhydrophobic residues of the TMD of E proteins. Taken together, these findings support a TMD-dependent sorting for viral E proteins along the secretory pathway. Moreover, similar mutations introduced into the TMD of DENV E protein resulted in the increased production of virus-like particles (VLPs), suggesting that modifications of TMD facilitate VLP production and have implications for utilizing flaviviral VLPs as serodiagnostic antigens and vaccine candidates.
The assembly of many enveloped viruses is known to take place at various locations along the secretory pathway. Members of the families Orthomyxoviridae, Paramyxoviridae, Rhabdoviridae, Retroviridae, and Togaviridae, such as influenza virus, parainfluenza virus, vesicular stomatitis virus, human immunodeficiency virus, and Sindbis virus, assemble at the plasma membrane (25, 30, 63). Members of the families Bunyaviridae and Coronaviridae assemble in the Golgi complex and endoplasmic reticulum-Golgi intermediate compartment (ERGIC), respectively, whereas members of the family Flaviviridae assemble in the membranous structure derived from the ER (2, 30, 36, 40). An important step in the morphogenesis of enveloped viruses involves the encounter and interaction between the viral nucleocapsid complex, which contains viral capsid (C) protein and nucleic acid derived from genome replication, and viral envelope (E) protein, as well as matrix protein in some cases at a particular organelle. As the default pathway for cellular membrane proteins goes from the ER to the plasma membrane, how the viral E protein, a membrane protein, retains in a particular intracellular organelle is critical for virus assembly.
Dengue viruses (DENV) belong to the genus Flavivirus of the family Flaviviridae. There are four serotypes of DENV (DENV1 to DENV4), which are the leading cause of arboviral diseases in tropical and subtropical areas, including a debilitating but self-limited disease, dengue fever, and the severe and potentially life-threatening diseases dengue hemorrhagic fever and dengue shock syndrome (21-24). DENV contains a positive-sense, single-stranded RNA genome. The single open reading frame encodes a polyprotein precursor, which is cleaved by cellular and viral protease into three structural proteins, C, precursor membrane (PrM), and E, at the N terminus, and seven nonstructural proteins, NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5, at the C terminus (38). Like most other flaviviruses, the replication cycle of DENV involves the binding of E protein to its cellular receptor and entry through receptor-mediated endocytosis, followed by uncoating, translation, and genome replication, which occurs in the membranous structures derived from the ER (38, 47). Assembly also occurs in these membranous structures, where immature virions are thought to bud into the lumen of the ER and transport through the secretory pathway (31, 38, 40, 68, 71, 73). Prior to release from the cells, PrM protein is cleaved to Pr and M proteins by furin or furin-like protease to generate mature virions, although the cleavage has been reported to be inefficient for DENV (32, 38, 50, 59, 65, 69).
The E protein of DENV is the major determinant of tropism and virulence and is the major target of neutralizing and enhancing antibodies (5, 21, 24, 38, 47). The N-terminal 395 amino acid residues of E protein contain three well-characterized domains as determined by crystallographic studies (45, 46, 61). The C-terminal 100 amino acid residues of E protein contain the stem and the anchor (transmembrane domain, or TMD), which crosses the two leaflets of the lipid bilayer and consists of two α-helices (T1 and T2) and a stretch of six mostly nonhydrophobic residues between them (see Table 1 and Fig. 5A) (1, 74). A common feature of flavivirus replication is the production of subviral particles, which are smaller and sediment slower than the mature virions (38). The coexpression of PrM and E proteins is sufficient to produce recombinant virus-like particles (VLPs). VLPs are similar to infectious virions in structural, biochemical, and antigenic properties and have been used to study the function of PrM/E proteins and the assembly of virus particles (17, 34, 62). VLPs also have been shown to be useful noninfectious serodiagnostic antigens and potential vaccine candidates for different flaviviruses, such as DENV, Japanese encephalitis virus, and West Nile virus (8, 9, 13, 29, 35, 41, 58).
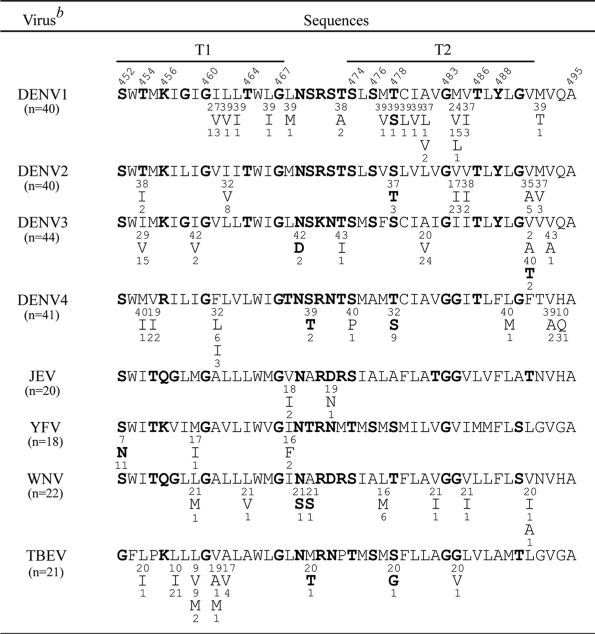
TABLE 1.
Comparison of the sequences of the TMD of E proteins of different flavivirusesa
The TMD of flaviviruses consists of two α-helices (T1 and T2), of which the boundaries are defined based on a recent cryo-EM study (74) and a stretch of six mostly nonhydrophobic residues between them. TMD sequences of multiple strains (numbers in parentheses) of each of these flaviviruses were obtained from GenBank and analyzed. Single-letter designations of amino acids are shown, with the numbers above indicating amino acid positions and the numbers beneath each amino acid indicating the numbers of strains containing that residue. The nonhydrophobic residues are in boldface.
DENV1, DENV2, DENV3, and DENV4, dengue virus type 1, 2, 3, and 4, respectively; JEV, Japanese encephalitis virus; YFV, yellow fever virus; WNV, West Nile virus; TBEV, tick-borne encephalitis virus.
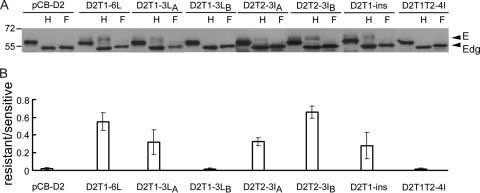
FIG. 5.
Glycosylation pattern of PrM/E expression constructs containing different mutations in the TMD. (A) Lysates derived from 293T cells transfected with each of these constructs were treated with endo H (H) or PNGase F (F) and subjected to Western blot analysis by using serum from a confirmed DENV2 case (70). Data are presented as described for Fig. 2C. (B) The ratio of the intensity of the endo H-resistant band to that of the endo H-sensitive band was determined for each construct. Data are means and standard errors from three experiments.
Chimeric experiments introducing different domains of flaviviral E protein into cellular proteins normally expressed on the surface, such as CD4 or CD8, have shown that the TMDs of both E1 and E2 proteins of hepatitis C virus (HCV) contain an ER retention (but not retrieval) signal (10, 11, 14, 19). Similarly, the TMDs of PrM and E proteins of yellow fever virus (YFV), another flavivirus, recently were reported to account for ER retention (54). We reported previously that the stem-TMD of DENV2 E protein contains a signal for retention in the ER (27).
In this study, we investigated the critical residues and elements in the TMD of DENV E protein responsible for the retention and assembly in the ER. Based on chimeric experiments and mutations introduced into the TMD, our results suggest that the length and certain nonhydrophobic residues of TMD are critical. Moreover, a trend of decreasing length and increasing nonhydrophobic residues of TMD of E proteins was found by comparing enveloped viruses assembled at the plasma membrane to those in the Golgi complex and ER. These findings support a TMD-dependent mechanism of sorting for viral E proteins. Furthermore, our observations that some TMD mutants increased the production of VLPs suggest modifications of TMD as a strategy to facilitate VLP production and have implications for employing flaviviral VLPs as serodiagnostic antigens and candidate vaccines.
MATERIALS AND METHODS
Plasmid constructs.
The constructs pCB-D2 and pCB-CD4 were as described previously, except that a unique restriction site (PstI) was introduced at amino acid residue 371 of CD4 (27, 28). To construct pCB-CD4D2A, a PCR fragment of the coding region of the anchor of DENV2 E protein was amplified by using the primer pair D2E452PstI-A and D2NotI-2402 with pCB-D2 as the template, digested with PstI and NotI, and cloned into the respective sites of pCB-CD4. To construct pCB-CD4D2SA, a PCR fragment containing the chimeric coding region, including a partial CD4 and the stem of DENV2 E protein, was amplified by two rounds of PCR with overlapping primers. The first-round PCR was done by using the primer pair CD4-Bsu36I 361A and CD4-D2stem-B1 with pCB-CD4 as the template and with primer pair CD4-D2stem-A2 and D2E451 PstI-B with pCB-D2 as the template. The second-round PCR used primer pair CD4-Bsu36I361A and d2E451PstI-B. After digestion with Bsu36I and PstI, the PCR product was cloned into the respective sites of pCB-CD4D2A.
To construct pCB-CD4D4SA and pCB-CD4D4A, PCR fragments were amplified by using primer pairs (D4E395PstI-A plus d4E495BNotI for pCB-CD4D4SA, D4E452PstI-A plus d4E495BNotI for pCB-CD4D4A) and pCB-D4 as the template, digested with PstI and NotI, and cloned into the respective sites of pCB-CD4.
To construct pCB-D2CD4, a PCR fragment of the chimeric coding region including partial DENV2 E protein plus the TMD and cytoplasmic (CY) domains of CD4 (residues 375 to 435) was amplified by two rounds of PCR. The first-round PCR was done by using the primer pair D2E200PstIA1 and D2Stem-CD4TM-B1 with pCB-D2 as the template and the primer pair D2Stem-CD4TM-A2 and NotICD4-435B with pCB-CD4 as the template. The second-round PCR used the primer pair D2E200PstIA1 and NotICD4-435B, digestion with PstI and NotI, and cloning into the respective sites of pCB-D2.
To construct pCB-D2T1-3LA, pCB-D2T1-3LB, pCB-D2T2-3IA, pCB-D2T1-ins, and pCB-D2T1T2-4I, PCR fragments containing corresponding mutations were amplified by two rounds of PCR. The first-round PCR was done by using primer pairs (for pCB-D2T1-3LA, D2E200PstIA1 and D2E452-454-456L-B1 plus D2E452-454-456L-A2 and D2NotI-2402; pCB-D2T1-3LB, D2E200PstIA1 and D2E460-464-467L-B1 plus D2E460-464-467L-A2 and D2NotI-2402; pCB-D2T2-3IA, D2E200PstIA1 and D2E474-476-478-B1 plus D2E474-476-478-A2 and D2NotI-2402; pCB-D2T1-ins, D2E200PstIA1 and D2ETM1ins-B1 plus D2ETM1insA2 and D2NotI-2402; pCB-D2T1T2-4I, D2E200PstIA1 and D2E469-470-472-473I-B1 plus D2E469-470-472-473I-A2 and D2NotI-2402) and pCB-D2 as the template. The second-round PCR used primer pair D2E200PstIA1 and D2NotI-2402, digestion with PstI and NotI, and cloning into the respective sites of pCB-D2.
To construct pCB-D2T1-6L, a PCR fragment containing corresponding mutations was amplified by two rounds of PCR. The first-round PCR was done by using primer pairs (D2E200PstIA1 and D2E460-464-467L-B1 plus D2E460-464-467L-A2 and D2NotI-2402) and pCB-D2T1-3LA as the template, followed by the second-round PCR using the primer pair D2E200PstIA1 and D2NotI-2402, digestion with PstI and NotI, and cloning into the respective sites of pCB-D2. To construct pCB-D2T2-3IB, a PCR fragment containing corresponding mutations was amplified by single-step PCR mutagenesis with primers D2E483-486-488-B1 and D2E200PstIA1 with pCB-D2 as the template, digested with PstI and NotI, and cloned into the respective sites of pCB-D2.
To construct pCB-CD4D2A mutants (pCB-CD4D2T1-3LB, pCB-CD4D2T2-3IA, pCB-CD4D2T2-3IB, and pCB-CD4D2T1-ins), PCR fragments were amplified by using the primer pair D2E452PstI-A and D2NotI-2402 with pCB-D2 mutants (pCB-D2T1-3LB, pCB-D2T2-3IA, pCB-D2T2-3IB, and pCB-D2T1-ins) as the template, digested with PstI and NotI, and cloned into pCB-CD4. To construct pCB-CD4D2T1-6L and pCB-CD4D2T1-3LA, the fragment of the coding region of the TMD of DENV2 E protein was amplified by using the primer pair D2E452-454-456LPstI-A and D2NotI-2402 with pCB-D2T1-6L and pCB-D2T1-3LA as the template, digested with PstI and NotI, and cloned into the respective sites of pCB-CD4. All constructs were confirmed by sequencing the entire inserts. The sequences of all of the primers used in this study will be provided upon request.
Cell lysates and VLPs.
293T cells were prepared in a 10 cm-culture dish at 5 × 105 cells per dish 1 day earlier and transfected with 10 μg of plasmid DNA by the calcium phosphate method. At 48 h posttransfection, culture supernatants were collected (see below) and cells were washed with 1× phosphate-buffered saline (PBS) and treated with 1% NP-40 lysis buffer (100 mM Tris [pH 7.5], 150 mM NaCl, 20 mM EDTA, 1% NP-40, 0.5% Na deoxycholate) containing protease inhibitors (Roche Diagnostics, Indianapolis, IN), followed by centrifugation at 20,000 × g at 4°C for 30 min to obtain cell lysates (27). Culture supernatants were clarified by centrifugation at 1,250 × g for 20 min, filtered through a 0.22-μm-pore-sized membrane (Sartorius, Goettingen, Germany), layered over a 20% sucrose buffer, and ultracentrifuged at 65,000 × g at 4°C for 5 h. The pellets were resuspended in 30 μl TNE buffer (50 mM Tris [pH 8.0], 100 mM NaCl, 10 mM EDTA) and subjected to Western blot analysis (27).
Antibodies.
Human DENV sera that can recognize E and PrM proteins were obtained from confirmed DENV2 cases as described previously (70). Anti-CD4 monoclonal antibodies (MAbs) RPA-T4 [IgG1(κ)] and OKT4, as well as a mouse isotype [IgG1(κ)] control, were purchased from eBioscience (San Diego, CA). Q4120 was from Sigma (St. Louis, MO) and SK3 was from Bio-Rad (Hercules, CA). Anti-CD4 (H-370) and anti-GM130 (a Golgi marker) rabbit polyclonal antibodies (PAbs) were purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA), anti-mannosidase II (Man II) mouse MAb 53FC3 was from Covance (Berkely, CA), and anti-PDI rabbit PAb SPA-890 was from Stressgen Bioreagents (Ann Arbor, MI). Anti-E mouse MAb FL0232 was purchased from Chance Biotechnology (Taipei, Taiwan), and 4G2 was from the American Type Culture Collection (Rockville, MD).
Western blot analysis.
Cell lysates or pellets from culture supernatants were subjected to 12% polyacrylaminde gel electrophoresis (PAGE), followed by transfer to a nitrocellulose membrane (Hybond-C; Amersham Biosciences, Hong Kong) as described previously (27). Membranes were blocked with 4% milk in wash buffer and incubated with primary (human sera or MAb) and secondary antibodies (horseradish peroxidase-conjugated anti-human or anti-mouse IgG) (Pierce, Rockford, IL) each at 37°C for 1 h. After a final wash, the signals were detected by enhanced chemiluminescence reagents (Perkin Elmer Life sciences, Boston, MA). The intensities of E, PrM, or CD4 bands were analyzed further by ImageQuant (GE Healthcare, United Kingdom), and the ratios of the intensities of different bands were determined (27).
Endoglycosidase digestion.
Aliquots from cell lysates or pellets were treated with 500 U of endo-β-N-acetylglucosaminidase H (endo H) or peptide N-glycosidase F (PNGase F) at 37°C for 1 h according to the manufacturer's instructions (New England Biolabs, Beverly, MA) and subjected to Western blot analysis.
Radioimmunoprecipitation.
293T cells prepared in six-well plates were transfected with plasmid DNA by the calcium phosphate method. At 20 h posttransfection, cells were washed, incubated with methionine-free Dulbecco's modified essential medium (DMEM) followed by 50 μCi [35S]methionine (Amersham Biosciences, Hong Kong) at 37°C for 6 h, and collected to obtain cell lysates as described previously (27). Following being precleared with beads, cell lysates were incubated with anti-CD4 MAb (RPA-T4, SK3, OKT4, and Q4120) (for CD4 chimeric constructs) or anti-E MAb FL0232 (for pCB-D2 constructs) at 4°C overnight and then with protein A Sepharose beads (Amersham Biosciences, Hong Kong) at 4°C for 6 h. After being washed with 1% NP-40 buffer, the beads were mixed with 2× sample buffer and heated, and the solubilized fraction was subjected to 12% PAGE as described previously (27).
Indirect immunofluorescence assay.
Plasmid DNA was transfected to 293T cells prepared in a six-well plate by the calcium phosphate method or to BHK-21 cells by Lipofectamine 2000 (Invitrogen, Carsblad, CA). At 48 h posttransfection, cells were resuspended in 1× PBS, spotted onto a slide, air dried, and fixed with 4% paraformaldehyde at room temperature for 30 min (27). Cells were permeabilized with or without 0.5% Triton X-100 at room temperature for 30 min and incubated with primary and secondary antibody, each at 37°C for 1 h. After a final wash and being dried and mounted, the slides were observed by a fluorescence microscope. For the visualization of the Golgi complex, pDsRed-monomer-Golgi (Clontech, Mountain View, CA), designated DsRed-Golgi, which expressed the red fluorescent protein targeting the trans-Golgi complex, was cotransfected with each of the plasmids to BHK-21 cells by Lipofectamine 2000 (Invitrogen, Carsblad, CA).
Flow cytometry.
293T cells prepared in a six-well plate were transfected with plasmid DNA by the calcium phosphate method. At 48 h posttransfection, cells were washed with 1× PBS, resuspended in 1× PBS, and treated with 2% paraformaldehyde at 4°C for 1 h, permeabilized with or without 0.5% Triton X-100 at room temperature for 30 min, and incubated with primary antibody (anti-CD4 MAb RPA-T4 or isotype control) and secondary antibody (fluorescein isothiocyanate [FITC]-conjugated anti-mouse IgG) each at 4°C for 1 h. Following the final wash, cells were resuspended in buffer containing 1% paraformaldehyde and subjected to FASCalibur (BD Bioscience, San Jose, CA) and CellQuest analyses (27).
Sucrose gradient sedimentation analysis.
Plasmid DNA was transfected to 293T cells prepared in a 10-cm culture dish by the calcium phosphate method. At 48 h posttransfection, cells were washed with 1× PBS, resuspended in 1× PBS, treated with 1% Triton X-100 on ice for 30 min, and then loaded into a 5 to 20% (wt/wt) sucrose gradient made with gradient buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 2 mM EDTA, 0.5% Triton X-100, 1 mM phenylmethylsulfonyl fluoride) (37). The gradient was ultracentrifuged at 247,606 × g at 15°C for 22 h, and each of the 13 fractions was collected and subjected to Western blot analysis.
Subcellular fractionation.
293T cells transfected with plasmid DNA were washed three times with 1× PBS 48 h later, resuspended in modified buffer B (10% sucrose, 20 mM Tris, 150 mM NaCl, 10 mM Mg acetate, 1 mM EGTA [pH 7.6]), and frozen-thawed eight times as described previously (27). After clearing the nuclei and debris by centrifugation at 1,000 × g for 5 min, the membrane fraction was pelleted by centrifugation at 20,000 × g for 30 min at 4°C and subjected to Western blot analysis. The resulting supernatants were layered over a 20% sucrose buffer and ultracentrifuged at 246,000 × g at 4°C for 1 h to obtain the pellets of the soluble fraction, which were resuspended in 30 μl TNE buffer and subjected to Western blot analysis.
RESULTS
TMD of E protein contains an ER retention signal.
To examine whether TMD alone is sufficient to account for the ER retention of DENV E protein, we generated chimeric CD4 constructs containing both the stem and TMD of DENV2 E protein (CD4D2SA) or TMD alone (CD4D2A) and examined their surface expression compared to that of wild-type CD4 (Fig. 1A). After transfection to 293T cells, cells were examined by immunofluorescence assay and flow cytometry analysis in the presence or absence of Triton X-100. As shown in Fig. 1B, CD4 was readily detected by an anti-CD4 MAb, RPA-T4, in the pCB-CD4-transfected cells in the presence or absence of Triton X-100, suggesting that CD4 did express on the surface of cells. In contrast, CD4D2SA and CD4D2A can be detected only in the presence of Triton X-100, suggesting that they did not express well on the surface of cells. Similarly, in the absence of Triton X-100, the percentage of positive cells stained with anti-CD4 MAb compared to that of the isotype control was higher in cells transfected with pCB-CD4 (27.13%) than in those transfected with pCB-CD4D2SA (13.67%) or pCB-CD4D2A (5.02%), suggesting that more CD4 was expressed on the surface than CD4D2SA or CD4D2A (Fig. 1C). To adjust different levels of expression, we calculated the relative surface expression of CD4, which was the ratio of the percentage of CD4-positive cells in the absence of Triton X-100 to that in the presence of Triton X-100; it was 0.57, suggesting that approximately 57% of CD4 was expressed on the surface of pCB-CD4-transfected cells. In contrast, the relative surface expression of CD4D2SA and CD4D2A were 0.22 and 0.13, respectively, suggesting that approximately 22 and 13% of chimeric CD4 was expressed on the surface of the pCB-CD4D2SA- and pCB-CD4D2A-transfected cells, respectively. To further examine the intracellular distribution of CD4, CD4D2SA, and CD4D2A, cell lysates were subjected to endo H or PNGase F digestion, followed by Western blot analysis.
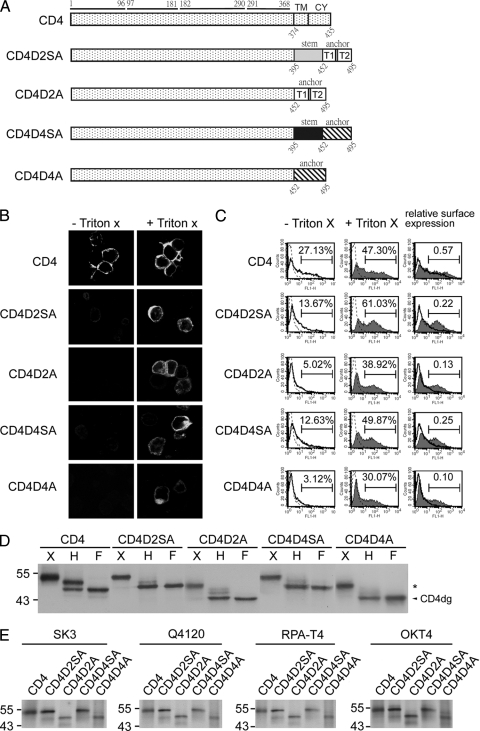
FIG. 1.
CD4 and chimeric CD4 constructs, surface expression, glycosylation pattern, and recognition by different MAbs. (A) Schematic diagram of the CD4 construct (pCB-CD4) and chimeric CD4 constructs containing the stem-TMD of DENV2 (CD4D2SA) or DENV4 (CD4D4SA) or TMD of DENV2 (CD4D2A) or DENV4 (CD4D4A). Open and gray bars indicate DENV2 sequences, and hatched and black bars indicate DENV4 sequences. The amino acid positions of the four domains of CD4 are shown above, and those of the stem, TMD, or CY domain are shown below. (B) 293T cells transfected with each of these constructs were subjected to indirect immunofluorescence assay using mouse anti-CD4 MAb RPA-T4 as described in Materials and Methods. (C) 293T cells transfected as described for panel A were permeabilized with or without Triton X-100 and subjected to flow cytometry analysis using MAb RPA-T4. Solid lines indicate MAb without Triton X-100, and solid lines with a shaded area indicate MAb with Triton X-100. Dotted lines indicate isotype controls, which overlap with those of mock-transfected cells (not shown). The relative surface expression of CD4 or chimeric CD4 was the ratio of the percentage of RPA-T4-positive cells in the absence of Triton X-100 to that in the presence of Triton X-100. (D) Lysates derived from 293T cells transfected as described for panel A were treated with endo H (H) or PNGase F (F) and subjected to Western blot analysis using RPA-T4. The arrowhead indicates deglycosylated CD4 (CD4dg), and asterisks indicate endo H-resistant bands. One representative experiment of three is shown. (E) 293T cells transfected as described for panel A were labeled with [35S]methionine at 20 h posttransfection and subjected to immunoprecipitation with anti-CD4 MAbs (RPA-T4, SK3, Q4120, and OKT4) and 12% PAGE as described in Materials and Methods. Molecular mass markers are shown in kDa.
As shown in Fig. 1D, two CD4 bands, which decreased in size by about 2 and 7 kDa, respectively, were found with the former as the predominant band. This is in agreement with previous reports that the band decreased in size by 2 kDa, which presumably represented the CD4 that normally had one of the two N-linked glycosylation sites converted to complex oligosaccharide and transported beyond the trans-Golgi complex (endo H resistant), was present in greater amounts than the band decreased by 7 kDa, which represented the CD4 remaining in the ER and sensitive to endo H digestion (10-12, 64). In contrast, the majority of CD4D2SA and CD4D2A bands were sensitive to endo H digestion, suggesting that CD4D2SA and CD4D2A were retained mainly in a compartment prior to the trans-Golgi complex (Fig. 1D). Taken together, these findings suggest that the TMD of DENV2 E protein is sufficient to retain CD4 in an intracellular compartment prior to the trans-Golgi complex, probably the ER.
To further investigate whether the TMD of another DENV serotype is sufficient to retain CD4 in the ER as well, we generated chimeric CD4 constructs containing both the stem and TMD of DENV4 E protein (CD4D4SA) or TMD alone (CD4D4A) (Fig. 1A), and we examined their surface expression and intracellular distribution. As shown in Fig. 1B, CD4D4SA and CD4D4A can be detected only in the presence of Triton X-100, suggesting that they did not express well on the cell surface. Consistently with this, the relative surface expression of CD4D4SA and CD4D4A, 0.25 and 0.10, respectively, was lower that of CD4 (Fig. 1C). After digestion with endo H, the majority of CD4D4SA and CD4D4A bands were sensitive to endo H digestion, suggesting that CD4D4SA and CD4D4A retained mainly in a compartment prior to the trans-Golgi complex (Fig. 1D). To exclude the possibility that the retention of the chimeric CD4 proteins (CD4D2SA, CD4D2A, CD4D4SA, and CD4D4A) is due to the misfolding of protein, cells transfected with each of these chimeric constructs were labeled metabolically and subjected to immunoprecipitation assay by using different anti-CD4 MAbs, including those targeting domain 1 of CD4 (SK3, Q4120, and RPA-T4) and domain 3 of CD4 (OKT4), some of which recognize conformational epitopes (OKT4, SK3, and Q4120) (4, 19). As shown in Fig. 1E, different chimeric CD4 proteins can be recognized by all four anti-CD4 MAbs tested, suggesting that introducing the stem-TMD or TMD into the C terminus of CD4 did not result in global conformational changes or the misfolding of the chimeric CD4 proteins. Taken together, these findings suggest that the TMD of DENV E protein alone is sufficient to retain CD4 in the ER.
Within the flavivirus-infected cells, E protein was found primarily in the ER (31, 38, 40, 68). To investigate the role of the TMD in retaining DENV2 E protein in the ER, a chimeric construct (pCB-D2CD4), in which the TMD of DENV2 E protein was replaced by the TMD and CY domain of CD4, was generated and examined (Fig. 2A). Flow cytometry analysis revealed that the majority of D2CD4 expressed on the cell surface with a relative surface expression of 0.85; that of the authentic DENV2 E protein is 0.13 (Fig. 2B). Consistently with this, the endo H digestion of lysates derived from pCB-D2CD4-transfected cells revealed almost exclusively an endo H-resistant band, whereas an endo H-sensitive band was noted in pCB-D2-transfected cells (Fig. 2C). These findings suggest that the TMD of DENV2 E protein plays an important role in retaining E protein in the ER.
FIG. 2.
PrM/E expression constructs, surface expression, and glycosylation pattern. (A) DENV2 PrM/E expression construct (pCB-D2) and chimeric construct containing the TMD and CY domain of CD4 in the TMD (D2CD4). ss, signal sequence. (B) 293T cells transfected with each of these constructs were permeabilized with or without Triton X-100 and subjected to flow cytometry analysis using anti-E MAb FL0232. Solid lines indicate anti-E MAb without Triton X-100, and solid lines with a shaded area indicate anti-E MAb with Triton X-100. Dotted lines indicate isotype controls, which overlap with those of mock-transfected cells (not shown). The relative surface expression of E protein was the ratio of the percentage of FL0232-positive cells in the absence of Triton X-100 to that in the presence of Triton X-100. (C) Lysates derived from 293T cells transfected with the constructs described for panel A were treated with endo H (H) or PNGase F (F) and subjected to Western blot analysis by using serum from a confirmed DENV2 case (70). Arrowheads indicate E or deglycosylated E protein (Edg). Molecular mass markers are shown in kDa. One representative experiment of three is shown.
Nonhydrophobic residues at the N terminus of T1, N or C terminus of T2, and the length of T1 contribute to the ER retention phenotype.
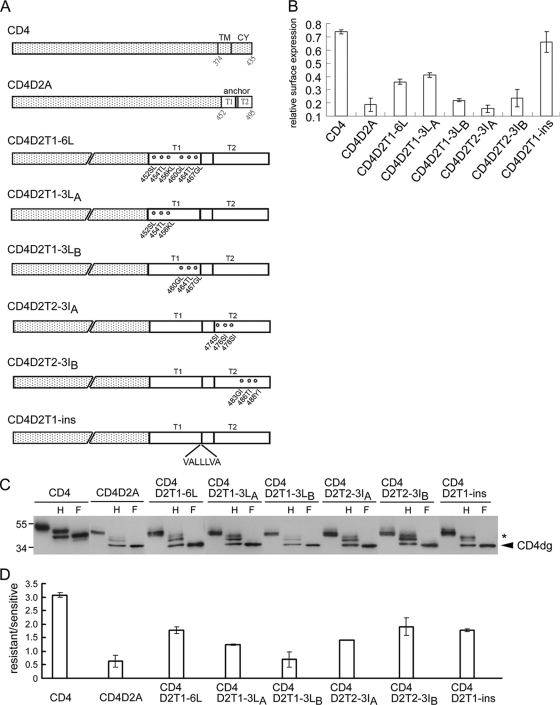
It was reported previously that an increase in the hydrophobicity or the length of the TMD of ER-resident proteins led to their release from the ER to the Golgi complex or plasma membrane (26, 60, 72). A closer examination of the sequences of the TMD of DENV2 E protein revealed six and seven nonhydrophobic residues at the T1 and T2 helices, respectively (Table 1). To investigate whether the six nonhydrophobic residues at T1 contribute to ER retention, site-directed mutagenesis was carried out to replace each of these six residues (serine at position 452, threonine at position 454, lysine at position 456, glycine at position 460, threonine at position 464, and glycine at position 467) with hydrophobic residues (leucines) in the backbone of pCB-CD4D2A to generate pCB-CD4D2T1-6L (Fig. 3A). After transfection to 293T cells, flow cytometry revealed that the relative surface expression of CD4D2T1-6L increased 2-fold (mean, 0.36) compared to that of CD4D2A (mean, 0.18) (Fig. 3B). To further examine whether the nonhydrophobic residues at the N or C terminus of T1 are sufficient to contribute to the ER retention phenotype, two mutants (pCB-CD4D2T1-3LA and pCB-CD4D2T1-3LB) containing mutatins of three nonhydrophobic residues to leucines at the N and C terminus, respectively, were generated (Fig. 3A). Flow cytometry analysis revealed that the relative surface expression of CD4D2T1-3LA increased (mean, 0.41) similarly to that of CD4D2T1-6L, whereas the relative surface expression of CD4D2T1-3LB only slightly increased (mean, 0.22) (Fig. 3B). This was supported by the enzyme digestion experiment, in which a nearly 2-fold or more increase in the amounts of the endo H-resistant band relative to that of the endo H-sensitive band was found in CD4D2T1-6L and CD4D2T1-3LA but not in CD4D2T1-3LB compared to results for CD4D2A (Fig. 3C and D). These findings suggest that the three nonhydrophobic residues at the N terminus of T1 contribute to the ER retention phenotype. To investigate whether the nonhydrophobic residues at the N terminus or C terminus of T2 also contribute to ER retention, another two mutants (pCB-CD4D2T2-3IA and pCB-CD4D2T2-3IB), which contained mutations of three nonhydrophobic residues to isoleucines at the N and C terminus of T2, respectively, were generated (Fig. 3A). Flow cytometry analysis revealed no significant increase in the relative surface expression of CD4D2T2-3IA (mean, 0.16) and CD4D2T2-3IB (mean, 0.24) (Fig. 3B); however, a nearly 2-fold or more increase in the amounts of the endo H-resistant band relative to that of the endo H-sensitive band was found for CD4D2T2-3IA and CD4D2T2-3IB compared to results for CD4D2A (Fig. 3C and D).
FIG. 3.
CD4 and chimeric constructs containing TMD and mutant TMD of DENV2, surface expression, and glycosylation pattern. (A) Schematic diagram of the CD4 construct (pCB-CD4) and chimeric CD4 constructs containing TMD and mutant TMD of DENV2. (B) 293T cells transfected with each of these constructs were subjected to flow cytometry analysis as described for Fig. 1C. Relative surface expression (means and standard errors) from three experiments is shown. (C) Lysates derived from 293T cells transfected with the constructs described above were treated with endo H (H) or PNGase F (F) and subjected to Western blot analysis by using MAb RPA-T4. Data were presented as described in the legend to Fig. 1D. (D) The ratio of the intensity of the endo H-resistant band to that of the endo H-sensitive band was determined for each construct. Data are means and standard errors of three experiments.
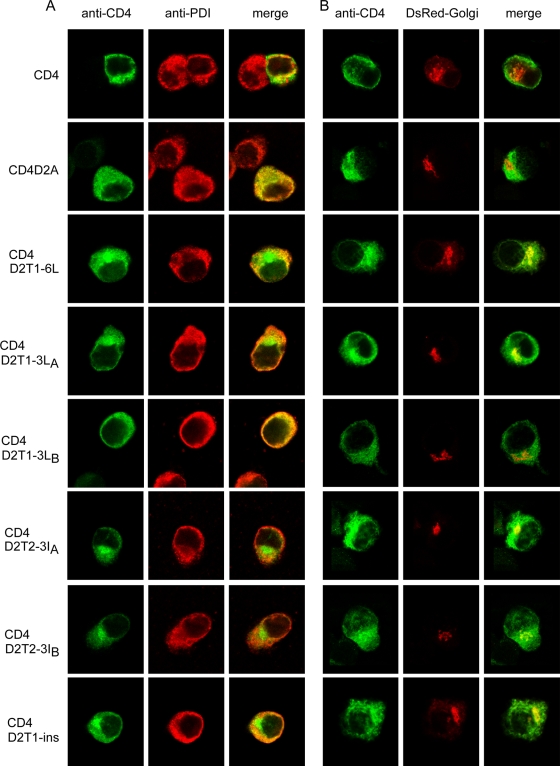
To examine the possibility that mutants CD4D2T2-3IA and CD4D2T2-3IB exit from the ER and transport to the trans-Golgi complex or beyond, as shown by the endo H-resistant band, but do not express on the cell surface, as revealed by flow cytometry, a double-label immunofluorescence assay was carried out to investigate the intracellular localization of these mutant proteins. As shown in Fig. 4A, CD4 was stained by MAb RPA-T4 both on the cell surface and in the cytoplasm, which had a staining pattern merging partially with that of PDI (an ER marker protein), suggesting that after being synthesized in the ER, CD4 transported beyond the Golgi complex and expressed on the cell surface (Fig. 4A). In contrast, CD4D2A and CD4D2T1-3LB were stained mainly intracellularly, with a staining pattern merging almost completely with that of PDI, suggesting that CD4D2A and CD4D2T1-3LB were retained mainly in the ER (Fig. 4A). Interestingly, CD4D2T1-6L, CD4D2T1-3LA, CD4D2T2-3IA, and CD4D2T2-3IB showed a different staining pattern, which merged primarily with that of PDI with a distinct perinuclear signal. Since the signal of anti-Golgi marker (Man II and GM130) antibodies tested was weak, DsRed-Golgi, which expressed the red fluorescent protein and was targeted to the trans-Golgi complex, was cotransfected with each of these CD4 mutant constructs and examined by fluorescent microscopy. Consistently with the findings for double-label staining shown in Fig. 4A, CD4D2T1-6L, CD4D2T1-3LA, CD4D2T2-3IA, and CD4D2T2-3IB were present mainly in the cytoplasm and had a distinct signal merging with that of DsRed-Golgi in the perinuclear region, whereas CD4D2A and CD4D2T1-3LB were present in the cytoplasm without merging with DsRed-Golgi (Fig. 4B). Of note, CD4 was present both on the surface and in the cytoplasm, which had very faint and scattered signal merging with that of DsRed-Golgi. Taken together, these findings suggest that a significant proportion of CD4D2T1-6L, CD4D2T1-3LA, CD4D2T2-3IA, and CD4D2T2-3IB has exited from the ER and transported to the Golgi complex or to the cell surface, whereas CD4D2A and CD4D2T1-3LB retained almost exclusively in the ER. The nonhydrophobic residues at the N terminus of T1 or the N or C terminus of T2 are likely to be critical for the ER retention phenotype.
FIG. 4.
Intracellular localization of CD4 and chimeric CD4 proteins containing the TMD of DENV2 with different mutations by double-label immunofluorescence assay. (A) BHK-21 cells were transfected with each of the constructs described in Fig. 3A, fixed at 48 h, and stained with mouse anti-CD4 MAb RPA-T4 (goat FITC-conjugated anti-mouse IgG as the secondary antibody) and rabbit anti-PDI Pab (goat rhodamine-conjugated anti-rabbit IgG as the secondary antibody) under an SP5 confocal laser-scanning microscope. (B) BHK-21 cells were cotransfected with DsRed-Golgi (a construct targeting to the trans-Golgi complex) and each of the constructs shown in Fig. 3A, fixed at 48 h, and stained with mouse anti-CD4 MAb RPA-T4 as above.
To further investigate the contribution of length to the ER retention phenotype, a strategy of inserting a stretch of seven artificially designed hydrophobic residues into the C terminus of T1 in the backbone of pCB-CD4D2A to generate pCB-CD4D2T1-ins was employed as described previously (Fig. 3A) (26). Flow cytometry analysis revealed its mean relative surface expression of 0.66 (Fig. 3B), and endo H digestion revealed an approximately 2-fold increase in the amount of the endo H-resistant band relative to that of the endo H-sensitive band compared to that for CD4D2A (Fig. 3C and 3D). Consistently with this, a staining pattern with a distinct perinuclear signal that did not merge with that of PDI was found by double-label immunofluorescence assay, and a signal merging with that of DsRed-Golgi perinuclearly was noted after cotransfection with DsRed-Golgi (Fig. 4A and B). These findings suggest that an increase in the length of T1 by hydrophobic residues results in the transport of CD4D2T1-ins to the Golgi complex and cell surface.
Nonhydrophobic residues at the N terminus of T1, N or C terminus of T2, and the length of T1 are involved in the retention of E protein in the ER.
To investigate the roles of the nonhydrophobic residues at the N terminus of T1, as well as the N or C terminus of T2, in retaining E protein in the ER, the mutations in the chimeric CD4 constructs were introduced into the DENV2 PrM/E-expressing construct, pCB-D2, and examined. The digestion of lysates derived from pCB-D2-transfected cells revealed a nearly completely endo H-sensitive pattern, suggesting that the E protein retains almost exclusively in the ER (Fig. 5A and B). In contrast, significant amounts of endo H-resistant bands were found in lysates derived from pCB-D2T1-6L-, pCB-D2T1-3LA-, pCB-D2T2-3IA-, or pCB-D2T2-3IB-transfected cells (Fig. 5A and B). Of note, lysates derived from pCB-D2T1-3LB-transfected cells showed an endo H-sensitive pattern similar to that of pCB-D2. These findings, generally in agreement with those of the chimeric CD4 experiment, suggest that substitutions of three nonhydrophobic residues at the N terminus of T1, or the N or C terminus of T2, affect the ability of the TMD to retain E protein in the ER. To further investigate the involvement of T1 length in retaining E protein in the ER, pCB-D2T1-ins was generated and examined. An endo H-resistant band was found in lysates derived from pCB-D2T1-ins-transfected cells (Fig. 5A and B), suggesting that an increase in the length of T1 by hydrophobic residues released the E protein from the ER.
Between the T1 and T2 helices there are six amino acid residues, most of which are nonhydrophobic (Table 1). To investigate the contribution of these residues to the retention of E protein in the ER, four nonhydrophobic residues (asparagine at position 469, serine at position 470, serine at position 472, and threonine at position 473) were replaced with isoleucines to generate the construct pCB-D2T1T2-4I. The digestion of lysates derived from pCB-D2T1T2-4I-transfected cells revealed an endo H-sensitive pattern, suggesting that these four nonhydrophobic residues do not contribute to the retention of E protein in the ER (Fig. 5A and B).
The effect of TMD mutations on PrM-E interaction and VLP production.
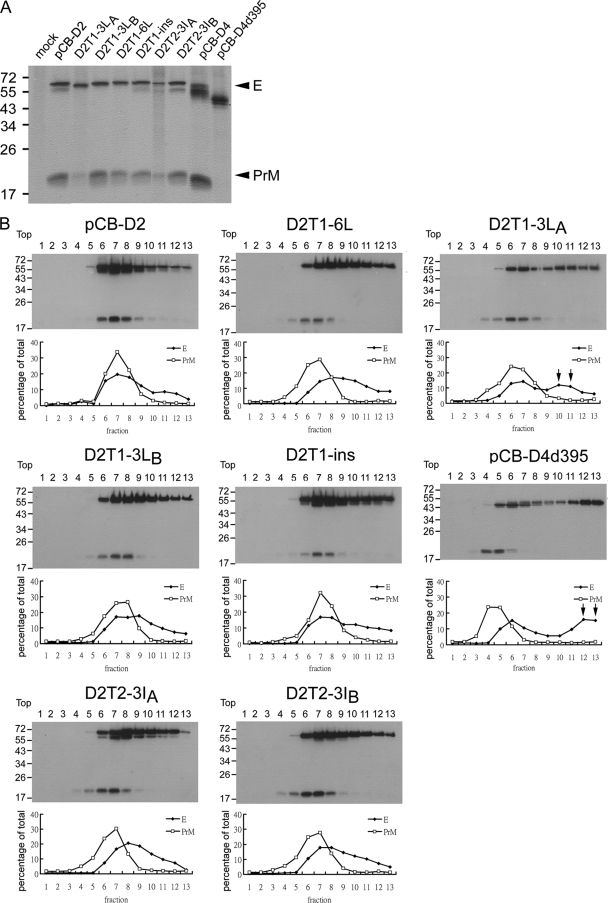
A previous study of the tick-borne encephalitis virus (TBEV) reported that deletion at the C terminus of E protein affects the heterodimeric interaction between PrM and E proteins as well as their assembly to form VLPs (1). We next examined whether the mutations introduced into the TMD affect the PrM-E interaction by performing sucrose gradient sedimentation analysis in cell lysates derived from each of these transfectants. As a control, the DENV4 PrM/E expression construct (pCB-D4) and its mutant (pCB-D4d395) containing the truncation of the stem-TMD, which has been shown to affect PrM-E heterodimerization, also were included in the analysis (1, 28). As shown in Fig. 6B, the majority of E protein of pCB-D2 and pCB-D2T1-ins was found in fractions 6 to 9 with a peak in fraction 7, which cosedimented with that of PrM protein, suggesting that mutant pCB-D2T1-ins does not affect the PrM/E heterodimerization. In contrast, the peak of the PrM protein (fraction 5) of mutant pCB-D4d395 did not cosediment with that of E protein, which had another peak in fractions 12 and 13 not seen with wild-type pCB-D4 or pCB-D2 (Fig. 6B and data not shown). Similarly, the E protein of mutant pCB-D2T1-3LA had another peak in fractions 10 and 11, suggesting that the PrM-E heterodimerization was affected. For mutants pCB-D2T1-6L, pCB-D2T1-3LB, pCB-D2T2-3IA, and pCB-D2T2-3IB, there was only a slight shift of the peak of E protein from that of PrM protein, suggesting that these mutations did not greatly affect the PrM-E heterodimerization (Fig. 6B). These findings were supported further by radioimmunoprecipitation with anti-E MAb, in which the intensity of the PrM protein band relative to that of the E protein band was reduced in mutants pCB-D4d395 (no PrM band detectable) and pCB-D2T1-3LA but not in other mutants (Fig. 6A).
FIG. 6.
Interaction between PrM and E proteins. (A) Radioimmunoprecipitation assay. 293T cells transfected with the PrM/E expression constructs were labeled with [35S]methionine at 20 h posttransfection, immunoprecipitated with anti-E MAb FL0232, and subjected to 12% PAGE as described in Materials and Methods. Arrowheads indicate E and PrM proteins. (B) Sucrose gradient sedimentation analysis. Cell lysates derived from 293T cells transfected with PrM/E expression constructs were subjected to 5 to 20% (wt/wt) sucrose gradient ultracentrifugation, and each of the 13 fractions was collected and subjected to Western blot analysis using serum from a confirmed DENV2 case (70). Molecular mass markers are shown in kDa. The intensities of the E and PrM bands in each fraction were determined and are presented as the percentage of total intensities of E and PrM bands, respectively, below each lane. Arrows indicate the shift of the peak of E protein in some mutants.
To further investigate whether these TMD mutations affect the production of VLPs, pellets derived from the ultracentrifugation of culture supernatants as well as cell lysates of each of these transfectants was examined by Western blot analysis. Compared to those of wild-type pCB-D2, the amounts of E protein in pellets relative to those in cell lysates were increased in mutants pCB-D2T1-6L, pCB-D2T1-3LA, pCB-D2T2-3IA, pCB-D2T2-3IB, and pCB-D2T1-ins, which showed a tendency to exit from the ER, but were not increased in mutant pCB-D2T1-3LB, which was retained strongly in the ER (Fig. 5 and 7). These findings suggested that several TMD mutations with an increase in the hydrophobicity or length not only resulted in less ER retention but also more VLP production. To examine whether intracellular PrM/E proteins of the TMD mutants that produced increased VLPs retain in the membrane-bound fraction or form VLPs in the soluble fraction of the ER or other compartments, 293T cells transfected with four such TMD mutants were subjected to a previously described subcellular fractionation experiment to obtain the membrane and soluble fractions (27). As shown in Fig. 8A, the amounts of PrM/E proteins in the pellets of the soluble fraction relative to those of the membrane fraction of these mutants were not greater than those of pCB-D2, suggesting that the VLPs produced by these TMD mutants, although greater in amount than those of wild-type pCB-D2 in the supernatants (Fig. 7), did not accumulate in the soluble fraction within the cells. As a control, calnexin, an integral ER membrane protein, was detected in the membrane fraction but not in the soluble fraction. The enzyme digestion of the membrane fraction revealed increased amounts of endo H-resistant bands relative to that of endo H-sensitive bands in the TMD mutants but not in pCB-D2 (Fig. 8B), suggesting that significant amounts of mutant E protein in the membrane fraction have released from the ER to the trans-Golgi complex or beyond. Similarly, the enzyme digestion of the soluble fraction showed that the endo H-resistant bands relative to the endo H-sensitive bands in these TMD mutants were stronger than those in pCB-D2, which had a faint endo H-resistant band (Fig. 8C), suggesting that significant amounts of mutant VLPs were present in the trans-Golgi complex or beyond compared to those of wild-type pCB-D2.
FIG. 7.
Production of VLPs by PrM/E expression constructs containing different mutations in the TMD (T1 and T2 helices). Forty-eight hours after transfection to 293T cells, cell lysates (A) and pellets (B) derived from culture supernatants by 20% sucrose cushion ultracentrifugation were subjected to Western blot analysis using serum from a confirmed DENV2 case (70). Arrowheads indicate PrM and E proteins recognized by the DENV2 serum, which did not react with lysates of mock-transfected cells (data not shown). The PrM bands of pellets after long exposure are shown below. Molecular mass markers are shown in kDa. One representative experiment of more than three is shown.
FIG. 8.
Subcellular fractionation experiment of TMD mutants with increased production of VLPs. (A) 293T cells transfected with mock and PrM/E expression constructs were resuspended in modified buffer B and frozen-thawed (27). After clearing the nuclei and debris, the membrane fraction and the pellets derived from the soluble fraction by 20% sucrose cushion ultracentrifugation were subjected to Western blot analysis by using serum from a confirmed DENV2 case (upper) (70) and then reprobed with anti-calnexin MAb (lower). Arrowheads indicate PrM, E, and calnexin. (B) Membrane fraction and (C) pellets of the soluble fraction of each transfectant were treated with endo H (H) or PNGase F (F) and subjected to Western blot analysis by using serum from a confirmed DENV2 case (70). The ratio of the intensity of the endo H-resistant band to that of the endo H-sensitive band is shown below the gels. Arrowheads indicate E or deglycosylated E protein (Edg). Molecular mass markers are shown in kDa. One representative experiment of two is shown.
DISCUSSION
A strategy for enveloped viruses that assemble at different cellular organelles is to develop signals in the E protein for localization to a specific organelle where morphogenesis takes place (2, 36, 54, 63). A recent study demonstrating the assembly and budding of DENV in the membranous structures derived from the ER suggests that an ER localization signal is present in its E protein (71). As deletional studies of the E protein of DENV4 and TBEV have shown the secretion of E protein containing a deletion of the TMD at the C terminus, the ER retention signal is unlikely to be present in the N-terminal ectodomain of E protein (1, 43). In agreement with these and previous studies of HCV and YFV, our chimeric experiment suggested that the TMD of DENV E protein contains an ER retention signal (10, 11, 14, 19, 54). Since there is no CY domain in the E protein (74) and no previously reported ER retention or retrieval motif based on sequence analysis (Table 1), the ER retention of DENV E protein most likely is due to the intrinsic properties of the TMD. By using the approach of site-directed mutagenesis, we demonstrated that the length and some nonhydrophobic residues of the TMD of DENV E protein are critical for its retention and assembly in the ER. To our knowledge, this is the first study reporting the determinants in the TMD of a viral E protein that are important for its intracellular localization in the ER. Moreover, our findings that modifications of the TMD led to the increased production of VLPs provided important information for the application of flaviviral VLPs as serodiagnostic antigens and candidate vaccines.
For ER-resident proteins, several specific signals for retention and retrieval in the ER have been identified, such as KDEL for the retrieval of ER-soluble proteins from the Golgi complex to ER, the dilysine motif K(X)KXX at the C terminus of the CY domain, and the arginine-based motif ΦΨ/RRXR at the N or C terminus of the CY domain for the retention and retrieval of certain ER membrane proteins (44, 56). For other ER membrane proteins, the properties of the TMD itself, including the relatively short length and charged residues or overall hydrophobicity, were found to be the key determinant for ER retention, such as cytochrome b5, UBC6, and Ufe1p (26, 60, 72). On the other hand, the TMDs of Golgi membrane proteins have been reported to play a major role for retention in the Golgi complex, such as galactosyltransferase and sialyltransferase (3, 39, 51, 52), and their length generally was shorter than that of the plasma membrane proteins. Since the concentration of membrane cholesterol, which is believed to increase the thickness and decrease the deformability of membrane, increases as one proceeds outward along the secretory pathway, it was proposed that cholesterol plays an important role in the lipid-based sorting of membrane proteins between the ER and Golgi complex, as well as between the Golgi complex and plasma membranes (6, 48).
The analysis of the locations of the assembly of different enveloped viruses and the length and number of nonhydrophobic residues in the TMD of E proteins revealed an interesting trend, namely, longer TMDs and relatively fewer nonhydrophobic residues were found in viruses that assemble at the plasma membrane, whereas shorter TMDs and more nonhydrophobic residues were found in those assembled in the ER (Table 2). Since flaviviruses contain two helices (T1 and T2) in the TMD and T2 is believed to be the signal sequence of NS1, reanalysis including the T1 helix only was performed and revealed virtually the same trend. These findings suggest that viral E proteins utilize a TMD-dependent and lipid-based sorting mechanism similar to that proposed for cellular proteins.
TABLE 2.
Comparison of the length and number of nonhydrophobic residues of TMD of E proteins of viruses assembled at different sitesa
| Virusb | Site of assemblyc | Length of TMD | No. of nonhydrophobic residues in TMD | Reference(s) |
|---|---|---|---|---|
| HIV-1 | PM | 22 | 5 | 18, 30 |
| Influenza A virus | PM | 27 | 8 | 7, 30, 63 |
| Ebola virus | PM | 24 | 4 | 20, 30, 42 |
| Sindbis virus | PM | 26 | 5 | 16, 25, 30 |
| Bunyaviridae | ||||
| UUKV | Golgi | 19-20 | 3-4 | 2, 15, 33, 49, 66 |
| RVFV | Golgi | 20 | 2 | |
| ANDV | Golgi | 19 | 4 | |
| BUNV | Golgi | 19 | 2 | |
| La Crosse virus | Golgi | 19 | 4 | |
| Coronavirus | ER-Golgi intermediate | E, 17 | E, 6 | 30, 36, 67 |
| DENV1 | ER | T1, 16; T2, 18 | T1, 7; T2, 7; T1-T2, 5 | Predicted from reference 74 |
| DENV2 | ER | T1, 16; T2, 18 | T1, 6; T2, 7; T1-T2, 5 | 74 |
| DENV3 | ER | T1, 16; T2, 18 | T1, 6; T2, 7; T1-T2, 5 | Predicted from reference 74 |
| DENV4 | ER | T1, 16; T2, 18 | T1, 4; T2, 6; T1-T2, 6 | Predicted from reference 74 |
| JEV | ER | T1, 16; T2, 18 | T1, 6; T2, 5; T1-T2, 4 | 68; predicted from reference 74 |
| YFV | ER | T1, 16; T2, 18 | T1, 5; T2, 5; T1-T2, 4 | 31; predicted from reference 74 |
| WNV | ER | T1, 16; T2, 18 | T1, 6; T2, 5; T1-T2, 4 | 40 |
| TBEV | ER | T1, 16; T2, 18 | T1, 4; T2, 6; T1-T2, 3 | Predicted from reference 74 |
The TMD of flaviviruses consists of two α-helices (T1 and T2) defined based on a recent cryo-EM study (74) and a stretch of six mostly nonhydrophobic residues between them (T1-T2). For coronavirus, only the E protein, which is the major determinant of localization in the ER-Golgi intermediate, is analyzed. Similarly, for bunyaviridae, only the Gn protein, the major determinant of localization in the Golgi complex, is analyzed.
HIV-1, human immunodeficiency virus type 1; UUKV, Uukuniemi virus; RVFV, Rift Valley fever virus; ANDV, Andes virus; BUNV, Bunyamwera virus; DENV1, DENV2, DENV3, and DENV4: dengue virus type 1, 2, 3 and 4, respectively; JEV, Japanese encephalitis virus; YFV, yellow fever virus; WNV, West Nile virus; TBEV, tick-borne encephalitis virus.
PM, plasma membrane; ER, endoplasmic reticulum.
A closer examination of the amino acid sequences of the TMD of different flaviviruses revealed that there were 13 to 19 nonhydrophobic residues in the entire TMD, including 4 to 7 in T1, 5 to 7 in T2, and 3 to 6 between T1 and T2 (Tables 1 and 2). With the exception of glycine residues at positions 460, 467, and 483, these nonhydrophobic residues are not absolutely conserved per se (Table 1). However, the number of nonhydrophobic residues in the entire TMD, more than 13, is much higher than that of the viruses that assemble at the plasma membrane, suggesting that it is the composition rather than the precise amino acid sequence that determines its localization in the ER. Notably, a similar finding has been reported for the TMD of a yeast ER membrane protein, Ufe1p (60). The distribution of the nonhydrophobic residues in the TMD appears to be scattered but tends to be more concentrated between T1 and T2 and at the N terminus of T1 than at the C terminus of T1 (Table 1). Interestingly, mutations to three nonhydrophobic residues at the N terminus of T1 (pCB-D2T1-3LA) but not to those at the C terminus of T1 (pCB-D2T1-3LB) affected the ER retention phenotype (Fig. 3 to 5), suggesting that the nonhydrophobic residues at the N terminus of T1 are more important for ER retention than those at the C terminus of T1. Of note, mutations introduced into nonhydrophobic residues at T2 (pCB-D2T2-3IA and pCB-D2T2-3IB) and between T1 and T2 (pCB-D2T1T2-4I) resulted in a smaller amount of PrM protein relative to that of E protein in the pellets (Fig. 7B and data not shown), suggesting that the nonhydrophobic residues at T2 or between T1 and T2 also are involved in maintaining the stability of PrM protein in pellets.
We also examined the effect of the TMD mutations on the PrM-E heterodimeric interaction and the production of VLPs and found that most of the mutants did not greatly affect the interaction between PrM and E proteins, except mutant pCB-D2T1-3LA, which contained mutations of three nonhydrophobic residues at the N terminus of T1 (positions 452, 454, and 456). This is generally in agreement with a recent study of the TMD of YFV E protein, in which alanines inserted into the C terminus of T1 (corresponding to DENV E positions 457, 461, and 465) or T2 (DENV E positions 479, 483, and 487) helix did not affect the PrM-E interaction (53) and suggests that the N terminus of T1 helix is involved in PrM-E heterodimerization. It is worth noting that the extent of reduced PrM-E interaction by mutant pCB-D2T1-3LA compared to that of pCB-D4d395 (Fig. 6) did not cause impaired VLP production. This is in contrast to the study of TBEV, in which deletional mutations at the C terminus of E protein resulted in the severe impairment of PrM-E interaction and VLP production (1). For the production of VLPs, interestingly we found that the TMD mutants that showed a tendency to release E protein from the ER, including pCB-D2T1-6L, pCB-D2T1-3LA, pCB-D2T2-3IA, pCB-D2T2-3IB, and pCB-D2T1-ins, produced greater amounts of VLPs than wild-type pCB-D2. Notably, the observation that two mutants at T2 (pCB-D2T2-3IA and pCB-D2T2-3IB) affected VLP production was in agreement with a recent report by Orlinger et al. that T2 of TBEV E protein has an essential function for virus assembly (55). Probably due to the rapid process of budding and transport of VLPs outside of cells as reported for several flaviviral particles (31, 38, 40, 68), subcellular fractionation experiments with four such TMD mutants did not reveal accumulation and thus increased amounts of VLPs in the soluble fraction. The enzyme digestion experiments with these TMD mutants revealed significant amounts of endo H-resistant bands in both membrane and soluble fractions (Fig. 8B and C), a finding consistent with that from the total cell lysates (Fig. 5B), and suggested that significant amounts of E protein of these TMD mutants and their VLPs were present in the trans-Golgi complex or beyond. Whether the PrM/E proteins of these TMD mutants, once released from ER, could produce VLPs at the trans-Golgi complex or beyond remains to be investigated. We have carried out a sucrose gradient subcellular membrane fractionation assay to separate the Golgi complex (fractions 1 and 2) and ER compartments (fractions 8 to 13). Compared with pCB-D2, increased levels of TMD mutant E proteins were found in the Golgi compartment; however, the amount was too little for further experiments to separate the membrane and soluble fractions and examine whether these TMD mutants produce VLPs in the Golgi compartment (data not shown). Future studies using electron microscopy (EM) might provide morphological information regarding the subcellular location of these TMD mutants and how they produce increased VLPs.
The cryo-EM study of DENV virions at high resolution revealed that the T1 and T2 helices of the TMD form an antiparallel coiled-coil structure in the membrane, leaving the C terminus end toward the lumen of the ER (74). Alanine scanning insertion mutagenesis, in which a single alanine inserted into a helix in the TMD displaces the residues on the N terminus of the insertion by 110° relative to those on the C terminus, has been utilized to study the helix-helix interaction in the membrane. Notably, two alanine insertional mutants in the TMD of YFV (corresponding to DENV E positions 465 and 479) greatly impaired the production of VLPs (53), whereas our TMD mutants did not, suggesting that the potential helix-helix packing, which could be disrupted by single alanine insertion but not by our mutants involving substitutions or insertion at the end of T1 helix, is important for the production of VLPs. In addition, previous study of DENV2 PrM/E-expressing constructs has shown that substitutions introduced into three residues (positions 398, 401, and 412) at the hydrophobic face of the stem helix enhanced the production of VLPs and suggested that an increase in the hydrophobicity and flexibility of the stem influences the curving and bending of the lipid membrane and lead to increased VLP production (57). In this regard, our study of TMD mutants suggests that an increase in the hydrophobicity at the N terminus of T1 or at either the N or C terminus of T2 as well as an increase in the length of T1 also may affect the curving and bending of the lipid membrane, thus leading to increased VLP production; nonetheless, the mechanisms remain to be investigated.
Acknowledgments
We thank Li-Kuang Chen at the Tzu-Chi University, Taiwan, for kindly providing mouse anti-E MAb FL0232 and Wei-Che Ko for technical assistance.
This work was supported by the National Science Council of Taiwan (NSC95-2320-B-002-084-MY3).
Footnotes
Published ahead of print on 24 February 2010.
REFERENCES
- 1.Allison, S. L., K. Stiasny, K. Stadler, C. W. Mandl, and F. X. Heinz. 1999. Mapping of functional elements in the stem-anchor region of tick-borne encephalitis virus envelope protein E. J. Virol. 73:5605-5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson, A. M., L. Melin, A. Bean, and R. F. Pettersson. 1997. A retention signal necessary and sufficient for Golgi localization maps to the cytoplasmic tail of a Bunyaviridae (Uukuniemi virus) membrane glycoprotein. J. Virol. 71:4717-4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aoki, D., N. Lee, N. Yamaguchi, C. Dubois, and M. N. Fukuda. 1992. Golgi retention of a trans-Golgi membrane protein, galactosyltransferase, requires cysteine and histidine residues within the membrane-anchoring domain. Proc. Natl. Acad. Sci. USA 89:4319-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bijlmakers, M. J., M. Isobe-Nakamura, L. J. Ruddock, and M. Marsh. 1997. Intrinsic signals in the unique domain target p56(lck) to the plasma membrane independently of CD4. J. Cell Biol. 137:1029-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bray, M., R. Men, I. Tokimatsu, and C. J. Lai. 1998. Genetic determinants responsible for acquisition of dengue type 2 virus mouse neurovirulence. J. Virol. 72:1647-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bretscher, M. S., and S. Munro. 1993. Cholesterol and the Golgi apparatus. Science 261:1280-1281. [DOI] [PubMed] [Google Scholar]
- 7.Bukrinskaya, A. G., A. I. Staroff, and K. Issayeffa. 1981. Influenza virus assembly and its defects. Biosystems 13:157-161. [DOI] [PubMed] [Google Scholar]
- 8.Chang, G. J., A. R. Hunt, and B. Davis. 2000. A single intramuscular injection of recombinant plasmid DNA induces protective immunity and prevents Japanese encephalitis in mice. J. Virol. 74:4244-4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, G. J., A. R. Hunt, D. A. Holmes, T. Springfield, T. S. Chiueh, J. T. Roehrig, and D. J. Gubler. 2003. Enhancing biosynthesis and secretion of premembrane and envelope proteins by the chimeric plasmid of dengue virus type 2 and Japanese encephalitis virus. Virology 306:170-180. [DOI] [PubMed] [Google Scholar]
- 10.Cocquerel, L., S. Duvet, J. C. Meunier, A. Pillez, R. Cacan, C. Wychowski, and J. Dubuisson. 1999. The transmembrane domain of hepatitis C virus glycoprotein E1 is a signal for static retention in the endoplasmic reticulum. J. Virol. 73:2641-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cocquerel, L., J. C. Meunier, A. Pillez, C. Wychowski, and J. Dubuisson. 1998. A retention signal necessary and sufficient for endoplasmic reticulum localization maps to the transmembrane domain of hepatitis C virus glycoprotein E2. J. Virol. 72:2183-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cocquerel, L., C. Wychowski, F. Minner, F. Penin, and J. Dubuisson. 2000. Charged residues in the transmembrane domains of hepatitis C virus glycoproteins play a major role in the processing, subcellular localization, and assembly of these envelope proteins. J. Virol. 74:3623-3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis, B. S., G. J. Chang, B. Cropp, J. T. Roehrig, D. A. Martin, C. J. Mitchell, R. Bowen, and M. L. Bunning. 2001. West Nile virus recombinant DNA vaccine protects mouse and horse from virus challenge and expresses in vitro a noninfectious recombinant antigen that can be used in enzyme-linked immunosorbent assays. J. Virol. 75:4040-4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duvet, S., L. Cocquerel, A. Pillez, R. Cacan, A. Verbert, D. Moradpour, C. Wychowski, and J. Dubuisson. 1998. Hepatitis C virus glycoprotein complex localization in the endoplasmic reticulum involves a determinant for retention and not retrieval. J. Biol. Chem. 273:32088-32095. [DOI] [PubMed] [Google Scholar]
- 15.Elliott, R. M. 1990. Molecular biology of the Bunyaviridae. J. Gen. Virol. 71(Pt 3):501-522. [DOI] [PubMed] [Google Scholar]
- 16.Feemster, R. F., R. E. Wheeler, J. B. Daniels, H. D. Rose, M. Schaeffer, R. E. Kissling, R. O. Hayes, E. R. Alexander, and W. A. Murray. 1958. Field and laboratory studies on equine encephalitis. N. Engl. J. Med. 259:107-113. [DOI] [PubMed] [Google Scholar]
- 17.Ferlenghi, I., M. Clarke, T. Ruttan, S. L. Allison, J. Schalich, F. X. Heinz, S. C. Harrison, F. A. Rey, and S. D. Fuller. 2001. Molecular organization of a recombinant subviral particle from tick-borne encephalitis virus. Mol. Cell 7:593-602. [DOI] [PubMed] [Google Scholar]
- 18.Fisher, A. G., E. Collalti, L. Ratner, R. C. Gallo, and F. Wong-Staal. 1985. A molecular clone of HTLV-III with biological activity. Nature 316:262-265. [DOI] [PubMed] [Google Scholar]
- 19.Flint, M., and J. A. McKeating. 1999. The C-terminal region of the hepatitis C virus E1 glycoprotein confers localization within the endoplasmic reticulum. J. Gen. Virol. 80(Pt 8):1943-1947. [DOI] [PubMed] [Google Scholar]
- 20.Geisbert, T. W., and P. B. Jahrling. 1995. Differentiation of filoviruses by electron microscopy. Virus Res. 39:129-150. [DOI] [PubMed] [Google Scholar]
- 21.Green, S., and A. Rothman. 2006. Immunopathological mechanisms in dengue and dengue hemorrhagic fever. Curr. Opin. Infect. Dis. 19:429-436. [DOI] [PubMed] [Google Scholar]
- 22.Gubler, D. J. 2002. Epidemic dengue/dengue hemorrhagic fever as a public health, social and economic problem in the 21st century. Trends Microbiol. 10:100-103. [DOI] [PubMed] [Google Scholar]
- 23.Guzmán, M. G., and G. Kouri. 2002. Dengue: an update. Lancet Infect. Dis. 2:33-42. [DOI] [PubMed] [Google Scholar]
- 24.Halstead, S. B. 1988. Pathogenesis of dengue: challenges to molecular biology. Science 239:476-481. [DOI] [PubMed] [Google Scholar]
- 25.Hernandez, R., C. Sinodis, M. Horton, D. Ferreira, C. Yang, and D. T. Brown. 2003. Deletions in the transmembrane domain of a sindbis virus glycoprotein alter virus infectivity, stability, and host range. J. Virol. 77:12710-12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Honsho, M., J. Y. Mitoma, and A. Ito. 1998. Retention of cytochrome b5 in the endoplasmic reticulum is transmembrane and luminal domain-dependent. J. Biol. Chem. 273:20860-20866. [DOI] [PubMed] [Google Scholar]
- 27.Hsieh, S. C., I. J. Liu, C. C. King, G. J. Chang, and W. K. Wang. 2008. A strong endoplasmic reticulum retention signal in the stem-anchor region of envelope glycoprotein of dengue virus type 2 affects the production of virus-like particles. Virology 374:338-350. [DOI] [PubMed] [Google Scholar]
- 28.Hu, H. P., S. C. Hsieh, C. C. King, and W. K. Wang. 2007. Characterization of retrovirus-based reporter viruses pseudotyped with the precursor membrane and envelope glycoproteins of four serotypes of dengue viruses. Virology 368:376-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunt, A. R., C. B. Cropp, and G. J. Chang. 2001. A recombinant particulate antigen of Japanese encephalitis virus produced in stably-transformed cells is an effective noninfectious antigen and subunit immunogen. J. Virol. Methods 97:133-149. [DOI] [PubMed] [Google Scholar]
- 30.Hunter, E. 2007. Virus assembly, p. 141-168. In D. M. Knipe, P. M.Howley, and D. E. Griffin (ed.), Fields virology. Lippincott William & Wilkins, Philadelphia, PA.
- 31.Ishak, R., D. G. Tovey, and C. R. Howard. 1988. Morphogenesis of yellow fever virus 17D in infected cell cultures. J. Gen. Virol. 69(Pt 2):325-335. [DOI] [PubMed] [Google Scholar]
- 32.Keelapang, P., R. Sriburi, S. Supasa, N. Panyadee, A. Songjaeng, A. Jairungsri, C. Puttikhunt, W. Kasinrerk, P. Malasit, and N. Sittisombut. 2004. Alterations of pr-M cleavage and virus export in pr-M junction chimeric dengue viruses. J. Virol. 78:2367-2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kinsella, E., S. G. Martin, A. Grolla, M. Czub, H. Feldmann, and R. Flick. 2004. Sequence determination of the Crimean-Congo hemorrhagic fever virus L segment. Virology 321:23-28. [DOI] [PubMed] [Google Scholar]
- 34.Konishi, E., and A. Fujii. 2002. Dengue type 2 virus subviral extracellular particles produced by a stably transfected mammalian cell line and their evaluation for a subunit vaccine. Vaccine 20:1058-1067. [DOI] [PubMed] [Google Scholar]
- 35.Kroeger, M. A., and P. C. McMinn. 2002. Murray Valley encephalitis virus recombinant subviral particles protect mice from lethal challenge with virulent wild-type virus. Arch. Virol. 147:1155-1172. [DOI] [PubMed] [Google Scholar]
- 36.Lim, K. P., and D. X. Liu. 2001. The missing link in coronavirus assembly. Retention of the avian coronavirus infectious bronchitis virus envelope protein in the pre-Golgi compartments and physical interaction between the envelope and membrane proteins. J. Biol. Chem. 276:17515-17523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin, Y. J., and S. C. Wu. 2005. Histidine at residue 99 and the transmembrane region of the precursor membrane PrM protein are important for the PrM-E heterodimeric complex formation of Japanese encephalitis virus. J. Virol. 79:8535-8544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindenbach, B. D., H. J. Thiel, and C. M. Rice. 2007. Flaviviridae: the viruses and their replication, p. 1101-1152. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott William & Wilkins, Philadelphia, PA.
- 39.Machamer, C. E. 1993. Targeting and retention of Golgi membrane proteins. Curr. Opin. Cell Biol. 5:606-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mackenzie, J. M., and E. G. Westaway. 2001. Assembly and maturation of the flavivirus Kunjin virus appear to occur in the rough endoplasmic reticulum and along the secretory pathway, respectively. J. Virol. 75:10787-10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin, J. E., T. C. Pierson, S. Hubka, S. Rucker, I. J. Gordon, M. E. Enama, C. A. Andrews, Q. Xu, B. S. Davis, M. Nason, M. Fay, R. A. Koup, M. Roederer, R. T. Bailer, P. L. Gomez, J. R. Mascola, G. J. Chang, G. J. Nabel, and B. S. Graham. 2007. A West Nile virus DNA vaccine induces neutralizing antibody in healthy adults during a phase 1 clinical trial. J. Infect. Dis. 196:1732-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Medina, M. F., G. P. Kobinger, J. Rux, M. Gasmi, D. J. Looney, P. Bates, and J. M. Wilson. 2003. Lentiviral vectors pseudotyped with minimal filovirus envelopes increased gene transfer in murine lung. Mol. Ther. 8:777-789. [DOI] [PubMed] [Google Scholar]
- 43.Men, R. H., M. Bray, and C. J. Lai. 1991. Carboxy-terminally truncated dengue virus envelope glycoproteins expressed on the cell surface and secreted extracellularly exhibit increased immunogenicity in mice. J. Virol. 65:1400-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Michelsen, K., H. Yuan, and B. Schwappach. 2005. Hide and run. Arginine-based endoplasmic-reticulum-sorting motifs in the assembly of heteromultimeric membrane proteins. EMBO Rep. 6:717-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2003. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl. Acad. Sci. USA 100:6986-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Modis, Y., S. Ogata, D. Clements, and S. C. Harrison. 2005. Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. J. Virol. 79:1223-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mukhopadhyay, S., R. J. Kuhn, and M. G. Rossmann. 2005. A structural perspective of the flavivirus life cycle. Nat. Rev. Microbiol. 3:13-22. [DOI] [PubMed] [Google Scholar]
- 48.Munro, S. 1995. An investigation of the role of transmembrane domains in Golgi protein retention. EMBO J. 14:4695-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murphy, F. A., A. K. Harrison, and S. G. Whitfield. 1973. Bunyaviridae: morphologic and morphogenetic similarities of Bunyamwera serologic supergroup viruses and several other arthropod-borne viruses. Intervirology 1:297-316. [DOI] [PubMed] [Google Scholar]
- 50.Murray, J. M., J. G. Aaskov, and P. J. Wright. 1993. Processing of the dengue virus type 2 proteins prM and C-prM. J. Gen. Virol. 74(Pt 2):175-182. [DOI] [PubMed] [Google Scholar]
- 51.Nilsson, T., J. M. Lucocq, D. Mackay, and G. Warren. 1991. The membrane spanning domain of beta-1,4-galactosyltransferase specifies trans Golgi localization. EMBO J. 10:3567-3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nilsson, T., P. Slusarewicz, M. H. Hoe, and G. Warren. 1993. Kin recognition. A model for the retention of Golgi enzymes. FEBS Lett. 330:1-4. [DOI] [PubMed] [Google Scholar]
- 53.Op De Beeck, A., R. Molenkamp, M. Caron, A. Ben Younes, P. Bredenbeek, and J. Dubuisson. 2003. Role of the transmembrane domains of prM and E proteins in the formation of yellow fever virus envelope. J. Virol. 77:813-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Op De Beeck, A., Y. Rouille, M. Caron, S. Duvet, and J. Dubuisson. 2004. The transmembrane domains of the prM and E proteins of yellow fever virus are endoplasmic reticulum localization signals. J. Virol. 78:12591-12602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Orlinger, K. K., V. M. Hoenninger, R. M. Kofler, and C. W. Mandl. 2006. Construction and mutagenesis of an artificial bicistronic tick-borne encephalitis virus genome reveals an essential function of the second transmembrane region of protein E in flavivirus assembly. J. Virol. 80:12197-12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pelham, H. R. 1995. Sorting and retrieval between the endoplasmic reticulum and Golgi apparatus. Curr. Opin. Cell Biol. 7:530-535. [DOI] [PubMed] [Google Scholar]
- 57.Purdy, D. E., and G. J. Chang. 2005. Secretion of noninfectious dengue virus-like particles and identification of amino acids in the stem region involved in intracellular retention of envelope protein. Virology 333:239-250. [DOI] [PubMed] [Google Scholar]
- 58.Purdy, D. E., A. J. Noga, and G. J. Chang. 2004. Noninfectious recombinant antigen for detection of St. Louis encephalitis virus-specific antibodies in serum by enzyme-linked immunosorbent assay. J. Clin. Microbiol. 42:4709-4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Randolph, V. B., G. Winkler, and V. Stollar. 1990. Acidotropic amines inhibit proteolytic processing of flavivirus prM protein. Virology 174:450-458. [DOI] [PubMed] [Google Scholar]
- 60.Rayner, J. C., and H. R. Pelham. 1997. Transmembrane domain-dependent sorting of proteins to the ER and plasma membrane in yeast. EMBO J. 16:1832-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature 375:291-298. [DOI] [PubMed] [Google Scholar]
- 62.Schalich, J., S. L. Allison, K. Stiasny, C. W. Mandl, C. Kunz, and F. X. Heinz. 1996. Recombinant subviral particles from tick-borne encephalitis virus are fusogenic and provide a model system for studying flavivirus envelope glycoprotein functions. J. Virol. 70:4549-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scheiffele, P., M. G. Roth, and K. Simons. 1997. Interaction of influenza virus haemagglutinin with sphingolipid-cholesterol membrane domains via its transmembrane domain. EMBO J. 16:5501-5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shin, J., R. L. Dunbrack, Jr., S. Lee, and J. L. Strominger. 1991. Signals for retention of transmembrane proteins in the endoplasmic reticulum studied with CD4 truncation mutants. Proc. Natl. Acad. Sci. USA 88:1918-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stadler, K., S. L. Allison, J. Schalich, and F. X. Heinz. 1997. Proteolytic activation of tick-borne encephalitis virus by furin. J. Virol. 71:8475-8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tischler, N. D., J. Fernandez, I. Muller, R. Martinez, H. Galeno, E. Villagra, J. Mora, E. Ramirez, M. Rosemblatt, and P. D. Valenzuela. 2003. Complete sequence of the genome of the human isolate of Andes virus CHI-7913: comparative sequence and protein structure analysis. Biol. Res. 36:201-210. [DOI] [PubMed] [Google Scholar]
- 67.Tooze, J., and S. A. Tooze. 1985. Infection of AtT20 murine pituitary tumour cells by mouse hepatitis virus strain A59: virus budding is restricted to the Golgi region. Eur. J. Cell Biol. 37:203-212. [PubMed] [Google Scholar]
- 68.Wang, J. J., C. L. Liao, Y. W. Chiou, C. T. Chiou, Y. L. Huang, and L. K. Chen. 1997. Ultrastructure and localization of E proteins in cultured neuron cells infected with Japanese encephalitis virus. Virology 238:30-39. [DOI] [PubMed] [Google Scholar]
- 69.Wang, S., R. He, and R. Anderson. 1999. PrM- and cell-binding domains of the dengue virus E protein. J. Virol. 73:2547-2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang, W. K., H. L. Chen, C. F. Yang, S. C. Hsieh, C. C. Juan, S. M. Chang, C. C. Yu, L. H. Lin, J. H. Huang, and C. C. King. 2006. Slower rates of clearance of viral load and virus-containing immune complexes in patients with dengue hemorrhagic fever. Clin. Infect. Dis. 43:1023-1030. [DOI] [PubMed] [Google Scholar]
- 71.Welsch, S., S. Miller, I. Romero-Brey, A. Merz, C. K. E. Bleck, P. Walther, S. D. Fuller, C. Antony, J. Krijnse-Locker, and R. Bartenschlager. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang, M., J. Ellenberg, J. S. Bonifacino, and A. M. Weissman. 1997. The transmembrane domain of a carboxyl-terminal anchored protein determines localization to the endoplasmic reticulum. J. Biol. Chem. 272:1970-1975. [DOI] [PubMed] [Google Scholar]
- 73.Yu, I. M., W. Zhang, H. A. Holdaway, L. Li, V. A. Kostyuchenko, P. R. Chipman, R. J. Kuhn, M. G. Rossmann, and J. Chen. 2008. Structure of the immature dengue virus at low pH primes proteolytic maturation. Science 319:1834-1837. [DOI] [PubMed] [Google Scholar]
- 74.Zhang, W., P. R. Chipman, J. Corver, P. R. Johnson, Y. Zhang, S. Mukhopadhyay, T. S. Baker, J. H. Strauss, M. G. Rossmann, and R. J. Kuhn. 2003. Visualization of membrane protein domains by cryo-electron microscopy of dengue virus. Nat. Struct. Biol. 10:907-912. [DOI] [PMC free article] [PubMed] [Google Scholar]