Abstract
A recombinant human monoclonal antibody, IgG1 b12 (b12), recognizes a conformational epitope on human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein (Env) gp120 that overlaps the CD4 binding domain. Although b12 is able to broadly neutralize HIV-1 subtype B, C, and D viruses, many HIV-1 CRF01_AE viruses are resistant to b12-mediated neutralization. In this report, we examined the molecular mechanisms underlying the low neutralization susceptibility of CRF01_AE viruses to b12, using recently established CRF01_AE Env recombinant viruses. Our results showed that two potential N-linked glycosylation (PNLG) sites in the V2 and C2 regions of Env gp120 played an important role in regulating the susceptibility of CRF01_AE Env to b12. The locations of these PNLG sites correspond to amino acid positions 186 and 197 in HXB2 Env gp120; thus, they are designated N186 and N197 in this study. Removal of N186 significantly conferred the b12 susceptibility of 2 resistant CRF01_AE Env clones, 65CC4 and 107CC2, while the introduction of N186 reduced the b12 susceptibility of a susceptible CRF01_AE Env clone, 65CC1. In addition, removal of both N186 and N197 conferred the b12 susceptibility of 3 resistant CRF01_AE Env clones, 45PB1, 62PL1, and 101PL1, whereas the removal of either N186 or N197 was not sufficient to confer the b12 susceptibility of these CRF01_AE Env clones. Finally, removal of N197 conferred the b12 susceptibility of 2 resistant CRF01_AE Env clones lacking N186, 55PL1 and 102CC2. Taken together, we propose that two PNLG sites, N186 and N197, in Env gp120 are important determinants of the b12 resistance of CRF01_AE viruses.
IgG1 b12 (b12) is a recombinant human monoclonal antibody established from a Fab (IgG1k) phage display library generated from bone marrow samples of human immunodeficiency virus type 1 (HIV-1)-infected patients (2, 4). The b12 antibody recognizes a conformational epitope on HIV-1 envelope glycoprotein (Env) gp120 that overlaps the CD4 binding domain (2, 5, 38) and is able to neutralize diverse strains of HIV-1 (3, 45). In addition, b12 protects hu-PBL-SCID mice (SCID mice given human peripheral blood lymphocytes) and macaque monkeys from infection with HIV-1 and SHIV, a virus combining parts of the HIV and simian immunodeficiency virus (SIV) genomes, respectively (31, 32, 48). Furthermore, it is demonstrated that serum antibodies specific to the CD4 binding domain of gp120 are responsible for the potent and broad neutralization of HIV-1 strains mediated by broadly reactive sera of HIV-1-infected patients (21); therefore, it is important to establish a vaccine strategy to elicit a broadly neutralizing antibody against the CD4 binding domain, such as b12 (12, 21). To this end, regulatory mechanisms underlying the different susceptibilities of various HIV-1 strains to b12 need to be clarified.
CRF01_AE is one of the major HIV-1 subtypes that dominate the global epidemic and is prevalent throughout Southeast Asia (13, 26). In particular, this subtype is responsible for more than 95% of infection cases in Thailand, Cambodia, and Viet Nam (13). Although b12 is able to broadly neutralize HIV-1 subtype B, C, and D clinical isolates, it poorly neutralizes many CRF01_AE strains (3, 47, 49); however, the mechanisms why CRF01_AE viruses show low susceptibility to b12-mediated neutralization are still not understood.
Recently, we established 35 infectious CRF01_AE Env recombinant viruses, and studied their neutralization susceptibility to neutralizing human monoclonal antibodies, patient serum and HIV-1 entry inhibitors (46, 47). Among them, a recombinant virus containing CRF01_AE Env, 65CC1 was susceptible to b12, while 34 remaining CRF01_AE Env recombinant viruses, including the virus containing CRF01_AE Env, 65CC4, were resistant to b12 (47). In this report, we examined the molecular mechanisms underlying the low b12 susceptibility of CRF01_AE Env, using these CRF01_AE Env recombinant viruses.
MATERIALS AND METHODS
Cells.
293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (10% FBS-DMEM). U87.CD4.CCR5 and U87.CD4.CXCR4 cells were obtained through the AIDS Research and Reference Reagent Program (Division of AIDS, NIAID, NIH) from HongKui Deng and Dan R. Littman and were maintained in 10% FBS-DMEM with puromycin (1 μg/ml) and G418 (300 μg/ml) (complete medium).
Preparation of recombinant proviral constructs.
cDNAs encoding CRF01_AE Env gp120 and gp41, 45PB1, 55PL1, 62PL1, 65CC1, 65CC4, 101PL1, 102CC2, and 107CC2 were cloned into pNL-envCT (14), luciferase reporter proviral DNA derived from pNL4-3 (1), to generate CRF01_AE Env recombinant proviral constructs as described previously (46). In addition, recombinant proviral constructs containing chimeric CRF01_AE Env, 65CC1N4C and 65CC4N1C (see Fig. 1A), were prepared as described previously (47). In order to generate N-linked glycosylation mutants of CRF01_AE Env, amino acid substitution(s) N186Q (amino acid substitution from asparagine [N] to glutamine [Q] at position 186), N187Q, S186N/N188S and/or N197Q, were introduced into the CRF01_AE Env, 45PB1, 55PL1, 62PL1, 65CC1, 65CC4, 101PL1, 102CC2, and 107CC2, by site-directed mutagenesis using the QuikChange site-directed mutagenesis kit (Stratagene, Cedar Creek, TX). In addition, an amino acid substitution, E302D, was introduced into a CRF01_AE Env, 65CC4, to generate 65CC4-E302D.
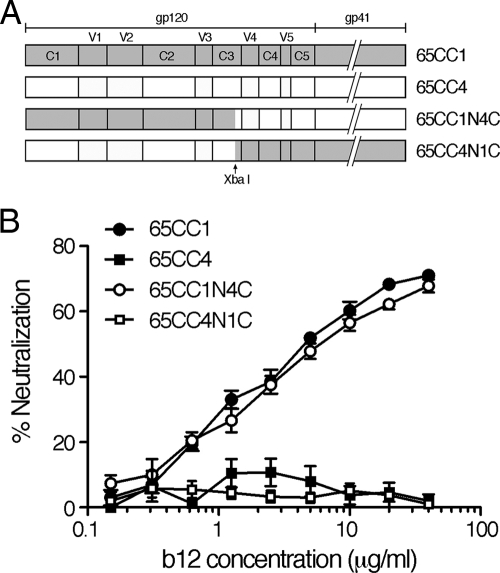
FIG. 1.
(A) Schematic illustration of variable (V) and conserved (C) regions of Env gp120 and gp41. Chimeric CRF01_AE Env containing partial fragments of 65CC1 and 65CC4, 65CC1N4C and 65CC4N1C, respectively, were constructed and subjected to neutralization tests. (B) N-terminal regions of gp120 contain the determinants of b12 resistance of 65CC4. Neutralization susceptibility of CRF01_AE Env recombinant viruses to b12 was evaluated as described in Materials and Methods. Results are expressed as percent neutralization, which was calculated by determining the reduction in viral infectivity in the presence of b12 compared to that in control experiments in the absence of b12. All data points represent the means and standard errors (error bars) of four independent experiments.
Preparation of recombinant virus.
Viral supernatants were prepared by transfecting 293T cells with the proviral construct using FuGENE HD transfection reagent (Roche, Basel, Switzerland). Forty-eight hours after transfection, the supernatants were cleared by centrifugation for 5 min at 8,000 rpm, and stored as aliquots at −85°C. The concentration of HIV-1 Gag p24 antigen in viral supernatants was measured by enzyme-linked immunosorbent assay (ELISA) (Vironostika HIV-1 antigen micro-ELISA system; bioMérieux, Boxtel, Netherlands).
Neutralization tests.
The neutralization susceptibility of the recombinant virus to the monoclonal antibody b12 (Polymun Scientific, Vienna, Austria) was examined as follows. Viral supernatants (10 ng of p24 antigen) were incubated with 2-fold serially diluted b12 (160 to 0.005 μg/ml) in 100 μl complete medium for 1 h at 37°C. In control experiments, viral supernatants were incubated with control human IgG. U87.CD4.CXCR4 or U87.CD4.CCR5 cells, which were seeded into the wells of a 96-well plate (5 × 103 cells per 100 μl per well) 24 h prior to neutralization tests, were then incubated with the mixture of viral supernatants and b12. U87.CD4.CXCR4 cells were used as target cells for the recombinant virus containing CRF01_AE Env, 45PB1, 65CC1, 65CC4, or 107CC2 as well as for pNL-envCT, whereas U87.CD4.CCR5 cells were used as target cells for the recombinant virus containing CRF01_AE Env, 55PL1, 62PL1, 101PL1, or 102CC2. Forty-eight hours after infection, luciferase activity in infected cells was measured using the Steady Glo luciferase assay kit (Promega, Madison, WI) with an LB960 microplate luminometer (Berthold, Bad Wildbad, Germany). Percent neutralization was calculated by determining the reduction in luciferase activity in the presence of b12 compared to that in control experiments in the absence of b12.
Viral infectivity assay.
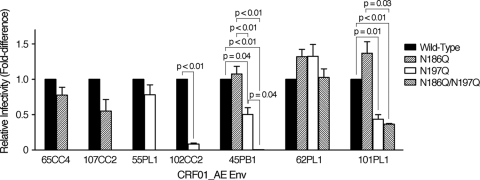
Viral supernatants were prepared as described above. U87.CD4.CCR5 or U87.CD4.CXCR4 cells, which were seeded into the wells of a 24-well plate (3 × 104 cells per 500 μl per well) 24 h prior to the tests, were infected with the recombinant virus (20 ng of p24). Forty-eight hours after infection, luciferase activity in infected cells was measured as described above. Relative infectivity was calculated by dividing the luciferase activity produced by the N-linked glycosylation mutant virus by that produced by the corresponding wild-type virus. Statistical analysis was carried out using the standard function of GraphPad Prism 5 software (GraphPad Software, San Diego, CA) with a paired t test.
Preparation of a structural model.
A model of the gp120 structure was constructed based on the following coordinates, the HIV-1 gp120 trimeric structure solved by tomographic reconstruction (Protein Data Bank identification code [accession number] [PDB ID] 3DNL) (22), the crystal structure of HIV-1 gp120 core complexed with b12 (PDB ID 2NY7) (52), and the crystal structure of the fully glycosylated SIV gp120 core (PDB ID 2BF1) (6). The two crystal structures (2NY7 and 2BF1) were fitted to each subunit of the trimeric structure (3DNL) using the secondary-structure matching method installed in the CCP4 suite (7, 16). Subsequent illustration of the model was prepared using the PyMOL molecular visualization system (DeLano Scientific, San Carlos, CA [http://www.pymol.org/]).
RESULTS
The N-terminal regions of Env gp120 contain the determinants of b12 resistance of the CRF01_AE Env 65CC4.
Two CRF01_AE Env clones, 65CC1 and 65CC4, showed distinct neutralization susceptibility to b12 (47), although these CRF01_AE Env clones had a close phylogenetic relationship (46). In addition, our previous results suggested that the determinants of b12 resistance of 65CC4 were located within the N-terminal regions (C1, V1, V2, C2, V3, and a 36-amino-acid sequence of C3 before the XbaI recognition site) of Env gp120 (47). First, we confirmed our previous results by performing neutralization tests using 4 CRF01_AE Env recombinant viruses. The results showed that 65CC4 and a chimeric Env, 65CC4N1C, containing N-terminal regions of 65CC4 gp120, were resistant to b12-mediated neutralization, while 65CC1 and a chimeric Env, 65CC1N4C, containing C-terminal regions of 65CC4 gp120, showed high neutralization susceptibility to b12 (Fig. 1). These results confirmed that the N-terminal regions of Env gp120 contained the determinants of b12 resistance of 65CC4.
Comparison of the amino acid sequences of the N-terminal regions of Env gp120.
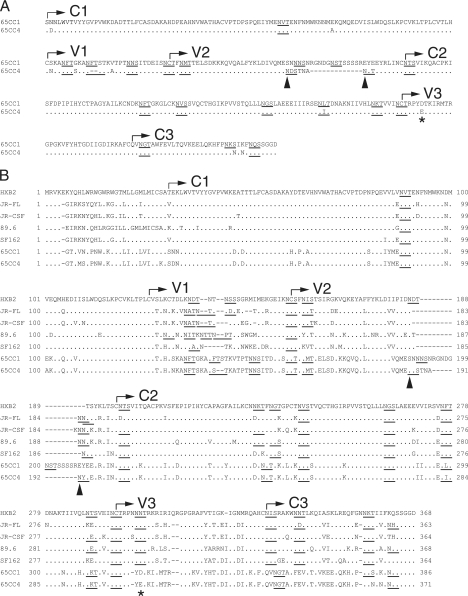
We next compared the amino acid sequences of the N-terminal regions of Env gp120 in 65CC1 and 65CC4. The results showed that several amino acid residues differed in 65CC1 and 65CC4 (Fig. 2A). In addition, the V1 and V2 regions of 65CC4 were 2 and 13 amino acids shorter than those of 65CC1, respectively (Fig. 2A). Furthermore, although 65CC1 and 65CC4 contained an equal number of potential N-linked glycosylation (PNLG) sites (N-X-S/T, where X is any amino acid except for proline) in N-terminal regions of Env gp120, the locations of a few PNLG sites differed in 65CC1 and 65CC4 (Fig. 2A). Recent studies on CRF01_AE (40, 47) and subtype C Env (39) showed that viral susceptibility to neutralizing antibodies was inversely correlated with the length of V1 and V2 regions of Env gp120. In addition, several CRF01_AE Env clones that contain a long V2 region, similar to 65CC1 (46), were resistant to b12-mediated neutralization (47). Thus, we considered that short V1 and V2 regions of 65CC4 might not account for the lower b12 susceptibility of 65CC4 compared to 65CC1. In contrast, it was demonstrated that N-linked glycosylation of a particular amino acid residue plays an important role as a glycan shield, reducing the susceptibility of HIV-1 Env to neutralizing antibodies (25, 50); therefore, we decided to examine the possible involvement of PNLG sites in regulating b12 susceptibility of 65CC1 and 65CC4. We found 2 PNLG sites located in the V2 region of 65CC4, but not in that of 65CC1 (Fig. 2A, arrowheads). Regulatory mechanisms of b12 susceptibility were well examined on HIV-1 subtype B laboratory strains (8, 20, 23, 28, 30, 34, 35); therefore, in order to define the locations of these PNLG sites, we next compared the amino acid sequences of N-terminal regions of Env gp120 among 5 subtype B strains, JR-FL, JR-CSF, 89.6, SF162, and HXB2, as well as 2 CRF01_AE Env clones, 65CC1 and 65CC4. The results showed that PNLG sites in the N-terminal regions of gp120 were relatively conserved among 5 subtype B and 2 CRF01_AE Env clones, with several exceptions (Fig. 2B). The locations of 2 PNLG sites found in the V2 region of 65CC4 corresponded to amino acid positions 186 and 187 in HXB2 Env gp120; thus, these PNLG sites were designated N186 and N187 by HXB2 numbering in this report (Fig. 2B, arrowheads).
FIG. 2.
Comparison of the N-terminal regions of gp120 between 2 CRF01_AE Env clones, 65CC1 and 65CC4 (A), or among 5 subtype B and 2 CRF01_AE Env clones (B). Amino acid sequences of the N-terminal regions (before the XbaI recognition site) of Env gp120 were compared among subtype B strains, HXB2 (GenBank accession no. K03455), JR-FL (U63632), JR-CSF (M38429), 89.6 (U39362) and SF162 (M65024), as well as CRF01_AE Env, 65CC1 (EU743779), and 65CC4 (EU743780). These amino acid sequences were aligned using the ClustalW algorithm (44) with slight manual adjustment. The positions of gp120 variable (V) and conserved (C) regions are denoted in the figure. Dots denote amino acid identity, while dashes represent gaps introduced to optimize alignment. PNLG sites are shown by underlining. In panel B, the numbering of amino acid residues is based on the sequence of HXB2 Env and is shown beside the aligned sequence. Arrowheads indicate two PNLG sites, N186 and N187, detected in 65CC4 but not 65CC1, whereas asterisks indicate the amino acid difference in the V3 region in 65CC1 and 65CC4.
In addition, since amino acid substitutions in the V3 region were reported to affect viral neutralization susceptibility to b12 (8, 15, 24), we decided to also examine the potential involvement of the amino acid difference at position 302 (by HXB2 numbering) (Fig. 2, asterisks) in regulating b12 susceptibility of CRF01_AE Env.
Neutralization susceptibility of 65CC4- and 65CC1-derived CRF01_AE Env mutants to b12.
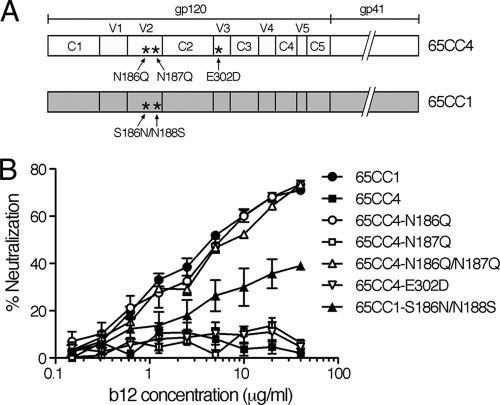
65CC4-derived N-linked glycosylation mutants lacking a PNLG site at N186 or N187 in the V2 region, 65CC4-N186Q or 65CC4-N187Q, respectively, the mutant lacking both N186 and N187, 65CC4-N186Q/N187Q, as well as the mutant with an amino acid substitution in the V3 region, 65CC4-E302D, were constructed and subjected to neutralization tests (Fig. 3A). The results showed that 65CC4-N186Q was highly susceptible to b12-mediated neutralization, while 65CC4-N187Q was resistant to b12 (Fig. 3B), whereas 65CC4-N186Q/N187Q showed similar b12 susceptibility to 65CC4-N186Q (Fig. 3B). In addition, 65CC4-E302D was resistant to b12 (Fig. 3B). These results suggested that removal of the PNLG site N186 conferred b12 susceptibility of 65CC4, whereas removal of the PNLG site N187, as well as the amino acid substitution, E302D, in the V3 region, had no effect on b12 susceptibility of 65CC4. To confirm the role of N186 in viral neutralization susceptibility to b12, we next constructed a 65CC1-derived mutant containing N186, 65CC1-S186N/N188S, and subjected it to neutralization tests (Fig. 3A). The results showed that 65CC1-S186N/N188S was notably less susceptible to b12-mediated neutralization than parental 65CC1 (Fig. 3B), suggesting that introduction of the PNLG sites N186 reduced b12 susceptibility of 65CC1. 65CC1-S186N/N188S was not completely resistant to b12, implying that other factors might be involved in determining b12 susceptibility of 65CC1. In order to introduce N186 into 65CC1, we had to remove the PNLG site, N188, from 65CC1, which might potentially affect the b12 susceptibility of 65CC1. Nevertheless, these results demonstrate that the PNLG site, N186, in the V2 region of Env gp120 plays an important role in regulating b12 susceptibility of CRF01_AE Env clones, 65CC1 and 65CC4.
FIG. 3.
(A) Schematic illustration of the locations of amino acid substitutions introduced into CRF01_AE Env, 65CC1 and 65CC4. (B) Removal of the PNLG site, N186, confers b12 susceptibility of CRF01_AE Env, 65CC4, while introduction of N186 reduces b12 susceptibility of 65CC1. Neutralization susceptibility of CRF01_AE Env recombinant viruses to b12 was evaluated as described in Materials and Methods. Results are expressed as percent neutralization, as described in the legend to Fig. 1. All data points represent the means and standard errors (error bars) of three independent experiments.
Removal of N186 significantly increases b12 susceptibility of particular, but not all, CRF01_AE Env recombinant viruses.
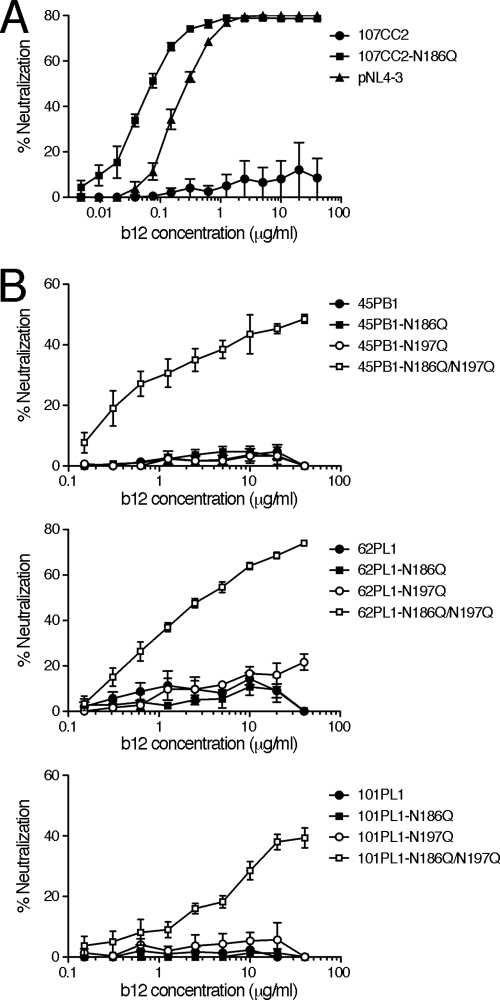
In our previous report, 65CC1 was susceptible to b12, while 34 remaining CRF01_AE Env clones, including 65CC4, were resistant to b12 (47). In addition, the PNLG site N186 was conserved in 13 of 35 CRF01_AE Env recombinant viruses studied (approximately 37%) (data not shown). In order to examine the role of N186 in the b12 susceptibility of other CRF01_AE Env clones, we selected 4 CRF01_AE Env clones, 45PB1, 62PL1, 101PL1, and 107CC2, which naturally contain N186. After mutant viruses lacking N186 were constructed, these mutants, as well as parental CRF01_AE Env recombinant viruses, were subjected to neutralization tests. The results showed that the CRF01_AE Env mutant lacking N186, 107CC2-N186Q, was highly susceptible to b12, while parental 107CC2 was resistant to b12-mediated neutralization (Fig. 4A). In addition, the b12 susceptibility of 107CC2-N186Q was higher than that of subtype B, pNL4-3 Env (Fig. 4A). Namely, removal of N186 markedly conferred the b12 susceptibility of 107CC2. In contrast, 3 CRF01_AE Env mutants lacking N186, 45PB1-N186Q, 62PL1-N186Q, and 101PL1-N186Q, as well as parental CRF01_AE Env, 45PB1, 62PL1, and 101PL1, respectively, were resistant to b12-mediated neutralization (Fig. 4B), suggesting the existence of other determinants of b12 resistance in these CRF01_AE Env viruses. These results demonstrate that removal of the PNLG site, N186, confers b12 susceptibility of particular, but not all, b12-resistant CRF01_AE Env. In other words, other determinants may regulate the b12 susceptibility of CRF01_AE Env.
FIG. 4.
(A and B) Removal of N186 significantly confers b12 susceptibility of CRF01_AE Env, 107CC2 (A), while removal of both N186 and N197 is required to confer b12 susceptibility of CRF01_AE Env, 45PB1, 62PL1, and 101PL1 (B). The susceptibility of CRF01_AE Env recombinant viruses, as well as pNL-envCT containing pNL4-3 Env, to b12-mediated neutralization was evaluated as described in Materials and Methods. Results are expressed as percent neutralization, as described in the legend to Fig. 1. All data points represent the means and standard errors (error bars) of three independent experiments.
N197 cooperates with N186 to regulate b12 susceptibility of CRF01_AE Env clones.
Removal of N186 from 3 CRF01_AE Env clones, 45PB1, 62PL1, and 101PL1, failed to confer their b12 susceptibility (Fig. 4B); thus, we searched for other determinants to regulate the b12 susceptibility of these CRF01_AE Env. The PNLG site at amino acid position 197 (by HXB2 numbering, designated N197) in the C2 region of Env gp120 plays an important role in regulating the b12 susceptibility of subtype B (20, 23, 30) and subtype C viruses (51). In our study, a b12-susceptible CRF01_AE Env, 65CC1, as well as 34 b12-resistant CRF01_AE Env, contained N197 (46, 47); therefore, it was unclear whether N197 plays a role in regulating the b12 susceptibility of CRF01_AE Env. However, a recent report showed that the susceptibility of CRF01_AE Env, DA5, to a neutralizing monoclonal antibody recognizing the CD4 binding domain, F105 (36, 43), is increased by removing N197 (42); thus, this PNLG site might potentially also play a role in regulating the b12 susceptibility of CRF01_AE Env. In order to address this possibility, we constructed mutant viruses lacking N197 and subjected them to neutralization tests. The results showed that 3 CRF01_AE Env mutants lacking N197, 45PB1-N197Q, 62PL1-N197Q, and 101PL1-N197Q were resistant to b12, similar to their parental CRF01_AE Env, 45PB1, 62PL1, and 101PL1, respectively (Fig. 4B). In stark contrast, 3 CRF01_AE Env mutants lacking both N186 and N197, 45PB1-N186Q/N197Q, 62PL1-N186Q/N197Q, and 101PL1-N186Q/N197Q, were susceptible to b12-mediated neutralization (Fig. 4B). These results demonstrate that N186 and N197 cooperate to regulate b12 susceptibility of CRF01_AE Env. Finally, approximately 63% of CRF01_AE Env did not naturally contain N186 in our study (data not shown); therefore, we examined the role of N197 in regulating the b12 susceptibility of CRF01_AE Env lacking N186. To this end, CRF01_AE mutants lacking N197 were constructed and subjected to a neutralization test. The results showed that removal of N197 from 2 CRF01_AE Env lacking N186, 55PL1 and 102CC2, conferred their b12 susceptibility (Fig. 5), indicating that N197 is a determinant of the b12 resistance of CRF01_AE Env in the absence of N186.
FIG. 5.
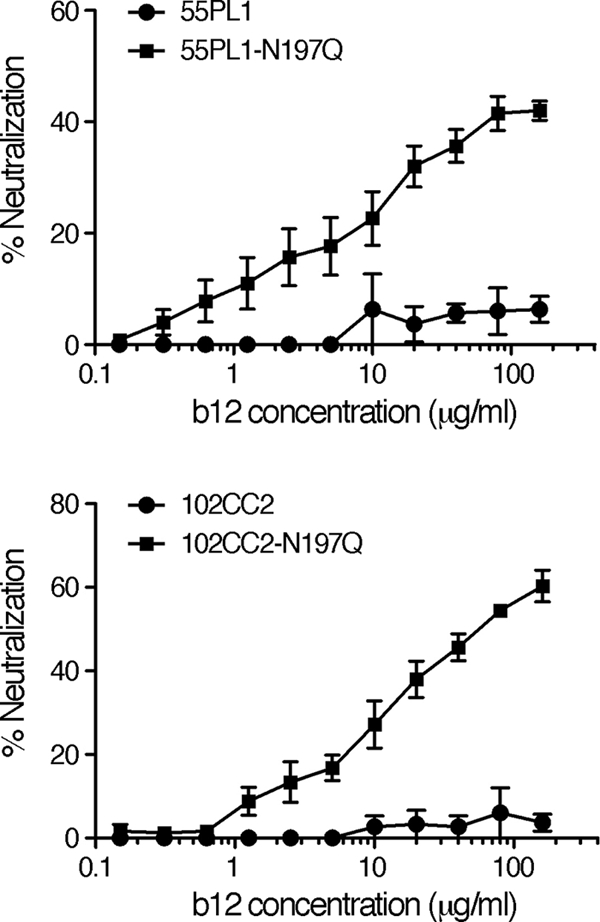
Removal of N197 confers b12 susceptibility of CRF01_AE Env clones lacking N186, 55PL1 and 102CC2. The susceptibility of CRF01_AE Env recombinant viruses to b12-mediated neutralization was evaluated as described in Materials and Methods. Results are expressed as percent neutralization, as described in the legend to Fig. 1. All data points represent the means and standard errors (error bars) of three independent experiments.
No strict correlation is observed between the infectivity and b12 susceptibility of CRF01_AE Env recombinant virus.
Finally, we examined the possible correlation between the infectivity and b12 susceptibility of CRF01_AE Env recombinant virus. For this purpose, the infectivity of the N-linked glycosylation mutant virus was compared with that of the corresponding wild-type virus. The results showed that removal of N186 did not statistically significantly affect (P values of >0.07) the level of single-round replication of CRF01_AE Env recombinant viruses (Fig. 6). In addition, removal of N197 reduced the replication capacity of the recombinant viruses containing 45PB1, 101PL1, and 102CC2, but not of the recombinant viruses containing 55PL1 and 62PL1 (Fig. 6). Furthermore, removal of both N186 and N197 drastically reduced the infectivity of the recombinant virus containing 45PB1, but not of the recombinant viruses containing 62PL1 and 101PL1 (Fig. 6). The recombinant virus containing the b12-susceptible CRF01_AE Env, 65CC4-N186Q, 55PL1-N197Q, 62PL1-N186Q/N197Q, or 101PL1-N186Q/N197Q showed a similar level of infectivity to the recombinant virus containing the b12-resistant CRF01_AE Env, 65CC4, 55PL1, 62PL1, or 101PL1-N197Q, respectively (Fig. 6); thus, we concluded that the increase in the b12 susceptibility of CRF01_AE Env recombinant viruses upon removal of the PNLG site(s) was not due to the decrease in the infectivity of the recombinant viruses.
FIG. 6.
No strict correlation is observed between the infectivity and susceptibility to b12 of CRF01_AE Env recombinant viruses. Viral infectivity of the N-linked glycosylation mutant virus lacking the PNLG site(s), N186Q, N197Q, or both N186 and N197 (N186Q/N197Q) was compared with that of the corresponding wild-type virus as described in Materials and Methods. Data are shown as fold difference in luciferase activity produced by the N-linked glycosylation mutant virus relative to luciferase activity produced by the wild-type virus. Results are presented as the means and standard errors (error bars) of three independent experiments. Differences among the viruses were analyzed by the paired t test and are reported when P < 0.05.
DISCUSSION
In this report, we examined the molecular mechanism underlying the low b12 susceptibility of CRF01_AE viruses. Env gp120 is the most variable HIV-1 protein with typical intersubtype and intrasubtype differences soaring to 35% and 20%, respectively (9). Such diversity in the amino acid sequence may affect the protein structure; thus, we cannot rule out the possibility that the structure of Env gp120 is somewhat different among diverse HIV-1 subtypes. Indeed, structural differences of the conserved and variable regions of gp120 are reported to exist between subtype B and C viruses (11, 33). The b12 antibody recognizes a conformational epitope; thus, b12 susceptibility of HIV-1 Env is necessarily affected by the protein structure of gp120. Taken together, it is conceivable that the mechanisms to determine b12 susceptibility potentially vary among different HIV-1 subtypes.
Amino acid mutations in V1 and V2 regions affect the susceptibility of subtype B viruses to neutralizing antibodies against the CD4 binding domain of gp120, including b12 (20, 23, 28, 29, 41). The results of structural analyses of Env gp120 suggest that part of the V1 and V2 regions contacts with the CD4 molecule when Env gp120 binds to CD4 (17, 22), and this may account for the role of V1 and V2 regions in viral susceptibility to neutralizing antibodies against the CD4 binding domain, including b12.
N-linked glycosylation of HIV-1 Env affects its protein structure and reduces viral susceptibility to neutralizing antibodies (25, 50). N-linked glycans, including the high-mannose type, complex type, and hybrid type, are attached to HIV-1 gp120 (10, 27, 53). In addition, high-mannose-type glycans with 5 to 9 mannose residues are the major type of N-linked glycans detected on gp120 (10, 27, 53). Several reports describe the role of N-linked glycosylation of particular amino acid residues in the V2, C2, and V3 regions of Env gp120 in regulating the b12 susceptibility of subtype B viruses as follows. Removal of the PNLG site, N197 (by HXB2 numbering), in the C2 region of Env gp120 significantly increases the b12 susceptibility of 89.6 and JR-CSF strains (20, 30). In addition, removal of N195 (197 by HXB2 numbering) also significantly increases the b12 susceptibility of the SF162 strain (23). These studies demonstrate that N197 plays an important role in regulating the b12 susceptibility of subtype B strains. Consistent with these reports, our results showed that N197 regulated the b12 susceptibility of CRF01_AE Env recombinant viruses by cooperating with N186 (Fig. 4B). Next, removal of the PNLG sites, N187 (by HXB2 numbering) and N301 (by HXB2 numbering) in the V2 and V3 regions of gp120, respectively, modestly increased the b12 susceptibility of the 89.6 strain (20). In addition, removal of N186 (188 by HXB2 numbering) moderately increased the b12 susceptibility of the SF162 strain, while removal of N154 (156 by HXB2 numbering) significantly increased the b12 susceptibility of the SF162 strain (23). Furthermore, introduction of the PNLG site with amino acid substitution D182N (D185N by HXB2 numbering) in the V2 region of Env gp120 moderately reduced the b12 susceptibility of the JR-CSF strain (28). Finally, JR-FL containing N187 (by HXB2 numbering) and SF162 lacking N187 (by HXB2 numbering) show similar susceptibility to b12-mediated neutralization (34). Taken together, these results show that the PNLG sites, N185, N187, and N188, moderately reduce the b12 susceptibility of JR-CSF, 89.6, and SF162 strains, respectively (20, 28), whereas N187 has no effect on the b12 susceptibility of JR-FL (23, 34). Partially consistent with these results, removal of N187 had no effect on the b12 susceptibility of a CRF01_AE Env, 65CC4 (Fig. 3B). In contrast, removal of N186 or removal of N186 and N197 significantly increased the b12 susceptibility of CRF01_AE Env recombinant viruses (Fig. 3B and 4).
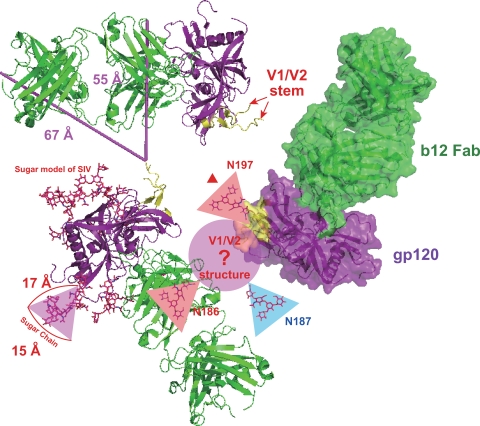
We constructed a structural model to understand the roles of N-linked glycans attached to the PNLG sites, N186, N187, and N197, on the interaction between gp120 and b12 (Fig. 7). Construction of a precise model is impossible because all available structures of gp120 do not include V1 and V2 regions, which may be due to the difficulty of crystallizing gp120 in the presence of these variable regions. For this reason, we have discussed only the relative arrangement and scale among sugar chains and proteins as follows. Part of the V1 and V2 regions (V1/V2 stem) remains in the gp120 molecule (PDB ID 2NY7), which was solved by Zhou et al. (52); thus, we could estimate the likely locations of V1 and V2 regions, as described previously (22). In this model, although V1 and V2 regions are located far from the b12 contact site in the monomeric structure of gp120, these regions are located close to the b12 molecule binding to the adjacent gp120 molecule in the trimeric complex. V1 and V2 regions consist of approximately 70 to 80 amino acid residues, whereas the molecular dimension of high-mannose-type glycan is roughly estimated to be 15 to 17 Å. Since N186 is located near the tip of the V2 region, we could estimate that V1 and V2 regions with an N-linked glycan attached to N186 may interfere with the binding between b12 and gp120. In addition, N197, which is located at the N terminus of the C2 region in close proximity to the V1/V2 stem region, is conserved in all CRF01_AE Env studied (45), implying the importance of an N-linked glycan attached to this position in stabilizing the tertiary structure of HIV-1 gp120. Indeed, removal of N197 tended to reduce viral infectivity (Fig. 6). Therefore, we consider that an N-linked glycan attached to N197 may contribute to stabilize the structure of V1 and V2 regions and that it plays a role in enhancing the interfering effect of these regions on the binding of gp120 and b12. Finally, although N186 and N187 should be located in close proximity, removal of N187 had no effect on the viral neutralization susceptibility to b12 (Fig. 3B), indicating that the direction of an N-linked glycan attached to N187 may not face the adjacent gp120 molecule in the trimeric complex (Fig. 7).
FIG. 7.
A model of the role of N-linked glycans at amino acid positions 186, 187, and 197 in the interaction between gp120 and b12. A structural model of glycosylated gp120 complexed with b12 was constructed on the basis of previously solved gp120 structures (PDB IDs 3DNL, 2NY7, and 2BF1) as described in Materials and Methods. The generated model has 3-fold rotational symmetry. Each complex of gp120 and b12 is represented by a different model, a ribbon model with distance measurements, a ribbon model with a surface, or a ribbon model with glycosylated gp120. The structures of gp120 and b12 are colored purple and green, respectively. In addition, the location of the V1/V2 stem is colored yellow. The unknown V1 and V2 structures and N-linked glycans are shown as a circle and triangles, respectively. Two bars indicate the distances in two different directions (55 and 67 Å) from the V1/V2 stem to b12 binding to the adjacent gp120 molecule.
Our results suggested that the determinants of the b12 resistance of CRF01_AE Env, 65CC4, were not located in the C-terminal regions of Env gp120, including the last 16 amino acid residues of C3 (C terminus of C3), V4, C4, V5, and C5 regions of gp120 (Fig. 1). In contrast, previous studies on subtype B viruses show that amino acid substitutions in the C terminus of the C3 region plays an important role in determining b12 susceptibility of the resistant variant or in the appearance of b12 escape mutants (8, 28). In addition, b12-resistant JR-CSF variants contain amino acid substitutions in the C terminus of C3 and V4 regions of gp120 (35). These results, when taken together with the reports described above as well as our results, indicate that multiple regions of gp120 may be responsible for regulating the b12 susceptibility of HIV-1 Env gp120.
Finally, the PNLG site, N186, in the V2 region of gp120 is conserved in approximately 16% and 11% of subtype B and C env clones, respectively, as described in recent reports (18, 19). However, no correlation has been observed between b12 susceptibility and the existence of N186 among subtype B and C env clones (18, 19), suggesting that N186 might not play an important role in regulating b12 susceptibility of subtype B and C viruses. Indeed, pNL4-3 Env, which naturally contains N186, was highly susceptible to b12-mediated neutralization in this study (Fig. 4A). In contrast, removal of N186 or of N186 and N197 conferred the b12 susceptibility of b12-resistant CRF01_AE Env (Fig. 3B and 4), demonstrating that N186 plays an important role, in cooperating with N197, in determining the b12 susceptibility of CRF01_AE Env. CRF01_AE is one of the major HIV-1 subtypes that dominate the global epidemic and is prevalent throughout southeastern Asian countries, including Thailand (13, 26). In addition, recent vaccine developments involve clinical trials using CRF01_AE Env as an immunogen (AIDS vaccine trials database, International AIDS Vaccine Initiative [http://www.iavireport.org/trials-db]). The results from a recently conducted vaccine trial in Thailand suggest that protective humoral immune responses to CRF01_AE viruses are not elicited by the immunogens used in this study (37); therefore, it may be required to further study the immunological properties of CRF01_AE Env to design new vaccine immunogens for the elicitation of neutralizing antibody responses. We believe that our results might provide important information to better understand the neutralization susceptibility of CRF01_AE viruses.
Acknowledgments
We are grateful to Yoshitake Nishimune (Research Institute for Microbial Diseases, Osaka University) for valuable help with this study. U87.CD4.CXCR4 and U87.CD4.CCR5 were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. The manuscript was proofread by Medical English Service (Kyoto, Japan).
This work was supported in part by a program of the Founding Research Center for Emerging and Re-emerging Infectious Diseases launched by a project commissioned by the Ministry of Education, Cultures, Sports, Science and Technology (MEXT) of Japan and a research grant from the Department of Medical Sciences, Ministry of Public Health of Thailand. RCC-ERI was established by the Research Institute for Microbial Diseases, Osaka University, Osaka, Japan, and the National Institute of Health, Department of Medical Sciences, Ministry of Public Health, Nonthaburi, Thailand.
Footnotes
Published ahead of print on 17 February 2010.
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbas, C. F., III, T. A. Collet, W. Amberg, P. Roben, J. M. Binley, D. Hoekstra, D. Cababa, T. M. Jones, R. A. Williamson, G. R. Pilkington, N. L. Haigwood, E. Cabezas, A. C. Satterthwait, I. Sanz, and D. R. Burton. 1993. Molecular profile of an antibody response to HIV-1 as probed by combinatorial libraries. J. Mol. Biol. 230:812-823. [DOI] [PubMed] [Google Scholar]
- 3.Binley, J. M., T. Wrin, B. Korber, M. B. Zwick, M. Wang, C. Chappey, G. Stiegler, R. Kunert, S. Zolla-Pazner, H. Katinger, C. J. Petropoulos, and D. R. Burton. 2004. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 78:13232-13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burton, D. R., C. F. Barbas III, M. A. Persson, S. Koenig, R. M. Chanock, and R. A. Lerner. 1991. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc. Natl. Acad. Sci. U. S. A. 88:10134-10137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burton, D. R., J. Pyati, R. Koduri, S. J. Sharp, G. B. Thornton, P. W. Parren, L. S. Sawyer, R. M. Hendry, N. Dunlop, P. L. Nara, M. Lamacchia, E. Garratty, E. R. Stiehm, Y. J. Bryson, Y. Cao, J. P. Moore, D. D. Ho, and C. F. Barbas III. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024-1027. [DOI] [PubMed] [Google Scholar]
- 6.Chen, B., E. M. Vogan, H. Gong, J. J. Skehel, D. C. Wiley, and S. C. Harrison. 2005. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433:834-841. [DOI] [PubMed] [Google Scholar]
- 7.Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760-763. [DOI] [PubMed] [Google Scholar]
- 8.Duenas-Decamp, M. J., P. Peters, D. Burton, and P. R. Clapham. 2008. Natural resistance of human immunodeficiency virus type 1 to the CD4bs antibody b12 conferred by a glycan and an arginine residue close to the CD4 binding loop. J. Virol. 82:5807-5814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaschen, B., J. Taylor, K. Yusim, B. Foley, F. Gao, D. Lang, V. Novitsky, B. Haynes, B. H. Hahn, T. Bhattacharya, and B. Korber. 2002. Diversity considerations in HIV-1 vaccine selection. Science 296:2354-2360. [DOI] [PubMed] [Google Scholar]
- 10.Geyer, H., C. Holschbach, G. Hunsmann, and J. Schneider. 1988. Carbohydrates of human immunodeficiency virus. Structures of oligosaccharides linked to the envelope glycoprotein 120. J. Biol. Chem. 263:11760-11767. [PubMed] [Google Scholar]
- 11.Gnanakaran, S., D. Lang, M. Daniels, T. Bhattacharya, C. A. Derdeyn, and B. Korber. 2007. Clade-specific differences between human immunodeficiency virus type 1 clades B and C: diversity and correlations in C3-V4 regions of gp120. J. Virol. 81:4886-4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haynes, B. F., and D. C. Montefiori. 2006. Aiming to induce broadly reactive neutralizing antibody responses with HIV-1 vaccine candidates. Expert Rev. Vaccines 5:347-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hemelaar, J., E. Gouws, P. D. Ghys, and S. Osmanov. 2006. Global and regional distribution of HIV-1 genetic subtypes and recombinants in 2004. AIDS 20:W13-W23. [DOI] [PubMed] [Google Scholar]
- 14.Kinomoto, M., M. Yokoyama, H. Sato, A. Kojima, T. Kurata, K. Ikuta, T. Sata, and K. Tokunaga. 2005. Amino acid 36 in the human immunodeficiency virus type 1 gp41 ectodomain controls fusogenic activity: implications for the molecular mechanism of viral escape from a fusion inhibitor. J. Virol. 79:5996-6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koch, M., M. Pancera, P. D. Kwong, P. Kolchinsky, C. Grundner, L. Wang, W. A. Hendrickson, J. Sodroski, and R. Wyatt. 2003. Structure-based, targeted deglycosylation of HIV-1 gp120 and effects on neutralization sensitivity and antibody recognition. Virology 313:387-400. [DOI] [PubMed] [Google Scholar]
- 16.Krissinel, E., and K. Henrick. 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60:2256-2268. [DOI] [PubMed] [Google Scholar]
- 17.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, M., F. Gao, J. R. Mascola, L. Stamatatos, V. R. Polonis, M. Koutsoukos, G. Voss, P. Goepfert, P. Gilbert, K. M. Greene, M. Bilska, D. L. Kothe, J. F. Salazar-Gonzalez, X. Wei, J. M. Decker, B. H. Hahn, and D. C. Montefiori. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79:10108-10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, M., J. F. Salazar-Gonzalez, C. A. Derdeyn, L. Morris, C. Williamson, J. E. Robinson, J. M. Decker, Y. Li, M. G. Salazar, V. R. Polonis, K. Mlisana, S. A. Karim, K. Hong, K. M. Greene, M. Bilska, J. Zhou, S. Allen, E. Chomba, J. Mulenga, C. Vwalika, F. Gao, M. Zhang, B. T. Korber, E. Hunter, B. H. Hahn, and D. C. Montefiori. 2006. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in southern Africa. J. Virol. 80:11776-11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, Y., B. Cleveland, I. Klots, B. Travis, B. A. Richardson, D. Anderson, D. Montefiori, P. Polacino, and S. L. Hu. 2008. Removal of a single N-linked glycan in human immunodeficiency virus type 1 gp120 results in an enhanced ability to induce neutralizing antibody responses. J. Virol. 82:638-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, Y., S. A. Migueles, B. Welcher, K. Svehla, A. Phogat, M. K. Louder, X. Wu, G. M. Shaw, M. Connors, R. T. Wyatt, and J. R. Mascola. 2007. Broad HIV-1 neutralization mediated by CD4-binding site antibodies. Nat. Med. 13:1032-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, J., A. Bartesaghi, M. J. Borgnia, G. Sapiro, and S. Subramaniam. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ly, A., and L. Stamatatos. 2000. V2 loop glycosylation of the human immunodeficiency virus type 1 SF162 envelope facilitates interaction of this protein with CD4 and CCR5 receptors and protects the virus from neutralization by anti-V3 loop and anti-CD4 binding site antibodies. J. Virol. 74:6769-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malenbaum, S. E., D. Yang, L. Cavacini, M. Posner, J. Robinson, and C. Cheng-Mayer. 2000. The N-terminal V3 loop glycan modulates the interaction of clade A and B human immunodeficiency virus type 1 envelopes with CD4 and chemokine receptors. J. Virol. 74:11008-11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mascola, J. R., and D. C. Montefiori. 2003. HIV-1: nature's master of disguise. Nat. Med. 9:393-394. [DOI] [PubMed] [Google Scholar]
- 26.McCutchan, F. E. 2006. Global epidemiology of HIV. J. Med. Virol. 78(Suppl. 1):S7-S12. [DOI] [PubMed] [Google Scholar]
- 27.Mizuochi, T., T. J. Matthews, M. Kato, J. Hamako, K. Titani, J. Solomon, and T. Feizi. 1990. Diversity of oligosaccharide structures on the envelope glycoprotein gp 120 of human immunodeficiency virus 1 from the lymphoblastoid cell line H9. Presence of complex-type oligosaccharides with bisecting N-acetylglucosamine residues. J. Biol. Chem. 265:8519-8524. [PubMed] [Google Scholar]
- 28.Mo, H., L. Stamatatos, J. E. Ip, C. F. Barbas, P. W. Parren, D. R. Burton, J. P. Moore, and D. D. Ho. 1997. Human immunodeficiency virus type 1 mutants that escape neutralization by human monoclonal antibody IgG1b12. J. Virol. 71:6869-6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nabatov, A. A., G. Pollakis, T. Linnemann, A. Kliphius, M. I. Chalaby, and W. A. Paxton. 2004. Intrapatient alterations in the human immunodeficiency virus type 1 gp120 V1V2 and V3 regions differentially modulate coreceptor usage, virus inhibition by CC/CXC chemokines, soluble CD4, and the b12 and 2G12 monoclonal antibodies. J. Virol. 78:524-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pantophlet, R., E. Ollmann Saphire, P. Poignard, P. W. Parren, I. A. Wilson, and D. R. Burton. 2003. Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. J. Virol. 77:642-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parren, P. W., H. J. Ditzel, R. J. Gulizia, J. M. Binley, C. F. Barbas III, D. R. Burton, and D. E. Mosier. 1995. Protection against HIV-1 infection in hu-PBL-SCID mice by passive immunization with a neutralizing human monoclonal antibody against the gp120 CD4-binding site. AIDS 9:F1-F6. [DOI] [PubMed] [Google Scholar]
- 32.Parren, P. W., P. A. Marx, A. J. Hessell, A. Luckay, J. Harouse, C. Cheng-Mayer, J. P. Moore, and D. R. Burton. 2001. Antibody protects macaques against vaginal challenge with a pathogenic R5 simian/human immunodeficiency virus at serum levels giving complete neutralization in vitro. J. Virol. 75:8340-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel, M. B., N. G. Hoffman, and R. Swanstrom. 2008. Subtype-specific conformational differences within the V3 region of subtype B and subtype C human immunodeficiency virus type 1 Env proteins. J. Virol. 82:903-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pinter, A., W. J. Honnen, Y. He, M. K. Gorny, S. Zolla-Pazner, and S. C. Kayman. 2004. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J. Virol. 78:5205-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poignard, P., R. Sabbe, G. R. Picchio, M. Wang, R. J. Gulizia, H. Katinger, P. W. Parren, D. E. Mosier, and D. R. Burton. 1999. Neutralizing antibodies have limited effects on the control of established HIV-1 infection in vivo. Immunity 10:431-438. [DOI] [PubMed] [Google Scholar]
- 36.Posner, M. R., L. A. Cavacini, C. L. Emes, J. Power, and R. Byrn. 1993. Neutralization of HIV-1 by F105, a human monoclonal antibody to the CD4 binding site of gp120. J. Acquir. Immune Defic. Syndr. 6:7-14. [PubMed] [Google Scholar]
- 37.Rerks-Ngarm, S., P. Pitisuttithum, S. Nitayaphan, J. Kaewkungwal, J. Chiu, R. Paris, N. Premsri, C. Namwat, M. de Souza, E. Adams, M. Benenson, S. Gurunathan, J. Tartaglia, J. G. McNeil, D. P. Francis, D. Stablein, D. L. Birx, S. Chunsuttiwat, C. Khamboonruang, P. Thongcharoen, M. L. Robb, N. L. Michael, P. Kunasol, and J. H. Kim. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N. Engl. J. Med. 361:2209-2220. [DOI] [PubMed] [Google Scholar]
- 38.Roben, P., J. P. Moore, M. Thali, J. Sodroski, C. F. Barbas III, and D. R. Burton. 1994. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. J. Virol. 68:4821-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rong, R., F. Bibollet-Ruche, J. Mulenga, S. Allen, J. L. Blackwell, and C. A. Derdeyn. 2007. Role of V1V2 and other human immunodeficiency virus type 1 envelope domains in resistance to autologous neutralization during clade C infection. J. Virol. 81:1350-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Samleerat, T., S. Thenin, G. Jourdain, N. Ngo-Giang-Huong, A. Moreau, P. Leechanachai, J. Ithisuknanth, K. Pagdi, P. Wannarit, S. Sangsawang, M. Lallemant, F. Barin, and M. Braibant. 2009. Maternal neutralizing antibodies against a CRF01_AE primary isolate are associated with a low rate of intrapartum HIV-1 transmission. Virology 387:388-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saunders, C. J., R. A. McCaffrey, I. Zharkikh, Z. Kraft, S. E. Malenbaum, B. Burke, C. Cheng-Mayer, and L. Stamatatos. 2005. The V1, V2, and V3 regions of the human immunodeficiency virus type 1 envelope differentially affect the viral phenotype in an isolate-dependent manner. J. Virol. 79:9069-9080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Teeraputon, S., S. Louisirirojchanakul, and P. Auewarakul. 2005. N-linked glycosylation in C2 region of HIV-1 envelope reduces sensitivity to neutralizing antibodies. Viral Immunol. 18:343-353. [DOI] [PubMed] [Google Scholar]
- 43.Thali, M., U. Olshevsky, C. Furman, D. Gabuzda, M. Posner, and J. Sodroski. 1991. Characterization of a discontinuous human immunodeficiency virus type 1 gp120 epitope recognized by a broadly reactive neutralizing human monoclonal antibody. J. Virol. 65:6188-6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trkola, A., A. B. Pomales, H. Yuan, B. Korber, P. J. Maddon, G. P. Allaway, H. Katinger, C. F. Barbas III, D. R. Burton, D. D. Ho, and J. P. Moore. 1995. Cross-clade neutralization of primary isolates of human immunodeficiency virus type 1 by human monoclonal antibodies and tetrameric CD4-IgG. J. Virol. 69:6609-6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Utachee, P., P. Jinnopat, P. Isarangkura-Na-Ayuthaya, U. C. de Silva, S. Nakamura, U. Siripanyaphinyo, N. Wichukchinda, K. Tokunaga, T. Yasunaga, P. Sawanpanyalert, K. Ikuta, W. Auwanit, and M. Kameoka. 2009. Genotypic characterization of CRF01_AE env genes derived from human immunodeficiency virus type 1-infected patients residing in central Thailand. AIDS Res. Hum. Retroviruses 25:229-236. [DOI] [PubMed] [Google Scholar]
- 47.Utachee, P., P. Jinnopat, P. Isarangkura-Na-Ayuthaya, U. C. de Silva, S. Nakamura, U. Siripanyaphinyo, N. Wichukchinda, K. Tokunaga, T. Yasunaga, P. Sawanpanyalert, K. Ikuta, W. Auwanit, and M. Kameoka. 2009. Phenotypic studies on recombinant human immunodeficiency virus type 1 (HIV-1) containing CRF01_AE env gene derived from HIV-1-infected patient, residing in central Thailand. Microbes Infect. 11:334-343. [DOI] [PubMed] [Google Scholar]
- 48.Veazey, R. S., R. J. Shattock, M. Pope, J. C. Kirijan, J. Jones, Q. Hu, T. Ketas, P. A. Marx, P. J. Klasse, D. R. Burton, and J. P. Moore. 2003. Prevention of virus transmission to macaque monkeys by a vaginally applied monoclonal antibody to HIV-1 gp120. Nat. Med. 9:343-346. [DOI] [PubMed] [Google Scholar]
- 49.Walker, L. M., S. K. Phogat, P. Y. Chan-Hui, D. Wagner, P. Phung, J. L. Goss, T. Wrin, M. D. Simek, S. Fling, J. L. Mitcham, J. K. Lehrman, F. H. Priddy, O. A. Olsen, S. M. Frey, P. W. Hammond, S. Kaminsky, T. Zamb, M. Moyle, W. C. Koff, P. Poignard, and D. R. Burton. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 51.Wu, X., T. Zhou, S. O'Dell, R. T. Wyatt, P. D. Kwong, and J. R. Mascola. 2009. Mechanism of human immunodeficiency virus type 1 resistance to monoclonal antibody B12 that effectively targets the site of CD4 attachment. J. Virol. 83:10892-10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou, T., L. Xu, B. Dey, A. J. Hessell, D. Van Ryk, S. H. Xiang, X. Yang, M. Y. Zhang, M. B. Zwick, J. Arthos, D. R. Burton, D. S. Dimitrov, J. Sodroski, R. Wyatt, G. J. Nabel, and P. D. Kwong. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445:732-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu, X., C. Borchers, R. J. Bienstock, and K. B. Tomer. 2000. Mass spectrometric characterization of the glycosylation pattern of HIV-gp120 expressed in CHO cells. Biochemistry 39:11194-11204. [DOI] [PubMed] [Google Scholar]