Abstract
The genomic RNA of retroviruses and retrovirus-like transposons must be sequestered from the cellular translational machinery so that it can be packaged into viral particles. Eukaryotic mRNA processing bodies (P bodies) play a central role in segregating cellular mRNAs from the translational machinery for storage or decay. In this work, we provide evidence that the RNA of the Saccharomyces cerevisiae Ty1 retrotransposon is packaged into virus-like particles (VLPs) in P bodies. Ty1 RNA is translationally repressed, and Ty1 Gag, the capsid and RNA binding protein, accumulates in discrete cytoplasmic foci, a subset of which localize to P bodies. Human APOBEC3G, a potent Ty1 restriction factor that is packaged into Ty1 VLPs via an interaction with Gag, also localizes to P bodies. The association of APOBEC3G with P bodies does not require Ty1 element expression, suggesting that P-body localization of APOBEC3G and Ty1 Gag precedes VLP assembly. Additionally, we report that two P-body-associated 5′ to 3′ mRNA decay pathways, deadenylation-dependent mRNA decay (DDD) and nonsense-mediated decay (NMD), stimulate Ty1 retrotransposition. The additive contributions of DDD and NMD explain the strong requirement for general 5′ to 3′ mRNA degradation factors Dcp1, Dcp2, and Xrn1 in Ty1 retromobility. 5′ to 3′ decay factors act at a posttranslational step in retrotransposition, and Ty1 RNA packaging into VLPs is abolished in the absence of the 5′ to 3′ exonuclease Xrn1. Together, the results suggest that VLPs assemble in P bodies and that 5′ to 3′ mRNA decay is essential for the packaging of Ty1 RNA in VLPs.
Exogenous retroviruses and endogenous long terminal repeat (LTR) retrotransposons have complex modes of replication involving reverse transcription of the genomic RNA in the cytoplasm and integration of the cDNA into the host genome. Consequently, the replication of these retroelements is modulated by a multitude of host facilitators and defense factors, some of which are conserved from Saccharomyces cerevisiae to humans (3, 41, 42, 61). APOBEC3 proteins are mammalian cytidine deaminases that mount a potent defense against exogenous and endogenous retroelements (25). The human genome encodes seven APOBEC3 proteins, including the prototypical APOBEC3G (A3G), which blocks replication of HIV-1 lacking the gene for the auxiliary factor Vif (77). A3G is incorporated into HIV-1 virions through an RNA-dependent interaction with the nucleocapsid domain of HIV-1 Gag (2, 13, 70, 82, 88). In the virion, A3G interferes with reverse transcription, at least partly by deaminating dC residues in the minus strand of HIV-1 cDNA, leading to cDNA degradation and to dG-to-dA mutations in the plus strand of the integrated provirus (44, 54, 58, 80, 89). A3G also restricts the replication of endogenous retroelements in other species, including the LTR retrotransposon Ty1 in Saccharomyces cerevisiae (31, 34, 74). A3G interacts with Ty1 Gag in an RNA-dependent fashion, is incorporated into cytoplasmic Ty1 virus-like particles (VLPs), and blocks Ty1 retrotransposition by a mechanism that is analogous to that of HIV-1 restriction (31, 74, 75).
In mammalian cells, A3G interacts with proteins involved in mRNA translational repression and decay and is localized to eukaryotic mRNA processing bodies (P bodies) and related ribonucleoprotein (RNP) granules known as stress granules (17, 37, 84). P bodies are dynamic cytoplasmic RNP granules at which components of the deadenylation-dependent mRNA decay (DDD) pathway and the nonsense-mediated decay (NMD) pathway are concentrated along with their mRNA substrates. P-body-associated mRNA degradation in eukaryotes is initiated by deadenylation of mRNA from the 3′ end by the Ccr4-Pop2-Not1-5 deadenylase complex, leading to translational repression by Dhh1/Rck and Pat1, removal of the 5′ cap by the essential Dcp1-Dcp2 complex, and 5′ to 3′ exonucleolytic degradation by Xrn1 (Kem1). Other components of the DDD pathway that are enriched in P bodies include the heptameric activator of decapping, Lsm1-7, and Edc3, which promotes P-body assembly (18, 35, 68, 78). Polyadenylated mRNAs can also be targeted to P bodies via the NMD pathway (79), which recognizes mRNAs containing premature stop codons. The NMD pathway includes Upf1, Upf2, Upf3, Dcp1, Dcp2, and Xrn1 but not Dhh1 (20, 45, 65). Despite the association of 5′ to 3′ mRNA decay factors with P bodies, 5′ to 3′ degradation of mRNA by either DDD or NMD can occur cotranslationally, and therefore degradation is not necessarily coupled to the localization of mRNA in P bodies (47, 48).
Work by the Sandmeyer and Garfinkel labs has uncovered distinct but significant roles for yeast P bodies in Ty3 and Ty1 VLP assembly, respectively (6, 14, 52). Ty3 Gag proteins, RNA, and VLPs localize to P bodies, and this localization requires the nucleocapsid domain of Ty3 Gag, which is essential for Ty3 RNA packaging in VLPs (6, 52). The P-body-associated proteins Xrn1, Dhh1, Pat1, and Lsm1 are required for efficient retrotransposition of Ty3 elements. Moreover, Ty3 Gag foci are increased in size in an xrn1Δ mutant and more diffuse in a dhh1Δ mutant, as are P bodies (6, 51). Together, these results provide compelling evidence for the assembly of Ty3 VLPs in P bodies. Interestingly, however, previous studies have suggested that this may not be the case with Ty1 VLPs. While Ty1 Gag and RNA colocalized in cytoplasmic foci, they were spatially distinct from P bodies (14, 57). However, formation of Ty1 Gag/RNA foci and retrotransposition were enhanced by the same P-body-associated proteins that are required for efficient Ty3 retrotransposition, including the 5′ to 3′ exonuclease Xrn1 and the DDD pathway components Dhh1, Lsm1, and Pat1 (8, 14, 43). In the xrn1Δ and dhh1Δ mutants, Ty1 RNA levels were not significantly altered, but the levels of Ty1 integrase, reverse transcriptase proteins, and cDNA were reduced, and the formation of VLP clusters as visualized by electron microscopy (EM) was diminished. Moreover, the levels of Ty1 antisense transcripts were increased to variable degrees in these mutants (14). These findings, together with the intriguing observation that Ty1 VLPs harbor a mixture of capped and decapped Ty1 RNA (16, 24), suggested that DDD components function outside P bodies to promote a posttranscriptional step in Ty1 retrotransposition.
Ty1 is a 5.9-kb element consisting of direct terminal repeats flanking two overlapping open reading frames, gag (TYA1) and pol (TYB1). Ty1 elements are transcribed from the 5′ LTR to 3′ LTR by RNA polymerase II, forming a 5.7-kb terminally redundant mRNA that is capped and polyadenylated. This mRNA is the template for the translation of all Ty1 proteins and for reverse transcription of the full-length cDNA. The following two primary translation products are synthesized, with the latter resulting from a programmed ribosomal frameshift from gag to pol: p49-TyA (Gag) and p199-TyA-TyB (Gag-Pol). A Ty1 RNA dimer is encapsulated into cytoplasmic VLPs consisting of Ty1 Gag and Gag-Pol (36). Inside the VLP, Gag and Gag-Pol are proteolytically processed into the following four proteins that are required for retrotransposition: p45-TyA (mature Gag), protease, integrase, and reverse transcriptase. Subsequently, Ty1 RNA is reverse transcribed into a linear, double-stranded cDNA. The cDNA, in association with integrase, is transported back to the nucleus, where integrase mediates nonhomologous insertion into chromosomal DNA.
The present study was undertaken to explore the role of P-body-associated 5′ to 3′ mRNA decay pathways in Ty1 retrotransposition and A3G-mediated restriction of Ty1. In contrast with the results from previous studies (14, 57), we report that a significant fraction of Ty1 Gag foci localize in P bodies. Moreover, the retrotransposition restriction factor A3G localizes in P bodies independently of its interaction with Ty1 RNA or Gag. Since A3G is packaged into Ty1 VLPs (31), our findings suggest that the localization of Ty1 Gag in P bodies is functionally significant. Consistent with the P-body localization of Ty1 Gag, we demonstrate that Ty1 RNA is translationally repressed in high-molecular-weight RNP complexes. We confirm and extend previous results implicating the P-body-associated DDD pathway in Ty1 retrotransposition and demonstrate that NMD also enhances Ty1 retromobility and that Upf1 is required for efficient restriction of Ty1 retrotransposition by A3G. Furthermore, we find that the DDD and NMD pathways play additive roles in enhancing Ty1 retrotransposition, since single and double mutations that block both of these pathways of 5′ to 3′ decay cause more severe retrotransposition defects than DDD- and NMD-specific mutations. In the absence of 5′ to 3′ decay factors, Ty1 cDNA levels, but not Ty1 Gag levels, are significantly reduced. In an xrn1Δ mutant, which lacks 5′ to 3′ decay, packaging of Ty1 RNA into VLPs is abolished. Together, our results indicate that 5′ to 3′ mRNA decay is essential for the assembly of Ty1 Gag and RNA into functional VLPs and suggest that P bodies are sites of RNA and A3G packaging into VLPs.
MATERIALS AND METHODS
Strains.
The Saccharomyces cerevisiae strains described below are derivatives of strains BY4741 and BY4742. Genotypes of the strains used are listed in Table 1. Derivatives of strains BY4741 and BY4743 in which individual yeast open reading frames (ORFs) are replaced by the kanMX marker gene (86) or fused at the 3′ end terminus to the TAP tag (40) were obtained from Open Biosystems. Derivatives of BY4741 harboring XRN1-GFP-HIS3, DCP2-GFP-HIS3, DHH1-GFP-HIS3, and NVJ1-GFP-HIS3 (50) were a generous gift from Roy Parker.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | 86 |
| JC3212 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 Ty1his3AI[Δ1]-3114 | 72 |
| JC3607 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 Ty1-GFP-3566 tec1Δ::URA3 | 72 |
| JC3688 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 Ty1-GFP-3566 fus3Δ::URA3 | 72 |
| JC3787 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 Ty1his3AI[Δ1]-3114 | 64 |
| JC3807 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 Ty1his3AI[Δ1]-3114 Ty1-GFP-3566 | 72 |
| JC3903 | MATahis3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 Ty1his3AI[Δ1]-3114 fus3Δ::kanMX | 72 |
| JC4684 | lsm1Δ::kanMX derivative of JC3212 | This study |
| JC4704 | MATahis3Δ1 leu2Δ0 ura3Δ0 Ty1his3AI[Δ1]-3114 tec1Δ::kanMX | 72 |
| JC4742 | dhh1Δ::MET15 derivative of JC3212 | This study |
| JC4746 | dhh1Δ::MET15 derivative of JC3807 | This study |
| JC4750 | pat1Δ::kanMX derivative of JC3212 | This study |
| JC4759 | xrn1Δ::kanMX derivative of JC3212 | This study |
| JC4762 | xrn1Δ::kanMX derivative of JC3807 | This study |
| JC4928 | his3Δ1 leu2Δ0 ura3Δ0 Ty1his3AI[Δ1]-3114 Ty1-GFP-3566 dcp2-DAmP-LEU2 | This study |
| JC4960 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 Ty1his3AI[Δ1]-3114 upf1Δ::kanMX | This study |
| JC4963 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 Ty1his3AI[Δ1]-3114 edc3Δ::kanMX | This study |
| JC4964 | ccr4Δ::kanMX derivative of JC3212 | This study |
| JC4965 | edc2Δ::kanMX derivative of JC3212 | This study |
| JC4969 | sbp1Δ::kanMX derivative of JC3212 | This study |
| JC5015 | upf3Δ::kanMX derivative of JC3212 | This study |
| JC5030 | upf1Δ::URA3 derivative of JC3807 | This study |
| JC5033 | upf1Δ::URA3 derivative of JC4964 | This study |
| JC5053 | his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 trp1:hisG Ty1his3AI[Δ1]-3114 dcp1Δ::kanMX [pCEN-DCP1] | This study |
| JC5057 | his3Δ1 leu2Δ0 ura3Δ0 trp1:hisG Ty1his3AI[Δ1]-3114 dcp1Δ::kanMX [pCEN-dcp1-34] | This study |
| JC5060 | ste12Δ::MET15 derivative of JC3212 | This study |
| JC5063 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ste12Δ::MET15 DCP2-GFP-HIS3 | This study |
| JC5066 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ste12Δ::MET15 DHH1-GFP-HIS3 | This study |
| JC5298 | spt3Δ::kanMX derivative of JC3212 | This study |
| JC5302 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 spt3Δ::MET15 DCP2-GFP-HIS3 | This study |
| JC5304 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 spt3Δ::MET15 DHH1-GFP-HIS3 | This study |
| JC5405 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 upf1Δ::kanMX DCP2-GFP-HIS3 | This study |
Strains JC3212, JC3787, and JC3807 have been described previously (64, 72). Primers used in strain construction are provided in Table S1 in the supplemental material. The lsm1Δ::kanMX, pat1Δ::kanMX, edc2Δ::kanMX, sbp1Δ::kanMX, upf3Δ::kanMX, and ccr4Δ::kanMX alleles were introduced into strain JC3212 by PCR-mediated gene disruption. A PCR product containing each allele was synthesized with primer pair PJ50 and PJ51, PJ47 and PJ48, PJ304 and PJ305, PJ322 and PJ323, PJ414 and PJ415, or PJ278 and PJ279 and genomic DNA from the lsm1Δ::kanMX, pat1Δ::kanMX, edc2Δ::kanMX, sbp1Δ::kanMX, upf3Δ::kanMX, or ccr4Δ::kanMX derivative of BY4741. The xrn1Δ::kanMX strains JC4759 and JC4762 are derivatives of strains JC3212 and JC3807, respectively, made by one-step gene disruption with a PCR product generated with primers PJ56 and PJ57 and genomic DNA from the xrn1Δ::kanMX derivative of BY4741. The upf1Δ::URA3 ccr4Δ::kanMX strain JC5033 was made by introducing the upf1Δ::URA3 disruption cassette from plasmid pPL65 (55) into strain JC4964. Strain JC4960 is a spore of a cross between the upf1Δ::kanMX derivative of BY4741 and JC3787. Strain JC5030 is a upf1Δ::URA3 derivative of JC3807 made by gene disruption with the upf1Δ::URA3 cassette from plasmid pPL65. Strain JC4963 is a spore of a cross between the edc3Δ::kanMX derivatives of strains BY4741 and JC3787.
Strains JC4742 and JC4746 were constructed by introducing a dhh1Δ::MET15 PCR product generated with primers PJ210 and PJ211 and plasmid pRS401 into strains JC3212 and JC3807, respectively. The ste12Δ::MET15 strains JC5060, JC5063, and JC5066 were constructed by PCR-mediated gene disruption of strain JC3212, the DHH1-GFP-HIS3 derivative of BY4741, and the DCP2-GFP-HIS3 derivative of BY4741, respectively. The ste12Δ::MET15 PCR product was generated with primers PJ144 and PJ145 and plasmid template pRS401. The strains JC5298, JC5302, and JC5304 were constructed by introducing a spt3Δ::kanMX allele into the strain JC3212, the DCP2-GFP-HIS3 derivative of BY4741, and the DHH1-GFP-HIS3 derivative of BY4741, respectively, by PCR-mediated gene disruption. The spt3Δ::kanMX allele was generated by PCR with primers SPT3F and SPT3R and genomic DNA from the spt3Δ::kanMX derivative of BY4741.
The dcp2::DAmP allele in strain JC4928 contains the LEU2 marker inserted into the 3′ untranslated region (UTR) of DCP2, which results in reduced expression of DCP2 (73). The strain was constructed by first generating a diploid strain by mating strain JC3807 to strain JC3787. A PCR product synthesized using primers PJ359 and PJ360 and plasmid pRS405 as a template was introduced into the diploid strain to generate a dcp2::DAmP/DCP2 diploid strain. Tetrads were dissected to obtain the spore JC4928.
Strains JC5053 and JC5057 are dcp1Δ::kanMX isolates harboring plasmids pRP783 (DCP1) and pRP896 (dcp1-34) (83), respectively. The strains were constructed by crossing a trp1::hisG derivative of strain JC3212 with a trp1::hisG derivative of JC3787. One copy of DCP1 was deleted in the resulting diploid by a one-step gene disruption that used two overlapping PCR products. One product was generated with primers PJ284 and DNTG1, and the second was generated with primers kanC and PJ334. Genomic DNA of the DCP1/dcp1Δ::kanMX derivative of BY4743 was used as a template in both PCRs. The resulting heterozygous dcp1Δ::kanMX/DCP1 strain was subsequently transformed with plasmids pRP783 and pRP896, a generous gift from Roy Parker. Individual transformants were sporulated, and G418r/Trp+ haploid spores were obtained by tetrad dissection.
Plasmid construction.
The plasmid pJC860 (pGAL1-A3G-RFP) is a LEU2-marked, CEN-based yeast plasmid containing an ORF encoding red fluorescent protein (RFP) fused to the A3G ORF under the control of the GAL1 promoter. The RFP ORF was amplified by PCR with primers PJ68 and PJ61 and the template plasmid pNB2265, kindly provided by Suzanne Sandmeyer (6). The A3G ORF was amplified with primers PJ69 and PJ70 and the template pcDNA-APOBEC3G, kindly provided by Bryan Cullen (31). The A3G and RFP ORFs were fused by overlap extension PCR and amplified using primers PJ70 and PJ61. The PCR product was digested with HindIII and XbaI and cloned into pJC837 (71) digested with HindIII and XbaI.
The plasmid pJC858 (pLTR-Gag-RFP) is a URA3-marked, 2μm-based yeast plasmid that contains an in-frame fusion of the RFP ORF to nucleotide 1203 of the Ty1-H3 TYA1 ORF driven by a Ty1 LTR promoter. The TYA1 ORF was amplified by PCR with primers PJ64 and PJ63 and the template plasmid pGTy1his3AI[Δ1] (71). The RFP ORF was amplified by PCR using primers PJ62 and PJ60 and the template plasmid pNB2265. The TYA1 and RFP ORFs were fused by overlap extension PCR with primers PJ64 and PJ60. The PCR product was digested with XhoI and BamHI and cloned into plasmid pBJC116 digested with XhoI and BglII. Plasmid BJC116 consists of a Yep24 vector harboring the XbaI-XhoI fragment of pNF4000 (69), which contains the 5′ end of the 5′ LTR of Ty1-588.
Microscopy.
Fluorescence microscopy was performed by using a Zeiss Axioskop (Carl Zeiss Inc.). Images were captured with an ORCA-ER digital camera (Hamamatsu) and Openlab software (Improvision). All images were obtained at a magnification of ×1,000 with a Zeiss Plan-Apochromat 100×/1.40 oil differential interference contrast (DIC) objective. Fluors were detected with the following exciter/emitter filter sets: 345/425, Hoechst 33342 dye; 480/535, green fluorescent protein (GFP); 560/645, RFP. Images were colored and merged in Adobe Photoshop 8.0.
Independent transformants of plasmid pGAL1-A3G-RFP were grown in synthetic complete medium lacking leucine and containing 2% glucose (SC-LEU 2% glucose broth) overnight at 30°C, diluted 1:50 into SC-LEU 2% galactose broth, and grown at 20°C to late log phase (optical density at 600 nm [OD600] of 0.8 to 1.1). Independent transformants of plasmid pLTR-Gag-RFP were grown in SC-URA 2% glucose broth overnight at 30°C, diluted 1:100 into SC-URA 2% glucose broth, and grown at 20°C to late log phase. Hoechst 33342 dye was added to 1 ml of cells to a final concentration of 50 μg/ml, and the cells were incubated at 20°C for 10 min. The cells were collected by centrifugation at 1,000 rpm, washed three times in 1 ml distilled water (dH2O), and suspended in 100 μl dH20. A 1-μl aliquot was spotted on a glass slide and visualized by fluorescence microscopy.
To assess the colocalization of GFP and RFP foci, the following criteria were used: (i) cells were selected for analysis if the Hoechst dye appeared concentrated in the nucleus and did not spread throughout the cell; (ii) characteristically bright GFP and RFP spots, above the background, were counted; and (iii) foci were categorized as overlapping when RFP and GFP foci produced yellow in the digitally merged image, separate when RFP and GFP foci were separated by the radius of one focus or a greater length, and adjacent when RFP and GFP foci were separated by less than the approximate radius of one focus in the digitally merged image.
Affinity purification of P-body proteins.
Derivatives of BY4741 harboring XRN1-TAP, PAT1-TAP, LSM1-TAP, or TDH3-TAP, or strain JC3212, which lacks a TAP tag, were transformed with pGAL1-A3G-3xHA plasmid DNA (31). Independent transformants were grown in SC-URA 2% sucrose/2% raffinose broth at 20°C to mid-log phase, when galactose was added to a 2% final concentration; the cultures were then grown at 20°C for 6 h. The cells were washed once in lysis/binding buffer (20 mM HEPES-NaOH at pH 7.8, 150 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 5% glycerol, 0.05% NP-40, 1 mM dithiothreitol [DTT]), and total cell extract was prepared by glass bead cell disruption in lysis/binding buffer plus the Complete Mini, EDTA-free protease inhibitor cocktail (Roche). IgG-Sepharose 6 Fast Flow (GE Healthcare) cross-linked by incubation with 20 mM dimethyl pimelimidate (Pierce) in 0.2 M Na-borate, pH 9.0, and quenched by incubation in 0.2 M ethanolamine, pH 8.0, was bound to cell lysates. Following incubation, the IgG-Sepharose was washed consecutively in 60, 40, and 40 volumes of TEV cleavage buffer (10 mM Tris-Cl at pH 8.0, 150 mM NaCl, 0.1% NP-40, 0.5 mM EDTA, 1 mM DTT) at 4°C. Tobacco etch virus protease (AcTEV; Invitrogen) was added to the settled IgG-Sepharose in a 100-μl volume, and the samples were incubated at 20°C for 6 h. An equal amount of supernatant was mixed with 4× sodium dodecyl sulfate (SDS) loading buffer, and the proteins were separated by polyacrylamide gel electrophoresis (PAGE) on 4 to 20% acrylamide gels. The proteins were analyzed by Western blotting with anti-HA monoclonal antibody or anti-calmodulin binding protein (CBP) polyclonal antibodies (Santa Cruz).
Polysome analysis.
Strains JC3212 and JC4742 were grown in yeast extract-peptone-dextrose (YPD) broth at 30°C to saturation, then diluted 1:100 into fresh YPD broth, and grown at 20°C to mid-log phase. Cells were harvested by centrifugation at 4°C and washed once with cold polysome lysis buffer (20 mM Tris-HCl at pH 8, 140 mM KCl, 5 mM MgCl2, 0.5 mM DTT, 1% Triton X-100, 1 mg/ml heparin). Cell extract was prepared from 12 to 15 OD600 units of cells by glass bead lysis in 500 μl polysome lysis buffer with or without 7.5 mM EDTA plus the Complete Mini, EDTA-free protease inhibitor cocktail (Roche) and 200 units/ml RNasin (Promega). The cell extract was centrifuged at 4,000 rpm for 5 min, then transferred to a fresh tube, and centrifuged at 10,000 rpm for 5 min. Cell extract at 18 A260 units was applied to 5% to 45% (wt/vol) sucrose gradients in polysome lysis buffer without Triton X-100 and with or without 7.5 mM EDTA. Sucrose gradients were formed using a BioComp gradient station (model 153; BioComp Instruments) set at an angle of 81.5° and a speed of 21 rpm for 84 s. The gradients were centrifuged at 39,000 rpm for 2.5 h at 4°C using an SW41 rotor. Twenty-four 450-μl fractions were collected into 1 ml of 8 M guanidine-HCl using the BioComp gradient station set at 0.5 mm/min and 3.10 mm per fraction. RNA was precipitated from each of the even-numbered fractions by the addition of an equal volume of 100% ethanol. RNA samples were subjected to Northern analysis as described previously using a 32P-labeled Ty1 riboprobe (71) and an ACT1 riboprobe. The template for the synthesis of the ACT1 riboprobe was a PCR fragment generated with primers PJ355 and PJ356 and yeast genomic template DNA. Ty1 mRNA and ACT1 mRNA bands were quantified by phosphorimage analysis.
Transposition frequency assays.
To measure the transposition of Ty1his3AI[Δ1]-3114, single-colony isolates of JC3212 and congenic derivatives were grown in YPD broth at 30°C to saturation. The cells were diluted 1:1,000 into fresh YPD broth and grown at 20°C for 3 days. A dilution of each culture was plated on YPD agar, and aliquots were plated on SC-HIS 2% glucose agar; plates were then incubated at 30°C for 3 days. The frequency of Ty1his3AI retrotransposition is the number of His+ prototrophs divided by the total number of CFU plated. The average frequency and standard error for each genotype tested were calculated using 4 to 9 cultures.
Independent colonies of BY4741 and congenic derivatives carrying plasmid pBJC838 (pGTy1his3AI[Δ1]-LEU2-CEN [81]) were grown in SC-LEU 2% glucose broth at 30°C to saturation. The cells were diluted 1:100 into SC-LEU 2% galactose/2% sucrose/2% raffinose broth and grown at 20°C for 3 days. A dilution of each culture was spread on SC-LEU 2% glucose agar, aliquots were plated on SC-LEU-HIS 2% glucose agar, and plates were incubated at 30°C for 3 days. The frequency of pGTy1his3AI retrotransposition is the number of His+ Leu+ colonies divided by the total number of Leu+ cells plated. The average frequency and standard error for each transformed strain were calculated using 3 or 4 cultures.
Strain BY4741 and congenic derivatives carrying pBJC838 were subsequently transformed with either pGAL1-A3G-3xHA or the pYES2 vector. Three independent colonies of each strain were grown in SC-LEU-URA 2% glucose broth at 30°C to saturation. The cells were diluted 1:100 into SC-LEU-URA 2% galactose/2% sucrose/2% raffinose broth and grown at 20°C for 3 days. A dilution of each culture was spread on SC-LEU-URA 2% glucose agar, aliquots were plated on SC-LEU-URA-HIS 2% glucose agar, and plates were incubated at 30°C for 3 days. The frequency of pGTy1his3AI retrotransposition is the number of His+ Leu+ Ura+ colonies divided by the total number Leu+ Ura+ cells plated. The median retrotransposition frequency was calculated using 6 to 13 independent cultures per strain. The ratio of the median pGTy1his3AI frequency in the strain harboring pGAL1-A3G-3xHA to that in the strain harboring the vector yielded the fold inhibition by APOBEC3G. One-sided hypothesis tests of whether APOBEC3G inhibition was significantly reduced in mutant strains were performed by conducting nonparametric bootstrap analyses (32) under the null hypothesis that the fold inhibition in each mutant is the same as that in the wild-type strain. For each test, the pGTy1his3AI transposition frequency data were resampled 10,000 times, and the ratios of median fold inhibition were recalculated. One-sided P values were obtained as the number of bootstrap samples with a ratio of the median fold inhibition statistic greater than 1. To account for the increased expectation of finding a significant result when performing multiple tests, false discovery rate (FDR) corrections were applied post hoc (7).
Ty1 cDNA levels.
Independent colonies of strain JC3212 and congenic derivatives were grown to saturation at 20°C. Total genomic DNA was digested with SphI and subjected to Southern blot analysis with a 32P-labeled TYB1 riboprobe as described previously (71).
Northern analysis.
Total RNA was prepared from cells grown to mid-log phase at 20°C, as described previously (60). Plasmids pGEM-TYA1 and pGEM-HIS3, the DNA templates for riboprobe synthesis to detect Ty1 mRNA and Ty1his3AI mRNA, respectively, have been described previously (29). Plasmid pDG512, a gift from David Garfinkel, was used as a template to synthesize a riboprobe to detect 18S ribosomal DNA (rDNA). Plasmid pJC940, which contains the XhoI-BglII fragment of Ty1-H3 (nucleotides 238 to 1702) cloned into the pSP70 vector (Promega), was used as a DNA template for the synthesis of a riboprobe to detect Ty1AS RNA. Bands were quantified by phosphorimaging.
Gag-GFP activity assay.
Strain JC3807 and derivatives, which harbor the Ty1-GFP-3566 (GAG-GFP) element (72), and strain JC3212 and derivatives, which lack Ty1-GFP-3566, were grown in YPD broth at 30°C overnight. The strains were diluted to an OD600 of 0.1 in YPD and grown at 20°C for 3 h. The cells were diluted 1:10 into dH2O. The geometric mean of the GFP activity in 10,000 cells of each strain was measured by flow cytometry using a FACSCalibur (Becton Dickinson). The difference between the mean GFP activity in each strain containing Ty1-GFP-3556 and the mean GFP activity in the isogenic strain lacking Ty1-GFP-3556 was calculated to correct for differences in autofluorescence arising from differences in cell size.
Western blot analyses.
Strain JC3212 and congenic derivatives were grown at 20°C to mid-log phase. Total cell lysates were separated on 10% SDS-PAGE gels, and proteins were transferred to polyvinylidene difluoride (PVDF) membranes. The membranes were incubated with affinity-purified anti-Gag polyclonal antibody in phosphate-buffered saline (PBS) plus 0.05% Tween 20 and 1% nonfat milk. Subsequently, the membrane was incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies and SuperSignal West Pico chemiluminescent substrate (Pierce), and then the blots were exposed to film. The films were photographed with a FluorChem 8900 (Alpha Innotech), and the intensity of the bands was quantified with AlphaEase FC (Alpha Innotech). The PVDF membranes were stripped of antibody in 50 mM Tris-HCl (pH 7), 2% SDS, and 50 mM DTT at 70°C for 30 min. The membrane was incubated with antitubulin polyclonal antibody (Chemicon International) as a loading control, and the protein band was visualized as described above. A series of 2-fold dilutions of whole-cell extracts were analyzed by SDS-PAGE to quantitate Ty1 Gag levels relative to antitubulin levels. The band intensities produced from samples in the linear range of each dilution series were used to calculate Gag levels relative to antitubulin levels. The average results and standard errors were calculated using three or four samples of each strain analyzed on two Western blots.
Benzonase protection assay.
Strain JC3212 and congenic derivatives were grown at 20°C to mid-log phase. Cells were harvested by centrifugation at 4°C and washed once with cold dH2O. Cell extract was prepared from 15 OD600 units by glass bead lysis in 350 μl extraction buffer (10 mM Tris-HCl at pH 7.5, 100 mM KCl, 10 mM EDTA) plus Complete Mini, EDTA-free protease inhibitor cocktail and 57 units/ml RNasin. All steps were conducted at 0 to 4°C unless otherwise indicated. The cell extract was centrifuged at 1,000 rpm for 5 min, then transferred to a fresh tube, and centrifuged at 10,000 rpm for 1 min. Three 65.75-μl aliquots of whole-cell extract were added to 7.5 μl of 10× Benzonase buffer (100 mM Tris-HCl at pH 7.5, 100 mM MgCl2, 500 mM NaCl), 0.25 μl in vitro-synthesized Ty1 control transcript (5 to 10 ng), and 1.5 μl Benzonase or dH2O. The Ty1 in vitro transcript was synthesized using pGEM-TYA1 as a template and T7 polymerase. Samples with and without Benzonase were transferred to a 24°C water bath for 6 min. A third set of samples lacking Benzonase were kept at 0°C. Addition of 5 μl of 0.5 M EDTA and 500 μl RNA isolation buffer (10 mM Tris-HCl at pH 7.5, 10 mM EDTA, 2% SDS) stopped the reaction. RNA was phenol-chloroform extracted, ethanol precipitated, and resuspended in dH2O. An aliquot of RNA (20 μg) from the control samples or the equivalent volume from Benzonase-treated samples was denatured with an equal volume of glyoxal, separated by agarose gel electrophoresis, and transferred to Hybond-XL (GE Healthcare). A 32P-labeled Ty1 riboprobe was synthesized using T7 polymerase. DNA templates for riboprobe synthesis were generated by PCR amplification with primers PJ357 and PJ358 and plasmid pGEM-TYA1 as a template.
RESULTS
Human A3G and Ty1 Gag localize to P bodies in yeast.
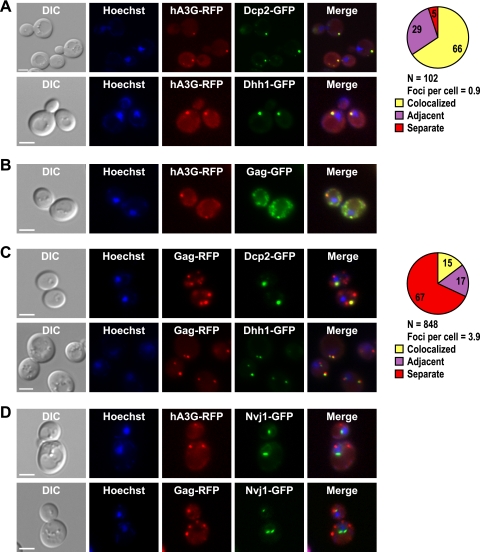
The A3G protein localizes to P bodies in human peripheral blood cells and cell lines (37, 84). To determine whether A3G localizes to P bodies when expressed in yeast cells, the ORF encoding RFP was fused to the 3′ end of the A3G ORF, which is driven from the inducible GAL1 promoter on a centromere-based plasmid (pGAL1-A3G-RFP). The A3G-RFP protein was expressed in strains harboring a genomic copy of either DCP2-GFP or DHH1-GFP (50). A3G-RFP localized in cytoplasmic foci in most cells, with a range of 0 to 4 foci per cell and an average of 0.9 focus per cell (Fig. 1A). (Cells that lack A3G foci may have lost the pGAL1-A3G-RFP plasmid, and therefore, the average number of foci per cell could be an underestimate.) In contrast, RFP expressed from the GAL1 promoter on a high-copy-number yeast plasmid does not form foci (6). Moreover, we found that 66% of A3G-RFP foci colocalized with P bodies, based on complete or partial overlap with Dcp2-GFP foci. An additional 29% of A3G-RFP foci were directly adjacent to a Dcp2-GFP focus, but no region of overlap was detected in the merged imaged. Similar extents of overlap and adjacency were also observed when Dhh1-GFP was used to visualize P bodies (Fig. 1A) (quantification not shown). The A3G foci did not overlap with the foci of the control fusion protein Nvj1-GFP (Fig. 1D). These data suggest that the A3G protein is closely associated with or contained in P bodies in yeast cells.
FIG. 1.
Human A3G and Ty1 Gag localize to P bodies. Individual yeast cells treated with Hoechst dye (Hoechst) to visualize cell nuclei were located by differential interference contrast (DIC) microscopy and fluorescence microscopy. Scale bar, 3 μm. Fluorescent images were merged using Adobe Photoshop. (A) Yeast strains harboring DCP2-GFP or DHH1-GFP and plasmid pGAL1-A3G-RFP were grown in galactose-containing broth, and live cells were visualized to determine the subcellular locations of P bodies and A3G-RFP and their joint subcellular location (Merge). The pie graph shows the percentage of A3G-RFP foci that colocalize with Dcp2-GFP foci or were adjacent to Dcp2-GFP foci (see Materials and Methods for complete description of quantitation). “N” represents the total number of A3G-RFP foci examined for colocalization with Dcp2-GFP. (B) Live cells harboring the chromosomal Ty1-GFP-3566 element and the pGAL1-A3G-RFP plasmid were grown in the presence of galactose to determine the subcellular locations of Gag-GFP and A3G-RFP. (C) Live cells harboring DCP2-GFP or DHH1-GFP and plasmid pLTR-GAG-RFP were visualized to determine the subcellular locations of P bodies and Ty1 Gag-RFP and their joint subcellular location. The pie graph shows the percentage of Gag-RFP foci that colocalize or are adjacent to Dcp2-GFP foci. “N” represents the number of Gag-RFP foci examined for colocalization with Dcp2-GFP. (D) The subcellular locations of Gag-RFP and A3G-RFP were visualized in live cells harboring NVJ1-GFP and plasmid pGAL1-A3G-RFP or pLTR-Gag-RFP. NVJ1 encodes a nuclear-vacuolar junction protein.
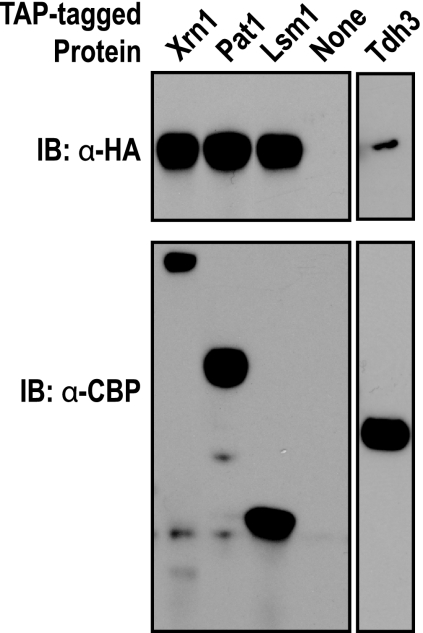
To determine whether the colocalization of A3G with P bodies was due to the formation of specific complexes containing A3G and P-body constituents, we expressed A3G with a C-terminal 3× hemagglutinin (3×HA) epitope tag in yeast strains containing an allele of XRN1, LSM1, or PAT1 in which the ORF was fused to the tandem affinity purification (TAP) tag (40). TAP-tagged proteins and biochemically associated complexes were purified from cell lysates by binding to IgG-Sepharose, and complexes were released with TEV protease. Western blot analysis of protein complexes demonstrated that A3G-3xHA copurified with Xrn1-TAP, Lsm1-TAP and Pat1-TAP (Fig. 2). However, all of these proteins can bind RNA, and therefore the interaction of A3G with Xrn1, Lsm1 and Pat1 could occur via nonspecific RNA-mediated bridging. To address the specificity of the interactions, we asked whether A3G copurifies with another RNA binding protein, Tdh3, which is not associated with P bodies. Only a very low level of A3G-3×HA copurified with Tdh3, supporting the idea that the presence of A3G in specific complexes with Xrn1, Lsm1, and Pat1 underlies its colocalization with P bodies.
FIG. 2.
Human A3G copurifies with core P-body proteins in yeast. Yeast strains expressing C-terminal fusions of the TAP tag to Xrn1, Pat1, Lsm1, or Tdh3 or expressing no TAP-tagged protein (None) and coexpressing A3G-3×HA were analyzed. TAP-tagged proteins and copurifying proteins were released from IgG-Sepharose by cleavage with TEV protease. (Top) Western blot reacted with anti-HA antibody (α-HA), which detects A3G-3×HA present in purified TAP-tagged protein complexes. (Bottom) Western blot reacted with anti-CBP antibodies (α-CBP), which detect the TAP-tagged protein in purified protein complexes. IB, immunoblot.
Because A3G interacts with Ty1 Gag in VLPs and potently inhibits retrotransposition (31), we asked whether Ty1 Gag also colocalizes with A3G in P bodies. To assess A3G colocalization with Gag, we expressed the pGAL1-A3G-RFP plasmid in a yeast strain that carries GFP fused to the 3′ end of the GAG ORF in a single chromosomal copy of Ty1 (Ty1-GFP-3566) (72). Consistent with previous results obtained using other methodologies (14, 57), we found that Gag-GFP expressed from a chromosomal Ty1 element was distributed in multiple cytoplasmic foci. Moreover, a significant proportion of Gag-GFP foci overlapped with A3G-RFP foci (Fig. 1B). To determine whether Gag localizes in P bodies, we constructed a GAG-RFP fusion gene driven from the LTR promoter on a high-copy-number plasmid (pLTR-GAG-RFP). Like Gag-GFP, Gag-RFP localized in multiple discrete cytoplasmic foci (Fig. 1C). Moreover, when pLTR-GAG-RFP was expressed in the strain harboring the chromosomal Ty1-GFP-3566 element, nearly complete colocalization of Gag-GFP and Gag-RFP was observed (see Fig. S1 in the supplemental material). When pLTR-GAG-RFP was expressed in a strain harboring DCP2-GFP, approximately 15% of Gag-RFP foci overlapped with Dcp2-GFP foci, while another 17% of Gag-RFP foci were directly adjacent to Dcp2-GFP foci (Fig. 1C). Comparable results were obtained when Gag-RFP colocalization with Dhh1-GFP was evaluated (Fig. 1C). The percentage of Gag-RFP foci that overlapped with Dcp2-GFP foci was not as high as the percentage of A3G foci that overlapped with Dcp2-GFP foci, and the average of 3.9 Gag-RFP foci per cell was substantially higher than the average of 0.9 A3G-RFP focus per cell. In summary, we observed a striking localization of A3G in P bodies, and a fraction of Ty1 Gag foci overlapped with P bodies and with A3G foci.
The localization of a fraction of Gag to P bodies could result from the formation of specific complexes between Gag and core P-body proteins. To explore this idea, expression of Gag was induced from a pGTy1 plasmid in strains expressing Xrn1-TAP, Lsm1-TAP, or Pat1-TAP (40). TAP-tagged proteins were successfully purified from cell lysates, but there was no detectable copurification of Ty1 Gag (data not shown). It remains possible that Gag interacts specifically with another component of P bodies aside from the three tested above. However, these results suggest that Ty1 Gag and A3G localize in P bodies by distinct mechanisms.
Localization of A3G to P bodies occurs independently of Ty retrotransposon expression.
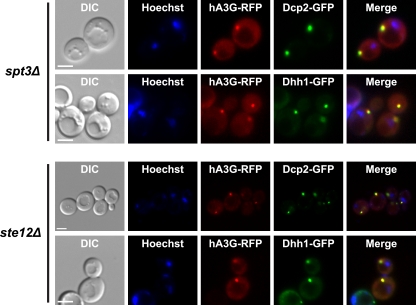
The results given above raise the possibility that A3G localizes in P bodies independently of Ty1 expression. To address this possibility, pGAL1-A3G-RFP was expressed in spt3Δ and ste12Δ derivatives of strains harboring the DCP2-GFP or DHH1-GFP allele. Spt3 and Ste12 are transcription factors that are required for expression of Ty1 elements (21, 23, 85). The A3G-RFP protein accumulated in cytoplasmic foci in spt3Δ and ste12Δ strains and colocalized with both Dcp2-GFP and Dhh1-GFP (Fig. 3). Control experiments in which pLTR-Gag-RFP was expressed in the spt3Δ or ste12Δ strain confirmed that Ty1 Gag-RFP foci are not observed in these mutants (see Fig. S2 in the supplemental material) (data not shown). Together, these results indicate that A3G localizes to P bodies in the absence of Ty1 RNA expression. In fact, there is likely to be very little expression of any retrotransposon in an spt3Δ mutant, since Ty2 expression is also abolished in this mutant, and the expression of Ty3, Ty4, and defective Ty5 elements is very low or repressed (9, 49, 85). Therefore, our data suggest that A3G associates with P bodies through its interactions with host proteins or RNAs that are not encoded by retroelements. The independent localization of A3G and Ty1 Gag in P bodies suggests that P bodies are sites of A3G interaction with Gag during VLP assembly.
FIG. 3.
Human A3G localizes to P bodies independently of Ty1 expression. Individual live yeast cells were treated with Hoechst dye and visualized by differential interference contrast (DIC) and fluorescence microscopies. Scale bar, 3 μm. The localization of human A3G-RFP foci and P bodies in strains lacking Ty1 expression was visualized by inducing expression of plasmid pGAL1-A3G-RFP in an spt3Δ strain harboring DCP2-GFP or DHH1-GFP (top) and an ste12Δ strain harboring DCP2-GFP or DHH1-GFP (bottom).
Ty1 RNA is sequestered from translation in high-molecular-weight nonpolysomal complexes.
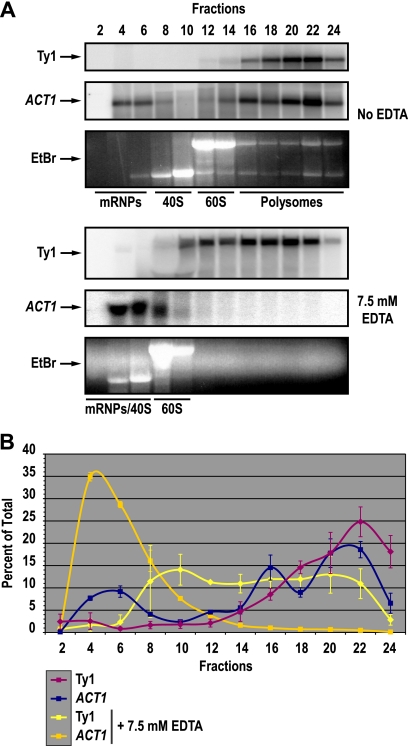
The presence of Ty1 Gag and the Gag-binding protein A3G in P bodies raises the possibility that some fraction of Ty1 RNA is also present in P bodies and therefore sequestered from translation. To determine whether there is a pool of translationally repressed Ty1 RNA, we detected Ty1 RNA across a polysome profile. Cell lysates were fractionated on 5 to 45% sucrose gradients in the absence and presence of 7.5 mM EDTA. Fractionation of lysates in the absence of EDTA allows actively translated mRNAs associated with polysomes to be separated from untranslated mRNPs, which typically sediment near the top of the density gradient. Fractionation of lysates in the presence of EDTA results in the dissociation of ribosomes to ribosomal subunits; therefore, Ty1 RNA on polysomes can be separated from Ty1 RNA in VLPs or other high-molecular-weight nonpolysomal RNPs that are stable in the presence of EDTA (33). Following fractionation on sucrose gradients, total RNA was extracted from even-numbered fractions of the gradients. Northern blot analysis was performed to detect Ty1 RNA and, as a control, the ACT1 mRNA (Fig. 4A). In the absence of EDTA, both Ty1 mRNA and the actively translated ACT1 mRNA comigrated with polysomes in fractions 16 to 24 (Fig. 4B). As expected, addition of EDTA to the sucrose gradient resulted in the dissociation of polysomes and a concomitant shift in ACT1 mRNA from heavy fractions of the sucrose gradient to fractions 4 to 8. Ty1 RNA, on the other hand, remained in high-molecular-weight RNPs in fractions 8 to 22. While Ty1 RNPs shifted modestly from fractions 16 to 24 in the absence of EDTA to fractions 8 to 22 in the presence of EDTA, 40S and 60S ribosomal subunits also shifted toward the top of the gradient, and the positions of Ty1 mRNPs relative to 40S and 60S subunits were not substantially altered by the addition of EDTA. The absence of Ty1 RNA that cosediments with ACT1 mRNA in the free mRNP/40S region of the gradient when polysomes are dissociated indicates that little if any Ty1 RNA is actively translated on polysomes. Instead, Ty1 RNA is located in high-molecular-weight RNPs that are resistant to dissociation by EDTA and therefore distinct from polysomes. A fraction of these large RNPs could be Ty1 VLPs. However, Ty1 RNA comprises up to 1% of total RNA (29), and yet, VLPs are barely detectable (22, 38, 63). Therefore, packaging of Ty1 RNA within VLPs cannot fully explain the strong translational repression of Ty1 RNA in high-molecular-weight RNPs. The sequestration of Ty1 RNA from translation is consistent with the localization of Ty1 Gag and the restriction factor A3G in P bodies.
FIG. 4.
Ty1 mRNA resides in high-molecular-weight nonpolysomal RNPs. (A) Northern blot analysis of Ty1 mRNA (Ty1) and ACT1 RNA (ACT1), the latter as a control for the sedimentation of an actively transcribed mRNA, in even-numbered fractions obtained by fractionation of cell lysates from wild-type strain JC3212 on 5% to 45% sucrose gradients lacking EDTA (top three panels) or containing 7.5 mM EDTA (bottom three panels). The positions of nontranslating mRNPs, 40S and 60S ribosomal subunits, and polysomes were determined by gel electrophoresis of RNA from each fraction on an agarose gel stained with ethidium bromide (EtBr). (B) The percentage of Ty1 or ACT1 mRNA in each fraction as a percentage of the total amount in every fraction assayed is indicated on the y axis. The positions of nontranslating mRNPs, the 40S and 60S ribosomal subunits, and the polysomes are indicated above the graph. Error bars indicate the standard errors of the average results obtained using three biological replicates with and without EDTA for the Ty1 RNA and using two biological replicates for the ACT1 RNA.
The P-body protein Dhh1 is a general repressor of translation that is required for efficient retrotransposition of Ty1 and Ty3 (6, 14, 19, 51). Therefore, we examined the possibility that Dhh1 is required for the translational repression of Ty1 RNA. When a dhh1Δ mutant strain was analyzed, sedimentation of Ty1 RNA in sucrose gradients in the presence and absence of EDTA was very similar to its sedimentation in the analysis of the wild-type strain (see Fig. S3 in the supplemental material). Therefore, Dhh1 plays no apparent role in the repression of Ty1 RNA translation.
A functional 5′ to 3′ mRNA decay pathway is required for efficient Ty1 retrotransposition.
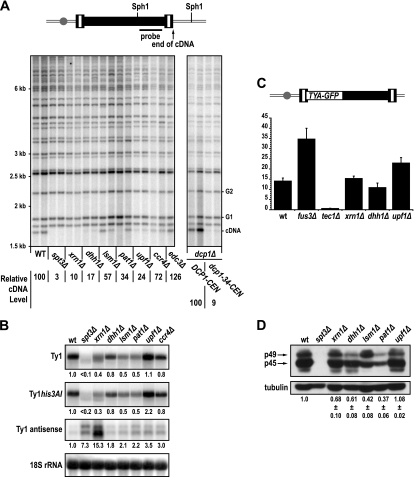
Four 5′ to 3′ mRNA decay proteins that are associated with P bodies—Xrn1, Dhh1, Lsm1, and Pat1—have been shown to promote Ty1 retrotransposition (8, 14). Both the DDD pathway, which includes Ccr4, Dhh1, Pat1, and Lsm1-Lsm7, and the NMD pathway, which includes Upf1, Upf2, and Upf3, target mRNAs for decapping by the Dcp1-Dcp2 complex and 5′ to 3′ degradation by Xrn1. To gain insight into the relative importance of P-body-associated DDD, NMD, and general 5′ to 3′ mRNA decay factors in Ty1 retrotransposition, we deleted the genes encoding these factors in a strain containing a single chromosomal Ty1his3AI element. The his3AI retrotransposition indicator gene allows cells in which the Ty1his3AI element retrotransposes to be detected as His+ prototrophs in a quantitative assay (28). In agreement with previous reports (8, 14), Ty1his3AI retrotransposition was dramatically reduced in the xrn1Δ mutant to 0.5% of the retrotransposition level in a wild-type strain (Table 2). DCP1 and DCP2 are essential genes, so we constructed a dcp1Δ strain carrying a plasmid-borne copy of the strong loss-of-function allele dcp1-34 (83) and a second strain carrying a hypomorphic DAmP allele of DCP2 (73). The strains with hypermorphic mutations in DCP1 or DCP2 each had a substantial reduction in the retrotransposition frequency to <9% of that in the corresponding wild-type strain (Table 2). Given that Dcp1 and Dcp2 are required to remove the 5′ cap from mRNA so that degradation by Xrn1 can occur, these findings suggest that exonucleolytic degradation of RNA by Xrn1 is necessary for Ty1 retrotransposition.
TABLE 2.
Frequency of Ty1his3AI retrotransposition in mRNA decay mutants
| Genotypea | Ty1his3AI retrotransposition frequency ± SE (10−7) | Relative Ty1his3AI transposition frequency (%) |
|---|---|---|
| WT | 4.55 ± 0.67 | 100 |
| xrn1Δ | 0.025 ± 0.005 | 0.5 |
| dcp2-DAmP | 0.258 ± 0.045 | 5.7 |
| dhh1Δ | 0.071 ± 0.001 | 1.6 |
| upf1Δ | 0.114 ± 0.033 | 2.5 |
| upf3Δ | 0.153 ± 0.040 | 3.4 |
| pat1Δ | 0.731 ± 0.133 | 16 |
| lsm1Δ | 0.473 ± 0.033 | 10 |
| edc2Δ | 3.93 ± 1.03 | 86 |
| edc3Δ | 2.29 ± 0.47 | 50 |
| sbp1Δ | 4.74 ± 0.44 | 104 |
| ccr4Δ | 0.905 ± 0.139 | 20 |
| upf1Δ ccr4Δ | <0.020 | <0.4 |
| dcp1Δ DCP1-CEN | 3.55 ± 0.66 | 100 |
| dcp1Δ dcp1-34-CEN | 0.316 ± 0.051 | 8.9 |
WT, wild type.
NMD has not previously been examined for a role in Ty1 retrotransposition, and we find that NMD is required to a large extent. The frequency of Ty1his3AI retrotransposition in the NMD-defective upf1Δ and upf3Δ mutants was <4% of that in a wild-type strain. An equivalent retrotransposition defect was also observed in a upf2Δ mutant (data not shown). The effect of NMD factors is as strong as or stronger than that of DDD factors. For example, in the DDD-defective dhh1Δ mutant, the frequency of retrotransposition was reduced to 1.6% of that in the wild-type strain. However, deletion of other DDD pathway-specific genes, including LSM1, PAT1, and CCR4, had a more modest effect on retrotransposition, yielding frequencies of 10%, 16%, and 20% of that in the wild-type strain, respectively. To determine whether the DDD and NMD pathways independently promote retrotransposition, we deleted both the DDD-specific gene CCR4 and the NMD-specific gene UPF1 in the Ty1his3AI strain. Retrotransposition in the upf1Δ ccr4Δ double mutant was significantly lower than that in either of the single mutants and comparable to that in the xrn1Δ mutant. This result indicates that Upf1 and Ccr4 have additive effects on retrotransposition and suggest that both DDD and NMD contribute to the strong requirement for 5′ to 3′ mRNA degradation in Ty1 retrotransposition.
In contrast to the core components of the DDD and NMD pathways, conditional enhancers of decapping EDC2 or SBP1 (66, 76) play no apparent role in promoting retrotransposition. Moreover, deletion of EDC3, whose gene product provides a scaffold for the decapping complex during P-body assembly (30), caused only a 50% reduction in retrotransposition. Since edc3Δ mutants have reduced levels of microscopically visible P bodies, this result suggests that P-body assembly is not as critical for retrotransposition as the 5′ to 3′ mRNA decay functions that are concentrated in P bodies.
Upf1 enhances A3G-mediated restriction of pGTy1 retrotransposition.
To explore the mechanisms by which 5′ to 3′ mRNA decay proteins promote retrotransposition, we examined the effect of xrn1Δ, dhh1Δ, lsm1Δ, and upf1Δ mutations on retrotransposition of a GAL1p:Ty1his3AI element on a centromere-based plasmid (pGTy1his3AI) (Table 3). Expression of Ty1his3AI from the GAL1 promoter increases not only the total amount of Ty1 RNA but also the efficiency of retrotransposition per Ty1 transcript (27, 39). Retrotransposition of the pGTy1his3AI element was not decreased in the lsm1Δ mutant, but in an xrn1Δ, dhh1Δ or upf1Δ mutant, it was reduced to 1.3%, 3.2%, or 8.7% of the frequency in a wild-type strain. The relative decrease in pGTy1his3AI retrotransposition in these mutants compared to that in the wild-type strain was similar to that of the chromosomal Ty1his3AI element (Table 2). However, the actual frequency of pGTy1his3AI element retrotransposition in the xrn1Δ mutant is almost 1,000-fold higher than that of a chromosomal Ty1his3AI element and is even higher than the retrotransposition frequency of the chromosomal Ty1his3AI element in a wild-type strain (Table 3). These results argue that Xrn1 affects the efficiency of a step in retrotransposition rather than performing an essential function.
TABLE 3.
Frequency of pGTy1his3AI retrotransposition in mRNA decay mutants
| Genotype | pGTy1his3AI retrotransposition frequency ± SE (10−4) | pGTy1his3AI transposition relative to WT strain (%) |
|---|---|---|
| WT | 1.60 ± 0.16 | 100 |
| xrn1Δ | 0.02 ± 0.005 | 1.3 |
| dhh1Δ | 0.05 ± 0.008 | 3.2 |
| lsm1Δ | 2.9 ± 0.5 | 184 |
| upf1Δ | 0.14 ± 0.02 | 8.7 |
Because the level of pGTy1his3AI retrotransposition in xrn1Δ, dhh1Δ, lsm1Δ, and upf1Δ mutants is well above the limit of detection of the retrotransposition assay, we were able to determine whether these P-body proteins are required for A3G-mediated restriction of Ty1 retrotransposition. Coinduction of pGAL1-A3G and pGTy1his3AI resulted in a 36.6-fold inhibition of Ty1his3AI retrotransposition in a wild-type strain (Table 4). This level of A3G-mediated inhibition was altered only slightly in the xrn1Δ, dhh1Δ, or lsm1Δ mutant background. However, A3G-mediated restriction was significantly reduced in the upf1Δ mutant. Therefore, Upf1 is required for the efficient restriction activity of A3G.
TABLE 4.
Inhibition of pGTy1his3AI retrotransposition by A3G in mRNA decay mutants
| Genotype | Fold inhibition of pGTy1his3AI retrotransposition by A3Ga | Relative A3G inhibition (P value)b |
|---|---|---|
| WT | 36.6 | 1.00 |
| xrn1Δ | 15.3 | 0.42 (0.11) |
| dhh1Δ | 75.0 | 2.05 (0.94) |
| lsm1Δ | 35.9 | 0.98 (0.54) |
| upf1Δ | 5.9 | 0.16 (0.0015)c |
Ratio of the median pGTy1his3AI retrotransposition frequency in the strain carrying an empty vector to that in the strain expressing pGAL1-A3G.
Ratio of the fold inhibition in the deletion strain to the fold inhibition in the wild-type strain. P values shown are for a one-sided test to determine whether inhibition is reduced in the deletion strain.
The P value was <0.01 after controlling for FDR.
Since A3G does not gain access to P bodies by interacting with Ty1 Gag or RNA (Fig. 3), we considered the possibility that Upf1's role in A3G-mediated restriction is to enhance the localization of A3G to P bodies. To address this possibility, pGAL1-A3G-RFP was expressed in a upf1Δ derivative of a strain harboring the DCP2-GFP allele. A3G-RFP is localized in cytoplasmic foci that overlap with Dcp2-GFP foci in the upf1Δ strain (see Fig. S4 in the supplemental material), and the degree of overlap was comparable to that in the wild-type strain (Fig. 1A). Moreover, Upf1 is not required for partial colocalization of Gag-RFP with Dcp2-GFP (see Fig. S4 in the supplemental material). This suggests that Upf1 is not necessary for the association of A3G or Gag with P bodies, and therefore, it might act at a later stage in the retrotransposition process, such as VLP assembly or reverse transcription.
Posttranslational regulation of Ty1 retrotransposition by Xrn1, Dhh1, and Upf1.
To investigate the role of 5′ to 3′ mRNA decay factors in retrotransposition, we first asked whether the accumulation of Ty1 cDNA was altered in decay mutants, as suggested previously for the xrn1Δ, lsm1Δ, and dhh1Δ mutants (14). Levels of unintegrated Ty1 cDNA were measured using a standard Southern blot assay (Fig. 5A). Ty1 cDNA levels in the xrn1Δ, dhh1Δ, and dcp1-34 mutants were reduced to ≤10% of the level in the wild-type strain, and the level in the upf1Δ mutant was 24% of the wild-type level. By comparison, cDNA levels were modestly reduced in the lsm1Δ, pat1Δ, and ccr4Δ mutants. No decrease in Ty1 cDNA was detected in the edc3Δ mutant, which had only a 2-fold decrease in retrotransposition. Given that cDNA is an essential intermediate in retrotransposition, these data, obtained in an assay that does not rely on a genetically marked element, provide independent confirmation that Xrn1, Dhh1, Dcp1, and Upf1 are activators of Ty1 retrotransposition. Moreover, they suggest that these mRNA decay factors promote a step in retrotransposition prior to the integration of Ty1 cDNA.
FIG. 5.
5′ to 3′ mRNA decay factors facilitate Ty1 retrotransposition posttranslationally. (A) Southern blot analysis of SphI-digested genomic DNA extracted from two independent isolates of strain JC3212 (wild type [WT]) and congenic mutant derivatives. The dcp1Δ strains complemented by a plasmid bearing DCP1 or dcp1-34 are also shown. The spt3Δ mutant is a negative control for Ty1 cDNA. A schematic of a chromosomal Ty1 element indicating the locations of SphI sites and sequences hybridizing to the probe is shown above the blot. The positions of molecular mass standards are indicated to the left of the blot. The position of the 1.7-kb unintegrated Ty1 cDNA fragment from the internal SphI site in Ty1 cDNA to the 3′ end of the cDNA (cDNA) is indicated to the right of the blot. The positions of two SphI fragments each containing the 3′ end of a genomic Ty1 elements and flanking genomic sequences (G1 and G2), which were used for normalization of Ty1 cDNA levels, are also indicated. The ratios of the signal in the Ty1 cDNA band to the average signal in two genomic Ty1 bands in each strain were averaged for each genotype to compute the relative Ty1 cDNA level, which is expressed as a percentage of the relative Ty1 cDNA level in a wild-type strain. (B) Northern blot analysis of Ty1, Ty1his3AI, and Ty1AS RNA in total yeast RNA prepared from strain JC3212 (wild type [wt]) or congenic derivatives, whose relevant genotypes are indicated. Ty1, Ty1his3AI, and Ty1AS RNA levels relative to the level in a wild-type strain after normalizing to the signal for 18S rRNA are provided below each image of the blot. (C) Histogram of the mean Gag-GFP activity in strain JC3807 (wt) and congenic mutant strains. Gag-GFP activity in the fus3Δ and tec1Δ mutants was measured to control for elevated or diminished levels of Gag, respectively (72). A diagram of the Ty1-GFP-3566 element containing the gag-GFP fusion is shown above the graph. The gray ball represents the centromere and indicates that Ty1-GFP-3566 is integrated into the chromosome. (D) Western blot analysis of Ty1 Gag levels. Total soluble yeast proteins from strain JC3212 or congenic mutant strains were separated by 10% SDS-PAGE, transferred to a PVDF membrane, and reacted with anti-Gag antibody (top). The spt3Δ strain that does not express Ty1 is a negative control. The positions of unprocessed p49-Gag and proteolytically processed p45-Gag are indicated. The α-tubulin protein (tubulin) was detected as a loading control (bottom). The average levels of p45-Gag and p49-Gag relative to α-tubulin were determined by serial dilution of independent samples from each strain (data not shown), and these values are provided below the blot images.
Northern blot analysis was performed to determine whether Ty1 or Ty1his3AI mRNA levels were altered by the deletion of XRN1, DHH1, and UPF1, since these mutants have strong effects on the levels of Ty1 retrotransposition and Ty1 cDNA. We also examined Ty1 RNA levels in the lsm1Δ, pat1Δ, and ccr4Δ mutants for comparison, since their effects on retrotransposition and cDNA are relatively minor. A modest decrease in total Ty1 RNA was observed in all the mutants except for the upf1Δ mutant (Fig. 5B). The amount of Ty1 RNA ranged from 40% of the wild-type level in the xrn1Δ mutant to 80% of that level in the dhh1Δ and ccr4Δ mutants. In addition, a smaller transcript running slightly below the full-length Ty1 transcript was observed in the xrn1Δ mutant. This shorter transcript has been described in previous work (8, 14) and is similar in size to the 5′-end truncated Ty1 transcript in the spt3Δ mutant (85). The levels of RNA from the single Ty1his3AI element were also modestly reduced in all of the mutants except upf1Δ. Since the effects of all of these mutations on Ty1 RNA and Ty1his3AI RNA levels are small, and since no correlation between the severity of the retrotransposition defect and the decrease in Ty1 or Ty1his3AI RNA was seen, the data indicated that Xrn1, Dhh1, and Upf1 affect a posttranscriptional step in retrotransposition.
Since Ty1AS RNA expression is negatively correlated with Ty1 cDNA and retrotransposition levels (59), we also measured the level of Ty1AS RNA in the mRNA decay mutants (Fig. 5B). The levels of the two major class sizes of Ty1AS RNA, which migrate between 0.5 and 1 kb, were substantially increased in the xrn1Δ mutant and the spt3Δ control strain. Modest increases in Ty1AS RNA of 1.8- to 3.5-fold were observed in the dhh1Δ, lsm1Δ, pat1Δ, ccr4Δ, and upf1Δ mutants. With the exception of the xrn1Δ mutant, which has very high levels of Ty1AS RNA and very low levels of Ty1 cDNA and retrotransposition, the mRNA decay mutants did not have increases in Ty1AS RNA that were correlated with the decreases in Ty1 cDNA or retrotransposition levels. Thus, it is not likely that elevated Ty1AS RNA is the sole cause of the retrotransposition defect in the mRNA decay mutants.
Next, we asked whether reduced efficiencies of Ty1 RNA translation could explain the low levels of Ty1 cDNA and retrotransposition in the xrn1Δ, dhh1Δ, and upf1Δ mutants. Synthesis of a Gag-GFP fusion protein expressed from the chromosomal reporter Ty1-GFP-3566 was measured by flow cytometry. The mean Gag-GFP activity in the xrn1Δ and dhh1Δ mutants was not significantly different than that in the wild-type strain. In the upf1Δ mutant, there was a 1.8-fold increase in Gag-GFP activity (Fig. 5C). Gag-GFP activity was increased in the fus3Δ control strain and decreased in the tec1Δ control strain, indicating that expression of the Ty1-GFP-3566 element is regulated appropriately. Therefore, the xrn1Δ, dhh1Δ, and upf1Δ mutants do not have major defects in Ty1 Gag synthesis, although the translational efficiency of Ty1 RNA might be increased modestly in the upf1Δ mutant. To confirm and extend these conclusions, quantitative Western blot analyses were performed to detect unprocessed p49-Gag and processed p45-Gag. In the wild-type strain, most of the Gag is present in the processed form (Fig. 5D). Small changes in the ratio of p49-Gag to p45-Gag were occasionally observed in some of the mutants, but these changes were not reproducible. The total amount of p49-Gag and p45-Gag relative to α-tubulin was determined by Western blot analysis of serial dilutions of total cell lysates. (The dilution series are not shown, but the quantification is shown in Fig. 5D.) In upf1Δ mutants, the total level of Ty1 Gag was equivalent to that in a wild-type strain. In the xrn1Δ and dhh1Δ mutants, the total level of Gag was 68% and 61% of that in the wild-type strain, respectively. Notably, these reductions in Gag protein were not as severe as the reductions in the pat1Δ and lsm1Δ mutants, both of which have relatively modest decreases in Ty1 cDNA synthesis and retrotransposition. Together, the results argue that neither changes in Gag protein levels nor a defect in Gag processing can explain the major defects in Ty1 cDNA accumulation in the xrn1Δ, dhh1Δ, and upf1Δ mutants.
Xrn1 is required for the packaging of Ty1 RNA in VLPs.
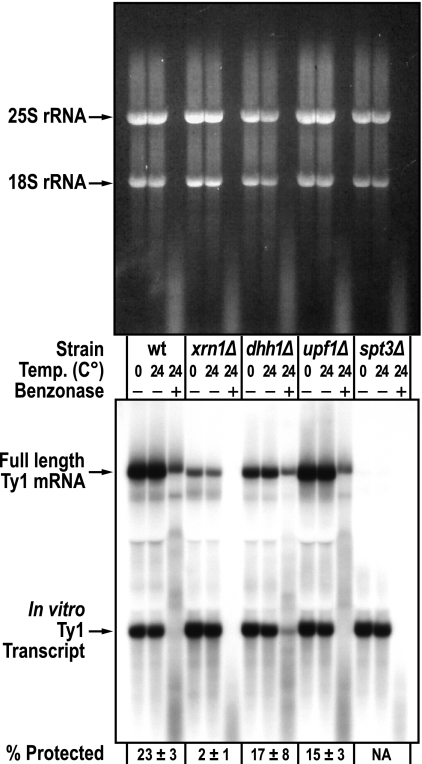
The above-described results suggest that P-body proteins Xrn1, Dhh1, and Upf1 affect Ty1 retrotransposition primarily at a step between Ty1 protein synthesis and the accumulation of Ty1 cDNA. Therefore, we examined the possibility that P-body-associated mRNA decay proteins are involved in the packaging of Ty1 RNA in VLPs. Genomic RNA from the Saccharomyces cerevisiae Ty3 and Schizosaccharomyces pombe Tf1 retrotransposons is partially protected from Benzonase digestion by packaging in VLPs (53, 56). We used the Benzonase protection assay to determine whether Xrn1, Dhh1, or Upf1 plays a role in the packaging of Ty1 RNA in VLPs (Fig. 6). Cell lysates prepared from wild-type and mutant strains were incubated with Benzonase at 24°C. To monitor endogenous nuclease activity under these conditions, in vitro Ty1 RNA was added to the cell lysate before addition of Benzonase. To monitor digestion by other nucleases present in the cell lysate, we incubated the lysate at 0°C or 24°C in the absence of Benzonase. RNA was extracted from each sample, and Ty1 RNA was detected by Northern blotting. No significant difference in the level of Ty1 RNA or the in vitro Ty1 transcript was seen when lysates were incubated at 0° or 24°C in the absence of Benzonase, indicating that cellular nucleases do not contribute to Ty1 RNA degradation at 24°C. In the wild-type strain, the average amount of Ty1 RNA remaining after Benzonase digestion at 24°C was 23% (±3%) of the amount of Ty1 RNA in the control sample incubated at 24°C without Benzonase. In contrast, rRNA (visualized on an ethidium-stained agarose gel) and the Ty1 in vitro transcript added to the wild-type cell lysate (visualized by Northern blotting) were completely digested. This fraction of endogenous Ty1 RNA packaging in wild-type cells is only slightly lower than that reported for Ty3 when transposition was induced by expression from the GAL1 promoter (52). In the dhh1Δ and upf1Δ mutants, the amount of Ty1 RNA that was protected from Benzonase was modestly reduced to 17% and 15%, respectively, of that present in the sample without Benzonase. In contrast, Ty1 RNA protection was abolished in the xrn1Δ mutant. Thus, while a defective DDD pathway (dhh1Δ) or NMD pathway (upf1Δ) reduces the level of Ty1 RNA packaging, it is completely eliminated in the absence of 5′ to 3′ mRNA degradation in the xrn1Δ mutant. Therefore, 5′ to 3′ mRNA decay appears to be essential for Ty1 packaging into VLPs.
FIG. 6.
Xrn1 promotes Ty1 mRNA packaging. Whole-cell extract from each strain was supplemented with an in vitro synthesized Ty1 transcript and divided into three samples of equal volume. Samples were incubated at 0°C or 24°C without Benzonase or at 24°C with Benzonase for 6 min. (Top) Ethidium bromide-stained agarose gel of total RNA prepared from each sample and denatured with glyoxal. The genotype of each strain, temperature of extract incubation, and addition (+) or deletion (−) of Benzonase are indicated below the image. The positions of the 18S and 25S rRNA bands are indicated to the left of the image. (Bottom) Northern blot analysis of the gel shown in the top panel. The positions of the endogenous Ty1 mRNA and the in vitro Ty1 transcript are indicated to the left of the image. The spt3Δ strain, lacking a transcriptional activator of Ty1 elements, serves as a negative control for the position of the endogenous Ty1 mRNA. The signals from the Ty1 mRNA bands were quantified by phosphorimage analysis. The ratio of endogenous Ty1 mRNA in the sample treated with Benzonase to that in the untreated sample at 24°C is expressed as a percentage. The average percentage of protected Ty1 mRNA and the standard error of the average percentage obtained using three biological replicates (one of which is shown) are provided below the image. NA, not applicable.
DISCUSSION
Colocalization of Ty1 Gag and the antiviral restriction factor A3G to P bodies.
We show that Ty1 Gag and the human antiviral restriction factor A3G colocalize with P bodies and that this colocalization is likely to have functional significance for Ty1 VLP formation and for A3G restriction of Ty1 retrotransposition. Several observations prompted us to determine whether Ty1 VLPs and A3G might be associated with cytoplasmic P bodies. First, A3G accumulates in P bodies in human cell lines and peripheral blood cells (37, 84). Second, P bodies are the probable sites of assembly or maturation of Ty3 VLPs (6, 52). Third, P-body components Xrn1, Dhh1, Pat1, and Lsm1 promote retrotransposition of both Ty3 and Ty1 elements (8, 14, 43). We found that 66% of A3G cytoplasmic foci localized in P bodies and that another 29% were directly adjacent to P bodies (Fig. 1A). Localization of A3G in P bodies is further supported by an interaction between A3G and TAP-tagged P-body proteins Xrn1, Pat1, and Lsm1 (Fig. 2). A3G and P-body constituents may interact indirectly by binding to common RNA molecules. In fact, binding of A3G to transcripts of endogenous retroelements has been suggested as a possible explanation for A3G accumulation in P bodies in mammalian cells (5); however, we found that A3G accumulates in yeast P bodies in spt3Δ mutants, which have very low levels of endogenous retroelement RNA (Fig. 3). Since A3G localizes in P bodies independently of Ty1 expression but interacts with Ty1 Gag in an RNA-dependent manner and is efficiently packaged into VLPs (31), we suggest that P bodies are sites of Ty1 RNA and A3G packaging into VLPs.
Ty1 Gag formed multiple, discrete cytoplasmic foci in most cells, but only 15% of these proteins overlapped with P bodies, while another 17% were adjacent to P bodies (Fig. 1C). Gag foci have been described in previous studies, including two that reported the detection of Gag foci containing Ty1 RNA (14, 57). In both of these reports, Ty1 RNA/Gag foci were spatially distinct from P bodies. Methodological differences between our study and the previous ones could explain the discrepancy in the results. In both previous studies, Ty1 RNA/Gag foci were detected in ∼30% of cells by combined RNA fluorescent in situ hybridization (FISH)/immunolocalization analyses. However, P bodies could not be visualized in exponentially growing cells unless the cells were deprived of glucose, and this treatment resulted in the disappearance of Ty1 Gag/RNA foci. In our study, Gag foci and P bodies were visualized in almost all exponentially growing cells in the presence of glucose. Therefore, imaging of live cells might be necessary to detect the ∼15% of Gag foci that colocalize with P bodies. A possible explanation for the small fraction of Gag foci that overlap with P bodies could be that Gag is transiently localized to P bodies. Gag may accumulate as foci within P bodies and then redistribute into distinct compartments. Perhaps this process is similar to the formation of yeast stress granules, which are RNP complexes that can overlap or be distinct from P bodies but are dependent on P-body formation (11, 12, 46).
Both Gag and Ty1 RNA may be present in P bodies, based on our demonstration that Ty1 RNA is sequestered from translation (Fig. 4). Polysome analysis revealed that virtually all detectable Ty1 RNA in the cell is translationally repressed in high-molecular-weight RNP complexes that cosediment with polysomes but are resistant to dissociation by EDTA. It is unlikely that all or most of these RNPs are VLPs, because yeast cells have very high levels of Ty1 RNA and barely detectable levels of VLPs (22, 38, 63). Moreover, the results of Benzonase protection assays indicated that only about one-quarter of Ty1 transcripts are packaged in VLPs (Fig. 6). Perhaps Gag associates with Ty1 RNA cotranslationally, leading to the formation of a translationally repressive Gag-RNA complex that seeds the formation of a high-molecular-weight complex. However, Ty1 proteins fail to display a cis preference for the RNA from which they are encoded, and VLPs package heterogeneous genomes (10, 26). Thus, there is no evidence that Ty1 RNA needs to be translated in order to be associated with Gag. Genomic HIV-1 RNA also lacks a cis preference for binding the proteins that it encodes, and there is a random distribution of HIV-1 virions containing RNA molecules from different genomes (15, 67). The unspliced genomic RNA of HIV-1 is transported to the cytoplasm by the RNA helicase DDX3, whose yeast ortholog Ded1 is a P-body protein (4, 87). This observation has led to the suggestion that unspliced genomic HIV-1 RNA is exported from the nucleus directly to P bodies (5). We propose that Ty1 RNA is translationally repressed by localization in P bodies soon after or concurrent with nuclear export. It would be interesting to test this idea by determining whether Ded1p is required for Ty1 retrotransposition. Future studies could also address the nature of the high-molecular-weight Ty1 RNPs and their relationship to Gag foci.
5′ to 3′ mRNA decay plays a central role in Ty1 retrotransposition at a posttranslational level.
Our suggestion that Ty1 Gag localization to P bodies is important for retrotransposition is supported by the observation that efficient retrotransposition of a chromosomal Ty1 element requires core components of P bodies, including Dcp1-Dcp2, Xrn1, Dhh1, Lsm1, Pat1, and Ccr4 (Table 2). NMD proteins, which accumulate in P bodies when decapping is inhibited (79), are also necessary for efficient retrotransposition. Furthermore, Xrn1, Dhh1, and Upf1 are required for retrotransposition of a pGTy1 element (Table 3), whose expression overcomes transpositional dormancy and copy number control by Ty1AS RNA (27, 39, 59). Therefore, Xrn1, Dhh1, and Upf1 may promote the efficiency of a step or steps in retrotransposition, rather than block a negative regulator of transposition such as Ty1AS RNA. The DDD and NMD pathways contribute additively to retrotransposition, since a mutant lacking both the DDD factor Ccr4 and the NMD factor Upf1 has a more severe retrotransposition defect than either of the single mutants (Table 2). The observed participation of both NMD and DDD in Ty1 retrotransposition explains why the xrn1Δ mutation, which abolishes 5′ to 3′ mRNA decay, has a more severe phenotype than NMD- and DDD-specific mutations. However, a mutation in EDC3, which promotes P-body assembly (30, 78), only modestly inhibits retrotransposition (Table 2), suggesting that the function of the 5′ to 3′ mRNA decay pathways in P bodies is more important for retrotransposition than for the structure of P bodies.
Analysis of Ty1 gene expression in the 5′ to 3′ decay mutants strongly suggests that these factors act at a step after the translation of Ty1 Gag (Fig. 5). Several of the mutants had modestly reduced levels of Gag, but there was no correlation between the level of Gag and the severity of the retrotransposition phenotype. In addition, all of the 5′ to 3′ decay mutants tested had elevated levels of Ty1AS RNA, but a correlation between the levels of Ty1AS RNA and the severity of the retrotransposition defect was also lacking. These data do not support a mechanism in which the accumulation of Ty1AS RNA in 5′ to 3′ decay mutants results in the retrotransposition defect. The fact that retrotransposition of a GAL1 promoter-driven Ty1his3AI element, whose expression overcomes the transpositional dormancy associated with Ty1AS RNA (59), is also dependent on Dhh1, Upf1, and Xrn1 (Table 3) also argues against a causative role for Ty1AS RNA in the retrotransposition defect. However, it remains possible that Ty1AS RNA has a unique inhibitory role in the xrn1Δ mutant, which has very high levels of Ty1AS RNA and very low levels of Ty1 retrotransposition compared to those of DDD-defective and NMD-defective mutants (Fig. 1B and Table 2). If increased Ty1AS RNA explains the severe retrotransposition defect in the xrn1Δ mutant, then the xrn1Δ mutant should have a phenotype similar to that of a strain producing high levels of Ty1AS RNA. Both strains do have reduced levels of reverse transcriptase and integrase (14, 59). However, the absence of Ty1 RNA packaging in the xrn1Δ mutant (Fig. 6) is not consistent with the robust exogenous reverse transcriptase activity that is observed in VLPs in a strain with high levels of Ty1AS RNA (59). Therefore, we favor the idea that the severe retrotransposition defect in the xrn1Δ mutant results directly from the complete lack of 5′ to 3′ mRNA decay rather than from a unique inhibitory role of Ty1AS RNA.
In addition to being required for Ty1 retrotransposition, the NMD protein Upf1 is required for efficient A3G-mediated restriction of Ty1 retrotransposition (Table 4). Apparently, A3G does not gain access to P bodies by interacting with retroelement RNA (Fig. 3) or by interacting with Upf1 (see Fig. S4 in the supplemental material), which functions independently of other NMD factors to shuttle targeted mRNAs from the translational machinery to P bodies (79). Therefore, Upf1 may promote a later step in retrotransposition, such as VLP assembly or RNA remodeling during reverse transcription. In fact, we did observe a modest defect in Ty1 RNA packaging in the upf1Δ mutant (Fig. 6). Notably, the human homolog of yeast Upf1 is a component of the intracellular HIV-1 RNP and plays a role in stabilization of HIV-1 RNA (1). Perhaps Upf1 can also associate with Ty1 Gag-RNA complexes in yeast, although Upf1 is clearly not required for Ty1 RNA stability (Fig. 5B).
A model: passage of Ty1 RNPs through P bodies is required for retrotransposition.
In light of the results presented here, we propose that Ty1 Gag and RNA are transiently targeted to P bodies and that their association in P bodies results in the formation of retrotransposition-competent VLPs. Ty1 RNA and Gag could be targeted to P bodies separately, since Ty1 proteins lack a cis preference for association with the RNA from which they are translated. A transient association in P bodies could serve to bring Ty1 Gag together with the translationally repressed pool of Ty1 RNA in the vicinity of 5′ to 3′ mRNA decay factors that are required for efficient retromobility. Once VLPs are formed, they may be relocated in cytoplasmic foci that are distinct from P bodies. The assembly of Ty1 Gag RNPs within P bodies is supported by the independent localization of the VLP component A3G to P bodies. P-body-associated Ty1 RNP assembly also provides an explanation for the fact that Ty1 RNA in VLPs is partially decapped (16, 24) and therefore is likely to be a substrate of the core P-body component Dcp1-Dcp2. The strongest evidence for Ty1 RNP assembly in P bodies is the observation that Ty1 RNA packaging into VLPs, as measured by protection from Benzonase digestion (Fig. 6), is abolished in the xrn1Δ mutant, which lacks 5′ to 3′ decay. The absence of Ty1 RNA packaging is correlated with a severe retrotransposition defect and a very low level of cDNA. Collectively, our results suggest that 5′ to 3′ mRNA decay may directly regulate the incorporation of Ty1 RNA into assembling VLPs. What is the role of mRNA decay in retrotransposition? Perhaps 5′ to 3′ decay pathways are necessary to degrade cellular mRNAs that could otherwise compete with Ty1 RNA for packaging into Ty1 VLPs and could therefore result in the formation of aberrant reverse transcripts within VLPs (62).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants AI065301 and GM52072.
We thank Suzanne Sandmeyer, Roy Parker, David Garfinkel, and Bryan Cullen for providing the plasmids and strains used in this study and Paul Masters and Patrick Maxwell for comments on the manuscript. We are grateful to Jason de Koning for statistical analysis of retrotransposition frequency results and to Dhanushki Samaranayake for construction of the dcp2::DAmP allele and the dcp1Δ/DCP1 heterozygote. Flow cytometry was performed by the Wadsworth Center Immunology Core.
Footnotes
Published ahead of print on 10 March 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Ajamian, L., L. Abrahamyan, M. Milev, P. V. Ivanov, A. E. Kulozik, N. H. Gehring, and A. J. Mouland. 2008. Unexpected roles for UPF1 in HIV-1 RNA metabolism and translation. RNA 14:914-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alce, T. M., and W. Popik. 2004. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J. Biol. Chem. 279:34083-34086. [DOI] [PubMed] [Google Scholar]
- 3.Beauregard, A., M. J. Curcio, and M. Belfort. 2008. The take and give between retrotransposable elements and their hosts. Annu. Rev. Genet. 42:587-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beckham, C., A. Hilliker, A. M. Cziko, A. Noueiry, M. Ramaswami, and R. Parker. 2008. The DEAD-box RNA helicase Ded1p affects and accumulates in Saccharomyces cerevisiae P-bodies. Mol. Biol. Cell 19:984-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beckham, C. J., and R. Parker. 2008. P bodies, stress granules, and viral life cycles. Cell Host Microbe 3:206-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beliakova-Bethell, N., C. Beckham, T. H. Giddings, Jr., M. Winey, R. Parker, and S. Sandmeyer. 2006. Virus-like particles of the Ty3 retrotransposon assemble in association with P-body components. RNA 12:94-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B. Stat. Methodol. 57:289-300. [Google Scholar]
- 8.Berretta, J., M. Pinskaya, and A. Morillon. 2008. A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S. cerevisiae. Genes Dev. 22:615-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bilanchone, V. W., J. A. Claypool, P. T. Kinsey, and S. B. Sandmeyer. 1993. Positive and negative regulatory elements control expression of the yeast retrotransposon Ty3. Genetics 134:685-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boeke, J. D., C. A. Styles, and G. R. Fink. 1986. Saccharomyces cerevisiae SPT3 gene is required for transposition and transpositional recombination of chromosomal Ty elements. Mol. Cell. Biol. 6:3575-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brengues, M., and R. Parker. 2007. Accumulation of polyadenylated mRNA, Pab1p, eIF4E, and eIF4G with P-bodies in Saccharomyces cerevisiae. Mol. Biol. Cell 18:2592-2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buchan, J. R., D. Muhlrad, and R. Parker. 2008. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J. Cell Biol. 183:441-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cen, S., F. Guo, M. Niu, J. Saadatmand, J. Deflassieux, and L. Kleiman. 2004. The interaction between HIV-1 Gag and APOBEC3G. J. Biol. Chem. 279:33177-33184. [DOI] [PubMed] [Google Scholar]
- 14.Checkley, M. A., K. Nagashima, S. Lockett, K. Nyswaner, and D. J. Garfinkel. 2010. P-body components are required for Ty1 retrotransposition during assembly of retrotransposition-competent virus-like particles. Mol. Cell. Biol. 30:382-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, J., O. Nikolaitchik, J. Singh, A. Wright, C. E. Bencsics, J. M. Coffin, N. Ni, S. Lockett, V. K. Pathak, and W. S. Hu. 2009. High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proc. Natl. Acad. Sci. U. S. A. 106:13535-13540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng, Z., and T. M. Menees. 2004. RNA branching and debranching in the yeast retrovirus-like element Ty1. Science 303:240-243. [DOI] [PubMed] [Google Scholar]
- 17.Chiu, Y. L., H. E. Witkowska, S. C. Hall, M. Santiago, V. B. Soros, C. Esnault, T. Heidmann, and W. C. Greene. 2006. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc. Natl. Acad. Sci. U. S. A. 103:15588-15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coller, J., and R. Parker. 2004. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 73:861-890. [DOI] [PubMed] [Google Scholar]
- 19.Coller, J., and R. Parker. 2005. General translational repression by activators of mRNA decapping. Cell 122:875-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coller, J. M., M. Tucker, U. Sheth, M. A. Valencia-Sanchez, and R. Parker. 2001. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA 7:1717-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Company, M., C. Adler, and B. Errede. 1988. Identification of a Ty1 regulatory sequence responsive to STE7 and STE12. Mol. Cell. Biol. 8:2545-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conte, D., Jr., E. Barber, M. Banerjee, D. J. Garfinkel, and M. J. Curcio. 1998. Posttranslational regulation of Ty1 retrotransposition by mitogen-activated protein kinase Fus3. Mol. Cell. Biol. 18:2502-2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conte, D., Jr., and M. J. Curcio. 2000. Fus3 controls Ty1 transpositional dormancy through the invasive growth MAPK pathway. Mol. Microbiol. 35:415-427. [DOI] [PubMed] [Google Scholar]
- 24.Coombes, C. E., and J. D. Boeke. 2005. An evaluation of detection methods for large lariat RNAs. RNA 11:323-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cullen, B. R. 2006. Role and mechanism of action of the APOBEC3 family of antiretroviral resistance factors. J. Virol. 80:1067-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Curcio, M. J., and D. J. Garfinkel. 1994. Heterogeneous functional Ty1 elements are abundant in the Saccharomyces cerevisiae genome. Genetics 136:1245-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Curcio, M. J., and D. J. Garfinkel. 1992. Posttranslational control of Ty1 retrotransposition occurs at the level of protein processing. Mol. Cell. Biol. 12:2813-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Curcio, M. J., and D. J. Garfinkel. 1991. Single-step selection for Ty1 element retrotransposition. Proc. Natl. Acad. Sci. U. S. A. 88:936-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Curcio, M. J., A. M. Hedge, J. D. Boeke, and D. J. Garfinkel. 1990. Ty RNA levels determine the spectrum of retrotransposition events that activate gene expression in Saccharomyces cerevisiae. Mol. Gen. Genet. 220:213-221. [DOI] [PubMed] [Google Scholar]
- 30.Decker, C. J., D. Teixeira, and R. Parker. 2007. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 179:437-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dutko, J. A., A. Schafer, A. E. Kenny, B. R. Cullen, and M. J. Curcio. 2005. Inhibition of a yeast LTR retrotransposon by human APOBEC3 cytidine deaminases. Curr. Biol. 15:661-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Efron, B. T., and R. J. Tibshirani. 1994. An introduction to the bootstrap. Chapman & Hall/CRC, Boca Raton, FL.
- 33.Eichinger, D. J., and J. D. Boeke. 1988. The DNA intermediate in yeast Ty1 element transposition copurifies with virus-like particles: cell-free Ty1 transposition. Cell 54:955-966. [DOI] [PubMed] [Google Scholar]
- 34.Esnault, C., O. Heidmann, F. Delebecque, M. Dewannieux, D. Ribet, A. J. Hance, T. Heidmann, and O. Schwartz. 2005. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature 433:430-433. [DOI] [PubMed] [Google Scholar]
- 35.Eulalio, A., I. Behm-Ansmant, and E. Izaurralde. 2007. P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 8:9-22. [DOI] [PubMed] [Google Scholar]
- 36.Feng, Y. X., S. P. Moore, D. J. Garfinkel, and A. Rein. 2000. The genomic RNA in Ty1 virus-like particles is dimeric. J. Virol. 74:10819-10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallois-Montbrun, S., B. Kramer, C. M. Swanson, H. Byers, S. Lynham, M. Ward, and M. H. Malim. 2007. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J. Virol. 81:2165-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garfinkel, D. J., J. D. Boeke, and G. R. Fink. 1985. Ty element transposition: reverse transcriptase and virus-like particles. Cell 42:507-517. [DOI] [PubMed] [Google Scholar]
- 39.Garfinkel, D. J., K. Nyswaner, J. Wang, and J. Y. Cho. 2003. Post-transcriptional cosuppression of Ty1 retrotransposition. Genetics 165:83-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghaemmaghami, S., W. K. Huh, K. Bower, R. W. Howson, A. Belle, N. Dephoure, E. K. O'Shea, and J. S. Weissman. 2003. Global analysis of protein expression in yeast. Nature 425:737-741. [DOI] [PubMed] [Google Scholar]
- 41.Goff, S. P. 2007. Host factors exploited by retroviruses. Nat. Rev. Microbiol. 5:253-263. [DOI] [PubMed] [Google Scholar]
- 42.Goff, S. P. 2008. Knockdown screens to knockout HIV-1. Cell 135:417-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Griffith, J. L., L. E. Coleman, A. S. Raymond, S. G. Goodson, W. S. Pittard, C. Tsui, and S. E. Devine. 2003. Functional genomics reveals relationships between the retrovirus-like Ty1 element and its host Saccharomyces cerevisiae. Genetics 164:867-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harris, R. S., K. N. Bishop, A. M. Sheehy, H. M. Craig, S. K. Petersen-Mahrt, I. N. Watt, M. S. Neuberger, and M. H. Malim. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803-809. [DOI] [PubMed] [Google Scholar]
- 45.He, F., and A. Jacobson. 2001. Upf1p, Nmd2p, and Upf3p regulate the decapping and exonucleolytic degradation of both nonsense-containing mRNAs and wild-type mRNAs. Mol. Cell. Biol. 21:1515-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoyle, N. P., L. M. Castelli, S. G. Campbell, L. E. Holmes, and M. P. Ashe. 2007. Stress-dependent relocalization of translationally primed mRNPs to cytoplasmic granules that are kinetically and spatially distinct from P-bodies. J. Cell Biol. 179:65-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu, W., C. Petzold, J. Coller, and K. E. Baker. 2010. Nonsense-mediated mRNA decapping occurs on polyribosomes in Saccharomyces cerevisiae. Nat. Struct. Mol. Biol. 17:244-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu, W., T. J. Sweet, S. Chamnongpol, K. E. Baker, and J. Coller. 2009. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature 461:225-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hug, A. M., and H. Feldmann. 1996. Yeast retrotransposon Ty4: the majority of the rare transcripts lack a U3-R sequence. Nucleic Acids Res. 24:2338-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425:686-691. [DOI] [PubMed] [Google Scholar]
- 51.Irwin, B., M. Aye, P. Baldi, N. Beliakova-Bethell, H. Cheng, Y. Dou, W. Liou, and S. Sandmeyer. 2005. Retroviruses and yeast retrotransposons use overlapping sets of host genes. Genome Res. 15:641-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Larsen, L. S., N. Beliakova-Bethell, V. Bilanchone, M. Zhang, A. Lamsa, R. Dasilva, G. W. Hatfield, K. Nagashima, and S. Sandmeyer. 2008. Ty3 nucleocapsid controls localization of particle assembly. J. Virol. 82:2501-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Larsen, L. S., M. Zhang, N. Beliakova-Bethell, V. Bilanchone, A. Lamsa, K. Nagashima, R. Najdi, K. Kosaka, V. Kovacevic, J. Cheng, P. Baldi, G. W. Hatfield, and S. Sandmeyer. 2007. Ty3 capsid mutations reveal early and late functions of the amino-terminal domain. J. Virol. 81:6957-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lecossier, D., F. Bouchonnet, F. Clavel, and A. J. Hance. 2003. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300:1112. [DOI] [PubMed] [Google Scholar]
- 55.Leeds, P., J. M. Wood, B. S. Lee, and M. R. Culbertson. 1992. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:2165-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lin, J. H., and H. L. Levin. 1998. Reverse transcription of a self-primed retrotransposon requires an RNA structure similar to the U5-IR stem-loop of retroviruses. Mol. Cell. Biol. 18:6859-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Malagon, F., and T. H. Jensen. 2008. The T body, a new cytoplasmic RNA granule in Saccharomyces cerevisiae. Mol. Cell. Biol. 28:6022-6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mangeat, B., P. Turelli, G. Caron, M. Friedli, L. Perrin, and D. Trono. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99-103. [DOI] [PubMed] [Google Scholar]
- 59.Matsuda, E., and D. J. Garfinkel. 2009. Posttranslational interference of Ty1 retrotransposition by antisense RNAs. Proc. Natl. Acad. Sci. U. S. A. 106:15657-15662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maxwell, P. H., C. Coombes, A. E. Kenny, J. F. Lawler, J. D. Boeke, and M. J. Curcio. 2004. Ty1 mobilizes subtelomeric Y′ elements in telomerase-negative Saccharomyces cerevisiae survivors. Mol. Cell. Biol. 24:9887-9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maxwell, P. H., and M. J. Curcio. 2007. Host factors that control long terminal repeat retrotransposons in Saccharomyces cerevisiae: implications for regulation of mammalian retroviruses. Eukaryot. Cell 6:1069-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maxwell, P. H., and M. J. Curcio. 2007. Retrosequence formation restructures the yeast genome. Genes Dev. 21:3308-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mellor, J., M. H. Malim, K. Gull, M. F. Tuite, S. McCready, T. Dibbayawan, S. M. Kingsman, and A. J. Kingsman. 1985. Reverse transcriptase activity and Ty RNA are associated with virus-like particles in yeast. Nature 318:583-586. [DOI] [PubMed] [Google Scholar]
- 64.Mou, Z., A. E. Kenny, and M. J. Curcio. 2006. Hos2 and Set3 promote integration of Ty1 retrotransposons at tRNA genes in Saccharomyces cerevisiae. Genetics 172:2157-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Muhlrad, D., and R. Parker. 1994. Premature translational termination triggers mRNA decapping. Nature 370:578-581. [DOI] [PubMed] [Google Scholar]
- 66.Neef, D. W., and D. J. Thiele. 2009. Enhancer of decapping proteins 1 and 2 are important for translation during heat stress in Saccharomyces cerevisiae. Mol. Microbiol. 73:1032-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nikolaitchik, O., T. D. Rhodes, D. Ott, and W. S. Hu. 2006. Effects of mutations in the human immunodeficiency virus type 1 Gag gene on RNA packaging and recombination. J. Virol. 80:4691-4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Parker, R., and U. Sheth. 2007. P bodies and the control of mRNA translation and degradation. Mol. Cell 25:635-646. [DOI] [PubMed] [Google Scholar]
- 69.Pure, G. A., G. W. Robinson, L. Naumovski, and E. C. Friedberg. 1985. Partial suppression of an ochre mutation in Saccharomyces cerevisiae by multicopy plasmids containing a normal yeast tRNAGln gene. J. Mol. Biol. 183:31-42. [DOI] [PubMed] [Google Scholar]
- 70.Schafer, A., H. P. Bogerd, and B. R. Cullen. 2004. Specific packaging of APOBEC3G into HIV-1 virions is mediated by the nucleocapsid domain of the gag polyprotein precursor. Virology 328:163-168. [DOI] [PubMed] [Google Scholar]
- 71.Scholes, D. T., M. Banerjee, B. Bowen, and M. J. Curcio. 2001. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics 159:1449-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Scholes, D. T., A. E. Kenny, E. R. Gamache, Z. Mou, and M. J. Curcio. 2003. Activation of a LTR-retrotransposon by telomere erosion. Proc. Natl. Acad. Sci. U. S. A. 100:15736-15741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schuldiner, M., S. R. Collins, N. J. Thompson, V. Denic, A. Bhamidipati, T. Punna, J. Ihmels, B. Andrews, C. Boone, J. F. Greenblatt, J. S. Weissman, and N. J. Krogan. 2005. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell 123:507-519. [DOI] [PubMed] [Google Scholar]
- 74.Schumacher, A. J., G. Hache, D. A. Macduff, W. L. Brown, and R. S. Harris. 2008. The DNA deaminase activity of human APOBEC3G is required for Ty1, MusD, and human immunodeficiency virus type 1 restriction. J. Virol. 82:2652-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schumacher, A. J., D. V. Nissley, and R. S. Harris. 2005. APOBEC3G hypermutates genomic DNA and inhibits Ty1 retrotransposition in yeast. Proc. Natl. Acad. Sci. U. S. A. 102:9854-9859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Segal, S. P., T. Dunckley, and R. Parker. 2006. Sbp1p affects translational repression and decapping in Saccharomyces cerevisiae. Mol. Cell. Biol. 26:5120-5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 78.Sheth, U., and R. Parker. 2003. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300:805-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sheth, U., and R. Parker. 2006. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell 125:1095-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shindo, K., A. Takaori-Kondo, M. Kobayashi, A. Abudu, K. Fukunaga, and T. Uchiyama. 2003. The enzymatic activity of CEM15/Apobec-3G is essential for the regulation of the infectivity of HIV-1 virion but not a sole determinant of its antiviral activity. J. Biol. Chem. 278:44412-44416. [DOI] [PubMed] [Google Scholar]
- 81.Stamenova, R., P. H. Maxwell, A. E. Kenny, and M. J. Curcio. 2009. Rrm3 protects the Saccharomyces cerevisiae genome from instability at nascent sites of retrotransposition. Genetics 182:711-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Svarovskaia, E. S., H. Xu, J. L. Mbisa, R. Barr, R. J. Gorelick, A. Ono, E. O. Freed, W. S. Hu, and V. K. Pathak. 2004. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J. Biol. Chem. 279:35822-35828. [DOI] [PubMed] [Google Scholar]
- 83.Tharun, S., and R. Parker. 1999. Analysis of mutations in the yeast mRNA decapping enzyme. Genetics 151:1273-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wichroski, M. J., G. B. Robb, and T. M. Rana. 2006. Human retroviral host restriction factors APOBEC3G and APOBEC3F localize to mRNA processing bodies. PLoS Pathog. 2:e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Winston, F., K. J. Durbin, and G. R. Fink. 1984. The SPT3 gene is required for normal transcription of Ty elements in S. cerevisiae. Cell 39:675-682. [DOI] [PubMed] [Google Scholar]
- 86.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, N. Liebundguth, D. J. Lockhart, A. Lucau-Danila, M. Lussier, N. M'Rabet, P. Menard, M. Mittmann, C. Pai, C. Rebischung, J. L. Revuelta, L. Riles, C. J. Roberts, P. Ross-MacDonald, B. Scherens, M. Snyder, S. Sookhai-Mahadeo, R. K. Storms, S. Veronneau, M. Voet, G. Volckaert, T. R. Ward, R. Wysocki, G. S. Yen, K. Yu, K. Zimmermann, P. Philippsen, M. Johnston, and R. W. Davis. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]
- 87.Yedavalli, V. S., C. Neuveut, Y. H. Chi, L. Kleiman, and K. T. Jeang. 2004. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev.-RRE export function. Cell 119:381-392. [DOI] [PubMed] [Google Scholar]
- 88.Zennou, V., D. Perez-Caballero, H. Gottlinger, and P. D. Bieniasz. 2004. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J. Virol. 78:12058-12061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang, H., B. Yang, R. J. Pomerantz, C. Zhang, S. C. Arunachalam, and L. Gao. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.