Abstract
In retroviruses and the double-stranded RNA totiviruses, the efficiency of programmed −1 ribosomal frameshifting is critical for ensuring the proper ratios of upstream-encoded capsid proteins to downstream-encoded replicase enzymes. The genomic organizations of many other frameshifting viruses, including the coronaviruses, are very different, in that their upstream open reading frames encode nonstructural proteins, the frameshift-dependent downstream open reading frames encode enzymes involved in transcription and replication, and their structural proteins are encoded by subgenomic mRNAs. The biological significance of frameshifting efficiency and how the relative ratios of proteins encoded by the upstream and downstream open reading frames affect virus propagation has not been explored before. Here, three different strategies were employed to test the hypothesis that the −1 PRF signals of coronaviruses have evolved to produce the correct ratios of upstream- to downstream-encoded proteins. Specifically, infectious clones of the severe acute respiratory syndrome (SARS)-associated coronavirus harboring mutations that lower frameshift efficiency decreased infectivity by >4 orders of magnitude. Second, a series of frameshift-promoting mRNA pseudoknot mutants was employed to demonstrate that the frameshift signals of the SARS-associated coronavirus and mouse hepatitis virus have evolved to promote optimal frameshift efficiencies. Finally, we show that a previously described frameshift attenuator element does not actually affect frameshifting per se but rather serves to limit the fraction of ribosomes available for frameshifting. The findings of these analyses all support a “golden mean” model in which viruses use both programmed ribosomal frameshifting and translational attenuation to control the relative ratios of their encoded proteins.
Viruses utilize programmed ribosomal frameshifting (PRF) to posttranscriptionally regulate the expression of multiple genes encoded on monocistronic viral mRNAs. In many RNA viruses that utilize programmed ribosomal frameshifting (e.g., most retroviruses, totiviruses, and Ty elements), the mRNAs transcribed from these viral templates contain two overlapping open reading frames (ORFs). In these viruses, the ORF encoding the major viral nucleocapsid proteins (e.g., Gag) is located at the 5′ end of the mRNA, whereas ORFs encoding proteins with enzymatic functions (typically Pro and Pol) are located 3′ of, and out of frame with, the Gag ORF. The enzymatic proteins are translated only as a result of PRF events that occur at frequencies of 1 to 40% depending on the specific virus and assay system employed (reviewed in reference 6). Thus, the majority of translational events result in the production of structural nucleocapsid proteins, while the intervention of frameshifting results in a decreased yield of enzymatic products (23). The importance of maintaining precise ratios of structural to enzymatic proteins on viral propagation has been demonstrated using two endogenous viruses of the yeast Saccharomyces cerevisiae and with two retroviruses (reviewed in reference 18). Small alterations in programmed frameshifting efficiencies promote the rapid loss of the yeast double-stranded RNA (dsRNA) L-A killer virus (13, 14, 17, 19, 38, 39, 40, 44, 49). Similarly, increasing or decreasing the efficiency of the +1 ribosomal frameshift in the Ty1 retrotransposable element of yeast results in reduced retrotranspostion frequencies (2, 17, 20, 27, 28, 33, 39). In L-A, Gag-pol dimerization nucleates the formation of the virus particles (10-12, 22). Increasing the amount of Gag-pol protein synthesized may cause too many particles to initiate nonproductively, while producing too little may prevent efficient dimerization (19). The proteolytic processing of the TyA-TyB (Gag-pol equivalent) polyprotein of Ty1 is more akin to the situation observed in retroviruses. In Ty1, increasing the amount of Gag-pol protein synthesized inhibited the proteolytic processing of the polyprotein (33). As a consequence, the formation of the mature forms of RNase H, integrase, and reverse transcriptase is blocked (33). Similarly, changing the ratio of Gag to Gag-pol proteins in retroviruses like HIV or Moloney murine leukemia virus interferes with virus particle formation (4, 24, 29, 32, 42, 53). In these cases, the overexpression of the Gag-pol protein results in the inefficient processing of the polyprotein and the inhibition of virus production. In sum, viral PRF efficiencies have been fine-tuned to deliver the precise ratios of proteins required for efficient viral particle assembly; too much or too little frameshifting alters this ratio, with detrimental consequences. Based on these studies, it has been proposed that −1 PRF is a viable target for the prevention of viral propagation (reviewed in 18).
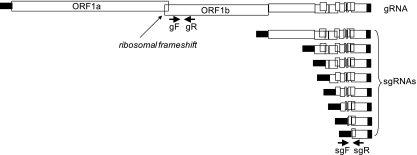
Coronaviruses are positive-strand RNA viruses with large genomes (∼30,000 nucleotides [nt]) that also utilize −1 PRF. They can cause enteric and respiratory tract infections with varying severity. For example, some genotypes affecting humans (HCoV-229E and HCoV-OC43) cause cold-like symptoms, while the coronavirus associated with severe acute respiratory disease (SARS-CoV) is associated with a high mortality rate. Similarly, the coronaviruses that affect other mammals have assorted phenotypes: the mouse hepatitis virus (MHV) enterotropic strains replicate initially in the intestinal epithelium and tend not to disseminate, whereas the neurotrophic MHV strains initially replicate in the respiratory tract and then disseminate to the liver, brain, and lymph nodes. The latter strains are used in models for acute and chronic central nervous system infection (54). While the SARS-CoV and MHV viruses have different pathologies, overall they are phylogenetically more similar to each other than SARS-CoV is to HCoV-229E (21). The genomic organization of coronaviruses is different from that of retroviruses and totiviruses: the structural proteins are encoded by subgenomic mRNAs, while the genes regulated by −1 PRF are involved in replicase/transcriptase function (56, 59). The genomic organization of SARS-CoV is shown in Fig. 1. The ORF1a-encoded polyprotein (pp1a) synthesizes nonstructural proteins. The −1 PRF signal is located at the 3′ end of ORF1a and redirects a fraction of the translating ribosomes into the ORF1b reading frame to synthesize the larger pp1ab polyprotein. The enzymatic functions required for viral replication are derived from pp1ab (1, 5, 55). Although frameshifting is an essential feature of the viral life cycle per se because it is required for the production of most of the replicase proteins, the consequences of changing −1 PRF efficiencies on the replication of this class of viruses have never been tested.
FIG. 1.
Schematic of genomic and subgenomic RNAs. The open reading frames of the SARS-CoV gRNA are shown as open boxes. The position of the frameshift signal where ORF1a and ORF1b overlap is indicated. The 5′ leader sequence and 3′ noncoding region common to the gRNA and all sgRNAs are shown as filled boxes. The positions of the primers gF and gR used to detect gRNA and the primers sgF and sgR used to dedect sgRNA are shown as arrows. Note that the detection of sgRNA requires a leader sequence proximal to the 3′ ORF.
The cis-acting signals that promote frameshifting consist of a heptameric slippery site and an strong mRNA structure separated by a short spacer. In general, the slippery site can be defined as N NNW WWH, where N is any three identical bases, W is AAA or UUU, and H is A, C, or U (the frame of the initiator AUG is indicated by the spacing) (8, 16). It appears that there is a preference within virus groups for certain slippery sites, and these preferences likely reflect the differences in the host ribosomes (3, 45). The second element is usually an mRNA pseudoknot that directs elongating ribosomes to pause with their A and P sites positioned over the slippery site (34, 51). The initial demonstration that a pseudoknot was required for efficient −1 PRF was for the avian infectious bronchitis coronavirus (IBV) (9). Subsequently, numerous pseudoknots have been described that facilitate frameshifting (reviewed in reference 25). Until recently, all of the frameshifting pseudoknots described contained two stems. However, structural analyses revealed that the SARS-CoV frameshift-stimulating pseudoknot contains three stems (47, 52). In addition, another cis-acting element affecting −1 PRF located immediately upstream of the SARS-CoV −1 PRF signal was suggested to attenuate the frameshifting efficiency of both the SARS-CoV and infectious bronchitis virus (IBV) signals (52). The availability of sequences from several new coronaviruses now allows more in-depth comparisons of regulatory sequences.
The current study begins by examining the question of the importance of synthesizing the correct ratios of viral proteins for SARS-CoV propagation and then addresses mechanisms through which these ratios may be controlled. Initially, a series of slippery-site mutants was introduced into an infectious clone to test the hypothesis that correct levels of −1 PRF are critical for the propagation of this virus. The viable mutant viruses produced less genomic RNA than subgenomic RNA. Furthermore, the infection of cells with equivalent amounts of wild-type and mutant genomic RNAs revealed that the mutants were significantly less infectious than the wild type, thus demonstrating an important role for −1 PRF in the viral life cycle. The hypothesis that the frameshift-stimulating mRNA pseudoknots have evolved in coronaviruses to promote frameshifting at specific levels so as to deliver the proper ratios of ORF1a and ORF1ab products was tested using a series of mutations that morphed the MHV −1 PRF signal into that from SARS-CoV. The results of this analysis reveal features of the coronavirus pseudoknots that are important for stimulating optimal levels of frameshifting. Lastly, the issue of an additional regulatory element, the so-called attenuator sequence (52), was examined. Phylogenetic analyses reveal that while there is little conservation of the sequence upstream of the various coronavirus −1 PRF signals, computational analyses show that they all are predicted to fold into strong secondary structures. Although prior findings suggested that the attenuator element reduced −1 PRF by ∼40%, the experimental design employed in that study did not preclude the hypothesis that strong secondary mRNA structures simply cause ribosomes to dissociate from the mRNA prior to encountering the frameshift signal, i.e., translational attenuation. Experiments presented in the current study support this hypothesis, suggesting that the function of the attenuator is to further help fine-tune the ratios of ORF1a and ORF1b viral products by limiting the number of ribosomes available to translate ORF1b. In sum, the current study shows that the ratios of ORF1a- and ORF1b-encoded proteins play a critical role for the coronaviruses, and that both −1 PRF and translational attenuation are employed to guarantee the production of a “golden mean” of viral proteins for optimal virus replication and viability.
MATERIALS AND METHODS
Sequence analysis.
The GenBank (http://www.ncbi.nlm.nih.gov/GenBank/) accession numbers for the sequences discussed in this paper are SARS-CoV (NC_004718), Bt-CoV Rp3 (NC_009693), Bt-CoV HKU3 (NC_009694), Bt-CoV Rf1 (NC_009695), Bt-CoV Rm1 (NC_009696), Bt-CoV HKU9-1 (NC_009021), Bt-CoV HKU5-1 (NC_009020), Bt-CoV HKU9 (NC_009019), Bt-CoV 133/2005 (NC_008315), IBV (NC_001451), TCoV (NC_010800), MuCoV (NC_011550), BuCoV (NC_011548), ThCoV (NC_011549), SW1-CoV (NC_010646), MHV-A59 (NC_001846), MHV-JHM (NC_006852), HCoV OC43 (NC_005147), BCoV (NC_003045), EqCoV (NC_010327), pigeon herpes encephalomyelitis virus (NC_007732), HCoV HKU1 (NC_006577), Bt-CoV HKU2 (NC_009988), feline infectious peritonitis virus (NC_007025), transmissible gastroenteritis virus (TGEV) (NC_002306), HCoV 229E (NC_002645), HCoV NL63 (NC_005831), porcine epidemic diarrhea virus (NC_003436), Bt-CoV 512/2005 (NC_009657), Bt-CoV HKU8 (NC_010438), Bt-CoV 1a (NC_010437), and Bt-CoV 1b (NC_010436). Sequences were aligned using ClustalW2 (35), and cladograms were constructed on the EMBL website (http://www.ebi.ac.uk/). Pairwise alignments were performed using the default Clustal settings in the Lasergene software (DNASTAR Inc., Madison, WI). RNA sequences were folded using mfold on the web server at Rensselaer Polytechnic Institute (36, 60).
Strains and genetic methods.
Escherichia coli strain DH5α was used to amplify plasmids, and high-efficiency transformations were performed using the method of Inoue et al. (30). Vero E6 cells were cultured at 37°C with 5% CO2 in Dulbecco's modified eagle medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS; HyClone, Logan, UT). Cells were transfected using FuGENE 6 (Roche, Indianapolis, IN) according to the manufacturer's protocol.
Plasmid constructs.
The parental plasmids pJD464 and pJD502, containing the Renilla and firefly luciferase genes flanking the wild-type SARS-CoV frameshift, have been described previously (47). pJD502 is the test construct (T) for measuring frameshifting efficiency, and pJD464 is a readthrough control plasmid (C) to normalize against any defects in overall translation that the introduced SARS sequence may cause. A PCR fragment corresponding to nucleotides 13057 to 14171 from MHV strain A59 (a kind donation from Paul Masters) was used as a template for our MHV studies. The region from nucleotides 13545 to 13681, which included 51 nucleotides upstream of the slippery site, the slippery site, and the predicted pseudoknot, was amplified by PCR. The 51 nucleotides upstream from the slippery site was the maximum amount that could be cloned while still maintaining two open reading frames such that both test and control vectors could be made. The primers (Table 1) included the restriction sites BamHI and SacI to allow for cloning. The PCR amplicon was digested with these two endonucleases and cloned into the similarly digested dual luciferase vector p2luci (26) to create the readthrough control plasmid pJD768. This then was subcloned as a BamHI/EcoRI fragment into the dual luciferase vector p2luc (26) to create the test construct pJD769. Site-directed mutagenesis was used to introduce mutations at various positions in the frameshift-stimulating pseudoknot on pJD768. Mutagenesis was performed using Stratagene's QuikChange II kit (La Jolla, CA) and the primers listed in Table 1. The mutations were confirmed by sequencing, and test constructs for each control were made by subcloning the BamHI/EcoRI fragment into p2luc.
TABLE 1.
Oligonucleotide primers used in this study
| Oligonucleotide and function | Sequence |
|---|---|
| Mutagenesis | |
| MHVf1 | 5′-AAGGATCCCTGTTCCTGTGTAGGCAC-3′ |
| MHVf2 | 5′-CCGGATCCCTTTTTAAACGGGTTCGG-3′ |
| MHVr3 | 5′-CCGAGCTCAAATGCCCTTAATTGAACATC-3′ |
| MHVstem2f | 5′-AAGTGTTGCCCGTCTTGTACCC-3′ |
| MHVstem2r | 5′-CGGGCAACACTTGTACCCCG-3′ |
| MHVloop3f | 5′-TGATGTCTTAAGGGCATTTGAGC-3′ |
| MHVloop3r | 5′-CCTTAAGACATCAGTGTCCAAGC-3′ |
| MHVloop3bulgef | 5′-TGATGTCTACAGGGCATTTGAGCTC-3′ |
| MHVloop3bulger | 5′-GCCCTGTAGACATCAGTGTCCAAG-3′ |
| MHVcomplexf | 5′-TGGCACTAGTACACATGGTGTCTACAGGGCATTTGAGC-3′ |
| MHVcomplexr | 5′-CCTGTAGACACCATGTGTACTAGTGCCACTGGCACAG-3′ |
| Attenuator analysis | |
| T7KozakSARS | 5′-TAATACGACTCACTATAGGGAGAGCCACCATGTTTATTGAATC-3′ |
| RevSARS | 5′-CCCTCATCTTAATGACGTAGAGC-3′ |
| T7SARS23kD | 5′-TAATACGACTCACTATAGGGAGAGCCACCATGGTTTTGGAGAATAAC-3′ |
| RevSAR23kd | 5′-CACCGGCTAAAAGAATTGAAG-3′ |
| T7Luc | 5′-TAATACGACTCACTATAGGGAGACCACCATGTTGCCATCAAAAATC-3′ |
| revLuc | 5′-TTTTTTTTTTTTTTTTTTTTGTTTTTCACTGCATACGACGTTC-3′ |
| T7forSHAPE | 5′-TAATACGACTCACTATAGGGAAGATGCACCTGATGAAATGG-3′ |
| revSHAPE | 5′-GCCCATATCGTTTCATAGCTTC-3′ |
| FlucFor | 5′-CCAATGCTATTGTTGAAGGTGC-3′ |
| SHAPEoligo | 5′-GCCGGGCCTTTCTTTATG-3′ |
| TaqMan analysis | |
| SARS13496 | 5′-TGCTGGTTTTGCAAAGTTCCT-3′ |
| SARS13564 | 5′-AAATTGCCTTCCTCATCCTTCTC-3′ |
| SARS30-50 | 5′-CCAACCAACCTCGATCTCTTG-3′ |
| SARS28539 | 5′-CAAGGCTCCCTCAGTTGCA-3′ |
SARS-CoV reverse genetics.
Briefly, a full-length cDNA clone of the SARS-CoV genome was constructed from six subclones (called SARS clones A through F), and SP6 RNA polymerase and a GTP cap analog were used to generate full-length infectious transcripts (58). The transfection of these into mammalian cell lines results in a productive, lytic viral infection. RNA was prepared by in vitro transcription, and the transcripts were transfected into Vero cells. The virus was allowed to grow for 5 days at 37°C. Viral supernatants were plated on Vero cells, and several clones were obtained by plaque purification. The plaque-purified viruses were expanded on Vero cells. Viral assays were conducted in a biosafety level 3 facility.
Quantitation of viral titer.
The abundance of viral genomic and subgenomic RNAs (gRNA and sgRNA, respectively) was determined by quantitative real-time PCR using SYBR green chemistry. RNA was extracted from infected cells using TRIzol (Invitrogen, Carlsbad CA) from which cDNA was produced using the Applied Biosystems high-capacity cDNA reverse transcription kit (Foster City, CA) according to the manufacturer's instructions. Primers complementary to nucleotides 13496 to 13516 and 13564 to 13542 (using the numbering of the Urbani SARS-CoV strain; GenBank number AY278741) were used to detect genomic transcripts, and primers complementary to nucleotides 30 to 50 and 28539 to 28521 were used to detect subgenomic RNA. Although the primers for detecting the sgRNA can anneal to the gRNA, only the smallest sgRNA has the 5′ leader and 3′ sequence in close enough proximity to allow amplification to proceed with normal PCR cycles (Fig. 1). To quantitate the ratio of subgenomic to genomic equivalents, we measured the ratio of genomic to subgenomic RNA from viral stocks, intracellular (unlysed cells) plus any secreted or membrane-bound viral RNA. We believe that this ratio most fairly describes replication events independent of viral infectivity.
For infection experiments, genomic RNA was quantitated in the viral stocks for the purpose of determining the amount of viral stock to use as inoculum. Equivalent amounts of genomic viral RNA were used to infect cells. Viral titers were determined by observing infected Vero E6 monolayers in 96-well plates by use of a 50% tissue culture infectious dose (TCID50) assay as previously described (15). Briefly, 10-fold serial dilutions of viral samples were incubated at 37°C for 4 days and then examined for cytopathic effect (CPE) in infected cells. The CPE of SARS-CoV-infected Vero E6 cells was determined by observing rounded, detached cells in close association with each other. The first dilution of viral sample was a 1:10 dilution, which set the limit of viral detection for this assay at 1 log10 TCID50. Error bars are the standard deviations from six measurements. Where infectivity was at the lower level of detection for the assay, error was not able to be calculated.
Translation assays.
PCR primers were designed to amplify the frameshift signal and the attenuator sequence when present from the dual luciferase plasmids. The forward primers included the T7 transcription promoter (Table 1). Small amplicons were generated so that differences in the proteins from transcripts with or without the attenuator sequence could be clearly resolved. Additionally, larger amplicons were generated that included the entire luciferase cassette. PCR products were made using the Fermentas 2× PCR Master Mix (Glen Burnie, MD). RNA was transcribed and translated from PCR amplicons using the Ambion mMESSAGE mMACHINE T7, MEGAScript T7, and Retic Lysate IVT kits (Austin, TX) or the Promega T7+ coupled transcription/translation kit (Madison, WI). Products labeled with [26S]methionine were separated by electrophoresis through Invitrogen 4 to 20% Tris-glycine gels (Carlsbad, CA), and autoradiographs were produced.
Dual luciferase assays.
Vero E6 cells were transfected with the dual luciferase plasmids and grown overnight in DMEM supplemented with 10% FBS. Cells were lysed using the passive lysis buffer (Promega, Madison, WI) per the manufacturer's instructions. Luminescence reactions were initiated by the addition of 10 to 20 μl of cell lysates to 100 μl of the Promega LAR II buffer and completed by the addition of 100 μl to the Stop'n'Glo reagent. Luminescence was measured using a Turner Design TD20/20. At least three readings were taken for each assay, and all assays were repeated (n = 3 to 12) until the data were normally distributed to enable statistical analyses both within and between experiments (31).
RNA structure probing.
The structures of the pseudoknots were probed using the SHAPE procedure of Wilkinson et al. (57). Briefly, DNA was amplified from each mutant plasmid using 2× PCR master mix (Fermentas, Glen Burnie, MD) and the primers T7forSHAPE, 5′-TAATACGACTCACTATAGGGAAGA TGCACCTGATGAAATGG-3′, and revSHAPE, 5′-GCCCATATCGTTTCATAGCTTC-3′. These primers correspond to the 3′ end of firefly luciferase and the 5′ end of Renilla luciferase coding sequences, respectively. RNA was transcribed from the PCR products using the Ambion T7 MEGAscript kit per the manufacturer's instructions. Two pmol of RNA in a total volume of 12 μl H2O was denatured at 95°C and cooled on ice. Six μl of a buffer containing 333 mM HEPES, pH 8.0, 20 mM MgCl2, 222 mM NaCl was added to each RNA sample, and the mixture was incubated at 37°C for 20 min. The reaction mixtures were split into two equal aliquots, one containing 1 μl dimethylsulfoxide (DMSO) and the other containing 1 μl of 30 mM N-methylisatoic anhydride (NMIA) in DMSO, and incubated at 37°C for 45 min. The RNAs were precipitated with 90 μl H2O, 4 μl 5 M NaCl, 1 μl 20 mg/ml glycogen, 2 μl 100 mM EDTA, pH 8.0, and 350 μl ethanol overnight at −80°C. After centrifugation, the RNA was resuspended in 7 μl of 0.5× Tris-EDTA. Fifty pmol of the oligonucleotide 5′-GCCGGGCCTTTCTTTATG-3′ (Integrated DNA Technology, Coralville, IA) was labeled with 30 μCi of [γ-24P]ATP using T4 kinase (Roche) and purified through a G-25 column (GE Healthcare; Piscataway, NJ). Seven μl of RNA and 3 μl of labeled oligonucleotide were annealed. Reverse transcription reactions were performed using SuperScript III enzyme (Invitrogen, Carlsbad, CA) at 52°C for 20 min. Products were separated through 8% polyacrylamide gels, dried, and exposed to a phosphorimager cassette for analysis.
RESULTS
Frameshift efficiency plays a critical role in SARS-CoV propagation.
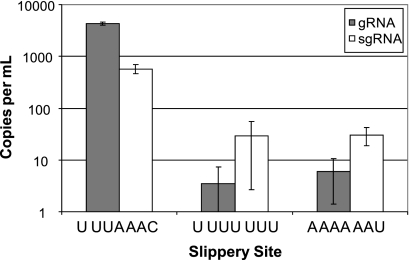
The composition of a slippery site can strongly influence its ability to promote −1 PRF (9, 19, 45). A series of slippery-site mutants was constructed in a dual luciferase-based −1 PRF reporter plasmid (pJD502), and the effects on −1 PRF efficiency were assayed as previously described (47). The plasmids are based on those described by Grentzmann et al. (26) and support transient expression. The activity of the second luciferase is dependent on −1 PRF and is normalized to the activity of the first. As shown in Table 2, the different slippery sites promoted a broad range of −1 PRF efficiencies from 14.4 to 0.15%. A reverse genetics system (58) then was employed to assess the effects of three of the mutant slippery sites (marked by an asterisk in Table 2) on SARS-CoV propagation. The three mutant viruses (and wild-type control) were constructed, recovered, and tested for infectivity using a tissue culture infectious dose assay. Initial visual inspection revealed that plaque numbers and diameters increased as −1 PRF efficiencies approached wild-type levels. Consistent with the hypothesis that −1 PRF efficiency is critical for coronavirus propagation, the U UUG AAC mutant, which does not alter the primary protein coding sequence of the slippery site but which almost completely abrogated frameshifting, was not able to support virus propagation. RNAs from the wild-type and remaining two mutant viruses were recovered and quantified using real-time PCR. Interestingly, the abundance of genomic RNA in infected cells from the mutant viruses was reduced by approximately three orders of magnitude relative to wild-type levels, while the subgenomic RNA levels only dropped by approximately one order of magnitude (Fig. 2). Equivalent amounts of virus (based on the amount of genomic viral RNA) subsequently were used to infect cells, after which TCID50 values were determined (Table 2). This analysis clearly shows that as little as a 3-fold decrease in −1 PRF efficiency essentially abrogated SARS-CoV propagation. Consistent with experimental data from retroviruses and dsRNA viruses, this demonstrates that maintaining the efficiency of frameshifting is essential for the optimal production of infectious viral particles for positive-sense single-stranded RNA (ssRNA) viruses as well.
TABLE 2.
Slippery-site SARS-CoV sequencesa
| Slippery-site sequence | % −1 PRF | SD | Plaque size | TCID50 |
|---|---|---|---|---|
| U UUA AAC (WT) | 14.40 | 2.35 | +++ | 4.53 |
| U UUU UUC | 12.24 | 2.41 | ||
| A AAT TTA | 4.93 | 1.56 | ||
| U UUU UUU* | 4.88 | 1.31 | ++ | <1 |
| A AAA AAC | 4.40 | 1.02 | ||
| G GGA AAC | 3.97 | 0.70 | ||
| G GGU UUA | 3.57 | 0.74 | ||
| U UUA AAU | 3.23 | 1.48 | ||
| U UUA AAA | 2.81 | 0.82 | ||
| A AAA AAU* | 2.33 | 0.55 | + | <1 |
| C CCA AAC | 2.21 | 0.81 | ||
| G GGU UUC | 1.94 | 0.83 | ||
| U UUA AAG | 1.72 | 1.24 | ||
| G GGU UUU | 1.67 | 0.67 | ||
| U UUG AAA | 0.70 | 0.19 | ||
| G GGU UUG | 0.58 | 0.13 | ||
| U UUG AAC* | 0.15 | 0.03 | − | ND |
Slippery-site SARS-CoV sequence mutants and incoming reading frames are indicated by spaces. *, slippery sites incorporated into infectious clones. −1 PRF efficiencies and standard deviations were measured in Vero cells as previously described (60). The plaque sizes of infectious clones are indicated by the number of plus signs. The TCID50 (log10/10 viral genomes) is the average of six measurements. The lower limit of the assay (1 log10) is 1.
FIG. 2.
Relative abundance of genomic and subgenomic RNA in viral stocks. Plaque-purified virus was used to infect Vero cells. Four days postinfection CPE was observed. Media and detached cells were removed and filtered. RNA was extracted from a 100-μl aliquot using TRIzol. TaqMan analysis was used to determine the total number of genomic and subgenomic RNA molecules compared to a reference RNA transcribed from a SARS replicon (1). The number of copies per ml of viral stock is shown with standard deviations.
Morphing the MHV frameshift-promoting pseudoknot into the SARS-CoV pseudoknot supports the hypothesis that viruses have evolved a golden mean for frameshift efficiency.
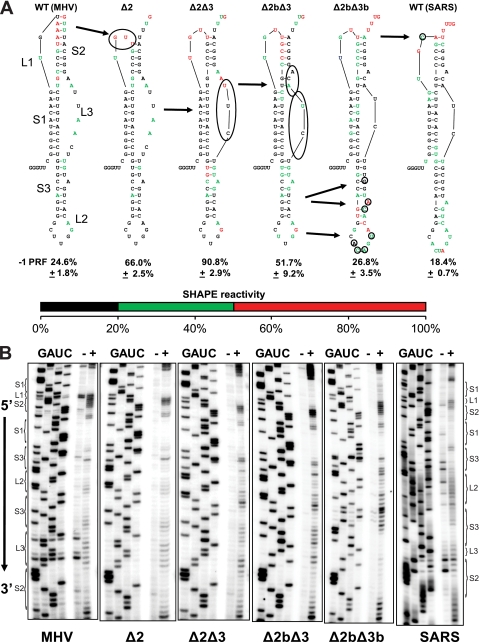
Although SARS-CoV and MHV belong to the same group of coronaviruses, their predicted −1 PRF-promoting pseudoknots differ significantly. Comparing MHV with SARS in Fig. 3A, the first stem of both pseudoknots is predicted to be 10 nucleotides long, i.e., one full helical turn. The second stem of the SARS pseudoknot is 7 nucleotides long with a bulged adenosine (47, 52), whereas the MHV stem is predicted to be 9 nucleotides long without a bulged residue. The loop joining stem 3 with stem 2 also differs significantly between the two pseudoknots: the loop in the MHV structure is predicted to be 8 nucleotides long, and that in the SARS structure is 2 nt. Finally, each pseudoknot has a bulged adenosine in the third stem but they differ slightly in placement, with the adenosine in the SARS pseudoknot being closer to the loop of stem 3. We hypothesized that the two viruses have similar functional requirements with regard to −1 PRF efficiency, and that the different −1 PRF signals have evolved over time toward a functionally equivalent golden mean, i.e., different structures have evolved to produce optimal levels of frameshifting for virus propagation.
FIG. 3.
Changing the MHV −1 PRF-promoting mRNA pseudoknot to the SARS-CoV pseudoknot: structural and functional analysis. (A) The predicted secondary structures of the MHV (left) and SARS-CoV (right) pseudoknots are shown, along with a series of mutants designed to sequentially change the MHV sequence into that of SARS-CoV. S and L denote stem and loop elements, respectively. Circled bases show the sequential mutations made to shorten stem 2 (Δ2) and loop 3 (Δ2Δ3) and to add and move the bulges in stems 2 (Δ2bΔ3) and 3 (Δ2bΔ3b). Frameshifting efficiency (percent) with standard errors is shown below each construct. Color coding indicates the extent of protection from modification by NMIA in the SHAPE reactions as indicated by the bar below. (B) Representative autoradiograms of SHAPE reactions. Dideoxynucleotide sequencing reactions (GUAC) are included for each mutant. The minus sign indicates control samples, and the plus sign represents NMIA-treated samples.
The predicted structural differences between the SARS-CoV and MHV frameshift-promoting pseudoknots provided natural starting points to probe for how differences in structure affect function. To this end, a series of four mutants was constructed that sequentially changed the MHV −1 PRF promoting pseudoknot into the SARS-CoV structure. Figure 3A shows the entire series of six constructs, from wild-type MHV to SARS-CoV, where the predicted secondary structures were computationally determined using pknots (48). The circles denote bases sequentially mutagenized to morph the MHV pseudoknot into the final SARS-CoV structure. To gain a better sense of their solution structures, SHAPE analysis of T7 RNA polymerase primer extension reactions was employed using NMIA (57). Representative autoradiograms of the SHAPE reactions for each construct are shown in Fig. 3B, and the results are color mapped onto the secondary structures in Fig. 3A, where black denotes fully protected bases, green means partial protection from modification, and red shows that the sugars were fully available for modification by NMIA. Most aspects of the predicted structures held true, with fully protected bases (black) mapping inside of the stems and partially protected bases (green) mapping to stem junctions and in loops. All of the stem 1 structures were highly stable. However, this analysis did reveal some important differences from the predicted structures. For example, although the MHV stem 2 is predicted to be 9 bp in length, the high A-U and G-U content at its distal end rendered it quite unstable, decreasing its actual length to a core of three to four G-C base pairs and consequentially increasing the length of loop 1 from the predicted 2 nt to approximately 7 nt. The SARS-CoV stem 2 was much more stable (5 to 6 bp plus the protected bulged A), and its actual loop 1 was shorter than that of MHV (4 nt). In contrast, while base pairing in the MHV stem 3 was very stable, progressive mutations toward the SARS-CoV sequence led base pairing in this region to become less stable. Further, while the repositioning of the bulged A in stem 3 helped to stabilize stem 2, this further destabilized stem 3. Similar patterns were observed in the loop 2 and loop 3 regions, although we note that loop changes simultaneous with moving the bulged A may have influenced stem 3 stability. For example, while the larger loop 2 of SARS-CoV was significantly more solvent accessible than its shorter counterpart in MHV, the longer loop 3 of MHV was more solvent accessible than the shorter loop 3 of SARS-CoV. Interestingly, increasing the stability of loop 3 by reducing its size progressively destabilized loop 2 (compare the series Δ2, Δ2Δ3, Δ2bΔ3 in Fig. 1A). In summary, while the MHV and SARS-CoV −1 PRF-promoting pseudoknots both contain stable stem 1 structures, they each appear to have exchanged solvent-accessible (stem 2/loop 3 for MHV and stem 3/loop 2 for SARS-CoV) and solvent-inaccessible modules (stem 3/loop 2 for MHV and stem 2/loop 3 for SARS-CoV). In contrast, the intermediate constructs appear to progress to less-solvent-accessible, and thus more compacted structures, culminating with Δ2 and Δ2Δ3, from which point they sequentially become more solvent accessible and, thus, less compacted.
The effects of these constructs on −1 PRF efficiency were assayed as previously described (47). The wild-type MHV and SARS-CoV sequences promoted roughly equivalent levels of −1 PRF (SARS-CoV, ∼18.4%; MHV, ∼24.6%) (Fig. 3A). Beginning with the MHV sequence, shortening loop 1 by one codon (Δ2) increased the stability of stem 2 and increased frameshifting efficiency to ∼66%. Reducing the length of loop 3 further compacted the structure (Δ2Δ3), increasing frameshifting to ∼90.8%. The insertion of a bulge into stem 2 (Δ2bΔ3) rendered loop 3 more solvent accessible (less compacted) and lowered the frameshifting frequency to ∼51.7%. A final mutant that repositioned the bulge in stem 3 made the loop more like that of the SARS-CoV pseudoknot (Δ2bΔ3b) lowered frameshifting efficiency back to a level closer to that of the wild-type SARS-CoV basal level (∼26.8%). These results demonstrate a relationship between pseukoknot stability/compactedness and −1 PRF efficiency and support the hypothesis that the pseudoknots of SARS-CoV and MHV evolved to promote the correct frequencies of −1 PRF required for the optimum synthesis of ORF1a- and ORF1b-encoded proteins.
The SARS-CoV attenuator sequence impedes ribosome processivity rather than inhibiting frameshifting.
A prior study had suggested the presence of a frameshift attenuator element located immediately 5′ of the SARS-CoV −1 PRF signal (52). This conclusion was based on the observation that apparent frameshifting efficiency was reduced by ∼40% when an additional 150 nucleotides of sequence located 5′ of the slippery site of the SARS −1 PRF signal was included in frameshift reporter constructs. The inclusion of this sequence upstream of the IBV frameshift signal resulted in a similar reduction in −1 PRF, leading the authors to conclude that the function of this element is to specifically attenuate frameshifting. An mfold analysis revealed that this sequence may assume very stable secondary structures, leading the authors to suggest that RNA-RNA interactions between the attenuator sequence and the −1 PRF signal promote decreased rates of −1 PRF. An alternative interpretation is that the stable RNA structure assumed by this sequence simply inhibits ribosome processivity, causing a significant fraction of ribosomes to dissociate from the mRNA before encountering the −1 PRF signal. This would result in an apparent, but not actual, change in −1 PRF efficiency.
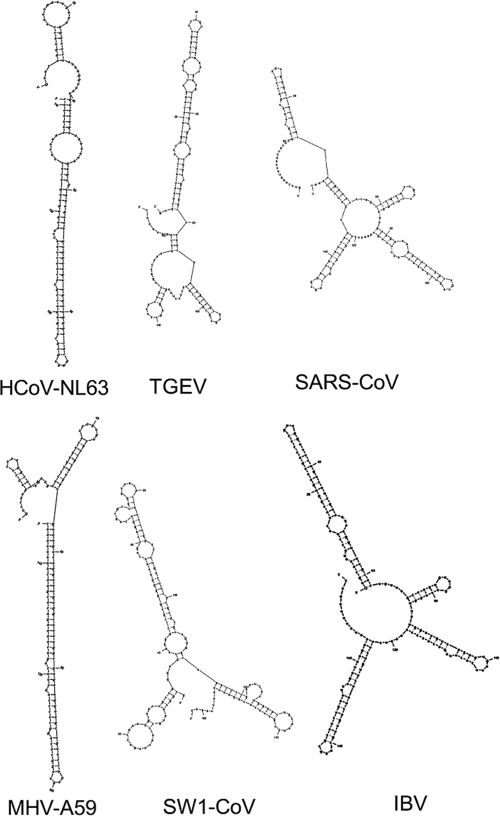
If such an element is functional, it should be conserved. In an initial survey, windows of 150 nt upstream of the −1 PRF signals from all 32 coronaviruses sequenced to date were extracted and compared. Multiple-alignment analyses using Clustal W2 (35) at both the peptide and nucleotide levels revealed that although there was good conservation of sequences within the different subgroups of viruses, there was no conservation of either primary nucleotide or peptide sequences between coronavirus subgroups (data not shown). However, since primary sequence information is not informative with regard to potential secondary- or tertiary-structure interactions, all of these sequences were folded in silico using mfold (36, 60) to address this issue. This analysis revealed that they all had the potential to form stable secondary structures, although there was no predicted consensus structure tying the different groups together. Figure 4 shows folding solutions from six representative viruses. Thus, it is possible that coronaviruses have evolved strong RNA structures to modulate frameshifting.
FIG. 4.
Coronavirus sequences upstream of the −1 PRF signal are predicted to fold into highly stable structures. mfold analyses of the predicted secondary structures of sequences 5′ of the slippery sites from six distantly related coronaviruses are shown.
To determine whether the effect of the SARS-CoV attenuator element on −1 PRF is direct or indirect, a series of dual luciferase reporters was constructed either with or without the attenuator element and/or the −1 PRF signal (slippery site plus pseudoknot) (Fig. 5A). The SARS-CoV attenuator sequence, slippery site, and pseudoknot (nucleotides 13224 to 13477) were cloned between the two Renilla and firefly luciferase genes into p2luci to create the test plasmid (T+). The insertion of an additional adenosine immediately 5′ of the slippery site created a readthrough control plasmid (C+), enabling the translation of the downstream firefly gene without frameshifting. Test and control constructs lacking the attenuator sequence and containing the −1 PRF signal (pJD502 and pJD464) were described previously (47) and were used to control for the absence of the attenuator sequence. These are designated T− and C−. pLuci provided the readthrough control (RT) lacking both the attenuator and −1 PRF signal.
FIG. 5.
Apparent effect of the attenuator sequence on frameshifting is due to its affect on ribosome processivity. (A) pLuci (no-insert readthrough, i.e., RT) was employed as the parent plasmid for all subsequent constructs. The SARS-CoV frameshift signal, including the slippery site (ss) and pseudoknot (Ψk), were cloned between the two luciferase genes (test construct T−). A readthrough plasmid was created by the addition of one nucleotide (n) upstream of the slippery site (control construct C−). Similar constructs containing the upstream attenuator sequence (att) also were made (T+ and C+). The reading frame of the firefly luciferase in each of the constructs is indicated. (B) Apparent frameshifting efficiencies were assayed in Vero cells transfected with each of the indicated constructs. Firefly activity was normalized to Renilla activity, and comparisons were made between the plasmids. The test constructs were compared to the control constructs to determine frameshifting efficiency. (C) Firefly/Renilla luciferase ratios generated by the readthrough constructs containing either the pseudoknot alone (C−) or the pseudoknot and the attenuator sequence (C+) were compared to those generated from pLuci (RT). (D) Full-length luciferase proteins produced using separate transcription and translation reactions were separated by SDS-PAGE. Two different dilutions of T− (i.e., T− and T−2) are shown. An increase in the abundance of smaller proteins from attenuator-containing transcripts is indicated with an asterisk. The expected sizes of the Renilla protein with or without the attenuator at 43 and 37 kDa, respectively, are marked on the gel. The frameshift or readthrough products (Firefly/Renilla) with or without the attenuator are predicted to be 101 and 107 kDa, respectively.
The apparent level of frameshifting promoted by the attenuator-containing construct (T+/C+) was 12.1%, i.e., ∼34% less than the 18.4% frameshifting promoted from the construct lacking this sequence (T+/C+) (Fig. 5B). This result compares favorably with results of the prior study (52). However, the comparison of firefly/Renilla luciferase ratios among the readthrough controls revealed that both the attenuator and −1 PRF signal inhibited ribosome processivity (Fig. 5C). Specifically, the addition of the slippery site plus pseudoknot reduced the firefly/Renilla luciferase ratios to 92% of the control plasmid lacking any inserts (compare RT/RT to C−/RT), i.e., the pseudoknot structure inhibited ribosome processivity by ∼8%. The addition of the attenuator reduced this ratio further to 62% of the no-insert readthrough control (C+/RT), i.e., the combination of the attenuator and pseudoknot inhibited ribosome processivity by 38%. This number is nearly identical to the apparent 34% reduction in −1 PRF in the presence of the attenuator. Thus, the attenuator element does not actually inhibit −1 PRF. Rather, we suggest that this element functions to reduce the fraction of ribosomes that encounter the −1 PRF signal, from which point they may shift the reading frame and translate ORF1b. In sum, we conclude that this element has evolved as an additional means to control the stoichiometric ratio of pp1a to pp1b.
If the attenuator functions to block elongating ribosomes, then its presence should result in the accumulation of truncated peptide products. To test this, PCR products containing T7 RNA polymerase transcription promoters were synthesized, T7 RNA polymerase runoff transcripts were synthesized in vitro, [26S]methionine-labeled peptides were translated in vitro, and the products were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The frameshift-promoting (T− and T−2) template lacking the attenuator primarily produced a product of 37 kDa (T− and T−2 in Fig. 5D), which is consistent with the presence of a 0-frame termination codon. The addition of the attenuator (T+) increased the size of this product to 43 kDa. Importantly, the presence of the attenuator also resulted in the production of a significant fraction of truncated peptides (indicated by an asterisk in Fig. 5D). A similar range of truncated products was produced from the readthrough reporter containing the attenuator (C+), and these were significantly less well represented in the attenuatorless readthrough control (C−). In addition, the presence of truncated products in the 37-kDa range in the C− sample is consistent with pseudoknot-induced ribosome dissociation. These results support the hypothesis that both the attenuator and the frameshift-stimulating pseudoknot cause a significant fraction of ribosomes to dissociate from the mRNA.
DISCUSSION
Maintaining the correct levels of coronavirus frameshifting efficiency is essential for viral infectivity.
The importance of maintaining precise ratios of Gag to Gag-pol has been demonstrated for the L-A dsRNA totivirus, the HIV and MMTV retroviruses, and the Ty1 retrotransposable element (reviewed in reference 18). In addition, the detrimental effects of altered frameshifting efficiency has been shown for the positive-sense ssRNA luteovirus barley yellow dwarf virus (43) and, more recently, with regard to the neuroinvasiveness of the Kunjin subtype of the positive-sense ssRNA flavivirus West Nile virus (37). Although one of the earliest frameshift signals identified was from a coronavirus (7), the importance of frameshifting has not been established formally for this group. The ORFs in which the coronavirus frameshift signals are found are very large and encode many different proteins, many of whose functions are uncharacterized. Altering the ORF1a/ORF1b protein ratios by reducing −1 PRF efficiency should result in fewer enzymes encoded by ORF1b compared to the level of proteases encoded by ORF1a.
The reduction of −1 PRF to 0.15% completely abrogated the production of infectious viral particles, whereas small amounts of infectious virus were produced at −1 PRF levels of 2.3%. These observations demonstrate that there is a lower limit of frameshifting below which coronaviruses cannot replicate to detectable levels. As anticipated, gRNA production was greatly reduced in the two viable −1 PRF mutants. After normalizing for virus levels in infected cells, the infectivity rates of the two viable slippery-site mutants remained at least 3.5 orders of magnitude less than that of the wild-type controls. One possible explanation for this dramatic effect consequent to mere 3- to 7-fold changes in −1 PRF is that decreased −1 PRF results in the decreased synthesis of ORF1ab-encoded replicase proteins, which are required for the replication of the virus. This species in turn serves as the enzyme for the synthesis of gRNA and sgRNA. In positive-sense ssRNA viruses, gRNA synthesis is greatly amplified from the negative strand, and thus the observation that the mutants decreased infectivity by >3.5 orders of magnitude, and gRNA synthesis by ∼3 orders of magnitude, may be accounted for by this amplification process. Alternatively, the decreased viral amplification may be due to a decrease in the abundance of the other nsp11 to nsp16 proteins relative to that of the nsp1 to nsp10 proteins. It is not clear from our analysis whether the loss in infectivity is directly the result of the altered RNA synthesis, protein levels, or a subsequent defect in viral particle production. Further study is needed to investigate these possibilities.
Interestingly, the slippery-site mutants had lesser effects on the accumulation of sgRNA (Fig. 2). Viral RNA synthesis occurs in the cytoplasm and requires only the ORF1a- and ORF1ab-encoded proteins (1, 55). The RNA-dependent RNA polymerase (RDRP) is responsible for the synthesis of both gRNAs and sgRNAs, and the ratio of these two species normally remains constant throughout the infectious cycle (50). The differences in this ratio observed between the wild-type virus and frameshift mutants demonstrate that alterations in the −1 PRF signal impact the utilization of the template RNA for the synthesis of these two RNA species. It has been shown that additional nsp12 and nucleocapsid proteins each enhance replication (41), as well as the presence of additional nsp3, a papainlike cysteine protease (50), suggesting that the processing of the polyprotein(s) is important for replication. However, those studies did not distinguish between the production of gRNA and sgRNA species. In the current study, reduced −1 PRF is expected to result in less nsp11 to nsp16 relative to the amount of nsp1 to nsp10 produced during infection. We observed a change in the ratio of gRNA and sgRNA when RNA from infected cells were analyzed by quantitative PCR. The link between these two points is, at present, unclear but likely is due to the inherent function of the nsp proteins. It is formally possible, however, that the defect in gRNA synthesis observed in the current study is due to changes in the RNA slippery-site sequence itself.
Coronavirus frameshift-promoting mRNA pseudoknot structures have evolved to fine-tune −1 PRF toward producing a golden mean of ORF1a- and ORF1b-encoded peptides.
While the structural analyses presented here show that both the SARS-CoV and MHV mRNA pseudoknots are three-stemmed structures, their primary sequences and basic structural elements are significantly different from one another. Specifically, although their stem 1 elements are highly stable, the long loop 1, short stem 2, and long loop 3 of MHV are less compact than those of their analogous SARS-CoV elements. Conversely, reverse relationships apply to the stem 3 and loop 2 elements. Stabilizing/compacting any of these elements promoted increased frameshifting efficiencies, which is consistent with the notion that highly stable mRNA secondary structures cause elongating ribosomes to pause longer over the slippery site. The critical finding here is that, despite having significantly different structures, both the MHV and SARS-CoV pseudoknots promoted equivalent rates of −1 PRF. Our results show that although a class of mRNA pseudoknots that promote very high levels of frameshifting can exist, the fact that all naturally occurring coronavirus frameshift signals assayed to date promote −1 PRF efficiencies in the 10 to 20% range (46) provides indirect support for the notion that high levels of frameshifting also are incompatible with coronavirus replication. This is consistent with the golden mean model of frameshifting, i.e., it appears that each virus has evolved the right balance between more and less stable structural elements to produce the optimum rates of −1 PRF.
Coronaviruses also use translational attenuation to obtain the correct ratios of upstream and downstream viral polypeptides.
The description of a sequence with a novel function, the attenuation of −1 PRF by a novel cis-acting element (52), warranted further inspection. Prior to that report, no other cis-acting elements affecting −1 PRF other than the slippery site and downstream stimulatory element had been described, and no concrete model for how the attenuator functioned was proposed. The presence of a large number of previously unavailable coronavirus genomes allowed an initial phylogenetic analysis. The alignment of these sequences revealed a large degree of diversity among the analogous regions in both homology and length; the smallest coronaviral genomes contained the shortest analogous sequences, and the most diverse genomes showed the least homology. This indicated that no specific attenuator sequence is conserved among coronaviruses, suggesting that the element does not actually directly affect the process of frameshifting. However, the computational prediction of highly stable secondary structures engendered the hypothesis that the attenuator element impedes the processivity of elongating ribosomes on the viral mRNA, causing them to dissociate prior to encountering the −1 PRF signal, i.e., a translational attenuation model. As supported by the experiments in the current study, the addition of the attenuator sequence upstream of the dual luciferase assay confirmed that its presence resulted in an apparent decrease in −1 PRF. However, the critical experiment, comparing the readthrough controls, revealed that while the −1 PRF signal itself promoted a small but significant decrease in the production of the full-length product (presumably due to the presence of the mRNA pseudoknot), the addition of the attenuator decreased this value by nearly 40%. Further, in vitro translation assays demonstrated that the attenuator sequence promoted the increased synthesis of prematurely terminated peptide products. Again, although indirect, this provides indirect support of the golden mean hypothesis, in that they show that coronaviruses have evolved a second cis-acting element to limit the fraction of ribosomes able to eventually translate ORF1b. In sum, the data presented here support a model in which a combination of translational attenuation and limited −1 PRF efficiency serve to fine-tune the fraction of elongating ribosomes able to translate the ORF1b mRNA sequence, thus ensuring that the pp1a and pp1b ratios are optimized for coronavirus propagation.
Acknowledgments
We thank members of the Dinman laboratory for comments and advice and give special thanks to Amy Sims and Ralph Baric for construction of the infectious clones and to Paul Masters for the MHV clone.
This work was supported by a grant from the National Institutes of Health to J.D.D. (AI064307).
Footnotes
Published ahead of print on 17 February 2010.
REFERENCES
- 1.Almazán, F., M. L. DeDiego, C. Galan, D. Escors, E. Alvarez, J. Ortego, I. Sola, S. Zuniga, S. Alonso, J. L. Moreno, A. Nogales, C. Capiscol, and L. Enjuanes. 2006. Construction of a severe acute respiratory syndrome coronavirus infectious cDNA clone and a replicon to study coronavirus RNA synthesis. J. Virol. 80:10900-10906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balasundaram, D., J. D. Dinman, R. B. Wickner, C. W. Tabor, and H. Tabor. 1994. Spermidine deficiency increases +1 ribosomal frameshifting efficiency and inhibits Ty1 retrotransposition in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 91:172-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barry, J. K., and W. A. Miller. 2002. A −1 ribosomal frameshift element that requires base pairing across four kilobases suggests a mechanism of regulating ribosome and replicase traffic on a viral RNA. Proc. Natl. Acad. Sci. USA 99:11133-11138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biswas, P., X. Jiang, A. L. Pacchia, J. P. Dougherty, and S. W. Peltz. 2004. The human immunodeficiency virus type 1 ribosomal frameshifting site is an invariant sequence determinant and an important target for antiviral therapy. J. Virol. 78:2082-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brian, D. A., and R. S. Baric. 2005. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 287:1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brierley, I. 1995. Ribosomal frameshifting viral RNAs. J. Gen. Virol. 76(Pt 8):1885-1892. [DOI] [PubMed] [Google Scholar]
- 7.Brierley, I., P. Digard, and S. C. Inglis. 1989. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell 57:537-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brierley, I., A. J. Jenner, and S. C. Inglis. 1992. Mutational analysis of the “slippery-sequence” component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 227:463-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brierley, I., N. J. Rolley, A. J. Jenner, and S. C. Inglis. 1991. Mutational analysis of the RNA pseudoknot component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 220:889-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bruenn, J. A. 1980. Virus-like particles of yeast. Annu. Rev. Microbiol. 34:49-68. [DOI] [PubMed] [Google Scholar]
- 11.Castón, J. R., B. L. Trus, F. P. Booy, R. B. Wickner, J. S. Wall, and A. C. Steven. 1997. Structure of L-A virus: a specialized compartment for the transcription and replication of double-stranded RNA. J. Cell Biol. 138:975-985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng, R. H., J. R. Caston, G. J. Wang, F. Gu, T. J. Smith, T. S. Baker, R. F. Bozarth, B. L. Trus, N. Cheng, R. B. Wickner, et al. 1994. Fungal virus capsids, cytoplasmic compartments for the replication of double-stranded RNA, formed as icosahedral shells of asymmetric Gag dimers. J. Mol. Biol. 244:255-258. [DOI] [PubMed] [Google Scholar]
- 13.Cui, Y., J. D. Dinman, T. G. Kinzy, and S. W. Peltz. 1998. The Mof2/Sui1 protein is a general monitor of translational accuracy. Mol. Cell. Biol. 18:1506-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui, Y., J. D. Dinman, and S. W. Peltz. 1996. Mof4-1 is an allele of the UPF1/IFS2 gene which affects both mRNA turnover and −1 ribosomal frameshifting efficiency. EMBO J. 15:5726-5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Darnell, M. E., E. P. Plant, H. Watanabe, R. Byrum, M. St Claire, J. M. Ward, and D. R. Taylor. 2007. Severe acute respiratory syndrome coronavirus infection in vaccinated ferrets. J. Infect. Dis. 196:1329-1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dinman, J. D., T. Icho, and R. B. Wickner. 1991. A −1 ribosomal frameshift in a double-stranded RNA virus of yeast forms a gag-pol fusion protein. Proc. Natl. Acad. Sci. USA 88:174-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dinman, J. D., and T. G. Kinzy. 1997. Translational misreading: mutations in translation elongation factor 1alpha differentially affect programmed ribosomal frameshifting and drug sensitivity. RNA 3:870-881. [PMC free article] [PubMed] [Google Scholar]
- 18.Dinman, J. D., M. J. Ruiz-Echevarria, and S. W. Peltz. 1998. Translating old drugs into new treatments: ribosomal frameshifting as a target for antiviral agents. Trends Biotechnol. 16:190-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dinman, J. D., and R. B. Wickner. 1992. Ribosomal frameshifting efficiency and gag/gag-pol ratio are critical for yeast M1 double-stranded RNA virus propagation. J. Virol. 66:3669-3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dinman, J. D., and R. B. Wickner. 1995. 5S rRNA is involved in fidelity of translational reading frame. Genetics 141:95-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Escutenaire, S., N. Mohamed, M. Isaksson, P. Thoren, B. Klingeborn, S. Belak, M. Berg, and J. Blomberg. 2007. SYBR green real-time reverse transcription-polymerase chain reaction assay for the generic detection of coronaviruses. Arch. Virol. 152:41-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esteban, R., and R. B. Wickner. 1986. Three different M1 RNA-containing viruslike particle types in Saccharomyces cerevisiae: in vitro M1 double-stranded RNA synthesis. Mol. Cell. Biol. 6:1552-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Farabaugh, P. J. 1996. Programmed translational frameshifting. Microbiol. Rev. 60:103-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Felsenstein, K. M., and S. P. Goff. 1988. Expression of the gag-pol fusion protein of Moloney murine leukemia virus without gag protein does not induce virion formation or proteolytic processing. J. Virol. 62:2179-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giedroc, D. P., and P. V. Cornish. 2009. Frameshifting RNA pseudoknots: structure and mechanism. Virus Res. 139:193-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grentzmann, G., J. A. Ingram, P. J. Kelly, R. F. Gesteland, and J. F. Atkins. 1998. A dual-luciferase reporter system for studying recoding signals. RNA 4:479-486. [PMC free article] [PubMed] [Google Scholar]
- 27.Harger, J. W., A. Meskauskas, J. Nielsen, M. C. Justice, and J. D. Dinman. 2001. Ty1 retrotransposition and programmed +1 ribosomal frameshifting require the integrity of the protein synthetic translocation step. Virology 286:216-224. [DOI] [PubMed] [Google Scholar]
- 28.Hudak, K. A., A. B. Hammell, J. Yasenchak, N. E. Tumer, and J. D. Dinman. 2001. A C-terminal deletion mutant of pokeweed antiviral protein inhibits programmed +1 ribosomal frameshifting and Ty1 retrotransposition without depurinating the sarcin/ricin loop of rRNA. Virology 279:292-301. [DOI] [PubMed] [Google Scholar]
- 29.Hung, M., P. Patel, S. Davis, and S. R. Green. 1998. Importance of ribosomal frameshifting for human immunodeficiency virus type 1 particle assembly and replication. J. Virol. 72:4819-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inoue, H., H. Nojima, and H. Okayama. 1990. High efficiency transformation of Escherichia coli with plasmids. Gene 96:23-28. [DOI] [PubMed] [Google Scholar]
- 31.Jacobs, J. L., and J. D. Dinman. 2004. Systematic analysis of bicistronic reporter assay data. Nucleic Acids Res. 32:e160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karacostas, V., E. J. Wolffe, K. Nagashima, M. A. Gonda, and B. Moss. 1993. Overexpression of the HIV-1 gag-pol polyprotein results in intracellular activation of HIV-1 protease and inhibition of assembly and budding of virus-like particles. Virology 193:661-671. [DOI] [PubMed] [Google Scholar]
- 33.Kawakami, K., S. Pande, B. Faiola, D. P. Moore, J. D. Boeke, P. J. Farabaugh, J. N. Strathern, Y. Nakamura, and D. J. Garfinkel. 1993. A rare tRNA-Arg(CCU) that regulates Ty1 element ribosomal frameshifting is essential for Ty1 retrotransposition in Saccharomyces cerevisiae. Genetics 135:309-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kontos, H., S. Napthine, and I. Brierley. 2001. Ribosomal pausing at a frameshifter RNA pseudoknot is sensitive to reading phase but shows little correlation with frameshift efficiency. Mol. Cell. Biol. 21:8657-8670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 36.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 37.Melian, E. B., E. Hinzman, T. Nagasaki, A. E. Firth, N. M. Wills, A. S. Nouwens, B. J. Blitvich, J. Leung, A. Funk, J. F. Atkins, R. Hall, and A. A. Khromykh. 2009. NS1′ of flaviviruses in the Japanese encephalitis serogroup is a product of ribosomal frameshifting and plays a role in viral neuro-invasiveness. J. Virol. 84:1641-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meskauskas, A., J. L. Baxter, E. A. Carr, J. Yasenchak, J. E. Gallagher, S. J. Baserga, and J. D. Dinman. 2003. Delayed rRNA processing results in significant ribosome biogenesis and functional defects. Mol. Cell. Biol. 23:1602-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Meskauskas, A., and J. D. Dinman. 2001. Ribosomal protein L5 helps anchor peptidyl-tRNA to the P-site in Saccharomyces cerevisiae. RNA 7:1084-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meskauskas, A., J. W. Harger, K. L. Jacobs, and J. D. Dinman. 2003. Decreased peptidyltransferase activity correlates with increased programmed −1 ribosomal frameshifting and viral maintenance defects in the yeast Saccharomyces cerevisiae. RNA 9:982-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pan, J., X. Peng, Y. Gao, Z. Li, X. Lu, Y. Chen, M. Ishaq, D. Liu, M. L. DeDiego, L. Enjuanes, and D. Guo. 2008. Genome-wide analysis of protein-protein interactions and involvement of viral proteins in SARS-CoV replication. PLoS One 3:e3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park, J., and C. D. Morrow. 1991. Overexpression of the gag-pol precursor from human immunodeficiency virus type 1 proviral genomes results in efficient proteolytic processing in the absence of virion production. J. Virol. 65:5111-5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paul, C. P., J. K. Barry, S. P. Dinesh-Kumar, V. Brault, and W. A. Miller. 2001. A sequence required for −1 ribosomal frameshifting located four kilobases downstream of the frameshift site. J. Mol. Biol. 310:987-999. [DOI] [PubMed] [Google Scholar]
- 44.Peltz, S. W., A. B. Hammell, Y. Cui, J. Yasenchak, L. Puljanowski, and J. D. Dinman. 1999. Ribosomal protein L3 mutants alter translational fidelity and promote rapid loss of the yeast killer virus. Mol. Cell. Biol. 19:384-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Plant, E. P., and J. D. Dinman. 2006. Comparative study of the effects of heptameric slippery site composition on −1 frameshifting among different eukaryotic systems. RNA 12:666-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Plant, E. P., and J. D. Dinman. 2008. The role of programmed-1 ribosomal frameshifting in coronavirus propagation. Front. Biosci. 13:4873-4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Plant, E. P., G. C. Perez-Alvarado, J. L. Jacobs, B. Mukhopadhyay, M. Hennig, and J. D. Dinman. 2005. A three-stemmed mRNA pseudoknot in the SARS coronavirus frameshift signal. PLoS Biol. 3:e172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rivas, E., and S. R. Eddy. 2000. The language of RNA: a formal grammar that includes pseudoknots. Bioinformatics 16:334-340. [DOI] [PubMed] [Google Scholar]
- 49.Ruiz-Echevarría, M. J., J. M. Yasenchak, X. Han, J. D. Dinman, and S. W. Peltz. 1998. The upf3 protein is a component of the surveillance complex that monitors both translation and mRNA turnover and affects viral propagation. Proc. Natl. Acad. Sci. USA 95:8721-8726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sawicki, S. G., and D. L. Sawicki. 2005. Coronavirus transcription: a perspective. Curr. Top. Microbiol. Immunol. 287:31-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Somogyi, P., A. J. Jenner, I. Brierley, and S. C. Inglis. 1993. Ribosomal pausing during translation of an RNA pseudoknot. Mol. Cell. Biol. 13:6931-6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Su, M. C., C. T. Chang, C. H. Chu, C. H. Tsai, and K. Y. Chang. 2005. An atypical RNA pseudoknot stimulator and an upstream attenuation signal for −1 ribosomal frameshifting of SARS coronavirus. Nucleic Acids Res. 33:4265-4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Telenti, A., R. Martinez, M. Munoz, G. Bleiber, G. Greub, D. Sanglard, and S. Peters. 2002. Analysis of natural variants of the human immunodeficiency virus type 1 gag-pol frameshift stem-loop structure. J. Virol. 76:7868-7873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Templeton, S. P., and S. Perlman. 2007. Pathogenesis of acute and chronic central nervous system infection with variants of mouse hepatitis virus, strain JHM. Immunol. Res. 39:160-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thiel, V., J. Herold, B. Schelle, and S. G. Siddell. 2001. Viral replicase gene products suffice for coronavirus discontinuous transcription. J. Virol. 75:6676-6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thiel, V., K. A. Ivanov, A. Putics, T. Hertzig, B. Schelle, S. Bayer, B. Weissbrich, E. J. Snijder, H. Rabenau, H. W. Doerr, A. E. Gorbalenya, and J. Ziebuhr. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 84:2305-2315. [DOI] [PubMed] [Google Scholar]
- 57.Wilkinson, K. A., E. J. Merino, and K. M. Weeks. 2006. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 1:1610-1616. [DOI] [PubMed] [Google Scholar]
- 58.Yount, B., K. M. Curtis, E. A. Fritz, L. E. Hensley, P. B. Jahrling, E. Prentice, M. R. Denison, T. W. Geisbert, and R. S. Baric. 2003. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA 100:12995-13000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ziebuhr, J. 2005. The coronavirus replicase. Curr. Top. Microbiol. Immunol. 287:57-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zuker, M. 2003. mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]