Abstract
The late expression factor 2 gene (lef-2) of baculovirus Autographa californica multiple nucleopolyhedrovirus (AcMNPV) has been identified as one of the factors essential for origin-dependent DNA replication in transient expression assays and has been shown to be involved in late/very late gene expression. To study the function of lef-2 in the life cycle of AcMNPV, lef-2 knockout and repair bacmids were generated by homologous recombination in Escherichia coli. Growth curve analysis showed that lef-2 was essential for virus production. Interestingly, a DNA replication assay indicated that lef-2 is not required for the initiation of viral DNA replication and that, rather, it is required for the amplification of DNA replication. lef-2 is also required for the expression of late and very late genes, as the expression of these genes was abolished by lef-2 deletion. Temporal and spatial distributions of LEF-2 protein in infected cells were also analyzed, and the data showed that LEF-2 protein was localized to the virogenic stroma in the nuclei of the infected cells. Analysis of purified virus particles revealed that LEF-2 is a viral protein component of both budded and occlusion-derived virions, predominantly in the nucleocapsids of the virus particles. This observation suggests that LEF-2 may be required immediately after virus entry into host cells for efficient viral DNA replication.
The Baculoviridae family consists of a group of invertebrate-specific viruses that contain a circular, double-stranded DNA genome with sizes between 82 and 180 kb (15). Among this group, Autographa californica multiple nucleopolyhedrovirus (AcMNPV) is the best-studied baculovirus. The size of the AcMNPV genome is ∼134 kb, and sequence analysis of the genome showed that it contains 155 open reading frames (ORFs) with protein-encoding potential (3). AcMNPV produces two forms of virus progeny during its infection cycle, budded viruses (BVs) and occlusion-derived viruses (ODVs). Although the two forms contain identical genetic materials, they are produced at different stages of virus infection and mediate baculovirus infection through different routes. BVs are produced during the early stage of baculovirus infection and mediate systematic infection within the host. BVs obtain their envelopes as they bud off of the plasma membranes of the infected cells, which contain baculovirus GP64 protein. GP64 is important for BV entry into neighboring cells (5). ODVs are produced during the very late stage of baculovirus infection and are embedded within a crystalline structure made up of polyhedrin proteins that form occlusion bodies (OBs). OBs are very stable in the natural environment and are disintegrated only under alkaline conditions, e.g., in the midguts of insects after ingestion, releasing ODVs for primary infection.
Based on the temporal expression patterns of the viral genes, the baculovirus infection cycle is divided into three phases: early, late, and very late. Early genes are transcribed by host RNA polymerase; late and very late genes are transcribed by a virus-encoded RNA polymerase during or after viral DNA replication. Most early gene products are involved in DNA replication, and the expression of late and very late genes depends on DNA replication and viral transactivators. Baculovirus late gene expression requires 20 virus-encoded factors, termed late expression factors (LEFs) (23, 33, 37). Six of these 20 factors, encoded by ie-1, lef-1, lef-2, lef-3, p143, and dnapol, were required for the replication of the origin-containing plasmid in previous transient expression experiments (16). They were also capable of indirectly triggering apoptosis through DNA replication events in virus-infected cells, leading to host translational shutoff (35). Because all the LEFs were transiently expressed during these experiments, it was possible to overlook the functional importance of some factors whose expression is regulated by other viral factors during the course of virus infection. The development of bacmid technology has enabled the generation of viral DNA lacking genes essential for virus viability in Escherichia coli, which was impossible to achieve in insect cells in the past. Based on this technique, the functional importance of viral genes in the virus life cycle can be studied in the context of the virus genome, providing new insights into the functions of the genes of interest. For example, analysis of viral DNA lacking lef-11, which was thought previously to be nonessential for DNA replication, demonstrated that lef-11 is in fact essential for viral DNA replication, as lef-11 deletion virus does not undergo viral DNA replication (17); hence, lef-11 became known as an essential replicative lef gene in addition to those identified by plasmid-based replication assays. It was also reported previously that ie-0 can functionally substitute for ie-1 (37), and therefore, there are a total of eight replicative LEF genes.
lef-2 was initially identified as one of the lef genes required for origin-dependent DNA replication based on findings from transient expression experiments (16). It was also demonstrated previously to be one of three genes essential for late and very late gene expression (32), and its expression level was found to have a positive effect on the strength of the very late polyhedrin promoter (41). A single mutation at the 3′ end of the lef-2 gene was also shown to abolish very late polyhedrin and p10 promoter activities (27). In addition, the lef-2 sequence is part of the polyhedrin upstream sequence (pu), which functions as a transcriptional enhancer (1, 22, 39). Through yeast two-hybrid analysis, LEF-2 was also demonstrated to interact with LEF-1 (10), another LEF required for viral DNA replication. The region encompassing amino acids 20 to 60 of LEF-2 was found to be required for the interaction of LEF-2 and LEF-1, and their interaction is a prerequisite for origin-dependent DNA replication. Because LEF-1 is a DNA primase (28), LEF-2 was thus termed a primase-associated factor. The data from these previous studies suggest that lef-2 is involved in at least two processes during baculovirus infection: DNA replication and late/very late gene expression. Nevertheless, the exact functions of LEF-2 in these two processes and its role in the life cycle of the virus are not yet clear.
In this study, we examined the effects of lef-2 deletion on the various stages of baculovirus infection by generation of a lef-2 knockout bacmid. The results showed that lef-2 deletion virus was deficient in BV production. Further dissection of the role of lef-2 in viral DNA replication showed that lef-2 is not essential for the initiation of viral DNA replication and that, rather, it is crucial for efficient viral DNA amplification. lef-2 deletion severely impaired late vp39 and very late p10 gene expression, but the onset of immediate early ie-1 gene expression was not compromised. Interestingly, LEF-2 was found to be incorporated into the nucleocapsids of BVs and ODVs, suggesting that the function of LEF-2 may be required immediately after virus entry into cells.
MATERIALS AND METHODS
Cell culture and viruses.
The Spodoptera frugiperda IPLB-Sf21 cell line (hereinafter referred to as Sf21) was cultured as a monolayer in TC-100 insect medium containing 10% heat-inactivated fetal bovine serum (FBS) (19). It was used for the generation and propagation of recombinant baculoviruses. Viral stocks were prepared, and their titers were determined by 50% tissue culture infective dose (TCID50) and quantitative PCR (Q-PCR) analyses (21).
Construction of lef-2 knockout and repair bacmids.
To generate the lef-2 deletion bacmid, a chloramphenicol acetyltransferase (CAT) cassette in which a chloramphenicol resistance gene was flanked by ∼500 nucleotides homologous to the upstream and downstream regions of the lef-2 coding sequence was first generated. This cassette was introduced into competent E. coli DH10Bac cells carrying wild-type AcMNPV genomic DNA (in parental bacmid bMON14272) for the generation of the lef-2 knockout bacmid, designated bAcΔlef2, by λ Red recombination as described previously (38). A reporter cassette containing the heat shock 70 promoter driving the expression of the enhanced green fluorescent protein (eGFP) was inserted into pFastBac1 (Invitrogen), yielding pFashE. pFashE was used to introduce the eGFP gene (egfp) into the wild-type AcMNPV bacmid bAcwt and into bAcΔlef2 by site-specific transposition, generating reporter bacmids bAcwt-hE and bAcΔlef2-hE. To introduce lef-2 back into bAcΔlef2, a lef-2 repair plasmid was constructed. The rabbit β-globin terminator sequence was amplified from pTriEx-3 (Novagen) with primers Glu-F and Glu-R (Table 1) and subsequently cloned into the pGL3-basic vector, which was digested with BamHI and NcoI, yielding pGlobin. The region containing the lef-2 gene sequence and its putative promoter was amplified from AcMNPV genomic DNA with orf4-F and lef-2-BamHI-R primers (Table 1). The PCR product was digested with XhoI and BamHI and cloned into pGlobin, which was linearized with SalI and BamHI, generating pLef2-globin. pLef2-globin was digested with SphI and NheI, and the resulting lef-2-globin gene fragment was inserted into pFashE, yielding pFhElef2-repair. pFhElef2-repair was used to insert egfp and lef-2 into the polyhedrin locus of bAcΔlef2, yielding repair bacmid bAcΔlef2-Rep-hE. Confirmation of deletion at the lef-2 locus was performed by PCR analysis with primers ko-A and ko-B (Table 1) and subsequent digestion of the PCR product by AscI, which has two unique cutting sites in the CAT cassette and none in lef-2. Intact lef-2 would yield a 945-bp product, and the replacement of lef-2 by the CAT reporter cassette would yield a 1,400-bp fragment (data not shown).
TABLE 1.
Primer sequences used in this study
| Primer name | Primer sequence (5′-3′) |
|---|---|
| orf5-F | CGCATCTCAACACGACTATGAT |
| orf603-R | TATGCGCAGCGGTACTATACAC |
| Glu-F | AATACCGGATCCATCGATCTTTTTCCCTCTG |
| Glu-R | TAGTTACCCATGGGCATATGTTGCCAAACTC |
| orf4-F | GTTATGACGCCTACAACTCCCCG |
| lef-2-BamHI-R | CGCGGATCCTCAATAATTACAAATAGG |
| orf4-KpnI-F | CGGGGTACCGTTGGGCATGTACGTCCGAA |
| orf5-SacI-R | GTCGAGCTCTTCGCGGCTTCTCGCACCA |
| lef1p-F | CAACCGTGGATTTCATTTGTGGT |
| lef1p-R | GTGAGCGCGTCCAAGTTTGAATC |
| ko-A | CATGACCCCCGTAGTGACAACGATC |
| ko-B | CGACAAGGCGTCTAGTTTATGTG |
| ie1-F | ATCACAACAGCAACATTTGCAC |
| ie1-R | TAATACGACTCACTATAGGGTCACAAATGTGGTTTGCACGTA |
| vp39-F | TAATACGACTCACTATAGGGGCAAACGATTGGGTTGACTTCT |
| vp39-R | CAACCAATTACCAAGACGTGTT |
| p10-F | ATAAGAATTATTATCAAATCAT |
| p10-R | TAATACGACTCACTATAGGGTTACTTGGAACTGCGTTTACCA |
To generate a hemagglutinin (HA)-tagged lef-2 repair bacmid, a fragment containing the lef-2 promoter region was amplified from AcMNPV genomic DNA with primers orf4-KpnI-F and orf5-SacI-R (Table 1) and inserted into an HA tag-encoding plasmid, pGL3HA, yielding pGLp3HA. The lef-2 coding sequence and β-globin terminator sequence were amplified from pLef2-globin with lef2-AvrII-F and RVprimer3, and the resulting PCR fragment was inserted downstream of the lef-2 putative promoter in pGLp3HA, in frame with the HA tag sequence, generating pG3HA-repair. The lef-2 repair fragment (the lef-2 promoter and coding region and the β-globin terminator sequence) was released from pG3HA-repair by digestion with SphI and NheI and inserted into pFashE to yield pFhElef2-HA. A fragment containing polyhedrin and its own promoter and transcriptional terminator was amplified with polh/SphI-F and polh/NotI-R. The PCR product was digested with SphI and NotI and inserted into pFhElef2-HA to yield pFhElef2-HApolh. This plasmid was used to insert the egfp reporter, lef-2 with a HA tag sequence, and polyhedrin into the polyhedrin locus of bAcΔlef2, yielding bAchE-lef2HA-polh.
Purification of bacmid DNA and transfection of cells.
Bacmid DNA was first purified with the PureLink HiPure plasmid DNA purification kit (Invitrogen), and ElectroMAX DH10B T1 phage-resistant cells (Invitrogen) were subsequently retransformed with the purified bacmid DNA. Bacterial colonies were selected on the basis of sensitivity to tetracycline and resistance to kanamycin and gentamicin, a pattern indicating that the bacterial clones contain only the desired bacmid DNA and no helper plasmid, and purified for subsequent experiments. Sf21 cells (2 × 106) were transfected with 2 μg of purified bacmid DNA by using Cellfectin as the transfection reagent according to the protocol of the manufacturer (Invitrogen).
Growth curve assay.
Sf21 cells were transfected with the bacmid DNA construct indicated below or infected with recombinant virus at a multiplicity of infection (MOI) of 5. Five hours after transfection or 1 h after infection, cells were washed with phosphate-buffered saline (PBS) and the medium was replenished with fresh TC-100 culture medium containing 10% FBS. This time point was designated time zero. Supernatants were collected from transfected or infected Sf21 cells at the time points indicated in Fig. 2 and cleared by centrifugation at 1,000 × g for 5 min. The titers were determined by TCID50 analysis and then double-checked by Q-PCR (21). The data points were plotted as averages of infection results from triplicate assays in three independent experiments.
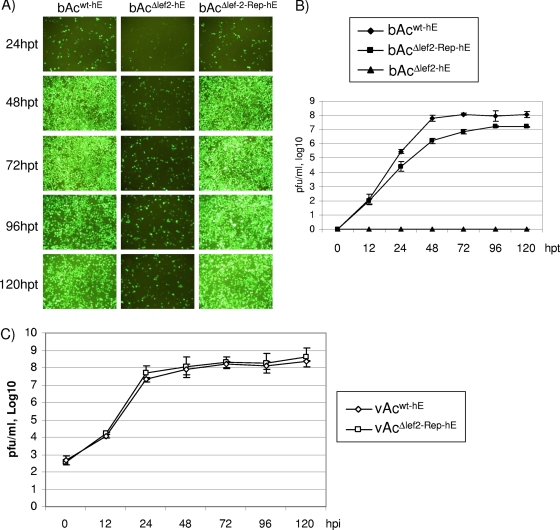
FIG. 2.
BV production by lef-2 knockout virus is impaired. (A) Fluorescence microscopic images of Sf21 cells transfected with bAcwt-hE, bAcΔlef2-hE, or bAcΔlef2-Rep-hE at 24, 48, 72, 96, and 120 h posttransfection (hpt). (B) Virus growth curve analysis. Sf21 cells were transfected with bAcwt-hE, bAcΔlef2-hE, or bAcΔlef2-Rep-hE, and virus titers were determined by TCID50 analysis at the indicated time points. (C) Growth curve analysis of vAcwt-hE and vAcΔlef2-Rep-hE in infected Sf21 cells. Supernatants of the infected cells were collected at time points up to 120 h postinfection (hpi), and virus titers were determined by TCID50 analysis.
DpnI replication assay.
Sf21 cells (106) were transfected with 1 μg of a gp64 knockout bacmid (bAcΔgp64), bAcΔlef2-hE, or a p143 knockout bacmid carrying pETLE, in which the egfp coding region is driven by the etl promoter (20) (bAcΔp143-ETLE), and at the time points indicated in Fig. 4, transfected cells were washed with PBS and collected. The cell pellets were lysed with 500 μl of lysis buffer (10 mM Tris, pH 8, 100 mM EDTA, 20 μg/ml RNase A, 0.5% sodium dodecyl sulfate [SDS], 80 μg/ml proteinase K) and incubated overnight at 37°C. Samples were extracted once with phenol, once with phenol-chloroform, and once with chloroform. Total cellular DNA was precipitated with a 0.5 volume of 7.5 M ammonium acetate and 2 volumes of ice-cold ethanol, washed with 70% ice-cold ethanol, and resuspended in 100 μl of Tris-EDTA buffer. Two micrograms of total DNA was treated with 20 U of EcoRI or EcoRI-DpnI, resolved on a 0.8% agarose gel, and subsequently transferred onto a Hybond-N+ nylon membrane (Amersham Biosciences). The membrane was probed with a digoxigenin (DIG)-labeled lef-1 gene sequence synthesized with a PCR DIG probe synthesis kit (Roche) using lef1p-F and lef1p-R (Table 1) and analyzed with a DIG detection kit (Roche).
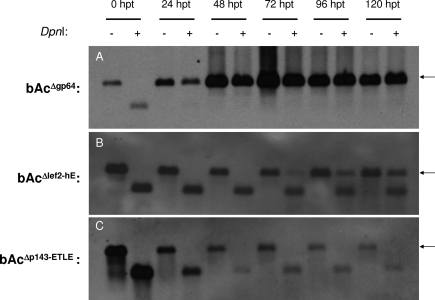
FIG. 4.
The timely initiation and efficiency of viral DNA replication were affected by lef-2 knockout. Sf21 cells were transfected with 1 μg of bAcΔgp64 (A), bAcΔlef2-hE (B), or bAcΔp143-ETLE (C). Total cellular DNA was extracted from transfected cells collected at 0, 24, 48, 72, 96, and 120 h posttransfection. Two micrograms of extracted DNA was digested with 20 U of EcoRI or 20 U of EcoRI-DpnI and resolved on a 0.8% agarose gel. Samples were hybridized with a DIG-labeled lef-1 gene sequence and analyzed with a DIG detection kit. −, digestion with EcoRI only; +, double digestion with EcoRI and DpnI. The upper bands (indicated by arrows) represent the DpnI-resistant signal, while the lower bands represent the DpnI-sensitive signal. Due to the active DNA replication in cells transfected with bAcΔgp64, the exposure time for panel A was much shorter than those for other panels.
Northern blot analysis.
Sf21 cells (106) were infected with virus produced in cells transfected with bAcwt-hE or bAcΔlef2-Rep-hE (vAcwt-hE or vAcΔlef2-Rep-hE virus) at an MOI of 10. Infected cells were harvested at the time points indicated below, and total RNA was extracted with an RNeasy minikit (Qiagen, Venlo, Netherlands). Five micrograms of total RNA was resolved on a 1% formaldehyde agarose gel and subsequently transferred onto a Hybond-N+ nylon membrane. The membrane was probed with DIG-labeled antisense strands of lef-2 transcripts and analyzed with a DIG detection kit (Roche). To study ie-1, vp39, and p10 expression, 5 × 105 Sf21 cells were transfected with 1 μg of bAcwt-hE, bAcΔlef2-hE, or bAcΔlef2-Rep-hE bacmid DNA. Transfected cells were harvested at 24, 48, and 72 h posttransfection. Total RNA was extracted, and 5 μg (for vp39 and p10 transcripts) or 15 μg (for ie-1 transcripts) of total RNA was resolved on a 1% formaldehyde agarose gel. RNA samples were transferred onto a Hybond-N+ nylon membrane and hybridized with DIG-labeled antisense RNA probes for ie-1, vp39, and p10 transcripts. The presence of target RNA was detected with a DIG detection kit (Roche) (42). The primers used to synthesize DIG-labeled lef-2, ie-1, vp39, and p10 RNA probes are listed in Table 1.
Localization of LEF-2 in the cell nucleus and cytoplasm.
Cells (2 × 106) were infected with HA-tagged repair virus vAchE-lef2HA-polh at an MOI of 10. Infected cells were harvested at 0, 6, 12, 16, 20, 24, 30, 36, and 48 h postinfection. Cytoplasmic and nuclear fractions of the infected cells were prepared as described by Fang et al. (11). One-fifth of each fraction was analyzed by SDS-10% PAGE and Western blotting with (i) primary mouse monoclonal antibody against HA (1:1,000; Cell Signaling) and secondary horseradish peroxidase (HRP)-conjugated antibody against mouse (1:5,000) and (ii) primary mouse monoclonal antibody against baculovirus GP64 (1:3,000; eBioscience) and HRP-conjugated antibody against mouse (1:5,000).
Immunofluorescence microscopy.
Sf21 cells were seeded onto chamber slides (Nalge Nunc International) and infected with vAchE-lef2HA-polh at an MOI of 20. At 16 h postinfection, infected cells were washed three times with PBS and fixed with 2% paraformaldehyde for 10 min at room temperature. Fixed cells were washed three times with PBS, permeabilized with 0.2% Triton X-100 for 15 min at room temperature, and rinsed three times with PBS-T (PBS containing 0.1% Tween 20). Cells were incubated in blocking buffer (1% normal goat serum in PBS-T) for 1 h at room temperature before incubation with primary mouse monoclonal anti-HA antibody (Covance) diluted 1:200 in blocking buffer at 4°C overnight. Cells were washed four times with PBS-T for 5 min each time and then incubated with Alexa Fluor 405 goat anti-mouse IgG antibody (Molecular Probes) diluted 1:200 in blocking buffer for 1 h at room temperature. Following four washes with PBS-T, cellular DNA and viral DNA were stained with propidium iodine (PI; diluted 1:10,000 in PBS [Molecular Probes]) for 5 min at room temperature. Cells were rinsed three times with PBS before the chamber slides were disassembled, covered with coverslips, and sealed with mounting medium. Cells were viewed with a 63× oil immersion lens objective on a confocal laser-scanning microscope (LSM510 META; Zeiss). Images were taken, viewed, and analyzed with LSM Image software (Zeiss).
Purification of virus particles from BVs and ODVs.
Sf21 cells were infected with recombinant baculovirus vAchE-lef2HA-polh at an MOI of 5. At 4 days postinfection, the cells were collected and used to extract ODVs as described previously (7). For BV purification, Sf21 cells were infected with vAchE-lef2HA-polh at an MOI of 0.5 and the supernatant was collected at 5 days postinfection and subjected to low-speed centrifugation at 3,000 × g for 10 min. The supernatant was then subjected to high-speed centrifugation at 80,000 × g (24,000 rpm) with an SW28 rotor (Beckman) for 90 min. The virus pellet was resuspended in protease inhibitor-containing PBS, and the suspension was loaded onto a 25 to 60% sucrose gradient. The sample was centrifuged at 96,000 × g (27,900 rpm) with an SW41 rotor at 4°C for 3 h. The virus particles were collected, diluted in PBS, and centrifuged again at 80,000 × g (21,600 rpm) with an SW41 rotor for 90 min to pellet the BVs. The purified BV particles were resuspended in Tris-HCl (pH 8.5).
Separation of nucleocapsid and envelope proteins of BVs and ODVs.
To separate nucleocapsids from the envelopes, the purified BVs and ODVs were incubated in separation buffer (1% Nonidet P-40 and 10 mM Tris-HCl, pH 8.5) on a rotating platform for 30 min at room temperature (11). A 4-ml solution of 30% (wt/vol) glycerol was overlaid with the mixture, and the preparation was centrifuged at 150,000 × g (34,000 rpm) with an SW60 rotor at 4°C for 1 h. The pellet obtained after centrifugation contained the nucleocapsids of the virus particles and was resuspended in 10 mM Tris-HCl, pH 7.5. The fraction above the interface at the top of the supernatant contained the envelope proteins and was collected and precipitated with 4 volumes of acetone. The precipitated envelope proteins were resuspended in 10 mM Tris-HCl, pH 7.5.
Western blot analysis.
To detect the presence of HA-tagged LEF-2 in BVs and ODVs, 10 μg of BV or ODV particles and 5-μg aliquots of the nucleocapsid and envelope fractions from purified virus particles were mixed with 2× SDS sample buffer and resolved by SDS-10% PAGE. The protein samples were probed with the following antibodies: (i) primary mouse monoclonal antibody against the HA tag (1:1,000) and HRP-conjugated secondary antibody against mouse (1:5,000), (ii) primary mouse monoclonal antibody against baculovirus GP64 (1:3,000; eBioscience) and HRP-conjugated antibody against mouse (1:5,000), and (iii) primary rabbit polyclonal antibody against baculovirus VP39 (1:2,500; Abnova) and HRP-conjugated antibody against rabbit (1:5,000). The membrane was analyzed with an enhanced chemiluminescence system (Immobilon Western; Millipore).
RESULTS
Generation of AcMNPV lef-2 knockout and repair bacmids.
To generate the lef-2 deletion bacmid DNA, the E. coli λ Red recombination system was exploited to replace the lef-2 gene with a CAT expression cassette in the lef-2 locus through homologous recombination in E. coli. Replacement at the lef-2 locus in the parental bacmid (bMON14272) was confirmed by PCR as mentioned in Materials and Methods.
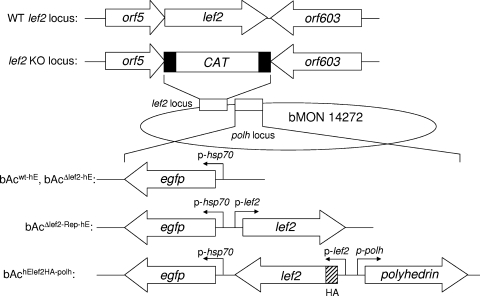
In order to monitor the progression of virus infection, the eGFP expression cassette was inserted into the polyhedrin locus in the wild-type bacmid (bAcwt) and the lef-2 knockout bacmid (bAcΔlef2) by site-specific transposition, yielding bAcwt-hE and bAcΔlef2-hE (Fig. 1). A lef-2 repair bacmid was constructed in which the lef-2 coding sequence and its own promoter region, along with the eGFP reporter cassette, were inserted into the polyhedrin locus of bAcΔlef2 to obtain bAcΔlef2-Rep-hE. To monitor the distribution of LEF-2 in infected cells, an additional lef-2 repair bacmid, bAchE-lef2HA-polh, was constructed in which the HA tag sequence was added to the 5′ end of the lef-2 coding sequence. All clones were checked by PCR at both lef-2 and polyhedrin loci (data not shown).
FIG. 1.
Schematic drawing of wild-type (WT), lef-2 knockout (lef2 KO), and repair bacmids used in this study. The lef-2 gene sequence was replaced by the CAT gene flanked by 50-nucleotide sequences homologous to the 5′- and 3′-end sequences of the lef-2 gene. Reporter cassettes containing the egfp gene under the control of the heat shock 70 promoter were inserted into the polyhedrin locus by site-specific transposition in E. coli to yield a bacmid backbone for further modifications. The first bacmid, bAcwt-hE, contains a wild-type bacmid backbone with the insertion of egfp; in the second bacmid, bAcΔlef2-hE, lef-2 was deleted; in the third bacmid, bAcΔlef2-Rep-hE, a lef-2 expression cassette was reinserted for repair; in the fourth bacmid, bAchE-lef2HA-polh, the HA tag sequence was inserted at the 5′ end of lef-2, followed by a complete polyhedrin expression cassette. p-hsp70, heat shock 70 promoter; p-lef2, putative lef-2 promoter; p-polh, polyhedrin promoter. Black boxes represent sequences homologous to the 5′ and 3′ ends of lef-2; the striped box represents the HA tag sequence.
Analysis of wild-type, lef-2 knockout, and repair bacmids in Sf21 cells.
In order to determine the effects of lef-2 deletion on virus growth, Sf21 cells were transfected with bAcwt-hE, bAcΔlef2-hE, or bAcΔlef2-Rep-hE. At 24 h posttransfection, green fluorescence could be observed in transfected cells (Fig. 2A). No obvious difference in the number of green fluorescent cells among samples transfected with bAcwt-hE, bAcΔlef2-hE, and bAcΔlef2-Rep-hE was observed, indicating equal transfection efficiencies even though the intensity of green fluorescence in cells transfected with bAcΔlef2-hE was noticeably lower than that in cells transfected with the other bacmids. At 48 h posttransfection, almost all cells transfected with bAcwt-hE fluoresced green, suggesting generation of BVs for secondary infections. The intensity of green fluorescence reached a maximum at 72 h posttransfection and then started to decline, due most likely to cell lysis as a result of virus infection. The number of green fluorescent cells and the intensity of green fluorescence among cells transfected with bAcΔlef2-hE also increased from 24 to 48 h posttransfection but to a much lesser extent than the corresponding indicators among cells infected with the wild-type virus. No further significant changes in the number of green fluorescent cells and the intensity of green fluorescence were observed after 48 h. Small aggregates of green fluorescent cells, which did not develop further into infection foci (data not shown), were occasionally observed after 72 h among cells transfected with bAcΔlef2-hE. bAcΔlef2-Rep-hE repair virus exhibited growth similar to that of the wild-type virus in transfected Sf21 cells. The similarity in infection pattern between wild-type and repair viruses suggested that the observed phenotype of lef-2 deletion virus was due to the absence of lef-2.
lef-2 deletion abolished BV production.
To quantify the effect of lef-2 deletion on BV production, growth curves for bAcwt-hE, bAcΔlef2-hE, and bAcΔlef2-Rep-hE viruses were determined. The supernatants from transfected cells were collected at the time points posttransfection indicated in Fig. 2B, and the titers were determined. The results showed that no BVs were produced in cells transfected with the lef-2 knockout construct up to 120 h posttransfection (Fig. 2B). On the other hand, bAcwt-hE and bAcΔlef2-Rep-hE yielded normal virus growth curves that reached plateaus at 48 and 96 h posttransfection, respectively (Fig. 2B). These results indicate that lef-2 deletion impaired BV production in transfected cells. This finding was in agreement with the green fluorescence patterns observed in transfected cells, where green fluorescence expression was restricted initially to cells transfected with bAcΔlef2-hE.
At 12 h posttransfection, the titer of bAcΔlef2-Rep-hE virus was consistently around 1 order of magnitude lower than that of bAcwt-hE virus. To confirm that the virus progeny produced by the repair virus and that produced by the wild-type virus were equally infectious, fresh Sf21 cells were infected with BVs collected from cells transfected with bAcwt-hE or bAcΔlef2-Rep-hE at an MOI of 5. No significant phenotypic difference was observed with a fluorescence microscope (data not shown). The growth curves for vAcwt-hE and vAcΔlef2-Rep-hE viruses in infected cells were determined (Fig. 2C). The virus titer in repair virus-infected cells was still lower than that in wild-type virus-infected cells at all time points, but by a much smaller margin than that between titers in transfection experiments, and the two viruses generated similar growth curves. This finding demonstrated that the repair virus was as infectious as the wild-type virus.
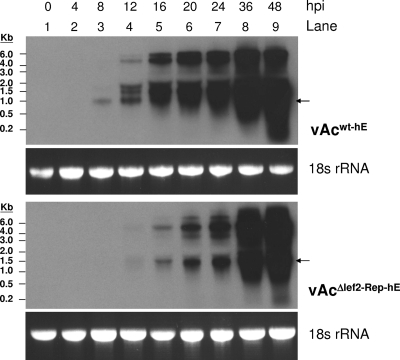
The slightly inferior growth of the repair virus may indicate that the temporal expression of lef-2 was altered in the repair virus since lef-2 was reinserted into the polyhedrin locus instead of its original locus. To compare the transcriptional profiles of lef-2 in wild-type and repair virus-infected cells, Northern blot analysis was performed with an antisense RNA probe for lef-2 transcripts. The onset of lef-2 expression in cells infected with repair virus was delayed by ∼4 h compared to that in cells infected with wild-type virus (Fig. 3). The lef-2-specific transcript from repair virus was somewhat larger than that from wild-type virus (bands corresponding to lef-2-specific transcripts are indicated by arrows in Fig. 3). This was due probably to the presence of the β-globin terminator sequence in place of the lef-2 putative terminator sequence in repair virus. lef-2-specific transcripts continued to accumulate and transcripts with higher molecular weights were detected in both wild-type and repair virus-infected cells as infection progressed. Detection of transcripts with high molecular weights corresponding to the lef-2 region of the AcMNPV genome during the late stage of virus infection has been reported previously (24) (see Discussion). We also noted that wild-type and repair viruses expressed different patterns lef-2, which may have contributed to the slower growth curve of the repair virus.
FIG. 3.
Temporal expression of lef-2 transcripts in infected cells. Sf21 cells were infected with either vAcwt-hE or vAcΔlef2-Rep-hE at an MOI of 5. Total cellular RNA was extracted from infected cells harvested at 0, 4, 8, 12, 16, 24, 36, and 48 h postinfection. Five micrograms of total RNA from each sample was resolved on a 1% formaldehyde gel and stained with ethidium bromide for visualization of 18S rRNA to ensure loading of equal sample amounts. The samples were then transferred onto a Hybond-N+ membrane (Amersham), hybridized with DIG-labeled lef-2 antisense RNA probes, and analyzed with a DIG detection kit. Arrows indicate bands corresponding to lef-2-specific transcripts.
DpnI sensitivity DNA replication assay.
lef-2 was identified previously as one of the lef genes required for origin-dependent DNA replication (16). To further dissect the role of lef-2 in viral DNA replication, the ability of lef-2 deletion virus to support viral DNA replication was investigated. A DpnI sensitivity DNA replication assay was performed to measure viral DNA replication in Sf21 cells. DpnI can recognize and digest viral DNA introduced by transfection, whereas viral DNA that replicates in transfected cells is resistant to DpnI digestion. Sf21 cells were transfected with bAcΔgp64, bAcΔp143-ETLE, or bAcΔlef2-hE, and the transfected cells were harvested at the time points indicated in Fig. 4. gp64 knockout virus can replicate viral DNA normally and is impaired only in cell-to-cell transmission of the virus particles (17, 29); therefore, bAcΔgp64 virus can serve as a positive control in a DNA replication assay. Total cellular DNA was extracted, and 2 μg of extracted DNA was digested with 20 U of EcoRI or EcoRI-DpnI and probed with a labeled lef-1 sequence (Fig. 4). Digestion with EcoRI would yield a 2.6-kb fragment containing lef-1, and double digestion with EcoRI and DpnI would yield a 1.1-kb fragment containing lef-1. The result demonstrated viral DNA replication in cells transfected with bAcΔgp64, and viral DNA continued to accumulate up to 120 h posttransfection (Fig. 4A). On the other hand, no viral DNA replication could be detected in cells transfected with bAcΔlef2-hE until 72 h posttransfection and there was slight and gradual accumulation of the DpnI-resistant bands from these cells from 72 h onward, indicating that initiation of DNA replication was possible, though greatly delayed and inefficient, in the absence of lef-2 (Fig. 4B).
To further confirm that the viral DNA replication detected in cells with lef-2 knockout virus was not resulting from nonspecific replication by host DNA polymerase, replication of p143 knockout virus DNA was also analyzed. Both lef-2 and p143 were previously demonstrated to be essential for viral DNA replication in transient expression assays (16). No DpnI-resistant band, an indication of viral DNA replication, could be detected for p143 knockout virus even at 120 h posttransfection (Fig. 4C). The absence of a DpnI-resistant band in p143 knockout virus samples suggested that viral DNA replication did occur, although at an extremely low level, in cells with lef-2 knockout virus. This result indicated that lef-2 is not absolutely required for the initiation of viral DNA replication; nevertheless, it is required for timely and efficient viral DNA amplification.
Analysis of early, late, and very late gene expression by lef-2 knockout baculovirus.
Ample evidence has implied that baculovirus late and very late gene expression depends on viral DNA replication (23, 32, 34). Origin-dependent DNA replication could not take place without lef-2 in transient expression assays (16), and as a consequence, the baculovirus late gene vp39 promoter was inactive (23). However, most of the factors in these prior experiments were transiently expressed and may not represent the real scenario in the context of the virus genome. Since lef-2 deletion virus can still initiate viral DNA replication, it was necessary to examine whether late and very late gene expression was turned on as a result of viral DNA replication.
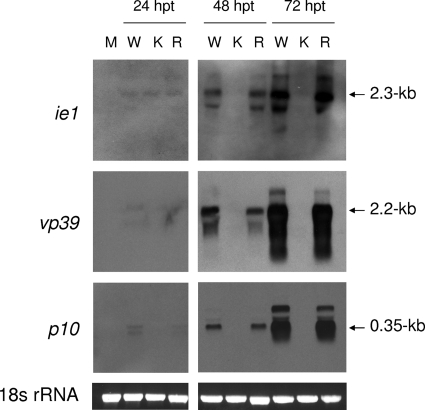
To estimate the transcriptional activities of early, late, and very late genes in lef-2 knockout virus, the expression of baculovirus early gene ie-1, late gene vp39, and very late gene p10 transcripts from wild-type, lef-2 knockout, and lef-2 repair viruses was measured by Northern blot analysis. Total RNA samples harvested from cells transfected with bacmids were hybridized with DIG-labeled antisense RNA probes specific for ie-1, vp39, and p10 transcripts. The data showed that vp39 or p10 transcripts could not be detected in lef-2 knockout virus samples even at 72 h posttransfection (Fig. 5), suggesting that late and very late gene expression was impaired upon lef-2 deletion. ie-1 transcripts could be detected in lef-2 knockout virus samples at 24 h posttransfection, due to the use of host transcription machinery. The Northern blot analysis showed that transcription of late gene vp39 and very late gene p10 in lef-2 knockout virus was severely impaired. These results indicate that, without amplification, initiation of viral DNA replication is not sufficient to support expression of late and very late genes from the baculovirus genome.
FIG. 5.
Late and very late gene expression was abolished by lef-2 deletion. Sf21 cells (106) were transfected with 1 μg of bAcwt-hE, bAcΔlef2-hE, or bAcΔlef2-Rep-hE DNA. Total cellular RNA was extracted from transfected cells collected at 24, 48, and 72 h posttransfection. Five micrograms (for vp39 and p10 transcripts) or 15 μg (for ie-1 transcripts) of total RNA was resolved on a 1% formaldehyde gel and transferred onto a Hybond-N+ membrane. The membrane was subsequently hybridized with DIG-labeled ie-1, vp39, or p10 antisense RNA probes and analyzed with a DIG detection kit. Ethidium bromide staining of 18S rRNA demonstrated loading of equal sample amounts. Arrows indicate the expected sizes of ie-1, vp39, and p10 transcripts. M, mock-transfected cells; W, cells transfected with bAcwt-hE; K, cells transfected with lef-2 knockout bacmid bAcΔlef2-hE; R, cells transfected with lef-2 repair bacmid bAcΔlef2-Rep-hE.
Spatial distribution of LEF-2.
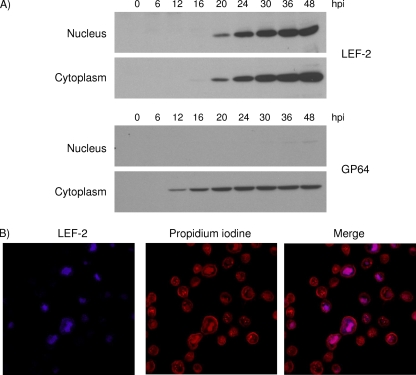
The temporal and spatial distributions of LEF-2 protein were analyzed by biochemical separation of infected cells into cytoplasmic and nuclear fractions and detection of LEF-2 by Western blot analysis. LEF-2 protein was initially detected in the cytoplasm at 16 h postinfection and was clearly detectable in both the nucleus and the cytoplasm starting at 20 h postinfection (Fig. 6A). In another experiment, we found that LEF-2 continued to accumulate in both the cytoplasm and the nucleus for up to 72 h postinfection and that the level of LEF-2 declined at 96 h postinfection (data not shown). About equal amounts of LEF-2 were detected in the cytoplasm and the nucleus at later time points postinfection; this pattern may be due to the relatively small size of LEF-2 and the lack of a known nuclear localization sequence in this protein (9, 13).
FIG. 6.
Localization of LEF-2 in infected Sf21 cells. (A) LEF-2 is localized in both cytoplasmic and nuclear regions in infected cells. Localization of GP64 is shown as a control. (B) LEF-2 colocalizes with the active viral DNA replication sites, i.e., virogenic stroma, in the nuclei of infected cells. Localization of LEF-2 was visualized by staining with Alexa 405 (blue), and DNA was stained with PI (red).
LEF-2 is one of the virally encoded proteins required for viral DNA replication; however, its localization relative to viral DNA replication sites has not yet been investigated. To study the localization of LEF-2 relative to the sites of viral DNA replication in vivo, localization of LEF-2 in infected cells was visualized by immunofluorescence staining. Cellular DNA and newly replicated viral DNA were visualized by PI staining. At 16 h postinfection, cellular chromatin lined the periphery of the cell nucleus (30) and the heavy PI staining in the central region of the nucleus represented the newly synthesized viral DNA and viral DNA replication foci (Fig. 6B). LEF-2 was shown to colocalize well with the newly synthesized viral DNA. This finding further indicates the involvement of LEF-2 in viral DNA replication processes.
Association of LEF-2 with nucleocapsids of both BVs and ODVs.
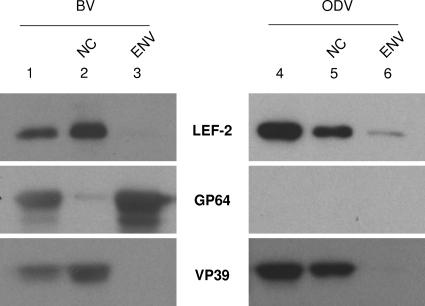
Previous analysis of purified ODVs revealed that five (IE-1, LEF-1, LEF-3, DNAPOL, and P143) of the viral proteins involved in DNA replication are associated with the nucleocapsids of ODVs (6). This arrangement gives the virus the ability to efficiently initiate viral DNA replication as soon as it enters host cells. Because LEF-2 was also required for viral DNA replication, it became a question whether LEF-2 was also associated with the nucleocapsid of the virus particles, along with the other replication LEFs. To this end, lef-2 repair virus vAchE-lef2HA-polh, in which a HA tag was added to the N terminus of LEF-2 along with the polyhedrin gene product, was generated. BVs were purified from the supernatant of vAchE-lef2HA-polh-infected cells, and ODVs were extracted from vAchE-lef2HA-polh-infected cells. LEF-2 protein could be detected in both purified BVs and ODVs, demonstrating that LEF-2 is a component of virus particles (Fig. 7). To further elucidate the localization of LEF-2 in virus particles, biochemical fractionation of the purified virus particles was undertaken, and Western blot analysis showed that LEF-2 protein was localized predominantly in the nucleocapsids of both BVs and ODVs (Fig. 7). To assess whether fractionation was successful, the localization patterns of GP64, an envelope protein, and VP39, a capsid protein, in the purified virus particles were also analyzed with antibodies specific for GP64 and VP39. GP64 protein was detected mainly in the envelope fraction of BVs, with a trace amount being detected in the nucleocapsid fraction. This level was considered to reflect acceptable envelope contamination (<10%) of the nucleocapsid preparation (4, 7). On the other hand, VP39 protein was detected primarily in the nucleocapsid fraction of the purified virus particles, indicating that the fractionation process was successful.
FIG. 7.
LEF-2 is incorporated into nucleocapsids of both BVs and ODVs. BVs and ODVs were purified from infected cells by using a sucrose gradient and separated into nucleocapsid and envelope fractions by using a glycerol cushion. Ten micrograms of purified BVs (lane 1) or ODVs (lane 4) and 5-μg samples of nucleocapsids (NC; lanes 2 and 5) and envelopes (ENV; lanes 3 and 6) were separated on a 10% polyacrylamide gel and transferred onto a polyvinylidene difluoride membrane (Millipore). The presence of LEF-2 protein was detected with mouse monoclonal antibody against the HA tag. The same blot was also probed with rabbit antiserum against baculovirus envelope protein GP64 and rabbit antiserum against baculovirus nucleocapsid protein VP39.
DISCUSSION
In this work, we studied the effects of lef-2 deletion on the baculovirus infection cycle. We found that in the absence of lef-2, no infectious virus particles could be produced. It is reasonable to attribute the defects in virus progeny production to lef-2 deletion, as the repair virus exhibited a wild-type virus growth curve (Fig. 2B and C). Findings of previous studies showed that LEF-2 was essential for origin-dependent DNA replication (16, 23); however, the results of our experiment showed that without LEF-2, DNA replication can still be initiated, though at an extremely low level (Fig. 4B). On the other hand, deletion of p143, which is an essential gene for viral DNA replication and encodes a helicase, completely abolishes viral DNA replication, as no viral DNA replication could be detected (Fig. 4C). This finding implies that the replicated DNA detected in cells with lef-2 knockout virus is not likely to be the result of nonspecific DNA replication. Therefore, in contrast to results from the transient expression assays (16), our results suggest that LEF-2 may be stimulatory rather than essential for viral DNA replication in the course of virus infection.
There are some possible explanations for this discrepancy in results. First, in previous studies, all the viral factors were expressed transiently, which may alter their temporal expression in comparison with that in the context of the virus. In addition, those viral factors were expressed from subgenomic fragments, which may accidently exclude the 5′ or 3′ end or cis-acting elements essential for gene regulation by other viral factors. The transient expression system may not have mimicked the real scenario during the normal course of virus infection. Second, the replication of a plasmid containing the standard replication origin, the homologous region (hr), was used as an indicator for DNA replication in those studies. However, plasmids containing viral sequences other than hr have been shown to replicate in the presence of virus (40), and their ability to initiate DNA replication may not rely on the copresence of replicative lef genes. Another possibility is that segments of the viral DNA were incorporated into the host genome and therefore replicated along with cellular DNA, which may also yield DpnI-resistant products. Integration of segments of baculoviral genomic DNA into the host genome in mammalian cells has been demonstrated previously (26). However, this possibility may not be the case because we also used a DIG-labeled DNA probe for the vp39 gene sequence and a DIG-labeled DNA probe for the lef-1 gene sequence. Since vp39 is located on the opposite end of the viral genome from lef-1, our results suggested that the whole genome, rather than segments, had replicated. Therefore, integration may not be the reason for the DpnI-resistant products observed in the DpnI replication assay in this study (Fig. 4).
Previous reports have suggested that baculovirus late and very late gene expression depends on viral DNA replication (23, 34). Most of the viral late and very late genes encode viral structural proteins, which are components of BVs and ODVs. As a consequence of low-level viral DNA replication, viral late and very late gene expression was abolished by lef-2 deletion (Fig. 5), indicating a total shutdown of the viral transcription system. The observation that low levels of viral DNA replication were not sufficient to turn on late and very late gene expression suggests that either vigorous viral DNA replication activity is required to turn on viral transcription or LEF-2 itself is involved in the transcription of late and very late genes, as suggested previously (32). We also noticed that the growth curve for the bAcΔlef2-Rep-hE repair virus indicated that it grew more slowly than the bAcwt-hE wild-type virus in transfected cells (Fig. 2B). This was possibly because lef-2 was located in a different context in the genome of the repair virus, which resulted in an altered profile of temporal transcription of lef-2 in the repair virus compared to that in the wild-type virus and subsequently slowed the virus growth rate (Fig. 3). Multiple transcripts corresponding to regions running through the lef-2 coding sequence in both repair and wild-type viruses were detected toward late time points postinfection. The presence of high-molecular-weight transcripts of the lef-2 coding region and other viral genomic regions has also been reported previously (12, 24), and the transcription of these transcripts was either initiated at regions farther upstream or terminated at regions farther downstream of the target sequence. Antisense probes recognizing the sequences upstream and downstream of lef-2 will help to elucidate the exact identities of the high-molecular-weight transcripts represented in Fig. 3.
In vivo immunofluorescence staining showed that LEF-2 colocalized with newly synthesized viral DNA (Fig. 6B). Several replicative LEFs, including IE-1, LEF-3, P143, and DBP, have been shown previously to localize to the sites of viral DNA replication in vivo (14, 25, 30, 31). Our results provide indirect evidence that LEF-2 does perform some important functions at the sites of viral DNA replication, i.e., virogenic stroma. Nevertheless, the exact functions of LEF-2 during viral DNA replication are yet to be elucidated. It was also observed that colocalization of newly synthesized viral DNA and LEF-2 was more readily detectible after the replication foci had passed the initial formation stage and enlarged (data not shown). Since late and very late gene expression also takes place in the virogenic stroma, the immunofluorescence staining data presented herein may also implicate LEF-2 in late and very late gene expression.
The incorporation of LEF-2 into the nucleocapsids of both BVs and ODVs is one of the important findings in the present study (Fig. 7). All the baculovirus replication LEFs except LEF-2 (IE-1, LEF-1, LEF-3, DNAPOL, and P143) have been shown previously to associate with nucleocapsids of the ODVs (6). Our present results add LEF-2 to the list, meaning that essentially all the replication LEFs are present in the nucleocapsids of the ODVs. This finding raises the possibility that viral DNA replication may be initiated and amplified by these capsid proteins even prior to the expression of these replication-related LEFs from the newly invading viral genome. More recently, LEF-11 was also found to be involved in viral DNA replication based on the results of genomic knockout experiments (17). Whether or not this gene product is also a component of the nucleocapsid of virus particles remains an interesting issue for further studies.
LEF-2 is generally viewed as a primase-associated factor due to its interaction with baculovirus-encoded DNA primase LEF-1 (10, 28). No known function of LEF-2 besides its requirement for viral DNA replication has been assigned, although LEF-2 has been suggested to possess single-stranded DNA (ssDNA) binding ability (28). The primase-associated factor of another double-stranded DNA virus, UL8 of herpes simplex virus type 1 (HSV-1), is a component of the helicase-primase complex, which forms part of the prereplicative sites (18). UL8 was also shown previously to be involved in efficient nuclear uptake of the other two proteins of the helicase-primase complex, UL5 and UL52, which function as a helicase and a primase, respectively (8). This is not quite the case for baculovirus, as LEF-2 is not required for the nuclear transport of baculovirus-encoded helicase P143. Instead, a virus-encoded ssDNA binding protein, LEF-3, is required for this process (2, 9). UL8 of HSV-1 is also required for efficient primer utilization during lagging-strand synthesis (36). LEF-2 may perform a function similar to that of UL8 in this regard if binding of LEF-2 to ssDNA and LEF-1 occurs during DNA synthesis on the lagging strand (28). The validation of this hypothesis may help further define the exact functions of LEF-2 during baculovirus replication processes. Moreover, elucidation of the mechanism by which LEF-2 affects late and very late gene expression would further our understanding of baculovirus transcriptional regulation.
Acknowledgments
We thank Gary Blissard and Oliver Lung for giving us the gp64 knockout bacmid as a necessary control and David Theilmann for valuable discussions and suggestions. We also thank Miranda Loney and Harry Wilson for revision.
This research is funded by grants 098-2811-B-001-025, 098-2811-B-001-036, and 98-2321-B-001-031-MY3 from the National Science Council and grant 94S-1303 from Academia Sinica, Taiwan.
Footnotes
Published ahead of print on 10 March 2010.
REFERENCES
- 1.Acharya, A., and K. P. Gopinathan. 2001. Identification of an enhancer-like element in the polyhedrin gene upstream region of Bombyx mori nucleopolyhedrovirus. J. Gen. Virol. 82:2811-2819. [DOI] [PubMed] [Google Scholar]
- 2.Au, V., M. Yu, and E. B. Carstens. 2009. Characterization of a baculovirus nuclear localization signal domain in the late expression factor 3 protein. Virology 385:209-217. [DOI] [PubMed] [Google Scholar]
- 3.Ayres, M. D., S. C. Howard, J. Kuzio, M. Lopez-Ferber, and R. D. Possee. 1994. The complete DNA sequence of Autographa californica nuclear polyhedrosis virus. Virology 202:586-605. [DOI] [PubMed] [Google Scholar]
- 4.Beniya, H., S. C. Braunagel, and M. D. Summers. 1998. Autographa californica nuclear polyhedrosis virus: subcellular localization and protein trafficking of BV/ODV-E26 to intranuclear membranes and viral envelopes. Virology 240:64-75. [DOI] [PubMed] [Google Scholar]
- 5.Blissard, G. W., and J. R. Wenz. 1992. Baculovirus gp64 envelope glycoprotein is sufficient to mediate pH-dependent membrane fusion. J. Virol. 66:6829-6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braunagel, S. C., W. K. Russell, G. Rosas-Acosta, D. H. Russell, and M. D. Summers. 2003. Determination of the protein composition of the occlusion-derived virus of Autographa californica nucleopolyhedrovirus. Proc. Natl. Acad. Sci. U. S. A. 100:9797-9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braunagel, S. C., and M. D. Summers. 1994. Autographa californica nuclear polyhedrosis virus, PDV, and ECV viral envelopes and nucleocapsids: structural proteins, antigens, lipid and fatty acid profiles. Virology 202:315-328. [DOI] [PubMed] [Google Scholar]
- 8.Calder, J. M., E. C. Stow, and N. D. Stow. 1992. On the cellular localization of the components of the herpes simplex virus type 1 helicase-primase complex and the viral origin-binding protein. J. Gen. Virol. 73(Pt. 3):531-538. [DOI] [PubMed] [Google Scholar]
- 9.Chen, Z., and E. B. Carstens. 2005. Identification of domains in Autographa californica multiple nucleopolyhedrovirus late expression factor 3 required for nuclear transport of P143. J. Virol. 79:10915-10922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans, J. T., D. J. Leisy, and G. F. Rohrmann. 1997. Characterization of the interaction between the baculovirus replication factors LEF-1 and LEF-2. J. Virol. 71:3114-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang, M., X. Dai, and D. A. Theilmann. 2007. Autographa californica multiple nucleopolyhedrovirus EXON0 (ORF141) is required for efficient egress of nucleocapsids from the nucleus. J. Virol. 81:9859-9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friesen, P. D., and L. K. Miller. 1985. Temporal regulation of baculovirus RNA: overlapping early and late transcripts. J. Virol. 54:392-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hicks, G. R., and N. V. Raikhel. 1995. Protein import into the nucleus: an integrated view. Annu. Rev. Cell Dev. Biol. 11:155-188. [DOI] [PubMed] [Google Scholar]
- 14.Ito, E., D. Sahri, R. Knippers, and E. B. Carstens. 2004. Baculovirus proteins IE-1, LEF-3, and P143 interact with DNA in vivo: a formaldehyde cross-linking study. Virology 329:337-347. [DOI] [PubMed] [Google Scholar]
- 15.Jehle, J. A., G. W. Blissard, B. C. Bonning, J. S. Cory, E. A. Herniou, G. F. Rohrmann, D. A. Theilmann, S. M. Thiem, and J. M. Vlak. 2006. On the classification and nomenclature of baculoviruses: a proposal for revision. Arch. Virol. 151:1257-1266. [DOI] [PubMed] [Google Scholar]
- 16.Kool, M., C. H. Ahrens, R. W. Goldbach, G. F. Rohrmann, and J. M. Vlak. 1994. Identification of genes involved in DNA replication of the Autographa californica baculovirus. Proc. Natl. Acad. Sci. U. S. A. 91:11212-11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin, G., and G. W. Blissard. 2002. Analysis of an Autographa californica nucleopolyhedrovirus lef-11 knockout: LEF-11 is essential for viral DNA replication. J. Virol. 76:2770-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liptak, L. M., S. L. Uprichard, and D. M. Knipe. 1996. Functional order of assembly of herpes simplex virus DNA replication proteins into prereplicative site structures. J. Virol. 70:1759-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, C. Y., C. H. Wang, W. K. Hsiao, H. R. Lo, C. P. Wu, and Y. C. Chao. 2009. RING and coiled-coil domains of baculovirus IE2 are critical in strong activation of the cytomegalovirus major immediate-early promoter in mammalian cells. J. Virol. 83:3604-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, Y. K., C. C. Chu, and T. Y. Wu. 2006. Baculovirus ETL promoter acts as a shuttle promoter between insect cells and mammalian cells. Acta Pharmacol. Sin. 27:321-327. [DOI] [PubMed] [Google Scholar]
- 21.Lo, H. R., and Y. C. Chao. 2004. Rapid titer determination of baculovirus by quantitative real-time polymerase chain reaction. Biotechnol. Prog. 20:354-360. [DOI] [PubMed] [Google Scholar]
- 22.Lo, H. R., C. C. Chou, T. Y. Wu, J. P. Yuen, and Y. C. Chao. 2002. Novel baculovirus DNA elements strongly stimulate activities of exogenous and endogenous promoters. J. Biol. Chem. 277:5256-5264. [DOI] [PubMed] [Google Scholar]
- 23.Lu, A., and L. K. Miller. 1995. The roles of eighteen baculovirus late expression factor genes in transcription and DNA replication. J. Virol. 69:975-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mainprize, T. H., K. Lee, and L. K. Miller. 1986. Variation in the temporal expression of overlapping baculovirus transcripts. Virus Res. 6:85-99. [DOI] [PubMed] [Google Scholar]
- 25.Mainz, D., I. Quadt, and D. Knebel-Morsdorf. 2002. Nuclear IE2 structures are related to viral DNA replication sites during baculovirus infection. J. Virol. 76:5198-5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merrihew, R. V., W. C. Clay, J. P. Condreay, S. M. Witherspoon, W. S. Dallas, and T. A. Kost. 2001. Chromosomal integration of transduced recombinant baculovirus DNA in mammalian cells. J. Virol. 75:903-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Merrington, C. L., P. A. Kitts, L. A. King, and R. D. Possee. 1996. An Autographa californica nucleopolyhedrovirus lef-2 mutant: consequences for DNA replication and very late gene expression. Virology 217:338-348. [DOI] [PubMed] [Google Scholar]
- 28.Mikhailov, V. S., and G. F. Rohrmann. 2002. Baculovirus replication factor LEF-1 is a DNA primase. J. Virol. 76:2287-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monsma, S. A., A. G. Oomens, and G. W. Blissard. 1996. The GP64 envelope fusion protein is an essential baculovirus protein required for cell-to-cell transmission of infection. J. Virol. 70:4607-4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagamine, T., Y. Kawasaki, A. Abe, and S. Matsumoto. 2008. Nuclear marginalization of host cell chromatin associated with expansion of two discrete virus-induced subnuclear compartments during baculovirus infection. J. Virol. 82:6409-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okano, K., V. S. Mikhailov, and S. Maeda. 1999. Colocalization of baculovirus IE-1 and two DNA-binding proteins, DBP and LEF-3, to viral replication factories. J. Virol. 73:110-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Passarelli, A. L., and L. K. Miller. 1993. Three baculovirus genes involved in late and very late gene expression: ie-1, ie-n, and lef-2. J. Virol. 67:2149-2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rapp, J. C., J. A. Wilson, and L. K. Miller. 1998. Nineteen baculovirus open reading frames, including LEF-12, support late gene expression. J. Virol. 72:10197-10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rice, W. C., and L. K. Miller. 1986. Baculovirus transcription in the presence of inhibitors and in nonpermissive Drosophila cells. Virus Res. 6:155-172. [DOI] [PubMed] [Google Scholar]
- 35.Schultz, K. L., and P. D. Friesen. 2009. Baculovirus DNA replication-specific expression factors trigger apoptosis and shutoff of host protein synthesis during infection. J. Virol. 83:11123-11132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sherman, G., J. Gottlieb, and M. D. Challberg. 1992. The UL8 subunit of the herpes simplex virus helicase-primase complex is required for efficient primer utilization. J. Virol. 66:4884-4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stewart, T. M., I. Huijskens, L. G. Willis, and D. A. Theilmann. 2005. The Autographa californica multiple nucleopolyhedrovirus ie0-ie1 gene complex is essential for wild-type virus replication, but either IE0 or IE1 can support virus growth. J. Virol. 79:4619-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vanarsdall, A. L., K. Okano, and G. F. Rohrmann. 2004. Characterization of a baculovirus with a deletion of vlf-1. Virology 326:191-201. [DOI] [PubMed] [Google Scholar]
- 39.Wu, C. P., J. Y. Wang, T. Y. Huang, H. R. Lo, and Y. C. Chao. 2008. Identification of baculoviral factors required for the activation of enhancer-like polyhedrin upstream (pu) sequence. Virus Res. 138:7-16. [DOI] [PubMed] [Google Scholar]
- 40.Wu, Y., and E. B. Carstens. 1996. Initiation of baculovirus DNA replication: early promoter regions can function as infection-dependent replicating sequences in a plasmid-based replication assay. J. Virol. 70:6967-6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu, Y. L., and Y. C. Chao. 2008. The establishment of a controllable expression system in baculovirus: stimulated overexpression of polyhedrin promoter by LEF-2. Biotechnol. Prog. 24:1232-1240. [DOI] [PubMed] [Google Scholar]
- 42.Wu, Y.-L., C. P. Wu, S.-T. Lee, H. Tang, C.-H. Chang, H.-H. Chen, and Y.-C. Chao. 2010. The early gene hhi1 reactivates Heliothis zea nudivirus 1 in latently infected cells. J. Virol. 84:1057-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]