Abstract
Human papillomaviruses (HPV) link their life cycles to epithelial differentiation and induce productive replication of viral DNA in suprabasal cells. Viral-DNA amplification requires cells to remain active in the cell cycle upon differentiation. This is in contrast to normal cells, which lose proliferative capability upon differentiation. One factor that negatively regulates proliferative capability upon differentiation is microRNA 203 (miR-203), which is expressed primarily in suprabasal epithelial cells. Although HPVs do not encode their own microRNAs (miRNAs), they modulate expression of cellular miRNAs to regulate the activities of cellular proteins. We show that the HPV E7 protein downregulates miR-203 expression upon differentiation, which may occur through the mitogen-activated protein (MAP) kinase/protein kinase C (PKC) pathway. One target of miR-203 is the p63 family of transcription factors, and we demonstrate that HPV-positive cells maintain significantly higher levels of these factors upon differentiation than do normal keratinocytes. Several downstream targets of p63, CARM-1, p21, and Bax, were also increased in E7-expressing cells, and their levels were inversely correlated with amounts of miR-203. Introduction of expression vectors for miR-203 into keratinocytes that stably maintain HPV episomes resulted in short-term elevation of HPV genome copy numbers, but these were rapidly lost upon subsequent passage. When HPV-positive cells expressing high levels of miR-203 were induced to differentiate in methylcellulose, impaired genome amplification was observed. We conclude that high levels of miR-203 are inhibitory to HPV amplification and that HPV proteins act to suppress expression of this microRNA to allow productive replication in differentiating cells.
Human papillomaviruses (HPV) infect stratified epithelia and link their productive life cycles to the differentiation state of the host cell (13, 15, 23, 42, 59). Following infection of basal epithelial cells, HPV genomes are established as multicopy episomes that replicate in synchrony with chromosomal DNA (9, 31, 37, 59). In normal epithelia, as cells leave the basal layer, they exit the cell cycle, stratify, and undergo differentiation in suprabasal layers. When HPV-positive cells leave the basal layer, they also stratify and differentiate but remain active in the cell cycle (25, 26, 31). In highly differentiated suprabasal layers, HPV-positive cells reenter S phase and use cellular replication proteins to amplify their genomes. This is coincident with expression of late viral genes and the assembly of virions (40, 42). The HPV E6 and E7 oncoproteins are the primary factors responsible for blocking cell cycle exit during differentiation (4, 12), while the E1 and E2 proteins mediate origin recognition and recruitment of cellular replication factors (6, 52). The E5 and E1^E4 fusion proteins contribute to activation of late viral functions, but their mechanisms of action are not fully understood (8, 16, 22). Given the linkage between HPV amplification and differentiation, it appears that viral proteins target cellular factors to control the proliferative ability of cells during differentiation.
MicroRNAs (miRNAs) are small noncoding RNAs that play key roles in differentiation and development by posttranscriptionally downregulating the functions of cellular genes (3, 34). Over 700 cellular miRNAs have been identified, and they bind to sequences in the 3′ untranslated regions (UTRs) of transcribed messages to inhibit translation, as well as to induce the degradation of mRNAs. miRNAs are 22 to 23 nucleotides in length and are generated by sequential processing of long mRNAs by two RNase II enzymes, Drosha and Dicer (28). The inactivation of Dicer in mouse skin results in inhibition of normal epithelial proliferation and differentiation, indicating the importance of miRNAs to epithelial development (19). miRNA 203 (miR-203) is expressed specifically in suprabasal layers of stratified epithelia, as well as in psoriatic plaques, implicating it as a regulator of epithelial maturation (45, 46, 57). The primary role of miR-203 is to suppress the proliferative capacity of epithelial cells upon differentiation (57). In mouse keratinocytes, the p63 family of transcription factors was identified as a primary target of miR-203 action (29). The p63 family, which is related to the p53 tumor suppressor group of transactivators, consists of six members, but the primary member expressed in epithelia is ΔNp63. The ΔNp63 isoform is generated by alternative splicing of the p63 gene and lacks the N-terminal transactivation domain. Despite deletion of a portion of the N-terminal transactivation domain, ΔNp63 retains the ability to transactivate a set of target genes, including several p53-responsive genes (39, 56). ΔNp63 is expressed at high levels in proliferating undifferentiated basal keratinocytes, and its expression is downregulated in normal differentiated nonproliferating cells. This downregulation of ΔNp63 has been proposed to regulate the balance between epithelial proliferation and differentiation (50).
A number of DNA viruses express their own miRNAs in infected cells and use them to regulate viral and cellular activities. Adenoviruses, simian virus 40 (SV40), and herpesviruses (cytomegalovirus [CMV], Kaposi's sarcoma-associated herpesvirus [KSHV], murine gammaherpesvirus 68 [MHV68], and Epstein-Barr virus [EBV]) have all been shown to express their own miRNAs (48, 51). In contrast, papillomaviruses do not express miRNAs but do alter the expression of cellular miRNAs (2, 33, 41, 53, 54). Since the HPV life cycle is linked to cellular differentiation and requires retention of the proliferative capability of suprabasal epithelial cells, we investigated if HPV proteins modulate expression of human miR-203 and its downstream target, ΔNp63. We demonstrate that high-risk HPV proteins downregulate miR-203 expression upon differentiation and that this correlates with increased levels of ΔNp63. Moreover, we show that the HPV oncoprotein E7 is sufficient for blocking upregulation of miR-203 expression and that this likely occurs by modulation of the mitogen-activated protein kinase (MAPK) pathway signaling.
MATERIALS AND METHODS
Cell types.
Normal human foreskin keratinocytes (NHKs) were isolated from neonatal human foreskin epithelium as previously described (17) and were cultured in keratinocyte basal medium supplemented with bovine pituitary extract, insulin, hydrocortisone, and epidermal growth factor (Lonza). To create cell lines stably expressing HPV E6, E7, and E6/E7, NHKs were transduced with retroviral particles from PT67 packaging lines for 6 h in the presence of 8 μg/ml Polybrene (Sigma-Aldrich, St. Louis, MO). To generate cells containing HPV episomes, we transfected NHKs with HPV type 31 (HPV-31) genomes released from the pBRmin-HPV31 wild-type plasmid backbone using the restriction enzyme HindIII. To isolate HPV-positive cells, transfected cells were selected using antibiotics, as described previously (20). The isolated HPV-positive cells were then cultured with mitomycin C-treated NIH 3T3 J2 fibroblast feeders in E medium supplemented with mouse epidermal growth factor (5 ng/ml; Collaborative Biomedical Products, Bedford, MA) (20). The creation of retroviral constructs has been described previously (18, 36). In addition to stably transfected cell lines, we used CIN612 cells that contained HPV-31 episomes. CIN612 cells were grown in E medium as described above.
Reagents.
MiR-203 inhibitor (Applied Biosystems) was used to transfect NHKs and CIN612 cells using FuGene6 transfection reagent (Roche) following the manufacturer's protocols. Phorbol 12-myristate 13-acetate (PMA) (Sigma-Aldrich, St. Louis, MO) was used to treat the cells at 100 ng/ml for 3, 6, and 9 h.
Differentiation of keratinocytes in semisolid media.
Keratinocyte differentiation was induced through suspension in 1.5% methylcellulose (Sigma, St. Louis, MO) as previously described (55). The methylcellulose solution was prepared by combining dry autoclaved methylcellulose with E medium. Keratinocytes were harvested and then suspended in a 10-cm-diameter petri dish with 25 ml of methylcellulose for either 24 or 48 h at 37°C. Following incubation, the cells were washed four times with phosphate-buffered saline (PBS) at 4°C. Total DNA, RNA, and protein were then extracted from the cell pellets as previously described (55).
qRT-PCR.
Total RNA was isolated using RNA Stat-60 reagent (Tel-Lab) according to the manufacturer's specifications. To assay for miRNAs, poly(A) tails were added to short RNAs with the QuantiMir reverse transcription kit from System Biosciences, using the manufacturer's protocol. Real-time quantitative PCR (qRT-PCR) was performed using SYBER green (Clonetech) with primers specific to the microRNAs of interest. The 5′ primers were single-stranded DNA (ssDNA) sequences of mature miRNAs, and unique 3′ primers were specific to the poly(A) tails described above. Each qRT-PCR assay was conducted in triplicate using cDNA derived from 50 ng total RNA from HPV-positive or -negative cell lines. The expression of miRNAs in HPV-positive cells relative to that in HPV-negative cells was calculated using the threshold cycle (CT) method (30). Samples were normalized using U6 RNA as an endogenous control. The ratios of miRNA amounts were compared among samples.
Northern blot analysis.
Northern analysis of miR-203 was performed on total RNA isolated using RNA Stat-60 as previously described (38). Briefly, 15 μg of total RNA was separated on a small 17% SDS gel containing 7 M urea for 2 h at 180 V in 0.5× Tris-borate-EDTA (TBE) buffer. The gels were stained with ethidium bromide after separation to confirm equal loading. The RNAs were then transferred onto a Gene Screen nylon membrane (Bio-Rad) and UV cross-linked. The nylon membranes were then placed in a prehybridization/hybridization solution (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 20 mM Na2HPO4, 7% SDS, 2× Denhardt's solution) for 30 min at 50°C. The cDNA sequence of mature miR-203 (CTAGTGGTCCTAAACATTTCAC) was used as a probe and radioactively labeled with [32P]ATP using T4 PNK-kinase (Invitrogen). Purified labeled probes were denatured, added to prehybridized nylon membranes in the fresh hybridization solution described above, and incubated at 50°C overnight. After hybridization, the nylon membranes were washed three times with 2× SSC-0.2% SDS for 20 min at 50°C and rinsed once with 3× SSC. RNA was visualized by autoradiography after −80°C exposure for 72 h, and quantitative analysis was performed with Image J software (NIH).
Luciferase assay.
A wild-type 60-bp fragment from the p63 3′ UTR containing an miR-203 consensus binding site (5′-GAGUCCUUGUGAUUUCAAAG-3′) (57) was synthesized (IDT DNA Technologies) and cloned, using PCR-created HindIII and SpeI sites, into the pMIR-REPORT miRNA Expression Reporter Vector System from Applied Biosystems. A pMIR-REPORT miRNA expression reporter vector carrying a mutated miR-203 binding site in the p63 3′-UTR fragment (5′-GAGUCCUUGUGCUCUCGAAAG-3′) was created using a QuikChange Site-Directed Mutagenesis Kit from Stratagene (La Jolla, CA). NHKs were plated in six-well plates the day prior to transfection and cotransfected with 1 μg of wild-type, mutant, or empty pMIR-REPORT miRNA expression reporter vector with 200 ng of the Renilla control vector pRL-TK (Promega) with or without miR-203 pre-miR miRNA precursor molecule (Applied Biosystems) and using FuGene6 transfection reagent (Roche Diagnostics) according to the manufacturer's instructions. After 48 h of incubation, cells were harvested for luciferase assay using the Dual Luciferase kit (Promega) according to the manufacturer's protocol. Transfection efficiencies were normalized for Renilla luciferase activity, and luciferase signals were normalized to the respective controls without the miR-203 pre-miR miRNA precursor molecule (set to 100%).
Western blot analysis.
Total cell protein extracts were obtained using RIPA lysis buffer supplemented with a cocktail of protease inhibitors (Roche). Prior to the harvesting of cells for Western analyses, J2 fibroblast feeders were removed via treatment with 0.5 mM EDTA in PBS. The protein concentration was measured by a Bradford protein assay (Bio-Rad). Protein samples (25 mg) were separated by electrophoresis on 12% sodium dodecyl sulfate-polyacrylamide gels. The separated proteins were then transferred from the SDS gels to polyvinylidene difluoride membranes (Immobolin-P; Millipore), followed by membrane blocking in a 5% nonfat milk solution (0.1% Tween 20 in PBS). The membranes were then incubated with one of the following primary antibodies: anti-p53 (Abcam), anti-p63, anti-p63a, anti-p73, anti-CARM-1 (Santa Cruz), anti-p21 (BD Pharmingen), anti-Bax (Cell Signaling), anti-involucrin, and anti-GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (Abcam). Enhanced chemiluminescence was used to visualize the proteins (Amersham Pharmacia).
Southern blot analysis.
Prior to harvesting cells for DNA isolation, J2 fibroblasts were removed by EDTA treatment. The cells were lysed in lysis buffer (400 mM NaCl, 10 mM Tris-HCl, and 10 mM EDTA), and extracts were then incubated with 50 μg/ml RNase A and proteinase K sequentially to remove residual RNA and proteins, followed by phenol-chloroform extraction. Ten micrograms of total DNA was digested with the restriction enzyme HindIII, and samples were run in a 0.8% agarose gel at 70 V overnight. DNA was transferred to Gene Screen nylon membranes (Bio-Rad) using vacuum transfer according to the manufacturer's protocols. The probe consisted of the full HPV-31 genome isolated from the pBRmin-HPV31 wild-type plasmid by restriction digestion with HindIII. The probe was labeled with [32P]dCTP using Ready-To-Go DNA-labeling beads (Amersham Biosciences) and column purified. The nylon membranes were placed in prehybridization/hybridization solution (4× SSC, 5× Denhardt's solution, 10% dextran sulfate, 1% SDS, 50% formamide, and 0.1 mg/ml denatured salmon sperm DNA) for 1 h at 42°C and hybridized to the labeled probe in the same buffer at 42°C overnight. After hybridization, the membranes were washed twice for 15 min each time in 2× SSC-0.1% SDS, 0.5× SSC-0.1% SDS, and 0.1× SSC-1% SDS and once for 30 min at 50°C in 0.1× SSC-1% SDS. The bands were visualized by autoradiography, and quantitative analysis was performed with Image J software (NIH).
RESULTS
HPV proteins block high-level induction of miR-203 upon epithelial differentiation.
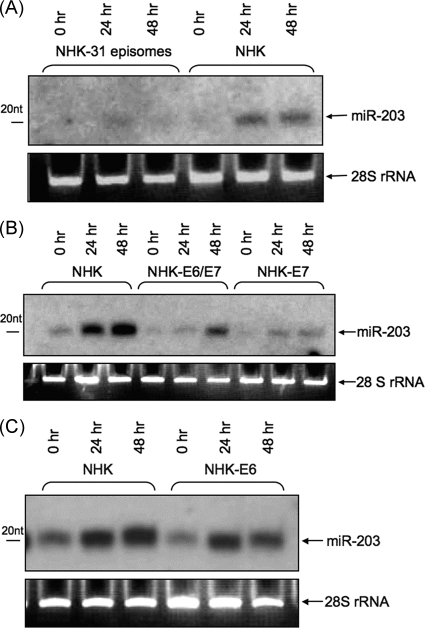
Studies in mouse, as well as human, cells have shown that miR-203 is expressed primarily in keratinocytes and that the levels of miR-203 increase significantly in suprabasal cells following differentiation (1, 29, 46, 57). Since HPVs link their life cycles to the differentiation state of the epithelial host, we investigated if HPV proteins altered miR-203 expression in either undifferentiated or differentiated keratinocytes. For these studies, we used primary NHKs derived from foreskin circumcisions and stable cell lines generated by transfection of keratinocytes from the same donor with recircularized cloned HPV-31 genomes (NHK-31). Both sets of cells were screened for miR-203 expression by Northern analysis using undifferentiated monolayer cultures, as well as following differentiation by suspension in methylcellulose. As shown in Fig. 1A, undifferentiated NHKs were found to express low levels of miR-203, and this increased 4- to 6-fold following differentiation in methylcellulose for 24 or 48 h. The level of miR-203 in undifferentiated NHK-31 cells was similar to that of NHKs. In contrast, upon differentiation, the level of miR-203 was reduced approximately 5-fold in HPV-positive cells from that seen in similarly differentiated NHKs. Identical results were seen in multiple independently derived NHK-31 cell lines and controls. We conclude that HPV proteins downregulate the levels of miR-203 expression upon differentiation.
FIG. 1.
miR-203 levels increase upon differentiation of normal keratinocytes but are downregulated in HPV-positive cells. Northern blot analysis of miR-203 levels in HPV-positive and normal keratinocytes differentiated in 1.5% methylcellulose for 24 and 48 h. (A) NHKs and keratinocytes stably expressing HPV-31 episomes (NHK-31). (B) NHKs and keratinocytes stably expressing HPV-31 E6/E7 and E7. (C) NHKs and keratinocytes expressing HPV-31 E6. 28S rRNA was a loading control. nt, nucleotides.
Next, it was important to identify the viral protein responsible for downregulating miR-203 expression upon differentiation. We reasoned that one or both of the viral oncoproteins E6 and E7 were good candidates, as they have been shown to control the proliferative capacity of differentiating cells (4, 12). For these studies, we used cell lines that had been generated by infecting NHKs with retroviruses expressing individual HPV-31 E6 (NHK-E6) and E7 (NHK-E7) oncoproteins or the combination of E6 and E7. Cell lines transduced with these retroviruses express comparable levels of E6 or E7, as seen in cells with viral episomes (J. Bodily and L. Laimins, unpublished data). As shown in Fig. 1B, cell lines expressing both E6 and E7 or E7 alone had significantly reduced levels of miR-203 following differentiation in methylcellulose compared to the corresponding NHKs. In contrast, cells expressing E6 alone exhibited levels of miR-203 similar to those seen in NHKs (Fig. 1C). This also demonstrated that the process of transduction itself had no effect on miR-203 expression. Similar results were seen with cell lines expressing E6 and E7 oncoproteins from another high-risk HPV type, HPV-16 (data not shown), implying that this effect is common to other high-risk HPV types. We conclude that the E7 protein is the primary viral factor responsible for blocking upregulation of miR-203 expression following differentiation.
HPV proteins interfere with downregulation of p63 expression upon differentiation.
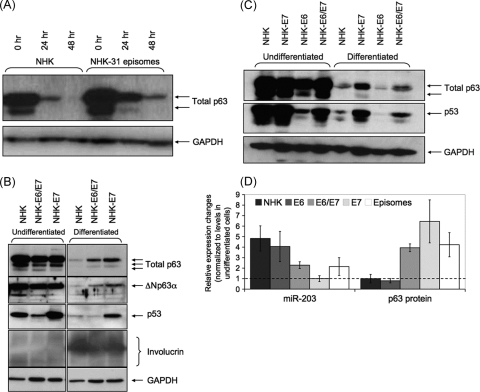
One target of miR-203 is the p63 transcription factor family, and we next examined if the presence of HPV proteins had any effect on the levels of these factors. We first compared the levels of total p63 proteins in NHK and NHK-31 cells using an antibody that detected all family members. Similar levels of total p63 proteins were detected in undifferentiated NHK and NHK-31 cells (Fig. 2A). Upon differentiation in methylcellulose, the levels of p63 decreased in both NHK and NHK-31 cells; however, significantly higher levels were retained in NHK-31 cells. In NHKs, the levels of p63 rapidly declined to low levels following suspension in methylcellulose for 48 h, while in NHK-31 cells, approximately 5-fold-higher levels of total p63 proteins were retained following differentiation. Identical effects were seen in cells expressing both HPV-31 E6 and E7 or E7 alone using antibodies specific for the most dominant isoform, ΔNp63 (Fig. 2B). Similarly, cells expressing HPV-16 E6 and E7 in combination or HPV-16 E7 alone retained high levels of p63, while cells expressing E6 alone exhibited reductions similar to those seen in NHKs (Fig. 2C). These results indicated that the HPV effect on p63 levels was common to several high-risk HPV types and that E7 mediates this activity. Consistent with published reports, the basal levels of p53 were reduced in cells expressing E6 and E7 but were increased in cells expressing E7 alone (Fig. 2C). In four independent experiments using different primary isolates and matched HPV-positive cells, we observed consistent upregulation of miR-203 expression and a corresponding decrease in p63 levels in differentiated cells. A graph showing the average quantitative changes in miR-203 and p63 levels upon differentiation of HPV-positive and -negative cells is shown in Fig. 2D. The inverse correlation of miR-203 with p63 levels indicates that miR-203 was negatively targeting p63 upon differentiation. Studies using mouse cells indicated that miR-203 regulates proliferative capacity upon cellular differentiation, but not expression of markers of differentiation (5, 57). Consistent with this, we observed a minimal effect of high-level expression of miR-203 on the levels of the differentiation marker involucrin (Fig. 2B). This indicates that differentiation is not delayed in miR-203-overexpressing cells. The multiple forms of involucrin seen in the Western blot are typically seen in analyses of this protein (5, 57). Similar effects were seen in E7-expressing cells or cells with complete HPV genomes (not shown), in agreement with the role of E7 in regulating miR-203 expression.
FIG. 2.
p63 protein levels are inversely correlated with miR-203 levels upon keratinocyte differentiation. (A) Total p63 protein levels in NHKs and NHK-31 cells upon differentiation. (B) Western blot analysis for total p63, ΔNp63α, p53, and involucrin in NHKs and keratinocytes stably expressing HPV-31 E6/E7 and E7. Levels in the undifferentiated state and following differentiation in methylcellulose for 48 h are shown. (C) Total p63 and p53 protein levels in undifferentiated and 48-h-differentiated NHKs and keratinocytes stably expressing HPV-16 E6/E7, E7, and E6. (D) Relative changes in miR-203 RNA and p63 protein levels in HPV-positive and -negative keratinocytes upon differentiation in methylcellulose for 48 h. miR-203 levels were normalized to NHK-E7 cell levels (set to 1) and p63 levels to those seen in NHKs. The results are from 4 independent experiments ± standard errors of the mean.
p63 is a target of miR-203 in human keratinocytes.
We next investigated if miR-203 directly regulates p63 protein levels in human keratinocytes. Using a luciferase reporter system, we tested if miR-203 targets sequences located in the 3′ UTR of the mature human p63 mRNA message and blocks p63 translation. A 60-bp DNA fragment containing the putative miR-203 consensus recognition site from the 3′ UTR of the human p63 gene (5′-GAGUCCUUGUGAUUUCAAAG-3′) (57) was synthesized and cloned into the 3′-UTR region of a firefly luciferase reporter gene. This reporter plasmid was cotransfected, along with a vector expressing miR-203 precursor molecule or an empty control vector, into NHKs. Luciferase expression was monitored after 48 h (Fig. 3A). In NHKs cotransfected with the miR-203 overexpression construct and the p63 UTR-luciferase construct, luciferase levels were approximately two-thirds lower than in cells cotransfected with empty vector and the p63 UTR-luciferase construct. This result implied that the presence of a miR-203 recognition sequence from p63 mRNA within the luciferase 3′ UTR rendered luciferase sensitive to high levels of miR-203 (Fig. 3A). In contrast, expression of a luciferase reporter containing a mutated miR-203 consensus recognition site was unchanged by the presence or absence of overexpressed miR-203 (Fig. 3A). These observations confirmed effects seen in mouse cells and indicated that human miR-203 targets the p63 3′ UTR and negatively regulates p63 levels in the human keratinocytes used in our study.
FIG. 3.
miR-203 targets p63 mRNA in human keratinocytes. (A) NHKs were cotransfected with a luciferase reporter containing a wild-type or mutated p63 3′-UTR fragment, along with an expression vector for the pre-miR-203 precursor, and analyzed after 48 h. The luciferase construct lacking the p63 3′-UTR fragment, as well as mutated 3′ UTR, served as negative controls. Relative luciferase levels were calculated by comparison to transfections without the pre-miR-203 precursor. Transfection efficiency was normalized using Renilla luciferase. The error bars indicate standard errors. (B) qRT-PCR analysis of miR-203 and ΔNp63α mRNA levels following transfection of NHKs with anti-miR-203 inhibitors after 24, 48, and 72 h. Levels were normalized to U6 mRNA. (C) NHKs were transfected with antisense miR-203 RNA for 24 and 48 h and analyzed for ΔNp63α and CARM-1 protein levels. NHKs transfected with nonspecific RNA served as negative controls. (D) NHKs were transfected with antisense miR-203 RNA, and the levels of ΔNp63α, p53, p21, and Bax were determined by Western blot analysis. Negative-control cells were transfected with nonspecific RNA. (E) Western blot analysis of NHKs and NHK-E6 and NHK-E7 cells for levels of ΔNp63α, p21, and Bax following differentiation in methylcellulose for 24 and 48 h.
We next investigated if the protein levels of p63 increased when the activity of miR-203 was blocked through the use of an antisense miR-203 RNA fragment as an inhibitor. Transfection of the antisense miR-203 for 24, 48, and 72 h in NHKs drastically reduced miR-203 levels but had a minimal effect on p63 mRNA (Fig. 3B). This is consistent with the mechanism of microRNA action, which is mediated at the posttranscriptional level (44). To investigate the effect of miR-203 inhibition on p63 protein levels, we transfected NHKs with antisense miR-203, induced cells to differentiate in methylcellulose, and screened for the levels of ΔNp63 by Western blot analysis. Consistent with our previous experiments, the levels of ΔNp63 declined rapidly following differentiation of NHKs. However, the levels of ΔNp63 remained unchanged upon differentiation of NHKs transfected with antisense miR-203 RNAs (Fig. 3C), again indicating that miR-203 has a negative effect on ΔNp63 protein levels.
To investigate how the function of ΔNp63 was altered in the presence of inhibitors of miR-203, we examined the levels of ΔNp63 downstream targets by Western blot analysis. Although the ΔNp63 isoform lacks the N-terminal transactivation domain, it retains the ability to activate several cellular genes, including the methyltransferase CARM-1 gene, and some p53 target genes, such as those encoding p21 and Bax (5, 58). CARM-1 is a suggested target of the ΔNp63 transcription factor and contains p63 binding sites in its promoter region (49). We observed little to no expression of CARM-1 in either undifferentiated or differentiated NHKs. In contrast, in cells in which miR-203 was inhibited using antisense miR-203, we detected increasing levels of CARM-1 upon differentiation (Fig. 3C). Consistent with E7's ability to block upregulation of miR-203, we observed induction of CARM-1 upon differentiation of E7-expressing keratinocytes (data not shown). Interestingly, we did not see CARM-1 expressed in undifferentiated cells despite high levels of ΔNp63, indicating that additional factors besides ΔNp63 contribute to CARM-1 activation upon cellular differentiation.
The levels of two additional ΔNp63 targets, p21 and Bax, were found to decrease following differentiation of NHKs (Fig. 3D). In contrast, in NHKs transfected with miR-203 antisense inhibitors, the levels of p21 and Bax remained high, which correlated with elevated ΔNp63 levels upon differentiation (Fig. 3D). These elevated levels of p21 and Bax, however, did not lead to increased apoptosis (data not shown). Interestingly, we also observed that inhibition of miR-203 resulted in retention of high levels of p53 following differentiation (Fig. 3D). We conclude that several ΔNp63 and p53 target genes, such as those encoding p21 and Bax, are regulated by miR-203 upon differentiation.
Consistent with E7's role in regulating miR-203 levels, we found that NHKs stably expressing HPV type 31 E7 oncoproteins exhibited the same p21 and Bax profile as NHKs with inhibited miR-203. The levels of p21 and Bax in NHK-E7 cells remained high upon differentiation, in contrast to the levels in HPV-negative NHKs (Fig. 3E). In normal keratinocytes, p53 levels declined upon differentiation, while in NHKs that expressed miR-203 inhibitors, no such decrease was seen (Fig. 3D). Similarly, in E7-expressing cells, a minimal decrease in p53 levels was detected upon differentiation (Fig. 2B and C). It is thus possible that the increase in p21 and Bax levels in E7-expressing cells is due to the actions of both p53 and ΔNp63.
E7 blocks the MAPK/PKC pathway-dependent activation of miR-203 expression.
Our studies indicated that the HPV E7 protein interferes with the normal upregulation of miR-203 expression upon differentiation, and we sought to determine the pathways targeted by E7 to mediate these effects. Our analysis of the promoter region on miR-203 (500 bp upstream from the miR-203 start codon) identified 9 strong and 13 weak AP-2α binding sites, along with approximately 20 Sp-1 binding sites, two NF-κB binding sites, and two NFAT binding sites. Interestingly, the MAPK pathways induce all these transcription factors. Since the MAPK/protein kinase C (PKC) pathway is also implicated in regulating keratinocyte differentiation (35, 43), we investigated if miR-203 levels were affected by the MAPK/PKC pathway signaling and if HPV E7 interfered with the activation of this pathway.
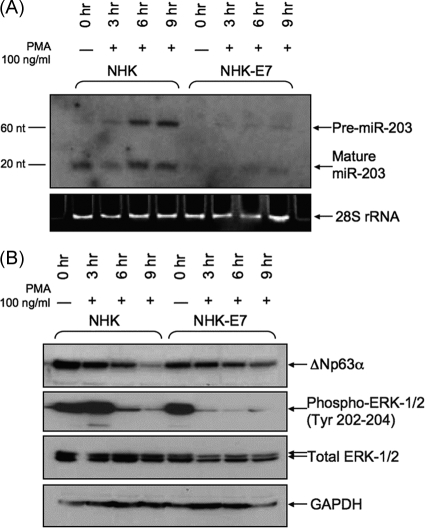
To induce the MAPK/PKC pathway, we treated NHKs and NHK-E7 cells with PMA, which initiates the MAPK pathway by activating PKC (7, 24). We treated both types of cells with 100 ng/ml PMA for 3, 6, and 9 h and then examined miR-203 levels using Northern blot analysis. In NHKs, the PMA treatment resulted in upregulation of a miR-203 precursor molecule, pre-miR-203, within 3 h and upregulation of mature miR-203 within 6 and 9 h (Fig. 4A). Since our experiments indicated an increase in the miR-203 precursor levels upon PMA treatment, we conclude that PMA most likely induces increased transcription of miR-203. In contrast to NHKs, PMA-treated NHK-E7 cells failed to upregulate pre-miR-203 or miR-203 to the same degree, suggesting that E7 acts to block the PMA-induced miR-203 upregulation. Moreover, the PMA treatment of NHK-31 cells, which stably maintain HPV type 31 episomes, induced a pattern of miR-203 expression similar to that observed in NHK-E7 cells (data not shown). We also observed that MAPK/PKC signaling had opposing effects on miR-203 and p63 protein levels. Figure 4B demonstrates that p63 protein levels decrease in NHKs treated with PMA. This PMA-induced p63 decrease was less prominent in NHK-E7 cells (Fig. 4B), suggesting that E7 has the ability to slow down the kinetics of MAPK/PKC pathway activation. This is evident from the lack of an increase in phospho-ERK (extracellular signal-regulated kinase), a marker of MAPK/PKC pathway activation, in NHK-E7 cells 3 h after PMA treatment that is observable in NHKs. This E7-mediated interference in the MAPK pathway signaling may contribute to blocking miR-203 transcription activation.
FIG. 4.
The MAP kinase pathway regulates miR-203 and p63 expression. (A) Northern blot analysis for pre-miR-203 in NHKs and NHK-E7 cells treated with 100 ng/ml PMA for 0, 3, 6, and 9 h. Shown is quantitation of the average fold increase of pre-miR-203 relative to 0 h in NHKs (1.0, 1.8, 3.1, and 3.7) and in NHK-E7 cells (1.0, 1.0,1.3, and 1.6). (B) Western blot analysis of p63, phospho-ERK-1/2, and total ERK-1/2 in NHKs and NHK-E7 cells following treatment with 100 ng/ml PMA for 0, 3, 6, and 9 h.
miR-203 overexpression modulates HPV gene expression and genome copy number.
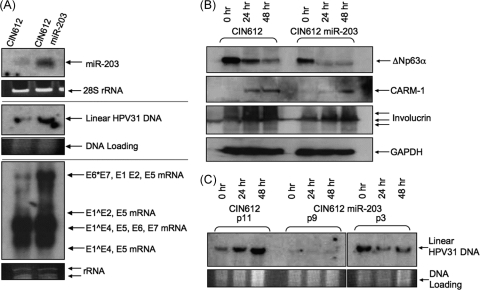
We next investigated what effect high-level expression of miR-203 had on the replication of HPV-31 genomes in undifferentiated and differentiated cells. For these studies, we created stable HPV-positive cell lines that expressed high levels of miR-203 from a heterologous promoter. We first generated recombinant retroviruses that expressed the miR-203 precursor along with a drug-selectable marker and used these to infect CIN612 cells. We used CIN612 cells for this analysis because they stably maintain HPV-31 genomes and do not express any drug resistance markers, as they were derived from a biopsy specimen. Following miR-203 retroviral infection, CIN612 lines stably expressing high levels of miR-203 (CIN612 miR-203 cells) were selected and expanded, and the miR-203 levels in undifferentiated cells were assessed by Northern blot analysis (Fig. 5A). We then used Southern blot analysis to measure HPV-31 DNA genome levels in CIN612 miR-203 cells. Interestingly, we observed that CIN612 miR-203 cells contained on average 2.6-fold-higher HPV genome levels than wild-type CIN612 cells (P < 0.005) (Fig. 5A). Both RNA and DNA were collected from CIN612 miR-203 cells passaged twice after drug selection was completed. Northern blot analysis of HPV mRNA transcripts demonstrated increased expression of E1- and E2-encoding messages in the CIN612 miR-203 cells, suggesting that miR-203 may directly or indirectly enhance HPV transcription. No change was observed in the levels of E6/E7 transcripts (Fig. 5A), indicating that the effect of miR-203 was specific for E1/E2 messages and not a general effect on all viral transcripts. CIN612 miR-203 cells also had reduced basal levels of p63, and these decreased more rapidly upon differentiation than p63 in wild-type CIN612 cells (Fig. 5B). The CIN612 miR-203 cells also exhibited delayed CARM-1 induction upon differentiation (Fig. 5B), consistent with our previous observations.
FIG. 5.
High levels of miR-203 induce increased HPV genomes and transcripts but block amplification. (A) Northern blot analysis of miR-203 levels in CIN612 cells expressing constitutive high levels of miR-203, Southern blot analysis of linearized HPV-31 genomes in CIN612 control cells and CIN612-miR-203 cells, and Northern blot analysis of HPV-31 transcripts in CIN612 cells and CIN612 cells expressing high levels of miR-203 (CIN612-miR-203). Loading controls indicated equal loading in each lane (not shown). E1^E4 is a viral fusion protein expressed from a spliced transcript. (B) Western blot analysis of ΔNp63α and CARM-1 lysates from the CIN612 cell line and CIN612 cells stably transfected with miR-203 overexpression vector. (C) Representative Southern blots showing levels of linearized HPV-31 DNA genomes in methylcellulose-differentiated CIN612 cells and CIN612 miR-203 cells three passages (p3) and nine passages (p9) after transfection.
Next, it was important to investigate the effects of miR-203 overexpression on genome amplification. Analysis of data from four independent experiments demonstrated that HPV-31 genome levels in wild-type CIN612 cells increased 2-fold on average upon 48 h of differentiation in methylcellulose (P < 0.005). Importantly, when CIN612 miR-203 cells were induced to differentiate by suspension in methylcellulose, they failed to amplify HPV genomes in these four experiments (Fig. 5C). This indicates that high-level expression of miR-203 may interfere with genome amplification and that its expression needs to be abrogated upon differentiation.
Upon further passaging of CIN612 miR-203 cells in monolayer culture (9 passages after transfection), we observed a loss of episomal HPV genomes in both undifferentiated and differentiated cells (Fig. 5C). This suggests that long-term overexpression of miR-203 is incompatible with stable maintenance of HPV episomes. No loss of episomes was observed in control CIN612 cells infected with control retroviruses. Identical effects were observed in several experiments using two independent miR-203 overexpression lines. We conclude that high-level expression of miR-203 interferes with HPV genome amplification upon differentiation and long-term stable maintenance of HPV genomes.
DISCUSSION
In HPV-infected epithelia, cells remain active in the cell cycle upon differentiation to allow productive HPV genome replication in suprabasal cells (31). This process is regulated in part by miR-203, which reduces the proliferative capacity of differentiating epithelial cells (46, 57). Our studies demonstrated that miR-203 is downregulated by high-risk HPV proteins upon keratinocyte differentiation. This HPV-mediated miR-203 downregulation is important, as high levels of miR-203 interfere with genome amplification, as well as long-term HPV genome maintenance.
miR-203 is a critical molecule for regulating the transition of keratinocytes from a proliferative state in undifferentiated basal cells to a nonproliferative status in differentiated suprabasal cells. It has even been suggested to promote epithelial maturation by suppressing the “stemness” potential (57). Reducing the expression of miR-203 thus appears to be important in facilitating the productive phase of the HPV life cycle in differentiating epithelia. In mouse and cell culture models, expression of miR-203 has a minimal effect on the synthesis of differentiation markers (5, 57). This is consistent with our observations that the levels of the differentiation marker involucrin did not significantly differ in HPV-positive and HPV-negative cells. Previous studies indicated that differentiation and proliferative capacity are separable activities, and our findings are consistent with this idea (25).
One major target of miR-203 is the p63 family of transcription factors that regulates the balance between epithelial proliferation and differentiation (29). p63 knockout mice are viable but exhibit abnormalities in ectodermal development and stem cell regeneration (27). In contrast to the frequent mutations detected in the p53 gene, p63 is rarely mutated in cancers; however, overexpression of the most dominant isoform, ΔNp63, is often observed in squamous cell carcinomas of the head and neck, skin, cervix, and lung (39). In our studies, the levels of p63 and ΔNp63 were inversely correlated with the amount of miR-203 expressed. In normal epithelia, miR-203 expression increases upon differentiation, while E7 blocks this increase in HPV-positive cells, resulting in retention of high levels of p63 proteins. We observed that in normal keratinocytes treated with antisense inhibitors of miR-203, high levels of p63 proteins were retained following differentiation. Since p63 promotes cellular proliferation, reduced levels of p63 are important for normal epithelial differentiation in which cells exit the cell cycle. We believe that one consequence of HPV E7 action on miR-203 in differentiating cells is the retention of high levels of p63 proteins and that this effect is potentially important for keeping cells active in the cell cycle.
ΔNp63 is the most abundantly expressed p63 isoform in epithelial cells. ΔNp63 lacks the N-terminal transactivation domain but still retains the ability to activate expression of a number of genes, some of which are also p53 responsive (56). Our studies demonstrated that abrogating miR-203 function with miR-203 antisense oligonucleotides resulted in elevated levels of ΔNp63 and a corresponding increase in expression of two ΔNp63 downstream targets, p21 and Bax. Consistent with E7's role in blocking increased expression of miR-203, high levels of p21 and Bax were observed in HPV E7 cells following differentiation in methylcellulose, while only low levels were seen in NHKs or cells expressing E6 alone. Our studies demonstrated an inverse correlation between miR-203 expression and that of the downstream targets of ΔNp63, p21 and Bax, in HPV-positive cells. Interestingly, E7 has been shown to block p21-mediated inhibition of Cdk2 function in differentiating cells, which also contributes to retaining proliferative capability (25). Inhibition of miR-203 also results in increased levels of p53, and it is possible that p53 also contributes to maintenance of high levels of p21 and Bax; however, p53-independent methods of activation have also been reported (5, 14). Importantly, no miR-203 consensus binding sites are present in the p53 mRNA, indicating that miR-203 acts indirectly to increase p53 protein levels. We also believe that miR-203 does not act directly on p21 or Bax messages. It is likely that, in addition to the p63 family, miR-203 targets additional as-yet-unidentified cellular genes whose activities are linked to regulating cell cycle progression in differentiating cells.
In our studies, the primary viral factor responsible for downregulation of miR-203 expression upon keratinocyte differentiation was E7, while the E6 oncoprotein had a minimal effect. E7 had been shown in previous studies to be responsible for keeping cells active in the cell cycle during differentiation, and we believe that targeting miR-203 may be critical to this process (4, 12). Additional targets of E7 are the retinoblastoma (Rb) family members, along with the histone deacetyltransferases (HDACs) (32). It is possible that association with these factors may contribute to E7's role in downregulating expression of miR-203. Alternatively, E7 binding to Rb and HDACs may function independently to complement the activities of miR-203 in maintaining the proliferative capacity of epithelial cells upon differentiation.
Further insights into how E7 acts to downregulate miR-203 expression upon differentiation are provided by our experiments, indicating that the MAPK/PKC pathway is linked to miR-203 expression. Treatment of NHKs with a known PKC activator, PMA, induced miR-203 expression to high levels, while no significant activation was seen in cells expressing E7. In NHKs, we observed increased phosphorylation of ERK, a downstream target of PKC, while no such activity was observed in E7-expressing cells. Since we observed an increase in the levels of pre-miR-203 in NHKs following PMA treatment, this activation most likely occurs at the level of transcription. We suspect that activation of the AP-1, AP-2, and Sp-1 transcription factor families by PMA is central to the mechanism by which miR-203 expression is increased, as the promoter region contains numerous AP-2 and Sp-1 sites. PMA is also an activator of differentiation, and this may also contribute to miR-203 activation. As our observations were being prepared for publication, another study was published that also demonstrated that miR-203 expression can be activated by phorbol esters like PMA, which is consistent with our observations (47). In our studies, we observed a decrease in the levels of downstream targets of miR-203, such as p63, upon PMA treatment, and this was correlated with an increase in the levels of miR-203. In E7-expressing cells, no significant decrease in p63 levels was observed upon PMA treatment, indicating that E7 interferes with downregulation of p63. The mechanism by which E7 specifically acts to block PMA induction is not clear and is an area for future study, including the potential relationship between Rb function, MAPK pathway signaling, and miR-203 (21).
Our findings indicate that high-level expression of miR-203 interferes with HPV genome amplification upon keratinocyte differentiation, as well as with long-term maintenance of HPV episomes. Immediately after infection of HPV-positive CIN612 cells with retroviruses overexpressing miR-203, we detected increased levels of HPV DNA episomes and HPV RNA transcripts that encoded E1 and E2, the major regulators of HPV genome replication. We suspect this may be due directly or indirectly to the action of ΔNp63. While no miR-203 or ΔNp63 binding sites are present in high-risk HPV types 31 and 16, ΔNp63 has been shown to bind to sequences in the upstream regulatory region of the cutaneous, low-risk HPV type 20 (10, 11). In HPV type 20, ΔNp63 binding resulted in transcriptional activation, but it is possible that in other HPV types, p63 may act as a negative regulator. Moreover, HPV genomes in CIN612 miR-203 cells also failed to amplify HPV genomes upon differentiation. Upon further passaging of the CIN612 miR-203-overexpressing cells, we consistently detected only integrated HPV genomes, in contrast to control CIN612 cells. We therefore conclude that high-level miR-203 expression is not compatible with HPV genome amplification and long-term HPV genome maintenance. This effect on HPV is most likely mediated by miR-203 and/or p63 altering the balance between cellular proliferation and differentiation. A model summarizing our findings is shown in Fig. 6.
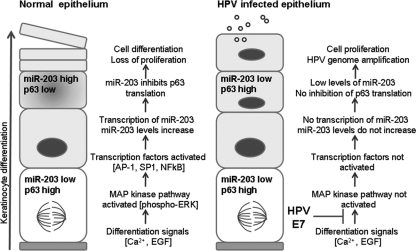
FIG. 6.
Model for the regulation of miR-203 expression in differentiating normal and HPV-positive epithelia. miR-203 expression is increased upon differentiation of normal keratinocytes, leading to suppression of p63 translation in suprabasal cells. In HPV-infected epithelia, E7 blocks miR-203 upregulation through the MAP kinase pathway, leading to increased levels of p63, cells remaining active in the cell cycle, and HPV genome amplification. EGF, epidermal growth factor.
HPV modulates the expression of numerous cellular microRNAs that likely contribute to viral pathogenesis. Our study identified one microRNA target, miR-203, which is responsible for downregulating proliferative capacity upon differentiation. MicroRNAs have many cellular targets, and we have identified human p63 as one target of miR-203. Further elucidation of the roles of other miR-203 targets and other p63 isoforms in the HPV life cycle will be the focus of future analyses.
Acknowledgments
We thank Kathy Rundell for helpful comments.
This work was supported by a grant to L.A.L. from the NCI (RO1CA 59655).
Footnotes
Published ahead of print on 10 March 2010.
REFERENCES
- 1.Bostjancic, E., and D. Glavac. 2008. Importance of microRNAs in skin morphogenesis and diseases. Acta Dermatovenerol Alp Panonica Adriat. 17:95-102. [PubMed] [Google Scholar]
- 2.Cai, X., G. Li, L. A. Laimins, and B. R. Cullen. 2006. Human papillomavirus genotype 31 does not express detectable microRNA levels during latent or productive virus replication. J. Virol. 80:10890-10893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, C. Z., L. Li, H. F. Lodish, and D. P. Bartel. 2004. MicroRNAs modulate hematopoietic lineage differentiation. Science 303:83-86. [DOI] [PubMed] [Google Scholar]
- 4.Cheng, S., D. C. Schmidt-Grimminger, T. Murant, T. R. Broker, and L. T. Chow. 1995. Differentiation-dependent up-regulation of the human papillomavirus E7 gene reactivates cellular DNA replication in suprabasal differentiated keratinocytes. Genes Dev. 9:2335-2349. [DOI] [PubMed] [Google Scholar]
- 5.De Laurenzi, V., A. Rossi, A. Terrinoni, D. Barcaroli, M. Levrero, A. Costanzo, R. A. Knight, P. Guerrieri, and G. Melino. 2000. p63 and p73 transactivate differentiation gene promoters in human keratinocytes. Biochem. Biophys. Res. Commun. 273:342-346. [DOI] [PubMed] [Google Scholar]
- 6.Dell, G., and K. Gaston. 2001. Human papillomaviruses and their role in cervical cancer. Cell Mol. Life Sci. 58:1923-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Efimova, T., P. LaCelle, J. F. Welter, and R. L. Eckert. 1998. Regulation of human involucrin promoter activity by a protein kinase C, Ras, MEKK1, MEK3, p38/RK, AP1 signal transduction pathway. J. Biol. Chem. 273:24387-24395. [DOI] [PubMed] [Google Scholar]
- 8.Fehrmann, F., D. J. Klumpp, and L. A. Laimins. 2003. Human papillomavirus type 31 E5 protein supports cell cycle progression and activates late viral functions upon epithelial differentiation. J. Virol. 77:2819-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fehrmann, F., and L. A. Laimins. 2003. Human papillomaviruses: targeting differentiating epithelial cells for malignant transformation. Oncogene 22:5201-5207. [DOI] [PubMed] [Google Scholar]
- 10.Fei, J. W., P. Angel, Q. X. Wei, and E. M. de Villiers. 2006. TAp63alpha indirectly regulates a cutaneous HPV promoter through complex formation with Jun family members. Oncogene 25:3914-3923. [DOI] [PubMed] [Google Scholar]
- 11.Fei, J. W., Q. X. Wei, P. Angel, and E. M. de Villiers. 2005. Differential enhancement of a cutaneous HPV promoter by DeltaNP63alpha, Jun and mutant p53. Cell Cycle 4:689-696. [DOI] [PubMed] [Google Scholar]
- 12.Flores, E. R., B. L. Allen-Hoffmann, D. Lee, and P. F. Lambert. 2000. The human papillomavirus type 16 E7 oncogene is required for the productive stage of the viral life cycle. J. Virol. 74:6622-6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frattini, M. G., H. B. Lim, and L. A. Laimins. 1996. In vitro synthesis of oncogenic human papillomaviruses requires episomal genomes for differentiation-dependent late expression. Proc. Natl. Acad. Sci. U. S. A. 93:3062-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallegos, J. R., J. Litersky, H. Lee, Y. Sun, K. Nakayama, K. Nakayama, and H. Lu. 2008. SCF TrCP1 activates and ubiquitylates TAp63gamma. J. Biol. Chem. 283:66-75. [DOI] [PubMed] [Google Scholar]
- 15.Galloway, D. A., G. W. Demers, S. A. Foster, C. L. Halbert, and K. Russell. 1994. Cell cycle checkpoint control is bypassed by human papillomavirus oncogenes. Cold Spring Harbor Symp. Quant. Biol. 59:297-306. [DOI] [PubMed] [Google Scholar]
- 16.Genther, S. M., S. Sterling, S. Duensing, K. Munger, C. Sattler, and P. F. Lambert. 2003. Quantitative role of the human papillomavirus type 16 E5 gene during the productive stage of the viral life cycle. J. Virol. 77:2832-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halbert, C. L., G. W. Demers, and D. A. Galloway. 1992. The E6 and E7 genes of human papillomavirus type 6 have weak immortalizing activity in human epithelial cells. J. Virol. 66:2125-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halbert, C. L., G. W. Demers, and D. A. Galloway. 1991. The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J. Virol. 65:473-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harris, K. S., Z. Zhang, M. T. McManus, B. D. Harfe, and X. Sun. 2006. Dicer function is essential for lung epithelium morphogenesis. Proc. Natl. Acad. Sci. U. S. A. 103:2208-2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hebner, C. M., R. Wilson, J. Rader, M. Bidder, and L. A. Laimins. 2006. Human papillomaviruses target the double-stranded RNA protein kinase pathway. J. Gen. Virol. 87:3183-3193. [DOI] [PubMed] [Google Scholar]
- 21.Hou, S. T., X. Xie, A. Baggley, D. S. Park, G. Chen, and T. Walker. 2002. Activation of the Rb/E2F1 pathway by the nonproliferative p38 MAPK during Fas (APO1/CD95)-mediated neuronal apoptosis. J. Biol. Chem. 277:48764-48770. [DOI] [PubMed] [Google Scholar]
- 22.Hubert, W. G., and L. A. Laimins. 2002. Human papillomavirus type 31 replication modes during the early phases of the viral life cycle depend on transcriptional and posttranscriptional regulation of E1 and E2 expression. J. Virol. 76:2263-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hummel, M., J. B. Hudson, and L. A. Laimins. 1992. Differentiation-induced and constitutive transcription of human papillomavirus type 31b in cell lines containing viral episomes. J. Virol. 66:6070-6080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaken, S., and S. H. Yuspa. 1988. Early signals for keratinocyte differentiation: role of Ca2+-mediated inositol lipid metabolism in normal and neoplastic epidermal cells. Carcinogenesis 9:1033-1038. [DOI] [PubMed] [Google Scholar]
- 25.Jones, D. L., R. M. Alani, and K. Munger. 1997. The human papillomavirus E7 oncoprotein can uncouple cellular differentiation and proliferation in human keratinocytes by abrogating p21Cip1-mediated inhibition of cdk2. Genes Dev. 11:2101-2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones, D. L., and K. Munger. 1996. Interactions of the human papillomavirus E7 protein with cell cycle regulators. Semin. Cancer Biol. 7:327-337. [DOI] [PubMed] [Google Scholar]
- 27.Koster, M. I., K. A. Huntzinger, and D. R. Roop. 2002. Epidermal differentiation: transgenic/knockout mouse models reveal genes involved in stem cell fate decisions and commitment to differentiation. J. Investig. Dermatol. Symp Proc. 7:41-45. [DOI] [PubMed] [Google Scholar]
- 28.Lee, Y., C. Ahn, J. Han, H. Choi, J. Kim, J. Yim, J. Lee, P. Provost, O. Radmark, S. Kim, and V. N. Kim. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425:415-419. [DOI] [PubMed] [Google Scholar]
- 29.Lena, A. M., R. Shalom-Feuerstein, P. Rivetti di Val Cervo, D. Aberdam, R. A. Knight, G. Melino, and E. Candi. 2008. miR-203 represses ‘stemness’ by repressing DeltaNp63. Cell Death Differ. 15:1187-1195. [DOI] [PubMed] [Google Scholar]
- 30.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 31.Longworth, M. S., and L. A. Laimins. 2004. Pathogenesis of human papillomaviruses in differentiating epithelia. Microbiol. Mol. Biol. Rev. 68:362-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Longworth, M. S., R. Wilson, and L. A. Laimins. 2005. HPV31 E7 facilitates replication by activating E2F2 transcription through its interaction with HDACs. EMBO J. 24:1821-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez, I., A. S. Gardiner, K. F. Board, F. A. Monzon, R. P. Edwards, and S. A. Khan. 2008. Human papillomavirus type 16 reduces the expression of microRNA-218 in cervical carcinoma cells. Oncogene 27:2575-2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mendell, J. T. 2005. MicroRNAs: critical regulators of development, cellular physiology and malignancy. Cell Cycle 4:1179-1184. [DOI] [PubMed] [Google Scholar]
- 35.Mitev, V., and L. Miteva. 1999. Signal transduction in keratinocytes. Exp. Dermatol. 8:96-108. [DOI] [PubMed] [Google Scholar]
- 36.Moody, C. A., A. Fradet-Turcotte, J. Archambault, and L. A. Laimins. 2007. Human papillomaviruses activate caspases upon epithelial differentiation to induce viral genome amplification. Proc. Natl. Acad. Sci. U. S. A. 104:19541-19546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moody, C. A., and L. A. Laimins. 2009. Human papillomaviruses activate the ATM DNA damage pathway for viral genome amplification upon differentiation. PLoS Pathog. 5:e1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakamura, M., J. M. Bodily, M. Beglin, S. Kyo, M. Inoue, and L. A. Laimins. 2009. Hypoxia-specific stabilization of HIF-1alpha by human papillomaviruses. Virology 387:442-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nylander, K., P. J. Coates, and P. A. Hall. 2000. Characterization of the expression pattern of p63 alpha and delta Np63 alpha in benign and malignant oral epithelial lesions. Int. J. Cancer. 87:368-372. [PubMed] [Google Scholar]
- 40.Ozbun, M. A., and C. Meyers. 1998. Temporal usage of multiple promoters during the life cycle of human papillomavirus type 31b. J. Virol. 72:2715-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reshmi, G., and M. R. Pillai. 2008. Beyond HPV: oncomirs as new players in cervical cancer. FEBS Lett. 582:4113-4116. [DOI] [PubMed] [Google Scholar]
- 42.Ruesch, M. N., F. Stubenrauch, and L. A. Laimins. 1998. Activation of papillomavirus late gene transcription and genome amplification upon differentiation in semisolid medium is coincident with expression of involucrin and transglutaminase but not keratin-10. J. Virol. 72:5016-5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seo, H. R., Y. W. Kwan, C. K. Cho, S. Bae, S. J. Lee, J. W. Soh, H. Y. Chung, and Y. S. Lee. 2004. PKCalpha induces differentiation through ERK1/2 phosphorylation in mouse keratinocytes. Exp. Mol. Med. 36:292-299. [DOI] [PubMed] [Google Scholar]
- 44.Soifer, H. S., J. J. Rossi, and P. Saetrom. 2007. MicroRNAs in disease and potential therapeutic applications. Mol. Ther. 15:2070-2079. [DOI] [PubMed] [Google Scholar]
- 45.Sonkoly, E., M. Stahle, and A. Pivarcsi. 2008. MicroRNAs: novel regulators in skin inflammation. Clin. Exp. Dermatol. 33:312-315. [DOI] [PubMed] [Google Scholar]
- 46.Sonkoly, E., T. Wei, P. C. Janson, A. Saaf, L. Lundeberg, M. Tengvall-Linder, G. Norstedt, H. Alenius, B. Homey, A. Scheynius, M. Stahle, and A. Pivarcsi. 2007. MicroRNAs: novel regulators involved in the pathogenesis of psoriasis? PLoS One 2:e610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sonkoly, E., T. Wei, E. Pavez Lorie, H. Suzuki, M. Kato, H. Torma, M. Stahle, and A. Pivarcsi. 2009. Protein kinase C-dependent upregulation of miR-203 induces the differentiation of human keratinocytes. J. Invest. Dermatol. 130:124-134. [DOI] [PubMed] [Google Scholar]
- 48.Sullivan, C. S., and D. Ganem. 2005. MicroRNAs and viral infection. Mol. Cell 20:3-7. [DOI] [PubMed] [Google Scholar]
- 49.Testoni, B., S. Borrelli, E. Tenedini, D. Alotto, C. Castagnoli, S. Piccolo, E. Tagliafico, S. Ferrari, M. A. Vigano, and R. Mantovani. 2006. Identification of new p63 targets in human keratinocytes. Cell Cycle 5:2805-2811. [DOI] [PubMed] [Google Scholar]
- 50.Truong, A. B., M. Kretz, T. W. Ridky, R. Kimmel, and P. A. Khavari. 2006. p63 regulates proliferation and differentiation of developmentally mature keratinocytes. Genes Dev. 20:3185-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Umbach, J. L., and B. R. Cullen. 2009. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 23:1151-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ustav, M., and A. Stenlund. 1991. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 10:449-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, X., S. Tang, S. Y. Le, R. Lu, J. S. Rader, C. Meyers, and Z. M. Zheng. 2008. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS One 3:e2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang, X., H. K. Wang, J. P. McCoy, N. S. Banerjee, J. S. Rader, T. R. Broker, C. Meyers, L. T. Chow, and Z. M. Zheng. 2009. Oncogenic HPV infection interrupts the expression of tumor-suppressive miR-34a through viral oncoprotein E6. RNA 15:637-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilson, R., F. Fehrmann, and L. A. Laimins. 2005. Role of the E1-E4 protein in the differentiation-dependent life cycle of human papillomavirus type 31. J. Virol. 79:6732-6740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu, G., S. Nomoto, M. O. Hoque, T. Dracheva, M. Osada, C. C. Lee, S. M. Dong, Z. Guo, N. Benoit, Y. Cohen, P. Rechthand, J. Califano, C. S. Moon, E. Ratovitski, J. Jen, D. Sidransky, and B. Trink. 2003. DeltaNp63alpha and TAp63alpha regulate transcription of genes with distinct biological functions in cancer and development. Cancer Res. 63:2351-2357. [PubMed] [Google Scholar]
- 57.Yi, R., M. N. Poy, M. Stoffel, and E. Fuchs. 2008. A skin microRNA promotes differentiation by repressing ‘stemness.’ Nature 452:225-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu, L., E. A. Rorke, and R. L. Eckert. 2007. DeltaNp63alpha promotes apoptosis of human epidermal keratinocytes. J. Investig. Dermatol. 127:1980-1991. [DOI] [PubMed] [Google Scholar]
- 59.zur Hausen, H. 2002. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer. 2:342-350. [DOI] [PubMed] [Google Scholar]