Abstract
A new class of N-(1-thia-4-azaspiro[4.5]decan-4-yl)carboxamide inhibitors of influenza virus hemagglutinin (HA)-mediated membrane fusion that has a narrow and defined structure-activity relationship was identified. In Madin-Darby canine kidney (MDCK) cells infected with different strains of human influenza virus A/H3N2, the lead compound, 4c, displayed a 50% effective concentration of 3 to 23 μM and an antiviral selectivity index of 10. No activity was observed for A/H1N1, A/H5N1, A/H7N2, and B viruses. The activity of 4c was reduced considerably when added 30 min or later postinfection, indicating that 4c inhibits an early step in virus replication. 4c and its congeners inhibited influenza A/H3N2 virus-induced erythrocyte hemolysis at low pH. 4c-resistant virus mutants, selected in MDCK cells, contained either a single D112N change in the HA2 subunit of the viral HA or a combination of three substitutions, i.e., R220S (in HA1) and E57K (in HA2) and an A-T substitution at position 43 or 96 of HA2. The mutants showed efficiency for receptor binding and replication similar to that of wild-type virus yet displayed an increased pH of erythrocyte hemolysis. In polykaryon assays with cells expressing single-mutant HA proteins, the E57K, A96T, and D112N mutations resulted in 4c resistance, and the HA proteins containing R220S, A96T, and D112N mutations displayed an increased fusion pH. Molecular modeling identified a binding cavity for 4c involving arginine-54 and glutamic acid-57 in the HA2 subunit. Our studies with the new fusion inhibitor 4c confirm the importance of this HA region in the development of influenza virus fusion inhibitors.
Currently available drugs for the prevention and treatment of seasonal influenza virus infections are the M2 ion channel blockers (amantadine and rimantadine) and the neuraminidase (NA) inhibitors (oseltamivir and zanamivir) (9). The clinical usefulness of amantadine and rimantadine is limited due to the increasing incidence of adamantane-resistant viruses in the population (3, 11). Moreover, the M2 ion channel blockers inhibit only influenza A virus replication and are associated with neurological side effects. NA inhibitors are favored clinically, since they are effective against all NA subtypes, are well tolerated, and have a higher barrier for resistance (27). However, drug-resistant isolates have been detected in A/H3N2- and A/H5N1-infected patients receiving oseltamivir treatment (10, 16). Even more reason for concern is the recent and worldwide isolation of oseltamivir-resistant A/H1N1 mutants, even among untreated patients (17, 46). Oseltamivir and, to a lesser extent, zanamivir have been stockpiled as part of pandemic preparedness plans and form the cornerstone of the response to the recent outbreak of the swine flu A/H1N1 virus (39, 40). However, it is unclear whether these antivirals will be sufficient to deal with larger influenza epidemics, so there is an urgent need to develop antivirals that act on a novel influenza virus target.
An attractive antiviral strategy is to block influenza virus entry into the host cell, a process in which the viral hemagglutinin (HA) plays a key role (42). HA is a trimeric envelope glycoprotein that contains two disulfide-linked polypeptide chains, HA1 and HA2. After attachment of the receptor binding domain in the HA1 subunit to sialic acid-containing cell surface glycans, the virion is internalized by endocytosis. The acidic pH of the endosome leads to an extensive and irreversible conformational change of the HA protein, resulting in exposure of the fusion peptide, which inserts into the endosomal target membrane of the host cell (18). After fusion of the viral and endosomal membranes, the viral ribonucleoproteins are released into the cytosol and transported into the nucleus, where replication occurs (6). Crystallographic studies have provided detailed insight into the processes of HA refolding and extrusion of the fusion peptide (4). The latter is a sequence of hydrophobic amino acids located at the N terminus of the HA2 subunit, which is, in the prefusogenic conformation, sequestered in a pocket of ionizable residues at the monomer interface of the HA trimer (48). In order to exploit the HA protein as an antiviral target, several small-molecule inhibitors that block the acid-induced conformational change of HA have been identified (2, 19, 21, 30, 49). For many of these, development has been hindered by their subtype-dependent activities. On the other hand, these diverse fusion inhibitors represent excellent tools to identify the HA amino acid residues involved in the fusion process and/or delineate the structural differences among HA subtypes (36).
We report here the identification of a new class of influenza A virus fusion inhibitors that have specific activity against the H3 subtype, but not the H1, H5, and H7 subtypes. A detailed analysis of their structure-activity relationships and mechanisms of action is described, with a focus on the interaction of these compounds with the HA protein.
MATERIALS AND METHODS
Chemical synthesis of the compounds.
Commercially available chemicals were obtained from Merck, Aldrich, or Fluka. Melting points (M.p.) were determined on a Buchi 530 capillary melting-point apparatus in open capillaries and were uncorrected. Compound purity was measured by thin-layer chromatography (TLC) on Silica gel 60 F254. Infrared (IR) spectra were recorded on a Perkin Elmer 1600 FT infrared spectrophotometer in potassium bromide pellets (υ in cm−1). 1H nuclear magnetic resonance (NMR) spectra were recorded on a VarianUnity 500-MHz NMR spectrophotometer using tetramethylsilane as an internal standard. Electron impact mass spectra (EIMS) were determined on a VG Zab Spec (70-eV) spectrometer. Elementary analyses were performed on a Carlo Erba 1106 and Elementar Micro Vario CHNS elemental analyzer at the Scientific and Technical Research Council of Turkey. 2a, 2b, and 3b were reported previously (45).
General synthesis of 2 (series II).
A mixture of 0.005 mol 1 (45), 0.01 mol of an appropriate ketone, a drop of concentrated H2SO4, and 10 ml of C2H5OH (96%) was refluxed for 6 h. The crude product that precipitated on cooling was filtered and recrystallized from a C2H5OH-H2O mixture.
6-Methyl-N′-(4-phenylcyclohexylidene)imidazo[2,1-b][1,3]thiazole-5-carbohydrazide (2c). M.p., 210 to 2°C; yield, 62.5%. IR, 3,280, 3,140 (NH), 1,625 (C=O). 1H-NMR (CDCl3) δ (ppm), 1.53-2.70 (9H, m, cyclohexane [cyclohex.]), 2.47 (3H, s, imidazothiazole [imidazothia.]-C6-CH3), 6.70 (1H, d, imidazothia.-C2-H), 7.00-7.14 (5H, m, phenyl), 8.05 (1H, d, imidazothia.-C3-H), 8.32 (1H, s, CONH). EIMS (70 eV) m/z (%), 352 (M+, 44), 165 (100). Analysis: calculated for C19H20N4OS (352.453): C, 64.75; H, 5.72; N, 15.90. Found: C, 64.97; H, 5.61; N, 15.66.
General synthesis of 3, 4, and 6 (series I and III).
To a solution of 1 or 5 (0.005 mol) in dry benzene (30 ml) was added an appropriate ketone (0.01 mol), and the mixture was refluxed for 1.5 h using a Dean-Stark trap. After the mixture was cooled at room temperature, sulfanylacetic acid or 2-sulfanylpropanoic acid (0.15 mol) was added dropwise to the solution, and the resulting solution was refluxed for 6 h. Excess benzene was evaporated in vacuo. The residue was triturated with saturated NaHCO3 until CO2 evaluation ceased and was allowed to stand overnight. The solid thus obtained was filtered, washed with H2O, and crystallized from the C2H5OH-H2O mixture.
6-Methyl-N-(8-methyl-3-oxo-1-thia-4-azaspiro[4.5]dec-4-yl)imidazo[2,1-b][1,3] thiazole-5-carboxamide (3a). M.p., 202 to 4°C; yield, 80.2%. IR, 3,153, 3,128 (NH), 1,698, 1,661 (C=O). 1H-NMR ([D6]DMSO [dimethyl sulfoxide]) δ (ppm), 0.9 (3H, d, C8-CH3), 1.15-2.00 (9H, m, C6-10-H), 2.59 (3H, s, imidazothia.-C6-CH3), 3.61 (2H, s, C2-H), 6.93 (1H, d, imidazothia.C2-H), 8.14 (1H, d, imidazothia.C3-H), 8.22 (1H, s, CONH). EIMS (70 eV) m/z (%), 364 (M+, 52), 165 (100). Analysis, calculated for C16H20N4O2S2 (364.48): C, 52.72; H, 5.53; N, 15.37. Found: C, 53.01; H, 5.59; N, 15.82.
N-(3-oxo-8-phenyl-1-thia-4-azaspiro[4.5]dec-4-yl)-6-methylimidazo[2,1-b][1,3]thiazole-5-carboxamide (3c). M.p., 238 to 9°C; yield, 86.9%. IR, 3,323, 3,110 (NH), 1,690, 1,658 (C=O). 1H-NMR (CDCl3) δ (ppm), 1.16-1.99 (8H, m, -C6,7,9,10-H), 2.26 (1H, m, -C8-H), 3.47 (2H, s, -C2-H), 2.50 (3H, s, imidazothia.-C6-CH3), 6.99-7.13 (5H, m, phenyl), 6.78 (1H, d, imidazothia.-C2-H), 7.87 (1H, s, CONH), 7.99 (1H, d, imidazothia.-C3-H). EIMS (70 eV) m/z (%), 426 (M+, 26), 165 (100). Analysis, calculated for C21H22N4O2S2 (426.55): C, 59.13; H, 5.20; N, 13.13. Found: C, 59.92; H, 4.55; N, 12.48.
6-Methyl-N-(2-methyl-3-oxo-1-thia-4-azaspiro[4.5]dec-4-yl)imidazo[2,1-b][1,3]thiazole-5-carboxamide (4a). M.p., 220 to 1°C; yield, 21.9%. IR, 3,251, 3,131 (NH), 1,700, 1,658 (C=O). 1H-NMR (CDCl3) δ (ppm), 0.80-1.97 (10H, m, C6-10-H), 1.57 (3H, d, C2-CH3), 2.56 (3H, s, imidazothia.-C6-CH3), 3.90 (1H, q, -C2-H); 6.92 (1H, d, imidazothia.-C2-H), 8.13 (1H, d, imidazothia.-C3-H), 8.61(1H, s, CONH). EIMS (70 eV) m/z (%), 364 (M+, 17), 165 (100). Analysis, calculated for C16H20N4O2S2 (364.48): C, 52.72; H, 5.53; N, 15.37. Found: C, 53.00; H, 5.42; N, 15.19.
6-Methyl-N-(2,7-dimethyl-3-oxo-1-thia-4-azaspiro[4.5]dec-4-yl)imidazo[2,1-b][1,3]thiazole-5-carboxamide (4b), M.p., 245 to 6°C; yield, 31.8%. IR, 3,250, 3,150 (NH), 1,700, 1,658 (C=O). 1H-NMR ([D6]DMSO) δ (ppm), 0.99 (3H, d, C7-CH3), 1.43 (3H, d, C2-CH3), 1.50-1.90 (9H, m, C6-10-H), 2.57 (3H, s, imidazothia.-C6-CH3), 3.94 (1H, q, -C2-H), 7.36 (1H, d, imidazothia.-C2-H), 8.04 (1H, d, imidazothia.-C3-H), 9.84 (1H, s, CONH). EIMS (70 eV) m/z (%), 378 (M+, 30), 181 (100). Analysis, calculated for C17H22N4O2S2 (378.5): C, 53.94; H, 5.86; N, 14.80. Found: C, 53.88; H, 5.68; N, 14.40.
6-Methyl-N-(2,8-dimethyl-3-oxo-1-thia-4-azaspiro[4.5]dec-4-yl)imidazo[2,1-b][1,3]thiazole-5-carboxamide (4c). M.p., 165 to 7°C; yield, 37.0%. IR, 3,261, 3,131 (NH), 1,698, 1,656 (C=O). 1H-NMR ([D6]DMSO) δ (ppm): 0.81 (3H, d, C8-CH3), 0.98-2.11 (9H, m, C6-10-H), 1.47 (3H, d, -C2-CH3), 2.57 (3H, s, imidazothia.-C6-CH3), 3.91 (1H, q, -C2-H), 7.35 (1H, d, imidazothia.-C2-H), 8.00 (1H, d, imidazothia.-C3-H), 9.89 (1H, s, CONH). EIMS (70 eV) m/z (%): 378 (M+, 42), 181 (100). Analysis, calculated for C17H22N4O2S2 (378.5): C, 53.94; H, 5.86; N, 14.80. Found: C, 54.17; H, 5.55; N, 15.01.
6-Methyl-N-(8-ethyl-2-methyl-3-oxo-1-thia-4-azaspiro[4.5]dec-4-yl)imidazo[2,1-b][1,3] thiazole-5-carboxamide (4d). M.p., 212 to 3°C; yield, 22.4%. IR, 3,246, 3,136 (NH), 1,701, 1,659 (C=O). 1H-NMR ([D6]DMSO) δ (ppm), 0.79 (3H, t, C8-CH2-CH3), 1.00 to 1.26 (5H, m, C8-CH2CH3 and C6-10-H), 1.46 (3H, d, -C2-CH3), 1.59-2.03 (6H, m, C6-10-H), 2.56 (3H, s, imidazothia.-C6-CH3), 3.94 (1H, q, C2-H), 7.38 (1H, d, imidazothia.-C2-H), 7.99 (1H, d, imidazothia.-C3-H), 9.87 (1H, s, CONH). EIMS (70 eV) m/z (%), 392 (M+, 40), 181 (100). Analysis, calculated for C18H24N4O2S2 (392.53): C, 55.08; H, 6.16; N, 14.27. Found: C, 55.19; H, 5.94; N, 14.16.
6-Methyl-N-(2-methyl-3-oxo-8-phenyl-1-thia-4-azaspiro[4.5]dec-4-yl)imidazo[2,1-b][1,3] thiazole-5-carboxamide (4e). M.p., 113 to 5°C; yield, 30.9%. IR, 3,283, 3,141 (NH); 1,702, 1,672 (C=O). 1H-NMR ([D6]DMSO) δ (ppm), 1.61-2.21 (9H, m, C6-10-H), 1.49 (3H, d, -C2-CH3), 2.62 (3H, s, imidazothia.-C6-CH3), 3.89 (1H, q, C2-H), 7.18-7.29 (5H, m, phenyl), 7.40(1H, d, imidazothia.-C2-H), 8.06 (1H, d, imidazothia.-C3-H), 9.96 (1H, s, CONH). EIMS (70 eV) m/z (%), 441 (M+,37), 252 (100). Analysis, calculated for C22H24N4O2S2 0.5 H2O (449.59): C, 58.77; H, 5.60; N, 12.46. Found: C, 58.85; H, 5.76; N, 12.46.
2-Hydroxy-N-(8-methyl-3-oxo-1-thia-4-azaspiro[4.5]dec-4-yl)benzamide (6a). M.p., 247 to 9°C; yield, 88.5%. IR, 3,240 (OH/NH), 1,687, 1,659 (C=O). 1H-NMR ([D6]DMSO) δ (ppm): 0.85 (3H, d, C8-CH3), 1.11-1.92 (9H, m, -C6-10-H), 3.62 (2H, s, -C2-H), 6.93-6.97 (2H, m, phenyl C3-H and C5-H), 7.45 (1H, t, phenyl C4-H), 7.86 (1H, d, phenyl C6-H), 10.5 (1H, s, CONH), 11.5 (1H, s, OH). Analysis, calculated for C16H20N2O3S. 0.5H2O (329.414): C, 58.33; H, 6.43; N, 8.50. Found: C, 58.73; H, 6.74; N, 8.40.
2-Hydroxy-N-(2,8-dimethyl-3-oxo-1-thia-4-azaspiro[4.5]dec-4-yl)benzamide (6b). M.p., 159 to 61°C; yield, 97.8%. IR, 3,230 (OH/NH), 1,692, 1,655 (C=O). 1H-NMR ([D6]DMSO) δ (ppm): 0.84 (3H, d, C8-CH3), 1.08-1.99 (9H, m, -C6-10-H), 1.43 (3H, d, -C2-CH3), 3.93 (1H, q, -C2-H); 6.93-6.98 (2H, m, phenyl C3-H and C5-H), 7.45 (1H, t, phenyl C4-H), 7.86 (1H, d, phenyl C6-H), 10.56 (1H, s, CONH), 11.5 (1H, s, OH). Analysis, calculated for C17H22N2O3S (334.433): C, 61.05; H, 6.63; N, 8.38. Found: C, 61.49; H, 6.78; N, 8.90.
Cells and virus strains.
Madin-Darby canine kidney (MDCK) cells (a kind gift from M. Matrosovich, Marburg, Germany) (22) were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS) (Integro), 1 mM sodium pyruvate, and 0.075% sodium bicarbonate. During virus experiments, the MDCK cells were maintained in Ultra MDCK medium (Lonza, Basel, Switzerland), supplemented with 0.0225% sodium bicarbonate, 2 mM l-glutamine, and 2 μg/ml tosylphenylalanylchloromethylketon (TPCK)-treated trypsin (Sigma, St. Louis, MO).
The human influenza virus strains A/PR/8/34 (A/H1N1) and B/HK/5/72 were purchased from the American Type Culture Collection (ATCC). The strains A/X-31 (A/H3N2) (A/Aichi/2/68 [H3N2] × A/PR/8/34 [H1N1]) and A/HK/7/87 (A/H3N2) were obtained from J. Neyts (Katholieke Universiteit Leuven, Leuven, Belgium), whereas the following strains were kindly provided by R. Fouchier (Rotterdam, Netherlands): A/FM/1/47 (A/H1N1), A/HK/2/68, and A/Victoria/3/75 (A/H3N2) and B/Lee/40. The A/H5N1 viruses A/Vietnam/1203/04 and A/chicken/Vietnam/C58/04 were generated by reverse genetics with plasmids kindly provided by R. Salomon and R. G. Webster (13, 32, 37). The A/H3N2 virus A/Aichi/2/68, used as a positive control in A/H5N1 plaque reduction assays, was obtained from the ATCC. A/chicken/Pennsylvania/143586/02 (A/H7N2) was kindly provided by R. G. Webster (14).
Virus stocks of these laboratory-adapted strains were prepared by passaging them in 10-day-old embryonated chicken eggs. In addition, two clinical isolates were included (A/Ned/378/05 [A/H1N1] and B/Ned/537/05; kind gifts from R. Fouchier), which underwent one passage in embryonated hen eggs.
Antiviral assays.
The following control compounds were included: ribavirin (Virazole), obtained from ICN Pharmaceuticals; rimantadine (Sigma); and oseltamivir carboxylate (GS-4071) (a kind gift from T. Cihlar, Gilead Sciences, Foster City, CA). The test compounds were evaluated for anti-influenza virus activity by a multicycle cytopathic effect (CPE) reduction assay (28). MDCK cells were seeded into 96-well plates at 7,500 cells per well 16 h prior to infection and incubated at 35°C. Serial dilutions of the compounds were added to the cells, together with the influenza virus (multiplicity of infection, 50 50% cell culture infective doses [CCID50] per well, which corresponds to 0.0004 PFU per cell). At 3 days postinfection (p.i.), microscopy was performed to determine the antiviral activity, expressed as the compound concentration producing 50% inhibition of virus-induced CPE (50% effective concentration [EC50]), as well as the cytotoxicity of the compounds, expressed as the compound concentration causing minimal changes in cell morphology (MCC). The data were confirmed by the formazan-based 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) cell viability assay (CellTiter 96 AQueous One Solution Cell Proliferation Assay from Promega, Madison, WI), and the spectrophotometric data were used to calculate the EC50 and 50% cytotoxic concentration (CC50).
In addition, the compound cytostatic activity was determined in uninfected MDCK cells, which were incubated with serial dilutions of the compounds for 72 h and then subjected to cell counting with a Z1 Coulter Counter apparatus (Beckman Coulter, Fullerton, CA). The data were expressed as the concentration causing 50% inhibition of cell proliferation (50% inhibitory concentration [IC50]).
To determine the amount of virus released into the supernatant, cells were incubated with virus and compounds as described above, and after 72 h, the culture supernatant was collected from selected wells. After 2 μl of the culture medium was mixed with 1 μl lysis enhancer and 10 μl resuspension buffer (both from the CellsDirect One-Step qRT-PCR kit; Invitrogen, Carlsbad, CA), the samples were incubated at 75°C for 10 min and diluted 5-fold. Then, quantification of viral-genome copies was performed with a one-step real-time reverse transcription (RT)-PCR assay (CellsDirect One-Step qRT-PCR kit; Invitrogen). The M1-FOR primer (5′-CCTGGTATGTGCAACCTGTG-3′) and M1-REV primer (5′-AGCCTGACTAGCAACCTCCA-3′) were derived from the influenza A virus M1 gene sequence and yielded an amplified fragment 193 bp long. The M1-probe (6-FAM [6-carboxyfluorescein]-5′-CTGACTCCCAGCATCGGTCTCATAGGC-3′-Black Hole Quencher [BHQ]; Eurogentec, Liège, Belgium) was chosen from within this sequence. Amplification was performed on an ABI Prism 7000 apparatus (Applied Biosystems, Foster City, CA) and consisted of 15 min of cDNA synthesis at 50°C and a 2-min initial activation at 95°C, followed by 40 thermal cycles of 15 s at 95°C and 90 s at 60°C. In each experiment, a standard curve (R2 > 0.99 within the range of 103 to 108 copies per reaction) was drawn to convert the respective cycle threshold (CT) values into the number of viral-genome copies. This standard consisted of a pCR2.1-TOPO plasmid construct in which was cloned a 473-bp sequence of influenza virus A/PR/8/34 segment 7. All samples were run in duplicate.
Plaque assay with influenza A/H5N1 and A/H7N2 viruses.
Confluent monolayers of MDCK cells were grown in 6-well dishes overnight at 37°C. Compound dilutions were added to the cells in the presence of A/H5N1 (A/Vietnam/1203/04 or A/Chicken/Vietnam/C58/04), A/H7N2 (A/Chicken/Pennsylvania/143586/02), or A/H3N2 (A/Aichi/2/68) viruses diluted to 200 PFU/ml. The virus was allowed to adsorb for 1 h at 37°C. The inoculum was removed, and the cells were washed with phosphate-buffered saline (PBS) prior to being overlaid with minimal essential medium (MEM) supplemented with 0.3% bovine serum albumin, 0.9% Bacto agar, and 1 μg/ml TPCK-treated trypsin. The infected cells were incubated for a further 72 h at 37°C prior to removal of the overlay medium and staining of the monolayer with crystal violet to visualize plaques. The EC50 was calculated as the compound concentration yielding 50% reduction of the number of plaques observed in untreated wells. Experiments with H5N1 and A/H7N2 viruses were conducted in a U.S. Department of Agriculture-approved biosafety level 3+ containment facility.
Time-of-addition experiment.
MDCK cells were seeded into 24-well dishes at 125,000 cells per well. After 16 h of incubation at 35°C, influenza virus A/X-31 was added at a multiplicity of infection of 0.0004 PFU per cell, and the compounds were added at −30 min, 0 h, 30 min, 1 h, 3 h, 5 h, or 8 h p.i. The reference compounds were rimantadine, ribavirin, chloroquine (from Sigma), and bisindolylmaleimide I (from Calbiochem). At 10 h p.i., the medium was discarded and total cellular RNA extracts were prepared with the RNeasy Mini Kit (Qiagen, Hilden, Germany). In order to quantify the viral RNA (vRNA), the samples were analyzed by two-step real-time RT-PCR, using the M1-specific primers, probe, and standard as described above. cDNA synthesis was performed on 0.5 μg of total RNA using Moloney murine leukemia virus (M-MLV) reverse transcriptase (Invitrogen) and 500 nM M1-FOR primer. Then, real-time PCR was performed, using the M1-FOR and M1-REV primers, the M1 probe (see above), and qPCR MasterMix (Eurogentec). The thermal profile consisted of a 10-min initial activation at 95°C, followed by 40 thermal cycles of 15 s at 95°C, 30 s at 60°C, and 40 s at 72°C. In each experiment, a standard curve (R2 > 0.99 within the range of 103 to 108 copies per reaction) was drawn to convert the respective CT values into the number of viral-genome copies. All samples were analyzed in duplicate.
Hemolysis inhibition assay.
The procedure to determine the inhibitory effects of the compounds on virus-induced hemolysis at low pH was slightly adapted from Luo et al. (20). After preincubation of 100 μl allantoic influenza virus stock with 100 μl compound (serially diluted in PBS) at 37°C for 1 h, an equal volume of chicken red blood cells (RBC) (2% in PBS) was added. The samples were incubated at 37°C for 10 min, and then the RBC (with adsorbed virus) were centrifuged (300 × g; 4°C; 8 min). The RBC pellets were resuspended in 450 μl PBS buffer containing the corresponding concentrations of the compounds, and the pH was lowered to 5.0 by addition of a predetermined amount of 1 N acetic acid. After incubation at 37°C for 25 min, the reaction was neutralized with NaOH, and intact chicken RBC were removed by centrifugation (400 × g; 8 min; 4°C). Then, 300 μl of the supernatant was transferred to a 96-well plate, and the optical density was measured at 540 nm with a Spectramax Plus apparatus (Molecular Devices, Union City, CA). Background values were derived from mock-infected samples that underwent identical treatment. The data were expressed as the EC50 relative to the value observed in the absence of compound.
Trypsin susceptibility assay.
To determine the inhibitory effect of 4c on the acid-induced conformational change of HA, which renders HA susceptible to tryptic digestion, influenza virus A/X-31 (allantoic stock diluted 1:10 in PBS) was incubated in the presence of various compound concentrations at 37°C for 15 min. The pH was lowered to 5.0 by addition of a predetermined amount of 1 M sodium acetate buffer (pH 4.8). After 10 min of incubation at 37°C, the mixture was neutralized to pH 7.2 by the addition of 0.5 M Tris-HCl buffer (pH 9.0). Then, samples were incubated in the presence of 10 mg per ml trypsin (Sigma) at 37°C for 1 h. The reaction was terminated by incubation (20 min at 37°C) with 5 mg per ml soybean trypsin inhibitor (Sigma). Subsequently, the samples were mixed with protein extraction buffer (20 mM Tris-HCl, pH 7.4, 0.137 M NaCl, 2 mM Na2EDTA, 1% Triton X-100, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride, 0.01 mg leupeptin per ml). The lysates were clarified (10 min at 23,000 × g) and analyzed by Bradford assay. After being mixed with loading dye containing 50 mM dithiothreitol and boiled for 3 min, aliquots containing 20 μg of total protein were loaded on 12% Tris-glycine SDS-polyacrylamide gels. Then, electrophoresis and electroblotting onto Hybond-P membranes (GE Healthcare) were performed, and the membranes were blocked overnight in 5% fat-free powdered milk in PBS. The blots were subjected to two washes with PBS containing 0.1% Tween 20 (PBS-T), 1 h of incubation with 5 μg per ml of primary antibody (anti-HA1 mouse monoclonal antibody clone InA246; HyTest), three washes in PBS-T, 45 min of incubation with secondary antibody (horseradish peroxidase-linked goat anti-mouse immunoglobulin polyclonal antibody; Dako), and six final washes in PBS-T. Protein bands were visualized with a ECL Plus Western blotting detection system (GE Healthcare) and exposed to X-ray film.
Selection and characterization of resistant mutants.
The influenza virus strain A/X-31 was passaged on MDCK cells in the presence of 20, 20, and 150 μM 4c for the first, second, and third passages, respectively. The final supernatant was harvested and plaque purified on 0.8% agar, and individual clones were incubated for 72 h on MDCK cells in the presence of 100 μM 4c. The cellular fractions were subjected to RNA extraction (RNeasy Mini Kit; Qiagen), followed by reverse transcription with M-MLV reverse transcriptase (Invitrogen) and a primer derived from the 3′ terminus of hemagglutinin, amplification of the cDNA by high-fidelity PCR (Expand High Fidelity PCR System; Roche, Mannheim, Germany), and cycle sequencing. The supernatant fractions were used as virus stocks in an antiviral CPE reduction assay as described above. In parallel, these stocks were inoculated in embryonated eggs, and the allantoic fluids were subjected to virus titration by cell culture and hemagglutination assay and virion quantification by two step real-time RT-PCR (as described above). Finally, these allantoic stocks were used to perform a hemolysis inhibition assay with the test compounds and to determine the pH of erythrocyte hemolysis for the mutant viruses.
Polykaryon assay for fusion of cells expressing wild-type or mutant HA.
The coding sequence for the A/X-31 HA was obtained by RNA extraction of A/X-31-infected MDCK cells, followed by reverse transcription and high- fidelity PCR, and was then cloned into the pcDNA3.1 D/V5-His-TOPO vector (Invitrogen). After generation of the mutant plasmids by site-directed mutagenesis (with the QuikChange Site-Directed Mutagenesis Kit; Stratagene), the wild-type and mutant HA sequences were amplified by high-fidelity PCR using primers extended with EcoRV and NotI sites to allow subcloning into the pCAGEN expression vector (kindly provided by C. Cepko [Boston, MA] via Addgene [plasmid no. 11160]) (23). The final plasmids were checked by cycle sequencing of the HA fragments.
To perform the polykaryon assay, HeLa cells were seeded into 12-well plates at 300,000 cells per well. One day later, transfection with the wild-type or mutant pCAGEN-HA plasmid (0.73 μg DNA per well) was performed using Fugene 6 reagent (Roche) and serum-free medium. After 3 h, the transfection medium was replaced by medium containing 2% FCS. Two days later, the cells were washed with DMEM, and the HA was cleaved by incubation with TPCK-treated trypsin (2.5 μg per 0.5 ml DMEM) for 15 min at 37°C. After two rinses with PBS containing Ca2+ and Mg2+ (PBS-CM), the cells were preincubated with 0.5 ml test compound (diluted in PBS-CM) for 15 min at 37°C. Then, the compound was removed, and the cells were incubated with 0.5 ml acidic buffer (PBS-CM adapted to pH 5.0 with 0.1 M citric acid) containing the corresponding concentration of test compound. After exactly 15 min of incubation at 37°C, the cells were rinsed with PBS-CM, and 1 ml medium containing 10% FCS was added, followed by 3 h of incubation at 37°C. Finally, the cells were fixed with 96% ethanol, stained with Giemsa (Sigma), and examined by microscopy at ×200 magnification. To score syncytium formation, the number of polykaryons (containing five or more nuclei) was counted in four randomly selected microscopic fields. To determine the fusion pH of wild-type and mutant HAs, the experiments were performed in the absence of compound, but the pH of the acidic buffer was varied (range, 4.9 to 6.0).
Molecular modeling. (i) X-ray structure.
In all docking experiments, the recently published X-ray structure of the H3 hemagglutinin trimeric protein in complex with tert-butyl hydroquinone (TBHQ) was used (Protein Data Bank [PDB] entry 3EYM [36]). The HA1 chains were denoted the A, C, and E chains, whereas B, D, and F are the HA2 chains. For the numbering of amino acid residues, a suffix 1 or 2 indicates their location in the HA1 or HA2 subunit, respectively.
(ii) Preparation of the compounds and the HA protein for docking.
The N-(1-thia-4-azaspiro[4.5]decan-4-yl)carboxamide compounds were drawn using Macromodel 5.0 (25). The Cambridge Structural Database (CSD) entry OFEGIQ was used as a template for the conformation of the aliphatic six-member ring (1). The molecular geometry was fed into Gamess for geometry optimization using the AM1 force field (38). Polar hydrogen atoms, as well as Gasteiger charges, were added to the protein and compound structures using autodock tools.
(iii) Docking of 4c in the HA structure.
For the different HA residues associated with 4c resistance, the calculated solvent accessibility was at least 100 Å2 for A432 and E572, whereas R2201, A962, and D1122 had values of 20 Å2 or less (33). Docking experiments with 4c (after removal of TBHQ) were therefore restricted to A432 and E572, using Autodock 4 software (26) and a grid size of 50 by 50 by 50 points and 0.375-Å resolution. When all protein residues were kept rigid, no specific interactions between 4c and E57(F) were observed. Superposition of chains B, D, and F (using the Dali server [http://www.ebi.ac.uk/Tools/dalilite/index.html]) showed that the orientation of the side chain of E57 differed among the three monomers, implying that this E57 residue is flexible and adopts different rotameric states. From these, we selected rotameric state (χ1 = 49,84, χ2 = 74.84, χ3 = 160.79), which orients the glutamic acid side chain in the direction of the cavity, for 4c docking. After docking, the top 50 of docked ligand conformations were examined, and the highest-score conformation was selected based on the following criteria: specific hydrogen bonding with the E57(F) side chain and hydrophobic interactions with the R and R2 methyl groups of 4c. Interactions (H bonds and hydrophobic) were calculated using Ligplot and HBPlus (24, 47), and the three-dimensional (3D) models were generated with UCSF Chimera (29).
RESULTS
Activities of N-(1-thia-4-azaspiro[4.5]decan-4-yl)carboxamide derivatives against human influenza A/H3N2 virus.
The anti-influenza virus activities of a series of structurally related N-(1-thia-4-azaspiro[4.5]decan-4-yl)carboxamide compounds (Fig. 1) were determined by measuring their inhibitory effects on virus replication in MDCK cells, using two strains of the influenza A/H3N2 virus subtype. The antiviral data obtained by microscopic evaluation of the virus-induced CPE were confirmed by the MTS cell viability assay and expressed as the EC50. The cytotoxicities of the compounds were expressed as the MCC (the compound concentration causing minimal alterations in cell morphology as estimated by microscopy) or the CC50 (the compound concentration causing 50% reduction in cell viability based on the MTS assay). Analysis of the structure-activity relationships revealed that the strongest antiviral activity was observed for the compounds 4c and 4d, in which the R substituent at position 8 of the spiro ring (Fig. 1) is a methyl and ethyl substituent, respectively. Both compounds had an average EC50 of 3.7 μM (Table 1). 4c showed minimal cytotoxicity at the highest concentration tested (100 μM), yielding a selectivity index (SI) (defined as the ratio of MCC to EC50) of 27. 4d had a lower SI of 14. The importance of the substituent at position 8 was evident from the data showing that 4b, which has a methyl group at position 7 (R1) (Fig. 1) instead of 8, was 11-fold less active than 4c, while the compounds having no substituent (4a) or an 8-phenyl group (4e) were devoid of antiviral activity. Likewise, the methyl substituent in the spiro ring (R2) (Fig. 1) was shown to play an important role, since the analogues of 4c and 4d lacking this function (3a and 3b, respectively) were inactive. Similarly, removal of the spiro ring was found to be deleterious (compounds 2a, 2b, and 2c). A few compounds were synthesized with variations in the aromatic system. Substitution of the imidazo[2,1-b]thiazole ring (4c) by an o-hydroxyphenyl ring (6b) resulted in a minimal increase in antiviral activity; however, 6b was also more cytotoxic. The analogue lacking the methyl in the spiro ring (6a) was inactive.
FIG. 1.
Strategy for chemical synthesis of the compounds.
TABLE 1.
Activity in influenza A/H3N2 virus-infected MDCK cellsa
| Compoundb | R | R1 | R2 | Antiviral EC50c (μM) |
Cytotoxicityd |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| A/X-31 |
A/HK/7/87 |
MCC (μM) | CC50 (μM) | IC50 (μM) | ||||||
| CPE | MTS | CPE | MTS | |||||||
| Series I | ||||||||||
| 3a | Me | H | H | >100 | >100 | >100 | >100 | >100 | ND | ND |
| 3b | Et | H | H | >100 | >100 | >100 | >100 | >100 | ND | ND |
| 3c | Phe | H | H | >100 | >100 | >100 | >100 | ≥100 | ND | ND |
| 4a | H | H | Me | 100 | 100 | >100 | >100 | >100 | ND | ND |
| 4b | H | Me | Me | 32 ± 17 | 44 | 42 ± 4 | 40 | >100 | ND | ND |
| 4c | Me | H | Me | 3.3 ± 1.3 | 3.2 ± 2.0 | 5.0 ± 2.3 | 3.2 ± 2.3 | ≥100 | >100 | 93 |
| 4d | Et | H | Me | 2.4 ± 2.1 | 4.0 ± 3.8 | 4.2 ± 3.3 | 4.3 ± 3.0 | 50 | >50 | 48 |
| 4e | Phe | H | Me | >100 | >100 | >100 | >100 | 100 | ND | ND |
| Series II | ||||||||||
| 2a | Me | >100 | >100 | >100 | >100 | ≥100 | ND | ND | ||
| 2b | Et | >100 | >100 | >100 | >100 | >100 | ND | ND | ||
| 2c | Phe | >100 | >100 | >100 | >100 | >100 | ND | ND | ||
| Series III | ||||||||||
| 6a | Me | H | >300 | >300 | >300 | >300 | ≥60 | >300 | ND | |
| 6b | Me | Me | 2.6 ± 2.3 | 5.9 ± 4.3 | 2.8 ± 1.8 | 1.4 ± 0.6 | >50 | >50 | 38 | |
| Oseltamivir carboxylate | 0.0042 ± 0.0035 | 0.020 ± 0.040 | 1.7 ± 1.9 | 0.63 ± 0.23 | >100 | >100 | 80 | |||
| Ribavirin | 9.9 ± 3.3 | 11 ± 4 | 7.4 ± 2.1 | 7.2 ± 3.8 | 61 ± 30 | >100 | 18 | |||
The data shown are the means ± standard deviations of 2 to 7 independent tests. ND, not done. MDCK cells, Madin-Darby canine kidney cells; Et, ethyl; Me, methyl; Phe, phenyl.
Figure 1 shows the basic structures of the N-(1-thia-4-azaspiro[4.5]decan-4-yl)carboxamide compounds, series I, II, and III.
Antiviral activity is expressed as the EC50, defined as the compound concentration producing 50% inhibition of virus replication, as estimated by microscopic scoring of the CPE or by measuring cell viability in the formazan-based MTS assay.
Cytotoxicity is expressed as the MCC, the compound concentration producing minimal changes in cell morphology, as estimated by microscopy; the CC50, estimated by the MTS cell viability assay; or the IC50, determined by cell counting.
In contrast to the promising activities of 4c, 4d, and 6b against influenza A/H3N2 virus, these compounds were completely inactive against A/H1N1 and B viruses (Table 2). In addition, 4c was shown to be inactive in a plaque assay with influenza A/H5N1 or A/H7N2 virus (highest concentration tested, 100 μM). The A/H3N2 (A/Aichi/2/68) virus, included as a control, displayed an EC50 of 7.5 μM, which is similar to the value obtained in the CPE reduction assay. This defined subtype dependency was consistent upon evaluation against a broad panel of human influenza virus strains (Table 2). Overall, 4c, 4d, and 6b had average EC50s for A/H3N2 of 9.6 μM, 3.7 μM, and 4.4 μM, respectively. A cell-counting assay was used to precisely estimate their cytostatic activities (expressed as the IC50), and from this their selectivity indexes (i.e., ratio of IC50 to EC50) values were calculated as 10, 13, and 9 for 4c, 4d, and 6b, respectively. The strong activities of 4c and 4d were also evident from a virus yield experiment, since both compounds (at a concentration of 20 μM) produced a 3- to 4-log-unit reduction in the number of virus particles released in the supernatant of infected MDCK cells at 72 h p.i. (data not shown).
TABLE 2.
Antiviral activities in influenza A/H1N1, A/H3N2, and B virus-infected MDCK cellsa
| Strain | Antiviral EC50b (μM) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 4c |
4d |
6b |
Ribavirin |
Oseltamivir carboxylate |
||||||
| CPE | MTS | CPE | MTS | CPE | MTS | CPE | MTS | CPE | MTS | |
| A/H1N1 | ||||||||||
| A/FM/1/47 | >50 | >50 | >50 | >50 | >50 | 38 ± 10 | 12 ± 3 | 13 ± 1 | 4.0 ± 2.9 | 4.0 ± 2.2 |
| A/Ned/378/05 | >50 | >50 | >50 | >50 | >50 | 21 ± 14 | 8.4 ± 2.2 | 6.1 ± 2.9 | 0.19 ± 0.06 | <0.20 |
| A/PR/8/34 | >50 | >50 | >50 | >50 | >50 | >50 | 10 ± 0 | 12 ± 5 | 12 ± 7 | 6.3 |
| A/H3N2 | ||||||||||
| A/X-31 | 3.3 ± 1.3 | 3.2 ± 2.0 | 2.4 ± 2.1 | 4.0 ± 3.8 | 2.6 ± 2.3 | 5.9 ± 4.3 | 9.9 ± 3.3 | 11 ± 4 | 0.0042 ± 0.0035 | 0.020 ± 0.040 |
| A/HK/2/68 | 7.4 ± 2.8 | 4.6 ± 3.0 | 3.4 ± 1.3 | 1.8 ± 1.4 | 5.4 ± 3.2 | 2.7 ± 1.1 | 11 ± 1 | 9.1 ± 1.3 | 1.2 ± 1.0 | <0.20 |
| A/Victoria/3/75 | 23 ± 17 | 27 ± 20 | 6.0 ± 1.5 | 3.7 ± 3.2 | 7.1 ± 3.9 | 7.2 ± 5.2 | 8.7 ± 3.7 | 7.7 ± 4.1 | <0.20 | <0.20 |
| A/HK/7/87 | 5.0 ± 2.3 | 3.2 ± 2.3 | 4.2 ± 3.3 | 4.3 ± 3.0 | 2.8 ± 1.8 | 1.4 ± 0.6 | 7.4 ± 2.1 | 7.2 ± 3.8 | 1.7 ± 1.9 | 0.63 ± 0.23 |
| A/Aichi/2/68 | 7.5 ± 2.7c | |||||||||
| A/H5N1 | ||||||||||
| A/Vietnam/1203/04 | >100c | |||||||||
| A/chicken/Vietnam/ C58/04 | >100c | |||||||||
| A/H7N2 | ||||||||||
| A/chicken/Pennsylvania/ 143586/02 | >100c | |||||||||
| B | ||||||||||
| B/HK/5/72 | >50 | >50 | >50 | >50 | >50 | >50 | 5.8 ± 2.9 | 11 ± 9 | 1.7 ± 0.9 | 2.2 ± 0.9 |
| B/Lee/40 | >50 | >50 | >50 | >50 | >50 | >50 | 15 ± 6 | 15 ± 9 | >20 | >20 |
| B/Ned/537/05 | >50 | >50 | >50 | >50 | >50 | >50 | 6.6 ± 3.1 | 7.9 ± 0.3 | 2.2 ± 2.3 | 1.7 ± 1.7 |
The data shown are the means ± standard deviations of 2 to 7 independent tests.
Antiviral activity is expressed as the EC50, defined as the compound concentration producing 50% inhibition of virus replication, as estimated by microscopic scoring of the CPE or by measuring cell viability in the formazan-based MTS assay.
Data obtained by plaque reduction assay.
4c acts within 30 min postinfection.
A one-cycle time-of-addition experiment was performed in which the antiviral compounds were added at different time points p.i. (range, −30 min to 8 h). As reference compounds, we included the M2 proton channel blocker rimantadine; the weak base chloroquine (which acts by increasing the endosomal pH); the protein kinase inhibitor bisindolylmaleimide I (which has been shown to inhibit the nuclear import of the viral ribonucleoprotein complex) (34); and ribavirin, an inhibitor of viral-RNA synthesis. In the absence of compounds, the increase in vRNA copies between time point zero (input virus) and 10 h p.i. was 170-fold (Fig. 2). No vRNA synthesis was detected when 4c was added at −30 min or 0 min p.i. In contrast, its inhibitory effect was considerably reduced when the compound was added at 30 min p.i. or later. The same time dependency was observed for rimantadine and chloroquine, while the time-of-addition curve was slightly shifted for bisindolylmaleimide I. Ongoing vRNA synthesis between 1 and 8 h p.i. was reflected in the gradual curve obtained with ribavirin treatment.
FIG. 2.
Inhibitory effect of 4c on vRNA synthesis as a function of the time of compound addition. MDCK cells were infected with A/X-31 influenza virus, and compounds were added at −30 min, 0 h, 30 min, 1 h, 3 h, 5 h, or 8 h p.i. At 10 h p.i., the cells were subjected to RNA extraction for subsequent analysis by real-time RT-PCR. The vRNA synthesis is presented as the fold increase in vRNA copies at 10 h p.i. relative to the number of added virus particles. The data shown are the means ± standard deviations (SD) (the numbers of experiments are in parentheses).
The N-(1-thia-4-azaspiro[4.5]decan-4-yl)carboxamide derivatives inhibit virus-mediated hemolysis.
As 4c was found not to influence the attachment of influenza virus to erythrocytes in a hemagglutination assay (data not shown), an inhibitory effect of 4c on the interaction between HA and cellular receptors was excluded. Next, a hemolysis inhibition assay was performed to determine whether 4c and its derivatives inhibit hemagglutinin-mediated interactions with target membranes. For several strains of A/H3N2 influenza virus, the hit compounds inhibited the acid-induced hemolysis with EC50s of ≥3 μM (4c), ≥3 μM (4d), and <1.6 μM (6b) (Table 3). Since 4c and 4d showed no inhibitory effect on hemolysis by influenza A/H1N1 and B viruses, the subtype-dependent activity, as observed in the CPE reduction experiment, was confirmed. Also, 6a and 3a showed much weaker inhibition of influenza A/H3N2 virus-induced hemolysis. For 4b and 4a, no inhibition was noted at a concentration of 100 μM. 4c proved to be highly inhibitory when the compound was present during both the 10-min virus-erythrocyte adsorption stage and the subsequent acid-induced hemolysis event. In contrast, no effect was observed when 4c was washed away after the adsorption stage, indicating that the binding of 4c to HA is reversible.
TABLE 3.
Inhibitory effects on virus-induced hemolysis
| Compound | EC50a (μM) |
||||||
|---|---|---|---|---|---|---|---|
| A/H1N1 |
A/H3N2 |
B (B/HK/5/72) | |||||
| A/PR/8/34 | A/FM/1/47 | A/X-31 | A/HK/7/87 | A/HK/2/68 | A/Vict/3/75 | ||
| 3a | ND | ND | 29 | ND | ND | ND | ND |
| 4a | ND | ND | >100 | ND | ND | ND | ND |
| 4b | ND | ND | >100 | ND | ND | ND | ND |
| 4c | >100 | >100 | 3.0 ± 0.9 | 30 ± 29 | 4.2 ± 0.9 | ≤2.6 | >100 |
| 4d | >100 | >100 | 3.2 ± 1.1 | 15 | 6.7 ± 4.2 | 2.5 ± 0.0 | >100 |
| 6a | ND | ND | 12 ± 1 | ND | ND | ND | ND |
| 6b | ND | ND | <1.6 | ND | ND | ND | ND |
The inhibitory effect, determined by hemolysis inhibition assay, was expressed as the EC50, i.e., the compound concentration producing 50% inhibition of virus-induced hemolysis relative to the value at pH 4.9. Means (± standard deviations) of 1 to 4 independent experiments are shown. ND, not done.
4c inhibits HA-mediated membrane fusion by prevention of the HA conformational change at low pH.
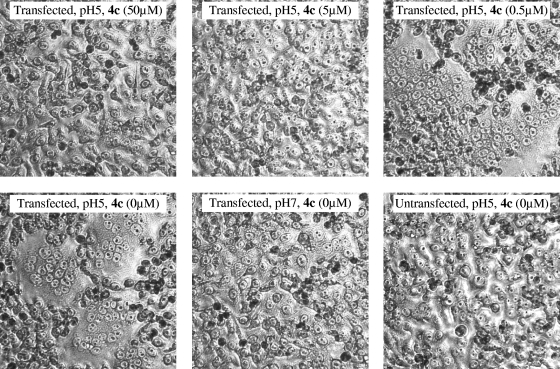
HeLa cells expressing the A/X-31 HA were exposed to pH 5.0 in the absence or presence of 4c, and the subsequent fusion of cell membranes and syncytium formation were evaluated by microscopy. As can be seen in Fig. 3, 4c produced dose-dependent inhibition of polykaryon formation, since no syncytia were formed when 4c was added at ≥5 μM 15 min prior to, as well as during, the acid stage.
FIG. 3.
4c inhibits low-pH-induced polykaryon formation. HeLa cells expressing wild-type A/X-31 HA were treated with trypsin to cleave HA0, washed, and incubated in the presence of various concentrations of 4c. Then, the pH was lowered to 5.0, and the cells were incubated for 15 min at 37°C in the presence of 4c. Following syncytium formation for 3 h at 37°C in medium, the cells were fixed, stained with Giemsa, and examined by microscopy (original magnification, ×200). Representative fields are shown.
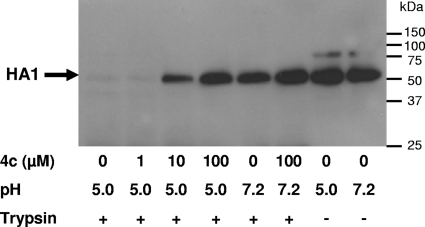
FIG. 4.
Effect of 4c on the conformational change of HA, as demonstrated by trypsin digestion. A/X-31 virus was incubated at 37°C for 15 min in the presence of various concentrations of 4c, and the pH was lowered to 5.0. After neutralization, the mixtures were treated with trypsin. The lysates were subjected to Western blot analysis under reducing conditions and using an anti-HA1 antibody. The low-pH-induced conformational change of the HA resulted in the disappearance of the HA1 band.
In order to investigate whether 4c exerts its fusion-inhibiting effect by directly preventing the conformational change of HA at acidic pH, a tryptic digestion assay was used, which is based on the exposure of trypsin cleavage sites when HA changes from its prefusogenic to its fusogenic conformation (41). As shown in Fig. 4, the HA1 subunit of the A/X-31 virus was clearly protected against trypsin cleavage when the virus was incubated in the presence of 4c at 10 or 100 μM.
Genotypic and phenotypic characterization of 4c-resistant mutants.
4c-resistant (4cr) mutants were selected during three passages of the A/X-31 strain in MDCK cells at 4c concentrations up to 100 μM. After clonal purification, three clones (designated 4cr-A, 4cr-B, and 4cr-C) were isolated. Three control clones were obtained from a parallel condition in which no compound was added. As the biological data for the control clones were identical, they were pooled. In CPE reduction experiments, replication of all three 4cr clones was not inhibited by 4c, 4d, and 6b at compound concentrations of 100 μM, whereas the EC50 of the control was in the same range observed previously (Table 4). The antiviral sensitivities of the 4cr clones to the control compounds ribavirin and oseltamivir carboxylate were unchanged, since the EC50s were similar to those obtained for the control. The 4cr clones were, however, resistant to the M2 blocker rimantadine. The 4c-resistant phenotype of the 4cr clones was confirmed by the hemolysis inhibition assay: none of the three clones was inhibited in its hemolytic activity at 4c and 4d concentrations of 100 μM.
TABLE 4.
Antiviral sensitivities of 4c-resistant mutants
| Clonea | EC50 (μM)b |
|||||||
|---|---|---|---|---|---|---|---|---|
| CPE assayc |
Hemolysis assayd |
|||||||
| 4c | 4d | 6b | Rimantadine | Ribavirin | Oseltamivir carboxylate | 4c | 4d | |
| 4cr-A | >100 | >100 | >100 | >500 | 12 ± 0 | 0.14 ± 0.15 | >100 | >100 |
| 4cr-B | >100 | >100 | >100 | >500 | 12 ± 1 | 0.046 ± 0.020 | >100 | >100 |
| 4cr-C | >100 | >100 | >100 | >500 | 13 ± 0 | 0.37 ± 0.36 | >100 | >100 |
| Control | 2.7 ± 1.1 | 1.8 ± 1.0 | 1.5 ± 0.9 | 58 ± 2 | 12 ± 1 | 0.10 ± 0.14 | 3.9 ± 1.1 | 4.0 ± 0.8 |
4cr-A, 4cr-B, and 4cr-C, clones selected under 4c pressure; control, pooled data from three clones selected in the absence of compound.
The data shown are the means (± standard deviations) of four independent tests.
Antiviral activity in Madin-Darby canine kidney cells was expressed as the EC50, as estimated by microscopic scoring of the CPE.
The inhibitory effect, determined by hemolysis inhibition assay, was expressed as the EC50 at low pH.
After isolation of the 4c-resistant and control clones, the HA gene was sequenced. Clones 4cr-A and 4cr-B were triple mutants, both of which contained the substitutions R2201S and E572K. A third substitution (Ala to Thr) was found at either of two positions: 4cr-A showed an A432T substitution and 4cr-B contained an A962T substitution. The 4cr-C clone was genotypically distinct, since it contained a single D1122N substitution. In addition, several amino acid changes compared to the published X-31 sequence were noted, but these were identical in the 4c-resistant and control clones.
To compare the binding efficiencies and replication fitness of the selected clones, the viruses were inoculated into hen eggs, and the resulting allantoic fluids were subjected to virus titration by cell culture, hemagglutination assay, and real-time RT-PCR. The results (Table 5) indicate that the fitness of the resistant mutants was not compromised, since the allantoic fluids contained comparable numbers of virions (assessed by RT-PCR) that were equally effective in binding to erythrocytes or replicating in cell culture.
TABLE 5.
Receptor binding and replication fitness of 4c-resistant virus mutantsa
| Cloneb | Amino acid substitution(s) | No. of viral copies (RT-PCR) | HAU | CCID50 | Ratio |
|
|---|---|---|---|---|---|---|
| HAU/no. of viral copies (×1,000,000) | CCID50/no. of viral copies | |||||
| 4cr-A | R2201S, A432T, E572K | 5.0E7 | 128 | 1.1E7 | 2.5 | 0.23 |
| 4cr-B | R2201S, E572K, A962T | 3.3E7 | 176 | 5.5E6 | 5.3 | 0.17 |
| 4cr-C | D1122N | 9.3E7 | 256 | 3.2E7 | 2.8 | 0.34 |
| Control | 9.3E7 | 341 | 3.6E7 | 3.7 | 0.39 | |
The individual clones were expanded in eggs, and the allantoic fluids were subjected to virus titration by cell culture (expressed as the CCID50) and hemagglutination assay (expressed as the number of hemagglutinating units [HAU]). Virion quantification was done by real-time RT-PCR. The data shown are the mean of two independent tests.
4cr-A, 4cr-B, and 4cr-C, clones selected under 4c pressure; control, pooled data from three clones selected in the absence of compound.
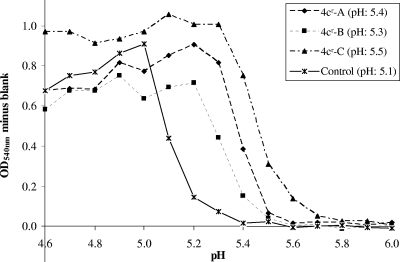
To determine the fusion pHs of the 4c-resistant and control clones, hemolysis assays were performed in which the pH during the acid-induced fusion stage was varied (pH range, 4.6 to 6.0). The fusion pHs, defined as the pH at which 50% hemolysis occurred, were 5.4, 5.3, and 5.5 for 4cr-A, 4cr-B, and 4cr-C, respectively, compared to 5.1 for the control (Fig. 5). Thus, the 4c-resistant viruses, whether they contained one (4cr-C) or three (4cr-A and 4cr-B) substitutions in the HA protein, displayed an elevated fusion pH. The increase was more pronounced for the 4cr-A and 4cr-C clones than for the 4cr-B clone. This increase in the fusion pH also explains the resistance of the 4cr clones to rimantadine. It is known that high concentrations of adamantane compounds have a pH-increasing effect in the endosomes, which can be counteracted by resistance mutations in the HA, leading to increased fusion pH (8). The relatively high antiviral EC50 of rimantadine for the wild-type virus suggests that the virus is insensitive to the specific M2 proton channel-blocking effect of the adamantanes.
FIG. 5.
pH dependence of hemolysis for wild-type and 4c-resistant mutants. A hemolysis assay was performed in which the pH of the acidic buffer was varied between 4.6 and 6.0. The y axis shows the degree of hemolysis, expressed as the optical density at 450 nm (OD540) minus the blank value obtained from a mock-infected condition. 4cr-A, 4cr-B, and 4cr-C, clones selected under 4c pressure; control, pooled data from three clones selected in the absence of compound. The values in parentheses are the pHs at which 50% hemolysis occurred relative to the value at pH 4.9.
Membrane fusion characteristics of mutant HA proteins containing 4c-associated substitutions.
To determine the phenotypic consequences of the 4c-associated HA protein mutations, wild-type and single-mutant HA genes were expressed in HeLa cells to perform the polykaryon fusion assay at low pH. The EC50s for inhibition of syncytium formation by 4c were 0.90, 1.8, 2.1, 7.9, and ≥23 μM for the wild-type and R2201S-, A432T-, E572K-, and D1122N-substituted HAs, respectively (Table 6). 4c produced no inhibition of membrane fusion in cells containing the A962T-substituted HA protein (highest concentration tested, 50 μM). Thus, the E572K, A962T, and D1122N substitutions resulted in resistance to the fusion-inhibiting effect of 4c. By varying the pH of the acidic buffer (range, 4.9 to 6.0) to which the HA-expressing cells were exposed, the fusion pH was determined. Strong increases in the fusion pH were observed for the R2201S, A962T, and D1122N mutants, whereas the A432T- and E572K-substituted HAs displayed the same fusion pH as wild-type HA protein.
TABLE 6.
Analysis of 4c-associated HA substitutions in a polykaryon assay of HA-transfected cells
| Substitution | Fusion pHa | EC50 for 4c (μM)b |
|---|---|---|
| R2201S | 5.5 ± 0.1c | 1.8 ± 0.7 |
| A432T | 5.2 ± 0.1 | 2.1 ± 1.5 |
| E572K | 5.2 ± 0.1 | 7.9 ± 0.6c |
| A962T | 5.5 ± 0.0c | >50c |
| D1122N | 5.6 ± 0.1c | ≥23c |
| Wild type | 5.2 ± 0.1 | 0.90 ± 0.40 |
The fusion pH was defined as the pH at which the number of syncytia was 50% of the number observed at pH 4.9.
The EC50 represents the compound concentration at which the number of syncytia was 50% of the number observed in the absence of compound.
P < 0.05 (two-sided Student's t test) for a statistically significant difference versus the value of the wild type.
Data shown are the mean (± standard deviations) of three independent tests.
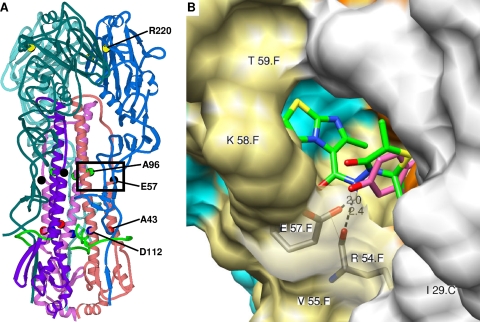
Locations of the HA residues associated with 4c resistance and molecular modeling for 4c binding.
Figure 6A shows the locations of the five amino acid residues associated with viral resistance to 4c within the available crystal structure of HA X-31 in complex with the H3-specific fusion inhibitor TBHQ. Each of the three HA regions suggested to be critically involved in the conformational change at low pH (43) appeared to be involved in 4c resistance. R2201 is located at an intersubunit interface in a coil region within the globular head of the HA trimer and is involved in hydrogen bonds and hydrophobic interactions. Residues A432 and D1122 are located in the same cavity, with A432 located at the entrance and D1122 at the bottom of this cavity. A432 shows local hydrophobic interactions, and D1122 forms hydrophobic interactions and hydrogen bonds with the nearby fusion peptide (8). The other two residues are situated at the entrance (E572) and bottom (A962) of the TBHQ binding pocket. E572 forms intrasubunit hydrogen bonds and hydrophobic interactions and is involved in TBHQ binding (36). With A962, local hydrophobic interactions were observed.
FIG. 6.
Positions of 4c-associated HA mutations and the predicted binding pocket of 4c in the HA protein. The modeling was based on the published crystal structure of the X-31 HA in complex with TBHQ (36). (A) Ribbon diagram showing the locations of the amino acid residues (R2201, A432, E572, A962, and D1122) involved in 4c resistance. The HA1 chains are colored in sky blue (A chain), royal blue (C chain), and slate blue (E chain); HA2 chains are in pink (B chain), flesh (D chain), and purple (F chain); and the fusion peptide is indicated in green. The frame indicates the position of the binding pocket of 4c. (B) Chimera model of the interactions of 4c in the binding pocket around glutamic acid-57. The numbering of the amino acid residues includes the polypeptide chain (HA1 chains, A, not visible; C, white; and E, cyan; HA2 chains, B, not visible; D, orange; and F, khaki). Predicted hydrogen bonds with the main chain carbonyl of R542 and the side chain carboxyl of E572 are shown as dashed lines, with the distance indicated in angstroms. Residues I291, R542, V552, K582, and T592, indicated on the protein structure, and residues P2931, K3071, Y942, E972, L982, L992, and A1012 (not visible) are involved in hydrophobic interactions with 4c (shown in green; note that the cyclohexane part is not displayed). The predicted position of the N-(1-thia-4-azaspiro[4.5]decan-4-yl)carboxamide moiety of 4c is similar to that of TBHQ (shown in purple). 4c-specific hydrophobic interactions were observed between its imidazo[2,1-b]thiazole part and residues P2931, K3071, K582, and T592.
Computer-aided docking (in the HA X-31 crystal structure after removal of TBHQ) was performed to predict the possible binding pocket for 4c in the HA trimer in the vicinity of any of the observed resistance mutations. R2201, A962, and D1122 were found to be solvent inaccessible. 4c was observed to fit within a cavity close to E572, and several hydrophobic interactions between the surrounding residues and 4c were observed in which the R and R2 methyl groups of 4c were also involved (Fig. 6B). The nitrogen atom of its carboxamide bridge forms hydrogen bonds with the side chain carboxyl of E572 and the main chain carbonyl of R542. Similar interactions within this E572 cavity were obtained with the 8-ethyl analogue 4d and the o-hydroxyphenyl analogue 6b (data not shown). Replacement of this E572 residue by lysine results in loss of the hydrogen bond between the glutamic acid carboxyl function and 4c. Alternatively, a change in charge could modify the local structure of the cavity, influencing the binding of 4c. Besides these hydrogen bonds, multiple hydrophobic interactions were observed between 4c and its binding pocket, namely, with residues I291, P2931, K3071, R542, V552, K582, T592, Y942, E972, L982, L992, and A1012 (Fig. 6B). Interestingly, the position of the aliphatic cyclic part of 4c within this hydrophobic cavity was clearly similar to that of TBHQ, which also forms a hydrogen bond with E572 (as well as with L982) and hydrophobic interactions with L291, R542, L552, E972, L982, L992, and A1012 (36). However, the imidazo[2,1-b]thiazole part of 4c allows the formation of several additional hydrophobic interactions.
DISCUSSION
Fusion between the viral and endosomal membranes is an essential event during influenza virus entry into the host cell. After endocytosis of the virion, the influenza virus HA protein undergoes an irreversible acid-induced structural rearrangement in which the highly hydrophobic fusion peptide, located at the amino terminus of HA2, is exposed and projected toward the endosomal membrane (42). Since this conformational change is crucial for the fusogenic activity of HA and viral entry, this event is an attractive target for the development of antiviral agents.
In the present work, we described a new series of N-(1-thia-4-azaspiro[4.5]decan-4-yl)carboxamide inhibitors that show marked antiviral activity against influenza A/H3N2 viruses in a CPE reduction assay. For the lead compound, 4c, the average EC50 for influenza A/H3 viruses was 9.6 μM and the IC50 was 93 μM, yielding a selectivity index of 10. Time-of-addition experiments with 4c revealed that the compound acts at an early step in virus replication, i.e., within 30 min p.i. The subtype-dependent activity of the N-(1-thia-4-azaspiro[4.5]decan-4-yl)carboxamide derivatives, and the time-of-addition curve for 4c, pointed to the influenza virus HA protein as the possible target. Whereas 4c did not influence the attachment of influenza virus to erythrocytes in a hemagglutination assay, the compound showed strong inhibition of virus-induced hemolysis. Moreover, the structure-activity relationship obtained in hemolysis inhibition studies agreed with the results of the CPE reduction assay. Hence, the available data are consistent with the mechanism of action for 4c and its congeners being the inhibition of HA protein-mediated membrane fusion. Several classes of small molecules that inhibit influenza virus fusion have been reported previously (2, 5, 30, 49). While most of these compounds inhibit the acid-induced conformational change, diiodofluorescein was shown to act in an irreversible manner by premature triggering of this HA conformational change (12). For 4c, the first mechanism of action was suggested by our observation that no inhibition of hemolysis occurred when the compound was washed away after the virus-erythrocyte adsorption stage and prior to acid-induced fusion. Definite proof of this mode of action was provided by the tryptic digestion assay, in which 4c prevented the acid-induced conversion of HA to its trypsin-sensitive conformation.
Most reported fusion inhibitors have subtype-dependent activities, and for the majority, the activity against the influenza A/H1 and A/H2 strains is superior to that against A/H3 strains (21, 30, 49). One exception is the antiviral drug arbidol, which displays antiviral activity against a broader range of subtypes and was recently reported to act as a fusion inhibitor (19). The small benzoquinones and hydroquinones, with TBHQ being the most potent, were, until now, the only known class of A/H3-specific fusion inhibitors (2). The class of N-(1-thia-4-azaspiro[4.5]decan-4-yl)carboxamide inhibitors we describe in this report shows specific activity against influenza A/H3 viruses. This is surprising, since the backbone structure of our inhibitors is related to that of some A/H1- and A/H2-specific inhibitors, consisting of an aromatic system linked to a nonaromatic cyclic system via an amide bridge (20, 30). To obtain a more detailed insight into this subtype dependency, we also evaluated the compounds against influenza A/H5 and A/H7 viruses, an issue which has not been investigated for the other reported fusion inhibitors. The inclusion of these emerging subtypes not only has implications for antiviral drug development, but also may aid us to understand the mode of action of 4c and its derivatives. Whereas the inactivity of 4c against A/H5 virus was not totally surprising, the insensitivity of the A/H7 virus was quite unexpected. Russell et al. performed a phylogenetic analysis of all 16 HAs based on the available crystal structures of representative HAs, including some regions involved in fusion (35). The H1 and H5 HAs are closely related and belong to the same group 1, while the H3 and H7 HAs are more distantly related and represent the two clades within group 2. The different activities of 4c against H3 and H7 viruses may thus reflect the fact that the region in H3 HA involved in binding of 4c differs from the corresponding region in H7 HA. We addressed this issue by characterizing 4c-resistant virus mutants and investigating the roles of the individual substitutions in a polykaryon assay in cells expressing mutant HA proteins. Experience with the other fusion inhibitors has shown that the HA mutations selected by these compounds may be explained by reduced binding of the inhibitor and/or decreased stability of the HA trimeric protein, and hence an increase in the fusion pH. Among the 4c-associated mutations, the A962T and D1122N substitutions were shown to reduce HA trimer stability, rendering HA resistant to the fusion-inhibiting effect of 4c. On the other hand, the observation that E572K-substituted HA displayed an unchanged fusion pH yet 9-fold-greater resistance to 4c is suggestive of a role of this residue in 4c binding, which was also proposed by our computer-aided docking. Interestingly, residue E572 is directly involved in binding of another H3-specific fusion inhibitor, TBHQ, as demonstrated by cocrystallization studies with the X-31 HA (36). We noticed a clear similarity between the position of TBHQ and that predicted for the N-(1-thia-4-azaspiro[4.5]decan-4-yl)carboxamide part of 4c. However, the aromatic imidazo[2,1-b]thiazole ring of 4c allows the formation of several additional hydrophobic interactions within this cavity. Although our modeling pointed to residue A432 as an alternative potential binding site for 4c (data not shown), we consider this to be less likely, since substitution of this residue did not result in significant resistance to 4c. In addition, A432 was changed in only one of the 4c-resistant viruses, whereas the E572K mutation was present in both triple-mutant clones. The finding that 4c and TBHQ have a stabilizing effect on HA upon binding to E572 and the nearby cavity is in agreement with the critical role played by the adjacent lysine-58, since the K582I mutation leads to stabilization of the HA proteins of the H3 and H5 subtypes (31, 32, 44). The single D1122N mutation observed in the third 4c-resistant virus clone has previously been identified in virus mutants with increased pH values for conformational changes and membrane fusion, including TBHQ-resistant H3 mutants (8, 12, 32). Thus, although 4c and TBHQ have very different chemical structures, there is a clear parallel in their interaction modes with the H3 HA.
The R2201S substitution is intriguing, since it is located in the globular-head domain of the HA trimer, in which mutations associated with resistance to fusion inhibitors (whether H1 or H3 specific) are not commonly observed. Residue 220 is located at the intersubunit interface (7), at some distance from the receptor binding site. The R2201S substitution does not impact the erythrocyte binding properties of the HA, as was evident from our hemagglutination experiments with the mutant viruses. The same substitution was recently identified during mouse adaptation of an H3 virus, a process that is known to select for viruses with increased fusion pH (15). Likewise, we demonstrated that the 4c-associated mutations, including R2201S, have no impact on viral fitness, since the virus mutants showed equal erythrocyte binding and replication efficiencies and all mutations were stable when the mutant viruses were expanded in eggs. On the other hand, an HA protein alignment showed that the 5 residues that were mutated in 4c-resistant viruses were conserved among the five H3N2 strains included in our study. Likewise, all 13 residues involved in hydrogen bonding and/or hydrophobic interactions of 4c within its predicted HA binding pocket were identical in these five H3N2 viruses, except for K3071 and V552, at which positions strain A/HK/7/87 contains R3071 and L552. Although this subtle difference could account for the lower sensitivity of this virus to 4c in the hemolysis test, its relevance is unclear, since all five H3N2 viruses displayed comparable sensitivities to 4c in the replication assay in MDCK cells.
Fusion inhibitors such as 4c are excellent tools to further delineate the structural changes during the HA-unfolding process. Our results show that the A962 and R2201 residues play important roles in HA stabilization, which was not previously demonstrated. The data we gained on the binding of 4c to the pocket around glutamic acid-57 confirm the relevance of this HA cavity in the structure-based design of fusion inhibitors. Computer-aided design of new derivatives of 4c could potentially lead to compounds with improved antiviral effect and less confined subtype dependency.
Acknowledgments
This study was supported by a Ph.D. grant from the Katholieke Universiteit Leuven (to E.V.), the Fonds voor Wetenschappelijk Onderzoek Vlaanderen (FWO no. 9.0188.07), the Geconcerteerde Onderzoeksacties (GOA/10/014), the International Consortium for Anti-Virals (ICAV), and the American Lebanese Syrian Associated Charities. This project has been funded in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract no. HHSN266200700005C.
We appreciate the dedicated assistance of Leentje Persoons, Frieda De Meyer, Vicky Broeckx, Wim van Dam, and Julie Borghÿs. We thank the Animal Resources Center at St. Jude and Scott Krauss, Heather Forrest, and Patrick Seiler for technical support in the biosafety level 3+ facility. We acknowledge Robert Webster, Scott Krauss, Rachelle Salomon, and Ron Fouchier for kindly providing influenza viruses and plasmids and Mikhail Matrosovich for the kind gift of the MDCK cells.
Footnotes
Published ahead of print on 24 February 2010.
REFERENCES
- 1.Akkurt, M., S. Karaca, E. Sahin, O. Guzel, A. Salman, and E. Ilhan. 2007. 2-Hydroxy-N-(3-oxo-1-thia-4-azaspiro[4.5]dec-4-yl)-2,2-diphenylacetamide. Acta Crystallogr. E. 63:o3379-o3380. [Google Scholar]
- 2.Bodian, D. L., R. B. Yamasaki, R. L. Buswell, J. F. Stearns, J. M. White, and I. D. Kuntz. 1993. Inhibition of the fusion-inducing conformational change of influenza hemagglutinin by benzoquinones and hydroquinones. Biochemistry 32:2967-2978. [DOI] [PubMed] [Google Scholar]
- 3.Bright, R. A., M. J. Medina, X. Xu, G. Perez-Oronoz, T. R. Wallis, X. M. Davis, L. Povinelli, N. J. Cox, and A. I. Klimov. 2005. Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: a cause for concern. Lancet 366:1175-1181. [DOI] [PubMed] [Google Scholar]
- 4.Bullough, P. A., F. M. Hughson, J. J. Skehel, and D. C. Wiley. 1994. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371:37-43. [DOI] [PubMed] [Google Scholar]
- 5.Cianci, C., K. L. Yu, D. D. Dischino, W. Harte, M. Deshpande, G. Luo, R. J. Colonno, N. A. Meanwell, and M. Krystal. 1999. pH-dependent changes in photoaffinity labeling patterns of the H1 influenza virus hemagglutinin by using an inhibitor of viral fusion. J. Virol. 73:1785-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cross, K. J., L. M. Burleigh, and D. A. Steinhauer. 2001. Mechanisms of cell entry by influenza virus. Exp. Rev. Mol. Med. 3:1-18. [DOI] [PubMed] [Google Scholar]
- 7.Daniels, P. S., S. Jeffries, P. Yates, G. C. Schild, G. N. Rogers, J. C. Paulson, S. A. Wharton, A. R. Douglas, J. J. Skehel, and D. C. Wiley. 1987. The receptor-binding and membrane-fusion properties of influenza virus variants selected using anti-haemagglutinin monoclonal antibodies. EMBO J. 6:1459-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daniels, R. S., J. C. Downie, A. J. Hay, M. Knossow, J. J. Skehel, M. L. Wang, and D. C. Wiley. 1985. Fusion mutants of the influenza virus hemagglutinin glycoprotein. Cell 40:431-439. [DOI] [PubMed] [Google Scholar]
- 9.De Clercq, E. 2006. Antiviral agents active against influenza A viruses. Nat. Rev. Drug Discov. 5:1015-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Jong, M. D., T. T. Tran, H. K. Truong, M. H. Vo, G. J. Smith, V. C. Nguyen, V. C. Bach, T. Q. Phan, Q. H. Do, Y. Guan, J. S. Peiris, T. H. Tran, and J. Farrar. 2005. Oseltamivir resistance during treatment of influenza A (H5N1) infection. N. Engl. J. Med. 353:2667-2672. [DOI] [PubMed] [Google Scholar]
- 11.Deyde, V. M., X. Xu, R. A. Bright, M. Shaw, C. B. Smith, Y. Zhang, Y. Shu, L. V. Gubareva, N. J. Cox, and A. I. Klimov. 2007. Surveillance of resistance to adamantanes among influenza A(H3N2) and A(H1N1) viruses isolated worldwide. J. Infect. Dis. 196:249-257. [DOI] [PubMed] [Google Scholar]
- 12.Hoffman, L. R., I. D. Kuntz, and J. M. White. 1997. Structure-based identification of an inducer of the low-pH conformational change in the influenza virus hemagglutinin: irreversible inhibition of infectivity. J. Virol. 71:8808-8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffmann, E., G. Neumann, Y. Kawaoka, G. Hobom, and R. G. Webster. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. U. S. A. 97:6108-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ilyushina, N. A., E. A. Govorkova, and R. G. Webster. 2005. Detection of amantadine-resistant variants among avian influenza viruses isolated in North America and Asia. Virology 341:102-106. [DOI] [PubMed] [Google Scholar]
- 15.Keleta, L., A. Ibricevic, N. V. Bovin, S. L. Brody, and E. G. Brown. 2008. Experimental evolution of human influenza virus H3 hemagglutinin in the mouse lung identifies adaptive regions in HA1 and HA2. J. Virol. 82:11599-11608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiso, M., K. Mitamura, Y. Sakai-Tagawa, K. Shiraishi, C. Kawakami, K. Kimura, F. G. Hayden, N. Sugaya, and Y. Kawaoka. 2004. Resistant influenza A viruses in children treated with oseltamivir: descriptive study. Lancet 364:759-765. [DOI] [PubMed] [Google Scholar]
- 17.Lackenby, A., O. Hungnes, S. G. Dudman, A. Meijer, W. J. Paget, A. J. Hay, and M. C. Zambon. 2008. Emergence of resistance to oseltamivir among influenza A(H1N1) viruses in Europe. Euro Surveill. 13:8026. [DOI] [PubMed] [Google Scholar]
- 18.Lai, A. L., and L. K. Tamm. 2007. Locking the kink in the influenza hemagglutinin fusion domain structure. J. Biol. Chem. 282:23946-23956. [DOI] [PubMed] [Google Scholar]
- 19.Leneva, I. A., R. J. Russell, Y. S. Boriskin, and A. J. Hay. 2009. Characteristics of arbidol-resistant mutants of influenza virus: implications for the mechanism of anti-influenza action of arbidol. Antiviral Res. 81:132-140. [DOI] [PubMed] [Google Scholar]
- 20.Luo, G., R. Colonno, and M. Krystal. 1996. Characterization of a hemagglutinin-specific inhibitor of influenza A virus. Virology 226:66-76. [DOI] [PubMed] [Google Scholar]
- 21.Luo, G., A. Torri, W. E. Harte, S. Danetz, C. Cianci, L. Tiley, S. Day, D. Mullaney, K. L. Yu, C. Ouellet, P. Dextraze, N. Meanwell, R. Colonno, and M. Krystal. 1997. Molecular mechanism underlying the action of a novel fusion inhibitor of influenza A virus. J. Virol. 71:4062-4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matrosovich, M., T. Matrosovich, J. Carr, N. A. Roberts, and H. D. Klenk. 2003. Overexpression of the alpha-2,6-sialyltransferase in MDCK cells increases influenza virus sensitivity to neuraminidase inhibitors. J. Virol. 77:8418-8425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuda, T., and C. L. Cepko. 2004. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc. Natl. Acad. Sci. U. S. A. 101:16-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDonald, I. K., and J. M. Thornton. 1994. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 238:777-793. [DOI] [PubMed] [Google Scholar]
- 25.Mohamadi, F., N. G. J. Richards, W. C. Guida, R. Liskamp, M. Lipton, C. Caufield, G. Chang, T. Hendrickson, and W. C. Still. 1990. MacroModel—an integrated software system for modeling organic and bioorganic molecules using molecular mechanics. J. Comput. Chem. 11:440-467. [Google Scholar]
- 26.Morris, G. M., D. S. Goodsell, R. S. Halliday, R. Huey, W. E. Hart, R. K. Belew, and A. J. Olson. 1998. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 19:1639-1662. [Google Scholar]
- 27.Moscona, A. 2008. Medical management of influenza infection. Annu. Rev. Med. 59:397-413. [DOI] [PubMed] [Google Scholar]
- 28.Naesens, L., E. Vanderlinden, E. Roth, J. Jeko, G. Andrei, R. Snoeck, C. Pannecouque, E. Illyes, G. Batta, P. Herczegh, and F. Sztaricskai. 2009. Anti-influenza virus activity and structure-activity relationship of aglycoristocetin derivatives with cyclobutenedione carrying hydrophobic chains. Antiviral Res. 82:89-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pettersen, E. F., T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, and T. E. Ferrin. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605-1612. [DOI] [PubMed] [Google Scholar]
- 30.Plotch, S. J., B. O'Hara, J. Morin, O. Palant, J. LaRocque, J. D. Bloom, S. A. Lang, Jr., M. J. DiGrandi, M. Bradley, R. Nilakantan, and Y. Gluzman. 1999. Inhibition of influenza A virus replication by compounds interfering with the fusogenic function of the viral hemagglutinin. J. Virol. 73:140-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reed, M. L., O. A. Bridges, P. Seiler, J. K. Kim, H. L. Yen, R. Salomon, E. A. Govorkova, R. G. Webster, and C. J. Russell. 2010. The pH of activation of the hemagglutinin protein regulates H5N1 influenza virus pathogenicity and transmissibility in ducks. J. Virol. 84:1527-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reed, M. L., H. L. Yen, R. M. DuBois, O. A. Bridges, R. Salomon, R. G. Webster, and C. J. Russell. 2009. Amino acid residues in the fusion peptide pocket regulate the pH of activation of the H5N1 influenza virus hemagglutinin protein. J. Virol. 83:3568-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richards, F. M. 1977. Areas, volumes, packing and protein structure. Annu. Rev. Biophys. Bioeng. 6:151-176. [DOI] [PubMed] [Google Scholar]
- 34.Root, C. N., E. G. Wills, L. L. McNair, and G. R. Whittaker. 2000. Entry of influenza viruses into cells is inhibited by a highly specific protein kinase C inhibitor. J. Gen. Virol. 81:2697-2705. [DOI] [PubMed] [Google Scholar]
- 35.Russell, R. J., S. J. Gamblin, L. F. Haire, D. J. Stevens, B. Xiao, Y. Ha, and J. J. Skehel. 2004. H1 and H7 influenza haemagglutinin structures extend a structural classification of haemagglutinin subtypes. Virology 325:287-296. [DOI] [PubMed] [Google Scholar]
- 36.Russell, R. J., P. S. Kerry, D. J. Stevens, D. A. Steinhauer, S. R. Martin, S. J. Gamblin, and J. J. Skehel. 2008. Structure of influenza hemagglutinin in complex with an inhibitor of membrane fusion. Proc. Natl. Acad. Sci. U. S. A. 105:17736-17741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salomon, R., J. Franks, E. A. Govorkova, N. A. Ilyushina, H. L. Yen, D. J. Hulse-Post, J. Humberd, M. Trichet, J. E. Rehg, R. J. Webby, R. G. Webster, and E. Hoffmann. 2006. The polymerase complex genes contribute to the high virulence of the human H5N1 influenza virus isolate A/Vietnam/1203/04. J. Exp. Med. 203:689-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmidt, M. W., K. K. Baldridge, J. A. Boatz, S. T. Elbert, M. S. Gordon, J. H. Jensen, S. Koseki, N. Matsunaga, K. A. Nguyen, S. Su, T. L. Windus, M. Dupuis, and J. A. Montgomery. 1993. General atomic and molecular electronic structure system. J. Comput. Chem. 14:1347-1363. [Google Scholar]
- 39.Schunemann, H. J., S. R. Hill, M. Kakad, R. Bellamy, T. M. Uyeki, F. G. Hayden, Y. Yazdanpanah, J. Beigel, T. Chotpitayasunondh, C. Del Mar, J. Farrar, T. H. Tran, B. Ozbay, N. Sugaya, K. Fukuda, N. Shindo, L. Stockman, G. E. Vist, A. Croisier, A. Nagjdaliyev, C. Roth, G. Thomson, H. Zucker, and A. D. Oxman. 2007. WHO rapid advice guidelines for pharmacological management of sporadic human infection with avian influenza A (H5N1) virus. Lancet Infect. Dis. 7:21-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shinde, V., C. B. Bridges, T. M. Uyeki, B. Shu, A. Balish, X. Xu, S. Lindstrom, L. V. Gubareva, V. Deyde, R. J. Garten, M. Harris, S. Gerber, S. Vagasky, F. Smith, N. Pascoe, K. Martin, D. Dufficy, K. Ritger, C. Conover, P. Quinlisk, A. Klimov, J. S. Bresee, and L. Finelli. 2009. Triple-reassortant swine influenza A (H1) in humans in the United States, 2005-2009. N. Engl. J. Med. 360:2616-2625. [DOI] [PubMed] [Google Scholar]
- 41.Skehel, J. J., P. M. Bayley, E. B. Brown, S. R. Martin, M. D. Waterfield, J. M. White, I. A. Wilson, and D. C. Wiley. 1982. Changes in the conformation of influenza virus hemagglutinin at the pH optimum of virus-mediated membrane fusion. Proc. Natl. Acad. Sci. U. S. A. 79:968-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skehel, J. J., and D. C. Wiley. 2002. Influenza haemagglutinin. Vaccine. 20(Suppl. 2):S51-S54. [DOI] [PubMed] [Google Scholar]
- 43.Steinhauer, D. A., J. Martin, Y. P. Lin, S. A. Wharton, M. B. Oldstone, J. J. Skehel, and D. C. Wiley. 1996. Studies using double mutants of the conformational transitions in influenza hemagglutinin required for its membrane fusion activity. Proc. Natl. Acad. Sci. U. S. A. 93:12873-12878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steinhauer, D. A., S. A. Wharton, J. J. Skehel, D. C. Wiley, and A. J. Hay. 1991. Amantadine selection of a mutant influenza virus containing an acid-stable hemagglutinin glycoprotein: evidence for virus-specific regulation of the pH of glycoprotein transport vesicles. Proc. Natl. Acad. Sci. U. S. A. 88:11525-11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ur, F., N. Cesur, S. Birteksoz, and G. Otuk. 2004. Synthesis of some new 6-methylimidazo[2,1-b]thiazole-5-carbohydrazide derivatives and their antimicrobial activities. Arzneimittelforschung 54:125-129. [DOI] [PubMed] [Google Scholar]
- 46.van der Vries, E. B. van den Berg, and M. Schutten. 2008. Fatal oseltamivir-resistant influenza virus infection. N. Engl. J. Med. 359:1074-1076. [DOI] [PubMed] [Google Scholar]
- 47.Wallace, A. C., R. A. Laskowski, and J. M. Thornton. 1995. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 8:127-134. [DOI] [PubMed] [Google Scholar]
- 48.Wilson, I. A., J. J. Skehel, and D. C. Wiley. 1981. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 289:366-373. [DOI] [PubMed] [Google Scholar]
- 49.Yoshimoto, J., M. Kakui, H. Iwasaki, T. Fujiwara, H. Sugimoto, and N. Hattori. 1999. Identification of a novel HA conformational change inhibitor of human influenza virus. Arch. Virol. 144:865-878. [DOI] [PubMed] [Google Scholar]