Abstract
The first fully sequenced papillomavirus (PV) of marsupials, tentatively named Bettongia penicillata papillomavirus type 1 (BpPV1), was detected in papillomas from a woylie (Bettongia penicillata ogilbyi). The circular, double-stranded DNA genome contains 7,737 bp and encodes 7 open reading frames (ORFs), E6, E7, E1, E2, E4, L2, and L1, in typical PV conformation. BpPV1 is a close-to-root PV with L1 and L2 ORFs most similar to European hedgehog PV and bandicoot papillomatosis carcinomatosis virus types 1 and 2 (BPCV1 and -2). It appears that the BPCVs arose by recombination between an ancient PV and an ancient polyomavirus more than 10 million years ago.
Papillomaviruses (PVs) are small, nonenveloped, icosahedral viruses with a circular double-stranded DNA (dsDNA) genome of approximately 8 kbp (12). These host-specific, epitheliotropic viruses may induce benign or malignant epithelial proliferations (12).
The woylie, Bettongia penicillata ogilbyi (Gray, 1837), is listed as a critically endangered marsupial species on the IUCN Red List of Threatened Species (www.iucnredlist.org). Woylies were the first Australian mammals to be delisted from Commonwealth and State endangered species lists, but they are currently experiencing a dramatic population decline related to predation and, possibly, disease (5, 24, 28).
Putative marsupial or monotreme PVs have been detected by electron microscopy in an American marsupial (Didelphis sp.) (14) and by molecular techniques in Australian species, including the koala, eastern grey kangaroo, echidna, and possum (1, 19). Molecular diagnosis of these putative PVs relied upon PCR to detect conserved regions within the L1 open reading frame (ORF); however, the bandicoot papillomatosis carcinomatosis virus types 1 and 2 (BPCV1 and -2), which infect two marsupial bandicoot species, also contain PV-like L1 and L2 ORFs but do not meet the criteria for inclusion within the Papillomaviridae (3, 6, 29). Rather, the BPCVs appear to be natural PV-polyomavirus hybrids (3, 29). The discovery of the BPCVs casts doubt on the classification of all PVs identified using partial sequence data from the late ORFs. Complete genomic analysis was conducted on the woylie isolate to definitively establish its taxonomic position.
An adult male Bettongia penicillata ogilbyi was initially trapped on 21 May 2008 in Western Australia (34°02′S, 116°36′E) (Department of Environment and Conservation Animal Ethics Committee approval, 2006/08). Multiple darkly pigmented, raised papillomatous lesions were noted on both the left and right eyelid margins and on the left muzzle. Lesions were biopsied with the animal under general anesthesia, and samples were placed in 10% neutral buffered formalin and in sterile cryovials. The animal was retrapped on another 6 occasions over the following 6 months, during which papillomatous lesions were not detected.
Fixed biopsy material was processed for histopathology (3), immunohistochemistry (3), and in situ hybridization (2). The epidermis was focally expanded by multiple projecting papillary folds, with marked orthokeratotic and parakeratotic hyperkeratosis. The basal layer was irregular but confined within the basement membrane. The stratum spinosum was thickened 10 to 15 cells deep, there were multiple, large keratinocytes with pale cytoplasm (koilocytosis), the stratum granulosum was thickened, and these lesions extended to the biopsy specimen margins (consistent with cutaneous papillomatosis). Indirect immunohistochemistry demonstrated scattered positive keratinocyte nuclear staining, and in situ hybridizations using a digoxigenin-labeled BpPV1 genomic DNA probe annealed to nuclei and nucleoli of keratinocytes within the epidermis, ruling out contamination. These findings provide a compelling temporal and spatial association between the papillomatous lesions and BpPV1. Based on the current understanding of PV infections in other species, it appears plausible that a causal relationship between clinical papillomatosis in B. penicillata and BpPV1 may exist.
Photographs and a histological slide from this case were deposited at the Australian Registry of Wildlife Health (ARWH), Taronga Zoo, Australia (ARWH accession number 6788/1).
Total DNA was extracted from the frozen sample using the DNeasy tissue kit (Qiagen). PCR using FAP59/64 primers (7) produced an ∼450-bp amplicon, and sequencing of this amplicon confirmed that an L1 ORF-containing novel virus was present. Multiply primed rolling circle amplification (RCA) using the TempliPhi 100 amplification kit (Amersham Biosciences) was performed on an aliquot of extracted DNA (21, 29). The RCA product was digested with restriction endonucleases: EcoRI produced 1 fragment that was ∼7.7 kb; SalI did not cut; BamHI produced 2 fragments that were ∼7.6 kb and ∼0.1 kb; HindIII produced 2 fragments that were ∼6.9 kb and ∼0.8 kb; and XbaI produced 2 fragments that were ∼7 kb and ∼0.7 kb. The RCA product, cut with EcoRI, XbaI, and HindIII, was cloned into linearized, dephosphorylated, and purified pGEM 3f(+) plasmids (Applied Biosystems) using the Roche Rapid DNA ligation kit (Roche Diagnostics) and transformed into competent Escherichia coli (Invitrogen; One Shot MAX Efficiency DH5α-T1), with permission from the Murdoch University Institutional Biosafety Committee. E. coli was subjected to blue/white colony screening; insert-containing colonies were grown in broth cultures, and plasmids were prepared using the QIAprep Spin miniprep kit (Qiagen). Purified BpPV1-containing plasmids were sequenced in triplicate in the forward and reverse directions with the version 3.1 BigDye Terminator kit (Applied Biosystems) (22). The sequence was determined using an ABI Prism Applied Biosystems 377 automatic DNA sequencer (Applied Biosystems). Chromatogram files were edited with Chromas Lite version 2.0 (Technelysium Pty. Ltd.) and sequences assembled and aligned in BioEdit version 7.0.9.0 (11). Putative ORFs were predicted manually and by using the National Center for Biotechnology Information online ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). The online Basic Local Alignment Search Tool (BLAST) was used to interrogate GenBank. Identified ORFs were translated using the ExPASy Translate tool, and theoretical isoelectric points and molecular masses were estimated using Compute pI/Mw (http://www.expasy.ch/tools).
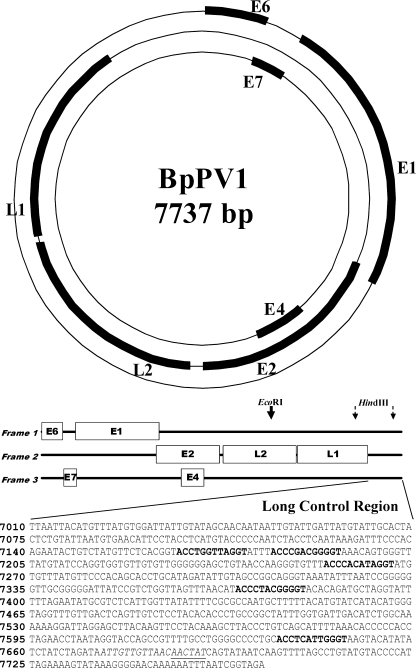
The BpPV1 genome was a circular dsDNA molecule, 7,737 bp around, with a GC content of 44.20%, and contained 2 classical PV late and 5 classical PV early protein ORFs separated by a long control region (LCR). All 7 ORFs were within the sense DNA strand (Fig. 1; Table 1). The putative E4 ORF did not include a recognized start codon.
FIG. 1.
Circular and linear genome maps of Bettongia penicillata papillomavirus type 1 (BpPV1), including a detail of the long control region, showing a putative origin of DNA replication (italics), predicted E2 binding sites (bold), and a predicted E1 binding site (underlined).
TABLE 1.
Percentage similarity of aligned nucleotide and amino acid sequences from BpPV1 compared to BPCV types 1 and 2 and selected papillomavirus types and predicted physicochemical properties of translated BpPV1 ORFsa
| ORF | Type | % similarity of BpPV1 to: |
ORF location | Length (aa) | Molecular mass (kDa) | pI | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EHPV | BPCV1 | BPCV2 | COPV | HPV1a | HPV16 | BPV1 | ||||||
| L1 | n | 62 | 61 | 60 | 55 | 53 | 52 | 52 | 5498-7009 | 503 | 57.3 | 7.51 |
| aa | 61 | 58 | 59 | 51 | 49 | 47 | 47 | |||||
| L2 | n | 39 | 39 | 36 | 40 | 40 | 36 | 33 | 3905-5476 | 523 | 56.5 | 4.93 |
| aa | 35 | 31 | 30 | 32 | 33 | 30 | 27 | |||||
| E6 | n | 36 | 36 | 38 | 38 | 32 | 28-472 | 147 | 17.2 | 4.94 | ||
| aa | 28 | 24 | 21 | 22 | 23 | |||||||
| E7 | n | 47 | 38 | 41 | 32 | 26 | 468-740 | 90 | 10.2 | 4.46 | ||
| aa | 33 | 25 | 26 | 14 | 15 | |||||||
| E1 | n | 54 | 53 | 53 | 49 | 49 | 730-2520 | 596 | 67.9 | 6.06 | ||
| aa | 45 | 45 | 43 | 37 | 39 | |||||||
| E2 | n | 33 | 39 | 33 | 27 | 2462-3826 | 454 | 50.1 | 7.83 | |||
| aa | 34 | 27 | 26 | 24 | 18 | |||||||
| E4b | n | 25 | 27 | 3006-3495 | 162 | 18.8 | 5.35 | |||||
| aa | 13 | 20 | ||||||||||
EHPV, European hedgehog PV (NC_011765); BPCV1, bandicoot papillomatosis carcinomatosis virus type 1 (NC_010107); BPCV2, bandicoot papillomatosis carcinomatosis virus type 2 (NC_010817); COPV, canine oral PV (NC_001619); HPV1a, human PV type 1a (NC_001356); HPV16, human PV type 16 (NC_001326); BPV1, bovine PV type 1 (NC_001522); pI, isoelectric point; n, percentage similarity based on aligned nucleotide sequence; aa, percentage similarity based on aligned amino acid sequence.
The E4 ORF does not contain a recognized start codon.
Pairwise sequence identity scores were calculated for BpPV1 and the late ORFs (L1 and L2) of the BPCVs and for the L1, L2, E6, E7, E1, E2, and E4 ORFs of selected PV types (Alphapapillomavirus human PV 16 [HPV16; GenBank accession number NC_001526], Deltapapillomavirus bovine PV 1 [BPV1; NC_001522], Lambdapapillomavirus canine oral PV [COPV; NC_001619], Mupapillomavirus human PV 1a [HPV1a; NC_001356], and European hedgehog PV [EHPV; NC_011765]) at the nucleotide and amino acid levels using the GAP program on the Sequence Analysis Server at Michigan Technological University (Table 1).
The time since BpPV1 and the BPCVs diverged from a theoretical common ancestor virus was estimated using the average mutation rate for the L1 ORF of lambdapapillomaviruses, which is 1.84 × 10−8 nucleotide substitutions per site per year (95% confidence interval [CI], 1.27 × 10−8 to 2.35 × 10−8 nucleotide substitutions per site per year) (20). Based upon the differences between BpPV1 and BPCV1 L1 ORFs (580 nucleotide substitutions over a 1,535-bp unambiguously aligned region), these viruses may have shared an ancestor ∼34.4 million years ago (mya) (95% CI, 26.9 to 49.9 mya).
Phylogenetic analysis.
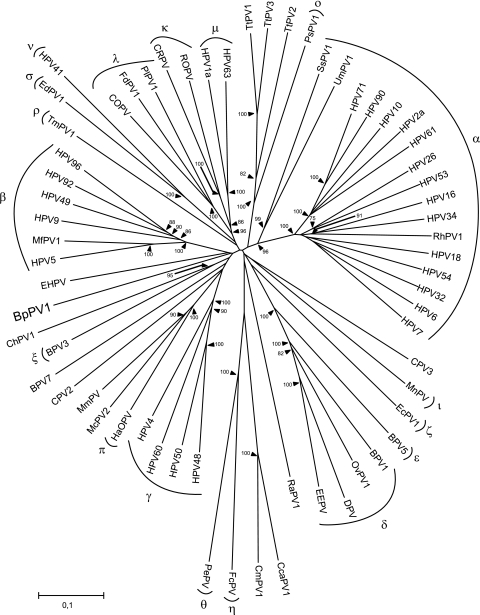
Multiple nucleotide sequence alignments were constructed in DAMBE version 5.0.52 (30). The sequence of BpPV1 and those of 62 other PV types were imported and aligned at the amino acid level using ClustalW (25). The nucleotide sequences were aligned according to the aligned amino acid sequences. This was performed separately for each ORF. Nucleotide alignments were corrected manually in GeneDoc Multiple Sequence Alignment Editor and Shading Utility version 2.6.003 (17). Only the unambiguously aligned parts of the E1, E2, L2, and L1 ORFs were used to form one concatenated alignment of 2,763 sites. Nucleotide positions included in the alignments were nucleotides (nt) 1195 to 1272, 1417 to 1533, 1627 to 1842, 1879 to 2271, and 2287 to 2424 in E1, nt 2507 to 2863, 2897 to 2941, and 3587 to 3640 in E2, nt 3917 to 4075 and 4874 to 4972 in L2, and nt 5525 to 5647, 5687 to 5893, 5927 to 6004, 6050 to 6301, 6374 to 6536, 6584 to 6712, 6746 to 6790, and 6842 to 6919 in L1, relative to the BpPV1 sequence. From these alignments, phylogenetic trees were constructed using the neighbor-joining method in MEGA version 4.1 (15). Trees were constructed using all 3 codon positions, as well as only the 1st and 2nd positions. Bootstrap support values were obtained for 10,000 replicates. In this tree, the PVs cluster in the previously defined genera, and high bootstrap values support the nodes joining PV congeners. According to this tree, BpPV1 is most closely related to European hedgehog papillomavirus (EHPV) but branches off very close to the root of the tree. This clustering is supported by a bootstrap value of 20% (Fig. 2).
FIG. 2.
Neighbor-joining phylogenetic tree, based on a combined concatenated E1/E2/L2/L1 nucleotide sequence alignment of BpPV1 and 62 other PVs, using only the 1st and 2nd codon positions. The PV types were, in the genus Alphapapillomavirus, HPV32 (GenBank accession number NC_001586), HPV10 (NC_001576), HPV61 (NC_001694), HPV2a (NC_001352), HPV26 (NC_001583), HPV53 (NC_001593), HPV18 (NC_001357), HPV7 (NC_001595), HPV16 (NC_001526), HPV6 (NC_001355), HPV34 (NC_001587), rhesus PV type 1 (RhPV1; NC_001678), HPV54 (NC_001676), HPV90 (NC_004104), and HPV71 (AY330621); in the genus Betapapillomavirus, HPV5 (NC_001531), HPV9 (NC_001596), HPV49 (NC_001591), HPV92 (NC_004500), HPV96 (NC_005134), and Macaca fascicularis PV type 1 (MfPV1; EF028290); in the genus Gammapapillomavirus, HPV4 (NC_001457), HPV48 (NC_001690), HPV50 (NC_001691), and HPV60 (NC_001693); in the genus Deltapapillomavirus, European elk PV (EEPV; NC_001524), deer PV (DPV; NC_001523), ovine PV type 1 (OvPV1; NC_001789), and BPV1 (NC_001522); in the genus Epsilonpapillomavirus, BPV5 (NC_004195); in the genus Zetapapillomavirus, Equus caballus PV type 1 (EcPV1; NC_003748); in the genus Etapapillomavirus, Fringilla coelebs PV (FcPV; NC_004068); in the genus Thetapapillomavirus, Psittacus erithacus timnesh PV (PePV; NC_003973); in the genus Iotapapillomavirus, Mastomys natalensis PV (MnPV; NC_001605); in the genus Kappapapillomavirus, cottontail rabbit PV (CRPV; NC_001541) and rabbit oral PV (ROPV; NC_002232); in the genus Lambdapapillomavirus, COPV (NC_001619), Felis domesticus PV type 1 (FdPV1; NC_004765), and Procyon lotor PV type 1 (PlPV1; NC_007150); in the genus Mupapillomavirus, HPV1 (NC_001356); in the genus Nupapillomavirus, HPV41 (NC_001354); in the genus Xipapillomavirus, BPV3 (NC_004197); in the genus Omicronpapillomavirus, Phocoena spinipinnis PV type 1 (PsPV1; NC_003348); in the genus Pipapillomavirus, hamster oral PV (HaOPV; E15111); in the genus Rhopapillomavirus, Trichechus manatus latirostris PV type 1 (TmPV1; NC_006563); in the genus Sigmapapillomavirus, Erethizon dorsatum PV type 1 (EdPV1; NC_006951); and unclassified types Rousettus aegyptiacus PV type 1 (RaPV1; NC_008298), BPV7 (NC_007612), canine PV type 2 (CPV2; NC_006564), CPV3 (NC_008297), Capra hircus PV type 1 (ChPV1; NC_008032), Mastomys coucha PV type 2 (McPV2; DQ664501), Micromys minutus PV (MmPV; NC_008582), Ursus maritimus PV type 1 (UmPV1; NC_010739), Sus scrofa PV type 1a (SsPV1a; EF395818), Chelonia mydas PV type 1 (CmPV1; EU257705), Caretta caretta PV type 1 (CcaPV1; EU257704), Tursiops truncatus PV type 1 (TtPV1; NC_011109), TtPV2 (NC_008184), TtPV3 (NC_011110), and EHPV (NC_011765). The established PV genera are indicated by their Greek symbols. The scale bar indicates the genetic distance (in nucleotide substitutions per site), and the numbers at the internal nodes represent the bootstrap probability percentages as determined for 10,000 iterations by the neighbor-joining method. Only bootstrap values of 75% or more are shown.
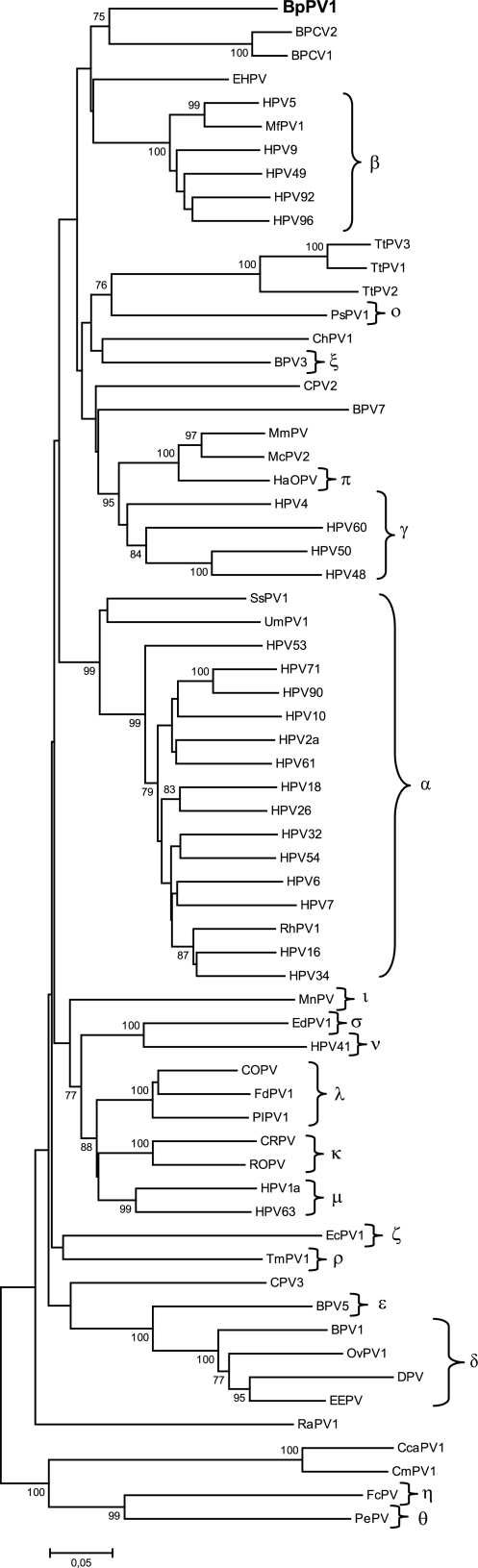
A second tree was constructed using a 1,503-site-long, concatenated alignment of the L1 and L2 ORFs. This alignment included the L1 and L2 sequences of BpPV1, BPCV1 (GenBank accession number NC_010107), BPCV2 (NC_010817), and the same 62 PV types used in the first tree. The nucleotide positions, relative to BpPV1, were the same as in the first tree, and bootstrap support values were obtained for 10,000 replicates. In this tree, the late region of BpPV1 appears to be most closely related to the late ORFs of BPCV1 and BPCV2. This clustering is supported by a bootstrap value of 75% (Fig. 3).
FIG. 3.
Evolutionary relationship of the BpPV1 L1 and L2 sequences to BPCV1, BPCV2, and selected members of the Papillomaviridae. A neighbor-joining phylogenetic tree based on a concatenated alignment of the L1 and L2 sequences of BpPV1, BPCV2 (GenBank accession number NC_010817), BPCV1 (NC_010107), and 62 PVs (defined in the legend to Fig. 2). The established PV genera are indicated by their Greek symbols. The scale bar indicates the genetic distance (in nucleotide substitutions per site), and the numbers at the internal nodes represent the bootstrap probability percentages as determined for 10,000 iterations by the neighbor-joining method. Only bootstrap values of 75% or more are shown.
Analysis of this close-to-root BpPV1 genome has provided insight into the evolutionary history of the BPCVs. It appears that the late ORFs of the BPCVs last shared a common PV ancestor with BpPV1 approximately 26.9 to 49.9 mya, while their marsupial hosts, Bettongia penicillata, Perameles bougainville, and Isoodon obesulus, are thought to have last shared a common ancestor during the Eocene (∼34 to 56 mya), based on mitochondrial DNA evidence (18).
At least 2 of the 4 postulated evolutionary mechanisms of PV diversification (10, 23), namely, codivergence between PVs and their hosts (20), infection across species boundaries (9), establishment of new ecological niches by adaptive radiation (8, 13), and recombination (16, 26, 27), appear to be relevant to BPCV evolutionary history. It seems most likely that recombination between an ancient marsupial PV and an ancient polyomavirus at least 10 mya, and potentially as much as ∼50 mya, gave rise to the BPCVs, which have since undergone virus-host codivergence in their respective hosts (3). If more sequence data become available from other novel PV types and polyomaviruses, the evolutionary histories of the marsupial PVs and BPCVs may be resolved with greater certainty (4). To that end, it is vital that novel isolates of circular dsDNA viruses, particularly those from marsupial hosts, be completely sequenced and analyzed (6, 8, 9, 16).
Nucleotide sequence accession number.
The sequence data for the BpPV1 genome were archived in GenBank, National Center for Biotechnology Information (accession number GU220391).
Acknowledgments
This project received funding from the Weston-Fernie Wildlife Research Endowment, Murdoch University, and the Department of Environment and Conservation.
Cloning and sequencing experiments were performed at the State Agriculture and Biotechnology Centre, Western Australia, with invaluable assistance from Frances Brigg, Moira Desport, Josh Lewis, and Tegan McNab.
Footnotes
Published ahead of print on 3 March 2010.
REFERENCES
- 1.Antonsson, A., and A. J. McMillan. 2006. Papillomavirus in healthy skin of Australian animals. J. Gen. Virol. 87:3195-3200. [DOI] [PubMed] [Google Scholar]
- 2.Bennett, M. D., L. Woolford, A. J. O'Hara, K. S. Warren, and P. K. Nicholls. 2008. In situ hybridization to detect bandicoot papillomatosis carcinomatosis virus type 1 in biopsies from endangered western barred bandicoots (Perameles bougainville). J. Gen. Virol. 89:419-423. [DOI] [PubMed] [Google Scholar]
- 3.Bennett, M. D., L. Woolford, H. Stevens, M. Van Ranst, T. Oldfield, M. Slaven, A. J. O'Hara, K. S. Warren, and P. K. Nicholls. 2008. Genomic characterization of a novel virus found in papillomatous lesions from a southern brown bandicoot (Isoodon obesulus) in Western Australia. Virology 376:173-182. [DOI] [PubMed] [Google Scholar]
- 4.Bernard, H.-U. 2008. Genome diversity and evolution of papillomaviruses, p. 417-429. In E. Domingo, C. Parish, and J. Holland (ed.), Origin and evolution of viruses, 2nd ed. Academic Press, London, England.
- 5.Christensen, P. 2002. Brush-tailed bettong, p. 292-293. In R. Strahan (ed.), The mammals of Australia, 2nd ed. Reed New Holland, Frenchs Forest, New South Wales, Australia.
- 6.de Villiers, E., C. Fauquet, T. R. Broker, H. Bernard, and H. zur Hausen. 2004. Classification of papillomaviruses. Virology 324:17-27. [DOI] [PubMed] [Google Scholar]
- 7.Forslund, O., A. Antonsson, P. Nordin, B. Stenquist, and B. G. Hansson. 1999. A broad range of human papillomavirus types detected with a general PCR method suitable for analysis of cutaneous tumours and normal skin. J. Gen. Virol. 80:2437-2443. [DOI] [PubMed] [Google Scholar]
- 8.García-Vallvé, S., Á. Alonso, and I. G. Bravo. 2005. Papillomaviruses: different genes have different histories. Trends Microbiol. 13:514-521. [DOI] [PubMed] [Google Scholar]
- 9.Gottschling, M., A. Köhler, E. Stockfleth, and I. Nindl. 2007. Phylogenetic analysis of beta-papillomaviruses as inferred from nucleotide and amino acid sequence data. Mol. Phylogenet. Evol. 42:213-222. [DOI] [PubMed] [Google Scholar]
- 10.Gottschling, M., A. Stamatakis, I. Nindl, E. Stockfleth, Á. Alonso, and I. G. Bravo. 2007. Multiple evolutionary mechanisms drive papillomavirus diversification. Mol. Biol. Evol. 24:1242-1258. [DOI] [PubMed] [Google Scholar]
- 11.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 12.Howley, P. M., and D. R. Lowy. 2007. Papillomaviruses, p. 2299-2354. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed., vol. 2. Lippincott Williams and Wilkins, Philadelphia, PA. [Google Scholar]
- 13.Jackson, A. 2005. The effect of paralogous lineages on the application of reconciliation analysis by cophylogeny mapping. Syst. Biol. 54:127-145. [DOI] [PubMed] [Google Scholar]
- 14.Koller, L. D. 1972. Cutaneous papillomas on an opossum. J. Natl. Cancer Inst. 49:309-313. [PubMed] [Google Scholar]
- 15.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinformatics 5:150-163. [DOI] [PubMed] [Google Scholar]
- 16.Narechania, A., Z. Chen, R. DeSalle, and R. D. Burk. 2005. Phylogenetic incongruence among oncogenic genital alpha human papillomaviruses. J. Virol. 79:15503-15510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicholas, K. B., H. B. Nicholas, and D. W. Deerfield II. 1997. GeneDoc: analysis and visualization of genetic variation. EMBnet.news 4:1-4. [Google Scholar]
- 18.Nilsson, M. A., U. Arnason, P. B. S. Spencer, and A. Janke. 2004. Marsupial relationships and a timeline for marsupial radiation in South Gondwana. Gene 340:189-196. [DOI] [PubMed] [Google Scholar]
- 19.Perrott, M. R. F., J. Meers, G. E. Greening, S. E. Farmer, I. W. Lugton, and C. R. Wilks. 2000. A new papillomavirus of possums (Trichosurus vulpecula) associated with typical wart-like lesions. Arch. Virol. 145:1247-1255. [DOI] [PubMed] [Google Scholar]
- 20.Rector, A., P. Lemey, R. Tachezy, S. Mostmans, S.-J. Ghim, K. Van Doorslaer, M. Roelke, M. Bush, R. J. Montali, J. Joslin, R. D. Burk, A. B. Jenson, J. P. Sundberg, B. Shapiro, and M. Van Ranst. 2007. Ancient papillomavirus-host co-speciation in Felidae. Genome Biol. 8:R57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rector, A., R. Tachezy, and M. Van Ranst. 2004. A sequence-independent strategy for detection and cloning of circular DNA virus genomes by using multiply primed rolling-circle amplification. J. Virol. 78:4993-4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U. S. A. 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schulz, E., M. Gottschling, I. G. Bravo, U. Wittstatt, E. Stockfleth, and I. Nindl. 2009. Genomic characterisation of the first insectivoran papillomavirus reveals an unusually long, second non-coding region and indicates a close relationship with Betapapillomavirus. J. Gen. Virol. 90:626-633. [DOI] [PubMed] [Google Scholar]
- 24.Start, T., A. Burbidge, and D. Armstrong. 1998. A review of the conservation status of the woylie, Bettongia penicillata ogilbyi (Marsupialia: Potoroidae) using IUCN criteria. CALMscience 2:277-289. [Google Scholar]
- 25.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Ranst, M., J. B. Kaplan, J. P. Sundberg, and R. D. Burk. 1995. Molecular evolution of papillomaviruses, p. 455-476. In A. Gibbs, C. H. Calisher, and F. García-Arenal (ed.), Molecular basis of virus evolution. Cambridge University Press, Cambridge, United Kingdom.
- 27.Varsani, A., E. van der Walt, L. Heath, E. P. Rybicki, A. L. Williamson, and D. P. Martin. 2006. Evidence of ancient papillomavirus recombination. J. Gen. Virol. 87:2527-2531. [DOI] [PubMed] [Google Scholar]
- 28.Wayne, A. F. 2008. Summary of progress and interim findings, p. 279-292. In A. F. Wayne (ed.), Diagnosis of recent woylie (Bettongia penicillata ogilbyi) declines in southwestern Australia: progress report of the woylie conservation research project. Western Australian Department of Environment and Conservation, Perth, Western Australia, Australia.
- 29.Woolford, L., A. Rector, M. Van Ranst, A. Ducki, M. D. Bennett, P. K. Nicholls, K. S. Warren, R. A. Swan, G. E. Wilcox, and A. J. O'Hara. 2007. A novel virus detected in papillomas and carcinomas of the endangered western barred bandicoot (Perameles bougainville) exhibits genomic features of both the Papillomaviridae and Polyomaviridae. J. Virol. 81:13280-13290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xia, X., and Z. Xie. 2001. DAMBE: software package for data analysis in molecular biology and evolution. J. Hered. 92:371-373. [DOI] [PubMed] [Google Scholar]