Abstract
Retroviruses and retrovirus-derived vectors integrate nonrandomly into the genomes of host cells with specific preferences for transcribed genes, gene-rich regions, and CpG islands. However, the genomic features that influence the transcriptional activities of integrated retroviruses or retroviral vectors are poorly understood. We report here the cloning and characterization of avian sarcoma virus integration sites from chicken tumors. Growing progressively, dependent on high and stable expression of the transduced v-src oncogene, these tumors represent clonal expansions of cells bearing transcriptionally active replication-defective proviruses. Therefore, integration sites in our study distinguished genomic loci favorable for the expression of integrated retroviruses and gene transfer vectors. Analysis of integration sites from avian sarcoma virus-induced tumors showed strikingly nonrandom distribution, with proviruses found prevalently within or close to transcription units, particularly in genes broadly expressed in multiple tissues but not in tissue-specifically expressed genes. We infer that proviruses integrated in these genomic areas efficiently avoid transcriptional silencing and remain active for a long time during the growth of tumors. Defining the differences between unselected retroviral integration sites and sites selected for long-terminal-repeat-driven gene expression is relevant for retrovirus-mediated gene transfer and has ramifications for gene therapy.
Retroviral replication requires integration of the provirus, the DNA intermediate of retroviral replication, into the chromosome of the host. Provirus integration occurs in most genomic regions with weak though statistically significant preference for specific target site DNA sequences (19, 49). Secondary DNA structures, DNA bending, nucleosome density, DNase hypersensitivity, and certain chromatin features represent other preferential target sites for retrovirus integration (see reference 20 for a review). The capture of dimeric retroviral integrase by host cell factors, which tether the whole preintegration complex with chromatin, has been described as a basic mechanism controlling retrovirus target site specificity. For example, LEDGF/p75 directs human immunodeficiency virus type 1 (HIV-1) integrase to chromatin, and its depletion from cells or alterations of its N-terminal domain impair both the efficiency and preferences of lentiviral integration (7, 31).
The availability of the assembled human genome sequence opened the chance to map and analyze retrovirus integration sites on a genome-wide scale with respect to the genomic features of the host DNA, such as GC content, gene density, and cytogenetic bands. In a small data set of unselected integration sites, it turned out that HIV-1 preferentially integrated into genes, GC-rich regions, and cytogenetic R bands (10). This approach has been rapidly applied to a wide variety of retroviruses, and several comparative studies have shown surprising differences between their integration preferences. While HIV-1 preferentially targets genes, particularly the transcriptionally active ones (10, 42, 33), avian sarcoma and leukosis viruses (ASLVs) integrate with only slight preference for genes (1, 33, 34, 38). The most random dispersion of integration without any tendency to integrate within genes was observed for mouse mammary tumor virus (12). Uniquely, murine leukemia virus (MLV) favors integration in close proximity to upstream or downstream transcription start sites (48).
Retrovirus-derived vectors retain the target site preferences of their parental retroviruses (33). Hence, the genome-wide integration results are relevant to the design of retroviral vectors for gene transfer and gene therapy. It is particularly important to know in which genomic locations the integrated retroviral vector retains transcriptional activity of the transduced gene and, vice versa, where it tends to be transcriptionally silenced by the inhibitory mechanisms of the host cell. In gene therapy trials, the selection of target cells with active retrovirus-transduced therapeutic genes can result in clones with trans-activated proto-oncogenes. This was demonstrated in child patients treated for X-linked severe combined immunodeficiency with MLV-based vectors, who exhibited four cases of B cell lymphoproliferative disorder with the vector integrated close to the promoter of the LMO2 proto-oncogene (15). Another example of clonal selection of retroviral integration sites can be seen in studies describing common integration sites (CIS) of chronically transforming retroviruses (4, 8, 35, 45).
The currently available genome-wide data sets of integration site preferences were obtained without any selection for or against the transcriptional activities of integrated proviruses. We have only limited data on the integration site distribution of silent lentiviral vectors or latent HIV-1 proviruses, which are very sensitive to the chromosomal environment (25, 26, 28) and the availability of required transcription factors in the target cell. The distributions of HIV-1 proviruses differ in resting and activated CD4+ cells infected in vitro. In activated cells, HIV integrations were found more often in gene-rich regions, close to CpG islands, and in GC-rich regions. Proviruses in regions with relatively low gene density may be more prone to be silenced, and therefore, the latent state is more frequently established in resting cells (2).
Here, we used the acutely transforming Rous sarcoma virus (RSV)-based retrovirus vector bearing the v-src oncogene for tumor induction in chickens. Subsequently, we cloned the junctions of proviral and chromosomal DNAs. Because the progressive growth of tumors requires high and long-term stable expression of the transduced v-src (37), in this way, we could select integration sites permitting high and stable expression of the integrated provirus and characterize the genomic features of these loci. We show that the distribution of integration sites from RSV-induced tumors is strikingly nonrandom, with a strong bias for GC-rich and gene-rich regions. Proviruses were found preferentially within or close to transcription units (TUs), particularly in genes broadly expressed in multiple tissues but only exceptionally in tissue-specifically expressed genes. We propose this approach for characterization of the genomic features of loci able to promote the expression of integrated retroviruses and gene therapy vectors.
MATERIALS AND METHODS
Construction of the AviPack packaging cell line.
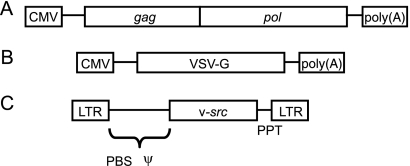
The packaging cell line AviPack was prepared by introduction of the packaging construct pcGagPol into the DF-1 chicken cell line (18). The packaging construct was generated by insertion of the gag-pol coding sequence into the pcDNA3 plasmid (Invitrogen, Carlsbad, CA). The replication-competent vector RCASBP(A) (13) was digested with SacI and PciI, and overhangs of the resulting 5,092-bp fragment were removed by treatment with mung bean nuclease. The 5,092-bp fragment was ligated into the multiple cloning site of pcDNA3 cleaved with EcoRV. The resulting pcGagPol vector, shown schematically in Fig. 1, was linearized by cleavage with PciI, purified by phenol-chlorophorm extraction, and transfected by calcium phosphate precipitation into the DF-1 cell line. The stably transfected cells were selected with 400 μg G418/ml. After 2 weeks of selection, the individual cell clones were isolated and expanded. Twelve fast-proliferating clones were selected and used for virus production (see below). One of these clones, dubbed AviPack, that provided the highest virus titer was expanded and used for subsequent preparation of virus stocks.
FIG. 1.
Schematic representations of the packaging construct pcGagPol in the AviPack line (A), the cotransfection envelope construct (B), and the transforming proviral construct pH19KE (C). CMV, promoter of human cytomegalovirus; VSV-G, gene encoding the envelope of vesicular stomatitis virus; PBS, primer binding site; Ψ, encapsidation signal; PPT, polypurine tract.
Cell culture and virus production.
The AviPack and DF-1 cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM) (Sigma, St. Louis, MO) supplemented with 5% calf serum, 2% fetal calf serum, 1% chicken serum (all sera from Gibco-BRL, Gaithersburg, MD), and antibiotics in a 3% CO2 atmosphere at 37°C. For virus production, the AviPack cells were cultivated on a 100-mm petri dish. The cells were cotransfected by calcium phosphate precipitation with 25 μg of the transforming and replication-defective vector pH19KE (17) and 5 μg of the plasmid pVSV-G (Clontech, Mountain View, CA), carrying the gene encoding the vesicular stomatitis virus envelope glycoprotein (VSV-G). The medium containing the virus was collected 36 h and 48 h after transfection. The virus stock was filtered through a 0.45-μm syringe cellulose-acetate filter, and the virus titer was assessed by a focus-forming assay in chicken embryo fibroblasts.
Tumor induction and monitoring.
Chickens of the close-bred line P free of ev loci, endogenous avian leukosis virus (ALV) sequences (14), were used in the experiments. These chickens are maintained at the Institute of Molecular Genetics and are free from exogenous avian leukosis viruses. Tumors were induced in 14-day-old chicks by subcutaneously inoculating 100 focus-forming units (FFU) in 0.2 ml of the virus stock into both the wing webs and the outer area of the pectoral muscle. To evaluate the progression of tumors objectively, we monitored the area of tumor that was prominent from the site of inoculation by placing transparent foil on the tumor and tracing its contours. The picture of the tumor was then transferred onto a sheet of millimeter paper. Estimates of the tumor area in square millimeters were calculated as half the sum of the outer and inner regular figures just fitting the picture of the tumor (46). Progressively growing tumors were harvested, and DNAs were isolated individually by phenol-chloroform extraction from tissue lysed in SDS buffer with proteinase K.
Cloning and sequencing of integration sites.
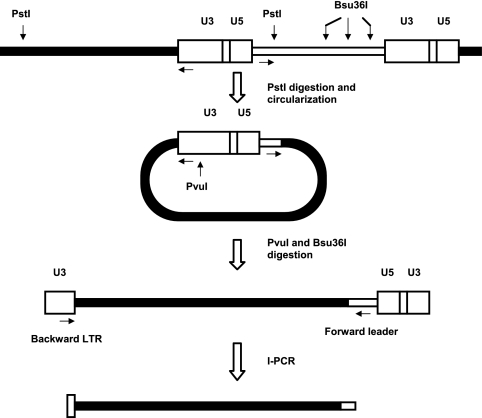
Amplification and cloning of sequences flanking the 5′ long terminal repeat (LTR) of pH19KE proviruses were done as described by Reinišová et al. (38) with slight modifications (Fig. 2). The inverse-PCR (I-PCR) strategy is schematically shown in Fig. 2. DNAs from the tumors were individually digested overnight with PstI at 37°C and circularized by self-ligation with 400 cohesive units of T4 DNA ligase (New England Biolabs, Ipswitch, MA) in 100 μl reaction mixture overnight at increasing temperatures, which started at 10°C and finished at 18°C. The product of self-ligation was subsequently cleaved overnight at 37°C with 10 U of PvuI and 10 U of Bsu36I (both enzymes from New England Biolabs, Ipswitch, MA) in 50 μl reaction mixture to eliminate the background from internal proviral PstI fragments and to increase the efficiency of I-PCR by linearization of the circles within the LTR, respectively. After being desalted, 150 ng of the resulting DNA was subjected to PCR amplification with Taq polymerase (TaKaRa Bio, Otsu, Japan) in a standard reaction according to the manufacturer's instructions, with the addition of betaine and dimethyl sulfoxide. The primers used were as follows: forward leader, 5′CCTCATCCGTCTCGCTTATTCG3′ (nucleotides 63 to 84 3′ to the end of the 5′LTR), and backward LTR, 5′CCTTACTACCACCAATCGGCA3′ (nucleotides 95 to 115 of the LTR). The conditions for I-PCR amplification were 95°C for 3 min, followed by 34 cycles, each consisting of 94°C for 20 s, 58°C for 50 s, and 72°C for 120 s, and finally 3 min at 72°C. The I-PCR products corresponding to sizes from 0.25 to 2.0 kbp were either treated with ExoSapIt (Affymetrix, Santa Clara, CA) or, in the case of multiple bands produced by I-PCR, extracted from the agarose gel using the Qiaex gel extraction kit (Qiagen, Hilden, Germany) and ligated into the pGEM-T Easy vector (Promega, Madison, WI). The ligation products were introduced into Escherichia coli XL1-Blue MRF′ bacteria, and the resulting colonies were screened for the presence of inserts longer than 250 bp by blue-white selection using X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and by PCR using forward and backward LTR primers. The selected clones were subverted to DNA sequencing using the Big Dye Terminator v. 3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA).
FIG. 2.
Scheme of the I-PCR procedure to clone the sequences flanking the 5′ LTR of avian sarcoma virus (ASV) provirus integrated into the chicken genomic DNA. PstI digest of DNA from infected cells was circularized by self-ligation, linearized with PvuI and Bsu36I, and subverted to I-PCR. The recognition sites of enzymes are depicted as solid vertical arrows and I-PCR primers as horizontal arrows. Proviral DNA is represented by open boxes, and flanking chicken DNA is shown as a filled box.
Identification of integration sites.
All the obtained sequences were mapped onto the chicken genome assembly using BLAT (27). We discarded sequences mapping to multiple locations in the genome or to unassembled contigs and the hits below the threshold of 99% identity over 90% or more of the length of the integration site. Targeted genes/TUs and TUs located within 100 kb of the integration sites were identified according to the annotation of the chicken genome in the Ensembl database (23). The Ensembl GeneID was used as the primary gene identifier. Several Perl scripts utilizing the Ensembl Perl API (http://www.ensembl.org/info/docs/api/) were written to obtain gene annotation and external references of Ensembl genes to the UniGene database.
EST data processing.
Seventy-two publicly available chicken expressed sequence tag (EST) libraries, including both normalized and nonnormalized, with at least 500 ESTs were used in the expression analysis. The EST data were obtained from the NCBI UniGene Chicken EST Library Browser (41) (http://www.ncbi.nlm.nih.gov/UniGene/lbrowse2.cgi?TAXID=9031&CUTOFF=500), sorted into 13 groups representing individual chicken organs or tissues, and ESTs from the same source were pooled (see Table S1 in the supplemental material). We enumerated the ESTs of genes targeted by provirus integration in each of the 13 tissues.
Database versions.
The following chicken genome databases were used for mapping of provirus integrations and analysis of insertion sites: the 2.1 (May 2006) release of the chicken genome assembly (24), Ensembl release 56 of September 2009 (23), and chicken UniGene build number 41 of October 2008 (41).
RESULTS
Production of transforming virus and tumor induction.
For tumor induction, we used the outbred chicken line P, free of ASLV-related endogenous viruses (14), in order to avoid the background in I-PCR cloning of integration sites. Because of mutations in tv receptor loci and inefficient entry of A, B, and C RSV subgroups in this breed (our unpublished observation), we constructed a cell line able to package replication-defective and transformation-competent ASLV-based vectors with pantropic VSV-G envelopes (50). The plasmid containing the gag and pol genes under the cytomegalovirus (CMV) promoter and the Neor gene as a selection marker was transfected into DF-1 cells. The AviPack clone with the highest capacity for packaging the LTR, v-src, and LTR proviral genomes with pantropic VSV-G envelope was used for virus production. Virus preparations were titrated and adjusted for monoclonal tumor induction in chicks. Tumors induced in 14-day-old chickens appeared at the site of inoculation with nearly 100% efficiency 2 weeks postinoculation. All induced tumors grew progressively with only minor differences in the growth kinetics, and chicks were sacrificed to take tissue samples within the subsequent 3 to 5 weeks.
The data set of integration sites.
We cloned and sequenced 201 DNA fragments produced by I-PCR from 311 harvested tumors. Only a few tumors provided two integration sites; on the other hand, from a reasonable fraction of chicken tumors, no integration site was cloned. Only 166 clones containing the PstI junction of the chicken genomic sequence with the defined part of the U3 region at the 5′ end and a part of the leader region at the 3′ end were regarded as bona fide 3′ proviral flanking sequence and included in further analysis. Another sign of regular integration sites was the absence of two terminal nucleotides of the LTRs. The lengths of the cloned 3′ flanking sequences varied from 22 to 1,741 bp. All 166 flanking sequences were examined by BLAT, and the genomic hits with the highest identity, mostly higher than 99% along the full length, were handled as integration sites. We further discarded 13 integration sites where the provirus landed in multiple repeats or unassembled genomic regions and integration sites that could not be localized into the current version of the chicken genome assembly. All 153 localized integration sites are listed in Table S2 in the supplemental material.
Genomic distribution of integration sites.
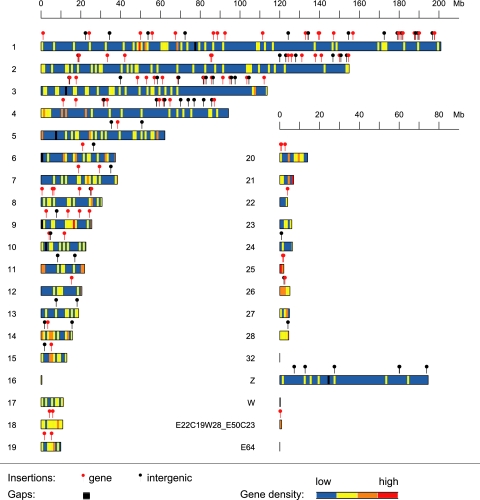
In total, 153 integration sites were unambiguously mapped to the draft chicken genome assembly (Fig. 3). Both macro- and microchromosomes were targets of provirus integration. We compared the observed numbers of provirus integrations in individual chromosomes with the numbers expected according to the chromosomal size (data not shown) and found the highest observed/expected ratio in macrochromosomes 3 and 1. In contrast, no integration sites were found on chromosomes 16, 17, 21, 23, 27, 32, and W. Chromosome W might have been underrepresented in our experiments, as we used chickens of both sexes for tumor induction. However, none of the discrepancies between observed and expected integrations is statistically significant (P > 0.1; χ2 test), and we conclude that integration sites are randomly distributed at the level of whole chromosomes.
FIG. 3.
Chromosomal locations of RSV integrations in virus-induced chicken tumors. In total, 153 out of 166 integration sites were unequivocally localized in the assembled chicken genome. Integrations into genes, defined as integrations within annotated Ensembl genes, are highlighted with red “lollipops.” Integrations outside of genes are shown as black lollipops. Chromosomes are depicted in colors to show gene density over 1-Mb-long windows of the chromosomal sequence (gene density increases from blue through yellow and orange to red); the black regions correspond to long gaps (often putative centromeres).
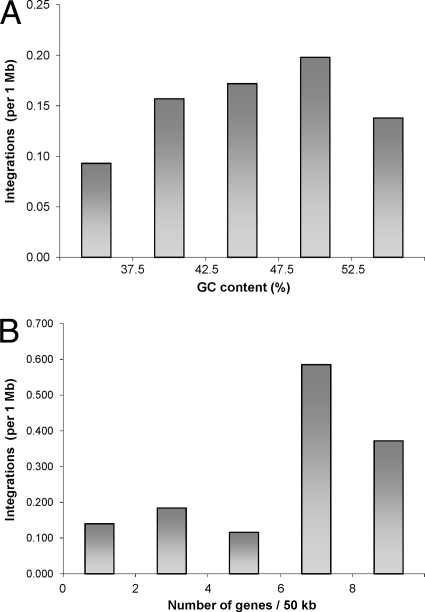
Regarding the GC content, proviruses tended to be localized in genomic regions with higher, but not the highest, percentages of GC (Fig. 4A). The highest density of integrations was observed in the fraction of the genome with a GC content between 47.5 and 52.5%. By analyzing the density of genes around the integration sites, we showed that proviruses localized preferentially in gene-rich regions (Fig. 4B). There was a low density of integration sites in gene deserts and in the regions with up to six genes per 50 kb. The highest density of integration sites was found in the genomic fraction containing seven or eight genes per 50 kb, and it dropped in fractions with even higher gene contents. Importantly, the frequency of integrations was not correlated with the increasing density of PstI recognition sites. The great majority of integration sites were found in genomic fractions with an average density of PstI sites, and only a few integrations went to microchromosomal PstI-rich regions (data not shown). We conclude that there was no detection bias caused by the restriction enzyme used for I-PCR.
FIG. 4.
GC and gene contents of integration sites. (A) GC content. The plot shows the density of integration according to the GC content in 50-kb-long nonoverlapping segments. (B) Gene content. The gene content was calculated as the number of annotated Ensembl genes in 50-kb-long nonoverlapping segments.
Proviruses in chicken tumors primarily localize in genes.
The striking feature of the provirus distribution in chicken tumors is the frequent targeting of TUs. Integrations within annotated chicken genes, either untranslated regions, exons, or introns, according to Ensembl v56, were regarded as targeting TUs. Out of 153 localized integrations, 88 (57.5%) were found in TUs, whereas only 65 (42.5%) targeted the intergenic regions. Provided that the annotated genes, including introns and all untranslated sequences, in Ensembl v56 span 40.4% of the sequenced chicken genome, there was significant preference for integration into TUs (P = 0.0002; binomial test) in comparison with intergenic regions. In addition, 45 integrations (29.4%) were found in regions 5 kb upstream and 5 kb downstream of CpG islands. Ensembl v56 registers 12,596 CpG islands spanning, together with sequences 5 kb upstream and 5 kb downstream, 139 Mb, i.e., 13.5% of the chicken genome. There was significant preference for integration into CpG islands (P = 0.0007; binomial test), obviously dependent on the preference for genes and GC-rich regions.
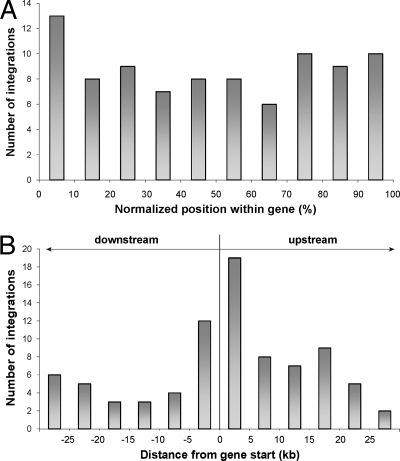
We also examined the specific positions of proviruses within the targeted genes. We calculated the normalized positions along the targeted genes for all 88 genic integrations (Fig. 5A) and showed only a slightly increased number of integrations within the first 10% of the gene length. Because the capacities of cellular promoters/enhancers to affect the transcription of adjacent integrated proviruses are correlated with the absolute distance between them, we calculated the distances of all integration sites from the transcription start sites of the neighboring genes. From this view, integration sites clustered within 10 kb around the transcription start sites, i.e., 5 kb upstream and 5 kb downstream of the transcription start site (Fig. 5B).
FIG. 5.
Distribution of integration sites around cellular genes. To avoid potential artifacts caused by selective removal of redundant records, all targeted genes were counted, even if one insertion was found within or near several transcription units. (A) Distribution of 88 integration sites within Ensembl genes. The gene lengths were normalized to a common length to facilitate the comparison. (B) Distribution of integration sites around transcription starts of Ensembl genes.
Proviruses in chicken tumors are overrepresented in genes expressed in multiple tissues.
In order to compare the transcription of previously analyzed unselected integration sites with our current data set of sites selected for high and long-term expression of integrated provirus, we checked the expression of 88 genes with provirus integrations and all genes found within 100 kb around integration sites, 50 kb upstream and 50 kb downstream of the site of integration. Gene expression was assessed according to the presence of ESTs in 72 chicken EST libraries representing 13 different organs or tissues (see Table S1 in the supplemental material). The genes examined were classified into two categories: (i) broadly expressed genes, whose ESTs were found in libraries from five and more tissues, and (ii) tissue-specific genes with ESTs in libraries exclusively from one tissue. Using this approach, we were able to examine 60 UniGene genes targeted by integration (see Table S3 in the supplemental material) and 236 genes within 100 kb around the integration sites (data not shown).
The summarized data on gene expression at integration sites are given in Table 1. Broadly expressed genes represent 51.7% of the genes targeted by integration and 52.5% of the genes within 100 kb of the integration sites. In contrast, barely 3.3% of the genes targeted by integration and 14% of the genes within 100 kb around integration sites are characterized as tissue specific. In comparison, similar fractions of genes in UniGene, 25.5% and 26.8%, are broadly expressed and tissue specific, respectively. The differences in comparison with all genes from UniGene are highly significant (P < 0.00002; Fischer's exact test). Even more stringent selection of genes expressed in seven or more tissues still indicated 36.7% of targeted genes and 34% of genes within 100 kb of the integration sites. Again, in comparison with 15.2% of all UniGene genes, the difference is highly significant (P < 0.00004; Fischer's exact test). We did not observe any striking pattern of expression in the targeted genes. These genes are mostly expressed in brain, ovary, cartilage, liver, and lymphoid tissues. Several organs, e.g., the pancreas, are underrepresented as to the expression of targeted genes. Based on these data, we conclude that broadly expressed or even housekeeping genes are preferential target sites for transcriptionally active proviruses.
TABLE 1.
Expression analysis of genes targeted by provirus integration and genes adjacent to the integration sites within 100 kb
| Category | No. (%) of genes targeted by integration | No. (%) of genes within 100 kb of the integration site | No. (%) of all genes from UniGene |
|---|---|---|---|
| Genes expressed in five or more tissues | 31 (51.7) | 124 (52.5) | 8,510 (25.5) |
| Tissue-specific genes expressed in one tissue | 2 (3.3) | 33 (14.0) | 8,949 (26.8) |
| Total | 60a | 236b | 33,383c |
Fifty-nine out of 89 Ensembl genes targeted by integration have UniGene reference to 60 UniGene genes.
Two hundred thirty-one out of 410 Ensembl genes targeted by integration have UniGene reference to 236 UniGene genes.
Twenty-eight thousand two hundred and eight genes out of all 33,383 UniGene genes are expressed in at least one tissue.
DISCUSSION
All retroviruses integrate with some preference into transcriptionally active genes, probably reflecting the accessibility of decondensed chromatin and tethering of retrovirus integrases with chromatin-associated proteins (reviewed in reference 6). In the present study, we have demonstrated a substantially increased proportion of retrovirus integration into or close to genes, particularly into genes broadly expressed in multiple tissues, in progressively growing chicken tumors. The selection for tumor formation and progressive growth allows the clonal expansion of cells, in which proviruses are integrated into genomic regions that are likely to favor strong LTR-driven expression of v-src. We infer that proviruses integrated into these genomic regions efficiently avoid transcriptional silencing and remain active for a long time during multiple cell cycles.
The selection of active proviral integrations through tumor induction, as proposed in our study, is based on the observation that the high expression of the transduced v-src oncogene is necessary for tumor growth in chickens and that the level of oncogene expression is correlated with tumor progression (37). In contrast, the epigenetic silencing of the provirus and loss of v-src expression trigger reversion of the transformed phenotype in cancer cell clones in vitro (16). We therefore consider this kind of selection to be very stringent and effective. One drawback of this approach might be, in rare cases, the presence of more than one provirus in one tumor. Under these conditions, it might be that only one provirus drives expression of the v-src oncogene whereas another is silent. We adjusted the efficiency of tumor induction to between 50 and 100% to minimize the probability of multiple integrations in single tumors. Nevertheless, we cannot completely exclude the possibility that our set of integration sites contains a small proportion of such silent bystanders.
The principal finding of this study is the extreme accumulation of transcriptionally active proviruses in genes, particularly in those with great transcriptional breadth. ASLVs were previously shown to integrate with a weak preference into transcription units (1, 34). Barr et al. (1) demonstrated a slightly but statistically insignificantly increased frequency of genic integration in chicken cells in comparison with HeLa cells. We observed 57.5% of integrations targeting TUs, which is highly significant in the small chicken genome with its high gene density. Statistically highly significant is the accumulation in the genes expressed in multiple tissues, potentially housekeeping genes. In a subset of the integration-targeted genes, where the data from EST libraries were available, the broadly expressed genes were overrepresented whereas only a marginal fraction of tissue-specifically expressed genes was found. This can be only partially explained by the weak correlation between ASLV de novo integration and the expression of target genes (1). We can assume that housekeeping genes are more frequent targets in a limited pool of target cells than the few tissue-specific genes expressed in these cells. Furthermore, the constitutively expressed genes are frequently found in GC-rich regions and clustered in gene-rich regions (40), so that whole chromosomal segments can associate with transcription factories (44). We could not precisely analyze the level of transcription in the targeted loci, as many EST libraries used in our study are normalized. Further analyses in human or mouse cells, where more representative microarray data are available, remain to be performed.
Similar results, i.e., increased frequency of integration into genes close to CpG islands and in gene-rich regions, were shown in a small number of transcriptionally active human T-cell leukemia virus type 1 (HTLV-1) provirus Tax+ clones (32). The same bias of provirus integration has been observed in peripheral blood mononuclear cells (PBMCs) from patients persistently infected with HTLV-1 and exhibiting the virus-associated inflammatory condition. The comparison with unselected in vitro HTLV-1 integration (9) suggests that the selection of transcriptionally active proviruses either in vitro or in vivo influences the distribution of integration sites in correlation with the outcome of HTLV-1 infection.
Although ASLVs preferentially integrate into transcribed genes (1), there are at least two observations providing evidence that the most active genes might not be the best targets for retrovirus integration. Maxfield et al. (30) demonstrated that the high level of transcription suppressed gene-specific integration of the Rous-associated virus 1 (RAV-1) retrovirus in the quail genome. In the inducible metallothionein gene, they showed that 100-fold induction of transcription reduced the frequency of integration events by six times. Similar results were obtained in inducible gene cassettes introduced into the quail genome artificially (47). Transcription-correlated inhibition could be explained by steric hindrance via the RNA polymerase II complex or by DNA duplex separation during transcription (11). Provided that the highly and broadly expressed genes cluster into so-called regions of increased density of gene expression (RIDGES) (5), characterized by the highest GC content, our observation of decreased frequency of integration into the most GC- and gene-rich genomic fractions can be explained by this interference. Independently, the RSV sequences in cell lines derived from hamster tumors were found in heavy isochores, but not in the most GC-rich DNA (39).
In several cases, we found proviral integration into genes potentially involved in tumor suppression, apoptosis, and cell transformation. Although the v-src oncogene was the immediate agent inducing sarcomas in our system, we can postulate additional promotion of the progressive tumor growth resulting from disruption/transactivation of these loci.
In our system, we selected for genomic locations permitting efficient expression of integrated loci. Endogenous retroviruses, in comparison, are subverted to the opposite selection forces during the evolution of their host genomes. Purifying selection removed human endogenous retroviruses (HERVs) with deleterious effects within transcription units. As a result, HERVs are enriched in intergenic regions and orientation biased when (rarely) found within transcription units. The antisense orientation of splicing and poly(A) signals does not disrupt the transcription of targeted units (29, 43). Reconstituted consensus HERV-Kcon, however, integrates with normal preferences of exogenous retroviruses (3), demonstrating experimentally that the distribution bias of HERVs does not arise during initial retrovirus integration. Similarly, the distribution of Alu retrotansposons evolves in time by virtue of negative selection (36).
Defining the differences between unselected retroviral integration sites and sites selected for long-terminal-repeat-driven gene expression is relevant for retrovirus-mediated gene transfer and has ramifications for gene therapy. As ASLV-based vectors are now being explored and optimized for gene therapy applications in hematopoietic cells (21, 22), it is highly important to study the clonal selection of integration sites in animal models.
Supplementary Material
Acknowledgments
This work was supported by Czech Science Foundation grant no. 204/07/1030 awarded to J.P. Construction of the AviPack cell line was supported by grant no. 301/09/P667 to F.Š.
We thank Jan Svoboda for helpful discussions and critical reading of the manuscript.
Footnotes
Published ahead of print on 10 February 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Barr, S. D., J. Leipzig, P. Shinn, J. R. Ecker, and F. D. Bushman. 2005. Integration targeting by avian sarcoma-leukosis virus and human immunodeficiency virus in the chicken genome. J. Virol. 79:12035-12044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brady, T., L. M. Agosto, N. Malani, C. C. Berry, U. O'Doherty, and F. Bushman. 2009. HIV integration site distributions in resting and activated CD4+ T cells infected in culture. AIDS 23:1461-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brady, T., Y. N. Lee, K. Ronen, N. Malani, C. C. Berry, P. D. Bieniasz, and F. D. Bushman. 2009. Integration target site selection by a resurrected human endogenous retrovirus. Genes Dev. 23:633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Callahan, R., and G. H. Smith. 2008. Common integration sites for MMTV in viral induced mouse mammary tumor. J. Mammary Gland Biol. Neoplasia 13:309-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caron, H., B. van Schaik, M. van der Mee, F. Baas, G. Riggins, P. van Sluis, M.-C. Hermus, R. van Asperen, K. Boon, P. A. Voûte, S. Heisterkamp, A. van Kampen, and R. Versteeg. 2001. The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science 291:1289-1292. [DOI] [PubMed] [Google Scholar]
- 6.Ciuffi, A., and F. Bushman. 2006. Retroviral DNA integration: HIV and the role of LEDGF/p75. Trends Genet. 22:388-395. [DOI] [PubMed] [Google Scholar]
- 7.Ciuffi, A., M. Llano, E. Poeschla, C. Hoffmann, J. Leipzig, P. Shinn, J. R. Ecker, and F. Bushman. 2005. A role of LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 11:1287-1289. [DOI] [PubMed] [Google Scholar]
- 8.de Ridder, J., A. Uren, J. Kool, M. Reinders, and L. Wessels. 2006. Detecting statistically significant common insertion sites in retroviral insertional mutagenesis screen. PLoS Comput. Biol. 2:e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Derse, D., B. Crise, Y. Li, G. Princler, N. Lum, C. Stewart, C. F. McGrath, S. H. Hughes, D. J. Munroe, and X. Wu. 2007. Human T-cell leukemia virus type 1 integration target sites in the human genome: comparison with those of other retroviruses. J. Virol. 81:6731-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elleder, D., A. Pavlíček, J. Pačes, and J. Hejnar. 2002. Preferential integration of human immunodeficiency virus type 1 into genes, cytogenetic R bands and GC-rich DNA regions: insight from the human genome sequence. FEBS Lett. 517:285-286. [DOI] [PubMed] [Google Scholar]
- 11.Engelman, A. 2005. The ups and downs of gene expression and retroviral DNA integration. Proc. Natl. Acad. Sci. U. S. A. 102:1275-1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faschinger, A., F. Rouault, J. Sollner, A. Lukas, B. Salmons, W. H. Günzburg, and S. Indik. 2008. Mouse mammary tumor virus integration site selection in human and mouse genomes. J. Virol. 82:1360-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Federspiel, M. J., and S. H. Hughes. 1994. Effects of the gag region on genome stability: avian retroviral vectors that contain sequences from the Bryan strain of Rous sarcoma virus. Virology 203:211-220. [DOI] [PubMed] [Google Scholar]
- 14.Gudkov, A. V., I. B. Obukh, S. M. Serov, and B. S. Naroditsky. 1981. Variety of endogenous proviruses in the genomes of chickens of different breeds. J. Gen. Virol. 57:85-94. [DOI] [PubMed] [Google Scholar]
- 15.Hacein-Bey-Abina, S., A. Garrigue, G. P. Wang, J. Soulier, A. Lim, E. Morillon, E. Clappier, L. Caccavelli, E. Delabesse, K. Beldjord, V. Asnafi, E. MacIntyre, L. Dal Cortivo, I. Radford, N. Brousse, F. Sigaux, D. Moshous, J. Hauer, A. Borkhardt, B. H. Belohradsky, U. Wintergerst, M. C. Velez, L. Leiva, R. Sorensen, N. Wulffraat, S. Blanche, F. D. Bushman, A. Fischer, and M. Cavazzana-Calvo. 2008. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Investig. 118:3132-3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hejnar, J., J. Svoboda, J. Geryk, V. J. Fincham, and R. Hák. 1994. High rate of morphological reversion in tumor cell line H-19 associated with permanent transcriptional suppression of the LTR, v-src, LTR provirus. Cell Growth Differ. 5:277-285. [PubMed] [Google Scholar]
- 17.Hejnar, J., P. Hájková, J. Plachý, D. Elleder, V. Stepanets, and J. Svoboda. 2001. CpG island protects Rous sarcoma virus-derived vectors integrated into nonpermissive cells from DNA methylation and transcriptional suppression. Proc. Natl. Acad. Sci. U. S. A. 98:565-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Himly, M., D. N. Foster, I. Bottoli, J. S. Iacovoni, and P. K. Vogt. 1998. The DF-1 chicken fibroblast cell line: transformation induced by diverse oncogenes and cell death resulting from infection by avian leukosis viruses. Virology 248:295-304. [DOI] [PubMed] [Google Scholar]
- 19.Holman, A. G., and J. M. Coffin. 2005. Symmetrical base preferences surrounding HIV-1, avian sarcoma/leukosis virus, and murine leukemia virus integration sites. Proc. Natl. Acad. Sci. U. S. A. 102:6103-6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holmes-Son, M. L., R. S. Appa, and S. A. Chow. 2001. Molecular genetics and target site specificity of retroviral integration. Adv. Genet. 43:33-69. [DOI] [PubMed] [Google Scholar]
- 21.Hu, J., G. Renaud, T. J. Gomes, A. Ferris, P. C. Hendrie, R. E. Donahue, S. H. Hughes, T. G. Wolfsberg, D. W. Russell, and C. E. Dunbar. 2008. Reduced genotoxicity of avian sarcoma leukosis virus vectors in rhesus long-term repopulating cells compared to standard murine retrovirus vectors. Mol. Ther. 16:1617-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu, J., A. Ferris, A. Larochelle, A. E. Krouse, M. E. Metzger, R. E. Donahue, S. H. Hughes, and C. E. Dunbar. 2007. Transduction of rhesus macaque hematopoietic stem and progenitor cells with avian sarcoma and leukosis virus vectors. Hum. Gene Ther. 18:691-700. [DOI] [PubMed] [Google Scholar]
- 23.Hubbard, T. J., B. L. Aken, S. Ayling, B. Ballester, K. Beal, E. Bragin, S. Brent, Y. Chen, P. Clapham, L. Clarke, G. Coates, S. Fairley, S. Fitzgerald, J. Fernandez-Banet, L. Gordon, S. Graf, S. Haider, M. Hammond, R. Holland, K. Howe, A. Jenkinson, N. Johnson, A. Kahari, D. Keefe, S. Keenan, R. Kinsella, F. Kokocinski, E. Kulesha, D. Lawson, I. Longden, K. Megy, P. Meidl, B. Overduin, A. Parker, B. Pritchard, D. Rios, M. Schuster, G. Slater, D. Smedley, W. Spooner, G. Spudich, S. Trevanion, A. Vilella, J. Vogel, S. White, S. Wilder, A. Zadissa, E. Birney, F. Cunningham, V. Curwen, R. Durbin, X. M. Fernandez-Suarez, J. Herrero, A. Kasprzyk, G. Proctor, J. Smith, S. Searle, and P. Flicek. 2009. Ensembl 2009. Nucleic Acids Res. 37(Database issue):D690-D697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.International Chicken Genome Sequencing Consortium. 2004. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 432:695-716. [DOI] [PubMed] [Google Scholar]
- 25.Jordan, A., P. Defechereux, and E. Verdin. 2001. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J. 20:1726-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jordan, A., D. Bisgrove, and E. Verdin. 2003. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 22:1868-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kent, W. J. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12:656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewinski, M. K., D. Bisgrove, P. Shinn, H. Chen, C. Hoffmann, S. Hannenhalli, E. Verdin, C. C. Berry, J. R. Ecker, and F. D. Bushman. 2005. Genome-wide analysis of chromosomal features repressing human immunodeficiency virus transcription. J. Virol. 79:6610-6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mager, D. L. 1999. Human endogenous retroviruses and pathogenesis: genomic considerations. Trends Microbiol. 7:431. [DOI] [PubMed] [Google Scholar]
- 30.Maxfield, L. F., C. D. Fraize, and J. M. Coffin. 2005. Relationship between retroviral DNA-integration-site selection and host cell transcription. Proc. Natl. Acad. Sci. U. S. A. 102:1436-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meehan, A. M., D. T. Saenz, J. H. Morrison, J. A. Garcia-Rivera, M. Peretz, M. Llano, and E. M. Poeschla. 2009. LEDGF/p75 proteins with alternative chromatin tethers are functional HIV-1 cofactors. PloS Pathog. 5:e1000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meekings, K. N., J. Leipzig, F. D. Bushman, G. P. Taylor, and C. R. M. Bangham. 2008. HTLV-1 integration into transcriptionally active genomic regions is associated with proviral expression and with HAM/TSP. PloS Pathog. 4:e1000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchell, R. S., B. F. Beitzel, A. R. W. Schroder, P. Shinn, H. Chen, C. C. Berry, J. R. Ecker, and F. D. Bushman. 2004. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PloS Biol. 2:1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narezkina, A., K. D. Taganov, S. Litwin, R. Stoyanova, J. Hayashi, C. Seeger, A. M. Skalka, and R. A. Katz. 2004. Genome-wide analyses of avian sarcoma virus integration sites. J. Virol. 78:11656-11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pajer, P., V. Pečenka, J. Králová, V. Karafiát, D. Průková, Z. Zemanová, R. Kodet, and M. Dvořák. 2006. Identification of potential human oncogenes by mapping the common viral integration sites in avian nephroblastoma. Cancer Res. 66:78-86. [DOI] [PubMed] [Google Scholar]
- 36.Pavlicek, A., K. Jabbari, J. Paces, V. Paces, J. Hejnar, and G. Bernardi. 2001. Similar integration but different stability of Alus and LINEs in the human genome. Gene 276:39-45. [DOI] [PubMed] [Google Scholar]
- 37.Plachý, J., K. Hála, J. Hejnar, J. Geryk, and J. Svoboda. 1994. src-specific immunity in inbred chickens bearing v-src DNA- and RSV-induced tumors. Immunogenetics 40:257-265. [DOI] [PubMed] [Google Scholar]
- 38.Reinišová, M., A. Pavlíček, P. Divina, J. Geryk, J. Plachý, and J. Hejnar. 2008. Target site preferences of subgroup C Rous sarcoma virus integration into the chicken DNA. Open Genomics J. 1:e6-12. [Google Scholar]
- 39.Rynditch, A., F. Kadi, J. Geryk, S. Zoubak, J. Svoboda, and G. Bernardi. 1991. The isopycnic, compartmentalized integration of Rous sarcoma virus sequences. Gene 106:165-172. [DOI] [PubMed] [Google Scholar]
- 40.Saccone, S., C. Federico, and G. Bernardi. 2002. Localization of gene-richest and the gene-poorest isochores in the interphase nuclei of mammals and birds. Gene 300:169-178. [DOI] [PubMed] [Google Scholar]
- 41.Sayers, E. W., T. Barrett, D. A. Benson, S. H. Bryant, K. Canese, V. Chetvernin, D. M. Church, M. DiCuccio, R. Edgar, S. Federhen, M. Feolo, L. Y. Geer, W. Helmberg, Y. Kapustin, D. Landsman, D. J. Lipman, T. L. Madden, D. R. Maglott, V. Miller, I. Mizrachi, J. Ostell, K. D. Pruitt, G. D. Schuler, E. Sequeira, S. T. Sherry, M. Shumway, K. Sirotkin, A. Souvorov, G. Starchenko, T. A. Tatusova, L. Wagner, E. Yaschenko, and J. Ye. 2009. Databse resources of the National Center for Biotechnology Information. Nucleic Acids Res. 37(Database issue):D5-D15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schroeder, A. R., P. Shinn, H. Chen, C. Berry, J. R. Ecker, and F. Bushman. 2002. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110:521-529. [DOI] [PubMed] [Google Scholar]
- 43.Smit, A. F. 1999. Interspersed repeats and other momentos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 9:657-663. [DOI] [PubMed] [Google Scholar]
- 44.Sutherland, H., and W. A. Bickmore. 2009. Transription factories: gene expression in unions? Nat. Rev. 10:457-466. [DOI] [PubMed] [Google Scholar]
- 45.Suzuki, T., H. Schen, K. Akagi, H. C. Morse, J. D. Malley, D. Q. Naiman, N. A. Jenkins, and N. G. Copeland. 2002. New genes involved in cancer identified by retroviral tagging. Nat. Genet. 32:166-174. [DOI] [PubMed] [Google Scholar]
- 46.Svoboda, J., J. Plachý, J. Hejnar, I. Karakoz, R. V. Guntaka, and J. Geryk. 1992. Tumor induction by the LTR, v-src, LTR DNA in four B (MHC) congenic lines of chickens. Immunogenetics 35:309-315. [DOI] [PubMed] [Google Scholar]
- 47.Weidhaas, J. B., E. L. Angelichio, S. Fenner, and J. M. Coffin. 2000. Relationship between retroviral DNA integration and gene expression. J. Virol. 74:8382-8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu, X., Y. Li, B. Crise, and S. M. Burgess. 2003. Transcription start regions in the human genome are favored targets for MLV integration. Science 300:1749-1751. [DOI] [PubMed] [Google Scholar]
- 49.Wu, X., Y. Li, B. Crise, S. M. Burgess, and D. J. Munroe. 2005. Weak palindromic consensus sequences are a common feature found at the integration target sites of many retroviruses. J. Virol. 79:5211-5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang, Y., E. F. Vanin, M. A. Whitt, M. Fornerod, R. Zwart, R. D. Schneiderman, G. Grosveld, and A. W. Nienhuis. 1995. Inducible, high-level production of infectious murine leukemia retroviral vector particles pseudotyped with vesicular stomatitis virus G envelope protein. Hum. Gene Ther. 6:1203-1213. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.