Abstract
Measles is a highly contagious human disease caused by measles virus (MeV) and remains the leading cause of death in children, particularly in developing countries. Wild-type MeV preferentially infects lymphocytes by using signaling lymphocytic activation molecule (SLAM), whose expression is restricted to hematopoietic cells, as a receptor. MeV also infects other epithelial and neuronal cells that do not express SLAM and causes pneumonia and diarrhea and, sometimes, serious symptoms such as measles encephalitis and subacute sclerosing panencephalitis. The discrepancy between the tissue tropism of MeV and the distribution of SLAM-positive cells suggests that there are unknown receptors other than SLAM for MeV. Here we identified CD147/EMMPRIN (extracellular matrix metalloproteinase inducer), a transmembrane glycoprotein, which acts as a receptor for MeV on epithelial cells. Furthermore, we found the incorporation of cyclophilin B (CypB), a cellular ligand for CD147, in MeV virions, and showed that inhibition of CypB incorporation significantly attenuated SLAM-independent infection on epithelial cells, while it had no effect on SLAM-dependent infection. To date, MeV infection was considered to be triggered by binding of its hemagglutinin (H) protein and cellular receptors. Our present study, however, indicates that MeV infection also occurs via CD147 and virion-associated CypB, independently of MeV H. Since CD147 is expressed in a variety of cells, including epithelial and neuronal cells, this molecule possibly functions as an entry receptor for MeV in SLAM-negative cells. This is the first report among members of the Mononegavirales that CD147 is used as a virus entry receptor via incorporated CypB in the virions.
Measles, a highly contagious and very serious human disease, remains a leading cause of death in children, particularly in developing countries, despite the availability of a safe and effective vaccine for the past 40 years (4). Measles is caused by measles virus (MeV), a member of the genus Morbillivirus in the family Paramyxoviridae. The hemagglutinin (H) protein of this virus binds to cellular receptors, while its fusion (F) protein mediates the fusion of the virion and plasma membrane, leading to viral infection.
Thus, far, 2 molecules—CD46 (9) and signaling lymphocyte activation molecule (SLAM) (33)—have been identified as MeV receptors. CD46 is expressed on all human nucleated cells; however, only vaccine strains such as the Edmonston strain utilize CD46 as a receptor. The major receptor for wild-type strains is SLAM. SLAM expression is restricted to hematopoietic cells such as lymphocytes and macrophages—target cells of MeV—resulting in leucopenia. However, MeV infection affects various tissues in the body, including the lungs, kidney, gastrointestinal tract, skin, vascular endothelium, and brain (13). MeV infection sometimes causes serious neuronal symptoms, such as acute demyelinating encephalomyelitis (ADEM) and subacute sclerosing panencephalitis (SSPE) (28), in the central nervous system. However, since the epithelial and neuronal cells involved do not express SLAM, it is unknown how MeV infects these cells and causes serious systemic symptoms. No candidate proteins acting as a receptor in epithelial and neuronal cells have yet been identified. MeV infectivity to epithelial and neuronal cells is 100- to 1,000-fold lower than that to SLAM-expressing immune cells (14); hence, the affinity of unknown receptors in the former cells for the MeV H protein is probably much lower than that of SLAM. This has made it difficult to identify the receptors.
Recently, it was reported that MeV N protein (MeV-N) associates with host cellular proteins, inducing immunological abnormalities and pathogenicity or mediating the immune response (27, 34). MeV-N binds to Fc receptors on B cells and dendritic cells and induces immunosuppression by inhibiting antibody production (27). Alternatively MeV-N binds to the interferon regulatory factor 3 (IRF3) and the virus-activated kinase and then activates signal cascades of innate immunity by phosphorylating IRF3 (34). In addition, we recently reported that MeV-N also binds to the p40 subunit of eukaryotic translation initiation factor 3 (eIF3-p40) and inhibits host translation (31). Since MeV-N is the most abundant among the viral proteins in infected cells, it must have more functions in MeV infection.
Cyclophilins A and B (CypA and CypB) (18, 24) are members of the immunophilin family of proteins, which are characterized by peptidyl-prolyl cis-trans isomerase (PPIase) activity and a tendency to bind with the immunosuppressant cyclosporine (CsA). CypA is a cytosolic protein that is partly secreted via the nonclassical secretion pathway (15). CypA interacts with apoptosis-inducing factor (AIF) and cooperates in apoptosis-associated chromatinolysis (5). CypB is mainly localized in the endoplasmic reticulum and is partly secreted and localized in the cytoplasm. CypB interacts with the somatolactogenic hormone prolactin and potentiates prolactin-induced proliferation, cell growth, and nuclear retrotransport of prolactin (29). In addition, extracellular CypA and CypB have been reported to be ligands for CD147, which is a multifunctional transmembrane protein involved in neural function, inflammation, and tumor invasion; they are also known to function as proinflammatory cytokines (26, 40).
Here we identified cyclophilins A and B as binding partners of MeV-N and showed that MeV utilizes CD147 (1) as a receptor on epithelial cells via CypB incorporated into virus particles.
MATERIALS AND METHODS
Antibodies.
Anti-CD46 mouse monoclonal antibody (Hycult-Biotechnologies), anti-SLAM mouse monoclonal antibody (Biolegend), anti-CD36 mouse monoclonal antibody (Abcam), anti-CD147 mouse monoclonal antibody (Ancell), anti-His6 rabbit polyclonal antibody (Santa Cruz), anti-CypA rabbit antiserum (Biomol), anti-CypB rabbit polyclonal antibody (Affinity BioReagents, Proteintech), and BD Living Colors A.V. mouse monoclonal antibody, which is an anti-enhanced green fluorescent protein (anti-EGFP) antibody (Clontech), were purchased from the indicated manufacturers. Anti-canine distemper virus N protein (anti-CDV-N) mouse monoclonal antibody 8G, which cross-reacts with MeV-N, was prepared in our laboratory (20).
Cells and viruses.
HEK293, HEK293-SLAM cells (32), which stably express marmoset SLAM, and CHO cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Sigma-Aldrich) with 5% fetal bovine serum (FBS). COBL-a (17) cells were cultured in RPMI (Sigma-Aldrich) with 10% FBS. B95a cells were cultured in RPMI with 5% FBS. Sf9 cells were cultured in Grace's insect cell culture medium (Sigma-Aldrich) with 10% FBS.
A wild-type strain, MeV-HL (16), and recombinant MeVs derived from MV-HL, MeV-EGFP (35) and MeV-luc, were propagated in B95a cells at 37°C in RPMI with 2% FBS. To prepare a high-titer virus solution, a 200-ml culture of B95a cells was infected with MeV at a multiplicity of infection (MOI) of <0.01, and at 18 h postinfection (hpi) for MeV-luc, 48 hpi for MeV-EGFP, and 32 hpi for MeV-HL, the medium of the infected cell culture was collected without disrupting cells and centrifuged with a JLA 10.500 rotor (Beckman Coulter) at 5,000 rpm at 4°C. The supernatant was ultracentrifuged with a type 19 rotor (Beckman Coulter) at 19,000 rpm at 4°C. The resultant pellet was dissolved with DMEM with 5% FBS. The viral titer was estimated using B95a cells.
Generation of luciferase-expressing MeV.
To create luciferase-expressing MeV, a previously established plasmid encoding the cDNA of the full-length genome of the wild-type HL strain of MeV was used (35). A firefly luciferase gene was inserted between the N and P genes; the result was designated pMV-LUC. The recombinant luciferase-expressing MeV-HL strain, namely, MeV-luc, was generated from pMV-LUC according to a previously described method (32). Briefly, HEK293 cells were placed in a 6-well culture dish, inoculated with a recombinant vaccinia virus (MVA-T7) for 1 hour, and then transfected with 1 μg of pMV-LUC per well and with 1 μg of pKSN1, 1 μg of pKSP, and 0.3 μg of pGEML per well, which expressed the N, P, and L proteins, respectively, under the control of the T7 promoter, using FuGENE6. After incubation for 3 days, the cells were cocultivated with B95a cells at a concentration of 2 × 106 cells per well and further incubated until extensive cytopathic effects were noted.
Expression and purification of His-tagged C-terminal fragment of MeV-N.
The tail region of MeV-N (N_TAIL) was amplified by PCR, using TaKaRa LA Taq polymerase (TaKaRa) with a specific primer pair corresponding to the N_TAIL region (5′-AAGCTAGCCACCATCATCATCATCATACTACTGAGGACAGGATCAGTA-3′ and 5′-AAGGATCCCTAGTCTAGAAGATCTCTGTCATTG-3′; restriction sites are underlined). The PCR fragment was inserted into the NheI/BamHI sites of pET21d vector (Novagen). The His-tagged N_TAIL construct was expressed in Rosetta Blue/pLysS (Stratagene) at 30°C for 4 h in the presence of 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) and purified with Ni-Sepharose 6 Fast Flow beads (GE Healthcare). Eluted protein was buffer exchanged with phosphate buffer (pH 7.4) and concentrated using Centricon Plus-20 devices (Millipore).
Proteomic analysis of MeV-N binding protein.
COBL-a cells were lysed with a lysis buffer (10 mM HEPES [pH 7.4], 0.15 M NaCl, 0.5% NP-40, protease inhibitor cocktails [BD Biosciences]). The lysate was incubated with 20 μg of His-tagged N_TAIL in 300 μl of binding buffer (10 mM HEPES [pH 7.4], 150 mM NaCl, 0.1% NP-40, protease inhibitor cocktails) at 4°C overnight. Next, 2 mg of the chemical cross-linker DTSSP (Pierce) was added to the mixture and incubated on ice for 2 h. Unreacted cross-linkers were inactivated by adding 30 μl of 1 M Tris-HCl (pH 8.0), and then NaCl and imidazole were added to the mixture at final concentrations of 0.5 M and 25 mM, respectively. Forty microliters of Ni-Sepharose 6 Fast Flow beads (GE Healthcare) was then added to the mixture to trap the complex of N_TAIL and cellular proteins. The beads were washed extensively with washing buffer 1 (10 mM HEPES [pH 7.4], 0.5 M NaCl, 25 mM imidazole, 1% Triton X-100) and then with washing buffer 2 (10 mM HEPES [pH 7.4], 0.5 M NaCl, 25 mM imidazole), followed by incubation with 100 μl of washing buffer 2 containing 200 mM dithiothreitol (DTT) at 4°C overnight. The supernatant, into which cellular proteins were released, was precipitated with trifluoroacetic acid. The resultant pellet was trypsinized and subjected to high-performance liquid chromatography (HPLC)-linked matrix-assisted laser desorption ionization-time-of-flight mass spectrometry (MALDI-TOF MS). Detected peptides were analyzed by the Mascot search engine.
GST pull-down assay.
The cDNA of human cyclophilin A (CypA) was obtained from total RNA of HEK293 cells by reverse transcription (RT) with Superscript II reverse transcriptase (Invitrogen), followed by PCR with TaKaRa LA Taq polymerase, using a specific primer pair corresponding to CypA cDNA (5′-AAGAATTCCCATGGTCAACCCCACCGTGTTCTTCGAC-3′ and 5′-AACTCGAGTCATTCGAGTTGTCCACAGTCAGCAATGGTGATC-3′; restriction sites are underlined). The cDNA of human cyclophilin B (CypB) was obtained from a human lymphocyte cDNA library (Clontech) by PCR with Phusion polymerase (New England Biolabs), using a specific primer pair corresponding to CypB cDNA (5′-TTGAATTCGGGATGAGAAGAAGAAGGGGCCCAAAG-3′ and 5′-TTCTCGAGCTACTCCTTGGCGATGGCAAAG-3′; restriction sites are underlined). The amplified cDNAs were inserted into the EcoRI/XhoI sites of pGEX-4T2 vector (GE Healthcare). Glutathione S-transferase (GST)-fused CypA and GST-fused CypB were expressed for 6 h at 30°C in the presence of 0.5 mM IPTG in Escherichia coli BL21(DE3) and then purified with glutathione-Sepharose 4B beads (GE Healthcare). GST was obtained by expressing empty pGEX-4T2 vector. The gene for the full-length MeV-N protein fused to a His6 tag at its C terminus was amplified by PCR with a specific primer pair (5′-AACTGCAGATGGCCACACTTTTGAGGAGCTTAGC-3′ and 5′-AAGCTAGCCTAATGATGATGATGATGATGATGATGGTCTAGAAGATCTCTGTCATTGTATAC-3′; restriction sites are underlined). The MeV-N gene was inserted into the XbaI/PstI sites of pVL1392 vector (Invitrogen). Sf9 cells were transfected with this plasmid and Baculogold linearized vector (BD Biosciences) by use of Lipofectin (Invitrogen) to obtain recombinant baculovirus. His-tagged full-length MeV-N protein was expressed by infecting Sf9 cells with the recombinant baculovirus and then was purified with Ni-Sepharose beads. Two micrograms of MeV-N was incubated with 2 μg of GST-CypA, GST-CypB, or GST in 50 mM phosphate-buffered saline (PBS; pH 7.4) containing 0.5% NP-40 on ice for 30 min, and then 40 μl glutathione-Sepharose beads was added to each sample. After another 30 min of incubation, each sample was washed five times with PBS containing 0.5% NP-40 and developed by SDS-PAGE. In assays using cyclosporine (Calbiochem), a 5 μM final concentration was used for CypA, and 5, 15, and 30 μM final concentrations were used for CypB. MeV-N was detected with anti-His6 antibody (Santa Cruz) by Western blotting.
Purification of virus particles by sucrose density gradient centrifugation.
The culture medium of MeV-infected B95a cells was collected without disrupting cells and centrifuged with a JLA 10.500 rotor at 5,000 rpm at 4°C. The supernatant was ultracentrifuged with a type 19 rotor (Beckman Coulter) at 19,000 rpm at 4°C, and the resultant pellet was resuspended with TN buffer (10 mM Tris-HCl [pH 8.0], 0.15 M NaCl). The suspension was ultracentrifuged with an SW-41Ti rotor (Beckman Coulter) at 25,000 rpm at 4°C on a 20% to 60% sucrose gradient (the volume of each sucrose layer was 1 ml). Visible bands were collected, or 600-μl fractions were collected sequentially from the bottom of the tube. Each fraction was analyzed by titration and Western blotting.
Western blot analysis.
Ten microliters of each fraction from the sucrose gradient was diluted with Laemmli buffer and electrophoresed in an SDS-12% polyacrylamide gel, followed by transfer to a polyvinylidene difluoride (PVDF) membrane (Millipore). The membrane was incubated with a primary antibody (1:1,000), followed by incubation with a horseradish peroxidase-conjugated secondary antibody (Dako). Signals were detected by enhanced chemiluminescence (Amersham ECL Advance Western blot detection kit; GE Healthcare) using an LAS-1000 image analyzer (Fujifilm) and were calculated by ImageGauge (Fujifilm).
Analysis of cytoplasmic fraction of MeV-infected B95a cells.
B95a cells (1.5 × 106 cells on a 6-mm dish) were infected with MeV-luc at an MOI of 0.2. At 18 hpi, cells were washed with PBS and then suspended in 1 ml of hypotonic buffer (10 mM HEPES [pH 7.4], 10 mM KCl). After incubation on ice for 30 min, NP-40 (final concentration, 0.6%; Sigma-Aldrich) was added and the sample was vortexed, followed by centrifugation. The supernatant was analyzed as the cytoplasmic fraction by Western blotting.
Indirect immunofluorescence staining.
B95a cells (3 × 105 cells/well in a 24-well plate) were infected with MeV-luc at an MOI of 0.2. At 18 hpi, cells were fixed with 2% paraformaldehyde at room temperature for 10 min and then permeabilized with PBS plus 0.5% Triton X-100 at room temperature for 5 min. After that, cells were incubated with anti-CDV-N mouse monoclonal antibody (1:100) and anti-CypA (1:100) or -CypB (1:50) (Proteintech) rabbit polyclonal antibody, followed by incubation with Alexa 568-conjugated goat anti-mouse IgG and Alexa 488-conjugated goat anti-rabbit IgG polyclonal antibodies (Invitrogen). Cells were observed by confocal fluorescence microscopy.
Flow cytometry.
HEK293 cells or CHO cell lines were stripped with 2 mM EDTA and then pelleted by centrifugation at 800 × g. The pelleted cells were resuspended with sample buffer (PBS plus 3% FBS plus 0.02% NaN3). A total of 1 × 106 cells were incubated with 1 μg of a mouse monoclonal primary antibody in 300 μl of sample buffer on ice for 1 hour and then washed three times with sample buffer, followed by incubation with 0.5 μl of Alexa 488-conjugated goat anti-mouse IgG polyclonal secondary antibody in 300 μl of sample buffer on ice for 1 hour. After being washed five times with sample buffer, cells were applied to a BD FACScan flow cytometer (BD Biosciences) and analyzed using Cellquest software (BD Biosciences).
Blocking experiments using viral infection with antibodies or recombinant CypB.
For blocking experiments with antibodies, HEK293 cells (6 × 104 cells/well in a 96-well plate) were preincubated with 50 μl of medium containing 50 μg/ml of a blocking antibody at room temperature for 30 min, and then 50 μl of medium containing 50 μg/ml of the blocking antibody and MeV-luc at an MOI of 1.0 was added to cells. The MOI corresponded to that estimated for B95a cells. After further incubation at 37°C for 1 hour, cells were washed three times with medium containing the fusion block peptide z-Phe-Phe-Gly (Sigma-Aldrich) to stop the infection and to remove viruses and the antibody. At 24 hpi, the luciferase activity of infected cells was measured using the Pikkagene system (Toyo Ink).
Blocking experiments with recombinant CypB were performed by the same procedure, using 0, 30, or 60 μg/ml of recombinant CypB (Abcam) instead of blocking antibodies.
Establishment of CHO cell line stably expressing CD147.
The zeocin resistance gene was amplified from pcDNA4/HisMax vector (Invitrogen) by PCR and replaced with the simian virus 40 (SV40) ori of pCAGGS vector (37) to construct pCAGzc, which is a pCAGGS derivative vector possessing zeocin resistance. The cDNA of human CD147 was obtained from total RNA of HEK293 cells by RT with Superscript II reverse transcriptase (Invitrogen), followed by PCR with Phusion polymerase, using a specific primer pair corresponding to human CD147 cDNA (5′-TTGAATTCACCATGGCGGCTGCGCTGTTCGTG-3′ and 5′-TTGGATCCTCAGGAAGAGTTCCTCTGGCGGA-3′; restriction sites are underlined). The PCR product was inserted into the EcoRI/BglII sites of pCAGzc to construct the expression plasmid pCAGzc-CD147. CHO-K1 cells were transfected with pCAGzc-CD147, using Lipofectamine LTX (Invitrogen), and selected by use of 300 μg/ml of zeocin (Invivogen). Several zeocin-resistant clones were picked, and CD147 expression was confirmed by immunohistochemistry using anti-CD147 antibody. In the same way, another CHO cell line carrying an empty vector, pCAGzc, was established as a negative control.
Infection assay on CHO cells.
CHO/CD147 and CHO/pCAG cells were seeded at 0.3 × 105 cells/well in a 96-well plate. Cells were incubated with 100 μl of medium containing MeV-luc at an MOI of 4.0 at 37°C for 1 hour. The MOI corresponded to that estimated for B95a cells. After incubation, cells were washed three times with medium containing fusion block peptide to stop the infection and to remove viruses. At 48 hpi, the luciferase activity of infected cells was measured using the Pikkagene system (Toyo Ink). To confirm that human CD147 supported MeV infection on CHO/CD147 cells, CHO/CD147 and CHO/pCAG cells (0.3 × 105 cells) were preincubated with 50 μg/ml of anti-CD147 antibody prior to MeV-luc infection at an MOI of 4.0. After infection, cells were washed and the luciferase activity was measured as described above.
RESULTS
Proteomic analysis of MeV-N binding proteins.
To identify cellular proteins that bind to MeV-N, the His-tagged C-terminal region of the N protein of a wild-type MeV-HL strain was incubated with the cell lysate of COBL-a cells, which are human umbilical blood cells susceptible to MeV infection (17). The complexes were trapped with Ni-Sepharose beads, and binding proteins were analyzed by MALDI-TOF MS. Among the many proteins assigned, 4 peptides derived from CypA and 1 peptide from CypB were detected, and CypA (Fig. 1A and C) and CypB (Fig. 1B and D) were identified based on their high total scores. Thus, CypA and CypB were considered candidate cellular binding proteins of MeV-N.
FIG. 1.
CypA and CypB are identified as MeV-N-interacting proteins by proteomic analysis. The C-terminal region of MeV-N, which is usually called the tail region, was used as a bait protein, and N_TAIL-binding proteins were pulled down from the COBL-a cell lysate. The binding proteins were trypsinized in solution without separation by SDS-PAGE and were subjected to MALDI-TOF MS. Detected ions were analyzed using the Mascot search engine. Amino acid sequences of CypA (A) and CypB (B) are shown. Peptides detected by MALDI-TOF MS are exhibited in red. Scores for each ion from CypA (C) and CypB (D) are shown, and individual ion scores of >36 indicate identity and extensive homology.
CypA and -B belong to the cyclophilin family, whose members are characterized as CsA binding proteins and possess a PPIase activity (18, 24).
Confirmation of direct binding of MeV-N and cyclophilins by GST pull-down assay.
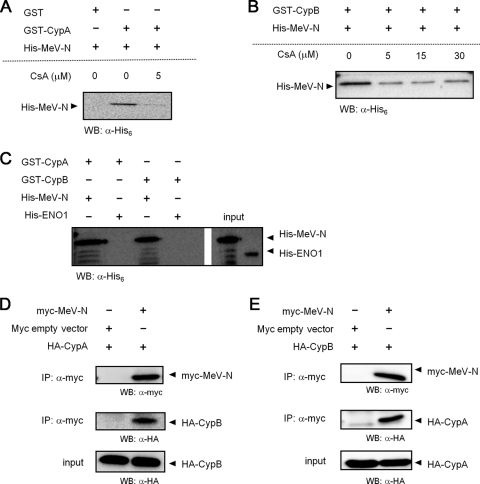
To confirm the binding of MeV-N, recombinant GST-CypA, GST-CypB, or GST alone was incubated with purified His-tagged full-length MeV-N. MeV-N was pulled down when it was incubated with GST-CypA (Fig. 2A, second lane) or GST-CypB (Fig. 2B, first lane); however, MeV-N was not pulled down when it was incubated with GST alone (Fig. 2A, first lane). When an irrelevant His-tagged protein, His-tagged human enolase 1 (His-ENO1), containing 15 proline residues, was incubated with GST-CypA or GST-CypB, His-ENO1 was not pulled down; this finding indicates that MeV-N specifically binds to CypA and CypB (Fig. 2C). Since CsA binds to the PPIase domain of cyclophilins and prevents cyclophilin binding to substrate proteins, the effects of CsA on the binding of CypA and CypB to MeV-N were examined. A concentration of 5 μM CsA completely abolished the CypA binding to MeV-N (Fig. 2A, third lane), which indicated that CypA bound to MeV-N on its PPIase domain. On the other hand, 5 μM CsA showed only a partial inhibition (about 60% inhibition) of the CypB binding to MeV-N, and higher concentrations of CsA (15 and 30 μM) did not show any additive inhibition of CypB binding compared with the 5 μM concentration of CsA (Fig. 2B, second to fourth lanes). While it is known that the binding affinity of CypB for CsA is almost equal to that of CypA, it is unclear why CypB bound with MeV-N in spite of the high concentration of CsA. We speculate that CypB possesses multiple binding sites for MeV-N, in addition to the PPIase domain.
FIG. 2.
GST pull-down and coimmunoprecipitation (IP) assays showing direct binding of CypA and CypB to MeV-N. We incubated 2 μg of His-tagged MeV-N with 2 μg of GST-CypA (A), GST-CypB (B), or GST (A) for 30 min on ice, and then glutathione-Sepharose beads were added to each sample. After washing of the beads, trapped proteins were analyzed by SDS-PAGE followed by Western blotting (WB). To detect MeV-N, anti-His6 antibody was used as the primary antibody. For assays using CsA, a 5 μM final concentration was used for CypA and 5, 15, and 30 μM final concentrations were used for CypB. The numbers in the figure show the final concentration of CsA in each sample. (C) We incubated 2 μg of His-tagged MeV-N or His-tagged ENO1 with 2 μg of GST-CypA or GST-CypB. GST pull-down assays were performed according to the procedure described above. (D and E) We transiently transfected 1 μg of pCMV-myc-MeV-N with 1 μg of pCMV-HA-CypA (E) or pCMV-HA-CypB (D) into HEK293 cells (1.5 × 106 cells). At 24 h posttransfection, the cells were lysed, and 0.5 μg of anti-myc monoclonal antibody and protein G Sepharose beads were added to the lysate. After washing of the beads, trapped proteins were analyzed by SDS-PAGE followed by Western blotting with anti-myc or anti-HA polyclonal antibody.
Binding of CypA and CypB to MeV-N was also confirmed by using a coimmunoprecipitation assay. HA-tagged CypA or CypB was coexpressed with myc-tagged MeV-N by transient transfection into HEK293 cells. At 24 h posttransfection, myc-MeV-N was immunoprecipitated from the cell lysate by using the anti-myc antibody. The immunoprecipitates were subjected to SDS-PAGE followed by Western blotting with anti-HA antibody. HA-CypA and -CypB were coimmunoprecipitated only when they were coexpressed with myc-MeV-N (Fig. 2D and E).
Incorporation of cyclophilins into MeV particles.
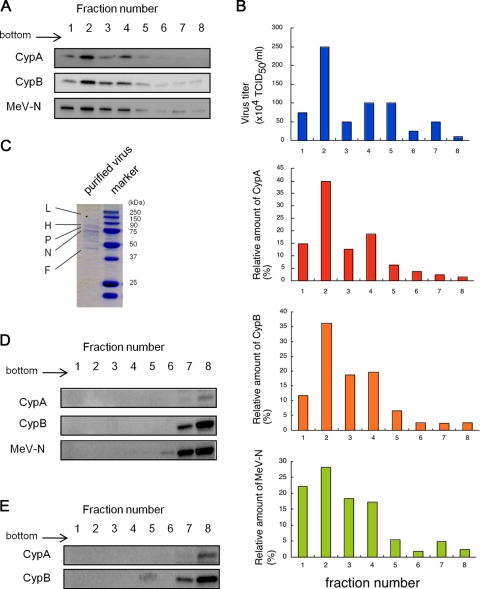
It was reported that CypA and CypB also bind to HIV-1 nucleocapsids (19). CypA is specifically incorporated into HIV-1 particles (11, 36) and then translocates to the virion surface (22). HIV-1 with exposed CypA binds to CD147, a cell surface transmembrane protein (26), and enters cells by using CD147 as a receptor (25). On the other hand, CypB is not incorporated into HIV-1 particles, despite its binding to HIV-1 nucleocapsids (19). Therefore, we analyzed whether CypA and/or -B exists on MeV virions and whether CD147 acts as a receptor for MeV via CypA and/or CypB, both of which are known as ligands of CD147 (26, 38). To reveal incorporation of cyclophilins into MeV particles, viral particles were purified. Culture medium of MeV-HL-infected cells was ultracentrifuged without lysis, and the resultant pellet was resuspended in a buffer and ultracentrifuged on a 20% to 60% sucrose gradient (21). Eight fractions were collected sequentially from the bottom of the tube, and the virus titer and relative amount of cyclophilins in each fraction were determined by titration using B95a cells and Western blotting, respectively. The distributions of CypA and -B exhibited similar patterns to those for virus titer and presence of MeV-N (Fig. 3A and B). To confirm the purity of the virus particles, trichloroacetic acid was added to fractions containing MeV-N, and the resultant pellet was subjected to SDS-PAGE and Coomassie brilliant blue staining (Fig. 3C). While several bands corresponding to the viral structural proteins were detected, only a few uncharacterized bands were detected; this finding suggested that the virus particles had sufficiently high purity. Furthermore, to exclude the possibility of contamination by free cyclophilins and released MeV-N in the fractions, cell lysates of mock-infected and MeV-infected B95a cells were subjected to 20% to 60% sucrose density gradient centrifugation, and 8 fractions were collected in the same manner. Free cyclophilins and released MeV-N were not detected in fractions 1 to 5, where most virus particles were distributed, but were detected in lower-density fractions (Fig. 3D and E). These results showed that CypA and CypB were actually incorporated into the MeV-HL virions.
FIG. 3.
Evidence of incorporation of CypA and CypB into MeV particles. Culture medium of MeV-infected B95a cells was ultracentrifuged to pellet down the virus. The resultant pellet was resuspended with TN buffer and placed on a 20% to 60% sucrose layer (the volume of each sucrose layer was 1 ml). After ultracentrifugation, 600-μl fractions were collected sequentially from the bottom of the tube in the case of MeV-HL. (A) CypA, CypB, and MeV-N in each fraction of MeV-HL were detected by Western blotting. (B) The virus titer of each fraction was estimated using B95a cells. The virus titer and relative amounts of CypA, CypB, and MeV-N in each fraction of MeV-HL were graphed. (C) Trichloroacetic acid was added to fraction 2, and the resultant pellet was analyzed by SDS-PAGE and Coomassie brilliant blue staining. MeV-infected or mock-infected B95a cells were lysed with PBS containing 0.5% NP-40, and the cell lysates were placed on a 20% to 60% sucrose layer (the volume of each sucrose layer was 1 ml). After ultracentrifugation, 600-μl fractions were collected sequentially from the bottom of the tube. CypA, CypB, and MeV-N in each fraction of the MeV-infected cell lysate (D) and CypA and CypB in each fraction of the mock-infected cell lysate (E) were detected by Western blotting.
CypA incorporation into MeV is less efficient than that of CypB.
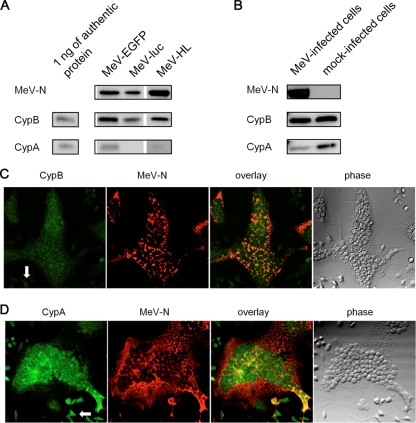
To facilitate the infection assay, we used a recombinant MeV expressing firefly luciferase (Luc), namely, MeV-luc, which was constructed using an MeV-HL-based reverse genetic system. When MeV-luc was subjected to sucrose density gradient centrifugation and then analyzed by Western blotting, CypA was rarely detected; this finding was different from that obtained in the case of MeV-HL. To confirm whether the difference was due to the insertion of a reporter gene into genomic RNA, we performed Western blotting and compared the incorporations of CypA and CypB into purified MeV-luc and MeV-EGFP, which was a recombinant MeV expressing EGFP (35). The incorporated CypA was observed in the particles of MeV-EGFP but not in those of MeV-luc, while the incorporated CypB was clearly observed in each of the viruses (Fig. 4A).
FIG. 4.
Incorporation of CypA or CypB into MeV. (A) MeV-HL, MeV-EGFP, and MeV-luc were purified by sucrose density gradient centrifugation. Virus particles were developed by SDS-PAGE and analyzed by Western blotting with anti-MeV-N, -CypA, and -CypB antibodies. (B) B95a cells were infected with MeV-luc at an MOI of 0.2. At 18 hpi, the cytoplasmic fraction of infected cells was extracted with hypotonic buffer. CypA, CypB, and MeV-N in the cytoplasmic fraction were detected by Western blotting. As a control, mock-infected B95a cells were used. (C and D) Localization of cyclophilins and MeV-N in MeV-luc-infected B95a cells was examined by immunofluorescence staining. CypB (C) and CypA (D) distributions (green; left panels), MeV-N distribution (red; second panels), their overlay (third panels), and phase-contrast images (right panels) are shown in striatal sections. Each panel represents sequential confocal scans of the same field. Arrows in the left panel indicate uninfected cells.
Since CypA is an abundant cytosolic protein, whereas CypB is localized mainly in the endoplasmic reticulum, the less-efficient incorporation of CypA than that of CypB was unpredicted. To elucidate the reason for this observation, the amounts of CypA and CypB in cytoplasmic fractions of MeV-infected and mock-infected B95a cells were examined by Western blotting. The cytoplasmic CypA level significantly decreased upon infection with MeV, although the cytoplasmic CypB level was not altered (Fig. 4B). Next, localization of CypA, CypB, and MeV-N in MeV-infected B95a cells was examined. MeV-N was localized mainly in the cytoplasm of syncytia (Fig. 4C and D, second panels). CypB was localized mainly in the perinuclear region and was also distributed almost equally in the cytoplasm and nuclei of syncytia (Fig. 4C, first panel) and colocalized with MeV-N in cytoplasm (Fig. 4C, third panel). In contrast, CypA was predominantly translocated into the nucleus (Fig. 4D, first panel). Furthermore, in the cytoplasm of syncytia, colocalization of CypA with MeV-N was not significant (Fig. 4D, third panel).
Cyclosporine inhibits CypB incorporation into MeV particles and reduces MeV infectivity to epithelial cells.
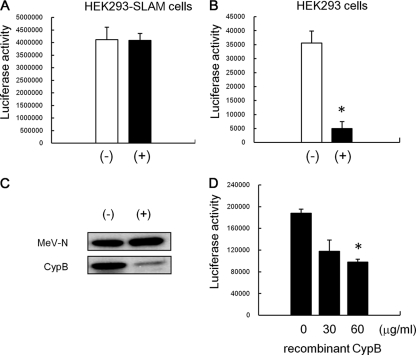
Since CsA inhibits incorporation of CypA into virions, HIV-1 treated with CsA significantly loses the ability to infect host cells (36). To examine the effects of cyclophilins on the infectivity of MeV, MeV-luc was propagated with and without 5 μg/ml of CsA, and then virions in culture medium of infected cells were purified by ultracentrifugation. The resultant pellets were rinsed and resuspended with fresh medium. The virus propagated without CsA was named MeV-luc (−), and that propagated with CsA was named MeV-luc (+). In this study, HEK293 cells, which are SLAM-negative cells, and HEK293-SLAM cells, which are HEK293-derivative cells stably expressing marmoset SLAM, were used in the infection assay. When HEK293 and HEK293-SLAM cells were infected with MeV-luc (+) or MeV-luc (−) at an MOI of 1.0, both MeV-luc (+) and MeV-luc (−) showed identical infectivities for the HEK293-SLAM cells (Fig. 5A), while the infectivity of MeV-luc (+) was significantly decreased, by 85%, compared with that of MeV-luc (−), on HEK293 cells (Fig. 5B). Since incorporation of CypB but not CypA was observed in MeV-luc, CypB was considered to be involved in the infection of SLAM-negative cells with MeV-luc.
FIG. 5.
Effect of CsA treatment on MeV infectivity to epithelial cells. MeV-luc was propagated with and without 5 μg/ml CsA, and then the culture medium of infected cells was ultracentrifuged to pellet down viruses. The resultant pellets were rinsed and resuspended with fresh medium. The virus propagated without CsA was named MeV-luc (−), and that propagated with CsA was named MeV-luc (+). MeV-luc (−) and MeV-luc (+) were titrated using B95a cells. HEK293-SLAM cells (A) and HEK293 cells (B) were infected with the viruses at an MOI of 1.0. At 24 hpi, the luciferase activity of infected cells was measured. The white and black bars represent the infectivities of MeV-luc (−) and MeV-luc (+), respectively. Luciferase assays were performed in triplicate. Data are means plus standard errors of the means (SEM). *, P = 0.003 (Student's t test). (C) MeV-luc (−) and MeV-luc (+) were further purified by sucrose gradient density centrifugation and were analyzed by Western blotting with anti-MeV-N, -CypA, and -CypB antibodies. (D) HEK293 cells (6 × 104 cells) were preincubated with 0, 30, or 60 μg/ml of recombinant CypB for 30 min at room temperature and then infected with MeV-luc at an MOI of 1.0 in the presence of 0, 30, or 60 μg/ml of recombinant CypB at 37°C for another hour. After incubation, cells were washed three times with medium containing fusion block peptide to stop infection and to remove viruses and recombinant CypB. The luciferase activity of infected cells was measured at 24 hpi. Luciferase assays were performed in triplicate. Data are means plus SEM. *, P = 0.0007 (Student's t test).
To confirm the effect of CsA on the incorporation of CypB into MeV particles, MeV-luc (−) and MeV-luc (+) were purified by sucrose density gradient centrifugation, and the purified MeV-luc (−) and MeV-luc (+) were analyzed by SDS-PAGE followed by Western blotting. CypB incorporation was significantly inhibited by CsA treatment (Fig. 5C).
Next, to examine further the involvement of CypB in MeV infection of epithelial cells, the effect of exogenous addition of CypB on MeV infection was examined. HEK293 cells were infected with MeV-luc at an MOI of 1.0 in the presence of 0, 30, or 60 μg/ml recombinant CypB. As a result, CypB inhibited MeV infection in a dose-dependent manner, by up to 50%, on HEK293 cells (Fig. 5D). These results indicated that CypB associating with virions plays a role in MeV infection of epithelial cells.
Contribution of CD147 to MeV infection of epithelial cells.
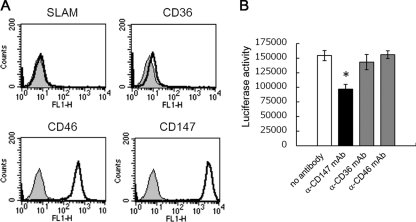
Since it has been reported that HIV-1 can infect target cells by using CD147 as a receptor via CypA on HIV-1 virions (25, 26) and that CypB also binds to CD147 (40), we analyzed the role of CD147 in MeV infection of epithelial cells. HEK293 cells express high levels of CD147 and CD46, while they do not express SLAM (Fig. 6A). HEK293 cells were infected with MeV-luc at an MOI of 1.0 in the presence of anti-CD147 or anti-CD46 monoclonal antibody or in the presence of anti-CD36 monoclonal antibody as a negative control. A total of 50 μg/ml of anti-CD147 antibody significantly reduced MeV-luc infectivity to HEK293 cells, by 40% (Fig. 6B), while the same concentration of anti-CD46 or -CD36 antibody showed no effect on MeV-luc infectivity. These results indicate that CD147 is involved in MeV infection of HEK293 cells.
FIG. 6.
Contribution of CD147 to MeV infection of epithelial cells. (A) HEK293 cells (1 × 106 cells) were incubated with 1 μg of anti-CD147, -CD36, -CD46, or -SLAM mouse monoclonal antibody for 1 hour. After being washed, cells were incubated with 0.5 μl of Alexa Fluor 468-conjugated goat anti-mouse IgG polyclonal antibody for 1 hour. After being washed, cells were analyzed by flow cytometry. (B) HEK293 cells (6 × 104 cells) were preincubated with 50 μg/ml of anti-CD147, -CD36, or -CD46 antibody for 30 min at room temperature and then infected with MeV-luc at an MOI of 1.0 in the presence of 50 μg/ml of each antibody at 37°C for another hour. After incubation, cells were washed three times with medium containing fusion block peptide to stop infection and to remove viruses and antibodies. The luciferase activity of infected cells was measured at 24 hpi. Luciferase assays were performed in triplicate. Data are means plus SEM. *, P = 0.020 (Student's t test).
Effect of overexpression of CD147 on MeV infection in CHO-K1 cells.
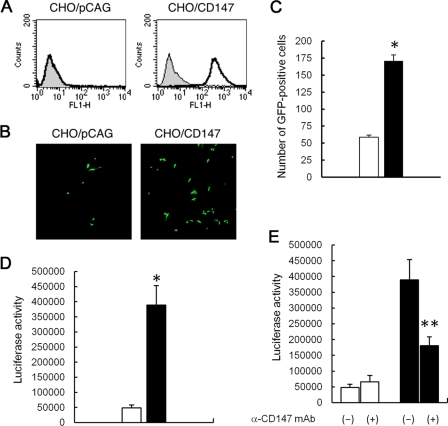
CHO-K1 cells are much less susceptible to MeV infection than are HEK293 cells. Thus, to confirm whether CD147 acts as an entry receptor for MeV, a CHO cell line that stably expresses human CD147 (CHO/CD147) was established by transfection of the human CD147 gene and subsequent selection. CHO/pCAG cells transfected with only the pCAGGS vector were used as negative controls. Flow cytometry analysis confirmed that CHO/CD147 cells expressed high levels of human CD147 (Fig. 7A). When CHO/CD147 and CHO/pCAG cells were infected with MeV-EGFP at an MOI of 4.0 and observed by confocal fluorescence microscopy, the number of GFP-positive cells significantly increased on CHO/CD147 cells (Fig. 7B and C). Similarly, when CHO/CD147 and CHO/pCAG cells were infected with MeV-luc at an MOI of 4.0, the luciferase activity of CHO/CD147 cells was eightfold higher than that of CHO/pCAG cells (Fig. 7D). Thus, stably expressed human CD147 on CHO cells facilitated MeV-luc infection. Furthermore, anti-CD147 antibody significantly inhibited MeV-luc infection of CHO/CD147 cells, while it had no effect on MeV-luc infection of CHO/pCAG cells (Fig. 7E). This evidence indicates that CD147 acted as an entry receptor for MeV.
FIG. 7.
Effect of overexpression of CD147 on MeV infection in CHO-K1 cells. (A) Expression of human CD147 on CHO/CD147 cells was confirmed by flow cytometry with anti-human CD147 antibody. (B) CHO/CD147 and CHO/pCAG cells (3 × 104 cells each) were incubated with MeV-EGFP at an MOI of 4.0 at 37°C for 1 hour. The MOI corresponded to that estimated for B95a cells. After the incubation, cells were washed three times with medium containing fusion block peptide. At 48 hpi, cells were observed by confocal fluorescence microscopy. (C) CHO/CD147 and CHO/pCAG cells (3 × 104 cells each) were incubated with MeV-EGFP at an MOI of 1.0 at 37°C for 1 hour. After incubation, the cells were washed thrice with a medium containing fusion block peptide. At 48 hpi, we counted the number of GFP-positive cells by using a confocal fluorescence microscope. Data are means plus SEM. *, P = 0.0004 (Student's t test). (D) CHO/CD147 and CHO/pCAG cells (3 × 104 cells each) were incubated with MeV-luc at an MOI of 4.0 at 37°C for 1 hour. After the incubation, cells were washed three times with medium containing fusion block peptide. The luciferase activity of infected cells was measured at 48 hpi. The white and black bars represent the infectivities to CHO/pCAG and CHO/CD147 cells, respectively. Luciferase assays were performed in triplicate. Data are means plus SEM. *, P = 0.022 (Student's t test). (E) CHO/CD147 and CHO/pCAG cells (3 × 104 cells each) were incubated with MeV-luc in the presence of anti-CD147 antibody at an MOI of 4.0 at 37°C for 30 min. After the incubation, cells were washed three times with medium containing fusion block peptide. The luciferase activity of infected cells was measured at 48 hpi. The white and black bars represent the infectivities to CHO/pCAG and CHO/CD147 cells, respectively. (−), infection without antibody, as a negative control; (+), infection with anti-CD147 antibody. Luciferase assays were performed in triplicate. Data are means plus SEM. **, P = 0.035 (Student's t test).
DISCUSSION
CypA and CypB belong to the cyclophilin family, whose members are known as CsA binding proteins and possess PPIase activity (18, 24). They are known as the most abundant cyclophilins in cells and have been implicated in the maturation of host cellular proteins. Recently, they were also reported to play important roles in the virus life cycle (3, 30, 38). CypA interacts with several proteins of HIV-1, vaccinia virus, and vesicular stomatitis virus (VSV) and is specifically incorporated into their particles (2, 7, 36). On the other hand, CypB is not incorporated into HIV-1 particles, although it interacts with the HIV-1 Gag protein, as does CypA (3, 19). In this study, we revealed that MeV-N also interacted with CypA and CypB (Fig. 2) and that the cyclophilin inhibitor CsA prevented CypA binding completely but CypB binding only partially, which indicated that CypA bound to MeV-N in a PPIase domain-dependent manner, while CypB bound in both a PPIase domain-dependent and -independent manner. Western blot analysis of MeV-HL particles purified by sucrose density gradient centrifugation showed that both CypA and CypB were incorporated into MeV particles, unlike the case for HIV-1 (Fig. 3). However, MeV-HL and other recombinant MeVs incorporated CypA less efficiently than CypB. Immunofluorescence staining of MeV-infected B95a cells showed that CypA was translocated into the nucleus and that MeV-N seemed colocalized more efficiently with CypB than with CypA in the cytoplasm of syncytia (Fig. 4C and D). In fact, the amount of CypA was much lower than that of CypB in the cytoplasmic fraction of MeV-infected B95a cells (Fig. 4B). A recent study reported that CypA interacted with AIF and was translocated into the nucleus at the early stage of apoptosis (5). Because MeV infection induces apoptosis in infected cells (10), it might trigger CypA translocation into the nuclei of MeV-infected cells. These pieces of evidence may be among the reasons that CypA incorporation into MeV was less efficient than that of CypB. In this study, we examined cyclophilin incorporation into strains MeV-luc, MeV-EGFP, and parental MeV-HL; interestingly, we found that the incorporation rates of CypA and CypB were different (Fig. 4A). Although the exact reason underlying this finding is still unclear, we speculate that this difference can be attributed to a difference in the growth rates of the three strains. Because of the differences in the growth rates of the three strains, the reasons for which are unknown, the time points of virus harvest were different. MeV-luc was harvested at 18 hpi, MeV-HL at 32 hpi, and MeV-EGFP at 48 hpi. As described above, CypA was translocated into the nucleus at 18 hpi in most syncytia (Fig. 4D); however, in older syncytia, nuclear accumulation of CypA was not significant, which may be attributed to redistribution into the cytoplasm (data not shown). Because of the localization of MeV-N in the cytoplasm of the syncytia and the delay in harvesting, CypA could have bound with MeV-N in the cytoplasm of the syncytia and could have been incorporated into the virus particles.
In the case of HIV-1, virus particles which are generated in the presence of the cyclophilin inhibitor CsA lack CypA incorporation because CsA abolishes the interaction between Gag protein and CypA (40). Similar to this, CypB incorporation into MeV-luc particles was significantly prevented by CsA treatment, which indicated that CypB was specifically incorporated into MeV particles by binding to MeV-N (Fig. 5C). Furthermore, MeV propagated in the presence of CsA significantly lost infectivity to HEK293 cells, by 90% (Fig. 5B), while the virus retained the same infectivity to HEK293-SLAM cells (Fig. 5A). SLAM-positive cells have a much higher susceptibility to MeV than do SLAM-negative cells. In fact, the ratio of the infectivity of MeV for SLAM-negative HEK293 cells to that for HEK293-SLAM cells was approximately 1 to 100. Thus, since SLAM is a strong receptor for MeV infection, the effect of CypB on SLAM-independent infection may not have appeared on HEK293-SLAM cells. In addition, recombinant CypB inhibited MeV infection by 50% on HEK293 cells (Fig. 5D). In these SLAM-negative cells, CypB is considered to play an essential role in MeV infection.
It is known that CypA incorporated into HIV-1 particles translocates to the surfaces of virions by an unknown mechanism (22). We tried to confirm the existence of CypB on MeV virions by immunoelectron microscopy, but commercially available anti-CypB antibody failed to detect CypB on the virions. This may have been due to the quality of the antibody or to the limited amount of CypB on the virions, because the sensitivity of immunoelectron microscopy is rather low.
CD147 is a transmembrane glycoprotein (1) expressed by various cell types, including epithelial, endothelial, and neuronal cells. CD147 is known to act as a signaling receptor for extracellular CypA and CypB, and it regulates lymphocyte responsiveness (39). It has been reported that the interaction between CypA and CD147 enables HIV-1 to infect target cells via CD147, independently of the binding of gp120 and CD4 (25). Additionally, severe acute respiratory syndrome coronavirus (SARS-CoV) is proposed to use CD147 as a receptor in the same manner as HIV-1, because its nucleocapsid binds to CypA and a CD147-blocking peptide inhibited SARS-CoV infection (8). In this study, although the neutralizing activity of the anti-CD147 antibody used in the present study was not high, showing only 50% inhibition of HIV-1 infection (25), it inhibited MeV infection by 40% on HEK293 cells (Fig. 6), showing that MeV also uses CD147 as a receptor, similar to HIV-1. However, unlike HIV-1 and SARS-CoV, MeV seems to use CypB instead of CypA for binding to CD147. This finding is the first among viruses belonging to the order Mononegavirales and shows a new infection mode of MeV which is independent of H protein. Furthermore, since the infection mode in this study has been reported for other viruses, it can be extended beyond families of enveloped viruses.
Although we recently reported that a heparin-like molecule is involved in morbillivirus infection in various cells (12, 35), thus far no specific protein on the surfaces of epithelial cells has been identified as a receptor for morbilliviruses. It has been reported that CypB interacts with two types of cell surface binding sites on T lymphocytes (6). The first site is CD147, and the second site is cell surface heparan sulfate, which was recently identified as syndecan-1 (23). It has been reported that syndecan-1 physically associates with CD147 and is required for CypB-mediated signaling, and it was proposed that CypB binding to the heparan sulfate moiety of syndecan-1 would complement a low-affinity interaction with CD147 (23). Syndecan-1 is also expressed by various cell types, including epithelial cells and endothelial cells. These reports strongly support the new infection mode of MeV, in which CypB and CD147 are involved.
Since CD147 is expressed on various cells, including epithelial and neuronal cells, our finding may provide a clue to understanding the mechanisms by which MeV causes severe symptoms of respiratory and gastrointestinal infections and central nervous system complications. Other mammals, such as mice, also express CD147; however, the CD147 proteins in mice and humans exhibit very little homology. The amino acid identity between mouse and human CD147 proteins is only 65%. Therefore, CD147-mediated infection may show species specificity. Establishment of a human CD147-expressing mouse model would be beneficial to reveal this point and to reveal the contribution of the infection mode to MeV pathogenicity.
Acknowledgments
We thank H. Fukuda and S. Ohmi, The Institute of Medical Sciences, The University of Tokyo, for MALDI-TOF MS analysis.
This study was supported by a grant-in-aid from the Ministry of Education, Science, Culture and Sports, Japan.
Footnotes
Published ahead of print on 10 February 2010.
REFERENCES
- 1.Biswas, C., Y. Zhang, R. DeCastro, H. Guo, T. Nakamura, H. Kataoka, and K. Nabeshima. 1995. The human tumor cell-derived collagenase stimulatory factor (renamed EMMPRIN) is a member of the immunoglobulin superfamily. Cancer Res. 55:434-439. [PubMed] [Google Scholar]
- 2.Bose, S., M. Mathur, P. Baltes, N. Joshi, and A. K. Banerjee. 2003. Requirement for cyclophilin A for the replication of vesicular stomatitis virus New Jersey serotype. J. Gen. Virol. 84:1687-1699. [DOI] [PubMed] [Google Scholar]
- 3.Braaten, D., E. K. Franke, and J. Luban. 1996. Cyclophilin A is required for an early step in the life cycle of human immunodeficiency virus type 1 before the initiation of reverse transcription. J. Virol. 70:3551-3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryce, J., C. Boschi-Pinto, K. Shibuya, and R. E. Black. 2005. WHO estimates of the causes of death in children. Lancet 365:1147-1152. [DOI] [PubMed] [Google Scholar]
- 5.Cande, C., N. Vahsen, I. Kouranti, E. Schmitt, E. Daugas, C. Spahr, J. Luban, R. T. Kroemer, F. Giordanetto, C. Garrido, and J. M. Penninger. 2004. AIF and cyclophilin A cooperate in apoptosis-associated chromatinolysis. Oncogene 23:1514-1521. [DOI] [PubMed] [Google Scholar]
- 6.Carpentier, M., F. Allain, B. Haendler, A. Denys, C. Mariller, M. Benaïssa, and G. Spik. 1999. Two distinct regions of cyclophilin B are involved in the recognition of a functional receptor and of glycosaminoglycans on T lymphocytes. J. Biol. Chem. 274:10990-10998. [DOI] [PubMed] [Google Scholar]
- 7.Castro, A. P., T. M. Carvalho, N. Moussatche, and C. R. Damaso. 2003. Redistribution of cyclophilin A to viral factories during vaccinia virus infection and its incorporation into mature particles. J. Virol. 77:9052-9068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, Z., L. Mi, J. Xu, J. Yu, X. Wang, J. Jiang, J. Xing, P. Shang, A. Qian, Y. Li, P. X. Shaw, J. Wang, S. Duan, J. Ding, C. Fan, Y. Zhang, Y. Yang, X. Yu, Q. Feng, B. Li, X. Yao, Z. Zhang, L. Li, X. Xue, and P. Zhu. 2005. Function of HAb18G/CD147 in invasion of host cells by severe acute respiratory syndrome coronavirus. J. Infect. Dis. 191:755-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorig, R. E., A. Marcil, A. Chopra, and C. D. Richardson. 1993. The human CD46 molecule is a receptor for measles virus (Edmonston strain). Cell 75:295-305. [DOI] [PubMed] [Google Scholar]
- 10.Esolen, L. M., S. W. Park, J. M. Hardwick, and D. E. Griffin. 1995. Apoptosis as a cause of death in measles virus-infected cells. J. Virol. 69:3955-3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franke, E. K., H. E. Yuan, and J. Luban. 1994. Specific incorporation of cyclophilin A into HIV-1 virions. Nature 372:359-362. [DOI] [PubMed] [Google Scholar]
- 12.Fujita, K., R. Miura, M. Yoneda, F. Shimizu, H. Sato, Y. Muto, Y. Endo, K. Tsukiyama-Kohara, and C. Kai. 2007. Host range and receptor utilization of canine distemper virus analyzed by recombinant viruses: involvement of heparin-like molecule in CDV infection. Virology 359:324-335. [DOI] [PubMed] [Google Scholar]
- 13.Griffin, D. E. 2001. Measles virus, p. 1401-1441. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 14.Hashimoto, K., N. Ono, H. Tatsuo, H. Minagawa, M. Takeda, K. Takeuchi, and Y. Yanagi. 2002. SLAM (CD150)-independent measles virus entry as revealed by recombinant virus expressing green fluorescent protein. J. Virol. 76:6743-6749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim, S.-H., S. M. Lessner, Y. Sakurai, and Z. S. Galis. 2004. Cyclophilin A as a novel biphasic mediator of endothelial activation and dysfunction. Am. J. Pathol. 164:1567-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kobune, F., H. Takahashi, K. Terao, T. Ohkawa, Y. Ami, Y. Suzaki, N. Nagata, H. Sakata, K. Yamanouchi, and C. Kai. 1996. Nonhuman primate models of measles. Lab. Anim. Sci. 46:315-320. [PubMed] [Google Scholar]
- 17.Kobune, F., Y. Ami, M. Katayama, M. Takahashi, R. Tuul, G. Korukluoglu, T. Kiyohara, R. Miura, H. Sato, M. Yoneda, and C. Kai. 2007. A novel monolayer cell line derived from human umbilical cord blood cells shows high sensitivity to measles virus. J. Gen. Virol. 88:1565-1567. [DOI] [PubMed] [Google Scholar]
- 18.Liu, J., M. W. Albers, C. M. Chen, S. L. Schreiber, and C. T. Walsh. 1990. Cloning, expression, and purification of human cyclophilin in Escherichia coli and assessment of the catalytic role of cysteines by site-directed mutagenesis. Proc. Natl. Acad. Sci. USA 100:1298-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luban, J., K. L. Bossolt, E. K. Franke, G. V. Kalpana, and S. P. Goff. 1993. Human immunodeficiency virus type I Gag protein binds to cyclophilins A and B. Cell 73:1067-1078. [DOI] [PubMed] [Google Scholar]
- 20.Masuda, M., H. Sato, H. Kamata, T. Katsuo, A. Takenaka, R. Miura, M. Yoneda, K. Tsukiyama-Kohara, K. Mizumoto, and C. Kai. 2006. Characterization of monoclonal antibodies directed against the canine distemper virus nucleocapsid protein. Comp. Immunol. Microbiol. Infect. Dis. 29:157-165. [DOI] [PubMed] [Google Scholar]
- 21.Miller, C. A., and C. S. Raine. 1979. Heterogeneity of virus particles in measles virus. J. Gen. Virol. 45:441-453. [DOI] [PubMed] [Google Scholar]
- 22.Misumi, S., T. Fuchigami, N. Takamune, I. Takahashi, M. Takama, and S. Shoji. 2002. Three isoforms of cyclophilin A associated with human immunodeficiency virus type 1 were found by proteomics by using two-dimensional gel electrophoresis and matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Virol. 76:10000-10008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pakula, R., A. Melchior, A. Denys, C. Vanpouille, J. Mazurier, and F. Allain. 2007. Syndecan-1/CD147 association is essential for cyclophilin B-induced activation of p44/42 mitogen-activated protein kinases and promotion of cell adhesion and chemotaxis. Glycobiology 17:492-503. [DOI] [PubMed] [Google Scholar]
- 24.Price, E. R., L. D. Zydowsky, M. J. Jin, C. H. Baker, F. D. McKeon, and C. T. Walsh. 1991. Human cyclophilin B: a second cyclophilin gene encodes a peptidyl-prolyl isomerase with a signal sequence. Proc. Natl. Acad. Sci. USA 88:1903-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pushkarsky, T., G. Zybarth, L. Dubrovsky, V. Yurchenko, H. Tang, H. Guo, B. Toole, B. Sherry, and M. Bukrinsky. 2001. CD147 facilitates HIV-1 infection by interacting with virus-associated cyclophilin A. Proc. Natl. Acad. Sci. USA 98:6360-6365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pushkarsky, T., V. Yurchenko, B. Sherry, and M. Bukrinsky. 2002. CD147 is a signaling receptor for extracellular cyclophilin A: role in HIV-1 infection. Int. Conf. AIDS 14:7-12. [Google Scholar]
- 27.Ravanel, K., C. Castelle, T. Defrance, T. F. Wild, D. Charron, V. Lotteau, and C. Rabourdin-Combe. 1997. Measles virus nucleocapsid protein binds to FcγRΙΙ and inhibits human B-cell antibody production. J. Exp. Med. 186:269-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rima, B. K., and W. P. Duprex. 2006. Morbilliviruses and human disease. J. Pathol. 208:199-214. [DOI] [PubMed] [Google Scholar]
- 29.Rycyzym, M. A., and C. V. Clevenger. 2002. The intranuclear prolactin/cyclophilin B complex as a transcriptional inducer. Proc. Natl. Acad. Sci. USA 99:6790-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saphire, A. C. S., D. Bobardt, and P. A. Gallay. 1999. Host cyclophilin A mediates HIV-1 attachment to target cells via heparans. EMBO J. 18:6771-6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sato, H., M. Masuda, M. Kanai, K. Tsukiyama-Kohara, M. Yoneda, and C. Kai. 2007. Measles virus N protein inhibits host translation by binding to eIF3-p40. J. Virol. 81:11569-11576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sato, H., R. Honma, M. Yoneda, R. Miura, K. Tsukiyama-Kohara, F. Ikeda, T. Seki, S. Watanabe, and C. Kai. 2008. Measles virus induces cell-type specific changes in gene expression. Virology 375:321-330. [DOI] [PubMed] [Google Scholar]
- 33.Tatsuo, H., N. Ono, K. Tanaka, and Y. Yanagi. 2000. SLAM (CDw150) is a cellular receptor for measles virus. Nature 406:893-897. [DOI] [PubMed] [Google Scholar]
- 34.tenOever, B. R., M. J. Servant, N. Grandvaux, R. Lin, and J. Hiscott. 2002. Recognition of the measles virus nucleocapsid as a mechanism of IRF-3 activation. J. Virol. 76:3659-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Terao-Muro, Y., M. Yoneda, T. Seki, A. Watanabe, K. Tsukiyama-Kohara, K. Fujita, and C. Kai. 2008. Heparin-like glycosaminoglycans prevent the infection of measles virus in SLAM-negative cell lines. Antiviral Res. 80:370-376. [DOI] [PubMed] [Google Scholar]
- 36.Thali, M., A. Bukovsky, E. Kondo, B. Rosenwirth, C. T. Walsh, J. Sodroski, and H. G. Göttlinger. 1994. Functional association of cyclophilin A with HIV-1 virions. Nature 372:363-365. [DOI] [PubMed] [Google Scholar]
- 37.Tokui, M., I. Takei, F. Tashiro, A. Shimada, A. Kasuga, M. Ishii, T. Ishii, K. Takatsu, T. Saruta, and J. Miyazaki. 1997. Intramuscular injection of expression plasmid DNA is an effective means of long-term systemic delivery of interleukin-5. Biochem. Biophys. Res. Commun. 233:527-531. [DOI] [PubMed] [Google Scholar]
- 38.Watashi, K., N. Ishii, M. Hijikata, D. Inoue, T. Murata, Y. Miyanari, and K. Shimotohno. 2005. Cyclophilin B is a functional regulator of hepatitis C virus RNA polymerase. Mol. Cell 19:111-122. [DOI] [PubMed] [Google Scholar]
- 39.Yurchenko, V., S. Constant, and M. Bukrinsky. 2006. Dealing with the family: CD147 interactions with cyclophilins. Immunology 117:301-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yurchenkoa, V., M. O'Connora, W. W. Daia, H. Guob, B. Tooleb, B. Sherrya, and M. Bukrinskya. 2001. CD147 is a signaling receptor for cyclophilin B. Biochem. Biophys. Res. Commun. 288:786-788. [DOI] [PubMed] [Google Scholar]