Abstract
The zinc finger antiviral protein (ZAP) is a host factor with potent antiviral activity when overexpressed in cells. ZAP blocks replication of the prototype alphavirus Sindbis virus (SINV) at a step at or before translation of the incoming viral genome. The mechanism of ZAP anti-SINV activity and the determinants of its antiviral function, however, have not been defined. Here, we have identified a dominant negative inhibitor of human ZAP. Rat ZAP with a cysteine-to-arginine mutation at position 88 (rZAPC88R), previously reported as a nonfunctional form of ZAP, increases SINV growth in cells. These results led us to discover a previously undetectable pool of endogenous functional ZAP within human cells. Investigation of the mechanism of dominant negative inhibition, combined with a comprehensive mutational analysis of the antiviral factor, revealed that homotypic associations are required for ZAP function in limiting SINV propagation.
Alphaviruses remain a serious global health concern. Human infection can cause symptoms including rash, fever, arthralgia, and polyarthritis and, in rare cases, fatal encephalitis (reviewed in reference 40). Currently, there is no specific treatment for alphavirus infection. Sindbis virus (SINV) is the type member of the Alphavirus genus and is an amenable study system that has provided valuable insights into viral replication and pathogen-host interactions (reviewed in reference 46). Replication of SINV takes place in the cytoplasm on modified endosomal and lysosomal membranes (9). Recent evidence has also implicated the plasma membrane as an initial site of replication (14). The positive-strand RNA genome encodes two open reading frames (ORFs) (reviewed in reference 44). Cap-dependent translation of the 5′-proximal ORF from the incoming viral genome produces a polyprotein that is processed to generate nonstructural proteins nsP1 to nsP4 (45). nsP1 possesses methyltransferase and guanylyltransferase activities (34, 42), nsP2 is the protease responsible for processing the nsP1-to-nsP4 region (6, 19) and also exhibits helicase activity (13), nsP3 is a phosphoprotein with an undefined but essential function in viral replication (5, 28), and nsP4 is the RNA-dependent RNA polymerase (20). The virus modulates replicase template preference by successive cleavage of the nonstructural polyproteins, providing temporal regulation of negative strand synthesis followed by genome amplification (24-26, 43). The 3′ ORF is translated from a subgenomic RNA and encodes the structural proteins, which include the capsid protein and glycoproteins E2, 6K, and E1 (45). The capsid protein has autoprotease activity (1) and cleaves itself from the remaining structural proteins before interacting with the viral RNA genome to form nucleocapsids (10). The glycoproteins are targeted via the host secretory pathway to the plasma membrane (37), where interaction between the cytoplasmic domain of E2 and the surface of the nucleocapsid drives virus budding (32).
We and others previously showed that an interferon (IFN)-stimulated host protein, zinc finger antiviral protein (ZAP), has strong anti-alphavirus activity (2, 22, 47). ZAP is also active against retroviruses and filoviruses (11, 36) but does not appear to induce a general antiviral state in cells (2). Based on the minimal overlap of the replication strategies of these viruses, it is hypothesized that ZAP mediates its antiviral effect in the cytoplasm (11); indeed, the protein shuttles between the cytoplasm and nucleus (31) and is found primarily in the cytoplasm when overexpressed in mammalian cells (31, 33). Association of ZAP with nucleic acid and with exosome components suggests a role in regulating RNA metabolism; interaction with target RNA and subsequent recruitment of exosome components for RNA degradation have been proposed (17). In addition, ZAP facilitates interaction of exosome factors and the host protein DEAD box helicase p72, which may unwind target RNA for degradation (4). Overexpression of ZAP enhances the degradation of retrovirus (11) and filovirus (36) RNA and blocks SINV at a step after uncoating and prior to translation of the incoming genome (2). In addition, retrovirus and SINV sequences have been shown to confer ZAP sensitivity on a reporter RNA (16). The interaction of ZAP with RNA is thought to be mediated by one or more zinc fingers (16). Indeed, the amino-terminal domain of ZAP (NZAP), which is sufficient to confer antiviral activity, contains four CCCH-type zinc-binding motifs. While the role of these sequences in zinc coordination is unclear, some of these residues are critical for ZAP function. Mutation of cysteine 88 to arginine (ZAPC88R) is predicted to disrupt the second zinc finger and was shown to impair the antiviral function and RNA-binding ability of ZAP (16).
ZAP was first identified in a rat cDNA library (11), and most published studies involve the rat isoform (rZAP). Recently, a human ZAP ortholog (hZAP) was reported (22). In the present study, we have identified rZAPC88R as a dominant negative inhibitor of wild-type hZAP. Overexpression of rZAPC88R augments SINV replication, leading us to discover a pool of functional endogenous ZAP existing within human cells. Further investigation of the dominant negative phenotype, combined with a comprehensive mutational analysis of ZAP, revealed that homotypic interactions are critical for the function of this antiviral factor.
MATERIALS AND METHODS
Cell lines.
Cell lines were maintained at 37°C in a humidified atmosphere containing 5% CO2. BHK-J cells (30), a derivative of BHK-21 (ATCC CCL-10; hamster kidney fibroblasts), were cultured in minimum essential medium (Invitrogen, Carlsbad, CA) supplemented with 7.5% fetal bovine serum (FBS). 293T cells (ATCC CRL-11268), a derivative of human embryonic kidney 293 cells expressing the simian virus 40 T antigen and selected for adherence and transfectability, were generously provided by Guangxia Gao (Chinese Academy of Sciences, Beijing, China) and Stephen P. Goff (Columbia University, New York, NY) and cultured in Dulbecco's modified Eagle medium (DMEM; Invitrogen) supplemented with 10% FBS. Human embryonic kidney 293 cells conditionally expressing rZAP, rZAPC88R, or LacZ have been previously described (16) and are designated T-REx-rZAP, T-REx-rZAPC88R, and T-REx-LacZ, respectively. T-REx lines were maintained in DMEM supplemented with 10% FBS, 5 μg/ml blasticidin, and 200 μg/ml zeocin. Expression of rZAP, rZAPC88R, or LacZ was induced by addition of doxycycline (1 μg/ml) to the growth medium for 12 h.
Viruses.
Wild-type SINV (Toto1101) (39), SINV expressing firefly luciferase as a fusion with nsP3 (Toto1101/Luc) (2), a SINV temperature-sensitive mutant expressing luciferase (Toto1101/Luc:ts6) (2), and SINV encoding the enhanced green fluorescent protein (EGFP) from a duplicated subgenomic promoter (TE/5′2J/GFP) (8) have been previously reported. Stocks were prepared and titers determined on BHK-J cells with 10-fold serial dilutions of sample, and then plaques were visually enumerated after crystal violet staining, as previously described (2); multiplicities of infection (MOI) were calculated based on BHK-J-derived titers. The temperature sensitivity of the Toto1101/Luc:ts6 virus stock in T-REx cells was verified by luciferase assay; no amplification of signal was detected when infection was carried out at 40°C.
Plasmid construction.
Plasmids were constructed by standard methods; clones were verified by restriction enzyme digestion and sequencing of PCR-generated fragments (Genewiz, Inc., South Plainfield, NJ). Descriptions of the cloning strategies are outlined below; additional primer sequences will be provided upon request.
pcDNA4/TO/rZAPHA and pcDNA4/TO/rZAPC88RHA.
Plasmids pcDNA4/TO/myc-rZAP and pcDNA4/TO/myc-rZAPC88R have been previously described (16) and express full-length rZAP or rZAP with the C88R mutation, respectively, each with a C-terminal myc epitope tag. Full-length rZAP was fused at its C terminus to the influenza virus hemagglutinin (HA) epitope tag (amino acid sequence YPYDVPDYA) by PCR amplification of the C terminus of the rZAP coding sequence with forward primer 5′-TATCGAACAGGCCTATTGTGATCC-3′ and a reverse primer encoding the HA tag followed by a stop codon and a NotI site (5′-TCGAGCGGCCGCCTCAAGCGTAGTCTGGGACGTCGTATGGGTACTCTGGACCTCTTCTCTTCTGC-3′). The NdeI/NotI-digested PCR products were used to replace the equivalent 321-bp fragment of pcDNA4/TO/myc-rZAP and pcDNA4/TO/myc-rZAPC88R to generate plasmids expressing HA-tagged rZAP (pcDNA4/TO/rZAPHA) and rZAPC88R (pcDNA4/TO/rZAPC88RHA).
Split-Gaussia constructs.
As previously described (38), Zip-GLuc1 and Zip-GLuc2 encode the GCN4 leucine zipper protein fused at its C terminus to a flexible linker, followed by the N (GLuc1)- or C (GLuc2)-terminal domain of Gaussia luciferase (GLuc); these constructs were generously provided by Stephen Michnick (Université de Montréal, Montreal, Quebec, Canada). To facilitate the expression of protein fragments as N- or C-terminal fusions with GLuc1 or GLuc2, we generated pGLuc1-GLuc2 and pGLuc2-GLuc1, which encode a fusion of the two GLuc domains with an intervening flexible linker; the first GLuc domain is flanked by NotI/ClaI sites, and the second GLuc fragment is flanked by BspEI/XbaI sites. pGLuc1-GLuc2 was created by amplification of the GLuc1 sequence with forward primer 5′-GCATGTCGGCGGCCGCACCATGAAGCCCACCGAGAACAACGAAG-3′ and reverse primer 5′-CTATCAGCATCGATGCCTATGCCGCCCTGTGCGGAC-3′, followed by digestion with NotI/ClaI and ligation to pZIP-GLuc2 digested with the same enzymes. pGLuc2-GLuc1 was created in the same manner, using forward primer 5′-GCATGTCGGCGGCCGCACCATGGAGGCGATCGTCGACATTCCTG-3′ and reverse primer 5′-CTATCAGCATCGATGTCACCACCGGCCCCCTTGATC-3′ to amplify GLuc2, followed by subcloning into pZIP-GLuc1.
phZAP-GLuc1 was constructed by amplifying the full-length hZAP coding sequence from hZAP (ATCC I.M.A.G.E. Consortium clone 752123) with forward primer 5′-ACAGTGGCGGCCGCACCATGGCGGACCCGGAGG-3′ and reverse primer 5′-GCCACCTTCGAACTCTGGCCCTCTCTTCATCTGC-3′ and ligation of the NotI/BstBI-digested PCR product to the compatible 5,323-bp NotI/ClaI fragment of pGLuc2-GLuc1. pGLuc2-Zip, pGLuc2-hZAP, pGLuc2-rZAPC88R, and pGLuc2-hNZAP were created by substitution of the 285-bp BspEI/XbaI fragment of pGLuc2-GLuc1 with the appropriate PCR-amplified sequence. Zip residues were derived from Zip-GLuc1 (38); hNZAP denotes the hZAP antiviral domain (N-terminal 252 residues). pHCV5A-GLuc2, generously provided by Shihyun You, was constructed by replacing the GLuc1 segment of pGLuc1-GLuc2 with the hepatitis C virus (HCV) nonstructural protein 5A (NS5A) coding sequence, amplified from J6/JFH (29) using forward primer 5′-GGCGGCCGCACCATGTCTAAATTGTTCCCCAAGCTGCCC-3′ and reverse primer 5′-CCACCATCGATGCAGCACACGGTGGTATCGTCCTC-3′; the N-terminal membrane-associated helix (residues 1 to 24) was deleted.
pGLuc2-hNZAP alanine mutant constructs.
Quintuple-alanine mutations were constructed in the context of pGLuc2-hNZAP using standard assembly PCR techniques as described previously (23). Briefly, the hNZAP coding sequence was amplified with primers containing five alanine substitutions at the desired positions, in conjunction with flanking primers. The primary PCR products were then assembled in a secondary PCR using flanking forward primer 5′-TCCTCCGGAATGGCGGACCCGGAGG-3′ and reverse primer 5′-ACTAGTTCTAGATTACCGATCTCTACTCTTGCTTCT-3′. Final PCR products were digested with BspEI/XbaI and ligated to pGLuc2-GLuc1 digested with the same enzymes.
pEBG-NZAP and alanine mutant constructs.
The N-terminal 254 residues of rZAP (rNZAP) were cloned into the mammalian expression vector pEBG (35) by subcloning the SwaI-and-NotI fragment from pGEX-rNZAP (33). The resulting construct, pEBG-rNZAP, encodes a glutathione S-transferase (GST)-rNZAP fusion protein. pEBG-hNZAP and the panel of alanine mutant constructs were generated by subcloning of the appropriate PCR-amplified segments into the EcoRI/NotI sites of pEBG-rNZAP, retaining the N-terminal GST fusion. Primers used for hNZAP amplification were forward primer 5′-AGGAATTCTGACCATGGCGGACCCGGAGG-3′ and reverse primer 5′-CTCGAGCGGCCGCTCACCGATCTCTACTCTTGCTTCTAGC-3′.
Virus infections and growth curves.
Cells were seeded at 7 × 105/9.6-cm2 dish and infected the following day with the appropriate wild-type or modified SINV; the MOI used is indicated in Results or the figure legends. After 1 h, the inoculum was removed, cells were washed twice with growth medium and fresh medium was added. Culture supernatants were collected at various times postinfection and stored at −80°C. Infectious titers were determined on BHK-J cells as described above. Replication of the reporter virus was detected by firefly luciferase assay of cell lysates harvested with 1× Passive Lysis Buffer (Promega, Madison, WI) according to the manufacturer's protocol. Luciferase activity was measured on a Centro LB960 luminometer (Berthold Technologies, Oak Ridge, TN).
DNA and siRNA transfection.
T-REx derivatives were seeded at 4 × 105/9.6-cm2 dish 24 h prior to DNA transfection using Fugene 6 (Roche, Indianapolis, IN) mixed with 1 μg of plasmid in Opti-MEM (Invitrogen), according to the manufacturer's protocol. For small interfering RNA (siRNA) transfection, T-REx-rZAPC88R cells were seeded the day before at 3.7 × 104/2.0-cm2 well. Cells were transfected using Lipofectamine RNAiMAX transfection reagent (Invitrogen) and 12 pmol of siRNA per well according to the manufacturer's protocol. siRNA duplexes were ZAP specific (5′-GGUAAAACCUGGACGGACUUU-3′, sense; 5′-P-AGUCCGUCCAGGUUUUACCUU-3′, antisense; catalogue no. J-017449-11; Dharmacon, Lafayette, CO) or irrelevant (catalogue no. D-001810-10). Knockdown experiments were conducted in triplicate. Where indicated, cell viability was determined using the CellTiter-Glo (Promega) assay according to the manufacturer's recommendations.
Affinity purification.
Transfected 293T cells expressing GST- and myc-tagged proteins were disrupted with lysis buffer (20 mM HEPES, 110 mM potassium acetate, 2 mM MgCl2, 0.1% Tween, 0.5% NP-40, 300 mM NaCl). Lysates were precleared by centrifugation (16,000 × g for 10 min at 4°C), and tagged proteins were affinity purified using 30 μl bovine serum albumin (BSA)-blocked glutathione agarose beads (Santa Cruz, Santa Cruz, CA) for 1 h at 4°C, followed by three washes in cold lysis buffer. Immunoprecipitated proteins were collected by boiling in sodium dodecyl sulfate gel loading buffer and analyzed by Western blotting.
Western blotting and antibodies.
Protein samples were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotted with the following antibodies as indicated in the figures: rabbit anti-Gaussia luciferase (1:10,000; NanoLight, Pinetop, AZ), mouse anti-myc (ATCC CRL 1792 hybridoma, 1.6 μg/ml), mouse anti-GST (1:3,000; Cell Signaling Technology, Boston, MA), and mouse anti-β-actin (1:5,000; Sigma-Aldrich, St. Louis, MO).
Split-Gaussia assay.
293T cells transfected with split-Gaussia plasmid DNA were lysed after 24 h using Renilla lysis buffer (Promega), and activity was measured using Renilla luciferase substrate according to the manufacturer's recommendations. Reconstituted luciferase activities normalized by protein expression (see Fig. 4A); for the raw data, see Fig. 3B. The levels of wild-type and mutant GLuc2-hNZAP protein expression were assessed by Western blotting using a rabbit polyclonal antibody specific for Gaussia luciferase (1:10,000; NanoLight, Pinetop, AZ). Enhanced chemiluminescence signal was captured by direct imaging using an Alphaimager (Alpha Innotech, San Leandro, CA), and the relative amounts of protein were quantified using the company's software. Of all 51 alanine mutant constructs, the mean protein expression was 52.5% relative to wild-type ZAP expression, with a standard deviation of 38%. Protein expression levels for 43 of the mutant constructs were within the first standard deviation of the mean. The cells expressing the least stable mutant construct (hNZAP66-70) had 5-fold less protein, and the cells expressing the most stable construct (hNZAP56-60) had 1.6-fold more protein, than wild-type ZAP.
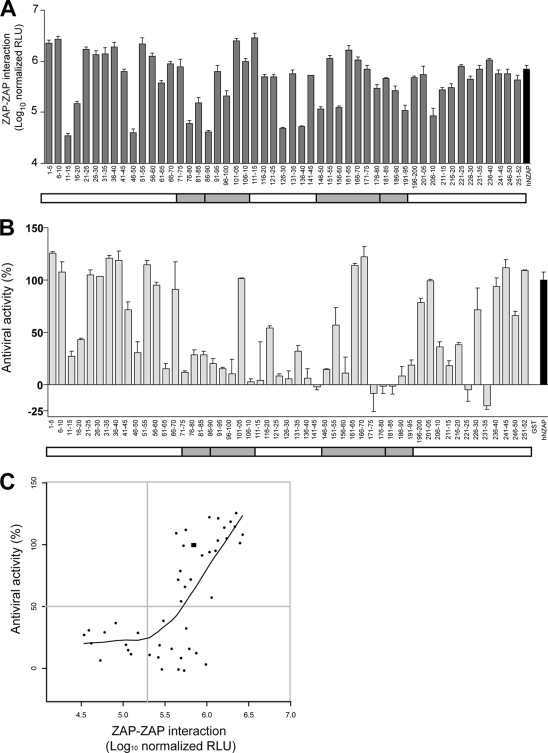
FIG. 4.
Identification of sequences important for ZAP-ZAP interaction and antiviral function. (A) hNZAP constructs containing quintuple-alanine mutations, expressed as a fusion with the carboxyl-terminal Gaussia luciferase fragment (GLuc2), were cotransfected with a plasmid encoding full-length wild-type hZAP fused to the amino-terminal Gaussia luciferase fragment (GLuc1). Reconstituted luciferase activity was normalized to mutant hNZAP protein levels (relative light units, RLU). Means and standard deviations of triplicate transfections are shown. For both panels A and B, a schematic of hNZAP is shown as a white box; gray shading indicates the position of the putative CCCH zinc finger motifs. Similar results were obtained in another independent experiment. (B) 293T cells were transfected with plasmids encoding alanine mutant hNZAP proteins fused to GST. Relative antiviral activity was determined by a flow cytometry based assay; GST-hNZAP inhibitory activity was defined as 100%, and GST was defined as having 0% antiviral activity. Means and standard deviations of duplicate experiments are shown. (C) The ZAP-ZAP interaction (A) and the antiviral activities (B) of the alanine mutant proteins were plotted. The black square represents the data point of wild-type hNZAP. Gray lines mark the activity level equivalent to 50% of that of wild type hNZAP in the antiviral activity and the ZAP-ZAP interaction. The fitted local regression curve (LOESS curve) is shown. Six mutant proteins (16-20, 111-15, 126-30, 171-75, 221-25, and 231-35) were omitted, as they showed <10% transfected cells in the antiviral assay, presumably due to reduced stability of the protein or poor transfection efficiency.
FIG. 3.
hZAP interacts with itself and with rZAPC88R. (A) 293T cells were cotransfected with plasmids expressing myc-tagged hZAP or rZAPC88R together with GST, GST-tagged hNZAP, or rNZAPC88R. Two days posttransfection, GST or GST fusion proteins were precipitated from cell lysates using glutathione agarose beads. The levels of GST or GST-tagged proteins were monitored by Western blotting; copurified myc-tagged proteins were detected by stripping the blot and reprobing with anti-myc antibodies. Expression of myc-tagged hZAP or rZAPC88R in lysates of transfected cells is shown. IP, affinity purification. (B) ZAP interaction was measured by determining reconstituted luciferase activity (relative light units, RLU) in 293T cells using the split-Gaussia assay. The assay was conducted in the absence (black) or presence (gray) of a competing construct encoding myc-tagged hZAP without a split-luciferase fragment. ZIP, leucine zipper GCN4. HCV5A, cytoplasmic fragment of HCV RNA-binding protein NS5A. Similar results were obtained in two other independent experiments. For each transfection pair, asterisks indicate statistically significant differences (P < 0.05, unpaired Student t test) obtained upon addition of the competing hZAPmyc construct.
Immunostaining and flow cytometry.
Cells were harvested and fixed with 1% paraformaldehyde in phosphate-buffered saline (PBS). Cells were blocked and permeabilized in PBS containing 0.1% saponin, 5% goat serum, and 0.1% sodium azide. Primary mouse antibodies specific for HA (16B12; Covance, Denver, PA) or GST (26H1; Cell Signaling, Beverly, MA) were diluted 1:500 (HA) or 1:1,000 (GST) in PBS containing 0.1% saponin, 0.5% BSA, and 0.1% sodium azide and incubated for 1 h at room temperature. Cells were washed three times in PBS, with centrifugation at 500 × g for 5 min. Alexa 633-conjugated anti-mouse secondary antibody (1:1,000; Invitrogen) was incubated for 30 min at room temperature, followed by three washes with PBS. Cells were analyzed using a FACScalibur cytometer (Becton Dickinson, Franklin Lakes, NJ) and Flowjo software (Tree Star, Inc., Ashland, OR), analyzing 10,000 events per sample. Gates were set with uninfected or untransfected cells such that less than 1% of the cells fell within the positive gate.
Determination of antiviral activity.
Relative antiviral activity was calculated by setting the inhibitory activity of GST-hNZAP to 100% and that of GST to 0% activity. The percent EGFP (infection) determined by flow cytometry is used to calculate the antiviral activity of ZAP and each alanine mutant construct (X) based on the formula [(A − X)/(A − B)] × 100, where A is percent EGFP (infection) in GST-expressing cells, B is percent EGFP (infection) in GST-hNZAP expressing cells, and X is percent EGFP (infection) in ZAP mutant X-expressing cells.
RESULTS
A nonfunctional mutant form of ZAP (rZAPC88R) enhances SINV replication.
Mutation of a residue contributing to a putative zinc finger motif (C88R) has previously been shown to abolish the antiviral activity of rZAP (16). Here, we observed that expression of this mutant protein not only failed to inhibit SINV replication but consistently enhanced infectious titers (Fig. 1A). Using human T-REx embryonic kidney 293 cell derivatives conditionally expressing rZAP or rZAPC88R (16), similar levels of virus replication were found in the uninduced state (filled symbols). As expected, induction of rZAP expression resulted in a marked inhibition of SINV replication. Induction of rZAPC88R expression, however, increased progeny virus production by >10-fold at 6 to 24 h postinfection, compared to that in uninduced cells. These cell lines expressed similar levels of rZAP or rZAPC88R protein after doxycycline treatment (Fig. 1B). Overexpression of another nonfunctional mutant form of rZAP, H191K, in which a putative zinc-coordinating histidine was replaced with lysine, did not result in enhancement (data not shown). We confirmed that the increased titers were not a clonal effect of our stable cell line, as transient overexpression of rZAPC88R in 293T cells similarly enhanced SINV replication (Fig. 2B and data not shown). Understanding the mechanism of rZAPC88R-mediated augmentation of SINV growth could provide clues as to the mechanism of ZAP antiviral activity.
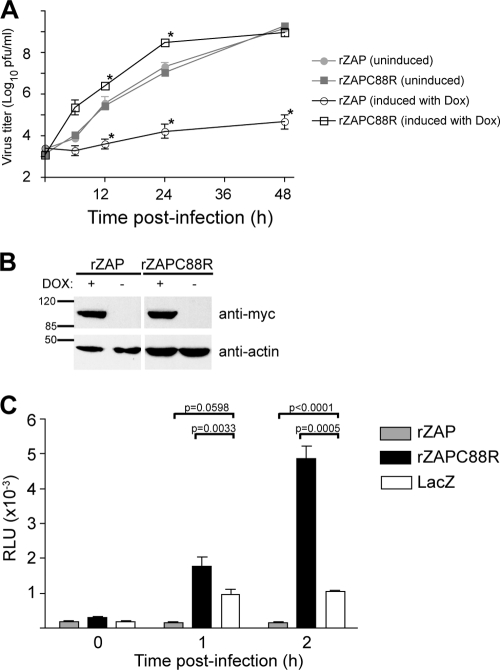
FIG. 1.
Expression of ZAP mutant construct rZAPC88R enhances SINV replication. (A) T-REx-rZAP or -rZAPC88R cells were left untreated or induced with doxycycline (Dox) for 12 h as indicated and then infected for 1 h with SINV Toto1101 (MOI = 0.1). At the indicated time points, the medium was harvested; separate wells were utilized for each time point. Virus titers were determined in duplicate by plaque assay on BHK-J cells. The mean and standard error of the mean of duplicate wells are shown. Asterisks indicate a significant difference (P < 0.05, unpaired Student t test) compared to the noninduced sample from the same time point. (B) Induction efficiency of T-REx-rZAP and -rZAPC88R cells. Cells were left untreated or induced with doxycycline (DOX) for 12 h and then Western blotted for myc-tagged rZAP or rZAPC88R, as well as β-actin. The values to the left are molecular sizes in kilodaltons. (C) rZAPC88R expression enhances translation of the incoming SINV genome. Induced T-REx-rZAP, -rZAPC88R, or -LacZ cells were infected with temperature-sensitive SINV encoding luciferase, Toto1101/Luc:ts6 (MOI = 10). Infection and incubation were carried out at the restrictive temperature (40°C). Translation was measured by luciferase activity. Means and standard deviations of triplicate infections are shown. Similar results were obtained in an independent experiment. P values (unpaired Student t test) for a subset of the data are shown. RLU, relative light units.
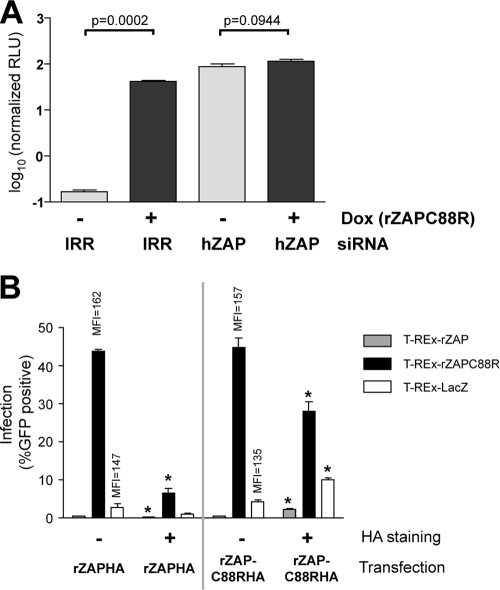
FIG. 2.
Wild-type and mutant ZAP levels dictate the cellular antiviral state. (A) T-REx-rZAPC88R cells were transfected with IRR siRNA or hZAP-specific siRNA, as indicated. Cells were then left uninduced (−) or treated with doxycycline (+) to induce expression of rZAPC88R and infected 12 h later with Toto1101/Luc (MOI = 10). Replication, as monitored by firefly luciferase activity, was normalized to cell viability at the time of infection. RLU, relative light units. Means and standard deviations of triplicate samples are shown. Similar results were obtained in an independent experiment. P values (unpaired Student t test) are shown. Dox, doxycycline. (B) T-REx cell lines were induced to express rZAP, rZAPC88R, or LacZ and subsequently transfected with plasmids expressing HA-tagged rZAP (rZAPHA) or rZAPC88R (rZAPC88RHA). After infection with EGFP-expressing SINV TE/5′2J/GFP (MOI = 10) for 12 h, cells were analyzed for HA (transfection) and EGFP (infection) levels using flow cytometry. HA− (nontransfected) and HA+ (transfected) populations of cells were separately analyzed. The percentage of cells infected (%GFP positive) is shown. Mean fluorescence intensities (MFI) of the infected populations are shown for nontransfected (HA−) cells in T-REx-rZAPC88R and -LacZ cells transfected with HA-tagged rZAPC88R (rZAPC88RHA). Similar results were obtained in three independent experiments. Asterisks indicate significant differences (P < 0.05, unpaired Student t test) between transfected cells and their nontransfected counterparts.
We were interested in characterizing the step at which rZAPC88R increases SINV replication. Since previous work has shown that ZAP prevents efficient translation of incoming SINV genomes (2), we used a temperature-sensitive virus to address the possibility that rZAPC88R mediates enhancement at this stage. Induced T-REx-rZAP, -rZAPC88R and -LacZ cells were infected (MOI = 10) with Toto1101/Luc:ts6, a luciferase-expressing SINV temperature sensitive for RNA synthesis (18). Infections were carried out at the restrictive temperature (40°C) so that luciferase activity results entirely from translation of the incoming virus genome. T-REx-rZAP, -rZAPC88R, and -LacZ cell lines supported similar levels of SINV translation under uninduced conditions (data not shown). Upon overexpression of rZAP, translation of the SINV genome was diminished compared to that in cells expressing the LacZ control (Fig. 1C). Overexpression of rZAPC88R, however, increased the translation of the incoming SINV genome, suggesting that this mutant construct augments the same step of the virus life cycle that is inhibited by wild-type ZAP.
The proviral effect of rZAPC88R is dependent on the level of wild-type ZAP.
Since ZAP was identified from a rat cDNA library screen for antiviral function (11), it is likely that some cells express endogenous ZAP capable of limiting virus replication. Indeed, ZAP mRNA has been detected in various human tissues and cultured cells (11, 22), although we have been unable to detect the endogenous ZAP protein (data not shown). We hypothesized that rZAPC88R acts as a dominant negative inhibitor of this previously undetectable pool of functional hZAP. To test this hypothesis, we examined the effect of reducing the levels of endogenous hZAP in T-REx-rZAPC88R cells. An irrelevant (IRR) siRNA or an siRNA specifically targeting hZAP, but not rZAP, was transfected into cells, which were then left untreated or induced with doxycycline to express rZAPC88R. Cell populations were subsequently infected for 12 h with a luciferase-encoding SINV, Toto1101/Luc (2). In IRR siRNA-treated cells, induction of rZAPC88R resulted in a marked enhancement of SINV replication, compared to levels in uninduced cells (Fig. 2A, IRR). Removal of endogenous ZAP by siRNA-mediated silencing resulted in a similar increase in SINV replication, even in the absence of rZAPC88R induction (Fig. 2A, hZAP, −Dox). Furthermore, induction of rZAPC88R in cells treated to silence endogenous ZAP did not further enhance SINV replication (Fig. 2A, hZAP, +Dox). These results indicate that knockdown of endogenous ZAP is beneficial to SINV growth and suggest that rZAPC88R mediates its proviral effects by targeting this pool of functional ZAP protein.
If rZAPC88R-mediated SINV enhancement is dependent on inhibition of the wild-type antiviral factor, overexpression of wild-type ZAP should limit the effect of rZAPC88R. Induced T-REx-rZAP, -rZAPC88R, or -LacZ cells were transfected with plasmids expressing HA-tagged rZAP or rZAPC88R. Cells were then infected with SINV encoding EGFP (TE/5′2J/GFP, MOI = 10) for 12 h before immunostaining and flow cytometric analysis of the HA tag (wild-type or mutant ZAP) and EGFP (virus) expression. This allowed direct comparison of virus replication in transfected (HA+) and nontransfected (HA−) cells in the same sample. As expected, in nontransfected cells, induction of wild-type rZAP (T-REx-rZAP) resulted in reduced levels of SINV replication (0.47% or 0.6% EGFP, Fig. 2B, HA−), compared to control LacZ-expressing cells (2.9% or 4.3% EGFP). Moreover, induced T-REx-rZAPC88R cells demonstrated high levels of infection (43.8% or 44.8% EGFP, Fig. 2B, HA−). In addition to the higher percentage of infected cells, the level of virus replication within the infected population expressing T-REx-rZAPC88R was increased 10% or 16% compared to LacZ-expressing cells (as measured by mean fluorescence intensity). In stable cell populations expressing rZAPC88R or LacZ, expression of HA-tagged rZAP correlated with a decrease in SINV infection (Fig. 2B, rZAPHA+). In T-REx-rZAPC88R cells, for example, expression of rZAPHA reduced the proportion of infected cells from 43.8% to 6.6%. In contrast, transient expression of rZAPC88RHA in T-REx-rZAP or -LacZ cells correlated with increased EGFP signal, indicative of higher SINV replication (Fig. 2B, rZAPC88RHA+). For example, overexpression of rZAPC88RHA increased the infection rate in T-REx-LacZ cells from 4.3% (rZAPC88RHA−) to 10.1% (rZAPC88RHA+). Together, these findings indicate that the relative levels of rZAPC88R and wild-type ZAP determine the permissivity of cells for SINV replication, consistent with the hypothesis that rZAPC88R acts a dominant negative inhibitor.
ZAP interacts with itself, forming higher-order complexes, and rZAPC88R retains this interaction.
One possible mechanism of dominant negative inhibition is a direct interaction with the wild-type protein. If homotypic multimerization is required for function, binding of a mutant partner may render the entire complex inactive. It is not known, however, if ZAP forms functional higher-order associations. Coprecipitation analysis of the tagged proteins suggested that ZAP interacts with itself and that the rZAPC88R mutant construct retains this interaction with the wild-type protein (Fig. 3A). To quantitatively investigate this ZAP-ZAP homotypic interaction, we employed a split-Gaussia luciferase interaction assay, also known as protein fragment complementation assay (38). In this system, two fragments of Gaussia luciferase are independently expressed as fusions with proteins of interest. GLuc1, the amino-terminal half of Gaussia luciferase, and GLuc2, the carboxyl-terminal portion of the luciferase protein, do not interact in isolation. An association between the fused proteins of interest, however, results in increased proximity of the Gaussia fragments and reconstituted luciferase activity. We observed robust luciferase signal for fusions to the GCN4 leucine zipper protein ZIP, which is known to interact with itself (38). We found that hZAP was similarly able to form homotypic interactions and that the rZAPC88R mutant construct associated well with the wild-type protein (Fig. 3B). In contrast, hZAP did not form a detectable interaction with an unrelated zinc- and RNA-binding domain of HCV NS5A (HCV5A). We further demonstrated the specificity of the ZAP-ZAP interactions through competition with a third plasmid expressing myc-tagged hZAP (hZAPmyc). Whereas hZAPmyc effectively inhibited hZAP-hZAP and hZAP-rZAPC88R interactions, the association of ZIP with itself was unaffected. These data suggest that ZAP self-associates and that the mutation of cysteine 88 to arginine does not affect this interaction.
Mapping the ZAP-ZAP interacting domain and the functional significance of self-association.
To identify the ZAP self-interacting region, we constructed a series of alanine-scanning mutations in the hZAP antiviral domain (residues 1 to 252, hNZAP). The antiviral domain of hZAP was identified based on its homology with the first 254 amino acid residues of rZAP, which were previously shown to be sufficient for antiviral activity (11). We substituted residues for alanine in groups of five, resulting in a total of 51 ZAP mutant constructs. Using the split-Gaussia luciferase assay, the series of alanine constructs fused with GLuc2 was cotransfected with full-length wild-type hZAP fused to GLuc1. Reconstituted luciferase activities were normalized to the level of mutant ZAP fusion protein expression, as determined by Western blotting using an antibody targeting Gaussia luciferase. These data revealed several regions of ZAP important for homotypic interactions (Fig. 4A). Only six mutations severely affected the ZAP-ZAP interaction, as defined by a >1-log reduction in luciferase signal. These mutations were distributed within the amino-terminal half of NZAP, and two of them overlapped with residues encompassing two of the putative zinc fingers.
To probe the functional relevance of the ZAP-ZAP association, the alanine mutations were introduced into a GST-hNZAP fusion construct and their antiviral phenotype was determined by a flow cytometry-based assay. 293T cells were transiently transfected with a mammalian expression vector expressing GST-hNZAP, GST-hNZAP containing each alanine mutation, or GST alone. Cells were infected with EGFP-encoding SINV TE/5′2J/GFP and then immunostained for GST, followed by flow cytometry analysis to determine the percentage of EGFP-positive cells in the transfected populations. The wild-type GST-hNZAP-transfected population showed about 40% GFP positivity at 12 h postinfection, which was defined as 100% antiviral activity (Fig. 4B); GST-transfected cells showed ∼90% infection at the same time point, which was defined as 0% antiviral activity. The mutant GST-hNZAP proteins showed a spectrum of antiviral activities ranging from undetectable to greater-than-wild-type efficacies (Fig. 4B). Twenty-eight of the 51 mutant ZAP proteins showed >50% reductions in antiviral activity, and half of them had mutations that overlapped regions comprising the putative zinc fingers. Six of the constructs exhibited <0% antiviral activity due to a higher number of infected cells than GST control cells. This suggests a potential proviral activity of these ZAP mutant constructs, similar to rZAPC88R, although this remains to be formally confirmed. In order to explore the relationship between the ZAP self-interaction and the antiviral activity, the effects of alanine replacement on the two phenotypes were plotted (Fig. 4C). In general, the constructs fall into three categories: (i) those that, like wild-type ZAP, show strong antiviral and interaction activities, (ii) those that do not self-associate and have lost antiviral activity, and (iii) those, like rZAPC88R, which continue to interact but do not inhibit SINV infection. Interestingly, we did not observe any mutant constructs displaying strong antiviral activity but with an impaired ZAP-ZAP interaction. We generated a locally weighted scatterplot smoothing (LOESS) curve to monitor the correlation of the two phenotypes (Fig. 4C) and observed a trend suggestive of higher-strength ZAP-ZAP interactions paralleling increased antiviral activity. These results suggest that ZAP self-association is critical for its antiviral function.
DISCUSSION
ZAP was first identified in a cDNA screen and shown to have antiviral activity in an overexpression context (11). The existence and importance of a functional endogenous pool of the factor in human cells, however, had not been demonstrated. ZAP mRNA is detected in cultured cell lines, as well as in various human tissues, including germ line cells and peripheral blood lymphocytes (11, 22); treatment of cells with IFN has been shown to increase ZAP mRNA levels (41). Attempts to detect ZAP protein by Western blotting and immunofluorescence, however, have been unsuccessful, even after induction with IFN (data not shown). Here, a nonfunctional mutant construct, rZAPC88R, led us to identify a pool of functional endogenous ZAP present in cultured cells. Elimination of endogenous ZAP function by the expression of dominant negative rZAPC88R or by siRNA-mediated silencing enhanced SINV replication (Fig. 1 and 2); native ZAP levels are, however, insufficient to completely inhibit SINV replication.
The significance of low-level stores of ZAP in human cells is unknown. Based on its strong antiviral activity, it is tempting to speculate that ZAP acts as a restriction factor to limit alphavirus infection. Alphaviruses exhibit a neurotropic pattern in infected animals (15) and, in rats and humans, ZAP has lower expression levels in the brain than in other tissues tested (11, 22). The relevance of ZAP expression to the tropism of SINV and other alphaviruses, however, has not been directly studied. ZAP plays an important role in innate immunity pathways, and the gene that encodes it is one of many IFN-stimulated genes (ISGs) (41). ISGs function to create an antiviral state within the cell, including halting transcription and translation, and must therefore be tightly regulated. Other ISGs with anti-SINV activity, such as those that encode ISG15, ISG20, ISG56, and viperin, are similar to ZAP in that they are quickly and strongly induced upon IFN treatment or virus infection (27, 47). Interestingly, these factors are capable of limiting virus replication under basal conditions in the absence of IFN (47). Moreover, silencing of both ZAP and ISG20 results in a greater enhancement of SINV replication than silencing of either alone, suggesting that multiple factors can contribute to virus inhibition (47). Of the factors encoded by ISGs, ZAP appears to be one of the dominant effectors in the IFN-mediated anti-SINV response, as various cell types with siRNA-mediated reduction of ZAP exhibit reduced sensitivity to the protective effect of IFN (33, 47 and data not shown). In addition, overexpression of ZAP synergizes with IFN treatment, likely due to cooperation with other induced effectors (33). While the remarkable antiviral activity of ZAP opens up the possibility that this protein may have additional signaling or regulatory roles, our preliminary studies show that ZAP is not critical for the induction of other early effectors of the IFN response (data not shown). However, roles for ZAP in modulating the activity or expression of particular IFN-induced factors or in modulating the innate pathways that initiate the production of IFN are still possible. The presence of low levels of ZAP and other IFN-induced factors may serve to prime cells for a rapid response to viral infection.
We suspected that rZAPC88R might act as a dominant negative inhibitor by direct interaction with endogenous ZAP. The discovery that wild-type ZAP shows a strong homotypic interaction, which is retained by the rZAPC88R mutant construct, is consistent with this hypothesis. Comprehensive mutational analysis of the ZAP antiviral domain revealed that all of the mutant constructs with strong anti-SINV activity retained self-interaction, suggesting that the homotypic-interaction and antiviral domains might coincide. Alternatively, we favor the possibility that a ZAP-ZAP interaction is critical for antiviral activity. A class of mutant constructs, including rZAPC88R, which retains the interaction but has low antiviral function supports this hypothesis. The role of the ZAP-ZAP interaction in the inhibitory process is not clear. Oligomerization of another CCCH zinc finger-containing protein, tristetraprolin, has been reported (3), and the in vitro RNA-binding activity of this protein was enhanced by higher-order complex formation. ZAP multimerization may increase points of contact with target RNA, enhancing interaction and subsequent inhibition. It is also possible that RNA may promote ZAP self-association by recruiting other factors necessary for SINV inhibition. For the anti-HIV protein APOBEC3G, oligomerization and RNA-binding activities are both linked to virion incorporation and the subsequent antiviral phenotype (7). Although the nature of the split-Gaussia system suggests a dimeric interaction between two ZAP molecules, we cannot exclude the formation of multimers or large aggregates. Functional higher-order interactions are seen for TIA-1, which aggregates during the formation of stress granules, organelles that perform triage and prevent the translation of certain RNAs (12, 21). Our preliminary data suggest that ZAP can also be targeted to stress granules (data not shown); whether this is a site of ZAP function is currently under investigation.
Based on our findings, we propose that rZAPC88R binding to endogenous ZAP renders the complex inactive, possibly as a result of the lower RNA-binding activity of rZAPC88R (16). It has been proposed that a ZAP-RNA interaction is critical for the antiviral phenotype (16). Although SINV sequences have been shown to confer ZAP sensitivity on a reporter construct, these regions are dispersed throughout the genome and primarily in the antisense orientation (16). A specific and direct interaction between ZAP and the incoming, positive-sense SINV RNA has so far not been demonstrated. Analysis of the series of alanine mutant constructs for general RNA-binding affinity, as well as interaction with the SINV genome and specific cellular RNAs, may reveal correlations indicative of ZAP function. Alternatively, we cannot rule out the possibility that the dominant negative phenotype of rZAPC88R is due to the sequestration of an additional host protein(s) critical for ZAP activity. Several ZAP interacting partners have been reported, including exosome components (17) and RNA helicases (4). Preliminary proteomic data have revealed similar global host-protein associations for wild-type ZAP and rZAPC88R (data not shown), but it would be of interest to compare binding affinities for specific candidate factors. Our discovery of a class of ZAP mutant constructs that, like rZAPC88R, abolish antiviral activity without disrupting multimerization suggests the possibility of additional ZAP functional domains.
Insights into the regulation and mechanism of ZAP are valuable in dissecting the complex interplay between host response and viral evasion. Many virus-specific restriction factors, similar to ZAP, are present at low levels within cells. Finding ways to boost the expression of these molecules without the damaging pleiotropic effects of IFN therapy could tip the balance toward protection or cure of the host cell. Studies of the ZAP-alphavirus interplay provide a valuable model for host antiviral strategies and may provide insights into how other host antiviral factors target additional viruses.
Acknowledgments
This work was supported by Public Health Service grant AI057905 from the National Institute of Allergy and Infectious Diseases (M.R.M.), by the Irma T. Hirschl/Monique Weill-Caulier Trust (M.R.M.), and by the Greenberg Medical Research Institute. This work was also supported by a postdoctoral fellowship from the National Sciences and Engineering Research Council of Canada (L.M.J.L.) and NRSA DK081193 (C.T.J.).
We thank Steve Goff and Guangxia Gao for sharing numerous ZAP-related reagents and Stephen Michnick for providing plasmids of the split-Gaussia system (protein fragment complementation assay). We thank Catherine Murray for critical reading of the manuscript and helpful discussions, Shihyun You for providing split-Gaussia plasmids encoding HCV-NS5A and helpful discussions, Suyan Tian for help with statistical analyses, and Sarah Wensley, Taylor Goodspeed, Arnella Webson, and Megan Holz for technical assistance. We are also grateful to Beate Kümmerer for sharing unpublished results.
Footnotes
Published ahead of print on 24 February 2010.
REFERENCES
- 1.Aliperti, G., and M. J. Schlesinger. 1978. Evidence for an autoprotease activity of Sindbis virus capsid protein. Virology 90:366-369. [DOI] [PubMed] [Google Scholar]
- 2.Bick, M. J., J.-W. N. Carroll, G. Gao, S. P. Goff, C. M. Rice, and M. R. MacDonald. 2003. Expression of the zinc finger antiviral protein inhibits alphavirus replication. J. Virol. 77:11555-11562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao, H., F. Dzineku, and P. J. Blackshear. 2003. Expression and purification of recombinant tristetraprolin that can bind to tumor necrosis factor-alpha mRNA and serve as a substrate for mitogen-activated protein kinases. Arch. Biochem. Biophys. 412:106-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, G., X. Guo, F. Lv, Y. Xu, and G. Gao. 2008. p72 DEAD box RNA helicase is required for optimal function of the zinc-finger antiviral protein. Proc. Natl. Acad. Sci. U. S. A. 105:4352-4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dé, I., C. Fata-Hartley, S. G. Sawicki, and D. L. Sawicki. 2003. Functional analysis of nsP3 phosphoprotein mutants of Sindbis virus. J. Virol. 77:13106-13116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding, M. X., and M. J. Schlesinger. 1989. Evidence that Sindbis virus NSP2 is an autoprotease which processes the virus nonstructural polyprotein. Virology 171:280-284. [DOI] [PubMed] [Google Scholar]
- 7.Friew, Y. N., V. Boyko, W. S. Hu, and V. K. Pathak. 2009. Intracellular interactions between APOBEC3G, RNA, and HIV-1 Gag: APOBEC3G multimerization is dependent on its association with RNA. Retrovirology 6:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frolova, E. I., R. Z. Fayzulin, S. H. Cook, D. E. Griffin, C. M. Rice, and I. Frolov. 2002. Roles of nonstructural protein nsP2 and Alpha/Beta interferons in determining the outcome of Sindbis virus infection. J. Virol. 76:11254-11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Froshauer, S., J. Kartenbeck, and A. Helenius. 1988. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J. Cell Biol. 107:2075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuller, S. D. 1987. The T=4 envelope of Sindbis virus is organized by interactions with a complementary T=3 capsid. Cell 48:923-934. [DOI] [PubMed] [Google Scholar]
- 11.Gao, G., X. Guo, and S. P. Goff. 2002. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science 297:1703-1706. [DOI] [PubMed] [Google Scholar]
- 12.Gilks, N., N. Kedersha, M. Ayodele, L. Shen, G. Stoecklin, L. M. Dember, and P. Anderson. 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 15:5383-5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorbalenya, A. E., and E. V. Koonin. 1989. Viral proteins containing the purine NTP-binding sequence pattern. Nucleic Acids Res. 17:8413-8440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorchakov, R., N. Garmashova, E. Frolova, and I. Frolov. 2008. Different types of nsP3-containing protein complexes in Sindbis virus-infected cells. J. Virol. 82:10088-10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffin, D. E. 2001. Alphaviruses, p. 917-962. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th edition. Lippincott Williams & Wilkins, Philadelphia, PA.
- 16.Guo, X., J. W. Carroll, M. R. Macdonald, S. P. Goff, and G. Gao. 2004. The zinc finger antiviral protein directly binds to specific viral mRNAs through the CCCH zinc finger motifs. J. Virol. 78:12781-12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo, X., J. Ma, J. Sun, and G. Gao. 2007. The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. Proc. Natl. Acad. Sci. U. S. A. 104:151-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hahn, Y. S., A. Grakoui, C. M. Rice, E. G. Strauss, and J. H. Strauss. 1989. Mapping of RNA− temperature-sensitive mutants of Sindbis virus: complementation group F mutants have lesions in nsP4. J. Virol. 63:1194-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hahn, Y. S., E. G. Strauss, and J. H. Strauss. 1989. Mapping of RNA− temperature-sensitive mutants of Sindbis virus: assignment of complementation groups A, B, and G to nonstructural proteins. J. Virol. 63:3142-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamer, G., and P. Argos. 1984. Primary structural comparison of RNA-dependent polymerases from plant, animal and bacterial viruses. Nucleic Acids Res. 12:7269-7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kedersha, N. L., M. Gupta, W. Li, I. Miller, and P. Anderson. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J. Cell Biol. 147:1431-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kerns, J. A., M. Emerman, and H. S. Malik. 2008. Positive selection and increased antiviral activity associated with the PARP-containing isoform of human zinc-finger antiviral protein. PLoS Genet. 4:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Law, L. M., R. Duncan, A. Esmaili, H. L. Nakhasi, and T. C. Hobman. 2001. Rubella virus E2 signal peptide is required for perinuclear localization of capsid protein and virus assembly. J. Virol. 75:1978-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemm, J. A., and C. M. Rice. 1993. Assembly of functional Sindbis virus RNA replication complexes: requirement for coexpression of P123 and P34. J. Virol. 67:1905-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lemm, J. A., and C. M. Rice. 1993. Roles of nonstructural polyproteins and cleavage products in regulating Sindbis virus RNA replication and transcription. J. Virol. 67:1916-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lemm, J. A., T. Rumenapf, E. G. Strauss, J. H. Strauss, and C. M. Rice. 1994. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus- and plus-strand RNA synthesis. EMBO J. 13:2925-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lenschow, D. J., N. V. Giannakopoulos, L. J. Gunn, C. Johnston, A. K. O'Guin, R. E. Schmidt, B. Levine, and H. W. Virgin III. 2005. Identification of interferon-stimulated gene 15 as an antiviral molecule during Sindbis virus infection in vivo. J. Virol. 79:13974-13983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, G. P., M. W. La Starza, W. R. Hardy, J. H. Strauss, and C. M. Rice. 1990. Phosphorylation of Sindbis virus nsP3 in vivo and in vitro. Virology 179:416-427. [DOI] [PubMed] [Google Scholar]
- 29.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 30.Lindenbach, B. D., and C. M. Rice. 1997. trans-Complementation of yellow fever virus NS1 reveals a role in early RNA replication. J. Virol. 71:9608-9617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, L., G. Chen, X. Ji, and G. Gao. 2004. ZAP is a CRM1-dependent nucleocytoplasmic shuttling protein. Biochem. Biophys. Res. Commun. 321:517-523. [DOI] [PubMed] [Google Scholar]
- 32.Lopez, S., J. S. Yao, R. J. Kuhn, E. G. Strauss, and J. H. Strauss. 1994. Nucleocapsid-glycoprotein interactions required for assembly of alphaviruses. J. Virol. 68:1316-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacDonald, M. R., E. S. Machlin, O. R. Albin, and D. E. Levy. 2007. The zinc finger antiviral protein acts synergistically with an interferon-induced factor for maximal activity against alphaviruses. J. Virol. 81:13509-13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mi, S., and V. Stollar. 1991. Expression of Sindbis virus nsP1 and methyltransferase activity in Escherichia coli. Virology 184:423-427. [DOI] [PubMed] [Google Scholar]
- 35.Mizushima, S., and S. Nagata. 1990. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 18:5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Müller, S., P. Möller, M. J. Bick, S. Wurr, S. Becker, S. Günther, and B. M. Kümmerer. 2007. Inhibition of filovirus replication by the zinc finger antiviral protein. J. Virol. 81:2391-2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pavan, A., L. V. Lotti, M. R. Torrisi, G. Migliaccio, and S. Bonatti. 1987. Regional distribution of Sindbis virus glycoproteins on the plasma membrane of infected baby hamster kidney cells. Exp. Cell Res. 168:53-62. [DOI] [PubMed] [Google Scholar]
- 38.Remy, I., and S. W. Michnick. 2006. A highly sensitive protein-protein interaction assay based on Gaussia luciferase. Nat. Methods 3:977-979. [DOI] [PubMed] [Google Scholar]
- 39.Rice, C. M., R. Levis, J. H. Strauss, and H. V. Huang. 1987. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J. Virol. 61:3809-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryman, K. D., and W. B. Klimstra. 2008. Host responses to alphavirus infection. Immunol. Rev. 225:27-45. [DOI] [PubMed] [Google Scholar]
- 41.Ryman, K. D., K. C. Meier, E. M. Nangle, S. L. Ragsdale, N. L. Korneeva, R. E. Rhoads, M. R. MacDonald, and W. B. Klimstra. 2005. Sindbis virus translation is inhibited by a PKR/RNase L-independent effector induced by alpha/beta interferon priming of dendritic cells. J. Virol. 79:1487-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scheidel, L. M., R. K. Durbin, and V. Stollar. 1987. Sindbis virus mutants resistant to mycophenolic acid and ribavirin. Virology 158:1-7. [DOI] [PubMed] [Google Scholar]
- 43.Shirako, Y., and J. H. Strauss. 1994. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J. Virol. 68:1874-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strauss, E. G., and J. H. Strauss. 1986. Structure and replication of the alphavirus genome, p. 35-90. In S. Schlesinger and M. J. Schlesinger (ed.), The Togaviridae and Flaviviridae. Plenum Publishing Corp., New York, NY.
- 45.Strauss, E. G., C. M. Rice, and J. H. Strauss. 1984. Complete nucleotide sequence of the genomic RNA of Sindbis virus. Virology 133:92-110. [DOI] [PubMed] [Google Scholar]
- 46.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, Y., C. W. Burke, K. D. Ryman, and W. B. Klimstra. 2007. Identification and characterization of interferon-induced proteins that inhibit alphavirus replication. J. Virol. 81:11246-11255. [DOI] [PMC free article] [PubMed] [Google Scholar]