Abstract
Human cytomegalovirus (HCMV) infection has been shown to activate the mTORC1 signaling pathway. However, the phosphorylation of mTORC1 targets is differentially sensitive to the mTORC1 inhibitor rapamycin, and the drug inhibits HCMV replication to a modest extent. Using Torin1, a newly developed inhibitor that targets the catalytic site of mTOR kinase, we show that HCMV replication requires both rapamycin-sensitive and rapamycin-resistant mTOR activity. The treatment of infected cells with Torin1 inhibits the phosphorylation of rapamycin-sensitive and rapamycin-resistant mTOR targets and markedly blocks the production of virus progeny. The blockade of mTOR signaling with Torin1, but not rapamycin, disrupts the assembly of the eIF4F complex and increases the association of the translational repressor 4EBP1 to the 7-methylguanosine cap-binding complex. Torin1 does not affect HCMV entry and only modestly reduces the accumulation of the immediate-early and early viral proteins that were tested despite the disruption of the eIF4F complex. In contrast, Torin1 significantly decreases the accumulation of viral DNA and the pUL99 viral late protein. Similar mTOR signaling events were observed during murine cytomegalovirus (MCMV) infection, and we utilized murine fibroblasts containing several different mutations to dissect the mechanism by which Torin1 inhibits MCMV replication. This approach demonstrated that mTORC2 and the Akt1 and Akt2 kinases are not required for the Torin1-mediated inhibition of cytomegalovirus replication. The inhibition of MCMV replication by Torin1 was rescued in cells lacking 4EBP1, demonstrating that the inactivation of 4EBP1 by mTORC1 is critical for cytomegalovirus replication. Finally, we show that Torin1 inhibits the replication of representative members of the alpha-, beta-, and gammaherpesvirus families, demonstrating the potential of mTOR kinase inhibitors as broad-spectrum antiviral agents.
As intracellular parasites with limited genetic resources, viruses must rely on the host cell machinery to perform tasks essential for viral replication, even as host cell defense mechanisms inactivate many processes most commonly hijacked by viruses. As a consequence, viruses have evolved mechanisms to maintain the function of these cellular processes and to subvert them for their own ends.
Viruses typically reprogram the host protein synthetic pathway to favor the translation of viral mRNAs (1, 3, 13). In response, the host cell has evolved multiple defenses to inhibit the translation of viral proteins, and viruses have evolved mechanisms to antagonize this response. For example, double-stranded RNA (dsRNA) produced during viral infection activates protein kinase R, which phosphorylates and inactivates the translation initiation factor eIF2α, blocking the initiation of translation. The activation of protein kinase R is an effective antiviral mechanism, so much so that multiple viruses, including members of all three subfamilies of herpesviruses, have evolved strategies to counteract the effects of PKR on viral replication (5, 17, 28, 33, 35, 36). By encoding proteins that disable the host cell control of translation, viruses maintain the capacity of the infected cell to translate viral proteins.
Viruses also induce cellular signaling pathways that activate translation and then reprogram the activated translational apparatus to promote the synthesis of viral proteins. The mammalian target of rapamycin (mTOR) kinase is a metabolic sensor that regulates translation (37). The mTOR serine/threonine kinase is the catalytic subunit of two complexes, mTORC1 and mTORC2, that control cell growth, proliferation, and survival. The activation of mTORC1 signaling results in the initiation of several processes required for the efficient translation of 7-methyl guanosine (m7G)-capped mRNAs. mTORC1 phosphorylates and induces the activity of the p70 S6 kinase (4), which in turn phosphorylates ribosomal protein S6 (rpS6) to promote ribosome biogenesis. At the same time, mTORC1 phosphorylates and inactivates the translational repressor 4EBP1 (4, 11, 15, 27). The translation of capped mRNAs requires the eIF4F complex, which is composed of eIF4E, eIF4A, and eIF4G (14, 43). The eIF4F complex binds to the m7G cap of mRNAs and facilitates their association with the ribosome. Hypophosphorylated 4EBP1 binds to the mRNA cap recognition protein eIF4E, preventing the formation of the eIF4F complex and thereby blocking translation (38). The phosphorylation of 4EBP1 by mTORC1 blocks its ability to bind to eIF4E, resulting in an increased translation of capped mRNAs (4).
Given its important role in the regulation of cap-dependent translation, it is not surprising that multiple viruses that rely on cap-dependent mRNA translation have evolved mechanisms to ensure that mTORC1 remains active during infection. As a case in point, human cytomegalovirus (HCMV), a widespread betaherpesvirus that causes life-threatening disease in immunologically immature or compromised individuals, utilizes cap-dependent mRNA translation for its protein synthesis (3), and it induces mTOR activity, presumably to increase the translation of viral proteins (21, 24, 53). Furthermore, to maintain mTORC1 activity, HCMV encodes a protein, pUL38, that binds to and inhibits the tuberous sclerosis protein complex (TSC1/TSC2) (31). The TSC1 and TSC2 proteins function as a heterodimer, which limits mTORC1 activity in response to cellular stresses such as limited nutrient availability (20, 45). pUL38 alone is sufficient to prevent the inhibition of mTORC1 under limiting nutrient conditions and when TSC1/TSC2 activity is induced by drug treatment. Under these conditions, the classical mTORC1 inhibitor rapamycin inhibits mTORC1 in the presence of the viral protein, demonstrating that pUL38 maintains the phosphorylation of mTORC1 targets by manipulating the canonical signaling pathway.
HCMV infection induces a unique set of events in the mTOR signaling pathway. While the phosphorylation of the mTORC1 effector rpS6 is inhibited by rapamycin during HCMV infection, the phosphorylation of the 4EBP1 protein becomes insensitive to the effects of rapamycin within 12 h after infection (24). The mechanism supporting the rapamycin-resistant phosphorylation of 4EBP1 is unclear. It was proposed previously that the other mTOR-containing complex, mTORC2, could replace mTORC1 during infection (23). While normally insensitive to rapamycin, mTORC2 may become rapamycin sensitive during HCMV infection. Another possibility is that a kinase other than mTORC1 is responsible for 4EBP1 phosphorylation during HCMV infection. This kinase could be of cellular or viral origin, as several cellular kinases have been shown to be capable of phosphorylating 4EBP1 in vitro (12, 18), and HCMV encodes a protein kinase (pUL97). A third possibility is that a modified mTORC1 activity is induced in infected cells, which is resistant to rapamycin.
Rapamycin blocks mTORC1 kinase activity through a unique mode of action. Rather than binding to mTOR directly, rapamycin first binds to its intracellular receptor FKBP12. The rapamycin-FKBP12 complex then binds mTOR, preventing its association with the essential mTORC1 component Raptor. Despite its widespread use as an inhibitor of mTORC1, rapamycin does not exert as profound an effect on cellular protein synthesis as would be predicted. In fact, many mammalian cell types are relatively resistant to the inhibition of protein synthesis by rapamycin. Recently, several groups have developed a new class of mTOR inhibitors that directly target the catalytic site of the mTOR kinase (41). One such drug, Torin1, has been utilized to identify a previously unrecognized rapamycin-resistant mTORC1 activity (47). In contrast to rapamycin, Torin1 profoundly inhibits protein synthesis by disrupting the formation of the eIF4F complex. The effects of Torin1 are independent of mTORC2 (47), as these events take place in cells lacking the essential mTORC2 component Rictor.
In this work we use Torin1 to show that rapamycin-resistant mTORC1 activity is required for efficient herpesvirus replication. We show that during cytomegalovirus infection, the regulation of 4EBP1 phosphorylation by rapamycin-resistant mTORC1 is a critical event required for viral replication.
MATERIALS AND METHODS
Cells, viruses, and reagents.
Primary human foreskin fibroblasts were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% normal calf serum and used between passages 6 and 14. 10.1 murine embryonic fibroblasts (MEFs) were grown in DMEM containing 10% fetal bovine serum, as were Rictor-null (40), Akt1- and Akt2-null (32), and 4EBP1-null MEFs (48). Cell viability was assessed by a trypan blue exclusion assay.
Phenotypically wild-type (WT) HCMV was prepared by the transfection of human fibroblasts with a bacterial artificial chromosome (BAC) clone of the AD169 strain modified to contain the green fluorescent protein (GFP) open reading frame (ORF) under the control of the simian virus 40 (SV40) early promoter (BADinGFP) (54). Murine cytomegalovirus (MCMV) was obtained by the transfection of a Smith strain BAC plasmid (50) into 10.1 MEFs. MCMV stocks were grown on 10.1 MEFs, and titers were determined. The KOS strain of herpes simplex virus type 1 (HSV-1) (10) was grown on 10.1 MEFs, and titers were determined, as was the WUMS strain of murine gammaherpesvirus 68 (γHV68) (49).
Unless otherwise noted, the following inhibitors were used at the indicated concentrations: rapamycin (20 nM; Cell Signaling Technology), Torin1 (250 nM; generously provided by N. Gray, Harvard Medical School), and LY294002 (20 μM; Cell Signaling Technology).
Multistep growth analysis of viruses.
Human fibroblasts or 10.1 MEFs were plated at confluence and serum starved for 48 h prior to infection. Cells were infected at a multiplicity of 0.05 PFU/cell with the indicated virus in serum-free DMEM. Cells in six-well plates were incubated with virus in 300 μl of medium for 1 h with rocking every 15 min. After adsorption, the inoculum was removed and replaced with fresh serum-free medium. The amount of virus present in cell-free supernatants was quantified by the 50% tissue culture infective dose (TCID50) method on primary human fibroblasts (HCMV) or 10.1 MEFs (MCMV, HSV-1, and γHV68).
m7GTP-Sepharose capture assay.
Cells were plated to achieve confluence in 10-cm dishes and were serum starved for 48 h prior to infection at a multiplicity of 3 PFU/cell. At the indicated times, cells were harvested by scraping them into medium and were stored as frozen cell pellets at −80°C until use. Cell pellets were lysed in 500 μl of cap lysis buffer {40 mM HEPES (pH 7.6), 120 mM NaCl, 1 mM EDTA, 10 mM β-glycerophosphate, 0.3% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS)}. The protein concentration of lysates was quantified by using the Bradford assay (Bio-Rad). Equal amounts of protein were used in each immunoprecipitation in a total volume of 1 ml of cap lysis buffer. Following the addition of 10 μl of 7-methyl GTP (m7GTP)-coupled Sepharose, lysates were incubated at 4°C with mixing. After 1 h, the m7GTP-Sepharose was washed three times with cap lysis buffer and then resuspended in 1× SDS sample buffer. The samples were boiled at 100°C for 5 min and then centrifuged at 14,000 × g for 3 min prior to electrophoresis in acrylamide gels for Western blot analysis.
Western blot analysis of proteins.
Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 7.4], 1% NP-40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA) containing protease inhibitors (complete EDTA free; Roche). Protein concentrations in each lysate were determined by the Bradford assay. Unless otherwise noted, 30 μg of protein was analyzed per sample. Proteins in cell lysates were resolved on 10% SDS-containing polyacrylamide gels, with the following exceptions: for 4EBP1 blots, lysates were resolved on 15% gels, and for eIF4G analysis, lysates were resolved on 8% gels. Proteins were transferred onto Protran membranes by using a semidry transfer apparatus. Membranes were blocked with phosphate-buffered saline containing 0.1% Tween 20 (PBS-T) and 5% fat-free dried milk for 1 h prior to incubation with primary antibody. When mouse monoclonal antibodies were used, the antibody was diluted in PBS-T containing 1% bovine serum albumin (BSA) and incubated with the membrane for 1 h at room temperature. Rabbit polyclonal antibodies were diluted in PBS-T containing 5% BSA overnight at 4°C. Following extensive washing with PBS-T, blots were incubated with either goat anti-mouse or goat anti-rabbit horseradish peroxidase (HRP)-coupled secondary antibodies diluted 1:5,000 in PBS-T containing 1% BSA. Membranes were then washed again in PBS-T, and proteins were visualized by chemiluminescence using ECL reagent (Amersham). The following antibodies were used in this study: tubulin (Sigma) and rpS6, phospho-rpS6 (S235/6), 4E-BP1, phospho-4E-BP1 (T37/46), Akt, eIF4G, eIF4E, and eIF4A (Cell Signaling Technology). Antibodies to the HCMV proteins IE1 (56) and pUL99 (42) were described previously. Antibody to the HCMV UL44 protein was obtained from Virusys.
Determination of viral DNA and transcript accumulation in infected cells.
The accumulation of viral DNA during HCMV infection was monitored by quantitative PCR (qPCR) as described previously (46). Briefly, primary human fibroblasts were infected with BADinGFP at a multiplicity of 0.05 PFU/cell. At the indicated times, cells were harvested by scraping them into medium and were stored as frozen cell pellets until analysis. Cell pellets were resuspended in 500 μl of a solution containing 400 mM NaCl, 10 mM Tris (pH 7.5), and 10 mM EDTA. Proteinase K (20 μg) was added together with 4 μl of a 20% SDS solution. The lysate was incubated overnight at 37°C. Lysates were phenol-chloroform extracted. RNase A was added (20 μg), and the lysates were incubated at 37°C for 1 h. Lysates were extracted with phenol-chloroform and then with chloroform. DNA was precipitated by the addition of 1 ml of 100% ethanol followed by centrifugation at 14,000 × g for 30 min. DNA was washed once in 70% ethanol prior to resuspension in 50 μl of 10 mM Tris (pH 7.5). For each sample, DNA was quantified by using a NanoDrop spectrophotometer (Thermo Scientific). Five hundred nanograms of DNA was added to 12.5 μl 2× SYBR green PCR master mix (Applied Biosystems) and 2 μM each primer in a total volume of 25 μl. As an additional control for equal loading, the amount of viral DNA in each sample was normalized to the amount of the cellular glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene in each sample.
The UL99 transcript levels following infection were determined as described previously (8). Briefly, total RNA was harvested at the indicated times by Trizol (Invitrogen) extraction. DNase-treated RNA (0.5 μg) was reverse transcribed with the TaqMan reverse transcription reagent kit (Applied Biosciences) using random hexamer primers. Two microliters of cDNA was added to SYBR green master mix (Applied Biosciences) together with primers specific for UL99 (5′-GTGTCCCATTCCCGACTCG-3′ and 5′-TTCACAACGTCCACCCACC-3′). Actin levels were measured in the same samples by using the following primers: 5′-TCCTCCTGAGCGCAAGTACTC-3′ and 5′-CGGACTCGTCATACTCCTGCTT-3′. Copy numbers for UL99 and actin transcripts were determined by comparing the threshold cycle for each sample to a standard curve, which consisted of serial dilutions of a recombinant HCMV BAC that contains the actin gene inserted into the UL21.5 locus. The standard curve for all experiments had an R value greater than 0.98.
RESULTS
Torin1 inhibits the production of HCMV progeny.
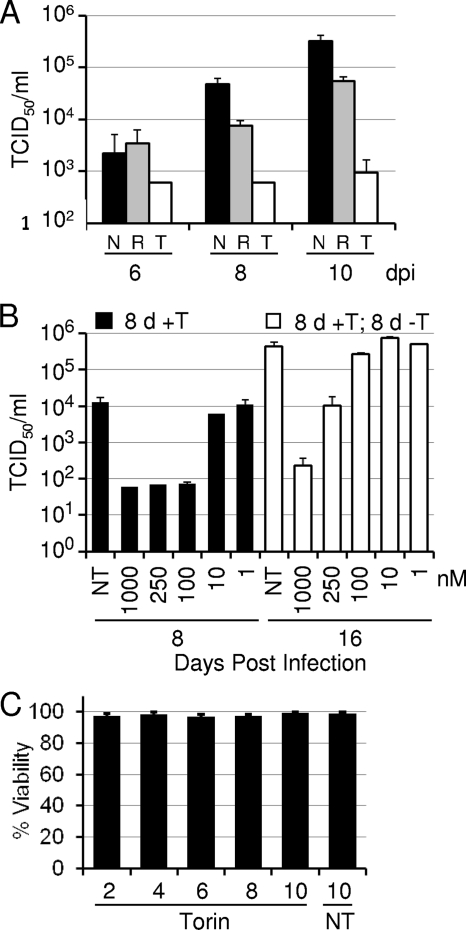
We first determined the effects of the newly developed mTOR inhibitor Torin1 on HCMV replication. Fibroblasts were growth arrested by serum starvation, infected with HCMV, and treated with either Torin1 or rapamycin, and growth was monitored over multiple rounds of viral replication. As shown in Fig. 1A, rapamycin treatment modestly inhibited HCMV replication, achieving an ∼8-fold effect on day 10, consistent with previous results (24). In contrast, Torin1 reduced the yield of HCMV by a factor of ∼160 on day 10. Torin1 was effective in blocking the production of HCMV progeny over a range of concentrations, with a 50% inhibitory concentration (IC50) of ∼60 nM (8 days postinfection) (Fig. 1B). This dose compares favorably with the IC50s of 2 to 10 nM at which Torin1 inhibits the kinase activities of mTORC1 and mTORC2 (47).
FIG. 1.
HCMV replication is inhibited by Torin1. Serum-starved confluent human fibroblasts were infected with HCMV at a multiplicity of 0.05 PFU/cell. Cell-free virus was quantified by a TCID50 assay, and error bars represent the standard errors of the means from two independent experiments, each performed in duplicate. (A) Torin1 inhibits HCMV replication to a greater extent than does rapamycin. Immediately following viral adsorption, cells were treated with vehicle alone (N) (black bars) (dimethyl sulfoxide [DMSO]), rapamycin (T) (gray bars) (20 nM), or Torin1 (T) (white bars) (250 nM). Supernatants were harvested every other day and replaced with fresh medium containing the appropriate treatment, and virus in the supernatant was assayed on the indicated days. (B) Inhibition of HCMV replication is dose dependent and does not result from cellular toxicity. Infected fibroblasts were treated with various doses of Torin1. Medium with drug was replaced every other day, and virus in the supernatant was assayed on day 8 postinfection (black bars). On day 8, a second set of cultures was washed twice, serum-free medium containing no drug was added to each well, and virus was assayed after an additional 8 days (16 days postinfection) (white bars). (C) Torin1 is not toxic to uninfected human fibroblasts. The viability of fibroblasts treated with Torin1 (250 nM) was monitored over a time course of 10 days by a trypan blue exclusion assay.
Previous reports have shown that although Torin1 substantially blocks cellular proliferation, it does not kill cells at concentrations of up to 500 nM (34, 47). We tested the effect of 250 nM Torin1 on the viability of growth-arrested fibroblasts. Torin1 treatment did not affect the viability of these cells, with more than 95% of the cells remaining viable over 10 days of Torin1 treatment (Fig. 1C). To further confirm that the viral growth defect was not the result of cytotoxicity, we performed a drug release experiment. Infected cells were treated with a range of concentrations of Torin1 for 8 days, after which the cells were maintained in medium lacking Torin1. Eight days later, virus in the supernatant was quantified by the TCID50 method (16 days postinfection) (Fig. 1C). Following the removal of the drug, HCMV replication partially recovered in cultures that had initially received 1 mM drug, substantially recovered in cells that had received 250 nM drug, and completely recovered in cells that had received 100 nM Torin1. The ≥100-fold increase in virus yield after the reversal of an 8-day Torin1 treatment further demonstrates that cells treated with ≤250 nM drug remained viable.
These results demonstrate that Torin1 is a potent inhibitor of HCMV replication. Given data from previous work demonstrating the selectivity of Torin1 for the mTOR kinase and its ability to inhibit rapamycin-resistant mTORC1 activity (47), it is very likely that this mTORC1 activity is critically important for HCMV lytic replication.
Torin1 blocks the accumulation of viral DNA and a late viral protein.
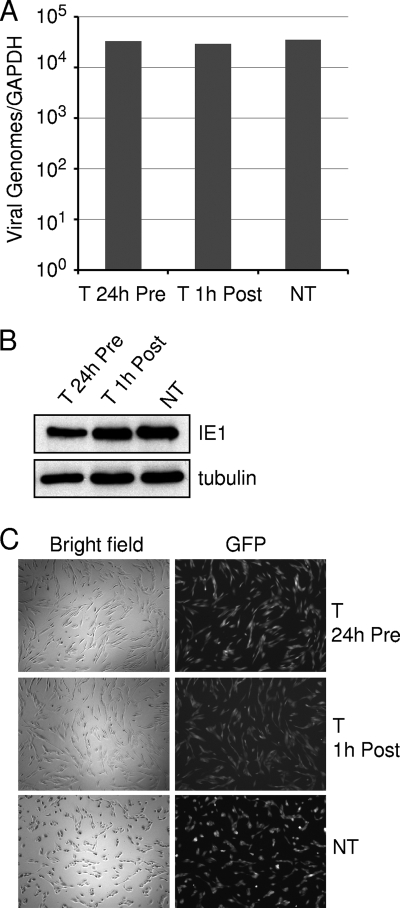
To determine the nature of the blockade in the viral life cycle imposed by Torin1, we initially examined the impact of drug treatment at a dose of 250 nM on HCMV entry. Cells were either pretreated for 24 h with Torin1 or treated with drug immediately following viral adsorption. The level of cell-associated viral DNA at 2 h postinfection (hpi) was not influenced by either drug treatment regimen (Fig. 2A). When the expression of the HCMV immediate-early IE1 protein was examined under these conditions, there was no appreciable difference in the amount of IE1 relative to cell-coded tubulin between the Torin1-treated and untreated cells at 6 hpi (Fig. 2B). Furthermore, drug treatment did not alter the percentage of cells expressing a GFP marker protein expressed from the virus genome at 24 hpi (Fig. 2C). Together, these results demonstrate that the initial steps of the HCMV life cycle, including the binding and entry of the virion and the expression of an immediate-early protein, are not affected by Torin1.
FIG. 2.
Torin1 does not affect HCMV entry into fibroblasts. Serum-starved confluent fibroblasts were infected with HCMV at a multiplicity of 3 PFU/cell. (A) Torin1 does not block the entry of viral DNA. Serum-free confluent fibroblasts were pretreated with Torin1 (T) (250 nM) for 24 h prior to infection (Pre) or beginning immediately after adsorption at 1 hpi (Post). Control cultures received the vehicle in which Torin1 was dissolved (NT). At 2 hpi cells were harvested, and cell-associated viral DNA was quantified by real-time PCR analysis. Error bars represent the standard errors of the means from two independent experiments performed in duplicate. (B) Torin1 does not alter the accumulation of the HCMV IE1 protein. The level of IE1 was determined at 6 hpi by a Western blot assay using an IE1-specific monoclonal antibody. The image is representative of two independent experiments. (C) Torin1 does not alter the percentage of infected cells. The expression of a GFP marker gene present in the viral genome was monitored at 24 h after infection in the presence or absence of drug.
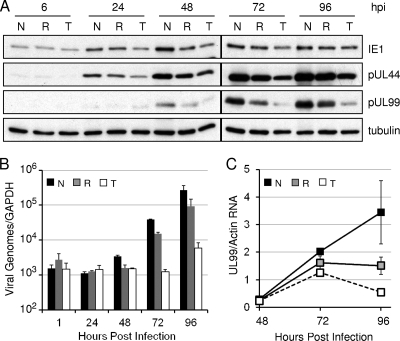
Having established that HCMV enters cells normally in the presence of Torin1, we next determined the effect of Torin1 compared to rapamycin on the accumulation of representative viral proteins from each kinetic class as after infection (Fig. 3A). Rapamycin had little effect on the accumulation of the immediate-early protein IE1 and the early protein pUL44, and it reduced the level of the late protein pUL99 to a modest extent. Torin1 inhibited the accumulation of IE1 and pUL44 to a limited extent, but it dramatically reduced the amount of pUL99. Since the expression of pUL99 is dependent on the initiation of viral DNA replication (9), we tested whether Torin1 inhibits viral DNA accumulation (Fig. 3B). Viral DNA accumulation was measured by quantitative real-time PCR of fibroblasts treated with rapamycin or Torin1. Rapamycin modestly inhibited viral DNA accumulation, consistent with its effect on the production of HCMV progeny. In contrast, Torin1 reduced viral DNA accumulation at 96 hpi by >150-fold. This finding suggested that the inhibition of viral late protein expression reflects a reduced transcription of viral late RNAs due to the inhibition of viral DNA accumulation. To test this hypothesis, we measured the levels of expression of UL99 mRNA in the presence of Torin1 and rapamycin. Both rapamycin and Torin1 decreased the levels of UL99 mRNA, and Torin1 had a greater effect than rapamycin (Fig. 3C). The decreased level of UL99 mRNA in Torin1-treated cells is consistent with the observed inhibition of viral DNA accumulation. The decrease in UL99 protein levels may be more severe than the decrease in UL99 mRNA levels, raising the possibility that mTOR activity might play a role in viral late protein synthesis specifically. However, an interpretation of these results in terms of an effect on late translation is confounded by the drug's effect on DNA accumulation.
FIG. 3.
Torin1 has little effect on the accumulation of an immediate-early protein and an early protein but inhibits the accumulation of HCMV DNA and a late protein. (A) Rapamycin-resistant mTOR activity is required for the accumulation of an some but not all HCMV proteins. Serum-starved confluent fibroblasts were infected with HCMV at a multiplicity of 3 PFU/cell and then incubated with vehicle (N) (DMSO), rapamycin (R) (20 nM), or Torin1 (T) (250 nM) immediately following adsorption. Cells were harvested at the indicated times, and the accumulation of the indicated proteins was analyzed by Western blotting. (B) Torin1 inhibits HCMV DNA accumulation. Serum-starved confluent human fibroblasts were infected with HCMV at a multiplicity of 0.05 PFU/cell and incubated with vehicle, rapamycin, or Torin1 as described above (A). At the indicated times DNA was isolated, and viral DNA was quantified by qPCR. Equivalent amounts of DNA were analyzed for each sample, and the results are normalized to the level of actin DNA per sample. (C) The levels of the viral late transcript UL99 are inhibited by Torin1 treatment. Fibroblasts were infected with HCMV at a multiplicity of 3 PFU/cell and treated with vehicle, rapamycin, or Torin1 as described above (A). At the indicated times the amount of UL99 RNA was determined by qPCR, and the results are normalized to the amount of actin RNA in each sample.
In sum, these results demonstrate that a rapamycin-insensitive mTOR activity is required for efficient HCMV DNA accumulation but is dispensable for the expression of viral immediate-early and early proteins.
Torin1 blocks the phosphorylation of 4EBP1 within HCMV-infected cells.
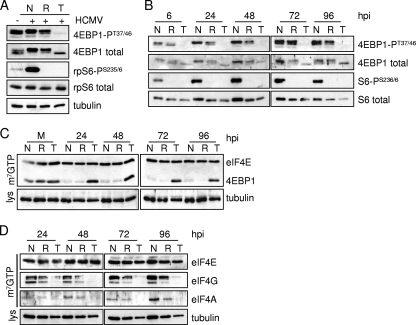
We next investigated the effect of Torin1 on the phosphorylation of mTORC1 targets during HCMV infection. HCMV infection induces mTORC1 activity, but the phosphorylation of mTORC1 targets is differentially sensitive to the mTORC1 inhibitor rapamycin (24, 53). While the mTORC1 phosphorylation of p70 S6 kinase, and its subsequent phosphorylation of rpS6, is inhibited by rapamycin during HCMV infection (21, 24, 53), the phosphorylation of another mTORC1 target, 4EBP1, is resistant to rapamycin (24). This differential effect on mTOR targets could indicate that a kinase other than mTOR is responsible for 4EBP1 phosphorylation during infection. To test this possibility, we treated infected fibroblasts with rapamycin or Torin1 and assessed the phosphorylation status of 4EBP1 and rpS6. Both drugs markedly inhibited the induction of rpS6 phosphorylation that is normally observed during HCMV infection, but only Torin1 substantially blocked the phosphorylation of 4EBP1 (Fig. 4A). This was evident both by the failure to detect phosphorylated 4EBP1-PT37/46 by using an antibody specific for the phosphoform and by the altered migration of total 4EBP1 in the presence of the drug. Total 4EBP1, rpS6, and tubulin levels were monitored to control for protein recovery. The differential effects of the drugs were observed throughout the course of infection (Fig. 4B). These results demonstrate that the rapamycin-resistant phosphorylation of 4EBP1 during HCMV infection is dependent on Torin1-sensitive mTOR activity rather than the action of another kinase.
FIG. 4.
Rapamycin-resistant mTOR activity is required for 4EBP1 phosphorylation and eIF4F complex integrity during HCMV infection. Serum-starved confluent human fibroblasts were infected with HCMV at a multiplicity of 3 PFU/cell. At 1 hpi, cultures were treated with the vehicle in which drugs were dissolved (N) (DMSO), rapamycin (R) (20 nM), or Torin1 (T) (250 nM). (A) At 48 hpi the phosphorylation status of mTORC1 targets was assessed by Western blot assay by using antibodies to phosphorylated targets (4EBP1-PT37/46 and rpS6-PS235/6) and total proteins. Tubulin was assayed as a loading control. (B) Same as above (A) except that cells were harvested at the indicated times. (C and D) After mock infection (M) or infection with HCMV (WT) at a multiplicity of 3 PFU/cell, cultures were harvested at the indicated times. Equivalent amounts of protein from each sample were incubated with m7GTP-Sepharose, and the isolated protein complexes were analyzed by Western blotting using the indicated antibodies to the eIF4F complex and 4EBP1. In all cases the results are representative of at least two independent experiments. lys, lysate.
The phosphorylation status of 4EBP1 regulates cap-dependent protein translation. Hypophosphorylated 4EBP1 binds to eIF4E and inhibits the formation of the eIF4F complex, while the phosphorylation of 4EBP1 inhibits its interaction with eIF4E (43). The ability of Torin1 to markedly inhibit 4EBP1 phosphorylation led us to examine the levels of the intact eIF4F complex in Torin1-treated cells. HCMV infection caused a decreased association of 4EBP1 with an analog of the m7G cap, m7GTP-Sepharose, throughout the course of infection (Fig. 4C), as was previously described (53). Rapamycin treatment did not increase the amount of 4EBP1 associated with the cap analog, consistent with its inability to block 4EBP1 phosphorylation during infection. In contrast, Torin1 treatment resulted in a substantially increased association of 4EBP1 with m7GTP-Sepharose throughout infection. HCMV infection did not alter the association of eIF4E with the cap analog, and this served as a loading control. In addition, the amount of tubulin in cell lysates was assayed to confirm that equal amounts of protein in each sample were loaded onto the cap analog. The increased level of 4EBP1 associated with m7GTP-Sepharose was consistent with the reduced association of eIF4G and eIF4A with the cap analog following Torin1 treatment (Fig. 4D). Rapamycin had minimal effects on the binding of eIF4G and eIF4A. Again, eIF4E levels were not affected by the drug and served as a loading control. These results indicate that the phosphorylation of 4EBP1 by rapamycin-resistant mTOR is required to maintain the integrity of the eIF4F complex during HCMV infection.
Torin1 does not block MCMV replication in 4EBP1-null cells.
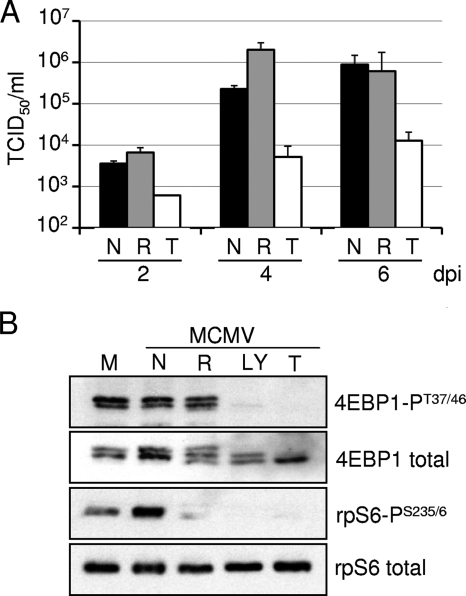
The identification of the functional roles of proteins in the mTOR signaling pathway has been facilitated by the generation of knockout mouse strains lacking individual mTOR components. For example, the availability of murine embryonic fibroblasts (MEFs) lacking the essential mTORC2 component Rictor led to the definitive identification of mTORC2 as the kinase complex responsible for the complete activation of Akt (16, 40). We used murine cytomegalovirus (MCMV) and several MEF lines deficient for mTOR signaling pathway components to test for a possible contribution of mTORC2 to rapamycin-resistant phosphorylation events. To confirm that MCMV behaves like HCMV and is a suitable model for the analysis of mTOR signaling events, we determined the effect of Torin1 and rapamycin on MCMV growth and mTOR-dependent phosphorylation events in MEFs. As was the case for HCMV, Torin1, but not rapamycin, inhibited MCMV replication (Fig. 5A). Indeed, although rapamycin reduced the yield of HCMV to a modest extent (Fig. 1A), it had no inhibitory effect on MCMV. Also as observed for HCMV (Fig. 4A), MCMV infection induced mTORC1 activity, as measured by the increased phosphorylation of rpS6. The phosphorylation of rpS6 was completely inhibited by rapamycin, Torin1, and LY294002, an inhibitor of class 1 phosphatidylinositol 3-kinase and mTOR (44), whereas the phosphorylation of 4EBP1 was inhibited by Torin1 and LY294002 but not rapamycin (Fig. 5B). Total rpS6 protein was assayed and served as a loading control. Like HCMV, MCMV induces the mTOR signaling pathway, and it depends on rapamycin-resistant mTOR activity to induce the phosphorylation of 4EBP1.
FIG. 5.
MCMV replication is inhibited by Torin1. (A) Torin1 but not rapamycin inhibits the production of MCMV progeny. MEFs were infected with MCMV at a multiplicity of 0.05 PFU/cell and treated with vehicle (black bars) (DMSO), rapamycin (gray bars) (20 nM), or Torin1 (white bars) (250 nM). Fresh serum-free medium containing drugs was added every other day. At the indicated times, cell-free supernatants were harvested, and the amount of virus in the supernatant was quantified by the TCID50 method. Error bars represent the standard errors of the means form two independent experiments performed in duplicate. (B) MEFs were infected with MCMV at a multiplicity of 3 PFU/cell and treated with vehicle (N), rapamycin (R), or Torin1 (T) as described above (A) or were treated with LY294002 (LY) (20 μM). At 48 hpi the phosphorylation state of the indicated mTORC1 targets was analyzed by a Western blot assay by using antibodies to phosphorylated targets (4EBP1-PT37/46 and rpS6-PS235/6) and total proteins. The results are representative of three independent experiments.
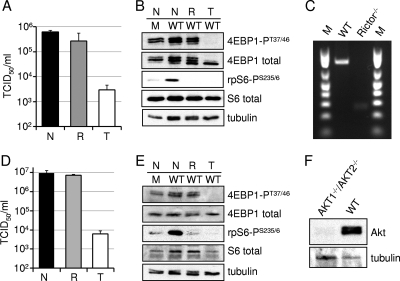
Having established that MCMV induces a set of mTOR signaling events similar to that of HCMV, we characterized the effect of Torin1 and rapamycin treatment on MEFs deficient for various effectors of mTOR action. We first investigated the requirement for mTORC2 for the replication of MCMV. Rictor-null MEFs (40) supported viral growth (no treatment) (Fig. 6A), demonstrating that mTORC2 is not required for efficient MCMV replication. Furthermore, Torin1 effectively inhibited MCMV replication (Fig. 6A) and 4EBP1 phosphorylation (Fig. 6B) in these cells, arguing that mTORC2 is not the target for Torin1 in MCMV-infected cells. Finally, the cells were confirmed to lack an intact Rictor locus when assayed by PCR (Fig. 6C). We also employed Akt1/Akt2-null MEFs (32) to evaluate a possible role for the Akt kinase, one of the targets of mTORC2. These cells supported Torin-sensitive MCMV replication (Fig. 6D), and Torin1 inhibited 4EBP1 phosphorylation in the absence of Akt (Fig. 6E), ruling out this kinase as the Torin1 target in MCMV-infected cells. Again, the cells were confirmed to lack Akt by Western blot assay (Fig. 6F).
FIG. 6.
mTORC2 and its target Akt are not the source of rapamycin-resistant mTOR activity. (A) MCMV growth is inhibited by Torin1 in Rictor-null MEFs. Confluent serum-starved cells were infected with MCMV at a multiplicity of 0.05 PFU/cell, and vehicle (black bars) (DMSO), rapamycin (gray bars) (20 nM), or Torin1 (white bars) (250 nM) was added at 1 hpi. At 6 days postinfection the amount of MCMV in cell-free supernatants was determined by the TCID50 method. (B) Torin1 blocks 4EBP1 phosphorylation in Rictor-null MEFs. MEFs were mock infected (M) or infected with MCMV (WT) at a multiplicity of 3 PFU/cell and treated with vehicle (N), rapamycin (R), or Torin1 (T) as described above (A). At 48 hpi, the phosphorylation state of mTORC1 targets was assessed by Western blotting using antibodies specific for the indicated proteins. (C) Confirmation of the genotype of Rictor-null MEFs. Total DNA was isolated from wild-type and Rictor-null MEFs, and the genotype was confirmed by use of PCR. (D) Same as above (A) except that Akt1- and Akt2-null MEFs were used. (E) Same as above (B) except that Akt1- and Akt2-null MEFs were used. For B and E, the error bars represent the standard errors of the means from at least two independent experiments, each performed in duplicate. For C and E, tubulin was assayed as a loading control. (F) Akt is not expressed in Akt1- and Akt2-null MEFs. Protein from wild-type or mutant MEFs was analyzed by Western blotting by use of an antibody specific for Akt.
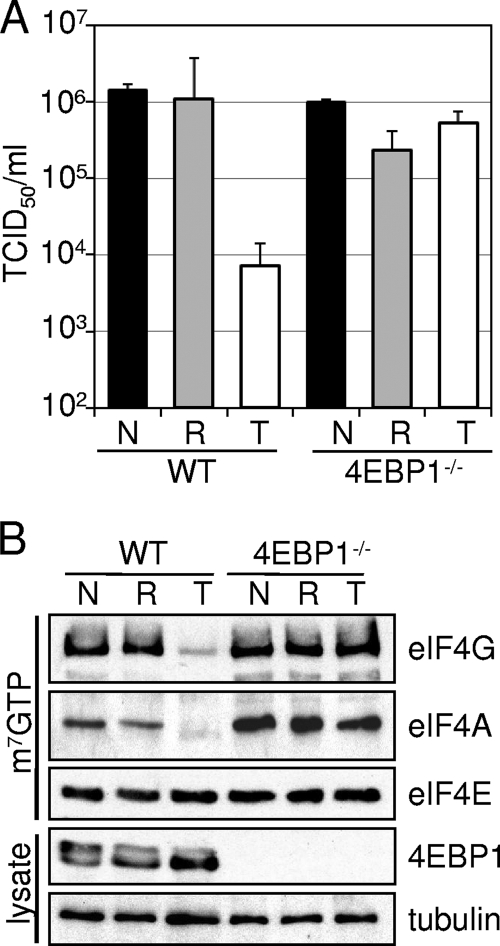
The inhibition of HCMV replication by Torin1 correlated with the hypophosphorylation of 4EBP1 (Fig. 3 and 4), suggesting that this phosphorylation event might be the critical Torin1 target. Accordingly, we tested the ability of Torin1 to inhibit MCMV replication in 4EBP1-null MEFs (48). MCMV replicated as well in these cells as in normal MEFs, indicating that 4EBP1 is not required for cytomegalovirus replication (4EBP1−/−, no treatment) (Fig. 7A). As in control cells, rapamycin had a minimal impact on MCMV replication in 4EBP1-null cells. Importantly, Torin1 was no longer capable of inhibiting MCMV replication in cells lacking 4EBP1 (Fig. 7A). As discussed above, 4EBP1 functions to inhibit eIF4F complex assembly unless inactivated by mTORC1-mediated phosphorylation (38). While Torin1 treatment inhibited the formation of the eIF4F complex in control cells, no such effect was observed for 4EBP1-null cells (Fig. 7B). Finally, our failure to detect 4EBP1 in lysates of these cells by Western blot assay confirmed the phenotype of the MEFs. We conclude that 4EBP1 is the critical target providing sensitivity to Torin1 during cytomegalovirus infection, and we propose that rapamycin-resistant mTORC1 is required for the maintenance of cap-dependent translation during the viral life cycle. This proposal is consistent with previous work showing that the phosphorylation of 4EBP1 in uninfected cells is resistant to rapamycin but inhibited by Torin1 (47).
FIG. 7.
Deletion of the mTORC1 target 4EBP1 rescues replication of MCMV in the presence of Torin1. (A) MCMV growth is not inhibited by Torin1 in 4EBP1-null MEFs. Confluent serum-starved cells were infected with MCMV at a multiplicity of 0.05 PFU/cell, and vehicle (black bars) (DMSO), rapamycin (gray bars) (20 nM), or Torin1 (white bars) (250 nM) was added at 1 hpi. At 6 days postinfection the amount of MCMV in cell-free supernatants was determined by the TCID50 method. The error bars represent the standard errors of the means from three independent experiments, each performed in duplicate. (B) Torin1 does not exclude eIF4G or eIF4A from the cap-binding complex in 4EBP1-null MEFs. Cells were infected with MCMV at a multiplicity of 3 PFU/cell and treated with vehicle (N), rapamycin (R), or Torin1 (T) as described above (A). At 48 hpi equal amounts of protein from cell lysates were incubated with m7G-Sepharose. The presence of eIF4F complex components bound by the cap analog was determined by Western blotting. The results are representative of two independent experiments.
Members of all three herpesvirus subfamilies are inhibited by Torin1.
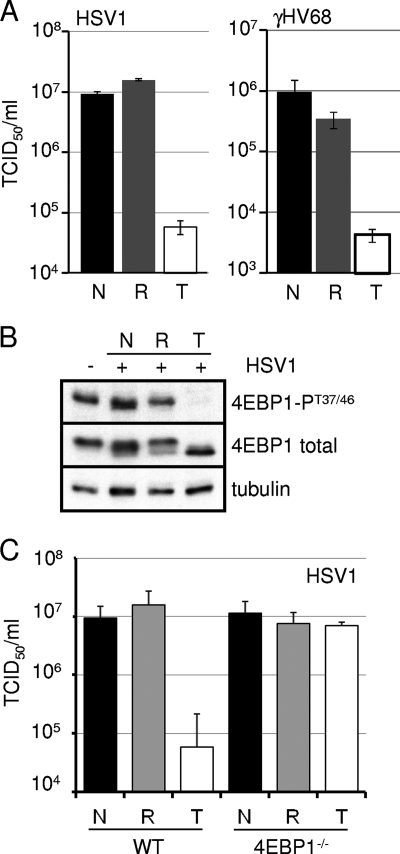
We were curious to determine if the requirement for rapamycin-resistant mTORC1 activity was unique to cytomegaloviruses, which are betaherpesviruses, or if it was a general feature of herpesvirus infection. We infected MEFs with the alphaherpesvirus herpes simplex virus type 1 (HSV-1) and the gammaherpesvirus murine gammaherpesvirus 68 (γHV68) (Fig. 8A). These viruses exhibited the same drug sensitivities as the cytomegaloviruses. While rapamycin was ineffective at preventing HSV-1 and γHV68 replication, Torin1 inhibited both viruses over multiple rounds of viral replication. In addition, Torin1, but not rapamycin, inhibited the phosphorylation of 4EBP1 during HSV-1 infection (Fig. 8B), and Torin1 failed to inhibit the production of HSV-1 in cells lacking 4EBP1 (Fig. 8C). We conclude that rapamycin-resistant mTOR activity is required for the replication of multiple herpesviruses.
FIG. 8.
Rapamycin-resistant mTOR activity is required for lytic replication by representative alpha- and gammaherpesviruses. (A) Confluent serum-starved MEFs were infected at a multiplicity of 0.05 PFU/cell with HSV-1 or γHV68. The amount of virus in cell-free supernatants was determined by the TCID50 method at 72 hpi for HSV-1 (left) and at 6 days postinfection for γHV68 (right). Black bars represent vehicle-treated samples (N) (DMSO), gray bars represent rapamycin-treated samples (R) (20 nM), and white bars represent Torin1-treated samples (T) (250 nM). The error bars represent the standard errors of the means from at least two independent experiments. (B) Confluent MEF monolayers were infected with HSV-1 at a multiplicity of 3 PFU/cell. Infected cell lysates were harvested at 8 hpi, and equal amounts of protein were analyzed by Western blotting. (C) WT or 4EBP1-null MEFs were infected with HSV-1 at a multiplicity of 0.05 PFU/cell. The amount of cell-free virus present in the supernatant at 72 hpi was quantified by the TCID50 method. The error bars represent the standard errors of the means from two independent experiments.
DISCUSSION
Using Torin1, a selective, ATP-competitive mTOR inhibitor (47), we show that rapamycin-resistant mTORC1 activity is required for herpesvirus growth in fibroblasts. The inhibition of mTOR kinase with Torin1 results in decreased HCMV DNA accumulation (Fig. 3C), and consequently, Torin1 inhibits the accumulation of at least some HCMV late proteins (Fig. 3A). Despite the disruption of the eIF4F complex (Fig. 4), the viral immediate-early protein IE1 and the early protein pUL44 were translated at nearly normal efficiencies during infection (Fig. 3A), suggesting that eIF4F- and mTOR-independent events mediate the translation of at least some viral proteins.
MCMV infection of MEFs induces the rapamycin-resistant phosphorylation of 4EBP1, and Torin1 inhibits 4EBP1 phosphorylation and MCMV growth (Fig. 5), demonstrating that MCMV is a suitable model system with which to study mTOR signaling events. Using MEFs deficient for Rictor or Akt1 and Akt2, we show that MCMV does not require mTORC2 signaling for viral replication and that neither mTORC2 nor Akt is required for the rapamycin-resistant phosphorylation of 4EBP1 (Fig. 6). Rather, we find that the regulation of 4EBP1 activity by rapamycin-resistant mTORC1 is critical for MCMV replication, as Torin1 was incapable of inhibiting MCMV growth in 4EBP1-null fibroblasts (Fig. 7). Using HSV1 and γHV68, representative members of the alpha- and gammaherpesvirus families, respectively (Fig. 8), we demonstrate that the requirement for rapamycin-resistant mTOR activity is conserved among representatives of all three herpesvirus subfamilies.
The activation of mTOR by herpesvirus infection was documented previously (2, 6, 21, 30, 52, 55). We show that the mTOR activity required for herpesvirus replication is mediated by mTORC1 and that it is relatively insensitive to rapamycin. This conclusion is based on several observations: (i) previous work has shown that Torin1 suppresses mTORC1 function more completely than rapamycin (47); (ii) Torin1 inhibits the rapamycin-resistant phosphorylation of 4EBP1, a well-known mTORC1 target, within HCMV-infected cells (Fig. 3); (iii) neither the mTORC2 complex nor its substrate Akt was required for rapamycin-resistant 4EBP1 phosphorylation in MCMV-infected cells (Fig. 6); and (iv) Torin1 did not inhibit MCMV replication or the assembly of the eIF4F cap-binding complex within cells lacking 4EBP1, a known target of mTORC1 (Fig. 7). Currently, the nature of this rapamycin-resistant mTORC1 activity is unknown. Rapamycin binds to its intracellular receptor FKBP12 (25), which then binds to the mTOR kinase within the mTORC1 complex (7, 22). There might be a population of mTORC1 in which the formation of the mTOR-rapamycin-FKBP12 trimer is unable to inhibit mTORC1 activity, perhaps due to the presence of additional proteins in the complex. Alternatively, herpesviruses might encode proteins that specifically act on or interact with the mTORC1 complex to generate a subset of mTORC1 complexes in which rapamycin is no longer effective. It is also possible that there is another uncharacterized mTOR complex that phosphorylates mTORC1 targets and does not require Raptor for its function.
We have found that rapamycin-resistant mTORC1 activity is required for HCMV DNA accumulation. How might mTORC1 regulate viral DNA synthesis? It is possible that a viral protein that is required for HCMV DNA replication requires rapamycin-resistant mTORC1 for its expression. While the viral immediate-early and early proteins assayed in this study are expressed in the absence of rapamycin-resistant mTORC1 activity, these results do not exclude the possibility that another viral protein required for the accumulation of viral DNA is sensitive to Torin1 and dependent upon eIF4F for its translation. Alternatively, the inhibition of eIF4F-dependent translation by Torin1 may result in the decreased expression of one or more cellular proteins necessary for viral DNA replication. This possibility is consistent with the observation that Torin1 treatment inhibited the replication of representative members of all three classes of herpesviruses (Fig. 8). Finally, it is conceivable that mTORC1 facilitates DNA replication by the direct phosphorylation of a viral or cellular protein. Further experiments to distinguish among these possibilities will be needed to precisely define the role of mTORC1 activity in promoting HCMV DNA replication.
It is interesting that the accumulation of the HCMV immediate-early protein IE1 and the early protein pUL44 was minimally affected by Torin1 treatment as late as 96 hpi (Fig. 3A) despite the disruption of the eIF4F complex beginning at 24 hpi (Fig. 4D). Since the eIF4F complex is required for the efficient translation of capped mRNAs, we had anticipated that Torin1 treatment would prevent the synthesis of viral proteins. pUL44 very clearly continues to accumulate after 24 hpi, when Torin1 has excluded a significant portion of eIF4A and eIF4G from the complex. Possibly, the eIF4F complex is not completely disrupted during the immediate-early and early phases of HCMV gene expression, and viral mRNAs compete effectively for the limited amount of complex that remains. It is also conceivable that the translation of HCMV proteins is mediated by another, as-yet-undefined, mechanism. How might HCMV achieve the eIF4F-independent synthesis of a portion of its proteins? The host response to viral infection typically includes the inhibition of protein synthesis (29). Viruses have evolved multiple means by which to maintain protein translation under these conditions to facilitate the expression of viral proteins (39). There is precedent in the herpesvirus family for the regulation of translational initiation. The ribonucleotide reductase homolog of herpes simplex virus, ICP6, binds to the eIF4F complex. ICP6 promotes eIF4F complex assembly by binding to its eIF4G subunit (51). Similar events may take place during HCMV infection, which could conceivably allow for the translation of viral proteins in the absence of a complete eIF4F complex. HCMV may also encode proteins that act to promote the translation of viral proteins in the complete absence of eIF4F. The finding that HCMV proteins are translated under conditions that disrupt eIF4F assembly provides a system with which to search for the cellular and viral proteins critical for continued protein translation in response to virus-induced stress.
Finally, we have discovered that rapamycin-resistant mTOR activity appears to be a common requirement for herpesvirus lytic replication (Fig. 1 and 8). This finding, together with previous work showing that rapamycin limits the scope and severity of beta- and gammaherpesvirus disease in immunocompromised patients (6, 19, 26), suggests that drugs like Torin1, which target the catalytic activity of mTOR, may be useful as broad-spectrum antiviral agents for the treatment of herpesvirus disease.
Acknowledgments
We thank members of our laboratory for helpful comments, criticisms, and discussions regarding this work. We are grateful to N. Gray (Harvard Medical School) for the gift of Torin1, N. Hay (University of Illinois at Chicago) for Akt1/2-null MEFs, M. Magnuson (Vanderbilt University) for Rictor-null MEFs, and N. Sonenberg (McGill University) for 4EBP1-null MEFs.
This work was supported by a grant from the U.S. National Institutes of Health to T.S. (CA85786) and an American Cancer Society postdoctoral fellowship (PF-07073-01-MBC) to N.J.M.
Footnotes
Published ahead of print on 24 February 2010.
REFERENCES
- 1.Alwine, J. C. 2008. Modulation of host cell stress responses by human cytomegalovirus. Curr. Top. Microbiol. Immunol. 325:263-279. [DOI] [PubMed] [Google Scholar]
- 2.Arias, C., D. Walsh, J. Harbell, A. C. Wilson, and I. Mohr. 2009. Activation of host translational control pathways by a viral developmental switch. PLoS Pathog. 5:e1000334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buchkovich, N. J., Y. Yu, C. A. Zampieri, and J. C. Alwine. 2008. The TORrid affairs of viruses: effects of mammalian DNA viruses on the PI3K-Akt-mTOR signalling pathway. Nat. Rev. Microbiol. 6:266-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burnett, P. E., R. K. Barrow, N. A. Cohen, S. H. Snyder, and D. M. Sabatini. 1998. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc. Natl. Acad. Sci. U. S. A. 95:1432-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cassady, K. A., M. Gross, and B. Roizman. 1998. The second-site mutation in the herpes simplex virus recombinants lacking the gamma134.5 genes precludes shutoff of protein synthesis by blocking the phosphorylation of eIF-2alpha. J. Virol. 72:7005-7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chaisuparat, R., J. Hu, B. C. Jham, Z. A. Knight, K. M. Shokat, and S. Montaner. 2008. Dual inhibition of PI3Kalpha and mTOR as an alternative treatment for Kaposi's sarcoma. Cancer Res. 68:8361-8368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choi, J., J. Chen, S. L. Schreiber, and J. Clardy. 1996. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science 273:239-242. [DOI] [PubMed] [Google Scholar]
- 8.Cuevas-Bennett, C., and T. Shenk. 2008. Dynamic histone H3 acetylation and methylation at human cytomegalovirus promoters during replication in fibroblasts. J. Virol. 82:9525-9536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Depto, A. S., and R. M. Stenberg. 1992. Functional analysis of the true late human cytomegalovirus pp28 upstream promoter: cis-acting elements and viral trans-acting proteins necessary for promoter activation. J. Virol. 66:3241-3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duff, R., and F. Rapp. 1973. Oncogenic transformation of hamster embryo cells after exposure to inactivated herpes simplex virus type 1. J. Virol. 12:209-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dufner, A., and G. Thomas. 1999. Ribosomal S6 kinase signaling and the control of translation. Exp. Cell Res. 253:100-109. [DOI] [PubMed] [Google Scholar]
- 12.Fadden, P., T. A. Haystead, and J. C. Lawrence, Jr. 1998. Phosphorylation of the translational regulator, PHAS-I, by protein kinase CK2. FEBS Lett. 435:105-109. [DOI] [PubMed] [Google Scholar]
- 13.Gale, M., Jr., S. L. Tan, and M. G. Katze. 2000. Translational control of viral gene expression in eukaryotes. Microbiol. Mol. Biol. Rev. 64:239-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gingras, A. C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913-963. [DOI] [PubMed] [Google Scholar]
- 15.Gingras, A. C., B. Raught, and N. Sonenberg. 2004. mTOR signaling to translation. Curr. Top. Microbiol. Immunol. 279:169-197. [DOI] [PubMed] [Google Scholar]
- 16.Guertin, D. A., D. M. Stevens, C. C. Thoreen, A. A. Burds, N. Y. Kalaany, J. Moffat, M. Brown, K. J. Fitzgerald, and D. M. Sabatini. 2006. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev. Cell 11:859-871. [DOI] [PubMed] [Google Scholar]
- 17.Hakki, M., E. E. Marshall, K. L. De Niro, and A. P. Geballe. 2006. Binding and nuclear relocalization of protein kinase R by human cytomegalovirus TRS1. J. Virol. 80:11817-11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haystead, T. A., C. M. Haystead, C. Hu, T. A. Lin, and J. C. Lawrence, Jr. 1994. Phosphorylation of PHAS-I by mitogen-activated protein (MAP) kinase. Identification of a site phosphorylated by MAP kinase in vitro and in response to insulin in rat adipocytes. J. Biol. Chem. 269:23185-23191. [PubMed] [Google Scholar]
- 19.Hill, J. A., M. Hummel, R. C. Starling, J. A. Kobashigawa, S. V. Perrone, J. M. Arizon, S. Simonsen, K. H. Abeywickrama, and C. Bara. 2007. A lower incidence of cytomegalovirus infection in de novo heart transplant recipients randomized to everolimus. Transplantation 84:1436-1442. [DOI] [PubMed] [Google Scholar]
- 20.Inoki, K., Y. Li, T. Zhu, J. Wu, and K. L. Guan. 2002. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 4:648-657. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, R. A., X. Wang, X. L. Ma, S. M. Huong, and E. S. Huang. 2001. Human cytomegalovirus up-regulates the phosphatidylinositol 3-kinase (PI3-K) pathway: inhibition of PI3-K activity inhibits viral replication and virus-induced signaling. J. Virol. 75:6022-6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, D. H., D. D. Sarbassov, S. M. Ali, J. E. King, R. R. Latek, H. Erdjument-Bromage, P. Tempst, and D. M. Sabatini. 2002. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110:163-175. [DOI] [PubMed] [Google Scholar]
- 23.Kudchodkar, S. B., Y. Yu, T. G. Maguire, and J. C. Alwine. 2006. Human cytomegalovirus infection alters the substrate specificities and rapamycin sensitivities of raptor- and rictor-containing complexes. Proc. Natl. Acad. Sci. U. S. A. 103:14182-14187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kudchodkar, S. B., Y. Yu, T. G. Maguire, and J. C. Alwine. 2004. Human cytomegalovirus infection induces rapamycin-insensitive phosphorylation of downstream effectors of mTOR kinase. J. Virol. 78:11030-11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lane, W. S., A. Galat, M. W. Harding, and S. L. Schreiber. 1991. Complete amino acid sequence of the FK506 and rapamycin binding protein, FKBP, isolated from calf thymus. J. Protein Chem. 10:151-160. [DOI] [PubMed] [Google Scholar]
- 26.Marty, F. M., J. Bryar, S. K. Browne, T. Schwarzberg, V. T. Ho, I. V. Bassett, J. Koreth, E. P. Alyea, R. J. Soiffer, C. S. Cutler, J. H. Antin, and L. R. Baden. 2007. Sirolimus-based graft-versus-host disease prophylaxis protects against cytomegalovirus reactivation after allogeneic hematopoietic stem cell transplantation: a cohort analysis. Blood 110:490-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mayer, C., and I. Grummt. 2006. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene 25:6384-6391. [DOI] [PubMed] [Google Scholar]
- 28.Mohr, I. 2004. Neutralizing innate host defenses to control viral translation in HSV-1 infected cells. Int. Rev. Immunol. 23:199-220. [DOI] [PubMed] [Google Scholar]
- 29.Mohr, I. 2006. Phosphorylation and dephosphorylation events that regulate viral mRNA translation. Virus Res. 119:89-99. [DOI] [PubMed] [Google Scholar]
- 30.Moody, C. A., R. S. Scott, N. Amirghahari, C. O. Nathan, L. S. Young, C. W. Dawson, and J. W. Sixbey. 2005. Modulation of the cell growth regulator mTOR by Epstein-Barr virus-encoded LMP2A. J. Virol. 79:5499-5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moorman, N. J., I. M. Cristea, S. S. Terhune, M. P. Rout, B. T. Chait, and T. Shenk. 2008. Human cytomegalovirus protein UL38 inhibits host cell stress responses by antagonizing the tuberous sclerosis protein complex. Cell Host Microbe 3:253-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng, X. D., P. Z. Xu, M. L. Chen, A. Hahn-Windgassen, J. Skeen, J. Jacobs, D. Sundararajan, W. S. Chen, S. E. Crawford, K. G. Coleman, and N. Hay. 2003. Dwarfism, impaired skin development, skeletal muscle atrophy, delayed bone development, and impeded adipogenesis in mice lacking Akt1 and Akt2. Genes Dev. 17:1352-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peters, G. A., D. Khoo, I. Mohr, and G. C. Sen. 2002. Inhibition of PACT-mediated activation of PKR by the herpes simplex virus type 1 Us11 protein. J. Virol. 76:11054-11064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peterson, T. R., M. Laplante, C. C. Thoreen, Y. Sancak, S. A. Kang, W. M. Kuehl, N. S. Gray, and D. M. Sabatini. 2009. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell 137:873-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poppers, J., M. Mulvey, D. Khoo, and I. Mohr. 2000. Inhibition of PKR activation by the proline-rich RNA binding domain of the herpes simplex virus type 1 Us11 protein. J. Virol. 74:11215-11221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poppers, J., M. Mulvey, C. Perez, D. Khoo, and I. Mohr. 2003. Identification of a lytic-cycle Epstein-Barr virus gene product that can regulate PKR activation. J. Virol. 77:228-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Proud, C. G. 2002. Regulation of mammalian translation factors by nutrients. Eur. J. Biochem. 269:5338-5349. [DOI] [PubMed] [Google Scholar]
- 38.Richter, J. D., and N. Sonenberg. 2005. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433:477-480. [DOI] [PubMed] [Google Scholar]
- 39.Schneider, R. J., and I. Mohr. 2003. Translation initiation and viral tricks. Trends Biochem. Sci. 28:130-136. [DOI] [PubMed] [Google Scholar]
- 40.Shiota, C., J. T. Woo, J. Lindner, K. D. Shelton, and M. A. Magnuson. 2006. Multiallelic disruption of the rictor gene in mice reveals that mTOR complex 2 is essential for fetal growth and viability. Dev. Cell 11:583-589. [DOI] [PubMed] [Google Scholar]
- 41.Shor, B., J. J. Gibbons, R. T. Abraham, and K. Yu. 2009. Targeting mTOR globally in cancer: thinking beyond rapamycin. Cell Cycle 8:3831-3837. [DOI] [PubMed] [Google Scholar]
- 42.Silva, M. C., Q. C. Yu, L. Enquist, and T. Shenk. 2003. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J. Virol. 77:10594-10605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sonenberg, N., and T. E. Dever. 2003. Eukaryotic translation initiation factors and regulators. Curr. Opin. Struct. Biol. 13:56-63. [DOI] [PubMed] [Google Scholar]
- 44.Takeuchi, H., Y. Kondo, K. Fujiwara, T. Kanzawa, H. Aoki, G. B. Mills, and S. Kondo. 2005. Synergistic augmentation of rapamycin-induced autophagy in malignant glioma cells by phosphatidylinositol 3-kinase/protein kinase B inhibitors. Cancer Res. 65:3336-3346. [DOI] [PubMed] [Google Scholar]
- 45.Tee, A. R., D. C. Fingar, B. D. Manning, D. J. Kwiatkowski, L. C. Cantley, and J. Blenis. 2002. Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc. Natl. Acad. Sci. U. S. A. 99:13571-13576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Terhune, S., E. Torigoi, N. Moorman, M. Silva, Z. Qian, T. Shenk, and D. Yu. 2007. Human cytomegalovirus UL38 protein blocks apoptosis. J. Virol. 81:3109-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thoreen, C. C., S. A. Kang, J. W. Chang, Q. Liu, J. Zhang, Y. Gao, L. J. Reichling, T. Sim, D. M. Sabatini, and N. S. Gray. 2009. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 284:8023-8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsukiyama-Kohara, K., F. Poulin, M. Kohara, C. T. DeMaria, A. Cheng, Z. Wu, A. C. Gingras, A. Katsume, M. Elchebly, B. M. Spiegelman, M. E. Harper, M. L. Tremblay, and N. Sonenberg. 2001. Adipose tissue reduction in mice lacking the translational inhibitor 4E-BP1. Nat. Med. 7:1128-1132. [DOI] [PubMed] [Google Scholar]
- 49.Virgin, H. W., IV, P. Latreille, P. Wamsley, K. Hallsworth, K. E. Weck, A. J. Dal Canto, and S. H. Speck. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 71:5894-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wagner, M., S. Jonjic, U. H. Koszinowski, and M. Messerle. 1999. Systematic excision of vector sequences from the BAC-cloned herpesvirus genome during virus reconstitution. J. Virol. 73:7056-7060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walsh, D., and I. Mohr. 2006. Assembly of an active translation initiation factor complex by a viral protein. Genes Dev. 20:461-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walsh, D., and I. Mohr. 2004. Phosphorylation of eIF4E by Mnk-1 enhances HSV-1 translation and replication in quiescent cells. Genes Dev. 18:660-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walsh, D., C. Perez, J. Notary, and I. Mohr. 2005. Regulation of the translation initiation factor eIF4F by multiple mechanisms in human cytomegalovirus-infected cells. J. Virol. 79:8057-8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang, D., W. Bresnahan, and T. Shenk. 2004. Human cytomegalovirus encodes a highly specific RANTES decoy receptor. Proc. Natl. Acad. Sci. U. S. A. 101:16642-16647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang, L., and B. Damania. 2008. Kaposi's sarcoma-associated herpesvirus confers a survival advantage to endothelial cells. Cancer Res. 68:4640-4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu, H., Y. Shen, and T. Shenk. 1995. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J. Virol. 69:7960-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]