Abstract
The entry mechanism of hepatitis B virus (HBV) has not been defined, and this impedes development of antiviral therapies aimed at an early step in the viral life cycle. HBV infection has both host and tissue specificities. For the related duck hepatitis B virus (DHBV), duck carboxypeptidase D (DCPD) has been proposed as the species-specific docking receptor, while glycine decarboxylase (DGD) may serve as a tissue-specific cofactor or secondary receptor. DGD binds to several truncated versions of the viral large envelope protein but not to the full-length protein, suggesting a need for proteolytic cleavage of the envelope protein by a furin-like proprotein convertase. In the present study, we found that transfected DCPD could confer DHBV binding to non-duck cell lines but that this was followed by rapid virus release from cells. Coexpression of furin led to DCPD cleavage and increased virus retention. Treatment of DHBV particles with endosome prepared from duck liver led to cleavage of the large envelope protein, and such viral preparation could generate a small amount of covalently closed circular DNA in LMH cells, a chicken hepatoma cell line resistant to DHBV infection. A furin inhibitor composed of decanoyl-RVKR-chloromethylketone blocked endosomal cleavage of the large envelope protein in vitro and suppressed DHBV infection of primary duck hepatocytes in vivo. These findings suggest that furin or a furin-like proprotein convertase facilitates DHBV infection by cleaving both the docking receptor and the viral large envelope protein.
Hepatitis B virus (HBV) infection has host and tissue specificities. Only humans and other higher primates are susceptible. The liver is the primary target, followed by the kidney and the pancreas (19). The tissue-specific factors and host barriers against HBV infection are still not fully understood, although transcription factors required for HBV gene transcription are enriched in the liver (46). HBV is an enveloped DNA virus. It expresses large (L), middle (M), and small (S) envelope proteins through alternative in-frame translation initiation sites within the envelope gene. Inside the envelope is the core particle that encloses the viral genome and DNA polymerase. Following virus entry into hepatocytes, the viral genome of 3.2 kb is translocated to the nucleus, where it is converted from a partially double-stranded molecule into a covalently closed circle, or the ccc DNA. The ccc DNA has a minichromosome structure and serves as the template for transcription of viral messenger RNAs, which direct the translation of core, envelope, and polymerase proteins. One particular transcript, the pregenomic RNA, is packaged into newly assembled core protein particles together with polymerase, where it serves as the template for reverse transcription into the minus DNA strand. Further degradation of RNA template and initiation of plus-strand-DNA synthesis lead to virion formation and secretion (15).
The late steps in the HBV life cycle, including transcription, translation, replication, and virus secretion, have been studied extensively, because these steps can be reproduced following transfection of human hepatoma cell lines with a functional equivalent of ccc DNA, such as vector-linked tandem dimers. However, the early events in the viral life cycle, including entry, uncoating, nuclear transport of the viral genome, and genome repair, have been difficult to study due to the lack of a convenient cell culture system of HBV infection. Indeed, the HBV receptor remains enigmatic despite decades of extensive research by numerous investigators. In this regard, HBV-like viruses have been found in ducks (34), woodchucks, and ground squirrels. These agents resemble the human virus with respect to genome organization, protein composition, replication strategy, and host/tissue specificities. Together with HBV, they form the family of hepatotropic DNA viruses, or hepadnaviridae (15). The duck hepatitis B virus (DHBV) represents a convenient small-animal model to study the early events in the hepadnavirus life cycle because of the ease of performance of infection studies both in vivo using ducklings and in vitro using primary duck hepatocytes (PDH). A chicken hepatoma cell line called LMH supports DHBV genome replication and virion formation upon transfection with tandem dimers of the DHBV genome, but it is resistant to DHBV infection. DHBV has just two envelope proteins, L and S, with the pre-S domain of the L protein involved in receptor binding. We and the Ganem group have independently identified and cloned duck carboxypeptidase D (DCPD), a Golgi network-resident protein that shuttles to and from the cell surface, as a binding partner for the pre-S domain of the L protein (4, 11, 24, 25, 48, 54). Consistent with the host specificity of DHBV infection, the DHBV L protein has no affinity for chicken or human carboxypeptidase D (43). On the other hand, DCPD distribution is not restricted to DHBV-susceptible tissues. Transfer of DCPD into human cell lines conferred DHBV binding and endocytosis, confirming the role of DCPD as a DHBV docking receptor (4, 47, 52). However, active viral replication did not occur, raising the possibility that additional cofactors are necessary for the establishment of productive DHBV infection.
While mapping the DCPD binding site using deletion mutants of the L protein, we accidentally identified a 120-kDa duck protein (p120) as a binding partner for the truncated L protein (29). Cloning and sequencing revealed p120 as the P protein of duck glycine decarboxylase (DGD) (28). In contrast to the selective binding of the full-length L protein to duck but not chicken carboxypeptidase D, truncated versions of the L protein could bind to the chicken glycine decarboxylase as well (29) (our unpublished observation). However, DGD is expressed only in tissues that can be infected by DHBV (i.e., liver, kidney, and pancreas) (19, 28, 29). Moreover, blocking DGD expression by antisense RNA or promoting its degradation via its binding to antibodies impaired productive DHBV infection in PDH (27). Thus, DGD represents a tissue-specific host factor essential for the establishment of productive DHBV infection. DGD binds with high affinity to several truncated forms of the pre-S domain, such as those at positions 92 to 161, 98 to 161, and 1 to 102. The minimal binding site was mapped to a linear sequence of 5 amino acids (aa), 98EAFRR102 (29). Interestingly, the sequence around residue Arg102 (97REAFRRY103) is compatible with the recognition motif for furin and PC7, members of calcium-dependent serine proteases that cleave protein precursors at positively charged amino acids to produce biologically active products (1, 7). Furin is ubiquitously expressed, with an intracellular localization in the trans-Golgi network (TGN), the endosome, and the cell surface (6, 9, 42, 53, 55). In the present study, we investigated the abilities of furin and duck liver endosomal fractions to cleave DCPD and/or L protein, as well as the consequence of such cleavage for DHBV retention or formation of the ccc DNA in LMH cells. The effect of a furin inhibitor on DHBV infection of primary duck hepatocytes (PDH) was also investigated.
MATERIALS AND METHODS
Reagents.
The DHBV genome F16 was cloned into the EcoRI site of the pUC18 vector as a tandem dimer. DCPD and DGD cDNA were cloned into the pcDNA3.1/Zeo vector as well as into the adenovirus vector (20, 27). To render DCPD resistant to furin cleavage, the RLGR sequence at positions 166 to 169 (47, 48) was converted to ALGA by overlap extension PCR (50). The PCR product was digested with XbaI and HindIII and used to replace the cognate fragment of the wild-type clone. The introduced mutations were confirmed by DNA sequencing. A fusion construct between glutathione S-transferase (GST) and the pre-S domain of DHBV L protein has been described previously (29, 48). The expression construct for chicken furin and the corresponding polyclonal rabbit antibody were kindly provided by Anke Feldmann, and the catalytic mutants were constructed by overlap extension PCR (12, 50). The human furin expression construct was purchased from Origene (insertion of 6 nt) with additional modifications (21). In-house polyclonal antibodies against the core protein, the pre-S domain of the DHBV L protein, DCPD, and DGD have been described previously (27, 28, 47, 48). The monoclonal β-actin antibody was purchased from Sigma. Proprotein convertase and carboxypeptidase D inhibitors were purchased from or customer synthesized by Bachem Americas, Inc. (Torrance, CA). They include H-RRRRRR-OH (6×Arg) (PC7), decanoyl-RVKR-chloromethylketone (furin), phenylmethylsulfonyl fluoride (PMSF) (serine protease), dynorphin A (carboxypeptidase D), and Met-enkephalin-KK (carboxypeptidase D).
Purification of DHBV particles.
A DHBV viremic duck serum containing approximately 1011 virus particles/ml was overlaid on top of 10% and 20% sucrose cushions and centrifuged with an SW41 rotor at 39,000 rpm for 18 h. The pellet was resuspended in TEN buffer (20 mM Tris-HCl, pH 7.4, 1 mM EDTA, 150 mM NaCl) in one-fourth of the original volume and stored at 4°C for endosomal cleavage or infection experiments.
Isolation of endosome.
Endosomal fractions were enriched using a one-step flotation gradient (16), with minor modifications. Briefly, duck liver (1 g) was homogenized in 5 ml of homogenization buffer (20 mM HEPES, pH 5.5, 0.25 M sucrose, 1 mM EDTA, and 1 mM dithiothreitol [DTT]). After centrifugation at 4,000 rpm for 10 min, the postnuclear supernatant was adjusted to 44% (1.3 M) sucrose by being mixed with 70% sucrose, 1 mM EDTA, pH 8.0, at a ratio of 2:3. The sample was loaded into a 12-ml SW41 centrifugation tube and overlaid with 3.6 ml of 37.6% (1.1 M) sucrose, 2.4 ml of 20.5% (0.6 M) sucrose, and 0.5 ml of homogenization buffer or 0.25 M sucrose. After centrifugation at 4°C and 30,000 rpm for 3 h, fractions were collected from the top. Endosome-enriched fractions were identified by Western blot analysis.
In vitro cleavage assay.
About 1 to 3 μl of purified virus particles (3 × 108/μl) was incubated at 37°C for 3 h with the endosomal fraction (5 to 12 μl) in a buffer containing 20 mM HEPES, pH 5.5, 5 mM MgCl2, and 3 mM CaCl2 or as indicated in the figure legends. A 1/10 volume of the sample was used to verify envelope protein cleavage by Western blot analysis, while the remainder was used for infection of one well of LMH cells (∼3 × 105) grown in 6-well plates. Alternatively, the pre-S domain of the DHBV L protein expressed as a GST fusion protein was purified from 40 ml of LB culture using a 50-μl bed volume of glutathione Sepharose beads as described previously (29). The immobilized proteins were digested at room temperature (RT) for 2 h with 3 U of thrombin, and the 161-aa pre-S peptide thus released was recovered from the supernatant following low-speed centrifugation. The pre-S peptide (2 μl of 1:50 diluted in cleavage buffer) was incubated with 2 μl of endosomal fraction F3 or cell lysate at 37°C for 3 h.
Gene transduction.
Bosc cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1% l-glutamine, and 1% penicillin-streptomycin and transfected by the calcium phosphate precipitation method or the polyamine method (TransIT; Mirus). LMH cells were cultured in a 1:1 mixture of DMEM and F-12 medium and transfected by the polyamine method. HepG2 cells were cultured in minimal essential medium (MEM) supplemented with 1% nonessential amino acids and transfected by either the calcium phosphate precipitation method or the polyamine method. Primary mouse hepatocytes (PMH) were prepared from 4-week-old mice (FVB) and cultured under the same conditions as were PDH. DCPD and DGD were introduced into PMH by adenovirus-mediated gene transduction (27).
Infection of primary duck hepatocytes.
PDH were prepared from 3-day-old DHBV-free ducklings by a two-step perfusion procedure as described previously (27, 48, 51). Cells were seeded in 6-well plates at approximately 90% confluence by using L15 medium supplemented with 5% fetal bovine serum, 5 μg/ml human recombinant insulin (Novo Nordisk), and 5 × 10−5 M hydrocortisone sodium succinate (Pharmacia). After an overnight incubation at 37°C, cells were maintained in serum-free medium supplemented with 1% dimethyl sulfoxide (DMSO). Cells were infected with 5 μl of viremic duck serum (∼5 × 108 virus particles) overnight in the presence or absence of the furin inhibitor and harvested 7 days later.
DHBV binding assay.
DCPD-transfected cells seeded in 6-well plates were incubated 2 days later in replicate with a 1:10 dilution of viremic duck serum for several hours, followed by an extensive wash with serum-free medium. One well of cells was harvested using cell scrapers, and cells were collected by low-speed centrifugation. The cell pellet was incubated on ice with 100 μl of lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate). Nuclei were removed by centrifugation, and the lysate was used for Southern blot analysis of DHBV DNA or Western blot analysis of the DHBV L protein. Other wells of cells were harvested 2, 4, or 6 days later, with interim medium changes for cells harvested at days 4 and 6. Culture medium for cells harvested at day 2 and combined culture medium for cells harvested at later time points were subjected to ultracentrifugation with an SW41 rotor at 39,000 rpm for 16 h. The pelleted virus particles were used for Southern and Western blot analyses.
Western blot analysis.
Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using a 12% or 15% separating gel for endosome digested DHBV envelope proteins, a 12% gel for undigested L protein and β-actin, and an 8% gel for DCPD, DGD, and furin. A shorter duration of gel electrophoresis facilitates the visualization of the cleavage products, whereas longer running helps the separation of the native forms of L protein (p35 and p28). Proteins were transferred to polyvinylidene difluoride (PVDF) membranes, which were blocked at RT for 3 h with 3% bovine serum albumin (BSA) dissolved in phosphate-buffered saline containing 0.05% Tween 20 (PBST). The blots were incubated at 4°C overnight with rabbit polyclonal antibodies against the viral pre-S domain, DCPD, or DGD (27, 28, 47) diluted 1:10,000 in 3% BSA-PBST, washed with PBST, and subsequently incubated at RT for 1 h with a 1:40,000 dilution of goat anti-rabbit Ig conjugated with horseradish peroxidase (HRP). After an additional wash with PBST, the blots were incubated with enhanced chemiluminescence substrate (PerkinElmer) for 1 min, followed by exposure to X-ray films. For detection of furin and β-actin, 3% skimmed milk instead of 3% BSA was used for blocking, and the antibody dilutions were as follows: 1:20,000 for the mouse monoclonal antibody against β-actin, 1:2,000 for the rabbit polyclonal antibody against furin, and 1:40,000 for HRP-conjugated rabbit anti-mouse Ig.
Southern blot analysis of total DHBV DNA.
Cells cultured in 6-well plates were harvested using cell scrapers and collected by low-speed centrifugation. Cell pellets were digested at 37°C for 3 h with proteinase K (0.5 mg/ml) in 400 μl of TEN buffer (10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 150 mM NaCl) supplemented with 0.5% SDS. Following phenol extraction and ethanol precipitation, DNA was separated in a 1.2% agarose gel. DHBV DNA was detected by Southern blot analysis using a random primed probe. Signals were revealed by autoradiography.
RCA of ccc DNA.
Protein-free DNA was extracted from the cell pellet or homogenized duck liver as described previously (56). Briefly, approximately 3 × 106 to 5 × 106 cells were incubated at 37°C for 5 min in 0.5 ml of ccc DNA isolation buffer (10 mM Tris-HCl, pH 7.5, 10 mM EDTA, 1% SDS). After the addition of 125 μl of 2.5 M KCl, the lysate was vortexed and chilled on ice for 5 min. The detergent-protein complex was removed by centrifugation at 10,000 rpm for 5 min. The supernatant was extracted with phenol, and DNA was precipitated with ethanol. The DNA pellet was dissolved in 10 μl of solution containing 1× Phi29 buffer and 10 pmol each of the sense primer TGTGTATGATCTACC and the antisense primer CCAGTGATTCCTCGT, with the two 3′ bases modified with phosphorothioate. The DNA/primer mixture was denatured at 95°C for 3 min, cooled down sequentially at 50°C for 15 s, 30°C for 15 s, and room temperature for 10 min, and placed on ice. The annealing product was combined with 10 μl of solution containing 1× Phi29 buffer, 1 mM deoxynucleoside triphosphate (dNTP), 10 pmol each of the sense and antisense primers, 0.5 mg/ml BSA, and 5 U of the Phi29 DNA polymerase (New England Biolabs, Worcester, MA), followed by DNA amplification at 30°C for 16 h. The reaction was terminated by heating the samples at 65°C for 10 min. The rolling circle amplification (RCA) product, consisting of concatemers of the full-length DHBV genome, was converted to the 3.0-kb monomeric form by digestion with EcoRI, which has a single recognition site in the DHBV genome. The digest was electrophoresed in a 1.2% agarose gel and detected by Southern blot analysis using randomly labeled DHBV DNA as a probe.
RESULTS
DHBV binding to and release from DCPD-reconstituted Bosc cells.
Reconstitution of several liver- or non-liver-derived cell lines with DCPD did not lead to productive DHBV infection despite efficient virus binding. Time course studies revealed that internalized (trypsin-resistant) DHBV particles maintained the same ratio of viral genome/L protein as did the inoculum, suggesting internalization of intact virions rather than core particles (data not shown). Moreover, as exemplified by experiments with Bosc cells, a human kidney cell line, cell-associated DHBV signals decreased rapidly following withdrawal of the inoculum. This reduction was accompanied by increased DHBV particles in the culture supernatant (Fig. 1A). Since DCPD shuttles between the TGN and the cell surface (4, 11), such a dynamic redistribution of DHBV signals raises the possibility that endocytosed DHBV particles were reexported during DCPD trafficking from the TGN back to the cell surface. Considering that furin, the basic endopeptidase, has a very similar subcellular localization and trafficking pattern (22, 35), we explored the effect of furin coexpression.
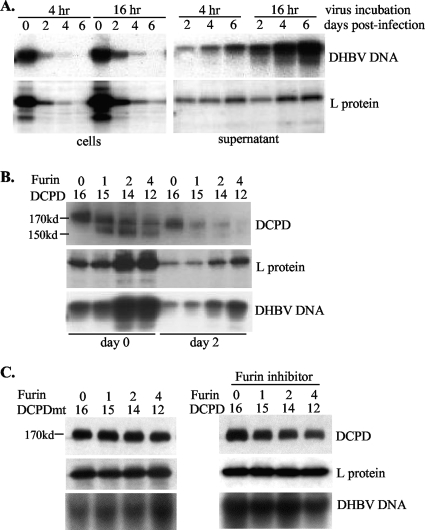
FIG. 1.
Kinetics of DHBV binding to and release from DCPD-reconstituted Bosc cells. (A) The rapid decline of cell-associated DHBV particles was due to their release to the culture supernatant. Bosc cells transiently transfected with DCPD cDNA (day 2 posttransfection) were incubated in quadruplicate with a 1:10 dilution of viremic duck serum for the indicated times. After an extensive wash, cells were harvested immediately (day 0) by scraping for analysis of DHBV DNA and viral L protein. For the remaining wells, cells and culture supernatant were harvested 2, 4, or 6 days later for DHBV DNA and protein analyses. Virus particles were concentrated from the culture supernatant by ultracentrifugation. (B) Cotransfection with chicken furin cDNA increased DHBV binding or retention. Bosc cells grown in 6-cm dishes were transfected in duplicate with furin and DCPD cDNAs at the indicated ratios and incubated with a 1:10 dilution of viremic duck serum for 15 h. After an extensive wash, cells were harvested immediately or 2 days later. Cell-associated DHBV DNA and L envelope protein were analyzed. (C) (Left) Furin failed to cleave a DCPD mutant (DCPDmt) in Bosc cells. A transfection/infection experiment similar to that described for panel B was performed, except that the 166RLGR169 motif of DCPD was mutated to ALGA. (Right) Impact of the furin inhibitor (decanoyl-RVKR-chloromethylketone) on DHBV binding/retention. The inhibitor (10 μM) was added immediately after transfection with chicken furin and wild-type DCPD. DCPD expression and viral L protein and DNA were analyzed.
Furin could cleave DCPD except in LMH cells.
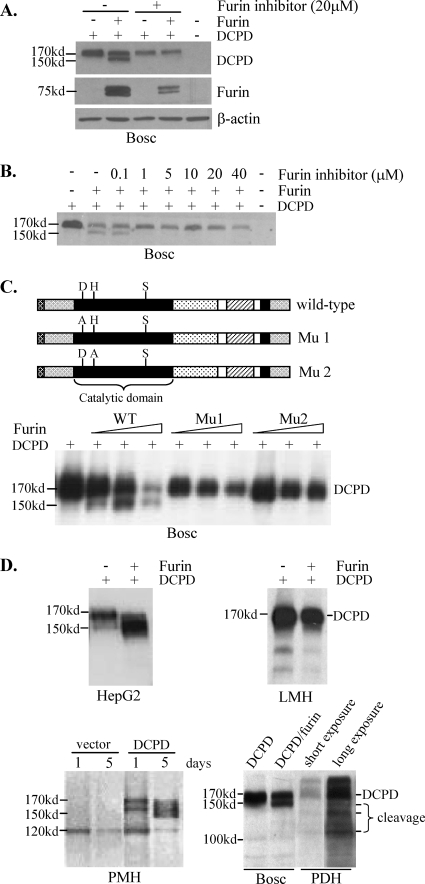
In Bosc cells transfected with DCPD cDNA alone, a full-length DCPD of about 170 kDa was detected. Cotransfection with chicken furin cDNA produced an additional band of around 155 kDa (Fig. 1B and 2A). This represented a DCPD cleavage product because it could be prevented by treatment of the transfected cells with decanoyl-RVKR-chloromethylketone, a potent furin inhibitor. As little as 1 μM inhibitor after transfection was sufficient to completely block the cleavage (Fig. 2B). Inspection of the deduced DCPD protein sequence (GenBank accession number AF039749) revealed a consensus sequence for furin cleavage (RXXR) at positions 166 to 169 (47, 48). Cleavage at this site would remove the N-terminal 169 residues to generate a truncated protein of approximately 151 kDa. Indeed, substitution of the two arginines with alanine completely abolished cleavage of DCPD (Fig. 1C, left). The ability of chicken furin to process DCPD requires its catalytic site (D154, H195, and S409), since either the D154A or the H195A substitution completely prevented cleavage of wild-type DCPD (Fig. 2C). In two human hepatoma cell lines, HepG2 and Huh7, the truncated form of DCPD was detectable even when DCPD was transfected alone, although coexpression of chicken furin further enriched the cleavage product at the expense of the full-length molecule (Fig. 2D, top left) (data not shown). In primary mouse hepatocytes (PMH) transduced with DCPD alone, a large fraction of DCPD protein was cleaved (Fig. 2D, bottom left). Similarly, significant cleavage of endogenous DCPD was also detected in PDH (Fig. 2D, bottom right) or duck liver (data not shown). These findings are consistent with the presence of endogenous furin in liver-derived cell lines and its abundance in normal hepatocytes. Surprisingly, no cleaved form of DCPD was detectable in LMH cells, even in the presence of exogenous chicken furin (Fig. 2D, top right). This observation suggests the presence of a furin inhibitor in the chicken hepatoma cell line.
FIG. 2.
DCPD cleavage by chicken furin or endogenous furin-like PC. (A) Ability of chicken furin to cleave DCPD in Bosc cells. Cells grown in 6-well plates were cotransfected with 0.5 μg each of DCPD cDNA and the pcDNA3.1/Zeo− vector or chicken furin cDNA. The furin inhibitor (decanoyl-RVKR-chloromethylketone) was added immediately after transfection. Cells were harvested 2 days later for Western blot analysis of DCPD, chicken furin, and β-actin. (B) Minimum concentration of the furin inhibitor required to block DCPD cleavage. (C) Critical role of catalytic sites of chicken furin for DCPD processing. D197, H235, and S409 in the catalytic domain (black box) form the catalytic triad. Mu1 and Mu2 contain D197A and H235A substitutions, respectively. Bosc cells in 6-well plates were either transfected with 1 μg of DCPD cDNA alone or cotransfected with DCPD and chicken furin cDNA (wild type [WT] or mutant) at a 1:16, 1:8, or 1:4 ratio, with the total amount of DNA kept at 1 μg. Cells were harvested 2 days later for DCPD detection. (D) DCPD cleavage in different cell types in the presence or absence of exogenous furin. DCPD and chicken furin cDNAs were delivered to HepG2, LMH, and Bosc cells by transient transfection and to primary mouse hepatocytes (PMH) by the adenovirus vector. The endogenous DCPD pattern in PDH was included for comparison (lower right).
DCPD cleavage enhances DHBV retention.
To determine the functional consequence of DCPD cleavage, we cotransfected Bosc and HepG2 cells with DCPD together with increasing doses of chicken furin cDNA, followed by incubation with DHBV viremic duck sera. Interestingly, furin coexpression increased DHBV binding and/or retention in both Bosc cells (Fig. 1B) and HepG2 cells (data not shown), despite the fact that the total DCPD levels (full-length plus cleaved) were actually reduced since less DNA was transfected. Considering that furin alone failed to confer any DHBV binding (data not shown), our findings suggest that furin-mediated DCPD cleavage improves DHBV binding or reduces viral exit. On the other hand, cotransfection of chicken furin with the DCPD mutant resistant to cleavage failed to increase DHBV DNA signals (Fig. 1C, left). Similarly, furin also failed to modulate DHBV signals when cleavage of the wild-type DCPD was prevented by its inhibitor (Fig. 1C, right).
Colocalization of DCPD, DGD, and DHBV large envelope protein in the endosomal fraction.
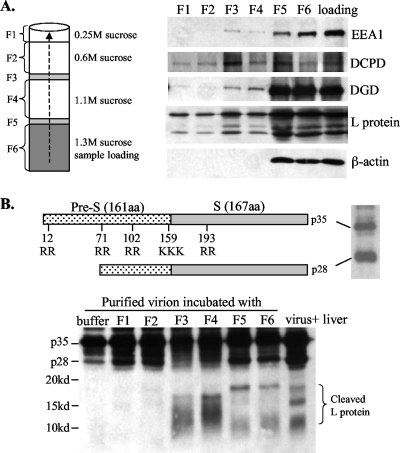
Furin is present on the cell surface, in the TGN, and in the endosome (1). Internalized DHBV particles were found translocated to the early and late endosomes of duck hepatocytes, as evidenced by colocalization with transferrin (8). Blocking endosomal trafficking abolished viral infectivity (8), suggesting a possible role of the endosome in membrane fusion and uncoating. To this end, endosome enriched fractions were isolated from Pekin duck liver using an established method. Briefly, duck liver homogenate was adjusted to a high concentration of sucrose and added to the bottom of the centrifugation tube. This was overlaid with three additional cushions with decreasing sucrose concentrations. After ultracentrifugation to form a sucrose gradient, individual fractions were collected from the top (Fig. 3A, left) (F3 and F5 showed visible protein bands). Western blot analysis revealed that EEA1, an early endosome marker, reached up to the F3 fraction (Fig. 3A). Thus, F3 contained relatively pure endosome, with less contamination with other proteins than F4 and F5. Interestingly, DCPD, DGD, and DHBV L protein were all detectable in this fraction, suggesting colocalization (Fig. 3A).
FIG. 3.
(A) Colocalization of DCPD, DGD, and viral envelope proteins in the endosomal fraction of infected duck liver. Duck liver was homogenized and mixed with sucrose solution to achieve a final concentration of 1.3 M. The mixture was loaded into the bottom of the centrifugation tube and overlaid with three additional layers of sucrose solutions. After ultracentrifugation, six fractions were collected from the bottom (note that F3 and F5 corresponded to two narrow visible protein bands). A 5-μl aliquot from each fraction was used for Western blot analysis of DCPD, DGD, and β-actin as well as EEA1, an early endosome marker (53). The sample prior to centrifugation (loading) was analyzed in parallel. (B) Cleavage of DHBV L protein by endosomal resident proteases. Shown at the top are the schematic representations of DHBV L protein (p35), its processed form (p28), and dibasic or tribasic residues. Approximately 3 × 107 purified virus particles were incubated at 37°C for 3 h with 2 μl of endosomal fractions (F1 to F6) derived from the liver of a DHBV-free duck or just with the digestion buffer. The cleavage product was resolved by 15% PAGE and detected by Western blot analysis with a polyclonal antibody against the pre-S domain. Liver lysate from a DHBV-infected duck served as a control. Note that short electrophoresis was performed to reveal <20-kDa cleavage products.
Cleavage of DHBV pre-S protein by a furin-like endosomal protease.
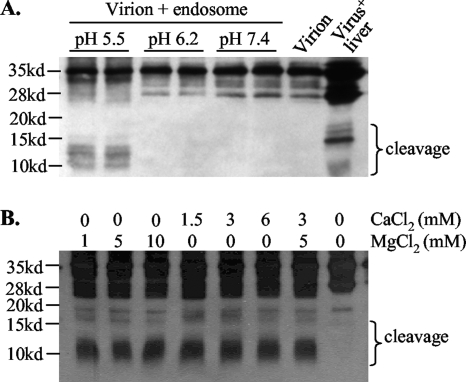
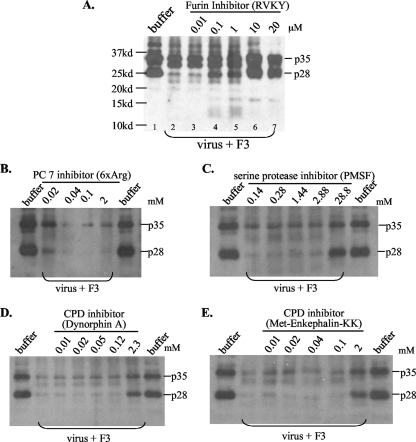
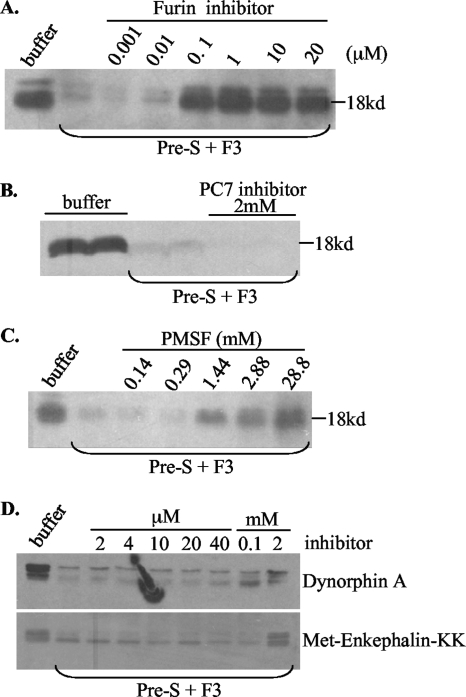
We next examined whether furin-like PCs present in the endosome could cleave DHBV envelope proteins. In this regard, the viremic duck serum contains not only the full-length L protein of 35 kDa (p35) but also a 28-kDa protein (p28), which probably represents a proteolytic cleavage product lacking the N terminus (13) (Fig. 3B, top). There are five clusters of dibasic or tribasic residues in p35 and four in p28 (Fig. 3B) (32, 49), with the sites in the pre-S domain probably exposed on the virion surface and accessible to proteases. Virus particles purified from viremic duck serum were incubated at 37°C for 3 h with various sucrose gradient fractions. Western blot analysis revealed protease activities from F3 to F6, as indicated by a reduction of p35 and p28 and the concomitant appearance of fast-migrating bands or smears (Fig. 3B). Further experiments were based on F3, the endosomal preparation less contaminated with other organelles. Consistent with the enzymatic properties of furin, cleavage of the DHBV L protein requires a low pH and a divalent cation, such as calcium or magnesium (Fig. 4). While both p35 and p28 could be processed, p28 seemed more accessible to the protease (Fig. 5A, compare lanes 1 and 2). Of the peptide-based protease inhibitors against furin, PC7, CPD, and serine proteases, only the furin inhibitor efficiently blocked cleavage of pre-S proteins. As little as 10 μM furin inhibitor completely prevented cleavage of both p35 and p28 (Fig. 5A). The effects seen at the high doses of PMSF (28.8 mM), dynorphin (2.3 mM), and M-enkephalin-KK (2 mM) (Fig. 5C to E) were probably due to the toxicity of DMSO used as the solvent. These data implicate furin or furin-like protease as a key endopeptidase responsible for cleavage of the DHBV L protein. To confirm the presence of a cleavage site(s) in the pre-S domain, we expressed the 161-aa pre-S domain as a GST fusion protein, followed by its release via thrombin digestion. Indeed, the endosome-associated enzyme(s) could efficiently cleave the pre-S domain (as suggested by reduced levels of the original 18-kDa polypeptide), which could be blocked by a low dose of the inhibitor for furin but not carboxypeptidase D or serine proteases (Fig. 6).
FIG. 4.
Requirement of low pH and divalent cations for endosomal cleavage of DHBV L protein. The F3 fraction was used as the source of the endosome. (A) The pH of the digestion buffer was adjusted to 5.5, 6.2, and 7.4. Untreated virions served as a negative control, while DHBV (+) liver lysate served as a positive control. The cleavage products are indicated. (B) The MgCl2 and CaCl2 concentrations were altered as indicated. Note that proteins were separated by 15% PAGE with short electrophoresis. For panel B, visualization of the cleavage products of <20 kDa was improved by increased protein loading.
FIG. 5.
Effects of protease inhibitors on endosomal cleavage of DHBV L protein. Purified virus particles (3 × 107) were incubated with 2 to 3 μl of endosome fraction F3 in 20 mM HEPES, pH 5.5, 5 mM MgCl2, and 3 mM CaCl2, with various protease inhibitors, at 37°C for 3 h. Proteins were separated by 12% PAGE. (A) Decanoyl-RVKR-chloromethylketone as the furin inhibitor. Note that p28 was more accessible to cleavage than p35. (B) H-RRRRRR-OH (6×Arg) as the PC7 inhibitor. (C) PMSF as a broad serine protease inhibitor. (D and E) Dynorphin A and Met-enkephalin-KK as carboxypeptidase (CPD) inhibitors. Note that only the furin inhibitor significantly suppressed cleavage of DHBV L protein at the low concentrations of 0.1 to 10 μM. While cleavage products of <20 kDa are visible in panel A, for the other panels a longer gel electrophoresis was taken and cleavage of L protein was judged by decreased intensities of p35 and p28.
FIG. 6.
Effects of protease inhibitors on cleavage of the pre-S domain by duck liver endosome. The entire 161-aa pre-S domain of the L protein was expressed as a GST fusion protein, purified using glutathione Sepharose beads, and separated from the GST tag by thrombin digestion. The pre-S domain thus released was incubated with the endosome fraction F3 at 37°C for 3 h in 20 mM HEPES, pH 5.5, 5 mM MgCl2, and 3 mM CaCl2, with various protease inhibitors. The 18-kDa pre-S domain was revealed by Western blot analysis after electrophoresis with a 12% gel. (A) Furin inhibitor. (B) PC7 inhibitor. (C) Broad serine protease inhibitor. (D) Two carboxypeptidase inhibitors, dynorphin A (top) and Met-enkephalin-KK (bottom). Note that cleavage of the 18-kDa pre-S peptide was judged by the intensity of the 18-kDa protein that survived digestion, because no cleavage products of <18 kDa could be detected.
Endosome-pretreated DHBV particles gained the ability to generate ccc DNA in LMH cells.
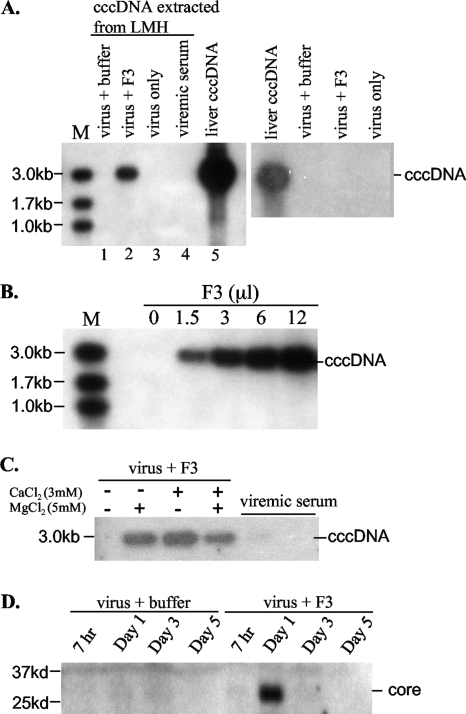
The biological relevance of endosomal cleavage of the DHBV envelope proteins was examined in the chicken hepatoma cell line LMH, which is resistant to DHBV infection but supports DHBV replication following transient transfection (no duck-derived hepatoma cell lines are available). Cultured LMH cells were incubated for 5 to 6 h with DHBV particles pretreated with the endosomal fraction F3. A pivotal event in hepadnaviral infection is the formation of ccc DNA, which is reliant on successful uncoating, capsid disassembly, and genome migration to the nucleus, followed by genome repair. This signals the transition from the early to the late stage of infection. Specific yet highly sensitive detection of ccc DNA was achieved by RCA, a method originally established for the amplification of chromosomal DNA or the circular genome and subsequently applied to the detection of HBV ccc DNA (33, 57). Protein-free DNA was extracted from infected duck liver or LMH cells and subjected to RCA using Phi29 polymerase. After digestion of the concatemeric product with EcoRI, the 3-kb DHBV genome was detected by Southern blot analysis. Control experiments revealed that liver ccc DNA preparation generated large amounts of the 3-kb hybridization signal, whereas purified virions or viremic duck serum did not (Fig. 7A, lanes 3 to 5). As expected, treatment of purified virus particles with endosome F3 or buffer failed to generate a positive RCA signal (Fig. 7A, right). However, incubation of endosome- but not buffer-pretreated virus particles with LMH cells generated the 3-kb product within 24 h (Fig. 7A, lanes 1 and 2). Omission of the divalent cation during endosomal digestion also prevented ccc DNA formation (Fig. 7C). Moreover, the amount of ccc DNA formed was dependent on the dose of endosomes used (Fig. 7B). It was estimated that 1 × 103 to 5 × 103 copies of ccc DNA were generated in 3 × 105 to 6 × 105 LMH cells incubated with 3 × 108 to 9 × 108 F3-pretreated DHBV particles, based on parallel RCA experiments of serially diluted duck liver ccc DNA (data not shown). Besides ccc DNA formation, we could detect viral core protein at day 1 after infection of LMH cells, indicating virus uncoating (Fig. 7D). However, no active viral replication was detected (data not shown), possibly due to the low efficiency of ccc DNA formation and the gradual decline of the ccc DNA signal over time (data not shown). At any rate, cleavage of viral envelope proteins by endosomal enzymes can lead to entry of a small number of DHBV particles into nonsusceptible LMH cells.
FIG. 7.
Incubation of LMH cells with endosome-pretreated DHBV particles led to ccc DNA formation and core protein detection. (A) ccc DNA formation requires endosome treatment and incubation with LMH cells. Purified DHBV particles were pretreated with F3 or just buffer. (Left) Such particles were incubated at 37°C for 5 h with LMH cells. Cells were harvested 24 h later. Protein-free DNA was extracted by phenol, and the ccc DNA was amplified by RCA. The RCA product was digested with EcoRI, and the 3-kb DHBV DNA was detected by Southern blot analysis. The ccc DNA extracted from the DHBV (+) liver served as a positive control, while DNA extracted from viremic duck serum served as a negative control. M, marker. (Right) F3- or buffer-treated virus particles were used directly for DNA extraction followed by RCA. (B) Effects of different doses of the endosome used for digestion of DHBV particles. The pretreated particles were used for incubation with LMH cells, followed by ccc DNA detection by RCA. (C) Effect of divalent cations on ccc DNA formation. Note that a lack of the divalent cation in the digestion buffer prevented ccc DNA formation. (D) Western blot analysis of DHBV core protein from LMH cells incubated with pretreated virus particles. Note that core protein detection in LMH cells requires endosome pretreatment of DHBV particles. The loss of signal at days 3 and 5 postinfection is probably due to degradation of the disassembled core protein.
A furin inhibitor reduced DHBV infection of primary duck hepatocytes.
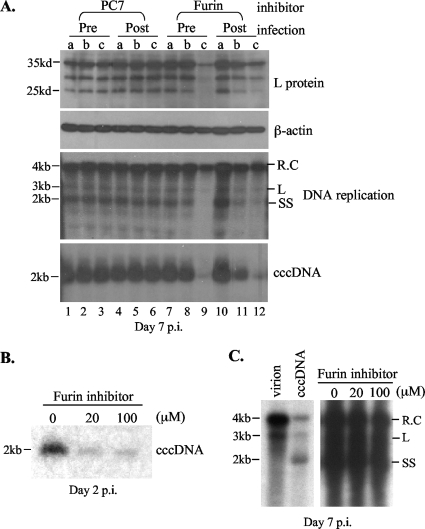
The data presented so far suggest that DCPD can be cleaved by furin and that cleavage of DHBV envelope proteins by a furin-like endosomal protease induces ccc DNA formation in LMH cells. If furin cleavage indeed plays a pivotal role in DHBV entry, then inhibition of its enzymatic activity should prevent DHBV infection. To test this hypothesis, we employed decanoyl-RVKR-chloromethylketone, the furin inhibitor capable of blocking furin cleavage of DCPD (Fig. 2A and B) and endosomal cleavage of the pre-S proteins (Fig. 5 and 6). The PC7 inhibitor was tested in parallel as a negative control. PDH were incubated with DHBV viremic duck serum overnight, washed, and cultured for one more week. Productive viral infection was determined by Southern blot analysis of viral ccc DNA and replicative DNA, as well as Western blot analysis of viral pre-S proteins. Levels of the pre-S proteins and ccc DNA were markedly diminished when 100 μM furin inhibitor was added throughout infection (virus incubation plus 7 days of culture) (Fig. 8A, lane 9). These levels were also affected when 100 or 20 μM inhibitor was added immediately after removal of the inoculum (Fig. 8, lane 12). Both 100 μM and 20 μM inhibitor reduced ccc DNA levels at an earlier stage of infection (day 2) (Fig. 8B). The inhibitor was ineffective if added 2 days after infection (Fig. 8C), suggesting an effect on the initiation of infection rather than virus replication/gene expression per se. Neither 100 μM nor 20 μM inhibitor had any toxic effect on duck hepatocytes, as measured by a cell viability assay (CCK-8 assay; data not shown) or by determination of the β-actin level (Fig. 8A).
FIG. 8.
The furin inhibitor diminished DHBV infection of primary duck hepatocytes. (A) PDH (7 × 105) were incubated with DHBV viremic serum (∼5 × 108 virus particles) overnight, followed by a wash with the medium. Cells were harvested 7 days later (day 7 postinfection [p.i.]). The furin inhibitor (decanoyl-RVKR-chloromethylketone) was added either beginning 12 h prior to incubation with the viremic serum (pre) or immediately following removal of the inoculum (post). DHBV L protein, replicative DNA, and ccc DNA were analyzed. β-Actin served as a control for possible cell toxicity. Lanes a, no inhibitor; lanes b, 20 μM inhibitor; lanes c, 100 μM inhibitor. (B) Southern blot analysis of the ccc DNA at day 2 postinfection. The furin inhibitor was added immediately following withdrawal of the inoculum (same as “post” for panel A). (C) The furin inhibitor was added 2 days after virus infection. Viral replication was detected by Southern blot analysis. The left panel shows the different forms of the viral DNA, including relaxed circular (R.C), linear (L), and single stranded (SS).
DISCUSSION
Envelope proteins of many viruses are cleaved by furin-like PCs. They include HIV gp160, cytomegalovirus glycoprotein B, mouse mammary tumor virus superantigen, influenza virus A hemagglutinin, measles virus F0, Newcastle disease virus F0, Sindbis virus gpE2, and parainfluenza virus F0 (see references 36 and 41 for reviews). In the case of HIV, the cleavage is mediated by the two ubiquitous PCs, furin and PC7 (18). The N-terminal cleavage product (surface, or SU) is involved in receptor binding, whereas the C-terminal part (transmembrane, or TM) mediates membrane fusion. The lack of processed envelope proteins in mature hepadnavirus particles could be explained by the unique mechanism of hepadnaviral morphogenesis. The pre-S domain of the large envelope protein is cytosolically oriented to serve as a matrix protein during capsid envelopment, thus precluding its accessibility to PCs (5, 17, 37, 38, 45). Only at a later stage would the pre-S domain of some large envelope proteins be translocated onto the virion surface, where it serves as the contact site for the viral receptor for the next round of infection. Thus, cleavage activation of the fusogenic peptide of viral envelope proteins may be postponed, such as after binding with a docking receptor, for this class of viruses. Identification of DGD as a binding partner for the truncated pre-S domain of the DHBV L envelope protein and demonstration of its requirement for productive DHBV infection of PDH reinforce this hypothesis (27-29). In the present study, we provide several pieces of evidence to suggest that enzymatic cleavage, possibly by a furin-like PC, plays a critical role in the DHBV life cycle.
First, introduction of DCPD into liver- or kidney-derived human cell lines led to virus binding and internalization in the form of intact virions. However, the bound and/or internalized virus particles were rapidly released to the culture supernatant (Fig. 1A). Furin could cleave DCPD and increase cell-associated DHBV signals (Fig. 1B and 2A). The cleavage site was located at residues 166 to 169 in the N terminus, because a double amino acid substitution prevented cleavage by chicken furin. In this regard, DCPD is composed of three domains, A, B, and C, each bearing similarity to carboxypeptidase N/E. Most of the DCPD molecule is extracellular, except for a single transmembrane segment followed by a cytoplasmic tail composed of the last 58 residues of domain C. Whereas the enzymatic activities reside in domains A and B, domain C mediates DHBV binding (10, 43). At present we do not know the subcellular location where DCPD is cleaved, whether this cleavage occurs prior to DCPD binding to DHBV particles, or whether it increases DHBV binding to domain C or rather slows down shedding of bound and/or internalized DHBV particles. It will be of interest to determine whether a DCPD mutant rendered resistant to furin cleavage acts in a dominant negative manner to interfere with DHBV infection of primary duck hepatocytes.
Second, we found endosomal colocalization of DCPD, DGD, and viral envelope proteins in the duck liver, suggesting the possible involvement of the endosome in switching DHBV binding from DCPD to DGD followed by membrane fusion. Recent studies by others also indicated that DHBV entry depends on trafficking to the endosomal compartment, where fusion is expected to occur (3, 8, 14, 44). Indeed, the endosomal fraction obtained from the susceptible duck liver could cleave DHBV L protein (Fig. 3 to 6), whereas the endosome prepared from the resistant LMH cell line could not (our unpublished observation). The requirements for low pH, divalent cations, and inhibition by decanoyl-RVKR-chloromethylketone are consistent with furin or a furin-like PC being the enzyme involved. At least some of the cleavage sites are located in the pre-S domain (Fig. 3B and Fig. 6), where four dibasic or tribasic residues are found. Since the cleavage product often showed a smearing pattern, possibly due to degradation of the initial cleavage product, the exact cleavage site(s) remains to be established. Generation of DHBV mutants with different sets of the basic residues mutated will help solve this issue. At any rate, such a cleavage event has biological significance, because DHBV particles pretreated with an endosomal enzyme(s) gained the ability to generate a small amount of ccc DNA in LMH cells (Fig. 7). This result suggests that a small proportion of the processed virus particles successfully entered LMH cells and removed their envelopes, followed by nuclear migration of the relaxed circular DNA, removal of the DNA polymerase, and DNA repair and ligation. The RCA method we used to amplify the ccc DNA is specific, as demonstrated by the negative results when virus particles, whether or not pretreated with endosome, were used directly for amplification. It is a major improvement over the previous method amplifying the gap region of the replicative DNA (23).
The relative low efficiency of ccc DNA formation by the endosome-treated DHBV particles in LMH cells could be attributed to many factors, such as the metastable nature of cleaved envelope proteins, trimming of the newly exposed basic residues by a carboxypeptidase such as DCPD (R101 and R102 are critical for DGD binding), lack of ccc DNA amplification, and additional restriction factors associated with the chicken hepatoma cell line. In this regard, it has been reported that Huh7 cells are permissive to HBV replication following lipid-mediated transfer of core particles to the cytosol (39), suggesting a block at or before uncoating. It will be of interest to perform similar experiments with LMH cells to determine whether there are additional defects subsequent to uncoating.
Finally, we demonstrated that a furin inhibitor markedly suppressed productive DHBV infection of primary duck hepatocytes if added immediately following virus incubation. The same inhibitor added 2 days later had no effect. This provided strong evidence for the involvement of furin or a furin-like protease in the initiation of DHBV infection. Since this inhibitor, decanoyl-RVKR-chloromethylketone, could block both furin cleavage of DCPD and endosomal cleavage of DHBV envelope proteins in vitro, the result obtained does not distinguish the contribution of DCPD cleavage relative to that of envelope protein cleavage in the initiation of infection. We also observed that a higher dose of this inhibitor (100 μM) is needed to efficiently inhibit DHBV infection, which could be associated with the wide intracellular distribution and stability of the inhibitor in vivo. Nevertheless, no cytotoxic effect was observed in PDH treated with 100 μM inhibitor. Use of RNA interference technology is necessary to further establish whether furin per se or another PC is required for productive DHBV infection.
The requirement of protease cleavage for hepadnavirus infection has been suggested by several previous studies. Lu et al. reported that in vitro treatment of HBV particles with bacterial V8 protease triggered infection of HepG2 cells, a human hepatoma cell line otherwise resistant to HBV infection (31). It was proposed that cleavage by V8 protease exposes the fusion peptide in the amino-terminal region of the HBV S protein (40). Another study from the same group revealed that a serine protease inhibitor Kazal (SPIK) was expressed at a 1,000-fold-higher concentration in HepG2 cells than in normal human liver, which may explain the lack of envelope protein cleavage in HepG2 cells (30). Stoeckl and colleagues reported that treatment of HBV particles with endosomal enzymes from HepG2 cells resulted in the exposure of the so-called translocation motif and viral infection of Huh7 cells (44). Similarly, treatment of DHBV particles with endosome derived from LMH cells enabled viral infectivity in LMH cells. However, we found that endosome prepared from LMH cells failed to cleave DHBV envelope proteins (our unpublished observation). Moreover, furin failed to cleave DCPD when introduced into LMH cells (Fig. 2D). These findings are more in line with the observations of Lu and colleagues regarding HBV and HepG2 cells. Furthermore, two recent reports demonstrated that the presumed translocation motif in HBV is dispensable for HBV infectivity (2, 26).
We have identified DCPD as a binding partner for the intact L protein and DGD as the binding partner for several truncated versions of the L protein. Moreover, we have demonstrated the importance of DGD in productive DHBV infection. The discovery that furin and an endosomal enzyme(s) could cleave DCPD and viral envelope proteins and the demonstration that such cleavages have functional consequences for ccc DNA formation and productive infection may provide the missing link between DCPD and DGD. We propose that DCPD serves as the DHBV docking receptor on the cell surface. Subsequent cleavage of DCPD and viral envelope proteins in the endosome switches the viral binding partner to DGD, which somehow leads to membrane fusion and uncoating. Further studies are needed to test this hypothesis and to elucidate the details of the early steps in the DHBV life cycle. Such studies will also shed light on the nature of the HBV receptor and cofactors.
Acknowledgments
We are grateful to Anke Feldman for the chicken furin cDNA construct and furin antibodies.
J. Li was a Liver Scholar of the American Liver Foundation. This work was supported by NIH grants CA109733, DK066950, and CA133976 and American Cancer Society grant RSG 06-059-01-MBC.
Footnotes
Published ahead of print on 24 February 2010.
REFERENCES
- 1.Bergeron, F., R. Leduc, and R. Day. 2000. Subtilase-like pro-protein convertases: from molecular specificity to therapeutic applications. J. Mol. Endocrinol. 24:1-22. [DOI] [PubMed] [Google Scholar]
- 2.Blanchet, M., and C. Sureau. 2007. Infectivity determinants of the hepatitis B virus pre-S domain are confined to the N-terminal 75 amino acid residues. J. Virol. 81:5841-5849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breiner, K. M., and H. Schaller. 2000. Cellular receptor traffic is essential for productive duck hepatitis B virus infection. J. Virol. 74:2203-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breiner, K. M., S. Urban, and H. Schaller. 1998. Carboxypeptidase D (gp180), a Golgi-resident protein, functions in the attachment and entry of avian hepatitis B viruses. J. Virol. 72:8098-8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruss, V., X. Lu, R. Thomssen, and W. H. Gerlich. 1994. Post-translational alterations in transmembrane topology of the hepatitis B virus large envelope protein. EMBO J. 13:2273-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruzzaniti, A., K. Goodge, P. Jay, S. A. Taviaux, M. H. Lam, P. Berta, T. J. Martin, J. M. Moseley, and M. T. Gillespie. 1996. PC8 [corrected], a new member of the convertase family. Biochem. J. 314:727-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan, S. J., A. A. Oliva, Jr., J. LaMendola, A. Grens, H. Bode, and D. F. Steiner. 1992. Conservation of the prohormone convertase gene family in metazoa: analysis of cDNAs encoding a PC3-like protein from hydra. Proc. Natl. Acad. Sci. U. S. A. 89:6678-6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chojnacki, J., D. A. Anderson, and E. V. Grgacic. 2005. A hydrophobic domain in the large envelope protein is essential for fusion of duck hepatitis B virus at the late endosome. J. Virol. 79:14945-14955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Constam, D. B., M. Calfon, and E. J. Robertson. 1996. SPC4, SPC6, and the novel protease SPC7 are coexpressed with bone morphogenetic proteins at distinct sites during embryogenesis. J. Cell Biol. 134:181-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eng, F. J., E. G. Novikova, K. Kuroki, D. Ganem, and L. D. Fricker. 1998. gp180, a protein that binds duck hepatitis B virus particles, has metallocarboxypeptidase D-like enzymatic activity. J. Biol. Chem. 273:8382-8388. [DOI] [PubMed] [Google Scholar]
- 11.Eng, F. J., O. Varlamov, and L. D. Fricker. 1999. Sequences within the cytoplasmic domain of gp180/carboxypeptidase D mediate localization to the trans-Golgi network. Mol. Biol. Cell 10:35-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feldmann, A., M. K. Schafer, W. Garten, and H. D. Klenk. 2000. Targeted infection of endothelial cells by avian influenza virus A/FPV/Rostock/34 (H7N1) in chicken embryos. J. Virol. 74:8018-8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernholz, D., G. Wildner, and H. Will. 1993. Minor envelope proteins of duck hepatitis B virus are initiated at internal pre-S AUG codons but are not essential for infectivity. Virology 197:64-73. [DOI] [PubMed] [Google Scholar]
- 14.Funk, A., M. Mhamdi, H. Hohenberg, H. Will, and H. Sirma. 2006. pH-independent entry and sequential endosomal sorting are major determinants of hepadnaviral infection in primary hepatocytes. Hepatology 44:685-693. [DOI] [PubMed] [Google Scholar]
- 15.Ganem, D., and R. Schneider. 2001. Hepadnaviridae: the viruses and their replication, p. 2923-2969. In D. M. Knipe, P. M. Howley, et al. (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 16.Gorvel, J. P., P. Chavrier, M. Zerial, and J. Gruenberg. 1991. rab5 controls early endosome fusion in vitro. Cell 64:915-925. [DOI] [PubMed] [Google Scholar]
- 17.Guo, J. T., and J. C. Pugh. 1997. Topology of the large envelope protein of duck hepatitis B virus suggests a mechanism for membrane translocation during particle morphogenesis. J. Virol. 71:1107-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hallenberger, S., M. Moulard, M. Sordel, H. D. Klenk, and W. Garten. 1997. The role of eukaryotic subtilisin-like endoproteases for the activation of human immunodeficiency virus glycoproteins in natural host cells. J. Virol. 71:1036-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halpern, M. S., J. M. England, D. T. Deery, D. J. Petcu, W. S. Mason, and K. L. Molnar-Kimber. 1983. Viral nucleic acid synthesis and antigen accumulation in pancreas and kidney of Pekin ducks infected with duck hepatitis B virus. Proc. Natl. Acad. Sci. U. S. A. 80:4865-4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He, T. C., S. Zhou, L. T. da Costa, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. U. S. A. 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito, K., K. H. Kim, A. S. Lok, and S. Tong. 2009. Characterization of genotype-specific carboxyl-terminal cleavage sites of hepatitis B virus e antigen precursor and identification of furin as the candidate enzyme. J. Virol. 83:3507-3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones, B. G., L. Thomas, S. S. Molloy, C. D. Thulin, M. D. Fry, K. A. Walsh, and G. Thomas. 1995. Intracellular trafficking of furin is modulated by the phosphorylation state of a casein kinase II site in its cytoplasmic tail. EMBO J. 14:5869-5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kock, J., and H. J. Schlicht. 1993. Analysis of the earliest steps of hepadnavirus replication: genome repair after infectious entry into hepatocytes does not depend on viral polymerase activity. J. Virol. 67:4867-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuroki, K., R. Cheung, P. L. Marion, and D. Ganem. 1994. A cell surface protein that binds avian hepatitis B virus particles. J. Virol. 68:2091-2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuroki, K., F. Eng. T. Ishikawa, C. Turck, F. Harada, and D. Ganem. 1995. gp180, a host cell glycoprotein that binds duck hepatitis B virus particles, is encoded by a member of the carboxypeptidase gene family. J. Biol. Chem. 270:15022-15028. [DOI] [PubMed] [Google Scholar]
- 26.Lepere, C., M. Regeard, J. Le Seyec, and P. Gripon. 2007. The translocation motif of hepatitis B virus envelope proteins is dispensable for infectivity. J. Virol. 81:7816-7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, J., S. Tong, H. B. Lee, A. L. Perdigoto, H. C. Spangenberg, and J. R. Wands. 2004. Glycine decarboxylase mediates a postbinding step in duck hepatitis B virus infection. J. Virol. 78:1873-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, J., S. Tong, and J. R. Wands. 1999. Identification and expression of glycine decarboxylase (p120) as a duck hepatitis B virus pre-S envelope-binding protein. J. Biol. Chem. 274:27658-27665. [DOI] [PubMed] [Google Scholar]
- 29.Li, J. S., S. P. Tong, and J. R. Wands. 1996. Characterization of a 120-kilodalton pre-S-binding protein as a candidate duck hepatitis B virus receptor. J. Virol. 70:6029-6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu, X., and T. Block. 2004. Study of the early steps of the hepatitis B virus life cycle. Int. J. Med. Sci. 1:21-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu, X., T. M. Block, and W. H. Gerlich. 1996. Protease-induced infectivity of hepatitis B virus for a human hepatoblastoma cell line. J. Virol. 70:2277-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mandart, E., A. Kay, and F. Galibert. 1984. Nucleotide sequence of a cloned duck hepatitis B virus genome: comparison with woodchuck and human hepatitis B virus sequences. J. Virol. 49:782-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Margeridon, S., S. Carrouee-Durantel, I. Chemin, L. Barraud, F. Zoulim, C. Trepo, and A. Kay. 2008. Rolling circle amplification, a powerful tool for genetic and functional studies of complete hepatitis B virus genomes from low-level infections and for directly probing covalently closed circular DNA. Antimicrob. Agents Chemother. 52:3068-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mason, W. S., G. Seal, and J. Summers. 1980. Virus of Pekin ducks with structural and biological relatedness to human hepatitis B virus. J. Virol. 36:829-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Molloy, S. S., L. Thomas, J. K. VanSlyke, P. E. Stenberg, and G. Thomas. 1994. Intracellular trafficking and activation of the furin proprotein convertase: localization to the TGN and recycling from the cell surface. EMBO J. 13:18-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakayama, K. 1997. Furin: a mammalian subtilisin/Kex2p-like endoprotease involved in processing of a wide variety of precursor proteins. Biochem. J. 327:625-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ostapchuk, P., P. Hearing, and D. Ganem. 1994. A dramatic shift in the transmembrane topology of a viral envelope glycoprotein accompanies hepatitis B viral morphogenesis. EMBO J. 13:1048-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prange, R., and R. E. Streeck. 1995. Novel transmembrane topology of the hepatitis B virus envelope proteins. EMBO J. 14:247-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rabe, B., D. Glebe, and M. Kann. 2006. Lipid-mediated introduction of hepatitis B virus capsids into nonsusceptible cells allows highly efficient replication and facilitates the study of early infection events. J. Virol. 80:5465-5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodriguez-Crespo, I., E. Nunez, B. Yelamos, J. Gomez-Gutierrez, J. P. Albar, D. L. Peterson, and F. Gavilanes. 1999. Fusogenic activity of hepadnavirus peptides corresponding to sequences downstream of the putative cleavage site. Virology 261:133-142. [DOI] [PubMed] [Google Scholar]
- 41.Seidah, N. G., and M. Chretien. 1999. Proprotein and prohormone convertases: a family of subtilases generating diverse bioactive polypeptides. Brain Res. 848:45-62. [DOI] [PubMed] [Google Scholar]
- 42.Seidah, N. G., J. Hamelin, M. Mamarbachi, W. Dong, H. Tardos, M. Mbikay, M. Chretien, and R. Day. 1996. cDNA structure, tissue distribution, and chromosomal localization of rat PC7, a novel mammalian proprotein convertase closest to yeast kexin-like proteinases. Proc. Natl. Acad. Sci. U. S. A. 93:3388-3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Spangenberg, H. C., H. B. Lee, J. Li, F. Tan, R. Skidgel, J. R. Wands, and S. Tong. 2001. A short sequence within domain C of duck carboxypeptidase D is critical for duck hepatitis B virus binding and determines host specificity. J. Virol. 75:10630-10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stoeckl, L., A. Funk, A. Kopitzki, B. Brandenburg, S. Oess, H. Will, H. Sirma, and E. Hildt. 2006. Identification of a structural motif crucial for infectivity of hepatitis B viruses. Proc. Natl. Acad. Sci. U. S. A. 103:6730-6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swameye, I., and H. Schaller. 1997. Dual topology of the large envelope protein of duck hepatitis B virus: determinants preventing pre-S translocation and glycosylation. J. Virol. 71:9434-9441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang, H., and A. McLachlan. 2001. Transcriptional regulation of hepatitis B virus by nuclear hormone receptors is a critical determinant of viral tropism. Proc. Natl. Acad. Sci. U. S. A. 98:1841-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tong, S., J. Li, and J. R. Wands. 1999. Carboxypeptidase D is an avian hepatitis B virus receptor. J. Virol. 73:8696-8702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tong, S., J. Li, and J. R. Wands. 1995. Interaction between duck hepatitis B virus and a 170-kilodalton cellular protein is mediated through a neutralizing epitope of the pre-S region and occurs during viral infection. J. Virol. 69:7106-7112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tong, S., F. Mattes, K. Teubner, and H. E. Blum. 1990. Complete nucleotide sequence of a Chinese duck hepatitis B virus. Nucleic Acids Res. 18:6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tong, S. P., J. S. Li, L. Vitvitski, D. Lepot, and C. Trepo. 1991. Generation of a 1.5-kb cDNA fragment of the hepatitis C virus genome by overlap extension. J. Med. Virol. 35:228-231. [DOI] [PubMed] [Google Scholar]
- 51.Tuttleman, J. S., J. C. Pugh, and J. W. Summers. 1986. In vitro experimental infection of primary duck hepatocyte cultures with duck hepatitis B virus. J. Virol. 58:17-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Urban, S., C. Schwarz, U. C. Marx, H. Zentgraf, H. Schaller, and G. Multhaup. 2000. Receptor recognition by a hepatitis B virus reveals a novel mode of high affinity virus-receptor interaction. EMBO J. 19:1217-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van de Loo, J. W., J. W. Creemers, N. A. Bright, B. D. Young, A. J. Roebroek, and W. J. Van de Ven. 1997. Biosynthesis, distinct post-translational modifications, and functional characterization of lymphoma proprotein convertase. J. Biol. Chem. 272:27116-27123. [DOI] [PubMed] [Google Scholar]
- 54.Varlamov, O., and L. D. Fricker. 1998. Intracellular trafficking of metallocarboxypeptidase D in AtT-20 cells: localization to the trans-Golgi network and recycling from the cell surface. J. Cell Sci. 111:877-885. [DOI] [PubMed] [Google Scholar]
- 55.Wouters, S., M. Leruth, E. Decroly, M. Vandenbranden, J. W. Creemers, J. W. van de Loo, J. M. Ruysschaert, and P. J. Courtoy. 1998. Furin and proprotein convertase 7 (PC7)/lymphoma PC endogenously expressed in rat liver can be resolved into distinct post-Golgi compartments. Biochem. J. 336:311-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu, M., and J. Summers. 1994. Multiple functions of capsid protein phosphorylation in duck hepatitis B virus replication. J. Virol. 68:4341-4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang, D. Y., M. Brandwein, T. Hsuih, and H. B. Li. 2001. Ramification amplification: a novel isothermal DNA amplification method. Mol. Diagn. 6:141-150. [DOI] [PubMed] [Google Scholar]