Abstract
The oncogenic human gammaherpesvirus Kaposi's sarcoma-associated herpesvirus (KSHV) expresses 12 viral microRNAs (miRNAs) in latently infected cells. Here, we report that cellular mRNAs encoding the cellular cyclin-dependent kinase inhibitor p21, a key inducer of cell cycle arrest, are direct targets for KSHV miR-K1. Ectopically expressed KSHV miR-K1 specifically inhibited the expression of endogenous p21 in KSHV-negative cells and strongly attenuated the cell cycle arrest that normally occurs upon p53 activation, yet miR-K1 did not prevent the induction of other p53-responsive genes. Stable knockdown of miR-K1 in latently KSHV-infected human primary effusion lymphoma (PEL) B cells revealed a derepression of p21 expression and enhanced cell cycle arrest following activation of p53. Our data demonstrate that miR-K1 represses the expression of p21, a protein with known tumor suppressor functions, and suggest that this KSHV miRNA is likely to contribute to the oncogenic potential of this opportunistic viral pathogen.
The human-pathogenic gammaherpesvirus Kaposi's sarcoma-associated herpesvirus (KSHV) is the etiological agent of Kaposi's sarcoma and B-cell primary effusion lymphoma (PEL) (5, 6, 14). PEL-derived B-cell lines maintain a latent infection by KSHV in culture, which is characterized by the episomal maintenance of multiple copies of the viral DNA genome and the expression of a small number of viral proteins and 12 viral microRNAs (miRNAs), miR-K1 to miR-K12 (4, 14, 17, 30, 35). The viral latent proteins have been extensively studied and mediate viral genome maintenance (LANA), activate cellular survival signaling (v-FLIP), and drive cell cycle progression (K-cyclin), among other functions (14). The KSHV miRNAs were only recently identified, and much less is known about their roles in KSHV replication and pathogenesis (4, 17, 30, 35). miRNAs are ∼22-nucleotide (nt)-long regulatory RNAs that repress the expression of mRNAs bearing partially complementary target sequences, most commonly in the 3′ untranslated region (UTR) (2). Based on published reports, and consistent with the finding that individual cellular miRNAs can directly repress multiple distinct mRNAs, the KSHV miRNAs may have evolved to target a number of cellular and viral transcripts and thereby regulate several different biological pathways (3, 16, 18, 22, 23, 26, 36, 39, 48). One important functional clue comes from the finding that miR-K11 functions as an ortholog of the cellular miRNA miR-155 (16, 39). The physiological expression of miR-155 is normally tightly regulated during activation of the immune response. Constitutive expression of miR-155 in B cells can lead to the development of B-cell lymphomas (8), and miR-K11 may play an analogous role in the development of KSHV-induced B-cell lymphomas. KSHV miR-K5 was shown to inhibit the expression of Bcl2-associated factor 1 (BCLAF1), possibly leading to reduced apoptosis, as well as an increase in the lytic reactivation of KSHV (48). KSHV miRNAs have also been reported to downregulate expression of thrombospondin 1, a cellular protein with antiangiogenic activity (36). KSHV miR-K7 has been reported to repress the expression of MICB, which mediates recognition of virus-infected cells by natural killer cells, and may thereby help KSHV-infected cells to avoid immune elimination (26). Multiple KSHV miRNAs target MAF in primary lymphatic endothelial cells, which may contribute to the transcriptional reprogramming of these cells observed upon KSHV infection (18). Recently, three reports have suggested a role of the KSHV miRNAs in stabilizing viral latency. KSHV miR-K9-5p directly targets the viral lytic transactivator RTA and thereby antagonizes lytic reactivation of the virus (3). Two groups have recently reported that the deletion of all KSHV miRNAs except miR-K10 and miR-K12 resulted in a modest increase in the spontaneous lytic reactivation of KSHV (22, 23).
Additional targets and functions of the KSHV miRNAs likely remain to be uncovered. In order to identify mRNAs directly regulated by the KSHV miRNAs, we performed microarray analyses using RNA samples derived from KSHV-negative B cells stably expressing physiological levels of individual KSHV miRNAs. The expression of KSHV miR-K1 resulted in the significant down- or upregulation of numerous cellular mRNAs (data not shown). During the course of this analysis, we noticed that the 3′ UTRs of mRNAs encoding the cyclin-dependent kinase inhibitor (CDKI) p21 contain two closely spaced matches to miR-K1. p21 functions as a key mediator of cell cycle arrest, in response to the activation of multiple tumor-suppressive pathways, by inhibiting cyclin-cyclin-dependent kinase (CDK) complexes containing Cdk2 or Cdk1 (1). While p21 is rarely mutated in tumors, its normal function is often disrupted by mutations that block pathways that depend upon p21 to effect cell cycle arrest, including the p53 tumor suppressor and the transforming growth factor beta (TGF-β) signaling pathways. p21 knockout mice show an increased incidence of spontaneous tumors late in life, and loss of p21 synergizes with the loss of other known tumor suppressors, causing an increase in cancer in vivo, thus arguing that p21 functions as a bona fide tumor suppressor (1, 24). Because of its involvement in several different pathways, the downregulation of p21 is expected to impact the response of cells to several biological stimuli, including DNA damage and signaling induced by interferon or TGF-β, as well as triggers of oncogene-induced senescence, such as K-cyclin expression (21). Here, we demonstrate that the KSHV miRNA miR-K1 indeed represses the expression of cellular p21 in latently KSHV-infected B cells, thus suggesting that miR-K1 may function to allow the progression of KSHV-infected cells through the cell cycle under conditions where this would normally be blocked by p21.
MATERIALS AND METHODS
Constructs.
Lentiviral indicator vectors based on pNL-SIN-CMV-RLuc, carrying no additional sequences or 2 perfect matches to miR-K1, miR-K3, or miR-K4 in the 3′ UTR of RLuc, as well as the internal-control vector pNL-SIN-CMV-FLuc, have been described previously (15). Indicator vectors expressing firefly luciferase (Fluc) (pL/SV40/GL3 [pLSG]) or Renilla luciferase (RLuc) (pL/SV40/RLuc [pLSR]) have also been described (16). The full-length p21 3′ UTR, ending just upstream of the poly(A) signal, was amplified from a cDNA clone (Open Biosystems) using primers 404 (AGAGATCTAGATCCGCCCACAGGAAGCCTGCAG) and 406 (AGAGAGCGGCCGCGAGCACCTGCTGTATATTCAGCATTG). The resulting fragment was cloned between the XbaI and NotI sites of pLSG and sequence verified [pLSG/p21(406)]. In order to achieve authentic polyadenylation, a fragment spanning the 3′ end of the p21 3′ UTR, from ∼300 bp 5′ of the poly(A) signal to ∼130 bp 3′ of the poly(A) signal, was amplified from BJAB cell genomic DNA using primers 719 (TGGGCTCATATGGGGCTGGG) and 720 (AGAGAGCGGCCGCTTGCTAAGTTTTCTGCATTC). The resulting fragment was digested using NdeI and NotI and used to replace the corresponding fragment in pLSG/p21(406) to yield pLSG/p21(720). The resulting vector contained the entire 3′ UTR of p21 and 130 nt of the authentic genomic sequence 3′ to the poly(A) signal and was used for Fig. 1. This approach was used because the miR-K1 binding sites are within 200 nt 5′ of the poly(A) signal. Seed matches to miR-K1 were mutated using mutant PCR primers 702 (CTTCCAGCTCCTATGACATACTGGCCTG)/703 (CAGGCCAGTATGTCATAGGAGCTGGAAG) (5′ site; M1) and 704 (CTCCACCTAGACTATGAACCTCTCGAGGGC)/705 (GCCCTCGAGAGGTTCATAGTCTAGGTGGAG) (3′ site; M2), as well as outer primers 719 and 720. For the combined mutation of both sites (DM), M2 was introduced into a DNA fragment already carrying M1. The resulting mutant PCR products were used to replace the corresponding NdeI and NotI fragment in pLSG/p21(720).
FIG. 1.
miR-K1 can directly repress gene expression via the p21 3′ UTR. (A) Schematic representation of human p21 mRNA (accession number NM_078467). (B) Seed matches to miR-K1 in the 3′ UTR of the p21 mRNA are shown in boldface. (C) FLuc indicator vectors carrying no additional sequences (none), the full-length 3′ UTR of p21 (p21), or p21 3′-UTR mutants (CUGUAA changed to CUAUGA) were cotransfected into 293T cells with an RLuc internal-control vector and empty vector (vector) or a miR-K1 expression plasmid (miR-K1). Dual luciferase assays were carried out as described previously (16). The FLuc-to-RLuc ratios observed were normalized to those observed for the control vector lacking additional 3′-UTR sequences. The values obtained in cells transfected with empty vector were set at 100%. n = 3; the error bars indicate standard deviations (SD). (D) RLuc indicator vectors carrying no additional sequences (none) or 2 perfect matches to miR-K1 (miR-K1) were cotransfected with the FLuc internal-control plasmid and empty vector (vector) or a miR-K1 expression plasmid (miR-K1). Dual luciferase assays were carried out as described previously (16). The RLuc-to-FLuc ratios observed for the miR-K1 indicator vector were normalized to those observed for the control vector lacking miRNA target sequences. The values obtained in cells transfected with empty vector were set at 100%. n = 3; the error bars indicate SD.
The lentiviral vector pL/CMV/EGFP (pLCE) has been described previously (47). The miR-K1 expression cassette used consisted of ∼250 nt from the KSHV genome present in BC-1 and was described and validated previously (15, 16). This cassette was amplified from the previously described vector pNL-SIN-CMV-AcGFP/miR-K1 (16) using primers 673F (AGAGACTCGAGGGGAGGAAGGATGTGGGGG) and 673R (AGAGAGCGGCCGCCCCATTATAATCTTAATC) and cloned between the XhoI and NotI sites of pLCE, 3′ to the enhanced green fluorescent protein (EGFP) open reading frame (ORF). The resulting vector was sequence verified. For the inducible expression of EGFP-miR-K1, the EGFP or EGFP-miR-K1 cassette was excised from pLCE or pLCE/miR-K1 using AgeI and EcoRI and used to replace the AgeI-EcoRI fragment of pTRIPZ (Open Biosystems). To express miR-K1 in BL40 cells, we constructed an murine stem cell virus (MSCV)-based retroviral vector derived from the previously described vector pMig-w (33). Primers 617 (GATCTAGAGACCTAGGAGCACTCGAGAGAGAGCGGCCGCAGAGAGAATTCTCTCTG) and 618 (TCGACAGAGAGAATTCTCTCTGCGGCCGCTCTCTCTCGAGTGCTCCTAGGTCTCTA) were annealed and ligated between the BglII and SalI sites of pMig-w to produce pM, which carried a BglII-AvrII-XhoI-NotI-EcoRI polylinker between the packaging signal and the woodchuck hepatitis virus posttranscriptional regulatory element (WRE) of the vector. pM was cut with AvrII and EcoRI and ligated with NheI-EcoRI fragments derived from pLCE or pLCE/K1 to produce pME and pME/K1. pTRIPZ expressing an artificial miRNA targeting p21 (sh-miR-p21) was purchased from Open Biosystems, and pTRIPZ/p21ORF was cloned by PCR amplification of the p21 ORF from a cDNA clone (Open Biosystems) using primers 706 (AGAGAACCGGTCGCCATGTCAGAACCGGCTGGGG) and 707 (AGAGAACGCGTTTAGGGCTTCCTCTTGGAGAAG). The resulting fragment was cloned between the AgeI and MluI sites of pTRIPZ. All PCR-amplified fragments were sequenced.
Lentiviral sponges directed against an artificial small interfering RNA (siRNA) specific for CXCR4 (sCX) or miR-K1 were based on the previously described lentiviral vector pLCE and designed essentially as described previously (10, 47). Oligonucleotides containing 3 imperfect matches each (CX_F [CTAGTAAGTTTTCAGAAAGCTAACAGTTGAAGTTTTCAGAAAGCTAACAGTTGAAGTTTTCAGAAAGCTAACATCTAGATTTGAATTC] and CX_R [AATTGAATTCAAATCTAGATGTTAGCTTTCTGAAAACTTCAACTGTTAGCTTTCTGAAAACTTCAACTGTTAGCTTTCTGAAAACTTA] for sCX and 627 [CTAGGCTTACACCCACAATCCTGTAATGTTTTGGCTTACACCCACAATCCTGTAATGTTTTGGCTTACACCCACAATCCTGTAATTCTAGATTTGAATTC]/628 [AATTGAATTCAAATCTAGAATTACAGGATTGTGGGTGTAAGCCAAAACATTACAGGATTGTGGGTGTAAGCCAAAACATTACAGGATTGTGGGTGTAAGC] for sK1) were annealed and inserted between XbaI and EcoRI 3′ of the EGFP ORF. Ligation of the oligonucleotides destroyed the 5′ XbaI site and introduced a new XbaI site 3′ of the matches. This allowed the ligation of two more copies of the annealed oligonucleotides to give 9 imperfect matches each. The resulting vectors, pLCE/sCX and pLCE/sK1, were sequence verified.
Indicator assays and primer extensions.
Indicator assays using pLSG/p21 constructs were performed by transfection of 293T cells as described previously (16). Indicator assays for miRNA activity (using pNL-SIN-CMV-RLuc and pNL-SIN-CMV-FLuc vectors) were performed either after transfection of indicators into U2OS or 293T cells or after transduction of B-cell lines, as described previously (15, 16). Primer extensions were carried out as described previously (16).
Cell culture and generation of viral transductants.
U2OS cells were grown in McCoy's 5A medium supplemented with 10% fetal bovine serum (FBS) and gentamicin. BC-3 cells were obtained from the ATCC and grown in RPMI 1640 medium containing 15% FBS, 10 mM HEPES, 1 mM sodium pyruvate, 0.05 mM β-mercaptoethanol, and gentamicin. The Burkitt's lymphoma cell line BL40 (a gift from Mark Wade) was grown in RPMI 1640 medium containing 10% FBS and gentamicin. All retroviral and lentiviral vectors were produced by cotransfection into 293T cells using Fugene6. pLCE-based vectors were produced by cotransfection with pMDLgpRRE, pRSV-Rev, and pVSV-G (9). pTRIPZ-based vectors were produced by cotransfection with pCMVΔR8.74 and pMD2.G (9). MSCV vectors were cotransfected with pMSCV-Gag/Pol and pHIT-G. Forty-eight hours after transfection, the filtered supernatants were used to infect logarithmically growing cells. Forty-eight hours after transduction, cells were selected with puromycin (1 μg/ml for U2OS; 3 μg/ml for BC-3) or sorted by fluorescence-activated cell sorter (FACS) as described previously (16).
Western blotting and qRT-PCR.
Cells were washed with ice-cold phosphate-buffered saline (PBS), incubated with lysis buffer (20 mM Tris, pH 7.4, 100 mM NaCl, 1% Triton X-100, 10% glycerol, 1 mM EDTA, 1 mM dithiothreitol [DTT], 0.1 mM sodium orthovanadate, 20 mM NaF, 10 mM pyrophosphate, and Complete EDTA-free protease inhibitors [Roche]) for 30 min on ice and centrifuged at 21,000 × g for 30 min at 4°C. Protein was quantified using the BCA Protein Assay Kit (Pierce), and equal amounts of total protein were analyzed by Western blotting. Anti-p21 (C-19) and anti-β-actin (C4) antibodies were from Santa-Cruz Biotechnology, Inc.; anti-p53 antibodies (DO-7) from Sigma; and anti-Mdm2 antibodies (2A9) from Calbiochem. Total RNA was prepared using TRIzol as instructed. Total RNA (500 ng) was reverse transcribed using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems). mRNA expression was measured using exon-spanning TaqMan probes (Applied Biosystems) against p21 (Hs00355782_m1), PIG3 (Hs00153280_m1), and Mdm2 (12) and normalized to beta-2 microglobulin mRNA expression.
Cell cycle experiments.
In all cases, equal cell numbers were used, and cells were subconfluent throughout the experiment. U2OS cells were treated with doxycycline (Dox) (1 μg/ml) for 24 h and then split into 0.7 × 106 cells/10-cm dish in medium with or without Dox; 24 h later, Nutlin-3 (Sigma) or solvent (dimethyl sulfoxide [DMSO]) was added for 24 h (final concentration, 5 μM Nutlin-3). During the last 30 min of Nutlin-3 treatment, the cells were pulse-labeled with 25 μM 5-bromodeoxyuridine (BrdU) (Calbiochem) and then trypsinized. Sponge-expressing BC-3 cells were plated at 2 × 105 cells/ml; 24 h later, Nutlin-3 (2.5 μM final concentration in DMSO) or DMSO was added. After 23.5 h, the cells were labeled with a final concentration of 25 μM BrdU for 30 min. pTRIPZ/p21(ORF) or pTRIPZ-transduced BC-3 cells were plated at 2 ×105 cells/ml; 24 h later, the cells were treated with 1 μg/ml Dox for 23.5 h and then labeled with 25 μM BrdU for 30 min. In all cases, the cells were washed once with ice-cold PBS and fixed with 70% ice-cold ethanol at −20°C for at least 1 h. The cells were costained with anti-BrdU-fluorescein isothiocyanate (FITC) antibodies (BD Pharmingen) and propidium iodide (PI)-RNase staining buffer (BD Pharmingen). Briefly, the fixed cells were washed with wash buffer (PBS-0.1% bovine serum albumin [BSA]) and treated with 3.5 M HCl for 30 min. The cells were washed, incubated with 0.1 M sodium borate (pH 8.5) for 2 min, washed, resuspended in 30 μl wash buffer and 20 μl anti-BrdU-FITC antibody solution, and stained for 30 min. The cells were washed and stained with 0.5 ml PI-RNase staining buffer for 15 min. Data were acquired and analyzed using a BD FACScan flow cytometer and Cellquest and FloJo software. A minimum of 10,000 cells were collected per sample.
qPCR analysis of KSHV genome content.
Genomic DNA was prepared from untransduced or sponge-expressing BC-3 cells using the DNeasy blood and tissue kit (Qiagen). Quantitative PCR (qPCR) was conducted using Power SYBR green PCR Master Mix (Applied Biosystems), 10 ng DNA template, and each primer at a concentration of 100 nM. The KSHV genome was detected using primers 766 (TTGCCACCCACGCAGTCT)/767 (GGACGCATAGGTGTTGAAGAGTCT) and normalized to the human β-globin DNA content (primers 764 [TGAGCCTTCACCTTAGGGTTGCCCA]/765 [GCCCTGGGCAGGTTGGTATCAAGGT]).
RESULTS AND DISCUSSION
miR-K1 represses p21 expression via the 3′ UTR.
p21 transcripts contain two closely spaced seed matches to miR-K1 within ∼100 nt of each other toward the 3′ end of their ∼1.5-kb 3′ UTR (Fig. 1A and B). Perfect complementarity of an mRNA target to the miRNA seed sequence, extending from nt 2 to 7 or 8 from the miRNA 5′ end, is seen in the majority of responsive mRNA species (2). We generated FLuc reporter plasmids by placing the entire 3′ UTR of p21, as well as 130 nt of flanking human genomic sequences located 3′ of the p21 poly(A) signal, 3′ of the FLuc ORF in the previously described indicator plasmid pLSG (16). This was expected to result in polyadenylation of the FLuc transcript at the authentic p21 poly(A) site. To control for specificity, 2-nt mutations were introduced into each miR-K1 seed match, individually (M1 and M2) or in combination (DM). The resulting indicator plasmids were cotransfected into 293T cells, along with an RLuc-based internal-control plasmid and either a miR-K1 expression plasmid or a control plasmid. Dual luciferase assays revealed that miR-K1 repressed expression of the FLuc indicator linked to the p21 3′ UTR by cooperatively engaging both seed matches, thereby confirming that p21 can be directly and specifically targeted by miR-K1 (Fig. 1C). miR-K1 expression was confirmed in parallel using a previously described artificial indicator plasmid for miR-K1 activity (Fig. 1D) (15).
Ectopic expression of miR-K1 antagonizes p21-mediated cell cycle arrest.
KSHV expresses a number of cell cycle regulatory proteins during viral latency (7, 13, 32, 34), and these have the potential to complicate the analysis of miR-K1 function in the context of latently KSHV-infected cells. We therefore decided to first address the regulation of p21 by miR-K1 in the KSHV-negative osteosarcoma cell line U2OS, which is commonly used to study p53 function (11). We established U2OS cell lines allowing the inducible expression of miR-K1 or, as a control for p21 function, of an inducible artificial miRNA designed to be perfectly complementary to the p21 mRNA (sh-miR-p21). Control cell lines transduced with the parental lentiviral vectors pTRIPZ and pTRIPZ/EGFP were also established. p53 expression and activity are tightly controlled by the ubiquitin ligase Mdm2, which represses the transcriptional activity of p53 and targets p53 for degradation (28, 46). These functions of Mdm2 can be disrupted by the small-molecule inhibitor Nutlin-3, which results in the stabilization of p53 and, consequently, the expression of p53-inducible genes, including the p21 gene (42). To evaluate the consequences of miR-K1 expression in untreated and Nutlin-3-treated cells, U2OS cells were plated at equal densities, and miR-K1 or sh-miR-p21 expression was induced using Dox. Twenty-four hours into Dox treatment, the cells were divided into multiple aliquots and, after an additional 24 h, treated with Nutlin-3 or an equivalent amount of solvent (DMSO). In parallel with Nutlin-3 treatment, small aliquots of the cells were transfected with indicator constructs for miR-K1 activity. After 24 h, cells were processed for analysis of miR-K1 activity, cellular protein and mRNA expression, and cell cycle properties. Indicator assays for miR-K1 activity confirmed the Dox-induced expression of miR-K1 (Fig. 2A). Western blotting showed that in DMSO-treated cells, a reduction in endogenous p21 expression was readily detectable in cells expressing either sh-miR-p21 or miR-K1 (Fig. 2B, top 2 gels). Upon Nutlin-3 treatment of cells, equivalent accumulation of p53 and its transcriptional target, Mdm2, was observed in all cell lines (Fig. 2B, bottom 3 gels). As expected, p21 expression under control conditions was induced following p53 activation. In contrast, p21 upregulation was strongly diminished in cells expressing either miR-K1 or sh-miR-p21 (Fig. 2B, bottom gels). Upon longer exposure, Nutlin-3-induced p21 expression was detectable in cells expressing either miR-K1 or sh-miR-p21, suggesting that the knockdown of p21 was incomplete under both conditions (data not shown). Results obtained using qRT-PCR confirmed that miR-K1 specifically reduced the abundance of p21 transcripts while the induction of other p53-responsive mRNAs (i.e., Mdm2 and PIG3) was not affected (Fig. 2C and D). Taken together, these results strongly suggest that miR-K1 directly targets p21 yet does not globally attenuate p53-dependent transcriptional activity.
FIG. 2.
Ectopic miR-K1 represses endogenous p21 protein and antagonizes p21-mediated cell cycle arrest. U2OS cells allowing the inducible expression of sh-miR-p21 or miR-K1 and control cells (TRIPZ and EGFP) were treated (ON) or not (OFF) with Dox. Forty-eight hours later, the cells were treated with 5 μM Nutlin-3 (N) or DMSO (D) for 24 h. (A) Cells were cotransfected with RLuc indicator vectors carrying no additional sequences (no target) or 2 perfect matches to miR-K1 (2 targets miR-K1) and an FLuc internal-control vector. Dual luciferase assays were carried out 24 h after transfection. The RLuc-to-FLuc ratios observed for the miR-K1 indicator vector were normalized to those observed for the control vector lacking miRNA target sequences. The values obtained in cells expressing only EGFP were set at 100%. The error bars indicate SD (n = 3). (B) Western blot analysis of p21, p53, Mdm2, and β-actin expression in DMSO-treated cells (top 2 gels) and Nutlin-3-treated cells (bottom 4 gels). One Nutlin-3- or DMSO-treated sample was included in each gel to demonstrate induction upon Nutlin-3 treatment. (C) Total RNA from all samples from one experiment was analyzed for the expression of p21, Mdm2, and PIG3 mRNA by TaqMan qRT-PCR. The data were normalized to the expression of β-2-microglobulin. (Z, pTRIPZ; sh, sh-miR-p21; E, EGFP; K1, miR-K1; ON, treated with 1 μg/ml Dox; OFF, not treated with Dox). The error bars indicate SD from three technical replicates. Expression levels are shown relative to the level of expression observed in the pTRIPZ-transduced, Dox-treated sample. (D) DMSO- or Nutlin-3-treated, EGFP- or miR-K1-expressing U2OS cells were analyzed by qRT-PCR as described for panel C. Data from three independent experiments are shown relative to the level of expression observed in the pTRIPZ/EGFP-transduced, Dox-treated sample. The error bars indicate SD. (E) Cells were costained with anti-BrdU-FITC and PI and analyzed using flow cytometry. S phase, upper gate; G1, lower left; G2, lower right. (F) Quantification of cell cycle distribution upon Nutlin-3 treatment (n = 5). The percentages obtained for sh-miR-p21-expressing cells were normalized to those from pTRIPZ-expressing cells and those from miR-K1-expressing cells to EGFP-expressing cells. The percentages obtained from Nutlin-3-treated cells were related to those obtained from DMSO-treated cells. The values obtained in TRIPZ- and EGFP-expressing cells were set at 100% (control). The error bars indicate SD. Only Dox-treated samples are shown.
To address the consequences of miR-K1 expression for p53-induced cell cycle arrest in U2OS cells, the cells were treated as described above and metabolically labeled with BrdU for 30 min. Cell cycle analysis confirmed that the Nutlin-3-induced cell cycle arrest observed in U2OS cells largely depended on p21 function, because expression of sh-miR-p21 resulted in a strong reduction of cell cycle arrest upon Nutlin-3 treatment (Fig. 2E; quantified in F). The incomplete rescue of sh-miR-p21-expressing cells from Nutlin-3-induced cell cycle arrest was probably due to the incomplete knockdown of p21 by sh-miR-p21 observed upon Nutlin-3 treatment (data not shown). This interpretation of our data is supported by the finding that HCT116 cells, in which both copies of the p21 gene are deleted, are resistant to p53-induced cell cycle arrest (45) and the observation that Nutlin has little effect on cell cycle progression in HCT116(p21−/−) cells (data not shown). Like sh-miR-p21-expressing U2OS cells, miR-K1-expressing U2OS cells, compared to control cells, also exhibited a larger percentage of cells in S phase upon Nutlin-3 treatment, thus indicating that miR-K1-expressing cells also partially escaped p53-induced cell cycle arrest (Fig. 2E and F). The increase of the percentage of cells in S phase upon Dox-induced miR-K1 expression appeared to primarily result from a reduction of the percentage of cells in the G2 phase of the cell cycle compared to Dox-induced control cells. Upon Nutlin treatment, however, cell cycle distributions also differed between Dox-induced and uninduced U2OS cells in the absence of miR-K1 expression (data not shown). To avoid possible artifacts in the cell cycle distribution arising due to Dox treatment, we evaluated the consequences of the constitutive expression of miR-K1 in U2OS cells (Fig. 3). U2OS cells were transduced with a control lentiviral vector (pLCE) or a miR-K1-expressing lentiviral vector (pLCE/miR-K1). Three days after transduction, aliquots of cells were transfected with indicator constructs for miR-K1 activity and treated with Nutlin-3 or DMSO. Twenty-four hours later, miR-K1 expression was confirmed using indicator assays (Fig. 3A), and the knockdown of p21 protein expression in miR-K1-expressing cells was confirmed by Western blotting (data not shown). The cells were pulse-labeled with BrdU and subjected to cell cycle analysis as described above (Fig. 3B). As in the experiment shown in Fig. 2, miR-K1-expressing cells exhibited a greater percentage of cells in S phase upon Nutlin-3 treatment than control cells, confirming that miR-K1 is indeed able to antagonize p53-induced cell cycle arrest. The observed increase in the percentage of cells in S phase correlated with a decrease in the number of cells in both the G1 and G2 phases of the cell cycle. This is consistent with the finding that the expression of p21 in U2OS cells causes cell cycle arrest at both G1 and G2 (data not shown) and argues against the hypothesis that miR-K1 preferentially inhibits cell cycle arrest in G2.
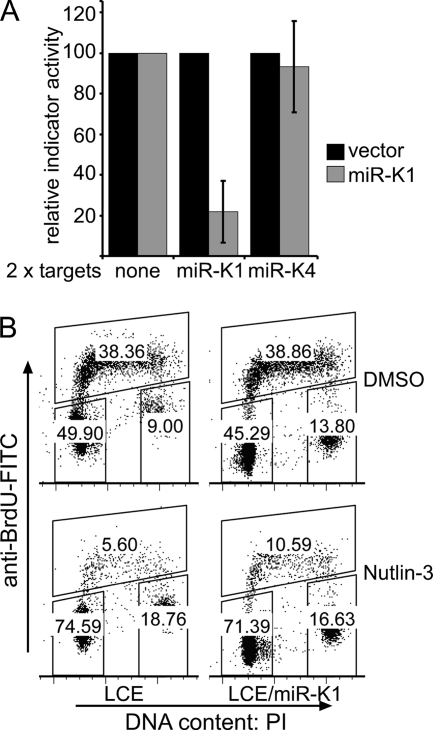
FIG. 3.
Constitutive expression of miR-K1 attenuates p21-induced cell cycle arrest. U2OS cells were transduced with a control lentiviral vector (pLCE) or a lentiviral vector expressing miR-K1 (LCE/miR-K1). (A) Three days after transduction, cells were cotransfected with RLuc indicator vectors carrying no additional sequences (no target) or 2 perfect matches to miR-K1 or miR-K4, as indicated, and an FLuc internal-control vector. Dual luciferase assays were carried out 24 h later. The RLuc-to-FLuc ratios observed for the miR-K1 and miR-K4 indicator vectors were normalized to those observed for the control vector lacking miRNA targets. The values obtained in cells expressing only EGFP were set at 100%. The error bars are from independently generated cell pools and indicate SD (n = 3). (B) Cells were treated for 24 h with 10 μM Nutlin-3 or DMSO, costained with anti-BrdU-FITC and PI, and analyzed using flow cytometry. S phase, upper gate; G1, lower left; G2, lower right. The data shown are representative of two experiments using independently generated cell pools. In experiments 1 and 2, miR-K1-expressing cells arrested ∼2.3-fold and ∼1.9-fold less efficiently than EGFP-expressing cells, respectively. The results from experiment 2 are shown in panel B.
Ectopically expressed miR-K1 represses p21 in a B-cell line.
In order to further strengthen our result showing that miR-K1 can directly repress p21 protein expression, we stably expressed miR-K1 in a human B-cell line, the KSHV-negative Burkitt's lymphoma cell line BL40. BL40 cells encode a wild-type (wt) p53 gene, and Nutlin-3 treatment of these cells is known to induce p21 expression (44). Primer extension confirmed that miR-K1 expression in these cells was close to the physiological levels of expression observed in the PEL cell lines BC1, BCBL-1, and BC-3 (Fig. 4A). As in U2OS cells, miR-K1 was able to strongly antagonize p21 expression following the treatment of BL40 cells with Nutlin-3 (Fig. 4B). mir-K1 also strongly antagonized p21 expression following the treatment of BL40 cells with doxorubicin, which causes p53 activation by inducing DNA damage without affecting the accumulation of p53 (Fig. 4C). This result demonstrated that physiological levels of miR-K1 expression can repress the expression of p21 in B cells.
FIG. 4.
miR-K1 attenuates p21 expression in B cells. BL40 cells were transduced with the control vector pME or pME expressing miR-K1 and sorted by FACS. (A) Primer extension for miR-K1 detection. miR-16 was used as a loading control. (B) Cells were plated at equal numbers and, 24 h later, treated with 2.5 μM Nutlin-3 for the indicated times. Protein lysates were analyzed for p21 expression by Western blotting. β-Actin served as a loading control. The asterisk marks a cross-reacting protein. (C) Cells were plated at equal numbers and, 24 h later, treated with 25 ng/ml doxorubicin for the indicated times. Protein lysates were analyzed for p21 and p53 expression by Western blotting. β-Actin served as a loading control. The asterisks mark cross-reacting proteins.
miR-K1 attenuates p21-mediated cell cycle arrest during KSHV latency.
Having established that miR-K1 can specifically repress p21 protein expression in KSHV-negative cells, we next addressed the phenotypic consequences of this interaction in the latently KSHV-infected PEL cell line BC-3, which retains wild-type p53 (29).
Nutlin-3 treatment of PEL cell lines results in the upregulation of p21 and cell cycle arrest (29, 38). In BC-3 cells, the induction of cell cycle arrest following p53 reactivation is less efficient than in KSHV-negative U2OS cells, possibly because this cell line has lost retinoblastoma protein (Rb) expression and/or due to the expression of viral proteins that repress p21 function (31). Nevertheless, we were able to confirm that expression of p21, lacking its authentic 3′ UTR, in BC-3 cells can indeed induce cell cycle arrest (Fig. 5). In order to achieve stable knockdown of miR-K1 function, BC-3 cells were transduced with a lentiviral “sponge” bearing 9 imperfect matches to miR-K1 (sK1), unmodified lentiviruses (EGFP only), or a control sponge consisting of 9 imperfect matches to a CXCR-4-directed artificial miRNA (sCX). Similar miRNA sponges have been previously shown to competitively inhibit miRNA function (10). Cell pools with matched mean green fluorescent intensities were sorted by flow cytometry, and two independently generated replicates were obtained. In each case, sorted cells represented a large percentage (between 48% and 92%) of the parental population and therefore did not represent small, possibly abnormal subpopulations. Functional knockdown of miR-K1 activity in sK1-transduced BC-3 cells was confirmed using lentiviral indicator assays for miR-K1 activity (Fig. 6A) (15). While high levels of miR-K1 activity were detected in control cell pools, miR-K1 activity was completely lost in cells expressing the miR-K1-directed sponge, i.e., indicator activity was comparable to that observed in the KSHV-negative B-cell line BJAB. In contrast, the activity of another KSHV miRNA (miR-K3), tested in parallel, was unaffected by the presence of the sK1 sponge. Since sK1-transduced cells grew with essentially wild-type kinetics (Fig. 6D and data not shown) and the KSHV genomic-DNA content was unaffected by the knockdown of miR-K1 (Fig. 6B), we conclude that miR-K1 activity is nonessential for the survival or growth of PELs or for the episomal maintenance of the viral genome.
FIG. 5.
Expression of p21 causes cell cycle arrest in BC-3 cells. BC-3 cells stably expressing a lentiviral vector allowing the inducible expression of p21 [p21(ORF)] or the parental vector (TRIPZ) were plated at equal densities and, 24 h later, treated with 1 μg/ml Dox for 24 h. The cells were labeled with 25 μM BrdU for 30 min. (A) Cells were analyzed for p21 expression by Western blotting. β-Actin served as a loading control. (B) Cells were fixed and stained with anti-BrdU-FITC antibodies and propidium iodide and analyzed using flow cytometry. One representative experiment (out of 3 independent experiments) is shown. The statistical analysis of all 3 replicates showed that, compared to untreated cells, in Dox-treated p21(ORF)-transduced cells, 32% ± 5% as many cells were in S phase and 176% ± 25% as many were in G1 phase (±SD).
FIG. 6.
Endogenous miR-K1 attenuates p21-mediated cell cycle arrest in the PEL cell line BC-3. BC-3 cells expressing EGFP only, a control sponge (sCX), or a miR-K1-directed sponge (sK1) were sorted by FACS. 1 and 2 refer to independently generated cell lines. (A) Lentiviral indicator assays for miR-K1 activity (2 targets K1) or miR-K3 activity (2 targets K3) were performed as previously described (15). The KSHV-negative B-cell line BJAB served as a negative control. n = 3; the error bars indicate SD. (B) The KSHV genomic-DNA (gDNA) content was quantified by qPCR of total DNA prepared from sponge-expressing BC-3 cell lines. Total DNA prepared from KSHV-negative BJAB cells was used as a negative control. The values were normalized to a β-globin endogenous control, and the values obtained for untransduced BC-3 cells (−) were set at 100%. The error bars indicate SD and are from 3 technical replicates. (C) Cell lines were treated for 24 h with 2.5 μM Nutlin-3 or solvent (DMSO) and analyzed by for p21, p53, Mdm2, and β-actin expression by Western blotting. (D) Cells were treated as for panel C, labeled with BrdU, costained with anti-BrdU-FITC and PI, and analyzed using flow cytometry. One representative experiment is shown. Statistical analysis (n = 5) indicated that Nutlin-3-induced cell cycle arrest was significantly enhanced in sK1-expressing BC-3 cells (P < 0.0004). Percentages (±SD) of cells in S phase compared to a DMSO-treated control cell line: EGFP/DMSO was set at 100; sK1/DMSO, 95% ± 5%; EGFP/Nutlin-3, 74% ± 2.5%; sK1/Nutlin-3, 60% ± 4%. Percentages (±SD) of cells in G1 phase compared to a DMSO-treated control cell line: EGFP/DMSO was set at 100; sK1/DMSO, 106% ± 4%; EGFP/Nutlin-3, 118% ± 3%; sK1/Nutlin-3, 132% ± 5%.
To assess whether miR-K1 represses p21 in PEL cells, sponge-expressing BC-3 cell lines were plated at equal densities and, 24 h later, treated with 2.5 μM Nutlin-3 for 24 h. We used this relatively low level of Nutlin-3 to avoid excessive p53-induced apoptosis during these experiments. The cells were labeled with BrdU and processed for analysis of protein expression and cell cycle progression. Western blotting showed that induction of endogenous p21 upon Nutlin-3 treatment was strongly enhanced in cells lacking miR-K1 activity, demonstrating that endogenous, virally encoded miR-K1 indeed represses p21 in PEL cells (Fig. 6C, top). In contrast, p53 stabilization and the induction of the p53 target Mdm2 gene were equivalent in all cell lines tested (Fig. 6C). These results mirror those obtained in U2OS and BL40 cells ectopically expressing miR-K1 (Fig. 2B and 4) and suggest that the overall transcriptional activity of p53 is not affected by miR-K1 but that miR-K1 instead directly targets p21 mRNAs in BC-3 cells. Nutlin-3-induced cell cycle arrest was equally efficient in cells expressing only EGFP and cells expressing the control sponge (sCX) (Fig. 6D). In contrast, BC-3 cells lacking miR-K1 activity were arrested significantly more efficiently upon Nutlin-3 treatment (P ≤ 0.0004), suggesting that miR-K1 activity attenuates p21-mediated cell cycle arrest in KSHV-infected cells.
In summary, we report that KSHV miR-K1 inhibits p21 expression through sites located within the 3′ UTR of p21 mRNAs and consequently attenuates p21-induced cell cycle arrest. The ectopic expression of miR-K1 strongly antagonized p21 expression in the osteosarcoma cell line U2OS and the B-cell line BL40 (Fig. 2B and 4), and miR-K1 attenuated the induction of cell cycle arrest observed following p53 activation in U2OS cells (Fig. 2 and 3). Conversely, the functional knockdown of endogenous miR-K1 in a latently KSHV-infected PEL cell line resulted in increased expression of p21 and increased the efficiency of cell cycle arrest following p53 activation (Fig. 6).
While several of the previously validated cellular targets of KSHV miRNAs are targeted by more than one viral miRNA (16, 18, 39), we currently have no evidence that this is the case for p21. The p21 3′ UTR also has potential 6-mer and 8-mer seed matches to KSHV miR-K11 and miR-K6-5p, respectively. Our preliminary data suggest that miR-K11 does not downregulate p21 indicator expression (data not shown), while we have not tested regulation by miR-K6-5p, which is expressed only at low levels in PEL cell lines (data not shown and reference 41).
The phenotype caused by inactivation of miR-K1 in the context of KSHV latency was relatively modest, which is not surprising, given that KSHV has evolved multiple ways to ensure cell cycle progression and to antagonize p21-mediated cell cycle arrest. While PEL cells commonly retain a wild-type p53 genotype (29), the viral LANA protein represses p53 activity (13, 37). The interaction of LANA with a complex containing both p53 and Mdm2 is disrupted by Nutlin-3, thereby allowing the reactivation of p53 upon Nutlin treatment (37). KSHV also expresses a viral cyclin (K-cyclin) with functional properties of cellular D- and E-type cyclins (7, 43). K-cyclin is resistant to inhibition by p21 and induces the phosphorylation of p21 at serine 130, resulting in the inactivation of p21 (7, 20, 40). We have independently confirmed that mutation of p21 serine 130 to alanine results in a p21 mutant protein that is a more potent inhibitor of cell cycle progression in BC-3 cells than wt p21 (data not shown). The fact that cell cycle progression in PEL is partially driven by a p21-resistant viral cyclin and that p21 function is attenuated by phosphorylation may help explain why the phenotype of miR-K1 inactivation is fairly subtle in the context of KSHV latency (Fig. 6D). Interestingly, repression of p21 by the oncogenic miR-17-92 cluster was shown to contribute to the deregulation of cell cycle properties seen upon miR-17-92 overexpression (19, 25). PEL cell lines consistently express the miR-17-92 cluster, and it is possible that these miRNAs could further contribute to the suppression of p21 expression and function in PEL (27). While miR-17-5p is detectable in BC-3 cells (data not shown), its level of expression may be too low to effectively inhibit p21 expression.
To our knowledge, the findings reported here document the first example of a viral miRNA that deregulates cell cycle progression. Infection of immunodeficient individuals by KSHV can lead to the transformation of endothelial cells to give rise to Kaposi's sarcoma and of B cells to give rise to primary effusion lymphoma, and we hypothesize that miR-K1 may play a key role in the oncogenic potential of this important viral pathogen.
Acknowledgments
We thank Neelanjan Mukherjee and Uwe Ohler for analysis of microarray data, Micah Luftig for helpful discussions, Irving Chung for technical assistance, and John Whitesides at the Duke Center for AIDS Research BSL3 Flow Cytometry Core Facility for cell sorting. Analytical FACS was performed through the Duke Comprehensive Cancer Center Flow Cytometry Shared Resource. BL40 cells were kindly provided by Mark Wade with permission from Martin Allday.
This work was supported by National Institutes of Health grant R01-AI067968 and AIDS-associated malignancy supplement P30-CA14236 to B.R.C. and NIH grant K99-CA137860 to E.G.
Footnotes
Published ahead of print on 10 March 2010.
REFERENCES
- 1.Abbas, T., and A. Dutta. 2009. p21 in cancer: intricate networks and multiple activities. Nat. Rev. Cancer 9:400-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel, D. P. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136:215-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bellare, P., and D. Ganem. 2009. Regulation of KSHV lytic switch protein expression by a virus-encoded microRNA: an evolutionary adaptation that fine-tunes lytic reactivation. Cell Host Microbe 6:570-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai, X., S. Lu, Z. Zhang, C. M. Gonzalez, B. Damania, and B. R. Cullen. 2005. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc. Natl. Acad. Sci. U. S. A. 102:5570-5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 6.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 7.Chang, Y., P. S. Moore, S. J. Talbot, C. H. Boshoff, T. Zarkowska, K. Godden, H. Paterson, R. A. Weiss, and S. Mittnacht. 1996. Cyclin encoded by KS herpesvirus. Nature 382:410. [DOI] [PubMed] [Google Scholar]
- 8.Costinean, S., N. Zanesi, Y. Pekarsky, E. Tili, S. Volinia, N. Heerema, and C. M. Croce. 2006. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc. Natl. Acad. Sci. U. S. A. 103:7024-7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dull, T., R. Zufferey, M. Kelly, R. J. Mandel, M. Nguyen, D. Trono, and L. Naldini. 1998. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 72:8463-8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ebert, M. S., J. R. Neilson, and P. A. Sharp. 2007. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 4:721-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Florenes, V. A., G. M. Maelandsmo, A. Forus, A. Andreassen, O. Myklebost, and O. Fodstad. 1994. MDM2 gene amplification and transcript levels in human sarcomas: relationship to TP53 gene status. J. Natl. Cancer Inst. 86:1297-1302. [DOI] [PubMed] [Google Scholar]
- 12.Forte, E., and M. A. Luftig. 2009. MDM2-dependent inhibition of p53 is required for Epstein-Barr virus B-cell growth transformation and infected-cell survival. J. Virol. 83:2491-2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friborg, J., Jr., W. Kong, M. O. Hottiger, and G. J. Nabel. 1999. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402:889-894. [DOI] [PubMed] [Google Scholar]
- 14.Ganem, D. 2007. Kaposi's sarcoma-associated herpesvirus, p. 2847-2888. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 15.Gottwein, E., X. Cai, and B. R. Cullen. 2006. A novel assay for viral microRNA function identifies a single nucleotide polymorphism that affects Drosha processing. J. Virol. 80:5321-5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gottwein, E., N. Mukherjee, C. Sachse, C. Frenzel, W. H. Majoros, J. T. Chi, R. Braich, M. Manoharan, J. Soutschek, U. Ohler, and B. R. Cullen. 2007. A viral microRNA functions as an orthologue of cellular miR-155. Nature 450:1096-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grundhoff, A., C. S. Sullivan, and D. Ganem. 2006. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA 12:733-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen, A., S. Henderson, D. Lagos, L. Nikitenko, E. Coulter, S. Roberts, F. Gratrix, K. Plaisance, R. Renne, M. Bower, P. Kellam, and C. Boshoff. 2010. KSHV-encoded miRNAs target MAF to induce endothelial cell reprogramming. Genes Dev. 24:195-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He, L., J. M. Thomson, M. T. Hemann, E. Hernando-Monge, D. Mu, S. Goodson, S. Powers, C. Cordon-Cardo, S. W. Lowe, G. J. Hannon, and S. M. Hammond. 2005. A microRNA polycistron as a potential human oncogene. Nature 435:828-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarviluoma, A., E. S. Child, G. Sarek, P. Sirimongkolkasem, G. Peters, P. M. Ojala, and D. J. Mann. 2006. Phosphorylation of the cyclin-dependent kinase inhibitor p21Cip1 on serine 130 is essential for viral cyclin-mediated bypass of a p21Cip1-imposed G1 arrest. Mol. Cell. Biol. 26:2430-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koopal, S., J. H. Furuhjelm, A. Jarviluoma, S. Jaamaa, P. Pyakurel, C. Pussinen, M. Wirzenius, P. Biberfeld, K. Alitalo, M. Laiho, and P. M. Ojala. 2007. Viral oncogene-induced DNA damage response is activated in Kaposi sarcoma tumorigenesis. PLoS Pathog. 3:1348-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lei, X., Z. Bai, F. Ye, J. Xie, C. G. Kim, Y. Huang, and S. J. Gao. 2010. Regulation of NF-kappaB inhibitor IkappaBalpha and viral replication by a KSHV microRNA. Nat. Cell Biol. 12:193-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu, F., W. Stedman, M. Yousef, R. Renne, and P. M. Lieberman. 2010. Epigenetic regulation of Kaposi's sarcoma-associated herpesvirus latency by virus-encoded microRNAs that target Rta and the cellular Rbl2-DNMT pathway. J. Virol. 84:2697-2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin-Caballero, J., J. M. Flores, P. Garcia-Palencia, and M. Serrano. 2001. Tumor susceptibility of p21(Waf1/Cip1)-deficient mice. Cancer Res. 61:6234-6238. [PubMed] [Google Scholar]
- 25.Mendell, J. T. 2008. miRiad roles for the miR-17-92 cluster in development and disease. Cell 133:217-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nachmani, D., N. Stern-Ginossar, R. Sarid, and O. Mandelboim. 2009. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe 5:376-385. [DOI] [PubMed] [Google Scholar]
- 27.O'Hara, A. J., W. Vahrson, and D. P. Dittmer. 2008. Gene alteration and precursor and mature microRNA transcription changes contribute to the miRNA signature of primary effusion lymphoma. Blood 111:2347-2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oliner, J. D., K. W. Kinzler, P. S. Meltzer, D. L. George, and B. Vogelstein. 1992. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature 358:80-83. [DOI] [PubMed] [Google Scholar]
- 29.Petre, C. E., S. H. Sin, and D. P. Dittmer. 2007. Functional p53 signaling in Kaposi's sarcoma-associated herpesvirus lymphomas: implications for therapy. J. Virol. 81:1912-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pfeffer, S., A. Sewer, M. Lagos-Quintana, R. Sheridan, C. Sander, F. A. Grasser, L. F. van Dyk, C. K. Ho, S. Shuman, M. Chien, J. J. Russo, J. Ju, G. Randall, B. D. Lindenbach, C. M. Rice, V. Simon, D. D. Ho, M. Zavolan, and T. Tuschl. 2005. Identification of microRNAs of the herpesvirus family. Nat. Methods 2:269-276. [DOI] [PubMed] [Google Scholar]
- 31.Platt, G., A. Carbone, and S. Mittnacht. 2002. p16INK4a loss and sensitivity in KSHV associated primary effusion lymphoma. Oncogene 21:1823-1831. [DOI] [PubMed] [Google Scholar]
- 32.Radkov, S. A., P. Kellam, and C. Boshoff. 2000. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat. Med. 6:1121-1127. [DOI] [PubMed] [Google Scholar]
- 33.Refaeli, Y., L. Van Parijs, S. I. Alexander, and A. K. Abbas. 2002. Interferon gamma is required for activation-induced death of T lymphocytes. J. Exp. Med. 196:999-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rivas, C., A. E. Thlick, C. Parravicini, P. S. Moore, and Y. Chang. 2001. Kaposi's sarcoma-associated herpesvirus LANA2 is a B-cell-specific latent viral protein that inhibits p53. J. Virol. 75:429-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Samols, M. A., J. Hu, R. L. Skalsky, and R. Renne. 2005. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi's sarcoma-associated herpesvirus. J. Virol. 79:9301-9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Samols, M. A., R. L. Skalsky, A. M. Maldonado, A. Riva, M. C. Lopez, H. V. Baker, and R. Renne. 2007. Identification of cellular genes targeted by KSHV-encoded microRNAs. PLoS Pathog. 3:e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarek, G., S. Kurki, J. Enback, G. Iotzova, J. Haas, P. Laakkonen, M. Laiho, and P. M. Ojala. 2007. Reactivation of the p53 pathway as a treatment modality for KSHV-induced lymphomas. J. Clin. Invest. 117:1019-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sarek, G., and P. M. Ojala. 2007. p53 reactivation kills KSHV lymphomas efficiently in vitro and in vivo: new hope for treating aggressive viral lymphomas. Cell Cycle 6:2205-2209. [DOI] [PubMed] [Google Scholar]
- 39.Skalsky, R. L., M. A. Samols, K. B. Plaisance, I. W. Boss, A. Riva, M. C. Lopez, H. V. Baker, and R. Renne. 2007. Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155. J. Virol. 81:12836-12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swanton, C., D. J. Mann, B. Fleckenstein, F. Neipel, G. Peters, and N. Jones. 1997. Herpes viral cyclin/Cdk6 complexes evade inhibition by CDK inhibitor proteins. Nature 390:184-187. [DOI] [PubMed] [Google Scholar]
- 41.Umbach, J. L., and B. R. Cullen. 2010. In-depth analysis of Kaposi's sarcoma-associated herpesvirus microRNA expression provides insights into the mammalian microRNA-processing machinery. J. Virol. 84:695-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vassilev, L. T., B. T. Vu, B. Graves, D. Carvajal, F. Podlaski, Z. Filipovic, N. Kong, U. Kammlott, C. Lukacs, C. Klein, N. Fotouhi, and E. A. Liu. 2004. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303:844-848. [DOI] [PubMed] [Google Scholar]
- 43.Verschuren, E. W., N. Jones, and G. I. Evan. 2004. The cell cycle and how it is steered by Kaposi's sarcoma-associated herpesvirus cyclin. J. Gen. Virol. 85:1347-1361. [DOI] [PubMed] [Google Scholar]
- 44.Wade, M., E. T. Wong, M. Tang, J. M. Stommel, and G. M. Wahl. 2006. Hdmx modulates the outcome of p53 activation in human tumor cells. J. Biol. Chem. 281:33036-33044. [DOI] [PubMed] [Google Scholar]
- 45.Waldman, T., K. W. Kinzler, and B. Vogelstein. 1995. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 55:5187-5190. [PubMed] [Google Scholar]
- 46.Wu, X., J. H. Bayle, D. Olson, and A. J. Levine. 1993. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 7:1126-1132. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, J., D. D. Jima, C. Jacobs, R. Fischer, E. Gottwein, G. Huang, P. L. Lugar, A. S. Lagoo, D. A. Rizzieri, D. R. Friedman, J. B. Weinberg, P. E. Lipsky, and S. S. Dave. 2009. Patterns of microRNA expression characterize stages of human B-cell differentiation. Blood 113:4586-4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ziegelbauer, J. M., C. S. Sullivan, and D. Ganem. 2009. Tandem array-based expression screens identify host mRNA targets of virus-encoded microRNAs. Nat. Genet. 41:130-134. [DOI] [PMC free article] [PubMed] [Google Scholar]