Abstract
The poliovirus 3′ noncoding region (3′ NCR) is necessary for efficient virus replication. A poliovirus mutant, PVΔ3′NCR, with a deletion of the entire 3′ NCR, yielded a virus that was capable of synthesizing viral RNA, albeit with a replication defect caused by deficient positive-strand RNA synthesis compared to wild-type virus. We detected multiple ribonucleoprotein (RNP) complexes in extracts from poliovirus-infected HeLa cells formed with a probe corresponding to the 5′ end of poliovirus negative-strand RNA (the complement of the genomic 3′ NCR), and the levels of these RNP complexes increased during the course of viral infection. Previous studies have identified RNP complexes formed with the 3′ end of poliovirus negative-strand RNA, including one that contains a 36-kDa protein later identified as heterogeneous nuclear ribonucleoprotein C (hnRNP C). We report here that the 5′ end of poliovirus negative-strand RNA is capable of interacting with endogenous hnRNP C, as well as with poliovirus nonstructural proteins. Further, we demonstrate that the addition of recombinant purified hnRNP C proteins can stimulate virus RNA synthesis in vitro and that depletion of hnRNP C proteins in cultured cells results in decreased virus yields and a correspondingly diminished accumulation of positive-strand RNAs. We propose that the association of hnRNP C with poliovirus negative-strand termini acts to stabilize or otherwise promote efficient positive-strand RNA synthesis.
Picornaviruses are a group of small, positive-sense RNA viruses that replicate in the host cell cytoplasm. Their genomes are characterized by highly structured 5′ noncoding regions (NCRs) containing a cruciform RNA structure (termed stem-loop I, or cloverleaf) necessary for viral-RNA replication and an internal ribosome entry site (IRES) that allows translation of a virus polyprotein from the single open reading frame of genomic RNA. The virus polyprotein is cleaved by encoded viral proteinases (30) to yield the mature structural and nonstructural proteins, including the RNA-dependent RNA polymerase 3D. Due to the limited size of the genome, picornaviruses have evolved to utilize host cell factors in concert with their own virus-encoded proteins and RNA secondary structures to efficiently drive the replication cycle. Poliovirus, an example of the viruses using such a combination of host and viral functions, is the causative agent of paralytic poliomyelitis and the most extensively studied picornavirus. Cellular proteins play an important role in the replication of the poliovirus RNA genome (19, 37). For example, poly(rC) binding protein 2 (PCBP2) binds to the stem-loop I RNA cruciform structure at the 5′ end of the genome. Binding of PCBP2 to this region is required for viral-RNA replication, and disruption of the capability of the protein to bind to stem-loop I disrupts RNA replication (55). During the synthesis of poliovirus negative-strand RNA intermediates, it has been proposed that the 5′ and 3′ ends of poliovirus positive-sense RNA communicate via interactions formed by host cell protein poly(A)-binding protein (PABP) bound to the poly(A) tract at the 3′ end of the genome and PCBP2 and the virus-encoded 3CD protein bound to the stem-loop I RNA structure (5, 20, 28).
The 3′ NCR of poliovirus genomic RNA is located immediately following the 3D polymerase coding region and is relatively short (71 nucleotides) compared to the 5′ NCR (742 nucleotides). For poliovirus, the 3′ NCR consists of two stem-loop structures (termed X and Y) that are formed by base-pairing interactions between the nucleotides of the 3′ terminus and the genetically encoded poly(A) tail. These stem-loop structures have been predicted to participate in tertiary “kissing” interactions between the RNA stem-loops themselves (31, 40) and/or to bind cellular factors (48, 53) to facilitate RNA synthesis. Previous studies by Brown et al. characterized a viable poliovirus mutant lacking the 3′ NCR (PVΔ3′NCR) that exhibited a growth defect in cultured human cell lines (12, 13). Due to the proximity of the 3′ NCR to the presumed site of negative-strand RNA synthesis initiation at the 3′ poly(A) tail, it was originally thought that the observed growth defect was due to a deficiency in negative-strand RNA synthesis owing to a lack of 3′ NCR stem-loop structures. However, further examination of the strand-specific synthesis during PVΔ3′NCR infection of HeLa cells revealed that while negative-strand RNA synthesis was largely unchanged compared to the wild type, positive-strand RNA synthesis was diminished significantly. This suggested either that a terminal RNA structure (e.g., the 3′ NCR) can act in trans to influence the initiation of RNA synthesis at the opposite end of an RNA strand of negative polarity or that the negative-strand complement of the 3′ NCR at the 5′ end of the replicative intermediate acts in cis to mediate positive-strand RNA synthesis. In addition, the observation that the PVΔ3′NCR positive-strand RNA synthesis defect was further exacerbated in a neuroblastoma cell line compared to HeLa cells indicated that a limiting cellular factor interacting with the 3′ NCR might play a role in RNA replication. Previous studies had identified the interaction of the nuclear protein nucleolin with the 3′ NCR (51, 53). It has also been reported that recombinant encephalomyocarditis virus (EMCV) 3Dpol, as well as the poliovirus polymerase precursor 3CD, bind this region of the genome (18, 26). As there has yet to be a function ascribed to the binding of these proteins to the poliovirus 3′ NCR, we hypothesized that the 5′ end of negative-strand RNA (the complement to the 3′ NCR) interacts with additional viral and/or cellular factors to promote efficient positive-strand RNA synthesis.
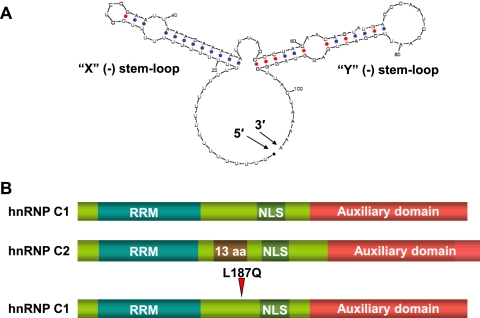
The 5′ end of poliovirus negative-strand RNA has been predicted by RNA-folding algorithms (59) to adopt stem-loop structures highly similar to those of its positive-strand equivalent (Fig. 1A), including base pairing with the poly(U) tract in negative-strand RNA. Initial RNA-protein binding assays carried out in our study using radiolabeled RNA probes corresponding to the 5′ end of negative-strand RNA, here denoted 5′(−), identified distinct ribonucleoprotein (RNP) complexes in extracts from poliovirus-infected HeLa cells. The appearance and pattern of these complexes were similar to those observed in previous studies using the opposite terminus of the poliovirus replicative intermediate, the 3′ end of negative-strand RNA [3′(−)]. Roehl and Semler first reported the appearance of these complexes using UV cross-linking approaches, and the identity of the 36-kDa protein component of such complexes was later determined by mass spectrometry to be the cellular protein heterogeneous nuclear ribonucleoprotein C (hnRNP C) by Brunner et al. (15, 43). hnRNP C is a predominantly nuclear protein that is highly abundant in human cells and exists as two alternatively spliced isoforms, C1 and C2 (22) (Fig. 1B). In the cell, hnRNP C forms a heterotetramer consisting of three C1 molecules and a single C2 molecule. This arrangement is thought to orient the amino-terminal high-affinity RNA recognition motifs (RRM) to bind RNA. hnRNP C functions in cellular mRNA biogenesis processes in the nucleus, including mRNA splicing and stabilization of pre-mRNA (22, 46).
FIG. 1.
The 5′ end of poliovirus negative-strand RNA and hnRNP C proteins. (A) Mfold-predicted structure of the 5′ end of poliovirus negative-strand RNA, the complement of the genomic-strand 3′ NCR. Shown are the X(−) stem-loop (formed by base pairing that includes a stretch of uridine residues) and the Y(−) stem-loop. The 5′ and 3′ ends of the RNA structure are indicated (59). (B) Scheme depicting the domains of wild-type hnRNP C1 and C2 proteins. NLS, nuclear localization signal; auxiliary domain, region implicated in protein-protein interactions. Indicated is the additional 13-amino-acid (aa) region of C2 proteins. The location of the CID mutation engineered to create hnRNP C1-L187Q is also indicated.
In this study, we used RNA binding assays in conjunction with the immunoprecipitation (IP) of RNP complexes by hnRNP C antibodies to demonstrate that hnRNP C binds to the 5′ end of poliovirus negative-strand RNA, providing evidence that hnRNP C can bind both termini of viral negative-strand RNA intermediates. We further show that addition of recombinant wild-type hnRNP C1 protein, but not C1 proteins harboring a mutation in the multimerization domain (56), to poliovirus in vitro translation/replication assays can stimulate poliovirus RNA synthesis, suggesting that hnRNP C binding as a multimer may be important for its participation in RNA replication during virus infection. In addition, we used retrovirus-mediated expression of hnRNP C-specific short hairpin RNAs (shRNAs) to significantly reduce the levels of hnRNP C proteins in HeLa cells. Poliovirus yields were decreased 5-fold in hnRNP C-specific shRNA-treated cells compared to infections of control shRNA-treated cells. Using quantitative real-time PCR analysis, we determined that the accumulation of positive-strand RNA is selectively decreased in hnRNP C-depleted cells during the early phases of poliovirus infection, implying a role for hnRNP C proteins in virus positive-strand RNA synthesis. Our results suggest that cellular hnRNP C proteins play an important role in the formation of positive-strand RNA replication complexes in infected cells.
MATERIALS AND METHODS
Cell culture.
HeLa cells and derivative cell lines were maintained in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 8% newborn calf serum. NGP cells were maintained in DMEM supplemented with 20% fetal calf serum. Phoenix-Ampho 293T cells (47) were maintained in DMEM supplemented with 10% fetal calf serum.
Generation of RNA probes used in this study.
To generate the plasmid pNCRRz5′−, a full-length poliovirus cDNA clone, pT7PV1 (25), was digested with StuI and BglII. The 4.2-kb portion of the plasmid was gel purified, treated with calf intestine phosphatase for 1 h at 37°C, and phenol-chloroform extracted twice to remove contaminating protein. The DNA vector fragment was precipitated with 2.5 volumes of 100% ethanol. Synthetic oligonucleotide pairs (Operon) were used in the generation of the plasmid (Table 1 lists all oligonucleotides used in this study).
TABLE 1.
Oligonucleotides used in this study
| No. | Name | Sequence (5′-3′) |
|---|---|---|
| 1 | NCRRz1-Fwd | CCTTAATACGAACTCACTATAGGAAAAAAAAAAAAAAACTGATGAGGCCGAAAGGCCG |
| 2 | NCRRz1-Rev | TTCGGCCTCATCAGTTTTTTTTTTTTTTTCCTATAGTGAGTCGTATTAAGG |
| 3 | NCRRz2-Fwd | AAAACCCGGTATCCCGGGTTCTTTTTTTTTTTTTTTCCTCCGAATTAAAGAAAAATTT |
| 4 | NCRRz2-Rev | TTCTTTAATTCGGAGGAAAAAAAAAAAAAAAGAACCCGGGATACCGGGTTTTCGGCCT |
| 5 | NCRRz3-Fwd | ACCCCTACAACAGTATGACCCAATCCAATTCGACTGAGGTAGGGTTACTA |
| 6 | NCRRz3-Rev | GATCTAGTAACCCTACCTCAGTCGAATTGGATTGGGTCATACTGTTGTAGGGGTAAATTT |
| 7 | NdeIC1pET22b | GGGGCATATGGCCAGCAACGTTACCAACAAG |
| 8 | EcoRIC1pET22b | GGGGGAATTCGGTCCTCCATTGGCGCTGTCTCT |
| 9 | CIDdisruptL187Q | CCTTCAGGCCATTAAGAAGGGGGAGACCCAGATAAAACAAAAAGTGG |
| 10 | CIDdisruptL187Q-r | CCACTTTTTGTTTTATCTGGGTCTCCCCCTTCTTAATGGCCTGAAGG |
| 11 | hcshRNAoligosense | GATCCCCCGTCAGCGTGTATCAGGAATTCAAGAGATTCCTGATACACGCTGACGTTTTTA |
| 12 | hcshRNAoligoanti | AGCTTAAAAACGTCAGCGTGTATCAGGAATCTCTTGAATTCCTGATACACGCTGACGGGG |
| 13 | hcshRNAScramfwd | GATCCCCCCGTAAGATCTGGGGCCTCCAAATACCTGTGGTGGAATAAATTCCAGTTTTTA |
| 14 | hcshRNAScramrev | AGCTTAAAAACTGGAATTTATTCCACCACAGGTATTTGGAGGCCCCAGATCTTACGGGGG |
| 15 | PV+Rev | CACTGTCCTGCTCTGGTTGG |
| 16 | PV-Rev | ACTCCTGACAACAACCAGACATC |
| 17 | β-actinRev | GGAGAAGCTGTGCTACGTCG |
| 18 | PV+Fwd | ATGTTCCTGTCGGTGCTGTG |
| 19 | PV-Fwd | GCGGGAACACAAAGGCATTC |
| 20 | β-actinFwd | GTGGATCAGCAAGCAGGAG |
| 21 | Linker 1F | CCGGAGGAAAAAAAAAAAAAAAA |
| 22 | Linker 1R | TTTTTTTTTTTTTTTTTTTTCCT |
| 23 | Linker 2F | AAAAAAAAACTATAGTGTCACCTAAATG |
| 24 | Linker 2R | AATTCATTTAGGTGACACTATAGTTTTT |
| 25 | HincIIBbsIF | TTAAAACAGCTCTGGGGTTGTACCCACCCCAGAGGCCCAC |
| 26 | HincIIBbsIR | CCCAGTGGGCCTCTGGGGTGGGTACAACCCCAGAGCTGTTTTAA |
| 27 | BspeIMut+ | GGGTCATACTGTTGTAGGGGTAAATTTTTCTTTAATCCGGAGGA |
| 28 | BspeIMut− | TCCTCCGGATTAAAGAAAAATTTACCCCTACAACAGTATGACCC |
| 29 | BspeIMutBck+ | GGGTCATACTGTTGTAGGGGTAAATTTTTCTTTAATTCGGAGGA |
| 30 | BspeIMutBck− | TCCTCCGAATTAAAGAAAAATTTACCCCTACAACAGTATGACCC |
DNA oligonucleotides were 5′ phosphorylated with T4 polynucleotide kinase (NEB) in the presence of 1 mM ATP. Following kinase treatment, oligonucleotide pairs 1 and 2, 3 and 4, and 5 and 6 were pooled, boiled for 3 min, and then allowed to cool to room temperature. Two hundred picomoles of each oligonucleotide pair (three in total) were incubated with the 4.2-kb vector fragment of pT7PV1 in the presence of T4 DNA ligase to generate pNCRRz5′−.
To synthesize the NCRRz5′− RNA probe, pNCRRz5′− plasmid was linearized with BglII and used in an RNA transcription reaction using T7 RNA polymerase in the presence of [32P]UTP. The transcription reaction mixture was incubated at 37°C for 2 h, followed by digestion with DNase I for 15 min. The probe was purified on a 7 M urea, 8% polyacrylamide gel for 3 h at 400 mV to allow separation of the probe and the hammerhead ribozyme RNA species. The NCRRz5′− RNA was excised from the urea gel and eluted using oligonucleotide elution buffer (0.5 M NH2OAc, 1 mM EDTA, 0.1% SDS) at room temperature overnight. The eluted probe was extracted twice with phenol-chloroform and precipitated with ethanol at −20°C for 1 h, and the probe was resuspended in diethyl pyrocarbonate (DEPC)-treated H2O and quantified using a scintillation counter (Beckman-Coulter).
Preparation of HeLa cell extracts.
The preparation of HeLa cytoplasmic S10 extracts has been described previously (33). Crude cytoplasmic extracts from mock-infected and poliovirus-infected HeLa and NGP cells were prepared as described previously (15).
UV cross-linking assays.
UV cross-linking assays were carried out as previously described (42). Briefly, 50 μg of either HeLa cell S10 extracts or crude cytoplasmic extracts was incubated in the presence of 1× binding buffer (5 mM HEPES-KOH [pH 7.8], 25 mM KCl, 10 mM MgCl2, 150 mM NaCl, 0.1 mM EDTA, and 3.8% [vol/vol] glycerol). One microgram of poly(I·C) RNA (Invitrogen) was included in reaction mixtures to reduce nonspecific protein binding. The reaction mixtures were incubated at 30°C for 10 min, followed by addition of the radiolabeled probe (approximately 0.2 pmol), and further incubated at 30°C for 15 min. For experiments in which competition for formation of RNP complexes was assayed, the competitor RNAs were added at the initial incubation step. Samples were exposed to 254-nm UV light for 8 min using a Stratalinker (Stratagene). The cross-linked complexes were treated with 0.5 μg of RNase A and 30 units of RNase T1 for 30 min at 37°C. An equal volume of Laemmli sample buffer was added to each reaction mixture prior to boiling it for 3 min, and samples were resolved on a 12.5% polyacrylamide-SDS-containing gel at 130 mV overnight. For relative size comparison, [35S]methionine-labeled in vitro translation of poliovirus virion RNA (vRNA) was run alongside sample reactions. The gels were dried and developed using a phosphorimager (Bio-Rad).
Immunoprecipitation of UV cross-linked RNA-protein complexes.
Immunoprecipitations of UV cross-linked proteins were carried out as a standard UV cross-linking assay except that reaction mixtures to be immunoprecipitated contained five times more total protein, poly(I·C) RNA, buffer, and RNA probe than reaction mixtures that were not precipitated. Following RNase treatment, 500 μl of IP buffer (50 mM Tris-HCl [pH 8.0], 5 mM EDTA, 150 mM NaCl, and 0.5% NP-40) was added to the reaction mixtures, along with 5 μl (approximately 5 μg) of either purified IgG (Zymed) or C1/C2 monoclonal antibody (Abcam). Samples were incubated with gentle rocking for 1.5 h at 4°C. Forty microliters of a prewashed 50% slurry of protein A-agarose beads (Roche) was added, followed by an additional incubation of 30 min at 4°C. Samples were pelleted and washed twice with ice-cold IP buffer before the addition of Laemmli sample buffer and resolution on a 12.5% polyacrylamide-SDS-containing gel. The gels were imaged using a phosphorimager.
Preparation and expression of recombinant proteins.
To generate pET22b-hnRNP C1, the plasmids pECE-FlagC1 and pECE-FlagC2 (generous gifts from M. Holcik) were PCR amplified using the synthetic oligonucleotides 7 and 8 to yield the C1 and C2 cDNA fragments flanked by NdeI and EcoRI restriction sites, respectively. The insert was digested with NdeI and EcoRI and cloned into the pET22b (EMD Biosciences) protein expression vector. The pET22bC1-L187Q plasmid was generated by site-directed mutagenesis of pET22b-C1 using the synthetic oligonucleotides 9 and 10. After verification by sequencing, the plasmids were transformed, and recombinant hnRNP C proteins were expressed in BL21(DE3) cells by induction with IPTG (isopropyl-β-d-thiogalactopyranosidase) at an optical density of 0.4 for 3 h at 18°C. The cells were pelleted, washed once with sucrose buffer (25% sucrose, 50 mM Tris-HCl [pH 8.0], 40 mM EDTA), and lysed with lysozyme buffer (25 mM Tris-HCl [pH 8.0], 2.0 × 105 U/ml chicken egg white lysozyme σ) at 0°C for 15 min, followed by sonication. The samples were then centrifuged at 10,000 rpm in a JA-17 (Beckman) rotor for 15 min. The supernatants were precipitated with ammonium sulfate (20% [wt/vol]) for 1 h, and the pellets were resuspended and dialyzed overnight in I30 buffer (20 mM Tris-HCl [pH 8.0], 250 mM NaCl, 30 mM imidazole, 10% glycerol, 0.5% NP-40). Following dialysis, samples were purified by Ni2+ ion-based affinity chromatography (GE Healthcare).
RNA electrophoretic mobility shift assays.
RNA electrophoretic mobility shift assays were carried out as described previously (55), except that the binding buffer consisted of 20 mM Tris-HCl (pH 8.0), 0.1 M NaCl, 1 mM dithiothreitol (DTT), 0.1% NP-40, and 3.6% glycerol. Incubations were carried out at room temperature for 20 min, with a final concentration of radiolabeled RNA probe of 0.1 nM. In reaction mixtures containing antibodies, 0.2 μg of antibody was added after the initial 20-min incubation for 10 min before they were loaded onto the glycerol-containing polyacrylamide gel.
In vitro translation/RNA replication.
In vitro translation/replication assays were carried out with HeLa S10 cytoplasmic extracts using poliovirus virion RNA as previously described (39), and labeled RNAs from replication reaction mixtures were isolated using RNaqueous spin columns (Ambion).
Recombinant retrovirus production and infection.
To deplete hnRNP C proteins from tissue culture cells, the pSuper RNA interference (RNAi) system (Oligoengine) was used to generate cell lines containing an integrated retrovirus element capable of constitutive or tetracycline-inducible production of hnRNP C-specific shRNAs. The pSuperior-retro-puro plasmid containing the retrovirus construct was digested with BglII and HindIII. The 7.0-kb vector fragment was excised from an agarose gel and treated with calf intestine alkaline phosphatase. The vector fragment was ligated with either the HC12KD oligonucleotide pair 11 and 12 or the HC12Scram oligonucleotide pair 13 and 14. Each oligonucleotide had been synthesized to contain a 5′ phosphate group (Operon). Following verification of the plasmids by sequencing, pHC12KD and pHC12Scram were transfected into 50% confluent monolayers of the packaging cell line Phoenix-Ampho 293T in 100-mm plates (47) using Fugene (Roche). Harvesting and concentration of retrovirus virion particles were carried out as described previously (6). Monolayers of HeLa cells in 100-mm plates (50% confluence) were infected with a resuspension of retrovirus particles (HC12KD or HC12Scram) consisting of 5 ml DMEM supplemented with 8% newborn calf serum and 4 μg/ml Polybrene. Following retrovirus infection, the cells were allowed to recover for 24 h and then passaged to 30-cm tissue culture plates, and infected cells were selected using DMEM supplemented with 8% newborn calf serum and 2 μg/ml puromycin. Knockdown of hnRNP C was verified by Western blotting with an hnRNP C-specific monoclonal antibody (Abcam).
One-step growth analysis.
One-step growth analysis was performed on HeLa, HeLaHC12KD, or HeLaHC12Scram cell line monolayers in 60-mm plates at 37°C as described previously (13). The monolayers were infected at a multiplicity of infection (MOI) of 25. Virus was adsorbed onto the monolayers for 30 min, and then they were washed twice with phosphate-buffered saline (PBS). Following infection, the cell monolayers were incubated in 4 ml of DMEM supplemented with 8% newborn calf serum. The cell monolayers and media were harvested at the indicated times postinfection, and virus was released from the cells by freezing/thawing them five times. Viral titers were determined by plaque assay on HeLa cells in triplicate and were reported as PFU per cell.
Total cellular-RNA extraction.
Total cellular RNA from HeLa cells or retrovirus-infected HeLa cells was harvested using Tri-reagent (Molecular Research Center, Inc.). Cell monolayers were infected with wild-type poliovirus or the virus mutant PVΔ3′NCR at an MOI of 25 at 37°C. At the indicated times postinfection, the medium was aspirated from the cell monolayers in 60-mm plates, and 1 ml of Tri-reagent was added. Total cellular RNA was removed from the lysates following chloroform extraction of the aqueous phase. The RNA was treated with DNase I for 30 min at 37°C and purified using RNaqueous spin columns.
Quantitative-PCR measurement of poliovirus RNA strand synthesis.
Purified total cellular RNA from HeLa cells was reverse transcribed with Moloney murine leukemia virus (M-MuLV) reverse transcriptase (New England Biolabs). Synthetic oligonucleotides used to prime cDNA synthesis were prepared based upon the Beacon Designer program's optimal predictions for quantitative PCR (Premier Biosoft International). The oligonucleotides used were as follows: PV+Rev (oligonucleotide 15) (used for poliovirus positive-strand RNA analysis), PV−Rev (oligonucleotide 16) (used for poliovirus negative-strand RNA analysis), and β-actinRev (oligonucleotide 17) (used for β-actin cellular-RNA analysis). PCR assays were carried out using IQ SYBR green supermix (Bio-Rad), the above-mentioned primers, and their counterparts, PV+Fwd (oligonucleotide 18), PV−Fwd (oligonucleotide 19), and β-actinFwd (oligonucleotide 20). Cesium-chloride-purified plasmid DNA of pT7PV1 (for positive-strand RNA), pT7NPV1 (for negative-strand RNA), or β-actin cDNA (Invitrogen) was used to generate standard curves. The real-time PCR conditions used consisted of an initial melting step at 95°C for 5 min. Samples were subjected to 40 amplification cycles consisting of 95°C for 10 s and 55°C for 13 s. Following PCR, a melting curve was performed at 55°C, increasing 0.5°C every 10 s for 80 cycles. The reaction mixtures were subjected to electrophoresis on a 2% Tris-acetate-EDTA (TAE)-agarose gel to check for any nonspecific DNA amplification that might contaminate the sample reactions. The β-actin gene was used as a reference gene for positive- and negative-strand RNA analysis in HeLa and derivative cell lines.
To generate the pT7NPV1 DNA plasmid, the full-length cDNA clone pT7PV1 and the vector pUC19 were digested with BamHI and EcoRI restriction endonucleases. The ∼2.9-kb fragment of pT7PV1 was ligated to the ∼2.6-kb vector portion of pUC19 to create pUC19pT7PV1. To facilitate subcloning, pUC19pT7PV1 was subjected to site-directed mutagenesis with oligonucleotides 27 and 28 to create pUC19pT7PV1BspE1 (for the conditions, see reference 7). The resulting plasmid, pUC19pT7PV1 BspE1, containing the BspE1 restriction site, was digested with BspE1 and EcoRI. The resulting ∼5.6-kb fragment was gel purified and incubated with oligonucleotide pairs 21-22 and 23-24 in the presence of T4 DNA ligase to generate the subclone pUC19pT7PV1BspE1SP6r. This subclone was then digested with BglII and EcoRI, and the ∼2.0-kb fragment was religated to the ∼7.9-kb portion of the poliovirus cDNA clone pT7PV1 digested with BglII and EcoRI to generate pT7PV1BspE1SP6r. To introduce a T7 promoter, pT7PV1BspE1SP6r was digested with StuI and PmlI, and the ∼9.9-kb portion of the plasmid was gel purified prior to additional ligation. The 242-bp vector fragment of the plasmid TVT7R digested with HincII and BbsI was incubated with the oligonucleotide pair 25 and 26 in the presence of the ∼9.9-kb pT7PV1BspE1SP6r DNA fragment and T4 DNA ligase, resulting in the plasmid pT7NPV1BspE1. Finally, pT7NPV1BspE1 was subjected to site-directed mutagenesis with the oligonucleotide pair 29 and 30 to remove the BspE1 cloning site. All quantitative-PCR assays were carried out in triplicate a minimum of three times.
RESULTS
RNP complex formation using the 5′(−) RNA probe and extracts generated from poliovirus-infected cells.
The terminal nucleotides of the 5′ end of poliovirus negative-strand RNA base pair to form two stem-loop structures similar to the X and Y stem-loops that have been determined to form in the complementary 3′ NCR of the poliovirus genomic RNA (40). In the same manner that the X stem-loop consists of residues from the poliovirus 3′ NCR and the poly(A) tail, the poliovirus poly(U) sequence at the 5′ end of the negative-strand RNA base pairs with the RNA sequence complementary to the 3′ NCR to form a stem-loop structure, referred here as X(−) (Fig. 1A). A virus engineered to harbor a deletion of the 3′ NCR, PVΔ3′NCR, is viable in cell culture; however, its ability to synthesize poliovirus positive-strand RNA is diminished, leading to a decrease in virus yield (13, 50). We hypothesized that the RNA synthesis defect is due to a lack of the 3′ NCR complement at the 5′ end of the negative-strand RNA. Brown and colleagues' initial characterization of the PVΔ3′NCR virus used HeLa cells, as well as the neuronal cell line SK-N-SH (13). The observation that the replication defect was even more pronounced in SK-N-SH cells suggested a cellular factor might be involved. To determine the ability of the 5′ end of poliovirus negative-strand RNA to interact with cellular (or viral factors), we carried out UV cross-linking assays using crude extracts generated from mock-infected or poliovirus-infected cells and a radiolabeled probe corresponding to the 5′-terminal 71 nucleotides of the negative strand, complete with a poly(U) “leading sequence” at the 5′ end. To generate a bona fide 5′ end, a cis-active hammerhead ribozyme sequence (27, 45) was included downstream of the T7 promoter sequence and upstream of the start of the poly(U) sequence. In this way, the two G residues included in in vitro RNA transcription using T7 polymerase would be removed, allowing the 5′ end to begin with the authentic poly(U) sequence (Fig. 2A).
FIG. 2.
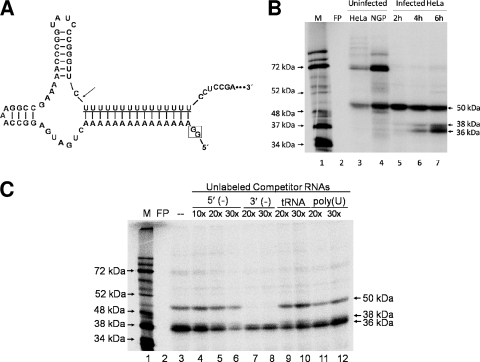
Binding of proteins from poliovirus-infected cells to the 5′ end of virus negative-strand RNA. (A) Mfold-predicted structure of the 5′ end of poliovirus negative-strand RNA with a 5′ hammerhead ribozyme. The “2-G start” (boxed) resulting from RNA transcription from the T7 promoter is indicated, followed by the nucleotide sequence forming the hammerhead ribozyme structure. The arrow indicates the site of autocatalytic cleavage of the ribozyme and the poly(U) sequence of poliovirus negative-strand RNA, followed by a portion of the 5′ end of the negative-strand RNA corresponding to the genomic 3′ NCR (underlined) (59). (B) UV cross-linking assays with poliovirus 5′(−) RNA probe and cytoplasmic extracts from mock-infected HeLa (lane 3) and mock-infected NGP (lane 4) cells. Lanes 5 to 7 show complexes from UV cross-linking assays carried out in the presence of 50 μg of cytoplasmic extracts generated from poliovirus-infected HeLa cells harvested at 2, 4, and 6 h postinfection, respectively. Samples were boiled in the presence of Laemmli sample buffer and resolved on a 12.5% polyacrylamide-SDS-containing gel. Analysis was carried out using a phosphorimager (Bio-Rad). Lane M, marker proteins ([35S]methionine-labeled in vitro translation of poliovirus virion RNA). FP, free probe cross-linked in the absence of any extract and treated with RNase as described in Materials and Methods. (C) UV cross-linking assay using extracts from poliovirus-infected cells in the presence of unlabeled competitor RNAs. These studies were carried out using cytoplasmic extracts generated from poliovirus-infected HeLa cells at 6 h postinfection. Competitor RNAs corresponding to nonradiolabeled 5′(−) RNA probe (lanes 4 to 6), nonradiolabeled 3′(−) RNA probe (lanes 7 and 8), tRNA (lanes 9 and 10), or nonradiolabeled transcript of 20 uridine residues (lanes 11 and 12) were added to reaction mixtures at the indicated molar excess compared to the amount of [32P]UTP-radiolabeled 5′(−) RNA probe. Analysis was carried out using a phosphorimager. Lane M, marker proteins ([35S]methionine-labeled in vitro translation of poliovirus virion RNA). FP, free probe in the absence of any extract.
The 5′(−) RNA probe was uniformly labeled with [32P]UTP and UV cross-linked in the presence of cytoplasmic extract generated from mock-infected cells (Fig. 2B, lanes 3 and 4) or poliovirus-infected cells (Fig. 2B, lanes 5 to 7). In mock-infected extracts of HeLa cells and the human neuronal cell line NGP, RNA-protein complexes migrating at approximately 72 and 50 kDa were observed (Fig. 2B, lanes 3 to 4). In crude cytoplasmic extracts generated from poliovirus-infected cells, we detected the formation of the 50-kDa complex and the disappearance of the 72-kDa complex. This may be due to cleavage of the 72-kDa protein(s) by the poliovirus proteinases, or in the case of a shuttling protein, these factors may be unable to bind to poliovirus RNA during the course of infection due to a disruption in cell trafficking and/or membrane rearrangement (16, 23, 24). In cell extracts harvested from cells at 4 h postinfection (Fig. 2B, lane 6), two lower-molecular-weight complexes appeared, with the association increasing at a later time postinfection (Fig. 2B, lane 7). The approximate molecular masses of these complexes are 38 and 36 kDa, and together with the 50-kDa complex, they bear a striking similarity to previous studies by Roehl and Semler in which UV cross-linking assays were carried out using the 3′ end of poliovirus negative-strand RNA (43). In that study, the 50-kDa complex was observed using extracts from mock- and poliovirus-infected cells UV cross-linked with the 3′-terminal 66 nucleotides of poliovirus negative-strand RNA; the 38- and 36-kDa complexes were detected in extracts harvested at approximately 3.5 h postinfection and later (43).
To determine if the 5′(−) probe forms the above-described RNP complexes in a specific manner, we performed UV cross-linking competition assays using the radiolabeled 5′(−) RNA probe and nonradiolabeled RNAs in molar excess as competitors (Fig. 2C). Nonradiolabeled 5′(−) RNA preincubated with the assay reaction mixtures before addition of radiolabeled probe competed for binding of proteins in a dose-dependent manner (Fig. 2C, lanes 4 to 6). tRNA was used as a nonspecific competitor RNA control with the additional advantage that, like the 5′(−) end, tRNA adopts a stable RNA secondary structure (Fig. 2C, lanes 9 and 10). Addition of tRNA in molar excess did not compete for binding with any of the complexes. Since the 3′ end of the poliovirus negative strand is also capable of binding RNP complexes of 50, 38, and 36 kDa in extracts from infected cells, we preincubated unlabeled 3′(−) RNA corresponding to the terminal 66 nucleotides of the 3′ end of the poliovirus negative strand. While the 3′(−) RNA was capable of completely competing for the 50-kDa complex (Fig. 2C, lanes 7 and 8), we observed only partial competition for the 38- and 36-kDa protein complexes. This suggested that while the 5′ and 3′ poliovirus negative-strand termini bind to some of the same proteins in the infected cell, they may not necessarily bind all of the same proteins or with the same affinity (17, 32). To rule out the possibility that the proteins interacting with the 5′(−) end were binding to the poly(U) sequence at the 5′ terminus of the probe and not the unique poliovirus RNA nucleotide sequence, we carried out UV cross-linking competition assays using T7 RNA polymerase transcripts consisting of 20 uridine residues (Fig. 2C, lanes 11 and 12). No significant competition was observed with the poly(U) competitor. These data indicate that the 5′ end of poliovirus negative-strand RNA interacts with the 38- and 36-kDa proteins in a specific manner.
Identification of hnRNP C binding to the 5′ end of negative-strand poliovirus RNA.
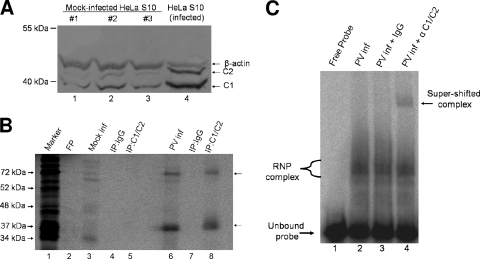
Based upon our initial RNA binding studies examining protein factors interacting with the 5′(−) end, we had evidence that suggested the 36-kDa RNP complex might be the cellular protein hnRNP C that had previously been identified to bind to the 3′(−) end of the RNA (15). Protein complexes UV cross-linked with the radiolabeled RNA probe were immunoprecipitated using either IgG or a monoclonal antibody for hnRNP C as a control. hnRNP C proteins could not be immunoprecipitated in cytoplasmic extracts generated from mock-infected cells (Fig. 3B, lanes 3 to 5), possibly due to the fact that the majority of cellular hnRNP C is localized in the nuclei of uninfected cells (35). Cytoplasmic extracts generated from mock-infected HeLa cells also contain some hnRNP C protein representing the basal levels of hnRNP C that have not yet translocated to the nucleus. However, S10 cytoplasmic extracts generated from poliovirus-infected HeLa cells contain increased amounts of cytoplasmic hnRNP C (Fig. 3A, compare lanes 1 to 3 to lane 4). hnRNP C and other hnRNP proteins have been shown to accumulate in the cell cytoplasm of picornavirus-infected cells (14, 15, 23). When crude extracts generated from poliovirus-infected HeLa cells at 6 h postinfection were cross-linked to the 5′(−) probe, hnRNP C proteins could be readily immunoprecipitated with the monoclonal antibody (Fig. 3B, lanes 6 and 8). Two separate protein complexes (arrows) were immunoprecipitated in our assay; a complex comigrating with the lower RNP complex observed in our initial UV cross-linking and a higher-molecular-weight complex that most likely represented a multimer of hnRNP C covalently linked to the RNA probe. In reactions using extracts from mock- or poliovirus-infected cells, the IgG control did not precipitate any of the detected cross-linked proteins (Fig. 3B, lanes 4 and 7).
FIG. 3.
The cellular protein hnRNP C binds the 5′ end of poliovirus negative-strand RNA. (A) Increased concentrations of hnRNP C proteins in HeLa S10 cytoplasmic extracts generated from poliovirus-infected cells. Cytoplasmic extracts of HeLa cells were prepared from either mock-infected cells (three separate extracts; lanes 1 to 3) or poliovirus-infected cells (lane 4) at 5 h postinfection. Equivalent amounts of total protein (10 μg) from each extract were resolved on a 12.5% polyacrylamide-SDS-containing gel. Western blot analysis was carried out using mouse monoclonal anti-C1/C2 as the primary antibody (Abcam) and an alkaline phosphatase-conjugated anti-mouse IgG (Zymed) as a secondary antibody. Molecular masses are indicated on the left. As a loading control, the blots were also probed with mouse monoclonal anti-β-actin antibody (Abcam). (B) Immunoprecipitation of UV cross-linked complexes confirms binding of hnRNP C to the 5′(−) RNA probe. UV cross-linking assays were carried out as described in Materials and Methods with the [32P]UTP-radiolabeled 5′(−) RNA probe and cytoplasmic extracts generated from mock-infected (Mock inf) (lanes 3 to 5) or poliovirus-infected (PV inf) (lanes 6 to 8) HeLa cells at 5 h postinfection. [32P]UTP-radiolabeled UV cross-linked samples from mock- or poliovirus-infected HeLa cells were immunoprecipitated by the inclusion of normal IgG (lanes 4 and 7) or hnRNP C1/C2 antibody (lanes 5 and 8) in reaction mixtures following RNase digestion. The immunoprecipitated complexes were resolved on a 12.5% polyacrylamide-SDS-containing gel. Analysis was carried out using a phosphorimager. Lane M, marker proteins ([35S]methionine-labeled in vitro translation of poliovirus virion RNA). FP, free probe in the absence of any extract. (C) Electrophoretic mobility shift assays using extracts generated from poliovirus-infected cells in the presence or absence of antibody to analyze the contents of RNP complexes. Cytoplasmic extracts (1 μg) from poliovirus-infected HeLa cells were incubated with radiolabeled poliovirus 5′(−) RNA probe. The reaction mixtures in lanes 3 and 4 contained 0.1 μg of either IgG or C1/C2 monoclonal antibody, respectively. The unbound probe is indicated at the bottom left, and the appearance of a supershifted complex in lane 4 is indicated by an arrow at the top right.
To confirm our identification of hnRNP C binding to the 5′(−) end, we carried out electrophoretic mobility shift assays of our radiolabeled RNA probe in the presence of extracts generated from poliovirus-infected HeLa cells. In Fig. 3C, incubation of extracts generated from poliovirus-infected HeLa cells resulted in the formation of multiple RNP complexes (Fig. 3C, lane 2). Inclusion of hnRNP C monoclonal antibody (Fig. 3C, lane 4) resulted in a supershifted complex compared to the use of IgG as a negative control (Fig. 3C, lane 3). Based upon these findings and our immunoprecipitation data, we conclude that hnRNP C is a participant in the formation of 5′(−) RNP complexes.
Multimerization of hnRNP C bound to poliovirus negative-strand RNA.
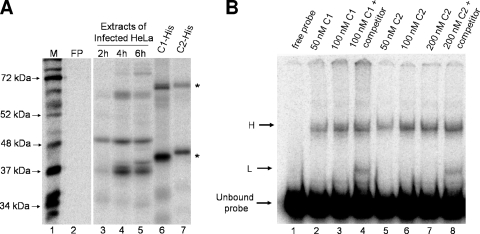
To examine the function of hnRNP C in poliovirus RNA replication, we engineered recombinant forms of histidine-tagged hnRNP C1 and C2 previously shown to be capable of binding cellular hnRNAs (56). Recombinant hnRNP C1-His proteins formed two separate complexes in our UV cross-linking reactions (Fig. 4A, lane 6, asterisks). These complexes were similar to those observed with endogenous protein in extracts from poliovirus-infected HeLa cells (compare Fig. 3B, lanes 6 and 8, to 4A, lane 6). Recombinant hnRNP C2 also bound the radiolabeled RNA probe, forming complexes similar to those seen with C1 (Fig. 4A, lane 7). Due to the presence of the histidine tag on recombinant hnRNP C1 and C2, the faster-migrating complexes in lanes 6 and 7 of Fig. 4A have slightly slower electrophoretic mobilities than the complexes formed with endogenous proteins in Fig. 4A, lanes 3 to 5, which migrated slightly faster than the 37-kDa molecular mass marker in lane 1. Electrophoretic mobility shift assays confirmed the ability of recombinant hnRNP C1-His (Fig. 4B, lanes 2 and 3) and hnRNP C2-His (Fig. 4B, lanes 5 to 7) to bind the 5′(−) RNA probe and form RNP complexes in a dose-dependent manner (designated H, for high-molecular-mass complex, in Fig. 4B). When nonradiolabeled 5′(−) RNA was used as an RNA competitor in molar excess (Fig. 4B, lanes 4 and 8), a faster-migrating complex (designated L, for low-molecular-mass complex) was observed compared to reactions lacking specific RNA competition, perhaps as a result of partial dissociation of the higher-molecular-mass complex. These data suggest that recombinant hnRNP C (and, by extrapolation, endogenous hnRNP C in our HeLa cell extracts) is capable of binding the 5′(−) terminus as a multimeric complex. Wan and colleagues have previously reported in vitro hnRNP C multimerization in complexes with cellular RNAs (56), providing additional support for our conclusions.
FIG. 4.
Binding of hnRNP C to the 5′ end of poliovirus negative-strand RNA under nondenaturing conditions. (A) Binding of recombinant hnRNP C to poliovirus negative-strand RNA. UV cross-linking assays were carried out using 50 μg of extracts generated from poliovirus-infected HeLa cells at 2 h (lane 3), 4 h (lane 4), and 6 h (lane 5) postinfection. Lanes 6 and 7 show UV cross-linked complexes with bacterially expressed His-tagged hnRNP C recombinant proteins containing a carboxyl-terminal hexahistidine tag. Lane 6, 0.5 μg hnRNP C1-His; lane 7, 0.5 μg of hnRNP C2-His. Image analysis was carried out using a phosphorimager. Lane M, marker proteins ([35S]methionine-labeled in vitro translation of poliovirus virion RNA). FP, free probe in the absence of any extract, as described in the legend to Fig. 2. (B) RNA mobility shift assays were carried out as described in Materials and Methods. Recombinant purified hnRNP C1 (lanes 2 to 4) or hnRNP C2 (lanes 5 to 8) was incubated with radiolabeled 5′(−) RNA probe. The competition reaction mixtures (lanes 4 and 8) contained nonradiolabeled 5′(−) RNA in molar excess. Following incubation, complexes were resolved on a 4% native polyacrylamide gel. The unbound probe is indicated on the left, as are the putative monomeric (L) and multimeric (H) complexes.
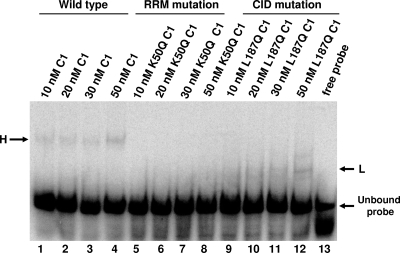
To further analyze the oligomeric state of hnRNP C binding to the 5′(−) probe, we carried out electrophoretic mobility shift assays using a version of hnRNP C1 with a leucine-to-glutamine mutation that disrupts the hydrophobic inner core of the C1-C1 interaction domain (CID), which has been determined to be important for hnRNP C multimerization (56). This mutated protein, here designated hnRNP C1-L187Q, had been shown previously to be defective in binding to a glutathione S-transferase (GST)-tagged recombinant hnRNP C1 (56). In our mobility shift assay, it formed a faster-migrating complex (L) than the complex (H) formed by wild-type recombinant hnRNP C1 (Fig. 5, compare lanes 9 to 12 with lanes 1 to 4). Although we have not confirmed their composition, the L and H complexes in Fig. 5 have electrophoretic mobilities similar to those shown in Fig. 4B and may represent equivalent species. These faster-migrating L complexes (but not the slower-migrating H complexes) formed with the 5′(−) RNA probe even at higher concentrations of recombinant hnRNP C1-L187Q protein ranging from 100 to 500 nM (data not shown), suggesting that the protein concentrations used in our assay were not limiting. The mutation of a leucine residue to glutamine was not predicted to disrupt the RRM of the hnRNP C protein, which is required for hnRNP C to bind RNA (22). This is in contrast to a previously described hnRNP C mutation, in which a lysine located in the RRM domain of hnRNP C1 was replaced with a glutamine. The mutated protein hnRNP C1-K50Q (produced as a recombinant GST fusion protein) could not bind poliovirus negative-strand RNA in a UV cross-linking assay (15), and we show here that a His-tagged version of hnRNP C1-K50Q was similarly incapable of binding to the 5′ end of poliovirus negative-strand RNA (Fig. 5, lanes 5 to 8). These data suggest that hnRNP C may bind the 5′ end of poliovirus negative-strand RNA as a multimeric protein complex.
FIG. 5.
Differential binding capabilities of wild-type and mutated hnRNP C proteins. Electrophoretic mobility shift assays were carried out as described in Materials and Methods. Recombinant purified hnRNP C1-His (lanes 1 to 4), hnRNP C1-K50Q (lanes 5 to 8), or hnRNP C1-L187Q (lanes 9 to 12) was incubated at the indicated concentrations with radiolabeled poliovirus 5′(−) RNA probe, and the resulting complexes were resolved on a 4% native polyacrylamide gel. Lane 13 contained free probe in the absence of any protein, with the unbound probe indicated at the lower right. L, lower-molecular-mass RNP complex; H, higher-molecular-mass RNP complex.
Interaction of poliovirus replication proteins with the 5′ end of negative-strand RNA.
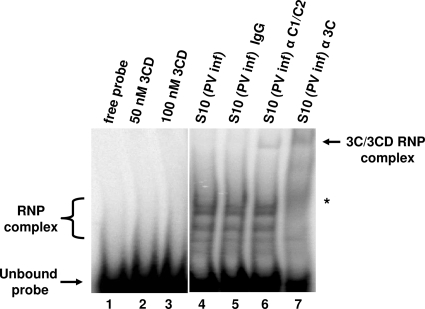
Previous studies have identified hnRNP C proteins as interacting with poliovirus nonstructural proteins, including 3CD (9, 15). Thus, viral proteins may interact with hnRNP C when it is bound to negative-strand RNA, either simultaneously or in a cooperative fashion, similar to what has been reported for the viral 3CD protein and cellular PCBP2 binding to poliovirus stem-loop I RNA (20, 37). Also, Banerjee et al. have previously identified viral protein 2C as binding the 3′ end of negative-strand RNA using assays similar to those employed in this study (2, 3). To determine whether PV replication proteins interact with the 5′ end of negative-strand RNA, we carried out mobility shift assays using the 5′(−) RNA probe and S10 cytoplasmic extracts generated from poliovirus-infected cells. As shown in Fig. 6, lanes 4 to 6, we observed several RNP complexes that formed with the 5′(−) RNA. At least one of these complexes could be supershifted by antibody to hnRNP C, but not by normal mouse IgG (Fig. 6, compare lane 5 to lane 6). In addition, inclusion of polyclonal antibodies capable of recognizing the viral proteins 3C and 3CD resulted in a supershifted complex (Fig. 6, lane 7). We also noted the disappearance of higher-mobility RNP complexes when 3C polyclonal antibody was added (Fig. 6, lane 7, asterisk). These may represent either 3C-containing proteins or binding partners indirectly interacting with the RNA probe via 3C/3CD. However, recombinant 3CD protein alone could not form a stable RNP complex with the 5′(−) RNA probe (Fig. 6, lanes 2 and 3). Recombinant 3CD proteins alone were similarly unable to bind to 5′(−) RNA probe in UV cross-linking assays (data not shown). Subsequent mobility shift assays using recombinant hnRNP C and 3C/3CD proteins did not result in a higher-molecular-weight complex than assays utilizing hnRNP C proteins alone (data not shown). These results suggest that, in order for 3C/3CD to stably interact with 5′(−) RNA, additional cellular proteins (such as hnRNP C) and/or viral factors may be required.
FIG. 6.
Interaction of infected-cell viral proteins 3C/3CD with the 5′ end of poliovirus negative-strand RNA. Electrophoretic mobility shift assays were performed as described in Materials and Methods. Recombinant purified 3CD protein containing a C147A mutation (a generous gift of R. Perera) was incubated with poliovirus 5′(−) RNA probe at the indicated concentrations. Supershift assays were carried out as described in Materials and Methods using a monoclonal antibody to hnRNP C1/C2 or a polyclonal antibody capable of recognizing virus proteins containing poliovirus 3C sequences (41). Cytoplasmic extracts from poliovirus-infected cells (1 μg) were incubated with the radiolabeled poliovirus 5′(−) RNA probe. The reaction mixtures in lanes 5, 6, and 7 contained 0.1 μg of normal mouse IgG (Zymed), mouse monoclonal hnRNP C1/C2 antibody (Abcam), or polyclonal antibody raised against recombinant poliovirus protein 3C, respectively. The presence of the supershifted complexes in lanes 6 and 7 is indicated by an arrow at the top right. An asterisk on the right indicates the disappearance of RNP complexes in lane 7.
Poliovirus RNA replication is stimulated by the addition of hnRNP C protein.
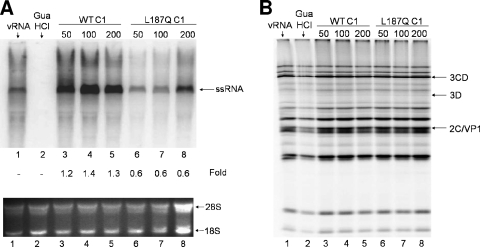
We next analyzed the abilities of our recombinant hnRNP C proteins to participate in poliovirus RNA replication. We carried out translation and RNA replication studies in vitro with wild-type hnRNP C1 protein and the recombinant hnRNP C1 harboring the L187Q mutation. Brunner et al. previously examined the activity of hnRNP C added to in vitro replication assays using a GST-hnRNP C fusion version of hnRNP C1 (15). However, the glutathione S-transferase tag is approximately 28 kDa, resulting in a combined protein molecule of ∼70 kDa that greatly exceeds the predicted 41-kDa size of endogenous hnRNP C1 (15). Since we have demonstrated the ability of these recombinant proteins to stably interact with the 5′ ends of negative-strand RNA probes, the His-tagged version of hnRNP C may better reflect in vitro the properties of hnRNP C in the infected cell. The addition of hnRNP C1-His to the RNA replication reaction increased RNA synthesis compared to negative-control reactions (Fig. 7A, compare lanes 3 to 5 to lane 1). The maximum increase in RNA synthesis occurred with the addition of approximately 100 nM hnRNP C1, whereas higher concentrations led to either decreased or no stimulation (Fig. 7A, lane 5, and data not shown). In contrast, our in vitro replication assays using the L187Q mutated hnRNP C1 resulted in a slight inhibition of poliovirus RNA synthesis at concentrations of 50 to 200 nM (Fig. 7A, compare lanes 6 to 8 to lane 1). Note that the fold increase value of 0.6 in lane 8 is based upon the normalized [32P]CTP-labeled single-stranded RNA (ssRNA) signal compared to rRNA (28S and 18S) loading controls shown at the bottom of Fig. 7A. These data suggest that poliovirus RNA synthesis stimulation requires hnRNP C proteins with an intact multimerization domain, perhaps reflecting the requirement for hnRNP C bound to poliovirus negative-strand RNA to recruit other protein binding partners. We also examined the effects of the addition of recombinant hnRNP C proteins on poliovirus translation. In assays using wild-type or the L187Q mutated protein, poliovirus translation was not affected (Fig. 7B). Based upon the increased levels of poliovirus RNA synthesis in vitro, we conclude that wild-type hnRNP C is capable of stimulating poliovirus RNA replication.
FIG. 7.
Stimulation of poliovirus RNA synthesis by the addition of wild-type (WT) hnRNP C1 proteins. Recombinant wild-type hnRNP C1 (lanes 3 to 5) or hnRNP C1-L187Q (lanes 6 to 8) protein or protein buffer alone (lane 1) was added to in vitro replication/translation reaction mixtures containing HeLa S10 cytoplasmic extracts from uninfected cells. Each reaction mixture was split into two fractions as described previously (15). (A) Replication of poliovirus virion RNA. Replication fractions containing purified poliovirus vRNA, HeLa S10 cytoplasmic extracts, and recombinant hnRNP C proteins were incubated at 30°C to allow translation of nonstructural proteins. Following addition of [32P]CTP to the reaction mixtures, RNA synthesis was allowed to proceed for 2 h. The replication reaction mixtures were then purified using an RNaqueous spin column and resolved on a 1.1% Tris-borate-EDTA-agarose gel containing ethidium bromide. As a negative control, 2 mM guanidine hydrochloride (GuaHCl) was added to the reaction mixture shown in lane 2 to inhibit negative-strand RNA synthesis. 18S and 28S rRNAs were used to confirm equal loading of samples. Comparative density values of the signals for RNA synthesis are depicted as fold increases compared to lane 1. (B) Translation fractions of the samples depicted in panel A were incubated with [35S]methionine and, following incubation at 30°C for 4 h, the reaction mixtures were boiled in Laemmli sample buffer and resolved on a 12.5% polyacrylamide-SDS-containing gel.
Knockdown of hnRNP C in HeLa cells delays poliovirus growth.
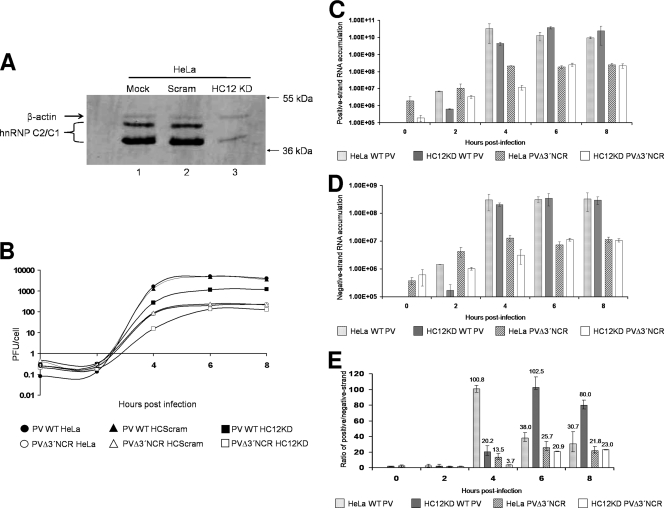
We next determined the contribution of hnRNP C to poliovirus replication by depleting the levels of hnRNP C proteins available to the virus replication complex. Efforts to deplete HeLa S10 cytoplasmic extracts of endogenous hnRNP C were unsuccessful, and transient transfection of small interfering RNAs (siRNAs) specific for hnRNP C mRNAs in HeLa cells only partially decreased hnRNP C expression (data not shown). To address the problem of significantly reducing a highly abundant cellular protein without disrupting normal cellular functions, we utilized a retrovirus-mediated shRNA expression system. A packaging cell line (Phoenix-Ampho 293T cells) was transfected with a retrovirus plasmid construct containing an integrated shRNA sequence under the control of an RNA polymerase III H1 promoter (34). These shRNAs were directed toward an mRNA sequence shared by the C1 and C2 isoforms of the hnRNP C protein. HeLa cells infected with the resulting retrovirus (HC12KD) were denoted HeLaHC12KD. These shRNA-producing retroviruses were collected from the packing cell line and used to infect HeLa cells prior to infection with poliovirus. As a control for any nonspecific effects of retrovirus infection, a retrovirus construct carrying a scrambled shRNA (HC12Scram) was also used. The incorporated retrovirus had the additional advantage of containing a puromycin gene, allowing the selection of HeLa cells containing the integrated vector. Whole-cell extracts from HeLa cells infected with the HC12KD retrovirus showed ∼70% knockdown of hnRNP C compared to extracts from cells infected with the HC12Scram retrovirus (Fig. 8A, compare lane 3 to lanes 1 and 2). To rule out possible pleiotropic effects from infection of cultured cells with the shRNA retrovirus, trypan blue staining of retrovirus-infected cell lines was used to assay for cell viability. Greater than 90% of cultured cell monolayers infected with either retrovirus stock (HC12KD or HC12Scram) were viable (data not shown). Therefore, the shRNA retrovirus system selectively knocked down hnRNP C protein without any major toxicity to treated cells. The establishment of a uniform population of HeLa cells depleted of hnRNP C allowed analysis of poliovirus replication kinetics without the caveat of the inefficiency of direct siRNA transfection. HeLa cells infected with the HC12KD or HC12Scram retrovirus were selected using puromycin and then infected with wild-type poliovirus to generate one-step growth curves (Fig. 8B). Compared to wild-type growth in either mock- or HC12Scram-infected cells, HC12KD-infected cells had approximately 5-fold-decreased poliovirus yields (measured in PFU/cell) at peak virus titer (approximately 6 h postinfection). The decrease in virus yield in cells depleted of hnRNP C proteins suggests a stimulatory role for these proteins during poliovirus infection of cultured cells.
FIG. 8.
Poliovirus growth kinetics and viral-RNA accumulation in hnRNP C-depleted HeLa cells. Knockdown of hnRNP C via retrovirus infection and selection of infected cells is described in Materials and Methods. Plaque assays were carried out as described previously (13) and in Materials and Methods. (A) Whole-cell lysates of mock-treated (lane 1), HCScram-treated (lane 2), and HC12KD-treated (lane 3) cells were generated as described in Materials and Methods. Ten micrograms of each cell lysate was resolved on a 12.5% polyacrylamide-SDS-containing gel. Western blot analysis was carried out with a mouse monoclonal anti-hnRNP C1/C2 antibody (Abcam) and an alkaline phosphatase-conjugated anti-mouse secondary antibody (Promega). As a loading control, mouse monoclonal antibodies recognizing β-actin were utilized (Abcam). (B) One-step growth analysis was carried out in HeLa, HeLaScram, or HeLaHC12KD monolayers in 60-mm plates at 37°C. The cells were infected at an MOI of 25 with wild-type or PVΔ3′NCR virus. Infected HeLa, HeLaScram, and HeLaHC12KD cells were harvested beginning at 0 h postinfection and every 2 h thereafter. The virus particles were released by freeze-thaw, and the total virus yield was determined from the plaque assay titer divided by the total initial cell count. (C) Quantitative real-time PCR of poliovirus positive-strand RNA accumulation was carried out using total-RNA harvests of monolayers from wild-type HeLa cells or retrovirus-infected HeLaHC12KD cells infected with either wild-type poliovirus or PVΔ3′NCR. The cells were infected at an MOI of 25, and total RNA was harvested as described in Materials and Methods. (D) Quantitative real-time PCR of poliovirus negative-strand RNA accumulation was carried out as described for panel C, except that DNA oligonucleotides annealing to poliovirus negative-strand RNA were used. (E) Viral-RNA synthesis is depicted as the ratio of detected positive- to negative-strand RNAs, with the mean ratio depicted above the bars for each sample. As an internal control for RNA content in individual samples, quantitative real-time PCR of cellular β-actin RNAs was carried out for each sample in positive-strand vRNA accumulation (C) and negative-strand vRNA accumulation (D) to normalize poliovirus RNA detection results. Note that for the quantitative real-time PCR assays, shRNA-mediated knockdown was compared to untreated HeLa cells because there were no significant differences in poliovirus growth curves when poliovirus infections of untreated HeLa cells were compared to HeLaScram-treated cells. The error bars indicate standard deviations.
The poliovirus mutant PVΔ3′NCR lacks the 3′ NCR and hence the 5′ negative-strand binding site for hnRNP C. This virus was previously reported to produce lower virus yields than the wild type due to an inability to synthesize high levels of positive-strand RNA (13). Our finding that hnRNP C binds to the 5′ terminus of poliovirus negative-strand RNA provides an explanation for the growth phenotype of this virus, i.e., lack of an hnRNP C binding site contributes to the positive-strand RNA synthesis defect. We therefore determined the effect of hnRNP C depletion in HeLa cell PVΔ3′NCR replication. The virus yield from PVΔ3′NCR infection of HC12KD-treated HeLa cells was approximately 2.5-fold lower than mutant virus growth levels in mock- or HCScram-treated HeLa cells, a slightly smaller decrease than we observed for wild-type poliovirus in hnRNP C-depleted cells (Fig. 8B). There was a significant difference in virus yield at 4 h postinfection (an approximately 5-fold decrease compared to controls). These data suggest that a decrease in hnRNP C available to virus replication complexes results in a decrease in virus yields, characterized by a significant delay in the kinetics of virus replication.
Strand-specific virus RNA synthesis effects of hnRNP C depletion in cultured cells.
Knockdown of hnRNP C in HeLa cells reduced the virus yield in infections by wild-type poliovirus and PVΔ3′NCR. Based upon the kinetics of our single-cycle growth analyses, we hypothesized that the deficiency in virus yield was caused by inefficient viral-RNA synthesis. In a typical poliovirus infection, the synthesis of RNA is asymmetric, with the ratio of positive- to negative-strand RNA ranging from approximately 30:1 to 70:1 during the course of an infection (21, 36). To determine the origin of the drop in virus production, we examined the ability of the wild type and PVΔ3′NCR to support RNA synthesis in HC12KD-treated cells using quantitative real-time PCR to analyze total RNA from infected cells. For these assays, β-actin was used as a control, since its expression is not affected as a consequence of retrovirus infection of HeLa cells (Fig. 8A). In agreement with our single-cycle growth data, our quantitative-PCR studies indicated that at 4 h postinfection, HC12KD-treated cells infected with wild-type poliovirus or PVΔ3′NCR accumulated decreased amounts of positive-strand RNAs compared to infections in HeLa cells (Fig. 8C). However, at later times postinfection, the positive-strand RNA accumulated to equivalent levels in PVΔ3′NCR infections of HeLa cells and HC12KD-treated HeLa cells or, in the case of wild-type poliovirus, to greater levels in infections of HC12KD cells than in non-retrovirus-infected HeLa cells, possibly due to lysis by wild-type virus of untreated HeLa cells at 6 h and 8 h postinfection. These data were confirmed by separate RNase protection assays (data not shown). Negative-strand poliovirus RNA synthesis in HC12KD or non-retrovirus-infected HeLa cells was not significantly affected, especially at later times after infections (Fig. 8D). By comparing the ratio of positive- to negative-strand RNAs detected by quantitative PCR, we observed that the maximal RNA synthesis of wild-type poliovirus in HeLa cells occurred between 4 h and 5 h postinfection (Fig. 8E), which is in agreement with previous studies (13, 36). In contrast, maximal RNA synthesis of poliovirus positive-strand RNAs in cells depleted of hnRNP C proteins was delayed, providing further evidence that the defect in the virus yield observed in Fig. 8B was a result of aberrant positive-strand RNA synthesis.
DISCUSSION
Previous studies of the role of the poliovirus 3′ NCR have shown that, while the genomic RNA 3′-terminal sequence is dispensable for overall virus replication, its deletion results in a decrease in the synthesis of positive-strand RNA and hence a drop in the total production of infectious particles by at least 1 order of magnitude (12, 13, 50). Similar observations have also been reported for other picornaviruses (44, 49, 52). Based on the strand specificity of the replication defect, we hypothesized that this was due to a lack of the authentic 5′ end RNA sequence in the negative-strand RNA intermediate. In this report, we provided evidence that the 5′ end of poliovirus negative-strand RNA can specifically bind the cellular protein hnRNP C in poliovirus-infected cells and that this cellular interaction has a role in virus RNA replication at the strand-specific level. The observation of cellular proteins binding to the 5′(−) end is not surprising, as earlier work by Brown and colleagues had hypothesized that a limiting cellular factor might explain the disparities in the replication phenotypes of the PVΔ3′NCR virus in different tissue culture cell lines (13). What was somewhat surprising was that proteins capable of binding to the 5′ end of poliovirus negative-strand RNA were remarkably similar in apparent size to RNP complexes previously shown to form with the 3′ termini at the opposite end of the replicative intermediate (43). One of these complexes was later identified as containing the cellular protein hnRNP C (15).
Our electrophoretic mobility shift data suggested that hnRNP C may bind poliovirus RNA as a multimer. In the cell nucleus, hnRNP C proteins interact as a heterotetramer (C13-C21) to bind cellular RNA transcripts as part of splicing complexes and also to stabilize pre-mRNAs (29, 57). However, hnRNP C1 has been shown to form multimers in the absence of hnRNP C2 (22). While recombinant hnRNP C lacking a functional C1-C1 interaction domain (L187Q) is still capable of binding cellular mRNA and poliovirus negative-strand RNA, the appearance of higher-molecular-weight complexes in extracts generated from poliovirus-infected cells suggests that binding of endogenous hnRNP C to poliovirus RNAs occurs as a multimeric complex. Binding of hnRNP C multimers to poliovirus RNAs may be important for additional protein-protein interactions within the viral replication complex. Indeed, we observed stimulation of poliovirus RNA synthesis when recombinant hnRNP C1 was added to coupled in vitro translation/replication assays using HeLa S10 cytoplasmic extracts. However, the L187Q mutated form of hnRNP C1 that is defective in multimerization could not stimulate viral-RNA synthesis in vitro, even at 200 nM. These data support our hypothesis that a cellular factor (i.e., hnRNP C) capable of binding to the 5′ end can promote RNA synthesis, and this factor may require multimerization for its function(s).
During poliovirus infection, hnRNP C and other predominantly nuclear factors partially relocalize to the cytoplasm due to the disruption of cellular trafficking (8, 23). Whether the contribution to viral-RNA synthesis is a direct effect of hnRNP C binding or an indirect effect of the interaction of additional cellular and/or viral cofactors is not yet known. However, RNP complexes formed in mobility shift assays using extracts containing poliovirus proteins were capable of forming a supershifted complex with the inclusion of polyclonal antibodies specific for the viral protein 3C/3CD (41). While our RNA binding studies have not been able to detect recombinant forms of hnRNP C and 3C/3CD interacting in a stable complex with poliovirus negative-strand RNA, previous studies have shown that poliovirus 3CD interacts with hnRNP C (9, 15). It is tempting to speculate that the binding of 3C/3CD to the 5′ terminus of negative-strand RNA, in conjunction with hnRNP C and other host cell factors, might influence the interaction of the negative-strand RNA termini with each other and the viral RNA polymerase to facilitate positive-strand RNA synthesis (see below).
We investigated the contribution of hnRNP C during poliovirus infection of HeLa cells by using a recombinant retrovirus capable of constitutive expression of hnRNP C mRNA-specific shRNAs. Wild-type poliovirus infection in shRNA-treated cells resulted in a decrease in virus yield of almost 1 log10 unit at 4 h postinfection compared to controls, while at later times after infection, the yield of wild-type poliovirus in hnRNP C-depleted cells was 5-fold lower than in mock or scrambled shRNA controls. Infection of hnRNP C knockdown HeLa cells with the PVΔ3′NCR virus resulted in a different growth profile in which the virus yield in hnRNP C-depleted cells was approximately 5-fold lower than in PVΔ3′NCR-infected control cells at 4 h postinfection, yet at later times after infection, there were only minimal differences in virus yields. It should be noted that even at late times postinfection, the yields from PVΔ3′NCR-infected cells (knockdown or controls) were still between 1 and 2 log10 units lower than those from wild-type poliovirus-infected cells (knockdown or controls). Given that PVΔ3′NCR lacks the binding site for hnRNP C at the 5′ ends of negative-strand intermediates, the levels of hnRNP C may not be limiting at late times during infection but would still not be predicted to overcome the RNA replication defect imparted by this genetic lesion.
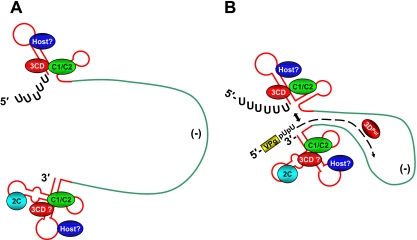
From the data presented in this study, as well as from prior observations, we propose an updated model (Fig. 9) for the role of hnRNP C in poliovirus RNA synthesis. For simplicity of presentation, we have depicted the negative-strand template as a single-stranded RNA, although it is normally present as a base-paired duplex with positive-strand RNA (referred to as the replicative form [RF]). Following synthesis of the negative-strand RNA, the positive-strand genomic RNA and the newly synthesized negative strand interact to form the double-stranded RF (38). In this form, the interaction of positive- and negative-strand viral RNAs is most likely contiguous. However, our structure predictions (Fig. 1) and those of others indicate that the 5′- and 3′-terminal regions of poliovirus RNA strands of both polarities contain a high degree of stable RNA secondary structure (3, 40, 41). It is reasonable to predict that the RNA termini would not be completely annealed to the complementary strand and may in fact “breathe” to allow the association of RNA binding proteins (Fig. 9A). In this case, interaction of poliovirus nonstructural proteins (e.g., 2C, 3C/3CD, and 3D) with the negative-strand RNA could be facilitated by close proximity due to their participation in the synthesis of the poliovirus negative-strand RNA (4, 20, 28, 58). While it is unclear how 3CD might translocate from the stem-loop I structure of the positive strand to the 5′ terminus of the negative strand, an explanation may be the presence of hnRNP C.
FIG. 9.
Model of the initiation of positive-strand RNA synthesis. Negative-strand template RNA, usually found as part of the double-stranded RF, is depicted in green as single stranded for simplicity. Nascent positive-strand RNA (B) is depicted by a black dashed arrow. (A) Prior to positive-strand RNA synthesis, the 5′ and 3′ termini associate with known (hnRNP C) and unknown (host) cellular proteins, in addition to viral proteins 3C/3CD and 2C. Binding of hnRNP C, possibly in conjunction with other viral/cellular proteins, allows interaction of 3C/3CD with the negative-strand RNA termini. (B) These interactions may allow contact of the 5′ and 3′ ends of the RF (indicated by the double-headed arrow), possibly facilitating initiation of positive-strand RNA synthesis by the RNA-dependent RNA polymerase (3Dpol). The activity of hnRNP C in the virus RNA replication cycle may be a rate-limiting step during the initial stages of poliovirus infection, where the hnRNP C cytoplasmic concentration is low. At later times postinfection, hnRNP C distribution shifts to the cytoplasm of the infected cell. VPg, the genome-linked protein of poliovirus; pUpU, the first two nucleotides of poliovirus genomic RNA.
We propose that hnRNP C binds the 5′ and 3′ ends of poliovirus negative-strand RNA to stabilize interactions between the two termini. Such interactions would result in a spatial organization of the positive-strand replication complex that allows the initiation of RNA synthesis at the 3′ end of the negative strand (Fig. 9B). This model could account for the RNA synthesis defect observed previously in PVΔ3′NCR, where positive-strand RNA synthesis was impaired. Lack of an authentic 5′ end of negative-strand RNA would result in a decrease in the ability of the 5′ and 3′ ends of negative-strand RNA to interact or otherwise communicate, causing an overall decrease in the efficiency of the polymerase to synthesize positive-strand RNAs. These 5′-3′-terminal interactions may exist only transiently to allow the initiation of positive-strand RNA synthesis, followed by dissociation to allow the 3D RNA polymerase to elongate newly initiated chains. This would differ from instances of other cellular factors binding poliovirus RNA, i.e., PCBP2, which has functions in both virus translation and replication (11, 20, 39, 54) and has a high affinity for two stem-loop structures within the poliovirus 5′ NCR (10, 20, 55). A caveat of this model is that although our investigations and those of others have provided evidence for hnRNP C and viral nonstructural proteins interacting with negative-strand RNA, there is a lack of direct evidence for hnRNP C and/or other proteins binding the 5′ and 3′ negative-strand RNA termini simultaneously. To address this issue, it will be necessary to determine the presence and identities of other cellular and virus factors participating in the formation of the replication complex. For instance, the 50-kDa and 38-kDa proteins that were originally described as interacting with the 3′ end of poliovirus negative-strand RNA have yet to be identified (15, 41, 43). Poliovirus protein 2C has been shown to associate with negative-strand viral RNA (2), in addition to its role in the induction of membranous vesicles during poliovirus infection (1). Further investigation will be required to understand how the different components of RNA replication complexes contribute to the exponential increase in the numbers of picornavirus genomic RNAs that will subsequently be packaged into virions. Components of the RNA replication complex may therefore prove to be effective targets for antiviral therapies directed toward stemming intracellular picornavirus replication and dissemination in an infected host.
Acknowledgments
We are grateful to Martin Holcik for the gift of FLAG-tagged hnRNP C expression constructs. We thank Andrea Cathcart, Sarah Daijogo, and Kerry Fitzgerald for critical comments on the manuscript. We also thank Rushika Perera for the gift of recombinant poliovirus 3CD.
This study was supported by Public Health Service grant AI 22693 from the National Institutes of Health. J.E.B. was a predoctoral trainee supported by Public Health Service training grant GM 07311.
Footnotes
Published ahead of print on 17 February 2010.
REFERENCES
- 1.Aldabe, R., and L. Carrasco. 1995. Induction of membrane proliferation by poliovirus proteins 2C and 2BC. Biochem. Biophys. Res. Commun. 206:64-76. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee, R., A. Echeverri, and A. Dasgupta. 1997. Poliovirus-encoded 2C polypeptide specifically binds to the 3′-terminal sequences of viral negative-strand RNA. J. Virol. 71:9570-9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerjee, R., W. Tsai, W. Kim, and A. Dasgupta. 2001. Interaction of poliovirus-encoded 2C/2BC polypeptides with the 3′ terminus negative-strand cloverleaf requires an intact stem-loop b. Virology 280:41-51. [DOI] [PubMed] [Google Scholar]
- 4.Barton, D. J., and J. B. Flanegan. 1997. Synchronous replication of poliovirus RNA: initiation of negative-strand RNA synthesis requires the guanidine-inhibited activity of protein 2C. J. Virol. 71:8482-8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barton, D. J., B. J. O'Donnell, and J. B. Flanegan. 2001. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 20:1439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartz, S. R., and M. A. Vodicka. 1997. Production of high-titer human immunodeficiency virus type 1 pseudotyped with vesicular stomatitis virus glycoprotein. Methods 12:337-342. [DOI] [PubMed] [Google Scholar]
- 7.Bedard, K. M., B. L. Walter, and B. L. Semler. 2004. Multimerization of poly(rC) binding protein 2 is required for translation initiation mediated by a viral IRES. RNA 10:1266-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belov, G. A., A. G. Evstafieva, Y. P. Rubtsov, O. V. Mikitas, A. B. Vartapetian, and V. I. Agol. 2000. Early alteration of nucleocytoplasmic traffic induced by some RNA viruses. Virology 275:244-248. [DOI] [PubMed] [Google Scholar]
- 9.Belov, G. A., C. Habbersett, D. Franco, and E. Ehrenfeld. 2007. Activation of cellular Arf GTPases by poliovirus protein 3CD correlates with virus replication. J. Virol. 81:9259-9267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blyn, L. B., K. M. Swiderek, O. Richards, D. C. Stahl, B. L. Semler, and E. Ehrenfeld. 1996. Poly(rC) binding protein 2 binds to stem-loop IV of the poliovirus RNA 5′ noncoding region: identification by automated liquid chromatography-tandem mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 93:11115-11120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blyn, L. B., J. S. Towner, B. L. Semler, and E. Ehrenfeld. 1997. Requirement of poly(rC) binding protein 2 for translation of poliovirus RNA. J. Virol. 71:6243-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown, D. M., C. T. Cornell, G. P. Tran, J. H. Nguyen, and B. L. Semler. 2005. An authentic 3′ noncoding region is necessary for efficient poliovirus replication. J. Virol. 79:11962-11973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown, D. M., S. E. Kauder, C. T. Cornell, G. M. Jang, V. R. Racaniello, and B. L. Semler. 2004. Cell-dependent role for the poliovirus 3′ noncoding region in positive-strand RNA synthesis. J. Virol. 78:1344-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brunner, J. E., K. J. Ertel, J. M. Rozovics, and B. L. Semler. 26 February 2010. Delayed kinetics of poliovirus RNA synthesis in a human cell line with reduced levels of hnRNP C proteins. Virology [Epub ahead of print.] [DOI] [PMC free article] [PubMed]
- 15.Brunner, J. E., J. H. C. Nguyen, H. H. Roehl, T. V. Ho, K. M. Swiderek, and B. L. Semler. 2005. Functional interaction of heterogeneous nuclear ribonucleoprotein C with poliovirus RNA synthesis initiation complexes. J. Virol. 79:3254-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho, M. W., N. Teterina, D. Egger, K. Bienz, and E. Ehrenfeld. 1994. Membrane rearrangement and vesicle induction by recombinant poliovirus 2C and 2BC in human cells. Virology 202:129-145. [DOI] [PubMed] [Google Scholar]
- 17.Choi, K. S., A. Mizutani, and M. C. Lai. 2004. SYNCRIP, a member of the heterogeneous nuclear ribonucleoprotein family, is involved in mouse hepatitis virus RNA synthesis. J. Virol. 78:13153-13162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui, T., S. Sankar, and A. G. Porter. 1993. Binding of encephalomyocarditis virus RNA polymerase to the 3′-noncoding region of the viral RNA is specific and requires the 3′-poly(A) tail. J. Biol. Chem. 268:26093-26098. [PubMed] [Google Scholar]
- 19.Gamarnik, A. V., and R. Andino. 1997. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA 3:882-892. [PMC free article] [PubMed] [Google Scholar]
- 20.Gamarnik, A. V., and R. Andino. 2000. Interactions of viral protein 3CD and poly(rC) binding protein with the 5′ untranslated region of the poliovirus genome. J. Virol. 74:2219-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giachetti, C., and B. L. Semler. 1991. Role of a viral membrane polypeptide in strand-specific initiation of poliovirus RNA synthesis. J. Virol. 65:2647-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorlach, M., M. Wittekind, R. A. Beckman, L. Mueller, and G. Dreyfuss. 1992. Interaction of the RNA-binding domain of the hnRNP C proteins with RNA. EMBO J. 11:3289-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gustin, K. E., and P. Sarnow. 2001. Effects of poliovirus infection on nucleo-cytoplasmic trafficking and nuclear pore complex composition. EMBO J. 20:240-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gustin, K. E., and P. Sarnow. 2002. Inhibition of nuclear import and alteration of nuclear pore complex composition by rhinovirus. J. Virol. 76:8787-8796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haller, A. A., and B. L. Semler. 1992. Linker scanning mutagenesis of the internal ribosome entry site of poliovirus RNA. J. Virol. 66:5075-5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris, K. S., W. Xiang, L. Alexander, W. S. Lane, A. V. Paul, and E. Wimmer. 1994. Interaction of poliovirus polypeptide 3CDpro with the 5′ and 3′ termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J. Biol. Chem. 269:27004-27014. [PubMed] [Google Scholar]
- 27.Herold, J., and R. Andino. 2000. Poliovirus requires a precise 5′ end for efficient positive-strand RNA synthesis. J. Virol. 74:6394-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Herold, J., and R. Andino. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holcik, M., B. W. Gordon, and R. G. Korneluk. 2003. The internal ribosome entry site-mediated translation of antiapoptotic protein XIAP is modulated by the heterogeneous nuclear ribonucleoproteins C1 and C2. Mol. Cell. Biol. 23:280-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leong, L. E. C., C. T. Cornell, and B. L. Semler. 2002. Processing determinants and functions of cleavage products of picornavirus polyproteins, p. 187-197. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, DC.
- 31.Lescrinier, E. M., M. Tessari, F. J. Van Kuppeveld, W. J. Melchers, C. W. Hilbers, and H. A. Heus. 2003. Structure of the pyrimidine-rich internal loop in the poliovirus 3′-UTR: the importance of maintaining pseudo-2-fold symmetry in RNA helices containing two adjacent non-canonical base-pairs. J. Mol. Biol. 331:759-769. [DOI] [PubMed] [Google Scholar]
- 32.McBride, A. E., A. Schlegel, and K. Kirkegaard. 1996. Human protein Sam68 relocalization and interaction with poliovirus RNA polymerase in infected cells. Proc. Natl. Acad. Sci. U. S. A. 93:2296-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Molla, A., A. V. Paul, and E. Wimmer. 1991. Cell-free, de novo synthesis of poliovirus. Science 254:1647-1651. [DOI] [PubMed] [Google Scholar]
- 34.Myslinski, E., J. C. Ame, A. Krol, and P. Carbon. 2001. An unusually compact external promoter for RNA polymerase III transcription of the human H1RNA gene. Nucleic Acids Res. 29:2502-2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakielny, S., and G. Dreyfuss. 1996. The hnRNP C proteins contain a nuclear retention sequence that can override nuclear export signals. J. Cell Biol. 134:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Novak, J. E., and K. Kirkegaard. 1991. Improved method for detecting poliovirus negative strands used to demonstrate specificity of positive-strand encapsidation and the ratio of positive to negative strands in infected cells. J. Virol. 65:3384-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parsley, T. B., J. S. Towner, L. B. Blyn, E. Ehrenfeld, and B. L. Semler. 1997. Poly(rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA 3:1124-1134. [PMC free article] [PubMed] [Google Scholar]
- 38.Paul, A. V. 2002. Possible unifying mechanism of picornavirus genome replication, p. 227-246. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, DC.
- 39.Perera, R., S. Daijogo, B. L. Walter, J. H. C. Nguyen, and B. L. Semler. 2007. Cellular protein modification by poliovirus: the two faces of poly(rC)-binding protein. J. Virol. 81:8919-8932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pilipenko, E. V., K. Poperechny, S. V. Maslova, W. J. G. Melchers, H. J. Bruins Slot, and V. I. Agol. 1996. Cis-element, oriR, involved in the initiation of (-) strand poliovirus RNA: a quasi-globular multi-domain RNA structure maintained by tertiary (′kissing') interactions. EMBO J. 15:5428-5436. [PMC free article] [PubMed] [Google Scholar]
- 41.Roehl, H. H., T. B. Parsley, T. V. Ho, and B. L. Semler. 1997. Processing of a cellular polypeptide by 3CD proteinase is required for poliovirus ribonucleoprotein complex formation. J. Virol. 71:578-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roehl, H. H., and B. L. Semler. 1994. In vitro biochemical methods for investigating RNA-protein interactions in picornaviruses, p. 169-182. In K. W. Adolph (ed.), Methods in molecular genetics, vol. 4. Academic Press, Orlando, FL. [Google Scholar]
- 43.Roehl, H. H., and B. L. Semler. 1995. Poliovirus infection enhances the formation of two ribonucleoprotein complexes at the 3′ end of viral negative-strand RNA. J. Virol. 69:2954-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saiz, M., S. Gomez, E. Martinez-Salas, and F. Sobrino. 2001. Deletion or substitution of the aphthovirus 3′ NCR abrogates infectivity and virus replication. J. Gen. Virol. 82:93-101. [DOI] [PubMed] [Google Scholar]
- 45.Scott, W. G., J. B. Murray, J. R. Arnold, B. L. Stoddard, and A. Klug. 1996. Capturing the structure of a catalytic RNA intermediate: the hammerhead ribozyme. Science 274:2065-2069. [DOI] [PubMed] [Google Scholar]
- 46.Swanson, M. S., and G. Dreyfuss. 1988. RNA binding specificity of hnRNP proteins: a subset bind to the 3′ end of introns. EMBO J. 7:3519-3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swift, S., J. Lorens, P. Achacoso, and G. P. Nolan. 2001. Rapid production of retroviruses for efficient gene delivery to mammalian cells using 293T cell-based systems. Curr. Protoc. Immunol. 31:10.17.14-10.17.29. [DOI] [PubMed] [Google Scholar]
- 48.Todd, S., J. H. Nguyen, and B. L. Semler. 1995. RNA-protein interactions directed by the 3′ end of human rhinovirus genomic RNA. J. Virol. 69:3605-3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Todd, S., and B. L. Semler. 1996. Structure-infectivity analysis of the human rhinovirus genomic RNA 3′ non-coding region. Nucleic Acids Res. 24:2133-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Todd, S., J. S. Towner, D. M. Brown, and B. L. Semler. 1997. Replication-competent picornaviruses with complete genomic RNA 3′ noncoding region deletions. J. Virol. 71:8868-8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tuteja, R., and N. Tuteja. 1998. Nucleolin: a multifunctional major nucleolar phosphoprotein. Crit. Rev. Biochem. Mol. Biol. 33:407-436. [DOI] [PubMed] [Google Scholar]
- 52.van Ooij, M. J., C. Polacek, D. H. Glaudemans, J. Kuijpers, F. J. Van Kuppeveld, R. Andino, V. I. Agol, and W. J. Melchers. 2006. Polyadenylation of genomic RNA and initiation of antigenomic RNA in a positive-strand RNA virus are controlled by the same cis-element. Nucleic Acids Res. 34:2953-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Waggoner, S., and P. Sarnow. 1998. Viral ribonucleoprotein complex formation and nucleolar-cytoplasmic relocalization of nucleolin in poliovirus-infected cells. J. Virol. 72:6699-6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walter, B. L., J. H. Nguyen, E. Ehrenfeld, and B. L. Semler. 1999. Differential utilization of poly(rC) binding protein 2 in translation directed by picornavirus IRES elements. RNA 5:1570-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walter, B. L., T. B. Parsley, E. Ehrenfeld, and B. L. Semler. 2002. Distinct poly(rC) binding protein KH domain determinants for poliovirus translation initiation and viral RNA replication. J. Virol. 76:12008-12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wan, L., J. K. Kim, V. W. Pollard, and G. Dreyfuss. 2001. Mutational definition of RNA-binding and protein-protein interaction domains of heterogeneous nuclear RNP C1. J. Biol. Chem. 276:7681-7688. [DOI] [PubMed] [Google Scholar]
- 57.Waterhouse, N., S. Kumar, Q. Song, P. Strike, L. Sparrow, G. Dreyfuss, E. S. Alnemri, G. Litwack, M. Lavin, and D. Watters. 1996. Heteronuclear ribonucleoproteins C1 and C2, components of the spliceosome, are specific targets of interleukin 1 beta-converting enzyme-like proteases in apoptosis. J. Biol. Chem. 271:29335-29341. [DOI] [PubMed] [Google Scholar]
- 58.Yang, Y., R. Rijnbrand, S. Watowich, and S. M. Lemon. 2003. Genetic evidence for an interaction between a picornaviral cis-acting RNA replication element and 3CD protein. J. Biol. Chem. 279:12659-12667. [DOI] [PubMed] [Google Scholar]
- 59.Zuker, M., D. H. Mathews, and D. H. Turner. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide, p. 11-43. In J. Barciszewski and B. F. C. Clark (ed.), RNA biochemistry and biotechnology. NATO Science Series, vol. 70. Kluwer Academic Publishers, Boston, MA. [Google Scholar]