Abstract
Most viral infections are self-limiting, resulting in either clearance of the pathogen or death of the host. However, a subset of viruses can establish permanent infection and persist indefinitely within the host. Even though persisting viruses are derived from various viral families with distinct replication strategies, they all utilize common mechanisms for establishment of long-lasting infections. Here, we discuss the commonalities between persistent infections with herpes-, retro-, flavi-, arena-, and polyomaviruses that distinguish them from acutely infecting viral pathogens. These shared strategies include selection of cell subsets ideal for long-term maintenance of the viral genome, modulation of viral gene expression, viral subversion of apoptotic pathways, and avoidance of clearance by the immune system.
Most viral pathogens cause acute, self-limiting infections whereby the virus replicates rapidly and disseminates to another organism prior to immune clearance or the death of the host. In contrast, some viruses are able to establish persistent infections (in many cases initiated by an acute infection) through adoption of sophisticated relationships with their hosts and manipulation of a wide array of cellular mechanisms for their own advantage. Persistence can take place through nonproductive infection (e.g., herpesvirus latency [21, 25, 42, 72]), proviral integration into the host genome (as in the case of retroviruses [13, 26]), and/or continuous viral replication (as in the cases of flaviviruses, arenaviruses, and polyomaviruses [11, 35, 50, 61, 65, 70]). While each virus has evolved distinct mechanisms to enable persistence, there are shared themes in the establishment and maintenance of permanent infection. These common threads linking persistent viral infections include (i) selection of cell subsets ideal for long-term maintenance of the viral genome, (ii) modulation of viral gene expression, (iii) viral subversion of cellular apoptotic pathways, and (iv) avoidance of clearance by the immune system. Amid the daunting complexity of host-virus interactions, these commonalities provide a point of focus for researchers hoping to treat these infections.
SELECTION OF CELL SUBSETS FOR LONG-TERM MAINTENANCE OF THE VIRAL GENOME
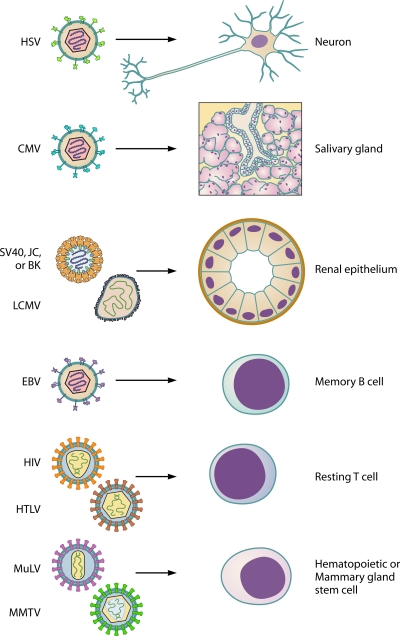
The first step in any viral invasion of a host is to gain access to and infect a susceptible cell. For acute infections, production of high viral titers by infected cells allows for easy dissemination and a high probability of infection of the subsequent host. On the other hand, viruses that establish persistent infections do not disseminate as easily and must often rely on close contact and direct exchange of tissues between host organisms in order to be efficiently transmitted. Persistence can thus be seen as a means for a virus to remain in the primary host until the opportunity to infect another organism arises. Many persistent infections are initiated through an acute phase of infection that provides the virus with an opportunity to establish an infection within the new host. Subsequent to this acute phase, a virus may be cleared from specific cell types, while particular cell subsets serve as sanctuaries for the retention of a permanent viral population (Fig. 1). In the case of herpes simplex virus (HSV), productive (lytic) infection occurs in epithelial cells; this lytic infection is subject to the actions of the immune system, and these sites are cleared in immunocompetent individuals (4, 21, 25, 72). However, a nonproductive (latent) infection is established in nearby sensory neurons (25, 72). Neurons are long-lived, terminally differentiated cells, providing the virus with a virtually everlasting home within the host. Another herpesvirus, mouse cytomegalovirus (MCMV), is able to reside within the cells of the salivary gland due to viral induction of immunosuppressive cytokine expression at this site (34) and the apparent refractive nature of the salivary gland epithelium to cytotoxic T-lymphocyte (CTL) effector functions (14).
FIG. 1.
Selection of cell subsets ideal for long-term maintenance of the viral genome. Particular cell subsets serve as sanctuaries for the retention of a permanent viral population. Herpes simplex virus (HSV) establishes latent infection in sensory neurons. Mouse cytomegalovirus (MCMV) maintains a viral reservoir in the salivary gland. The renal epithelium is the site of permanent infection for lymphocytic choriomeningitis virus (LCMV), simian virus 40 (SV40), and JC and BK viruses. Epstein Barr Virus (EBV) resides in the memory B-cell compartment. Human immunodeficiency virus (HIV) and human T-lymphotropic virus (HTLV) persist in resting T cells. Stem cells serve as a reservoir for both mouse mammary tumor virus (MMTV) and murine leukemia virus (MuLV).
Specific organ anatomy can also provide a virus with the opportunity to establish a sanctuary site, such as the kidney in the case of lymphocytic choriomeningitis virus (LCMV), wherein a robust antiviral T-cell response is able to clear the virus from most organs but actually facilitates viral replication in the renal tubular epithelium (71) (Fig. 1). The renal epithelium is also the site of persistence for several polyomaviruses, including simian virus 40 (SV40), the K virus of mice, and the JC and BK viruses of humans (12, 23, 24, 30, 69, 76), suggesting that the specific architecture of the kidney may provide a particularly attractive environment for establishment of a long-term infection.
Long-lived cells of the immune system can provide an ideal shelter for persistent viral infections, as in the case of Epstein-Barr virus (EBV), which resides in the resting memory B-cell compartment (2, 57) (Fig. 1). In some instances, the differentiation state of the infected cell can determine whether it is a target for productive replication or establishment of persistence. An example is infection by human immunodeficiency virus (HIV), in which activated CD4+ T cells support productive viral replication (55), while resting CD4+ T cells maintain a dormant reservoir of the virus (16) (Fig. 1).
Since retroviruses integrate into the host genome, any infected cell retains the viral genome throughout its lifetime. If the infected cell belongs to the stem cell population, then the virus will persist for an indefinite period, providing a continuous supply of infectious virions. This strategy is employed by retroviruses such as murine leukemia virus (MuLV), which establishes infection in hematopoietic progenitor cells in the bone marrow (73, 74), and by mouse mammary tumor virus (MMTV), which infects and persists in mammary epithelial stem cells (28, 44) (Fig. 1).
MODULATION OF VIRAL GENE EXPRESSION
As already mentioned, patterns of persistent infections can be quite different; some viruses continue to replicate (arenaviruses, flaviviruses), while others produce no infectious particles (herpesviruses during the latency stage). The need for a virus to shut down its gene expression program depends on the virus life cycle. Whereas some infections yield virions without causing host cell death, others induce cytopathic effects in target cells. In general, noncytopathic viruses do not need to modify their gene expression program but require attenuation of host responses to prevent clearance. In contrast, cytopathic viruses need to modulate expression of their genes to ensure survival of the host cell and also to avoid immune recognition. To achieve this, many cytopathic viruses utilize host mechanisms such as chromatin modification for the regulation of gene expression.
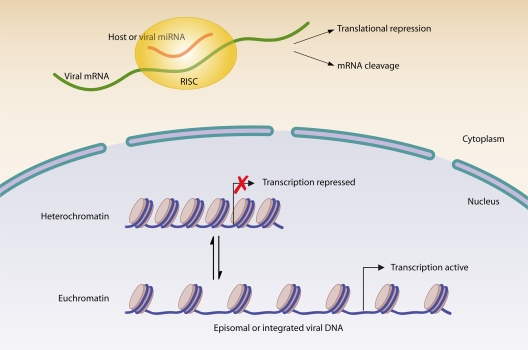
In the host cell nucleus, DNA is wrapped around a core of histone proteins to form the nucleosome; the higher-order folding of the DNA in the nucleosome results in either the transcriptionally active, less-condensed euchromatin form or the transcriptionally silent, highly condensed heterochromatin form (36) (Fig. 2). Modification of histones alters their interaction with DNA and thus influences gene expression (36). Viruses can utilize and induce changes in chromatin structure to aid in the establishment of a persistent infection. HSV latency provides an excellent example of viral interplay with host chromatin.
FIG. 2.
Modulation of viral gene expression. Viruses utilize reversible chromatin modifications to control transcription of viral genes. Viral transcription is active when associated with euchromatin and repressed when coupled to heterochromatin. Alternatively, host and/or viral miRNAs can be used to control production of viral proteins through translational repression or cleavage of viral mRNA. RISC, RNA-induced silencing complex.
The HSV replication cycle is cytopathic (25, 42); thus, establishment of a quiescent latent infection state is required for the virus to persist. During latent infection of neurons, HSV DNA exists in the nucleus in a circular episomal form associated with histones (42, 72) (Fig. 2). During this time, lytic genes are associated with heterochromatin, preventing their transcription, while only the latency-associated transcript (LAT) gene is associated with active euchromatin, allowing for its expression (42, 46). In fact, this appears to be a virally directed process, as LAT gene expression alone is sufficient to promote the assembly of heterochromatin on lytic gene promoters, thereby limiting lytic gene expression (91).
Retroviral proviruses are also subject to the effects of chromatin modification. The site of integration is a critical factor in the level of proviral gene expression (38); a provirus may integrate into euchromatin and be expressed or integrate into heterochromatin and become repressed (59). However, provirus integrated into a silent locus may later be reactivated (59) and can thus serve as a hidden reservoir of infection. One of the most well-characterized dynamic viral interactions with host chromatin proteins is that of the human T-lymphotropic virus type 1 (HTLV-1) Tax protein. Tax activity is a major factor contributing to HTLV-1 persistence, and its interactions with host chromatin-modifying enzymes are known to selectively activate or repress both viral and host genes to facilitate viral persistence (8).
While viruses that do not use the host cell nucleus for replication cannot establish latent infections or utilize chromatin modifications to regulate viral gene expression, this does not prohibit them from establishing persistent infections. One such example is hepatitis C virus (HCV), which persists in the liver of most infected individuals but does not establish latency (77). Furthermore, virus production remains high during the persistent stage of infection (65). In this case, the virus must rely on its numerous immune evasion mechanisms, which are discussed later and reviewed in reference 80.
In addition to maintaining persistent infection by limiting viral gene transcription, viruses often prevent translation of already-transcribed genes. The eukaryotic cell contains a complex system to regulate the expression of transcribed genes. This system includes the use of RNA interference, whereby noncoding RNAs inhibit translation of mRNAs (15). Persistent pathogens have not failed to coopt RNA interference pathways during infection, as they use both host and viral microRNAs (miRNAs) to control gene expression and promote long-term infection (Fig. 2). Examples include negative regulation of the HSV type 2 (HSV-2) lytic proteins ICP34.5 and ICP0 in neurons by LAT-encoded miR-I, -II, and -III (83, 84). The LAT of HSV-1 has also been revealed as a miRNA precursor for miR-H2-3p and miR-H6, serving to inhibit expression of ICP0 and ICP4 (88). SV40 has also been shown to encode miRNAs, which accumulate later in infection and reduce expression of early viral genes, in particular that of the highly immunogenic large T antigen, thereby reducing CTL activation while maintaining infectious virion production (82). Inhibition of viral gene expression by miRNAs is not necessarily directed by the virus; host miRNAs can also play a role in limiting viral gene expression, allowing the virus to persist in cells with a specific miRNA profile. For example, HIV proviral silencing in resting CD4+ T cells is attributed at least in part to host miRNA-mediated suppression (33) and thus contributes to the establishment of a latent reservoir of the virus. In this case, it appears that establishment of the latent reservoir is not the result of virus subversion of the immune response but rather an indirect result of host suppression of the virus.
VIRAL SUBVERSION OF CELLULAR APOPTOTIC PATHWAYS
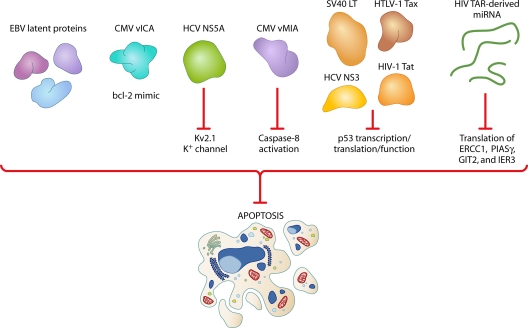
Apoptosis is perhaps the most primordial response of a virally infected cell, designed to thwart the spread of infection and protect the organism as a whole. Whereas an acutely infecting virus needs to prevent apoptosis of infected cells only long enough to produce progeny virions, a persisting virus must have a means to suppress apoptosis for a much longer time in order to maintain a compartment of infected cells (Fig. 3). For instance, EBV latent proteins have been shown to prevent apoptosis of infected B cells, promoting survival of the latently infected resting B-cell compartment (31). Other examples from the herpesvirus family include the CMV proteins vMIA and vICA, which are shown to inhibit apoptosis by mimicking the host antiapoptotic protein bcl-2 and inhibiting caspase-8 activation, respectively (27, 79). The tumor suppressor protein p53 is an attractive target for many viruses; most notably, the SV40 large T antigen (LT) is known to bind and inactivate p53, thereby preventing apoptosis (47, 52, 54). Among retroviruses, the HLTV-1 Tax protein prevents apoptosis through activation of the NF-κB and Akt cellular prosurvival pathways and negative regulation of p53 (85), while the HIV accessory protein Tat thwarts apoptosis through inhibition of p53 transcription (29). The antiapoptotic HCV protein NS3 has also been shown to target p53 and inhibit its function (22). An additional novel HCV antiapoptotic mechanism wherein the viral protein NS5A inhibits the Kv2.1 K+ channel, preventing hepatoma cells from apoptosis in response to oxidative stress, has recently been described (53).
FIG. 3.
Viral subversion of cellular apoptotic pathways. Persistent viruses must have a means to suppress apoptosis in order to maintain a compartment of infected cells. The latent proteins of EBV prevent apoptosis of infected B cells. The CMV protein vICA inhibits apoptosis by mimicking the host protein bcl-2. HCV NS5A suppresses apoptosis by inhibiting the Kv2.1 K+ channel, while CMV vMIA prevents caspase-8 activation. The cellular proapoptotic tumor suppressor p53 is inhibited at the transcriptional, translational, or functional level by SV40 large T antigen (LT), HCV NS3, HTLV-1 Tax, and HIV Tat. miRNAs derived from the HIV Tar transcript inhibit translation of the cellular proapoptotic proteins ERCC1, PIASγ, GIT2, and IER3.
Host and viral miRNAs are also utilized by persistent viruses to inhibit translation of cellular proapoptotic genes. For example, a recent report demonstrated that miRNAs derived from the HIV-1 trans-acting responsive element (TAR) suppress the expression of several cellular genes involved in apoptosis, including ERCC1, PIASγ, GIT2, and IER3 (41). Additionally, HTLV-1 Tax transactivates promoters of the host miRNAs that target the p53 transcript, thereby inhibiting apoptosis (7, 68, 93). These examples are by no means exhaustive, and for each virus, the manipulation of the apoptotic pathway is quite complex. However, an in-depth discussion is beyond the scope of this review.
AVOIDANCE OF CLEARANCE BY THE IMMUNE SYSTEM
Though mammals have developed an intricate and highly protective immune system, viruses have evolved mechanisms in order to avoid recognition and clearance by their hosts. Just as the signaling networks involved in the host immune response are varied and complex, viral evasion of the effects of these networks is also quite diverse. Acutely infecting viruses avoid immune elimination (or death of the host) just long enough to transmit to a subsequent organism, and they can accomplish this through universal disruption of the immune response. In contrast, persistent viruses must prevent recognition and elimination by both the innate and the adaptive immune systems for long periods of time. For this reason, they utilize a few common strategies to manipulate the host response. Here, we will focus on the common mechanisms employed by distinct viruses to avoid immune clearance. For an in-depth discussion of modulation of the immune response during persistent viral infections, we recommend the recently published review by Virgin et al. (89).
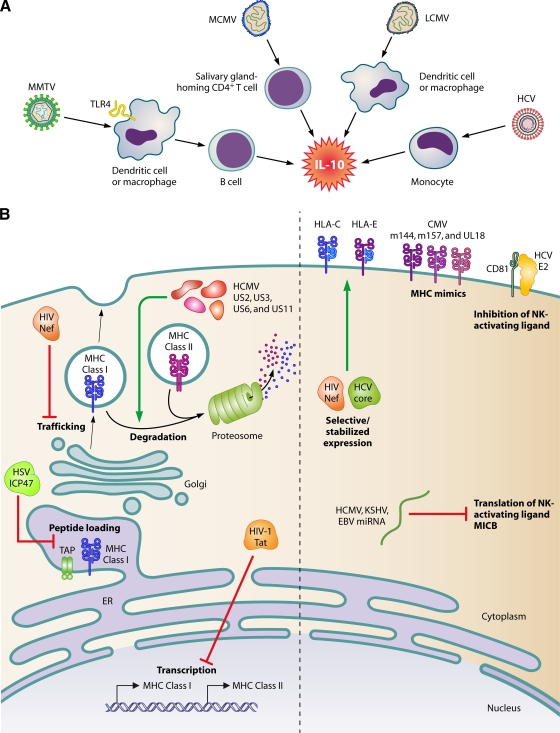
One way to avoid immune clearance is by modification of host cytokine production and signaling. Examples in persistent infections include inhibition of type I interferon (IFN) production by HCV (5), modification of immune cell migration through chemokine mimicry by cytomegalovirus (CMV) and HIV (62), and dysregulation of expression of cell adhesion molecules necessary for immune cell migration, such as LFA-3 and ICAM-1, by EBV and CMV (6, 56, 58). However, the most common theme in viral subversion of immune signaling is stimulation of interleukin-10 (IL-10) production (Fig. 4A). IL-10 is a well-characterized immunosuppressive cytokine that inhibits a broad spectrum of immune responses, including the suppression of stimulatory cytokine production, T-cell proliferation, and B-cell responses (60). Thus, it is not surprising that successful viral infections can take advantage of IL-10 in their evasion of the immune response. CMV appears to exploit elevated production of IL-10 by CD4+ T cells homed to the salivary glands to establish persistent infection at this site (34). An increase in IL-10 production by antigen-presenting cells in LCMV-infected mice impairs T-cell responses, contributing to chronic/persistent infection (10). The NS4 protein of HCV stimulates IL-10 production by monocytes and thereby inhibits CD4+ T-cell responses against the virus (9). Similarly, increased IL-10 production in HIV-infected patients has been observed (18), and reduced expression of IL-10 has been correlated with a slower progression to AIDS (78). Another retrovirus, MMTV, subverts the toll-like receptor 4 (TLR4) signaling on dendritic cells and/or macrophages to stimulate production of IL-10 by B cells (39). IL-10 homologues with immunosuppressive activity are also encoded by some viruses, including EBV and HCMV (45, 75, 81). In addition to the stimulation of IL-10 production to suppress the antiviral immune response, there is also evidence that many persistent viral infections result in the upregulation of regulatory T-cell function, which inhibits virus-specific CTL responses. This topic was recently reviewed in reference 51.
FIG. 4.
Avoidance of clearance by the immune system. (A) Viral stimulation of IL-10 production. MMTV subverts TLR4 signaling on dendritic cells or macrophages to stimulate IL-10 production by B cells, while MCMV elicits IL-10 production by salivary gland-homed CD4+ T cells. LCMV stimulate dendritic cells, whereas HCV stimulates macrophages and monocytes to secrete IL-10. (B, left) Viral proteins counteract cell surface antigen presentation by MHC molecules. The HIV Tat protein inhibits transcription of MHC class I and II. Peptide loading onto MHC class I is repressed by HSV ICP47 binding to the transporter associated with antigen processing (TAP) complex. HIV Nef inhibits trafficking of MHC class I to the cell surface. HCMV proteins US2, US3, US6, and US11 target MHC class I and II for proteasomal degradation. (B, right) Inhibition of the NK cell response. Translation of the NK cell-activating ligand, MICB, is inhibited by HCMV-, KSHV-, and EBV-produced miRNAs. HIV Nef and HCV core proteins stabilize expression of HLA-C and -E, inhibitory ligands of NK receptors. CMV inhibits NK activation by expressing MHC mimics. HCV E2 binds to CD81, inhibiting its NK-activating function.
In order to persist, any virus must avoid presentation of viral antigens and subsequent activation of CTLs and/or B cells. Persistent viruses utilize a variety of mechanisms to inhibit the presentation of viral peptides to T cells by major histocompatibility complex (MHC) molecules (Fig. 4B). HSV ICP47 binds to the transporter associated with antigen processing (TAP) complex, preventing peptide loading onto the MHC class I heavy chain (32). Human cytomegalovirus (HCMV) encodes four proteins, US2, US3, US6, and US11, to downregulate MHC class I (and class II) molecules by targeting them to proteasomal degradation (3). Finally, the HIV Tat protein has been shown to inhibit transcription of MHC class I and class II molecules in infected cells (40, 92), while the Nef protein modifies the intracellular trafficking of MHC class I molecules to prevent their surface expression (49). In addition to downregulation of MHC expression, persistent RNA viruses such as HIV and HCV can also avoid CTL recognition through the continued production of immune escape viral variants due to their low-fidelity polymerases (37, 86).
The downregulation of MHC class I molecules, while limiting viral antigen presentation, can also lead to recognition and killing of infected cells by NK cells. Persistent viruses have thus evolved mechanisms to specifically inhibit the NK cell response (Fig. 4B). A virus can take advantage of the varied specificity of MHC molecules to prevent NK cell activation. For example, HLA-A and -B molecules are known to effectively present HIV peptides to CTLs (17, 43, 66, 90), while HLA-C and -E serve as ligands for inhibitory NK cell receptors (48). The Nef protein of HIV-1 specifically downregulates HLA-A and -B molecules but leaves expression of HLA-C and -E unaffected, providing protection from NK-mediated cell lysis (19, 49). HCV inhibits the NK cell response through the stabilization of HLA-E expression by the HCV core protein (64) and binding of the envelope protein E2 to the cell surface molecule CD81 (20, 64, 87).
MicroRNAs can also be an effective tool to avoid NK cell recognition; miRNAs expressed by HCMV, Kaposi's sarcoma-associated herpesvirus (KSHV), and EBV have recently been shown to target the stress-induced NK cell ligand MICB to prevent elimination of infected cells (63). CMV has developed another elegant mechanism to prevent NK cell killing of infected cells—the production of MHC mimics, such as the m144 and m157 proteins of MCMV and the UL18 protein of HCMV (67). In fact, the use of cytokine and chemokine mimics is a common strategy employed by viruses to evade the immune system, the vast majority of which are not discussed here but are reviewed in reference 1.
SUMMARY
The common threads that appear among diverse persistent viral infections are evidence of the long-term evolutionary relationships that exist between organisms and their viral invaders. Despite the complex cell-intrinsic and intercellular antivirus mechanisms available at the host's disposal, viruses have developed various means that counteract these mechanisms and allow for virus persistence. Evaluation of these common mechanisms provides us not only with a greater understanding of the complex biology of host-virus interactions but also with the hope of treating and eliminating these infections.
Acknowledgments
We thank Glenn Randall and Alexander Chervonsky for helpful discussions.
Biography
 Tatyana Golovkina received her M.S. in biochemistry from Moscow State University, Moscow, former Soviet Union. She pursued graduate studies at the USSR Academy of Medical Sciences Cancer Research Center in Moscow. Her doctoral thesis described studies on evolution of endogenous retroviruses in the mammalian genome. She then joined Susan Ross’ laboratory at the Department of Biochemistry, University of Illinois at Chicago, and later moved with the laboratory to the Department of Microbiology, University of Pennsylvania, where her research focused on mechanisms of mouse mammary tumor virus-host interactions. She joined the Jackson Laboratory in Bar Harbor, ME, in 1997 and began studying the genetics of resistance to retroviral infection. In 2005, she relocated her laboratory to the Department of Microbiology at the University of Chicago. The major interest of her laboratory is the utilization of genetically resistant inbred mice to understand how the genetic makeup of the host influences susceptibility to retroviral infection and pathogenesis.
Tatyana Golovkina received her M.S. in biochemistry from Moscow State University, Moscow, former Soviet Union. She pursued graduate studies at the USSR Academy of Medical Sciences Cancer Research Center in Moscow. Her doctoral thesis described studies on evolution of endogenous retroviruses in the mammalian genome. She then joined Susan Ross’ laboratory at the Department of Biochemistry, University of Illinois at Chicago, and later moved with the laboratory to the Department of Microbiology, University of Pennsylvania, where her research focused on mechanisms of mouse mammary tumor virus-host interactions. She joined the Jackson Laboratory in Bar Harbor, ME, in 1997 and began studying the genetics of resistance to retroviral infection. In 2005, she relocated her laboratory to the Department of Microbiology at the University of Chicago. The major interest of her laboratory is the utilization of genetically resistant inbred mice to understand how the genetic makeup of the host influences susceptibility to retroviral infection and pathogenesis.
 Melissa Kane received her B.S. in animal sciences from Cornell University, Ithaca, NY, in 2005. She then worked as a research assistant studying B-cell development and autoreactivity in the laboratory of Roberta Pelanda at the University of Colorado Health Sciences/National Jewish Medical and Research Center. She is currently in her third year of graduate studies in microbiology in the laboratory of Tatyana Golovkina at the University of Chicago. Her thesis research focuses on the mechanisms of retroviral resistance inherited by I/LnJ mice.
Melissa Kane received her B.S. in animal sciences from Cornell University, Ithaca, NY, in 2005. She then worked as a research assistant studying B-cell development and autoreactivity in the laboratory of Roberta Pelanda at the University of Colorado Health Sciences/National Jewish Medical and Research Center. She is currently in her third year of graduate studies in microbiology in the laboratory of Tatyana Golovkina at the University of Chicago. Her thesis research focuses on the mechanisms of retroviral resistance inherited by I/LnJ mice.
Footnotes
Published ahead of print on 2 December 2009.
REFERENCES
- 1.Alcami, A. 2003. Viral mimicry of cytokines, chemokines and their receptors. Nat. Rev. Immunol. 3:36-50. [DOI] [PubMed] [Google Scholar]
- 2.Babcock, G. J., L. L. Decker, M. Volk, and D. A. Thorley-Lawson. 1998. EBV persistence in memory B cells in vivo. Immunity 9:395-404. [DOI] [PubMed] [Google Scholar]
- 3.Basta, S., and J. R. Bennink. 2003. A survival game of hide and seek: cytomegaloviruses and MHC class I antigen presentation pathways. Viral Immunol. 16:231-242. [DOI] [PubMed] [Google Scholar]
- 4.Bloom, D. C. 2004. HSV LAT and neuronal survival. Int. Rev. Immunol. 23:187-198. [DOI] [PubMed] [Google Scholar]
- 5.Bode, J. G., E. D. Brenndorfer, and D. Haussinger. 2008. Hepatitis C virus (HCV) employs multiple strategies to subvert the host innate antiviral response. Biol. Chem. 389:1283-1298. [DOI] [PubMed] [Google Scholar]
- 6.Boldogh, I., T. Albrecht, and D. D. Porter. 1996. Persistent viral infections, p. 585-596. In S. Baron (ed.), Medical microbiology, 4th ed. University of Texas Medical Branch at Galveston, Galveston, TX. [PubMed]
- 7.Bouzar, A. B., and L. Willems. 2008. How HTLV-1 may subvert miRNAs for persistence and transformation. Retrovirology 5:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boxus, M., J. C. Twizere, S. Legros, J. F. Dewulf, R. Kettmann, and L. Willems. 2008. The HTLV-1 Tax interactome. Retrovirology 5:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brady, M. T., A. J. MacDonald, A. G. Rowan, and K. H. Mills. 2003. Hepatitis C virus non-structural protein 4 suppresses Th1 responses by stimulating IL-10 production from monocytes. Eur. J. Immunol. 33:3448-3457. [DOI] [PubMed] [Google Scholar]
- 10.Brooks, D. G., M. J. Trifilo, K. H. Edelmann, L. Teyton, D. B. McGavern, and M. B. Oldstone. 2006. Interleukin-10 determines viral clearance or persistence in vivo. Nat. Med. 12:1301-1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buchmeier, M. J., J. de la Torre, and C. J. Peters. 2007. Arenaviridae: the viruses and their replication, p. 1791-1828. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 12.Buffett, R. F., and J. D. Levinthal. 1962. Polyoma virus infection in mice. Arch. Pathol. 74:513-526. [PubMed] [Google Scholar]
- 13.Bushman, F., M. Lewinski, A. Ciuffi, S. Barr, J. Leipzig, S. Hannenhalli, and C. Hoffmann. 2005. Genome-wide analysis of retroviral DNA integration. Nat. Rev. Microbiol. 3:848-858. [DOI] [PubMed] [Google Scholar]
- 14.Campbell, A. E., V. J. Cavanaugh, and J. S. Slater. 2008. The salivary glands as a privileged site of cytomegalovirus immune evasion and persistence. Med. Microbiol. Immunol. 197:205-213. [DOI] [PubMed] [Google Scholar]
- 15.Carthew, R. W., and E. J. Sontheimer. 2009. Origins and mechanisms of miRNAs and siRNAs. Cell 136:642-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chun, T. W., D. Finzi, J. Margolick, K. Chadwick, D. Schwartz, and R. F. Siliciano. 1995. In vivo fate of HIV-1-infected T cells: quantitative analysis of the transition to stable latency. Nat. Med. 1:1284-1290. [DOI] [PubMed] [Google Scholar]
- 17.Claverie, J. M., P. Kourilsky, P. Langlade-Demoyen, A. Chalufour-Prochnicka, G. Dadaglio, F. Tekaia, F. Plata, and L. Bougueleret. 1988. T-immunogenic peptides are constituted of rare sequence patterns: use in the identification of T epitopes in the human immunodeficiency virus gag protein. Eur. J. Immunol. 18:1547-1553. [DOI] [PubMed] [Google Scholar]
- 18.Clerici, M., C. Balotta, A. Salvaggio, C. Riva, D. Trabattoni, L. Papagno, A. Berlusconi, S. Rusconi, M. L. Villa, M. Moroni, and M. Galli. 1996. Human immunodeficiency virus (HIV) phenotype and interleukin-2/interleukin-10 ratio are associated markers of protection and progression in HIV infection. Blood 88:574-579. [PubMed] [Google Scholar]
- 19.Cohen, G. B., R. T. Gandhi, D. M. Davis, O. Mandelboim, B. K. Chen, J. L. Strominger, and D. Baltimore. 1999. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity 10:661-671. [DOI] [PubMed] [Google Scholar]
- 20.Crotta, S., A. Stilla, A. Wack, A. D'Andrea, S. Nuti, U. D'Oro, M. Mosca, F. Filliponi, R. M. Brunetto, F. Bonino, S. Abrignani, and N. M. Valiante. 2002. Inhibition of natural killer cells through engagement of CD81 by the major hepatitis C virus envelope protein. J. Exp. Med. 195:35-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Decman, V., M. L. Freeman, P. R. Kinchington, and R. L. Hendricks. 2005. Immune control of HSV-1 latency. Viral Immunol. 18:466-473. [DOI] [PubMed] [Google Scholar]
- 22.Deng, L., M. Nagano-Fujii, M. Tanaka, Y. Nomura-Takigawa, M. Ikeda, N. Kato, K. Sada, and H. Hotta. 2006. NS3 protein of Hepatitis C virus associates with the tumour suppressor p53 and inhibits its function in an NS3 sequence-dependent manner. J. Gen. Virol. 87:1703-1713. [DOI] [PubMed] [Google Scholar]
- 23.Dorries, K., and V. ter Meulen. 1983. Progressive multifocal leucoencephalopathy: detection of papovavirus JC in kidney tissue. J. Med. Virol. 11:307-317. [DOI] [PubMed] [Google Scholar]
- 24.Dubensky, T. W., and L. P. Villarreal. 1984. The primary site of replication alters the eventual site of persistent infection by polyomavirus in mice. J. Virol. 50:541-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Efstathiou, S., and C. M. Preston. 2005. Towards an understanding of the molecular basis of herpes simplex virus latency. Virus Res. 111:108-119. [DOI] [PubMed] [Google Scholar]
- 26.Goff, S. 2007. Retroviridae: the viruses and their replication, p. 1999-2070. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 27.Goldmacher, V. S., L. M. Bartle, A. Skaletskaya, C. A. Dionne, N. L. Kedersha, C. A. Vater, J. W. Han, R. J. Lutz, S. Watanabe, E. D. Cahir McFarland, E. D. Kieff, E. S. Mocarski, and T. Chittenden. 1999. A cytomegalovirus-encoded mitochondria-localized inhibitor of apoptosis structurally unrelated to Bcl-2. Proc. Natl. Acad. Sci. U. S. A. 96:12536-12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Golovkina, T. V., J. P. Dudley, and S. R. Ross. 1998. B and T cells are required for mouse mammary tumor virus spread within the mammary gland. J. Immunol. 161:2375-2382. [PubMed] [Google Scholar]
- 29.Gougeon, M. L. 2003. Apoptosis as an HIV strategy to escape immune attack. Nat. Rev. Immunol. 3:392-404. [DOI] [PubMed] [Google Scholar]
- 30.Greenlee, J. E., R. C. Phelps, and W. G. Stroop. 1991. The major site of murine K papovavirus persistence and reactivation is the renal tubular epithelium. Microb. Pathog. 11:237-247. [DOI] [PubMed] [Google Scholar]
- 31.Gregory, C. D., C. Dive, S. Henderson, C. A. Smith, G. T. Williams, J. Gordon, and A. B. Rickinson. 1991. Activation of Epstein-Barr virus latent genes protects human B cells from death by apoptosis. Nature 349:612-614. [DOI] [PubMed] [Google Scholar]
- 32.Hill, A., P. Jugovic, I. York, G. Russ, J. Bennink, J. Yewdell, H. Ploegh, and D. Johnson. 1995. Herpes simplex virus turns off the TAP to evade host immunity. Nature 375:411-415. [DOI] [PubMed] [Google Scholar]
- 33.Huang, J., F. Wang, E. Argyris, K. Chen, Z. Liang, H. Tian, W. Huang, K. Squires, G. Verlinghieri, and H. Zhang. 2007. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat. Med. 13:1241-1247. [DOI] [PubMed] [Google Scholar]
- 34.Humphreys, I. R., C. de Trez, A. Kinkade, C. A. Benedict, M. Croft, and C. F. Ware. 2007. Cytomegalovirus exploits IL-10-mediated immune regulation in the salivary glands. J. Exp. Med. 204:1217-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Imperiale, M. J., and E. O. Major. 2007. Polyomaviruses, p. 2263-2298. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 36.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 37.Johnson, W. E., and R. C. Desrosiers. 2002. Viral persistance: HIV's strategies of immune system evasion. Annu. Rev. Med. 53:499-518. [DOI] [PubMed] [Google Scholar]
- 38.Jordan, A., P. Defechereux, and E. Verdin. 2001. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J. 20:1726-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jude, B. A., Y. Pobezinskaya, J. Bishop, S. Parke, R. M. Medzhitov, A. V. Chervonsky, and T. V. Golovkina. 2003. Subversion of the innate immune system by a retrovirus. Nat. Immunol. 4:573-578. [DOI] [PubMed] [Google Scholar]
- 40.Kanazawa, S., T. Okamoto, and B. M. Peterlin. 2000. Tat competes with CIITA for the binding to P-TEFb and blocks the expression of MHC class II genes in HIV infection. Immunity 12:61-70. [DOI] [PubMed] [Google Scholar]
- 41.Klase, Z., R. Winograd, J. Davis, L. Carpio, R. Hildreth, M. Heydarian, S. Fu, T. McCaffrey, E. Meiri, M. Ayash-Rashkovsky, S. Gilad, Z. Bentwich, and F. Kashanchi. 2009. HIV-1 TAR miRNA protects against apoptosis by altering cellular gene expression. Retrovirology 6:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knipe, D. M., and A. Cliffe. 2008. Chromatin control of herpes simplex virus lytic and latent infection. Nat. Rev. Microbiol. 6:211-221. [DOI] [PubMed] [Google Scholar]
- 43.Koenig, S., T. R. Fuerst, L. V. Wood, R. M. Woods, J. A. Suzich, G. M. Jones, V. F. de la Cruz, R. T. Davey, Jr., S. Venkatesan, B. Moss, et al. 1990. Mapping the fine specificity of a cytolytic T cell response to HIV-1 nef protein. J. Immunol. 145:127-135. [PubMed] [Google Scholar]
- 44.Kordon, E. C., and G. H. Smith. 1998. An entire functional mammary gland may comprise the progeny from a single cell. Development 125:1921-1930. [DOI] [PubMed] [Google Scholar]
- 45.Kotenko, S. V., S. Saccani, L. S. Izotova, O. V. Mirochnitchenko, and S. Pestka. 2000. Human cytomegalovirus harbors its own unique IL-10 homolog (cmvIL-10). Proc. Natl. Acad. Sci. U. S. A. 97:1695-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kubat, N. J., A. L. Amelio, N. V. Giordani, and D. C. Bloom. 2004. The herpes simplex virus type 1 latency-associated transcript (LAT) enhancer/rcr is hyperacetylated during latency independently of LAT transcription. J. Virol. 78:12508-12518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lane, D. P., and L. V. Crawford. 1979. T antigen is bound to a host protein in SV40-transformed cells. Nature 278:261-263. [DOI] [PubMed] [Google Scholar]
- 48.Lanier, L. L. 1998. NK cell receptors. Annu. Rev. Immunol. 16:359-393. [DOI] [PubMed] [Google Scholar]
- 49.Le Gall, S., L. Erdtmann, S. Benichou, C. Berlioz-Torrent, L. Liu, R. Benarous, J. M. Heard, and O. Schwartz. 1998. Nef interacts with the mu subunit of clathrin adaptor complexes and reveals a cryptic sorting signal in MHC I molecules. Immunity 8:483-495. [DOI] [PubMed] [Google Scholar]
- 50.Lemon, S. M., C. Walker, M. J. Alter, and M. Yi. 2007. Hepatitis C virus, p. 1253-1304. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 51.Li, S., E. J. Gowans, C. Chougnet, M. Plebanski, and U. Dittmer. 2008. Natural regulatory T cells and persistent viral infection. J. Virol. 82:21-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Linzer, D. I., and A. J. Levine. 1979. Characterization of a 54K Dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell 17:43-52. [DOI] [PubMed] [Google Scholar]
- 53.Mankouri, J., M. L. Dallas, M. E. Hughes, S. D. Griffin, A. Macdonald, C. Peers, and M. Harris. 2009. Suppression of a pro-apoptotic K+ channel as a mechanism for hepatitis C virus persistence. Proc. Natl. Acad. Sci. U. S. A. [DOI] [PMC free article] [PubMed]
- 54.McCormick, F., and E. Harlow. 1980. Association of a murine 53,000-dalton phosphoprotein with simian virus 40 large-T antigen in transformed cells. J. Virol. 34:213-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McDougal, J. S., A. Mawle, S. P. Cort, J. K. Nicholson, G. D. Cross, J. A. Scheppler-Campbell, D. Hicks, and J. Sligh. 1985. Cellular tropism of the human retrovirus HTLV-III/LAV. I. Role of T cell activation and expression of the T4 antigen. J. Immunol. 135:3151-3162. [PubMed] [Google Scholar]
- 56.Middeldorp, J. M., and D. M. Pegtel. 2008. Multiple roles of LMP1 in Epstein-Barr virus induced immune escape. Semin. Cancer Biol. 18:388-396. [DOI] [PubMed] [Google Scholar]
- 57.Miyashita, E. M., B. Yang, G. J. Babcock, and D. A. Thorley-Lawson. 1997. Identification of the site of Epstein-Barr virus persistence in vivo as a resting B cell. J. Virol. 71:4882-4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mocarski, E. S., Jr. 2002. Immunomodulation by cytomegaloviruses: manipulative strategies beyond evasion. Trends Microbiol. 10:332-339. [DOI] [PubMed] [Google Scholar]
- 59.Mok, H. P., and A. M. Lever. 2007. Chromatin, gene silencing and HIV latency. Genome Biol. 8:228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moore, K. W., R. de Waal Malefyt, R. L. Coffman, and A. O'Garra. 2001. Interleukin-10 and the interleukin-10 receptor. Annu. Rev. Immunol. 19:683-765. [DOI] [PubMed] [Google Scholar]
- 61.Moser, J. M., and A. E. Lukacher. 2001. Immunity to polyoma virus infection and tumorigenesis. Viral Immunol. 14:199-216. [DOI] [PubMed] [Google Scholar]
- 62.Murphy, P. M. 2001. Viral exploitation and subversion of the immune system through chemokine mimicry. Nat. Immunol. 2:116-122. [DOI] [PubMed] [Google Scholar]
- 63.Nachmani, D., N. Stern-Ginossar, R. Sarid, and O. Mandelboim. 2009. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe 5:376-385. [DOI] [PubMed] [Google Scholar]
- 64.Nattermann, J., H. D. Nischalke, V. Hofmeister, G. Ahlenstiel, H. Zimmermann, L. Leifeld, E. H. Weiss, T. Sauerbruch, and U. Spengler. 2005. The HLA-A2 restricted T cell epitope HCV core 35-44 stabilizes HLA-E expression and inhibits cytolysis mediated by natural killer cells. Am. J. Pathol. 166:443-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Neumann, A. U., N. P. Lam, H. Dahari, D. R. Gretch, T. E. Wiley, T. J. Layden, and A. S. Perelson. 1998. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science 282:103-107. [DOI] [PubMed] [Google Scholar]
- 66.Nixon, D. F., A. R. Townsend, J. G. Elvin, C. R. Rizza, J. Gallwey, and A. J. McMichael. 1988. HIV-1 gag-specific cytotoxic T lymphocytes defined with recombinant vaccinia virus and synthetic peptides. Nature 336:484-487. [DOI] [PubMed] [Google Scholar]
- 67.Orange, J. S., M. S. Fassett, L. A. Koopman, J. E. Boyson, and J. L. Strominger. 2002. Viral evasion of natural killer cells. Nat. Immunol. 3:1006-1012. [DOI] [PubMed] [Google Scholar]
- 68.Pichler, K., G. Schneider, and R. Grassmann. 2008. MicroRNA miR-146a and further oncogenesis-related cellular microRNAs are dysregulated in HTLV-1-transformed T lymphocytes. Retrovirology 5:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Prywes, R., J. G. Foulkes, N. Rosenberg, and D. Baltimore. 1983. Sequences of the A-MuLV protein needed for fibroblast and lymphoid cell transformation. Cell 34:569-579. [DOI] [PubMed] [Google Scholar]
- 70.Rawls, W. E., M. A. Chan, and S. R. Gee. 1981. Mechanisms of persistence in arenavirus infections: a brief review. Can. J. Microbiol. 27:568-574. [DOI] [PubMed] [Google Scholar]
- 71.Recher, M., K. S. Lang, A. Navarini, L. Hunziker, P. A. Lang, K. Fink, S. Freigang, P. Georgiev, L. Hangartner, R. Zellweger, A. Bergthaler, A. N. Hegazy, B. Eschli, A. Theocharides, L. T. Jeker, D. Merkler, B. Odermatt, M. Hersberger, H. Hengartner, and R. M. Zinkernagel. 2007. Extralymphatic virus sanctuaries as a consequence of potent T-cell activation. Nat. Med. 13:1316-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Roizman, B., D. M. Knipe, and R. J. Whitley. 2007. Herpes simplex viruses, p. 2501-2602. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 73.Rosenberg, N., and P. Jolicoeur. 1997. Retroviral pathogenesis, p. 475-586. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- 74.Ruddle, N. H., M. K. Armstrong, and F. F. Richards. 1976. Replication of murine leukemia virus in bone marrow-derived lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 73:3714-3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Salek-Ardakani, S., J. R. Arrand, and M. Mackett. 2002. Epstein-Barr virus encoded interleukin-10 inhibits HLA-class I, ICAM-1, and B7 expression on human monocytes: implications for immune evasion by EBV. Virology 304:342-351. [DOI] [PubMed] [Google Scholar]
- 76.Sheffield, W. D., J. D. Strandberg, L. Braun, K. Shah, and S. S. Kalter. 1980. Simian virus 40-associated fatal interstitial pneumonia and renal tubular necrosis in a rhesus monkey. J. Infect. Dis. 142:618-622. [DOI] [PubMed] [Google Scholar]
- 77.Shepard, C. W., L. Finelli, and M. J. Alter. 2005. Global epidemiology of hepatitis C virus infection. Lancet Infect. Dis. 5:558-567. [DOI] [PubMed] [Google Scholar]
- 78.Shin, H. D., C. Winkler, J. C. Stephens, J. Bream, H. Young, J. J. Goedert, T. R. O'Brien, D. Vlahov, S. Buchbinder, J. Giorgi, C. Rinaldo, S. Donfield, A. Willoughby, S. J. O'Brien, and M. W. Smith. 2000. Genetic restriction of HIV-1 pathogenesis to AIDS by promoter alleles of IL10. Proc. Natl. Acad. Sci. U. S. A. 97:14467-14472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Skaletskaya, A., L. M. Bartle, T. Chittenden, A. L. McCormick, E. S. Mocarski, and V. S. Goldmacher. 2001. A cytomegalovirus-encoded inhibitor of apoptosis that suppresses caspase-8 activation. Proc. Natl. Acad. Sci. U. S. A. 98:7829-7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sklan, E. H., P. Charuworn, P. S. Pang, and J. S. Glenn. 2009. Mechanisms of HCV survival in the host. Nat. Rev. Gastroenterol. Hepatol. 6:217-227. [DOI] [PubMed] [Google Scholar]
- 81.Spencer, J. V., K. M. Lockridge, P. A. Barry, G. Lin, M. Tsang, M. E. Penfold, and T. J. Schall. 2002. Potent immunosuppressive activities of cytomegalovirus-encoded interleukin-10. J. Virol. 76:1285-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sullivan, C. S., A. T. Grundhoff, S. Tevethia, J. M. Pipas, and D. Ganem. 2005. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature 435:682-686. [DOI] [PubMed] [Google Scholar]
- 83.Tang, S., A. S. Bertke, A. Patel, K. Wang, J. I. Cohen, and P. R. Krause. 2008. An acutely and latently expressed herpes simplex virus 2 viral microRNA inhibits expression of ICP34.5, a viral neurovirulence factor. Proc. Natl. Acad. Sci. U. S. A. 105:10931-10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tang, S., A. Patel, and P. R. Krause. 2009. Novel less-abundant viral microRNAs encoded by herpes simplex virus 2 latency-associated transcript and their roles in regulating ICP34.5 and ICP0 mRNAs. J. Virol. 83:1433-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Taylor, J. M., and C. Nicot. 2008. HTLV-1 and apoptosis: role in cellular transformation and recent advances in therapeutic approaches. Apoptosis 13:733-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Thimme, R., C. Neumann-Haefelin, T. Boettler, and H. E. Blum. 2008. Adaptive immune responses to hepatitis C virus: from viral immunobiology to a vaccine. Biol. Chem. 389:457-467. [DOI] [PubMed] [Google Scholar]
- 87.Tseng, C. T., and G. R. Klimpel. 2002. Binding of the hepatitis C virus envelope protein E2 to CD81 inhibits natural killer cell functions. J. Exp. Med. 195:43-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Umbach, J. L., M. F. Kramer, I. Jurak, H. W. Karnowski, D. M. Coen, and B. R. Cullen. 2008. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 454:780-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Virgin, H. W., E. J. Wherry, and R. Ahmed. 2009. Redefining chronic viral infection. Cell 138:30-50. [DOI] [PubMed] [Google Scholar]
- 90.Walker, B. D., C. Flexner, T. J. Paradis, T. C. Fuller, M. S. Hirsch, R. T. Schooley, and B. Moss. 1988. HIV-1 reverse transcriptase is a target for cytotoxic T lymphocytes in infected individuals. Science 240:64-66. [DOI] [PubMed] [Google Scholar]
- 91.Wang, Q. Y., C. Zhou, K. E. Johnson, R. C. Colgrove, D. M. Coen, and D. M. Knipe. 2005. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc. Natl. Acad. Sci. U. S. A. 102:16055-16059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Weissman, J. D., J. A. Brown, T. K. Howcroft, J. Hwang, A. Chawla, P. A. Roche, L. Schiltz, Y. Nakatani, and D. S. Singer. 1998. HIV-1 tat binds TAFII250 and represses TAFII250-dependent transcription of major histocompatibility class I genes. Proc. Natl. Acad. Sci. U. S. A. 95:11601-11606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yeung, M. L., J. Yasunaga, Y. Bennasser, N. Dusetti, D. Harris, N. Ahmad, M. Matsuoka, and K. T. Jeang. 2008. Roles for microRNAs, miR-93 and miR-130b, and tumor protein 53-induced nuclear protein 1 tumor suppressor in cell growth dysregulation by human T-cell lymphotrophic virus 1. Cancer Res. 68:8976-8985. [DOI] [PMC free article] [PubMed] [Google Scholar]