Abstract
Although the herpes simplex virus type 1 (HSV-1) tegument is comprised of a large number of viral and cellular proteins, how and where in the cell these proteins are recruited into the virus structure is poorly understood. We have shown previously that the immediate-early gene product ICP0 is packaged by a mechanism dependent on the major tegument protein VP22, while others have shown a requirement for ICP27. We now extend our studies to show that ICP0 packaging correlates directly with the ability of ICP0 to complex with VP22 in infected cells. ICP27 is not, however, present in this VP22-ICP0 complex but is packaged into the virion in a VP22- and ICP0-independent manner. Biochemical fractionation of virions indicated that ICP0 associates tightly with the virus capsid, but intranuclear capsids contained no detectable ICP0. The RING finger domain of ICP0 and the N terminus of VP22 were both shown to be essential but not sufficient for ICP0 packaging and complex formation. Strikingly, however, the N-terminal region of VP22, while unable to form a complex with ICP0, inhibited its translocation from the nucleus to the cytoplasm. PML degradation by ICP0 was efficient in cells infected with this VP22 mutant virus, confirming that ICP0 retains activity. Hence, we would suggest that VP22 is an important molecular partner of ICP0 that controls at least one of its activities: its assembly into the virion. Moreover, we propose that the pathway by which VP22 recruits ICP0 to the virion may begin in the nucleus prior to ICP0 translocation to its final site of assembly in the cytoplasm.
The herpesvirus tegument is the virion compartment located between the DNA-containing capsid and the virus envelope (6). The tegument is the equivalent of the matrix of a range of other enveloped viruses but is unusual in that it comprises a very large number of proteins. A recent proteomic study of the composition of herpes simplex virus type 1 (HSV-1) virions has shown that at least 26 virus-encoded components are recruited into the HSV-1 tegument (32). Some of these proteins, such as VP1/2, VP13/14, VP16, and VP22, are classified as major, structurally significant components (23, 44), while some, such as vhs and the protein kinase UL13, are minor but nonetheless important components (38, 42). In addition to virus-encoded proteins, human cytomegalovirus (hCMV), Epstein-Barr virus (EBV), Kaposi's sarcoma-associated herpesvirus (KSHV), and HSV-1 have all been shown to package a wide range of cellular proteins into their teguments (25, 32, 49, 54).
It is proposed that after fusion of the virus envelope with the cellular membrane, the contents of the tegument are released into the cytoplasm of the cell. Hence, the tegument is believed to deliver a range of factors involved in the initiation of virus infection. The roles of a few tegument proteins, such as the immediate-early (IE) gene transactivator VP16 and the host shutoff protein Vhs, are well established at early stages of infection (2, 29, 39, 41). A number of years ago, several studies suggested that the IE protein ICP0 was also packaged into the HSV-1 particle (50, 52), but these results have remained somewhat controversial, with the suggestion that the ICP0 detectable in HSV-1 virion preparations may be a consequence of contamination. Evidence for specific ICP0 assembly into the virus was strengthened by our own more recent studies, which showed that while we could easily detect ICP0 in wild-type (Wt) virus particles, it was not detected with virion preparations lacking the major tegument protein VP22, suggesting that VP22 was somehow involved in the assembly of ICP0 (9). The case for ICP0 as a virion component was further enhanced by recent proteomic studies in which ICP0 was clearly identified as a component of highly purified HSV-1 virions (32). Nonetheless, a role for ICP0 at very early stages of infection prior to IE gene expression is yet to be determined.
ICP0 has been characterized as a general transactivator of gene expression and has recently been shown to dissociate the genome silencing complex consisting of REST-CoREST-histone deacetylase 1/2 (HDAC1/2) during infection (19, 20). In addition, ICP0 has been shown to function as an E3 ubiquitin ligase (5, 46). This activity directs a number of cellular proteins, including components of nuclear ND10 domains and centromeres for proteasomal degradation (4, 14, 15, 30). ICP0 also binds a cellular ubiquitin-specific protease, USP7, which contributes to the activities of ICP0 in infection by preventing its degradation (3, 17). Consistent with its role in the disruption of nuclear complexes and its role in gene expression, ICP0 localizes to the nucleus early in infection (1, 16). However, during the course of infection it relocalizes to the cytoplasm, where it is located in discrete domains (28, 31). Moreover, in some cell lines, such as primary human fibroblasts, ICP0 is predominantly cytoplasmic throughout infection (28). Once in the cytoplasm, ICP0 is believed to associate dynamically with proteasomes and has been shown to induce the accumulation of colocalizing conjugated ubiquitin, suggesting that its E3 ubiquitin ligase activity is functional in the cytoplasm as well as the nucleus (13). A recent study has shown that another IE protein, ICP27, is required for the translocation of ICP0 from the nucleus to the cytoplasm (43). Interestingly, these authors showed that in cells infected with HSV-1 expressing an ICP27 mutant that was unable to relocalize ICP0 to the cytoplasm, ICP0 was not assembled into the virion (43). Hence, the cytoplasmic localization of ICP0 would seem to be fundamental for its packaging. Nonetheless, cytoplasmic localization of ICP0 is not sufficient for its assembly into the virion, as our own previous data have shown that VP22 is also required for ICP0 packaging, even though ICP0 localizes efficiently to the cytoplasm in the absence of VP22 (9). In this case, we have proposed that VP22 recruits ICP0 from the cytosol, once it has translocated from the nucleus, to ICP0-specific domains in the cytoplasm. We have postulated that these cytoplasmic domains represent a step on the virus assembly pathway (9, 40).
In this current study we set out to determine the mechanism by which VP22 is involved in the recruitment of ICP0 into the virus structure. We show that ICP0 and VP22 are present in a complex in infected cells and that VP22-ICP0 complex formation requires the N terminus of VP22. We also show that ICP0 is assembled into a region of the tegument that is tightly associated with the capsid and that its assembly into the virus correlates with its ability to complex with VP22. Localization of ICP0 to our previously characterized cytoplasmic domains is not, however, sufficient for ICP0 assembly. Interestingly, the RING finger of ICP0 is required for complex formation and assembly into the virion, but it is not yet clear if this is a physical or functional requirement. While VP22 is not required for ICP0 translocation to the cytoplasm per se, the expression of the N terminus alone inhibits translocation, resulting in nuclear retention of ICP0. We conclude that the HSV-1 tegument protein VP22 is an important molecular partner of the IE protein ICP0 that recruits it to the virion but may also influence its activities in other ways.
MATERIALS AND METHODS
Cells and viruses.
Vero, U2OS, and BHK cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% newborn calf serum and antibiotics. Viruses were routinely grown in BHK or Vero cells and titrated on Vero cells. The parental virus strain used in this study was strain 17 (s17) of HSV-1. Strains sc16 and HFEM were kindly provided by Helena Browne (Department of Pathology, University of Cambridge) and Peter O'Hare (Marie Curie Research Institute, Surrey, United Kingdom), respectively. The recombinant virus expressing green fluorescent protein (GFP)-tagged VP22 (166v) and the VP22 deletion mutant (169v) that expresses GFP in place of VP22 have been described previously (9, 11). Viruses expressing residues 108 to 301, 160 to 301, 1 to 212, and 1 to 165 as GFP fusion proteins have also been described before (22). Virus mutants in ICP0 FXE, E52X, M1, D12, and dl1403 (12, 34) or expressing yellow fluorescent protein (YFP)-tagged ICP0 (18) were kindly provided by Roger Everett (MRC Virology Unit, University of Glasgow).
Virion and capsid purification.
Extracellular virions were purified on Ficoll gradients from the infected cell medium of 5 × 108 BHK cells, as described previously (11). Intranuclear capsid preparations were carried out from nuclear extracts made from 5 × 108 BHK cells infected at a multiplicity of infection of 0.05. After 3 days, cell pellets were solubilized in phosphate-buffered saline (PBS)/1% NP-40 containing protease inhibitors, and nuclei were pelleted at low speed following a 30-min incubation on ice. Nuclei were washed in PBS, then resuspended in PBS, and subjected to 3 rounds of freeze-thawing. Cell debris was removed by centrifugation, and the supernatant was purified by centrifugation for 1 h at 25,000 × g through a 30% sucrose cushion in an SW41 rotor. The resulting pellet was resuspended in 1 ml PBS and fractionated by centrifugation for 1 h at 25,000 × g through a 15 to 50% sucrose gradient. Fractions were collected across the gradient and precipitated in 10% trichloroacetic acid (TCA).
Virion stripping experiments.
Equivalent amounts of virions were pelleted at 2,300 × g for 5 min at 4°C and resuspended in 50 μl of 10 mM Tris-HCl (pH 7.5), 1 mM EDTA, 1% NP-40, and NaCl ranging from 0.1 to 1 M. The virions were then incubated for 15 min at room temperature and pelleted again to separate soluble from insoluble proteins.
Antibodies.
Monoclonal antibodies to GFP and HSV-1 major capsid protein VP5 were obtained from Clontech and Autogen Bioclear, respectively. The monoclonal anti-VP16 (LP1) and anti-gD (LP14) antibodies were kindly provided by Helena Browne (Department of Pathology, University of Cambridge). The anti-VP22 polyclonal antibodies AGV031 (N-terminal specific) and AV600 (C-terminal specific) have been described previously (10). Monoclonal anti-ICP0 (monoclonal antibody 11060) and anti-ICP27 antibodies were kindly provided by Roger Everett (MRC Virology Unit, University of Glasgow) and Steve Rice (University of Minnesota), respectively. Anti-PML polyclonal antibody DB75 was provided by Peter O'Hare (MCRI, Surrey, United Kingdom). The anti-conjugated ubiquitin antibody FK2 was obtained from Enzo Life Sciences.
Immunoprecipitation assay.
Vero cells grown in 6-cm dishes were infected with the relevant viruses at a multiplicity of infection of 1. After 20 to 24 h, the cells were washed twice with PBS, solubilized in 1 ml RIPA buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.1% SDS, 1% Na deoxycholate, 1% NP-40) and incubated on ice for 20 min. The cells were then centrifuged at 12,000 × g for 30 min at 4°C, and the supernatant was collected. The appropriate antibody was added to between 400 and 800 μl of lysate and incubated for 3 h at 4°C with rotation. Forty microliters of protein A Sepharose beads were added and incubated overnight at 4°C with rotation, and the resulting protein A-antibody complexes were washed five times with PBS. The immunoprecipitated proteins were then analyzed by SDS-PAGE followed by Western blotting.
SDS-polyacrylamide gel electrophoresis and Western blotting.
Protein samples were analyzed on 10% polyacrylamide gels and electrophoresed in SDS-Tris-glycine buffer. Following electrophoresis, gels were either stained with Coomassie blue (Thermo Scientific), silver stained (Perbio), or transferred to nitrocellulose for analysis by Western blotting. Western blots were developed using enhanced chemiluminescence (Pierce).
Immunofluorescence.
Cells for immunofluorescence were grown on 16-mm coverslips in individual wells of a six-well plate. Cells were fixed for 20 min in 4% paraformaldehyde, permeabilized for 10 min in PBS containing 0.5% Triton X-100, and blocked by incubation for 10 min in PBS containing 10% newborn calf serum. Primary antibody was added for 15 min in the same solution, and after the cells were washed extensively in PBS, the appropriate secondary antibody (all from Vector Labs) was added in a block solution and incubated for a further 10 min. The coverslips were then washed extensively in PBS and mounted in Vectashield or Mowiol containing 4′,6-diamidino-2-phenylindole (DAPI) (Vector Labs). Images were acquired using a Zeiss LSM510 Meta confocal microscope. Resulting images were processed using Adobe Photoshop software.
RESULTS
ICP0 is packaged into the tegument of the HSV-1 virion.
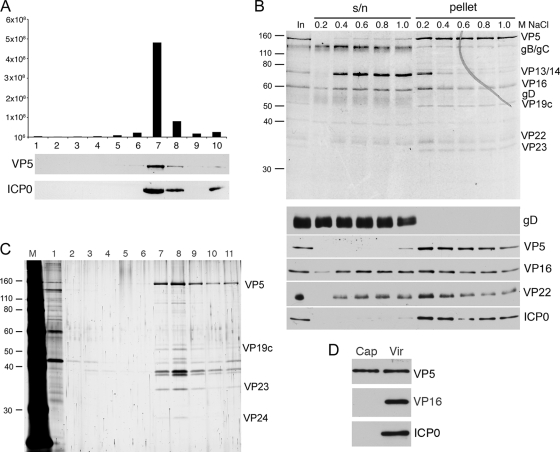
Our previous results have indicated that ICP0 is present in gradient-isolated preparations of extracellular HSV-1 virions that have been purified from the supernatant of infected BHK21 cells. To confirm that ICP0 is a true virion component and not simply a contaminant of our virion preparations, we first analyzed the concentration of ICP0 across a Ficoll gradient used to isolate virions. A crude preparation of extracellular s17 virions was produced by pelleting virus at high speed from infected cell supernatant. The resulting pellet was resuspended in PBS and centrifuged through a 5 to 15% Ficoll gradient. Ten fractions were collected across the gradient, and the content of each was analyzed by titration on Vero cells for the presence of infectious virus, showing that the peak of infectivity was present in fraction 7 (Fig. 1A). Samples of the fractions were also analyzed by SDS-PAGE and Western blotting for the major capsid protein VP5 and ICP0. These blots revealed that both ICP0 and VP5 also peaked in fraction 7, confirming that ICP0 copurifies with capsids and infectivity.
FIG. 1.
ICP0 is an HSV-1 tegument protein. (A) Extracellular s17 virions were centrifuged through a 5 to 15% Ficoll gradient, and fractions were taken across the gradient. Each sample was analyzed for infectivity (top) or subjected to Western blotting for the major capsid protein VP5 or ICP0. (B) Purified extracellular s17 virions were fractionated using detergent and increasing concentrations of NaCl. Soluble (s/n) and insoluble (pellet) fractions were separated by pelleting and then analyzed by SDS-PAGE followed by Coomassie blue staining (top panel) or Western blotting for a range of virus structural proteins. The sizes of the molecular weight markers (in thousands) are indicated on the left. (C) Intranuclear capsids from s17-infected cells were centrifuged through a 15 to 50% sucrose gradient, and fractions were collected across the gradient. Each sample was analyzed by SDS-PAGE followed by silver staining. (D) Equivalent amounts of intact s17 virions and s17 capsids from fraction 8, based on their VP5 content, were analyzed by Western blotting for VP5, VP16, and ICP0.
We next wished to determine in which compartment of the virus ICP0 is packaged. To this end, we used detergent treatment to fractionate extracellular virions into envelope and tegument/capsid fractions. Equal aliquots of virions were extracted in buffer containing salt concentrations ranging from 0.2 to 1 M NaCl to discriminate between loosely and tightly associated tegument proteins. The resulting fractions were analyzed by SDS-PAGE and then by Coomassie blue staining or Western blotting with antibodies specific for the capsid protein VP5, the envelope protein gD, and the tegument proteins VP16 and VP22. As would be expected, the envelope glycoprotein gD was completely removed from the virions at the lowest concentration of NaCl (Fig. 1B, gD). In contrast, the capsid protein VP5 remained in the pellet fraction even in NaCl concentrations as high as 1 M (Fig. 1B, VP5). Both tegument proteins VP16 and VP22 can be seen dividing into two populations; one is removed with 0.4 M NaCl, but the second one remains tightly associated with the capsid in the pellet (Fig. 1B, VP16 and VP22). Interestingly, ICP0 behaves like the capsid protein and remains entirely in the pellet fraction of the virions, even at 1 M NaCl (Fig. 1B, ICP0). These results suggest that ICP0 is tightly associated with the capsid within the virus structure.
HSV-1 capsids are assembled in the nucleus of the cell. Therefore, we wished to determine if ICP0 was present on intranuclear capsids. Nuclear extracts from Wt virus-infected BHK21 cells were subjected to centrifugation on a sucrose gradient, and fractions were collected across the gradient and analyzed by SDS-PAGE and then by silver staining. The capsid population was seen to peak in fraction 8 of the gradient (Fig. 1C), which was then analyzed by Western blotting beside an equivalent concentration of purified extracellular virions, based on VP5 content (Fig. 1D, VP5). Blotting for the tegument protein VP16, which has been shown to be present on primary virions in the perinuclear space (35) and which is present in the nucleus at high concentrations, revealed that these intranuclear capsids contained no VP16 and could therefore be considered highly purified. Furthermore, blotting for ICP0 showed that ICP0 was also absent from the intranuclear capsids, suggesting that ICP0 is not added to capsids within the nucleus but at a later stage downstream in the virus assembly pathway.
ICP0 coimmunoprecipitates with VP22 throughout infection.
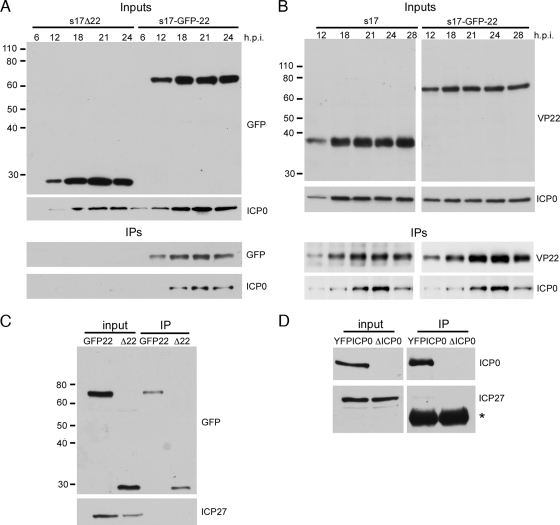
Previously, we have shown that ICP0 requires the presence of VP22 to be assembled into the HSV-1 virion (9). Hence, we wished to determine if this requirement reflected the presence of a VP22-ICP0 complex within the infected cell. Vero cells were infected with HSV-1 (s17) expressing GFP-tagged VP22 at a multiplicity of infection of 1, and lysates harvested in RIPA buffer at various times throughout infection. VP22 was then immunoprecipitated with an anti-VP22 antibody, and resulting immunocomplexes were analyzed by SDS-PAGE and Western blotting. As a control for nonspecific binding of ICP0 to the VP22 antibody, the same experiment was also carried out with cells infected with our Δ22 mutant virus expressing GFP in place of VP22. Western blotting of the input samples with a GFP antibody showed that the GFP-22 and Δ22 mutant infections progressed with similar efficiencies through the time courses (Fig. 2A, Inputs, GFP). As we have noted before, ICP0 levels for the Δ22 mutant infection were consistently lower than those for the GFP-22 infection (Fig. 2A, Inputs, ICP0). Blotting of the resulting complexes with a GFP antibody indicated that VP22 was successfully immunoprecipitated from 12 h onward (Fig. 2A, IPs, GFP). Interestingly, blotting of these complexes for ICP0 showed that ICP0 was efficiently coimmunoprecipitated with VP22 at these times of infection (Fig. 2A, IPs, ICP0). No ICP0 was detected in the immunoprecipitation samples from the Δ22 mutant-infected cells, confirming that pulldown of ICP0 with the VP22 antibody was specific to the presence of GFP-VP22 in the extract.
FIG. 2.
VP22 and ICP0 form a complex in infected cells. (A) Vero cells infected with s17 expressing GFP in place of VP22 (s17Δ22) or expressing GFP-tagged VP22 (s17-GFP-22) at a multiplicity of infection of 1 were harvested at times ranging between 6 and 24 h after infection, and immunoprecipitations carried out with a VP22 polyclonal antibody. The original input samples and resulting complexes were analyzed by Western blotting for GFP or ICP0. The sizes of the molecular weight markers (in thousands) are indicated on the left. (B) As described in the legend for panel A, but using s17 and s17-GFP-22 viruses. (C) Vero cells infected with the same viruses as those shown in panel A were harvested 20 h after infection and immunoprecipitated with a polyclonal anti-GFP antibody. The resulting complexes were analyzed by Western blotting for GFP and ICP27. (D) Vero cells infected with either s17 expressing YFP-tagged ICP0 or the ICP0 deletion virus dl1403 (ΔICP0) were treated as described in the legend for panel C and analyzed by Western blotting for ICP0 and ICP27. Asterisk denotes polyclonal heavy chain.
To confirm that the VP22-ICP0 pulldown occurred in wild-type virus-infected cells and was not specific to the expression of GFP-tagged VP22, a similar experiment was carried out comparing time courses of Vero cells infected with HSV-1-GFP-22 or HSV-1 s17. Western blotting of input samples again showed that both infections had progressed at similar rates and that in this case, ICP0 was efficiently expressed in s17 and GFP-22 infections (Fig. 2B, Inputs). Immunoprecipitation of VP22 also resulted in the pulldown of ICP0 in both infections, confirming that VP22 and ICP0 are present in an infected cell complex (Fig. 2B, IPs).
ICP27 is not present in the ICP0-VP22 complex.
It has recently been published that, like VP22, ICP27 is required for the virion incorporation of ICP0 (43), and therefore, we hypothesized that ICP27 may be present in the same infected cell complex as VP22 and ICP0. Hence, an extract of cells infected with the GFP22-expressing virus was immunoprecipitated with an anti-GFP antibody to pull down VP22-containing complexes, and the resulting complexes were tested for the presence of ICP27. At the same time, an extract from Δ22 mutant-infected cells expressing GFP in place of VP22 was used as a control for potential nonspecific binding of ICP27 to the antibody. As shown in Fig. 2C, although ICP27 expression was readily detectable in the infected cell extracts, it was not present in the GFP-22 pulldown complex (Fig. 2C, IP). As ICP27 has a more defined relationship with ICP0, we also immunoprecipitated ICP0 and tested for the presence of ICP27 in the resulting complexes. In this case, Vero cells were infected with s17 expressing YFP-tagged ICP0 or s17 lacking ICP0 and immunoprecipitated with an anti-GFP antibody (Fig. 2D). Once again, although ICP0 was efficiently precipitated and ICP27 was present in the input extract, no ICP27 was pulled down with ICP0 (Fig. 2D, IPs). Taken together, these results suggest that ICP27 does not form a complex with ICP0 or VP22 in the infected cell, in spite of being required for efficient ICP0 recruitment into the tegument.
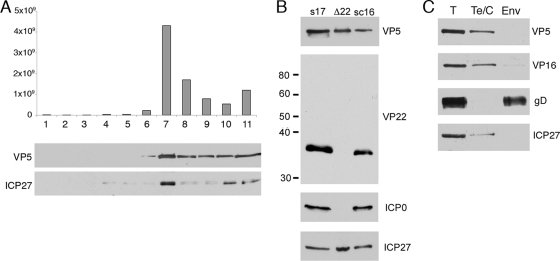
It has been published by others that ICP27 is not associated with virions of HSV-1 (43, 52), a result that would correlate with our inability to detect ICP27 in the VP22-ICP0 complexes in infected cells. To confirm that ICP27 was absent from virions, we fractionated extracellular virions across a Ficoll gradient, in this case from the HFEM strain of HSV-1, as described in the legend for Fig. 1A, and analyzed the resulting samples for infectivity and the presence of the capsid protein VP5 and ICP27 by Western blotting (Fig. 3A). Surprisingly, we were able to detect ICP27 in the peak infectious sample, in which it copurified with the capsid protein (Fig. 3A, fraction 7). These results were somewhat unexpected, and therefore, we next looked for ICP27 in extracellular virions from two other strains of HSV-1, s17 and sc16, and were able to readily detect the protein in these particles too, confirming that ICP27 is a component of virions (Fig. 3B, s17 and sc16). Furthermore, detergent stripping of virions revealed that ICP27 was present in the tegument/capsid fraction of the virions (Fig. 3C).
FIG. 3.
ICP27 is packaged into the HSV-1 tegument in a VP22-independent mechanism. (A) Extracellular HFEM virions were centrifuged through a 5 to 15% Ficoll gradient, and fractions taken across the gradient. Each sample was analyzed for infectivity (top) or subjected to Western blotting for the major capsid protein VP5 or ICP27. (B) Approximately equivalent amounts of s17, s17Δ22, and sc16 virions were analyzed by Western blotting for VP5, VP22, ICP0 and ICP27. The sizes of the molecular weight markers (in thousands) are indicated on the left. (C) Strain HFEM virions were fractionated into envelope (Env) and tegument-capsid (Te/C) fractions using detergent and 0.1 M NaCl. Soluble and insoluble fractions were separated by pelleting and then analyzed by Western blotting for the capsid protein VP5, the tegument protein VP16, the envelope protein gD, and ICP27.
Because it has been shown that ICP27 is required for ICP0 assembly into the virion, these results raised the possibility that although we could not detect ICP27 in the ICP0-VP22 complex, ICP0 and ICP27 may enter the virion structure by the same VP22-specific mechanism. Therefore, we tested Δ22 mutant virions for the presence of ICP27 but found that ICP27 packaging was unaltered in the absence of VP22 and ICP0 packaging (Fig. 3B, Δ22). Taken together, these results suggest that ICP27 is recruited into the virion by an ICP0- and VP22-independent mechanism.
Virion recruitment of ICP0 requires the N terminus of VP22.
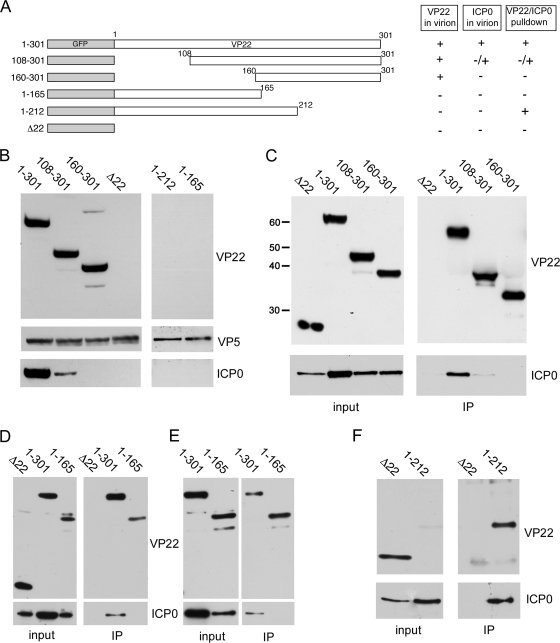
To further investigate the role of VP22 in the recruitment of ICP0 into the virion, we used viruses expressing truncation mutants of VP22 fused to GFP (Fig. 4A) that have been characterized previously (22). Extracellular virions were purified from the viruses shown in Fig. 4A, together with the Δ22 mutant virus, and analyzed by Western blotting for the presence of ICP0. As previously shown, assembly of VP22 did not require the N-terminal half of the protein (Fig. 4B, VP22, compare 1 to 301 with 160 to 301) but did require the C-terminal region (Fig. 4B, VP22, compare 1 to 301 with 1 to 212). Also, as we have shown before, assembly of ICP0 was seen to be dependent on VP22, as the Δ22 mutant virions contained no detectable amounts of ICP0 (Fig. 4B, ICP0, compare 1 to 301 with Δ22). Interestingly, the level of ICP0 present in virions that packaged residues 108 to 301 of VP22 was greatly reduced and was undetectable in virions that packaged residues 160 to 301 of VP22, in spite of the assembly of the VP22 variants being unaffected (Fig. 4B, ICP0). ICP0 was also absent from virions made from cells expressing the 1-to-212 and 1-to-165 variants of VP22, which themselves are not packaged (Fig. 4B, ICP0).
FIG. 4.
The N terminus of VP22 is required for ICP0 assembly into the virion. (A) Line drawing of the VP22 truncation mutants used in this study. (B) Equivalent amounts of virions from the viruses described in the legend for panel A were analyzed by Western blotting for VP22, VP5, and ICP0. (C) Immunoprecipitations using a polyclonal GFP antibody were carried out on Vero cells infected with HSV-1 expressing full-length GFP-22 (1 to 301), the two N-terminal VP22 truncation mutants (108 to 301 and 160 to 301), or GFP in place of VP22 (Δ22), 20 h after infection at a multiplicity of infection of 1. The resulting complexes were analyzed by Western blotting for the C terminus of VP22 or ICP0. The sizes of the molecular weight markers (in thousands) are indicated on the left. (D and E) As described in the legend for panel C, using cells infected with HSV-1 expressing full-length VP22 (1 to 301), the N-terminal half of VP22 (1 to 165), or GFP in place of VP22 (Δ22). Western blots were carried out using antibodies for the N terminus of VP22 or ICP0. (F) As described in the legend for panel D, using cells infected with HSV-1 expressing GFP in place of VP22 (Δ22) or the C-terminal truncation (1 to 212).
We next addressed the ability of these VP22 truncation mutants to form a complex with ICP0. First, the N-terminal truncation viruses were used in coimmunoprecipitation assays on extracts made at 20 hours postinfection (h.p.i), in which VP22 was immunoprecipitated using an antibody specific for the C terminus of VP22, and the resulting complexes blotted for ICP0. In this case, while full-length and N-terminally truncated VP22 viruses were precipitated in equivalent amounts (Fig. 4C, IP, VP22), the content of ICP0 present in the immunocomplexes was quite different, with deletion of the N-terminal third of the protein greatly reducing the pulldown of ICP0 and deletion of the N-terminal half of VP22 abolishing the pulldown of ICP0 (Fig. 4C, IP, ICP0). These results correlate directly with the level of ICP0 assembly into the respective virions (Fig. 4B), confirming that VP22 has a major role in the assembly pathway of ICP0.
To assess if the N-terminal half of VP22 was sufficient for complex formation with ICP0, the mutant virus expressing only residues 1 to 165 of VP22 was used in a coimmunoprecipitation assay, in this case using an antibody specific to the N terminus of VP22. Western blotting of the resulting pulldown complexes revealed that ICP0 was not present in the complex bound to the VP22 antibody, a result that was highly reproducible (Fig. 4D and E). This implies that this region of VP22 may be necessary but is not sufficient for complex formation with ICP0. In contrast, a similar pulldown carried out on cells expressing residues 1 to 212 of VP22 resulted in the efficient coprecipitation of ICP0, indicating that this region of the protein is sufficient to form a complex with ICP0 (Fig. 4F). Although this mutant expresses a low level of the VP22 1-to-212 mutant protein (Fig. 4F, input), as we have shown before (22), the amount of VP22 present in the immunoprecipitation complex was comparable to those of the other samples (Fig. 4F, IP). As this region of VP22 is not itself assembled into the virion (Fig. 4B), the fact that ICP0 is not assembled, even though ICP0 can complex with VP22, is consistent with a physical requirement for VP22 in the assembly of ICP0 (Fig. 4B).
ICP0 localization in VP22 mutant virus infections.
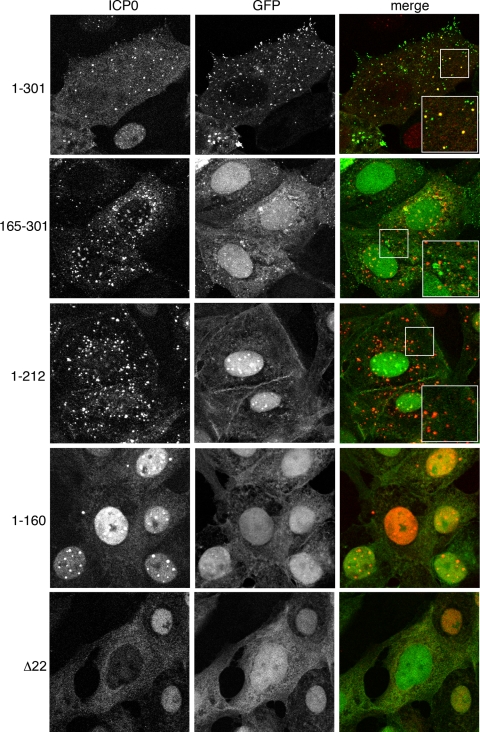
We have previously shown that, following translocation of ICP0 from the nucleus to the cytoplasm of HSV-1-infected cells, VP22 and ICP0 colocalize in discrete cytoplasmic domains (9). Furthermore, while ICP0 was able to translocate to the cytoplasm of cells infected with our Δ22 mutant virus, its localization to these cytoplasmic domains was shown to be greatly reduced, a result that led us to postulate that these cytoplasmic domains were on the virus assembly pathway for HSV-1 (9). As the colocalization of VP22 and ICP0 may be representative of their presence in the same complex as well as their recruitment into the virus tegument, we wished to determine if expression of any of the VP22 truncation mutants affected the cytoplasmic localization of ICP0. Vero cells were infected with HSV-1 expressing GFP-tagged full-length or truncated VP22 or GFP in place of VP22 (Δ22 mutant) at a multiplicity of infection of 10. Six hours after infection, cells were fixed and analyzed by immunofluorescence with anti-ICP0. As described before, ICP0 colocalized with a subset of GFP-positive cytoplasmic domains containing full-length VP22 (Fig. 5, 1 to 301), while in the absence of VP22, ICP0 was predominantly in a diffuse cytoplasmic localization (Fig. 5, Δ22). In cells infected with HSV-1 expressing the N-terminal 212 residues of VP22, which we have shown above to complex with ICP0, ICP0 was also present in cytoplasmic domains (Fig. 5, 1 to 212). However, in this case, VP22 was predominantly nuclear and in spite of their ability to coimmunoprecipitate, there was little evidence of colocalization at the cytoplasmic domains or elsewhere in the cell. Likewise, in cells infected with HSV-1 expressing the C-terminal half of VP22, which we have shown above does not complex with ICP0 but is packaged into the virus, ICP0 localized efficiently to its cytoplasmic domains. Hence, interaction with VP22 is not required for ICP0 to localize to these domains. Furthermore, a presence in these domains is not sufficient for ICP0 assembly into the virus. Strikingly, in cells infected with HSV-1 expressing the N-terminal half of VP22, shown above to be necessary but not sufficient for complex formation with ICP0, ICP0 localization is retained in its nuclear punctate pattern (Fig. 5, 1 to 160). Even at later time points up to 16 h after infection, there is little evidence that ICP0 can translocate to the cytoplasm in cells infected with this virus (data not shown). We would suggest then that this region of VP22 functions as a dominant negative inhibitor of ICP0 translocation to the cytoplasm.
FIG. 5.
ICP0 localizes to cytoplasmic domains in the absence of interacting VP22. Vero cells on coverslips were infected with the GFP-expressing viruses described in the legend for Fig. 4A at a multiplicity of infection of 10 and fixed with 4% paraformaldehyde 6 h later. Immunofluorescence was carried out for ICP0 (red).
ICP0 is biologically active in cells infected with VP22 truncation mutants.
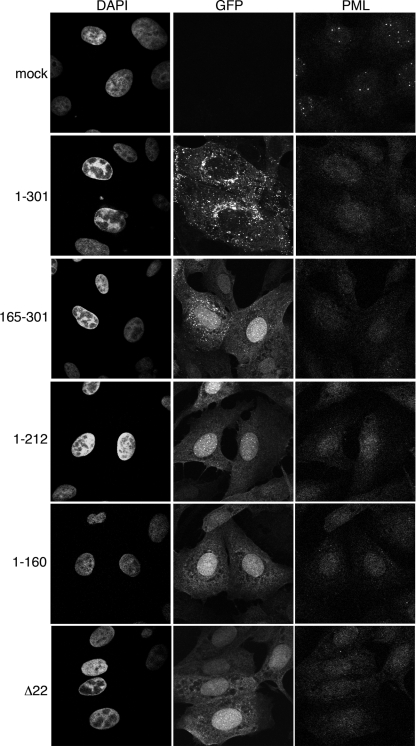
A major characterized role of ICP0 during infection is the localization to ND10 domains and subsequent degradation of PML and other components localized in those domains. We wanted to ensure that ICP0 was functioning correctly in our VP22 truncation mutant viruses, and therefore, Vero cells infected with each of the viruses, as described in the legend for Fig. 5, were fixed at 6 h after infection and stained for immunofluorescence using a polyclonal anti-PML antibody. Uninfected cells showed the characteristic pattern for PML staining in punctate nuclear domains (Fig. 6, mock). In contrast, infection with wild-type virus had efficiently dispersed PML at the time of fixation, and no ND10 domains in the infected cell nuclei were obvious (Fig. 6, 1 to 301). Likewise, all mutant virus infections, including the Δ22 mutant infection, caused the dispersal of PML from ND10 domains (Fig. 6), confirming that ICP0 was functional in all these infections. Interestingly, in cells infected with virus expressing only the N-terminal half of VP22, which we have shown above to result in the retention of ICP0 in nuclear domains, PML behavior was identical to that in the other infections, suggesting that the retention of ICP0 in nuclear domains is not a consequence of inefficient PML degradation (Fig. 6, 1 to 212).
FIG. 6.
PML is dispersed from ND10s in all VP22 mutant virus infections. Vero cells on coverslips were either uninfected (mock) or infected with the GFP-expressing viruses described in the legend for Fig. 4A at a multiplicity of infection of 10 and fixed with 4% paraformaldehyde 6 h later. Immunofluorescence was carried out for PML, and nuclei were stained with DAPI.
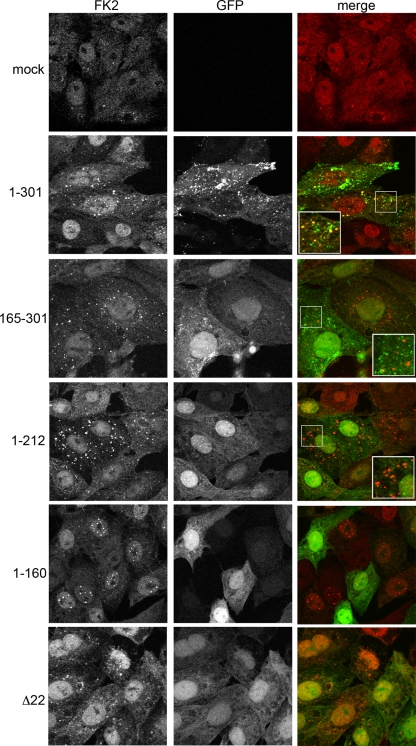
Another measure of ICP0 activity is its ability to induce the accumulation of colocalizing conjugated ubiquitin in the infected cell (13). Hence, we next measured this activity by carrying out the same infections described in the legend for Fig. 6 and then staining for immunofluorescence with the anti-ubiquitin antibody FK2 that detects both poly- and monoubiquitinated species. In uninfected cells, ubiquitin was predominantly nuclear in a speckled and diffuse pattern (Fig. 7, mock). In contrast, in cells infected with Wt virus expressing GFP-22, FK2 staining was present in cytoplasmic punctate domains that colocalized with a subset of GFP-positive domains (Fig. 7, 1 to 301), in a pattern similar to that seen for ICP0 in wild-type infection (Fig. 5, 1 to 301). Indeed, in all mutant virus infections, FK2 staining reflected the respective ICP0 staining (compare Fig. 7 and 5). Of note is the fact that in cells infected with the virus expressing the N-terminal half of VP22, which retains ICP0 in nuclear domains, ubiquitin accumulates to high levels in nuclear domains, thereby again reflecting the relative ICP0 localization (Fig. 7, 1 to 212). Taken together with the PML degradation results, these results imply that ICP0 is functional in all our VP22 mutant viruses, and therefore, the lack of assembly is not due to the presence of inactive ICP0.
FIG. 7.
ICP0 induces colocalizing, conjugated ubiquitin in all VP22 mutant virus infections. Vero cells on coverslips were either uninfected (mock) or infected with the GFP-expressing viruses described in the legend for Fig. 4A at a multiplicity of infection of 10 and fixed with 4% paraformaldehyde 6 h later. Immunofluorescence was carried out for conjugated ubiquitin using antibody FK2 (red).
The RING finger of ICP0 is necessary but not sufficient for recruitment into the virion.
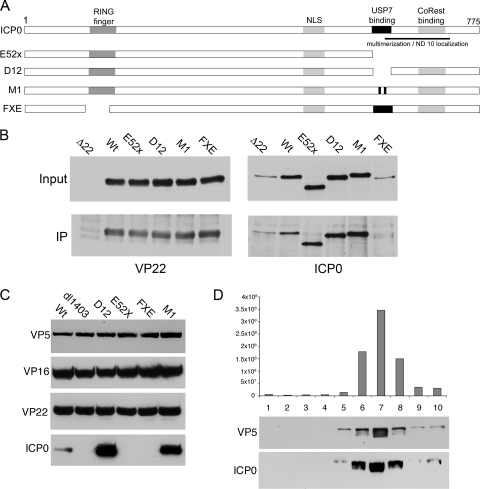
To investigate the domains of ICP0 required for interaction with VP22, we carried out coimmunoprecipitations from cells infected with a number of ICP0 mutant viruses kindly provided to us by Roger Everett, Glasgow (Fig. 8A). In this case, infections were carried out at a multiplicity of infection of 1 in U2OS cells, as ICP0 mutations are known to have little effect on virus growth in this cell line (53), and harvested 20 h later. Resulting immunocomplexes were analyzed by Western blotting for VP22 and ICP0, with a Δ22 mutant-infected cell lysate being used as a negative control. VP22 was expressed and precipitated in equivalent amounts in all infections, indicating that mutation of ICP0 has no effect on VP22 expression levels at least in U2OS cells (Fig. 8B, VP22). Levels of ICP0 were more variable in the infected cell lysates, with the Δ22 and FXE (Δ RING finger) mutant viruses consistently expressing less ICP0 than the other viruses (Fig. 8B, ICP0, input). Nonetheless, the coimmunoprecipitations indicated that ICP0 lacking its RING finger did not form a complex with VP22 during infection (Fig. 8B, ICP0, IP). Longer exposures of this blot showed no evidence of an ICP0 Δ RING finger mutant pulldown with VP22 (data not shown). In contrast, ICP0 lacking its C terminus (E52X) or mutated in its USP7 binding domain within the C terminus (D12 and M1) precipitated efficiently with VP22 (Fig. 8B, ICP0, IP). These results suggest that the RING finger but not the C terminus of ICP0 is required for complex formation with VP22.
FIG. 8.
Virion assembly of ICP0 requires its RING finger and its C terminus. (A) Line drawing of the ICP0 mutants used in this study. (B) U2OS cells were infected with all the viruses shown in panel A together with the Δ22 mutant virus at a multiplicity of infection of 1 and harvested 20 h later. Immunoprecipitations were carried out using a VP22 antibody, and the resulting complexes were analyzed by Western blotting for VP22 and ICP0. (C) Equivalent amounts of extracellular virions from the viruses shown in panel A, together with the ICP0 deletion mutant (dl1403), were analyzed by Western blotting for VP5, VP16, VP22, and ICP0. (D) Extracellular virions from the USP7 binding site mutant D12 were centrifuged through a 5 to 15% Ficoll gradient, and fractions were taken across the gradient. Each sample was analyzed for infectivity (top) or subjected to Western blotting for the major capsid protein VP5 or ICP0.
We next wished to correlate the ability of these ICP0 mutants to form VP22 complexes with their relative assemblies into virions. Western blotting was carried out on extracellular virions from the mutant viruses shown in Fig. 8A, together with the ΔICP0 mutant virus dl1403, and ICP0 and VP22 levels were compared to that present in Wt (s17) virions. Blotting for the capsid protein VP5 confirmed that all virions had been loaded in equivalent amounts (Fig. 8C, VP5), while blotting for VP22 in the ΔICP0 mutant virus indicated that VP22 packaging was not dependent on ICP0 (Fig. 8C, VP22, dl1403). ICP0 lacking its RING finger was not detected in virions, consistent with our failure to pull down this variant of ICP0 with VP22 (Fig. 8C, FXE). However, ICP0 lacking its C terminus was also not packaged into virions, in spite of its ability to interact efficiently with VP22 (Fig. 8C, E52X). In contrast, the ICP0 variants that were mutated specifically in their USP7 binding domains were packaged at levels higher than Wt levels (Fig. 8C, D12 and M1). Blotting virions for VP22 showed that this increase in ICP0 content was not a result of increased VP22 levels in the virion (Fig. 8C, VP22, D12, and M1). Likewise, the lack of packaging of the ICP0 RING finger and C-terminal mutants was also not a reflection of reduced VP22 levels in these virions (Fig. 8C, VP22, FXE, and E52X).
To confirm that the increased ICP0 level present in virions made from D12-infected cells was real, a crude preparation of virions was fractionated on a 5 to 15% Ficoll gradient, and 1-ml fractions were collected. Each fraction was then analyzed by titration for the presence of infectivity and by Western blotting for the presence of ICP0. The ICP0 content across the gradient was seen to cofractionate with both the major capsid protein VP5 and infectivity (Fig. 8D). Hence, these virions truly package increased levels of ICP0, albeit a variant of ICP0 mutated in its USP7 binding domain.
DISCUSSION
The HSV-1 IE protein ICP0 is expressed early in infection and is involved in establishing the correct environmental conditions within the cell to allow optimal lytic virus replication. As such, it counteracts several antiviral pathways that the cell may use to inhibit virus replication, including genome silencing and the interferon response. With such activities in its repertoire, the packaging of ICP0 into the virus structure, so that it is delivered to the cell immediately upon virus entry, would be a logical mechanism for the virus to utilize. However, although it has been some years since ICP0 was originally proposed to be a virion component (52), it is only recently that the concept has been explored in more detail. Here we provide further compelling evidence for the presence of ICP0 in the virus and, together with a number of other recent publications, prove that the presence of ICP0 in extracellular virion preparations is not due to sample contamination but is the result of specific assembly of the protein into the tegument compartment of the virus structure (7, 9, 32). In this paper we have also shown for the first time that the IE protein ICP27 is additionally packaged into the tegument of the virus. As ICP4 has also been shown to be present in the HSV-1 virion (9, 43, 51), a unifying theme of assembly of apparent nonstructural regulatory proteins seems to be emerging.
VP22-specific assembly of ICP0.
In our previous studies we have shown that virions made from HSV-1 lacking the tegument protein VP22 fail to package ICP0, suggesting an important relationship between these two proteins (9). Nonetheless, in this situation the complete deletion of the VP22 open reading frame from the HSV-1 genome may have had an indirect effect on the packaging of ICP0. Indeed, the fact that intracellular ICP0 levels are reduced in the absence of VP22 has led others to suggest that it may be the relative level of ICP0 that dictates its assembly into the virion rather than a specific interaction and/or localization (8). Importantly, however, we now show that VP22 and ICP0 are present in a complex within the infected cell that is dependent on the N terminus of VP22. Moreover, the ability of ICP0 to complex with VP22, in both VP22 and ICP0 mutant viruses, directly reflects its level of recruitment into the virion with VP22 (Table 1), implying that this interaction is the main determinant of ICP0 assembly. Taking these results together with our previous work, we conclude that, in our hands at least, an immunoprecipitated VP22-specific complex from infected cells contains gE, gM, VP16, and ICP0 but not gD or VP13/14 (45). We and others have previously identified binding domains for VP16 and gE within the C-terminal half of VP22 (22, 36, 37, 45), whereas, as we have shown here, the ICP0 interaction domain requires the N-terminal half of VP22. However, this region is not sufficient for complex formation with additional sequences extending into the VP16/gE binding domains being required (summarized in Table 1). Hence, it is possible that VP16 and/or gE is essential in stabilizing the VP22-ICP0-containing complex.
TABLE 1.
Binding and assembly properties of VP22 truncation mutantsf
| VP22 residues | gE bindinga | VP16 co-IPb | ICP0 co-IPc | Virion VP22d | Virion ICP0e |
|---|---|---|---|---|---|
| 1-301 | + | + | + | + | + |
| 108-301 | + | + | −/+ | + | −/+ |
| 160-301 | + | + | − | + | − |
| 194-301 | − | − | − | −/+ | − |
| 1-212 | − | + | + | − | − |
| 1-165 | − | − | − | − | − |
Binding of VP22 truncation mutants to the cytoplasmic tail of gE as described previously (45).
Infected cell coimmunoprecipitation of VP16 with VP22 mutants as shown previously (22).
Infected cell coimmunoprecipitation of ICP0 with VP22 mutants as shown here.
Virion assembly of VP22 truncation mutants as described previously (22).
Virion assembly of ICP0 in viruses expressing VP22 truncation mutants as described here.
gE and VP16 assembly is unaffected by the absence of VP22. co-IP, coimmunoprecipitation.
When considering molecular interactions involved in herpesvirus tegument assembly, the tegument is often referred to as having an inner and outer compartment. This designation is based on the ease with which tegument proteins can be extracted from the capsid structure and is often used to invoke an order of assembly of the tegument: inner proteins added first and outer proteins added later. The biochemical data presented here indicate that ICP0 is tightly associated with the capsid of the virion, to the extent that its extraction profile is akin to a true capsid protein rather than a tegument protein. Early work showed that there were around 150 copies of ICP0 in the virion (52), a number that would fit with a location proximal to the capsid. Furthermore, while the manuscript was in preparation, Delboy and coworkers published a similar result demonstrating that ICP0 is tightly associated with the capsid (7). The two tegument proteins that we looked at, VP16 and the ICP0 partner VP22, both separated into two populations, one that was readily extracted at a lower salt concentration and one that remained tightly associated with capsids under all conditions. Thus, we would propose that the second, tightly associated population of VP22 would be the one involved in recruiting ICP0 into the tegument. It is not yet clear what the differences between these two tegument protein populations are, but the differential extractability may be dictated by the respective binding partners of the two, the presence of different posttranslationally modified forms, and/or their cellular location of addition. Importantly, ICP0, VP16, and VP22 were all absent from our intranuclear capsids, confirming that recruitment of even the tightly associated populations of these molecules occurs outside the nucleus.
Cellular location of ICP0 assembly into the tegument.
Current data suggest that a pathway of events must occur in infection for ICP0 to assemble into the virus. When ICP0 is first expressed, it localizes to the nucleus, where it is involved in the proteasomal degradation of ND10 and centromere components and the inhibition of genome silencing. Recent studies from the Roizman laboratory suggest that both of these events must be complete before ICP0 translocates into the cytoplasm (21, 26). Translocation requires, among other things, a late gene product(s), as inhibition of DNA synthesis results in nuclear retention of ICP0 (31). Translocation also requires the IE protein ICP27, and failure of ICP0 to translocate in this ICP27-dependent manner abrogates its assembly into the virion (43). Hence, cytoplasmic localization would seem to be a prerequisite for ICP0 assembly (43). Once in the cytoplasm, ICP0 requires VP22 for physical recruitment into the virion, but where and how this recruitment takes place is not yet clear. Like others, we have observed ICP0 in specific punctate domains in the cytoplasm. In addition, in cells infected with virus lacking the VP22 gene, ICP0 failed to localize to these puncta to any degree (9). It is of note then that some full-length GFP-VP22 is concentrated in these domains with ICP0 (Fig. 5), a result that led us to initially propose that VP22 brought ICP0 to these sites where the proteins were then assembled into the virus (9). However, several results from our fluorescence data have led us to modify this model. First, these studies have clearly indicated that VP22 can be assembled into the virus at Wt levels without exhibiting steady-state localization to these cytoplasmic puncta, as indicated by the mutant protein 160 to 301. Second, ICP0 can localize efficiently to these sites without interacting with VP22 and without being assembled, again exemplified by the virus expressing mutant protein 160 to 301. Last, ICP0 and VP22 can form a complex without VP22 appearing at these sites, as is the case with VP22 mutant protein 1 to 212. Hence, our data now suggest that the ICP0 puncta may be specific sites through which VP22 and other tegument proteins transit prior to assembly but that final assembly occurs at a different site. Variants of VP22 that are assembled but do not interact with ICP0 may transit through these ICP0 puncta but would not accumulate there because ICP0 would not retain them, whereas variants that can interact with ICP0 but are not assembled would not be targeted to these sites in the first place. Therefore, VP22 localization to the ICP0-specific puncta would be dictated by one of its other binding partners rather than ICP0 itself. The failure of ICP0 to accumulate in these puncta in the absence of VP22 may reflect a more widespread phenotype of the Δ22 mutant virus, as discussed below.
The role of the E3 ligase activity in ICP0 assembly.
Our studies of a number of ICP0 mutants implied that the RING finger of the protein containing the E3 ligase activity was required for assembly into the virus, a result that has recently been corroborated by Delboy and coworkers (7). We have further shown that the ICP0 RING finger is necessary for complex formation with VP22, suggesting that the RING finger may be the physical domain of ICP0 involved in the formation of the VP22 complex. VP22 may therefore regulate the activity of the ICP0 E3 ligase during infection. Nonetheless, we have shown that all our VP22 mutants express functional ICP0, with regard to PML degradation, and any role that VP22 may play in ICP0 activity would presumably be subsequent to translocation to the cytoplasm. Alternatively, the ICP0 RING finger mutant may simply be folded incorrectly for complex formation with VP22. In the case of the C-terminal truncation mutant of ICP0 (E52X), this mutant forms a complex efficiently with VP22 but is not recruited to the virion, in spite of Wt levels of VP22 packaging. The C-terminal region of ICP0 absent from the E52X mutant incorporates USP7 binding, multimerization, ND10 localization, and CoREST binding (Fig. 8A) (20, 33, 34), and any or a combination of these activities may be required for ICP0 assembly into the virion. Given the results presented here that ICP0 mutated specifically in the USP7 binding domain is packaged to levels higher than Wt ICP0 (ICP0 mutants D12 and M1), it is unlikely that USP7 binding is involved in recruiting ICP0 to the virion. This higher level of ICP0 packaging in these mutants is counterintuitive, as USP7 binding mutants of ICP0 have been shown to be less stable in infected cells than in Wt ICP0 (3). Hence, the reason for such enhanced packaging is yet to be determined but may be related to the apparently enhanced levels of these mutants seen by others in the cytoplasm of infected cells (13, 17). Interestingly, the E52X mutant remains in the nucleus throughout infection (34) and, like the previously described ICP27 mutant which retains ICP0 in the nucleus (43), would be predicted to be absent from the virion. The absence of both the ND10 localization and the CoREST binding domains from E52X means that this protein is unable to locate to and disrupt ND10s or dissociate HDAC from the silencing complex (34). Consistent with the model that both events are a prerequisite for ICP0 translocation to the cytoplasm, the E52X protein is retained in the nucleus.
A role for VP22 in the translocation of ICP0 from the nucleus to the cytoplasm.
The ICP0 translocation pathway would suggest that VP22 does not have an involvement in the compartmentalization of ICP0. Furthermore, ICP0 translocates to the cytoplasm normally in cells infected with our Δ22 mutant virus (9). However, in this current study we have shown that the N-terminal half of VP22, which in itself is unable to stably complex with ICP0, retains ICP0 in the nucleus. Hence, the N terminus alone seems to be functioning as a dominant-negative inhibitor of ICP0 translocation. There are several situations known in which Wt ICP0 is retained in the nucleus, such as exposure to proteasomal inhibitors, failure to degrade PML (21), treatment with HDAC inhibitors (27), inhibition of DNA replication (31), or the presence of an ICP27 mutant (43). Our immunofluorescence studies of PML in infected cells showed clearly that ND10s were efficiently disrupted in this and all VP22 mutant virus infections (Fig. 6), implying that the block to ICP0 translocation occurs downstream of ND10 disruption. Furthermore, DNA replication is not obviously affected in this mutant, and although it is possible that the N terminus of VP22 inhibits the role of ICP27 in ICP0 translocation, our pulldown experiments from infected cells revealed that ICP27 is not an obvious binding partner of VP22. Unlike full-length VP22, the dominant-negative N-terminal region of VP22 has a predominantly nuclear steady-state localization. One potential model would be that this region of VP22 interacts with and inhibits a third partner in the VP22-ICP0 complex, be it cellular or viral, that is involved normally in the ICP0 translocation pathway from ND10s to the cytoplasm. Thus, it is noteworthy that this VP22 mutant virus exhibits an ICP0 phenotype similar to that of a point mutant of ICP0 (with a D199A mutation) described by the Roizman laboratory. This mutant is unable to bind cyclin D3, a cellular factor that this group has characterized as being involved in ICP0 translocation (26, 47, 48).
Taken together, these results may imply that VP22 is actively involved in ICP0 translocation from the nucleus. While the majority of full-length VP22 localizes to the cytoplasm of infected cells as described above, a small population localizes to discrete nuclear domains around the time that ICP0 begins to leave the nucleus. We have previously characterized these VP22-containing nuclear domains as being juxtaposed to ICP0-containing ND10s (24). It is conceivable that this nuclear population plays a role in directing ICP0 into its translocation pathway by forming the complex identified in our pulldown assays, with the N terminus dictating the formation of this transport complex by interacting with a third partner. If VP22 is truly involved in ICP0 translocation, one question that remains to be addressed concerns the cytoplasmic localization of ICP0 in Δ22 mutant-infected cells. However, this situation is similar to that of a ΔICP4 mutant virus infection in which ICP0 is efficiently transported, even in the absence of β and γ gene expression (31). Observations with the latter virus have led to the proposal that an additional, post-α gene is actively involved in retaining ICP0 in the nucleus. Hence, as deletion of the entire VP22 reading frame has more widespread effects than simply an absence of ICP0 from virus particles (8, 9), it is possible that overall ICP0 behavior is quite different in Δ22 mutant-infected cells and that any activity for nuclear retention is not functional.
In summary, we have shown that ICP0 is a true virion component that is assembled into the virion via a pathway that is absolutely dependent on VP22. Furthermore, VP22 may play an active role in determining the subcellular localization of ICP0 during infection. The next stage of this work will be to determine if virion-incorporated ICP0 has a function at early times of infection. Such studies will be experimentally challenging but will provide invaluable information on the ever-increasing complexity of ICP0 biology.
Acknowledgments
We thank Chris Boutell and Roger Everett for kindly providing reagents and discussions. We also thank Peter O'Hare, Helena Browne, and Steve Rice for antibodies and viruses.
This work was funded by the Medical Research Council.
Footnotes
Published ahead of print on 17 February 2010.
REFERENCES
- 1.Ackermann, M., D. K. Braun, L. Pereira, and B. Roizman. 1984. Characterization of herpes simplex virus 1 alpha proteins 0, 4, and 27 with monoclonal antibodies. J. Virol. 52:108-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batterson, W., and B. Roizman. 1983. Characterization of the herpes simplex virion-associated factor responsible for the induction of alpha genes. J. Virol. 46:371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boutell, C., M. Canning, A. Orr, and R. D. Everett. 2005. Reciprocal activities between herpes simplex virus type 1 regulatory protein ICP0, a ubiquitin E3 ligase, and ubiquitin-specific protease USP7. J. Virol. 79:12342-12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boutell, C., A. Orr, and R. D. Everett. 2003. PML residue lysine 160 is required for the degradation of PML induced by herpes simplex virus type 1 regulatory protein ICP0. J. Virol. 77:8686-8694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boutell, C., S. Sadis, and R. D. Everett. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and its isolated RING finger domain act as ubiquitin E3 ligases in vitro. J. Virol. 76:841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dargin, D. 1986. The structure and assembly of herpes viruses, p. 359-437. In J. R. Harris and R. W. Horne (ed.), Electronmicroscopy of proteins, vol. 5. Virus structure. Academic Press, London, United Kingdom. [Google Scholar]
- 7.Delboy, M. G., C. R. Siekavizza-Robles, and A. V. Nicola. 2010. Herpes simplex virus tegument ICP0 is capsid associated, and its E3 ubiquitin ligase domain is important for incorporation into virions. J. Virol. 84:1637-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duffy, C., E. F. Mbong, and J. D. Baines. 2009. VP22 of herpes simplex virus 1 promotes protein synthesis at late times in infection and accumulation of a subset of viral mRNAs at early times in infection. J. Virol. 83:1009-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elliott, G., W. Hafezi, A. Whiteley, and E. Bernard. 2005. Deletion of the herpes simplex virus VP22-encoding gene (UL49) alters the expression, localization, and virion incorporation of ICP0. J. Virol. 79:9735-9745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elliott, G., and P. O'Hare. 1997. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell 88:223-233. [DOI] [PubMed] [Google Scholar]
- 11.Elliott, G., and P. O'Hare. 1999. Live-cell analysis of a green fluorescent protein-tagged herpes simplex virus infection. J. Virol. 73:4110-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Everett, R. D. 1989. Construction and characterisation of herpes simplex virus type 1 mutants with defined lesions in immediate early gene 1. J. Gen. Virol. 70:1185-1202. [DOI] [PubMed] [Google Scholar]
- 13.Everett, R. D. 2000. ICP0 induces the accumulation of colocalizing conjugated ubiquitin. J. Virol. 74:9994-10005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Everett, R. D., W. C. Earnshaw, J. Findlay, and P. Lomonte. 1999. Specific destruction of kinetochore protein CENP-C and disruption of cell division by herpes simplex virus immediate-early protein Vmw110. EMBO J. 18:1526-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Everett, R. D., P. Freemont, H. Saitoh, M. Dasso, A. Orr, M. Kathoria, and J. Parkinson. 1998. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J. Virol. 72:6581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Everett, R. D., and G. G. Maul. 1994. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 13:5062-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Everett, R. D., M. Meredith, and A. Orr. 1999. The ability of herpes simplex virus type 1 immediate-early protein Vmw110 to bind to a ubiquitin-specific protease contributes to its roles in the activation of gene expression and stimulation of virus replication. J. Virol. 73:417-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Everett, R. D., G. Sourvinos, and A. Orr. 2003. Recruitment of herpes simplex virus type 1 transcriptional regulatory protein ICP4 into foci juxtaposed to ND10 in live, infected cells. J. Virol. 77:3680-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu, H., Y. Liang, G. Mandel, and B. Roizman. 2005. Components of the REST/CoREST/histone deacetylase repressor complex are disrupted, modified, and translocated in HSV-1-infected cells. Proc. Natl. Acad. Sci. U. S. A. 102:7571-7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gu, H., and B. Roizman. 2007. Herpes simplex virus-infected cell protein 0 blocks the silencing of viral DNA by dissociating histone deacetylases from the CoREST-REST complex. Proc. Natl. Acad. Sci. U. S. A. 104:17134-17139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gu, H., and B. Roizman. 2009. The two functions of herpes simplex virus 1 ICP0, inhibition of silencing by the CoREST/REST/HDAC complex and degradation of PML, are executed in tandem. J. Virol. 83:181-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hafezi, W., E. Bernard, R. Cook, and G. Elliott. 2005. Herpes simplex virus tegument protein VP22 contains an internal VP16 interaction domain and a C-terminal domain that are both required for VP22 assembly into the virus particle. J. Virol. 79:13082-13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heine, J. W., R. W. Honess, E. Cassai, and B. Roizman. 1974. Proteins specified by herpes simplex virus XII. The virion polypeptides of type 1 strains. J. Virol. 14:640-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hutchinson, I., A. Whiteley, H. Browne, and G. Elliott. 2002. Sequential localization of two herpes simplex virus tegument proteins to punctate nuclear dots adjacent to ICP0 domains. J. Virol. 76:10365-10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johannsen, E., M. Luftig, M. R. Chase, S. Weicksel, E. Cahir-McFarland, D. Illanes, D. Sarracino, and E. Kieff. 2004. Proteins of purified Epstein-Barr virus. Proc. Natl. Acad. Sci. U. S. A. 101:16286-16291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalamvoki, M., and B. Roizman. 2009. ICP0 enables and monitors the function of D cyclins in herpes simplex virus 1 infected cells. Proc. Natl. Acad. Sci. U. S. A. 106:14576-14580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalamvoki, M., and B. Roizman. 2008. Nuclear retention of ICP0 in cells exposed to HDAC inhibitor or transfected with DNA before infection with herpes simplex virus 1. Proc. Natl. Acad. Sci. U. S. A. 105:20488-20493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawaguchi, Y., R. Bruni, and B. Roizman. 1997. Interaction of herpes simplex virus 1 alpha regulatory protein ICP0 with elongation factor 1delta: ICP0 affects translational machinery. J. Virol. 71:1019-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwong, A. D., and N. Frenkel. 1989. The herpes simplex virus virion host shutoff function. J. Virol. 63:4834-4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lomonte, P., K. F. Sullivan, and R. D. Everett. 2001. Degradation of nucleosome-associated centromeric histone H3-like protein CENP-A induced by herpes simplex virus type 1 protein ICP0. J. Biol. Chem. 276:5829-5835. [DOI] [PubMed] [Google Scholar]
- 31.Lopez, P., C. Van Sant, and B. Roizman. 2001. Requirements for the nuclear-cytoplasmic translocation of infected-cell protein 0 of herpes simplex virus 1. J. Virol. 75:3832-3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loret, S., G. Guay, and R. Lippe. 2008. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J. Virol. 82:8605-8618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maul, G. G., and R. D. Everett. 1994. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J. Gen. Virol. 75:1223-1233. [DOI] [PubMed] [Google Scholar]
- 34.Meredith, M., A. Orr, M. Elliott, and R. Everett. 1995. Separation of sequence requirements for HSV-1 Vmw110 multimerisation and interaction with a 135-kDa cellular protein. Virology 209:174-187. [DOI] [PubMed] [Google Scholar]
- 35.Naldinho-Souto, R., H. Browne, and T. Minson. 2006. Herpes simplex virus tegument protein VP16 is a component of primary enveloped virions. J. Virol. 80:2582-2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Regan, K. J., M. A. Bucks, M. A. Murphy, J. W. Wills, and R. J. Courtney. 2007. A conserved region of the herpes simplex virus type 1 tegument protein VP22 facilitates interaction with the cytoplasmic tail of glycoprotein E (gE). Virology 358:192-200. [DOI] [PubMed] [Google Scholar]
- 37.O'Regan, K. J., M. A. Murphy, M. A. Bucks, J. W. Wills, and R. J. Courtney. 2007. Incorporation of the herpes simplex virus type 1 tegument protein VP22 into the virus particle is independent of interaction with VP16. Virology 369:263-280. [DOI] [PubMed] [Google Scholar]
- 38.Overton, H. A., D. J. McMillan, L. S. Klavinskis, L. Hope, A. J. Ritchie, and P. Wong-kai-in. 1992. Herpes simplex virus type 1 gene UL13 encodes a phosphoprotein that is a component of the virion. Virology 190:184-192. [DOI] [PubMed] [Google Scholar]
- 39.Post, L. E., S. Mackem, and B. Roizman. 1981. Regulation of alpha genes of herpes simplex virus: expression of chimeric genes produced by fusion of thymidine kinase with alpha gene promoters. Cell 24:555-565. [DOI] [PubMed] [Google Scholar]
- 40.Potel, C., and G. Elliott. 2005. Phosphorylation of the herpes simplex virus tegument protein VP22 has no effect on incorporation of VP22 into the virus but is involved in optimal expression and virion packaging of ICP0. J. Virol. 79:14057-14068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Read, G. S., B. M. Karr, and K. Knight. 1993. Isolation of a herpes simplex virus type 1 mutant with a deletion in the virion host shutoff gene and identification of multiple forms of the vhs (UL41) polypeptide. J. Virol. 67:7149-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schek, N., and S. L. Bachenheimer. 1985. Degradation of cellular mRNAs induced by a virion-associated factor during herpes simplex virus infection of Vero cells. J. Virol. 55:601-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sedlackova, L., and S. A. Rice. 2008. Herpes simplex virus type 1 immediate-early protein ICP27 is required for efficient incorporation of ICP0 and ICP4 into virions. J. Virol. 82:268-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spear, P. G., and B. Roizman. 1972. Proteins specified by herpes simplex virus. V. Purification and structural proteins of the herpesvirion. J. Virol. 9:143-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stylianou, J., K. Maringer, R. Cook, E. Bernard, and G. Elliott. 2009. Virion incorporation of the herpes simplex virus type 1 tegument protein VP22 occurs via glycoprotein E-specific recruitment to the late secretory pathway. J. Virol. 83:5204-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Van Sant, C., R. Hagglund, P. Lopez, and B. Roizman. 2001. The infected cell protein 0 of herpes simplex virus 1 dynamically interacts with proteasomes, binds and activates the cdc34 E2 ubiquitin-conjugating enzyme, and possesses in vitro E3 ubiquitin ligase activity. Proc. Natl. Acad. Sci. U. S. A. 98:8815-8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Sant, C., Y. Kawaguchi, and B. Roizman. 1999. A single amino acid substitution in the cyclin D binding domain of the infected cell protein no. 0 abrogates the neuroinvasiveness of herpes simplex virus without affecting its ability to replicate. Proc. Natl. Acad. Sci. U. S. A. 96:8184-8189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Sant, C., P. Lopez, S. J. Advani, and B. Roizman. 2001. Role of cyclin D3 in the biology of herpes simplex virus 1 ICP0. J. Virol. 75:1888-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Varnum, S. M., D. N. Streblow, M. E. Monroe, P. Smith, K. J. Auberry, L. Pasa-Tolic, D. Wang, D. G. Camp II, K. Rodland, S. Wiley, W. Britt, T. Shenk, R. D. Smith, and J. A. Nelson. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 78:10960-10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang, T. Y., and R. J. Courtney. 1995. Influence of the host cell on the association of ICP4 and ICP0 with herpes simplex virus type 1. Virology 211:209-217. [DOI] [PubMed] [Google Scholar]
- 51.Yao, F., and R. J. Courtney. 1989. A major transcriptional regulatory protein (ICP4) of herpes simplex virus type 1 is associated with purified virions. J. Virol. 63:3338-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yao, F., and R. J. Courtney. 1992. Association of ICP0 but not ICP27 with purified virions of herpes simplex virus type 1. J. Virol. 66:2709-2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yao, F., and P. A. Schaffer. 1995. An activity specified by the osteosarcoma line U2OS can substitute functionally for ICP0, a major regulatory protein of herpes simplex virus type 1. J. Virol. 69:6249-6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu, F. X., J. M. Chong, L. Wu, and Y. Yuan. 2005. Virion proteins of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79:800-811. [DOI] [PMC free article] [PubMed] [Google Scholar]