Abstract
Protease inhibitors (PIs) of hepatitis C virus (HCV) provide an additional or alternative therapy for chronic infection. However, assessment of their efficacy and ability to inhibit replication of different genotypes is hampered by the lack of a convenient animal model or a method for in vitro culture of HCV other than the type 1/2-based replicons and the infectious genotype 2a clone JFH1. To address this problem, we constructed a panel of replication-competent chimeric Jc1 (pFK JFH1/J6/C-846) clones containing protease and NS4A coding sequences from all six major genotypes, enabling the determination of replication and the susceptibility to PIs. Chimeras showed substantial variability in replication kinetics, attributable in part to naturally occurring polymorphisms and differing requirements for adaptive mutations in NS3 and NS4A. Through calculation of 50% inhibitory concentrations (IC50s) of BILN 2061, measuring reduction in the number of focus-forming units/ml (FFU/ml) and replication inhibition, consistent genotype-associated differences in antiviral susceptibilities were observed. IC50s for genotype 1b, 4a, and 6a-derived chimeras (1 to 3 nM) were approximately 100-fold lower than those for genotypes 2a, 3a, and 5a (range, 80 to 720 nM), implying major differences in response to therapy. In vitro passage in increasing concentrations of BILN 2061 rapidly induced resistance-associated mutations at position 168 in chimeras of all 6 genotypes and at position 156 in genotypes 1b and 4a, each with substantial variability in the identity of substituted amino acids. The system will allow future comprehensive phenotypic characterization of naturally occurring and treatment-induced mutations for PIs in trial or entering clinical use.
Worldwide, about 170 million individuals are estimated to be infected with hepatitis C virus (HCV) (1, 48). Chronic HCV infection is a leading cause of chronic liver diseases, such as cirrhosis and hepatocellular carcinoma (6). HCV has a positive-sense, single-stranded RNA genome of approximately 9,600 nucleotides, belonging to the family Flaviviridae (7). A single polyprotein of around 3,000 amino acids (53) is translated and processed by cellular and viral proteases to generate 10 different structural and nonstructural proteins (16, 18, 19).
The error-prone RNA-dependent RNA polymerase (RdRp) NS5B, and the resulting high mutation frequencies during replication, contributes to the substantial genetic and antigenic heterogeneity of HCV, with seven major genotypes showing >30% nucleotide sequence divergence from each other and numerous subtypes identified to date (5, 50-52). The distribution of genotypes varies by geographical location and risk groups for infection; the predominant genotypes within the United States, Europe, Australia, and East Asia (Japan, Taiwan, Thailand, and China) are 1, 2, and 3. Genotype 4 is largely confined to the Middle East, Egypt, and Central Africa, whereas genotypes 5 and 6 are found predominantly in South Africa and Southeast Asia, respectively (49).
The current treatment of pegylated interferon and ribavirin has limited efficacy and serious side effects; infections with genotype 1 in particular respond poorly even to prolonged treatment, with 48% failing to clear infections after 48 weeks of combined therapy (33, 39). To address this problem, several direct antiviral inhibitors of the NS3/4A serine protease and the RNA-dependent RNA polymerase have been developed. Among the former are the noncovalent inhibitor BILN 2061 (24) and the covalent inhibitors SCH 503034 (30) and VX-950 (37). In ongoing trials, encouraging results have been reported for the covalent inhibitors (12, 17, 42, 44), whereas the noncovalent inhibitor BILN 2061 development has been halted due to cardiotoxicity in laboratory animals (58) (reviewed recently by de Bruijne et al. [9]).
Research into antiviral drugs and vaccines has been hampered by the lack of a full viral life cycle cell culture system. Only recently, a full-length HCV cell culture system in which infectious virus can be generated in Huh7 cells from transfection of complete HCV genomic RNA sequences has been described (26, 59). Viable JFH1-based intergenotypic recombinants containing genotype-specific structural proteins (core, E1, and E2), p7, and NS2 have been developed for all seven genotypes (14, 15, 21, 38, 45, 65), which allow the study of vaccines and entry inhibitors for all genotypes. However, full-length HCV cell culture systems allowing the study of the NS3 protease are currently available only for genotypes 2a (JFH1 and HC-J6) (26, 34, 59) and 1a (H77), which needs adaptive mutations to replicate efficiently (64). The limited number of replication-competent full-length reference sequences limits the assessment of how genetic variation between the different genotypes and within subtypes influences susceptibility to antiviral therapy and development of resistance.
The aim of the current study was to develop effective cell culture systems for the six major genotypes of HCV, to compare the susceptibility of each to the protease inhibitor (PI), BILN 2061, and through passaging in subinhibitory concentrations of the drug, to compare the ability and mechanism of antiviral resistance development between genotypes. The use of replicon vectors, where protease genes of different genotypes were inserted into the genotype 1b replicon, has been demonstrated in several reports (3, 40). Other approaches involve the release of reporter molecules upon NS3/4A cleavage (29). None of these methods, however, construct infectious virus that would allow the whole replication cycle of HCV to be analyzed.
Recently, the full-length replication-competent clone Jc1 (pFK JFH1/J6/C-846) has been developed. This clone comprises the HCJ6 core and envelope coding sequences and a portion of the NS2 gene, with the remainder of the polyprotein derived from JFH1 (26, 38). It replicates autonomously and yields high infectious titers in the Huh7.5 cells. It was therefore chosen as a backbone for the construction of the intra- and intergenotypic recombinants in the current study and enabled the activities of HCV protease inhibitors against different genotypes and diverse natural isolates to be directly assessed. The development of these replication-competent intergenotype chimeras will improve the ability to predict clinical doses, efficacy, and development of drug resistance mutations in a diverse range of HCV variants circulating worldwide.
MATERIALS AND METHODS
HCV plasmids, sequences, and clinical specimens.
pJFH1 and pJFH1-GND (AB047639), used in the construction of the intra- and intergenotypic recombinants, were provided by T. Wakita (Tokyo Metropolitan Institute for Neuroscience, Tokyo, Japan) (59), and pFK JFH1/J6/C-846_dg (Jc1) was provided by R. Bartenschlager (Department of Molecular Virology, University of Heidelberg, Heidelberg, Germany) (38). pJ6CF (GenBank accession no. AF177036) and pH 77* (differs from pH 77 [GenBank accession no. AF011751] at M1205T) were provided by J. Bukh (NIH, Hepatitis Viruses Section, National Institutes of Health, Bethesda, MD) (62, 63), and HCV3a-Gla (p3a) was provided by E. A. McCruden (Division of Virology, Institute of Biomedical and Life Sciences, University of Glasgow, Glasgow, United Kingdom) (47). HC-J4 (p1b, D10750) was described by Okamoto et al. (35). ED43* (p4a) differs from ED43 (GenBank accession no. NC_009825) by 4 amino acids (T1048A, T1064I, I1160T, and R1176A), EUH1480* (p5a) differs from EUH1480 (GenBank accession no. NC_009826) by 9 amino acids (L1045V, F1061V, I1072T, L1081V, K1117T, G1118R, R1122P, I1694V, and T1695I), and EUHK2* (p6a) differs from EUHK2 (GenBank accession no. Y12083) by 6 amino acids (D1065A, V1070L, P1085A, F1087S, K1094R, and I1196V). Plasmids p4a, p5a, and p6a were provided by Richard Elliot (Centre for Biomolecular Sciences, University of St. Andrews, St. Andrews, United Kingdom). Differences between clones and prototype sequences were present in received clones and likely arose during cloning.
Sequences of HCV isolates used for sequence diversity analysis were retrieved from the HCV sequence database (22) and the NCBI GenBank. A total of 570 NS3 and NS4A sequences of genotype 1a, 459 NS3 sequences of genotype 1b, 242 NS3 and 180 NS4A sequences of genotype 3a, 39 NS3 sequences of genotype 4a, and 15 NS3 sequences of genotype 6a were analyzed. Anonymized research samples of residual plasma from previous investigations of HCV epidemiology and treatment response were used for estimation of sequence diversity in NS3.
Cell culture.
Huh7.5 cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 4,500 mg/liter glucose, 2 mM l-glutamine, 10% heat-inactivated fetal calf serum (FCS; Harlan Sera-Lab), nonessential amino acids, 20 mM HEPES, 100 U/ml penicillin, and 100 μg/ml streptomycin and incubated at 37°C, 5% CO2, and 100% relative humidity. Cells were split every second to third day at a ratio of 1:2 to 1:3.
Construction of pJFH1- and Jc1-based intra- and intergenotypic recombinants. (i) Replacement of the NS3 protease gene (Fx/Jx).
All nucleotide positions are referred to according to their position in the H77 reference position, AF009606. To construct F1a, F1b, F2a, F3a, F4a, F5a, and F6a, a BstBI restriction site was generated at the junction of NS2 and NS3 in the pJFH1 plasmid, and a BglII restriction enzyme site was generated at the junction of the NS3 protease and NS3 helicase domain. To reduce the possibility of PCR errors, the NS3 gene was subcloned into Zero Blunt TOPO using the naturally occurring restriction sites NotI and SpeI. A QuikChange site-directed mutagenesis kit (Stratagene) was used to introduce two silent point mutations to generate a BstBI site (C3398T and C3401G) and one to create a BglII restriction site (G3993A). Each of the introduced base changes was verified by DNA sequence analysis. The NS3 protease region was amplified from pH 77* with primers 1aBstBI and 1aBglII, of pJ6CF with primers 2aBstBI and 2aBglII, and so on. A list of all the primers used can be found in the supplemental material (http://www.virus-evolution.org/Downloads/JVI02698-09/). Primers were designed to include the JFH1 sequence of the NS2 region and the corresponding intergenotypic sequence of the NS3 protease region. Products were digested with BstBI and BglII (all restriction enzymes were obtained from New England Biolabs), gel purified (gel extraction kit; Qiagen) and ligated into pJFH1. pJFH1 has similarly been digested with BstBI and BglII, dephosphorylated using alkaline phosphatase (New England Biolabs), and gel purified. Each introduced NS3 protease gene was verified by DNA sequence analysis. To generate the corresponding Jc1-based chimeras (Jx), recombinant plasmids were digested with NotI and SfiI, gel purified, and ligated into Jc1, which was digested with NotI and SfiI and dephosphorylated.
(ii) Replacement of NS3 protease and NS4A cofactor gene (Fxx/Jxx).
To generate F1a1a, F1b1b, F2a2a, F3a3a, F4a4a, F5a5a, and F6a6a, two new restriction sites were introduced into pJFH1. To reduce the possibility of PCR errors, the NS4A was subcloned into Zero Blunt TOPO using the naturally occurring restriction sites NsiI and XbaI. A BlpI restriction site was generated by introducing a silent point mutation (C5297G) at the NS3 helicase and NS4A junction, and a MluI site was generated by introducing 4 point mutations (T5478A, T5480G, A5481C, G5483T) at the NS4A and NS4B junction, in which one was nonsilent. The NS4A region was amplified from pH 77* with primers 1aSapI and 1aMluI, introducing SapI (5′-end) and MluI (3′-end) restriction sites. The same strategy was used to amplify NS4A from genotype 1a to 6a prototype plasmids, using the corresponding genotype-specific primers (primer sequences can be found in the supplemental material at http://www.virus-evolution.org/Downloads/JVI02698-09/). Products were digested with SapI and MluI, gel purified, and ligated into the TOPO vector containing the JFH1 insert digested with BlpI and MluI and dephosphorylated. A5478 was mutated back to T by site-directed mutagenesis to recreate a native JFH1 amino acid sequence (Ala to Thr) outside the NS4A region. To generate Fxx and Jxx, TOPO vector, including the modified insert, was digested with NotI and SfiI, gel purified, and ligated into pJHF1 and Jc1, respectively, which were digested with NotI and SfiI and dephosphorylated.
Amplification of the NS3 protease gene from study subject plasma.
HCV RNA was isolated from 150 μl study subject plasma using the Qiagen RNeasy kit, according to the manufacturer's guidelines. Extracted RNA was eluted into 30 μl of RNase-free water. To generate cDNA and the primary PCR product, 0.75-μM genotype-specific outer primers (NS3p1a/1b/3a/4a/6aOS and NS3p1a/1b/3a/4a/6aOA; for sequences, see the supplemental material at http://www.virus-evolution.org/Downloads/JVI02698-09/) and 10 μl of extracted RNA were used in a 50-μl reaction using the SuperScript III one-step reverse transcriptase PCR (RT-PCR) system with Platinum Taq DNA polymerase (Invitrogen), as recommended by the manufacturer. The PCR protocol consisted of an RT step at 43°C for 1 h, followed by 20 cycles of 53°C for 1 min, 55°C for 1 min, and final extension at 70°C for 15 min. Subsequent to a denaturation step at 94°C for 2 min, 35 cycles of 94°C for 30 s; 54°C for 30 s; 68°C for 2 min; and a final extension at 68°C for 5 min were performed. The first-round PCR products were used as templates in a nested secondary PCR using genotype-specific primers (1a/1b/3a/4a/6aBstBIs and 1a/1b/3a/4a/6aBglIIas) introducing BstBI (5′-end) and BglII (3′-end) restriction sites. Nested PCR was performed using the high-fidelity KOD Hot Start polymerase (Novagen), as recommended by the manufacturer. Each PCR product was sequenced to obtain the consensus sequence of the corresponding NS3 protease gene. The nested PCR products were treated with BstBI and BglII and gel purified. To remove the NS3 protease gene, Jxxs including prototype NS3 protease genes were likewise digested with BstBI and BglII, dephosphorylated, and gel purified. Purified study subject-derived NS3 protease genes were then ligated into the corresponding Jxx, creating intergenotypic chimeras where the NS3 protease gene is study subject derived and where the NS4A cofactor has prototype sequence.
Construction of adapted genomes.
RNA was extracted from infectious supernatant using the QIAamp viral RNA minikit (Qiagen), according to the manufacturer's protocol. If no infectious supernatant was generated, RNA was alternatively extracted from cell pellets using Qiagen shredder columns, followed by a Qiagen RNeasy kit used as described above. A PCR product was generated using SuperScript III as described above. JFH-NotI and JFH-SpeI were used to amplify a 1.2-kb-long fragment encompassing the HCV NS3 protease, and JFH-5230 and JFH-5536 were used to amplify a 300-bp-long fragment encompassing the NS4A cofactor. The PCR product was then subjected to bulk sequence determination. To generate J2a2a-C3538G (J2a2a-T1066S), J5a5a-C3416G, T3968C, A4081T (J5a5a-Q1247L), and J6a6a-A3458G, G3459T (J6a6a-V1040L), the fragment encompassing the HCV NS3 protease was digested with SpeI and NotI and cloned into the corresponding (J2a2a, J5a5a, or J6a6a) recombinant plasmids. To generate J3a3a-3-C5328G, T5329C (J3a3a-3-L1663A) and J3a3a-8-C5328G, T5329C (J3a3a-8-L1663A), point mutations were introduced by site-directed mutagenesis and cloning. Modified fragments were verified by sequencing.
RNA synthesis and transfection.
Plasmid templates were linearized by XbaI digestion (for pJFH1 and pJFH1 chimeras) or MluI digestion (for Jc1 and Jc1 chimeras) and treated with mung bean nuclease (New England Biolabs) to remove 5′-end overhangs. The linearized DNA template was cleaned by phenol-chloroform extraction following ethanol precipitation. RNA was synthesized from 1 μg DNA template with T7 RNA polymerase (Promega) for 1 h at 37°C. Following treatment with RNase-free DNase, RNA was cleaned up using the RNeasy kit (Qiagen), and the integrity of the RNA was analyzed by nondenaturing agarose gel electrophoresis. RNA concentrations were determined using spectrophotometry, and 10-μg aliquots were stored at −80°C.
RNA was transfected into Huh7.5 cells by electroporation. Huh7.5 cells were washed with phosphate-buffered saline (PBS) and detached with trypsin. Cells were pelleted by centrifugation (1,600 rpm for 7 min at 4°C), then resuspended in 10 ml chilled diethyl pyrocarbonate (DEPC)-treated PBS, counted, washed three times with chilled DEPC-treated PBS (1,600 rpm for 4 min at 4°C), and then chilled on ice for at least 5 min. A total of 10 μg of RNA was mixed with 5 × 106 cells suspended in 400 μl of PBS and transferred to an electroporation cuvette (0.4-cm gap width; Bio-Rad, Munich, Germany). Electroporation consisted of one square wave pulse for 25 ms of current delivered by the Bio-Rad Gene Pulser Xcell electroporation device, set at 150 V. Transfected cells were immediately resuspended in 4.5-ml 50:50 dilution mix of conditioned and fresh media (containing 10% FCS) and then transferred into T25 flasks containing 10 ml complete growth medium or seeded into 24-well plates for NS5A immunostaining and incubated at 37°C, 5% CO2, and 100% relative humidity. Cells were passaged every 3 to 4 days by trypsinization and reseeding with a 1:3 to 1:4 split ratio into fresh culture vessels or 24-well plates for NS5A immunostaining. Virus-containing supernatant was collected, cleared of cell debris by centrifugation, and stored at 4°C overnight or at −80°C for the long term.
Immunohistochemistry staining for HCV NS5A.
Viral replication was assessed by NS5A staining. Electroporated cells were seeded into 24-well plates containing coverslips and immunostained for NS5A when they were subconfluent. Following fixation of cells in 4% paraformaldehyde for 20 min, cells were washed three times with PBS and then permeabilized using 0.1% Triton X-100 in PBS for 7 min. Following 2 further washes with PBS, cells were incubated with an in-house-derived polyclonal sheep anti-NS5A serum (provided by Mark Harris, University of Leeds) diluted 1:5,000 in 10% FCS-PBS for 1 h. After cells were washed two times with PBS, bound NS5A-specific antibody was detected by 1 h of incubation with Alexa Fluor 488 donkey anti-sheep IgG (Invitrogen) diluted 1:1,000 in PBS. Cells were further washed two times before NS5A-positive cells were detected using a fluorescence microscope. Three images per coverslip were taken from two coverslips per sample, and the percentage of HCV-positive cells was determined using AxioVision 4.8 software, where 0% shows that no cells are infected and 100% shows that all cells are infected. Jc1 served as a positive control, and JFH1-GND served as a negative control.
Determination of TCID50 in HCV cultures.
Naïve Huh7.5 cells were seeded in 96-well plates the previous day at a concentration of 6 × 103 cells/well. The sample was serially diluted in complete growth medium and inoculated onto the cells at 10-fold dilutions, with 6 replicates at each dilution. After 3 days of incubation, cells were stained for NS5A as described above. Viral titers were determined by calculating the tissue culture infectious dose at which 50% of the wells were positive for viral antigen (TCID50). Wells were scored positive if at least 1 positive cell was detected. TCID50 was calculated according to the method of Reed and Muench (41).
Drug inhibition studies.
The HCV-specific protease inhibitor (PI) BILN 2061 was resuspended at 5 mM in dimethyl sulfoxide (DMSO). Naïve Huh7.5 cells were seeded in 96-well plates the previous day at a concentration of 6 × 103 cells/well. The next day, cells were infected for 8 h with the corresponding infectious supernatant at a multiplicity of infection (MOI) of 0.015 and washed with PBS, and the medium was replaced with complete growth medium containing 0.1% DMSO, as a carrier control, with or without the indicated concentrations of the PI. Cultures were incubated for 3 days and then stained for NS5A as described above. Each concentration was assayed in triplicate. The percent inhibition of supernatant infectivity was calculated from the reduction in the number of FFU/ml after antiviral addition compared to the number of FFU/ml in virus control without antiviral addition.
Alternatively, 10 μg synthetic RNA was electroporated into Huh7.5 cells as described above and seeded into 24-well plates. The next day, the medium was replaced with complete growth medium containing 0.1% DMSO with or without the indicated concentrations of PI. Cultures were incubated for a further 3 days and then stained for NS5A as described above. Antiviral efficacy was measured as the inhibition of RNA replication by staining for NS5A and determining the percentage of HCV-positive cells. Each concentration was assayed in triplicate. For J2a2a, 1 μg instead of 10 μg was used to achieve an infection frequency of around 40%. For J6a6a, cells were first passaged for 24 days to allow 40% of the culture to be infected before BILN 2061 was added.
Dose-response curves were fitted to a four-parameter logistic function to obtain 50% maximal inhibitory concentrations (IC50s) using SigmaPlot.
In vitro selection of BILN 2061-resistant recombinant viruses.
Synthetic RNA of the recombinant viruses J1b1b, J2a2a-T1066S, J3a3a, J4a4a-19, J5a5a-V1040L, and J6a6a was electroporated into Huh7.5 cells as described above and seeded into 12-well plates. At 3 (J1b1b, J3a3a, and J4a4a-19) or 4 (J2a2a-T1066S and J5a5a-Q1247L) days postelectroporation, BILN 2061 was added, and the cells serially passaged under increasing BILN 2061 concentrations (from 23 nM to 230 nM for J1b1b, from 500 nM to 1000 nM for J2a2a-T1066S, from 350 nM to 7000 nM for J3a3a, from 10 nM to 100 nM for J4a4a-19, 500 nM for J5a5a-Q1247L, and from 1 nM to 10 nM for J6a6a; see Table S5 in the supplemental material at http://www.virus-evolution.org/Downloads/JVI02698-09/). J6a6a was first passaged for 35 days before antiviral addition. During the course of selection, infected cells were split when 70 to 80% confluence was reached. Fresh medium and BILN 2061 were added every 3 or 4 days, regardless of whether the cell culture was split. Whenever the cell culture was split, cell pellets were collected, and total cellular RNA was extracted and subjected to RT-PCR as described above. To determine the frequency of BILN 2061 resistance mutations, the 1.2-kb-long RT-PCR products of HCV RNA were ligated into the TOPO cloning vector, and individual bacterial colonies were subjected to colony PCR and sequenced. Each passaging experiment was carried out in duplicate and with a parallel passage containing control without antiviral.
Nucleotide sequence accession numbers.
Sequences obtained in the course of the study have been submitted to GenBank and have been assigned accession no. GU945424 to GU945462.
RESULTS
Design and construction of intra- and intergenotypic recombinants containing heterologous NS3 protease.
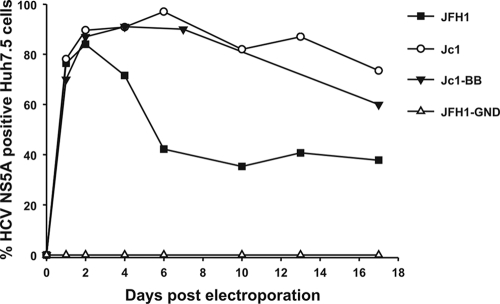
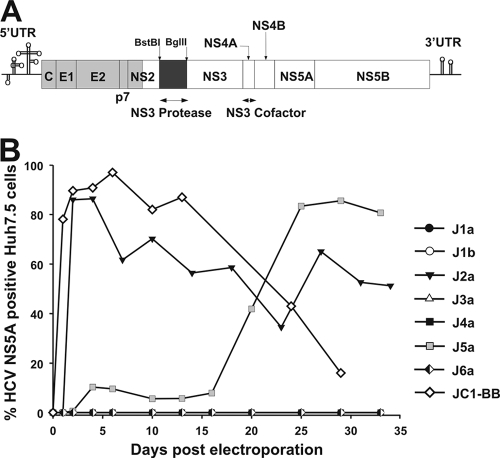
Intergenotype chimeras containing the NS3 protease gene of heterologous genotypes were created through ligation of protease domain sequences from different genotypes into the Jc1 backbone sequence. This necessitated creation of two unique restriction sites, a BstBI site at the 5′ end and a BglII site at the 3′ end of the protease-encoding region, creating the Jc1-BB clone. This showed the same replication kinetics as the parental strain Jc1 (Fig. 1). Protease genes from genotypes 1a (H77*), 1b (HC-J4), 2a (J6CF), 3a (HCV3a-Gla), 4a (ED43*), 5a (EUH1480*), and 6a (EUHK2*) were amplified by PCR and cloned into Jc1-BB. Recombinants containing heterologous proteases were termed Jx, where “x” identifies the genotype of the protease domain (Fig. 2A). RNAs transcribed from the recombinant plasmids and the replication-defective pJFH1-GND and Jc1 were electroporated into the highly permissive Huh7.5 cells, and virus replication was assessed by NS5A immunostaining. Besides J2a, which showed replication kinetics similar to that of the parental Jc1, only J5a replicated to detectable levels, and it took 20 days to spread to 50% of the cell culture (Fig. 2B). As the cell culture was split through the course of the experiment, the virus would have had to spread to neighboring cells to achieve this infection frequency during passaging.
FIG. 1.
Replication kinetics in Huh7.5 cells of the parental JFH1 and Jc1 full-length RNA transcripts and comparison with that of Jc1-BB containing introduced BstBI and BglII restriction sites in NS3 for protease gene insertion. The replication-defective mutant JFH1-GND was used as a negative control. The y axis records the percentage of HCV NS5A-positive cells scored by fluorescence microscopy.
FIG. 2.
Jx recombinants and their viability in Huh7.5 cells. (A) Genome map of cDNA clones (pJ6CF, gray; pJFH1, white). The NS3 protease was replaced with the corresponding intra- or intergenotypic gene (replaced region, black). UTR, untranslated region. (B) RNA transcripts from J1a, J1b, J2a, J3a, J4a, J5a, and J6a were electroporated into Huh7.5 cells, and replication was assessed by immunostaining against NS5A. Jc1 and pJFH1-GND served as positive and negative controls, respectively. Transcripts from the J1a, J1b, J3a, J4a, and J6a clones showed no detectable replication (all values lying on the x axis line).
Design and construction of intra- and intergenotypic recombinants containing heterologous NS3 protease and NS4A cofactor.
Since the NS4A cofactor is an indispensable part of the NS3 protease function and highly variable in sequence between genotypes, we investigated whether inclusion of the homologous NS4A gene in the intragenotypic/intergenotypic recombinants improved their replication ability (2, 11, 54).
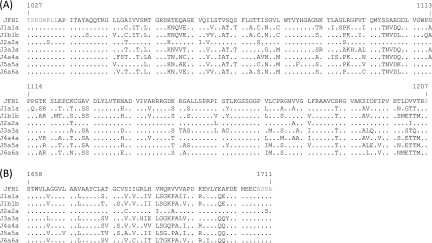
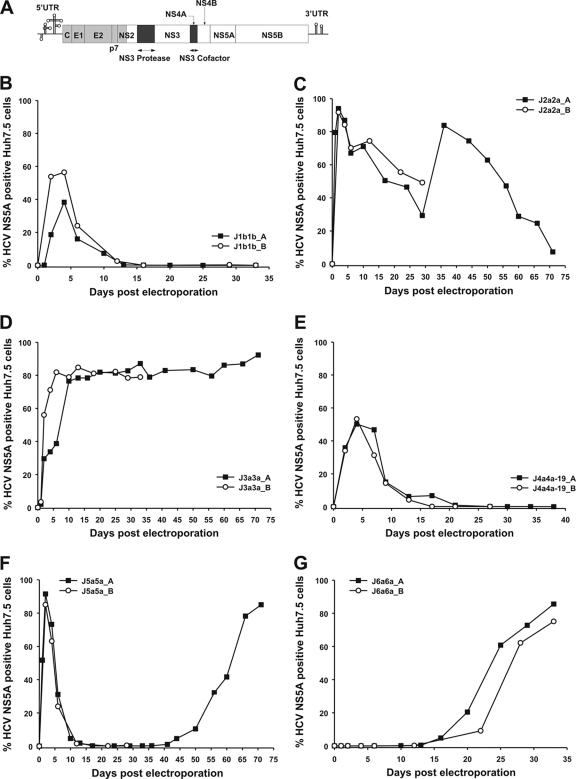
Two unique restriction sites were introduced at the 5′ end (BlpI) and at the 3′ end (MluI) of NS4A, and the corresponding NS4A genes from genotypes 1a, 1b, 2a (pJ6CF), 3a, 4a, 5a, and 6a were amplified by PCR using primers with SapI and MluI restriction sites. The primers were designed to include the JFH1 sequence up to the start of the NS4A gene (nucleotide [nt] 5313) and then the corresponding genotype sequence to the 3′ end of NS4A (nt 5474). As the introduction of the MluI restriction site was not synonymous, it was reversed to the JFH1 amino acid sequence after NS4A gene insertion by site-directed mutagenesis. The corresponding intragenotypic/intergenotypic recombinants containing the NS3 protease and NS4A gene sequences of hetero- or homologous genotypes were termed Jxx, where “xx” stands for the corresponding genotype in the protease and NS4A regions, respectively (Fig. 3 and 4A).
FIG. 3.
Comparison of genotype 1a, 1b, 2a, 3a, 4a, 5a, and 6a NS3 protease and NS4A cofactor sequences. (A) Alignment of NS3 protease residues (in black) from recombinants J1a1a, J1b1b, J2a2a, J3a3a, J4a4a, J5a5a, and J6a6a to JFH1 (NCBI accession no. AB047639). (B) Alignment of NS4A protease cofactor residues (in black) from recombinants J1a1a, J1b1b, J2a2a, J3a3a, J4a4a, J5a5a, and J6a6a to JFH1. Dots indicate amino acid sequence identity.
FIG. 4.
Jxx recombinants and their viability in Huh7.5 cell results from two independent experiments (Jxx_A and Jxx_B). (A) Genome map of cDNA clones (pJ6CF, gray; pJFH1, white). The NS3 protease and NS4A were replaced with intra- and intergenotypic genes (replaced region, black). (B to G) RNA transcripts from J1a1a, J1b1b, J2a2a, J3a3a, J4a4a-19, J5a5a, and J6a6a were electroporated into Huh7.5 cells. Jc1 and pJFH1-GND served as positive and negative controls, respectively.
Replication-competent recombinants could be created with genotypes 1b, 2a, 3a, 5a, and 6a (Fig. 4B to G). Four days after transfection, J1b1b was detected in 50% of the cell culture but was then cleared from the cell culture, likely reflecting an inability to infect Huh7.5 cells de novo beyond the transfection stage. In marked contrast, J2a2a and J5a5a replication was detected in almost all cells 2 days after transfection, comparable to that of Jc1. The immediate spread of these recombinants and Jc1 was accompanied by increased cell death, followed by proliferation of HCV-NS5A-negative Huh7.5 cells, as described in previous studies (14). Both, however, start to respread in the cell culture after this initial decrease in infected cell frequency, indicating sequence changes that reduce the cytopathic effect of the virus and promote its survival in cell culture. We have termed these changes as “attenuating” to make clear the difference with conventional “adaptive” mutations that enhance replication ability.
Replicating J3a3a and J6a6a viruses were found in 80% of the cell culture after an eclipse phase of 6 and 33 days, respectively. Compared to other Jxx recombinants, where the percentage of NS5A-positive cells was reduced again after an eclipse phase, J3a3a still infected 80% of the cell culture 70 days postelectroporation, indicating continuous spread to uninfected cells. Supernatant infectivity was measured at the peak of the infection by determining TCID50s (Table 1). The highest infectivity titer was measured for J2a2a, with a TCID50/ml of 104.2. J1b1b did not secrete any detectable infectious virus into the supernatant (32, 45, 66). J1a1a and J4a4a were not viable or their replication was not efficient enough to be detected with our assay. However, replacement of the NS3 protease gene in J4a4a with that of study subject-derived protease genes generated the replication-competent recombinants J4a4a-7, -8, and -19 (see next section) (Fig. 4E and 5C). J4a4a-7 and J4a4a-19 showed a replication profile similar to that of J1b1b, with NS5A-positive cells initially detected in 50% of the cell culture but then cleared out as the culture was passaged. As with J1b1b, no infectious virus was secreted into the supernatant. J1a1a and J4a4a both differ from the prototype sequences of H77 and ED43 by 1 (H77-M1205T) and 4 (ED43-T1048A, T1064I, I1160T, R1176A) amino acids, respectively, in the protease gene, whereas J1a1a and J4a4a are identical to H77 and ED43, respectively, in the NS4A gene (see Fig. S2 in the supplemental material at http://www.virus-evolution.org/Downloads/JVI02698-09/). M1205 is predominant among genotype 1a isolates, although one of the 601 available sequences of other genotype 1a isolates contained the Thr residue at this site, one contained Val, and one contained Ile. A total of 4 of 459 genotype 1b isolates in the HCV database contained the Thr residue at this position as well, largely discounting this substitution as a cause of the poor replication ability of the J1a1a clone.
TABLE 1.
Supernatant infectivity titers determined by TCID50 assay
| Viral isolate | Days postelectroporation | TCID50/ml |
|---|---|---|
| J1b1b | 5 | <10 |
| J2a2a | 5 | 104.2 |
| J3a3a | 15 | 103.4 |
| J4a4a-19 | 5 | <10 |
| J5a5a | 5 | 101.9 |
| J6a6a | 33 | 102.6 |
FIG. 5.
J1b1b, J3a3a, J4a4a, and J6a6a study subject-derived protease recombinants and their viability in Huh7.5 cells. The protease region of J1b1b, J3a3a, J4a4a, and J6a6a was replaced with that of two or three study subject-derived protease genes, and their replication capacities were assessed in the Huh7.5 cells.
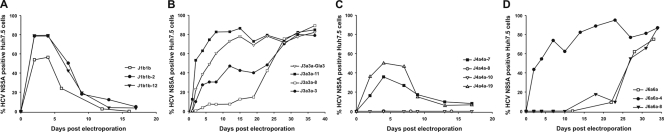
Replication kinetics of J1a1a, J1b1b, J3a3a, J4a4a, and J6a6a containing NS3 protease genes derived from HCV-infected study subjects.
We investigated whether the differing replication kinetics of chimeras constructed from different genotypes were a consistent genotype-associated property or whether it originated from naturally occurring variability between HCV variants within a genotype. For this purpose, we constructed chimeras from protease genes amplified from multiple study subjects infected with genotypes 1a, 1b, 3a, 4a, and 6a that showed markedly different replication kinetics and abilities to generate infectious virus (Fig. 4; Table 1). For each genotype, chimeras were created using PCR-amplified protease sequences from epidemiologically unlinked subjects infected with genotypes 1a, 1b, 3a, 4a, and 6a.
In an initial survey of 33 HCV variants recovered from genotype 3a-infected individuals, amplified protease gene sequences showed 6.9 and 2.7% nucleotide and amino acid sequence divergence, respectively, from each other, consistent with previous analyses of genotype 3a variability within this genome region (27, 60) (see Fig. S1B in the supplemental material at http://www.virus-evolution.org/Downloads/JVI02698-09/). From these 33 consensus sequences, the protease genes from three study subjects were used to replace the protease domain of J3a3a-Gla. Clones containing amino acid polymorphisms not found in any other sequence in the Los Alamos HCV sequence database, GenBank, or the consensus sequences in our data set were discarded (see Fig. S2B [http://www.virus-evolution.org/Downloads/JVI02698-09/]), ensuring representation of naturally occurring polymorphism. All three constructs yielded replicating virus but with different replication kinetics (Fig. 5B). J3a3a-11 replication was detected in most cells on day 9, slightly earlier than that with J3a3a-Gla. J3a3a-8 and J3a3a-3 spread occurred after an eclipse phase of 28 days.
It was unclear whether the differences in the replication kinetics of the J3a3a chimeras represented different degrees of compatibility between the subject-derived protease sequences with the NS4A cofactor sequence derived from the reference strain or whether there were specific compatibility problems with the genotype 2a backbone sequence. To investigate this, we compared the NS4A sequences of the prototype (HCV3a-Gla) with those of the three subjects (see Fig. S2B in the supplemental material at http://www.virus-evolution.org/Downloads/JVI02698-09/). All J3a3a recombinants' NS4A cofactor sequences were identical at the amino acid level, except for an L1670I amino acid substitution in the J3a3a-11 recombinant. L1670 is dominant among genotype 3a isolates, although 14 sequences with Val and 3 with Phe were identified among the 180 isolates analyzed. These observations indicate that incompatibility between the NS3 protease and the prototype HCV3a-Gla NS4A is most likely not a determinant for the observed differences in the replication kinetics of the J3a3a recombinants.
By the same methods, J1a1a recombinants with subject-derived proteases were created. NS3 protease sequences obtained from 31 subjects infected with genotype 1a showed 7.6 and 2.5% nucleotide and amino acid sequence divergence, respectively, from each other, again representing the naturally occurring diversity of this genotype (20, 27, 57, 60) (see Fig. S1A in the supplemental material at http://www.virus-evolution.org/Downloads/JVI02698-09/). The replacement of the NS3 protease of J1a1a-H77* with that of genotype 1a-infected subject-derived proteases resulted in replicating virus in 7 (J1a1a-1, -2, -3, -4, -5, -6, and -8) out of 9 recombinant viruses. None, however, spread to more than 0.08% of the cell culture, and all cells were NS5A negative after 30 days (data not shown). Except for J1a1a-1, all J1a1a recombinants contained one or more naturally occurring amino acid polymorphisms represented among sequences of the 570 isolates available from GenBank and the 31 isolates analyzed in this study (see Fig. S1 in the supplemental material at http://www.virus-evolution.org/Downloads/JVI02698-09/). In contrast, J1a1a-1, which was the recombinant showing the best replication kinetics (data not shown), contained one amino acid substitution (G1056) that does not occur in any other isolate analyzed. To investigate whether the H77*-NS4A cofactor was responsible for the severally impaired replication fitness of the J1a1a recombinants, NS4A amino acid sequences obtained from subjects whose proteases were used in the recombinants were compared with that of H77*. The membrane segment and the NS3 cofactor region showed amino acid identity (see Fig. S2A [http://www.virus-evolution.org/Downloads/JVI02698-09/]) (4). Some polymorphism occurred in the C-terminal domain of the NS4A cofactor, which, however, is not involved in the direct interaction with the NS3 protease. Differences in replication kinetics between the different J1a1a recombinants are therefore most likely due to the protease itself. Whether the overall poor replication kinetics of the J1a1a recombinants can be attributed to the reduced enzymatic function of the 1a proteases or to their incompatibility with the remaining type 2a sequence remains to be investigated.
The generation of J4a4a subject-derived protease recombinants resulted in a replicating virus in three (J4a4a-7, -8, and -9) out of four (J4a4a-7, -8, -9, and -10) recombinant viruses (Fig. 5C). A total of 40 to 50% of all cells were positive for NS5A at 4 days posttransfection with J4a4a-7 and J4a4a-19 RNA, whereas J4a4a-8 replicated only to very low levels. J4a4a-ED43* (A1048 and I1064), J4a4a-7 (R1056), J4a4a-10 (P1169), and J4a4a-19 (I1153) all showed amino acid polymorphisms which do not exist in any other genotype 4a isolate analyzed (n = 39) (see Fig. S2 in the supplemental material at http://www.virus-evolution.org/Downloads/JVI02698-09/). Only recombinant J4a4a-8 shows an amino acid polymorphism that also exists within the 4a isolates from the database. It is unclear which amino acid polymorphism contributes to the impaired replication phenotype in J4a4a-ED43* and J4a4a-10.
The generation of J1b1b subject-derived protease recombinants resulted in replicating virus for both of the generated recombinants (J1b1b-2 and J1b1b-12), with both showing replication kinetics similar to that of the prototype recombinant J1b1b (Fig. 5A). The same was true for J6a6a subject-derived protease recombinants (Fig. 5D). J6a6a-4 immediately infected 70 to 80% of the cell culture, whereas J6a6a-8 took 30 days to spread to the rest of the cell culture, similar to the prototype recombinant J6a6a.
Identification of adaptive and attenuating mutations in recovered Jx and Jxx viruses.
J5a, J6a6a, and the subject-derived chimeras J3a3a-3 and J3a3a-8 spread only in the cell culture after an eclipse phase. To investigate whether the delay in replication reflected a requirement for adaptive mutations to allow efficient replication, we determined the nucleotide sequence from the protease and NS4A regions amplified from cell culture supernatants after the chimera had spread to 80% of the cell culture (Table 2). The gene regions from nt position 2863 to 4178, which included the 3′ end of NS2 and the 5′ end of the NS3 helicase, and nt position 5230 to 5536, including NS4A, were sequenced at peak infectivity of the individual viruses. In the recovered genomes of passaged J3a3a-8 and J3a3a-3, no substitutions were detected, except in NS4A, where all 5 clones analyzed had the C5328G and T5329C substitutions, among others. The resulting L1663A amino acid substitution only became dominant in the J3a3a-Gla recombinant after 43 days of cell culture passaging. J5a had two substitutions within the NS3 protease domain (A3649G and C3854G), leading to the amino acid substitutions N1103S and C1171W. Within the type 2a NS4A, one nucleotide substitution (G5430A) led to the amino acid change D1679N. The J6a6a recombinant virus had two substitutions (A3558G and G3439T), resulting in a V1040L amino acid change within the N-terminal part of the NS3 protease. This amino acid is highly conserved among all genotypes, allowing only Leu, Val, or Ile (4).
TABLE 2.
Mutations of Jxs and Jxxs during passaging in Huh7.5 cells
| HCV chimera | Mutationsa |
||
|---|---|---|---|
| NS3p | NS3h | NS4A | |
| J5a | A3649G | G5430A | |
| C3854G | |||
| J2a2a | C3538G | ||
| J3a3a | C5328G+T5329C | ||
| J3a3a-3 all | C5328G+T5329C | ||
| Clones 1, 4, and 5 | C5328G+T5329C | ||
| Clone 2 | C5328G+T5329C+T5365C | ||
| Clone 3 | C5328G+T5329C+T5447C | ||
| Clone 6 | C5328G+T5329C+T5358C | ||
| J3a3a-8 | |||
| Clones 1, 4, and 5 | C5328G+T5329C | ||
| Clone 2 | C5328G+T5329C | ||
| T5389C+A5434G | |||
| Clone 3 | C5328G+T5329C+A5472G | ||
| J5a5a | C3416G | T3968C | |
| A4081T | |||
| J6a6a | A3458G | ||
| G3459T | |||
Abbreviations: NS3p, NS3 protease; NS3h, NS3 helicase. Nucleotide positions are numbered according to H77 (GenBank accession no. AF009606) reference positions.
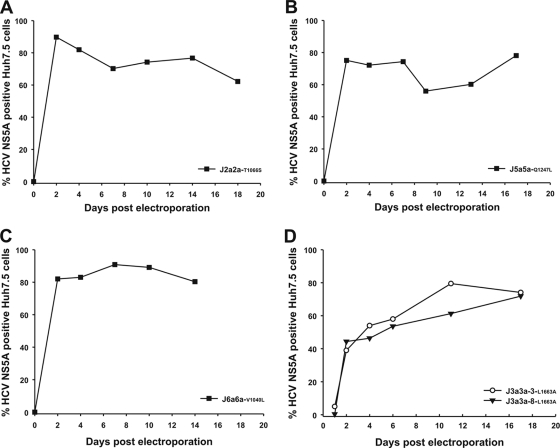
Both J2a2a and J5a5a replicated to high levels in cell culture, accompanied by increased cell death. After an initial clearance of the virus, both spread in cell culture again, indicating the acquisition of attenuating mutations (Fig. 4C and F). A C3538G nucleotide change within the protease domain of J2a2a leads to a T1006S amino acid change. Within J5a5a, three nucleotide changes (C3416G, T3968C, and A4081T), leading to a Q1247L amino acid change, were identified (Table 2).
Recombinant adapted/attenuated viruses efficiently infect Huh7.5 cells.
To identify whether the substitutions occurring in NS4A accounted for the differences in replication kinetics of the J3a3a-3 and -8 recombinant viruses, mutations were introduced into the original plasmids, and their viability was tested in Huh7.5 cells. Both J3a3a-3-L1663A (J3a3a-3-mt) and J3a3a-8-L1663A (J3a3a-8-mt) spread directly within the cell culture, and after 11 and 17 days, respectively, almost all cells were infected, with replication kinetics similar to those of J3a3a-Gla and J3a3a-11 (Fig. 5B and 6D). The L1663A amino acid (C5328G and T5329C) change at the 5′ end of NS4A was therefore sufficient to restore efficient replication. Reintroducing the V1040L amino acid change into J6a6a generated a recombinant (J6a6a-V1040L) which showed improved replication kinetics as well (Fig. 5C). At 2 days postelectroporation, J6a6a-V1040L replicated in 80% of the cell culture and continued to infect most of the cell culture at 14 days postelectroporation. Both J2a2a and J5a5a replicated to high levels, but replication was accompanied by increased cell death. Reintroduction of the identified mutations (Table 2) generated two attenuated viruses (Fig. 6A and B). Both J2a2a-T1066S and J5a5a-Q1247L were immediately able to infect about 80% of the cell culture and continued to infect the majority of cells at 14 days postelectroporation.
FIG. 6.
Viability of J2a2a, J5a5a, J6a6a, and J3a3a recombinants with study subject-derived NS3 protease genes, including adaptive/attenuating mutations observed in clonal analysis.
Susceptibility of different HCV genotypes to protease inhibitors.
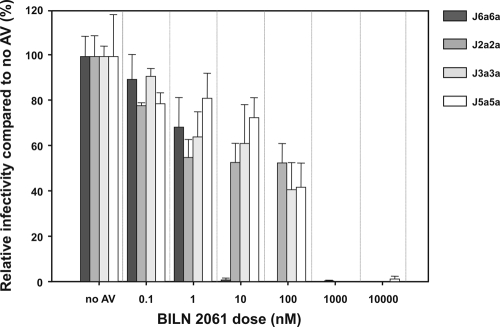
Intra- and intergenotypic recombinants derived from genotypes 2a, 3a, 5a, and 6a that produced infectious virus were evaluated for protease inhibitor sensitivity. We wished to determine how the activity of BILN 2061 was affected by the sequence differences of enzymes from various genotypes (Fig. 3). Huh7.5 cells were infected with virus containing supernatant (MOI of 0.015), and then the reduction in the number of FFU/ml upon BILN 2061 treatment was assessed (Fig. 7). Supernatant infectivities of all recombinant viruses were inhibited but to different extents. J2a2a, J3a3a, and J5a5a showed similar dose responses (IC50 = 210 nM, 80 nM, and 110 nM, respectively), whereas J6a6a (IC50 = 2 nM) was reproducibly 100-fold more susceptible to BILN 2061.
FIG. 7.
Antiviral inhibition of Jxxs. Reduction in supernatant infectivity. After 8 h of inoculation with Jxx (MOI, 0.015), Huh7.5 cells were washed and incubated in media containing 0.1% DMSO, as a carrier control, with or without the indicated doses of BILN 2061. Inhibition was calculated at 72 h postinfection as a reduction in supernatant infectivity (the number of FFU/ml; mean ± SEM; n = 3) after antiviral (AV) addition compared to infectivity of the control without antiviral.
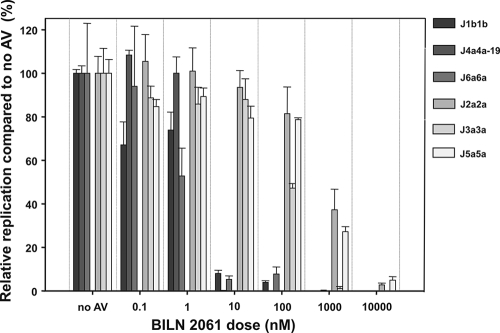
Since no infectious virus could be generated for J1b1b and J4a4a-19, protease inhibitor susceptibility was also assessed after synthetic RNA had been electroporated into fresh Huh7.5 cells (Fig. 8). After 24 h, protease inhibitor was added, and the reduction in the frequency of NS5A-positive cells was assessed at 96 h postelectroporation. J1b1b, J4a4a-19, and J6a6a showed 100- to 1,000-fold-greater susceptibility (IC50 = 3 nM, 1 nM, and 1 nM, respectively) than J2a2a, J3a3a, and J5a5a (IC50 = 720 nM, 105 nM, and 480 nM, respectively).
FIG. 8.
Antiviral inhibition of Jxxs. Reduction in viral replication. RNA (1 to 10 μg) was electroporated into Huh7.5 cells and incubated for 24 h. Cells were then washed and incubated in media containing 0.1% DMSO, as a carrier control, with or without the indicated doses of BILN 2061 for a further 72 h. The percent inhibition of replication was determined at 96 h postelectroporation (mean ± SEM; n = 3) and calculated as the ratio of NS5A-positive cells in BILN 2061-treated cells to those of the control without antiviral.
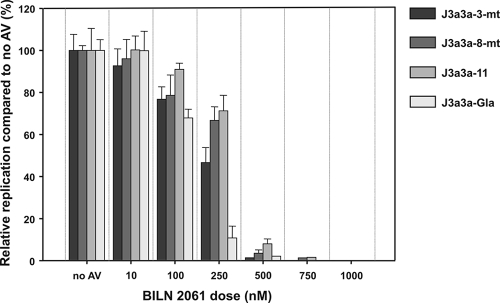
To investigate whether naturally occurring sequence variability within a genotype led to differences in antiviral susceptibilities, the J3a3a recombinants with study subject-derived proteases (J3a3a-3-mt, -8-mt, and -11) were subjected to BILN 2061 treatment as described above. The J3a3a-Gla chimera generated from the HCV3a-Gla prototype sequence showed an IC50 of 130 nM, comparable to previous assays (Fig. 8) but 2 to 3-fold lower than the IC50s of the subject-derived sequences (310 nM for J3a3a-11, 300 nM for J3a3a-8-mt, and 240 nM for J3a3a-3-mt) (Fig. 9). Although requiring further evaluation of more replicates' antiviral dilutions to establish formal statistical significance, these small but reproducible differences in apparent susceptibility suggest that some of the naturally occurring sequence variability within a subtype or genotype might have a direct and potentially clinically significant effect on response to antiviral therapy.
FIG. 9.
Antiviral inhibition of J3a3a recombinants with study subject-derived proteases. RNA (10 μg) was electroporated into Huh7.5 cells and incubated for 24 h. Cells were then washed and incubated in media containing 0.1% DMSO, as a carrier control, with or without the indicated doses of BILN 2061 for a further 72 h. The percent inhibition of replication was determined at 96 h postelectroporation (mean ± SEM; n = 3) and calculated as the ratio of NS5A-positive cells in BILN 2061-treated cells to those of the control without antiviral.
In vitro selection of BILN 2061-resistant recombinant viruses.
Chimeras constructed from HCV genotypes 1 to 6 were passaged in initially subinhibitory but increasing concentrations of BILN 2061 beyond the IC50s determined for each genotype to investigate whether antiviral resistance could be induced in vitro. After replicate transfection with chimeras of each genotype, BILN 2061 was added initially at concentrations approximately 10% of the IC50 and increased to final concentrations greater than the IC50 recorded for each genotype (individual schedules are provided in Table S1 in the supplemental material at http://www.virus-evolution.org/Downloads/JVI02698-09/). Attenuated clones of genotypes 2a (J2a2a-T1066S) and 5a (J5a5a-Q1247L) were used to allow long-term passage. For each genotype, control passages without addition of BILN 2061 were performed. Every 3 or 4 days, cells were split, the medium was replenished, and fresh BILN 2061 was added. At the end of passaging, HCV RNA was extracted from cell pellets of each replicate and control and subjected to RT-PCR to amplify the coding region of the HCV NS3 serine protease domain. To delineate the identity and frequency of substitutions, the RT-PCR product from each genotype passage was subcloned into the TOPO vector, and 10 individual colonies of two replicates and 10 clones from the control were subjected to sequencing.
All 6 genotypes showed substitutions at position 168, although this position differed in both the identity of the wild type-encoded amino acid and the substitutions that arose on passaging (Table 3). In contrast, this site remained invariant in each control passage experiment performed in parallel without addition of BILN 2061. Genotypes 2, 3, and 5 showed complete replacement of the wild-type codon in both replicate passages, while replacement frequencies of genotypes 1b, 4a, and 6a ranged from 70% to 90%. Genotypes 1b and 4a additionally showed a further substitution of the Ala residue at position 156 for Val and Gly in a proportion of clones (Table 3). One of the two replicates from the genotype 2a chimera showed a change at position 195 in half of the clones analyzed. All other substitutions recorded among clones in either BILN 2061 or control passages occurred infrequently (0 to 4 among each set of 10 clones), at variable positions, and were equally frequent among BILN 2061-passaged and control virus populations (data not shown).
TABLE 3.
Acquisition of mutations in NS3 during passaging in BILN 2061
| Clone | BILN 2061 |
% remaining wild type (mutation[s])c |
||||||
|---|---|---|---|---|---|---|---|---|
| Genotype | Conc/nMa | Day | Position | WTb | Replicate 1 | Replicate 2 | Control | |
| J1b1b | 1b | 230 | 8 | 168 | D | 0 (7G, 1A, 1E, 1V) | 10 (3G, 1A, 5E) | 100 |
| 156 | A | 60 (4V) | 90 (1V) | 100 | ||||
| J2a2a-T1066S | 2a | 1,000 | 22 | 168 | D | 0 (8V, 2Y) | 0 (4V, 6Y) | 100 |
| 195 | Q | 100 | 40 (6H) | 100 | ||||
| J3a3a | 3a | 7,000 | 21 | 168 | Q | 0 (4L, 5R, 1K) | 0 (8L, 2K) | 100 |
| J4a4a-19 | 4a | 100 | 13 | 168 | D | 20 (6G, 2V) | 20 (4G, 2A, 2E) | 100 |
| 156 | A | 80 (2V) | 80 (2T) | 100 | ||||
| J5a5a-Q1240L | 5a | 500 | 21 | 168 | E | 0 (7A, 1V, 2G) | 10 (9A) | 100 |
| J6a6a | 6a | 10 | 28 | 168 | D | 30 (4V, 2E, 1H) | 30 (3V, 1E, 3N) | 100 |
Final passage concentration of BILN 2061.
WT, wild type.
Proportion of clones retaining the original amino acid results from two replicate passage experiments (10 clones analyzed in each) and in a control passaged without BILN 2061 (10 clones analyzed).
DISCUSSION
Genetic variability of HCV proteins of different genotypes influences the molecular structure of protease and polymerase enzymatic sites and potentially limits the effectiveness of antiviral therapy targeting viral replication proteins (20). For example, BILN 2061, a protease inhibitor specifically developed for genotype 1, shows a nearly 2 log-weaker binding affinity to genotype 2 and 3 proteases than to genotype 1 protease (55), a difference that translates to much weaker antiviral efficacy of BILN 2061 among subjects with HCV genotype 2 or 3 infection (43) than those with HCV genotype 1 infection (24). The reduced effectiveness of antiviral drugs on certain genotypes also potentially facilitates the development of resistance mutations. Another PI, VX-950, showed similar efficacy in genotypes 1a, 1b, and 2a but lower efficacy in genotype 3a (37, 42).
Current drug discovery and optimization are mostly dependent on laboratory-optimized standard replicons. However, the standard replicon system allows only the evaluation of a limited number of laboratory strains that do not reflect the great genetic inter- and intragenotypic diversity of HCV. Different replicon-based vector approaches have to date been limited to the investigation of the activity of compounds against different HCV isolates from genotypes 1 to 4 (3, 40). The replicon-based system, however, does not include structural genes and therefore does not represent the full viral life cycle of HCV. We describe here the development of a full-length HCV cell culture system, allowing the study of the NS3 protease of all 6 genotypes. Recombinant viruses were constructed from Jc1, an intragenotypic genotype 2a recombinant, which replicates efficiently in the Huh7.5 cells (38). The resulting recombinant viruses encode the NS3 protease or the NS3 protease and the NS4A gene from genotype 1a, 1b, 2a, 3a, 4a, 5a, or 6a prototype strains or different clinical isolates, respectively.
The original attempt to replace the NS3 protease alone was not successful in creating replication-competent intergenotypic viruses, except for genotype 5a (Fig. 2B), which spread only within the cell culture after acquiring adaptive mutations (Table 2). Since the NS3 protease forms a stable complex with NS4A and is required for its trans-cleavage activity (10, 11, 54, 56), we reasoned that the poor replication ability of most Jx constructs may have originated through incompatibilities between these two protease components. It has, however, been shown that the NS3 protease domain can functionally cross-interact with the NS4A cofactor from another genotype (13, 61), and NS3 protease genes from different genotypes cloned into 1b/2a reference strain replicons do produce replication-competent replicons. Attempts by other groups to include full-length NS3/4A genes of other genotypes, including the helicase, in the 1b or 2a chimeric replicon system generally failed to create efficiently replicating replicons, although in the latter case, this may reflect a further compatibility restriction for the helicase to be of the same genotype as the RNA polymerase encoded by NS5B (3, 40).
The NS4A of genotype 2 has been shown to be much less efficient in heterologous combinations than those of other genotypes (61) and might explain why most of our recombinant viruses containing the type 2a cofactor did not replicate efficiently enough for detection. NS4As of genotypes 1a, 1b, 3a, 4a, 5a, and 6a are more similar to each other on the amino acid sequence level than to genotype 2a (Fig. 3). Why, however, genotype 5a protease forms a viable complex with the 2a backbone and the others do not remains unclear.
To resolve this compatibility issue, we included the corresponding homologous genotype-specific NS4A cofactor in the construction of the recombinant viruses (Fig. 3). This led to viable chimeras for genotypes 1b, 2a, 3a, 5a, and 6a (Fig. 4). J1a1a and J4a4a were impaired in their replication, but replacement of the NS3 protease gene in J4a4a with that of study subjects allowed the generation of replication-competent 4a recombinants (Fig. 4E and 5C). As expected, J2a2a replicated most efficiently and immediately spread within the cell culture (Fig. 4C), and J5a5a also spread to most cells within 2 days (Fig. 4F), both comparable with the parental Jc1. Similarly, the spread of Jc1 and these two recombinant viruses was also accompanied by increased cell death, followed by proliferation of HCV-NS5A-negative Huh7.5 cells, as described in previous studies (14, 32, 45, 66). Reintroduction of attenuating mutations (Table 2) allowed the generation of two recombinants, which immediately and continuously infected the cell culture (Fig. 6A and B). In contrast, the observed clearance of J1b1b and J4a4a-19 after transfection is the likely consequence of their failure to generate any infectious virus (Fig. 4B and E). Unlike the other recombinant viruses, where spread is reduced after an eclipse phase, J3a3a continuously infected about 80% of the cell culture, with high frequencies of infected cells even after 70 days of passaging in cell culture (Fig. 4D). As it has been shown that the catalytic efficiencies of genotype 1a, 3a, and 4a proteases are similar (13), the inability of genotype 1a and some genotype 4a chimeras to replicate efficiently likely is a consequence of the incompatibility between the NS3 protease/NS4A and the remaining type 2 sequence. The J6a6a recombinant spread within the cell culture after an eclipse phase, indicating adaptive mutations (Fig. 4F). The identified mutation within the NS3 protease (V1040L) is a change at a position with a highly conserved L/V/I polymorphism, and reintroduction of it into the original recombinant rescued its impaired replication kinetic (Fig. 6C) (4).
Even though the NS3 protease is considered one of the more conserved proteins encoded by the HCV genome, different genotypes do show substantial amino acid sequence variability that potentially influences its structure and function (20, 27, 57, 60). Furthermore, it has been demonstrated that catalytic efficiencies within a subtype can vary widely, especially within genotype 1b, whereas genotype 3a proteases showed the most homogenous range of activities (13). Cloning of subject-derived NS3 protease genes of genotype 1 into the 1b replicon construct by Qi et al. has shown 2- to 7-fold differences in the replication capacities of the constructs (40). To investigate the influence of these intrasubtype sequence differences on chimera viability, we directly amplified and cloned NS3 protease gene sequences from study subject plasma samples into the corresponding J1a1a, J1b1b, J3a3a, J4a4a, and J6a6a recombinant viruses.
All three J3a3a recombinants constructed from the original J3a3a-Gla and subject-derived protease genes generated viable chimeras (Fig. 5B). Recombinants with diminished replication kinetics could be rescued by introducing an adaptive L1663A amino acid change within the membrane segment of NS4A (Fig. 6D). The L1663A amino acid change likely is crucial for efficient and continuous spread of the J3a3a recombinants, as it was observed within the J3a3a-Gla recombinant as well after 43 days in cell culture (Table 2). Furthermore, L1663A is an amino acid change toward the consensus genotype 2a sequence, suggesting adaptation to the Jc1 backbone sequence.
Similarly, NS3 proteases from study subjects infected with genotypes 1b, 4a, and 6a were cloned into the corresponding J1b1b, J4a4a, and J6a6a recombinants. Both J1b1b recombinants replicated to the same extent as the prototype J1b1b, indicating that patient-derived sequences can be easily swapped and analyzed (Fig. 5A). Replacement of the NS3 protease gene of the J4a4a prototype recombinant with those amplified from genotype 4a-infected subjects reversed the impaired replication phenotype in three (J4a4a-7, -8, and -19) out of four cases (Fig. 5C). To which amino acid polymorphism this discrepancy is due is unclear, as not only J4a4a and J4a4a-10 but also J4a4a-19 have an amino acid polymorphism that does not occur in any other sequence of our analyzed data set. Replacement of the J6a6a NS3 protease with that of subject-derived genes resulted in replicating recombinants in both cases (J6a6a-4 and -8) (Fig. 5D). J6a6a-4 immediately spread within the cell culture and maintained a continuous infection, whereas J6a6a-8 showed a replication profile similar to that of prototype J6a6a. V1040L has been identified to be a crucial adaptive change for J6a6a. J6a6a-8 also contains a Val at position 1040, whereas J6a6a-4 contains an Ile, a bulkier amino acid similar to Leu and which may underlie its improved replication kinetics.
In a similar manner, NS3 proteases from study subjects infected with genotype 1a were cloned into the corresponding J1a1a recombinants. The replication kinetics of the resulting chimeras were very low or below detection limits, and none of the viruses were able to acquire adaptive mutations; all Huh7.5 cell cultures were NS5A negative after 30 days. It has been reported that the in vitro catalytic efficiency of the genotype 1a NS3 protease is similar to those of 3a and 4a, both of whose chimeras were replication competent in vitro (13). It is therefore unlikely that the J1a1a recombinants reproduce less efficiently than those of J3a3a, J4a4a-7, and J4a4a-19 because of differences in the enzymatic activity of the proteases. This suggests that genotype 1a NS3 protease/NS4A are less compatible with the genotype 2a backbone than genotypes 1b, 3a, 4a, 5a, and 6a.
Susceptibility and resistance development of different HCV genotypes to PIs.
The HCV-NS3 serine protease is essential for viral replication and therefore an attractive target for HCV-specific antiviral therapy (36). One of the first PIs developed, BILN 2061, was highly optimized for genotype 1 enzymes, making its binding more sensitive to sequence differences than substrate binding. However, it still shows Ki values below 100 nM for nongenotype 1 NS3-4A proteins (55). Using the intergenotype chimeras developed in the current study, we showed J1b1b and J4a4a-19 to be more susceptible to BILN 2061 than J2a2a and J3a3a, as has been previously demonstrated in vitro and in vivo (43, 55). Interestingly, J4a4a-19 and J6a6a were equally as susceptible to BILN 2061 as J1b1b and J5a5a, similarly to J2a2a and J5a5a. Experiments using expressed proteins showed similar IC50s for genotypes 1a, 1b, 4a, 5a, and 6a, whereas we observed a lower susceptibility level for J5a5a toward BILN 2061 (31, 46). Equivalent differences in susceptibility to BILN 2061 sensitivity were observed whether determined by supernatant infectivity reduction or inhibition of replication (Fig. 7 and 8). Differences in susceptibility extended even to variants within a genotype, with IC50s ranging from around 130 nM to 310 nM in a panel of subject-derived and reference genotype 3a protease sequences (Fig. 9). This 2- to 3-fold difference may influence the effectiveness of antiviral therapy.
The rapid selection of viral variants displaying drug-resistant phenotypes is a major concern in HCV treatment. Several resistant phenotypes have been observed in patients experiencing viral rebound during therapy and in replicon experiments (8, 23, 25, 28). Resistant mutations arising under BILN 2061 pressure have been described at the three main positions, Arg155, Ala156, and Asp168 (8, 25, 28). In this study, we identified several new in vitro resistance mutations in the NS3 protease gene of all 6 major genotypes. The dominant resistance mutations observed against BILN 2061 occurred at position 168, where the Asp residue in genotypes 1b, 2a, 4a, and 6a changed to Gly/Glu/Val/Ala (genotypes 1b and 4a), Val/Tyr (2a), and Val/Asn/Glu/His (6a). In the genotype 3a protease, a replacement of Gln168 by Leu/Arg/Lys was detected, and in the genotype 5a protease, Glu168 was replaced by Ala/Val/Gly. Although conserved in position, a striking feature of this pattern of mutation was the variable nature of the substituted amino acid, suggesting great flexibility in the nature of the structural disruption to the protease induced by these resistance-associated changes. The observation that resistance-associated mutations in one genotype (such as D168E substitution in genotypes 1b and 6a) may occur as the wild-type amino acid in another (genotype 5) is consistent with significant structural differences in the protease gene implied by the differences between genotypes in their susceptibility to inhibition by BILN 2061.
Although further mutations at positions 155 and 156 have been associated with BILN 2061 resistance (25, 28), substitutions at position 156 were observed only in minority populations of genotype 1b and in genotype 4a. The exact phenotypic effects of substitutions here and of further mutations potentially associated with resistance, such as the Q195H mutation in genotype 2a (Table 3), will require specific site-directed mutagenesis and IC50 determinations, along with comparisons of resistance profiles of the many different substitutions and genotype backgrounds at position 168. Progress toward characterization of these mutations and combinations will in the long term provide the necessary data for the development of genotypic drug resistance databases analogous to those developed for human immunodeficiency virus type 1.
In summary, we have developed a full-length HCV cell culture system, which allows the investigation of protease gene function in all six genotypes, a much greater range than the genotypes 2a and 1a full-length replication-competent clones described previously (26, 34, 59, 64). The developed system represents a powerful tool to study the NS3 protease within the full viral life cycle and adds to the currently available JFH1-based systems for the study of the nonstructural genes (14, 15, 21, 38, 45, 65). Overall, this system has allowed us to demonstrate that different NS3 proteins react differently to the same protease inhibitor. Any protease inhibitor identified in high-throughput screening can be evaluated for its efficacy on different genotypes and treatments designed according to the outcome. Passaging the recombinants in subinhibitory concentrations of antiviral drugs allowed us to identify potential resistance mutations to BILN 2061 and will therefore be of considerable value in preclinical assessment of resistance induction and genotypic variability for newly developed compounds.
The ease with which protease gene sequences amplified directly from clinical specimens can be inserted in the expression vector of the appropriate genotypes allows for the first time direct monitoring of antiviral drug inhibition for specific proteases through assessment of reduction in both supernatant infectivity and replication kinetics. Protease genes from study subjects naïve to treatment can then easily be assessed for sensitivity to a range of antiviral drugs and screened for preoccurring resistance mutations, as well as providing a phenotypic assay for rapid assessment of emerging resistance during therapy and the influence of specific mutations on treatment outcome.
Acknowledgments
We are grateful to David Evans for help in designing new restriction sites. We thank Mark Harris for providing Huh7.5 cells and the sheep anti-NS5A serum. Furthermore, we thank T. Wakita, R. Bartenschlager, J. Bukh, E. A. McCruden, and R. Elliot for providing us with HCV plasmids.
This work was supported by a studentship to I.I. from the BBSRC.
Footnotes
Published ahead of print on 17 February 2010.
REFERENCES
- 1.Anonymous. 1999. Global surveillance and control of hepatitis C. Report of a WHO Consultation organized in collaboration with the Viral Hepatitis Prevention Board, Antwerp, Belgium. J. Viral Hepat. 6:35-47. [PubMed] [Google Scholar]
- 2.Bartenschlager, R., V. Lohmann, T. Wilkinson, and J. O. Koch. 1995. Complex formation between the NS3 serine-type proteinase of the hepatitis C virus and NS4A and its importance for polyprotein maturation. J. Virol. 69:7519-7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binder, J., S. Tetangco, M. Wick, K. Maegley, L. Lingardo, A. Patick, and G. Smith. 2007. Development of hepatitis C virus (HCV) chimeric replicons for identifying broad spectrum NS3 protease inhibitors. Antiviral Res. 74:A38. [DOI] [PubMed] [Google Scholar]
- 4.Brass, V., J. M. Berke, R. Montserret, H. E. Blum, F. Penin, and D. Moradpour. 2008. Structural determinants for membrane association and dynamic organization of the hepatitis C virus NS3-4A complex. Proc. Natl. Acad. Sci. U. S. A. 105:14545-14550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukh, J. 1995. Genetic heterogeneity of hepatitis C virus: quasispecies and genotypes. Semin. Liver Dis. 15:41-63. [DOI] [PubMed] [Google Scholar]
- 6.Chen, S. L., and T. R. Morgan. 2006. The natural history of hepatitis C virus (HCV) infection. Int. J. Med. Sci. 3:47-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choo, Q. L., K. H. Richman, J. H. Han, K. Berger, C. Lee, C. Dong, C. Gallegos, D. Coit, R. Medina Selby, P. J. Barr, A. J. Weiner, D. W. Bradley, G. Kuo, and M. Houghton. 1991. Genetic organization and diversity of the hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 88:2451-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cubero, M., J. I. Esteban, T. Otero, S. Sauleda, M. Bes, R. Esteban, J. Guardia, and J. Quer. 2008. Naturally occurring NS3-protease-inhibitor resistant mutant A156T in the liver of an untreated chronic hepatitis C patient. Virology 370:237-245. [DOI] [PubMed] [Google Scholar]
- 9.de Bruijne, J., C. J. Weegink, P. L. Jansen, and H. W. Reesink. 2009. New developments in the antiviral treatment of hepatitis C. Vox Sang. 97:1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Failla, C., L. Tomei, and R. Defrancesco. 1994. Both NS3 and NS4A are required for proteolytic processing of hepatitis C virus nonstructural proteins. J. Virol. 68:3753-3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Failla, C., L. Tomei, and R. Defrancesco. 1995. An amino-terminal domain of the hepatitis C virus NS3 protease is essential for interaction with NS4A. J. Virol. 69:1769-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forestier, N., H. W. Reesink, C. J. Weegink, L. McNair, T. L. Kieffer, H. M. Chu, S. Purdy, P. L. Jansen, and S. Zeuzem. 2007. Antiviral activity of telaprevir (VX-950) and peginterferon alfa-2a in patients with hepatitis C. Hepatology 46:640-648. [DOI] [PubMed] [Google Scholar]
- 13.Franco, S., B. Clotet, and M. A. Martinez. 2008. A wide range of NS3/4A protease catalytic efficiencies in HCV-infected individuals. Virus Res. 131:260-270. [DOI] [PubMed] [Google Scholar]
- 14.Gottwein, J. M., T. K. H. Scheel, A. M. Hoegh, J. B. Lademann, J. Eugen-Olsen, G. Lisby, and J. Bukh. 2007. Robust hepatitis C genotype 3a cell culture releasing adapted intergenotypic 3a/2a (S52/JFH1) viruses. Gastroenterology 133:1614-1626. [DOI] [PubMed] [Google Scholar]
- 15.Gottwein, J. M., T. K. Scheel, T. B. Jensen, J. B. Lademann, J. C. Prentoe, M. L. Knudsen, A. M. Hoegh, and J. Bukh. 2009. Development and characterization of hepatitis C virus genotype 1-7 cell culture systems: role of CD81 and scavenger receptor class B type I and effect of antiviral drugs. Hepatology 49:364-377. [DOI] [PubMed] [Google Scholar]
- 16.Grakoui, A., C. Wychowski, C. Lin, S. M. Feinstone, and C. M. Rice. 1993. Expression and identification of hepatitis C virus polyprotein cleavage products. J. Virol. 67:1385-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hezode, C., N. Forestier, G. Dusheiko, P. Ferenci, S. Pol, T. Goeser, J. P. Bronowicki, M. Bourliere, S. Gharakhanian, L. Bengtsson, L. McNair, S. George, T. Kieffer, A. Kwong, R. S. Kauffman, J. Alam, J. M. Pawlotsky, and S. Zeuzem. 2009. Telaprevir and peginterferon with or without ribavirin for chronic HCV infection. N. Engl. J. Med. 360:1839-1850. [DOI] [PubMed] [Google Scholar]
- 18.Hijikata, M., N. Kato, Y. Ootsuyama, M. Nakagawa, and K. Shimotohno. 1991. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc. Natl. Acad. Sci. U. S. A. 88:5547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hijikata, M., H. Mizushima, Y. Tanji, Y. Komoda, Y. Hirowatari, T. Akagi, N. Kato, K. Kimura, and K. Shimotohno. 1993. Proteolytic processing and membrane association of putative nonstructural proteins of hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 90:10773-10777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holland-Staley, C. A., L. C. Kovari, E. M. Golenberg, K. J. Pobursky, and D. L. Mayers. 2002. Genetic diversity and response to IFN of the NS3 protease gene from clinical strains of the hepatitis C virus. Arch. Virol. 147:1385-1406. [DOI] [PubMed] [Google Scholar]
- 21.Jensen, T. B., J. M. Gottwein, T. K. Scheel, A. M. Hoegh, J. Eugen-Olsen, and J. Bukh. 2008. Highly efficient JFH1-based cell-culture system for hepatitis C virus genotype 5a: failure of homologous neutralizing-antibody treatment to control infection. J. Infect. Dis. 198:1756-1765. [DOI] [PubMed] [Google Scholar]
- 22.Kuiken, C., K. Yusim, L. Boykin, and R. Richardson. 2005. The Los Alamos hepatitis C sequence database. Bioinformatics 21:379-384. [DOI] [PubMed] [Google Scholar]
- 23.Kuntzen, T., J. Timm, A. Berical, N. Lennon, A. M. Berlin, S. K. Young, B. Lee, D. Heckerman, J. Carlson, L. L. Reyor, M. Kleyman, C. M. McMahon, C. Birch, W. J. Schulze Zur, T. Ledlie, M. Koehrsen, C. Kodira, A. D. Roberts, G. M. Lauer, H. R. Rosen, F. Bihl, A. Cerny, U. Spengler, Z. Liu, A. Y. Kim, Y. Xing, A. Schneidewind, M. A. Madey, J. F. Fleckenstein, V. M. Park, J. E. Galagan, C. Nusbaum, B. D. Walker, G. V. Lake-Bakaar, E. S. Daar, I. M. Jacobson, E. D. Gomperts, B. R. Edlin, S. M. Donfield, R. T. Chung, A. H. Talal, T. Marion, B. W. Birren, M. R. Henn, and T. M. Allen. 2008. Naturally occurring dominant resistance mutations to hepatitis C virus protease and polymerase inhibitors in treatment-naive patients. Hepatology 48:1769-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamarre, D., P. C. Anderson, M. Bailey, P. Beaulieu, G. Bolger, P. Bonneau, M. Bos, D. R. Cameron, M. Cartier, M. G. Cordingley, A. M. Faucher, N. Goudreau, S. H. Kawai, G. Kukolj, L. Lagace, S. R. LaPlante, H. Narjes, M. A. Poupart, J. Rancourt, R. E. Sentjens, R. St. George, B. Simoneau, G. Steinmann, D. Thibeault, Y. S. Tsantrizos, S. M. Weldon, C. L. Yong, and M. Llinas-Brunet. 2003. An NS3 protease inhibitor with antiviral effects in humans infected with hepatitis C virus. Nature 426:186-189. [DOI] [PubMed] [Google Scholar]
- 25.Lin, C., K. Lin, Y. P. Luong, B. G. Rao, Y. Y. Wei, D. L. Brennan, J. R. Fulghum, H. M. Hsiao, S. Ma, J. P. Maxwell, K. M. Cottrell, R. B. Perni, C. A. Gates, and A. D. Kwong. 2004. In vitro resistance studies of hepatitis C virus serine protease inhibitors, VX-950 and BILN 2061: structural analysis indicates different resistance mechanisms. J. Biol. Chem. 279:17508-17514. [DOI] [PubMed] [Google Scholar]
- 26.Lindenbach, B. D., M. J. Evans, A. J. Syder, B. Wolk, T. L. Tellinghuisen, C. C. Liu, T. Maruyama, R. O. Hynes, D. R. Burton, J. A. McKeating, and C. M. Rice. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623-626. [DOI] [PubMed] [Google Scholar]
- 27.Lodrini, S., S. Bagaglio, F. Canducci, M. S. De Mitri, P. Andreone, E. Loggi, A. Lazzarin, M. Clementi, and G. Morsica. 2003. Sequence analysis of NS3 protease gene in clinical strains of hepatitis C virus. J. Biol. Regul. Homeost. Agents 17:198-204. [PubMed] [Google Scholar]
- 28.Lu, L., T. J. Pilot-Matias, K. D. Stewart, J. T. Randolph, R. Pithawalla, W. He, P. P. Huang, L. L. Klein, H. Mo, and A. Molla. 2004. Mutations conferring resistance to a potent hepatitis C virus serine protease inhibitor in vitro. Antimicrob. Agents Chemother. 48:2260-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ludmerer, S. W., D. J. Graham, M. Patel, K. Gilbert, M. Stahlhut, and D. B. Olsen. 2008. A transient cell-based phenotype assay for hepatitis C NS3/4A protease: application to potency determinations of a novel macrocyclic inhibitor against diverse protease sequences isolated from plasma infected with HCV. J. Virol. Methods 151:301-307. [DOI] [PubMed] [Google Scholar]
- 30.Malcolm, B. A., R. Liu, F. Lahser, S. Agrawal, B. Belanger, N. Butkiewicz, R. Chase, F. Gheyas, A. Hart, D. Hesk, P. Ingravallo, C. Jiang, R. Kong, J. Lu, J. Pichardo, A. Prongay, A. Skelton, X. Tong, S. Venkatraman, E. Xia, V. Girijavallabhan, and F. G. Njoroge. 2006. SCH 503034, a mechanism-based inhibitor of hepatitis C virus NS3 protease, suppresses polyprotein maturation and enhances the antiviral activity of alpha interferon in replicon cells. Antimicrob. Agents Chemother. 50:1013-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Massariol, M. J., S. Zhao, M. Marquis, D. Thibeault, and P. W. White. 2010. Protease and helicase activities of hepatitis C virus genotype 4, 5, and 6 NS3-NS4A proteins. Biochem. Biophys. Res. Commun. 391:692-697. [DOI] [PubMed] [Google Scholar]
- 32.Mateu, G., R. O. Donis, T. Wakita, J. Bukh, and A. Grakoui. 2008. Intragenotypic JFH1 based recombinant hepatitis C virus produces high levels of infectious particles but causes increased cell death. Virology 376:397-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McHutchison, J. G., S. C. Gordon, E. R. Schiff, M. L. Shiffman, W. M. Lee, V. K. Rustgi, Z. D. Goodman, M. H. Ling, S. Cort, and J. K. Albrecht. 1998. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. N. Engl. J. Med. 339:1485-1492. [DOI] [PubMed] [Google Scholar]
- 34.Murayama, A., T. Date, K. Morikawa, D. Akazawa, M. Miyamoto, M. Kaga, K. Ishii, T. Suzuki, T. Kato, M. Mizokami, and T. Wakita. 2007. The NS3 helicase and NS5B-to-3′X regions are important for efficient hepatitis C virus strain JFH-1 replication in Huh7 cells. J. Virol. 81:8030-8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okamoto, H., M. Kojima, S.-I. Okada, H. Yoshizawa, H. Iizuka, T. Tanaka, E. E. Muchmore, Y. Ito, and S. Mishiro. 1992. Genetic drift of hepatitis C virus during an 8.2 year infection in a chimpanzee: variability and stability. Virology 190:894-899. [DOI] [PubMed] [Google Scholar]
- 36.Pawlotsky, J. M., and J. G. McHutchison. 2004. Hepatitis C. Development of new drugs and clinical trials: promises and pitfalls. Summary of an AASLD hepatitis single topic conference, Chicago, IL, February 27-March 1, 2003. Hepatology 39:554-567. [DOI] [PubMed] [Google Scholar]
- 37.Perni, R. B., S. J. Almquist, R. A. Byrn, G. Chandorkar, P. R. Chaturvedi, L. F. Courtney, C. J. Decker, K. Dinehart, C. A. Gates, S. L. Harbeson, A. Heiser, G. Kalkeri, E. Kolaczkowski, K. Lin, Y. P. Luong, B. G. Rao, W. P. Taylor, J. A. Thomson, R. D. Tung, Y. Wei, A. D. Kwong, and C. Lin. 2006. Preclinical profile of VX-950, a potent, selective, and orally bioavailable inhibitor of hepatitis C virus NS3-4A serine protease. Antimicrob. Agents Chemother. 50:899-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pietschmann, T., A. Kaul, G. Koutsoudakis, A. Shavinskaya, S. Kallis, E. Steinmann, K. Abid, F. Negro, M. Dreux, F. L. Cosset, and R. Bartenschlager. 2006. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. U. S. A. 103:7408-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poynard, T., P. Marcellin, S. S. Lee, C. Niederau, G. S. Minuk, G. Ideo, V. Bain, J. Heathcote, S. Zeuzem, C. Trepo, and J. Albrecht. 1998. Randomised trial of interferon alpha 2b plus ribavirin for 48 weeks or for 24 weeks versus interferon alpha 2b plus placebo for 48 weeks for treatment of chronic infection with hepatitis C virus. Lancet 352:1426-1432. [DOI] [PubMed] [Google Scholar]
- 40.Qi, X., A. Bae, S. Liu, H. Yang, S. C. Sun, J. Harris, W. Delaney, M. Miller, and H. Mo. 2009. Development of a replicon-based phenotypic assay for assessing the drug susceptibilities of HCV NS3 protease genes from clinical isolates. Antiviral Res. 81:166-173. [DOI] [PubMed] [Google Scholar]
- 41.Reed, L. J., and H. A. Muench. 1938. Simple method of estimating fifty per cent endpoints. Am. J. Hyg. (Lond.) 27:493-497. [Google Scholar]
- 42.Reesink, H. W., S. Zeuzem, C. J. Weegink, N. Forestier, A. van Vliet, J. van de Wetering de Rooij, L. McNair, S. Purdy, R. Kauffman, J. Alam, and P. L. Jansen. 2006. Rapid decline of viral RNA in hepatitis C patients treated with VX-950: a phase Ib, placebo-controlled, randomized study. Gastroenterology 131:997-1002. [DOI] [PubMed] [Google Scholar]
- 43.Reiser, M., H. Hinrichsen, Y. Benhamou, H. W. Reesink, H. Wedemeyer, C. Avendano, N. Riba, C. L. Yong, G. Nehmiz, and G. G. Steinmann. 2005. Antiviral efficacy of NS3-serine protease inhibitor BILN-2061 in patients with chronic genotype 2 and 3 hepatitis C. Hepatology 41:832-835. [DOI] [PubMed] [Google Scholar]
- 44.Sarrazin, C., R. Rouzier, F. Wagner, N. Forestier, D. Larrey, S. K. Gupta, M. Hussain, A. Shah, D. Cutler, J. Zhang, and S. Zeuzem. 2007. SCH 503034, a novel hepatitis C virus protease inhibitor, plus pegylated interferon alpha-2b for genotype 1 nonresponders. Gastroenterology 132:1270-1278. [DOI] [PubMed] [Google Scholar]
- 45.Scheel, T. K., J. M. Gottwein, T. B. Jensen, J. C. Prentoe, A. M. Hoegh, H. J. Alter, J. Eugen-Olsen, and J. Bukh. 2008. Development of JFH1-based cell culture systems for hepatitis C virus genotype 4a and evidence for cross-genotype neutralization. Proc. Natl. Acad. Sci. U. S. A. 105:997-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seiwert, S. D., S. W. Andrews, Y. Jiang, V. Serebryany, H. Tan, K. Kossen, P. T. Rajagopalan, S. Misialek, S. K. Stevens, A. Stoycheva, J. Hong, S. R. Lim, X. Qin, R. Rieger, K. R. Condroski, H. Zhang, M. G. Do, C. Lemieux, G. P. Hingorani, D. P. Hartley, J. A. Josey, L. Pan, L. Beigelman, and L. M. Blatt. 2008. Preclinical characteristics of the hepatitis C virus NS3/4A protease inhibitor ITMN-191 (R7227). Antimicrob. Agents Chemother. 52:4432-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shaw, M. L., J. McLauchlan, P. R. Mills, A. H. Patel, and E. A. McCruden. 2003. Characterisation of the differences between hepatitis C virus genotype 3 and 1 glycoproteins. J. Med. Virol. 70:361-372. [DOI] [PubMed] [Google Scholar]
- 48.Shepard, C. W., L. Finelli, and M. J. Alter. 2005. Global epidemiology of hepatitis C virus infection. Lancet Infect. Dis. 5:558-567. [DOI] [PubMed] [Google Scholar]
- 49.Simmonds, P. 1997. Clinical relevance of hepatitis C virus genotypes. Gut 40:291-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simmonds, P. 1998. Variability of the hepatitis C virus genome. Curr. Stud. Hematol. Blood Transfus. 62:38-63. [DOI] [PubMed] [Google Scholar]
- 51.Simmonds, P. 2000. Hepatitis C virus genotypes, p. 53-70. In T. J. Liang and J. H. Hoofnagle (ed.), Hepatitis C. Academic Press, San Diego, CA.
- 52.Simmonds, P., J. Bukh, C. Combet, G. Deleage, N. Enomoto, S. Feinstone, P. Halfon, G. Inchauspe, C. Kuiken, G. Maertens, M. Mizokami, D. G. Murphy, H. Okamoto, J. M. Pawlotsky, F. Penin, E. Sablon, I. Shin, L. J. Stuyver, H. J. Thiel, S. Viazov, A. J. Weiner, and A. Widell. 2005. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42:962-973. [DOI] [PubMed] [Google Scholar]
- 53.Takamizawa, A., C. Mori, I. Fuke, S. Manabe, S. Murakami, J. Fujita, E. Onishi, T. Andoh, I. Yoshida, and H. Okayama. 1991. Structure and organization of the hepatitis C virus genome isolated from human carriers. J. Virol. 65:1105-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanji, Y., M. Hijikata, S. Satoh, T. Kaneko, and K. Shimotohno. 1995. Hepatitis C virus-encoded nonstructural protein NS4A has versatile functions in viral protein processing. J. Virol. 69:1575-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thibeault, D., C. Bousquet, R. Gingras, L. Lagace, R. Maurice, P. W. White, and D. Lamarre. 2004. Sensitivity of NS3 serine proteases from hepatitis C virus genotypes 2 and 3 to the inhibitor BILN 2061. J. Virol. 78:7352-7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ueno, T., S. Inuzuka, M. Sata, H. Koh, S. Tamaki, M. Kin, H. Sugawara, R. Sakata, T. Torimura, and K. Tanikawa. 1995. Serum hyaluronate predicts response to interferon-alpha therapy in patients with chronic hepatitis C. Hepatogastroenterology 42:522-527. [PubMed] [Google Scholar]
- 57.Vallet, S., S. Gouriou, J. B. Nousbaum, M. C. Legrand-Quillien, A. Goudeau, and B. Picard. 2005. Genetic heterogeneity of the NS3 protease gene in hepatitis C virus genotype 1 from untreated infected patients. J. Med. Virol. 75:528-537. [DOI] [PubMed] [Google Scholar]
- 58.Vanwolleghem, T., P. Meuleman, L. Libbrecht, T. Roskams, R. De Vos, and G. Leroux-Roels. 2007. Ultra-rapid cardiotoxicity of the hepatitis C virus protease inhibitor BILN 2061 in the urokinase-type plasminogen activator mouse. Gastroenterology 133:1144-1155. [DOI] [PubMed] [Google Scholar]
- 59.Wakita, T., T. Pietschmann, T. Kato, T. Date, M. Miyamoto, Z. J. Zhao, K. Murthy, A. Habermann, H. G. Krausslich, M. Mizokami, R. Bartenschlager, and T. J. Liang. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Winters, M. A., S. L. Welles, and M. Holodniy. 2006. Hepatitis C virus protease gene diversity in patients coinfected with human immunodeficiency virus. J. Virol. 80:4196-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wright-Minogue, J., N. Yao, R. Zhang, N. J. Butkiewicz, B. M. Baroudy, J. Y. Lau, and Z. Hong. 2000. Cross-genotypic interaction between hepatitis C virus NS3 protease domains and NS4A cofactors. J. Hepatol. 32:497-504. [DOI] [PubMed] [Google Scholar]
- 62.Yanagi, M., R. H. Purcell, S. U. Emerson, and J. Bukh. 1997. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc. Natl. Acad. Sci. U. S. A. 94:8738-8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yanagi, M., R. H. Purcell, S. U. Emerson, and J. Bukh. 1999. Hepatitis C virus: an infectious molecular clone of a second major genotype (2a) and lack of viability of intertypic 1a and 2a chimeras. Virology 262:250-263. [DOI] [PubMed] [Google Scholar]
- 64.Yi, M., R. A. Villanueva, D. L. Thomas, T. Wakita, and S. M. Lemon. 2006. Production of infectious genotype 1a hepatitis C virus (Hutchinson strain) in cultured human hepatoma cells. Proc. Natl. Acad. Sci. U. S. A. 103:2310-2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yi, M. K., Y. H. Ma, J. Yates, and S. M. Lemon. 2007. Compensatory mutations in el, p7, NS2, and NS3 enhance yields of cell culture-infectious intergenotypic chimeric hepatitis C virus. J. Virol. 81:629-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhong, J., P. Gastaminza, J. Chung, Z. Stamataki, M. Isogawa, G. Cheng, J. A. McKeating, and F. V. Chisari. 2006. Persistent hepatitis C virus infection in vitro: coevolution of virus and host. J. Virol. 80:11082-11093. [DOI] [PMC free article] [PubMed] [Google Scholar]