Abstract
Clevudine (CLV) is a nucleoside analog with potent antiviral activity against chronic hepatitis B virus (HBV) infection. Viral resistance to CLV in patients receiving CLV therapy has not been reported. The aim of this study was to characterize CLV-resistant HBV in patients with viral breakthrough (BT) during long-term CLV therapy. The gene encoding HBV reverse transcriptase (RT) was analyzed from chronic hepatitis B patients with viral BT during CLV therapy. Sera collected from the patients at baseline and at the time of viral BT were studied. To characterize the mutations of HBV isolated from the patients, we subjected the HBV mutants to in vitro drug susceptibility assays. Several conserved mutations were identified in the RT domain during viral BT, with M204I being the most common. In vitro phenotypic analysis showed that the mutation M204I was predominantly associated with CLV resistance, whereas L229V was a compensatory mutation for the impaired replication of the M204I mutant. A quadruple mutant (L129M, V173L, M204I, and H337N) was identified that conferred greater replicative ability and strong resistance to both CLV and lamivudine. All of the CLV-resistant clones were lamivudine resistant. They were susceptible to adefovir, entecavir, and tenofovir, except for one mutant clone. In conclusion, the mutation M204I in HBV RT plays a major role in CLV resistance and leads to viral BT during long-term CLV treatment. Several conserved mutations may have a compensatory role in replication. Drug susceptibility assays reveal that adefovir and tenofovir are the most effective compounds against CLV-resistant mutants. These data may provide additional therapeutic options for CLV-resistant patients.
Chronic hepatitis B virus (HBV) infection is a major health problem worldwide and leads to chronic hepatitis, cirrhosis, and hepatocellular carcinoma (13). Antiviral treatment for chronic hepatitis B improves the outcome of the disease and prevents the development of hepatocellular carcinoma (14). Currently, several oral antiviral agents, including lamivudine (LMV), adefovir (ADV), and entecavir (ETV), have been approved for the treatment of chronic HBV infections (8). However, oral antiviral treatment does not provide a cure or durable remission and it has limited long-term efficacy due to the emergence of resistance (12). Long-term treatment with nucleos(t)ide analogs is associated with an increased risk of drug resistance. Antiviral drug resistance in patients infected with HBV is associated with subsequent virologic breakthrough (BT), viral rebound, and biochemical BT.
Clevudine [1-(2-deoxy-2-fluoro-β-arabinofuranosyl)thymine, L-FMAU] (CLV) is a pyrimidine analog with potent antiviral activity against HBV (4). CLV inhibits the DNA-dependent DNA activity of HBV polymerase, as well as reverse transcription and priming (1, 16). Phase III clinical trial results have shown that CLV therapy for 24 weeks has a potent and sustained antiviral effect in both HBeAg-positive and -negative chronic hepatitis B patients (23, 24). Clinical evidence of viral resistance has not been shown in phase III trials following 24 weeks of CLV treatment (23, 24).
CLV was approved for the treatment of chronic hepatitis B patients in South Korea in 2006. Studies evaluating the long-term efficacy of CLV are now ongoing. However, the incidence of and genotypic mutations that cause CLV resistance in chronic hepatitis B patients have not been reported. Although there has been no report of CLV resistance in chronic hepatitis B patients, a methionine-to-isoleucine (YIDD) mutation, but not a methionine-to-valine (YVDD) mutation, in the YMDD motif of woodchuck HBV reverse transcriptase (RT) was identified during week 32 of CLV therapy for woodchuck HBV infection (22). In vitro studies using site-directed mutagenesis have also demonstrated that CLV is active against HBV harboring a single YVDD mutation but that it is not effective against YIDD mutants (3, 18). However, viral resistance associated with a specific HBV mutation during CLV therapy has not been reported.
Here we report the sequence of the complete HBV RT gene and the drug susceptibility of mutants isolated from patients with CLV resistance during long-term CLV treatment.
MATERIALS AND METHODS
Patients.
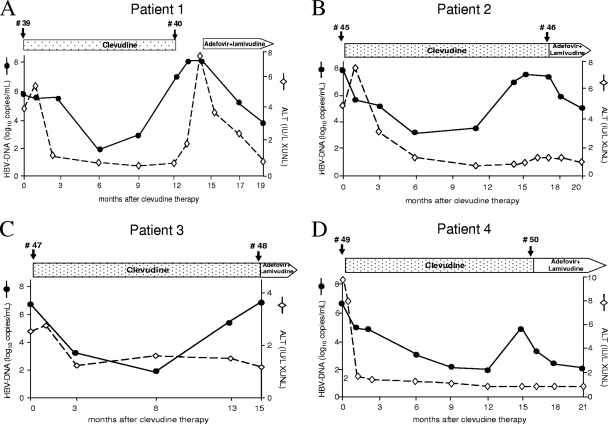
Patient 1 was a 48-year-old female with a 1-year history of hepatitis B e antigen (HBeAg)-positive chronic hepatitis B and no history of antiviral treatment. The patient was given 30 mg CLV/day due to an increased alanine aminotransferase (ALT) level and viremia. CLV therapy reduced the patient's serum HBV DNA level below the detection limit (300 copies/ml) during the first 6 months. However, viral BT occurred 9 months after the start of therapy and the patient's HBV DNA level increased to 1.1 × 107 copies/ml after 12 months. The patient's HBV DNA level decreased after a switch to combination therapy with ADV and LMV (Fig. 1A). Patient 2 was a 39-year-old man with an HBV DNA level of 5.2 × 108 copies/ml; the patient's HBV DNA level dropped to 1.3 × 103 copies/ml after 6 months of CLV therapy. The level of viremia increased to 2.6 × 108 after 14 months of CLV therapy; thus, the patient was switched to ADV and LMV (Fig. 1B). Patients 3 (a 29-year-old male) and 4 (a 52-year-old female) showed a viral response following CLV therapy; however, viral BT developed after 13 and 15 months of treatment, respectively (Fig. 1C and D). All of the patients were treatment naïve and had been compliant with their medications. They were all HBeAg positive, and there was no HBeAg loss or seroconversion during the treatment period. The patients' HBV DNA levels were determined by PCR using the Roche COBAS Amplicor system (detection limit, 300 copies/ml). Viral BT was defined as an increase in the HBV DNA level to greater than 1 log10 copy/ml from the lowest point. Serum was collected from each patient at the start of CLV therapy and at the time of viral BT. All of the patients provided written informed consent for HBV sequence analysis.
FIG. 1.
Clinical courses of four chronic hepatitis B patients treated with CLV. Serum samples were analyzed for HBV DNA and ALT. The HBV DNA levels were measured by real-time PCR (Roche). Panels A to D represent patients 1 to 4, respectively. The time points of serum sampling are indicated by arrowheads. The numbers indicate the series of HBV clones isolated from the patients. The clinical course of patient 1 was used in our previous paper (10).
Construction of the HBV1.2mer and sequence analysis of the polymerase RT domains.
To analyze the complete sequence of HBV RT from four chronic hepatitis B patients with viral BT during CLV treatment, we isolated HBV DNA from the patients' sera prior to CLV therapy and at the time of viral BT. HBV DNA was extracted using a QIAamp MinElute virus spin kit (Qiagen) according to the manufacturer's protocol. To generate HBV1.2mers harboring patient-derived RT mutations, we amplified the RT region of the HBV genome using the following primers: forward, 5′-AAT CTT CTC GAG GAC TGG GGA CCC TGC ACC-3′ (the XhoI site is underlined); reverse, 5′-GAG CAG CCA TGG GAA GGA GGT GTA TTT CCG-3′ (the NcoI site is underlined). The PCR conditions were 95°C for 5 min, followed by 32 cycles of 95°C for 50 s, 62°C for 50 s, and 72°C for 1 min 20 s, with a final extension at 72°C for 10 min. To avoid mutations during amplification, we used the FastStart High Fidelity PCR system (Roche). The purified products were digested with XhoI and NcoI, after which the wild-type (WT) RT sequence of the HBV1.2mer (9) was swapped for the patient-derived sequence. Sequence analysis of the cloned RT mutants was done using cloning primers specific for each side. To characterize the role of each point mutation in HBV replication and resistance, we generated chimeric clones (e.g., CLV-1, -2, -3, and -4) by overlap PCR using clones 39-8 and 40-3 as templates. CLV-5, -6, and -7 were generated by site-directed mutagenesis using a WT RT clone. All clones were confirmed by sequencing and further purified using phenol-chloroform before transfection.
Antiviral drugs and treatment.
LMV and CLV were provided by GlaxoSmithKline (Brentford, United Kingdom) and Bukwang Pharmaceutical Co. (Seoul, South Korea), respectively. Tenofovir (TDF) and ADV were provided by Gilead Sciences (Foster City, CA). ETV monophosphate was purchased from Moravek (Brea, CA). All antiviral drugs were suspended in sterile water and filtered using a 0.20-μm filter. Huh7 human hepatoma cells were transfected with 2 μg of the WT and mutant HBV1.2mers using Lipofectamine 2000 (Invitrogen). At 6 h posttransfection, the cells were treated with antiviral drugs in fresh media; the culture media were supplemented with fresh drugs every day for 4 days. The final concentration of each drug was as follows unless otherwise mentioned: 20 μM CLV, 20 μM LMV, 1 μM ETV, 20 μM ADV, and 20 μM TDF. Transfected cells were harvested at 4 days posttransfection; HBV replication was analyzed using Southern blotting. A 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide assay showed no significant cell viability differences between drug treatments.
In vitro drug susceptibility assay.
A susceptibility assay was performed using Southern blotting as described previously (7). HBV replication was quantified using a phosphorimager and Multi-Gauge v3.2 software (Fujifilm, Tokyo, Japan). To normalize the transfection yield of the HBV1.2mers, we determined the levels of HBV core protein and β-actin using Western blot analysis (9). In vitro drug susceptibility data are representative of at least three independent experiments.
RESULTS
Mutation profiles of the HBV polymerase RT domains isolated from CLV-resistant patients.
HBV DNA was isolated from the patients' sera before and after CLV treatment to identify mutations in RT associated with viral BT (Fig. 1). After construction of the HBV1.2mers, 5 to 15 clones were obtained from each serum sample to analyze the quasispecies of the entire RT domain. The sequences were compared against the sequence from genotype C HBV (NCBI GenBank accession no. GQ872210), a WT HBV genome isolated from the serum of a 25-year-old HBeAg-positive asymptomatic HBV carrier enrolled in our hospital.
Four conserved mutations were identified in patient 1 (M204I, L229V, N238H, and K333N) after viral BT. It is interesting that the mutations M204I, L229V, and K333N were not detected in all of the clones isolated from the patient's serum prior to CLV treatment, indicating that they may have been associated with CLV resistance (Table 1). In patient 2, 8 of 10 clones isolated before CLV treatment had the same sequence in the RT domain, indicating that a dominant HBV clone existed in the serum of patient 2 prior to CLV treatment. However, after viral BT, HBV triple mutants harboring M204I, V207I, and L269I appeared, suggesting an association between these mutations and CLV resistance (Table 2). In patient 3, only one mutation, M204I, was conserved after viral BT (Table 3). In patient 4, HBV quadruple mutants harboring L129M, V173L, M204I, and H337N appeared (80%, four out of five clones) after viral BT (Table 4). The mutation L80I, which is associated with LMV resistance (21), was also accompanied by the mutation M204I in one clone. For a summary of the conserved HBV RT and corresponding surface mutations from representative clones in CLV-resistant patients, see Fig. 4A.
TABLE 1.
HBV RT mutations in CLV-resistant patient 1
| CLV treatment point and strain(s) | Amino acid at RT position: |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 27 | 35 | 45 | 70 | 123 | 189 | 191 | 204 | 212 | 229 | 235 | 238 | 274 | 302 | 313 | 319 | 333 | 336 | 344 | |
| Baselinea | |||||||||||||||||||
| WTc (GQ872210) | V | H | D | T | N | S | V | M | K | L | L | N | R | Q | A | Q | K | L | Q |
| 39-3 | I | R | P | ||||||||||||||||
| 39-4 | D | I | R | ||||||||||||||||
| 39-5 | I | R | |||||||||||||||||
| 39-6 | R | ||||||||||||||||||
| 39-7 | R | H | |||||||||||||||||
| 39-8 | |||||||||||||||||||
| 39-9 | R | H | |||||||||||||||||
| Viral BTb | |||||||||||||||||||
| 40-2 | I | V | H | W | N | ||||||||||||||
| 40-3, 40-4 | I | V | H | N | |||||||||||||||
| 40-5 | F | R | I | V | H | N | |||||||||||||
| 40-6 | I | V | N | ||||||||||||||||
| 40-7 | G | I | V | H | R | N | |||||||||||||
| 40-8 | I | V | H | N | R | ||||||||||||||
| 40-9 | P | I | I | V | R | N | |||||||||||||
| 40-10 | S | I | V | T | N | ||||||||||||||
HBV1.2mers harboring RT domains isolated from the patient's serum prior to CLV treatment. The mutations T38A, Y124H, M129L, Q267L, L269I, Q271E, V291A, Y305F, and S317A were present even before therapy.
HBV1.2mers harboring RT domains isolated from the patient's serum at the time of viral BT during CLV treatment.
Conserved mutations that appeared after viral BT are in boldface.
TABLE 2.
HBV RT mutations in CLV-resistant patient 2
| CLV treatment point and strain(s) | Amino acid at RTc position: |
|||||||
|---|---|---|---|---|---|---|---|---|
| 35 | 122 | 148 | 204 | 207 | 229 | 238 | 269 | |
| Baselinea | ||||||||
| WT (GQ872210) | H | I | Y | M | V | L | N | L |
| 45-1, 45-2, 45-3, 45-5, 45-6, 45-7, 45-8, 45-10 | ||||||||
| 45-4, 45-9 | N | |||||||
| Viral BTb | ||||||||
| 46-1 | H | I | I | K | I | |||
| 46-2, 46-4 | I | I | I | |||||
| 46-3, 46-8, 46-10 | I | I | K | I | ||||
| 46-5 | T | I | I | I | ||||
| 46-6 | R | I | I | I | ||||
| 46-7 | T | I | I | I | ||||
| 46-9 | I | I | V | I | ||||
HBV1.2mers harboring RT domains isolated from the patient's serum prior to CLV treatment. The mutations S106C, H126Y, and D134E were present even before therapy.
HBV1.2mers harboring RT domains isolated from the patient's serum at the time of viral BT during CLV treatment.
Conserved mutations that appeared after viral BT are in boldface.
TABLE 3.
HBV RT mutations in CLV-resistant patient 3
| CLV treatment point and strain(s) | Amino acid at RTc position: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 80 | 129 | 131 | 135 | 138 | 142 | 191 | 204 | 207 | 238 | 267 | |
| Baselinea | |||||||||||
| WT (GQ872210) | L | M | D | S | R | V | V | M | V | N | Q |
| 47-1 | G | M | H | L | |||||||
| 47-2 | I | H | |||||||||
| 47-3 | V | I | D | L | |||||||
| 47-4 | I | I | H | L | |||||||
| 47-5 | F | G | I | L | |||||||
| Viral BTb | |||||||||||
| 48-1, 48-2 | I | I | I | H | |||||||
| 48-3, 48-5 | I | I | H | L | |||||||
| 48-4 | K | I | I | H | L | ||||||
HBV1.2mers harboring RT domains isolated from the patient's serum prior to CLV treatment. The mutation L269I was present even before therapy.
HBV1.2mers harboring RT domains isolated from the patient's serum at the time of viral BT during CLV treatment.
Conserved mutations that appeared after viral BT are in boldface.
TABLE 4.
HBV RT mutations in CLV-resistant patient 4
| CLV treatment point and strain(s) | Amino acid at RTc position: |
||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 40 | 55 | 80 | 91 | 124 | 126 | 128 | 129 | 164 | 173 | 186 | 204 | 213 | 221 | 223 | 225 | 232 | 267 | 269 | 285 | 295 | 317 | 319 | 329 | 337 | |
| Baselinea | |||||||||||||||||||||||||
| WT | S | H | L | I | Y | H | T | M | L | V | A | M | S | F | S | T | G | Q | L | K | G | A | Q | A | H |
| 49-1 | A | M | Y | M | A | ||||||||||||||||||||
| 49-2 | H | A | M | Y | A | M | I | ||||||||||||||||||
| 49-3, 49-7, 49-12, 49-15 | A | M | Y | M | I | ||||||||||||||||||||
| 49-4 | R | A | M | T | Y | M | I | ||||||||||||||||||
| 49-5 | A | M | T | Y | M | I | R | ||||||||||||||||||
| 49-6 | A | M | T | Y | M | I | E | D | |||||||||||||||||
| 49-8 | R | A | M | V | T | Y | M | I | |||||||||||||||||
| 49-9 | V | A | M | Y | M | I | |||||||||||||||||||
| 49-10 | P | A | M | Y | M | I | |||||||||||||||||||
| 49-11, 49-13 | A | M | T | Y | M | I | |||||||||||||||||||
| 49-14 | A | M | Y | A | M | I | S | ||||||||||||||||||
| Viral BTb | |||||||||||||||||||||||||
| 50-1 | I | A | M | I | Y | M | I | ||||||||||||||||||
| 50-2,50-3,50-5 | L | L | I | I | N | ||||||||||||||||||||
| 50-4 | L | L | I | S | I | N | |||||||||||||||||||
HBV1.2mers harboring RT domains isolated from the patient's serum prior to CLV treatment.
HBV1.2mers harboring RT domains isolated from the patient's serum at the time of viral BT during CLV treatment.
Conserved mutations that appeared after viral BT are in boldface.
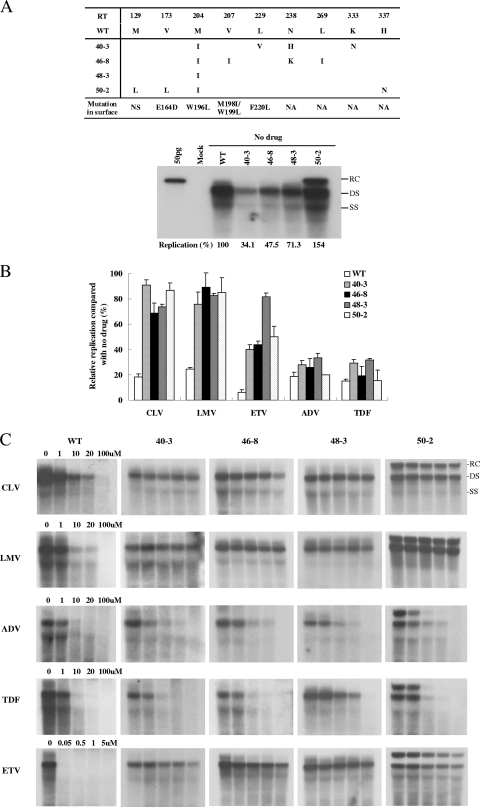
FIG. 4.
Susceptibility of CLV-resistant RT mutants to other antiviral agents. (A, top) Conserved HBV RT and corresponding surface mutations from representative clones in CLV-resistant patients. NS, no substitution; NA, not applicable. (Bottom) Comparison of the replicative abilities of representative CLV-resistant RT mutants without drug treatment. The percent replication shown in the lower part of the panel represents the relative replicative ability compared to that of the WT without drug treatment. Values represent the mean of at least three independent experiments. (B) Relative replicative abilities of the mutants following alternative antiviral treatment were obtained after normalization by the replicative ability without drug exposure. Transfected cells were treated with a single drug concentration (CLV, 20 μM; LMV, 20 μM; ETV, 1 μM; ADV, 20 μM; TDF, 20 μM). Replication was quantified using a phosphorimager. Data are the mean values of at least three independent Southern blots. (C) IC50s of other antiviral agents for the WT and representative CLV-resistant clones. A susceptibility assay was performed after the transfection of 2 μg of the HBV1.2mers into Huh7 cells at the indicated drug concentrations. The replicative ability of each mutant was quantified using a phosphorimager; the amount of replication without drug treatment was set at 100%. The IC50s were obtained by at least three independent experiments and are summarized in Table 5.
Taken together, our sequence analysis results indicate the existence of quasispecies before and after CLV treatment. M204I, which confers LMV resistance, was detected in all of the serum samples from four patients after viral BT. However, other mutations related to LMV resistance, such as M204V and L180M, were not detected.
Susceptibility of the cloned RT mutants to CLV.
To determine whether the RT mutants isolated from the serum samples were resistant to CLV, we performed drug susceptibility assays using representative RT mutants obtained from the four patients. After transfection of the HBV1.2mers into Huh7 cells, the replicative capacity of each mutant virus was examined using Southern blotting. The levels of HBV core protein and β-actin were measured using Western blotting to show the transfection yield and amount loaded (Fig. 2).
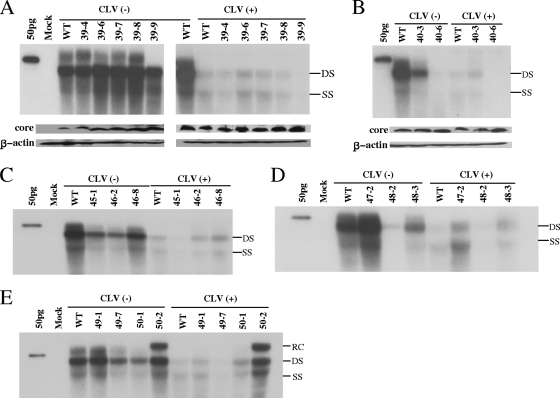
FIG. 2.
In vitro susceptibilities of representative RT mutants isolated from patients with viral BT. The 1.2mers of the HBV constructs (2 μg) were transfected into Huh7 cells grown in six-well plates and harvested 4 days later. The same batch of lysate was used for Southern and Western blotting. Western blotting was performed with polyclonal anti-core antibodies; after stripping, the membrane was reblotted with anti-β-actin antibodies. (A and B) Replicative abilities of RT mutants isolated from patient 1 at the time point indicated in Fig. 1. (C to E) Replicative abilities of the RT mutants cloned from patients 2 to 4, respectively. Each panel shows representative autoradiograms from at least three independent Southern blots with or without CLV treatment. The linearized whole genome of HBV was loaded in lane 1 as a marker. RC, relaxed circular; DS, double strand; SS, single strand.
The HBV clones obtained from patient 1 at baseline (series 39) showed a replicative capacity similar to that of the WT in the absence of antiviral drugs, suggesting that the quasispecies that appeared before CLV therapy (Table 1) did not affect viral replication. The replication of the HBV clones was greatly reduced with CLV treatment commensurate with the extent in the WT, indicating that the RT quasispecies at baseline were susceptible to CLV and did not harbor primary resistance mutations (Fig. 2A). However, 40-3, a representative clone isolated after viral BT bearing the mutations M204I, L229V, N238H, and K333N, did not show a significant decrease in viral replication following CLV treatment compared to the WT (Fig. 2B and 3A). In patient 2, 45-1, the dominant clone obtained at baseline, was susceptible to CLV, whereas 46-2 and 46-8, the major clones at the time of viral BT, were CLV resistant (Fig. 2C). It is interesting that the replicative ability of clone 46-2, which harbored the mutations M204I, V207I, and L269I, was much lower than that of the WT. However, clone 46-8, which carried an additional point mutation (N238K), exhibited significantly greater replicative ability. In patient 3, the level of replication of clone 47-2 in baseline serum was similar to that of the WT in the absence of antiviral drugs. However, clone 48-2, which was obtained after viral BT and which carried the additional mutations L80I and M204I, exhibited defective replication without antiviral drugs (Fig. 2D). Clones 48-2 and 48-3, which were obtained upon viral BT, showed clear CLV resistance as determined by the ratio of replication in the absence versus the presence of CLV. In patient 4, the dominant HBV mutants during viral BT (50-2), which harbored the mutations L129M, V173L, M204I, and H337N, showed strong CLV resistance, whereas the clones isolated at baseline were susceptible to CLV (Fig. 2E). It is interesting that clone 50-2 showed an enhanced replicative ability compared to the WT (154% of the WT level; Fig. 4A).
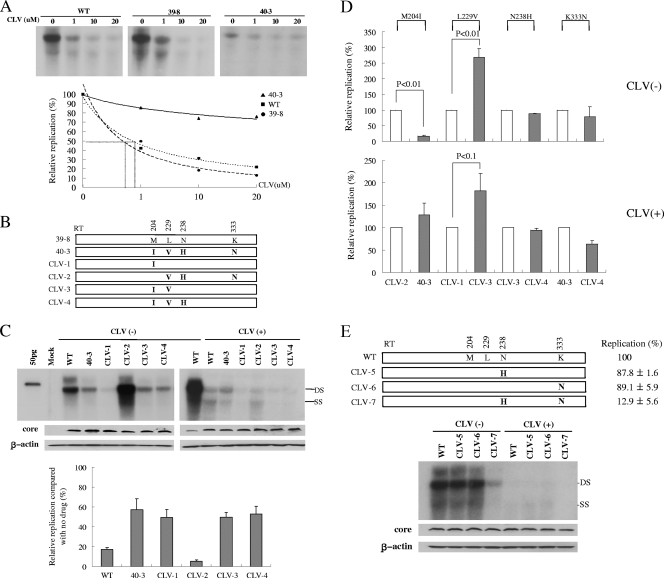
FIG. 3.
Characterization of RT mutations associated with viral BT during CLV treatment. (A) IC50s for the WT and mutant HBV clones. A CLV susceptibility assay was performed after the transfection of 2 μg of the HBV1.2mers into Huh7 cells following Southern blot analysis. The replicative ability of each mutant was quantified using a phosphorimager; the amount of replication without drug treatment was set at 100%. The IC50s were obtained by interpolation of the data. (B) Schematic representation of the mutant constructs. Clone 39-8 has WT residues at positions 204, 229, 238, and 333, as shown in Table 1. Each residue that differs from that in the WT sequence is shown. (C) Functional characterization of each conserved mutation in terms of replicative ability and susceptibility to CLV. Each value obtained by phosphorimager analysis was divided by the WT value [CLV(−)] on the same blot to normalize the intensity. The levels of replication without CLV treatment were set at 100% for each clone, and the relative amount of replication in the presence of CLV was calculated. Western blotting was performed with polyclonal anti-core antibodies; after stripping, the membrane was reblotted with anti-β-actin antibodies. (D) Relative replication of the mutant clones with or without CLV treatment. Empty bars represent the WT sequence at the indicated positions, whereas dotted bars indicate a point mutation, as shown in the upper part of the panel. Replication of the WT sequence at the indicated position (empty bars) was set at 100%, and the relative replication of the point mutants (dotted bars) was calculated. The graph includes data from at least three independent Southern blots with or without CLV treatment; each part of the panel shows data from a representative autoradiogram. (E) Schematic representation of the mutant constructs and their effects on genome replication and CLV susceptibility. Values represent the mean of at least three independent experiments. DS, double strand; SS, single strand.
Characterization of the conserved mutations that appeared after CLV treatment.
As shown in Fig. 2, the replicative ability of the drug-resistant RT mutants was generally much lower than that of the WT; therefore, it may not be accurate to judge the level of resistance by Southern blotting using a fixed dose of the test drugs. To clarify this issue, we determined the 50% inhibitory concentrations (IC50s) for representative clones 39-8 and 40-3 at baseline and after viral BT. After treatment with CLV at four different concentrations, the replicative abilities of the clones were quantified using Southern blotting and phosphorimager analysis. Figure 3A shows the IC50s for 40-3, the WT, and 39-8 (>20, 0.89, and 0.73 μM, respectively). These values are consistent with the value reported previously for WT HBV (0.83 μM) (3). These data indicate that clone 40-3, which was obtained after viral BT, was strongly resistant to CLV.
The clones obtained after viral BT (series 40) possessed several conserved mutations (M204I, L229V, N238H, and K333N). To characterize the role of each conserved mutation in HBV replication and CLV resistance, we constructed several mutant clones, as depicted in Fig. 3B, and tested the effect of each mutation on that clone's replicative ability and CLV susceptibility. As shown in Fig. 3C, all of the clones harboring the M204I mutation (40-3, CLV-1, CLV-3, and CLV-4) were resistant to CLV; the effect of M204I on replication and CLV resistance is shown clearly in Fig. 3D. CLV-2 exhibited a greatly enhanced replicative ability compared to the WT in the absence of CLV; however, it was susceptible to CLV due to the absence of the M204I mutation (Fig. 3C). It is interesting that the increase in replication was attributed to the point mutation L229V. L229V enhanced the replication of the mutant 2.7-fold in the absence of CLV and 1.8-fold during CLV treatment (CLV-1 versus CLV-3; Fig. 3D). The N238H (40-3 versus 40-6) and N238K (46-2 versus 40-8) mutations seem to be responsible for the enhanced replication observed (Fig. 2B and C). However, this result is inconsistent with that of Fig. 3C and D (CLV-3 versus CLV-4), which shows no effect on replication or CLV susceptibility. To settle this issue, we constructed three additional mutants using the WT backbone as depicted in Fig. 3E. Neither the N238H nor the K333N single mutation affected replication in the WT context. However, the N238H N333N double mutant showed significantly reduced replication (Fig. 3E). These results suggest that the effect of the N238 mutation on replication is context dependent. The consistent levels of core protein through the CLV mutant series indicate that the differences in genome replication between mutants are core protein independent. Taken together, our data show that M204I is a major mutation associated with CLV resistance and that, among the four conserved mutations, L229V acts as a compensatory mutation regardless of CLV treatment, whereas N238H and K333N have no effect on CLV susceptibility.
Susceptibility of the CLV-resistant HBV mutants to other antiviral agents.
To test the susceptibility of the CLV-resistant mutants to other antiviral agents, we performed antiviral drug susceptibility assays using LMV, ETV, ADV, and TDF with representative CLV-resistant clones. The mutation patterns and replication abilities of representative CLV-resistant clones are shown in Fig. 4A. To determine the profiles of resistance to other antiviral agents of CLV-resistant clones, we performed the drug susceptibility assay using a single concentration of each drug (CLV, 20 μM; LMV, 20 μM; ETV, 1 μM; ADV, 20 μM; TDF, 20 μM). TDF and ADV were effective against the replication of the CLV-resistant mutants, whereas these clones showed cross-resistance to LMV. However, all of the mutant clones were less susceptible to ETV (Fig. 4B). To demonstrate the clear pattern of cross-resistance of CLV-resistant clones to other HBV agents, we determined the IC50s for the WT and representative clones using increasing doses of CLV, LMV, ETV, ADV, and TDF. Representative autoradiograms of Southern blots and The IC50s are shown in Fig. 4C and Table 5, respectively. Our data indicate that the CLV-resistant clones were also resistant to LMV and ETV. All of the CLV-resistant clones were susceptible to ADV and TDF.
TABLE 5.
In vitro susceptibility of CLV-resistant clones to other antiviral agents
| Clone | Avg IC50 (fold resistance) ± SDa |
||||
|---|---|---|---|---|---|
| CLV | LMV | ADV | TDF | ETV | |
| WT | 0.9 (1) | 3.3 (1) | 6.3 (1) | 2.5 (1) | 0.01 (1) |
| 40-3 | >100 (>111) | >100 (>30) | 2.5 ± 0.6 (0.4 ± 0.1) | 5.2 ± 0.8 (2.1 ± 0.4) | 0.7 ± 0.04 (69 ± 4.2) |
| 46-8 | >100 (>111) | >100 (>30) | 7.8 ± 0.4 (1.25 ± 0.1) | 6.6 ± 0.5 (2.7 ± 0.2) | 0.7 ± 0.1 (69 ± 12) |
| 48-3 | >100 (>111) | >100 (>30) | 8.5 ± 0.8 (1.4 ± 0.1) | 4.7 ± 0.8 (1.9 ± 0.4) | >5 (>500) |
| 50-2 | >100 (>111) | >100 (>30) | 3.5 ± 0.8 (0.6 ± 0.1) | 5.9 ± 0.4 (2.3 ± 0.1) | 0.7 ± 0.1 (70 ± 12) |
SDs were calculated on the basis of at least three independent experiments. Fold resistance = (mutant IC50)/(WT IC50).
DISCUSSION
In this study, we identified a number of mutations in HBV RT conferring CLV resistance from patients with viral BT during long-term CLV therapy. M204I plays a major role in CLV resistance, whereas several additional mutations may have a compensatory function in HBV replication.
Previous phase III studies showed that CLV is safe, is tolerated well for 24 weeks, and is not associated with serious adverse events. Thus, CLV was recently approved as an anti-HBV agent in Korea. Clinical trials of CLV showed no emergence of drug resistance over a 24-week period (16, 24). However, the long-term use of nucleos(t)ide analogs leads to the emergence of antiviral-resistant HBV mutants. We recently reported one case of viral BT among 45 patients (2.2%) during the first year of CLV treatment (10). In this study, the four treatment-naïve patients treated with CLV showed a significant decrease in their respective HBV DNA levels. However, viral BT occurred after 9 to 15 months of CLV treatment. The CLV-resistant HBV mutants isolated from the patients were subjected to phenotypic assays for antiviral susceptibility. The complete HBV RT region was cloned and tested for in vitro susceptibility to antiviral agents, including CLV, LMV, ETV, ADV, and TDF. In vitro phenotyping of HBV for antiviral resistance is useful for the identification and confirmation of genotypic resistance mutations and cross-resistance to other antiviral agents.
Viral quasispecies of the gene encoding HBV polymerase were observed both at baseline and after viral BT. The changes identified in the RT sequence at baseline did not confer CLV resistance. The mutation M204I was identified in all of the HBV RT clones isolated from patient sera during viral BT after CLV therapy. In vitro susceptibility assays also demonstrated that M204I, which has been characterized as an HBV variation that confers resistance to LMV, telbivudine, and ETV, has a major role in CLV resistance. Among the CLV-resistant patients, no other YMDD mutation (e.g., M204V or L180M) was detected. These findings are similar to the pattern of genotypic resistance to telbivudine. Lai et al. (11) reported that telbivudine resistance was associated only with the mutation M204I, whereas the M204V and/or L180M mutations, which commonly develop after LMV treatment, were not observed.
Additional amino acid substitutions, including those at positions 129, 173, 207, 229, 238, 333, and 337, in the HBV RT region were detected in the sera of the four patients during viral BT. Additional studies are needed to characterize the role of each mutation, in isolation and in combination, in HBV replication and CLV resistance. Substitutions at amino acids 181, 191, and 245 in the HBV RT region were previously reported in patients treated with CLV; however, no viral BT occurred during the 24-week study period (15, 24). None of these mutations were detected in the HBV clones isolated from the patients in this study.
The compensatory mutations that restored the replicative ability of the M204V/I mutants are important with respect to the enhanced-replication phenotype. Compensatory mutations, including L180M, that enhance the viral replication of drug-resistant HBV have been reported (6, 18). It has also been reported that the mutation V173L exerts a slight compensatory effect on L180M M204V mutants (5). Our results demonstrate that additional substitutions (L229V [Fig. 3D] or L129M, V173L, and H337N [Fig. 2E]) work with M204I to compensate for the impaired viral replication of the M204I mutant. It is surprising that HBV clone 50-2, which harbored the mutations L129M, V173L, and H337N in addition to M204I, showed strong replicative ability (154% of the WT level) even though it contained mutation M204I, which attenuates HBV replication (17). More importantly, treatment with CLV or LMV did not inhibit the enhanced viral replication of clone 50-2, a quadruple mutant carrying L129M, V173L, M204I, and H337N (Fig. 4). These additional sequence changes might restore replication and potentiate drug resistance. In vitro studies on the contribution of each mutation in isolation and in combination to CLV resistance using a WT HBV backbone are under way.
CLV has potent and sustained antiviral effects; thus, it is suitable for use as a first-line agent against HBV (23, 24). However, our present data indicate that long-term CLV treatment causes the M204I mutation, which has cross-resistance to LMV, in HBV RT. A previous in vitro study showed that mutations that confer resistance to LMV (L180M and M204V) and to LMV and ADV (L180M, M204V, and N236T) confer cross-resistance to CLV (2). Therefore, CLV is inadequate for the secondary treatment of LMV resistance. There are no data on the treatment of CLV resistance. In this regard, controlling CLV-resistant mutants by other drugs or determining the cross-resistance of CLV is of great clinical concern. On the basis of our phenotypic analysis (Fig. 4), we propose that ADV, TDF, or a combined approach that includes ADV could be the best choice for patients with CLV resistance. The patients with CLV resistance in this study were treated with ADV and LMV in combination. Follow-up data after treatment with ADV and LMV were available in three cases (patients 1, 2, and 4), and they showed a decrease in serum HBV and ALT levels 3 months after treatment. These results suggest that combination therapy including ADV is effective in cases of CLV resistance. According to our in vitro study, while TDF may also be used as rescue therapy for CLV resistance, ETV is not sufficient for the control of CLV-resistant HBV. All of the CLV-resistant mutants displayed approximately 70-fold or higher resistance to ETV (Table 5). Recently, consistent with our data, studies involving the genotypic analysis of ETV-resistant HBV isolated from long-term ETV therapy have been reported (19, 20). They showed that most of the patients developed an M204I/V substitution with or without an L180M substitution with or without additional mutations such as L80I, V173L, T184L, S202G, and M250L. Therefore, long-term evaluation of such secondary treatment is needed to assess antiviral efficacy in patients with CLV resistance.
In conclusion, we characterized the first cases of CLV resistance in treatment-naïve patients with chronic hepatitis B. The mutation M204I is mainly associated with CLV resistance and leads to viral BT during long-term CLV therapy. Additional conserved mutations may play a compensatory role in HBV replication. Our in vitro drug susceptibility results suggest that ADV and TDF are useful for rescue therapy in cases of CLV-resistant HBV.
Acknowledgments
This work was supported by the Institute of Biomedical Science and Technology, Konkuk University, and the Second-Phase of BK (Brain Korea) 21 Project (to Y.K.P.).
No conflicts of interest exist.
Footnotes
Published ahead of print on 17 February 2010.
REFERENCES
- 1.Balakrishna Pai, S., S. H. Liu, Y. L. Zhu, C. K. Chu, and Y. C. Cheng. 1996. Inhibition of hepatitis B virus by a novel l-nucleoside, 2′-fluoro-5-methyl-beta-l-arabinofuranosyl uracil. Antimicrob. Agents Chemother. 40:380-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brunelle, M. N., A. C. Jacquard, C. Pichoud, D. Durantel, S. Carrouée-Durantel, J. P. Villeneuve, C. Trépo, and F. Zoulim. 2005. Susceptibility to antivirals of a human HBV strain with mutations conferring resistance to both lamivudine and adefovir. Hepatology 41:1391-1398. [DOI] [PubMed] [Google Scholar]
- 3.Chin, R., T. Shaw, J. Torresi, V. Sozzi, C. Trautwein, T. Bock, M. Manns, H. Isom, P. Furman, and S. Locarnini. 2001. In vitro susceptibilities of wild-type or drug-resistant hepatitis B virus to (−)-beta-d-2,6-diaminopurine dioxolane and 2′-fluoro-5-methyl-beta-l-arabinofuranosyluracil. Antimicrob. Agents Chemother. 45:2495-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chu, C. K., T. Ma, K. Shanmuganathan, C. Wang, Y. Xiang, S. B. Pai, G. Q. Yao, J. P. Sommadossi, and Y. C. Cheng. 1995. Use of 2′-fluoro-5-methyl-beta-l-arabinofuranosyluracil as a novel antiviral agent for hepatitis B virus and Epstein-Barr virus. Antimicrob. Agents Chemother. 39:979-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delaney, W. E., IV, H. Yang, C. E. Westland, K. Das, E. Arnold, C. S. Gibbs, M. D. Miller, and S. Xiong. 2003. The hepatitis B virus polymerase mutation rtV173L is selected during lamivudine therapy and enhances viral replication in vitro. J. Virol. 77:11833-11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu, L., and Y. C. Cheng. 1998. Role of additional mutations outside the YMDD motif of hepatitis B virus polymerase in L(−)SddC (3TC) resistance. Biochem. Pharmacol. 55:1567-1572. [DOI] [PubMed] [Google Scholar]
- 7.Guarnieri, M., K. H. Kim, G. Bang, J. Li, Y. Zhou, X. Tang, J. Wands, and S. Tong. 2006. Point mutations upstream of hepatitis B virus core gene affect DNA replication at the step of core protein expression. J. Virol. 80:587-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoofnagle, J. H., E. Doo, T. J. Liang, R. Fleischer, and A. S. Lok. 2007. Management of hepatitis B: summary of a clinical research workshop. Hepatology 45:1056-1075. [DOI] [PubMed] [Google Scholar]
- 9.Kim, K. H., and B. L. Seong. 2003. Pro-apoptotic function of HBV X protein is mediated by interaction with c-FLIP and enhancement of death-inducing signal. EMBO J. 22:2104-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ko, S. Y., S. Y. Kwon, W. H. Choe, B. K. Kim, K. H. Kim, and C. H. Lee. 2009. Clinical and virological responses to clevudine therapy in chronic hepatitis B patients: results at 1 year of an open-labelled prospective study. Antivir. Ther. 4:585-590. [PubMed] [Google Scholar]
- 11.Lai, C. L., E. Gane, Y. F. Liaw, C. W. Hsu, S. Thongsawat, Y. Wang, Y. Chen, E. J. Heathcote, J. Rasenack, N. Bzowej, N. V. Naoumov, A. M. Di Bisceglie, S. Zeuzem, Y. M. Moon, Z. Goodman, G. Chao, B. F. Constance, and N. A. Brown for the Globe Study Group. 2007. Telbivudine versus lamivudine in patients with chronic hepatitis B. N. Engl. J. Med. 357:2576-2588. [DOI] [PubMed] [Google Scholar]
- 12.Lai, C. L., J. Dienstag, E. Schiff, N. W. Leung, M. Atkins, C. Hunt, N. Brown, M. Woessner, R. Boehme, and L. Condreay. 2003. Prevalence and clinical correlates of YMDD variants during lamivudine therapy for patients with chronic hepatitis B. Clin. Infect. Dis. 36:687-696. [DOI] [PubMed] [Google Scholar]
- 13.Lee, W. M. 1997. Hepatitis B virus infection. N. Engl. J. Med. 337:1733-1745. [DOI] [PubMed] [Google Scholar]
- 14.Liaw, Y. F., J. J. Sung, W. C. Chow, G. Farrell, C. Z. Lee, H. Yuen, T. Tanwandee, Q. M. Tao, K. Shue, O. N. Keene, J. S. Dixon, D. F. Gray, and J. Sabbat. 2004. Lamivudine for patients with chronic hepatitis B and advanced liver disease. N. Engl. J. Med. 351:1521-1531. [DOI] [PubMed] [Google Scholar]
- 15.Lim, S. G., N. Leung, H. W. Hann, G. K. Lau, C. Trepo, H. Mommeja-Marin, C. Moxham, J. Sorbel, A. Snow, M. R. Blum, F. Rousseau, and P. Marcellin. 2008. Clinical trial: a phase II, randomized study evaluating the safety, pharmacokinetics and anti-viral activity of clevudine for 12 weeks in patients with chronic hepatitis B. 2008. Aliment. Pharmacol. Ther. 27:1282-1292. [DOI] [PubMed] [Google Scholar]
- 16.Liu, S. H., K. L. Grove, and Y. C. Cheng. 1998. Unique metabolism of a novel antiviral l-nucleoside analog, 2′-fluoro-5-methyl-beta-l-arabinofuranosyluracil: a substrate for both thymidine kinase and deoxycytidine kinase. Antimicrob. Agents Chemother. 42:833-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melegari, M., P. P. Scaglioni, and J. R. Wands. 1998. Hepatitis B virus mutants associated with 3TC and famciclovir administration are replication defective. Hepatology 27:628-633. [DOI] [PubMed] [Google Scholar]
- 18.Ono, S. K., N. Kato, Y. Shiratori, J. Kato, T. Goto, R. F. Schinazi, F. J. Carrilho, and M. Omata. 2001. The polymerase L528M mutation cooperates with nucleotide binding-site mutations, increasing hepatitis B virus replication and drug resistance. J. Clin. Invest. 107:449-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tenney, D. J., S. M. Levine, R. E. Rose, A. W. Walsh, S. P. Weinheimer, L. Discotto, M. Plym, K. Pokornowski, C. F. Yu, P. Angus, A. Ayres, A. Bartholomeusz, W. Sievert, G. Thompson, N. Warner, S. Locarnini, and R. J. Colonno. 2004. Clinical emergence of entecavir-resistant hepatitis B virus requires additional substitutions in virus already resistant to lamivudine. Antimicrob. Agents Chemother. 48:3498-3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tenney, D. J., R. E. Rose, C. J. Baldick, K. A. Pokornowski, B. J. Eggers, J. Fang, M. J. Wichroski, D. Xu, J. Yang, R. B. Wilber, and R. J. Colonno. 2009. Long-term monitoring shows hepatitis B virus resistance to entecavir in nucleoside-naïve patients is rare through 5 years of therapy. Hepatology 49:1503-1514. [DOI] [PubMed] [Google Scholar]
- 21.Warner, N., S. Locarnini, M. Kuiper, A. Bartholomeusz, A. Ayres, L. Yuen, and T. Shaw. 2007. The L80I substitution in the reverse transcriptase domain of the hepatitis B virus polymerase is associated with lamivudine resistance and enhanced viral replication in vitro. Antimicrob. Agents Chemother. 51:2285-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamamoto, T., S. Litwin, T. Zhou, Y. Zhu, L. Condreay, P. Furman, and W. S. Mason. 2002. Mutations of the woodchuck hepatitis virus polymerase gene that confer resistance to lamivudine and 2′-fluoro-5-methyl-beta-l-arabinofuranosyluracil. J. Virol. 76:1213-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoo, B. C., J. H. Kim, T. H. Kim, K. C. Koh, S. H. Um, Y. S. Kim, K. S. Lee, B. H. Han, C. Y. Chon, J. Y. Han, S. H. Ryu, H. C. Kim, K. S. Byun, S. G. Hwang, B. I. Kim, M. Cho, K. Yoo, H. J. Lee, J. S. Hwang, Y. S. Kim, Y. S. Lee, S. K. Choi, Y. J. Lee, J. M. Yang, J. W. Park, M. S. Lee, D. G. Kim, Y. H. Chung, S. H. Cho, J. Y. Choi, Y. O. Kweon, H. Y. Lee, S. H. Jeong, H. W. Yoo, and H. S. Lee. 2007. Clevudine is highly efficacious in hepatitis B e antigen-negative chronic hepatitis B with durable off-therapy viral suppression. Hepatology 46:1041-1048. [DOI] [PubMed] [Google Scholar]
- 24.Yoo, B. C., J. H. Kim, Y. H. Chung, K. S. Lee, S. W. Paik, S. H. Ryu, B. H. Han, J. Y. Han, K. S. Byun, M. Cho, H. J. Lee, T. H. Kim, S. H. Cho, J. W. Park, S. H. Um, S. G. Hwang, Y. S. Kim, Y. J. Lee, C. Y. Chon, B. I. Kim, Y. S. Lee, J. M. Yang, H. C. Kim, J. S. Hwang, S. K. Choi, Y. O. Kweon, S. H. Jeong, M. S. Lee, J. Y. Choi, D. G. Kim, Y. S. Kim, H. Y. Lee, K. Yoo, H. W. Yoo, and H. S. Lee. 2007. Twenty-four-week clevudine therapy showed potent and sustained antiviral activity in HBeAg-positive chronic hepatitis B. Hepatology 45:1172-1178. [DOI] [PubMed] [Google Scholar]