Abstract
Human T-lymphotropic virus type 1 (HTLV-1) encodes the viral protein Tax, which is believed to act as a viral transactivator through its interactions with a variety of transcription factors, including CREB and NF-κB. As is the case for all retroviruses, the provirus is inserted into the host DNA, where nucleosomes are deposited to ensure efficient packaging. Nucleosomes act as roadblocks in transcription, making it difficult for RNA polymerase II (Pol II) to proceed toward the 3′ end of the genome. Because of this, a variety of chromatin remodelers can act to modify nucleosomes, allowing for efficient transcription. While a number of covalent modifications are known to occur on histone tails in HTLV-1 infection (i.e., histone acetyltransferases [HATs], histone deacetylases [HDACs], and histone methyltransferases [HMTs]), evidence points to the use of chromatin remodelers that use energy from ATP hydrolysis to remodel nucleosomes. Here we confirm that BRG1, which is the core subunit of eight chromatin-remodeling complexes, is essential not only for Tax transactivation but also for viral replication. This is especially evident when wild-type infectious clones of HTLV-1 are used. BRG1 associates with Tax at the HTLV-1 long terminal repeat (LTR), and coexpression of BRG1 and Tax results in increased rates of transcription. The interaction of BRG1 with Tax additionally recruits the basal transcriptional machinery and removes some of the core histones from the nucleosome at the start site (Nuc 1). When using the BRG1-deficient cell lines SW13, C33A, and TSUPR1, we observed little viral transcription and no viral replication. Importantly, while these three cell lines do not express detectable levels of BRG1, much of the SWI/SNF complex remains assembled in the cells. Knockdown of BRG1 and associated SWI/SNF subunits suggests that the BRG1-utilizing SWI/SNF complex PBAF is responsible for HTLV-1 nucleosome remodeling. Finally, HTLV-1 infection of cell lines with a knockdown in BRG1 or the PBAF complex results in a significant reduction in viral production. Overall, we concluded that BRG1 is required for Tax transactivation and HTLV-1 viral production and that the PBAF complex appears to be responsible for nucleosome remodeling.
Human T-lymphotropic virus type 1 (HTLV-1) was the first human retrovirus discovered, in 1980 (18, 54, 55, 73). Upon infection with HTLV-1, patients can develop adult T-cell leukemia (ATL) or HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP). HTLV-1 immortalizes and activates human T lymphocytes, leading to polyclonal proliferation of infected cells and finally to oligoclonal or monoclonal growth. The virus relies on Tax, a 40-kDa phosphoprotein that localizes primarily in the nucleus, as a transactivator that can regulate a number of viral and cellular factors (52). Tax can immortalize cells in two different ways. HTLV-1-infected cells can express many early-response genes, such as erg-1, erg-2, c-fos, IL-2Rα, and cyclin D (16, 23). Tax can additionally induce immortalization by interacting with a number of cell cycle regulators via protein-protein interactions (23). Tax transactivates the HTLV-1 viral long terminal repeat (LTR) through the Tax-responsive elements (TREs), which are found in the U3 region of the LTR. Tax does not act as an enzyme, nor does it directly bind to DNA to assist in transcription. Instead, it acts on a variety of viral and cellular host factors influencing a number of pathways, such as the cell cycle and apoptosis (46). The three most probable mechanisms for trans-activation are transcriptional induction of TREs, posttranslational modifications of TRE-binding factors, and complex formation with transcriptional factors, allowing indirect binding of Tax to the TRE. Tax is known to interact with a number of transcriptional factors, including CREB, serum-responsive factor (SRF), and NF-κB; however, interactions also occur with cyclins D2 and D3, mitotic checkpoint regulators (MAD1), cyclin-dependent kinases (Cdks), Cdk inhibitors (p16INK4a and p21/waf1), and the tumor suppressor p53 (5-7, 17, 21, 29, 49, 61, 69, 71, 72). HTLV-1- and/or Tax-expressing cells display differential expression of cell-cycle-associated genes, such as the p53, p21/waf1, cyclin D2, cyclin D3, and p16 genes (2, 9, 10, 31, 32, 44, 48).
Tax activates HTLV-1 transcription through CREB and three cis-acting replication element (CRE) enhancer sequences on the viral promoter. CREB and Tax additionally interact with CBP/p300, which is believed to be involved in the formation of the preinitiation complex (19, 30, 37, 68). A number of histone acetyltransferases (HATs), including p300, CREB binding protein (CBP), and p300/CBP-associated factor (P/CAF), play roles in activating HTLV-1 gene transcription, while overexpression of histone deacetylases 1 (HDAC1) leads to the repression of Tax transactivation (19, 20, 26, 37, 41, 42). Methylation of histones also plays a role in HTLV-1 transcription; for instance, histone H3 methylated at lysine 9 (methylated H3K9) results in transcriptionally inactive chromatin, and when H3K9 is demethylated, transcription is promoted (36). Histone H3 is methylated by coactivator-associated arginine methyltransferase 1 (CARM1), leading to an increase in transcription (24). Cdks also play a role in HTLV-1 transcription; Cdk9, along with TFIIH, phosphorylates the RNA polymerase II (Pol II) carboxyl-terminal domain (CTD), allowing transcriptional elongation to occur (39). Despite all of these apparent interactions, we still do not have a clear picture of how Tax is involved in the transcription of chromatin templates through either viral or cellular promoters. Other chromatin-modifying enzymes have been shown to regulate transcription in HTLV-1. Tax interacts with a number of HATs, including p300, CBP, and the P/CAF.
Both cellular gene expression and viral gene expression are based on the state of chromatin structure. When chromatin is in the loosely packed euchromatin state, transcription can proceed with few interruptions. When chromatin is in the more-condensed heterochromatin form, transcription cannot proceed as efficiently, and thus, the level of transcription is reduced. Upon infection, the HTLV-1 provirus inserts itself into the host genome; as a result, nucleosomes are deposited along the viral chromatin, leading to a roadblock in transcription. In order to overcome this barrier, chromatin remodeling must take place to modify nucleosomes. This can be done in a number of ways, either by covalent modification of histone tails (via HATs or HDAC) or by the formation of complexes that use the energy of ATP hydrolysis to remodel chromatin. HATs such as CBP/p300 and P/CAF are associated with the HTLV-1 LTR, signifying that histone tail modifications are an important step in transactivation (31). CBP/p300 can acetylate histones H2A, H2B, H3, and H4; however, histones H3 and H4 are preferred substrates (3, 50).
There are three types of complexes that use ATP in chromatin remodeling: Brahma, ISWI, and Mi-2 (also referred to as NuRD [nucleosome remodeling and histone deacetylase]) (11). The Brahma complex uses either BRG1 (Brahma-related gene 1) or BRM (Brahma) as the ATPase subunit. BRG1 and BRM display similar biochemical activities and share a high degree of sequence identity (74%); however, the two ATPase subunits play different roles in processes such as proliferation and differentiation (33, 53, 56). The ATPase subunit BRG1 is utilized in six chromatin-remodeling complexes, including SWI/SNF's BAF (BRG1-associated factor) and PBAF complexes, as well as NCoR (nuclear receptor corepressor), WINAC (Williams syndrome transcription factor including nucleosome assembly complex), NUMAC (nucleosome methylation activator complex), and mSin3A/HDAC. These complexes can act either to repress or to activate chromatin remodeling. NUMAC activates transcription, and NCoR, mSin3A/HDAC, and WINAC are transcriptional repressors (35, 60, 64, 70). BAF and PBAF can function either as transcriptional activators or as repressors (63). BRG1 can act independently to remodel chromatin; however, the addition of core subunits (Baf155, Baf170, and Ini1) allows remodeling to proceed at a faster pace (53). In HIV-1, the remodeling complex SWI/SNF is the BRG1-utilizing complex needed for Tat transactivation (1, 45, 62).
Previous studies using immunoprecipitation from Tax-expressing cells, 2-dimensional gel electrophoresis, and mass spectrometry analysis have identified the ATPase subunit BRG1 as a Tax-interacting protein (68). Additionally, immunoprecipitation using antibodies against BRG1 and other BAFs showed interaction with Tax. Recently, the requirement for BRG1 in HTLV-1 LTR transcription has come into question; some believe that the need for BRG1 is bypassed when Tax forms a complex with a number of transcription factors (i.e., CREB/ATF-1 and CBP/p300) in activating transcription (75). Here, using a wild-type infectious clone of HTLV-1 (ACH.WT), we show that BRG1 interacts with Tax, coelutes with components of the basal transcription machinery (Pol II and CBP/p300), and is required for efficient nucleosome removal and Tax transactivation. We determined that nucleosome removal occurred at the HTLV-1 LTR, but not downstream (Gag region), in immortalized peripheral blood mononuclear cells (PBMCs) transfected with a full-length infectious ACH.WT plasmid. Additionally, we found that three BRG1 mutant cell lines, SW13, C33A, and TSUPR1, are deficient in viral production relative to that of wild-type cells and that the addition of BRG1 to infected mutant cells dramatically increases viral production. Interestingly, while the parent cells do not express BRG1 in quantifiable amounts, they are still able to assemble SWI/SNF complexes (47, 67).
MATERIALS AND METHODS
Cell culture.
HEK293T, C33A, SW13, TSUPR1, and H1GFP (25) cells were grown in Dulbecco's modified Eagle's medium supplemented with fetal bovine serum (FBS) (10%), 2 mM l-glutamine, and antibiotics (penicillin at 100 U/ml and streptomycin at 100 mg/ml) (cDMEM). H1GFP cells were additionally supplemented with 150 μg of G418/μl. H1GFP, HC1143, and C8166 cells were grown in RPMI 1640 supplemented with FBS, l-glutamine, and antibiotics as described above (cRPMI). All cell lines were maintained at 37°C under 5% CO2.
ChIP assay.
Chromatin immunoprecipitation (ChIP) assays were performed using antibodies as described below. Cells were seeded to 60% confluence in 10-cm-diameter dishes, transfected with an ACH.WT plasmid (20 μg) or pBluescript KS (20 μg of empty vector) (data not shown) using Lipofectamine (Life Technologies, Gaithersburg, Md.) in Opti-MEM (Gibco), and incubated for 48 h with cDMEM. After transfection, the cells were maintained for 1 week, and the remaining live cells (latent cells) were treated with tumor necrosis factor (TNF) to activate the virus. Samples were processed every 6 h following TNF treatment for ChIP analysis with appropriate antibodies. After proteins were cross-linked to DNA by 1.0% formaldehyde, chromatin was sonicated five times for 20 s each time, generating DNA fragments. The supernatants were diluted with ChIP dilution buffer (0.01% sodium dodecyl sulfate [SDS], 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl [pH 8.1], and 167 mM NaCl) to a total volume of 5.5 ml and were precleared by rotation for 1 h at 4°C with ChIP-prepared protein A/G beads (beads had been washed twice with 1 ml TNE50 (100 mM Tris, 50 mM NaCl, and 1 mM EDTA) plus NP-40 and had been resuspended in 650 μl with the addition of 40 μl of 10-mg/ml single-stranded DNA [ssDNA] and 75 μl of 10-mg/ml bovine serum albumin [BSA]). The extract was centrifuged at 3,000 rpm for 10 min at 4°C, and the cleared lysate was transferred to a fresh tube. Supernatants were used for immunoprecipitation (IP) with 10 μg of each antibody. After overnight rotation at 4°C, the immune complexes were collected by the addition of ChIP-prepared protein A/G beads. After extensive washes, the immune complexes were eluted with a 1% SDS-NaHCO3 solution for 30 min at room temperature. The eluted complexes were treated with an NaCl solution and were reverse cross-linked overnight. DNA was extracted using 1:1 phenol-chloroform (500 μl), followed by the addition of 1 ml of absolute ethanol and 3 M sodium acetate (50 μl) incubated at −20°C for at least 20 min. The solution was spun for 20 min at 14,000 rpm and 4°C, followed by a 70% ethanol wash and a 5-min spin. The DNA pellet was resuspended in 1× Tris-EDTA (TE) and was stored at 4°C. Afterwards, DNA was purified with a PCR purification kit (Bioneer) and was amplified by PCR. PCR was performed using primers against the HTLV-1 LTR promoter (−350 to +250) or the beta-globin promoter (5′-TCACACACTTGACCCTGTGCCATA/TTATATGCCCTGTCCTGGCTCCT T-3′).
Antibodies.
Antibodies against acetylated H4 (ac-H4) (at K8) (06-760), ac-H3 (at K9) (06-942), methylated H3 (met-H3) (at K9) (07-450), H1 (05-457), H2A (07-146), H2B (07-371), H3 (4-801), and H4 (05-858) were obtained from Upstate Biotech. Antibodies against p300 (sc584), CBP (sc1211), and BRG1 (sc10768) were obtained from Santa Cruz. The antibodies against Pol II (8WG16) and TATA box-binding protein (TBP) (a polyclonal rabbit antibody) have been used by us previously (13, 76). The anti-c-Myb antibodies were raised against wild-type c-Myb and acetylated (K471, K480, K485) peptides (58).
CAT assays.
SW13 or C33A BRG1 mutant cells were transfected with HTLV-1-LTR-CAT with pcTax or pcTax plus BRG1. After 48 h, cells were lysed, and chloramphenicol acetyltransferase (CAT) activity was determined. A standard reaction was performed by adding the cofactor acetyl coenzyme A to a microcentrifuge tube containing the cell extract and radiolabeled [14C]chloramphenicol in a final volume of 50 μl and then incubating the mixture at 37°C for 1 h. The reaction mixture was then extracted with ethyl acetate and was separated by thin-layer chromatography on Bakerflex silica gel plates in a chloroform-methanol (19:1) solvent. The resolved reaction products were then detected by exposing the plate to a PhosphorImager cassette.
Western blotting.
Cell extracts were resolved by SDS-polyacrylamide gel electrophoresis (PAGE) on a 4 to 20% Tris-glycine gel (Invitrogen). Proteins were transferred to Immobilon membranes (Millipore) at 200 mA for 2 h. Membranes were blocked with Dulbecco's phosphate-buffered saline (PBS)-0.1% Tween 20 plus 5% BSA. A primary antibody against specified antibodies was incubated with the membrane in PBS plus 0.1% Tween 20 overnight at 4°C. Membranes were washed three times with PBS plus 0.1% Tween 20 and were then incubated with a horseradish peroxidase (HRP)-conjugated secondary antibody for 1 h. The presence of the secondary antibody was detected by a SuperSignal West Dura extended-duration substrate (Pierce). Luminescence was visualized on a Kodak 1D image station.
Conventional chromatography of C8166 nuclear extracts.
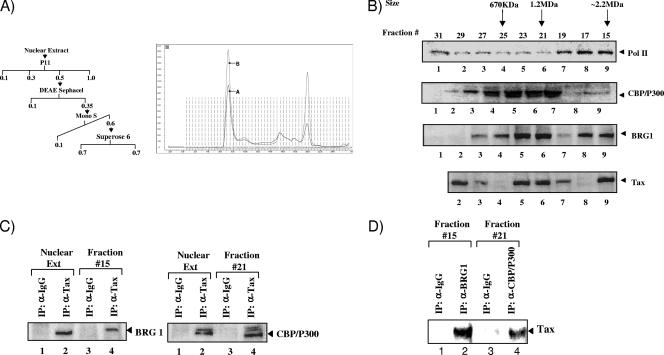
The nuclear extract (from 10 liters of log-phase growing cultures) was loaded onto a 250-ml column of phosphocellulose (P11; Whatman) and was fractionated stepwise by the indicated KCl concentrations (see Fig. 1A) in buffer A (20 mM Tris-HCl [pH 7.9], 0.2 mM EDTA, 10 mM β-mercaptoethanol [βME], 10% glycerol, 0.2 mM phenylmethylsulfonyl fluoride [PMSF]). The P11 0.5 M KCl fraction (250 mg) was loaded onto a 45-ml DEAE-Sephacel column (Pharmacia) and eluted with 0.35 M KCl. The 0.35 M KCl elution (140 mg) was dialyzed to 700 mM NH4SO4 in buffer HB (20 mM HEPES [pH 7.6], 4 mM dithiothreitol [DTT], 0.5 mM EDTA, 10% glycerol, 0.5 mM PMSF, 1 μg/ml aprotinin, 1 μg/ml leupeptin, and 1 μg/ml pepstatin) and was loaded onto a Mono S column. The column was resolved using a linear 10-column-volume gradient of 600 to 100 mM NH4SO4 in buffer HB. The 0.6 M fraction was loaded onto a Superose 6 sizing column at a salt concentration of 0.7 M. Fractions were collected, dialyzed to 50 mM KCl buffer, and Western blotted for the presence of Tax, BRG1, CBP/p300, and Pol II.
FIG. 1.
Association of Tax with CBP/p300 and SWI/SNF complexes. (A) (Left) Schematic representation of chromatographic steps using C8166 nuclear extracts. Nuclear extracts were passed through P11, DEAE Sephacel, Mono S, and Superose 6 sizing columns to obtain Tax and associated factors. (Right) Chromatogram of the fast protein liquid chromatography (FPLC) fractions, where peak A shows Superose 6 at 0.7 M salt and peak B shows 0.15 M salt. Fractions 13 to 17, which contain a high-molecular-weight complex, are fairly stable at either salt concentration. (B) Western blot analysis of Superose 6 column fractions using antibodies against Pol II, CBP/p300, Tax, and BRG1. Fractions were loaded and isolated at 0.7 M salt. Tax eluted at three distinct locations, including a small complex (120 kDa), a medium complex (1 MDa), and a large complex (∼2.2 MDa). (C) Immunoprecipitation using anti-Tax or control antibodies (IgG), followed by Western blot analysis for antibodies against BRG1 and CBP/p300. p300 is slightly larger than CBP (molecular weight, 265,000 [265K]) (68). A total of 330 μg of each fraction (fractions 15 and 21) was used for immunoprecipitation with 1.0 μg of the primary antibody. (D) Reciprocal IP of fraction 15 with anti-BRG1, as well as IP with anti-CBP/p300 from fraction 21, followed by Western blotting for Tax.
siRNA treatment.
BRG1 (29827), BRM (29831), and Baf250 (sc-43628) small interfering RNAs (siRNAs) were purchased from Santa Cruz Biotechnology, Inc. The siGENOME SMART pool for PB1 (Baf180) (M-008692-01-0005) was purchased from Dharmacon. siRNA (75 nM) was transfected into cells by using Attractene (Qiagen) according to the manufacturer's recommendations.
Transfections and RT assays.
Cells were seeded into 6-well plates at 400,000 cells/well in cDMEM or cRPMI (HCC1143 only). The following day, the cells were transfected with an ACH.WT plasmid (20 μg) (a generous gift from Lee Ratner, Washington University, St. Louis, MO) using Attractene (Qiagen) lipid reagent. Cells were harvested on days 0, 1, 4, and 6 posttransfection for determination of protein concentrations and reverse transcriptase (RT) analysis.
Viral supernatants (10 μl) were incubated in a 96-well plate with an RT reaction mixture containing 1× RT buffer (50 mM Tris-HCl, 1 mM DTT, 5 mM MgCl2, 20 mM KCl), 0.1% Triton, poly(A) (1 U/ml), poly(dT) (1 U/ml), and [3H]TTP. The mixture was incubated overnight at 37°C, and 10 μl of the reaction mixture was spotted onto a DEAE Filtermat paper, washed four times with 5% Na2HPO4 and three times with water, and then dried completely. RT activity was measured in a Betaplate counter (Wallac, Gaithersburg, MD).
Immunoprecipitation.
Cells were harvested at 4°C, and cell pellets were washed with PBS. Cell lysates were prepared as previously described (74). pcTax (5 μg) or control antibodies (5 μg) (anti-IgG) were incubated with whole-cell lysates overnight at 4°C with rotation. The overnight-incubated mixture was then cleared by centrifugation, and protein A/G beads (30% slurry) were added for 2 h at 4°C. The immunoprecipitated complex was washed with buffer K (150 mM KCl, 20 mM HEPES [pH 7.4], 5 mM MgCl2) and was then resuspended in SDS-PAGE loading Laemmli buffer. Samples were separated on a 4 to 20% SDS-PAGE gel and were subjected to Western blotting.
Restriction enzyme assay.
A glycerol gradient-purified 5S DNA array (2 nM) was exposed to SalI (5,000 U/ml) in the presence of either the SWI/SNF complex (4 nM) and ATP (1 mM) or SWI/SNF, ATP, and purified Tax protein (10 nM). The reaction mixtures were incubated at 37°C. The reactions were stopped after 5, 15, and 30 min, and the products were then run on an agarose gel. At the indicated time points, an aliquot of the reaction product was vigorously mixed for 10 s with water (25 μl) and 1:1 phenol-chloroform (50 μl). The purified DNA fragments were resolved by nondenaturing agarose gel electrophoresis in the presence of ethidium bromide. The gels were then dried onto 3M Whatman paper. The fraction of uncut DNA template on the gel was obtained by phosphorimager analysis by using ImageQuant software and taking the ratio of uncut signal to the sum of the cut and uncut signals present in the lane.
Glycerol gradient sedimentation of arrays was carried out on a 10 to 30% linear glycerol gradient in 10 mM Tris (pH 7.5), 125 mM NaCl, 2.5 mM MgCl2, and 1 mM DTT. Sedimentation was performed for 16 h at 33,000 × g in a SW28 rotor (Beckman). Sedimentation velocity studies were performed on a Beckman XL-A analytical ultracentrifuge equipped with scanner options as described previously (43).
DNase I treatment of H1GFP cells.
Cells were grown to 70% confluence in T-150 culture flasks and were either left untransfected (no DNA); transfected with either pcTax alone (10 μg), five wild-type BRG1-specific siRNAs (20 μg) (68) plus pcTax (10 μg), or mutant BRG1 siRNAs (20 μg) (68) plus pcTax (10 μg); or treated with NaB (10 μM). Forty-eight hours later, cells were trypsinized, combined in a 50-ml conical tube, and gently pelleted at 500 × g in a tabletop centrifuge. Cells were then washed gently once in solution A (150 mM sucrose, 80 mM KCl, 35 mM HEPES [pH 7.4], 5 mM K2HPO4, 5 mM MgCl2, 0.5 mM CaCl2) and once in solution B (150 mM sucrose, 80 mM KCl, 35 mM HEPES [pH 7.4], 5 mM K2HPO4, 5 mM MgCl2, 2 mM CaCl2) and were then resuspended at 500 μl per T-150 flask in solution B. Then 500 μl of solution B containing 0.4% NP-40 and 20 or 40 μg of DNase I (1 mg/ml) was added to the cell suspension, which was then mixed and incubated for 2 min at 37°C. The digestion was stopped with 4 ml of lysis buffer, and the suspension was incubated overnight at room temperature. DNA was subsequently isolated by standard techniques, resuspended in TE (1 μg/μl), size fractionated on a 1.4% agarose gel at 60 V for 12 h, stained with ethidium bromide, and transferred to a Hybond N+ membrane by capillary transfer. The nick-translated H1GFP fragment was used as a hybridization probe for Southern blotting.
PBMC purification.
Human PBMCs from two healthy donors were purified by Ficoll-Paque centrifugation and activated for 72 h with phytohemagglutinin (PHA) (10 μg/ml) and recombinant human interleukin-2 (IL-2) (50 U/ml) in cRPMI medium. The cells were transfected with double-CsCl-purified ACH.WT plasmids (40 μg) using the Amaxa reagent (Lonza). The cells were then cultured in cRPMI supplemented with PHA (5 μg/ml) and IL-2 (50 U/ml) for ∼6 weeks, after which they were cultured in the same medium without PHA. At various time points, cell culture supernatants were collected for p19 determination (data not shown), and relative cellular viability was assayed by 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) conversion assays on 100-μl aliquots of cells. Cells that survived for 70 days were ∼90% infected with HTLV-1.
RESULTS
Interaction of Tax with SWI/SNF and CBP/p300 complexes.
Chromatin-remodeling complexes that use ATPase units to alter chromatin structure allow cellular machinery access to proviral DNA, making it possible for transcription to proceed with minimal roadblocks. Using conventional chromatography, we have isolated a large complex containing both Tax and the SWI/SNF ATPase component BRG1. This complex was purified from nuclear extracts obtained from the Tax-expressing C8166 cell line. Figure 1A shows a diagram of chromatographic steps, in addition to fractions that were obtained from a Superose 6 sizing column. Using these fractions, we performed Western blotting for components of the basal transcription machinery (Pol II and CBP/p300) to determine the association of Tax/BRG1 and Tax/CBP/p300 (Fig. 1B). We observed that Tax coelutes with BRG1 in two different places on the sizing column: fractions 21 to 23 (1.2 MDa) and fraction 15 (∼2.2 MDa). To further confirm the association of Tax with CBP/p300 and SWI/SNF, we performed immunoprecipitation using pcTax or control antibodies (IgG), followed by Western blot analysis for BRG1 and CBP/p300 (Fig. 1C). Importantly, Tax immunoprecipitated with CBP/p300 from fraction 21, as well as with a control nuclear extract; Tax also immunoprecipitated with BRG1 in fraction 15, the same fraction that contains Pol II. Consistent with these results, we have previously shown that CBP activates Tax-dependent transcription by promoting transcriptional initiation and reinitiation (30). Finally, as seen in Fig. 1D, when performing reciprocal IP and Western blotting against BRG1 and CBP/p300, followed by Western blotting for Tax, we detected binding to Tax in both complexes. Collectively, these results indicate that the Tax/SWI/SNF complex and the Tax/CBP/p300 complex are tightly associated in HTLV-1-infected cells, suggesting the involvement of Tax in chromatin remodeling and the initiation of transcription.
Functional significance of the Tax/SWI/SNF interaction in vitro.
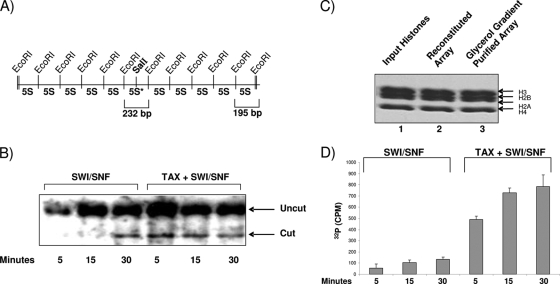
We next asked if Tax binding to SWI/SNF could accelerate nucleosome remodeling in vitro. We used a well-established procedure from the Peterson lab that utilizes the 5S nucleosome positioning plasmid, which bears a unique SalI site that is close to the dyad axis of a reconstituted nucleosome in the middle of an array of 10 5S nucleosome positioning sequences (Fig. 2A) (43). To determine the extent to which the SWI/SNF complex increases the accessibility of the SalI site in the central nucleosome, the array was exposed to SalI in the presence of either SWI/SNF and ATP or SWI/SNF, ATP, and purified Tax protein (Fig. 2B). Under these conditions, SalI digestion of the array DNA with SWI/SNF alone started at 30 min post-enzyme addition. However, in the presence of Tax, SalI cleavage was enhanced after 5 min, indicating that the Tax/SWI/SNF complex could possibly increase the remodeling of the nucleosome. The amplification of SalI cleavage required both SWI/SNF and ATP. All core histones were present throughout the process of obtaining the glycerol gradient-purified array (Fig. 2C). Finally, we performed a similar set of experiments using an LTR luciferase construct in which the plasmid was assembled into chromatin and further processed as seen in Fig. 2C. We performed time course experiments followed by restriction enzyme digestion with SmaI and SacI after the addition of Tax or the Tax/SWI/SNF complex. Digested fragments were run on an agarose gel and Southern blotted with a probe covering the LTR area from −52 to +1. The results (Fig. 2D) show that in the presence of Tax, cleavage of the native LTR is enhanced when SWI/SNF is present. Collectively, these data indicate that Tax/SWI/SNF increases chromatin remodeling on DNA at a higher rate than that with SWI/SNF alone.
FIG. 2.
Functional interaction of Tax with the chromatin-remodeling complex in vitro. (A) Diagram of the 5S nucleosome positioning plasmid, bearing a unique SalI site close to the dyad axis of a reconstituted nucleosome in the middle of an array of 10 5S nucleosome positioning sequences. EcoRI sites are 195 bp apart, while the SalI 5S repeat is located on a 232-bp EcoRI fragment. Modified from reference 43. (B) Glycerol gradient-purified 5S chromatinized DNA was used in a SalI restriction enzyme accessibility assay using either SWI/SNF alone or Tax plus SWI/SNF. The reaction mixtures were incubated at 37°C. Reactions were stopped after 5, 15, and 30 min before samples were processed and run on an agarose gel. The DNA was run on a 1.2% gel and was then stained with ethidium bromide. The gels were then dried onto 3MM Whatman paper. The fraction of uncut DNA template on the array was obtained by phosphorimager analysis using ImageQuant software, by taking the ratio of uncut signal to the sum of cut and uncut signals present in the lane. (C) Presence of histones on DNA after gradient purification as shown by Coomassie blue staining. Glycerol gradient sedimentation of arrays was carried out on 10 to 30% linear gradients. Sedimentation velocity studies were performed on a Beckman XL-A analytical ultracentrifuge equipped with scanner options as described previously (43). Lane 1, input histones used for reconstitution arrays (2.5 μg); lanes 2 and 3, reconstituted DNA molecules before and after exposure to the glycerol gradient. (D) Similar to panel C, but the plasmid used contained an LTR luciferase construct, and following nucleosome assembly, samples were treated with SmaI and SacI and were run on an agarose gel (2.5%), followed by hybridization with a nick-translated probe spanning the area of the LTR from −52 to +1. Data represent the results of three experiments with the hybridized cut fragment and SWI/SNF or Tax/SWI/SNF.
Chromatin remodelers and histone acetyltransferases are recruited sequentially to the HTLV-1 LTR in vivo.
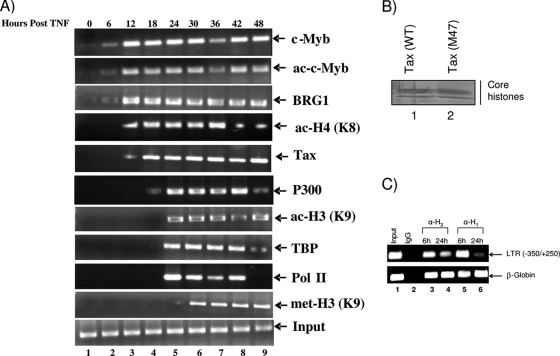
Since both histone tail modifications and an ordered recruitment of chromatin remodelers are required for the initiation of transcription of cellular and viral genes, we next asked whether the HTLV-1 promoter and coding sequences were regulated through histone modifications and whether p300 or SWI/SNF had a role in virus regulation in vivo. One test of the histone code hypothesis for HTLV-1-infected cells is to determine whether a specific pattern of histone acetylations can be observed upon gene activation. To test this, we carried out ChIP experiments to determine whether proteins such as SWI/SNF and CBP/p300 bound to DNA in vivo and whether the lysine residues on histones H3 and H4 were acetylated when transcription of the HTLV-1 gene was induced following transfections. To assess the in vivo transcription factor occupancy of the infectious ACH.WT clone, 293T cells were transfected with cloned viral DNA. ACH.WT was the first infectious HTLV-1 molecular clone containing the full-length HTLV-1 proviral genome from the CH strain and a wild-type C91/PL tax gene, and it is able to express a functional Tax protein, as assayed by CAT experiments for transactivation capability (34). After 1 week, the remaining live cells (latent cells) were treated with TNF for 2 h. Next, samples were processed for both ChIP and viral particle production. DNA was amplified by PCR using primers from position −350 to +250 (U3/R/U5). Figure 3A demonstrates the overall pattern of protein recruitment on the HTLV-1 LTR and histone H3 and H4 modifications in vivo. Tax, BRG1, and c-Myb (acetylated) were recruited to the promoter within the first 6 to 12 h, followed by the recruitment of p300 and TBP within 18 to 24 h. The presence of TBP is a hallmark of activated transcription and recruitment of Pol II. Among the histone modifications, we observed H4 (K8) and H3 (K9) acetylation, which are needed for the recruitment and stabilization of BRG1 and p300, respectively. Also, H3 (K9) was methylated, which is a hallmark of transcriptional elongation. We also isolated core histones from cells transfected with either a wild-type vector or the M47 mutant vector, and we saw no changes in histone levels after 48 h of ACH transfections (Fig. 3B). More specifically, when assaying for total histone H3 loaded onto the LTR, we observed a drop (∼50%) in the total histone H3 level after 24 h (when Tax is expressed); however, this drop was not apparent on the beta-globin promoter (Fig. 3C). A similar pattern was also observed with the linker histone protein, where the 24-h sample showed more than a 90% drop in the level of histone H1 on the LTR but not in the β-globin gene, indicating an “opening up” of the chromatin DNA that is responsive to Tax. Finally, it is noteworthy that these ChIP assays scored for factors that were needed for activated transcription by Tax. The assay was not sensitive enough for the detection of basal transcription, which would have been needed for initial transcription of the Tax mRNA (doubly spliced message) (data not shown). Collectively, these data indicate that factors such as acetylated c-Myb, BRG1, p300, and Tax are recruited to the promoter prior to the activated transcription required for elongation on the HTLV-1 promoter.
FIG. 3.
HTLV-1 clone transfection and in vivo ChIP analysis. (A) Human 293T kidney fibroblasts were seeded to 60% confluence in 10-cm-diameter dishes and were transfected with ACH.WT (20 μg) or pBluescript KS (20 μg of empty vector) (data not shown). After transfection, the cells were maintained for 1 week, and the remaining live cells (latent cells) were treated with TNF to activate the virus. Samples were processed every 6 h following TNF treatment for ChIP analysis with appropriate antibodies. Cell culture supernatants were collected at various time points posttransfection, and virus particle production was monitored by a p19 enzyme-linked immunosorbent assay (ELISA) (data not shown). The input panel (positive-control PCR) shows the presence of HTLV-1 DNA in total chromosomal DNA prior to the ChIP assay. The primers for the ChIP assay spanned the region from −350 to +250 (U3/R/U5). The 24-h sample represents the start of activated transcription (recruitment of the nucleating factor TBP) by Tax. ac, acetylation; met, methylation. (B) Isolation of total histones from cells transfected with ACH.WT (20 μg) or an ACH.M47 (20 μg) mutant clone. Histones were isolated after 48 h and were run on a 4 to 20% gel, which was stained with Coomassie blue. (C) 293T cells were transfected with ACH.WT (20 μg) as for panel A. Samples were processed after either 6 or 24 h for ChIP analysis. Antibodies against histone H1 and H3 were used for ChIP, and recovered DNA was subsequently used for PCR with primers against the LTR (from −350 to +250) or the beta-globin promoter.
Determination of translational positioning of HTLV-1 nucleosomes in vivo.
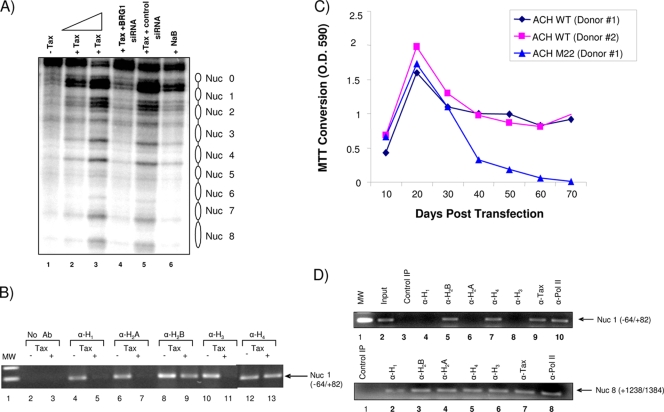
Active and inactive genes can be distinguished based on chromatin structure and accessibility at the promoter. Active genes are accessible to factors that allow for effortless transcription compared to inactive genes; this can be determined using nuclease sensitivity assays. Promoters of active genes are especially hypersensitive to DNase I, and this is more evident in nucleosome-free regions of a promoter. To examine the nucleosomal organization of the HTLV-1 promoter, we examined the translational positioning of nucleosomes in active and inactive HTLV-1 promoter regions in permeabilized cells. We used the H1GFP cell line, which contains a wild-type HTLV-1 LTR-green fluorescent protein (GFP) incorporated into the cellular chromosome, where the LTR can easily be activated by the addition of exogenous Tax (25). Cells were transfected with wild-type Tax either alone or with a siRNA against BRG1 (siBRG1) or control siRNAs (68), or NaB (a histone deacetylase inhibitor) was added as a positive control to activate chromatin DNA. Following transfection, cells were prepared for a DNase I hypersensitivity assay. Figure 4A shows the Southern blot analysis of the DNase I cleavage patterns on the inactive and active HTLV-1 LTR-GFP promoters. Surprisingly, there were no clear DNase I-hypersensitive sites on the HTLV-1 promoter in the absence of Tax (Fig. 4A, lane 1). However, upon the addition of increasing amounts of Tax (Fig. 4A, lanes 2 and 3), there was a clear indication of DNase I-hypersensitive sites. The pattern largely disappeared after treatment with siBRG1 but not after treatment with the control siRNA (Fig. 4A, lanes 4 and 5). Because the inactive promoter region in permeabilized cells was more nuclease resistant than the active promoter, it is likely that nucleosomes are present on the inactive promoter. However, the lack of specific 200-bp periodicity in the DNase I cleavage suggests that nucleosomes on the inactive promoter region are not translationally positioned. Instead, this cleavage pattern is consistent with random translational positioning or inaccessible nucleosomes in the inactive promoter.
FIG. 4.
Determination of translational positioning of HTLV-1 nucleosomes and absence of various histones in transfected H1GFP cells and PBMCs. (A) DNase I treatment of permeabilized H1GFP (HTLV-1 LTR-GFP) cells for mapping of DNase I-hypersensitive sites. Cells were grown to 70% confluence and were then either left untransfected (lane 1); transfected with increasing amounts of Tax alone (lane 2, 5 μg; lane 3, 10 μg), five wild-type BRG1-specific siRNAs (20 μg) plus Tax (10 μg) (lane 4), or five mutant BRG1 siRNAs (20 μg) plus Tax (10 μg) (lane 5); or treated with NaB (10 μM). Forty-eight hours later, cells were prepared for a DNase I hypersensitivity assay, size fractionated on an agarose gel, and transferred to a membrane. The nick-translated H1GFP fragment (5 μg) was used as a hybridization probe for Southern blotting. (B) Effect of Tax on histone eviction after Tax treatment. H1GFP cells were either left untreated or transfected with a Tax plasmid using Lipofectamine. Forty-eight hours later, samples were processed for ChIP using various antibodies against histones. Primers for the +1 start site (Nuc 1 area) spanned the area from −64 to +82. (C) Human PBMCs from two healthy donors were first purified by Ficoll-Paque centrifugation and then activated with PHA (10 μg/ml) and recombinant human IL-2 (50 U/ml). The cells were transfected with double-CsCl-purified ACH.WT (40 μg) using the Amaxa reagent. The cells were then cultured in cRPMI supplemented with PHA (5 μg/ml) and IL-2 for ∼6 weeks, after which they were cultured in the same medium without PHA. At various time points, cell culture supernatants were collected for p19 determination (data not shown), and relative cellular viability was assayed by MTT conversion assays on 100-μl aliquots of cells. Cells that survived for 70 days were ∼90% infected with HTLV-1. O.D. 590, optical density at 590 nm. (D) ChIP assays with samples from donor 1 PBMCs transfected with the wild-type clone ACH.WT. ChIPs were performed with 70-day samples using various antibodies. PCR amplification was performed for both the Nuc 1 (+1 area) and Nuc 8 (Gag) regions.
We performed ChIP assays from the HTLV-1 nucleosome that spanned the transcription start site (+1) and then used LTR primers covering a portion of the Nuc 1 area (−64 to +82). Figure 4B shows that histones H1, H2A, H2B, H3, and H4 were present on the transcription start site of an inactive promoter (−Tax lanes). However, upon the addition of Tax, three of the five histones—histones H1, H2A, and H3—were removed from the start site (Fig. 4B, +Tax lanes). These data indicate that the promoter may have nucleosomes loaded randomly on the LTR, and upon the addition of Tax, some of the histones are removed, resulting in phased DNA-accessible DNase I-hypersensitive sites (Fig. 4A, lanes 1 to 3). These results are also consistent with previous reports on mouse mammary tumor virus (MMTV), Hsp70, and hypoxanthine phosphoribosyltransferase (HPRT) genes, where histones H1, H2A, and H3 were preferentially removed at the initiation site and not on the actual open reading frame (ORF) DNA downstream of the promoter-proximal site (4, 15, 51, 59, 65).
We next asked if histones were differentially removed in PBMCs transfected with an HTLV-1 clone. We used the ACH.WT clone, with an NF-κB mutant HTLV-1 clone (ACH.M22) as a control. PBMCs from two donors were transfected with either the wild-type clone or the ACH.M22 clone. Wild-type clones showed immortalization after 40 days and continued to grow for as long as 70 days; however, transfections with the ACH.M22 clone failed to immortalize cells (Fig. 4C). These results are consistent with previously published results from the Ratner lab (57). ChIP assays with samples from one of the ACH.WT-transfected PBMC clones indicated that histones H1, H2A, and H3 were absent from the Nuc 1 area (Fig. 4D); however, when we used primers spanning Nuc 8 (+1238 to +1384), which covers the Gag ORF, we observed only partial removal of histone H1. Finally, it is noteworthy that the results in Fig. 4A show the presence of a few nucleosomes (−350 to +250). However, when using primers that cover only Nuc 1, we consistently observed removal of the H3 from the transcription start site in the presence of Tax. Collectively, our results using HTLV-1 integrated chromatin indicate that the Tax-activated promoter was assembled into an ordered array of translationally positioned nucleosomes, which was interrupted over a 350-bp region that showed increased accessibility to DNase I and that contained the entire functional promoter. In contrast, the inactive promoter was relatively inaccessible to nuclease and was not assembled into a translationally positioned nucleosomal array.
Effect of BRG1 on HTLV-1 LTR Tax-activated transcription.
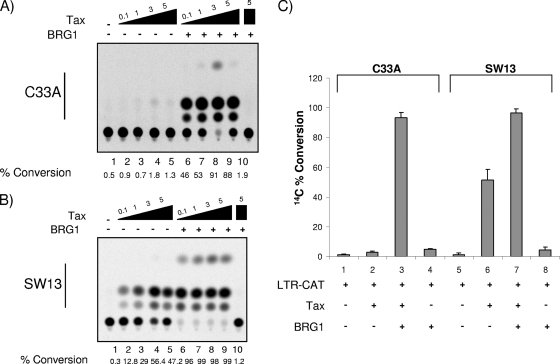
To further determine if BRG1 was needed for optimal Tax-activated transcription, we transfected HTLV-1-LTR-CAT with Tax or Tax plus BRG1 into one of two BRG1 mutant cell lines, SW13 or C33A. The SW13 cell line does not express detectable levels of either BRG1 or BRM (47, 67), although much of SWI/SNF remains assembled in SW13 cells (66, 67). The C33A cell line does not express BRM but expresses low levels of BRG1 (47). Results from these experiments can be seen in Fig. 5. Figure 5A shows transfection with increasing amounts of Tax in lanes 2 to 5 and then addition of BRG1 in lanes 6 to 10. C33A cells transfected with Tax alone showed almost no Tax transactivation, while the addition of BRG1 led to efficient Tax-activated transcription. When performing similar experiments with SW13 cells (Fig. 5B), we observed low levels of Tax-activated transcription in the presence of low concentrations of Tax; however, the addition of BRG1 enhanced the level of activated transcription dramatically (lanes 2 and 3 versus lanes 6 and 7). A BRG1 dominant-negative mutant was used as a negative control (Fig. 5B, lane 10). Furthermore, the results in Fig. 5C show that BRG1 alone was not able to activate the LTR in the absence of Tax (lanes 4 and 8) in either cell type. Collectively, these experiments indicate that BRG1 is required and necessary for optimal Tax-activated transcription.
FIG. 5.
Effect of BRG1 on HTLV-1 LTR Tax-activated transcription in mutant cells. (A and B) Titration of Tax (0.1, 1, 3, and 5 μg) either alone (lanes 2 to 5) or with a BRG1 plasmid (5 μg) (lanes 6 to 9). A total of 5 × 106 C33A (A) or SW13 (B) cells were transfected for 3 days and were subsequently processed for CAT assays (50 μg of total extract). Lanes 1 serve as a negative control (HTLV-1-LTR-CAT and no Tax), and lanes 10 also serve as a control for BRG1 (5 μg of a BRG1 dominant-negative mutant [38]). BRG1 enhanced the level of activation dramatically even with minimal concentrations of Tax (i.e., lanes 6 in both panels). (C) LTR-CAT was transfected either with BRG1 alone or with BRG1 plus Tax (3 μg) into either C33A or SW13 cells, as for panels A and B. The data represent three independent replicates. %Conversion, %conversion from unacetylated to mono- or diacetylated chloramphenicol.
Lack of HTLV-1 viral replication in BRG1 mutant cells.
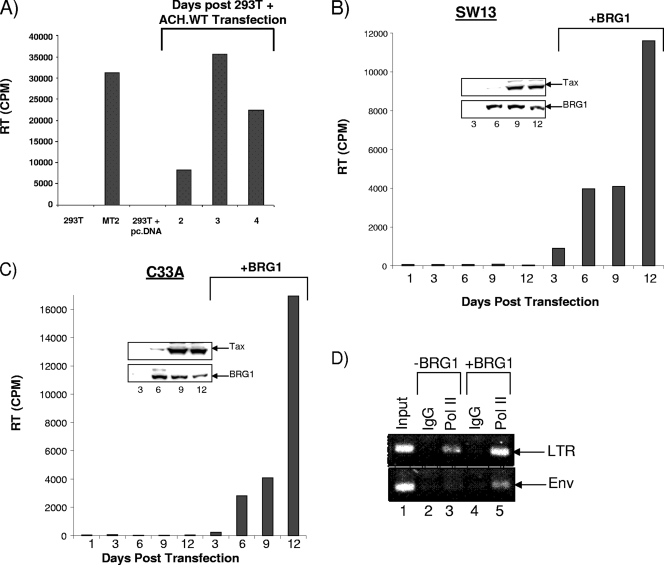
We next transfected both cell lines with the wild-type HTLV-1 clone. The results in Fig. 6A show that when ACH.WT is transfected into 293T cells, there are ample viral particles in the supernatants after days 3 and 4. 293T cells alone and 293T cells transfected with pcDNA served as negative controls, and MT2 was a positive-control supernatant from HTLV-1-infected MT2 cells treated with TNF. We next performed a similar transfection in both SW13 and C33A cells and carried out the experiment for 12 days. The results of these two transfections are shown in Fig. 6B and C. Supernatants were collected at days 1, 3, 6, 9, and 12 and were assayed for the presence of RT. Neither of these cell types showed viral replication in this experiment, which was performed in triplicate. However, when a BRG1 vector was transfected into both of these BRG1 mutant cell lines, RT was present in the supernatants. Both Fig. 6B and C show the presence of viral RT; the largest amount occurs 12 days after BRG1 transfection. Currently, it is not clear why SW13 cells showed less viral production, when activated transcription was much more pronounced than that in C33A cells (Fig. 5B). Finally, we performed ChIP assays to determine if Pol II was capable of loading and transcribing from the LTR as well as the Env gene in the presence or absence of BRG1. The results in Fig. 6D (C33A) show that Pol II was capable of being loaded onto the LTR but not the Env gene in the absence of BRG1 in C33A cells. This result indicates that BRG1-associated SWI/SNF is needed for Pol II to move through the entire length of the HTLV-1 DNA. Collectively, these results show that the HTLV-1 full-length infectious clone can replicate in cells that contain wild-type BRG1 protein.
FIG. 6.
Lack of HTLV-1 viral replication in BRG1 mutant cells. (A) 293T cells, used as a control, were transfected with the HTLV-1 wild-type clone, ACH.WT. Supernatants from cells infected with HTLV-1 MT2 (20 μl) and treated with TNF or from 293T cells transfected with an empty vector (supernatants after 4 days) served as positive and negative controls, respectively. Optimal HTLV-1 replication occurred on day 3. (B and C) Both SW13 and C33A BRG1 mutant cells were transfected with either the pcDNA3 vector (data not shown) or the ACH.WT plasmid (20 μg), and experiments were carried out to day 12. Day 1 is a negative control with no transfection. Viral RT was present only in the supernatants of BRG1-transfected cells and peaked at day 12 in either cell type. All experiments in panels B and C were performed in triplicate. Western blotting for BRG1 and actin of samples from SW13 or C33A cells transfected with a mock plasmid (left lanes 1, 3, 9, and 12) or a BRG1 plasmid on days 3, 6, 9, and 12 (right lanes and insets). (D) C33A cells from panel C at day 12 were processed for ChIP experiments using either an IgG control or the anti-Pol II antibody 8WG16. The LTR primers spanned the region from −350 to +250 (U3/R/U5), and the Env primers spanned the region from +8990 to +9450.
The BAF/PBAF complex is the BRG1-associated complex responsible for Tax-activated transcription.
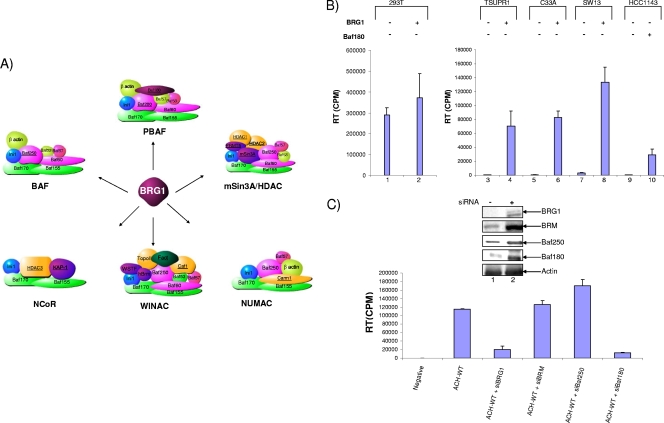
BRG1 uses energy derived from ATP hydrolysis to activate a number of complexes toward chromatin remodeling. The human SWI/SNF complex (BAF/PBAF) is one of these complexes; however, four additional complexes, WINAC, NUMAC, NCoR, and mSin3A/HDAC, also utilize BRG1 as their ATPase subunit. These complexes generally act either to repress or to activate chromatin remodeling. NUMAC acts as a transcriptional activator, while NCoR and mSin3A/HDAC act as transcriptional repressors (35, 60, 64, 70). Figure 7A shows the six complexes that remodel chromatin through BRG1 and further shows the specific subunits needed in each complex. These complexes share a number of subunits, including both core subunits (Baf170, Baf155, and Ini1) and those with other functional roles, such as Baf250, Baf60, and Baf53, which are known to be associated with nuclear receptors. While this report focuses on BRG1, we additionally examined SWI/SNF as a potential BRG1-utilizing complex in Tax transactivation. The BAF and PBAF complexes are very similar; they differ only in three subunits, of which Baf250 is specific to BAF and Baf180 and Baf200 are PBAF specific. The subunits seen in Fig. 7A show the general components needed for the BAF and PBAF complexes; however, the makeup of these complexes can vary. For example, in embryonic stem cells, the SWI/SNF complexes are functional but have low levels of BRM, Baf170, and Baf250 (28). Additionally, the BAF complex can function with either BRG1 or BRM as its ATPase subunit (27).
FIG. 7.
Effects of various siRNAs against SWI/SNF components on the wild-type HTLV-1 LTR. (A) BRG1-associated complexes. Brahma-related gene-1 (BRG1) is a catalytic subunit found in a variety of chromatin-remodeling complexes, including the SWI/SNF complexes BAF and PBAF, as well as WINAC, NCoR, mSin3A/HDAC, and NUMAC. The various complexes share other subunits that have a variety of activities, color coded as follows: green, core components; pink, nuclear receptor association; gold, DNA replication; light green, actin-related complexes; blue, Ini1/Baf47/SNF5; purple, other functions (63). As chromatin remodelers, these complexes can act either as transcriptional activators, as transcriptional repressors, or as both. For example, NUMAC acts as a transcriptional activator, while the mSin3A/HDAC complex acts as a transcriptional repressor. SWI/SNF complexes can act as either repressors or activators. (B) RT assay of supernatants (10 μl) from 293T cells (positive control), BRG1 mutant (TSUPR1, C33A, and SW13) cells, and Baf180 mutant (HCC1143) cells. Cells were transfected with the ACH.WT plasmid (20 μg) and were grown for 6 days. Supernatants were collected at day 6 and were then processed for the RT assay. BRG1 (10 μg), along with ACH.WT, was also transfected into five cell types (lanes 2, 4, 6, 8, and 10). (C) 293T cells were transfected with various siRNAs, including siBRG1, siBRM, siBaf250, and siBaf180 (300 nM), along with ACH.WT (20 μg). Samples were processed for RT at day 6. (Inset) Cell pellets were processed for Western blotting with antibodies against BRG1 and various other components. The actin panel represents cells transfected with siBRG1. Data represent one replicate of three independent experiments.
We asked if the wild-type HTLV-1 clone could replicate in either BRG1 mutant or PBAF mutant cell lines. For this purpose, we transfected the 293T, SW13, C33A, TSUPR1 (BRG1 mutant), and HCC1143 (Baf180 mutant) cell lines with an ACH.WT plasmid and collected supernatants at various time points up to 6 days. Transfection of ACH.WT in 293T cells produced increasing amounts of RT after 6 days and served as a positive control for these experiments. We further added a wild-type BRG1 construct and observed a slight increase in RT activity in the supernatant (Fig. 7B, lane 2). Like SW13 cells, TSUPR1 cells are deficient in both BRG1 and BRM; however, they do express core components of SWI/SNF complexes (67). As expected from previous results, the BRG1 mutant cell lines produced undetectable levels of RT when transfected with a full-length HTLV-1 plasmid (Fig. 7B, lanes 3, 5, 7, and 9). Interestingly, the Baf180 mutant cell line also produced no detectable levels of RT. All of these experiments produced similar results and were reproduced in three independent experiments. However, once wild-type BRG1 was transfected along with the ACH.WT clone, we observed an increase in RT levels in the supernatants (although these were never as high as the levels seen in the 293T cells). Furthermore, addition of the supernatants from transfected BRG1 mutant cells or BAF mutant cells to uninfected 293T cells produced no detectable virus after 5 to 6 days of culture (data not shown). Also, we tested the BRG1 mutant cells for the presence of the HTLV-1 proviral clone and observed integration of the viral DNA in these cells (data not shown). Overall, these results indicate that one or both of the BRG1 complexes (BAF or PBAF) are required for HTLV-1 production in cells.
BAF and PBAF complexes have been known to act as antagonists against each other: one complex activates transcription by remodeling chromatin, while the other ensures that chromatin positioning remains unchanged (28). We decided to compare the impacts of the BAF and PBAF complexes by observing the effects of knockdown on subunits that are expressed only in either the BAF complex (Baf250) or the PBAF complex (Baf180). To do this, we transfected various siRNAs into 293T cells along with the ACH.WT clone. The siRNAs included siBRG1, siBRM, siBaf250, and siBaf180. The results of such an experiment are shown in Fig. 7C: RT levels in the supernatants of siBRG1- or siBaf180-transfected cells dropped after 6 days. Western blot analysis of these transfected cells showed a >75% drop in the level of BRG1 or various associated Bafs. Collectively, these results indicate that the SWI/SNF complex PBAF is required both for HTLV-1 transcriptional activation and for viral production.
DISCUSSION
In the present report we show not only that BRG1 is required for Tax-activated transcription but also that the SWI/SNF complex PBAF is most likely the BRG1-utilizing complex for HTLV-1 Tax transactivation. We observed that Tax/SWI/SNF and Tax/CBP/p300 associate tightly in HTLV-1-infected cells, suggesting that BRG1 is involved in both chromatin remodeling and transcriptional initiation (Fig. 1). Although we have previously observed the presence of Tax in a 1.8-MDa complex by size exclusion chromatography of a C8166 total-cell lysate (68), here our chromatography technique allowed for the dissociation of weak complexes from the nuclear extracts so that only strong complexes that are salt resistant would remain intact. Also, this is the first time that we have seen the presence of Tax in such a high-molecular-weight fraction with BRG1. We are currently performing proteomic assays on fractions 15 and 21 of the Tax-associated complexes, and preliminary data indicate that fraction 15 may be a large complex partly due to the presence of phosphorylated Pol II with components of the RNA splicing machinery complexed with Tax (data not shown).
Using reconstituted nucleosome arrays, we determined that the combination of Tax and SWI/SNF increases the rate of chromatin remodeling over that with SWI/SNF alone (Fig. 2). We determined that Tax, BRG1, and acetylated c-Myb (ac-c-Myb) were recruited to the LTR, followed by the recruitment of p300 and TBP (a hallmark of activated transcription and Pol II recruitment), and finally other indicators of activated transcription were seen, including Pol II, methylated H3 (met-H3) (K9), and ac-H3 (K9). Furthermore, without Tax, there are no obvious DNase I-hypersensitive sites on the HTLV-1 promoter; however, the addition of Tax clearly indicates nucleosome positioning (Fig. 4). We also found that histones H1, H2A, and H3 were seemingly removed at the initiation site. Experiments using cells deficient in BRG1 showed that the ATPase subunit is required for both Tax transactivation and viral replication (Fig. 5). Finally, we observed that the SWI/SNF complex PBAF appears to be the complex required for nucleosome remodeling when either Baf180 mutant cells or a siRNA against the specific BAF (Baf250) or PBAF (Baf180 and Baf200) complex is used. Collectively, our data indicate that Tax requires BRG1 chromatin remodelers for activation of the HTLV-1 LTR through the modification of nucleosomes.
A 2006 paper by Zhang et al. states that BRG1 is not required for HTLV-1 transactivation by Tax (75). We believe that these results inaccurately describe the need for BRG1 in Tax activation, due to a number of discrepancies in these investigators' methods. First, the group used the lentivirus vector SMPU-18x21-EGFP, which contains 18 repeats of the Tax-responsive 21-bp repeats on the promoters. While this group showed that the vector is more Tax-GFP responsive than similar vectors that contain the natural 21 repeats of the Tax-responsive repeats, we believe that this does not accurately resemble the HTLV-1 genome, and therefore, results obtained by using these vectors probably do not produce useful information about viral transcription. They also utilized transient reporters in addition to a chromatinized template in their assays. Additionally, the group used the BRG1-deficient cell line SW13 to determine the necessity of BRG1 in Tax transactivation. As previously stated, the SW13 cell line produces no detectable levels of either BRM or BRG1, although the SWI/SNF complex still remains assembled in the SW13 cell line (47, 67). Therefore, results obtained from SW13 cells cannot be regarded as results from SWI/SNF-deficient cells. While the group additionally used another BRG1-deficient cell line, TSUPR1 (data not shown), similar studies have shown that this cell line also has intact core components of the SWI/SNF complex (67). Furthermore, we have used another BRG1 mutant cell line, ALAB, which has a defined mutation (C → T) at nucleotide 1630 in exon 10 of BRG1 (a stop codon), resulting in the absence of detectable protein expression (68). This cell line was transfected with either the HTLV-1 LTR promoter or the cyclin D2 promoter (with or without Tax) with BRG1 and was found to be activated 8.7-fold (HTLV-1 promoter) and 6.2-fold (cyclin D2 promoter) (data not shown). Interestingly, the Lees lab and others, which initially characterized these cells, also showed the downregulation of a number of cellular genes that are activated by Tax (12, 22). Furthermore, Zhang et al. failed to titrate their vectors in many of their experiments. It is well known in the field that viral activators such as Tax follow a “bell-shaped” curve for their activated transcription. Low concentrations of Tax would show low levels of activated transcription; medium levels of Tax show high, optimal transcription; and high levels of Tax show suppression of transcription. This is evident for many viral activators, including HIV-1 Tat, simian virus 40 (SV40) T, hepatitis B virus (HBV) X, and herpes simplex virus (HSV) VP16, among others (F. Kashanchi, unpublished results).
Additionally, Lemasson et al. showed that BRG1 is removed from the promoter when wild-type Tax is used in a ChIP assay (40). Furthermore, they showed that both histone H1 and histone H3 levels were lowered when Tax was added to the cells. In agreement with the latter results, we have also seen a drop in levels of both histone H1 and histone H3 from the HTLV-1 promoter when we used a wild-type virus construct in 293T cells and PBMCs (Fig. 4) (i.e., conversion of an unorganized, ordered array containing multiple nucleosomes to an ordered array at the transcription start site). However, we have consistently observed lower levels of BRG1 when Tax is transfected into cells. This is observed in a number of different cell types, including 293T and CHOK1-Luc cells, among others (data not shown). Interestingly, Lemasson et al. did not look for BRG1 expression after Tax transfection; however, Zhang et al. did notice (although they did not describe) a drop in the level of BRG1 after Tax transfection. Therefore, we believe that the ChIP results of Lemasson et al. showing the presence of lower levels of BRG1 on the HTLV-1 promoter after Tax transfection reflect the overall lower levels of BRG1 in CHOK1-Luc cells. When one accounts for the loss of BRG1 levels, one can see no change in the level of BRG1 uploaded onto the HTLV-1 promoter in the presence of Tax. We are currently investigating a possible binding of Tax/RB/BRG1 to the proteasome complex in the cytoplasm and the degradation of one or more BRG1-associated complexes in HTLV-1-infected cells. Previously published data indicate the presence of a classical LXCXE motif in the BRG1 protein (8, 14).
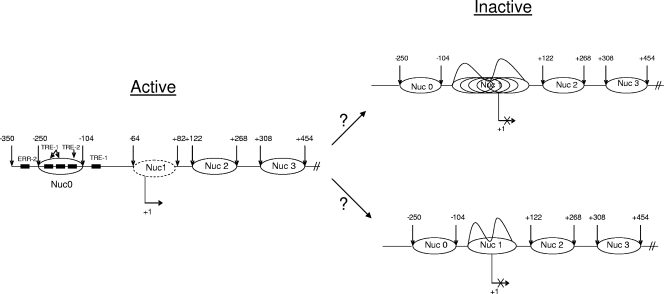
Our data on the inactive and Tax-activated HTLV-1 LTR have pointed toward the following model of nucleosomal arrays. The hypoxanthine phosphoribosyltransferase (HPRT) gene is subject to X chromosome inactivation, a process that leads to the transcriptional silencing of genes on one of the two X chromosomes in each female somatic cell. This results in the presence of both a transcriptionally active and a transcriptionally inactive HPRT allele within each female nucleus. On the active allele, the promoter region in vivo is unmethylated, is relatively sensitive to DNase I, and contains a DNase I-hypersensitive site that maps to the 5′ flanking region. In contrast, the inactive promoter is densely methylated, is resistant to DNase I, and does not exhibit DNase I hypersensitivity or detectable transcription factor binding in vivo. Our results using HTLV-1 integrated chromatin indicate that the Tax-activated promoter was assembled into an ordered array of translationally positioned nucleosomes, which was interrupted over a 350-bp region that showed increased accessibility to DNase I and that contained the entire functional promoter. In contrast, the inactive promoter was relatively inaccessible to nuclease and was not assembled into a translationally positioned nucleosomal array (Fig. 8). Collectively, our results (although no DNA methylation experiments were performed here) point to similarities with cellular genes such as the HPRT gene, where inefficient access to DNA binding sites may be responsible for little to no activated transcription. Once a viral activator, such as Tax, is recruited to the viral promoter, there appear to be removal of some of the histones and a more-ordered array where basal factors along with Pol II can transcribe through DNA. Further experiments will determine which factor(s) is responsible for the removal of the histones at the transcription start site. Finally, our results clearly show that BRG1 and SWI/SNF complexes are needed for activated transcription and viral replication. This is especially evident when wild-type infectious clones of HTLV-1 are used. It would therefore be interesting to determine which mutant viral clones are responsible for the observed BRG1 effects. Current experiments in our lab are aimed at addressing these related questions.
FIG. 8.
Model of nucleosomal arrays on the Tax-activated HTLV-1 LTR in vivo. (Left) In the presence of Tax, there are well-defined translationally positioned nucleosomal arrays on the active promoter, as seen by the presence of DNase I-hypersensitive sites and the removal of histones H1, H2A, and H3 at the transcriptional start site region. (Right) DNase I cleavage analysis indicates that the inactive promoter does not contain translationally positioned nucleosomal arrays. This is indicated by the lack of DNase I-hypersensitive sites and the presence of all core histones as well as H1. Currently it is not clear whether the Nuc 1 area at the transcriptional start site contains multiple arrays that are not phased (top right) or whether there is only one nucleosome that makes the DNA inaccessible for transcription factor and/or Pol II occupancy (bottom right). The arrows at both ends of each nucleosome represent the beginning of the hypersensitive site. The removal of some of the histones is shown as a dotted oval around Nuc1.
Acknowledgments
We thank Jin-Tang Dong of Emory University for the TSU-PR1 cell line. We also thank Jennifer Nyborg of Colorado State University for the CHOK1-Luc cell line. Additionally, we are grateful to Lee Ratner (Washington University, St. Louis, MO) for the donation of the ACH.WT plasmid. The initial in vitro transactivation experiments with Tax and SWI/SNF were performed in the lab of John N. Brady (deceased) at NIH/NCI by Meishing Zhou (NIH/NCI), who graciously shared the unpublished data with us.
This work was supported by grants from the George Washington University (REF funds to F.K. and Akos Vertes) and by NIH grants AI44357, AI43894, and AI46459 to F.K.
Footnotes
Published ahead of print on 17 February 2010.
REFERENCES
- 1.Agbottah, E., L. Deng, L. O. Dannenberg, A. Pumfery, and F. Kashanchi. 2006. Effect of SWI/SNF chromatin remodeling complex on HIV-1 Tat activated transcription. Retrovirology 3:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akagi, T., H. Ono, and K. Shimotohno. 1996. Expression of cell-cycle regulatory genes in HTLV-I infected T-cell lines: possible involvement of Tax1 in the altered expression of cyclin D2, p18Ink4 and p21Waf1/Cip1/Sdi1. Oncogene 12:1645-1652. [PubMed] [Google Scholar]
- 3.Bannister, A. J., and T. Kouzarides. 1996. The CBP co-activator is a histone acetyltransferase. Nature 384:641-643. [DOI] [PubMed] [Google Scholar]
- 4.Brower-Toland, B., D. A. Wacker, R. M. Fulbright, J. T. Lis, W. L. Kraus, and M. D. Wang. 2005. Specific contributions of histone tails and their acetylation to the mechanical stability of nucleosomes. J. Mol. Biol. 346:135-146. [DOI] [PubMed] [Google Scholar]
- 5.Caron, C., R. Rousset, C. Beraud, V. Moncollin, J. M. Egly, and P. Jalinot. 1993. Functional and biochemical interaction of the HTLV-I Tax1 transactivator with TBP. EMBO J. 12:4269-4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clemens, K. E., G. Piras, M. F. Radonovich, K. S. Choi, J. F. Duvall, J. DeJong, R. Roeder, and J. N. Brady. 1996. Interaction of the human T-cell lymphotropic virus type 1 Tax transactivator with transcription factor IIA. Mol. Cell. Biol. 16:4656-4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colgin, M. A., and J. K. Nyborg. 1998. The human T-cell leukemia virus type 1 oncoprotein Tax inhibits the transcriptional activity of c-Myb through competition for the CREB binding protein. J. Virol. 72:9396-9399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dahiya, A., M. R. Gavin, R. X. Luo, and D. C. Dean. 2000. Role of the LXCXE binding site in Rb function. Mol. Cell. Biol. 20:6799-6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de La Fuente, C., L. Deng, F. Santiago, L. Arce, L. Wang, and F. Kashanchi. 2000. Gene expression array of HTLV type 1-infected T cells: up-regulation of transcription factors and cell cycle genes. AIDS Res. Hum. Retrovir. 16:1695-1700. [DOI] [PubMed] [Google Scholar]
- 10.de La Fuente, C., F. Santiago, S. Y. Chong, L. Deng, T. Mayhood, P. Fu, D. Stein, T. Denny, F. Coffman, N. Azimi, R. Mahieux, and F. Kashanchi. 2000. Overexpression of p21waf1 in human T-cell lymphotropic virus type 1-infected cells and its association with cyclin A/cdk2. J. Virol. 74:7270-7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de la Serna, I. L., and A. N. Imbalzano. 2002. Unfolding heterochromatin for replication. Nat. Genet. 32:560-562. [DOI] [PubMed] [Google Scholar]
- 12.de la Serna, I. L., Y. Ohkawa, C. A. Berkes, D. A. Bergstrom, C. S. Dacwag, S. J. Tapscott, and A. N. Imbalzano. 2005. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol. Cell. Biol. 25:3997-4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng, L., C. de la Fuente, P. Fu, L. Wang, R. Donnelly, J. D. Wade, P. Lambert, H. Li, C. G. Lee, and F. Kashanchi. 2000. Acetylation of HIV-1 Tat by CBP/P300 increases transcription of integrated HIV-1 genome and enhances binding to core histones. Virology 277:278-295. [DOI] [PubMed] [Google Scholar]
- 14.Dunaief, J. L., B. E. Strober, S. Guha, P. A. Khavari, K. Alin, J. Luban, M. Begemann, G. R. Crabtree, and S. P. Goff. 1994. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell 79:119-130. [DOI] [PubMed] [Google Scholar]
- 15.Flavin, M., L. Cappabianca, C. Kress, H. Thomassin, and T. Grange. 2004. Nature of the accessible chromatin at a glucocorticoid-responsive enhancer. Mol. Cell. Biol. 24:7891-7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujii, M., T. Niki, T. Mori, T. Matsuda, M. Matsui, N. Nomura, and M. Seiki. 1991. HTLV-1 Tax induces expression of various immediate early serum responsive genes. Oncogene 6:1023-1029. [PubMed] [Google Scholar]
- 17.Gachon, F., S. Thebault, A. Peleraux, C. Devaux, and J. M. Mesnard. 2000. Molecular interactions involved in the transactivation of the human T-cell leukemia virus type 1 promoter mediated by Tax and CREB-2 (ATF-4). Mol. Cell. Biol. 20:3470-3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallo, R. C. 2005. History of the discoveries of the first human retroviruses: HTLV-1 and HTLV-2. Oncogene 24:5926-5930. [DOI] [PubMed] [Google Scholar]
- 19.Giebler, H. A., J. E. Loring, K. van Orden, M. A. Colgin, J. E. Garrus, K. W. Escudero, A. Brauweiler, and J. K. Nyborg. 1997. Anchoring of CREB binding protein to the human T-cell leukemia virus type 1 promoter: a molecular mechanism of Tax transactivation. Mol. Cell. Biol. 17:5156-5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrod, R., Y. L. Kuo, Y. Tang, Y. Yao, A. Vassilev, Y. Nakatani, and C. Z. Giam. 2000. p300 and p300/cAMP-responsive element-binding protein associated factor interact with human T-cell lymphotropic virus type-1 Tax in a multi-histone acetyltransferase/activator-enhancer complex. J. Biol. Chem. 275:11852-11857. [DOI] [PubMed] [Google Scholar]
- 21.Harrod, R., Y. Tang, C. Nicot, H. S. Lu, A. Vassilev, Y. Nakatani, and C. Z. Giam. 1998. An exposed KID-like domain in human T-cell lymphotropic virus type 1 Tax is responsible for the recruitment of coactivators CBP/p300. Mol. Cell. Biol. 18:5052-5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hendricks, K. B., F. Shanahan, and E. Lees. 2004. Role for BRG1 in cell cycle control and tumor suppression. Mol. Cell. Biol. 24:362-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Höllsberg, P. 1999. Mechanisms of T-cell activation by human T-cell lymphotropic virus type I. Microbiol. Mol. Biol. Rev. 63:308-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeong, S. J., H. Lu, W. K. Cho, H. U. Park, C. Pise-Masison, and J. N. Brady. 2006. Coactivator-associated arginine methyltransferase 1 enhances transcriptional activity of the human T-cell lymphotropic virus type 1 long terminal repeat through direct interaction with Tax. J. Virol. 80:10036-10044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jewell, N. A., and L. M. Mansky. 2005. Construction and characterization of deltaretrovirus indicator cell lines. J. Virol. Methods 123:17-24. [DOI] [PubMed] [Google Scholar]
- 26.Jiang, H., H. Lu, R. L. Schiltz, C. A. Pise-Masison, V. V. Ogryzko, Y. Nakatani, and J. N. Brady. 1999. PCAF interacts with Tax and stimulates Tax transactivation in a histone acetyltransferase-independent manner. Mol. Cell. Biol. 19:8136-8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kadam, S., and B. M. Emerson. 2003. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol. Cell 11:377-389. [DOI] [PubMed] [Google Scholar]
- 28.Kaeser, M. D., A. Aslanian, M. Q. Dong, J. R. Yates III, and B. M. Emerson. 2008. BRD7, a novel PBAF-specific SWI/SNF subunit, is required for target gene activation and repression in embryonic stem cells. J. Biol. Chem. 283:32254-32263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kashanchi, F., and J. N. Brady. 2005. Transcriptional and post-transcriptional gene regulation of HTLV-1. Oncogene 24:5938-5951. [DOI] [PubMed] [Google Scholar]
- 30.Kashanchi, F., J. F. Duvall, R. P. Kwok, J. R. Lundblad, R. H. Goodman, and J. N. Brady. 1998. The coactivator CBP stimulates human T-cell lymphotropic virus type I Tax transactivation in vitro. J. Biol. Chem. 273:34646-34652. [DOI] [PubMed] [Google Scholar]
- 31.Kehn, K., R. Berro, C. de la Fuente, K. Strouss, E. Ghedin, S. Dadgar, M. E. Bottazzi, A. Pumfery, and F. Kashanchi. 2004. Mechanisms of HTLV-1 transformation. Front. Biosci. 9:2347-2372. [DOI] [PubMed] [Google Scholar]
- 32.Kehn, K., L. Deng, C. de la Fuente, K. Strouss, K. Wu, A. Maddukuri, S. Baylor, R. Rufner, A. Pumfery, M. E. Bottazzi, and F. Kashanchi. 2004. The role of cyclin D2 and p21/waf1 in human T-cell leukemia virus type 1 infected cells. Retrovirology 1:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khavari, P. A., C. L. Peterson, J. W. Tamkun, D. B. Mendel, and G. R. Crabtree. 1993. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature 366:170-174. [DOI] [PubMed] [Google Scholar]
- 34.Kimata, J. T., F. H. Wong, J. J. Wang, and L. Ratner. 1994. Construction and characterization of infectious human T-cell leukemia virus type 1 molecular clones. Virology 204:656-664. [DOI] [PubMed] [Google Scholar]
- 35.Kitagawa, H., R. Fujiki, K. Yoshimura, Y. Mezaki, Y. Uematsu, D. Matsui, S. Ogawa, K. Unno, M. Okubo, A. Tokita, T. Nakagawa, T. Ito, Y. Ishimi, H. Nagasawa, T. Matsumoto, J. Yanagisawa, and S. Kato. 2003. The chromatin-remodeling complex WINAC targets a nuclear receptor to promoters and is impaired in Williams syndrome. Cell 113:905-917. [DOI] [PubMed] [Google Scholar]
- 36.Kouzarides, T. 2007. Chromatin modifications and their function. Cell 128:693-705. [DOI] [PubMed] [Google Scholar]
- 37.Kwok, R. P., M. E. Laurance, J. R. Lundblad, P. S. Goldman, H. Shih, L. M. Connor, S. J. Marriott, and R. H. Goodman. 1996. Control of cAMP-regulated enhancers by the viral transactivator Tax through CREB and the co-activator CBP. Nature 380:642-646. [DOI] [PubMed] [Google Scholar]
- 38.Lee, D., J. W. Kim, T. Seo, S. G. Hwang, E. J. Choi, and J. Choe. 2002. SWI/SNF complex interacts with tumor suppressor p53 and is necessary for the activation of p53-mediated transcription. J. Biol. Chem. 277:22330-22337. [DOI] [PubMed] [Google Scholar]
- 39.Lee, T. I., and R. A. Young. 2000. Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 34:77-137. [DOI] [PubMed] [Google Scholar]
- 40.Lemasson, I., N. J. Polakowski, P. J. Laybourn, and J. K. Nyborg. 2006. Tax-dependent displacement of nucleosomes during transcriptional activation of human T-cell leukemia virus type 1. J. Biol. Chem. 281:13075-13082. [DOI] [PubMed] [Google Scholar]
- 41.Lemasson, I., N. J. Polakowski, P. J. Laybourn, and J. K. Nyborg. 2004. Transcription regulatory complexes bind the human T-cell leukemia virus 5′ and 3′ long terminal repeats to control gene expression. Mol. Cell. Biol. 24:6117-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lenzmeier, B. A., H. A. Giebler, and J. K. Nyborg. 1998. Human T-cell leukemia virus type 1 Tax requires direct access to DNA for recruitment of CREB binding protein to the viral promoter. Mol. Cell. Biol. 18:721-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Logie, C., and C. L. Peterson. 1997. Catalytic activity of the yeast SWI/SNF complex on reconstituted nucleosome arrays. EMBO J. 16:6772-6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Low, K. G., L. F. Dorner, D. B. Fernando, J. Grossman, K. T. Jeang, and M. J. Comb. 1997. Human T-cell leukemia virus type 1 Tax releases cell cycle arrest induced by p16INK4a. J. Virol. 71:1956-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mahmoudi, T., M. Parra, R. G. Vries, S. E. Kauder, C. P. Verrijzer, M. Ott, and E. Verdin. 2006. The SWI/SNF chromatin-remodeling complex is a cofactor for Tat transactivation of the HIV promoter. J. Biol. Chem. 281:19960-19968. [DOI] [PubMed] [Google Scholar]
- 46.Matsuoka, M., and K. T. Jeang. 2007. Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation. Nat. Rev. Cancer 7:270-280. [DOI] [PubMed] [Google Scholar]
- 47.Muchardt, C., and M. Yaniv. 1993. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 12:4279-4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neuveut, C., K. G. Low, F. Maldarelli, I. Schmitt, F. Majone, R. Grassmann, and K. T. Jeang. 1998. Human T-cell leukemia virus type 1 Tax and cell cycle progression: role of cyclin D-cdk and p110Rb. Mol. Cell. Biol. 18:3620-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nicot, C., R. Mahieux, R. Opavsky, A. Cereseto, L. Wolff, J. N. Brady, and G. Franchini. 2000. HTLV-I Tax transrepresses the human c-Myb promoter independently of its interaction with CBP or p300. Oncogene 19:2155-2164. [DOI] [PubMed] [Google Scholar]
- 50.Ogryzko, V. V., R. L. Schiltz, V. Russanova, B. H. Howard, and Y. Nakatani. 1996. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87:953-959. [DOI] [PubMed] [Google Scholar]
- 51.Park, Y. J., J. V. Chodaparambil, Y. Bao, S. J. McBryant, and K. Luger. 2005. Nucleosome assembly protein 1 exchanges histone H2A-H2B dimers and assists nucleosome sliding. J. Biol. Chem. 280:1817-1825. [DOI] [PubMed] [Google Scholar]
- 52.Peloponese, J. M., Jr., T. Kinjo, and K. T. Jeang. 2007. Human T-cell leukemia virus type 1 Tax and cellular transformation. Int. J. Hematol. 86:101-106. [DOI] [PubMed] [Google Scholar]
- 53.Phelan, M. L., S. Sif, G. J. Narlikar, and R. E. Kingston. 1999. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell 3:247-253. [DOI] [PubMed] [Google Scholar]
- 54.Poiesz, B. J., F. W. Ruscetti, A. F. Gazdar, P. A. Bunn, J. D. Minna, and R. C. Gallo. 1980. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc. Natl. Acad. Sci. U. S. A. 77:7415-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poiesz, B. J., F. W. Ruscetti, J. W. Mier, A. M. Woods, and R. C. Gallo. 1980. T-cell lines established from human T-lymphocytic neoplasias by direct response to T-cell growth factor. Proc. Natl. Acad. Sci. U. S. A. 77:6815-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Randazzo, F. M., P. Khavari, G. Crabtree, J. Tamkun, and J. Rossant. 1994. brg1: a putative murine homologue of the Drosophila brahma gene, a homeotic gene regulator. Dev. Biol. 161:229-242. [DOI] [PubMed] [Google Scholar]
- 57.Robek, M. D., and L. Ratner. 1999. Immortalization of CD4(+) and CD8(+) T lymphocytes by human T-cell leukemia virus type 1 Tax mutants expressed in a functional molecular clone. J. Virol. 73:4856-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sano, Y., and S. Ishii. 2001. Increased affinity of c-Myb for CREB-binding protein (CBP) after CBP-induced acetylation. J. Biol. Chem. 276:3674-3682. [DOI] [PubMed] [Google Scholar]
- 59.Schwartz, B. E., and K. Ahmad. 2005. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev. 19:804-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sif, S., A. J. Saurin, A. N. Imbalzano, and R. E. Kingston. 2001. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 15:603-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suzuki, T., H. Hirai, and M. Yoshida. 1994. Tax protein of HTLV-1 interacts with the Rel homology domain of NF-κB p65 and c-Rel proteins bound to the NF-κB binding site and activates transcription. Oncogene 9:3099-3105. [PubMed] [Google Scholar]
- 62.Tréand, C., I. du Chene, V. Bres, R. Kiernan, R. Benarous, M. Benkirane, and S. Emiliani. 2006. Requirement for SWI/SNF chromatin-remodeling complex in Tat-mediated activation of the HIV-1 promoter. EMBO J. 25:1690-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Trotter, K. W., and T. K. Archer. 2008. The BRG1 transcriptional coregulator. Nucl. Recept. Signal. 6:e004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Underhill, C., M. S. Qutob, S. P. Yee, and J. Torchia. 2000. A novel nuclear receptor corepressor complex, N-CoR, contains components of the mammalian SWI/SNF complex and the corepressor KAP-1. J. Biol. Chem. 275:40463-40470. [DOI] [PubMed] [Google Scholar]
- 65.Wang, H., L. Wang, H. Erdjument-Bromage, M. Vidal, P. Tempst, R. S. Jones, and Y. Zhang. 2004. Role of histone H2A ubiquitination in Polycomb silencing. Nature 431:873-878. [DOI] [PubMed] [Google Scholar]
- 66.Wang, W., Y. Xue, S. Zhou, A. Kuo, B. R. Cairns, and G. R. Crabtree. 1996. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 10:2117-2130. [DOI] [PubMed] [Google Scholar]
- 67.Wong, A. K., F. Shanahan, Y. Chen, L. Lian, P. Ha, K. Hendricks, S. Ghaffari, D. Iliev, B. Penn, A. M. Woodland, R. Smith, G. Salada, A. Carillo, K. Laity, J. Gupte, B. Swedlund, S. V. Tavtigian, D. H. Teng, and E. Lees. 2000. BRG1, a component of the SWI-SNF complex, is mutated in multiple human tumor cell lines. Cancer Res. 60:6171-6177. [PubMed] [Google Scholar]
- 68.Wu, K., M. E. Bottazzi, C. de la Fuente, L. Deng, S. D. Gitlin, A. Maddukuri, S. Dadgar, H. Li, A. Vertes, A. Pumfery, and F. Kashanchi. 2004. Protein profile of tax-associated complexes. J. Biol. Chem. 279:495-508. [DOI] [PubMed] [Google Scholar]
- 69.Xiao, G., M. E. Cvijic, A. Fong, E. W. Harhaj, M. T. Uhlik, M. Waterfield, and S. C. Sun. 2001. Retroviral oncoprotein Tax induces processing of NF-κB2/p100 in T cells: evidence for the involvement of IKKα. EMBO J. 20:6805-6815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu, W., H. Cho, S. Kadam, E. M. Banayo, S. Anderson, J. R. Yates III, B. M. Emerson, and R. M. Evans. 2004. A methylation-mediator complex in hormone signaling. Genes Dev. 18:144-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yin, M. J., L. B. Christerson, Y. Yamamoto, Y. T. Kwak, S. Xu, F. Mercurio, M. Barbosa, M. H. Cobb, and R. B. Gaynor. 1998. HTLV-I Tax protein binds to MEKK1 to stimulate IκB kinase activity and NF-κB activation. Cell 93:875-884. [DOI] [PubMed] [Google Scholar]
- 72.Yin, M. J., E. J. Paulssen, J. S. Seeler, and R. B. Gaynor. 1995. Protein domains involved in both in vivo and in vitro interactions between human T-cell leukemia virus type I Tax and CREB. J. Virol. 69:3420-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yoshida, M. 2005. Discovery of HTLV-1, the first human retrovirus, its unique regulatory mechanisms, and insights into pathogenesis. Oncogene 24:5931-5937. [DOI] [PubMed] [Google Scholar]
- 74.Zhang, J., D. T. Scadden, and C. S. Crumpacker. 2007. Primitive hematopoietic cells resist HIV-1 infection via p21. J. Clin. Invest. 117:473-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang, L., M. Liu, R. Merling, and C. Z. Giam. 2006. Versatile reporter systems show that transactivation by human T-cell leukemia virus type 1 Tax occurs independently of chromatin remodeling factor BRG1. J. Virol. 80:7459-7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou, M., L. Deng, F. Kashanchi, J. N. Brady, A. J. Shatkin, and A. Kumar. 2003. The Tat/TAR-dependent phosphorylation of RNA polymerase II C-terminal domain stimulates cotranscriptional capping of HIV-1 mRNA. Proc. Natl. Acad. Sci. U. S. A. 100:12666-12671. [DOI] [PMC free article] [PubMed] [Google Scholar]