Abstract
The matrix protein (M1) of influenza A virus is generally viewed as a key orchestrator in the release of influenza virions from the plasma membrane during infection. In contrast to this model, recent studies have indicated that influenza virus requires expression of the envelope proteins for budding of intracellular M1 into virus particles. Here we explored the mechanisms that control M1 budding. Similarly to previous studies, we found that M1 by itself fails to form virus-like-particles (VLPs). We further demonstrated that M1, in the absence of other viral proteins, was preferentially targeted to the nucleus/perinuclear region rather than to the plasma membrane, where influenza virions bud. Remarkably, we showed that a 10-residue membrane targeting peptide from either the Fyn or Lck oncoprotein appended to M1 at the N terminus redirected M1 to the plasma membrane and allowed M1 particle budding without additional viral envelope proteins. To further identify a functional link between plasma membrane targeting and VLP formation, we took advantage of the fact that M1 can interact with M2, unless the cytoplasmic tail is absent. Notably, native M2 but not mutant M2 effectively targeted M1 to the plasma membrane and produced extracellular M1 VLPs. Our results suggest that influenza virus M1 may not possess an inherent membrane targeting signal. Thus, the lack of efficient plasma membrane targeting is responsible for the failure of M1 in budding. This study highlights the fact that interactions of M1 with viral envelope proteins are essential to direct M1 to the plasma membrane for influenza virus particle release.
The late phase of the influenza A virus replication cycle is marked by the occurrence of assembly and budding at the plasma membrane of infected cells, which leads to the separation of virion and host cell membranes and ultimately results in the production of infectious virus particles. This critical step is a highly concerted process driven largely by protein-protein, protein-lipid, and protein-nucleic acid interactions (34, 40). It has been established for many years that four viral structural components, namely, the matrix protein (M1), hemagglutinin (HA), neuraminidase (NA), and M2, are actively involved in the assembly and budding process (34, 35, 40), although the identities of these inter- and intramolecular interactions and regulatory mechanisms for influenza A virus assembly and budding are unclear. It has also been suggested that interactions of M1 with various cytoplasmic tails (CTs) of HA, NA, and M2 are critical to drive the assembly and release of influenza A virions from the surface of infected cells (1, 5, 10, 18, 25, 29, 30, 68). To date, these interactions have been largely speculative because direct interactions have been demonstrated only for M1 and M2 (5, 18, 29).
Early investigations into the budding machinery of influenza A virus using vaccinia virus- and baculovirus-based expression systems indicated that M1 was the only viral protein absolutely required for the assembly of virus particles (14, 15, 26, 31). This assumption seemed reasonable because M1, like the retroviral Gag protein, is the most abundant protein in the virion and lies directly underneath the lipid membrane, structurally forming a bridge between viral envelope proteins and the soluble viral RNA nucleoprotein (vRNP) complex (34, 35, 40). Observations that M1 provided the major driving force for influenza A virus budding were consistent with other findings showing that neither HA nor NA is absolutely essential for influenza virus budding (27, 42). However, a recent study involving the use of a plasmid-based transfection system demonstrated that HA and NA, not M1, were required for influenza A virus assembly and budding (6). Surprisingly, the latter study discovered that M1 expressed in transfected cells lacking HA or NA could not form virus-like particles (VLPs). Therefore, it was concluded that HA and NA glycoproteins, rather than M1 (6), are the driving force in influenza virus assembly and budding. A follow-up study further demonstrated that an interaction between M2 and M1 is important for virion incorporation of M1, as well as for productive virus assembly at virus budding sites (5). Consistent with these reports using influenza H3N2 virus as a model system, a study analyzing neutralizing antibodies present in survivors of the 1918 influenza pandemic showed that H1N1 VLPs can be produced from expression of HA and NA proteins only (65). Despite these recent advances in the understanding of influenza A virus budding, little is known about the underlying mechanism of why the M1 protein is incapable of forming extracellular VLPs. This information is paramount for the resolution of the issue at hand: is there an intrinsic deficiency in the initiation of the M1 budding process, or are results simply dependent on the various expression systems used in each particular study (24, 34)?
In this study, we sought to explore the mechanism underlying the apparent M1 defect in producing extracellular VLPs. Our results showed that the lack of plasma membrane targeting is responsible for the failure of M1 to produce extracellular particles when M1 is expressed alone. However, this M1 budding deficiency can be overcome through either the addition of a heterologous membrane targeting signal or coexpression of the M2 envelope protein, which in both cases directs M1 to the plasma membrane and results in the efficient production of M1 VLPs. Our findings help to elucidate the mechanisms responsible for the failure of influenza A virus M1 to bud from cells as well as offering important new insights into fundamental aspects of influenza A virus assembly and budding.
MATERIALS AND METHODS
Plasmid construction.
To express influenza A viral proteins, the eukaryotic expression vector pPRE was used under the control of both the cytomegalovirus (CMV) immediate-early promoter and the bovine growth hormone polyadenylation signal (9). This specific plasmid contains the hepatitis B virus posttranscriptional regulatory element (PRE) to facilitate the export of mRNA into the cytoplasm for optimal protein expression and has been used in characterization of the budding machinery for various retroviruses, including HIV-1 (8, 22, 23, 41, 54). The influenza H1N1 A/WSN/33 virus reverse genetics system used in this study was a gift from Eric Hoffmann at St. Jude Children's Research Hospital, Memphis, TN (16). For the construction of expression plasmids, DNA sequences corresponding to the influenza virus M1 and M2 proteins were subcloned into the pPRE expression vector, resulting in pM1 and pM2. For direct comparison of M1 protein expression levels between pPRE and pCAGGS (an expression vector driven by the chicken β-actin promoter, widely used for expression of influenza viral proteins), the M1 gene was also subcloned into pCAGGS (Addgene plasmid 11160; Addgene, Cambridge, MA), producing pCAGGS-M1. To facilitate the detection of protein expression, an HA epitope tag was placed at the C terminus of both M1 and M2. An M2 cytoplasmic tail mutant was generated by overlapping PCR in which the M2 tail was completely deleted from the C terminus, and the resultant construct was designated pM2ΔCT.
Various membrane-targeted M1 expression plasmids, pFyn-M1 and pLck-M1, were derived from pM1 in the context of a PRE backbone by inserting annealed oligonucleotides encoding the peptides Fyn (MGCVQCKDKE) and Lck (MGCGCSSHPE) into the N terminus of the M1 protein in frame (66). A cysteine-free pFyn-M1 expression plasmid was generated by exchanging two cysteine residues for serine residues in the Fyn peptide by PCR-directed mutagenesis, and the resultant construct was termed pFynSS-M1. In addition, two membrane targeting green fluorescent protein (GFP) expression plasmids, pFyn-GFP and pLck-GFP, were generated following the same methodology as was used in the production of pFyn-M1 or pLck-M1.
For the construction of bimolecular fluorescence complementation (BiFC) plasmids, sequences encoding either amino-terminal fragments (residues 1 to 173, VN) or carboxyl-terminal fragments (residues 155 to 238, VC) of the Venus fluorescence protein were fused to both the N and C termini of M1 via a protein linker (YPYDVPDYAASDIAAA) (22), which also included an HA tag sequence. The BiFC plasmids were termed M1-VN, VN-M1, M1-VC, and VC-M1, respectively. The use of a flexible linker facilitates complementation of the split fragments when they are brought into close proximity by the interacting proteins. Detailed protocols, including primers and sequence information for each construct, are available upon request. All plasmids were isolated using a Qiagen Midiprep kit (Valencia, CA), and the specific mutations were confirmed by DNA sequencing.
Cells and viruses.
COS-1, 293T, and MDCK cells were cultured in Dulbecco's minimal essential medium (DMEM) supplemented with 10% fetal bovine serum. Influenza viruses (H1N1 A/WSN/33) were generated using the reverse genetics system developed by Hoffmann et al. (16). Viruses were harvested 72 h posttransfection, and stock viruses were generated in MDCK cells.
Antibodies.
Monoclonal mouse anti-HA antibody was purchased from Sigma-Aldrich Co. (St. Louis, MO), and a monoclonal mouse anti-H1N1 M1 antibody was obtained from AbD Serotec of MorphoSys US Inc. (Raleigh, NC). M2-specific monoclonal antibody 14C2-S1-4 was kindly provided by Walter Gerhard and Jan Erikson through the Wistar Institute. Goat anti-H1N1 M1 IgG conjugated to fluorescein isothiocyanate (FITC) was purchased from Virostat Co. (Portland, ME). Polyclonal rabbit anti-GFP antibody, mouse anti-β-actin monoclonal antibody, donkey anti-goat IgG-horseradish peroxidase (HRP), donkey anti-rabbit IgG-HRP, goat anti-mouse IgG-HRP, and goat anti-mouse IgG-FITC were obtained from Abcam Inc. (Cambridge, MA).
Immunofluorescence assay (IFA) and confocal microscopy.
MDCK cells infected with influenza H1N1 A/WSN/33 virus or COS-1 cells transfected with the plasmid DNAs as indicated above were fixed with 2% (wt/vol) paraformaldehyde (Electron Microscopy Sciences, Hatfield, PA) followed by permeabilization with 0.2% (vol/vol) Triton X-100. Cells were further incubated with the appropriate primary antibody directly conjugated with FITC or with the primary antibody followed by secondary antibody conjugated to FITC and were then washed briefly in phosphate-buffered saline (PBS) before being mounted on a slide with Vectashield mounting medium (Vector Laboratories, Burlingame, CA). Sytox orange (Invitrogen, Carlsbad, CA) and DAPI (4′,6-diamidino-2-phenylindole; Invitrogen) were used for staining nuclei of infected MDCK cells and transfected COS-1 cells, respectively. Fluorescent imaging of fixed cells was done using a FluoView FV300 confocal system (Olympus America Inc., Melville, NY) equipped with an IX81 microscope. Digital images were processed with Adobe Photoshop (version 6). All the images were taken under similar experimental conditions (i.e., exposure time, magnification, and intensification), and processing was also the same for all the images shown in this study.
For confocal microscopy analysis of M1 intracellular localization in MDCK cells, the BiFC approach was used. MDCK cells grown on 12-well plates were transfected with either M1-VN/M1-VC BiFC plasmids or individual BiFC plasmids (0.5 μg of each plasmid). At 48 h posttransfection, transfected cells were fixed, permeabilized, stained with DAPI (Invitrogen), and examined by the procedure described above.
Virus budding assay.
COS-1 cells in 60-nm plates were transfected with 3 μg of pM1 (or pCAGGS-M1) and its derivatives (pFyn-M1, pLck-M1, and pFynSS-M1). For cotransfection, equal amounts of pM1 and pM2 or its mutant (pM2ΔCT) were used (3 μg of each plasmid). The total amount of transfected DNA in each sample was held constant using an empty vector lacking a coding region to compensate for any variations. Transfection was performed by using TransIT-LT1 (Mirus, Madison, WI) following the procedure outlined by the manufacturer. For the preparation of VLPs, at 48 h or 72 h after transfection, the culture medium was harvested and pelleted by centrifugation at 2,000 × g for 20 min. Clarified supernatants were then layered through a 20% sucrose cushion (PBS) and centrifuged at 100,000 × g for 2 h at 4°C in a Beckman Ti rotor (Beckman Coulter, Fullerton, CA). The pellet was resuspended on ice in 60 μl of PBS for 1 h followed by centrifugation at 2,000 × g for 10 min at 4°C. Clarified supernatants as VLPs were subject to further analysis. For the preparation of cell lysates, cells were harvested at 48 h posttransfection and lysed in 600 μl of lysis buffer (25 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% deoxycholic acid, 1% Triton X-100, 1× protease inhibitor cocktail). The cell lysates were prepared by centrifugation at 2,000 × g at 4°C for 10 min.
VLPs and cell lysates were separated on a 12.5% sodium dodecyl sulfate (SDS)-polyacrylamide gel and transferred to a nitrocellulose membrane (Pierce, Rockford, IL) followed by blocking in a PBS buffer containing 0.5% Tween 20 and 5% dry milk powder. Influenza virus proteins were detected by standard Western blotting procedure coupled with the ECL system (Pierce, Rockford, IL) with specific antibodies and corresponding secondary antibodies as described above.
Two pharmacological inhibitors of protein palmitoylation, 2-bromopalmitate (2BP) (Sigma-Aldrich) and cerulenin (Sigma-Aldrich), were used to address the effect of palmitoylation on VLP release by palmitoylated M1 budding. COS-1 cells were transfected with pFyn-M1 as described above. 2BP at a concentration of 50 μM, cerulenin at a concentration of 1 μg/ml, or dimethyl sulfoxide (DMSO; no-drug control) was maintained throughout the period of the culture, and SDS-PAGE/Western blot analysis of either M1 proteins or the endogenous actin derived from these cultures was performed as described above.
Protease protection assay.
Purified VLPs derived from COS-1 cells transfected with Fyn-M1 were split into three equivalent aliquots. Two aliquots were treated with TPCK (tosylsulfonyl phenylalanyl chloromethyl ketone)-trypsin (Sigma-Aldrich) in the presence or absence of 1% Triton X-100 for 30 min at 37°C. The third aliquot was treated with only 1% Triton X-100 under the same experimental conditions. All samples were analyzed by SDS-PAGE and Western blotting with anti-M1 monoclonal antibody.
RESULTS
Failure of M1 to produce extracellular VLPs.
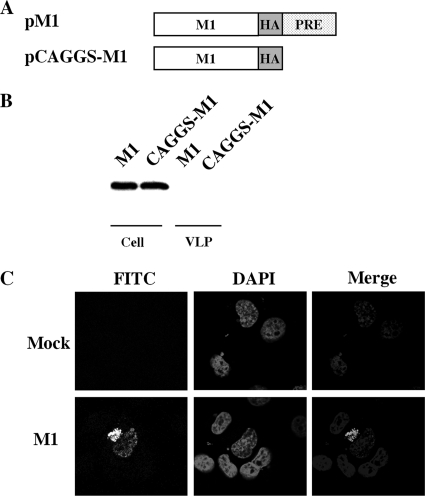
Transient transfection coupled with Western blot analysis was used to address whether the expression of M1 protein could drive VLP formation without additional viral proteins present. Transfection of COS-1 by the two M1 expression plasmids, pCAGGS and PRE (9), resulted in similar levels of M1 protein expression, respectively, but failed to produce any detectable VLPs (Fig. 1B). Our experiments with pCAGGS-M1 in COS-1 cells replicated the M1 protein budding deficiency that was described previously by Chen et al. under their experimental conditions involving 293T and HeLa cells (6). Thus, results from our studies support the previous observation that the M1 protein alone is insufficient to bud and form VLPs (6). It also suggests that the M1 budding deficiency observed is not cell type dependent. Notably, the quantities of M1 expression in the two vectors were indistinguishable; thus, we used only M1 in the PRE vector for further experiments.
FIG. 1.
M1 alone is not sufficient for extracellular VLP formation. (A) Schematic diagram of M1 expression constructs. An HA epitope tag (YPYDVPDYA) was appended in frame to the C terminus of the M1 protein in pPRE and pCAGGS vectors, producing pM1 and pCAGGS-M1, respectively. (B) VLP production by COS-1 cells transfected with pM1 and pCAGGS-M1 plasmids. VLPs and cell lysates were analyzed by Western blotting with an anti-HA monoclonal antibody. (C) Representative M1 subcellular localization images in transfected COS-1 cells expressing M1 protein. At 48 h posttransfection, COS-1 cells transfected with either pM1 or empty vector (mock) plasmids were fixed, permeabilized, and stained with goat anti-M1 IgG-FITC. Nuclei were stained with DAPI. M1 localization was then examined using confocal microscopy at an ×60 magnification.
M1 localizes in the nucleus and perinuclear region of the transfected cells in the absence of other viral proteins.
During the late stage of viral replication in HIV-1 and other enveloped viruses, e.g., Ebola virus and vesicular stomatitis virus (VSV), newly synthesized Gag or matrix proteins are targeted to the plasma membrane of infected cells, where they self-oligomerize and colocalize at lipid rafts assembling into immature virions (19, 33, 38, 49, 52, 53, 58, 62, 69). To determine if M1's inability to release into VLPs is caused by inappropriate targeting of M1 in the absence of other viral proteins, we stained permeabilized COS-1 cells expressing M1 at 48 h posttransfection with an FITC-conjugated antibody for M1. The results showed that M1 protein localizes primarily to the nucleus and perinuclear region with no apparent accumulation at the plasma membrane (Fig. 1C), much like budding-defective retroviral Gag mutants that lack the plasma membrane-binding domain required for Gag assembly and release (36, 37, 39, 50, 64). A similar M1 localization pattern without the plasma membrane staining was also observed at both 24 h and 72 h posttransfection (data not shown).
We also characterized the subcellular localization of M1 in MDCK cells, as they are widely used for studying influenza virus replication and infectivity. In this experiment, we employed a BiFC assay to study M1 subcellular localization. We used the BiFC approach rather than an indirect immunofluorescence assay (IFA) to locate M1 in MDCK cells because repeated transfection of MDCK cells using M1 expression plasmids failed to reveal any detectable signals when attempted in IFA (using anti-M1 antibody conjugated with FITC). We feel that this is probably due to a low transfection efficiency of MDCK cells. The BiFC assay by nature has greater sensitivity, exhibiting detection of fluorescence complementation at extremely low levels of protein expression (as low as 25 to 100 copies per cell) (17). This sensitive technique is designed for visualizing protein interactions and localization in living cells and as such seemed to us a wise choice for experimentation (17). BiFC M1-VN and M1-VC fusion constructs were generated in which VN (N-terminal 173 residues of Venus) and VC (C-terminal 83 residues of Venus) segments were each fused to the carboxy terminus of the M1 protein (Fig. 2A). Presumably as an oligomeric complex, M1 in MDCK cells transfected with BiFC M1-VN/M1-VC plasmids primarily localized to the nucleus and perinuclear region with no significant accumulation at the plasma membrane (Fig. 2B). We also generated VN-M1 and VC-M1 constructs in which VN and VC segments were each fused to the amino terminus of the M1 protein. Transfection of MDCK cells with BiFC VN-M1/VC-M1 resulted in a similar M1 localization in the nucleus/perinuclear region (data not shown). However, in contrast to the pattern of M1 localization observed when M1 was expressed alone, M1 in the context of H1N1 virally infected MDCK cells, which showed significant accumulation of M1 at the plasma membrane (Fig. 2C), in addition to some nuclear and perinuclear localization as observed previously in our transfection experiment involving only M1. M1 localization to the plasma membrane in virally infected cells reinforces the idea that factors such as viral envelope proteins (HA, NA, and M2) are needed for transportation of M1 to the cell surface as suggested previously (1, 5, 10, 18, 25, 29, 30, 68). Based on these data, it is evident that M1 alone cannot be trafficked to the plasma membrane.
FIG. 2.
Comparative studies of M1 localization patterns between transient transfection and virus infection in MDCK cells. (A) Schematic representation of M1 BiFC constructs (M1-VN and M1-VC). Venus fragments VN (N-terminal 173 residues) and VC (C-terminal 83 residues) were fused in frame to the C terminus of M1, which resulted in pM1-VN and pM1-VC, respectively. “L” indicates a linker sequence (YPYDVPDYAASDIAAA) inserted between VN or VC fragments and M1. A portion of the linker is an HA tag sequence. (B) M1 subcellular localization images in MDCK cells expressing BiFC M1-VN and M1-VC fusion proteins. At 48 h posttransfection, MDCK cells transfected with BiFC M1-VN/M1-VC plasmids or M1-VC plasmids (negative control) were fixed, stained with DAPI, and imaged with a confocal microscope at an ×60 magnification. Two representative BiFC images of M1 localization are shown (M1-M1a and M1-M1b). (C) M1 subcellular localization images of virus-infected MDCK cells. At 48 h postinfection, MDCK cells infected with influenza H1N1 A/WSN/33 virus were fixed, permeabilized, and stained with goat anti-M1 IgG-FITC. Sytox orange was used for staining nuclei of infected cells. M1 localization was examined with a confocal microscopy at an ×100 magnification.
In summary, data indicate that M1, in the absence of other viral proteins, localizes in the nucleus and the perinuclear region, not the plasma membrane where the whole influenza virus assembles and buds. Thus, a deficiency in plasma membrane targeting by M1 best explains the failure of M1 to produce extracellular VLPs as shown here and elsewhere (6). This could be the result of a defective membrane targeting domain or the lack of an inherent membrane targeting signal for influenza virus M1 protein.
Production of extracellular VLPs by rational targeting of M1 to the plasma membrane.
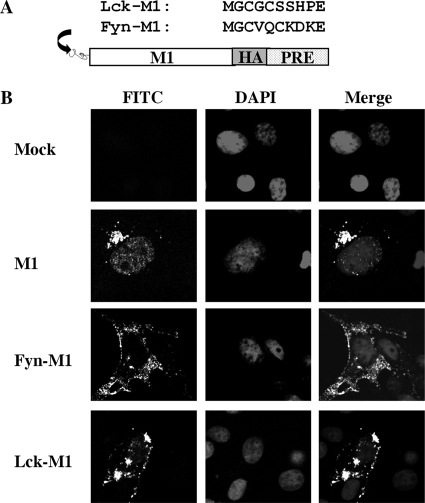
Based on our results, we further hypothesized that targeting of M1 to the plasma membrane could result in M1 budding into VLPs. We were particularly interested in the possibility of VLP formation through either M1 interactions with other viral proteins or nonviral factors such as heterologous membrane targeting signals. To test our hypothesis, we took advantage of two well-characterized plasma membrane targeting domains (66): a 10-residue myristoylated and palmitoylated peptide derived from the Fyn oncoprotein and a 10-residue myristoylated and palmitoylated peptide derived from the Lck oncoprotein. For the purpose of this test, each 10-residue membrane targeting peptide was fused to the M1 protein at the N terminus, resulting in two constructs which we designated Fyn-M1 and Lck-M1 (Fig. 3A).
FIG. 3.
Membrane targeting domains redirect M1 to the plasma membrane. (A) Schematic representation of two membrane targeting domains derived from Fyn and Lck oncoproteins that were appended to the M1 N terminus. (B) Representative M1 subcellular localization images in COS-1 cells expressing M1 or Fyn-M1 or Lck-M1. At 48 h posttransfection, COS-1 cells individually transfected with pM1, pFyn-M1, pLck-M1, and empty vector (mock) were fixed, permeabilized, and stained with goat anti-M1 IgG-FITC. Nuclei of COS-1 cells were stained with DAPI. M1 localization among these cells was then examined by confocal microscopy at an ×60 magnification.
Once our constructs were completed, we first determined whether or not our targeting strategy had the intended effect. Remarkably, both Fyn-M1 and Lck-M1 efficiently trafficked to the plasma membrane in transfected COS-1 cells. This contrasted greatly with earlier transfections of wild-type (WT) M1, where a nuclear/perinuclear localization pattern had been observed (Fig. 3B). We next asked whether shifting M1 to the plasma membrane through the use of Fyn or Lck targeting domains had the ability to result in extracellular VLP formation from M1 budding. Both Fyn-M1 and Lck-M1 generated VLPs, with Fyn-M1 VLP formation being the more prevalent (Fig. 4A). Again, WT M1 failed to produce VLPs under the same experimental conditions as those used for Fyn-M1 and Lck-M1, which supports our previous results (Fig. 1B). Our data clearly indicate that the rational targeting of M1 to the plasma membrane can result in the production of extracellular VLPs. The results also suggest that various 10-residue membrane targeting domains can functionally replace the role of viral envelope proteins for the targeting of M1 to the plasma membrane. In summary, results of these experiments further support our hypothesis that the lack of a membrane targeting signal within M1 is responsible for M1 budding failure.
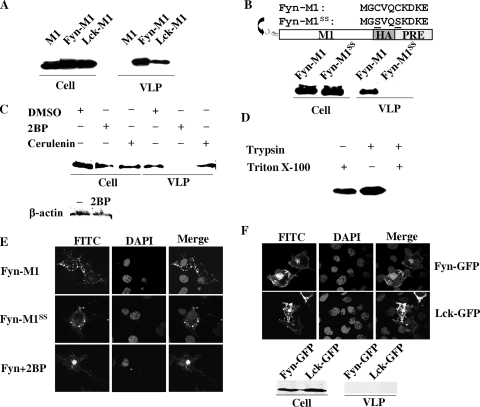
FIG. 4.
Production of extracellular M1 particles by membrane-targeted M1 proteins. (A) VLP production by COS-1 cells transfected with pM1 or pFyn-M1 or pLck-M1 plasmid. M1 VLPs and cell lysates were analyzed in a Western blot assay with anti-M1 monoclonal antibody. (B) VLP production by COS-1 cells transfected with Fyn-M1 or FynSS-M1 mutant defective in protein palmitoylation. VLPs and cell lysates were analyzed by Western blotting with anti-M1 monoclonal antibody. (C) VLP production by COS-1 cells transfected with Fyn-M1 in the absence or presence of protein palmitoylation inhibitors, 2-bromopalmitate (2BP; Sigma-Aldrich) and cerulenin (Sigma-Aldrich). VLPs and cell lysates were analyzed by Western blotting with anti-HA monoclonal antibody (Sigma-Aldrich). Cell lysates derived from the 2BP treatment and the DMSO control were also analyzed for the expression of endogenous β-actin proteins with antiactin monoclonal antibody (Abcam). (D) Trypsin digestion of VLPs. Purified VLPs derived from COS-1 cells transfected with Fyn-M1 were treated with TPCK-trypsin (Sigma-Aldrich) in the presence or absence of 1% Triton X-100 for 30 min at 37°C. Fyn-M1 VLPs were also treated with 1% Triton X-100 without TPCK-trypsin under the same experimental conditions. M1 protein was then visualized by Western blotting with anti-HA monoclonal antibody. (E) Representative M1 subcellular localization images in COS-1 cells expressing Fyn-M1 with or without 2BP treatment or Fyn-M1SS. At 48 h posttransfection, COS-1 cells individually transfected with pFyn-M1 (in the presence or absence of 2BP) and pFyn-M1SS were fixed, permeabilized, and stained with goat anti-M1 IgG-FITC. Nuclei of COS-1 cells were stained with DAPI. M1 localization among these cells was then examined by confocal microscopy at an ×60 magnification. (F) Representative GFP subcellular localization images in COS-1 cells expressing Fyn-GFP and Lck-GFP and VLP production by COS-1 cells transfected with pFyn-GFP or pLck-GFP plasmid. GFP VLPs and cell lysates were analyzed in a Western blot assay with anti-GFP polyclonal antibody (Abcam).
We next chose to examine whether the VLPs assembled by Fyn-M1 or Lck-M1 were specifically regulated by fatty acylation of the M1 protein (myristoylation and palmitoylation) through the introduced 10-residue membrane targeting peptides. We focused our study on the potential contribution of palmitoylation in induced M1 budding because the viral envelope proteins (HA, NA, and M2) that interact with M1 are not myristoylated (40), whereas palmitoylation of M2 and HA has been reported previously (55, 56, 59-61). Several early studies have also suggested a role for palmitoylation in influenza virus assembly (1, 7, 10, 20). Because Fyn-M generates VLPs with greater efficiency than does Lck-M1, it was selected for further experimentation. We constructed Fyn-M1 proteins devoid of palmitoylation acceptors by replacing two N-terminal cysteine residues with serine residues, and the resultant construct was designated FynSS-M1 (Fig. 4B). The absence of cysteine in the Fyn membrane targeting domain should inhibit the addition of the 16-carbon fatty acid palmitate (palmitoylation), as demonstrated in previous studies (44-46). Remarkably, FynSS-M1 failed to generate extracellular VLPs, although M1 intracellular protein production levels were comparable to those observed in the Fyn-M1 system (Fig. 4B). In addition to this site-directed mutagenesis approach, we also utilized chemical inhibitors of protein palmitoylation to determine the functional significance of palmitoylation in driving Fyn-M1 budding (46). Two pharmacological inhibitors of protein palmitoylation, 2BP and cerulenin, were used. As demonstrated in Fig. 4C, 2BP showed no obvious effect on the expression levels of intracellular M1 protein as well as endogenous actin protein compared to DMSO control but instead completely blocked Fyn-M1 particle release. This indicates that palmitoylation specifically is involved in the regulation of Fyn-M1 particle release. In contrast, cerulenin exerted no obvious effect on Fyn-M1 particle budding. At present, we do not know how to explain the different activities exhibited by these two inhibitors but speculate that it may reflect their mechanisms of action: 2BP is a nonmetabolizable palmitate analog that blocks palmitate incorporation into proteins, while cerulenin inhibits protein palmitoylation by targeting some groups of palmitoyl acyltransferase, which catalyzes the addition of palmitate to cysteine residues of protein through thioester linkages (46).
As an additional verification of the authenticity of Fyn-M1 assembly, we performed a protease digestion experiment on Fyn-M particles in the presence or absence of a detergent (Triton X-100). It has been well established that extracellular VLPs completely enveloped by the lipid bilayer are resistant to protease digestion unless the bilayer is disrupted with a detergent (4, 11, 13, 34, 63). To examine whether the Fyn-M1 VLPs released from our experimental conditions were encased in the plasma membrane, purified Fyn-M1 particles were digested with trypsin in the presence or absence of 1% Triton X-100 and analyzed by Western blotting using an anti-M1 monoclonal antibody (Fig. 4D). The M1 proteins were completely digested with trypsin in the presence of the detergent. In contrast, no digestion of M1 protein was observed in the absence of the detergent (Fig. 4D). These results indicate that extracellular VLPs produced by Fyn-M1 in COS-1 cells were completely surrounded by the lipid membrane, suggesting that Fyn-M1 mimics WT M1 in the presence of viral envelope proteins in that it is directed to the plasma membrane, assembled, and released.
We also performed a series of control experiments that enabled us to examine whether a mutated Fyn membrane targeting peptide or the administration of 2BP inhibitors could block M1 transportation to the plasma membrane, thereby inhibiting the formation of M1 VLPs. As demonstrated in Fig. 4E, neither M1 directed by the mutated Fyn peptide nor M1 in the presence of 2BP resulted in a significant accumulation of M1 at the cell surface compared to WT Fyn-M1 proteins. This result coupled with the above data (Fig. 3B and C) further highlights the specificity of palmitoylation in directing M1 to the plasma membrane and subsequently causing M1 VLP release.
Lastly, we were interested in determining whether any soluble protein such as GFP has the ability to bud from cells when directed specifically to the plasma membrane. For this examination, we generated two GFP fusion constructs in which Fyn or Lck membrane targeting peptide was fused to the N terminus of GFP. Our resulting constructs were named Fyn-GFP and Lck-GFP, respectively. Transfection of COS-1 cells with Fyn-GFP and Lck-GFP plasmids shifted a significant portion of the GFPs to the plasma membrane; however, none of them produced any detectable GFP particles (Fig. 4F). The results further support the idea that M1 has an inherent budding ability as well as our hypothesis that M1 fails to bud into VLPs primarily due to the lack of an inherent membrane targeting signal.
Coexpression of the M2 protein, not M2 cytoplasmic tail (CT) deletion, redirects M1 to the plasma membrane and facilitates M1 budding.
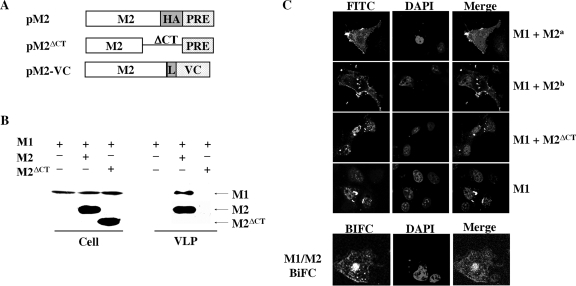
It has been shown that two viral envelope proteins (HA and NA) are required for budding of M1 from the cell, and the coexpression of HA and NA can efficiently incorporate M1 into extracellular VLPs (6, 24, 43). Knowing this to be the case, we asked whether coexpression of the M2 protein could independently promote M1 budding into VLPs without HA and NA proteins. We were particularly interested in determining whether the established M1-M2 interaction (5, 18, 29, 30) was capable of shifting M1 to the plasma membrane; subsequently resulting in the formation of M1 particles. As such, an M2 expression construct was generated in the PRE vector with an HA epitope tag appended to the C terminus. The same strategy was also used to generate an M2 mutant with complete deletion of the CT, termed M2ΔCT. This M2 mutant served as a negative control, as previous studies showed that the deletion of the CT in M2 disrupted normal M1-M2 interactions and inhibited virus assembly (5, 18, 29, 30). We also generated a BiFC M2-VC construct in which the VC segment of Venus was fused to the C terminus of our M2 protein, and this construct was used in combination with M1-VN in BiFC to localize the M1-M2 complex in living cells (Fig. 5A).
FIG. 5.
Coexpression of M2 shifts M1 to the plasma membrane and allows M1 particle budding. (A) A schematic diagram of the expression constructs of M2 (pM2) and M2 mutant (pM2ΔCT) lacking the entire CT domain as well as our BiFC M2-VC fusion construct. The experimental approach for the generation of these constructs has been detailed in Materials and Methods. (B) VLP production by COS-1 cells transfected with pM1 plasmid, either alone or in combination with pM2 or pM2ΔCT. At 72 h following transfection, supernatant medium and cellular lysates from each culture were individually collected and VLPs were prepared by ultracentrifugation. Equal amounts of VLP and lysate samples were run on different gels with the same concentration (12.5% SDS-PAGE) under similar experimental conditions, followed by transferring of proteins to different nitrocellulose membranes. Membranes were then probed separately with M1- or M2-specific antibody and exposed to different films for the visualization of protein expression. Note that Ml and M2/M2CTΔ protein bands were placed together arbitrarily according to their molecular weights by comparison with protein markers. Similar results were also obtained in experiments involving the separation of equal amounts of VLPs and cellular lysates on the same gel/membrane followed by cutting of the membrane into two different pieces corresponding to the migration distances of Ml and M2/M2CTΔ proteins according to the position of molecular weight markers. The separated membrane pieces were then individually analyzed in Western blotting via protein-specific antibodies (Ml and M2), respectively. (C) Subcellular localization patterns of M1 protein in transfected COS-1 cells expressing M1, M1 and M2, and M1 and M2ΔCT and subcellular localization patterns of M1-M2 complex in transfected COS-1 cells expressing M1-VN and M1-VC. At 48 h posttransfection, COS-1 cells were fixed, permeabilized, and stained with goat anti-M1 IgG-FITC. For the BiFC experiment, staining with anti-M1 IgG-FITC was not needed. Nuclei of COS-1 cells were stained with DAPI. Two representative images of M1 localization, M1+M2a and M1+M2b, are shown from cells coexpressing M1 and M2 proteins.
We first determined if M2 could rescue M1 budding into extracellular VLPs independently of HA and NA proteins. As demonstrated in Fig. 5B, coexpression of M2 protein with M1 efficiently generated VLPs. In contrast, VLP release was not detected in cells transfected with both M2ΔCT and M1, although comparable amounts of M1 and M2ΔCT proteins at the intracellular level were observed. These results indicated that the M2 protein is capable of rescuing M1 to form extracellular VLPs, which is likely mediated through a specific M1-M2 interaction. It should be noted that the expression of M2 protein alone did not result in any detectable M2 particles during our experimentation (data not shown). We next compared M1 localization patterns among COS-1 cells transfected with either M1, M1 and M2, or M1 and M2ΔCT plasmids by using an indirect immunofluorescence assay featuring the use of an anti-M1 monoclonal antibody. When M1 and M2 were cotransfected, the M1 intracellular localization pattern was greatly altered compared to cells transfected with M1 only or cells cotransfected with M1 and CT-deleted M2 (M2ΔCT) (Fig. 5C). Two representative images of M1 localization patterns from the M1/M2 cotransfection experiment are shown in Fig. 5C. These images demonstrate a shift in M1 localization from the nucleus/perinuclear region to the plasma membranes as well as to some internal cellular compartments. In contrast, coexpression of M2ΔCT with M1 failed to redirect M1 out of the nucleus/perinuclear region, resulting in an M1 localization pattern that is similar to that observed in cells expressing M1 alone. The Western blot analysis of duplicate samples used for the indirect immunofluorescence assay (Fig. 5C) revealed that M1 and M2 or M2 mutant proteins are expressed (data not shown). To complement our cotransfection experiments, we also used the BiFC approach to monitor M1 localization as a function of M1-M2 complex formation. Notably, BiFC data (Fig. 5C) largely recapitulate the M1 localization pattern observed in indirect immunofluorescence experiments involving coexpression of M1 and M2 proteins (Fig. 5C): M1 is shifted out of the nucleus to the plasma membrane as well as other cellular compartments. These results indicate that M2 protein can efficiently target M1 through specific protein-protein interactions to the assembly site (e.g., plasma membrane), which in turn promotes M1 incorporation into virus particles. Thus, the data continuously support our hypothesis that the lack of a membrane targeting signal within M1 accounts for its failure to redirect M1 to the plasma membrane to produce extracellular VLPs.
DISCUSSION
Previous studies from Chen and colleagues have demonstrated that influenza A virus requires expression of the envelope proteins for budding of intracellular M1 from the cells (6). The current studies support these findings and further demonstrate for the first time that the lack of plasma membrane targeting is responsible for the failure of M1 to generate extracellular particles in the absence of other viral proteins. The present study also reveals for the first time that redirecting M1 to the plasma membrane, either by a heterologous membrane targeting domain or through interaction with the envelope protein (M2), can result in the production of M1 particles. Taken together, the results discussed here and elsewhere (5, 6) collectively highlight the importance of viral envelope proteins in directing M1 to the plasma membrane and indicate that multiple interactions involving M1 and viral envelope proteins are essential to drive influenza virus particle budding.
Results of our experiments reveal several interesting aspects of influenza virus assembly and budding. First, the correlation between the efficiency of membrane targeting and M1 budding competency may help to resolve the current debate regarding the role of M1 in influenza virus budding. Previous studies using recombinant baculovirus or vaccinia virus systems solely expressing M1 showed that M1 was capable of assembly and production of extracellular VLPs (14, 15, 26). Taking into account the current observations, we can speculate that baculovirus and vaccinia virus expression systems may produce some unknown factors that target M1 to the plasma membrane of cells, which in turn promotes M1 particle budding. However, we cannot rule out the other possibilities, such as M1 “leaking” from infected cells as they undergo necrosis, as proposed previously in the vaccinia virus expression system (6). Second, the observation that M1 is incorporated into VLPs by M2 in the absence of HA and NA proteins appears to continue a theme established by previous studies revealing that multiple and often redundant pathways occur in the influenza virus budding process and that only one envelope protein is required for influenza virus assembly and budding (1, 12, 20, 21, 27, 32, 42). Specifically, these studies showed that none of the three viral envelope proteins (HA, NA, and M2) are absolutely required for influenza A virus budding, and virus particles lacking either viral envelope proteins or the cytoplasmic tails of both HA and NA can be formed and released, although such particles are not infectious. Our observations presented in this paper support these earlier studies regarding considerable redundancy in influenza virus budding and further highlight the fact that only a single interaction between M1 and one viral envelope protein such as M2 is required to direct intracellular M1 to the plasma membrane and to produce virus particles. Finally, our finding that the M1 protein assembles and forms extracellular particles, when aided by a 10-residue targeting peptide, suggests that M1 may have the ability to induce membrane curvature, undergo envelopment, and achieve budding into extracellular VLPs once transported to the plasma membrane. Although our hypothesis needs to be tested experimentally in the future, there are several lines of evidence in support of this hypothesis. Previous studies of diverse M1 mutants have demonstrated that M1 is critical in influenza virus budding and particle release, given that the introduced mutations did not disrupt its interactions with viral envelope proteins (2, 3, 28, 34, 47). A recent study of the foamy virus (a retrovirus) Gag demonstrated that the Gag particle budding process was still dependent on the intact PSAP L domain within foamy virus Gag and the associated recruitment of the class E vacuolar protein sorting (VPS) machinery to the budding site (66), even though Fyn and Lck membrane targeting peptides had been added to Gag in this study. Furthermore, it has been shown in various studies that the two peptides used in our study direct various fusion proteins with different biological features exclusively to the lipid rafts of the plasma membrane, not the extracellular environment (44, 45). The results showing that Fyn-GFP fusion proteins efficiently transport to the plasma membrane but fail to produce any detectable GFP particles also support the statement of M1 having an inherent budding ability.
Our observation that M1, in the absence of other viral proteins, was primarily targeted to the nucleus/perinuclear region initially came as a surprise but seems logical since M1 contains a well-documented nuclear signal, as demonstrated by several previous studies involving different approaches (1, 51, 57, 67). It should be noted that our documented findings on M1 localization differ substantially from those reported previously using the M1-expressing vaccinia virus system (10, 25, 67). These previous studies showed that M1 protein, when expressed alone, was associated with cellular membranes, including the plasma membrane. The association of M1 with the plasma membrane that was observed in the vaccinia virus-based expression system seems to indicate that the M1 protein in this system has the ability to produce extracellular VLPs as reported previously (14, 15). As discussed above, we reason that membrane targeting by M1 in the vaccinia virus expression system was not likely mediated by the M1 protein itself. Rather, we believe that some unknown factors produced by vaccinia viruses transport M1 to the plasma membrane, causing release of the M1 particle, although this hypothesis has not yet been tested. It should be pointed out that our data do not argue against the presence of multiple membrane-binding domains within the M1 protein as other previous studies have suggested (25, 48, 57). Instead, our results suggest that when M1 is targeted to the plasma membrane via envelope protein interactions, M1 depends on these distinctive membrane-binding domains to synergistically induce membrane curvature, undergo envelopment, and release M1 VLPs as we demonstrated in our Fyn-M and Lck-M1 systems.
In summary, we have shown that the lack of plasma membrane targeting is responsible for the failure of M1 in generating extracellular VLPs. Our results strongly indicate that M1 may not possess an inherent membrane targeting signal, as has been reported for retroviral Gags and other viral matrix proteins (19, 33, 38, 49, 52, 53, 58, 62, 69). This deficiency highlights interactions of M1 with influenza virus envelope proteins as being essential for the directing of M1 to the plasma membrane for particle release. By overcoming the defects for trafficking and VLP formation through membrane targeting peptides, M1 can now be characterized in this relatively simple system to identify individual functional domains responsible for various steps in the influenza virus budding process.
Acknowledgments
We thank members of the Li lab for helpful discussions and critical reviews of the manuscript. We also thank Michael Hildreth for his expertise in confocal microscopy.
We acknowledge use of the SDSU-Functional Genomics Core Facility supported in part by NSF/EPSCoR grant no. 0091948, the Center of Excellence in Drought Tolerance through the South Dakota 2010 Initiative, and the South Dakota Agricultural Experiment Station. This work was supported by the South Dakota Agricultural Experiment Station (3AH203 to F.L.), South Dakota 2010 Initiative through the Center for Infectious Disease Research and Vaccinology (CIDRV), Public Health Service grant 5K02AI076125-02 from NIAID to F.L., and a subaward of the Rocky Mountain Regional Center of Excellence for Biodefense and Emerging Infectious Diseases at Colorado State University (U54 AI065357) to F.L.
Footnotes
Published ahead of print on 24 February 2010.
REFERENCES
- 1.Ali, A., R. T. Avalos, E. Ponimaskin, and D. P. Nayak. 2000. Influenza virus assembly: effect of influenza virus glycoproteins on the membrane association of M1 protein. J. Virol. 74:8709-8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bourmakina, S. V., and A. Garcia-Sastre. 2003. Reverse genetics studies on the filamentous morphology of influenza A virus. J. Gen. Virol. 84:517-527. [DOI] [PubMed] [Google Scholar]
- 3.Burleigh, L. M., L. J. Calder, J. J. Skehel, and D. A. Steinhauer. 2005. Influenza A viruses with mutations in the M1 helix six domain display a wide variety of morphological phenotypes. J. Virol. 79:1262-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chazal, N., and D. Gerlier. 2003. Virus entry, assembly, budding, and membrane rafts. Microbiol. Mol. Biol. Rev. 67:226-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, B. J., G. P. Leser, D. Jackson, and R. A. Lamb. 2008. The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding. J. Virol. 82:10059-10070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, B. J., G. P. Leser, E. Morita, and R. A. Lamb. 2007. Influenza virus hemagglutinin and neuraminidase, but not the matrix protein, are required for assembly and budding of plasmid-derived virus-like particles. J. Virol. 81:7111-7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, B. J., M. Takeda, and R. A. Lamb. 2005. Influenza virus hemagglutinin (H3 subtype) requires palmitoylation of its cytoplasmic tail for assembly: M1 proteins of two subtypes differ in their ability to support assembly. J. Virol. 79:13673-13684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, C., O. Vincent, J. Jin, O. A. Weisz, and R. C. Montelaro. 2005. Functions of early (AP-2) and late (AIP1/ALIX) endocytic proteins in equine infectious anemia virus budding. J. Biol. Chem. 280:40474-40480. [DOI] [PubMed] [Google Scholar]
- 9.Donello, J. E., A. A. Beeche, G. J. Smith III, G. R. Lucero, and T. J. Hope. 1996. The hepatitis B virus posttranscriptional regulatory element is composed of two subelements. J. Virol. 70:4345-4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enami, M., and K. Enami. 1996. Influenza virus hemagglutinin and neuraminidase glycoproteins stimulate the membrane association of the matrix protein. J. Virol. 70:6653-6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freed, E. O. 2004. Mechanisms of enveloped virus release. Virus Res. 106:85-86. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Sastre, A., and P. Palese. 1995. The cytoplasmic tail of the neuraminidase protein of influenza A virus does not play an important role in the packaging of this protein into viral envelopes. Virus Res. 37:37-47. [DOI] [PubMed] [Google Scholar]
- 13.Garoff, H., R. Hewson, and D. J. Opstelten. 1998. Virus maturation by budding. Microbiol. Mol. Biol. Rev. 62:1171-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomez-Puertas, P., C. Albo, E. Perez-Pastrana, A. Vivo, and A. Portela. 2000. Influenza virus matrix protein is the major driving force in virus budding. J. Virol. 74:11538-11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomez-Puertas, P., I. Mena, M. Castillo, A. Vivo, E. Perez-Pastrana, and A. Portela. 1999. Efficient formation of influenza virus-like particles: dependence on the expression levels of viral proteins. J. Gen. Virol. 80:1635-1645. [DOI] [PubMed] [Google Scholar]
- 16.Hoffmann, E., G. Neumann, Y. Kawaoka, G. Hobom, and R. G. Webster. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. U. S. A. 97:6108-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu, C. D., Y. Chinenov, and T. K. Kerppola. 2002. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9:789-798. [DOI] [PubMed] [Google Scholar]
- 18.Iwatsuki-Horimoto, K., T. Horimoto, T. Noda, M. Kiso, J. Maeda, S. Watanabe, Y. Muramoto, K. Fujii, and Y. Kawaoka. 2006. The cytoplasmic tail of the influenza A virus M2 protein plays a role in viral assembly. J. Virol. 80:5233-5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jayakar, H. R., E. Jeetendra, and M. A. Whitt. 2004. Rhabdovirus assembly and budding. Virus Res. 106:117-132. [DOI] [PubMed] [Google Scholar]
- 20.Jin, H., G. P. Leser, and R. A. Lamb. 1994. The influenza virus hemagglutinin cytoplasmic tail is not essential for virus assembly or infectivity. EMBO J. 13:5504-5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin, H., G. P. Leser, J. Zhang, and R. A. Lamb. 1997. Influenza virus hemagglutinin and neuraminidase cytoplasmic tails control particle shape. EMBO J. 16:1236-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin, J., T. Sturgeon, C. Chen, S. C. Watkins, O. A. Weisz, and R. C. Montelaro. 2007. Distinct intracellular trafficking of equine infectious anemia virus and human immunodeficiency virus type 1 Gag during viral assembly and budding revealed by bimolecular fluorescence complementation assays. J. Virol. 81:11226-11235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin, J., T. Sturgeon, O. A. Weisz, W. Mothes, and R. C. Montelaro. 2009. HIV-1 matrix dependent membrane targeting is regulated by Gag mRNA trafficking. PLoS One 4:e6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang, S. M., J. M. Song, F. S. Quan, and R. W. Compans. 2009. Influenza vaccines based on virus-like particles. Virus Res. 143:140-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kretzschmar, E., M. Bui, and J. K. Rose. 1996. Membrane association of influenza virus matrix protein does not require specific hydrophobic domains or the viral glycoproteins. Virology 220:37-45. [DOI] [PubMed] [Google Scholar]
- 26.Latham, T., and J. M. Galarza. 2001. Formation of wild-type and chimeric influenza virus-like particles following simultaneous expression of only four structural proteins. J. Virol. 75:6154-6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, C., M. C. Eichelberger, R. W. Compans, and G. M. Air. 1995. Influenza type A virus neuraminidase does not play a role in viral entry, replication, assembly, or budding. J. Virol. 69:1099-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, T., and Z. Ye. 2002. Restriction of viral replication by mutation of the influenza virus matrix protein. J. Virol. 76:13055-13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCown, M. F., and A. Pekosz. 2006. Distinct domains of the influenza a virus M2 protein cytoplasmic tail mediate binding to the M1 protein and facilitate infectious virus production. J. Virol. 80:8178-8189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCown, M. F., and A. Pekosz. 2005. The influenza A virus M2 cytoplasmic tail is required for infectious virus production and efficient genome packaging. J. Virol. 79:3595-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mena, I., A. Vivo, E. Perez, and A. Portela. 1996. Rescue of a synthetic chloramphenicol acetyltransferase RNA into influenza virus-like particles obtained from recombinant plasmids. J. Virol. 70:5016-5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitnaul, L. J., M. R. Castrucci, K. G. Murti, and Y. Kawaoka. 1996. The cytoplasmic tail of influenza A virus neuraminidase (NA) affects NA incorporation into virions, virion morphology, and virulence in mice but is not essential for virus replication. J. Virol. 70:873-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Money, V. A., H. K. McPhee, J. A. Mosely, J. M. Sanderson, and R. P. Yeo. 2009. Surface features of a Mononegavirales matrix protein indicate sites of membrane interaction. Proc. Natl. Acad. Sci. U. S. A. 106:4441-4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nayak, D. P., R. A. Balogun, H. Yamada, Z. H. Zhou, and S. Barman. 2009. Influenza virus morphogenesis and budding. Virus Res. 143:147-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nayak, D. P., E. K. Hui, and S. Barman. 2004. Assembly and budding of influenza virus. Virus Res. 106:147-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ono, A., D. Demirov, and E. O. Freed. 2000. Relationship between human immunodeficiency virus type 1 Gag multimerization and membrane binding. J. Virol. 74:5142-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ono, A., and E. O. Freed. 1999. Binding of human immunodeficiency virus type 1 Gag to membrane: role of the matrix amino terminus. J. Virol. 73:4136-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ono, A., and E. O. Freed. 2001. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc. Natl. Acad. Sci. U. S. A. 98:13925-13930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ono, A., J. M. Orenstein, and E. O. Freed. 2000. Role of the Gag matrix domain in targeting human immunodeficiency virus type 1 assembly. J. Virol. 74:2855-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palese, P., and M. Shaw. 2007. Orthomyxoviridae: the viruses and their replication, p. 1647-1689. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 41.Patnaik, A., V. Chau, F. Li, R. C. Montelaro, and J. W. Wills. 2002. Budding of equine infectious anemia virus is insensitive to proteasome inhibitors. J. Virol. 76:2641-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pattnaik, A. K., D. J. Brown, and D. P. Nayak. 1986. Formation of influenza virus particles lacking hemagglutinin on the viral envelope. J. Virol. 60:994-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perrone, L. A., A. Ahmad, V. Veguilla, X. Lu, G. Smith, J. M. Katz, P. Pushko, and T. M. Tumpey. 2009. Intranasal vaccination with 1918 influenza virus-like particles protects mice and ferrets from lethal 1918 and H5N1 influenza virus challenge. J. Virol. 83:5726-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Resh, M. D. 2006. Palmitoylation of ligands, receptors, and intracellular signaling molecules. Sci. STKE 2006:re14. [DOI] [PubMed] [Google Scholar]
- 45.Resh, M. D. 2006. Trafficking and signaling by fatty-acylated and prenylated proteins. Nat. Chem. Biol. 2:584-590. [DOI] [PubMed] [Google Scholar]
- 46.Resh, M. D. 2006. Use of analogs and inhibitors to study the functional significance of protein palmitoylation. Methods 40:191-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roberts, P. C., R. A. Lamb, and R. W. Compans. 1998. The M1 and M2 proteins of influenza A virus are important determinants in filamentous particle formation. Virology 240:127-137. [DOI] [PubMed] [Google Scholar]
- 48.Ruigrok, R. W., A. Barge, P. Durrer, J. Brunner, K. Ma, and G. R. Whittaker. 2000. Membrane interaction of influenza virus M1 protein. Virology 267:289-298. [DOI] [PubMed] [Google Scholar]
- 49.Ruigrok, R. W., G. Schoehn, A. Dessen, E. Forest, V. Volchkov, O. Dolnik, H. D. Klenk, and W. Weissenhorn. 2000. Structural characterization and membrane binding properties of the matrix protein VP40 of Ebola virus. J. Mol. Biol. 300:103-112. [DOI] [PubMed] [Google Scholar]
- 50.Sandefur, S., V. Varthakavi, and P. Spearman. 1998. The I domain is required for efficient plasma membrane binding of human immunodeficiency virus type 1 Pr55Gag. J. Virol. 72:2723-2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sato, Y., K. Yoshioka, C. Suzuki, S. Awashima, Y. Hosaka, J. Yewdell, and K. Kuroda. 2003. Localization of influenza virus proteins to nuclear dot 10 structures in influenza virus-infected cells. Virology 310:29-40. [DOI] [PubMed] [Google Scholar]
- 52.Schmitt, A. P., G. P. Leser, D. L. Waning, and R. A. Lamb. 2002. Requirements for budding of paramyxovirus simian virus 5 virus-like particles. J. Virol. 76:3952-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scianimanico, S., G. Schoehn, J. Timmins, R. H. Ruigrok, H. D. Klenk, and W. Weissenhorn. 2000. Membrane association induces a conformational change in the Ebola virus matrix protein. EMBO J. 19:6732-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shehu-Xhilaga, M., S. Ablan, D. G. Demirov, C. Chen, R. C. Montelaro, and E. O. Freed. 2004. Late domain-dependent inhibition of equine infectious anemia virus budding. J. Virol. 78:724-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stegmann, T. 2000. Membrane fusion mechanisms: the influenza hemagglutinin paradigm and its implications for intracellular fusion. Traffic 1:598-604. [DOI] [PubMed] [Google Scholar]
- 56.Sugrue, R. J., R. B. Belshe, and A. J. Hay. 1990. Palmitoylation of the influenza A virus M2 protein. Virology 179:51-56. [DOI] [PubMed] [Google Scholar]
- 57.Thaa, B., A. Herrmann, and M. Veit. 2009. The polybasic region is not essential for membrane binding of the matrix protein M1 of influenza virus. Virology 383:150-155. [DOI] [PubMed] [Google Scholar]
- 58.Timmins, J., R. W. Ruigrok, and W. Weissenhorn. 2004. Structural studies on the Ebola virus matrix protein VP40 indicate that matrix proteins of enveloped RNA viruses are analogues but not homologues. FEMS Microbiol. Lett. 233:179-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Veit, M., H. D. Klenk, A. Kendal, and R. Rott. 1991. The M2 protein of influenza A virus is acylated. J. Gen. Virol. 72:1461-1465. [DOI] [PubMed] [Google Scholar]
- 60.Veit, M., E. Kretzschmar, K. Kuroda, W. Garten, M. F. Schmidt, H. D. Klenk, and R. Rott. 1991. Site-specific mutagenesis identifies three cysteine residues in the cytoplasmic tail as acylation sites of influenza virus hemagglutinin. J. Virol. 65:2491-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Veit, M., and M. F. Schmidt. 1993. Timing of palmitoylation of influenza virus hemagglutinin. FEBS Lett. 336:243-247. [DOI] [PubMed] [Google Scholar]
- 62.Verderame, M. F., T. D. Nelle, and J. W. Wills. 1996. The membrane-binding domain of the Rous sarcoma virus Gag protein. J. Virol. 70:2664-2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Waheed, A. A., and E. O. Freed. 2009. Lipids and membrane microdomains in HIV-1 replication. Virus Res. 143:162-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wills, J. W., R. C. Craven, R. A. Weldon, Jr., T. D. Nelle, and C. R. Erdie. 1991. Suppression of retroviral MA deletions by the amino-terminal membrane-binding domain of p60src. J. Virol. 65:3804-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu, X., T. Tsibane, P. A. McGraw, F. S. House, C. J. Keefer, M. D. Hicar, T. M. Tumpey, C. Pappas, L. A. Perrone, O. Martinez, J. Stevens, I. A. Wilson, P. V. Aguilar, E. L. Altschuler, C. F. Basler, and J. E. Crowe, Jr. 2008. Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature 455:532-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhadina, M., M. O. McClure, M. C. Johnson, and P. D. Bieniasz. 2007. Ubiquitin-dependent virus particle budding without viral protein ubiquitination. Proc. Natl. Acad. Sci. U. S. A. 104:20031-20036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang, J., and R. A. Lamb. 1996. Characterization of the membrane association of the influenza virus matrix protein in living cells. Virology 225:255-266. [DOI] [PubMed] [Google Scholar]
- 68.Zhang, J., A. Pekosz, and R. A. Lamb. 2000. Influenza virus assembly and lipid raft microdomains: a role for the cytoplasmic tails of the spike glycoproteins. J. Virol. 74:4634-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou, W., L. J. Parent, J. W. Wills, and M. D. Resh. 1994. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J. Virol. 68:2556-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]