Abstract
Cauliflower mosaic virus (CaMV) is transmitted from plant to plant through a seemingly simple interaction with insect vectors. This process involves an aphid receptor and two viral proteins, P2 and P3. P2 binds to both the aphid receptor and P3, itself tightly associated with the virus particle, with the ensemble forming a transmissible viral complex. Here, we describe the conformations of both unliganded CaMV P3 protein and its virion-associated form. X-ray crystallography revealed that the N-terminal domain of unliganded P3 is a tetrameric parallel coiled coil with a unique organization showing two successive four-stranded subdomains with opposite supercoiling handedness stabilized by a ring of interchain disulfide bridges. A structural model of virus-liganded P3 proteins, folding as an antiparallel coiled-coil network coating the virus surface, was derived from molecular modeling. Our results highlight the structural and biological versatility of this coiled-coil structure and provide new insights into the molecular mechanisms involved in CaMV acquisition and transmission by the insect vector.
Cauliflower mosaic virus (CaMV) is the type member of the plant virus family Caulimoviridae. This family is grouped together with hepadnaviruses into the pararetrovirus group due to its mode of replication via reverse transcription of a pregenomic RNA intermediate (17). The CaMV genome is a double-stranded circular DNA of approximately 8,000 bp, comprising seven major open reading frames (ORFs), only six of which have clearly identified biological functions. The product of ORF VI (P6) is expressed early in infection from the monocistronic 19S RNA. In addition to its recently reported role in the suppression of RNA silencing (14), P6 rapidly self-aggregates into the so-called electron-dense inclusion bodies, demonstrated to be the viral factory (17). Within these structures, P6 then trans-activates the translation of ORFs I to V from the polycistronic pregenomic 35S RNA according to a complex reinitiation process (39). ORFs I to V encode the major capsid protein (P4, encoded by ORF IV), reverse transcriptase (P5, encoded by ORF V), and three auxiliary proteins: P1 (ORF I) is involved in cell-to-cell and long-distance within-plant movement, P2 (ORF II) is involved in aphid transmission, and P3 (ORF III) is tightly associated with the virus particles and has a complex regulatory function in the virus infection cycle, particularly during plant-to-plant vector transmission.
The CaMV viral particle is roughly spherical, 520 Å in diameter, with an icosahedral T7 symmetry. It is constituted of three concentric shells, built from 420 capsid protein subunits, surrounding a large inner cavity with a diameter of approximately 250 Å (4, 37). The physical association between P3 and viral particles has been consistently demonstrated in mature CaMV virions by copurification, immunolabeling experiments, and in vitro interactions (7, 23-25). Recently, cryo-electron microscopy (cryo-EM) image reconstruction further showed that P3 molecules decorate virions, with their N-terminal ectodomains forming an antiparallel α-helical coiled-coil network at the surface and their C-terminal domains interacting with the coat protein and reaching inside the virus particle. Although direct evidence is still lacking, the C terminus of P3 may be attached to the genome, which is packed between the intermediate and inner shells (37), through its previously characterized DNA binding domain (33).
The transmission of CaMV by insect vectors is categorized as “noncirculative” and is reminiscent of that of hundreds of plant virus species (35). The virus is quickly acquired from an infected host during feeding by the aphid vector, where it is retained in the mouthparts for a short time. From there, infectious CaMV units are simply released and inoculated into a new, healthy plant after movement of the vector, with no requirement for replication, or even cycling of the virus, within the vector body. The attachment of CaMV to the aphid is mediated by P2, the N-terminal domain of which specifically recognizes a cuticular protein receptor located at the extreme tip of the maxillary stylets (32, 47), whereas the C-terminal α-helix binds, via predicted coiled-coil structures, to the ectodomain of the P3 decorating the virions (16, 25, 37).
This antiparallel coiled-coil motif of P3 supporting P2 attachment at the virion surface has also been demonstrated to establish another, perhaps competitive, interaction with the α-helical coiled-coil segment of the C-terminal domain of P1, an association that has been shown to be mandatory for CaMV cell-to-cell movement (43). In addition to the two-stranded antiparallel conformation when anchored to virions (37), the N-terminal α-helical domain of P3 has been demonstrated to form a distinct coiled-coil structure as a parallel tetramer when free in solution (22). Consistently, a tetrameric form of P3 has been extracted from infected plants (45, 46) and also shown to exist in other species of the family Caulimoviridae (42), suggesting a different biological role for this conformation. The functional promiscuity of the P3 protein, correlated with its configurational heterogeneity, has attracted attention in recent years. However, the molecular bases underlying its different biological properties remain largely obscure, due partly to a lack of structural data.
Here, we report the first X-ray crystal structure of a CaMV protein, the virus-associated protein P3, at 2.6-Å resolution. The unliganded crystal structure reveals a novel segmented tetrameric helix bundle comprising two successive coiled-coil subdomains with opposite twist, stabilized by four interchain disulfide bridges. We also derive a model for the two-stranded antiparallel coiled-coil network of P3 decorating the viral particle compatible with our earlier cryo-EM reconstruction data (37). The results presented and discussed here show that the conformational versatility and adaptability of ubiquitous coiled-coil domains (28) can play a central regulatory role in the relationship between viruses and their insect vectors.
MATERIALS AND METHODS
Crystallization and data collection.
The recombinant P3 protein was expressed in the Escherichia coli BL21(DE3)/pLysS host strain using a pET3a vector and purified as described previously (37). The protein was further purified by fast-flow gel filtration chromatography on a Superdex 200 column. The crystallization conditions for the P3 protein were screened at room temperature using several commercially available crystallization screens (Emerald BioSystems, Hampton Research) in hanging-drop vapor diffusion setups. The optimal crystallization conditions were 16 to 18% polyethylene glycol (PEG) 2000, 0.1 mol·liter−1 Tris-HCl buffer at pH 6.8, and NaCl at 0.1 mol·liter−1; 0.1 ml of reservoir solution was dispensed in each of the 96-well plates over a 1-μl protein drop at 5 mg/ml mixed with 1 μl of reservoir buffer. These crystals diffract at low resolution (∼6 Å) and belong to space group P41212 (a = b = 105.3 Å, c = 79.9 Å). Efforts to obtain better-diffracting crystals under these conditions were unsuccessful. Two other crystal forms belonging to monoclinic and hexagonal systems were also successively identified with slightly different crystallization conditions: 25% PEG 1000 as a precipitating agent in 0.1 mol·liter−1 morpholineethanesulfonic acid (MES)-NaOH buffer (pH = 6.5), 0.1 mol·liter−1 NaCl, and a 1.2 molar excess of DNA oligonucleotide [poly(AT); 14 bp]. These crystals belonged to space groups P64 (a = b = 104.94 Å, c = 72.53 Å) and P21 (a = 69.30 Å, b = 28.82 Å, c = 75.96 Å, and β = 92.08°). They diffracted to resolutions of 3.1 Å and 2.6 Å, respectively, using European Synchrotron Radiation Facility (ESRF) (Grenoble, France) beam lines ID23-2 and ID14-3 and a Q4-R Quantum charge-coupled device (CCD) detector. The crystals were cryoprotected by being either soaked in glycerol or coated by oil, and the data were collected at 0.93-Å wavelength under cryogenic conditions. Image data were processed and scaled using the programs MOSFLM (26) and SCALA of the CCP4 suite (3). The 180 images of the hexagonal crystal collected were also processed into 3 separate zones in order to evaluate the strength of the signal using the radiation damage-induced phasing (RIP) method (53).
Structure determination and refinement.
As our attempts to prepare selenium-substituted protein and heavy-atom derivatives failed, the structure of the P3 protein was determined using molecular replacement. This protein has no known structural homologs. However, sequence analyses revealed the presence of coiled-coil motifs, subsequently confirmed by circular-dichroism measurements of the purified protein (data not shown). The parallel tetrameric organization was also proposed from molecular-modeling studies (22). Initial phases of the hexagonal crystal form were determined by exploratory molecular replacement using different canonical coiled-coil models (5). The search models were short parallel coiled-coiled structural motifs: tetramers (Sendai virus phosphoprotein [Protein Data Bank {PDB} entry 1ezj], NSP4 [1g1i], tetrabrachion [1fe6], and VASP [1use]) and dimers (geminin [1t6f] and hepatitis delta antigen [1a92]). The length of the coiled-coil poly-Ser models was limited to 32 residues, and no Cα steric clashes were allowed during packing validation. All models were searched against the P3 data set in both P62 and P64 enantiomorphic space groups. The best solution was obtained for space group P64, with two 1t6f template models using the program PHASER (31) and data in the resolution range 20 to 3.1 Å (translation function Z [TFZ] score = 10.1; log likelihood gain [LLG] = 228). This partial model represented ∼38% of the scattering of the nonhydrogen atoms of the refined P3 structure. Density modification methods, including solvent flattening, histogram matching, and noncrystallographic symmetry (NCS) averaging, were performed in the program DM (3). The quality of the improved density map (see Fig. S1 in the supplemental material) was sufficient to trace a significant part of the missing polypeptide chain and to identify side chain residues. The final model was obtained with iterative cycles of model building using the program COOT (8) and density modification and refinement with REFMAC5 (34), incorporating NCS restraints and translation-libration-screw (TLS) anisotropic components. This model refined to an R factor of 22.2% (Rfree = 28.7%) at 3.1-Å resolution. The radiation-induced-damage signal was too weak to start the phasing process. Nevertheless, using the final phases from the refined model, inspection of the Fourier difference RIP map showed significant peaks corresponding to the disulfide bridges and carboxyl groups of acidic residues (see Fig. S2 in the supplemental material).
The structure of the monoclinic crystal form was determined by molecular replacement with PHASER using the final P64 crystal structure of the P3 tetramer as the search model (TFZ score = 18.4; LLG = 898). The coordinates were refined against data up to 2.6-Å resolution using REFMAC5. Later cycles included overall anisotropic refinement with four TLS groups. The final model refined to an R factor of 21.9% (Rfree = 28.9% for a test set of 7% randomly chosen reflections). Structure quality was monitored with the PROCHECK and SFCHECK programs (3, 48). Table 1 summarizes the data collection, refinement statistics, and model quality. It was found that 98.1% and 1.9% of the residues were in the most favored and allowed regions of the Ramachandran plot, respectively.
TABLE 1.
Data collection and refinement statistics of the CaMV P3 crystals
| Parameter | Hexagonal form | Monoclinic form |
|---|---|---|
| Data collection | ||
| Space group | P64 | P21 |
| Unit cell dimensions | ||
| a, b, c (Å) | 104.94, 104.94, 72.53 | 69.30, 28.82, 75.96 |
| α, β, γ (°) | 90, 90, 120 | 90, 92.08, 90 |
| Resolution range (Å)a | 20.85-3.10 (3.10-3.27) | 18.98-2.59 (2.59-2.74) |
| Rmerge on I (%)b | 0.065 (0.306) | 0.074 (0.302) |
| Average I /σI | 16.5 (4.6) | 14.4 (3.6) |
| Completeness (%) | 99.9 (99.9) | 96.1 (99.2) |
| Redundancy | 5.4 (5.5) | 4.3 (4.4) |
| Refinement | ||
| Unique reflections (free) | 8,336 (918) | 8,660 (657) |
| Rwork/Rfreec | 0.222/0.287 | 0.219/0.289 |
| No. of protein atoms | 2,203 | 2,161 |
| No. of water molecules/ions | 30 | |
| Average B factor (Å2) | 34.6 | 69.3 |
| Rmsd of bond lengths (Å) | 0.008 | 0.010 |
| Rmsd of bond angles (°) | 1.06 | 1.25 |
| Ramachandran plot (%) (favored/allowed/ disallowed) | 97.7/2.3/0.0 | 98.1/1.9/0.0 |
The values in parentheses are the statistics for the highest-resolution shell.
Rmerge = ΣhΣi |Ii(h) − <I(h)> |/ΣhΣi Ii(h), where Ii(h) and <I(h)> are the ith and mean measurements of the intensity of reflection, h, respectively.
Rwork = Σh ||Fo(h)| − |Fc(h)||/Σh |Fo(h)|, where Fo(h) and Fc(h) are the observed and calculated structure factors, respectively. No I/σ cutoff was used in the calculations of R factors. Rfree = Rwork calculated using randomly chosen reflection data and omitted from the start of refinement.
Structure analysis.
Coiled-coil parameters were calculated with the programs TWISTER (44) and SOCKET (49). Knobs-into-holes interactions were revealed with packing cutoffs of 7 Å and 8 Å for coiled-coil subdomains 1 and 2, respectively. P3 oligomer interfaces were analyzed with the program AREAIMOL (3) and using the PISA service (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html). Cavity calculations were performed with VOIDOO (19). The figures were generated using CHIMERA (http://www.cgl.ucsf.edu/chimera/).
Molecular modeling and fitting of the P3 model to the cryo-EM reconstruction.
The molecular models of the antiparallel coiled coil of P3 (residues 4 to 59) were built by homology modeling using a canonical two-stranded coiled coil (PDB entry 1aqt) as a template. The GP segment (residues 33 to 37) joining the two α-helices was modeled and optimized with Modloop (10), and molecular models were energy minimized using the MODELLER program (9). The antiparallel dimeric coiled-coil ectodomain P3 models were placed manually into a triskelion of the P3-decorated CaMV cryo-EM map published previously at 26-Å resolution (37) in the arbitrarily chosen T=7l icosahedral lattice. At this resolution, the elongated N-terminal domain of P3 is not sensitive to chiral structural features. The fit of these models was improved by using rigid-body refinement (the fit option in CHIMERA). The cryo-EM map and models were visualized using the program CHIMERA.
Protein structure accession numbers.
The atomic coordinates and structure factors have been deposited in the Protein Data Bank (http://www.pdb.org) (PDB identifier [ID] codes 3F6N and 3K4T for the P64 and P21 crystal forms, respectively).
RESULTS
Overview of the P3 crystal structure.
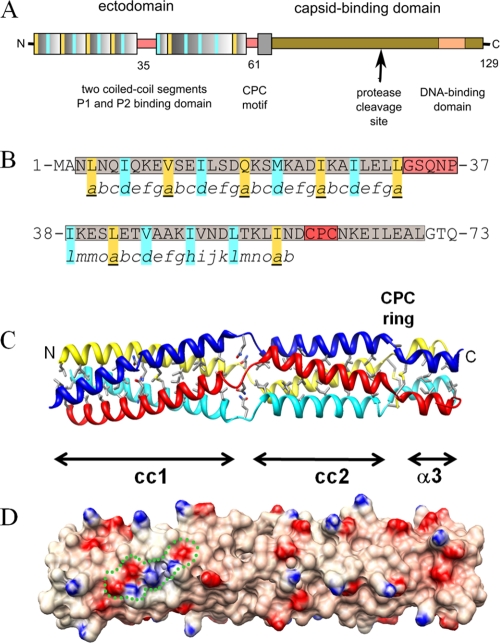
Sequence-based secondary-structure predictions indicate that P3 is composed of two amphipathic α-helices, with residues 3 to 32 and 38 to 57, respectively, having high and moderate propensities to form coiled-coil structures—a widespread protein-protein interaction and oligomerization motif (1)—thought to be assembled as a parallel tetramer (22, 42). The domain organization of the P3 protein (Fig. 1A) is characterized by an N-terminal region containing two coiled-coil segments, assigned as the P1- and/or P2-binding domain, and by a C-terminal region that corresponds to the anchoring domain in the viral capsid and that contains a DNA binding motif (residues 76 to 129) characterized by a proline-rich segment (23).
FIG. 1.
Overall structure of the CaMV P3 protein. (A) Schematic diagram of P3 domain architecture. Coiled-coil repeats are represented as wide colored boxes, and key residues in the hydrophobic coiled-coil core are highlighted in yellow and blue. (B) Structure-based assignments of coiled-coil heptads seen in the P3 sequence. Helical segments are shaded in gray and nonhelical segments in red. Yellow and blue residues are defined as in panel A. (C) Ribbon diagram of the unliganded crystal structure of P3. The overall length of the tetramer is ∼120 Å, with a diameter of 25 Å. The hydrophobic residues and the Gln18 layer in the 4-helix bundle, the Gln35 layer, and the cysteine ring within the GP and CPC motifs are displayed in stick representation. (D) Electrostatic potential rendered at ±10 kT/e, e being the unit electron charge (blue, positive; red, negative), and mapped on the molecular surface of P3 shown in the same orientation as in panel C. A cluster of residues involved in interchain electrostatic interactions is delineated by a green dotted line.
The X-ray structure of the multifunctional N-terminal domain of the P3 protein was determined at 3.1 Å and 2.6 Å resolution in space groups P64 and P21, respectively, using molecular replacement. In both cases, the asymmetric unit contains a tetrameric P3 assembly that folds into a parallel four-helix bundle subdivided into the two coiled-coil subdomains mediating oligomerization. The two crystal structures were almost identical (root mean square deviations [RMSD] for 280 Cα atoms, 0.75 Å) and, unless otherwise specified, the higher-resolution structure is described here. The structure-based assignment of the heptad registers to the coiled-coil sequence is shown in Fig. 1B. The molecular model has a cylindrical overall shape with a length of 120 Å and a cross-sectional diameter of 25 Å (Fig. 1C). Each chain folds into 3 consecutive α-helical segments (α1, residues 3 to 32; α2, 38 to 59; and α3, 63 to 70). The root mean square deviations between the individual chains in the P3 tetramer vary from 0.93 Å (chains A and C) to 1.31 Å (chains B and D). The model comprises solely residues 2 to 74, because the four C-terminal domains (residues 75 to 129) were disordered or absent from the electron density map. In fact, this C-terminal region of the P3 protein is susceptible to partial proteolysis both in infected cells (6, 12) and in crystallization drops (see Fig. S3 in the supplemental material). Here, the proteolysis generated a fragment of 11 kDa, resulting in the deletion of about 30 residues.
The original feature of this cylinder-shaped structure is that it is packed into 3 successive subdomains (Fig. 1C): a canonical left-handed coiled-coil domain (residues 3 to 32) is followed by a noncanonical coiled-coil domain (residues 38 to 59) characterized by right-handed supercoiling and, finally, by a short four-helix bundle (residues 63 to 70). The three α-helices of the P3 protomer are connected by two nonhelical segments. The first segment contains a Gly-Ser-X-X-Pro motif (GP linker; residues 33 to 37), and the slightly increased B factors in this region suggest some flexibility providing a local switch from the first left-handed to the second right-handed coiled-coil subdomain. The second short segment is characterized by a Cys-Pro-Cys sequence (CPC motif; residues 60 to 62) involving four vicinal interchain disulfide bridges forming an annular ring that stabilizes the distal part of this tetrameric assembly. As expected for a long coiled-coil structure, the large buried surface (∼12,500 Å2) represents 43.8% of the total solvent-accessible area of the four helical monomers. The radius of the four-helix bundle (r = 7.6 ± 0.4 Å) does not change significantly over the length of the molecule, and the exterior of this tetrameric stalk displays a highly charged surface in which the positive and negative electrostatic potentials follow the ridges of the superhelical backbone (Fig. 1D).
Canonical coiled-coil subdomain 1.
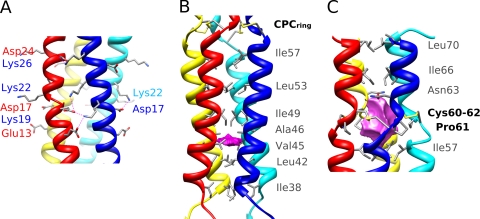
The overall structure of the N-terminal coiled-coil subdomain 1 is that of a twisted left-handed four-stranded coiled coil (Fig. 1C). The conserved sequence motif of 4 heptad repeats (abcdefg)n observed in the P3 protein (22, 42) is characteristic of coiled coils. Overall, the left-handed superhelix has a pitch of 144 Å and a radius of 7.7 Å (see Table S1 in the supplemental material). The a and d positions of the repeat are commonly occupied by hydrophobic residues that interact by classical “knobs-into-holes” packing (28) to form alternate layers. Here, Ile is the most represented side chain in d, while Leu, Val, and Ile are observed in position a. Much of the oligomerization energy of coiled-coil motifs derives from van der Waals packing interactions between the buried hydrophobic residues located at these a and d heptad positions (Fig. 1C). The buried surface area in this coiled-coil subdomain is 6,450 Å2, thus representing half of the total buried surface in tetrameric P3. The hydrophobic core is interrupted by a polar layer made of Gln18 residues occupying the a-heptad position, a feature frequently observed in coiled-coil interfaces. Here, the amide side chains of Gln18 residues form a buried layer stabilized by an interchain network of hydrogen bonds with backbone carbonyl oxygen of Ile14. Several salt bridges and hydrogen bonds between residues flanking the interface (positions b, c, e, and g) provide further stabilization energy to the coiled-coil assembly; such interchain stabilizing interactions are represented in Fig. 2A by Glu13-Lys19, Asp17-Lys19, Asp17-Lys22, and Asp24-Lys26. These locally clustered ionic interactions, combined with a tight hydrophobic core, are a key feature of the “trigger sequences” that promote α-helical coiled-coil formation. An earlier study showed that mutation of the conserved Lys22 to Asn22 in a peptide corresponding to this coiled-coil subdomain 1 impaired its association as a tetramer (22)—this is consistent with our structural data, where Lys22 makes a major contribution to the network of interhelical salt bridges.
FIG. 2.
Detailed view of P3 subdomains. (A) Detailed view of the intermolecular salt bridges in the canonical coiled-coil subdomain 1 of P3. Salt bridges are indicated as dotted lines. (B) Lateral view of P3 subdomain 2 revealing the small cavity (magenta) and the hydrophobic layers of the right-handed coiled-coil core, rendered as gray sticks. (C) View of the P3 structure in the region of the 4 cysteine bridges. The CPC ring motif delineates a large cavity (magenta) along the tetramer axis, and the lining residues are shown as sticks.
Noncanonical right-handed coiled-coil subdomain 2.
Subdomain 2 of the P3 structure is a four-helix bundle with a noncanonical coiled-coil topology. Interestingly, this part of the P3 structure has a unique pattern of hydrophobic residues (Fig. 1B), homologous to the 15-residue repeat of the VASP protein (21), and also displays a right-handed supercoiling geometry (Fig. 1C). In this pattern, positions a (Leu42 and Ile57), d (Val45), e (Ala46), h (Ile49), and l (Ile38 and Leu53) are occupied by hydrophobic residues forming a single pentadecad repeat motif in the central core of the segment (Fig. 2B). The a, d/e, h, and l layers at the tetramer interface exhibit characteristic meshing into a knobs-into-holes packing scheme (see Fig. S4 in the supplemental material). The superhelix has a pitch of 205 Å and a radius of 7.6 Å; the helical parameters are compared to those of subdomain 1 in Table S1 in the supplemental material. The tightness of the hydrophobic side chain packing is less pronounced, and hydrophobic core residues are more exposed to solvent, with a buried surface area of 3,350 Å2 (representing 38.1 Å2 per residue), i.e., much less than that found in the canonical coiled-coil subdomain 1 (52.4 Å2 per residue). This coiled-coil subdomain 2 is also remarkable in lacking any surface interhelical H bonds or salt bridges (16 and 8, respectively, in subdomain 1). In addition, a small cavity centered on the d/e layer and occupying a volume of 24 Å3 is lined by the hydrophobic side chains Leu42, Val45, Ala46, and Ile49 (Fig. 2B), further suggesting that the four-stranded coiled-coil subdomain 2 forms a less stable structure. Such a relatively unstable coiled-coil domain has been described previously in the scallop myosin rod, where the bending and unwinding of the α-helices were shown to play important roles in the functional properties of this structure (27). A similar functional significance of the two coiled-coil domains of P3 is discussed further below.
The CPC cystine ring motif.
The CPC motifs form a ring of four interchain disulfide bridges between Cys60 and Cys62 of two neighboring chains. This unique disulfide bridge pattern, which is localized at the C-terminal junction of the right-handed coiled-coil subdomain 2 and the short downstream 4-helix-bundle subdomain 3 (residues 63 to 70), stabilizes the C-terminal part of the P3 oligomer. The disulfide bridges have stereochemically optimal conformations for χ1 and χ2 angles, and the formation of such an unusual four-cystine arrangement requires the appropriate parallel registration of the four polypeptide chains. A few additional interchain polar interactions further anchor this CPC ring motif to the Asn63 and Glu65 residues in the short C-terminal subdomain 3 and to Ile57 in the coiled-coil subdomain 2. The eight cysteine residues from the CPC motif encircle a large cavity centered on the axial-symmetry axis, occupying a volume of 128 Å3 and lined almost exclusively by hydrophobic side chains: Ile57, Cys60, Pro61, Cys62, Asn63, and Ile66 (Fig. 2C). An analogous CXXC disulfide ring motif involving interchain bridges has been described in cartilage oligomeric matrix protein (COMP), which assembles as a five-stranded coiled coil (29). In this case, the redox state dictates interchain bridging and is critical for controlling the entrance of the vitamin D3 binding cavity delineated by the five coiled-coil helices (36). How the CPC ring in P3 could similarly induce functionally relevant conformational changes is discussed further below.
In summary, although the overall architecture of P3 may appear similar at first sight to those of the other parallel tetrameric coiled-coil proteins reported to date (see Fig. S5 in the supplemental material), it harbors remarkable peculiarities, highlighting the structural diversity that can be obtained with various combinations of superhelical parameters (radius of curvature, supercoil pitch, radius, and handedness). P3 is the first segmented tetrameric fold displaying a combination of a left-handed canonical and a right-handed noncanonical coiled-coil domain, thus underlining the versatility of this new structural motif.
Molecular model of the P3 lattice decorating CaMV particles.
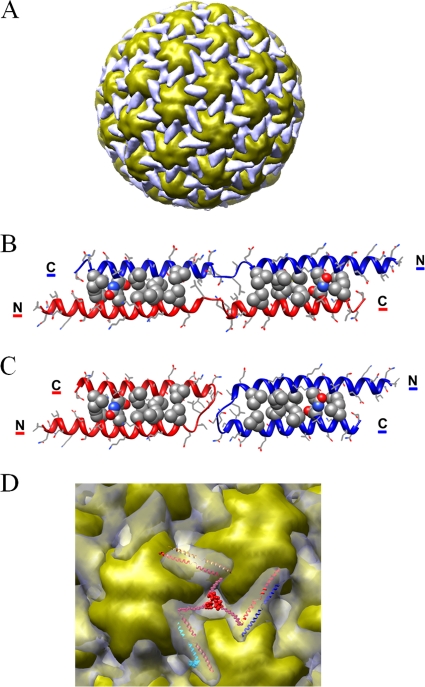
The CaMV virion capsid is composed of 420 P4 proteins assembled as 60 hexavalent and 12 pentavalent capsomers. The locations of P3 proteins were proposed based on cryo-EM reconstruction of free and P3-decorated virions at 18-Å and 26-Å resolution, respectively, combined with secondary-structure predictions (37). P3 binding on CaMV virion particles was demonstrated to form a triskelion structure that cements three hexavalent or pentavalent capsomers together (Fig. 3A). Although at low resolution (∼26 Å), the difference map clearly indicated that the number of digitations and the volume of each triskelion arm (∼15,000 Å3) are compatible with a stoichiometry of 1:1 for the P3/P4 ratio. Moreover, the volume and the size of these rod-like domains (∼56 by 22 Å) suggest that the 60 N-terminal residues of adjacent P3 proteins can form a complex through the formation of a two-stranded antiparallel coiled-coil (37). The C-terminal parts are deeply anchored inside the pores surrounding the capsomers and traverse the concentric layers formed by the coat proteins to reach the genomic DNA packaged in the inner shells (37).
FIG. 3.
Structural model of the P3 ectodomain on the surface of the P3-decorated CaMV particle. (A) Overall view of the cryo-EM reconstruction of the P3-decorated CaMV particle (37). The coiled-coil network formed by P3 ectodomains is colored gray and the capsid yellow. (B) A model of the swapped antiparallel dimeric coiled-coil P3 is displayed as a ribbon diagram with side chains as stick models. The residues involved in knob-and-holes packing (Ile14-Leu56, Gln18-Asp52, Met21-Ile49, Ile25-Val45, and Ile28-Leu42) are displayed as compact models. (C) The same model of the N-terminal domain of P3 as in panel B in the nonswapped conformation. (D) Enlarged view of a P3 triskelion motif (gray) in the cryo-EM difference map (37) surrounded by three adjacent capsid hexamers (yellow). The ectodomains of 3 antiparallel dimeric coiled-coil P3 molecules are displayed as ribbons and superimposed on the digitations of a triskelion motif. The anchoring domain at the center of the triskelion is modeled as dummy Cα atoms corresponding to the C-terminal segments.
The assembly and modeling rules of α-helical coiled-coil motifs are now well understood (51). In order to establish a plausible model for this P3 ectodomain arranged around the viral particles, we applied molecular-modeling techniques to build a two-stranded coiled coil using the two N-terminal helical segments of the crystal structure of P3 and canonical antiparallel coiled-coil structures as templates. As illustrated in Fig. 3B, this model comprises the N-terminal α1 (residues 4 to 32) and α2 (residues 38 to 59) helices separated by the flexible GP linker (residues 33 to 37). The interfacial positions compatible with a knobs-into-holes arrangement are occupied by residues Ile14, Gln18, Met21, Ile25, and Ile 28 in the α1 helix a and d positions and residues Leu42, Val45, Ile49, Asp52, and Leu56 of the α2 helix in a 3-4-3-4 spacing. The potentially favorable electrostatic interactions between Asp24 and Lys48, and Asp17 and Lys55, also favor this antiparallel coiled-coil alignment. Thus, this N-terminal α1-α2 region of P3 has a significant potential to adopt an antiparallel dimeric coiled-coil conformation. A second model is also compatible with the current rules for coiled-coil folding and assembly. As illustrated in Fig. 3C, it is constituted of two antiparallel coiled coils forming helical hairpins having the same core packing geometry. The minor difference between these two models lies in the GP linker, which forms a loop rather than an extended flexible segment linking swapped helices.
A reasonable fit of these models, which may coexist on the viral capsid surface, is obtained in the low-resolution cryo-EM difference density corresponding to the ectodomain of the P3 protein. As displayed in Fig. 3D, the shape and size of the antiparallel coiled-coil models (∼65 by 20 Å) match the difference density in the triskelion arms. The 2-fold axis relating two of the P3 polypeptide chains is coincident with the pseudo-2-fold symmetry axis located in the middle of each digitation. The salient feature of this model is that the swapped dimeric arrangement of adjacent α1-α2 helical segments generates an intricate coiled-coil lattice covering the whole viral particle. P3 thus could act as a capsid cement, filling holes and canyons around capsomers. This topological arrangement could be of significant importance, especially during transport and under the various stringent conditions encountered in the aphid buccal apparatus.
DISCUSSION
The crystal structure of the tetrameric P3 protein supports previous biochemical characterizations and provides detailed information on the interactions of this homotetramer. The structure displays several fundamental differences from other tetrameric coiled-coil structures described to date. The most striking originality is the existence of two consecutive coiled-coil subdomains with opposite handedness of supercoiling, the whole stalk being locked by an interchain cystine ring. To our knowledge, this is the first crystal structure of a protein containing a combination of coiled-coil segments with opposite handedness. Such a structural feature has been suggested by sequence analysis and molecular-modeling studies of the stalk domain of the nonfimbrial adhesin YadA from Yersinia (20). In that trimeric protein, 10 pentadecad repeats forming a right-handed coiled coil are followed by 4 heptads specifying a left-handed coiled coil.
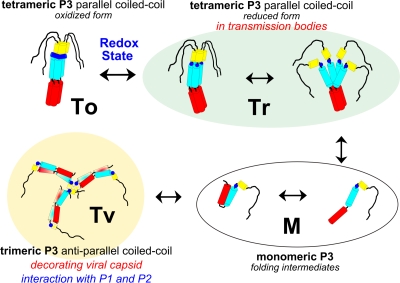
The two coiled-coil subdomains in the tetrameric P3 protein have different structural properties. The N-terminal canonical subdomain is stable and can be considered a nucleating coiled coil, while subdomain 2 is comparatively less stable, and bending or unwinding of the helices could determine the functional properties of P3. As displayed in Fig. 4, the versatility of the quaternary structure of the N-terminal coiled-coil segments of the P3 protein is also illustrated by the plausible organization in the antiparallel coiled-coil network decorating the virus particle, derived from molecular modeling and cryo-EM data (37). However, a higher-resolution cryo-EM reconstruction is needed for further improvement of the fit and detailed analysis of this capsid-bound P3 conformation.
FIG. 4.
Schematic representations of the various conformations of the CaMV P3 protein. Parallel coiled-coil P3 tetramers in the reduced form (Tr) are localized in TBs as partially destabilized structures or more compact stalks with reduced cysteines. The P3 oxidized form (To) stabilized by the cystine ring is displayed on the left; monomeric P3 folding intermediates are labeled as M forms. The trimeric P3 proteins (Tv) decorate the surface of the CaMV viral particle: α-helical segments form an antiparallel coiled-coil network as either α-helical hairpin monomers or swapped antiparallel coiled-coil dimers. The α1, α2, and α3 helices are schematized as red, blue, and yellow cylinders, respectively. The CPC motif is represented as a blue circle or line for the reduced and oxidized forms, respectively; the black segment corresponds to the disordered C-terminal region.
Different conformers of the same coiled-coil sequence adopting various quaternary structures and playing different regulatory roles have been described previously for the SNARE complex (50), the transcription factor GCN4 (52), and viral glycoproteins (41). While it is clear that P3 plays a central regulatory role in the biology of CaMV and exists in different conformational states depending on its involvement in protein-protein interactions and its cellular location, the accumulated data that have appeared in the literature over the last 2 decades have remained very difficult to unravel into an intelligible, comprehensive image. With the help of the structural information elucidated in the present study, we can now derive a putative scheme of the infection cycle of CaMV that accommodates most P3-related data available.
P3 is produced initially in the viral factories (30), where replication, encapsidation, and accumulation of around 95% of virions also occur (7, 17). From studies implicating virion-bound P3 in aphid transmission and cell-to-cell movement, it was proposed that P3 is an auxiliary protein whose role is to “functionalize” the preformed virus particle (43). This possibility is consistent with experimental data showing that P3 readily associates, with very high affinity, with previously formed and purified virions (7, 37). Once associated with virions as trimeric assemblies through the formation of an antiparallel coiled-coil network (Tv in Fig. 4), P3 likely acts as capsid cement, filling holes and canyons around capsomers. There are many examples of domain swapping (13), especially in the case of viral capsid proteins, where this frequent phenomenon provides long-range stabilizing interactions throughout icosahedral networks (11, 38) or may facilitate viral assembly (18).
One important prediction inherent in this analysis is that the CPC cystine ring of the P3 produced in the viral factories would be in a reduced form, as dictated by the prevailing reducing conditions of the intracellular environment (40). The conformation of the reduced form of P3 could be heterogeneous (Fig. 4, Tr forms), with partially unfolded parallel tetramers solely stabilized by the N-terminal canonical parallel coiled-coil domain, in equilibrium with monomeric folding intermediates (Fig. 4, M forms), and α1-α2 helices optionally forming an α-helical hairpin. While the canonical coiled-coil α1 segment alone has the propensity to associate as a parallel tetramer (22), the second, noncanonical coiled-coil α2 segment is shown here to be less stable, and the absence of stabilization by the cystine ring could facilitate the dissociation of the P3 N terminus and its reassembling as an antiparallel coiled-coil network around the virus particle (Fig. 4, Tv conformers). This network also provides functional arms protruding from the surface of the virion and mediating multivalent interactions with trimeric P1 and/or P2. Such P3-decorated virions can thus move to and traverse the plasmodesmata through the lumen of the tubules formed by the movement protein P1 (43). Alternatively, for plant-to-plant transmission, they can be taken up by aphids, where they attach to P2 already associated with the insect receptor (7).
A prominent feature of the P3 structure relies on the presence of the CXC ring motif conserved in all members of the genus Caulimovirus. The four intermolecular disulfide bonds stabilizing the P3 protein into a rigid extended conformation (Fig. 4, form To) exist only in an oxidizing environment, a situation that does not seem to apply to the P3 stored within viral factories. Recently, Martinière and collaborators (30) have demonstrated that the fraction of P3 that is not associated with the virions is rapidly exported from the viral factories, together with P2, and finally accumulates as P2/P3 aggregates within a single large inclusion body per infected cell, designated the transmission body (TB). In an earlier study, we showed that, when a TB is ingested by an aphid vector, P2 is efficiently retained within its mouthparts whereas P3 is set free and lost. The preformed P3-virion complexes are acquired later by the aphid, during subsequent intracellular probing, and attach to the aphid-bound P2 (7). A way to understand this switch from P2/P3 coaggregation within the TB and their mutual release in the aphid mouthparts would be to imagine that the cysteine residues in P3 act as a redox-sensitive cluster, reminiscent of the property of a comparable cystine ring described in the COMP protein referred to above (36). A change in redox potential could be induced by cell disruption by aphid stylets or, alternatively, by oxidizing enzymes that have previously been identified within the aphid saliva (2, 15). The formation of disulfide bridges between P3 protomers could thus trigger the structural rearrangement of this protein from the Tr to the To conformer (Fig. 4). This switch from the reduced to the oxidized form of P3 could strongly stabilize the N terminus stalk, reducing its flexibility, masking a significant fraction of its solvent-accessible interfaces, and abolishing the interactions with other molecular partners. It is interesting that the P3 expressed and purified from bacteria, shown here to be in the form of oxidized tetramers, is totally lacking any detectable affinity for both P2 (7) and P1 (43). This “switch” hypothesis is also consistent with the report of the oxidized tetrameric form of P3 in extracts from infected plants (45, 46). It will be particularly interesting to investigate whether this oxidized P3 conformation exists within live infected cells and to further validate its role in the release of P2 from TBs during aphid acquisition.
Conclusion.
Overall, the information reported in this study provides a unique illustration of how the structural and biological versatility of coiled coils can be used in sophisticated ways by viruses. A canonical parallel tetrameric coiled-coil subdomain with a left-handed twist is linked to a less stable noncanonical right-handed subdomain, which might destabilize the tetramer and favor distinct interactions with various partners unless the whole stalk is locked into a rigid conformation by a cystine ring. The CPC motif forming this P3 interchain disulfide ring may also serve as a redox-sensitive cluster triggering the corresponding conformational changes in different environments. The accumulation of all these features in a single molecule makes P3 a central regulator of the virus infection cycle, particularly during transmission by insect vectors, a complex phenomenon that involves versatile associations with various molecular partners both inside living cells and outside in the insect stylets.
Supplementary Material
Acknowledgments
We thank Joanna McCarthy and Ganesh Natrajan, staff members of the European Synchrotron Radiation Facility (ESRF) in Grenoble, for technical support during data collection. M.U., M.D., and S.B. acknowledge the financial support from the INRA SPE department and ANR. We are grateful to Andrey Kajava and Helen Rothnie for critical reading of the manuscript and English editing.
The work at ESRF was supported by the European Union.
Footnotes
Published ahead of print on 24 February 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Burkhard, P., J. Stetefeld, and S. V. Strelkov. 2001. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 11:82-88. [DOI] [PubMed] [Google Scholar]
- 2.Carolan, J. C., C. I. Fitzroy, P. D. Ashton, A. E. Douglas, and T. L. Wilkinson. 2009. The secreted salivary proteome of the pea aphid Acyrthosiphon pisum characterised by mass spectrometry. Proteomics 9:2457-2467. [DOI] [PubMed] [Google Scholar]
- 3.CCP4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760-763. [DOI] [PubMed] [Google Scholar]
- 4.Cheng, R. H., N. H. Olson, and T. S. Baker. 1992. Cauliflower mosaic virus: a 420 subunit (T=7), multilayer structure. Virology 186:655-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conners, R., D. J. Hill, E. Borodina, C. Agnew, S. J. Daniell, N. M. Burton, R. B. Sessions, A. R. Clarke, L. E. Catto, D. Lammie, T. Wess, R. L. Brady, and M. Virji. 2008. The Moraxella adhesin UspA1 binds to its human CEACAM1 receptor by a deformable trimeric coiled-coil. EMBO J. 27:1779-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dautel, S., T. Guidasci, M. Pique, J. L. Mougeot, G. Lebeurier, P. Yot, and J. M. Mesnard. 1994. The full-length product of cauliflower mosaic virus open frame III is associated with the viral particle. Virology 202:1043-1045. [DOI] [PubMed] [Google Scholar]
- 7.Drucker, M., R. Froissart, E. Hebrard, M. Uzest, M. Ravallec, P. Esperandieu, J. C. Mani, M. Pugniere, F. Roquet, A. Fereres, and S. Blanc. 2002. Intracellular distribution of viral gene products regulates a complex mechanism of cauliflower mosaic virus acquisition by its aphid vector. Proc. Natl. Acad. Sci. U. S. A. 99:2422-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 9.Fiser, A., and A. Sali. 2003. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 374:461-491. [DOI] [PubMed] [Google Scholar]
- 10.Fiser, A., and A. Sali. 2003. ModLoop: automated modeling of loops in protein structures. Bioinformatics 19:2500-2501. [DOI] [PubMed] [Google Scholar]
- 11.Garriga, D., J. Querol-Audi, F. Abaitua, I. Saugar, J. Pous, N. Verdaguer, J. R. Caston, and J. F. Rodriguez. 2006. The 2.6-angstrom structure of infectious bursal disease virus-derived T=1 particles reveals new stabilizing elements of the virus capsid. J. Virol. 80:6895-6905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giband, M., J. M. Mesnard, and G. Lebeurier. 1986. The gene III product (P15) of cauliflower mosaic virus is a DNA-binding protein while an immunologically related P11 polypeptide is associated with virions. EMBO J. 5:2433-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gronenborn, A. M. 2009. Protein acrobatics in pairs-dimerization via domain swapping. Curr. Opin. Struct. Biol. 19:39-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haas, G., J. Azevedo, G. Moissiard, A. Geldreich, C. Himber, M. Bureau, T. Fukuhara, M. Keller, and O. Voinnet. 2008. Nuclear import of CaMV P6 is required for infection and suppression of the RNA silencing factor DRB4. EMBO J. 27:2102-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harmel, N., E. Letocart, A. Cherqui, P. Giordanengo, G. Mazzucchelli, F. Guillonneau, E. De Pauw, E. Haubruge, and F. Francis. 2008. Identification of aphid salivary proteins: a proteomic investigation of Myzus persicae. Insect. Mol. Biol. 17:165-174. [DOI] [PubMed] [Google Scholar]
- 16.Hebrard, E., M. Drucker, D. Leclerc, T. Hohn, M. Uzest, R. Froissart, J. M. Strub, S. Sanglier, A. van Dorsselaer, A. Padilla, G. Labesse, and S. Blanc. 2001. Biochemical characterization of the helper component of Cauliflower mosaic virus. J. Virol. 75:8538-8546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hohn, T., and J. Fütterer. 1997. The proteins and functions of plant pararetroviruses: knowns and unknowns. Crit. Rev. Plant Sci. 16:133-167. [Google Scholar]
- 18.Ivanov, D., O. V. Tsodikov, J. Kasanov, T. Ellenberger, G. Wagner, and T. Collins. 2007. Domain-swapped dimerization of the HIV-1 capsid C-terminal domain. Proc. Natl. Acad. Sci. U. S. A. 104:4353-4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kleywegt, G. J., and T. A. Jones. 1994. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr. D Biol. Crystallogr. 50:178-185. [DOI] [PubMed] [Google Scholar]
- 20.Koretke, K. K., P. Szczesny, M. Gruber, and A. N. Lupas. 2006. Model structure of the prototypical non-fimbrial adhesin YadA of Yersinia enterocolitica. J. Struct. Biol. 155:154-161. [DOI] [PubMed] [Google Scholar]
- 21.Kuhnel, K., T. Jarchau, E. Wolf, I. Schlichting, U. Walter, A. Wittinghofer, and S. V. Strelkov. 2004. The VASP tetramerization domain is a right-handed coiled coil based on a 15-residue repeat. Proc. Natl. Acad. Sci. U. S. A. 101:17027-17032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leclerc, D., L. Burri, A. V. Kajava, J. L. Mougeot, D. Hess, A. Lustig, G. Kleemann, and T. Hohn. 1998. The open reading frame III product of cauliflower mosaic virus forms a tetramer through a N-terminal coiled-coil. J. Biol. Chem. 273:29015-29021. [DOI] [PubMed] [Google Scholar]
- 23.Leclerc, D., L. Stavolone, E. Meier, O. Guerra-Peraza, E. Herzog, and T. Hohn. 2001. The product of ORF III in cauliflower mosaic virus interacts with the viral coat protein through its C-terminal proline rich domain. Virus Genes 22:159-165. [DOI] [PubMed] [Google Scholar]
- 24.Leh, V., E. Jacquot, A. Geldreich, M. Haas, S. Blanc, M. Keller, and P. Yot. 2001. Interaction between the open reading frame III product and the coat protein is required for transmission of cauliflower mosaic virus by aphids. J. Virol. 75:100-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leh, V., E. Jacquot, A. Geldreich, T. Hermann, D. Leclerc, M. Cerrutti, P. Yot, M. Keller, and S. Blanc. 1999. Aphid transmission of cauliflower mosaic virus requires the viral PIII protein. EMBO J. 18:7077-7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leslie, A. G. 1999. Integration of macromolecular diffraction data. Acta Crystallogr. D Biol. Crystallogr. 55:1696-1702. [DOI] [PubMed] [Google Scholar]
- 27.Li, Y., J. H. Brown, L. Reshetnikova, A. Blazsek, L. Farkas, L. Nyitray, and C. Cohen. 2003. Visualization of an unstable coiled coil from the scallop myosin rod. Nature 424:341-345. [DOI] [PubMed] [Google Scholar]
- 28.Lupas, A. N., and M. Gruber. 2005. The structure of alpha-helical coiled coils. Adv. Protein Chem. 70:37-78. [DOI] [PubMed] [Google Scholar]
- 29.Malashkevich, V. N., R. A. Kammerer, V. P. Efimov, T. Schulthess, and J. Engel. 1996. The crystal structure of a five-stranded coiled coil in COMP: a prototype ion channel? Science 274:761-765. [DOI] [PubMed] [Google Scholar]
- 30.Martinière, A., D. Gargani, M. Uzest, N. Lautredou, S. Blanc, and M. Drucker. 2009. A role for plant microtubules in the formation of transmission-specific inclusion bodies of Cauliflower mosaic virus. Plant J. 58:135-146. [DOI] [PubMed] [Google Scholar]
- 31.McCoy, A. J., R. W. Grosse-Kunstleve, P. D. Adams, M. D. Winn, L. C. Storoni, and R. J. Read. 2007. Phaser crystallographic software. J. Appl. Crystallogr. 40:658-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moreno, A., E. Hebrard, M. Uzest, S. Blanc, and A. Fereres. 2005. A single amino acid position in the helper component of cauliflower mosaic virus can change the spectrum of transmitting vector species. J. Virol. 79:13587-13593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mougeot, J. L., T. Guidasci, T. Wurch, G. Lebeurier, and J. M. Mesnard. 1993. Identification of C-terminal amino acid residues of cauliflower mosaic virus open reading frame III protein responsible for its DNA binding activity. Proc. Natl. Acad. Sci. U. S. A. 90:1470-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240-255. [DOI] [PubMed] [Google Scholar]
- 35.Ng, J. C., and B. W. Falk. 2006. Virus-vector interactions mediating nonpersistent and semipersistent transmission of plant viruses. Annu. Rev. Phytopathol. 44:183-212. [DOI] [PubMed] [Google Scholar]
- 36.Ozbek, S., J. Engel, and J. Stetefeld. 2002. Storage function of cartilage oligomeric matrix protein: the crystal structure of the coiled-coil domain in complex with vitamin D(3). EMBO J. 21:5960-5968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plisson, C., M. Uzest, M. Drucker, R. Froissart, C. Dumas, J. Conway, D. Thomas, S. Blanc, and P. Bron. 2005. Structure of the mature P3-virus particle complex of cauliflower mosaic virus revealed by cryo-electron microscopy. J. Mol. Biol. 346:267-277. [DOI] [PubMed] [Google Scholar]
- 38.Qu, C., L. Liljas, N. Opalka, C. Brugidou, M. Yeager, R. N. Beachy, C. M. Fauquet, J. E. Johnson, and T. Lin. 2000. 3D domain swapping modulates the stability of members of an icosahedral virus group. Structure 8:1095-1103. [DOI] [PubMed] [Google Scholar]
- 39.Ryabova, L. A., M. M. Pooggin, and T. Hohn. 2006. Translation reinitiation and leaky scanning in plant viruses. Virus Res. 119:52-62. [DOI] [PubMed] [Google Scholar]
- 40.Sevier, C. S., and C. A. Kaiser. 2002. Formation and transfer of disulphide bonds in living cells. Nat. Rev. Mol. Cell Biol. 3:836-847. [DOI] [PubMed] [Google Scholar]
- 41.Skehel, J. J., and D. C. Wiley. 1998. Coiled coils in both intracellular vesicle and viral membrane fusion. Cell 95:871-874. [DOI] [PubMed] [Google Scholar]
- 42.Stavolone, L., E. Herzog, D. Leclerc, and T. Hohn. 2001. Tetramerization is a conserved feature of the virion-associated protein in plant pararetroviruses. J. Virol. 75:7739-7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stavolone, L., M. E. Villani, D. Leclerc, and T. Hohn. 2005. A coiled-coil interaction mediates cauliflower mosaic virus cell-to-cell movement. Proc. Natl. Acad. Sci. U. S. A. 102:6219-6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strelkov, S. V., and P. Burkhard. 2002. Analysis of alpha-helical coiled coils with the program TWISTER reveals a structural mechanism for stutter compensation. J. Struct. Biol. 137:54-64. [DOI] [PubMed] [Google Scholar]
- 45.Tsuge, S., K. Kobayashi, H. Nakayashiki, K. Mise, and I. Furusawa. 1999. Cauliflower mosaic virus ORF III product forms a tetramer in planta: its implication in viral DNA folding during encapsidation. Microbiol. Immunol. 43:773-780. [DOI] [PubMed] [Google Scholar]
- 46.Tsuge, S., T. Okuno, I. Furusawa, Y. Kubo, and O. Horino. 2001. Stabilization of cauliflower mosaic virus P3 tetramer by covalent linkage. Microbiol. Immunol. 45:365-371. [DOI] [PubMed] [Google Scholar]
- 47.Uzest, M., D. Gargani, M. Drucker, E. Hebrard, E. Garzo, T. Candresse, A. Fereres, and S. Blanc. 2007. A protein key to plant virus transmission at the tip of the insect vector stylet. Proc. Natl. Acad. Sci. U. S. A. 104:17959-17964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vaguine, A. A., J. Richelle, and S. J. Wodak. 1999. SFCHECK: a unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr. D Biol. Crystallogr. 55:191-205. [DOI] [PubMed] [Google Scholar]
- 49.Walshaw, J., and D. N. Woolfson. 2001. Socket: a program for identifying and analysing coiled-coil motifs within protein structures. J. Mol. Biol. 307:1427-1450. [DOI] [PubMed] [Google Scholar]
- 50.Weninger, K., M. E. Bowen, S. Chu, and A. T. Brunger. 2003. Single-molecule studies of SNARE complex assembly reveal parallel and antiparallel configurations. Proc. Natl. Acad. Sci. U. S. A. 100:14800-14805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woolfson, D. N. 2005. The design of coiled-coil structures and assemblies. Adv. Protein Chem. 70:79-112. [DOI] [PubMed] [Google Scholar]
- 52.Yadav, M. K., L. J. Leman, D. J. Price, C. L. Brooks III, C. D. Stout, and M. R. Ghadiri. 2006. Coiled coils at the edge of configurational heterogeneity. Structural analyses of parallel and antiparallel homotetrameric coiled coils reveal configurational sensitivity to a single solvent-exposed amino acid substitution. Biochemistry 45:4463-4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zwart, P. H., S. Banumathi, M. Dauter, and Z. Dauter. 2004. Radiation-damage-induced phasing with anomalous scattering: substructure solution and phasing. Acta Crystallogr D Biol. Crystallogr. 60:1958-1963. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.