Abstract
A new pathogenic R5-tropic simian/human immunodeficiency virus (SHIV) was generated following serial passaging in rhesus macaques. All 13 animals inoculated with SHIVAD8 passaged lineages experienced marked depletions of CD4+ T cells. Ten of these infected monkeys became normal progressors (NPs) and had gradual losses of both memory and naïve CD4+ T lymphocytes, generated antiviral CD4+ and CD8+ T cell responses, and sustained chronic immune activation while maintaining variable levels of plasma viremia (102 to 105 RNA copies/ml for up to 3 years postinfection [p.i.]). To date, five NPs developed AIDS associated with opportunistic infections caused by Pneumocystis carinii, Mycobacterium avium, and Campylobacter coli that required euthanasia between weeks 100 and 199 p.i. Three other NPs have experienced marked depletions of circulating CD4+ T lymphocytes (92 to 154 cells/μl) following 1 to 2 years of infection. When tested for coreceptor usage, the viruses isolated from four NPs at the time of their euthanasia remained R5 tropic. Three of the 13 SHIVAD8-inoculated macaques experienced a rapid-progressor syndrome characterized by sustained plasma viremia of >1 × 107 RNA copies/ml and rapid irreversible loss of memory CD4+ T cells that required euthanasia between weeks 19 and 23 postinfection. The sustained viremia, associated depletion of CD4+ T lymphocytes, and induction of AIDS make the SHIVAD8 lineage of viruses a potentially valuable reagent for vaccine studies.
Simian immunodeficiency virus (SIV)/macaque models of AIDS have been extensively used as surrogates for human immunodeficiency virus type 1 (HIV-1) in studies of virus-induced immunopathogenesis and vaccine development. As is observed for the HIVs recovered from a majority of individuals during the asymptomatic phase of their infections, pathogenic SIVs utilize the CCR5 coreceptor to enter their CD4+ T lymphocyte targets in vivo (36). This leads to the elimination of memory CD4+ T cells circulating in the blood and residing at effector sites (gastrointestinal [GI] tract, mucosal surfaces, and lung), particularly during acute HIV and SIV infections (5, 29, 32, 49). In contrast to naturally occurring SIVs and HIVs, SIV/HIV chimeric viruses (simian/human immunodeficiency viruses [SHIVs]) were constructed in the laboratory by inserting a large segment of the HIV genome, including the env gene, into the genetic backbone of the molecularly cloned SIVmac239 (44). SHIVs were developed because they expressed the HIV envelope glycoprotein and could be used in vaccine experiments to evaluate neutralizing antibodies (NAbs) elicited by HIV-1 gp120 immunogens. The commonly used pathogenic SHIVs generated high levels (107 to 108 RNA copies/ml) of plasma viremia and induced an extremely rapid, systemic, and nearly complete depletion of the entire CD4+ T cell population, resulting in death from immunodeficiency beginning at 3 months postinoculation (23, 26, 41). Unlike SIVs, however, these pathogenic SHIVs exclusively targeted CXCR4-expressing CD4+ T cells during infections of rhesus monkeys (36). Despite their extraordinary virulence, most vaccine regimens (naked DNA, peptides, proteins, inactivated virions, recombinant modified vaccinia virus Ankara (MVA), and DNA prime/recombinant viral-vector boosting) were effective in controlling intravenous (i.v.) and mucosal X4-tropic SHIV challenges (1, 3, 33, 42, 46). When it became apparent that the same vaccination strategies that were effective in suppressing pathogenic SHIVs failed to control SIV infections, concerns were raised about whether X4 SHIVs were appropriate surrogates for HIV in vaccine experiments (13).
The unusual biological properties of the X4 SHIVs plus the discrepant outcomes of SIV and X4 SHIV vaccine experiments have become a driving force for developing CCR5-utilizing (R5) SHIVs. Although several clade B and clade C R5-tropic SHIVs have been constructed (7, 15, 21, 30, 38), the SHIVSF162 lineage viruses are the best-characterized and most widely used R5 SHIVs (20). They have been employed in microbicide (10), neutralizing monoclonal antibody (MAb) passive-transfer (16, 17), and vaccination (2) studies.
In the aftermath of the failed STEP HIV vaccine trial, there was general consensus that additional SIVs and SHIVs should be developed, particularly for use as heterologous challenge viruses in vaccine studies (12). With this goal in mind, we report the generation of a new pathogenic R5-tropic SHIV bearing the env gene from the HIV-1Ada isolate (14). HIV-1Ada was selected because it is a prototypical macrophage-tropic strain (8), uses CCR5 for cell entry (53), and has the potential for eliciting NAbs against HIV-1 gp120, and we had previously constructed a full-length infectious molecular clone (pHIV-1AD8) (48). Based on previous experience in obtaining pathogenic X4-tropic SHIVs, serial passaging in macaques, treated with an anti-CD8 MAb at the time of virus inoculation, was used to expedite the adaptation of R5-SHIV sequences in a nonhuman primate host. Of the 13 animals inoculated with in vivo-passaged SHIVAD8#2 (see below) and its immediate derivatives, 10 exhibited a normal-progressor (NP) phenotype, sustaining gradual depletions of both memory and naïve CD4+ T cells from the circulation and memory CD4+ T cells at an effector site (lung) while maintaining variable viral-RNA loads (102 to 105 RNA copies/ml) for up to 3 years postinfection (p.i.). Five of these monkeys developed immunodeficiency with associated opportunistic infections requiring euthanasia. Three other NPs currently have total CD4+ T cell counts of 92 to 154 cells/μl plasma after 1 to 2 years of infection. The remaining 3 of the 13 SHIVAD8-inoculated macaques experienced a rapid-progressor (RP) clinical course and were euthanized between weeks 19 and 23 p.i. because of intractable diarrhea and marked weight loss. The sustained viremia, associated depletion of CD4+ T lymphocytes, and induction of AIDS make the SHIVAD8 lineage of viruses a potentially valuable reagent for vaccine studies.
MATERIALS AND METHODS
Construction of SHIVAD8.
SHIVAD8 contains the env gene from the R5-tropic HIV-1Ada (14)-derived molecular clone pHIVAD8 (48). A 3.04-kb segment from pHIVAD8, including a portion of the vpr gene and the entire tat, rev, vpu, and env genes, was PCR amplified using the forward primer TGAAACTTATGGGGATACTTGGGC, which begins at nucleotide 141 of the AD8 vpr gene, allowing the incorporation of a unique EcoRI site, located 21 nucleotides downstream from the primer, into the PCR product. The reverse PCR primer (TCCACCCATAAGCTTATAGCAAAGTCCTTTCCAAGCCC) generated a HindIII site adjacent to and encompassing the last 2 nucleotides of the env reading frame, as well as a substitution of a Thr for a Leu 3 codons upstream from the env termination codon. PCRs were performed using 10 pmol each of the forward and reverse primers, Platinum PCR SuperMix High Fidelity (Invitrogen), and 1 μl of pHIVAD8 in a final volume of 50 μl. The reaction mixtures were heated to 94°C for 2 min, followed by 30 cycles of 94°C for 20 s, 59°C for 30 s, and 70°C for 3 min and a 7-min extension at 70°C. The PCR product was gel extracted using a Qiaquick gel extraction kit (Qiagen) and digested with EcoRI and HindIII, and the resulting 3.04-kb restriction fragment was cloned directly into the previously described and similarly digested pSHIVDH12 (45) to generate pSHIVAD8. DNA sequencing of the entire 3.04-kb insert in pSHIVAD8 was conducted to verify that no spurious changes had been introduced during the PCR amplification and cloning.
Preparation of SHIVAD8 virus stocks.
HeLa cells were transfected with 25 μg of pSHIVAD8, and the virus present in the supernatant at 48 h was pelleted in an ultracentrifuge and resuspended in RPMI 1640 medium as previously described (52). Stocks of the cloned SHIVAD8 were prepared by infecting PM1 cells (31) or concanavalin A (ConA)-activated rhesus monkey peripheral blood mononuclear cells (PBMC) with the HeLa-derived SHIVAD8, as previously described (23, 24), and pooling the supernatant media at the times of peak reverse transcriptase (RT) production from both infections.
SHIVAD8 stock 2 (SHIVAD8#2) was prepared from PBMC and bone marrow (BM), spleen, and lymph node (LN) samples collected from macaque CK1G on day 6 p.i. Cell suspensions from axillary, inguinal, iliac, and mesenteric LNs, PBMC, and BM were cocultivated with PBMC from uninfected animals; the culture supernatants were monitored daily for RT activity, pooled, and designated SHIVAD8#2. The infectious titer of SHIVAD8#2 was 1.5 × 103 tissue culture infective doses (TCID50)/ml, as determined in rhesus macaque PBMC.
SHIVAD8 lymph node virus (SHIVAD8LN) was prepared from supernatant medium collected from cocultures of lymph node suspensions plus PBMC recovered from animal CJ8B at week 59 p.i. (Table 1) and PBMC from uninfected rhesus monkeys. The infectious titer of SHIVAD8LN was 6.4 × 103 TCID50/ml, as determined in rhesus macaque PBMC.
TABLE 1.
Infection of rhesus macaques with SHIVAD8#2 and immediate derivatives
| Animal | Inoculum |
|---|---|
| CJ8B | SHIVAD8#2 |
| CK15 | Blood transfusion from CJ8B (wk 60) |
| CJ58 | Blood transfusion from CJ8B (wk 60) |
| CE8J | Lymph node virusa (SHIVAD8#2LN, 3.2 × 105 TCID50) from CJ8B (wk 59) |
| CJ35 | Lymph node virus (SHIVAD8#2LN, 3.2 × 105 TCID50) from CJ8B (wk 59) |
| CJ3V | PBMC virusb (SHIVAD8#2PBMC, 5.9 × 104 TCID50) from CK15 + CJ58 (wk 4) |
| CK5G | PBMC virus (SHIVAD8#2PBMC, 5.9 × 104 TCID50) from CK15 + CJ58 (wk 4) |
| DB99 | Blood transfusion from CJ8B (wk 117) + CK15 (wk 57) + CJ58 (wk 57) |
| DA1Z | Blood transfusion from CJ8B (wk 117) + CK15 (wk 57) + CJ58 (wk 57) |
| A4E008 | Blood transfusion from DA1Z (wk 1) + DB99 (wk 1) |
| DA4W | Blood transfusion from DA1Z (wk 1) + DB99 (wk 1) |
| CL5A | SHIVAD8#2 passaged in vitro for 30 days (SHIVAD8#2.d30, 4.3 × 105 TCID50) |
| CL98 | SHIVAD8#2 passaged in vitro for 30 days (SHIVAD8#2.d30, 4.3 × 105 TCID50) |
Lymph node virus: SHIVAD8 derivative prepared from the supernatant medium collected from cocultures of lymph node suspensions plus PBMC, recovered from animal CJ8B at week 59 p.i., and PBMC from uninfected rhesus monkeys.
PBMC virus: SHIVAD8 derivative prepared from the supernatant medium collected from cocultures of PBMC, recovered from the indicated infected animals at week 4 p.i., and PBMC from uninfected rhesus monkeys.
SHIVAD8 PBMC virus (SHIVAD8PBMC) was prepared from supernatant medium collected from cocultures of PBMC recovered and pooled from animals CK15 and CJ58 at week 4 p.i. (Table 1) and PBMC from uninfected rhesus monkeys. The infectious titer of SHIVAD8PBMC was 1.1 × 104 TCID50/ml, as determined in rhesus macaque PBMC.
SHIVAD8#2.d30 was prepared by infecting ConA-stimulated pig-tailed macaque (PT) PBMC with SHIVAD8#2. Fresh ConA-stimulated PT PBMC were added to the infected cultures on days 10 and 20, and the supernatant medium collected on day 30, designated SHIVAD8#2.d30, had an infectious titer of 8.5 × 104 TCID50/ml, as determined in rhesus macaque PBMC.
Virus replication assay in rhesus monkey PBMC.
The preparation and infection of rhesus monkey PBMC have been described previously (25). Briefly, PBMC stimulated with concanavalin A and cultured in the presence of recombinant human interleukin-2 (IL-2) were spinoculated (1,200 × g for 1 h) (37) with virus normalized for RT activity. Virus replication was assessed by RT assay of the culture supernatant as described above.
Animal experiments.
Rhesus macaques (Macaca mulatta) were maintained in accordance with the guidelines of the Committee on Care and Use of Laboratory Animals (9) and were housed in a biosafety level 2 facility; biosafety level 3 practices were followed. Phlebotomies, i.v. virus inoculations, euthanasia, and tissue sample collections were performed as previously described (11). Bronchoalveolar lavage (BAL) fluid lymphocytes were prepared from uninfected and infected animals using a pediatric bronchoscope (Olympus BF3C40; Olympus America, Inc., Melville, NY), as previously described (22).
Serial in vivo passaging of SHIVAD8 was initiated by transferring whole blood (10 ml) and BM (2 ml) to a recipient animal previously treated with the anti-CD8+ T cell-depleting MAb cM-T807 (10 mg/kg of body weight) on days −1 and +3 p.i. In subsequent passages, spleen, LN (axillary, inguinal, iliac, and mesenteric), PBMC, and BM cell suspensions were prepared from infected donors at the time of necropsy and transferred (1 × 108 to 3 × 108 mononuclear cells and 1 × 108 to 10 × 108 BM cells) to a new recipient by the i.v., intraperitoneal (i.p.), and BM routes.
Quantitation of proviral-DNA and plasma viral-RNA levels.
The number of viral-DNA copies in PBMC was measured by quantitative DNA PCR (45). Viral-RNA levels in plasma were determined by real-time reverse transcription-PCR (ABI Prism 7700 sequence detection system; Applied Biosystems, Foster City, CA) as previously reported, using reverse-transcribed viral RNA in plasma samples from SIVmac239-inoculated rhesus macaques (11).
Lymphocyte immunophenotyping and data analysis.
EDTA-treated blood samples and BAL fluid lymphocytes were stained for flow cytometric analysis as described previously (34, 36), using combinations of the following fluorochrome-conjugated MAbs: CD3 (fluorescein isothiocyanate [FITC] or phycoerythrin [PE]), CD4 (PE, peridinin chlorophyll protein-Cy5.5 [PerCP-Cy5.5], or allophycocyanin [APC]), CD8 (PerCP or APC), CD28 (FITC or PE), CD95 (APC), and Ki-67 (FITC or PE). All antibodies were obtained from BD Biosciences (San Diego, CA), and samples were analyzed by four-color flow cytometry (FACSCalibur; BD Biosciences Immunocytometry Systems). Data analysis was performed using CellQuest Pro (BD Biosciences) and FlowJo (TreeStar, Inc., San Carlos, CA). For Ki-67 staining, cells were fixed with fluorescence-activated cell sorter (FACS) lysing solution (Becton Dickinson), treated with FACS permeabilization buffer 2 (Becton Dickinson), and stained with Ki-67 MAb or a control isotype IgG1. In this study, naïve CD4+ T cells were identified by their CD95low CD28high phenotype, whereas memory CD4+ T cells were CD95high CD28high or CD95high CD28low in the CD4+ small lymphocyte gate (36, 39).
Intracellular-cytokine assays.
Stimulation was performed on frozen lymphocytes as described previously (40). Freshly thawed lymphocytes were resuspended (106/ml) in RPMI medium supplemented with antibiotics and glutamine. Anti-CD28 conjugated to Alexa 594-PE was used for costimulation. Staphylococcus enterotoxin B (1 μg/ml; Sigma-Aldrich, St. Louis, MO) was used to stimulate T cells mitogenically through the T cell receptor as a positive control. A negative control (cells treated only with costimulatory anti-CD28) was included in every experiment. Peptides used to stimulate SIV-specific T cells were 15 amino acids (aa) in length, overlapping by 11 amino acids, and encompassed SIVmac239 Gag (New England Peptide, Gardner, MA). The concentration of each peptide was 2 μg/ml for stimulations, which were performed in the presence of brefeldin A (BFA) (1 μg/ml; Sigma-Aldrich, St. Louis, MO) for 16 h at 37°C. All cells were surface stained with the dead-cell exclusion dye Aqua Blue (Invitrogen Corp., Carlsbad CA), followed by staining with anti-CD3 Alexa 700 (BD Biosciences), anti-CD4 Cy5.5-PE (eBioscience Inc., San Diego, CA), anti-CD8 Pacific Blue (BD Biosciences), and anti-CD95 Cy5-PE (BD Biosciences). The cells were then fixed, permeabilized, and stained with anti-gamma interferon (IFN-γ) Cy7-PE (BD Biosciences), anti-IL-2 APC (BD Biosciences), tumor necrosis factor (TNF) FITC (BD Biosciences), and Mip1-β PE (BD Biosciences). SIV-specific CD8 T cell responses are reported as the frequency of memory CD8 T cells, gated by characteristic light scatter properties; then as Aqua Blue−, CD3+, CD8+, CD4−, or CD95+; and by production of either TNF or Mip-1β. All data are reported after background subtraction.
Virus neutralization assays.
Autologous plasma samples (1:20 dilution) from SHIVAD8-infected macaques were incubated with (i) the same uncloned SHIV-AD8 derivative used for inoculation or (ii) the SHIVAD8 isolated from PBMC at week 4 p.i. (for monkeys CJ58 and CK15) in quadruplicate in 96-well flat-bottom culture plates in a total volume of 50 μl for 1 h at 37°C. Prechallenge plasma samples from each animal served as controls. Freshly trypsinized TZM-bl cells (50) (1.5 × 104 in 150 μl Dulbecco's modified Eagle's medium [DMEM] containing 20 μg/ml DEAE dextran) were added to each well, and the cultures were maintained in a 37°C incubator for 28 h. The amount of virus-induced luciferase activity, measured as relative light units (RLU), present in cell lysates was determined as previously described (51), and the average neutralization activity for each plasma sample was determined. The average number of RLU for the prechallenge plasma controls ranged from 1 × 105 to 2 × 105. Any sample resulting in a 50% reduction of luciferase activity compared to that obtained with the uninfected control sample was considered positive for NAbs. To determine neutralizing-antibody titers, 40 μl of diluted virus, sufficient to generate the desired numbers of RLU, was mixed with 10 μl of appropriately diluted plasma samples in a 96-well plate and incubated for 1 h at 37°C. TZM-bl cells were added, cultures were maintained for an additional 28 h, and intracellular luciferase activity was measured as described above.
Coreceptor utilization assays.
Freshly trypsinized TZM-bl cells (1 × 104 per well) in 135 μl DMEM containing 10% fetal calf serum (FCS) and DEAE dextran (15 μg/ml) were seeded in flat-bottom 96-well plates. Twenty-five microliters of coreceptor antagonists (AD101 against CCR5, AMD3100 against CXCR4, or both, at final concentrations ranging from 0.1 nM to 1,000 nM) was added to each well. Following incubation for 1 h at 37°C, 10 TCID50 of replication-competent virus, determined in TZM-bl cells as previously described, in 40 μl was added to each well. After 24 h of incubation at 37°C, luciferase activity was determined. The percent infectivity reported was derived from the mean of quadruplicate assays.
To generate 293T cell-derived SHIVAD8 pseudotyped viruses, two separate plasmids were constructed. The first (pNLenv1) contained a frameshifted mutation in the leader peptide region of gp120 (43). Plasmids expressing the SHIVAD8(RIG+) and SHIVAD8(RIG-) env genes [pCMV-AD8(RIG+) and AD8(RIG−)] were generated by reverse transcription-PCR of plasma viral RNA, collected from macaque DB99 at the time of euthanasia, and subcloning into NotI and (newly created) XbaI sites of the pCMVbeta expression plasmid (Clonetech, Palo Alto, CA). Both plasmids [pNLenv1 and pCMV-AD8(RIG) in a 5:1 ratio] were cotransfected into 293T cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). The titers of pseudotyped-virus preparations were determined, and they were assayed for coreceptor usage 48 h following infection of TZM-bl cells, as described for replication-competent virus.
RESULTS
Construction of a CCR5-tropic SHIV.
We previously reported the construction of a full-length infectious HIV-1 molecular clone (pHIV-1AD8) derived from the prototypical macrophage-tropic CCR5-utilizing HIV-1Ada isolate (14, 48). A SHIV expressing the env gene from pHIVAD8 was obtained by inserting the 3.04-kb EcoRI-to-HindIII DNA fragment (including a portion of vpr and the entire tat, rev, vpu, and env genes) into the genetic background of pSHIVDH12 (45), as described in Materials and Methods. The resulting molecular clone, pSHIVAD8, directed the production of progeny virions following the transfection of HeLa cells. Virus stocks were prepared by infecting PM1 cells or ConA-stimulated rhesus PBMC with virions pelleted from HeLa cell transfection culture supernatants.
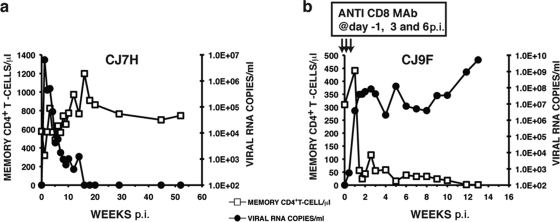
It is not generally appreciated how daunting it is to generate an R5-tropic SHIV able to maintain detectable levels of set-point viremia, exclusively target memory CD4+ T cells, and induce immunodeficiency in inoculated rhesus monkeys. Simply replacing orthologous SIV sequences with a DNA segment including a CCR5-utilizing HIV-1 env gene does not usually result in a SHIV exhibiting robust replication kinetics in vivo and a disease-inducing phenotype. This was, in fact, the case for SHIVAD8: levels of plasma viremia following virus inoculation (1 ml of undiluted virus by the i.v., i.p., and BM routes) were promptly and durable suppressed, and the numbers of memory CD4+ T lymphocytes did not change appreciably, as shown for a representative infected animal (CJ7H) in Fig. 1a. To be certain that we were on the right track with respect to the targeting and elimination of memory, not naïve, CD4+ T cells in vivo, a second macaque (CJ9F) was treated with the CD8+ T lymphocyte-depleting MAb cM-T807 24 h prior to SHIVAD8 inoculation, as well as on days 3 and 6 post-virus infection, to promote a vigorous in vivo infection. Unlike untreated macaque CJ7H, the levels of plasma viral RNA in monkey CJ9F rapidly rose to 3.8 × 107 copies/ml by day 10 p.i. and were associated with a rapid and irreversible decline of circulating memory CD4+ T cells (Fig. 1b). In contrast, the numbers of naïve CD4+ T lymphocytes in animal CJ9F were maintained in the 600- to 800-cell/μl range during this period. This result, therefore, confirmed that SHIVAD8 could sustain high virus loads and preferentially target the memory CD4+ T cell subset in vivo, but only in an animal with a compromised immune system.
FIG. 1.
Infectivity of the original SHIVAD8 in rhesus monkeys. Macaques CJ7H (a) and CJ9F (b) were inoculated with 1 ml of undiluted SHIVAD8 by the i.v., i.p., and BM routes. Macaque CJ9F was treated with the depleting anti-CD8 MAb cM-T807 as indicated.
In vivo passaging of SHIVAD8.
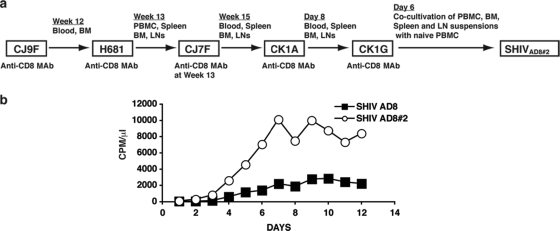
The prompt control of plasma viremia and the nonpathogenic phenotype of SHIVAD8 observed in untreated macaques were reminiscent of the infectivity patterns observed with first-generation X4-tropic SHIVs (28, 44). We therefore initiated serial animal-to-animal passaging of SHIVAD8 with macaque CJ9F as the “founder” infected monkey (Fig. 2a). This approach had previously been used to generate X4 SHIVs exhibiting more robust replicative and pathogenic properties (26, 41). Unfortunately, in vivo serial passaging of virus to optimize infectivity is an empirical and stochastic process. One never knows when or if an R5 SHIV has acquired an augmented replicative phenotype. The ultimate proof that such a change has occurred requires the inoculation of additional animals and waiting several months to assess the resultant viral replication kinetics and CD4+ T cell dynamics.
FIG. 2.
Serial animal-to-animal passage of SHIVAD8. (a) Passage history of SHIVAD8 and origin of SHIVAD8#2. (b) Rhesus monkey PBMC were infected with SHIVAD8 or the passaged SHIVAD8#2 virus stock, normalized for RT activity. CPM, counts per minute.
The strategy employed was to maximize the emergence of disease-inducing SHIV variants, putatively present in an increasingly genetically diverse virus population, by serially transferring large numbers of infected cells by i.v., i.p., and BM routes into recipient animals previously treated with an anti-CD8 depleting MAb. As indicated in Fig. 2a, whole blood and bone marrow cells were transferred from macaque CJ9F to macaque H681 by these three routes. In subsequent passages, cell suspensions were prepared from spleen, LN (axillary, inguinal, iliac, and mesenteric), PBMC, and BM cells collected at the time of necropsy, as described in Materials and Methods. With one exception (macaque CJ7F), the depleting anti-CD8 MAb was administered to a recipient animal on days −1 and +3 p.i. to facilitate unrestricted replication in vivo. Animal CJ7F did not receive anti-CD8 MAb at the time of virus transfer to investigate the possibility that SHIVAD8 had acquired improved replication properties in vivo following the initial two in vivo passages. Because this was not the case (its plasma viral-RNA loads had declined to 360 RNA copies/ml at week 10 p.i.), macaque CJ7F was treated with anti-CD8 MAb at week 13 p.i. and sustained an immediate burst of virus production that reached 1.4 × 106 RNA copies/ml of plasma at week 14 p.i. CJ7F was euthanized at week 15 p.i., and cell suspensions were prepared as described above and transferred by the i.v., i.p., and BM routes into macaque CK1A, previously treated with anti-CD8 MAb (Fig. 2a). Following the fifth in vivo passage, macaque CK1G was euthanized on day 6 p.i., and cell suspensions, prepared at the time of necropsy, were cocultivated with ConA-stimulated PBMC from uninfected rhesus monkeys as described in Materials and Methods; the culture supernatants were monitored for the presence of reverse transcriptase activity, pooled, and designated SHIVAD8#2.
Inoculation of rhesus macaques with SHIVAD8#2 and its immediate derivatives resulted in sustained plasma viremia and loss of CD4+ T lymphocytes.
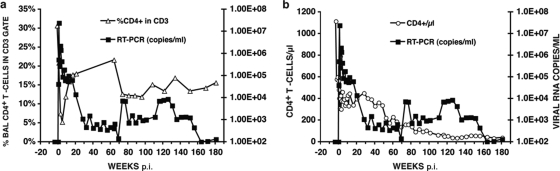
To ascertain whether serial passaging of SHIVAD8 in vivo had resulted in the acquisition of improved replicative properties, ConA-stimulated rhesus monkey PBMC were infected with SHIVAD8#2 or the starting SHIVAD8 virus preparation, both normalized for RT activity. As shown in Fig. 2b, SHIVAD8#2 replicated to much higher levels in cultured macaque PBMC than the original SHIVAD8. To determine whether this improved infectivity of SHIVAD8#2 for rhesus PBMC was correlated with augmented replication in an animal not treated with the depleting anti-CD8 MAb, macaque CJ8B was inoculated i.v. with 1.5 × 104 TCID50 of SHIVAD8#2. As shown in Fig. 3a, this monkey experienced a marked but transient depletion of memory CD4+ T cells in BAL specimens during the acute infection and maintained detectable levels of plasma viremia. Because animal CJ8B subsequently experienced a decline in the total circulating CD4+ T lymphocyte population from 565 to 175 cells/μl at week 56 p.i. (Fig. 3b), whole blood or virus propagated ex vivo from CJ8B lymph node suspensions (lymph node virus [SHIVAD8LN]) was inoculated into four additional macaques (CK15, CJ58, CE8J, and CJ35) (Fig. 4). Four other animals (DB99, DA1Z, A4E008, and DA4W) received blood transfusions, and two (CJ3V and CK5G) were inoculated with PBMC coculture virus (SHIVAD8PBMC) derived from monkeys CK15 and CJ58 (Fig. 4). In addition, because it was unknown at the time of its preparation whether SHIVAD8#2 had acquired augmented in vivo infectivity properties, SHIVAD8#2 was propagated for an additional 30 days ex vivo in macaque PBMC as described in Materials and Methods. Because the resulting derivative, designated SHIVAD8#2.d30, exhibited robust infectivity in both pigtailed and rhesus macaque PBMC (data not shown), it was inoculated intravenously into two rhesus monkeys (CL5A and CL98) (Fig. 4). The inocula used to infect rhesus monkeys with SHIVAD8#2 and its immediate derivatives are listed in Table 1. None of these monkeys received the depleting anti-CD8 MAb.
FIG. 3.
SHIVAD8#2 induces sustained plasma viremia and loss of CD4 T cells in an inoculated rhesus macaque. Plasma viremia and the percentage of BAL fluid CD4+ T cells (a) or the absolute numbers of circulating CD4+ T cells (b) in rhesus macaque CJ8B inoculated intravenously with SHIVAD8#2 are shown. RT-PCR, reverse transcription-PCR.
FIG. 4.
SHIVAD8#2 and its immediate derivatives cause immunodeficiency in rhesus macaques. The dashed arrows indicate virus transfer by blood transfusion. The thick arrows indicate LN or PBMC specimens used to generate virus stocks by coculturing with PBMC from uninfected donors. †, euthanized animals.
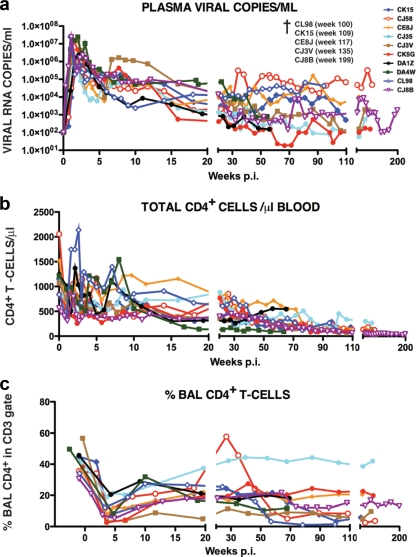
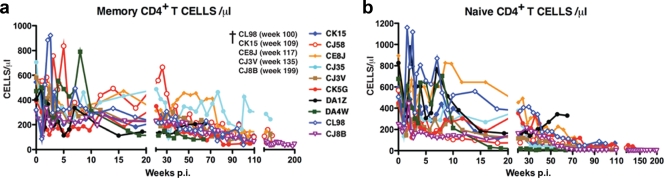
Ten of the 13 animals infected with SHIVAD8#2 or its immediate derivatives experienced an NP clinical course characterized by set-point virus loads that varied widely (from less than 103 to more than 105 RNA copies/ml) and a gradual depletion of circulating CD4+ T lymphocytes (Fig. 5a and b). Transient, and in some cases quite significant, losses of memory CD4+ T cells in BAL samples was a common finding during the acute infection (Fig. 5c). The loss of circulating CD4+ T lymphocytes in the 10 SHIVAD8#2-infected NPs affected both memory and naïve subsets (Fig. 6). With one exception (monkey CJ35), these animals sustained depletions of circulating memory CD4+ T cells to the 200-cell/μl level by week 100. NPs also experienced increased memory CD4+ T lymphocyte turnover, as monitored by Ki-67 expression, particularly during the first 10 weeks and the final stages of the infection (see Fig. S1 in the supplemental material). The loss of naïve CD4+ T lymphocytes in NP monkeys was even more profound. By week 80 p.i., this subset had declined to below 100 cells/μl in all of the animals (Fig. 6b). At the time of their euthanasia, five NPs (CJ8B, CE8J, CJ3V, CK15, and CL98) had only 1, 3, 6, 12, and 68 circulating naïve CD4+ T cells/μl, respectively. We previously reported that SIVsmE543-infected NPs had also experienced a marked loss of naïve CD4+ T cells as early as 20 weeks p.i. (35). It was therefore not unexpected that NP SHIVAD8-infected monkeys might also sustain a depletion of their naïve CD4+ T cell subset.
FIG. 5.
Total CD4+ T lymphocytes are gradually lost in normal progressors following infection with SHIVAD8#2 and its immediate derivatives. The levels of plasma viremia (a), absolute numbers of peripheral CD4+ T cells (b), and percentages of BAL fluid CD4+ T cells (c) are shown. The five normal progressors that developed AIDS and were euthanized are indicated (†).
FIG. 6.
Marked depletion of naïve and memory CD4+ T lymphocytes characterizes long-term SHIVAD8 infection in NP rhesus monkeys. Absolute numbers of memory CD4+ T cells (a) and naive CD4+ T cells (b) in 10 normal-progressor macaques during 200 weeks of SHIVAD8 infection are shown. †, euthanized animals.
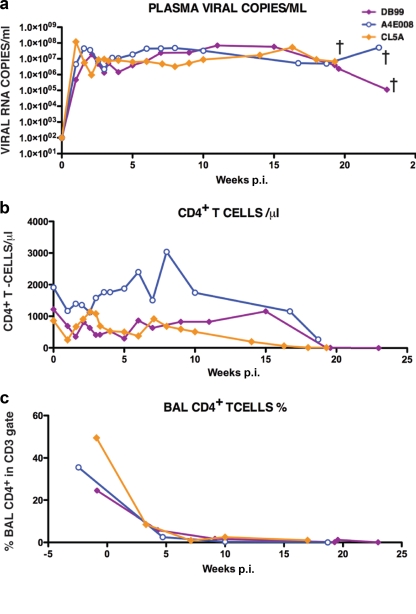
Three of the 13 macaques inoculated with SHIVAD8#2 and its immediate derivatives became RPs, requiring euthanasia between weeks 19 and 23 p.i. because of anorexia, intractable diarrhea, and marked weight loss (Fig. 7). Virus set points in the RPs exceeded 107 RNA copies/ml, memory CD4+ T cells in BAL specimens rapidly and irreversibly declined, and at the time of death, all of the animals had sustained marked losses of circulating CD4+ T cells.
FIG. 7.
Patterns of virus replication and CD4+T cell dynamics in SHIVAD8 rapid progressors. The levels of plasma viremia (a), absolute numbers of peripheral CD4+ T cells (b), and percentages of BAL fluid CD4+ T cells (c) are shown. †, euthanized animals.
Immune responses to SHIVAD8.
In the context of its use as a challenge virus in vaccine experiments, it was important to show that SHIVAD8 elicited both cellular and humoral immune responses during infections of rhesus monkeys. Therefore, anti-SHIVAD8 Gag-specific CD8+ T lymphocyte responses were measured by flow cytometry for 6 of the 10 NPs by intracellular staining of cells expressing TNF-α and/or IFN-γ following stimulation with a 15-mer peptide pool spanning SIVmac239 Gag. The levels of virus-specific CD8+ T cells in this group of rhesus monkeys ranged from 0.33 to 1.68% during the second year of their infection (see the table in the supplemental material). A similar analysis of Gag-specific responses in memory CD4+ T cells at these times in the same animals indicated that 0.90 to 2.90% expressed TNF-α and/or IFN-γ (see the table in the supplemental material).
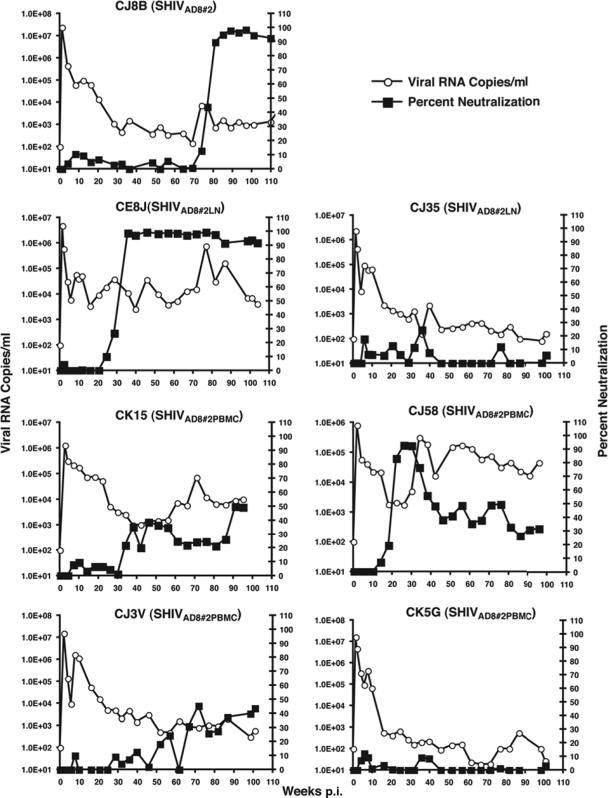
NAbs were detected in several of the NPs during the course of their infections (Fig. 8). The seven macaques evaluated had been inoculated with SHIVAD8#2 or two immediate derivatives (SHIVAD8#2LN and SHIVAD8#2PBMC). Plasma neutralizing activity directed against the same virus used for animal challenge was evaluated in monkeys CJ8B, CE8J, CJ35, CJ3V, and CK5G. The neutralization sensitivity of autologous virus (SHIVAD8#2PBMC) was monitored using plasma collected from PBMC of macaques CK15 and CJ58 (Fig. 4). The time of appearance of neutralization activity varied widely (week 20 to week 78 p.i.) and was generally correlated with levels of set-point viremia. In the three macaques producing the highest levels of anti-SHIVAD8 NAbs, the actual 50% inhibitory concentration (IC50) neutralization titers determined by limiting plasma dilution were 1:159 (CJ8B at week 89), 1:102 (CJ58 at week 30), and 1:143 (CE8J at week 52).
FIG. 8.
Neutralizing-antibody activities detected in normal-progressor macaques following infection with SHIVAD8#2 or its immediate derivatives. Plasma samples (1:20 dilution) from the indicated SHIVAD8-infected macaques were incubated in quadruplicate for 1 h at 37°C with the virus isolates shown in parentheses and then used as an inoculum to infect TZM-bl cells. The luciferase activity present in cell lysates at 28 h p.i. was measured, and the average percent neutralization activity in plasma at each time point was determined. Prechallenge plasma samples served as negative controls and baselines for zero neutralizing-antibody activity.
Coreceptor usage by SHIVAD8 lineage viruses.
The env gene of SHIVAD8 was derived from the prototypical macrophage-tropic HIV-1Ada, previously shown to use CCR5 for cell entry (53). When tested in a TZM-bl entry assay with inhibitors that specifically target CXCR4 or CCR5, the original SHIVAD8, SHIVAD8#2 (data not shown), and SHIVAD8#2LN exclusively utilized CCR5 (see Fig. S2 in the supplemental material). The marked depletion of circulating naïve CD4+ T cells in all SHIVAD8 NPs (Fig. 6b) raised the possibility that a coreceptor switch had occurred, enabling these viruses to enter and eliminate naïve CD4+ T cells, which express high levels of surface CXCR4, but not CCR5. Accordingly, virus was recovered from three NPs (CK15, CE8J, and CL98) immediately prior to euthanasia. When tested for coreceptor usage, the viruses isolated from all three NPs remained R5 tropic (see Fig. S2 in the supplemental material), indicating that the loss of naïve CD4+ T cells was not due to direct virus-induced cell killing.
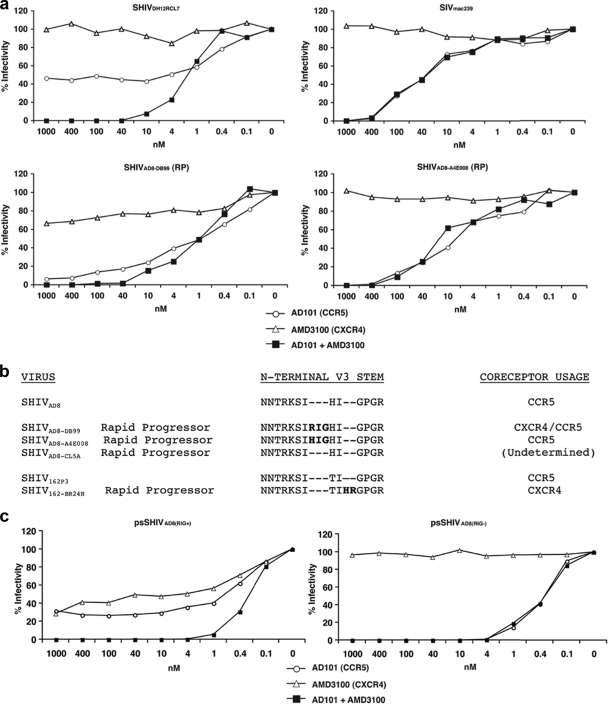
As noted earlier, three monkeys infected with SHIVAD8#2 derivatives exhibited an RP phenotype. By week 10 p.i., these macaques (DB99, A4E008, and CL5A) had experienced massive loss of memory CD4+ T cells in samples collected by BAL (Fig. 7c) but had little change in the number of circulating naive CD4+ T lymphocytes (data not shown). However, by week 19 p.i., the levels of total CD4+ T cells in the blood had declined significantly in all three RPs (Fig. 7b), raising again the possibility that coreceptor usage might have changed. To assess a possible coreceptor switch, virus was collected from RP monkeys DB99 and A4E008 at the time of euthanasia and evaluated in the TZM-bl assay with specific CXCR4 and CCR5 inhibitors. As shown in Fig. 9a, blocking the entry of SHIVAD8-DB99 required both inhibitors, whereas SHIVAD8-A4E008 was inhibited only by the CCR5 inhibitor. This result indicates that SHIVAD8-DB99 had acquired the capacity to use CXCR4 during its infection of macaque DB99 and that SHIVAD8-A4E008 had remained R5 tropic.
FIG. 9.
Coreceptor utilization of SHIVAD8 derivatives isolated from rapid progressors. (a) TZM-bl cells were infected in quadruplicate with viruses (SHIVAD8-DB99 and SHIVAD8-A4E008) recovered from rapid progressors DB99 and A4E008, respectively, in the presence of the indicated amounts of the small-molecule coreceptor inhibitors AD101 (CCR5), AMD 3100 (CXCR4), or both. SIVmac239 and SHIVDH12RCL-7 were also analyzed as representative R5-tropic and dual-tropic viruses, respectively. The luciferase activities present in cell lysates 24 h p.i. were measured, and percent infectivities were determined in the absence or presence of coreceptor inhibitors. (b) gp120 sequences from the N-terminal V3 regions of SHIVAD8 variants, recovered from three RP animals, were aligned with the starting SHIVAD8 V3 loop. The V3 regions of the R5-tropic SHIVSF162P3 and its SHIVSF162-BR24N derivative, which also emerged in an RP, are included in the alignment. (c) Coreceptor utilization of virus, pseudotyped with Envs present in RP DB99 at the time of necropsy, containing or lacking the 3-aa RIG V3 loop insertion.
Reverse transcription-PCR cloning and sequencing of env genes amplified from the plasma of macaque DB99 at the time of its euthanasia revealed that 28 of 29 recovered clones contained a 3-aa insertion (RIG) located 2 residues upstream of the GPGR sequence in the crown of the gp120 V3 region (Fig. 9b). A similar analysis of the env gene from virus circulating in monkey A4E008 revealed a different 3-aa insertion (HIG) at the same location in its V3 loop. The V3 loop sequences amplified from the plasma of both animals at week 2 p.i. did not contain any insertion. The gp120 region amplified from the third RP (macaque CL5A) at the time of euthanasia contained no insertion (Fig. 9b).
One of the 28 viral-DNA clones amplified from macaque DB99 plasma at the time of euthanasia containing the RIG insertion in V3 and the single clone simultaneously obtained from this animal lacking the V3 insertion were used to prepare pseudotyped virus for testing in the entry assay, as described in Materials and Methods. As shown in Fig. 9c, the V3 RIG insertion conferred usage of both CCR5 and CXCR4 coreceptors on SHIVpsAD8(RIG+) compared to the exclusive utilization of CCR5 by SHIVpsAD8(RIG-), which lacks the gp120 V3 insertion.
SHIVAD8-infected macaques developed immunodeficiency.
The clinical statuses and disease outcomes of all 13 animals inoculated with SHIVAD8#2 and its immediate derivatives during a 2- to 3-year observation period are presented in Table 2. As noted above, 10 of these 13 macaques were NPs and experienced gradual and irreversible depletions of both memory and naïve CD4+ T lymphocyte subsets (Fig. 6). Five of these animals were euthanized with symptoms of AIDS, and 3 additional NPs currently have CD4+ T cell counts ranging from 92 to 154 cells/μl plasma (Table 2). Histopathological studies performed on specimens collected at the time of necropsy revealed the presence of Pneumocystis carinii, Mycobacterium avium, and Campylobacter coli infections in individual macaques (see Fig. S3 in the supplemental material). In addition, 3 of the 13 R5-SHIV-infected monkeys experienced an RP syndrome characterized by sustained plasma viremia of >1 × 107 RNA copies/ml; rapid and irreversible loss of memory CD4+ T cells in the blood and at an effector site (BAL); and intractable diarrhea, anorexia, and weight loss requiring euthanasia between weeks 19 and 23 p.i.
TABLE 2.
Clinical and pathological findings in rhesus monkeys infected with SHIVAD8#2 and its immediate derivatives
| Animal | Clinical data/pathological findings |
|---|---|
| CJ8B | Euthanized (wk 199); uncontrolled diarrhea; wt loss |
| CK15 | Euthanized (wk 112); P. carinii pneumonia |
| CJ58 | Total CD4+ T cells, 154/mm3 (wk 111) |
| CE8J | Euthanized (wk 117); uncontrolled diarrhea, C. coli enteritis |
| CJ35 | Total CD4+ T cells, 270/mm3 (wk 129) |
| CJ3V | Euthanized (wk 135); uncontrolled diarrhea; typhlocolitis |
| CK5G | Total CD4+ T cells: 101/mm3 (wk 101) |
| DB99 | Euthanized (wk 23); rapid progressor |
| DA1Z | Total CD4+ T cells, 545/mm3 (wk 65) |
| A4E008 | Euthanized (wk 20); rapid progressor |
| DA4W | Total CD4+ T cells, 92/mm3 (wk 64) |
| CL5A | Euthanized (wk 19); rapid progressor |
| CL98 | Euthanized (wk 100); disseminated M. avium |
DISCUSSION
The results presented clearly show that the generation of a pathogenic R5-SHIV was not a trivial undertaking. Animal-to-animal passaging eventually gave rise to SHIVAD8#2, possessing greatly augmented infectivity for rhesus PBMC compared to the starting SHIVAD8 construct. Although it was not appreciated at the time, SHIVAD8#2 had also acquired improved in vivo properties, as evidenced by its and its immediate derivatives' capacity to cause fatal immunodeficiency in 8 of 13 inoculated rhesus monkeys (Fig. 4 and Table 2). The most consistent and distinguishing property of the passaged SHIVAD8 family of viruses during infections of rhesus macaques was the slow and unremitting loss of both memory and naïve CD4+ T cells (Fig. 6), a pattern of depletion observed in all 10 NPs. Surprisingly, and in contrast to both SIVmac and SIVsmE lineages, the pace of CD4+ T lymphocyte decline was not correlated with plasma virus loads. Although the geometric mean plasma viral-RNA level at week 50 in the SHIVAD8-infected monkey cohort was 1.7 × 103 RNA copies/ml, the set-point virus loads varied widely in the 10 infected animals (1.6 × 102 to 1.5 × 105 RNA copies/ml). This variability was also observed in pairs of animals inoculated with identical SHIVAD8#2 derivatives (viz. CK15 and CJ58, and DB99 and DA1Z). An extreme example of the nonlinkage between viral-RNA levels and CD4+ T cell loss with SHIVAD8 occurred with animal CK5G, which had 43 and 42 circulating naïve and memory CD4+ T cells/μl, respectively, at week 86 p.i. and a plasma viral load of only 5.4 × 102 RNA copies/ml. During the chronic phase of SHIVAD8 infections, the loss of naïve CD4+ T cells was more rapid and more marked than the depletion of the memory subset, as was previously observed in SIVsmE543-infected animals (35) (Fig. 6). By week 80, for example, NPs had sustained an 87 to 93% loss of naïve CD4+ T cells from their preinoculation levels, whereas the depletion of memory cells was significant, but not as pronounced. The dissociation of plasma virus loads and CD4+ T cell loss is reminiscent of the previously reported infection of pig-tailed macaques with SIVl'hoest and SIVsun (4). In that study, 8 of 12 infected animals developed immunodeficiency over a 5-year period while maintaining set-point viremia between 102 and 103 RNA copies/ml.
We do not presently understand why naïve CD4+ T lymphocytes are lost in SHIVAD8 NPs. Based on coreceptor expression, this T cell subset expresses CXCR4, not CCR5, on its surface and should therefore be refractory to infection by R5-tropic SHIVs and virus-induced cell killing. An assessment of the coreceptor utilization status of late-stage viruses recovered from SHIVAD8 NPs, in fact, revealed that a coreceptor switch had not occurred in these animals (see Fig. S2 in the supplemental material). Although a dissociation between viral-RNA levels and memory/naïve CD4+ T cell loss was observed, the NPs did experience increased memory CD4+ T lymphocyte turnover (see Fig. S1 in the supplemental material), even in animals with very low plasma virus loads. Activation-induced proliferation and killing of memory CD4+ T cells during the lengthy chronic SHIVAD8 infection might therefore be responsible for driving the differentiation of naïve CD4+ lymphocytes into memory cells and impose an unsustainable drain on this CD4+ T cell subset. It is also possible that SHIVAD8 infection of rhesus macaques negatively affects naïve CD4+ T lymphocyte homeostasis in the thymus, thereby impeding the differentiation or emigration of this T cell subset. It has also recently been reported that the loss of naive CD4+ T cells during SIVsmE543 infections was associated with the presence of autoreactive antibodies to CD4+ T lymphocytes, platelets, double-stranded DNA, and phospholipid (27). Increased numbers of circulating IgG-coated CD4+ T cells were observed in that study, and the levels of autoreactive antibodies were correlated with the extent of naïve CD4+ T cell depletion.
Approximately 20% of rhesus monkeys infected with SIVmac/SIVsm lineage viruses become RPs, experiencing persistently high virus set points, rapid and complete losses of memory CD4+ T cells, undetectable or transient antiviral antibody responses, and early onset (3 to 6 months p.i.) of symptomatic disease (6). Despite losing virtually all of their memory CD4+ T lymphocytes, SIV RPs, at the time of death, usually maintain preinoculation levels of naïve CD4+ T cells (35). This was not the case for SHIVAD8 RPs. Although all three experienced early and massive depletions of memory CD4+ T cells, two of the infected macaques had lost virtually all of their naïve CD4+ T cells at the time of euthanasia. In one of these animals (DB99), the virus recovered at the time of euthanasia, as well as a virus pseudotyped with an Env possessing the RIG insertion in the V3 loop, had acquired the capacity to infect cells expressing CXCR4 (Fig. 9a and c). Interestingly, coreceptor switching has been previously reported to occur during RP infections of macaques inoculated with a different R5-tropic SHIV, SHIVSF162P3 (18, 19, 47). In one of the SHIVSF162P3 coreceptor-switching events, the insertion of two positively charged amino acids (HR) immediately upstream of the V3 loop GPGR crown (Fig. 9b) was shown to confer X4 tropism (18). In the case of SHIVAD8-DB99, a 3-aa (RIG) insertion, also located in the N-terminal V3 stem and which increased the net charge of the V3 loop from +3 to +5, was responsible for the acquisition of CXCR4 usage. The insertion of HIG at the same location of the SHIVAD8-A4E008 V3 region did not affect the net charge and did not confer tropism for CXCR4-expressing cells.
Independent and unrecognized cross-species transmissions and spread of SIVsm at different U.S. primate facilities during the 1970s contributed to the emergence of SIVmac and SIVsmE660 lineages with distinctive replicative and pathogenic phenotypes. The serial passaging of SHIVAD8 in rhesus monkeys described here also resulted in an AIDS-inducing primate lentivirus with its own characteristic properties. First, in contrast to commonly used pathogenic SIVs, SHIVAD8#2 and its immediate derivatives generated sustained but, as previously noted, highly variable set-point virus loads in NPs. Similarly variable viral loads were also observed in eight rhesus monkeys inoculated with four independent SHIVAD8 stocks prepared from macaques CK15, CE8J, CL98, and CJ58 at the time of their euthanasia (data not shown). Profound depletions of both memory and naïve CD4+ T cells, which accompany relatively low virus set points (geometric mean level, 1.7 × 103 RNA copies/ml) in NPs, is a second property that distinguishes the R5-tropic SHIVAD8 from pathogenic SIVs. Finally, unlike SIVs, SHIVAD8 RPs experience an initial loss of memory CD4+ T lymphocytes and a later rapid deletion of naïve CD4+ T cells prior to death, which in one animal occurred following a CCR5-to-CXCR4 coreceptor switch. Based on the results shown in Fig. 4 and Table 2, we plan to use and distribute SHIVAD8#2LN, SHIVAD8#2PBMC, or the SHIVs recovered from NPs at the time of euthanasia (SHIVAD8-CL98, SHIVAD8-CK15, or SHIVAD8-CE8J) as challenge viruses in vaccine experiments. Animals inoculated with cell-free preparations of the last group of viruses have experienced variable but sustained plasma viremia associated with a gradual but significant CD4+ T cell loss during 30 weeks of infection. Some of these macaques have developed a rapid-progressor clinical course.
Supplementary Material
Acknowledgments
We are indebted to Keith Reimann and the NIH Nonhuman Primate Reagent Resource for providing cM-T807; to the NIH AIDS Research and Reference Reagent Program for providing AMD3100; and to Julie Strizki, Schering-Plough, for providing AD101. We thank John Mascola for TZM-bl cells and instructions for performing virus neutralization assays, Robin Kruthers and Ranjini Iyengar for determining viral RNA levels, and Vanessa Hirsch for critical comments during the preparation of this paper. We appreciate the contributions of Boris Skopits in diligently assisting in the care and maintenance of our animals.
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
Published ahead of print on 10 February 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Amara, R. R., F. Villinger, J. D. Altman, S. L. Lydy, S. P. O'Neil, S. I. Staprans, D. C. Montefiori, Y. Xu, J. G. Herndon, L. S. Wyatt, M. A. Candido, N. L. Kozyr, P. L. Earl, J. M. Smith, H. L. Ma, B. D. Grimm, M. L. Hulsey, J. Miller, H. M. McClure, J. M. McNicholl, B. Moss, and H. L. Robinson. 2001. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science 292:69-74. [DOI] [PubMed] [Google Scholar]
- 2.Barnett, S. W., I. K. Srivastava, E. Kan, F. Zhou, A. Goodsell, A. D. Cristillo, M. G. Ferrai, D. E. Weiss, N. L. Letvin, D. Montefiori, R. Pal, and M. Vajdy. 2008. Protection of macaques against vaginal SHIV challenge by systemic or mucosal and systemic vaccinations with HIV-envelope. AIDS 22:339-348. [DOI] [PubMed] [Google Scholar]
- 3.Barouch, D. H., S. Santra, J. E. Schmitz, M. J. Kuroda, T. M. Fu, W. Wagner, M. Bilska, A. Craiu, X. X. Zheng, G. R. Krivulka, K. Beaudry, M. A. Lifton, C. E. Nickerson, W. L. Trigona, K. Punt, D. C. Freed, L. Guan, S. Dubey, D. Casimiro, A. Simon, M. E. Davies, M. Chastain, T. B. Strom, R. S. Gelman, D. C. Montefiori, M. G. Lewis, E. A. Emini, J. W. Shiver, and N. L. Letvin. 2000. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science 290:486-492. [DOI] [PubMed] [Google Scholar]
- 4.Beer, B. E., C. R. Brown, S. Whitted, S. Goldstein, R. Goeken, R. Plishka, A. Buckler-White, and V. M. Hirsch. 2005. Immunodeficiency in the absence of high viral load in pig-tailed macaques infected with simian immunodeficiency virus SIVsun or SIVlhoest. J. Virol. 79:14044-14056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brenchley, J. M., T. W. Schacker, L. E. Ruff, D. A. Price, J. H. Taylor, G. J. Beilman, P. L. Nguyen, A. Khoruts, M. Larson, A. T. Haase, and D. C. Douek. 2004. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J. Exp. Med. 200:749-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, C. R., M. Czapiga, J. Kabat, Q. Dang, I. Ourmanov, Y. Nishimura, M. A. Martin, and V. M. Hirsch. 2007. Unique pathology in simian immunodeficiency virus-infected rapid progressor macaques is consistent with a pathogenesis distinct from that of classical AIDS. J. Virol. 81:5594-5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, Z., X. Zhao, Y. Huang, A. Gettie, L. Ba, J. Blanchard, and D. D. Ho. 2002. CD4+ lymphocytopenia in acute infection of Asian macaques by a vaginally transmissible subtype-C, CCR5-tropic simian/human immunodeficiency virus (SHIV). J. Acquir. Immune Defic. Syndr. 30:133-145. [DOI] [PubMed] [Google Scholar]
- 8.Collman, R., N. F. Hassan, R. Walker, B. Godfrey, J. Cutilli, J. C. Hastings, H. Friedman, S. D. Douglas, and N. Nathanson. 1989. Infection of monocyte-derived macrophages with human immunodeficiency virus type 1 (HIV-1). Monocyte-tropic and lymphocyte-tropic strains of HIV-1 show distinctive patterns of replication in a panel of cell types. J. Exp. Med. 170:1149-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Committee on the Care and Use of Laboratory Animals. 1985. Guide for the care and use of laboratory animals. Department of Health and Human Services publication no. NIH 85-23. National Institutes of Health, Bethesda, MD.
- 10.Dudley, D. M., J. L. Wentzel, M. S. Lalonde, R. S. Veazey, and E. J. Arts. 2009. Selection of a simian-human immunodeficiency virus strain resistant to a vaginal microbicide in macaques. J. Virol. 83:5067-5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Endo, Y., T. Igarashi, Y. Nishimura, C. Buckler, A. Buckler-White, R. Plishka, D. S. Dimitrov, and M. A. Martin. 2000. Short- and long-term clinical outcomes in rhesus monkeys inoculated with a highly pathogenic chimeric simian/human immunodeficiency virus. J. Virol. 74:6935-6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fauci, A. S., M. I. Johnston, C. W. Dieffenbach, D. R. Burton, S. M. Hammer, J. A. Hoxie, M. Martin, J. Overbaugh, D. I. Watkins, A. Mahmoud, and W. C. Greene. 2008. HIV vaccine research: the way forward. Science 321:530-532. [DOI] [PubMed] [Google Scholar]
- 13.Feinberg, M. B., and J. P. Moore. 2002. AIDS vaccine models: challenging challenge viruses. Nat. Med. 8:207-210. [DOI] [PubMed] [Google Scholar]
- 14.Gendelman, H. E., J. M. Orenstein, M. A. Martin, C. Ferrua, R. Mitra, T. Phipps, L. A. Wahl, H. C. Lane, A. S. Fauci, and D. S. Burke, et al. 1988. Efficient isolation and propagation of human immunodeficiency virus on recombinant colony-stimulating factor 1-treated monocytes. J. Exp. Med. 167:1428-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harouse, J. M., A. Gettie, R. C. Tan, J. Blanchard, and C. Cheng-Mayer. 1999. Distinct pathogenic sequela in rhesus macaques infected with CCR5 or CXCR4 utilizing SHIVs. Science 284:816-819. [DOI] [PubMed] [Google Scholar]
- 16.Hessell, A. J., P. Poignard, M. Hunter, L. Hangartner, D. M. Tehrani, W. K. Bleeker, P. W. Parren, P. A. Marx, and D. R. Burton. 2009. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nat. Med. 15:951-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hessell, A. J., E. G. Rakasz, P. Poignard, L. Hangartner, G. Landucci, D. N. Forthal, W. C. Koff, D. I. Watkins, and D. R. Burton. 2009. Broadly neutralizing human anti-HIV antibody 2G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS Pathog. 5:e1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho, S. H., S. Tasca, L. Shek, A. Li, A. Gettie, J. Blanchard, D. Boden, and C. Cheng-Mayer. 2007. Coreceptor switch in R5-tropic simian/human immunodeficiency virus-infected macaques. J. Virol. 81:8621-8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho, S. H., N. Trunova, A. Gettie, J. Blanchard, and C. Cheng-Mayer. 2008. Different mutational pathways to CXCR4 coreceptor switch of CCR5-using simian-human immunodeficiency virus. J. Virol. 82:5653-5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu, M., J. M. Harouse, A. Gettie, C. Buckner, J. Blanchard, and C. Cheng-Mayer. 2003. Increased mucosal transmission but not enhanced pathogenicity of the CCR5-tropic, simian AIDS-inducing simian/human immunodeficiency virus SHIV(SF162P3) maps to envelope gp120. J. Virol. 77:989-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Humbert, M., R. A. Rasmussen, R. Song, H. Ong, P. Sharma, A. L. Chenine, V. G. Kramer, N. B. Siddappa, W. Xu, J. G. Else, F. J. Novembre, E. Strobert, S. P. O'Neil, and R. M. Ruprecht. 2008. SHIV-1157i and passaged progeny viruses encoding R5 HIV-1 clade C env cause AIDS in rhesus monkeys. Retrovirology 5:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Igarashi, T., O. K. Donau, H. Imamichi, M. J. Dumaurier, R. Sadjadpour, R. J. Plishka, A. Buckler-White, C. Buckler, A. F. Suffredini, H. C. Lane, J. P. Moore, and M. A. Martin. 2003. Macrophage-tropic simian/human immunodeficiency virus chimeras use CXCR4, not CCR5, for infections of rhesus macaque peripheral blood mononuclear cells and alveolar macrophages. J. Virol. 77:13042-13052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Igarashi, T., Y. Endo, G. Englund, R. Sadjadpour, T. Matano, C. Buckler, A. Buckler-White, R. Plishka, T. Theodore, R. Shibata, and M. Martin. 1999. Emergence of a highly pathogenic simian/human immunodeficiency virus in a rhesus macaque treated with anti-CD8 mAb during a primary infection with a nonpathogenic virus. Proc. Natl. Acad. Sci. U. S. A. 96:14049-14054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Igarashi, T., Y. Endo, Y. Nishimura, C. Buckler, R. Sadjadpour, O. K. Donau, M. J. Dumaurier, R. J. Plishka, A. Buckler-White, and M. A. Martin. 2003. Early control of highly pathogenic simian immunodeficiency virus/human immunodeficiency virus chimeric virus infections in rhesus monkeys usually results in long-lasting asymptomatic clinical outcomes. J. Virol. 77:10829-10840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imamichi, H., T. Igarashi, T. Imamichi, O. K. Donau, Y. Endo, Y. Nishimura, R. L. Willey, A. F. Suffredini, H. C. Lane, and M. A. Martin. 2002. Amino acid deletions are introduced into the V2 region of gp120 during independent pathogenic simian immunodeficiency virus/HIV chimeric virus (SHIV) infections of rhesus monkeys generating variants that are macrophage tropic. Proc. Natl. Acad. Sci. U. S. A. 99:13813-13818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joag, S. V., Z. Li, L. Foresman, E. B. Stephens, L. J. Zhao, I. Adany, D. M. Pinson, H. M. McClure, and O. Narayan. 1996. Chimeric simian/human immunodeficiency virus that causes progressive loss of CD4+ T cells and AIDS in pig-tailed macaques. J. Virol. 70:3189-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuwata, T., Y. Nishimura, S. Whitted, I. Ourmanov, C. R. Brown, Q. Dang, A. Buckler-White, R. Iyengar, J. M. Brenchley, and V. M. Hirsch. 2009. Association of progressive CD4(+) T cell decline in SIV infection with the induction of autoreactive antibodies. PLoS Pathog. 5:e1000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, J. T., M. Halloran, C. I. Lord, A. Watson, J. Ranchalis, M. Fung, N. L. Letvin, and J. G. Sodroski. 1995. Persistent infection of macaques with simian-human immunodeficiency viruses. J. Virol. 69:7061-7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, Q., L. Duan, J. D. Estes, Z. M. Ma, T. Rourke, Y. Wang, C. Reilly, J. Carlis, C. J. Miller, and A. T. Haase. 2005. Peak SIV replication in resting memory CD4+ T cells depletes gut lamina propria CD4+ T cells. Nature 434:1148-1152. [DOI] [PubMed] [Google Scholar]
- 30.Luciw, P. A., E. Pratt-Lowe, K. E. Shaw, J. A. Levy, and C. Cheng-Mayer. 1995. Persistent infection of rhesus macaques with T-cell-line-tropic and macrophage-tropic clones of simian/human immunodeficiency viruses (SHIV). Proc. Natl. Acad. Sci. U. S. A. 92:7490-7494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lusso, P., F. Cocchi, C. Balotta, P. D. Markham, A. Louie, P. Farci, R. Pal, R. C. Gallo, and M. S. Reitz, Jr. 1995. Growth of macrophage-tropic and primary human immunodeficiency virus type 1 (HIV-1) isolates in a unique CD4+ T-cell clone (PM1): failure to downregulate CD4 and to interfere with cell-line-tropic HIV-1. J. Virol. 69:3712-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mehandru, S., M. A. Poles, K. Tenner-Racz, A. Horowitz, A. Hurley, C. Hogan, D. Boden, P. Racz, and M. Markowitz. 2004. Primary HIV-1 infection is associated with preferential depletion of CD4+ T lymphocytes from effector sites in the gastrointestinal tract. J. Exp. Med. 200:761-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nehete, P. N., S. Chitta, M. M. Hossain, L. Hill, B. J. Bernacky, W. Baze, R. B. Arlinghaus, and K. J. Sastry. 2001. Protection against chronic infection and AIDS by an HIV envelope peptide-cocktail vaccine in a pathogenic SHIV-rhesus model. Vaccine 20:813-825. [DOI] [PubMed] [Google Scholar]
- 34.Nishimura, Y., C. R. Brown, J. J. Mattapallil, T. Igarashi, A. Buckler-White, B. A. Lafont, V. M. Hirsch, M. Roederer, and M. A. Martin. 2005. Resting naive CD4+ T cells are massively infected and eliminated by X4-tropic simian-human immunodeficiency viruses in macaques. Proc. Natl. Acad. Sci. U. S. A. 102:8000-8005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishimura, Y., T. Igarashi, A. Buckler-White, C. Buckler, H. Imamichi, R. M. Goeken, W. R. Lee, B. A. Lafont, R. Byrum, H. C. Lane, V. M. Hirsch, and M. A. Martin. 2007. Loss of naive cells accompanies memory CD4+ T-cell depletion during long-term progression to AIDS in simian immunodeficiency virus-infected macaques. J. Virol. 81:893-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishimura, Y., T. Igarashi, O. K. Donau, A. Buckler-White, C. Buckler, B. A. Lafont, R. M. Goeken, S. Goldstein, V. M. Hirsch, and M. A. Martin. 2004. Highly pathogenic SHIVs and SIVs target different CD4+ T cell subsets in rhesus monkeys, explaining their divergent clinical courses. Proc. Natl. Acad. Sci. U. S. A. 101:12324-12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Doherty, U., W. J. Swiggard, and M. H. Malim. 2000. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 74:10074-10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pal, R., B. Taylor, J. S. Foulke, R. Woodward, M. Merges, R. Praschunus, A. Gibson, and M. Reitz. 2003. Characterization of a simian human immunodeficiency virus encoding the envelope gene from the CCR5-tropic HIV-1 Ba-L. J. Acquir. Immune. Defic. Syndr. 33:300-307. [DOI] [PubMed] [Google Scholar]
- 39.Pitcher, C. J., S. I. Hagen, J. M. Walker, R. Lum, B. L. Mitchell, V. C. Maino, M. K. Axthelm, and L. J. Picker. 2002. Development and homeostasis of T cell memory in rhesus macaque. J. Immunol. 168:29-43. [DOI] [PubMed] [Google Scholar]
- 40.Pitcher, C. J., C. Quittner, D. M. Peterson, M. Connors, R. A. Koup, V. C. Maino, and L. J. Picker. 1999. HIV-1-specific CD4+ T cells are detectable in most individuals with active HIV-1 infection, but decline with prolonged viral suppression. Nat. Med. 5:518-525. [DOI] [PubMed] [Google Scholar]
- 41.Reimann, K. A., J. T. Li, R. Veazey, M. Halloran, I. W. Park, G. B. Karlsson, J. Sodroski, and N. L. Letvin. 1996. A chimeric simian/human immunodeficiency virus expressing a primary patient human immunodeficiency virus type 1 isolate env causes an AIDS-like disease after in vivo passage in rhesus monkeys. J. Virol. 70:6922-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rose, N. F., P. A. Marx, A. Luckay, D. F. Nixon, W. J. Moretto, S. M. Donahoe, D. Montefiori, A. Roberts, L. Buonocore, and J. K. Rose. 2001. An effective AIDS vaccine based on live attenuated vesicular stomatitis virus recombinants. Cell 106:539-549. [DOI] [PubMed] [Google Scholar]
- 43.Schubert, U., and K. Strebel. 1994. Differential activities of the human immunodeficiency virus type 1-encoded Vpu protein are regulated by phosphorylation and occur in different cellular compartments. J. Virol. 68:2260-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shibata, R., M. Kawamura, H. Sakai, M. Hayami, A. Ishimoto, and A. Adachi. 1991. Generation of a chimeric human and simian immunodeficiency virus infectious to monkey peripheral blood mononuclear cells. J. Virol. 65:3514-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shibata, R., F. Maldarelli, C. Siemon, T. Matano, M. Parta, G. Miller, T. Fredrickson, and M. A. Martin. 1997. Infection and pathogenicity of chimeric simian-human immunodeficiency viruses in macaques: determinants of high virus loads and CD4 cell killing. J. Infect. Dis. 176:362-373. [DOI] [PubMed] [Google Scholar]
- 46.Shiver, J. W., T. M. Fu, L. Chen, D. R. Casimiro, M. E. Davies, R. K. Evans, Z. Q. Zhang, A. J. Simon, W. L. Trigona, S. A. Dubey, L. Huang, V. A. Harris, R. S. Long, X. Liang, L. Handt, W. A. Schleif, L. Zhu, D. C. Freed, N. V. Persaud, L. Guan, K. S. Punt, A. Tang, M. Chen, K. A. Wilson, K. B. Collins, G. J. Heidecker, V. R. Fernandez, H. C. Perry, J. G. Joyce, K. M. Grimm, J. C. Cook, P. M. Keller, D. S. Kresock, H. Mach, R. D. Troutman, L. A. Isopi, D. M. Williams, Z. Xu, K. E. Bohannon, D. B. Volkin, D. C. Montefiori, A. Miura, G. R. Krivulka, M. A. Lifton, M. J. Kuroda, J. E. Schmitz, N. L. Letvin, M. J. Caulfield, A. J. Bett, R. Youil, D. C. Kaslow, and E. A. Emini. 2002. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature 415:331-335. [DOI] [PubMed] [Google Scholar]
- 47.Tasca, S., S. H. Ho, and C. Cheng-Mayer. 2008. R5X4 viruses are evolutionary, functional, and antigenic intermediates in the pathway of a simian-human immunodeficiency virus coreceptor switch. J. Virol. 82:7089-7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Theodore, T. S., G. Englund, A. Buckler-White, C. E. Buckler, M. A. Martin, and K. W. Peden. 1996. Construction and characterization of a stable full-length macrophage-tropic HIV type 1 molecular clone that directs the production of high titers of progeny virions. AIDS Res. Hum. Retroviruses 12:191-194. [DOI] [PubMed] [Google Scholar]
- 49.Veazey, R. S., M. DeMaria, L. V. Chalifoux, D. E. Shvetz, D. R. Pauley, H. L. Knight, M. Rosenzweig, R. P. Johnson, R. C. Desrosiers, and A. A. Lackner. 1998. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science 280:427-431. [DOI] [PubMed] [Google Scholar]
- 50.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 51.Willey, R., M. C. Nason, Y. Nishimura, D. A. Follmann, and M. A. Martin. 2010. Neutralizing antibody titers conferring protection to macaques from a simian/human immunodeficiency virus challenge using the TZB-bl assay. AIDS Res. Hum. Retroviruses 26:89-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Willey, R. L., T. Klimkait, D. M. Frucht, J. S. Bonifacino, and M. A. Martin. 1991. Mutations within the human immunodeficiency virus type 1 gp160 envelope glycoprotein alter its intracellular transport and processing. Virology 184:319-329. [DOI] [PubMed] [Google Scholar]
- 53.Wu, L., G. LaRosa, N. Kassam, C. J. Gordon, H. Heath, N. Ruffing, H. Chen, J. Humblias, M. Samson, M. Parmentier, J. P. Moore, and C. R. Mackay. 1997. Interaction of chemokine receptor CCR5 with its ligands: multiple domains for HIV-1 gp120 binding and a single domain for chemokine binding. J. Exp. Med. 186:1373-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.