Abstract
The herpes simplex virus type 1 (HSV-1) UL25 gene encodes a minor capsid protein, pUL25, that is essential for packaging the full-length viral genome. Six regions which contain disordered residues have been identified in the high-resolution structure of pUL25. To investigate the significance of these flexible regions, a panel of plasmids was generated encoding mutant proteins, with each member lacking the disordered residues in one of the six regions. In addition, UL25 constructs were produced, which specified proteins that contained missense mutations individually affecting two of the four regions on the surface of pUL25 predicted from evolutionary trace analysis to be important in protein-protein interactions. The impacts of these mutations on viral DNA packaging, virus assembly, and growth were examined. Of the nine mutant proteins analyzed, five failed to complement the growth of a UL25 deletion mutant in Vero cells. These noncomplementing proteins fell into three classes. Proteins in one class did not alter the DNA packaging phenotype of an HSV-1 UL25 deletion mutant, whereas proteins from the other two classes allowed the UL25 null mutant to package full-length viral DNA. Subsequent analysis of the latter classes of mutant proteins demonstrated that one class enabled the null virus to release enveloped virus particles from U2OS cells, whereas the other class prevented egress of mature HSV-1 capsids from the nucleus. These findings reveal a new role for pUL25 in virion assembly, consistent with its flexible structure and location on the capsid.
Herpes simplex virus type 1 (HSV-1) has a large, complex virion structure that is composed of four morphologically distinct components. The central core comprises the linear double-stranded DNA genome, which is compactly organized inside an icosahedral capsid containing four different capsid shell proteins. The pUL6 DNA packaging protein forms the portal at the unique vertex. Two additional DNA packaging proteins, pUL25 and pUL17, are found on the capsid, and these proteins are present as heterodimers at multiple sites on the surface, adjacent to the vertices (20, 33, 35). A protein matrix containing more than 20 different proteins, referred to as the tegument, surrounds the capsid (15). One of the tegument proteins, pUL36, interacts with pUL25 and probably with VP5, the major capsid shell protein (5, 19, 23), and these associations are likely to be important for anchoring the tegument to the capsid (5). Finally, the outermost layer consists of a lipid membrane embedded with the viral glycoproteins.
Viral DNA synthesis and the early stages of virion assembly take place in the nucleus, within replication compartments (8, 25). The viral DNA is replicated as head-to-tail concatemers, and cleavage of the viral DNA into unit-length molecules is tightly associated with the packaging of the DNA into a preformed precursor capsid, the procapsid, through the portal. After the viral DNA is encapsidated, the mature C capsid rapidly buds into the inner nuclear membrane and is released into the cytoplasm by fusion of the enveloped particles with the outer nuclear membrane. In the cytoplasm, the capsids acquire the tegument and envelope.
pUL25 is one of seven viral proteins that are essential for packaging the viral DNA into the procapsid (10, 14, 18, 28). In contrast to the other DNA packaging proteins, pUL25 is not required for initiation of DNA packaging but is important at a later stage in the process. Mutant viruses lacking pUL25 cleave and package viral DNA in nonpermissive cells, but most of the encapsidated DNA is shorter than the full-length genome (4, 32). On the basis of the HSV-1 and pseudorabies virus (PRV) null mutant phenotypes, it has been suggested that pUL25 is important in stabilizing the capsid during and after DNA packaging (12, 13, 32). Recently, the N-terminal region of pUL25 was shown to be essential for the binding of pUL25 to capsids, since a mutant virus lacking the first 50 residues of pUL25 produced capsids that had reduced amounts of pUL25 under restrictive conditions and failed to package full-length viral DNA (4). pUL17 has been shown to be important for the efficient binding of pUL25 to capsids (33). However, analysis of pUL25 mutants for the ability to interact with pUL17 in an immunoprecipitation assay indicated that the first 50 amino acids of pUL25 are not required for the association of pUL25 with pUL17 (29). Although pUL25 was initially identified as a DNA packaging protein, several studies have suggested that it has a role both at a later stage in virion maturation and at very early times in infection, during uncoating of the viral genome (12, 13, 24, 32).
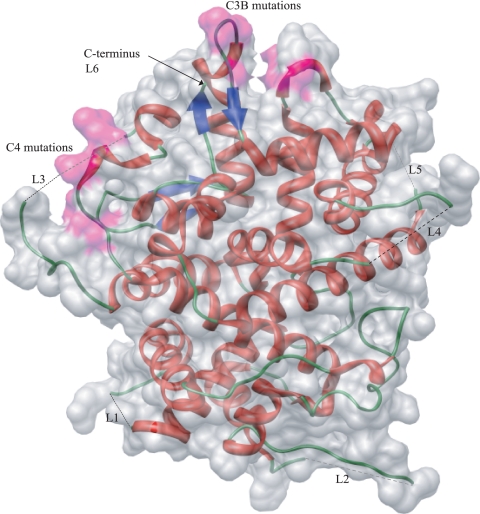
The crystallographic structure of an N-terminally truncated pUL25 protein (residues 134 to 580) has been determined and shown to have a distinctive architecture, with numerous loops radiating from the surface of a box-shaped core (3). Five of the extended loops and the C-terminal end of pUL25 contain disordered residues, indicative of a plastic and dynamic protein (Fig. 1). Using the structural information together with evolutionary trace analysis, Bowman et al. (3) identified four clusters of class-specific conserved residues on the surface of the protein (C1 to C4), which were predicted to be involved in protein-protein interactions.
FIG. 1.
Ribbon diagram of pUL25 (residues 134 to 580) showing the positions of the unstructured residues in loops L1 to L5 superimposed on a space-filling model and indicating the positions of the C3 and C4 residues that were mutated (colored pink). L6 unstructured residues are at the C terminus. Light-colored dashed lines represent unstructured residues at the back of the molecule, and black dashed lines represent those at the front.
We used a genetic approach to relate the structure of pUL25 to its function. Site-specific mutagenesis of UL25 was carried out, targeting unstructured amino acids and two of the four functional clusters of amino acids, C3 and C4, on the surface of the protein. The resulting mutant proteins were examined for the ability to alter the phenotype of an HSV-1 UL25 deletion mutant to obtain insight into the roles of specific regions on the surface of pUL25.
MATERIALS AND METHODS
Virus and cells.
Vero cells, Vero-derived 8-1 cells (18), and the human osteosarcoma cell line U2OS were grown in Dulbecco's medium supplemented with 10% fetal calf serum, penicillin (100 U/ml), and streptomycin (100 μg/ml). Sf21 cells were cultivated in TC100 containing 5% fetal calf serum, penicillin, and streptomycin. The wild-type (wt) HSV-1 strain used was strain 17.
Plasmids.
The HSV-1 strain 17 UL25 gene (nucleotides 48,813 to 50,553) (17) was derived from the plasmid pGX294 (34) and cloned as an EcoRI-XbaI fragment into the vector pFBpCI (1), cleaved with the corresponding enzymes, to create the plasmid pFBpCI-UL25. The UL25 gene in this vector was under the control of the human cytomegalovirus (HCMV) major immediate-early (IE) promoter, and its DNA sequence was identical to that previously reported (17). The plasmid pGX2 contains the 5.9-kbp BamHI fragment spanning the junction of the HSV-1 strain 17 L and S segments, which was inserted into the BamHI site of the vector pAT153 and used as a probe in Southern blot experiments. The bacterial artificial chromosome (BAC) containing the HSV-1 strain 17 genome used in this study was fHSVΔpac, which lacks all copies of the a sequence required for cleavage and packaging of the viral genome (27).
Construction of a UL25 deletion mutant.
A virus mutant was created by in vivo Red/ET recombination, using a Counter-Selection BAC modification kit supplied by Gene Bridges. Briefly, the recombination enzymes were induced in a culture of Escherichia coli DH10 carrying the HSV-1 strain 17 BAC fHSVΔpac and the plasmid pSC101-BAD-gbaA-tet by the addition of l-arabinose to a final concentration of 1%. After incubation of the culture at 37°C for 45 min, electrocompetent bacteria were prepared, and a 1,420-bp fragment, formed using the primers PCR1-F and PCR1-R (Table 1) in a PCR with an rpsL-neo cassette as the DNA template, was electroporated into the bacteria. Kanamycin-resistant, streptomycin-sensitive bacteria were isolated, and the BAC isolates were screened by Southern blotting for the presence of the rpsL-neo cassette. To obtain infectious virus, BAC DNA containing the rpsL-neo cassette in place of UL25 was cotransfected into 8-1 cells along with BamHI-cleaved pGX2 (containing the HSV-1 strain 17 BamHI K fragment) in order to repair the deleted a sequences of the parental BAC. Transfections were performed using Lipofectamine and Plus reagents (Life Technologies) according to the manufacturer's recommendations. The cells were harvested after 2 to 3 days of incubation at 37°C, and the virus, ΔUL25MO, was plaque purified.
TABLE 1.
Oligonucleotides used for generation of ΔUL25MO and mutated UL25 genesa
The sequences of oligonucleotides used in PCRs or as annealed pairs are shown. Relevant restriction endonuclease sites are underlined. Lowercase letters represent the template pEP-Kan-S sequence. Uppercase letters represent UL25 sequences. Letters in bold italics indicate mutations resulting in amino acid substitutions. Letters in bold represent silent mutations. _, deletion.
Construction of baculoviruses.
Baculoviruses expressing the wt or mutant UL25 gene under the control of the major IE promoter of HCMV and a control virus lacking the recombinant gene were constructed by recombining the plasmid pFBpCI or pFBpCI derivatives containing the UL25 gene into the bacmid bMON14272 in DH10Bac, using the Bac-to-Bac baculovirus expression system (Life Technologies). The recombinant bacmid DNA was transfected into Sf21 cells, and high-titer baculovirus stocks were produced.
Construction of UL25 mutant plasmids.
Regions of UL25 were mutated either by PCR, using pFBpCI-UL25 as the template and primers containing the mutations, or by annealing overlapping oligonucleotides possessing the mutations. Each PCR product containing mutations was subcloned into pGEM-T Easy and sequenced, and a restriction endonuclease fragment containing the mutations was cloned into pFBpCI-UL25, replacing the corresponding wt sequences in UL25. The annealed oligonucleotides possessing the required mutations and suitable 5′ and 3′ cohesive ends were cloned directly into pFBpCI-UL25 cleaved with the relevant restriction endonucleases or, in the case of cluster 3B mutations, into cleaved pFBpCI-UL25-C3A. The mutated UL25 genes were all sequenced to confirm their identities. Table 1 lists the PCR primers and oligonucleotides used in the construction of mutant UL25 genes, with the restriction endonuclease sites used in cloning or identification of constructs underlined. The annealed oligonucleotide used to create mutations in cluster 4 residues also contained a novel internal NotI site. UL25 sequences encoding residues that formed unstructured regions were deleted using either overlapping oligonucleotides containing the deletion or PCR primers with the deletion (Table 1). In some cases, new restriction endonuclease sites were generated, and for the construction of the loop 4 deletion, a PvuII site was removed, without affecting the encoded amino acids. Tables 2 and 3 list the UL25 mutant proteins and the encoded UL25 amino acid substitutions and deletions.
TABLE 2.
Substitution mutant proteins generated in this study
| Cluster | Protein | Amino acid substitutions and residues in clustera |
|---|---|---|
| 3 | pUL25-C3A | G169A, S170A, G172A, R180, G202, R203, K206, P413 |
| pUL25-C3B | G169A, S170A, G172A, R180, G202V, R203A, K206A, P413 | |
| 4 | pUL25-C4 | R390, N396A, Y398A, D400A, L402A, R552 |
The residues present in clusters 3 and 4 are shown, with the amino acid alterations in the mutant proteins indicated in bold.
TABLE 3.
Deletion mutant proteins generated in this study
| Protein | Deleted residues in pUL25 |
Total no. of deleted residues | |
|---|---|---|---|
| Start of deletion | End of deletion | ||
| pUL25-L1 | A249 | D254 | 6 |
| pUL25-L2 | R335 | G345 | 11 |
| pUL25-L3 | P417 | A425 | 9 |
| pUL25-L4 | P479 | T483 | 5 |
| pUL25-L5 | R511 | N513 | 3 |
| pUL25-L6 | S578 | V580 | 3 |
Transient complementation assay.
Duplicate Vero cell monolayers (2 × 105 cells per well) were transfected with plasmid DNA, using Lipofectamine and Plus reagents according to the manufacturer's instructions. At 5 hours posttransfection, the cell monolayers were infected with the HSV-1 deletion mutant ΔUL25MO at a multiplicity of infection of 2 PFU/cell. After the cells had been incubated for 1 h at 37°C, the virus inoculum was removed and the virus remaining on the cell surface was inactivated by treating the cells with 0.1 M glycine, 0.14 M NaCl, pH 3.0, for 1 min as described previously (32). The cells were then incubated at 37°C. The following day, the cells were harvested, the progeny virus yields on 8-1 cells were determined from one set of samples, and Western blot analysis was carried out on the other set.
Transient DNA packaging assay.
U2OS cell monolayers (1.6 × 106 cells/dish) were infected with 50 PFU of recombinant baculovirus per cell and incubated for 1 h at room temperature. The cells were washed twice with medium and infected with 2 PFU of ΔUL25MO per cell. After 1 h of incubation at 37°C, the cells were washed twice and overlaid with culture medium, and incubation was continued at 37°C. Twenty-four hours after infection with ΔUL25MO, the cells were harvested and total cellular DNA and DNase-resistant DNA samples were prepared as described previously (32). An aliquot of DNA was digested with BamHI, and the fragments were separated in agarose gels, blotted onto Hybond XL membranes (GE Healthcare), and hybridized to 32P-labeled plasmid pBE1, containing HSV-1 sequences corresponding to HSV-1 nucleotides 596 to 2905 within the long repeat sequence, or pST17, containing HSV-1 sequences corresponding to nucleotides 148825 to 151857 within the terminal short repeat (17, 32). The DNA was labeled with [α-32P]dCTP and [α-32P]dGTP by use of a random primer extension kit (Roche). Phosphorimages of the Southern blots were obtained using an FX personal molecular imager (Bio-Rad), and the relative amounts of radioactivity in the DNA bands were calculated using the software package Quantity One (Bio-Rad).
PFGE.
DNase-resistant DNA samples were prepared from U2OS cells infected with recombinant baculovirus and ΔUL25MO in essentially the same manner as that for the Southern blot samples, except that samples were concentrated 2-fold and the DNAs were neither phenol extracted nor ethanol precipitated. The DNA molecules were separated in a DR-II pulsed-field gel electrophoresis (PFGE) apparatus (Bio-Rad) as described previously (32) and analyzed by Southern blotting.
Electron microscopy.
U2OS cell monolayers on 35-mm-diameter petri dishes, infected first with recombinant baculovirus and subsequently with ΔUL25MO, were prepared for electron microscopy essentially as described by Roberts et al. (26).
Fluorescent in situ hybridization.
Infection of U2OS cells was carried out in the same manner as that described for the transient DNA packaging assay. Fluorescent in situ hybridization was performed as outlined previously, using cosmid 56, containing HSV-1 strain 17 sequences from bp 79442 to 115152, labeled by nick translation with Cy3-dCTP, as the probe (7, 17, 24). The cell nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole) after in situ hybridization. Samples were visualized using a Zeiss LSM 510 confocal microscope with 405- and 543-nm laser lines, with each channel scanned separately, and also by phase-contrast microscopy.
RESULTS
Generation of UL25 mutant plasmids and complementation analysis.
When this study was initiated, the only HSV-1 UL25 null mutant available was an insertion mutant with an in-frame stop codon terminating translation of pUL25 after codon 105 (18). For this reason, Red/ET recombination was used to construct an HSV-1 UL25 deletion mutant, ΔUL25MO, containing an rpsL-neo cassette in place of the UL25 gene (see Materials and Methods). DNA analysis showed that this virus had a similar DNA packaging defect in noncomplementing cells to that of the UL25 null mutant KUL25NS reported previously (32). Although most of the encapsidated DNA was shorter than the full-length genome, DNA-containing capsids were readily observed in the nuclei of both KUL25NS- and ΔUL25MO-infected Vero cells (data not shown), consistent with a recent study characterizing an HSV-1 UL25 deletion mutant (13). The mutant virus, ΔUL25MO, was used in subsequent experiments to characterize the mutant pUL25 proteins.
Since disordered portions of a protein are often highly flexible areas that are involved in the molecule's function, we initially targeted the unstructured regions of pUL25 for mutagenesis. Six UL25 mutant gene constructs were made in the expression plasmid pFBpCI, each of which contained a deletion removing the codons specifying the unstructured residues in one of the five flexible loops or at the extreme C terminus of pUL25 (loops L1 to L6) (Fig. 1). Three more mutant UL25 genes were generated, encoding substitutions of groups of residues in two of the four functionally important clusters, C3 and C4, on the surface of pUL25 (Fig. 1). Initially, three amino acids (G169, S170, and G172) in C3 were changed, but since preliminary complementation analysis suggested that these alterations did not significantly impair the function of pUL25, another construct was made, encoding three further substitutions, at residues G202, R203, and K206. Each target amino acid was replaced with alanine, with one exception, at a position (G202) where alanine was predicted by SIFT analysis to be tolerated, where a valine was used instead (21, 22). The mutant pUL25s generated are shown in Tables 2 and 3.
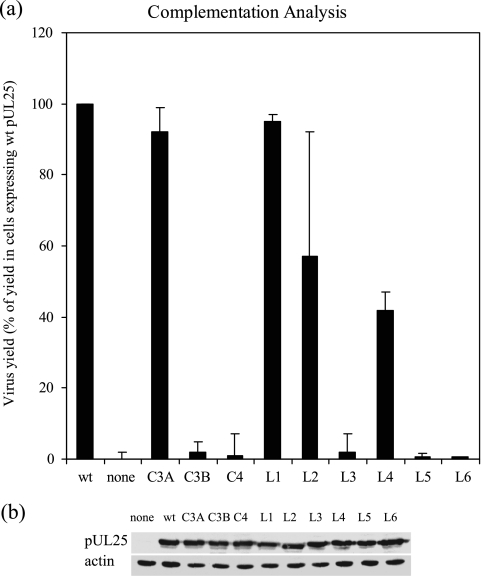
To examine the effects of the mutations on the function of pUL25, transient complementation analysis was carried out. Vero cells were transfected with a mutant UL25 expression plasmid, the wt UL25 expression plasmid, or empty plasmid and subsequently infected with ΔUL25MO. After incubation at 37°C for a further 24 h, the cells were harvested and the virus yield on the UL25-complementing cell line 8-1 was determined. The amount of mutant virus produced from ΔUL25MO-infected Vero cells expressing the pUL25-C3B, pUL25C4, pUL25-L3, pUL25-L5, or pUL25-L6 mutant protein was low. The yield was <5% of the yield from mutant virus-infected cells expressing wt pUL25 and was similar to the yield from virus-infected cells transfected with the empty vector pFBpCI (Fig. 2). These results suggested that the mutations in these positions impaired the function of pUL25. The other four mutant proteins, pUL25C3A, pUL25-L1, pUL25-L2, and pUL25-L4, complemented the growth of ΔUL25MO in Vero cells, and yields of >40% of the level obtained in the presence of the wt protein were obtained. Western blot analysis (Fig. 2b) confirmed that each of the mutant proteins was expressed at a similar level to that of wt pUL25 in the transfected cells.
FIG. 2.
Complementation analysis. (a) Vero cells were transfected with individual plasmids expressing wt or mutated versions of pUL25, as indicated, and subsequently infected with ΔUL25MO. The virus yield was determined in 8-1 cells and was expressed as a percentage of the yield obtained from cells transfected with the plasmid pFBpCI-UL25, encoding wt pUL25. Control cells received the empty vector pFBpCI and are labeled “none.” The results shown are the averages for three independent experiments. The error bars represent the standard deviations of the means. (b) Western blot analysis of transfected cells. Western blots of cells transfected with plasmids expressing the indicated UL25 proteins were probed with monoclonal antibodies MAb166 (34) and AC-40 (Sigma) to detect pUL25 (upper panel) and actin (lower panel), respectively.
Effects of mutant pUL25s on packaging phenotype of null mutant.
The noncomplementing UL25 mutant proteins were screened in a transient packaging assay for the ability to alter the phenotype of the UL25 null mutant. To enhance the sensitivity of the assay by increasing the number of cells expressing pUL25, wt and mutant UL25 genes under the control of the HCMV IE promoter in the pFBpCI vector were transferred into the bacmid bMON14272, and recombinant baculoviruses were generated. As a control, a recombinant baculovirus containing the HCMV IE promoter but lacking the UL25 gene was also constructed. U2OS cells were used instead of Vero cells in the packaging assay, because U2OS cells have been shown to have a high level of transduction efficiency with baculoviruses (31). To confirm that the infection efficiencies of the recombinant baculoviruses in U2OS cells were similar, the cells were infected with 50 PFU of each virus per cell for 24 h. The virus-infected cells were identified using pUL25 MAb166 in an immunofluorescence assay. All of the pUL25-expressing baculoviruses infected 41 to 47% of the cells, and this concentration of baculovirus was used in all subsequent experiments. Western blot analysis performed under these conditions additionally showed that wt and mutant UL25 proteins accumulated to similar levels (Fig. 3e).
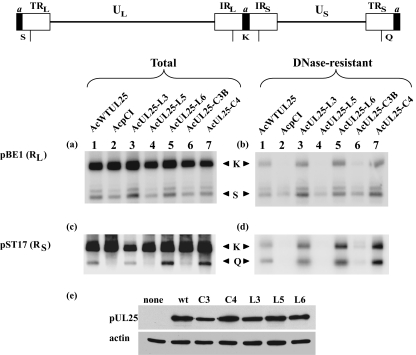
FIG. 3.
Analysis of DNA packaging in ΔUL25MO-infected U2OS cells expressing different pUL25s. U2OS cells were coinfected with ΔUL25MO and the indicated recombinant baculovirus. After 24 h of incubation at 37°C, the cells were harvested and total and DNase-resistant DNA samples were prepared. The DNAs were digested with BamHI, and the separated fragments were transferred to a membrane. The total (a and c) and DNase-resistant (b and d) DNA fragments were hybridized to 32P-labeled pBE1 (a and b) and pST17 (c and d), specific for the RL and RS regions, respectively. The positions of the BamHI K, Q, and S fragments are indicated. The locations of the BamHI fragments recognized by plasmids pBE1 (BamHI K and S) and pST17 (BamHI K and Q) are shown at the top. (e) Western blots of U2OS cells infected with baculoviruses expressing the indicated UL25 proteins were probed with monoclonal antibodies MAb166 (34) and AC-40 (Sigma) to detect pUL25 (upper panel) and actin (lower panel), respectively.
In the transient DNA packaging assay, U2OS cells were infected with individual recombinant baculoviruses expressing wt or mutant protein or with the negative-control virus, together with ΔUL25MO. Twenty-four hours after infection with the null mutant, the cells were harvested and total cellular DNA and DNase-resistant DNAs were prepared. The DNA samples were digested with BamHI, separated by gel electrophoresis, and analyzed by Southern blotting using a 32P-labeled probe specific for the short or long repeat sequences (Fig. 3). Packaged full-length viral genomes digested with BamHI produce two terminal BamHI fragments, S and Q, recognized by the pBE1 and pST17 probes, respectively (32). Both probes also hybridize to the joint-spanning BamHI K fragment. In nonpermissive cells infected with the UL25 null mutant KUL25NS, the concatemeric viral DNA is cleaved and encapsidation is initiated, starting with the L segment, but fails to go to completion (32). As a consequence, the L terminus is overrepresented with respect to the internal repeat, and the S terminus is underrepresented.
The pattern of DNA packaging reported for KUL25NS in Vero and BHK cells (32) was also observed in U2OS cells infected with ΔUL25MO and the negative-control baculovirus AcpCI (Fig. 3b and d, lanes 2). The amount of the BamHI K fragment was reduced compared to that of the L-terminal fragment, S, suggesting that in most instances DNA packaging did not proceed as far as the junction between the L and S segments. Incomplete packaging was observed in ΔUL25MO-infected U2OS cells expressing pUL25-L5 (Fig. 3b and d, lanes 4) or pUL25-C3B (Fig. 3b and d, lanes 6). In contrast, in mutant-infected cells expressing wt pUL25, pUL25-L3, pUL25-L6, or pUL25-C4, the relative amount of the BamHI S fragment present in packaged viral DNA was similar to that of the BamHI K fragment, and the S-terminal fragment Q was similarly abundant, suggesting that the complete genome had been packaged (Fig. 3b and d, lanes 1, 3, 5, and 7). It should be noted that in some experiments, and when longer exposures of the phosphorimage were performed, small amounts of fragments K and Q could be detected in cells that received AcpCI, AcUL25-L5, or AcUL25-C3B. We believe that this represents residual input ΔUL25MO that may not have been removed effectively by the washing procedure.
As expected, the BamHI S fragment, from the L terminus, was also detected in each of the total cell DNA samples (Fig. 3a). The S-terminal fragment Q was detected in total DNA of cells expressing wt pUL25, pUL25-L3, pUL25-L6, or pUL25-C4 (Fig. 3c, lanes 1, 3, 5, and 7). However, DNA from cells infected with ΔUL25MO and AcpCI, AcUL25-L5, or AcUL25-C3B contained only background levels of this fragment, suggesting that the free S-terminal end generated after cleavage of the genome is unstable.
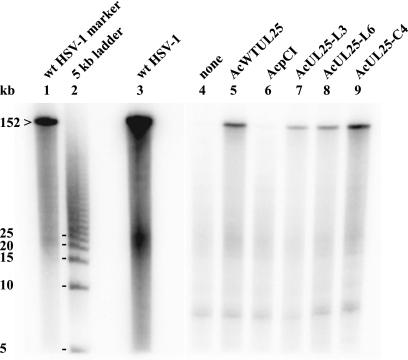
To investigate whether the genomic S termini present in ΔUL25MO-infected U2OS cells expressing pUL25-C4, -L3, or -L6 were derived from packaged full-length viral genomes, the DNA packaging experiment was repeated. DNase-resistant DNA samples were prepared, taking care to minimize shearing of DNA. The molecules were resolved by PFGE and analyzed by Southern blotting, using 32P-labeled plasmid pGX2 containing the HSV-1 BamHI K fragment as a probe (Fig. 4). A high-molecular-weight fragment which comigrated with the wt HSV-1 genomic DNA was detected in ΔUL25MO-infected U2OS cells expressing wt pUL25, pUL25-L3, pUL25-L6, or pUL25-C4, confirming that the mutant proteins enabled the UL25 null mutant to overcome the defect in packaging of full-length genomes (Fig. 4, lanes 5, 7, 8, and 9). Very low levels of full-length encapsidated genomes were detected in U2OS cells singly infected with ΔUL25MO or dually infected with ΔUL25MO and AcpCI, possibly derived from input particles that had not been uncoated (Fig. 4, lanes 4 and 6).
FIG. 4.
PFGE analysis of packaged ΔUL25MO DNA. DNase-resistant DNAs were prepared from ΔUL25MO-infected U2OS cells coinfected with the indicated recombinant baculovirus. The DNAs were separated by PFGE, transferred to a membrane, and hybridized to 32P-labeled pGX2, which contains the genomic BamHI K fragment. The position of the HSV-1 full-length genome (152 kb) and the sizes of markers within the 5-kb ladder are shown on the left.
Electron microscopic examination of mutant-infected cells expressing mutant pUL25.
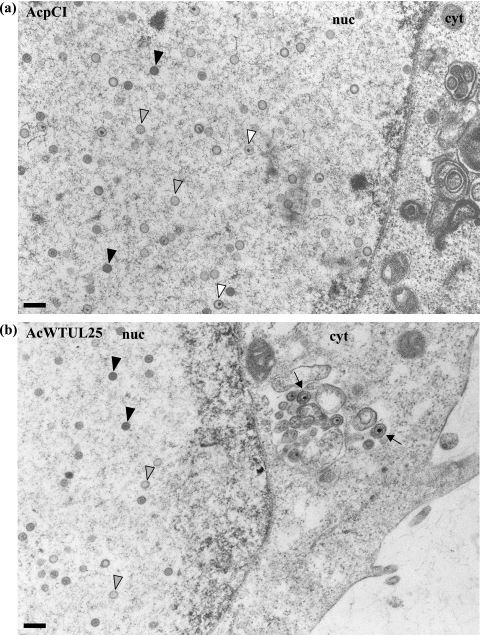
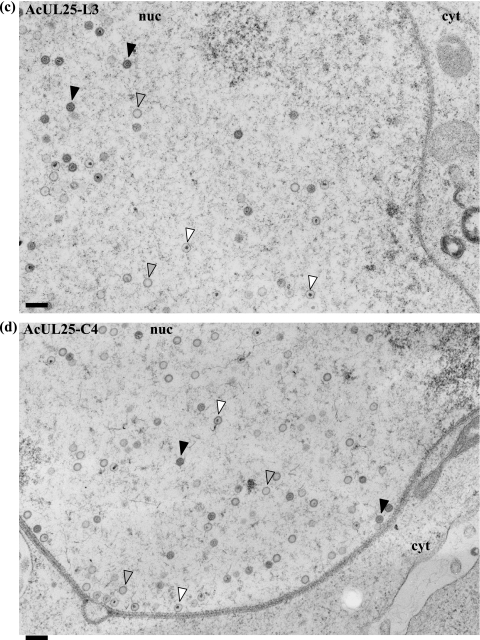
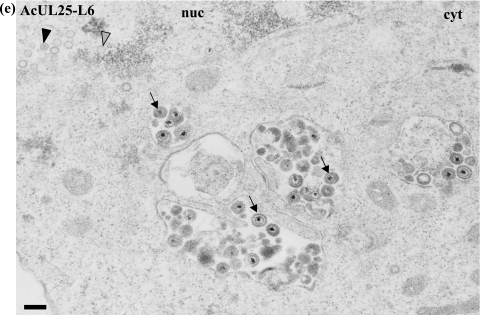
To determine the stage(s) within the HSV-1 life cycle disrupted by the UL25 mutations, samples were prepared for electron microscopic analysis. U2OS cells were infected with baculoviruses expressing either C4, L3, or L6 mutant protein or with the positive- or negative-control virus and subsequently infected with ΔUL25MO. After incubation at 37°C for 24 h, the virus-infected cells were embedded in Epon resin and the stained thin sections examined under an electron microscope. In cells coinfected with ΔUL25MO and the control baculovirus AcpCI, DNA-containing capsids were seen in the nuclei (Fig. 5a), consistent with the findings of our electron microscopic analysis of cells singly infected with either ΔUL25MO or KUL25NS (data not shown). Capsids were rarely observed in the cytoplasm of null mutant virus-infected cells, and in the few cases where cytoplasmic capsids were seen, the nuclear membrane did not appear to be intact. The pattern of virus assembly in ΔUL25MO-infected cells expressing the pUL25-L3 or pUL25-C4 mutant protein closely resembled that of ΔUL25MO-infected cells (Fig. 5c and d). Thus, although these two mutants were capable of supporting the packaging of full-length viral genomes (Fig. 3 and 4), the resulting C capsids remained defective in egress from the nucleus. In addition, inspection of multiple independent micrographs did not reveal any significant differences in the accumulation of different capsid types in the nuclei of cells infected with the null mutant alone or in the presence of the C4 or L3 mutant protein.
FIG. 5.
Analysis of virus assembly in ΔUL25MO-infected U2OS cells expressing different pUL25s. U2OS cells were coinfected with ΔUL25MO and the recombinant baculovirus AcpCI (a), AcWTUL25 (b), AcUL25-L3 (c), AcUL25-C4 (d), or AcUL25-L6 (e). After 24 h of incubation at 37°C, the cells were harvested, fixed, and embedded in Epon resin. Stained thin sections were examined under an electron microscope. Gray arrowheads indicate A (empty) capsids, black arrowheads denote B capsids, which lack viral DNA but contain the scaffolding protein, and white arrowheads indicate DNA-containing capsids. The arrows indicate enveloped capsids. nuc, nucleus; cyt, cytoplasm. Bar = 250 nm.
In contrast, in ΔUL25MO-infected cells expressing the L6 mutant protein, DNA-containing capsids were found in both the nuclei and the cytoplasm of the cells. In addition, enveloped virus particles were observed in the cytoplasm and on the cell surface (Fig. 5e). This pattern of virus assembly resembled that in ΔUL25MO-infected cells expressing wt pUL25 (Fig. 5b).
Location of viral DNA within the cell.
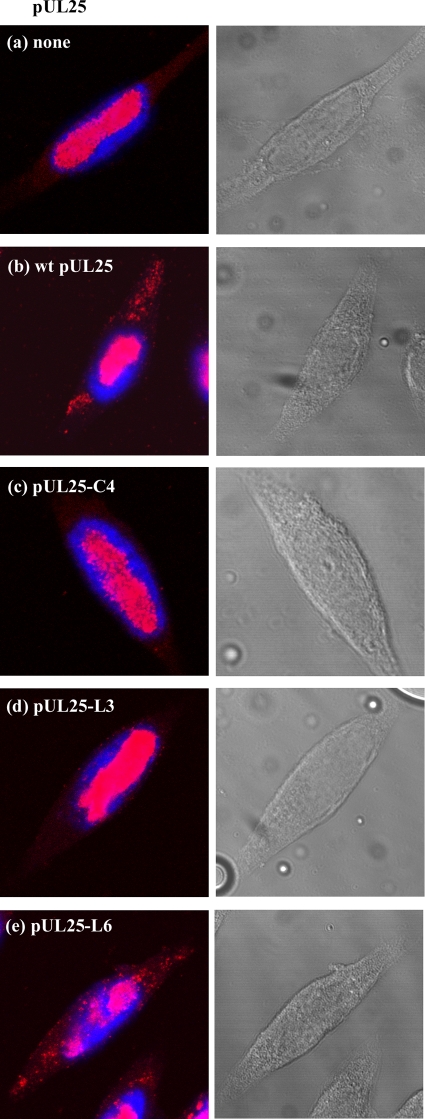
Fluorescence in situ hybridization was carried out on ΔUL25MO-infected cells expressing pUL25-C4, pUL25-L3, or pUL25-L6 protein to determine the location of the viral DNA within the cells. In ΔUL25MO-infected cells expressing either pUL25-C4 or pUL25-L3, the viral DNA was restricted to the nuclei (Fig. 6c and d), whereas in cells expressing wt pUL25 or pUL25-L6, viral DNA was also observed in the cytoplasm (Fig. 6b and e). This finding is consistent with the results from the electron microscopy analysis (Fig. 5), and importantly, it extends the observations from ultrathin sections, in which C particles present at low abundance might easily have been missed, to the whole-cell situation. The data from both the electron microscopic and fluorescence in situ hybridization analyses suggest that the residues in cluster C4 or the unstructured residues in loop L3 are important for egress of capsids from the nucleus and that the L6 residues at the carboxy terminus of UL25 are involved at a later stage, after the capsids have been released into the cytoplasm.
FIG. 6.
Localization of virus DNA in U2OS cells coinfected with ΔUL25MO and the recombinant baculoviruses AcpCI (a), AcWTUL25 (b), AcUL25-C4 (c), AcUL25-L3 (d), and AcUL25-L6 (e), which express the indicated pUL25 proteins. Virus DNA was detected by in situ hybridization and confocal microscopy (left). Nuclei were stained with DAPI. Phase-contrast images are shown on the right.
Complementation analysis of ΔUL25MO by pUL25-L6 in U2OS cells.
The UL25-L6 deletion mutant failed to complement the growth of ΔUL25MO in Vero cells. Since U2OS cells coinfected with ΔUL25MO and AcUL25-L6 produced DNA-containing capsids in the cytoplasm and enveloped virus particles in the cytoplasm and on the cell surface, the ability of the mutant L6 protein to complement ΔUL25MO was examined in Vero and U2OS cells. U2OS and Vero cells were transfected with pFBpCI-UL25 (expressing wt pUL25), pFBpCI-UL25-L6 (expressing pUL25-L6), or the empty vector pFBpCI and subsequently infected with ΔUL25MO for 24 h at 37°C. The cells were then harvested, and the progeny virus titers were determined in the complementing cell line 8-1. The level of complementation of ΔUL25MO in Vero cells expressing pUL25-L6 was similar to the level obtained in cells transfected with the empty vector, consistent with earlier results (Table 4 and Fig. 2). In U2OS cells expressing pUL25-L6, slightly higher levels of complementation of the null mutant virus were observed, but the mutant protein was still severely impaired in the ability to support virus growth.
TABLE 4.
Comparison of yields of ΔUL25MO in U2OS and Vero cells transfected with pFBpCI-UL25-L6, pFBpCI-UL25, or empty vector
| Plasmid | Virus yield (%)a |
|
|---|---|---|
| U2OS cells | Vero cells | |
| pFBpCI-UL25-L6 | 3.5 | 1.3 |
| 4.3 | 0.4 | |
| pFBpCI | 0.1 | 0.06 |
| 0.4 | 0.13 | |
| pFBpCI-UL25 | 100 | 100 |
| 100 | 100 | |
Data are the results of two independent experiments, expressed as percentages of the virus yield obtained in cells transfected with the wt pUL25-expressing plasmid, pFBpCI-UL25.
DISCUSSION
The functions of the six unstructured regions and two of the four conserved clusters that were previously defined by Bowman et al. (3) were investigated by site-directed mutagenesis, taking advantage of the information on the structure of HSV-1 pUL25. The findings are summarized in Table 5.
TABLE 5.
Summary of results obtained with mutant proteins
| Mutant protein | Complementation of UL25 null mutanta | Full-length DNA packagingb | Virions in cytoplasmc |
|---|---|---|---|
| wt pUL25 | + | + | + |
| None | − | − | − |
| pUL25-C3A | + | ND | − |
| pUL25-C3B | − | − | − |
| pUL25-C4 | − | + | − |
| pUL25-L1 | + | ND | − |
| pUL25-L2 | + | ND | − |
| pUL25-L3 | − | + | − |
| pUL25-L4 | + | ND | − |
| pUL25-L5 | − | − | − |
| pUL25-L6 | − | + | + |
Complementation of growth of ΔUL25MO in Vero cells.
The ability to package full-length virus DNA in U2OS cells coinfected with ΔUL25MO and the recombinant baculovirus expressing the mutant protein. ND, not determined.
Detection of virions in the cytoplasm in U2OS cells coinfected with ΔUL25MO and the recombinant baculovirus expressing the mutant protein, based on electron microscopic and fluorescence in situ hybridization results.
The results from DNA packaging experiments suggested that the wt residues mutated in pUL25-C3B and the unstructured residues (511 to 513) absent in pUL25-L5 are critical for packaging the full-length viral genome. Using the imaging program Chimera, a deep cleft was identified in the C3 region of pUL25, with five of the six wt residues mutated in pUL25-C3B located within this crevice (Fig. 1). Three of these residues, G169, S170, and G172, are situated on one side of the cleft, whereas the two other residues, G202 and R203, are located on the opposing face. Characterization of the pUL25-C3A protein revealed that replacement of residues G169, S170, and G172 with alanine did not significantly impair the function of the protein, suggesting that the other two C3 residues in this region are more important. It is possible, however, that mutations of residues on both sides of the cleft are required to abolish the function of the protein. The clustering of the critical residues around a crevice suggests that this region may be the site of an essential interaction between UL25 and another DNA packaging protein. The protein pUL17 is a strong candidate, but it is also possible that capsid shell proteins VP5 and VP19C or the portal protein pUL6 might be involved. Further analysis is required to determine the contributions of the individual mutated residues in pUL25-C3B to the functional C3 interface.
Since the unstructured residues in L5 are located on the opposing side of the molecule, approximately 180° from the C3B mutated residues, the deleted residues in L5 may affect a different protein interaction. A third region that is important for DNA packaging, near the N terminus, was reported earlier (4), but further investigation is required to determine whether these sites interact with different protein partners. An additional site that affects the binding of pUL25 to capsids, and hence virus DNA packaging, was identified through the characterization of the temperature-sensitive (ts) mutant ts1249, which has the missense mutation E233K (24). Although this lesion does not lie close to any of the regions mutated in the current study, it is situated in a pocket on the surface of the protein, located on the opposite face from that shown in the lower portion of the molecule in Fig. 1.
The C4 and L3 regions are important after DNA packaging, probably in the envelopment of C capsids at the inner nuclear membrane, but a role in the transport of the mature capsids to the site of primary envelopment cannot be excluded. Previously, pUL25 was speculated to have a function in the nucleus after packaging of the full-length genome, based on two observations. First, wt C capsids contain more pUL25 than do either the dead-end B capsids, which retain the cleaved scaffold proteins, or the precursor capsids, the procapsids (30, 34). Second, HSV-1 and PRV UL25 null mutant viruses produce DNA-containing capsids that remain in the nuclei (12, 13). In contrast to the UL25 null mutants, all of which had a reduced capacity to encapsidate full-length virus DNA, ΔUL25MO did not exhibit any obvious DNA packaging defect in U2OS cells expressing either the pUL25-C4 or pUL25-L3 mutant protein, and therefore our study is the first direct evidence for a role of pUL25 in the egress of mature capsids from the nucleus. Interestingly, the HSV-1 UL25 mutant 143i contains an insertion of five residues after residue 143 (4), which is located close to the cluster 4 residues mutated in pUL25-C4. This mutant, however, exhibits a different phenotype from that of the C4 mutant and, like the null mutant, packages shorter-than-full-length viral DNA (4). Since the insertion is adjacent to a well-conserved residue and lies between two beta sheets, it is possible that rather than having a localized effect on the function of pUL25, the mutation causes a more drastic disruption of the protein's function.
The failure of three of the six deletion mutants, each lacking a disordered region of pUL25, to complement the UL25 null mutant suggests that these disordered regions are essential for the function of pUL25. Although it is possible that deletion of disordered amino acids disrupted the overall architecture of the protein, we think that this is unlikely because the disordered amino acids are on the surface of the protein. Furthermore, at least in the case of pUL25-L3 and pUL25-L6, the function affected was at a later stage of virus assembly than DNA packaging. Interestingly, there was no correlation between the number of amino acids deleted and the effect on pUL25 function. The deletion of the 11 unstructured amino acids R335 to G345 (L2) did not impair the function of the mutant protein, whereas the loss of 3 amino acids, R511 to N513 (L5) or S578 to V580 (L6), had a drastic effect. None of the unstructured regions are highly conserved. Residues in L1 and L4 showed the least conservation in alignments of alphaherpesvirus pUL25s, and removal of these amino acids did not markedly affect the function of pUL25, although pUL25-L4 showed some impairment (Fig. 2).
The removal of unstructured residues in a region may compromise the flexibility of the protein and, as a consequence, affect the ability of the protein to respond to changes in its local environment. In the case of pUL25-L3, for example, it is possible that alterations in the capsid shell induced by the encapsidation of the HSV-1 genome could trigger a subtle conformational change in pUL25, exposing a binding site required for the egress of the capsid from the nucleus. Interestingly, the unstructured residues mutated in pUL25-L3 lie in very close proximity to the residues mutated in pUL25-C4, along a loop on the surface of pUL25. Since both regions are situated in the same area of pUL25 and since ΔUL25MO-infected U2OS cells expressing either mutant protein had similar patterns of DNA packaging and capsid assembly, it is possible that the L3 and C4 mutations disrupted the same functional interface of pUL25 (Fig. 1). The amino acids comprising C4 are more highly conserved than those of L3, suggesting that one or more of the residues mutated in pUL25-C4 may be involved in direct interactions that are required for primary envelopment, while L3 may act less directly.
Although mutant pUL25-L6, lacking the last three amino acid residues of pUL25, was unable to complement virus growth, the pattern of virion assembly of the UL25 null mutant in U2OS cells expressing this protein was similar to that of the wt virus, with enveloped virus particles readily detected in the cytoplasm, often in vesicles, and on the cell surface. For this reason, the major effect of the L6 mutation was probably at an early stage in infection, perhaps during uncoating of the virus genome. Recently, a UL25 ts mutant, ts1249, was shown to have an uncoating defect, but since the E233K mutation does not lie close to the C terminus of pUL25, where the L6 residues lie, it may affect a different ligand (24). Two different regions of the large tegument protein pUL36 have been shown to interact with pUL25, and like pUL25 (5, 23), pUL36 has been implicated in uncoating the virus genome (2, 6, 11). It is possible that pUL25-L6 is impaired in its interaction with this protein at either of these binding sites (5, 23). In the absence of pUL36, no envelopment of cytoplasmic C capsids takes places (9, 26). Therefore, since envelopment of C capsids in the cytoplasm occurs in ΔUL25MO-infected U2OS cells coinfected with AcUL25-L6, these virus particles must have a tegument and some pUL36. However, the enveloped ΔUL25MO C capsids in these cells may have reduced amounts of pUL36. Alternatively, pUL25-L6 could affect a different function that is unrelated to pUL36. For example, it is possible that pUL25-L6 fails to interact with the nucleoporins CAN/Nup214 and hCG1, which were recently found to interact with pUL25 (23) and are components of the nuclear pore complex through which the viral DNA is released into the nucleus.
Intriguingly, a sequence alignment of alphaherpesvirus pUL25 homologs showed that 75% contained potential class I or II PDZ domain binding motifs at the C terminus. PDZ domains are modular protein interaction domains that are approximately 90 amino acids in length and bind the C termini of target proteins in a sequence-specific manner. It is interesting that the three C-terminal residues from one of the head stabilization proteins of bacteriophage λ, gpW, which are thought to be unstructured, display sequence specificity for a class II PDZ domain binding motif and are critical for the protein's activity (16). However, further analysis is required to determine whether the putative PDZ domain binding motif in pUL25 has any functional significance.
In conclusion, we have identified specific regions of pUL25 that are essential for different functions of the protein. The mutant proteins generated were disrupted in one of three functions, namely, DNA packaging, nuclear egress of mature capsids, and possibly uncoating of the virus genome, and moreover, the relevant mutations all mapped within approximately one-third of the protein. This represents the first report of a pUL25 mutant that affects the exit of mature capsids from the nucleus.
Acknowledgments
We thank Cornel Fraefel (University of Zurich) for supplying fHSVΔpac and Chris Preston for helpful comments.
M. O'Hara was supported by an MRC studentship.
Footnotes
Published ahead of print on 24 February 2010.
REFERENCES
- 1.Adamson, W. E., D. McNab, V. G. Preston, and F. J. Rixon. 2006. Mutational analysis of the herpes simplex virus triplex protein VP19C. J. Virol. 80:1537-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Batterson, W., D. Furlong, and B. Roizman. 1983. Molecular genetics of herpes simplex virus. VIII. Further characterization of a temperature-sensitive mutant defective in the release of viral DNA and in other stages of the viral reproductive cycle. J. Virol. 45:397-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowman, B. R., R. L. Welschhans, H. Jayaram, N. D. Stow, V. G. Preston, and F. A. Quiocho. 2006. Structural characterization of the UL25 DNA-packaging protein from herpes simplex virus type 1. J. Virol. 80:2309-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cockrell, S. K., M. E. Sanchez, A. Erazo, and F. L. Homa. 2009. Role of the UL25 protein in herpes simplex virus DNA encapsidation. J. Virol. 83:47-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coller, K. E., J. I.-H. Lee, A. Ueda, and G. A. Smith. 2007. The capsid and tegument of the alphaherpesviruses are linked by an interaction between the UL25 and VP1/2 proteins. J. Virol. 81:11790-11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Copeland, A. M., W. W. Newcomb, and J. C. Brown. 2009. Herpes simplex virus replication: roles of viral proteins and nucleoporins in capsid-nucleus attachment. J. Virol. 83:1660-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cunningham, C., and A. Davison. 1993. A cosmid-based system for constructing mutants of herpes simplex virus type 1. Virology 197:116-124. [DOI] [PubMed] [Google Scholar]
- 8.de Bruyn Kops, A., S. L. Uprichard, M. Chen, and D. M. Knipe. 1998. Comparison of the intranuclear distributions of herpes simplex virus proteins involved in various viral functions. Virology 252:162-178. [DOI] [PubMed] [Google Scholar]
- 9.Desai, P. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 74:11608-11618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Homa, F. L., and J. C. Brown. 1997. Capsid assembly and DNA packaging in herpes simplex virus. Rev. Med. Virol. 7:107-122. [DOI] [PubMed] [Google Scholar]
- 11.Jovasevic, V., L. Liang, and B. Roizman. 2008. Proteolytic cleavage of VP1-2 is required for release of herpes simplex virus 1 DNA into the nucleus. J. Virol. 82:3311-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klupp, B. G., H. Granzow, G. M. Keil, and T. Mettenleiter. 2006. The capsid-associated UL25 protein of the alphaherpesvirus pseudorabies virus is nonessential for cleavage and encapsidation of genomic DNA but is required for nuclear egress of capsids. J. Virol. 80:6235-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuhn, J., T. Leege, B. G. Klupp, H. Granzow, W. Fuchs, and T. C. Mettenleiter. 2008. Partial functional complementation of a pseudorabies virus UL25 deletion mutant by herpes simplex virus type 1 pUL25 indicates overlapping functions of alphaherpesvirus pUL25 proteins. J. Virol. 82:5725-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lamberti, C., and S. K. Weller. 1998. The herpes simplex virus type 1 cleavage/packaging protein, UL32, is involved in efficient localization of capsids to replication compartments. J. Virol. 72:2463-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loret, S., G. Guay, and R. Lippé. 2008. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J. Virol. 82:8605-8618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maxwell, K. L., A. Davidson, H. Murialdo, and M. Gold. 2000. Thermodynamic and functional characterization of protein W from bacteriophage lambda. The three C-terminal residues are critical for activity. J. Biol. Chem. 275:18879-18886. [DOI] [PubMed] [Google Scholar]
- 17.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 18.McNab, A. R., P. Desai, S. Person, L. L. Roof, D. R. Thomsen, W. W. Newcomb, J. C. Brown, and F. L. Homa. 1998. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J. Virol. 72:1060-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McNabb, D. S., and R. J. Courtney. 1992. Characterization of the large tegument protein (ICP1/2) of herpes simplex virus type 1. Virology 190:221-232. [DOI] [PubMed] [Google Scholar]
- 20.Newcomb, W. W., F. L. Homa, and J. C. Brown. 2006. Herpes simplex virus capsid structure: DNA packaging protein UL25 is located on the external surface of the capsid near the vertices. J. Virol. 80:6286-6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ng, P. C., and S. Henikoff. 2001. Predicting deleterious amino acid substitutions. Genome Res. 11:863-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng, P. C., and S. Henikoff. 2003. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 31:3812-3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pasdeloup, D., D. Blondel, A. L. Isidro, and F. J. Rixon. 2009. Herpesvirus capsid association with the nuclear pore complex and viral DNA release involve the nucleoporin CAN/Nup214 and the capsid protein pUL25. J. Virol. 83:6610-6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Preston, V. G., J. Murray, C. M. Preston, I. M. McDougall, and N. D. Stow. 2008. The UL25 gene product of herpes simplex virus type 1 is involved in uncoating of the viral genome. J. Virol. 82:6654-6666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quinlan, M. P., L. B. Chen, and D. M. Knipe. 1984. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell 36:857-868. [DOI] [PubMed] [Google Scholar]
- 26.Roberts, A. P. E., F. Abaitua, P. O'Hare, D. McNab, F. J. Rixon, and D. Pasdeloup. 2009. Differing roles of inner tegument proteins pUL36 and pUL37 during entry of herpes simplex virus type 1. J. Virol. 83:105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saeki, Y., T. Ichikawa, A. Saeki, E. A. Chiocca, K. Tobler, M. Ackermann, X. O. Breakefield, and C. Fraefel. 1998. Herpes simplex virus type 1 DNA amplified as bacterial artificial chromosome in Escherichia coli: rescue of replication-competent virus progeny and packaging of amplicon vectors. Hum. Gene Ther. 9:2787-2794. [DOI] [PubMed] [Google Scholar]
- 28.Salmon, B., C. Cunningham, A. J. Davison, W. J. Harris, and J. D. Baines. 1998. The herpes simplex virus type 1 UL17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J. Virol. 72:3779-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scholtes, L., and J. D. Baines. 2009. Effects of major capsid proteins, capsid assembly, and DNA cleavage/packaging on the pUL17/pUL25 complex of herpes simplex virus 1. J. Virol. 83:12725-12737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheaffer, A. K., W. W. Newcomb, M. Gao, D. Yu, S. K. Weller, J. C. Brown, and D. J. Tenney. 2001. Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J. Virol. 75:687-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Song, S. U., S.-H. Shin, S.-K. Kim, G.-S. Choi, W.-C. Kim, M.-H. Lee, S.-J. Kim, I.-H. Kim, M.-S. Choi, Y.-J. Hong, and K.-H. Lee. 2003. Effective transduction of osteogenic sarcoma cells by a baculovirus vector. J. Gen. Virol. 84:697-703. [DOI] [PubMed] [Google Scholar]
- 32.Stow, N. D. 2001. Packaging of genomic and amplicon DNA by the herpes simplex virus type 1 UL25-null mutant KUL25NS. J. Virol. 75:10755-10765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thurlow, J. K., M. Murphy, N. D. Stow, and V. G. Preston. 2006. Herpes simplex virus type 1 DNA-packaging protein UL17 is required for efficient binding of UL25 to capsids. J. Virol. 80:2118-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thurlow, J. K., F. J. Rixon, M. Murphy, P. Targett-Adams, M. Hughes, and V. G. Preston. 2005. The herpes simplex virus type 1 DNA packing protein UL17 is a virion protein that is present in both the capsid and tegument compartments. J. Virol. 79:150-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trus, B. L., W. W. Newcomb, N. Cheng, C. Giovanni, L. Marekov, F. L. Homa, J. C. Brown, and A. C. Steven. 2007. Allosteric signaling and a nuclear exit strategy: binding of UL25/UL17 heterodimers to DNA filled HSV-1 capsids. Mol. Cell 26:479-489. [DOI] [PMC free article] [PubMed] [Google Scholar]