Abstract
Coronaviruses encode multifunctional proteins that are critical for viral replication and for blocking the innate immune response to viral infection. One such multifunctional domain is the coronavirus papain-like protease (PLP), which processes the viral replicase polyprotein, has deubiquitinating (DUB) activity, and antagonizes the induction of type I interferon (IFN). Here we characterized the DUB and IFN antagonism activities of the PLP domains of human coronavirus NL63 and severe acute respiratory syndrome (SARS) coronavirus to determine if DUB activity mediates interferon antagonism. We found that NL63 PLP2 deconjugated ubiquitin (Ub) and the Ub-line molecule ISG15 from cellular substrates and processed both lysine-48- and lysine-63- linked polyubiquitin chains. This PLP2 DUB activity was dependent on an intact catalytic cysteine residue. We demonstrated that in contrast to PLP2 DUB activity, PLP2-mediated interferon antagonism did not require enzymatic activity. Furthermore, addition of an inhibitor that blocks coronavirus protease/DUB activity did not abrogate interferon antagonism. These results indicated that a component of coronavirus PLP-mediated interferon antagonism was independent of protease and DUB activity. Overall, these results demonstrate the multifunctional nature of the coronavirus PLP domain as a viral protease, DUB, and IFN antagonist and suggest that these independent activities may provide multiple targets for antiviral therapies.
The front-line defense of a host cell against virus infection is the innate immune system, which utilizes multiple membrane and cytoplasmic sensors, such as toll-like receptors (TLRs) and RNA helicases, to detect pathogen-associated molecular patterns like viral RNA (3, 9, 31, 47, 54). Activation of these sensors by viral RNA intermediates sets off a cascade of signaling events that ultimately turn on transcription factors, such as NF-κB, ATF2/c-Jun, IRF-7, and IRF-3. These activated transcription factors translocate to the nucleus and upregulate transcription of interferon (IFN) mRNAs. The translation and subsequent secretion of IFNs activates cells to upregulate interferon-stimulated genes (ISGs) to establish an antiviral state hostile to viral replication. Of importance for this study, many of the signaling events that link the sensors to the transcription factors are mediated by the activities of kinases and ubiquitinating enzymes that modify and activate critical intermediates in the cascade (7, 8, 20, 25). For example, signaling from RIG-I can proceed through MAVS/TRAF3/TANK to TBKI and inducible IκB kinase (IKKi), which ultimately phosphorylate IRF-3. Recent studies indicate that both TRAF3 and TANK are modified by lysine-63-linked polyubiquitination and can be inactivated by DUBA, a cellular deubiquitinating (DUB) enzyme (28). Thus, ubiquitinating enzymes and DUBs are critical players in modulating the innate immune response.
For positive-strand RNA viruses that replicate in the cytoplasm of the cell using double-stranded RNA intermediates, the cytoplasmic innate immune sensors and subsequent signaling cascades represent a minefield that must be either neutralized, navigated by stealth, or both. Recent studies have revealed that viral proteases can act as “multitaskers” during viral replication by not only processing viral polyproteins but also cleaving/inactivating key players in the innate immune response. For example, the hepatitis C virus NS3-4A protease cleaves the viral replicase polyprotein and inactivates the key signaling proteins TRIF and MAVS (36, 38, 42, 46). Picornavirus 3C protease is essential for processing the replicase polyprotein and inactivating NF-κB and RIG-I (2, 49). In these studies, the catalytic function of the viral proteases was essential for the inactivation of host factors involved in signaling the innate immune response. Recently the coronavirus papain-like protease domains have also been identified as modulators of the innate immune response; however, the mechanisms of inhibition are not entirely clear (13, 16, 70).
Coronaviruses are enveloped viruses with large RNA genomes (28 to 32 kb) that cause disease in humans ranging from common colds (human coronavirus [CoV] 229E [HCoV-229E] and OC43) to croup and pneumonia, seen mostly in very young and old populations (HCoV-NL63 and -HKU1), to severe acute respiratory syndrome (SARS) coronavirus (SARS-CoV) with 10% mortality (53). Upon entry, coronavirus genomic RNA is translated to produce two large polyproteins, pp1a and pp1ab. These polyproteins are processed by viral cysteine proteases, papain-like (PLPs/PLpro) and picornavirus 3C-like (3CLpro), to generate mature nonstructural proteins (nsp's) that assemble with host cell membranes to form double membrane vesicles (DMVs) (18, 19, 61). These DMVs are the site of viral RNA synthesis producing double-stranded intermediates and genomic/subgenomic mRNAs. Interestingly, robust replication of SARS-CoV was shown to trigger low but detectable levels of beta interferon (IFN-β) (13, 62, 63). The low-level IFN response to a vigorously replicating RNA virus suggests that SARS-CoV either evades or inactivates the innate immune response. Indeed, recent studies indicate that SARS-CoV encodes multiple proteins, such as nsp1, protein 3b, protein 6, and the nucleocapsid protein that modulate multiple pathways of the innate immune response (17, 27, 30, 48, 68). In addition, we showed that the SARS-CoV papain-like protease (PLpro) domain acts as an interferon antagonist that blocks the phosphorylation and subsequent nuclear translocation of IRF-3 (13). We also showed via X-ray structural studies that the SARS-CoV PLpro domain is similar to cellular deubiquitinating enzymes (57), and we and others demonstrated that PLpro is both a protease and a DUB (4, 39, 40). Initially, we hypothesized that either the protease or DUB activity would be required for modulating the innate immune response, but we found that inactivation of the catalytic cysteine residue of PLpro, which ablates both proteolysis and deubiquitinating activity, decreased but did not abrogate PLpro's ability to block activation of interferon (13). These results are consistent with a protease/DUB-independent mechanism that contributes to interferon antagonism. Frieman and coworkers also showed that catalytic mutants of PLpro retained interferon antagonism, and they reported that deletion of the PLP ubiquitin-like (Ubl) domain upstream of the catalytic site resulted in a loss of antagonism (16). Studies by Zheng et al. of the PLP domain of murine hepatitis virus (MHV) suggested that viral DUB activity may be required for interferon antagonism, although they reported that MHV PLP2 catalytic cysteine mutants became less efficient at, but did not eliminate, blockade of the interferon response (70). Therefore, further studies are required to clarify the role of coronavirus protease/DUB activity in PLP-mediated interferon antagonism.
In this study, we analyzed the DUB and IFN antagonism profiles of the papain-like proteases of human coronavirus NL63 and SARS-CoV (group 1 and group 2 coronaviruses, respectively). We show that HCoV-NL63 PLP2 is a deubiquitinating and deISGylating (ISG15-removing) enzyme. HCoV-NL63 infection, like that of SARS-CoV, triggers a weak type I IFN response in human airway epithelial cell cultures. We also evaluated the role of PLP2 and PLpro enzymatic activity in interferon antagonism. By generating dose-response profiles of IFN antagonism, we found that the papain-like proteases do not require enzymatic activity to inhibit type I IFN induction. Using a pharmacological approach, we found that the inhibition of PLpro did not alter the antagonism of IRF-3-dependent reporters but did affect an NF-κB- dependent reporter. Overall, we show that multifunctional coronavirus PLPs target the activities of key transcription factors involved in the induction of type I interferons and thereby hinder the activation of the innate immune system.
MATERIALS AND METHODS
Cells and HCoV-NL63.
HeLa cells, HEK293 cells, and HEK293-TLR3 (stable expression of human TLR3 receptor) cells were cultured using Dulbecco's modified Eagle's medium containing 10% (vol/vol) fetal calf serum, supplemented with penicillin (100 U/ml) and streptomycin (100 μg/ml). The HCoV-NL63 (P8) virus and LLC-MK2 cells were kindly provided by Lia van der Hoek (University of Amsterdam, Amsterdam, Netherlands) and propagated as described previously (12). A plaque-purified isolate of HCoV-NL63 was kindly provided by Christian Drosten and propagated in CaCo2-TC7 cells (21). This virus stock was used to infect human airway epithelial cells as described previously (1).
Plasmid DNA.
pcDNA3.1-Flag-Ub was kindly provided by Adriano Marchese (Loyola University Medical Center). IFN-β-Luc was kindly provided by John Hiscott (Jewish General Hospital, Montreal, Canada). pISRE-Luc has been previously described (35). pRL-TK Renilla luciferase reporter was purchased from Promega. N-RIG-Flag, NF-κB-Luc, and nsp2/3-GFP were kindly provided by Ralph Baric (University of North Carolina). pcDNA3-myc6-mISG15 was kindly provided by Min-Jung Kim (Pohang University of Science and Technology, Pohang, Republic of Korea). pcDNA3-Ube1L and pcDNA3-UbcH8 were kind gifts from Robert M. Krug (University of Texas).
PLP1 and PLP2 core domain synthesis, cloning, and site-directed mutagenesis.
To obtain high expression in eukaryote cells, the codon usage of the HCoV-NL63 PLP1 core domain (amino acids 1018 to 1277 of HCoV-NL63) and PLP2 core domain (amino acids 1570 to 1884) were optimized based on human codon usage frequency, and the potential splicing sites and poly(A) signal sequences were removed and cloned into pcDNA3.1-V5/HisB at the BamHI and EcoRI sites as an in-frame fusion with the V5 peptide. The native viral sequence for the remainder of nsp3 (including the transmembrane domain downstream of PLP2) was cloned into pcDNA3.1-PLP2(N) using the existing EcoRI site and XhoI to generate transmembrane (TM)-containing PLP2 (PLP2-TM) in frame with the V5 peptide. To generate specific mutations in the catalytic residues (C1062 and H1212 in PLP1 and C1678 and H1836 in PLP2), mutagenic primers were incorporated into newly synthesized DNA using the QuikChange II XL site-directed mutagenesis protocol (Stratagene, La Jolla, CA) according to the manufacturer's instruction. Mutated nucleotides are indicated in bold. PLP1 C1062A was amplified using the forward primer 5′ AAC AAC GCC TGG ATC AGC ACC ACC CTG GTG CAA CTG 3′ and reverse primer 5′ GAT CCA GGC GTT GTT GTC GCT CTG GTC CAG CAC CCG 3′. PLP1 H1212A was amplified using the forward primer 5′ AGC GGC GCC TAC CAG ACC AAC CTG TAC AGC TTC AAC 3′ and reverse primer 5′ CTG GTA GGC GCC GCT GCC CTT CAC GCC CAG GTA CAC 3′. PLP2 C1678A was amplified using the forward primer 5′ AAC AAC GCC TGG GTG AAC GCC ACC TGC ATC ATC CTG 3′ and reverse primer 5′ CAC CCA GGC GTT GTT GTC GGT GGT GCC CAG CAC CCG 3′. PLP2 H1836A was amplified using the forward primer 5′ AAC GGC GCC TAC GTG GTG TAC GAC GCC GCC AAC AAC 3′ and reverse primer 5′ CAC GTA GGC GCC GTT GTC GAA GCT GCC GCT GAA GG 3′. The primers used for mutagenesis were designed according to the modified methods of Zheng et al. (71). All introduced mutations were confirmed by DNA sequencing.
Assay of deubiquitinating activity in cultured cells.
The effect of HCoV-NL63 PLP1 and PLP2 on ubiquitinated proteins in cultured cells was assessed as described previously (14). Briefly, HeLa cells cultured in 60-mm dishes were cotransfected with 0.4 μg of pcDNA3.1-Flag-Ub plus appropriate amounts of constructs containing PLP1, PLP2, or the corresponding catalytic mutants. Transfection was performed with Lipofectamine 2000 per the manufacturer's instructions. The empty vector pcDNA3.1/V5-HisB was used to standardize the total amount of DNA used for transfection. After 48 h, cells were harvested by adding 250 μl 2× LBA, containing 20 mM N-ethylmaleimide (NEM) (Calbiochem) and 20 mM iodoacetamine (Sigma). Cell lysates were then analyzed for ubiquitin (Ub)-conjugated proteins by Western blotting with anti-Flag M2 antibody (1:10,000) (Sigma). Proteins were separated via SDS-PAGE, followed by transfer to a polyvinylidene difluoride (PVDF) membrane in transfer buffer (0.025 M Tris, 0.192 M glycine, and 20% methanol) for 2 h at 4°C. The membrane was blocked using 5% dried skim milk in Tris-buffered saline (TBS) (0.9% NaCl, 10 mM Tris-HCl, pH 7.5) plus 0.1% Tween 20 (TBST) for 2 h at room temperature. The blot was probed with the indicated antibody overnight at 4°C. The membrane was washed in TBST three times for 20 min (each). Following the washes, the membrane was incubated with peroxidase-conjugated secondary antibody (donkey anti-rabbit or goat anti-mouse IgG horseradish peroxidase [HRP]) (Amersham) at a dilution of 1:10,000 for 2 h at room temperature. The membrane was then washed three times with TBST and detected with Western Lightning Chemoluminescence Reagent Plus (PerkinElmer LAS Inc.). To confirm the expression levels of PLP1, PLP2, and the mutants, anti-V5 antibody (Invitrogen) was used to detect the V5-tagged proteins. Calnexin was detected with anti-calnexin monoclonal antibody (MAb) (BD Transduction Lab) as a protein loading control.
NL63 PLP2 cleavage of K48- and K63-linked ubiquitin chains.
The NL63 PLP2 wild-type protein was purified as previously described (4, 12), and ubiquitin chains were purchased from Boston Biochem (K48-Ub6 [catalog no. UC-217] and K63-Ub6 [catalog no. UC-317]). Proteolytic cleavage of K48-linked or K63-linked ubiquitin chains was carried out under the following conditions: 0.01 μg of purified PLP2 was incubated with 2.5 to 5 μg of K48-Ub6 or K63-Ub6 at 25°C in a 10- to 20-μl volume containing 50 mM HEPES, pH 7.5, 0.1 mg/ml bovine serum albumin (BSA), 100 mM NaCl, and 2 mM dithiothreitol (DTT). A control reaction was incubated under identical conditions with the exclusion of enzyme. At specified time points, the reactions were quenched with the addition of SDS-PAGE sample loading dye to a 1× concentration (25 mM Tris, pH 6.8, 280 mM β-mercaptoethanol, 4% glycerol, 0.8% SDS, 0.02% bromophenol blue) and heat treated at 95°C for 5 min. The samples were analyzed by electrophoresis on a 15% SDS-PAGE gel and stained with Coomassie dye.
Assay of deISGylating activity in cultured cells.
The effect of HCoV-NL63 PLP2 on ISGylated proteins in cultured cells was assessed as described previously (69). Briefly, HEK293 cells cultured in 60-mm dishes were cotransfected with 0.5 μg of pcDNA3-myc6-mISG15, 0.25 μg of pcDNA3-Ube1L, and 0.25 μg of pcDNA3-UbcH8 plus appropriate amounts of constructs containing PLP2 or the corresponding catalytic mutant with a total of 2 μg of plasmid DNA for each transfection. The empty vector pcDNA3.1/V5-HisB was used to standardize the total amount of DNA used for transfection. Transfection was performed with Lipofectamine 2000 per the manufacturer's instructions. After 30 h, cells were harvested by adding 250 μl 2× LBA containing 20 mM N-ethylmaleimide (Calbiochem) and 20 mM iodoacetamine (Sigma). Cell lysates were then analyzed for ISG-conjugated proteins by Western blotting with monoclonal anti-myc antibody (1:2,000; MBL Companies, Japan) as described above. To confirm the expression levels of PLP2 and the mutants, anti-V5 antibody (1:5,000; Invitrogen) was used to detect the V5-tagged proteins. Actin was detected with antiactin MAb (Beyotime Institute of Biotechnology, China) as a protein loading control.
Enzyme-linked immunosorbent assay for IFN-β secretion in HAE culture supernatants.
Human airway epithelial (HAE) cultures were generated as previously described (1). Cultures were inoculated with 100 μl of 105 PFU/ml HCoV-NL63, 2,000 hemagglutinating units (HAU)/ml Sendai virus (SeV), or medium for 1 h at 37°C, after which the inoculum was removed and apical washes with 200 μl of F12 medium (Gibco) were performed at indicated times. The IFN-β concentration was determined by a commercial enzyme-linked immunosorbent assay (ELISA) (PBL Biomedical Laboratories) according to the manufacturer's instructions. Data were analyzed and plotted using the GraphPad Prism 5.0 software program.
Luciferase reporter gene assay.
To determine if PLpro and/or PLP2 modulates IFN expression in host cells, the IFN-β-Luc reporter, consisting of the IFN-β promoter upstream of firefly luciferase, was transfected into HEK293 cells along with PLpro, ΔUbl-PLpro-Sol, ΔUbl-PLpro-TM, PLP2, PLP2-TM, or the C1678A and H1836A PLP2 catalytic mutants in the soluble or transmembrane versions. pRL-TK, encoding Renilla luciferase under the control of the herpes simplex thymidine kinase promoter (low to moderate Renilla expression), was used for normalization of transfection efficiency. HEK293 cells were transfected by Lipofectamine 2000 or LT1 transfection reagent (MirusBio) according to the manufacturer's instructions and incubated for 24 h. Cells were then mock infected or infected with Sendai virus (Cantell strain; Charles River Laboratories) at the dose of 100 HAU/ml for 16 h or transfected with N-RIG-Flag for 24 h to activate the RIG-I-dependent IFN pathway. To detect TLR3-dependent IFN expression, HEK293-TLR3 cells were transfected with IFN-β-Luc and PLP2 for 24 h. Cells were then treated with poly(IC) for 6 h as described previously (13). Firefly luciferase and Renilla luciferase activities were assayed using the Dual-Luciferase reporter assay kit (Promega) and a luminometer.
Drug inhibition of SARS-CoV PLpro.
HEK293 cells were transfected with plasmids encoding PLpro-TM (13), IFN-β-Luc, ISRE-Luc, NF-κB-Luc, pRL-TK, nsp2/3-GFP substrate, and/or N-RIG-Flag. The pcDNA3.1/V5-HisB vector was used to standardize the DNA concentration for transfection. Dimethyl sulfoxide (DMSO) vehicle control or 100 μM GRL-0617S (56) was added at the time of transfection. Tumor necrosis factor alpha (TNF-α) (10 ng/ml) from Roche was used to stimulate the NF-κB-Luc reporter (6 h of stimulation). Cell lysates were prepared per the manufacturer's instruction using 1× passive lysis buffer (Promega), and luciferase activity was measured using the Dual-Luciferase reporter assay kit (Promega) and a luminometer. Cell lysates were also mixed 1:1 with 2× sample buffer and subjected to Western blotting as described above. PLpro-TM was detected with anti-V5 (Invitrogen), and nsp2/3-GFP and nsp3-GFP were detected with rabbit anti-green fluorescent protein (anti-GFP) (Invitrogen).
RESULTS
NL63 PLP2 but not PLP1 has deubiquitinating activity.
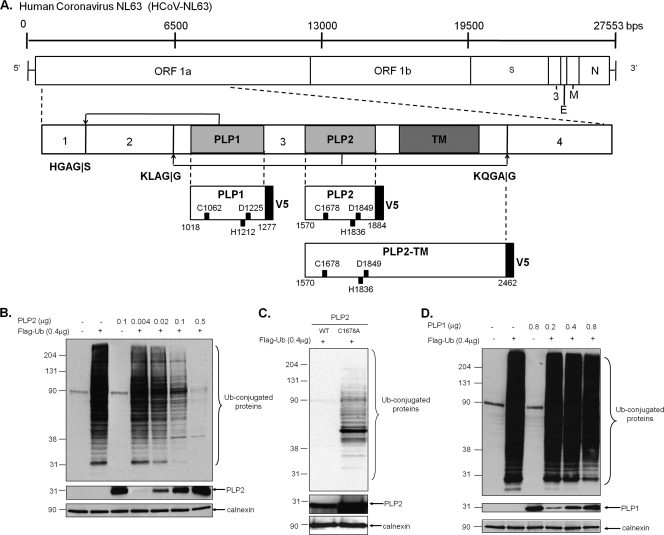
Previously we showed that HCoV-NL63 replicase nonstructural protein 3 (nsp3) encodes two papain-like proteases, PLP1 and PLP2, that process the amino-terminal end of the replicase polyprotein (Fig. 1A). In addition, we have shown that PLP2 can process K48-linked polyubiquitin chains, suggesting that this protease has deubiquitinating activity (12). Both the polyprotein cleavage and K48-linked polyubiquitin chain processing are dependent on a cysteine residue in the catalytic triad of this cysteine protease (12). To determine if one or both of these NL63 PLPs can remove ubiquitin conjugated to cellular proteins, HeLa cells were transfected with plasmid DNA encoding PLP1 or PLP2 along with pcDNA3.1-3×Flag-Ub, and the effect of expression of PLP1 and PLP2 on the extent of ubiquitinated cellular proteins was assessed via Western blotting with anti-Flag antibodies. We found that increasing expression of PLP2 resulted in a dramatic reduction in the level of Ub-conjugated proteins (Fig. 1B). We noted that PLP2 appears to have global DUB activity, since no particular cellular substrates were spared. As expected, this PLP2 DUB activity is dependent on an intact catalytic cysteine residue 1678, and mutation of this residue to alanine resulted in the loss of DUB activity (Fig. 1C). In contrast, PLP1 did not show any significant reduction of Flag-Ub conjugates at the concentrations tested (Fig. 1D). These results indicate that NL63 PLP2, like SARS-CoV PLpro, has potent DUB activity that can remove ubiquitin conjugates from many cellular substrates.
FIG. 1.
HCoV-NL63 PLP2, but not PLP1, has a dose-dependent global deubiquitinating activity in cultured cells. (A) Schematic diagram of the NL63 genomic RNA and the resulting polyprotein 1ab, which contains three viral proteases. PLP1 and PLP2 cleavage sites are indicated, as are the resulting nonstructural proteins. The V5-tagged constructs of PLP2 used in this study are listed, and the catalytic residues numbering from ORF 1a are shown. DNA encoding HCoV-NL63 PLP2 (B), PLP2 C1678A (C), or PLP1 (D) was transfected into HeLa cells along with pcDNA3.1-3×Flag-Ub. Cell lysates were prepared at 24 h posttransfection and analyzed for Flag-Ub-conjugated proteins by Western blotting with an anti-Flag antibody. Mouse anti-V5 was used to confirm the expression of PLP1 and PLP2, and anticalnexin antibody was used to detect calnexin, which serves as a protein loading control. Molecular weight markers shown on the left of each gel are in thousands.
NL63 PLP2 processes lysine-63-linked in addition to lysine-48-linked polyubiquitin.
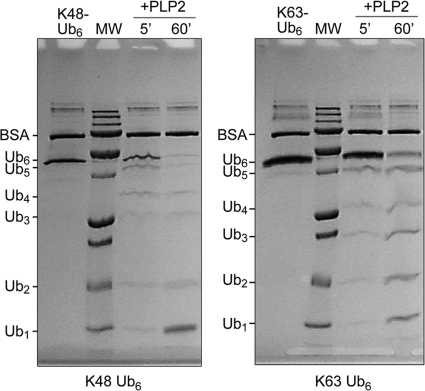
Cellular proteins can be covalently modified with ubiquitin at one or multiple lysines through an isopeptide bond that links the carboxy terminus of ubiquitin to a lysine on the target protein. Ubiquitin itself contains seven lysines that can be further conjugated to the C terminus of another ubiquitin molecule, forming different types of polyubiquitin-linked chains on the targeted protein. The two most common types of polyubiquitin chains are linked through ubiquitin lysine 48 (K48) and lysine 63 (K63). These modifications play key regulatory roles in protein degradation and pathway signaling and have been associated with controlling several pathways of innate and adaptive immunity (7). Previous studies indicated that SARS-CoV PLpro processes both K48- and K63-linked ubiquitin (39, 40). To assess if HCoV-NL63 PLP2 has isopeptidase activity that will deconjugate K63-linked ubiquitin in addition to K48-linked isopeptidase activity, purified PLP2 enzyme was incubated with hexameric K48-linked and K63-linked polyubiquitin chains. SDS-PAGE analysis of the cleavage products shows that PLP2 can cleave the substrates in vitro (Fig. 2), and with prolonged incubation times, both chains can be reduced to monoubiquitin (data not shown). These data show that both major forms of polyubiquitin can be recognized and degraded by HCoV-NL63 PLP2.
FIG. 2.
Processing of K48- and K63-linked ubiquitin chains by PLP2. NL63 PLP2 was incubated with K48-linked (left) or K63-linked (right) Ub6 chains for the indicated time points before being analyzed by SDS-PAGE. Uncleaved Ub6 is run in the first lane of each gel. The expected sizes of the Ub species are indicated to the left of all gels. Molecular weight (MW) markers include 250,000-, 100,000-, 75,000-, 50,000-, 37,000-, 25,000-, 20,000-, 15,000-, and 10,000-molecular-weight bands.
PLP2 possesses deISGylating activity.
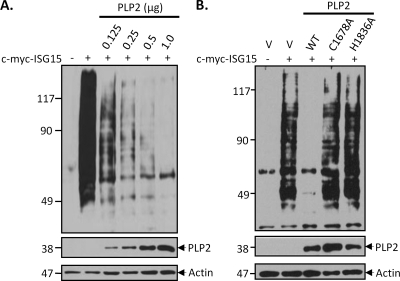
Several viral DUBs, including SARS-CoV PLpro, can also deconjugate Ub-like moieties such as ISG15 (39, 40). Conjugation of ISG15 has been shown to be important for protection against viral infection (15, 33, 34). We assessed whether HCoV-NL63 PLP2 can deISGylate cellular c-myc-tagged ISG15 (c-myc-ISG15) conjugates. HeLa cells were transfected with increasing amounts of plasmid DNA encoding PLP2 along with c-myc-ISG15 and the ISG15 conjugation machinery Ube1L and UbcH8 to enhance ISGylation of host cell proteins. The ability of PLP2 to deISGylate cellular proteins was then assayed via Western blotting with anti-myc antibody. We found that expression of increasing amounts of PLP2 was associated with a dramatic reduction in the levels of ISGylated cellular proteins (Fig. 3A), in agreement with a previous report (50). The deISGylating activity was dependent on intact catalytic residues C1678 and H1836, since mutation of these residues to alanine resulted in the loss of deISGylating activity (Fig. 3B). Thus, HCoV-NL63 PLP2 is a potent DUB/deISGylating enzyme that acts on many modified cellular substrates.
FIG. 3.
NL63 PLP2 has global deISGylating activity in cultured cells. HEK293 cells were transfected with pcDNA3-myc6-mISG15, pcDNA3-Ube1L, and pcDNA3-UbcH8 plus indicated amounts of the PLP2 expression construct (A) or PLP2 expression construct and the corresponding catalytic mutants (B). To ensure that the total amount (2 μg/transfection) of plasmids for transfection was equal under every condition, empty vector pcDNA3.1/V5-HisB (v) was used to standardize the total amount of DNA. After 30 h, cells were harvested, and cell lysates were analyzed for ISG-conjugated proteins by Western blotting with monoclonal anti-myc antibody. Expression levels of V5-tagged PLP2 and mutant enzymes were detected with anti-V5 antibody. Actin was detected with antiactin MAb antibody as a protein loading control.
IFN-β release from human airway epithelial (HAE) cell cultures.
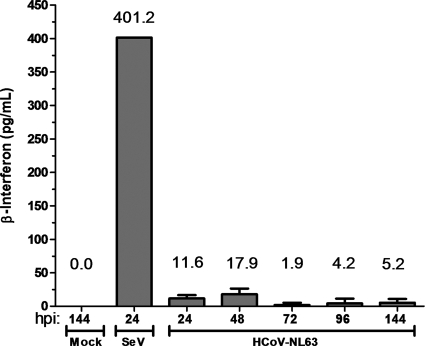
HAE cultures are a useful model system for studying human respiratory viruses, including HCoV-NL63, since they mimic human bronchial epithelium, which is the primary site of infection (1, 60). HAE cell cultures were infected with HCoV-NL63 or Sendai virus, and apical wash samples were collected at 24, 48, 72, 96, and 144 h postinfection. The presence of secreted IFN-β in the apical wash was measured by ELISA. Mock-infected cultures released no detectable IFN-β, whereas cultures inoculated with a potent IFN inducer, Sendai virus, released more than 400 pg/ml of IFN-β. In contrast, inoculation of HAE cultures with HCoV-NL63 stimulated low but detectable levels of IFN-β from 24 to 144 h postinfection (Fig. 4). These results are reminiscent of the reports of low but detectable levels of IFN-β produced from SARS-CoV-infected cells (13, 63) and indicate that either HCoV-NL63 is a weak inducer of the IFN-β response or, like SARS-CoV, HCoV-NL63 encodes potent IFN antagonists.
FIG. 4.
Evaluating interferon-β secretion from human airway epithelial (HAE) cell cultures following HCoV-NL63 infection. Apical washes were collected from HAE cultures at 24, 48, 72, 96, and 120 h postinfection, and secreted IFN-β was measured by ELISA. Supernatant from HAE cells infected with Sendai virus (SeV) for 24 h was used as a positive control.
PLP2 inhibits both RIG-I- and TLR3-dependent IFN-β expression.
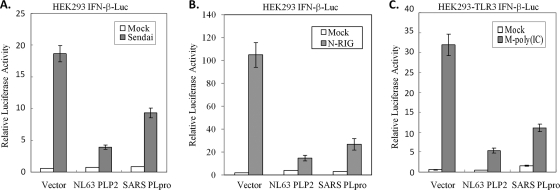
To determine if PLP2 is an IFN antagonist, we transfected HEK293 cells with plasmids encoding HCoV-NL63 PLP2 or SARS-CoV PLpro along with IFN-β-luciferase and Renilla luciferase reporters for 24 h. Then, the RIG-I-dependent pathway leading to IFN-β expression was activated by Sendai virus infection for 16 h or by a dominant active N-terminal portion of RIG-I (N-RIG). We found that activation of the IFN-β promoter by Sendai virus (Fig. 5A) or N-RIG (Fig. 5B) was inhibited in the presence of either NL63 PLP2 or SARS-CoV PLpro. To determine if HCoV-NL63 PLP2 inhibits TLR3-mediated activation of IFN-β production, PLP2 and the reporters were transfected into HEK293-TLR3 cells, and then the TLR3-mediated pathway was activated by addition of poly(IC) to the cell culture medium. We found that activation of the IFN-β promoter by the TLR3-mediated pathway was significantly inhibited by HCoV-NL63 PLP2 and SARS-CoV PLpro (Fig. 5C). These results demonstrate that the IFN antagonism mediated by coronavirus PLPs is conserved in two distinct viruses, although there is only 19% amino acid identity between the catalytic domains of HCoV-NL63 PLP2 and SARS-CoV PLpro in this region of nsp3 (4).
FIG. 5.
PLP2 inhibits both RIG-I- and TLR3-dependent IFN-β induction. (A) HEK293 cells were transfected with IFN-β-Luc, pRL-TK, and either 300 ng of HCoV-NL63 PLP2 or 300 ng of SARS-CoV PLpro. At 24 h posttransfection, cells were either mock infected or infected with Sendai virus for 16 h. Following infection, cell lysates were prepared and assayed using the Dual-Luciferase reporter assay. (B) HEK293 cells were transfected with IFN-β-Luc, pRL-TK, 200 ng N-RIG, and either 300 ng of HCoV-NL63 PLP2 or 300 ng of SARS-CoV PLpro. At 24 h posttransfection, cell lysates were prepared and assayed using the Dual-Luciferase reporter assay. (C) HEK293-TLR3 cells were transfected with IFN-β-Luc, pRL-TK, and either 300 ng of HCoV-NL63 PLP2 or 300 ng of SARS-CoV PLpro. At 24 h posttransfection, cells were either mock treated or treated with poly(IC) for 6 h. Following poly(IC) treatment, cell lysates were prepared and assayed using the Dual-Luciferase reporter assay. Error bars indicate standard deviations from the means for triplicates.
Mutation of the catalytic residues does not abolish HCoV-NL63 PLP2 IFN antagonism.
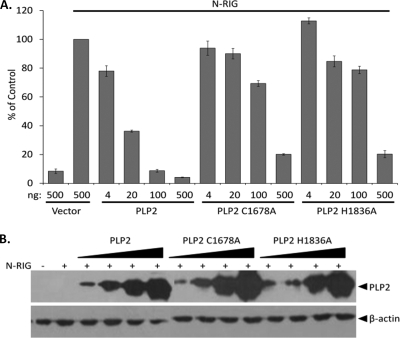
To further elucidate the interferon antagonism profile of PLP2, a dose-dependent IFN antagonism profile was generated. Concurrently, to determine if IFN antagonism is dependent on catalytic activity, plasmid DNAs encoding PLP2 or the C1678A (devoid of enzymatic activity) and H1836A (severely reduced) catalytic mutants were transfected with the IFN-β and pRL-TK reporters into HEK293 cells, and IFN-β reporter activity was assessed. N-RIG was used to stimulate IFN-β induction. We found that, like the PLP2 wild type, both PLP2 C1678A and PLP2 H1836A exhibit dose-dependent inhibition of IFN-β promoter activity; however, the catalytic mutants were less efficient than wild-type PLP2 (Fig. 6A). Expression of the proteases was verified by Western blotting (Fig. 6B). These results indicate that enzymatic activity of PLP2 is not strictly required for inhibition of antiviral IFN expression.
FIG. 6.
HCoV-NL63 PLP2 and the catalytic mutants inhibit RIG-I-mediated IFN induction in a dose-dependent manner. PLP2 and the C1678A and H1836A catalytic mutants were cotransfected with IFN-β-Luc and pRL-TK reporters into HEK293 cells. A dominant active N-terminal portion of RIG-I was used to stimulate IFN-β induction. At 24 h posttransfection, cell lysates were harvested and assayed for luciferase activity via the Dual-Luciferase reporter assay. Values are expressed as percentages of N-RIG-stimulated luciferase controls set to 100. Error bars indicate standard deviations from the means for triplicates. (B) The cell lysates described above were mixed with 2× sample buffer and subjected to 12.5% SDS-PAGE. Following transfer to nitrocellulose, the membrane was blotted with mouse anti-V5 to detect the proteases and antiactin as a loading control.
The transmembrane (TM) form of PLP2 is also an IFN antagonist.
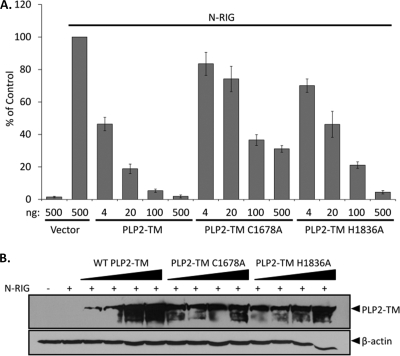
The PLP domains are part of a larger nsp3 protein in SARS-CoV and HCoV-NL63 that is membrane bound. Previously we showed that the biologically relevant transmembrane-containing form of SARS-CoV PLpro, termed PLpro-TM, is a potent IFN antagonist (13). To determine if the membrane-tethered version of HCoV-NL63 PLP2 can function as an interferon antagonist, the NL63 TM sequence was cloned into the PLP2 construct in frame with the V5 epitope tag, and the resulting construct was designated PLP2-TM. HEK293 cells were transfected with PLP2-TM or the catalytic mutant PLP2-TM C1678A or PLP2-TM H1836A, along with the IFN-β and pRL-TK reporters. N-RIG was used to stimulate IFN-β induction. We found that PLP2-TM and the catalytic mutants were able to inhibit N-RIG-induced IFN-β reporter activity in a dose-dependent manner, although like the soluble version of PLP2, the catalytic mutants were less efficient than the wild type (Fig. 7A). Expression of the proteases was verified by Western blotting (Fig. 7B). These data corroborate our previous results indicating that the catalytic site is not essential for IFN-β antagonism by HCoV-NL63 PLP2.
FIG. 7.
The transmembrane form of HCoV-NL63 PLP2 and catalytic mutants inhibit RIG-I-mediated IFN-β induction in a dose-dependent manner. The native downstream hydrophobic domain was cloned into the PLP2 plasmid, and the catalytic cysteine or histidine residue was mutated to alanine. (A) HEK293 cells were transfected with the indicated amounts of PLP2-TM, PLP2-TM C1678A, or PLP2-TM H1836A along with the IFN-β-Luc and pRL-TK reporters. N-RIG was used to stimulate IFN-β induction. At 24 h posttransfection, cell lysates were harvested and assayed for luciferase activity via the Dual-Luciferase reporter assay. Values are expressed as percentages of N-RIG-stimulated luciferase controls set to 100. Error bars indicate standard deviations from the means for triplicates. (B) The cell lysates described above were mixed with 2× sample buffer and subjected to 12.5% SDS-PAGE. Following transfer to nitrocellulose, the membrane was blotted with mouse anti-V5 to detect the proteases and antiactin as a loading control.
PLpro IFN antagonism is not dependent on the ubiquitin-like domain.
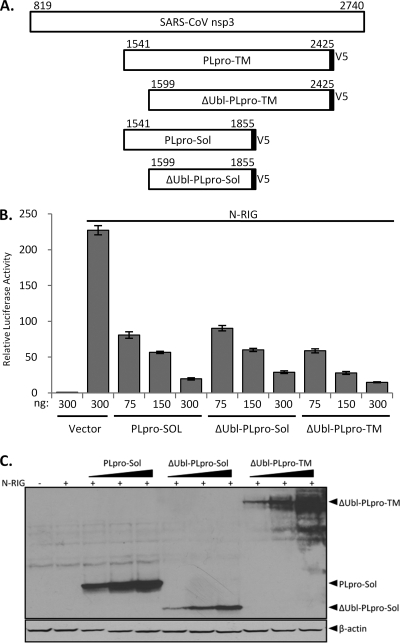
In addition to the downstream TM domain, a previous study suggested that the upstream ubiquitin-like domain (Ubl) plays a role in the IFN antagonism of SARS-CoV PLpro, since deletion of this domain in the soluble version of PLpro results in a loss of IFN antagonism (16). Currently, no analogous domain has been identified in HCoV-NL63 PLP2. To determine the role of the Ubl domain in the more biologically relevant transmembrane form of PLpro, we generated identical Ubl deletions (ΔUbl) of PLpro in both the soluble and transmembrane forms and performed a dose-response profile of IFN antagonism. In contrast to results of the study by Frieman and coworkers, we found that ΔUbl-PLpro-Sol was as potent as wild-type PLpro in inhibiting N-RIG-induced IFN-β reporter activity, as was ΔUbl-PLpro-TM (Fig. 8B). Expression of the proteases was verified by Western blotting (Fig. 8C). These results indicate that the Ubl domain of PLpro has no effect on antagonism of type I IFN induction.
FIG. 8.
The ubiquitin-like domain of SARS-CoV PLpro is not required for IFN antagonism. (A) Schematic diagram of nsp3 and the various V5-tagged deletion constructs. Numbers above the constructs indicate the amino acid numbers counting from ORF 1a. (B) HEK293 cells were transfected with the indicated amounts PLpro-Sol, ΔUbl-PLpro-Sol, or ΔUbl-PLpro-TM along with the IFN-β-Luc and pRL-TK reporters. A dominant active N-terminal portion of RIG-I was used to stimulate IFN-β induction. At 24 h posttransfection, cell lysates were harvested and assayed for luciferase activity via the Dual-Luciferase reporter assay. Values expressed are relative to results for cells transfected with the reporters alone. Error bars indicate standard deviations from the means for triplicates. (C) The cell lysates described above were mixed with 2× sample buffer and subjected to 12.5% SDS-PAGE. Following transfer to nitrocellulose, the membrane was blotted with mouse anti-V5 to detect the proteases and antiactin as a loading control.
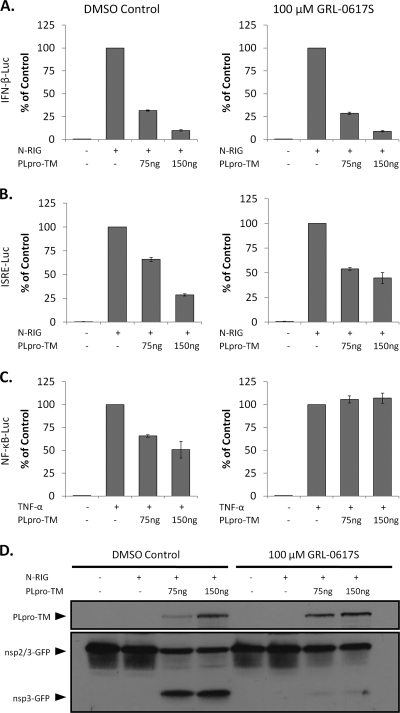
Inhibition of SARS-CoV PLpro by protease inhibitor GRL-0617S has no effect on IFN-β or IFN-stimulated response element (ISRE) reporter activity but abrogates inhibition of NF-κB reporter activity.
The mutagenesis data for PLP2 suggest that the catalytic residues (and thus catalytic activity) are not required for interferon antagonism, and Devaraj et al. reached a similar conclusion for SARS-CoV PLpro (13). To further evaluate the role of protease/DUB activity in interferon antagonism of wild-type PLpro, we added a protease inhibitor (GRL-0617S) that has been developed and shown to specifically and selectively block protease and DUB activity of SARS-CoV PLpro (56) and assessed the ability of PLpro to inhibit activation of IFN-β-Luc, ISRE-Luc, or NF-κB-Luc reporter activity. HEK293 cells were transfected with plasmid DNA encoding the transmembrane form of PLpro-TM (amino acids 1541 to 2425 of SARS-CoV ORF 1a), previously shown to be a potent IFN antagonist (13), and pRL-TK along with the IFN-β, ISRE, or NF-κB reporter, and an nsp2/3-GFP substrate. At the time of transfection, the cells were treated with 100 μM GRL-0617S (10 times the 50% effective concentration [EC50]) or DMSO (vehicle control). At 24 h after transfection, cell lysates were generated and evaluated for induction of the reporters and proteolytic processing of the nsp2/3 substrate. We found that inhibition of protease activity by GRL-0617S had no effect on the IFN-β reporter (Fig. 9A) and little to no effect on the ISRE reporter, which is dependent on IRF-3 (Fig. 9B). We did detect an alleviation of PLpro-mediated inhibition of the NF-κB reporter by GRL-0617S (compare Fig. 9A and 9B with 9C). These results indicate that protease/DUB activity may be important for PLpro-mediated inhibition of NF-κB activity but not essential for inhibition of IRF-3 activity. To demonstrate efficacy of the protease inhibitor, cell lysates used in the reporter assay were assessed for nsp2/3-GFP substrate cleavage. The nsp2/3-GFP substrate contains a region of nsp2/nsp3 (including the cleavage site) fused in-frame with GFP and was previously shown to be a substrate for PLpro (16). As shown in Fig. 9D, the nsp3-GFP cleavage product was readily detected in cell lysates that contain PLpro in the absence of GRL-0617S. In contrast, the processing of nsp2/3-GFP and liberation of nsp3-GFP was almost completely abrogated in the presence of the drug. Overall, these data provide further support that there is a catalysis-independent component to type I IFN antagonism by the papain-like proteases of human coronaviruses.
FIG. 9.
SARS-CoV PLpro inhibits IFN-β and ISRE but not NF-κB reporter activity in a dose-dependent manner in the presence or absence of a protease inhibitor. HEK293 cells were transfected with the indicated amounts of PLpro-TM, pRL-TK, nsp2/3-GFP, and either IFN-β-Luc (A), ISRE-Luc (B), or NF-κB-Luc (C). N-RIG was used to stimulate IFN-β and ISRE. TNF-α (10 ng/ml) was used to stimulate the NF-κB-Luc reporter. DMSO vehicle control or 100 μM GRL-0617S was added at the time of transfection. At 24 h posttransfection, cell lysates were harvested and assayed for luciferase activity via the Dual-Luciferase reporter assay. Values are expressed as percentages of N-RIG- or TNF-α-stimulated luciferase controls, set to 100. Error bars indicate standard deviations from the means from triplicates. (D) The cell lysates described above were mixed with 2× sample buffer and subjected to 12.5% SDS-PAGE. Following transfer to nitrocellulose, the membrane was blotted with mouse anti-V5 to detect PLpro-TM and rabbit anti-GFP to detect the nsp2/3-GFP substrate and the nsp3-GFP cleavage product.
DISCUSSION
In this study, we describe the multifunctional nature of the papain-like protease domains of NL63 and SARS coronaviruses. These coronavirus PLP domains act as viral proteases, deubiquitinating/deISGylating enzymes, and are able to antagonize innate immune induction of type I interferon. We found that PLP interferon antagonism is enhanced by, but is not strictly dependent on, the catalytic activity of the enzyme. Inhibition of coronavirus protease and DUB activity by mutagenesis or pharmacological means did not abrogate interferon antagonism. Therefore, these distinct PLP activities provide multiple targets for antiviral therapies.
The recognition of the DUB/deISGylating activity of SARS-CoV PLpro and HCoV-NL63 PLP2 provides new opportunities to investigate how the virus is modifying the host cell environment. Posttranslational modification of proteins by ubiquitin and ubiquitin-like (Ubl) molecules, such as SUMO, ISG15, and Nedd8, plays a critical role in the regulatory processes of virtually all aspects of cell biology (14, 15, 20, 22, 25, 29, 34, 41, 55). These modifications, though covalent, are highly reversible. Deubiquitinating enzymes can deconjugate Ub and Ub-like moieties and thus modulate the activities of ubiquitinated proteins. There are about 100 DUBs encoded in the human genome, and most of the known DUBs are cysteine proteases, characterized by a Cys-His-Asp catalytic triad (51). Several RNA viruses encode cysteine proteases to generate mature viral proteins necessary for replication, and many have been found to be multifunctional proteins. Like SARS-CoV PLpro and HCoV-NL63 PLP2, the protease of a nairovirus, Crimean Congo hemorrhagic fever virus (CCHFV), and the proteases of arteriviruses, including equine arteritis virus (EAV) and porcine respiratory and reproductive syndrome virus (PRRSV), have DUB and deISGylase activity (4, 15, 39). The contribution of these enzymatic activities to inhibition of type I IFN induction is currently poorly understood. A vast array of proteins involved in the type I IFN signaling cascade are activated by ubiquitination. Induction of IFN-β, for example, requires the activation of IRF-3 and NF-κB (45, 55). Ubiquitination is known to be intimately involved in the activation of NF-κB. Polyubiquitination of receptor-interacting protein (RIP), TNF receptor-associated factor 6 (TRAF6), and TNF receptor-associated factor 2 (TRAF2) activates these signaling intermediates, which leads to the polyubiquitination of IκB. IκB, which binds to and holds NF-κB inactive in the cytoplasm, is degraded via the proteasome, thereby freeing NF-κB to translocate to the nucleus and induce IFN-β transcription (20). Here we have shown that HCoV-NL63 PLP2 has a profound and global deconjugation effect on ubiquitinated cellular conjugates, suggesting that the DUB activity of PLP2 may modulate NF-κB activation. In addition, we demonstrate that protease inhibitors, which block coronavirus DUB activity, abrogate the moderate inhibition of NF-κB reporter activity imposed by transient, ectopic expression of PLpro. These results support a role for coronavirus DUBs in modulating the NF-κB response during coronavirus replication. However, further investigation is needed to delineate the physiological effect of coronavirus PLPs on NF-κB signaling, since SARS-CoV PLpro did not inhibit virus- or double-stranded RNA (dsRNA)-induced activation of two well-characterized NF-κB-dependent genes, encoding interleukin 6 (IL-6) and A20, when it was stably expressed in HeLa cells in a tetracycline-regulated fashion (13).
The effect of ISGylation of cellular proteins on the antiviral response is far less understood. It is known that ISG15 conjugation is required for protection against lethal Sindbis virus infection of IFN-α/β receptor knockout mice (15). Also, ISGylation has been shown to influence the activation of the JAK/STAT pathway, involved in type I IFN signaling (44). Intriguingly, IRF-3 also undergoes ISGylation during viral infections, which was found to enhance innate antiviral responses by inhibiting virus-induced IRF-3 degradation (43). We found that HCoV-NL63 PLP2 globally deconjugates ISG15 similarly to its deconjugation of Ub, and this activity depends on the catalytic sites of PLP2. Although the contribution of the deISGylation activity of PLP2 to IFN antagonism remains to be further investigated, deISGylation of IRF-3 is unlikely to be a significant contributing factor, since the PLP catalytic mutants devoid of deISGylation activity still effectively inhibited IFN induction.
Using human airway epithelial cells, which represent a cell culture model for respiratory infection, we found that IFN-β release induced by HCoV-NL63 was weak but measurable. This finding is similar to weak IFN induction by the far more pathogenic human coronavirus SARS-CoV, suggesting that antagonism of type I IFN is a common trait of coronavirus infection (13, 58, 63). In addition to PLpro, SARS-CoV encodes several other IFN antagonists. ORF 3b, ORF 6, and nucleocapsid inhibit type I IFN induction via inhibition of IRF-3 phosphorylation and its subsequent nuclear translocation (30). ORF 6 and nsp1 have been shown to inhibit IFN signaling by interfering with the activity of STAT1 (17, 27, 48). Mouse hepatitis virus also encodes several IFN antagonists, including nsp1, nucleocapsid, and PLP2 (58, 68, 70). Thus, there is clear evolutionary pressure to encode and maintain multiple IFN antagonists. It has yet to be determined if nsp1 and nucleocapsid from HCoV-NL63 are IFN antagonists as well.
Using reporter assays, we found that HCoV-NL63 PLP2 can antagonize type I IFN induction independently of catalytic activity. Catalytic mutants of PLP2 can dose-dependently inhibit IFN-β induction; however, this inhibition is reduced compared to equivalent amounts of wild-type PLP2. In addition, we note that the presence of the transmembrane domain confers enhanced IFN antagonism, particularly in the catalytic mutants. We speculate that the TM domain may facilitate either protein folding or interaction with cellular protein partners. Overall, the antagonism profile of HCoV-NL63 PLP2 is remarkably similar to that of PLpro of SARS-CoV. In fact, using a known specific inhibitor of SARS-CoV PLpro (GRL-0617S), we found that PLpro can effectively inhibit type I IFN induction despite a profound reduction in proteolysis, which corroborates the notion of a catalysis-independent mechanism for type I IFN antagonism. Previously we showed that SARS-CoV PLpro is able to inhibit the phosphorylation and nuclear accumulation of IRF-3 following Sendai virus infection (13). We found that HCoV-NL63 PLP2 was also able to inhibit the translocation of IRF-3 to the nucleus (data not shown); however, the mechanism of this inhibition is not yet clear. We are actively searching for cellular factors that associate with HCoV-NL63 PLP2 as well as SARS-CoV PLpro.
The crystal structure of SARS-CoV PLpro has identified a unique domain that has remarkable similarity to ubiquitin (57). Frieman and coworkers reported that removal of the Ubl domain in the soluble version of PLpro resulted in a loss of IFN antagonism (16). However, we note that the authors of that study assessed IFN antagonism using one concentration of PLpro. In this study, by performing a dose-response profile of IFN antagonism, we found that deletion of the Ubl domain from both the soluble and the transmembrane version of PLpro had no effect on IFN antagonism. Thus, it is critical to assess the effect of IFN antagonism across a range of protein concentrations to fully evaluate the activity of these proteases.
Many viruses have been shown to inhibit the transcriptional activity of IRF-3 in a wide variety of ways (9, 24, 52). Some viruses inhibit IRF-3 phosphorylation, dimerization, and/or translocation to the nucleus. Others can induce IRF-3 degradation or sequester the transcription factor (11, 23). The mechanism of inhibition that can lead to these phenotypes can occur directly on IRF-3 or may affect any of the vast array of proteins upstream of IRF-3 in the type I IFN induction cascade. For example, the VP35 protein of Ebola Zaire virus (EBOV) has been shown to impact IRF-3 activity by binding dsRNA, thus preventing detection by RIG-I (5, 6). In addition, VP35 was shown to interact with Ubc9 (SUMO E2 enzyme), PIAS1 (SUMO E3 ligase), and IRF-7, leading to SUMOylation of IRF-7 and transcriptional repression of the IFN-β promoter (10). Respiratory syncytial virus (RSV) encodes two proteins, NS1 and NS2, that act individually or cooperatively to inhibit the activity of IRF-3. It was reported that these proteins reduce the expression of key kinases involved in IRF-3 phosphorylation (TRAF3 and IκB kinase epsilon [IKKɛ]), but how NS1 and NS2 induce TRAF3 and IKKɛ degradation is still unclear (64). Coronavirus PLPs could be acting on an as yet unidentified cellular factor involved in the IFN induction cascade. For example, in a recent study by Schröder et al., a new protein, DEAD box protein 3 (DDX3), was found to be involved in the type I IFN cascade. DDX3 was identified by coimmunoprecipitation with an IFN antagonist from vaccinia virus (VACV) protein K7 (59). Alternatively, it is intriguing to speculate that coronavirus PLPs may function by sequestering polyubiquitin complexes or membrane-associated factors, such as STING/MITA (26, 67, 72). Previously we showed that the block must be downstream of TBK1 but at or upstream of IRF-3 since a constitutively active form of IRF-3 is not blocked by PLpro (13). Further studies are needed to elucidate the mechanism by which coronavirus PLPs modulate IRF-3 activity.
The multiple enzymatic activities of SARS-CoV PLpro and HCoV-NL63 PLP2 may all influence the host cell type I IFN response. The DUB activity of these proteins could modulate the activity of key players in the signaling cascade that are known to be activated by lysine-48- or lysine-63-linked polyubiquitination. Our data suggest that catalytic activity may contribute to IFN antagonism but ablation of proteolysis does not abrogate IFN antagonism. Thus, coronavirus PLPs also possess a catalytic activity-independent mechanism that acts to inhibit IFN induction. We are currently working to delineate both the DUB and catalytic activity-independent mechanism by which coronavirus PLPs inhibit type I IFN induction.
The data presented in this study draw significant parallels between the single papain-like protease of SARS-CoV PLpro and the second papain-like protease of HCoV-NL63 PLP2. Despite modest sequence identity (∼19%), these two proteases have similar enzymatic activities and can inhibit type I IFN induction independently of catalytic activity. Since coronavirus PLpro/PLP2 domains are required for viral replication, they are attractive targets for antiviral therapeutics. Indeed, inhibitors of SARS-CoV PLpro have been shown to block virus replication (56). Though less pathogenic than SARS-CoV, HCoV-NL63 causes significant morbidity in children, the elderly, and immune-compromised individuals and has been shown to be an etiological agent causing croup (65, 66). In addition, we now recognize that bats and other mammals can serve as reservoirs for potentially emerging pandemic coronaviruses (32, 37). Thus, further studies of these multifunctional coronavirus PLPs are needed to determine if both protease inhibitors and blockers of interferon antagonism can be developed to reduce replication and pathogenesis of human and zoonotic coronaviruses.
Acknowledgments
We thank members of the Baker lab for helpful discussions.
The work was supported by NIH grant P01AI060915 (to S.C.B., A.K.G., and A.D.M.), DoD grant W81XWH-09-01-0391 (to Gerald Byrne), and grants from the National Natural Science Foundation of China (30870536 and 30972761 to Z.C.) and Beijing Municipal Natural Science Foundation (7092075 to Z.C.). M.A.C. and B.S.B. were supported by NIH training grant T32AI007508.
Footnotes
Published ahead of print on 24 February 2010.
REFERENCES
- 1.Banach, B., J. M. Orenstein, L. M. Fox, S. H. Randell, A. H. Rowley, and S. C. Baker. 2009. Human airway epithelial cell culture to identify new respiratory viruses: coronavirus NL63 as a model. J. Virol. Methods 156:19-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barral, P. M., D. Sarkar, P. B. Fisher, and V. R. Racaniello. 2009. RIG-I is cleaved during picornavirus infection. Virology 391:171-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barral, P. M., D. Sarkar, Z. Z. Su, G. N. Barber, R. DeSalle, V. R. Racaniello, and P. B. Fisher. 2009. Functions of the cytoplasmic RNA sensors RIG-I and MDA-5: key regulators of innate immunity. Pharmacol. Ther. 124:219-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barretto, N., D. Jukneliene, K. Ratia, Z. Chen, A. D. Mesecar, and S. C. Baker. 2005. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 79:15189-15198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basler, C. F., A. Mikulasova, L. Martinez-Sobrido, J. Paragas, E. Muhlberger, M. Bray, H. D. Klenk, P. Palese, and A. Garcia-Sastre. 2003. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J. Virol. 77:7945-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basler, C. F., X. Wang, E. Muhlberger, V. Volchkov, J. Paragas, H. D. Klenk, A. Garcia-Sastre, and P. Palese. 2000. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc. Natl. Acad. Sci. U. S. A. 97:12289-12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhoj, V. G., and Z. J. Chen. 2009. Ubiquitylation in innate and adaptive immunity. Nature 458:430-437. [DOI] [PubMed] [Google Scholar]
- 8.Bibeau-Poirier, A., and M. J. Servant. 2008. Roles of ubiquitination in pattern-recognition receptors and type I interferon receptor signaling. Cytokine 43:359-367. [DOI] [PubMed] [Google Scholar]
- 9.Bowie, A. G., and L. Unterholzner. 2008. Viral evasion and subversion of pattern-recognition receptor signalling. Nat. Rev. Immunol. 8:911-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, T. H., T. Kubota, M. Matsuoka, S. Jones, S. B. Bradfute, M. Bray, and K. Ozato. 2009. Ebola Zaire virus blocks type I interferon production by exploiting the host SUMO modification machinery. PLoS Pathog. 5:e1000493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, Z., R. Rijnbrand, R. K. Jangra, S. G. Devaraj, L. Qu, Y. Ma, S. M. Lemon, and K. Li. 2007. Ubiquitination and proteasomal degradation of interferon regulatory factor-3 induced by Npro from a cytopathic bovine viral diarrhea virus. Virology 366:277-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, Z., Y. Wang, K. Ratia, A. D. Mesecar, K. D. Wilkinson, and S. C. Baker. 2007. Proteolytic processing and deubiquitinating activity of papain-like proteases of human coronavirus NL63. J. Virol. 81:6007-6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Devaraj, S. G., N. Wang, Z. Chen, Z. Chen, M. Tseng, N. Barretto, R. Lin, C. J. Peters, C. T. Tseng, S. C. Baker, and K. Li. 2007. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J. Biol. Chem. 282:32208-32221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans, P. C., H. Ovaa, M. Hamon, P. J. Kilshaw, S. Hamm, S. Bauer, H. L. Ploegh, and T. S. Smith. 2004. Zinc-finger protein A20, a regulator of inflammation and cell survival, has de-ubiquitinating activity. Biochem. J. 378:727-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frias-Staheli, N., N. V. Giannakopoulos, M. Kikkert, S. L. Taylor, A. Bridgen, J. Paragas, J. A. Richt, R. R. Rowland, C. S. Schmaljohn, D. J. Lenschow, E. J. Snijder, A. Garcia-Sastre, and H. W. Virgin IV. 2007. Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe 2:404-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frieman, M., K. Ratia, R. E. Johnston, A. D. Mesecar, and R. S. Baric. 2009. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-kappaB signaling. J. Virol. 83:6689-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frieman, M., B. Yount, M. Heise, S. A. Kopecky-Bromberg, P. Palese, and R. S. Baric. 2007. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 81:9812-9824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldsmith, C. S., K. M. Tatti, T. G. Ksiazek, P. E. Rollin, J. A. Comer, W. W. Lee, P. A. Rota, B. Bankamp, W. J. Bellini, and S. R. Zaki. 2004. Ultrastructural characterization of SARS coronavirus. Emerg. Infect. Dis. 10:320-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gosert, R., A. Kanjanahaluethai, D. Egger, K. Bienz, and S. C. Baker. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 76:3697-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haglund, K., and I. Dikic. 2005. Ubiquitylation and cell signaling. EMBO J. 24:3353-3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herzog, P., C. Drosten, and M. A. Muller. 2008. Plaque assay for human coronavirus NL63 using human colon carcinoma cells. Virol. J. 5:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hicke, L., and R. Dunn. 2003. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu. Rev. Cell Dev. Biol. 19:141-172. [DOI] [PubMed] [Google Scholar]
- 23.Higgs, R., J. Ni Gabhann, N. Ben Larbi, E. P. Breen, K. A. Fitzgerald, and C. A. Jefferies. 2008. The E3 ubiquitin ligase Ro52 negatively regulates IFN-beta production post-pathogen recognition by polyubiquitin-mediated degradation of IRF3. J. Immunol. 181:1780-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Honda, K., and T. Taniguchi. 2006. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat. Rev. Immunol. 6:644-658. [DOI] [PubMed] [Google Scholar]
- 25.Isaacson, M. K., and H. L. Ploegh. 2009. Ubiquitination, ubiquitin-like modifiers, and deubiquitination in viral infection. Cell Host Microbe 5:559-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishikawa, H., and G. N. Barber. 2008. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455:674-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kamitani, W., K. Narayanan, C. Huang, K. Lokugamage, T. Ikegami, N. Ito, H. Kubo, and S. Makino. 2006. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc. Natl. Acad. Sci. U. S. A. 103:12885-12890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kayagaki, N., Q. Phung, S. Chan, R. Chaudhari, C. Quan, K. M. O'Rourke, M. Eby, E. Pietras, G. Cheng, J. F. Bazan, Z. Zhang, D. Arnott, and V. M. Dixit. 2007. DUBA: a deubiquitinase that regulates type I interferon production. Science 318:1628-1632. [DOI] [PubMed] [Google Scholar]
- 29.Kirkin, V., and I. Dikic. 2007. Role of ubiquitin- and Ubl-binding proteins in cell signaling. Curr. Opin. Cell Biol. 19:199-205. [DOI] [PubMed] [Google Scholar]
- 30.Kopecky-Bromberg, S. A., L. Martinez-Sobrido, M. Frieman, R. A. Baric, and P. Palese. 2007. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J. Virol. 81:548-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koyama, S., K. J. Ishii, C. Coban, and S. Akira. 2008. Innate immune response to viral infection. Cytokine 43:336-341. [DOI] [PubMed] [Google Scholar]
- 32.Lau, S. K., P. C. Woo, K. S. Li, Y. Huang, H. W. Tsoi, B. H. Wong, S. S. Wong, S. Y. Leung, K. H. Chan, and K. Y. Yuen. 2005. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U. S. A. 102:14040-14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lenschow, D. J., N. V. Giannakopoulos, L. J. Gunn, C. Johnston, A. K. O'Guin, R. E. Schmidt, B. Levine, and H. W. Virgin IV. 2005. Identification of interferon-stimulated gene 15 as an antiviral molecule during Sindbis virus infection in vivo. J. Virol. 79:13974-13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lenschow, D. J., C. Lai, N. Frias-Staheli, N. V. Giannakopoulos, A. Lutz, T. Wolff, A. Osiak, B. Levine, R. E. Schmidt, A. Garcia-Sastre, D. A. Leib, A. Pekosz, K. P. Knobeloch, I. Horak, and H. W. Virgin IV. 2007. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc. Natl. Acad. Sci. U. S. A. 104:1371-1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, K., Z. Chen, N. Kato, M. Gale, Jr., and S. M. Lemon. 2005. Distinct poly(I-C) and virus-activated signaling pathways leading to interferon-beta production in hepatocytes. J. Biol. Chem. 280:16739-16747. [DOI] [PubMed] [Google Scholar]
- 36.Li, K., E. Foy, J. C. Ferreon, M. Nakamura, A. C. Ferreon, M. Ikeda, S. C. Ray, M. Gale, Jr., and S. M. Lemon. 2005. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc. Natl. Acad. Sci. U. S. A. 102:2992-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li, W., Z. Shi, M. Yu, W. Ren, C. Smith, J. H. Epstein, H. Wang, G. Crameri, Z. Hu, H. Zhang, J. Zhang, J. McEachern, H. Field, P. Daszak, B. T. Eaton, S. Zhang, and L. F. Wang. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310:676-679. [DOI] [PubMed] [Google Scholar]
- 38.Li, X. D., L. Sun, R. B. Seth, G. Pineda, and Z. J. Chen. 2005. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc. Natl. Acad. Sci. U. S. A. 102:17717-17722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindner, H. A., N. Fotouhi-Ardakani, V. Lytvyn, P. Lachance, T. Sulea, and R. Menard. 2005. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J. Virol. 79:15199-15208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindner, H. A., V. Lytvyn, H. Qi, P. Lachance, E. Ziomek, and R. Menard. 2007. Selectivity in ISG15 and ubiquitin recognition by the SARS coronavirus papain-like protease. Arch. Biochem. Biophys. 466:8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu, Y. C., J. Penninger, and M. Karin. 2005. Immunity by ubiquitylation: a reversible process of modification. Nat. Rev. Immunol. 5:941-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loo, Y. M., D. M. Owen, K. Li, A. K. Erickson, C. L. Johnson, P. M. Fish, D. S. Carney, T. Wang, H. Ishida, M. Yoneyama, T. Fujita, T. Saito, W. M. Lee, C. H. Hagedorn, D. T. Lau, S. A. Weinman, S. M. Lemon, and M. Gale, Jr. 2006. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc. Natl. Acad. Sci. U. S. A. 103:6001-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu, G., J. T. Reinert, I. Pitha-Rowe, A. Okumura, M. Kellum, K. P. Knobeloch, B. Hassel, and P. M. Pitha. 2006. ISG15 enhances the innate antiviral response by inhibition of IRF-3 degradation. Cell Mol. Biol. (Noisy-le-grand) 52:29-41. [PubMed] [Google Scholar]
- 44.Malakhova, O. A., and D. E. Zhang. 2008. ISG15 inhibits Nedd4 ubiquitin E3 activity and enhances the innate antiviral response. J. Biol. Chem. 283:8783-8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maniatis, T., J. V. Falvo, T. H. Kim, T. K. Kim, C. H. Lin, B. S. Parekh, and M. G. Wathelet. 1998. Structure and function of the interferon-beta enhanceosome. Cold Spring Harb. Symp. Quant. Biol. 63:609-620. [DOI] [PubMed] [Google Scholar]
- 46.Meylan, E., J. Curran, K. Hofmann, D. Moradpour, M. Binder, R. Bartenschlager, and J. Tschopp. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167-1172. [DOI] [PubMed] [Google Scholar]
- 47.Meylan, E., and J. Tschopp. 2006. Toll-like receptors and RNA helicases: two parallel ways to trigger antiviral responses. Mol. Cell 22:561-569. [DOI] [PubMed] [Google Scholar]
- 48.Narayanan, K., C. Huang, K. Lokugamage, W. Kamitani, T. Ikegami, C. T. Tseng, and S. Makino. 2008. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J. Virol. 82:4471-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neznanov, N., K. M. Chumakov, L. Neznanova, A. Almasan, A. K. Banerjee, and A. V. Gudkov. 2005. Proteolytic cleavage of the p65-RelA subunit of NF-kappaB during poliovirus infection. J. Biol. Chem. 280:24153-24158. [DOI] [PubMed] [Google Scholar]
- 50.Nicholson, B., C. A. Leach, S. J. Goldenberg, D. M. Francis, M. P. Kodrasov, X. Tian, J. Shanks, D. E. Sterner, A. Bernal, M. R. Mattern, K. D. Wilkinson, and T. R. Butt. 2008. Characterization of ubiquitin and ubiquitin-like-protein isopeptidase activities. Protein Sci. 17:1035-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nijman, S. M., M. P. Luna-Vargas, A. Velds, T. R. Brummelkamp, A. M. Dirac, T. K. Sixma, and R. Bernards. 2005. A genomic and functional inventory of deubiquitinating enzymes. Cell 123:773-786. [DOI] [PubMed] [Google Scholar]
- 52.Ozato, K., P. Tailor, and T. Kubota. 2007. The interferon regulatory factor family in host defense: mechanism of action. J. Biol. Chem. 282:20065-20069. [DOI] [PubMed] [Google Scholar]
- 53.Perlman, S., and J. Netland. 2009. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 7:439-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pichlmair, A., and C. Reis e Sousa. 2007. Innate recognition of viruses. Immunity 27:370-383. [DOI] [PubMed] [Google Scholar]
- 55.Platanias, L. C. 2005. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 5:375-386. [DOI] [PubMed] [Google Scholar]
- 56.Ratia, K., S. Pegan, J. Takayama, K. Sleeman, M. Coughlin, S. Baliji, R. Chaudhuri, W. Fu, B. S. Prabhakar, M. E. Johnson, S. C. Baker, A. K. Ghosh, and A. D. Mesecar. 2008. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc. Natl. Acad. Sci. U. S. A. 105:16119-16124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ratia, K., K. S. Saikatendu, B. D. Santarsiero, N. Barretto, S. C. Baker, R. C. Stevens, and A. D. Mesecar. 2006. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. U. S. A. 103:5717-5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roth-Cross, J. K., L. Martinez-Sobrido, E. P. Scott, A. Garcia-Sastre, and S. R. Weiss. 2007. Inhibition of the alpha/beta interferon response by mouse hepatitis virus at multiple levels. J. Virol. 81:7189-7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schroder, M., M. Baran, and A. G. Bowie. 2008. Viral targeting of DEAD box protein 3 reveals its role in TBK1/IKKepsilon-mediated IRF activation. EMBO J. 27:2147-2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sheahan, T., B. Rockx, E. Donaldson, A. Sims, R. Pickles, D. Corti, and R. Baric. 2008. Mechanisms of zoonotic severe acute respiratory syndrome coronavirus host range expansion in human airway epithelium. J. Virol. 82:2274-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Snijder, E. J., Y. van der Meer, J. Zevenhoven-Dobbe, J. J. Onderwater, J. van der Meulen, H. K. Koerten, and A. M. Mommaas. 2006. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 80:5927-5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Spiegel, M., A. Pichlmair, L. Martinez-Sobrido, J. Cros, A. Garcia-Sastre, O. Haller, and F. Weber. 2005. Inhibition of Beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J. Virol. 79:2079-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spiegel, M., K. Schneider, F. Weber, M. Weidmann, and F. T. Hufert. 2006. Interaction of severe acute respiratory syndrome-associated coronavirus with dendritic cells. J. Gen. Virol. 87:1953-1960. [DOI] [PubMed] [Google Scholar]
- 64.Swedan, S., A. Musiyenko, and S. Barik. 2009. Respiratory syncytial virus nonstructural proteins decrease multiple members of the cellular interferon pathways. J. Virol. 83:9682-9693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van der Hoek, L., K. Sure, G. Ihorst, A. Stang, K. Pyrc, M. F. Jebbink, G. Petersen, J. Forster, B. Berkhout, and K. Uberla. 2005. Croup is associated with the novel coronavirus NL63. PLoS Med. 2:e240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van der Hoek, L., K. Sure, G. Ihorst, A. Stang, K. Pyrc, M. F. Jebbink, G. Petersen, J. Forster, B. Berkhout, and K. Uberla. 2006. Human coronavirus NL63 infection is associated with croup. Adv. Exp. Med. Biol. 581:485-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xia, Z. P., L. Sun, X. Chen, G. Pineda, X. Jiang, A. Adhikari, W. Zeng, and Z. J. Chen. 2009. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature 461:114-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ye, Y., K. Hauns, J. O. Langland, B. L. Jacobs, and B. G. Hogue. 2007. Mouse hepatitis coronavirus A59 nucleocapsid protein is a type I interferon antagonist. J. Virol. 81:2554-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao, C., C. Denison, J. M. Huibregtse, S. Gygi, and R. M. Krug. 2005. Human ISG15 conjugation targets both IFN-induced and constitutively expressed proteins functioning in diverse cellular pathways. Proc. Natl. Acad. Sci. U. S. A. 102:10200-10205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng, D., G. Chen, B. Guo, G. Cheng, and H. Tang. 2008. PLP2, a potent deubiquitinase from murine hepatitis virus, strongly inhibits cellular type I interferon production. Cell Res. 18:1105-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zheng, L., U. Baumann, and J. L. Reymond. 2004. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 32:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhong, B., Y. Yang, S. Li, Y. Y. Wang, Y. Li, F. Diao, C. Lei, X. He, L. Zhang, P. Tien, and H. B. Shu. 2008. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29:538-550. [DOI] [PubMed] [Google Scholar]