Abstract
Poliovirus infection requires that the particle undergo a series of conformational transitions that lead to cell entry and genome release. In an effort to understand the conformational changes associated with the release of the RNA genome, we have used cryo-electron microscopy to characterize the structure of the 80S “empty” particles of poliovirus that are thought to represent the final product of the cell entry pathway. Using two-dimensional classification methods, we show that preparations of 80S particles contain at least two structures, which might represent snapshots from a continuous series of conformers. Using three-dimensional reconstruction methods, we have solved the structure of two distinct forms at subnanometric resolution, and we have built and refined pseudoatomic models into the reconstructions. The reconstructions and the derived models demonstrate that the two structural forms are both slightly expanded, resulting in partial disruption of interprotomer interfaces near their particle 2-fold axes, which may represent the site where RNA is released. The models demonstrate that each of the two 80S structures has undergone a unique set of movements of the capsid proteins, associated with rearrangement of flexible loops and amino-terminal extensions that participate in contacts between protomers, between pentamers, and with the viral RNA.
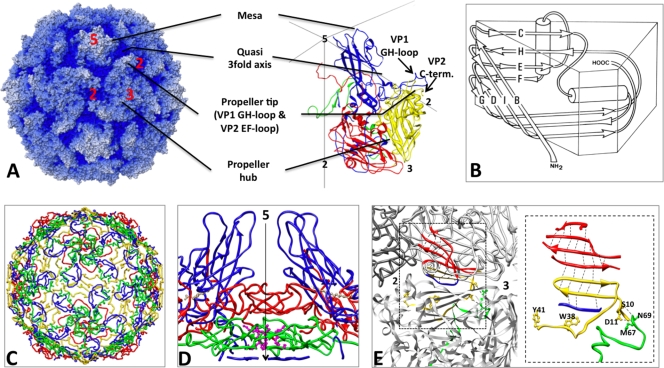
Like all members of the picornavirus family, poliovirus has an icosahedral capsid (300 Å in diameter) consisting of 60 copies of each of four coat proteins, VP1, VP2, VP3, and myrVP4, which encapsidates a plus-sense single-stranded RNA genome (Fig. 1A). The poliovirus capsid proteins are responsible for several functions along the infection pathway, including programmed assembly, encapsidation of the viral genome, protection of the genome, host cell recognition, cell entry (involving both membrane binding and penetration), and release and delivery of the viral genome. This variation in function requires both individual capsid proteins and the capsid as a whole to undergo a series of triggered dynamic structural changes.
FIG. 1.
Structural features of poliovirus. (A, left) Radial-depth-cued (regions closer to the particle center are darker) space-filling model of the native virus particle (accession number 1HSX [23]); (right) a ribbon representation of one protomer, shown enlarged over a portion of an icosahedral framework. VP1 is blue, VP2 is yellow, VP3 is red, and myrVP4 is green. The axes of icosahedral symmetry are labeled using the numbers 2, 3, and 5. (B) Geometric representation of the core structure shared by the major capsid proteins (VP1, VP2, and VP3). The major coat proteins are structurally similar to one another; each has a central core consisting of two β-sheets arranged in an eight-stranded, wedge-shaped β-barrel motif. Individual β-strands are labeled using the letters B through I (in an amino-to-carboxyl direction) resulting in two sheets named BIDG (front) and CHEF (back) for the participating strands. Connecting loops are denoted by two letters (e.g., the GH loop connects β-strands G and H). Structurally conserved α-helices are represented by cylinders. The narrow ends of 5-fold-symmetry-related VP1 proteins are clustered together around each 5-fold axis of symmetry, while the narrow ends of the VP2 and VP3 β-barrels alternate around the 3-fold axes (28). (C) Internal network formed by the minor coat protein myrVP4 and by the amino-terminal extensions of the major coat proteins. (D) A twisted β-tube serves as a “plug” by separating a low-density region below the 5-fold VP1 mesa (blue) from the space interior to the capsid. This intrapentameric structure is made from 5-fold-symmetry-related copies of the extreme amino-terminal peptides from VP3 (red), myrVP4 (green), and VP1 (blue). The amino terminus of myrVP4 is covalently linked to a myristic acid moiety (magenta), which mediates the interactions of myrVP4 with the VP3 plug. During viral assembly, formation of this structure stabilizes the assembly of protomers into pentamers. (E) A seven-stranded β-sheet with contributions from two different pentamers. The uppermost four strands are contributed by the CHEF sheet from VP3 (red) and the lowermost, seventh strand is contributed by VP1 (blue) from the same protomer. Strands 4 and 7 thus form a clamp around strands 5 and 6, which are contributed by a β-hairpin from the amino-terminal extension of VP2 (yellow) from a neighboring pentamer. Formation of this extended sheet stabilizes the association of pentamers to form a closed icosahedral shell. Dashed lines represent electrostatic interactions. VP2 residues (Y2041 and W2038, shown as yellow ball-and-stick models), which flank the β-hairpin, appear to interact with encapsidated RNA and with the extreme carboxyl terminus of myrVP4. VP2 residues S2010 and D2011 interact with myrVP4 residues (shown as green ball-and-stick structures) N4069 and M4067, respectively, tethering the products of myrVP0 cleavage together.
Several structurally unique assembly and disassembly intermediates have been described for poliovirus, which can be distinguished by sedimentation properties on a sucrose gradient and by antigenicity (16, 35, 42, 48). During assembly, all four capsid protein sequences are translated together as a single polypeptide that is cotranslationally myristoylated. This precursor form of the protomer, called myrP1, sediments at 7S. Once the protomer has been cleaved by virally encoded protease to yield myrVP0, VP3, and VP1, subsequent rearrangements of the freed amino and carboxyl termini allow protomers to assemble into pentamers that sediment at 14S. In turn, pentamers can assemble into an empty-capsid intermediate that sediments at 73S, which contains 60 copies of myrVP0, VP3, and VP1. Encapsidation of the viral genome is associated with the autocatalytic cleavage of myrVP0 to yield myrVP4 and VP2. This final cleavage event results in a significant increase in the stability of the resulting mature virus particle, which sediments at 160S, making assembly irreversible.
During cell entry, viral attachment to the host cell receptor CD155/Pvr catalyzes marked conformational changes, including the externalization of myrVP4 and of the amino terminus of VP1 (both normally internal components), during the production of an intermediate that sediments at 135S. After the 135S particle has been internalized by the host cell, unknown triggers result in the externalization of the viral RNA and the translocation of the RNA across a vesicle membrane into the cytoplasm, leaving the protein shell as an 80S empty capsid intermediate. RNA release from seemingly intact mature virus particles and formation of cell entry intermediates can be recapitulated in vitro by heat treatment (9, 49).
The peptides that are externalized during the virus-to-135S transition (myrVP4 and the amino terminus of VP1) insert into lipid bilayers, resulting in a direct association of the 135S particles with membranes (20, 46). Electrophysiology experiments revealed that the externalization of myrVP4 results in the formation of ion channels in membranes (15, 44, 45). Specific mutations in myrVP4 result in a virus that is defective in both RNA release and channel formation (15, 44, 45). This information supports a cell entry model in which the insertion of myrVP4 and regions from the amino-terminal extension of VP1 into the host cell membrane results in the formation of channels that serve to facilitate the translocation of the viral RNA into the cytoplasm (10).
Structure of the virus.
In the mature virion, VP1, VP2, and VP3 each include a wedge-shaped, eight-stranded β-sandwich core structure, consisting of two four-stranded β-sheets (Fig. 1B). Although the β-barrel cores of the subunits are very similar in size and shape, each of the proteins has a unique set of loops connecting the β-strands, carboxyl-terminal extensions, and long amino-terminal extensions. The β-barrel cores of VP1, VP2, and VP3 pack together in a T=1 (pseudo T=3) icosahedrally symmetric pattern to form a closed shell. The narrow ends of the VP1 β-barrels are clustered around the 5-fold axes, and the narrow ends of VP2 and VP3 alternate around the 3-fold axes (Fig. 1A). The outer surface of the virion is decorated by the carboxyl termini of VP1, VP2, and VP3 and by loops that connect the core β-strands, resulting in a surface that is dominated by star-shaped mesas at the 5-fold axes, with the loops at the narrow end of VP1 forming the hub, and three-bladed propeller-shaped features at 3-fold axes, with the loops at the narrow ends of the β-barrels of VP2 and VP3 forming the hub and the EF loops of VP2, the GH loop of VP1, and the carboxyl terminus of VP2 forming the blades of the propellers (Fig. 1A). VP1 is unique in this regard: buried between the two β-sheets of the core lies a hydrophobic pocket, occupied by electron density for a hydrocarbon that has variously been modeled as sphingosine or palmitate in various poliovirus crystal structures. This is the same pocket that a group of antipicornaviral agents bind within, and it has been suggested to play a role in regulating the structural dynamics required for viral infection (31, 32, 37).
myrVP4 and the amino-terminal extensions of the major VPs line the interior surface of the viral protein shell, thus forming a network that governs assembly and stabilizes the mature 160S poliovirion (Fig. 1C). A key feature of this network is a plug at the 5-fold axes that blocks a channel through the capsid that has been proposed (perhaps mistakenly) to be the route for RNA egress from the protein shell (Fig. 1D) (33). The plug includes a parallel β-tube formed by five intertwined amino termini of VP3, seated on top of five copies of a three-stranded β-sheet contributed by the amino termini of myrVP4 and VP1. The myristoyl group at the amino terminus of myrVP4 mediates the interactions between the VP3 and myrVP4 portions of the plug. Another important assembly-dependent feature of the internal network is a seven-stranded β-sheet that stabilizes the interaction between neighboring pentamers (Fig. 1E) (19). To form this extended sheet, the two-stranded β-hairpin from the amino-terminal extension of VP2 (that belongs to one of the pentamers) is sandwiched between the four-stranded CHEF sheet of VP3 and a peptide from the amino-terminal extension of VP1 (both of which belong to a neighboring pentamer). In this seven-stranded pattern, the C strand of VP3 lies on the outer surface of the capsid and the amino-terminal peptide from VP1 lies on the inner surface. The formation of this extended structure is dependent on the assembly of pentamers into an icosahedral shell.
The only density for packaged RNA that appears in the native virus structures is associated with residues W2038 and Y2041 from the amino-terminal extension of VP2. Density for nucleotides stacked against one or both of these highly conserved aromatic residues is seen in the crystal structures of several picornavirus genera, suggesting a common role for this peptide in stabilizing the interaction between the capsid and the packaged RNA (2, 19, 21, 25, 41).
Structures of assembly and cell entry intermediates.
For some time, we have been interested in understanding the nature of the conformational changes that take place during assembly and cell entry. Our first success was the determination of the crystal structure of the 73S assembly intermediate (4). The outer surface of the 73S particle is nearly identical to that of the native virion. This was expected, because the two forms of the virus share antigenic markers. However, except for the amino terminus of VP3 and the extreme amino terminus of myrVP4 (which together form the “plug” at the 5-fold axes), most of the internal network is either disordered or rearranged dramatically in 73S. These changes affect the entire amino-terminal extension of VP1, the carboxyl-terminal half of myrVP4, and the amino-terminal extension of VP2. Notably, a peptide that includes the scissile bond of myrVP0 is clearly visible in the 73S structure, and its environment has suggested a plausible mechanism for the RNA-mediated autocatalytic cleavage.
In the past, we have attempted to prepare crystals of cell entry intermediates, but we have not been able to obtain crystals suitable for high-resolution studies. We have therefore turned to cryo-electron microscopy (cryo-EM) to investigate the structures of these intermediates. In early studies, we reported cryo-EM structures for a complex of the virus with the ectodomain of the receptor (6) and for the 135S particle and 80S particle (5), all at 22 to 23 Å resolution. The resolution of the structure of the virus receptor complex has since been extended to subnanometer resolution (52) (D. N. Belnap, D. J. Filman, and J. M. Hogle, unpublished data). Additionally, we have solved the structure of a virus-receptor-liposome complex at 30 Å resolution, both by cryo-electron microscopy (11) and by cryo-electron tomography (7), and extended the resolution of the structure of the 135S particle to 10 Å (10). The virus-receptor-liposome complex showed that receptor binding brings a particle 5-fold axis into close proximity with the membrane and suggests that the virus-receptor complex induces an outward-projecting distortion of the outer leaflet of the lipid bilayer of the liposome. This observation was consistent with previous models, which suggested that the peptide myrVP4 and the amino terminus of peptide VP1, which is believed to form a structurally conserved amphipathic helix, become externalized at or near a 5-fold axis. Once externalized, these two peptides were expected to combine in some way to form pores in the membrane contiguous with the solvent-filled channel at the 5-fold axis of the viral capsid, and the viral genome was proposed to be released through these pores to gain access to the cytoplasm during cell entry. However, the cryo-EM structure of the 135S particle demonstrated both that the amino-terminal extension of VP1 exits from the particle at the base of the canyon and that the amphipathic helix, which is formed by the amino terminus of VP1, is located at the tip of the propeller near the 3-fold axis (10). This has caused us to rethink our hypothesis on routes of egress from the capsid interior.
In the present paper, we report cryo-EM studies characterizing the 80S particles. We show that the 80S preparations contain at least three different kinds of particles, and we report the structures of the two most highly populated alternative conformers (from a particle population that may contain a continuum of structures) at subnanometer resolution. (Similarly, two distinct classes of particles were observed previously by Hewat et al. in lower-resolution reconstructions of 80S particles of rhinovirus 2 and rhinovirus 14 [27, 28].) Elsewhere, we described cryo-EM and cryo-electron tomography characterization of the third structural state, in which the particles have been caught in the act of releasing their viral RNA (D. M. Bostina, H. C. Levy, D. J. Filman, and J. M. Hogle, submitted for publication). Together, these studies suggest that the viral RNA also exits through holes at the base of the canyon (not far from the site at which the amino terminus of VP1 exits in 135S particles).
MATERIALS AND METHODS
Virus propagation and purification and preparation of RNA-releasing particles.
Native Mahoney strain poliovirus was grown in a HeLa cell suspension, harvested by centrifugation, and released from the cell pellets by freeze-thaw lysis. The virus was purified from the cell lysate by CsCl density gradient fractionation, as previously described (14, 43). Purified native particles were converted by heat treatment at 56°C in 20 mM Tris buffer (pH 7.4), 50 mM NaCl, 2 mM CaCl2. We performed three separate experiments, heating the specimens for 4, 20, and 60 min, respectively.
Electron microscopy.
Samples of converted particles at 0.1 to 0.2 mg/ml were vitrified on glow-discharged, Quantifoil holey carbon grids. Images were recorded using a Gatan cryoholder on a Tecnai F30 microscope at 200 kV, with a magnification of ×59,000 and a dose of 15 to 20 e-/Å2, and were recorded under focus with a range of 0.9 to 3.0 μm on Kodak ISO153 films. A total of 138 micrographs from all three experiments were scanned using a Zeiss SCAI microdensitometer with a raster size of 7 μm and binned twice to a final pixel size of 2.32 Å/pixel. In total, about 10,000 individual particles were windowed using the program SIGNATURE (12). Visual inspection indicated a variable amount of RNA present within individual capsids.
Image processing.
The images were corrected for the contrast transfer function (CTF), for both phase reversal and envelope decay, using the program BSHOW (29, 30). Orientation search was done by projection matching, assuming icosahedral symmetry, using the program PFT2 (3). The reconstructions were performed with a Fourier-Bessel algorithm, as implemented in the program EM3DR2 (22). Resolution was determined using the Fourier shell correlation (FSC) method, with a cutoff threshold of 0.5. A soft Gaussian mask was used for delimiting the icosahedral capsid. All image manipulations were done using the BSOFT program package (29, 30) and with scripts written in Python.
As an initial model, we used an atomic model of poliovirus (50), low-pass filtered to a resolution of 60 Å. Orientation parameters were refined in a series of iterations but did not improve beyond 16 Å resolution. In order to determine the degree of variability between virus particles, we used a bootstrap method (53), wherein a series of 1,000 reconstructions was calculated, each based on a randomly chosen 5,000-image subset of the data, and a variance map was calculated from the reconstructions. In order to compensate for the bias introduced by the icosahedral reconstruction algorithm and for noise present in the images, we randomly shuffled the pixel values in each image and recalculated the reconstructions using the same orientation parameters as were used for the authentic images. The values of the variance map thus obtained were subtracted from those of the previous one. Areas of high and low variance suggested that the heterogeneity of the data set was the most probable cause for the modest resolution obtained.
We decided to investigate whether distinct classes of structures might be present in our specimens. Initially, aligned particles were divided into six equally populated classes, according to the ratio between the densities of the interior region and the capsid region of each projection. Then, separate refinements and reconstructions were performed on each group. The resulting maps did show the expected variation in RNA content but no distinguishable structural features in the capsid. We then chose the maps with the highest and the lowest RNA content as initial references for a sequential classification process. Individual particle images were compared with each of the two maps and assigned to the one with the highest phase agreement, with the comparison being restricted to the capsid area. In order to avoid the spurious migration of single images to the class having the greatest number of particles, new references were calculated using the same number of particles in each class. This process was halted after several rounds of dual-reference refinement, when no further improvement in resolution was observed. The resolution for the final maps obtained for each class was estimated to be 9.5 Å. An FSC curve calculated between the two final maps indicated that detectable differences occur only at resolutions higher than 16 Å.
To analyze our results, we then looked at the series of heating times, to see if the proportion of particles from each of the two classes differed according to how long the sample was heated. To facilitate visual comparisons of the capsid surfaces, we also generated electron density maps from atomic coordinates, based on crystal structures of the Mahoney strain of type 1 poliovirus. Coordinate-based maps were produced both for the mature 160S poliovirion (33, 51) and for the native-antigenic 73S empty capsid assembly intermediate (4) and filtered to 10 Å resolution using the BSF function from the BSOFT suite of electron microscopy programs (29, 30).
To address the question of whether the particle is indeed detectably larger when RNA is in the process of exiting, we conducted an experiment wherein we sorted the population of 10,000 projection images into three groups. In one group of 500 boxed particles, RNA was clearly visible in the immediate vicinity, which is expected to be a population of particles that is enriched in particles “caught in the act” of RNA release. The remainder of the boxed images had previously been assigned to the “early empty” or “late empty” groups, based on correlation statistics. The “early empty” group is expected to be more greatly enriched in “caught in the act” particles than the “late empty” group is but not as enriched as the 500-particle subset that has in-plane RNA density features visible just outside the capsid. For each group, a radially averaged density plot was calculated directly from the two-dimensional (2D) projections.
Fitting pseudoatomic models to cryo-EM maps.
Model building and automated refinement of the rigid-body models were initiated from a series of plausible-looking starting points created by interactive model-building using the Swiss-PdbViewer (SPDBV) (24) and COOT (18). The refinement explicitly included nine of the icosahedral-symmetry-related protomers moving in tandem, chosen in such a way that the central protomer (whose fit to the electron density was being optimized) was completely surrounded. Density lying outside this cluster of protomers was excluded by a generous mask. Refinement was carried out by the INSOUT protocol, as described in reference 10. The refinement residual, which was calculated in reciprocal space, was designed to optimize the Fourier-amplitude-weighted average of the cosines of the phase discrepancies between the model-based Fourier coefficients and the Fourier transform of the corresponding portion of the experimental map. That statistic is somewhat analogous to a crystallographic figure-of-merit (ranging from 1, for perfect phase agreement, to 0, for a complete lack of correspondence) and assesses how well the model-based electron density agrees with the experimental density, without the problems of systematic differences between the crystallographer's solvent-free atom representation and the electron microscope's contrast transfer function or the problem of density distortions due to limited resolution. Simultaneously, unfavorable contacts (i.e., steric clashes between neighboring capsid proteins) were avoided, in part, by maintaining the sum of the squares of the model-based Fourier amplitudes at a value similar to that calculated from the same rigid bodies, arranged as seen in the crystal structure of 73S particles. The actual values of the refinement statistics that we report have no absolute meaning because they depend on the resolution range chosen, the details of masking, the relative strength of the rigid-body contact restraints, and the completeness of the model. However, when several similar refinements were run in parallel, differing only in their starting parameters and their reference reconstructions, we assumed that a higher figure-of-merit value on output would indicate that the refined model did a better job of accounting for the observed electron density.
Models refined by this procedure have a gratifying tendency to resemble the shapes of their experimental density envelopes. However, at the current low resolution, neither the rigid-body assumptions, the assignment of specific protein sequences to each rigid body, nor the final arrangement of bodies is provably the correct one. Therefore, we regard the rigid-body pseudoatomic models as subjectively based aids to the interpretation of the current experimental density maps.
The poliovirus 73S protomer (4), which is one-sixtieth of the icosahedral capsid, was divided into five rigid bodies. Three of the bodies, which were movable in the refinement, each consisted of one of the β-barrels from the major capsid proteins, VP1, VP2 (myrVP0), or VP3, plus whichever additional polypeptide sequences (from amino termini, carboxyl termini, or loops) were either covalently or noncovalently attached to that barrel in the mature 160S poliovirion, and which we subjectively judged (based on their extensive intermolecular contacts and inspection of the electron density map) seemed likely to remain associated with that β-barrel when it shifts during the structural transitions of the virus. As we have come to understand, deciding which otherwise-flexible chains belong with which large β-barrels is one of the most important steps in our current modeling process. The two additional rigid bodies were manually placed in the density and not moved after that, due to the impossibility of shifting them automatically in a sensible way: these consisted of the 5-fold-symmetric β-tube formed by the intertwined amino termini of VP3 and the α-helical segment that we have tentatively identified as being comprised of residues 1042 to 1052 of the capsid protein VP1. For computational efficiency, side chain atoms were excluded. Multiple starting points were tried, as a way to compensate for the nonlinearity of the refinement space.
Atomic model parts were obtained from the crystal structure of poliovirus native empty capsids (RCSB Protein Data Bank [PDB] entry 1POV) (4). The first rigid body consisted of residues 1068 to 1207 and 1234 to 1290 (including all of VP1 except for the amino terminus, the GH loop, and the carboxyl terminus), plus 3013 to 3049 (that portion of the VP3 amino-terminal extension that wraps underneath the VP1 barrel). (By convention, we use the leftmost digit of the four-digit residue number as the chain identifier.) The second rigid body consisted of residues 0097 to 0113 and 0127 to 0341 (in the myrVP0 numbering, corresponding to the main barrel of VP2, except for one loop of VP2 on the inner surface of the capsid that is disordered in 73S particles) plus 1208 to 1233 (the GH loop of VP1). The third rigid body consisted of residues 3050 to 3231 (the main body of VP3) plus 0083 to 0096 (the two-stranded β-sheet from myrVP0 that extends the β-sheet hydrogen bonding pattern of the back sheet of VP3 in a symmetry-related pentamer) and 1291 to 1302 (the extreme carboxyl terminus of VP1). The fourth body (model built but not refined) is a single helix tentatively numbered 1042 to 1052, located on the outer surface of the capsid, and extending across the “canyon” between the 5-fold mesa and the 3-fold plateau. This helix, first identified by Bubeck et al. (10), is one of the most distinctive features of the 135S particle that distinguishes it from native virus and is believed to correspond to residues 1042 to 1052 of the amino-terminal extension of VP1, which are known to become externalized during the 160S-to-135S transition. The fifth rigid body (model built but not refined) consists of residues 3001 to 3012 from the extreme amino terminus of VP3. Five copies of this polypeptide intertwine around the 5-fold axis, forming a β-sheet tube on the inner surface of the capsid, which serves to block the hole at the viral 5-fold.
All of the atomic coordinates are expressed in a Cartesian coordinate system whose x, y, and z axes correspond to three mutually perpendicular 2-fold axes of the viral symmetry. We have calculated the major and minor axes of the ellipsoid of rotation for each of the β-barrels (belonging to each of the major capsid proteins) in each refined model, in order to more easily compare their shifts and tilts.
Accession numbers.
The 80S reconstructions and models have been deposited with the Electron Microscopy Database (EMDB) and the RCSB Protein Data Bank (PDB). 80S.e has EMDB accession number 5123 and PDB accession number 3IYB. 80S.l has EMDB accession number 5122 and PDB accession number 3IYC.
RESULTS
Cryo-electron microscopy.
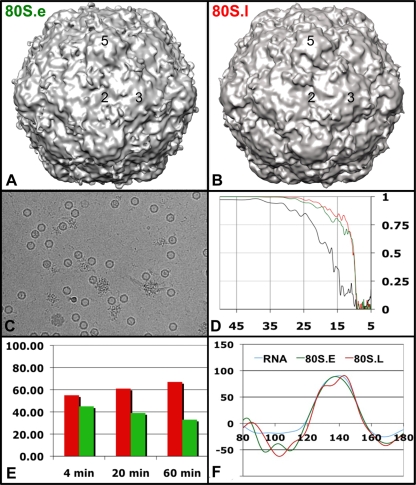
Initial attempts to solve the cryo-EM structure of the 80S particles resulted in reconstructions that converged at a resolution no better than 16 to 17 Å (Fig. 2D). Subsequent visual inspection of the micrographs revealed that heat-treated virus samples are mixtures of particle forms that vary in the amount of RNA density in their capsid interiors. Unlike mature virus particles, which appeared as filled spheres, the RNA-releasing “empty” particles appeared to be more hexagonal in profile and varied between partially full and nearly empty in the amounts of density that were visible within them. In micrographs taken at high defocus, it was also clear that a small subset of the particles (perhaps 5%) had clusters of strong density on both the inside and outside surfaces of the particles, which suggested that those particles had been caught in the act of releasing their RNA (Fig. 2C).
FIG. 2.
Cryo-EM reconstructions of poliovirus 80S empty capsids (early and late RNA-releasing forms) to ∼9.5 Å resolution. 3D reconstructions of the early form, 80S.e (A), and the late form, 80S.l (B), are shown as isocontour surfaces, showing the outer surface of the capsid, each contoured at 1.5σ and viewed along a 2-fold axis of symmetry. To recapitulate the RNA release process, purified samples of mature (160S) poliovirions were heated to 56°C. (C) Electron micrograph of vitrified, heat-treated poliovirus particles at various stages of RNA release imaged at ×59,000 magnification and ∼7 μm underfocus. (D) Fourier shell correlation plots for 80S reconstructions calculated using the entire data set (black line) or using only those images classified as early (green) or late (red) in the RNA release process. Vertical axis: FSC; horizontal axis: resolution in angstroms. Classification improved the resolution of the reconstructions from about 16 Å to 9.5 Å, indicating that the differences are real. (E) The proportion of 80S particles that partition into the early (green) and late (red) classifications varies, depending on the length of the heat treatment time. (F) Radially averaged density plots were determined from three groups of 2D images: green, particles from the early class; red, particles from the late class; blue, particles that appeared to be attached to expelled RNA. The radial location of the inner and outer protein-solvent boundaries (∼125 to 155 Å) does not appear to differ significantly between the three groups, suggesting that the particles are not expanded to different extents overall.
A data set of ∼10,300 boxed particle images was compiled from 138 micrographs. These particles were ultimately partitioned into two distinct data classes automatically, based on structural differences in the capsid region. Particles in this data set had been prepared by heating for various times. In all cases the preparations contained a mixture of the two classes. However, particles from preparations heated for shorter times were more heavily represented in one class, and particles from preparations heated for longer times were more heavily represented in the other (Fig. 2E). The two classes were therefore designated 80S.e (for early) and 80S.l (for late), respectively. In the assembled data set, the particles classified as 80S.l accounted for 59% of particles and the particles classified as 80S.e accounted for 41% of the particles. On average, particles in the 80S.e class had significantly higher density levels on the inside (which we believe corresponds to RNA) than particles in the 80S.l class did. A relatively small number of particles in the data set (∼500) appeared to have strong localized density for RNA on both the inside and the outside of the particle. Almost all of these had originally been classified in the 80S.e class. Radial-density plots, calculated from subsets of the particles classified as 80S.e, 80S.l, or “caught in the act,” demonstrated that the rotationally averaged inner and outer radii of the three classes of particles were indistinguishable (125 Å and 155 Å, respectively) (Fig. 2F).
Independent three-dimensional (3D) reconstructions were computed for each class (Fig. 2A and B). The resolution of each cryo-EM map was to 9.5 Å (as determined by Fourier shell correlation of half data sets, using a correlation criterion of 0.5) (Fig. 2D). Fourier shell correlations that included both classes indicated that the particles were similar to about 16 Å resolution (Fig. 2D). Consistent with our previously published low-resolution reconstructions of the 80S particle, the 80S.e and 80S.l reconstructions show that the particle has expanded ∼4%, relative to mature 160S virions. This expansion is accompanied by a pronounced thinning of the density in the interpentamer interface (along the boundary defined by the line from 3-fold axis to 2-fold axis to 3-fold axis) and also at the base of the canyon in an interprotomer interface. (The latter site lies roughly along the vertical plane joining a particle 2-fold axis and 5-fold axis and corresponds approximately to the site of the quasi-3-fold axis in a T=3 virus particle.)
Surface features of EM maps.
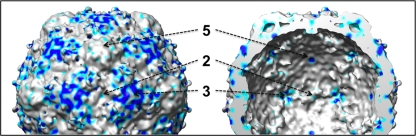
The two 80S density maps share common exterior surface features: depressions around their 2-fold symmetry axes, propeller-like protrusions surrounding the 3-fold axes, and a star-shaped 5-fold mesa surrounded by a moat-like depression (Fig. 2A and B). In the 80S.l reconstruction, the tips of the 5-fold mesa and the tips of the propellers are strongly connected by a rod-shaped density feature, which was previously noted in the 135S reconstruction at 10 Å resolution (10), where it was tentatively identified as an α-helical segment. The presence of this feature creates the impressions that the mesa is larger in diameter than it is in the 160S particle and that the tips of the mesa have been rotated clockwise from their positions in the native virions. The apparent broadening and the rotation of the 5-fold mesas were previously noted in our low-resolution reconstruction of the 80S particle and in the reconstructions of 80S particles from rhinoviruses. The rod-shaped density feature is less prominent when the 80S.e reconstructions are contoured at similar density levels but becomes visible when the contour level is dropped, suggesting that the density feature is partially occupied.
Interior features of EM maps.
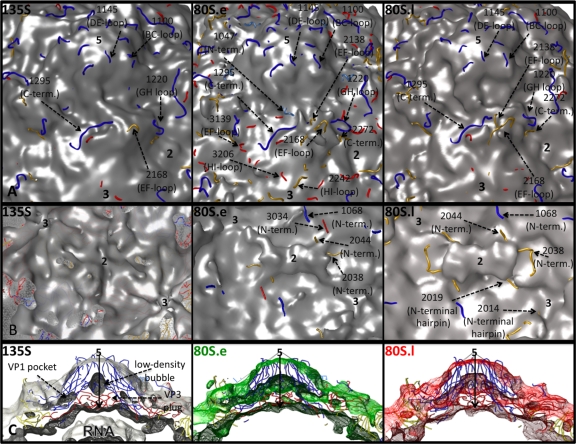
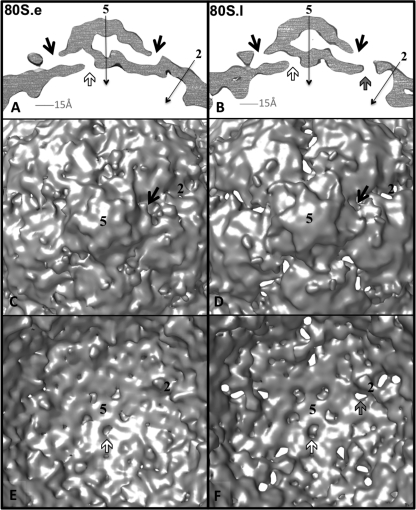
On the inner surface of the capsid, near their 3-fold axes, both 80S forms (Fig. 3B) closely resemble the prepackaging 73S empty capsids (Fig. 4C), having deep, trefoil-shaped depressions. 80S.l has less density in the depressions than 80S.e does. 80S.e and 80S.l share a feature on the capsid interior that distinguishes them from the other known structures of poliovirus intermediates, namely, a large density bridge that lies across the 2-fold axes and spans neighboring pentamers (Fig. 3B). In 135S (and 73S), the corresponding region appears to be smaller, with two discrete protrusions flanking a deep oblong depression at the 2-fold axes (Fig. 3B and 4C), a feature that was also seen in the empty human rhinovirus 2 (HRV2) cryo-EM structure (28). When the density maps for 135S particles are superimposed on 80S maps, it is clear that the 2-fold protrusions of 135S and 73S particles correspond in location to the two ends of the 80S bridge (not shown). 80S.e and 80S.l differ from each other in this region, in that the bridge is suspended over a patch of low density at the 2-fold axis in 80S.l, which is filled with a higher level of density adjacent to the bridge in 80S.e (at slightly higher radius). The two 80S structures also differ in the amount of density found in their 5-fold plugs, which appears to be reduced in 80S.e (Fig. 3C).
FIG. 3.
Close-up views of the inner and outer isocontour surfaces of the 135S and 80S capsids, and the fit of pseudoatomic models, after refinement. (A and B) Views of the outer surface (A) and inner surface (B) of the capsid, shown at contour levels of 1σ, 1.5σ, and 1.5σ, respectively, for the 135S (left), 80S.e (middle), and 80S.l (right) maps (a different contour level is used for the 135S density map because it contains density for the RNA genome and is compared to 80S at similar capsid protein volumes). Refined coordinates for pseudoatomic models of the poliovirus capsid proteins VP1 (blue), VP2 (yellow), and VP3 (red) are shown as main chain traces. Polypeptides that lie outside the chosen contours and representative residue numbers within them are labeled and indicated by dashed arrows. Symmetry axes are labeled with the numbers 2, 3, and 5. The chosen contour level is low enough that the protein shell appears to be closed, with none of the holes extending completely through the capsid, but high enough that unattached noise density is not visible. (C) 30-Å-thick, central slice of the cryo-EM density maps, 135S (gray, left), 80S.e (green, middle), and 80S.l (red, right), showing the enclosed pseudoatomic models docked into them, using density contour levels of 1.5σ, 3σ, and 3σ, respectively. In the 135S map, black arrows indicate the 5-fold axis, and labels indicate the VP3 plug and low-density bubble lying along that axis, the VP1 lipid-binding pocket, and RNA density on the interior.
FIG. 4.
Density maps made from atomic models of the native-antigenic empty capsid assembly intermediate (73S) and the mature, native capsid (160S). These two structures are nearly identical on their exterior surfaces but differ dramatically at the interior surface. The transition from 73S to 160S is marked by cleavage of the precursor protein myrVP0 into myrVP4 and VP2 and by encapsidation of viral RNA. The density maps (shown superimposed on an icosahedral framework) were generated by filtering the atomic models from crystal structures (73S from PDB entry 1POV [4]; 160S from PDB entry1HSX [23]) to 10 Å resolution and applying a temperature factor of 300. (A) Outer surface of 160S. (B) Central sections from the density maps are shown in gray mesh, with 73S on the left and 160S on the right, superimposed on a ribbon representation of the 160S atomic model. Certain interior features and the 5-fold-symmetry axes are labeled. (C) The inner surface of the capsid, viewed along the 2-fold-symmetry axis, in 73S (left) and 160S (right). To provide landmarks, ribbon representations of the 160S atomic model are provided, with VP1 in blue, VP2 in yellow, VP3 in red, and myrVP4 in green. Symmetry axes are labeled 2, 3, and 5. On its inner surface, 73S (left) displays an oblong depression at each 2-fold axis and a deep trefoil depression at each 3-fold axis. In 160S (right), these depressions have been filled in by ordered polypeptides: the trefoil depression is filled by the products of myrVP0 cleavage (the carboxyl terminus of myrVP4 and amino terminus of VP2), together with 1028 to 1054 of VP1, and 3160 to 3161 of VP3; the 2-fold depression is filled by polypeptides 1048 to 1067 and 2045 to 2056. Several of these dynamic peptides become rearranged further during cell entry.
Variance maps.
In order to more objectively evaluate the structural heterogeneity across the population of 2D particle images, we calculated a variance map (Fig. 5). The variance map identifies the areas in the 80S capsid where the density is either the most dissimilar or the most conformationally variable (53). Among the variance map peaks, we can identify the places where the two classes of particles differ most significantly: at the interpentamer interface near the 2-fold symmetry axis, at the hub and tips of the propellers surrounding the 3-fold axis on the outer surface, at the VP3 plug near the 5-fold axis, at the trefoil-shaped depressions at the 3-fold axis on the inner surface, and at the protrusions bridging the 2-fold axis on the inner surface. The two classes of 80S particles also differ in the average amount of density in the center of their protein shells, midway between the inner and outer capsid surfaces. This point is further illustrated by radial-density plots (Fig. 2F), which reveal that 80S.e has more density on average than 80S.l does in the radial range between 125 Å and 155 Å radius.
FIG. 5.
Variance maps, calculated from the population of heat-treated particles. An isocontour surface, showing a portion of the 80S.e reconstruction, has been colored to reflect the degree of voxel-wise variability seen in the heat-treated data set as a whole. The outer surface of the capsid is shown on the left and the inner surface on the right, and the symmetry axes are labeled 2, 3, and 5. Regions colored in gray have the lowest variance (<1σ), those colored in cyan have a higher variance (1 to 2σ), and those colored in dark blue have the highest variance (2 to 3σ).
Pseudoatomic modeling refinement results.
Because of the limited resolution of the reconstructions, we are constrained to build and refine models using well-defined rigid bodies (such as the well-folded β-barrel domains) previously shown to be stable in high-resolution crystal structures (Fig. 1A and B). Where warranted, we have included select well-ordered segments of the loops and carboxyl- and amino-terminal extensions of VP1, VP2, and VP3. In several cases, we have associated these segments with the β-barrels that they bind noncovalently (in 160S), whenever their covalent attachment points appeared to be insufficient to account for the stability of their conformations. Our previous attempts to derive such “pseudoatomic” models for the 80S and 135S particles were based on rigid bodies generated from 160S crystal structures. These modeling efforts resulted in generally satisfactory fits for VP1 and VP3, but we were unable to derive models of VP2 that simultaneously fit the contours on the inside and outside surfaces of the capsid density. In particular, we often found portions of the VP2 model protruding through its outer contour, not far from two “unfilled” density features that the model failed to explain. This clearly told us that something must be wrong with our rigid-body assumptions, but it gave no indication of how we could fix our errors.
Our breakthrough occurred after we noted two things. First, we observed that the inner surface of the 135S and 80S capsids (Fig. 3B) more closely matched the appearance of the inner surface of the 73S particle (Fig. 4C) than that of the 160S particle (Fig. 4D). Second, we saw that the atomic model of 160S that we were using for rigid-body modeling included an ordered loop on the inner surface of VP2 that protruded badly through the isocontour surface of the 80S map. Interestingly, the corresponding loop is disordered in the 73S crystal structure. With those points in mind, we performed parallel modeling and refinement studies using rigid bodies generated from both the 160S atomic model (1HXS) and the 73S atomic model (1POV). We observed that models for both the 80S.e and 80S.l reconstructions derived from the 73S structure produced superior fits and refinement statistics. Based on this observation, we also used rigid-body models derived from the 73S structure to build and refine new models for the 135S particle to better account for the density in our previously reported 135S reconstruction. Atomic models were refined with the program INSOUT following the method of Bubeck et al. (10), using one single, subjectively chosen weighting scheme that allowed the agreement of the model with the experimental density, the restriction of unfavorable steric clashes between rigid bodies, and the requirement for strict icosahedral symmetry to all have an impact on the refinement at once (see Materials and Methods) (10).
For the three refined models (80S.e, 80S.l, and 135S particles), Fourier amplitude-weighted phase agreement statistics were 0.322, 0.417, and 0.557, respectively. We surmise that the ability of a single atomic model to better account for the 135S density may reflect greater structural homogeneity in preparations of this particle and that the poorer statistics for the 80S structures (and in particular the 80S.e particle) reflect either errors in classification, greater flexibility of these structures, or possibly additional structural states in the population that are not distinguishable by classification methods at this resolution. For the three models, the sum-of-Fcalc-squared statistic that tracks protein overlap was indistinguishable (data not shown). These statistic values were all very similar to that calculated from the 73S model (1POV) in its original unexpanded arrangement of rigid-body parts. Correspondingly, only one contact between the model parts seems subjectively to be slightly too close, which is the interaction between the EF loop of VP3 and the carboxyl-terminal chain of VP2, occurring between adjacent protomers within the same pentamer. This observation, together with the mutational data discussed below, raises the possibility that some of the loops involved in this contact area may have been rearranged.
Fit of models into density maps.
Overall, the models fit well into the density maps (when viewed at a sigma range from 1 to 2), with only a few peptides falling outside the chosen isocontour surface of the map (Fig. 3). Most of these polypeptides become enclosed once the contour level is lowered, and we usually consider the result to be plausible as long as the resulting contour remains similar in shape to the model. Weakness in the density can be due to conformational variability, partial occupancy, the tendency of sharp features to disappear at low resolution, or a localized breakdown of the rigid-body modeling assumptions. The variance map constructed from our data set (see Fig. 5) was often useful for confirming that the discrepancies are likely to be real.
On the outer surface of the capsid, the peptides that protrude beyond the isocontour surface (in all three maps) include VP1 BC-loop residues 1091 to 1102, VP1 DE-loop residues 1144 to 1146, VP1 GH-loop residues 1216 to 1226, VP1 extreme-carboxyl-terminal residues 1288 to 1302, VP2 EF-loop residues 2160 to 2171, and VP2 extreme-carboxyl-terminal residues 2266 to 2272 (Fig. 3A). Additional peptides lying outside the isocontour surface for the 80S density maps include VP2 EF-loop residues 2137 to 2139 in both maps and VP3 EF-loop residues 3138 to 3142 in 80S.e only (Fig. 3A). This region of the VP3 EF loop makes electrostatic contacts with 5-fold-symmetry-related copies of the VP2 carboxyl terminus in 160S that are likely to be broken in our expanded 135S and 80S models. Recall that these two peptides make up the one contact in our pseudoatomic models that seems to be too close and therefore may rearrange.
At the interior surface of the capsid, all peptides from the 135S model were enclosed within the map contour. However, in the 80S.e and 80S.l structures, certain residues from the rigid-body model were not enclosed. These included 1068 to 1071 (from the portion of the VP1 amino-terminal extension that is not externalized) and VP2 2036 to 2044 (corresponding to the 0114-to-0126 loop in the amino-terminal extension of myrVP0 that is disordered in 73S particles and that is RNA associated in 160S). Additionally, VP3 residues 3032 to 3035 (in 80S.e), and VP2 amino-terminal hairpin residues 2014 to 2019 (in 80S.l) were all located outside the map isocontour (Fig. 3B). It is significant that these differences on the interior surface, seen between maps and between maps and models, involve residues with a demonstrated ability to rearrange (as previously seen in crystallographic or biochemical studies). Furthermore, the pseudoatomic modeling places most of these residues just adjacent to (at a slightly higher radius from) the density bridge across the 2-fold axis. It is tempting to speculate that these nearby residues from the VP2 and VP1 amino-terminal extensions may rearrange to form this density bridge, but the density is not sufficiently well defined to warrant further modeling at this resolution.
Vector shifts of the β-barrels.
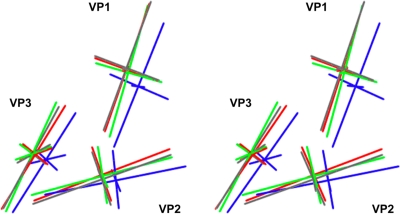
The rigid-body model building and refinement resulted in small translations and rotations to the individual core β-barrels, relative to mature 160S virus particles (Fig. 6; see Movies S1 and S2 in the supplemental material). To simplify the display, each of the β-barrels is approximated by showing the major and minor axes of the ellipsoid of rotation of its α-carbon coordinates. To simplify our description of the shifts and rotations of the β-barrels, we focus on the major capsid proteins of one particular protomer (Fig. 1A), using a Cartesian coordinate frame wherein the x, y, and z axes are coincident with icosahedral 2-fold axes of symmetry of the virion.
FIG. 6.
How the position and orientation of the major capsid proteins differ in the refined pseudoatomic models. Each capsid protein is represented by the major and minor axes of its ellipsoid of rotation, and displayed as a stereo pair. The axes for 160S (blue), 135S (gray), 80S.e (green), and 80S.l (red) are overlaid to illustrate relative positions and tilts. Axes for VP1, VP2, and VP3 are labeled.
Essentially, all three of the major capsid proteins (VP1, VP2, and VP3) in all three intermediate models (135S, 80S.e, and 80S.l) shifted most significantly along the z axis due to the expansion, with the shift in z being most pronounced for VP1 and VP3 and less so for VP2. The shift of VP1 outward along the z axis is a bit further in 135S than in the two 80S structures. In all three intermediates, VP1 shifts slightly in the x and y directions and appears to rotate slightly about both the x and z axes in the 80S.e structure. VP2 from 80S.l shifts slightly in both the x and y directions, while those in 80S.e and 135S shift significantly only in the y direction. Notable tilting of VP2 around the x axis occurs in both the 80S.l and 135S models but not in the 80S.e model. This tilting results in elevating the wider end of the VP2 wedge-shaped β-barrel while lowering the narrow end, near the 3-fold axis. VP3 does not exhibit significant rotations of the subunit or translations in the x and y directions.
Taken together, these shifts and rotations of the β-barrels result in significant alterations in the interprotomer and interpentamer interfaces, including the creation of gaps between the subunits along these interfaces, thereby accounting for the previously noted thinning of the density at these interfaces. The contacts between subunits in the interpentamer interface (which is disrupted during cell entry of aphthoviruses, sometimes leading to the formation of discrete pentamers) appear to be particularly tenuous. For example, near the 3-fold axis, the seven-stranded β-sheet that holds pentamers together tightly in 160S virions (Fig. 1E) has lost its innermost strand in both 73S and 80S particles, and attachment of the VP2 β-hairpin in 73S particles (in the crystal structure) is known to be stabilized by fewer hydrogen bonds than in 160S. In 80S.l particles (as noted above), the VP2 β-hairpin is no longer convincingly enclosed by the density contour, so disorder, rearrangement, or partial occupancy must be suspected. Meanwhile, in the immediate vicinity of the 2-fold axis, the gap between models has widened to be about 30 Å, and it corresponds to a patent opening in the density, seen at high contour levels. Thus, to a significant extent, the continued association of pentamers with each other appears to depend on the presence of the newly formed 2-fold bridging density features on the inner surface.
DISCUSSION
Improvements to our modeling procedure.
Rigid-body pseudoatomic modeling involves many subjective choices, particularly about which parts of the atomic model to include, which parts to group together, what positions to start a refinement with, and what residuals to optimize. At the current resolution limit, we have no objective test for the correctness of the result, but we believe that the latest models are more plausible than the previous ones, and we present them as testable hypotheses, providing landmarks that make it easier to navigate through the maps.
Over the past 9 years, our efforts to make pseudoatomic models by rigid-body fitting have gradually improved, partly due to the increased resolution of the reconstructions. In particular, the current reconstructions at ∼9.5 Å resolution include low-density areas within the capsid that correspond to the centers of the β-barrels and to gaps between β-barrels and which help us to identify unique positions for the rigid bodies. Additionally, we now have a better understanding of how to handle the polypeptides whose conformations (in the 160S crystal structures) depend entirely on the binding contacts that they are seen to make with neighboring proteins. These are polypeptides that extend across the inner and outer surfaces of the capsid to make interactions with the β-barrel cores of neighboring proteins. Such contacts are largely responsible for holding the proteins of the capsid together, and the polypeptides in question would have “no visible means of support” if their protein neighbors were removed. In rigid-body refinement, grouping these segments together with their own β-barrel cores would obviously not have been a sensible choice. Instead, whenever possible, we include these segments and group them with the neighboring barrel to which they are bound. Partly owing to the clarity of the protein-solvent boundary in 80S reconstructions, we can incorporate a greater number of the loops and amino- and carboxyl-terminal extensions into our pseudoatomic models. Overall, the result is more similar in shape to the isocontour surfaces of the density maps than our previous cryo-EM models were. These more-inclusive models fill the available density more completely, thus reducing the number of alternative models that give similar refinement scores. Most importantly, they permit a better understanding of those portions of the structure that affect the structural transitions of the virus and change during the transitions.
Thus, when the density map for 135S particles was originally calculated to 9.5 Å resolution, it became obvious that the amino terminus of VP3 (residues 3014 to 3050) travels with the main VP1 rigid body, as that chain wraps around the lower surface of the VP1 barrel in mature virus. Similarly, it became clear that the β-hairpin at the amino terminus of VP2 (residues 2010 to 2018) continued to extend the back β-sheet of a VP3 β-barrel that belongs to a neighboring pentamer (10), as it does in the crystal structures of 160S and 73S particles (Fig. 1E). However, in the Bubeck et al. (10) model of 135S, the presence of RNA on the interior of the capsid made it impossible to tell exactly where the protein ended. Despite this limitation, we were able to build and refine models for VP1 and VP3 that fit the inner and outer density contours well. In contrast, building a satisfactory model for VP2 based on rigid-body fitting and refinement of the VP2 coordinates from mature 160S virus particles was problematic. We could be sure that the modeling was wrong from the fact that the large EF loop of VP2, located at the tip of a 3-fold propeller, protruded through the outer contour of the reconstruction while, at that same contour, the loop was flanked by two empty density peaks that the model failed to account for. We speculated that VP2 might deform in some way or that the two empty peaks might correspond to VP2 in two or more partially occupied conformational arrangements. As we pointed out at the time, conservative rigid-body modeling had not provided a good description of the VP2 density, but the limited resolution restricted the number of parameters that we were entitled to use.
The recent reconstructions of 80S particles have allowed us to solve several of the earlier problems, partly because the absence of strong RNA density near the capsid shell has made the limits of the protein more obvious than in 135S. Moreover, when we docked the VP2 model (from 160S) to fit the current contour of the outer solvent boundary, we could see clearly that residues 2044 to 2057 were sticking outside the density contour on the inner surface. This was very reminiscent of the crystal structure of myrVP0, the precursor protein that is cleaved into myrVP4 and VP2 upon capsid maturation, in the atomic model of 73S empty capsids (4). As we showed in Basavappa et al. (4), this loop in myrVP0 is disordered, presumably correlated (both in 73S and in the present 80S density maps) with the disorder or absence of the amino terminus of VP1 and with the absence of RNA (Fig. 4C). We therefore initiated modeling and refinement with the VP2 portion of myrVP0 from the 73S structures, and we found that it yielded a significantly improved fit to both the inner and outer solvent boundaries in both the 80S and 135S reconstructions.
Once the myrVP0 β-barrel was reliably placed, it also became apparent that additional flexible peptides previously omitted from the model should be assigned to specific rigid bodies and included. These parts were the GH loop of VP1, which remains associated with the outer surface of VP2 and accounts for one of the “empty” density peaks that previously caused a problem in modeling 135S, and the extreme carboxyl terminus of VP2, which makes extensive contacts with the GH loop of VP1. Following this approach, we have been able to go back to the Bubeck et al. reconstruction of the 135S infectious intermediate, as well as the two current 80S reconstructions, and obtain more satisfactory models to describe the cryo-EM density maps for all three uncoating intermediates. Furthermore, now that the obvious problems with VP2 placement have been solved, we feel that we are on much firmer ground in describing which aminoacyl residues are located in the interfaces that change during the conformational transitions of the virus and allow a much more accurate and detailed analysis of the mutational data.
The current model indicates that much of the GH loop of VP1 remains attached noncovalently to the large EF loop of VP2 and to the VP2 β-barrel during the 160S-to-135S transition, even though VP1 and VP2 are shifted and tilted differently from one another during the expansion of the virus. It is hard to imagine how the GH loop could remain associated with VP2, unless some of the GH-loop polypeptide functions as a slack linker to provide an expansion joint. This is a biologically important result because a number of residues of the VP1 GH loop (including 1226, 1228, and 1234) lie within the footprint of the poliovirus receptor (Pvr/CD155) and several residues (1225, 1226, 1228, 1231, 1234, and 1236) (6, 13, 52) have been shown to affect receptor binding. Mutants containing single-amino-acid substitution at residues of the VP1 GH loop (including 1226, 1228, 1231, and 1236) result in a virus particle with a reduced propensity to undergo the 160S-to-135S transition. Interestingly, residue 1236 is flanked by residues 1235 and 1237, which both contact the pocket factor directly in the native 160S crystal structure (33). In addition, both the amino and carboxyl ends of the VP1 GH loop (residues 1199 and 1237, respectively) flank the opening of the pocket (the so-called “pore”) and are also in direct contact with the “pocket factor” in the native 160S crystal structure. Notably, the “pocket factor” appears to be either reduced or missing in 135S, 80S.e, and 80S.l (Fig. 3C). The relative shifts of VP2 and VP1, together with the loss of pocket factor and conformational rearrangement of the GH loop of VP1 during the 160S-to-135S transition, are collectively likely to be responsible for the fact that poliovirus loses its affinity for the receptor once the 135S transition has taken place. Taken together, these data suggest that the GH loop of VP1, along with peptides that interact with it, is involved in transmitting the receptor-binding signal to regions of the capsid that are responsible for the structural transitions.
The only modeled peptide that now remains outside our map contours when viewed at a generous contour level (0.5σ) is the extreme carboxyl terminus of VP1 (residues 1293 to 1302) in the 135S model. Regions from the carboxyl-terminal extension of VP1 have previously been shown to influence structural transitions. Thus, single mutations at residue 1290 lower the stability of 135S particles (17), and monoclonal antibodies raised against a peptide that corresponds to residues 1280 to 1286 are able to immunoprecipitate 80S but not 135S particles (36). In mature 160S virions, the loop of VP1 that includes residues 1280 to 1286 makes extensive contacts with a portion of the EF loop of VP2 that includes residue 2142 from the receptor-binding footprint (6, 52). Viruses with single mutations at 2142 have been shown to influence receptor-binding affinity, to expand host range and receptor specificity, and to become more labile to heat conversion from 160S to 135S (13, 51). Our conservative rigid-body modeling procedure and the resolution of our density maps do not permit us to make a more specific suggestion for how the carboxyl-terminal extension of VP1 may have changed. However, taken together with the mutagenesis and antibody-binding data, our maps and models both suggest that this polypeptide is unlikely to maintain its native (160S) conformation in the cell entry intermediates.
As we noted previously, VP2 has historically been the most problematic of the major capsid proteins to fit into our 135S and 80S maps; it appears to undergo the most dramatic changes in the normally well-ordered portions of the protein, and its interfaces with neighboring proteins are changed most extensively. In our 135S and 80S.l models, and to a lesser extent 80S.e, VP2 is the only one of the capsid proteins that tilts appreciably, relative to its orientation in 160S and 73S particles (Fig. 6). This tilting would move the broader end of the VP2 wedge outward and the narrow end (the end closer to the 3-fold axes) inward. This rearrangement might disrupt the interaction (seen in mature 160S virions) between the carboxyl-terminal extension of VP1 and the EF loop of VP2. The difference in amount of tilting of VP2 seen between the two 80S structures may also account for differences in the density maps surrounding the 3-fold axes (VP3 tilting also influences changes in this region, although to a lesser extent) and at the 3-fold propeller tips, as illustrated by the variance map (Fig. 5; see Movies S1 and S2 in the supplemental material).
Common structural changes associated with both RNA packaging and release.
The major structural differences that we observe between 135S and the 80S reconstructions occur on the inner surface of the capsid. Some of these structural changes, which may be associated with the RNA-release transition, are located in areas that also are seen to change during assembly (i.e., during the packaging of RNA and the cleavage of myrVP0), and are evident when the structures of 73S and 160S particles are compared (Fig. 4). For example, we observe a 73S-like trefoil depression in our 80S structures (Fig. 3B), while the resemblance of the 135S trefoil to that of 73S particles is less close. In contrast, on the inner surface of 135S particles, along the 3-fold axis, there is instead a 160S-like arrangement of density, which we attribute to the amino-terminal residues 2006 to 2013 and which caps off a low-density bubble in the capsid (Fig. 3C). This structural difference suggests that the extreme amino terminus of VP2 becomes rearranged during the initiation of RNA release (in the 135S-to-80S.e transition). Note that this polypeptide terminus is the same one that is liberated by myrVP0 cleavage and that it moves by more than 20 Å during the 73S-to-160S transition. Indications of where the largest changes are taking place in the 80S.e-to-80S.l transition can be obtained by noting which regions have the highest variance between 80S particle forms (Fig. 5). One region of high variability occurs in the floor of the trefoil and corresponds to the DE loop from the narrow end of the VP2 β-barrel, which exhibits a high degree of sequence conservation among picornaviruses. A second region of high variability occurs at the 5-fold plug, corresponding to five intertwined copies of the amino terminus of VP3. A third area occurs around the 2-fold axis, at the bridge of density (Fig. 3B), which we suspect is likely to have been formed by a rearrangement of the amino-terminal extension of VP2. Interestingly, this suggests that peptides at all three symmetry axes become altered during the 80S.e-to-80S.l transition.
The most notable difference between the 135S and 80S structures occurs on the inner surface of the capsid, in the area that surrounds the 2-fold symmetry axis. This region is 73S-like in the 135S structure (Fig. 3B), having a 2-fold depression flanked by protrusions, but becomes bridged across the 2-fold axis, with the addition of an oblong density feature in 80S particles. In 73S the flanking protrusions occur near the transition point where the amino-terminal polypeptide of VP1 becomes disordered (around residue 1067) and near the disordered loop in the amino-terminal extension of myrVP0 (corresponding to the neighborhood of residue 2043). Both of these dynamic peptides become well ordered in 160S, making numerous electrostatic interactions that stabilize them, including contacts between the 1063-to-1069 and 2045-to-2051 polypeptides in adjacent protomers. We are limited by the resolutions of our reconstructions of the cell entry intermediates, and by our rigid-body modeling, and therefore cannot yet model the specifics of the structural changes that we see near the 2-fold axis and which we are attributing to the rearrangement of these two polypeptides.
One interesting additional candidate for a participant in forming the bridging structure is the two-stranded β-hairpin from the amino-terminal extension of VP2, which forms the fifth and sixth strands of the interpentameric seven-stranded β-sheet (Fig. 1E), since the hairpin is located close to the 2-fold bridge and because the density for it in the reconstruction, as noted above, is weaker than expected. As an aside, it is probably relevant that the corresponding residues of uncoated equine rhinitis A virus (ERAV) were shown to rearrange during RNA release (47), though the specifics of the rearrangement are different. Thus, changes in atomic coordinates for ERAV before and after RNA release (47) did not correspond to the density changes that we see in the poliovirus intermediates.
Holes in the capsid at the interface between 5-fold-symmetry-related protomers may serve as routes for RNA egress.
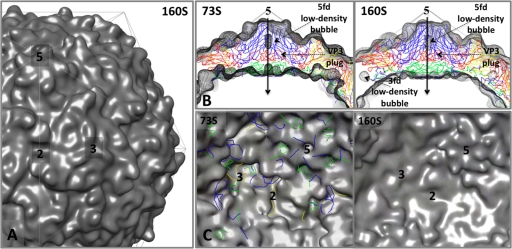
The 80S.e and 80S.l reconstructions were each made from a common, CTF-corrected data set of 2D images, classified into two distinct groups, using a structure-based method. Additionally, the current relatively high resolution of the reconstructions was impossible to achieve without carrying out the classification. For those reasons, the differences in density that we observe between these maps at a single contour level can be believed. Our radial-density plots illustrate that between the inner and outer capsid surfaces, there is less total protein density in the 80S.l than the 80S.e density map (Fig. 2F). Additionally, when viewed at a stringent contour level (3σ) both maps have holes that span the capsid shell (Fig. 7). These twisting channels start on the inner surface at openings close to the 5-fold plug, continue through the large low-density bubble on the 5-fold axis, run out through the VP1 lipid-binding pocket, and terminate where the pocket opens to the outer surface of the capsid, at the bottom of the “canyon,” near the quasi-3-fold axes (Fig. 7A and B). The 80S.l map has additional, more direct holes, running through the capsid shell, which are located at points on the surface that lie between the 2-fold and quasi-3-fold axes of symmetry (Fig. 7B, D, and F).
FIG. 7.
Holes in the 80S density maps may serve as routes of egress through the capsid. Cryo-EM density maps for 80S.e (A, C, and E) and 80S.l (B, D, and F) are contoured at 3σ to visualize regions of lowest density. (A and B) 5-Å-thick central slices of maps show the locations of low-density pores through the capsid shell. In each structure, openings at the interior surface (white arrows), near the 5-fold plug, are connected, via the low-density bubble along the 5-fold axis, to the lipid-binding pocket of VP1, which connects to the outer surface via a pore (black arrows) that is located near the quasi-3-fold axis. Thus, a twisted, continuous channel through the shell is formed. 80S.l has an additional more direct path through the shell (gray arrow). Symmetry axes are labeled with thin black arrows. (C and D) 80S.e (C) and 80S.l (D). The outer surface of each map is viewed along a 5-fold axis of symmetry. At the present contour level, the direct holes in the 80S.l map are evident. At a higher contour level, holes in the 80S.e map appear in similar positions (not shown). (E and F) 80S.e (E) and 80S.l (F). Shown are close-up views of a portion of the inner surface. All arrow colors and axis labels correspond to those in panels A and B.
The 80S and 135S density maps are generally displayed at a contour level between 1σ and 2σ (Fig. 2, 3, and 5), but when viewed at a slightly higher, stringent contour level (3.5σ, not shown), the 80S.l map displays large oblong holes along the 2-fold axis that provide straight pathways from inside to outside. In their longer dimension, these holes run from one quasi-3-fold axis to the other; in their shorter dimension, the holes measure about 15 Å in width, roughly corresponding to the width of the hole in the pseudoatomic model (Fig. 8). In the 80S.e density map at 3.5σ, a similar direct hole can be seen, but instead of one large hole across the 2-fold axis there are two smaller holes separated by the 2-fold-bridge of density. Differences between 80S.e and 80S.l in this region suggest that rearrangement of the peptides that make up the 2-fold bridges facilitates the 80S.e-to-80S.l transition during RNA release. The bridging region seen in 80S.e at 3.5σ is part of the more extensive bridge seen in both 80S reconstructions at lower contour levels (Fig. 3B), which is broader and extends further inward, to a lower radius. 80S.e and 80S.l thus differ from one another in the contour levels required to see specific features, and the majority of the bridge occurs at a distinctly lower contour level than most of the rest of the capsid, which suggests partial occupancy, whose extent differs between the two 80S populations and most likely increases over time.
FIG. 8.
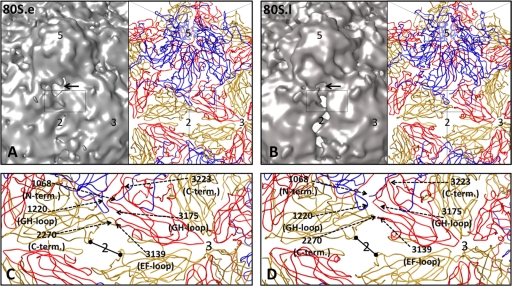
Pseudoatomic models reveal peptides that may rearrange for RNA egress. Density maps (contoured at 3σ) for 80S.e (A) and 80S.l (B) are shown on the left, side by side with their respective pseudoatomic models (right), to reveal low-density regions between protomers. Some of the symmetry axes are labeled 2, 3, and 5. The region indicated by the black rectangle includes residues previously implicated by mutagenesis to have an effect on assembly, receptor binding, capsid stability and structural transitions (see text). (C and D) Close-up views of a portion of the pseudoatomic models boxed in panels A and B, respectively. Representative residues, labeled using dashed arrows, lie in the interface between 5-fold-symmetry-related protomers. Black bars with dots mark the shortest dimension across the hole at the 2-fold axis, which is 15.3 Å in 80S.e and 15.0 Å in 80S.l. The longer dimension of the hole is 27.3 Å for 80S.e and 31.6 Å for 80S.l.
The holes through the 80S density maps are located between 5-fold-symmetry-related protomers and extend between the 2-fold and quasi-3-fold axes of symmetry (Fig. 8B). The holes in the density maps are blocked in the immediate vicinity of the 2-fold axes by a bridge of density that we attribute to residues of the amino-terminal extension of VP2. The holes in the 80S pseudoatomic models (Fig. 8C and D) appear to extend all the way across the 2-fold axes because the residues that contribute to the bridge density have not been included in the atomic models. Note that the holes in the 80S pseudoatomic models are easily large enough for single-stranded RNA to pass through them and that the low density levels for the bridges that block these holes in the density maps suggests that the bridges are only partially occupied. The partial occupancy raises the possibility that at least some number of the large holes at the 2-fold axes may be open during RNA release. In the native 160S structure, there is continuous density in these interface regions. Thus, in 160S, portions of the VP1 GH loop, the VP3 EF loop and GH loop, the VP2 and VP3 carboxyl-terminal polypeptides, and portions of the N-terminal peptide of VP2 all interact with one another across interfaces between symmetry-related protomers, in places where holes exist in 80S. Mutations to residues in this particular interface have been shown to influence the formation of pentamers (38), receptor binding (13), and the ability of viruses to undergo both the 73S-to-160S transition (1, 40) and the 160S-to-135S transition (13, 51) (Fig. 9). Based on these data and the symmetrized reconstructions, the locus of RNA exit could be anywhere along the intersubunit interface that extends from the pseudo-3-fold to the 2-fold.
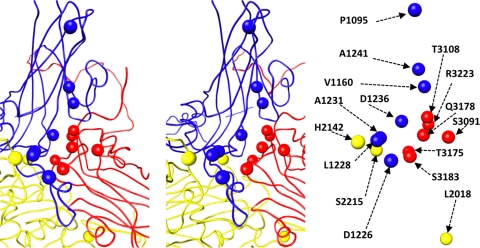
FIG. 9.
Mutations that influence assembly, receptor binding, and uncoating are clustered in the interface between 5-fold-symmetry-related protomers. Main chain traces of a portion of VP1 (blue), VP2 (yellow), and VP3 (red) are shown in stereo (left). The locations of specific residues are shown as spheres (left) and labeled (right). Residues from VP1 and VP2 on the left side of the interface come from one protomer. On the right side of the interface, residues from VP3 belong to a second (5-fold-symmetry-related) protomer, and residues from VP2 belong to a third (3-fold-symmetry-related) protomer. When mutated, the indicated residues influence assembly (2018, 2215, 3091, 3108, 3175, 3178, and 3223) (1, 19, 38, 39), receptor binding (1226, 1228, 1231, 1241, 2215, and 3178) (13), and the ability of particles to transition from 160S to 135S (1095, 1160, 1226, 1228, 1231, 1241, 2215, and 3178) (13, 51). Residues 2215 and 3178 fall into all three categories.
In order to elucidate the exact egress point from the capsid shell of the single copy of the RNA genome and to provide details of the interaction with RNA, we would need asymmetric information. In the present study, we have used icosahedral averaging across multiple particles to determine these structures, which provides greater resolution but limits what we can learn about the unique site. In a related study of this same heat-treated virus sample, we have used cryo-tomography and asymmetric single-particle reconstruction procedures to map the footprint of the exiting RNA in a subset of the particles that have density for RNA inside and outside the capsid, and confirmed that the RNA does indeed exit at the interface between 5-fold-symmetry-related protomers, in an area between the 2-fold symmetry axis and the quasi-3-fold axis (Bostina et al., submitted).
Why are the structural changes so small?
We had anticipated that the structural changes associated with the 160S-to-135S and 135S-to-80S transitions would be much larger than the changes that we have observed. Those expectations had been based on the large changes in sedimentation in the 135S particle and on the need to externalize large peptides and RNA. In our previous reports of the 80S (5) and 135S (5, 10) structures, we rationalized the small magnitude of the changes by suggesting that there might be additional transient intermediates in both structural transitions that are significantly expanded, creating openings for the externalization of peptides or RNA. This argument was based in part on an analogy with the expansion of T=3 plant viruses, which is induced by exposing the virus to slightly alkaline pH in the presence of chelators of divalent cations and which (as in poliovirus) results in the externalization of the amino-terminal extensions of the capsid proteins. Furthermore, when the expansion of tomato bushy stunt virus (TBSV) (26) or turnip crinkle virus (TCV) (34) is quenched by lowering the pH rapidly, the particles are trapped in a partially expanded state with their amino-terminal extensions caught on the outside of the particle. According to this analogy, the more fully expanded state of TBSV or TCV would correspond to the imagined transient fully expanded intermediate of poliovirus, and their quenched state would correspond to the 135S particle.
However, our current cryo-EM studies of poliovirus 80S particles provide no direct evidence for the existence of a form that is more expanded than the ones we have observed. In contrast to 135S particles (which each contain a full complement of RNA), there is good experimental support for the idea that much of the population of 80S particles has been “caught in the act,” with RNA trapped partway through the capsid. Supporting evidence includes the existence of at least two states of the 80S particle, with a variable amount of density on the inside that corresponds to residual RNA, and the finding that a small but significant percentage of the 80S.e particles have putative RNA density features both inside and outside the particle at the same time. Nevertheless, 135S particles and both forms of 80S particles all appear to be very similar in overall size (Fig. 2F). Plausible explanations for this include the possibility that the holes observed at the particle 2-fold axes (perhaps in symmetry copies where the bridging density is absent) are large enough to allow passage of single-stranded regions of the RNA. Alternatively, if the observed holes are not large enough, and a still greater icosahedrally symmetric expansion of the capsid is required (as in TBSV and TCV), then it remains possible that the RNA externalization steps are tightly coupled with a transient expansion, with recontraction to the observed structure occurring during pauses. As a final alternative, we cannot rule out the possibilities that the icosahedral symmetry of the capsid is broken only locally at the site of RNA externalization and that the capsid expansion (or hole creation) at that site is too limited in its extent to have a detectable impact on the icosahedrally symmetrized cryo-EM reconstructions that we report here. In an effort to address those possibilities, we are currently attempting to extend the resolution of our asymmetric reconstruction of “caught in the act” particles (work in progress), though at present, its resolution is not yet high enough to answer the question.
Why are there two different forms of the 80S particle, and what does it mean for there to be residual RNA density inside the shell?
The presence of two structural classes of 80S particles, each containing a variable amount of residual RNA, was first observed in cryo-EM studies of 80S particles of rhinovirus (27). However, the significance of the residual RNA and of the existence of two classes was not clear. Our observations that similar classes of 80S particles are seen in poliovirus argue strongly that the existence of these two forms is an inherent property of the RNA-release process, at least in the enterovirus and rhinovirus genera of the picornavirus family. In thinking about the significance of these observations, it is important to note that the RNA genome, which is extensively folded into complex double-stranded structures in the virion, must be locally unfolded into single-stranded regions during RNA release. Thus, our experimental protocols for producing the 80S particles in vitro call for the virus to be heated to temperatures which are close to the bulk melting temperature of the RNA (49), and we have previously demonstrated that the dyes that bind double-stranded RNA are released during the release of RNA from virions, implying that RNA unfolding must be taking place in natural infections as well (8).
To account for the observation that 80S particles contain variable levels of RNA on the inside, we postulate that unfolding of secondary structure is more difficult in some regions of the genome than at others. Thus, RNA release could take place in stages, with pauses occurring at points where the secondary structure is more stable. As an interesting aside, note that unfolding, followed by externalization, and then refolding of the externalized segments of the RNA provide a ratchet that could help to drive the externalization of the RNA to completion once initiated. We can further postulate that the failure to find externalized RNA in the majority of particles that have residual RNA density on the inside is due to hydrolysis by RNases in the sample during pauses. The existence of two classes of particles might then be explained by whether or not RNA continued to be engaged by the exit machinery. Thus, the 80S.l class might include both the particles that had finished releasing their genomes and those whose RNA had disengaged after hydrolysis, whereas the 80S.e class might represent those particles that have partially released RNA but in which the RNA remains engaged with the exit site even after hydrolysis.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants AI20566, 1F32AI081427-01, and 2P01GM062580-07.
We thank Nikolaus Grigorieff and Chen Xu for their assistance with data collection, David Belnap for valuable conversations about image processing, Doryen Bubeck for helpful discussions and sharing her expertise, and the members of the Hogle lab for creating a great work environment.
Footnotes
Published ahead of print on 24 February 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Ansardi, D. C., M. Luo, and C. D. Morrow. 1994. Mutations in the poliovirus P1 capsid precursor at arginine residues VP4-ARG34, VP3-ARG223, and VP1-ARG129 affect virus assembly and encapsidation of genomic RNA. Virology 199:20-34. [DOI] [PubMed] [Google Scholar]
- 2.Arnold, E., and M. G. Rossmann. 1990. Analysis of the structure of a common cold virus, human rhinovirus 14, refined at a resolution of 3.0 Å. J. Mol. Biol. 211:763-801. [DOI] [PubMed] [Google Scholar]
- 3.Baker, T. S., and R. H. Cheng. 1996. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol. 116:120-130. [DOI] [PubMed] [Google Scholar]
- 4.Basavappa, R., R. Syed, O. Flore, J. P. Icenogle, D. J. Filman, and J. M. Hogle. 1994. Role and mechanism of the maturation cleavage of VP0 in poliovirus assembly: structure of the empty capsid assembly intermediate at 2.9 Å resolution. Protein Sci. 3:1651-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belnap, D. M., D. J. Filman, B. L. Trus, N. Cheng, F. P. Booy, J. F. Conway, S. Curry, C. N. Hiremath, S. K. Tsang, A. C. Steven, and J. M. Hogle. 2000. Molecular tectonic model of virus structural transitions: the putative cell entry states of poliovirus. J. Virol. 74:1342-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belnap, D. M., B. M. McDermott, Jr., D. J. Filman, N. Cheng, B. L. Trus, H. J. Zuccola, V. R. Racaniello, J. M. Hogle, and A. C. Steven. 2000. Three-dimensional structure of poliovirus receptor bound to poliovirus. Proc. Natl. Acad. Sci. U. S. A. 97:73-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bostina, M., D. Bubeck, C. Schwartz, D. Nicastro, D. J. Filman, and J. M. Hogle. 2007. Single particle cryoelectron tomography characterization of the structure and structural variability of poliovirus-receptor-membrane complex at 30 Å resolution. J. Struct. Biol. 160:200-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brandenburg, B., L. Y. Lee, M. Lakadamyali, M. J. Rust, X. Zhuang, and J. M. Hogle. 2007. Imaging poliovirus entry in live cells. PLoS Biol. 5:e183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Breindl, M. 1971. The structure of heated poliovirus particles. J. Gen. Virol. 11:147-156. [DOI] [PubMed] [Google Scholar]
- 10.Bubeck, D., D. J. Filman, N. Cheng, A. C. Steven, J. M. Hogle, and D. M. Belnap. 2005. The structure of the poliovirus 135S cell entry intermediate at 10-angstrom resolution reveals the location of an externalized polypeptide that binds to membranes. J. Virol. 79:7745-7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bubeck, D., D. J. Filman, and J. M. Hogle. 2005. Cryo-electron microscopy reconstruction of a poliovirus-receptor-membrane complex. Nat. Struct. Mol. Biol. 12:615-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, J. Z., and N. Grigorieff. 2007. SIGNATURE: a single-particle selection system for molecular electron microscopy. J. Struct. Biol. 157:168-173. [DOI] [PubMed] [Google Scholar]
- 13.Colston, E., and V. R. Racaniello. 1994. Soluble receptor-resistant poliovirus mutants identify surface and internal capsid residues that control interaction with the cell receptor. EMBO J. 13:5855-5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Curry, S., M. Chow, and J. M. Hogle. 1996. The poliovirus 135S particle is infectious. J. Virol. 70:7125-7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Danthi, P., M. Tosteson, Q. H. Li, and M. Chow. 2003. Genome delivery and ion channel properties are altered in VP4 mutants of poliovirus. J. Virol. 77:5266-5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Sena, J., and B. Mandel. 1977. Studies on the in vitro uncoating of poliovirus. II. Characteristics of the membrane-modified particle. Virology 78:554-566. [DOI] [PubMed] [Google Scholar]
- 17.Duncan, G., and F. Colbere-Garapin. 1999. Two determinants in the capsid of a persistent type 3 poliovirus exert different effects on mutant virus uncoating. J. Gen. Virol. 80(part 10):2601-2605. [DOI] [PubMed] [Google Scholar]
- 18.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 19.Filman, D. J., R. Syed, M. Chow, A. J. Macadam, P. D. Minor, and J. M. Hogle. 1989. Structural factors that control conformational transitions and serotype specificity in type 3 poliovirus. EMBO J. 8:1567-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fricks, C. E., and J. M. Hogle. 1990. Cell-induced conformational change in poliovirus: externalization of the amino terminus of VP1 is responsible for liposome binding. J. Virol. 64:1934-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fry, E. E., N. J. Knowles, J. W. Newman, G. Wilsden, Z. Rao, A. M. King, and D. I. Stuart. 2003. Crystal structure of swine vesicular disease virus and implications for host adaptation. J. Virol. 77:5475-5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuller, S. D., S. J. Butcher, R. H. Cheng, and T. S. Baker. 1996. Three-dimensional reconstruction of icosahedral particles—the uncommon line. J. Struct. Biol. 116:48-55. [DOI] [PubMed] [Google Scholar]
- 23.Grant, R. A., C. N. Hiremath, D. J. Filman, R. Syed, K. Andries, and J. M. Hogle. 1994. Structures of poliovirus complexes with anti-viral drugs: implications for viral stability and drug design. Curr. Biol. 4:784-797. [DOI] [PubMed] [Google Scholar]
- 24.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 25.Hadfield, A. T., W. Lee, R. Zhao, M. A. Oliveira, I. Minor, R. R. Rueckert, and M. G. Rossmann. 1997. The refined structure of human rhinovirus 16 at 2.15 Å resolution: implications for the viral life cycle. Structure 5:427-441. [DOI] [PubMed] [Google Scholar]
- 26.Harrison, S. C. 1980. Protein interfaces and intersubunit bonding. The case of tomato bushy stunt virus. Biophys. J. 32:139-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hewat, E. A., and D. Blaas. 2004. Cryoelectron microscopy analysis of the structural changes associated with human rhinovirus type 14 uncoating. J. Virol. 78:2935-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hewat, E. A., E. Neumann, and D. Blaas. 2002. The concerted conformational changes during human rhinovirus 2 uncoating. Mol. Cell 10:317-326. [DOI] [PubMed] [Google Scholar]
- 29.Heymann, J. B. 2001. Bsoft: image and molecular processing in electron microscopy. J. Struct. Biol. 133:156-169. [DOI] [PubMed] [Google Scholar]
- 30.Heymann, J. B., and D. M. Belnap. 2007. Bsoft: image processing and molecular modeling for electron microscopy. J. Struct. Biol. 157:3-18. [DOI] [PubMed] [Google Scholar]
- 31.Hindiyeh, M., Q. H. Li, R. Basavappa, J. M. Hogle, and M. Chow. 1999. Poliovirus mutants at histidine 195 of VP2 do not cleave VP0 into VP2 and VP4. J. Virol. 73:9072-9079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hiremath, C. N., D. J. Filman, R. A. Grant, and J. M. Hogle. 1997. Ligand-induced conformational changes in poliovirus-antiviral drug complexes. Acta Crystallogr. D Biol. Crystallogr. 53:558-570. [DOI] [PubMed] [Google Scholar]
- 33.Hogle, J. M., M. Chow, and D. J. Filman. 1985. Three-dimensional structure of poliovirus at 2.9 Å resolution. Science 229:1358-1365. [DOI] [PubMed] [Google Scholar]
- 34.Hogle, J. M., A. Maeda, and S. C. Harrison. 1986. Structure and assembly of turnip crinkle virus. I. X-ray crystallographic structure analysis at 3.2 Å resolution. J. Mol. Biol. 191:625-638. [DOI] [PubMed] [Google Scholar]
- 35.Jacobson, M. F., and D. Baltimore. 1968. Polypeptide cleavages in the formation of poliovirus proteins. Proc. Natl. Acad. Sci. U. S. A. 61:77-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ketterlinus, R., and K. Wiegers. 1994. Mapping of antigenic domains in poliovirus VP1 involved in structural rearrangements during virus morphogenesis and antigenic alterations of the virion. Virology 204:27-37. [DOI] [PubMed] [Google Scholar]
- 37.Kim, K. H., P. Willingmann, Z. X. Gong, M. J. Kremer, M. S. Chapman, I. Minor, M. A. Oliveira, M. G. Rossmann, K. Andries, G. D. Diana, et al. 1993. A comparison of the anti-rhinoviral drug binding pocket in HRV14 and HRV1A. J. Mol. Biol. 230:206-227. [DOI] [PubMed] [Google Scholar]
- 38.Macadam, A. J., G. Ferguson, C. Arnold, and P. D. Minor. 1991. An assembly defect as a result of an attenuating mutation in the capsid proteins of the poliovirus type 3 vaccine strain. J. Virol. 65:5225-5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minor, P. D., G. Dunn, D. M. Evans, D. I. Magrath, A. John, J. Howlett, A. Phillips, G. Westrop, K. Wareham, J. W. Almond, et al. 1989. The temperature sensitivity of the Sabin type 3 vaccine strain of poliovirus: molecular and structural effects of a mutation in the capsid protein VP3. J. Gen. Virol. 70(part 5):1117-1123. [DOI] [PubMed] [Google Scholar]
- 40.Minor, P. D., A. J. Macadam, D. M. Stone, and J. W. Almond. 1993. Genetic basis of attenuation of the Sabin oral poliovirus vaccines. Biologicals 21:357-363. [DOI] [PubMed] [Google Scholar]
- 41.Muckelbauer, J. K., M. Kremer, I. Minor, L. Tong, A. Zlotnick, J. E. Johnson, and M. G. Rossmann. 1995. Structure determination of coxsackievirus B3 to 3.5 Å resolution. Acta Crystallogr. D Biol. Crystallogr. 51:871-887. [DOI] [PubMed] [Google Scholar]
- 42.Phillips, B. A., and R. Fennell. 1973. Polypeptide composition of poliovirions, naturally occurring empty capsids, and 14S precursor particles. J. Virol. 12:291-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rueckert, R. R., and M. A. Pallansch. 1981. Preparation and characterization of encephalomyocarditis (EMC) virus. Methods Enzymol. 78:315-325. [PubMed] [Google Scholar]
- 44.Tosteson, M. T., and M. Chow. 1997. Characterization of the ion channels formed by poliovirus in planar lipid membranes. J. Virol. 71:507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tosteson, M. T., H. Wang, A. Naumov, and M. Chow. 2004. Poliovirus binding to its receptor in lipid bilayers results in particle-specific, temperature-sensitive channels. J. Gen. Virol. 85:1581-1589. [DOI] [PubMed] [Google Scholar]
- 46.Tuthill, T. J., D. Bubeck, D. J. Rowlands, and J. M. Hogle. 2006. Characterization of early steps in the poliovirus infection process: receptor-decorated liposomes induce conversion of the virus to membrane-anchored entry-intermediate particles. J. Virol. 80:172-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tuthill, T. J., K. Harlos, T. S. Walter, N. J. Knowles, E. Groppelli, D. J. Rowlands, D. I. Stuart, and E. E. Fry. 2009. Equine rhinitis A virus and its low pH empty particle: clues toward an aphthovirus entry mechanism? PLoS Pathog. 5:e10000620. [DOI] [PMC free article] [PubMed]
- 48.Watanabe, Y., K. Watanabe, S. Katagiri, and Y. Hinuma. 1965. Virus-specific proteins produced in HeLa cells infected with poliovirus: characterization of subunit-like protein. J. Biochem. 57:733-741. [DOI] [PubMed] [Google Scholar]
- 49.Wetz, K., and T. Kucinski. 1991. Influence of different ionic and pH environments on structural alterations of poliovirus and their possible relation to virus uncoating. J. Gen. Virol. 72(part 10):2541-2544. [DOI] [PubMed] [Google Scholar]
- 50.Wien, M. W., M. Chow, and J. M. Hogle. 1996. Poliovirus: new insights from an old paradigm. Structure 4:763-767. [DOI] [PubMed] [Google Scholar]
- 51.Wien, M. W., S. Curry, D. J. Filman, and J. M. Hogle. 1997. Structural studies of poliovirus mutants that overcome receptor defects. Nat. Struct. Biol. 4:666-674. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, P., S. Mueller, M. C. Morais, C. M. Bator, V. D. Bowman, S. Hafenstein, E. Wimmer, and M. G. Rossmann. 2008. Crystal structure of CD155 and electron microscopic studies of its complexes with polioviruses. Proc. Natl. Acad. Sci. U. S. A. 105:18284-18289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, W., M. Kimmel, C. M. Spahn, and P. A. Penczek. 2008. Heterogeneity of large macromolecular complexes revealed by 3D cryo-EM variance analysis. Structure 16:1770-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.