Abstract
Extensive cell culture passage of the virulent Bucyrus (VB) strain of equine arteritis virus (EAV) to produce the modified live virus (MLV) vaccine strain has altered its tropism for equine CD3+ T lymphocytes and CD14+ monocytes. The VB strain primarily infects CD14+ monocytes and a small subpopulation of CD3+ T lymphocytes (predominantly CD4+ T lymphocytes), as determined by dual-color flow cytometry. In contrast, the MLV vaccine strain has a significantly reduced ability to infect CD14+ monocytes and has lost its capability to infect CD3+ T lymphocytes. Using a panel of five recombinant chimeric viruses, we demonstrated that interactions among the GP2, GP3, GP4, GP5, and M envelope proteins play a major role in determining the CD14+ monocyte tropism while the tropism for CD3+ T lymphocytes is determined by the GP2, GP4, GP5, and M envelope proteins but not the GP3 protein. The data clearly suggest that there are intricate interactions among these envelope proteins that affect the binding of EAV to different cell receptors on CD3+ T lymphocytes and CD14+ monocytes. This study shows, for the first time, that CD3+ T lymphocytes may play an important role in the pathogenesis of equine viral arteritis when horses are infected with the virulent strains of EAV.
Equine arteritis virus (EAV) is a small enveloped virus with a positive-sense, single-stranded RNA genome of 12.7 kb that belongs to the family Arteriviridae (genus Arterivirus, order Nidovirales), which also includes porcine reproductive and respiratory syndrome virus, simian hemorrhagic fever virus, and lactate dehydrogenase-elevating virus of mice (12, 55). The EAV genome includes nine known functional open reading frames (ORFs) (57). ORFs 1a and 1b encode two replicase polyproteins (pp1a and pp1ab) that are posttranslationally processed by three ORF1a-encoded proteinases (nsp1, -2, and -4) to yield at least 13 nonstructural proteins (nsp1 to -12, including nsp7α and -7β) (55, 61, 68), and the remaining seven ORFs (2a, 2b, and 3 to 7) encode structural proteins of the virus. These include four minor envelope proteins (E, GP2, GP3, and GP4, encoded by ORFs 2a, 2b, 3, and 4, respectively), two major envelope proteins (GP5 and M, encoded by ORFs 5 and 6) and the highly conserved nucleocapsid (N) protein (encoded by ORF7) (56). The two major envelope proteins GP5 and M form a disulfide-linked heterodimer (18, 54), and the three minor envelope proteins GP2, GP3, and GP4 form a covalently associated heterotrimeric complex in the virion (63, 65). By independently knocking out the expression of each structural protein, it was shown that all major and minor structural proteins are required for the production of infectious progeny virus (46).
EAV is the causative agent of equine viral arteritis (EVA) (10, 11). Geographically and temporally different strains of EAV vary in the severity of the clinical disease they induce and in their abortigenic potential (1, 45, 49, 58). While most EAV infections are asymptomatic, some infected horses can exhibit clinical manifestations such as an influenza-like illness, abortion in pregnant mares, and pneumonia or pneumoenteritis in neonatal foals (17, 22, 23, 28, 43, 44, 58, 60). Following experimental infection of horses by the respiratory route, the virus initially replicates in both macrophages and endothelial cells in the lung (14, 42). EAV is then transported to the draining lymph nodes, where it replicates and is released into the blood and lymphatic system for transport throughout the body. The virus infects the smaller blood vessels, especially the arterioles, causing a panvasculitis (16, 35, 42). A number of studies have demonstrated that different strains of EAV vary significantly in their pathogenicity, with very different clinical outcomes upon experimental inoculation of horses (2, 5, 6, 21, 43). The horse-adapted virulent Bucyrus (VB) strain of EAV is highly velogenic and causes severe clinical disease with a case fatality rate of 50 to 60% under experimental conditions (35, 51). Horses inoculated with virulent strains of EAV (e.g., the VB, recombinant VB [rVBS], and KY84 strains) develop severe lymphopenia with a high-titer viremia (6 × 103 to 1 × 105 PFU/ml) (2, 5, 35, 44). In contrast, horses inoculated with the attenuated modified live virus (MLV) vaccine strain or other avirulent strains of EAV (e.g., 030H and CA95G) (6, 49) develop a mild, transient lymphopenia with only a very-low-titer viremia (≤1 × 101 PFU/ml) (20, 24, 37-40).
The MLV vaccine (ARVAC; Fort Dodge Animal Health, Fort Dodge, IA) that is in current use in the United States and Canada (20, 37-40) was developed by serial passage of the VB strain of EAV in primary horse kidney (131 passages [HK131]) cells, primary rabbit kidney (111 passages [RK111]) cells, and equine dermis (24 passages [ED24]) cells, which resulted in attenuation of the virus (24). Previously, it has been shown that the VB strain was fully attenuated for horses by 116 passages in primary horse kidney cells (EAV HK116) (37, 38, 41). Subsequently, EAV HK116 was further passaged in HK cells an additional 15 times (HK131) and in two other cell lines (RK111 and ED24) to obtain the vaccine MLV (HK131, RK111, ED24) used in this study. There are 23 additional amino acid substitutions that accumulated in the GP2, GP3, GP4, GP5, and M envelope proteins during cell culture passage of the HK116 virus to generate the current MLV strain (ARVAC; Table 1) which may have contributed to further attenuation of the vaccine strain of EAV and increased the safety of the vaccine for horses. Recently, using reverse genetics, we showed that a chimeric virus containing the nonstructural proteins of the VB virus and the structural proteins of the HK116 strain (rVBS/HK116 S) (66) has an attenuated phenotype in horses. The data showed that critical amino acid substitutions in structural protein genes of the HK116 virus were responsible for attenuation of the VB strain.
TABLE 1.
Comparative amino acid analysis of rVBS, HK116, and rMLV envelope proteinsa
| ORF | Protein (length [amino acids]) | Position | Amino acid in: |
||
|---|---|---|---|---|---|
| rVBS | HK116 | rMLV | |||
| 2a (9751-9954) | E (67) | ||||
| 2b (9824-10507) | GP2 (227) | 62 | Tyr | Tyr | His |
| 92 | Ile | Thr | Thr | ||
| 158 | Gly | Gly | Glu | ||
| 223 | Arg | Pro | Pro | ||
| 3 (10306-10797) | GP3 (rVBS and HK116, 163; rMLV, 168) | 80 | Leu | Leu | Val |
| 123 | Leu | Leu | Ser | ||
| 160 | Cys | Cys | Tyr | ||
| 164 | Stop | Stop | Gln | ||
| 165 | Phe | ||||
| 166 | Tyr | ||||
| 167 | Leu | ||||
| 168 | His | ||||
| 168 | Stop | ||||
| 4 (10700-11158) | GP4 (152) | 4 | Tyr | Tyr | Ser |
| 8 | Leu | Ser | Ser | ||
| 29 | Ala | Ala | Thr | ||
| 37 | Ile | Ile | Thr | ||
| 5 (11146-11913) | GP5 (255) | 69 | Leu | Leu | Pro |
| 72 | Gln | Gln | Lys | ||
| 81 | Asn | Asp | Asp | ||
| 100 | Ser | Gly | Gly | ||
| 104 | Asn | Gly | Gly | ||
| 106 | Met | Met | Val | ||
| 170 | Ala | Ala | Ser | ||
| 214 | Gly | Gly | Glu | ||
| 6 (11901-12389) | M (162) | 38 | Leu | Leu | Ser |
| 49 | Phe | Phe | Leu | ||
| 71 | Val | Val | Ala | ||
| 81 | Met | Thr | Thr | ||
| 122 | Ile | Val | Val | ||
| 150 | Phe | Phe | Cys | ||
| 154 | Ala | Thr | Met | ||
Amino acid substitutions located in the predicted ectodomain of each protein are in bold. All potential N-glycosylation sites are conserved in GP2 (amino acid 155), GP3 (amino acids 28, 29, 49, 96, 106, and 118), and GP4 (amino acids 33, 55, and 90).
To enhance fundamental understanding of the pathobiology of EAV infections, it is important to identify those peripheral blood mononuclear cells (PBMCs) that are most closely associated with the virus during vascular transport and how genetic changes associated with an attenuated phenotype alter the dynamics of virus-host cell relationships in blood. Thus, we hypothesized that the VB and MLV strains differ in the ability to infect PBMCs and that the altered tropism of the attenuated MLV strain of EAV in PBMCs is associated with amino acid changes in the viral proteins. To test this hypothesis, we used three previously described recombinant viruses, rVBS (5), rVBS/HK116 S (66), and rMLV (J. Zhang et al., unpublished data), as well as four newly generated chimeric viruses (rVBS/MLV S, rMLV/VBS S, rMLV/VBS 234, and rMLV/VBS 56), and infected ex vivo preparations of PBMCs collected from horses. The data suggested that the difference in cellular tropism and virulence phenotype between the VB and MLV strains is associated with the collective interactions of both major (GP5 and M) and minor (GP2, GP3, and GP4) envelope proteins of EAV. Furthermore, this study also demonstrated that CD3+ T-lymphocyte tropism is primarily determined by amino acid substitutions in the GP2, GP4, GP5, and M envelope proteins but not in the GP3 minor envelope protein. However, the macrophage tropism is mainly determined by intricate interactions among the GP2, GP3, GP4, GP5, and M envelope proteins of the virus.
MATERIALS AND METHODS
Cell lines.
Equine pulmonary artery endothelial cells (EECs) were maintained in Dulbecco's modified essential medium (Mediatech, Herndon, VA) with sodium pyruvate, 10% fetal bovine serum (FBS; HyClone Laboratories, Inc., Logan, UT), 100 U/ml penicillin/streptomycin, and 200 mM l-glutamine (2, 5, 35). Rabbit kidney cells (RK-13, ATCC CCL-37; American Type Culture Collection, Manassas, VA) were grown in Eagle's minimum essential medium with 10% ferritin-supplemented bovine calf serum (HyClone Laboratories, Inc., Logan, UT) and 100 U/ml penicillin/streptomycin (Gibco, Carlsbad, CA).
Horses.
A total of 10 horses of different breeds (thoroughbreds, n = 4; standardbreds, n = 2; mixed breeds, n = 4) were used in this study. The horses were maintained on pasture at the Department of Veterinary Science's Maine Chance Farm, Lexington, KY. All horses were confirmed seronegative for EAV-neutralizing antibodies using previously described protocols (53). The horses were clinically evaluated prior to collection of blood samples for isolation of PBMCs. Furthermore, complete blood counts and differential counts were performed to establish that all of the animals had normal blood cell values. Blood samples were collected according to a protocol approved by the Institutional Animal Care and Use Committee at the University of Kentucky, Lexington, KY.
Antibodies.
To determine the phenotype of EAV-infected mononuclear cells, the following monoclonal antibodies (MAbs) directed against different cell-type-specific surface molecules were used in this study: anti-equine CD3+ T lymphocytes (UC F6G; kindly provided by Jeff Stott, University of California at Davis) (7), anti-equine CD4+ T lymphocytes, and anti-equine CD8+ T lymphocytes (CVS4 and CVS8, respectively; Serotec, Raleigh, NC) (33, 34). A MAb specific for human CD14 (Alexis Biochemicals, Lausen, Switzerland) that cross-reacts with equine CD14 (27) was used to identify equine monocytes, and R-phycoerythrin (R-PE)-conjugated anti-human CD21 (B-ly4; BD Pharmingen, San Jose, CA), previously shown to cross-react with equine B cells, was used (36). The R-PE-conjugated F(ab′)2 fragment of goat anti-mouse IgG1 (Southern Biotech, Birmingham, AL) was used as the secondary antibody.
To detect EAV antigens in infected cells, MAbs against EAV nonstructural protein 1 (nsp1; MAb 12A4) and N protein (MAb 3E2) were used (35, 62). Mouse ascitic fluid containing MAbs 12A4 and 3E2 was purified using a Melon Gel IgG Spin Purification kit (Pierce, Rockford, IL). Purified IgG was directly conjugated to Alexa Fluor 488 (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. Briefly, purified IgG from MAbs 12A4 and 3E2 was diluted with phosphate-buffered saline (PBS) to a concentration of 2 mg/ml and 1 M sodium bicarbonate (pH 8.3) was added. Subsequently, diluted IgG solution was incubated with Alexa Fluor 488 reactive dye for 1 h at room temperature in the dark with slow stirring. The reaction mixture was loaded onto a fine-size exclusion purification resin column for separation of labeled antibodies from unincorporated dye. Fractions of Alexa Fluor 488-labeled antibodies were collected and stored at 4°C. To detect nonspecific binding, mouse IgG1 (MOPC; Sigma, St. Louis, MO) and Alexa Fluor 488-conjugated mouse IgG1 isotype controls (Invitrogen, Carlsbad, CA) were used.
Viruses.
Two strains of EAV, the VB strain (ATCC VR-796) (10, 11) and the MLV vaccine strain (ARVAC; Fort Dodge Animal Health, Fort Dodge, IA) (24, 59), were used for in vitro studies to identify the EAV target cell population in PBMCs. Each virus was propagated once in EECs to prepare high-titer working stocks as previously described (26, 47). Briefly, EECs infected with each virus were frozen at −80°C when a 90 to 100% cytopathic effect (CPE) was observed. Cell lysates were clarified by centrifugation (500 × g) at 4°C for 15 min and then ultracentrifuged (Beckman Coulter, Miami, FL) at 121,600 × g through a 20% sucrose cushion in NET buffer (150 mM NaCl, 5 mM EDTA, 50 mM Tris-HCl, pH 7.5) at 4°C for 4 h to pellet the virus. Purified preparations of each strain of EAV were resuspended in PBS (pH 7.4) and frozen at −80°C. Virus stocks were titrated by standard plaque assay in RK-13 cells, and titers were expressed as PFU/ml (41).
Construction of chimeric cDNA clones of EAV and recovery of recombinant viruses.
Infectious cDNA clones of the VB strain of EAV and the MLV vaccine strain of EAV (pEAVrVBS and pEAVrMLV, respectively) were recently constructed in our laboratory (5; Zhang et al., unpublished). In vitro and in vivo characterization of these viruses showed that they are phenotypically indistinguishable from the respective wild-type parental viruses. Five chimeric infectious cDNA clones, pEAVrVBS/HK116 S, pEAVrVBS/MLV S, pEAVrMLV/VBS S, pEAVrMLV/VBS 234, and pEAVrMLV/VBS 56, were constructed by using standard molecular biological techniques (5, 66, 67). Recombinant chimeric infectious cDNA clone pEAVrVBS/HK116 S was generated by replacing the genes (ORFs 2a to 7) encoding the structural proteins of pEAVrVBS with the corresponding genes from the HK116 virus (66). Recombinant chimeric infectious cDNA clone pEAVrVBS/MLV S was constructed by replacing the structural protein genes (ORFs 2a to 7) of infectious cDNA clone pEAVrVBS (5), which was derived from the VB strain of EAV, with the corresponding genes of the pEAVrMLV clone (Zhang et al., unpublished). A similar approach was used to construct recombinant infectious cDNA clone pEAVrMLV/VBS S, where the structural protein genes (ORFs 2a to 7) of infectious cDNA clone pEAVrMLV were replaced with the corresponding genes of the pEAVrVBS clone, the pEAVrMLV/VBS 234 clone, in which the minor envelope protein genes (ORFs 2a to 4) of the rMLV were replaced with the corresponding genes of pEAVrVBS, and the pEAVrMLV/VBS 56 clone, in which the major envelope protein genes (ORFs 5 and 6) of the pEAVrMLV were replaced with the corresponding genes of pEAVrVBS.
Recombinant viruses rVBS, rMLV, rVBS/HK116 S, rVBS/MLV S, rMLV/VBS S, rMLV/VBS 234, and rMLV/VBS 56 were generated by in vitro transcription of infectious viral RNAs from XhoI-linearized full-length infectious cDNA clones and electroporated into EECs following previously described protocols (5). Virus particles were harvested from cell culture supernatant at 48 to 72 h after electroporation when a CPE was evident, clarified of cell debris by centrifugation, and stored at −80°C in single-use aliquots (passage 0 [P0]). Recombinant viruses (P0) harvested from transfected EECs were used to prepare high-titer working stocks by one more passage in EECs (P1) as described above, and purified viruses were used for in vitro infection studies.
Comparative amino acid sequence analysis and membrane topology prediction of EAV envelope proteins.
The published sequences of rVBS, rMLV, and HK116 were downloaded from the GenBank database (http://www.ncbi.nlm.nih.gov/nucleotide; accession numbers DQ846751 [5], FJ798195 [Zhang et al., unpublished], and EU586274 [66], respectively). ORFs 2a, 2b, 3, 4, 5, and 6 were translated into amino acid sequences and aligned with the Vector NTI Advance 11 software (Invitrogen, Carlsbad, CA). Prediction of membrane topology for viral envelope proteins was performed using the four most commonly used topology prediction methods available on the Internet: PSIPRED (http://www.psipred.net/psiform.html) (29, 30), TMHMM (http://www.cbs.dtu.dk/services/TMHMM/) (31), TMPred (http://www.ch.embnet.org/software/TMPRED_form.html), and TOPPRED (http://mobyle.pasteur.fr/cgi-bin/portal.py) (13). All user-adjustable parameters were left at their default values.
Preparation of PBMC cultures.
Blood (150 to 200 ml) was collected aseptically from each of 10 horses using Vacutainer tubes containing 15% EDTA solution (Kendall Healthcare, Mansfield, MA). PBMCs were isolated from the buffy-coat fraction by centrifugation through Ficoll-Paque Plus (Amersham Biosciences, Piscataway, NJ) at 500 × g for 30 min at 25°C. The PBMC layer was collected and washed twice with PBS (pH 7.4) by centrifugation at 100 × g for 10 min to eliminate the platelets. The cells were resuspended in complete RPMI (c-RPMI) 1640 medium (Gibco, Carlsbad, CA) supplemented with 10% heat-inactivated FBS (HyClone Laboratories, Logan, UT), 2 mM glutamine, 100 U/ml penicillin/streptomycin, and 55 μM 2-mercaptoethanol and counted using a Vicell Counter-XR (Beckman Coulter, Miami, FL). The adherent and nonadherent cell populations were separated by plating the PBMCs on 100-mm tissue culture dishes (Corning, Corning, NY). Briefly, the cells were incubated at 37°C for 4 h to allow adherent cells to attach, and after incubation, the unattached cells were removed, centrifuged, and resuspended to be counted and plated as a nonadherent cell population in 100-mm tissue culture dishes containing 5 ml of c-RPMI medium. The adherent cells were washed twice with PBS to remove the remaining nonadherent cells and replaced with 5 ml of fresh c-RPMI medium. Both adherent and nonadherent cells were incubated at 37°C until infected with EAV.
Sorting of lymphocytes and monocytes.
Positive selection of five different cell populations (CD3+, CD4+, CD8+, CD21+, and CD14+ cells) was performed using aseptically obtained horse blood and a MoFlo Cell Sorter (Dako Cytomation, Glostrup, Denmark). Briefly, 5 × 107 PBMCs were incubated with specific MAbs against equine CD3, CD4, or CD8 for T lymphocytes and human CD14 that cross-react with equine monocytes. Equine B lymphocytes were sorted by two-way sorting using an R-PE-conjugated anti-human CD21 antibody that cross-reacts with equine B lymphocytes on cells already incubated with an anti-equine CD8 MAb. All antibodies were incubated on ice in sorting buffer (1% FBS in Hanks' balanced salt solution, pH 7.4; Gibco, Carlsbad, CA) for 20 min. After incubation with the primary antibody, cells were washed and stained with goat anti-mouse IgG1-R-PE (Southern Biotechnology Inc., Birmingham, AL) or goat anti-mouse IgG-fluorescein isothiocyanate (Caltag Laboratories, Burlingame, CA) on ice for 20 min.
In vitro infection of lymphocytes and monocytes.
Cultures of lymphocytes and monocytes were infected with either wild-type (VBS or MLV) or recombinant (rVBS, rMLV, rVBS/HK116 S, rVBS/MLV S, rMLV/VBS S, rMLV/VBS 234, or rMLV/VBS 56) EAV at a multiplicity of infection (MOI) of 2. As negative controls, mock-infected lymphocytes and monocytes were cultured under identical conditions.
Dual-color immunofluorescence staining and flow cytometry.
For two-color immunofluorescence staining of wild-type or recombinant virus-infected cells, lymphocytes (1 × 106) were incubated on ice for 30 min with MAbs specific for equine CD3+, CD4+, or CD8+ T cells and R-PE-conjugated anti-human CD21 antibody for B cells. Blood-derived monocytes (1 × 106) were stained with anti-human CD14 MAb. After washing, cells were incubated with secondary R-PE-conjugated goat anti-mouse IgG1 (Southern Biotech, Birmingham, AL) for 30 min on ice. After washing to remove unbound secondary antibody, cells were fixed with 4% paraformaldehyde and then washed once in PBS-saponin buffer (PBS [pH 7.4] supplemented with 1% FBS, 0.1% saponin, and 0.1% sodium azide). Intracellular staining for EAV antigen was performed using Alexa Fluor 488-conjugated anti-EAV nsp1 MAb 12A4 or anti-EAV N MAb 3E2 in PBS-saponin buffer and incubation on ice for 30 min. After incubation, washed cells were resuspended in PBS containing 0.5% paraformaldehyde for two-color cytometric acquisition using a FACScalibur (Becton Dickinson, San Jose, CA). Lymphocytes and monocytes were gated and selected based on forward and side scatter parameters of analysis. Cells were evaluated by a two-color plot of anti-EAV antigen (FL-1) versus cell surface antigen (FL-2), and the percentage of CD3+, CD4+, CD8+, CD21+, or CD14+ and EAV antigen-positive cells was determined by CellQuest quadrant statistics. Results were expressed as the percentage of lymphocytes or monocytes infected with EAV after subtraction of the nonspecific staining of mock-infected cells.
Virus growth curve and quantitative EAV real-time TaqMan RT-PCR assay.
Sorted CD3+, CD4+, and CD8+ T lymphocytes, CD21+ B lymphocytes, and CD14+ monocytes were infected with the VB or MLV strain of EAV at an MOI of 2. Tissue culture supernatants were collected at 0, 1, 6, 12, 18, 24, 36, 48, 60, and 72 h postinfection (hpi) for one-step growth curve and quantitative EAV real-time TaqMan reverse transcription (RT)-PCR assays. For one-step growth curve analysis, virus titers in tissue culture supernatants were determined according to the method of Reed and Muench (52) and expressed as 50% tissue culture infective doses (TCID50)/50 μl. To detect EAV nucleic acids, a one-tube real-time TaqMan RT-PCR assay was performed using the TaqMan One-Step RT-PCR Master Mix (Applied Biosystems, Foster City, CA) in an Applied Biosystems 7500 Fast real-time PCR system according to a previously published protocol (3, 32). The copy numbers of EAV molecules were determined by absolute quantification with an IVT ORF7 RNA standard curve as described previously (3, 32).
Statistical analysis.
The Student t test was used to establish significant differences among infected lymphocyte subpopulations and between monocytes from group A and B horses. Statistical analysis was performed using Sigma Plot 11 (Systat Inc., Richmond, CA).
Nucleotide sequence accession numbers.
The nucleotide sequences of rVBS/HK116 S, rVBS/MLV S, rMLV/VBS S, rMLV/VBS 56, and rMLV/VBS 234 were deposited in GenBank under accession numbers GU732202, GU732201, GU732200, GU732199, and GU732198, respectively.
RESULTS
Differences in susceptibility of equine PBMCs to infection with virulent and attenuated strains of EAV.
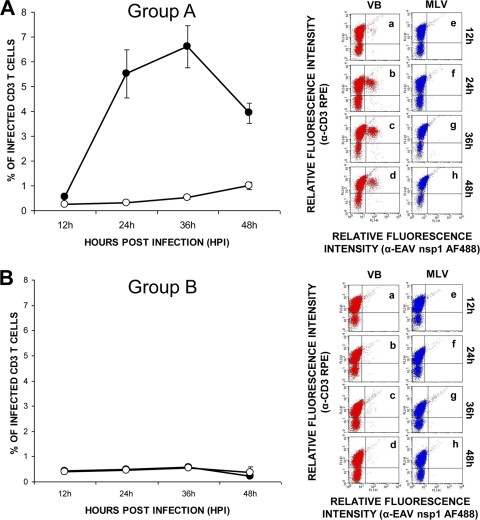
To investigate the susceptibility of equine PBMCs to virulent and attenuated strains of EAV, PBMCs collected from 10 randomly selected horses were infected with either the VB or the MLV strain of the virus. Dual-fluorescent-antibody staining of PBMC cultures was performed using a panel of leukocyte differentiation antigen-specific MAbs specific for pan-CD3+ T lymphocytes, CD4+ helper T lymphocytes, CD8+ cytotoxic T lymphocytes, and CD21+ B lymphocytes, as well as a MAb specific for EAV nsp1. Dual-immunofluorescence labeling with a CD3-specific MAb and an antibody against EAV nsp1 indicated that CD3+ T lymphocytes recovered from 6 of the 10 horses tested could be infected with the VB strain of EAV (Fig. 1A). At 12 hpi, the mean percentage of CD3+ T lymphocytes double labeled with cell surface antibody and MAb to the EAV nsp1 antigen was 0.5% ± 0.02%. The mean percentage of double-labeled CD3+ T lymphocytes increased to 5.5% ± 1.0% by 24 hpi. EAV nsp1 antigen expression peaked with a mean percentage of 6.6% ± 0.8% of double-labeled CD3+ T cells at 36 hpi and decreased to 4% ± 0.4% at 48 hpi. Interestingly, none of the CD3+ T lymphocytes from the remaining four horses were infected with the VB strain of EAV (Fig. 1B). Based on the phenotype of CD3+ T-lymphocyte subpopulations upon in vitro infection with the VB strain of EAV, the 10 horses could be divided into susceptible (group A; 6/10) and resistant (group B; 4/10) groups. Contrary to infection with the VB strain, T lymphocytes from none of the 10 donor horses were susceptible to infection with the MLV vaccine strain of EAV (Fig. 1A and B).
FIG. 1.
Differences in susceptibility of CD3+ T lymphocytes to infection with the VB and MLV strains of EAV. Dual-color immunofluorescence flow cytometric analysis was performed at various times postinfection. (A) Values represent the mean percentages (± the standard error of the mean) of CD3+ T cells infected with VB (black circles) and MLV (white circles) for six horses with T lymphocytes susceptible to EAV VB infection (group A). Percentages of infected cells at the indicated times (data points on the curve) were determined by using dot plots derived from dual-fluorescence flow cytometric analyses of cells (representative dot plots of VB infection, insets a through d; MLV infection, insets e through h). (B) Values represent the mean percentages (± the standard error of the mean) of CD3+ T cells infected with VB (black circles) and MLV (white circles) for four horses with T lymphocytes resistant to EAV VB infection (group B) as determined by using dot plots derived from dual-fluorescence flow cytometric analyses of cells (representative dot plots of VB infection, insets a through d; MLV infection, insets e through h).
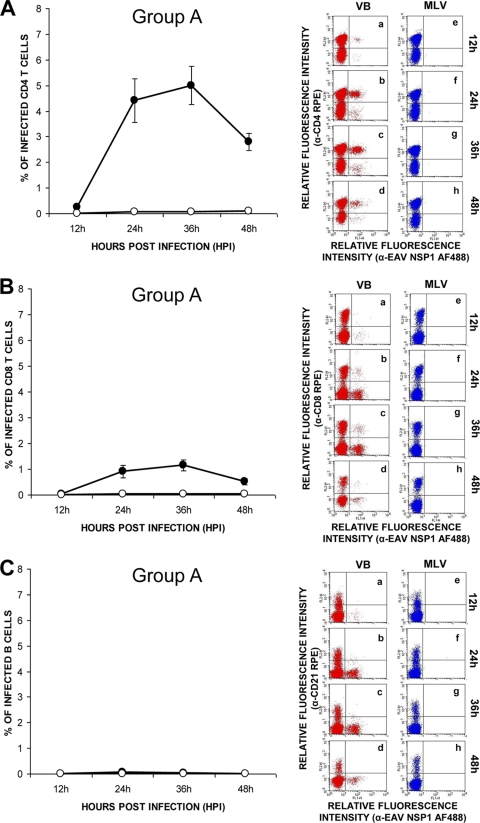
In an attempt to define the T-lymphocyte subpopulation susceptible to VB infection, lymphocytes from group A horses were infected with the VB strain and stained with either a CD4- or a CD8-specific MAb and an antibody to EAV nsp1. Similar to CD3+ T lymphocytes, the mean percentage of double-labeled CD4+ T cells increased to 4.4% ± 0.8% by 24 hpi. EAV nsp1 antigen expression peaked at 36 h after inoculation with a mean percentage of 5% ± 0.7% of double-labeled CD4+ T cells, which decreased subsequently to 2.8% ± 0.3% at 48 hpi (Fig. 2A). The mean percentage of double-labeled CD8+ T cells ranged from 0.03% to 1.1% ± 0.03% to 0.2% (Fig. 2B) through to the completion of the experiment. These findings indicated that the majority of CD3+ T lymphocytes infected with the VB strain were CD4+ T lymphocytes rather than CD8+ T lymphocytes. Not surprisingly, in a similar experiment, neither CD4+ nor CD8+ T lymphocytes from the four horses in group B could be infected with the VB strain of EAV (data not shown). EAV nsp1 antigen expression was not detected in the CD21+ B-lymphocyte cultures from either group A or B horses inoculated with either the VB or the MLV vaccine strain (Fig. 2C), indicating that B lymphocytes are not susceptible to EAV infection. In summary, viral antigen could be detected primarily in CD4+ T-lymphocyte subpopulations from group A horses infected with the VB strain of EAV. These data showed that only CD4+ T lymphocytes from some horses are susceptible to infection with the VB strain but none of them are susceptible to infection with the MLV strain, indicating that attenuation of the VB strain has altered its cellular tropism.
FIG. 2.
Susceptibility differences between CD4+ and CD8+ T lymphocytes and CD21+ B lymphocytes to infection with the VB and MLV strains of EAV. Dual-color immunofluorescence flow cytometric analysis was performed at various times postinfection. Values represent the mean percentages (± the standard error of the mean) of (A) CD4+ T cells, (B) CD8+ T cells, and (C) CD21+ B cells infected with the VB (black circles) and MLV (white circles) strains of EAV. Percentages of infected cells at the indicated times (data points on the curve) were determined by using dot plots derived from dual-fluorescence flow cytometric analyses of cells (representative dot plots of VB infection, insets a through d; MLV infection, insets e through h).
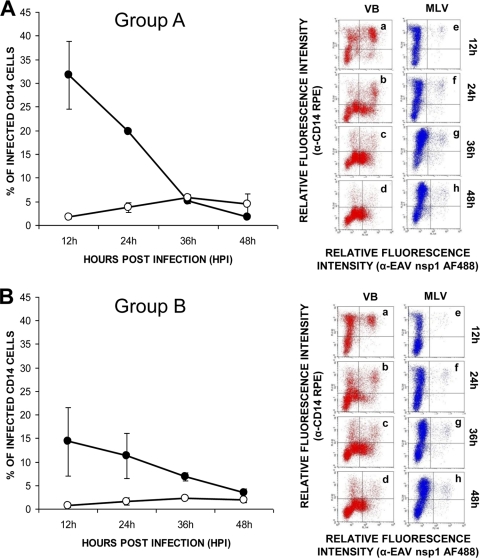
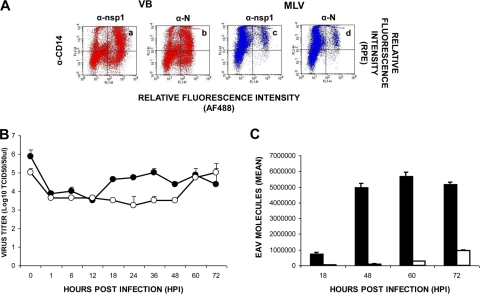
To investigate whether monocytes are equally susceptible to infection with the VB and MLV strains of EAV, dual-immunofluorescence staining was performed using MAbs specific for CD14+ monocytes and a MAb specific for EAV nsp1. Using the adherent cells obtained from PBMCs derived from two horses of each group, the percentage of CD14+ monocytes ranged from 60 to 75%, depending on the preparation. Double-labeled flow cytometric analysis showed that monocytes from all of the horses tested could be infected with the VB and MLV strains of EAV (Fig. 3). However, the mean percentage of MLV-infected cells was significantly lower and remained near the lower limit of detection throughout the experimental time course compared to those detected in VB-infected monocytes (Fig. 3A and B) from both groups of horses. In the case of cultures infected with the VB strain, the maximum mean percentage of monocytes from group A horses (n = 2) expressing the EAV nsp1 antigen was approximately 31.8% ± 7.2% at 12 hpi, after which it declined sharply (1.8% ± 0.2%) by 48 hpi (Fig. 3A, group A). Similar patterns were noted for monocytes purified from the group B horses (n = 2; Fig. 3B, group B). However, the overall mean percentage of cells expressing EAV antigen was significantly lower in monocytes from the group B horses (14.3% ± 7.3% at 12 hpi and 11.3% ± 4.7% at 24 hpi) than in monocytes from the susceptible horses (group A) at 12 and 24 hpi (Fig. 3A and B). The mean percentage of infected monocytes in MLV-inoculated cultures was minimal at 12 hpi, after which it gradually increased. Although similar patterns of infection were shown for cells derived from horses in groups A and B, the mean percentage of infected cells in the case of the former was significantly higher than in the case of the latter (Fig. 3A and B). Furthermore, these findings confirm that not only T lymphocytes but also monocytes differ in their susceptibility to infection with the VB and MLV strains of EAV.
FIG. 3.
Infection of monocytes with the VB and MLV strains of EAV. (A and B) Dual-color immunofluorescence flow cytometric analysis was performed at various times postinfection. Values represent the mean percentages (± the standard error of the mean) of monocytes infected with VB (black circles) and MLV (white circles) for (A) two horses with T lymphocytes susceptible to EAV VB infection (group A) and (B) two horses with T lymphocytes resistant to EAV VB infection (group B). Percentages of infected cells at the indicated times (data points on the curve) were determined by using dot plots derived from dual-fluorescence flow cytometric analyses of cells (representative dot plots of VB infection, insets a through d; MLV infection, insets e through h).
Expression of the structural proteins of EAV in PBMCs infected with virulent and attenuated strains of EAV.
One of the key findings of this study is the susceptibility of CD3+ T lymphocytes (predominantly CD4+ T lymphocytes) of group A horses to VB infection as determined by the expression of the nsp1 protein. To investigate whether viral structural protein genes are also expressed in infected cultures, two-color flow cytometric analysis was performed using MAbs specific for cell surface markers and a MAb specific for the EAV N protein. When T lymphocytes isolated from horses in group A were infected with the VB strain, both nsp1 expression and N protein expression were detected in CD3+, CD4+, and CD8+ T lymphocytes (Fig. 4a through c [nsp1] and d through f [N]). In contrast, CD3+, CD4+, and CD8+ T lymphocytes were completely refractory to infection with the MLV strain since neither nsp1 nor N protein expression could be observed in inoculated cultures (Fig. 4g through i [nsp1] and j through l [N]). The data indicated that a virus replication cycle, at least up to expression of structural proteins, occurred in VB-infected T lymphocytes but not in MLV-infected T lymphocytes of group A horses. However, release of progeny virus particles could not be detected in VB-inoculated cultures of sorted CD3+, CD4+, and CD8+ T lymphocytes from any of the horses tested based on virus one-step growth curve and quantitative real-time RT-PCR (qrRT-PCR) results (data not shown).
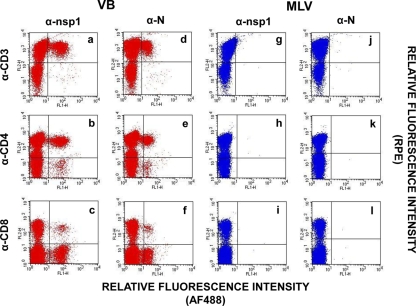
FIG. 4.
Detection of EAV N protein expression in T lymphocytes infected with the VB strain. CD3+, CD4+, and CD8+ T lymphocytes from group A horses infected with the VB and MLV strains of EAV were examined by dual-color immunofluorescence flow cytometric analysis using MAbs against the EAV nsp1 (MAb 12A4) and N (MAb 3E2) proteins and MAbs for cell-specific cell surface antigens at 24 hpi. (a through c) Lymphocytes infected with the VB strain were stained with anti-EAV nsp1 MAb AF488 and one of the specific antibodies for T lymphocytes, including MAbs to CD3, CD4, and CD8, respectively. (d through f) Dot plots derived from the same cultures stained with anti-EAV N MAb AF488 and antibodies specific for CD3+, CD4+, and CD8+ T lymphocytes, respectively. (g through i) Lymphocytes infected with the MLV strain were stained with anti-EAV nsp1 MAb and antibodies specific for T lymphocytes, including MAbs to CD3, CD4, and CD8, respectively. (j through l) Dot plots derived from the same cultures stained with anti-EAV N MAb AF488 and antibodies specific for CD3+, CD4+, and CD8+ T lymphocytes, respectively.
Furthermore, we also investigated the expression of N protein in monocytes infected with the VB and MLV strains of EAV (Fig. 5A). In contrast to T lymphocytes, expression of both nsp1 and N protein was detected in monocytes infected with the VB and MLV strains. Not surprisingly, the percentage of monocytes expressing the nsp1 and N proteins was significantly greater in VB-infected cultures (24.8% ± 4.4% and 18.3% ± 1.8%, respectively [panels a and b]) than the corresponding percentage of cultures infected with the MLV strain (5.5% ± 1.9% and 3.7% ± 1.8%, respectively; Fig. 5A, panels c and d). Additionally, viral titers and viral nucleic acid copy numbers from sorted CD14+ monocytes inoculated with the VB and MLV strains of EAV confirmed the findings from double-staining flow cytometric analysis (Fig. 5B and C, respectively). In monocyte cultures inoculated with the VB strain, viral titers increased gradually by 18 hpi, reaching a maximum titer of approximately 105 TCID50/50 μl at 36 hpi. After reaching this peak, titers gradually declined to 104 TCID50/50 μl by the final sample collection time at 72 hpi. Although productive viral replication also occurred in MLV-inoculated monocytes, viral titers were lower than those detected in VB-inoculated cells (Fig. 5B). Interestingly, while MLV titers remained low until 48 hpi, they showed a significant increase by 72 hpi. The trends in the viral growth curves were similar to those observed in nucleic acid copy numbers quantified by qrRT-PCR (Fig. 5C). Monocytes infected with the VB strain had increased viral nucleic acid copy numbers from 18 hpi, reaching a maximum after 48 hpi in both groups of horses. In contrast to VB-infected monocytes, viral nucleic acid copy numbers in monocytes infected with the MLV strain remained low throughout the time course of the experiment, though they had started to increase at 72 hpi, which was consistent with the viral growth curve results (Fig. 5C). These data confirmed that the MLV strain replicates more slowly and to lower titers in monocytes than does the VB strain of EAV in group A horses.
FIG. 5.
Replication of the VB and MLV strains of EAV in blood-derived monocytes of group A horses. (A) The CD14+ monocytes infected with the VB and MLV strains of EAV were examined by dual-color immunofluorescence flow cytometric analysis using MAbs against the EAV nsp1 (12A4) and N (3E2) proteins and MAbs for cell-specific cell surface antigens at 24 hpi. (a, b) Monocytes infected with the VB strain were stained with anti-EAV nsp1 MAb (panel a) or anti-EAV N MAb (panel b) and a MAb specific to cell surface antigen CD14. (c, d) Monocytes infected with the MLV strain were stained with anti-EAV nsp1 MAb (panel c) or anti-EAV N MAb (panel d) and a MAb specific to cell surface antigen CD14. (B) Replication of the VB (black circles) and MLV (white circles) strains in purified monocytes. Tissue culture fluids were collected at the indicated times, and viral titers were determined as the log10 TCID50/50 μl. The values shown are the mean viral titers ± the standard error of the mean. (C) Quantification of viral RNA copy number in cell culture fluid from purified monocytes infected with the VB (black bars) and MLV (white bars) strains using qrRT-PCR. Results are expressed as mean values ± the standard error of the mean.
Infection of CD3+ T lymphocytes and CD14+ monocytes with recombinant chimeric EAV strains.
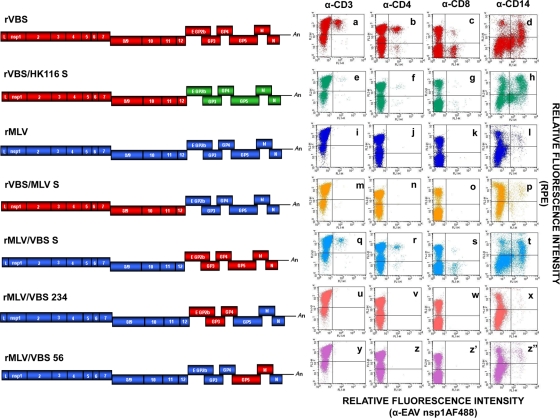
A marked disparity in the ability of the VB and MLV strains to infect subpopulations of susceptible CD3+ T lymphocytes and monocytes (group A horses) suggested that there is a clear difference in cellular tropism between the two strains. In our preliminary studies, the recombinant viruses, rVBS and rMLV, had similar growth kinetics in EECs (data not shown) and an equivalent capacity for lymphocyte infectivity, as confirmed by flow cytometry analysis (Fig. 6a through d [rVBS] and i through l [rMLV]) compared to the respective wild-type parental viruses, VB and MLV. To identify the viral proteins responsible for the differential cellular tropism between VB and MLV, we attempted to assess the impact of amino acid changes accumulated during cell culture passage of the HK116 strain, which is fully attenuated for horses compared to the VB strain. Therefore, we used the previously described rVBS/HK116 S chimeric virus (66) containing the structural proteins of EAV HK116 in the backbone of the rVBS genome to infect PBMCs from group A horses. When susceptible T lymphocytes were inoculated with chimeric rVBS/HK116 S, the number of T lymphocytes expressing the EAV nsp1 antigen decreased significantly compared to rVBS (2.3% to 0.2%; Fig. 6e through g). In contrast, the percentage of infected monocytes was comparable to that obtained using rVBS, indicating that the tropism of the HK116 strain had changed for lymphocytes but not for monocytes following 116 passages in primary horse kidney cells (Fig. 6h). In summary, these data showed that amino acid substitutions in the structural protein genes of strain HK116 changed the CD3+ T-lymphocyte tropism of the virus without any significant effect on its CD14+ monocyte tropism.
FIG. 6.
Infection of lymphocytes and monocytes with recombinant EAV strains. The genome of the infectious full-length cDNA clone of rVBS (red boxes) and the genome of the rMLV clone (blue boxes) are depicted. The genes encoding structural proteins of EAV HK116 are shown in green. The four chimeric viruses containing nonstructural and structural protein genes of either rVBS or rMLV are also depicted. L, leader; An, poly(A). CD3+, CD4+, and CD8+ T lymphocytes and CD14+ monocytes infected with recombinant viruses rVBS (a through d), rVBS/HK116 S (e through h), rMLV (i through l), rVBS/MLV S (m through p), rMLV/VBS S (q through t), rMLV/VBS 234 (u through x), and rMLV/VBS 56 (y through z″) were examined by dual-color immunofluorescence flow cytometric analysis using MAbs against EAV nsp1 (12A4) and MAbs for cell-specific cell surface antigens at 24 hpi.
In an attempt to further identify the specific viral proteins responsible for the differential tropism among the VB, HK116, and MLV strains in equine PBMCs, we generated a new panel of four recombinant chimeric viruses using the infectious cDNA clones of the VB and MLV strains (rVBS and rMLV, respectively). The panel consisted of reciprocal chimeric viruses containing interchanged structural and nonstructural protein genes of the VB and MLV strains (Fig. 6). When susceptible lymphocytes and monocytes were infected with the rVBS/MLV S and rMLV/VBS S viruses, rVBS/MLV S did not infect any CD3+ T lymphocytes and only replicated in CD14+ monocytes at a very low level which was identical to that observed in rMLV infection (Fig. 6m through p). In contrast, rMLV/VBS S infected and replicated in both CD3+ T lymphocytes and CD14+ monocytes, similar to rVBS, but there was a reduction in viral protein expression in lymphocytes compared with that observed in cells infected with rVBS (Fig. 6q through t). These results suggest that the structural proteins of the VB strain are responsible for determining its tropism for lymphocytes and monocytes. Furthermore, comparison of dual-color flow cytometric data on PBMCs infected with rVBS/HK116 S and rVBS/MLV S showed significant differences in CD14+ monocyte infectivity, indicating that amino acid substitutions that occurred during further cell culture passage of HK116 may have contributed to the change in monocyte tropism. Taken together, the data suggest that the tropism of EAV for CD3+ T lymphocytes and CD14+ monocytes was altered by amino acid changes in structural proteins of EAV.
To further investigate the role of minor and major envelope proteins of EAV in cellular tropism, additional recombinant viruses, rMLV/VBS 234 and rMLV/VBS 56, were generated with the MLV infectious cDNA clone as the viral backbone and used for in vitro infection of susceptible lymphocytes and monocytes. rMLV/VBS 234 had a genome sequence identical to that of rMLV, except for ORFs 2a, 2b, 3, and 4 (encoding minor envelope proteins E, GP2, GP3, and GP4), which were replaced with the corresponding regions of rVBS. In the case of rMLV/VBS 56, ORFs 5 and 6 (encoding major envelope proteins GP5 and M) were replaced with the corresponding genes of rVBS in the rMLV backbone (Fig. 6). In contrast to expectations, neither the rMLV/VBS 234 nor the rMLV/VBS 56 chimera was able to infect T lymphocytes (Fig. 6u through w [rMLV/VBS 234] and y through z′ [rMLV/VBS 56]). However, comparisons in monocytes showed that while the percentage of cells infected with rMLV/VBS 56 was similar to that of rMLV and rVBS/MLV S, rates of infection with rMLV/VBS 234 were significantly lower (Fig. 6z″ and x, respectively). Therefore, the higher relative fluorescence intensity values observed in monocytes with rMLV/VBS 56 (Fig. 6z″) than in those with rMLV/VBS 234 (Fig. 6x) suggest that the GP5 and M amino acid sequences may play a more critical role than those of E, GP2, GP3, and GP4 in facilitating monocyte infections. Taken together, these data demonstrate that the minor envelope glycoproteins, as well as the two major envelope proteins, play a critical role in determining monocyte tropism.
Comparative amino acid sequence analysis of the GP2, GP3, GP4, GP5, and M proteins of rVBS, HK116, and rMLV.
The reverse genetic studies using recombinant chimeric viruses clearly indicated that collective interactions among all EAV envelope proteins are required for efficient infection of susceptible subpopulations of CD3+ T lymphocytes and CD14+ monocytes, and these interactions were disabled or altered by the amino acid changes which had occurred during extensive serial passage of the VB strain resulting in the HK116 and MLV vaccine strains of EAV. Accordingly, the nucleotide sequences of ORFs 2 to 6 of the rVBS, HK116, and rMLV strains have been translated, aligned, and analyzed in the context of the predicted topography of each viral structural protein. The E minor envelope protein encoded by ORF2a was conserved (100% identity) among the rVBS, HK116, and rMLV strains, and therefore it is almost certainly not responsible for the differences in tropism among the three EAV strains (Table 1). Compared to the GP2, GP4, GP5, and M envelope proteins of the rVBS strain, those of the HK116 strain had several amino acid substitutions (two, one, three, and three, respectively; Table 1). With the exception of amino acid substitutions in the M protein and one amino acid substitution in the GP2 protein (223R→P) of the HK116 virus, all of the amino acid changes were located in the ectodomain of these proteins (Table 1 and Fig. 7). Interestingly, in contrast to the GP2, GP4, GP5, and M envelope proteins, the amino acids of the GP3 protein of the HK116 strain were identical to those of the parental VB strain. The data indicated that loss of CD3+ T-lymphocyte tropism of the HK116 virus is primarily due to substitutions involving the GP2, GP4, GP5, and M envelope proteins rather than the GP3 minor envelope protein. However, further cell culture passage of this HK116 strain resulted in numerous other nonsynonymous amino acid substitutions in ORFs 2b and 3 to 6 (Table 1 and Fig. 7). All of these substitutions appear to be nonconservative and therefore are likely to affect secondary and tertiary structure, which may change the interactions between these envelope proteins (Fig. 7). Analysis of ORF3 demonstrated a U→C substitution at position 10,795 in rMLV, resulting in the removal of the normal stop codon present in rVBS and HK116 and permitting the addition of five amino acids to the carboxyl terminus of GP3. Furthermore, there are three other nonsynonymous nucleotide substitutions between rMLV and rVBS (as well as the intermediate HK116 strain) in ORF3, of which the leucine-to-serine change at amino acid position 123 and the cysteine-to-tyrosine change at position 160 appear to be nonconservative (Table 1 and Fig. 7). In common with ORF3, all four nonsynonymous nucleotide substitutions in ORF4 are predicted to occur in the ectodomain of GP4, a trend also observed in ORF5, where six of the eight nonsynonymous substitutions occur within the same predicted domain for GP5 (Table 1 and Fig. 7). Therefore, it appears that most of the variation in amino acid content between the structural proteins of rVBS and rMLV occurs in those regions that could interact directly with host cell receptor molecules. An exception to this is in ORF6, where all seven nonsynonymous nucleotide substitutions between rMLV and rVBS are predicted to occur in the endodomain or transmembrane-spanning domain of the M protein. During extensive cell culture passage, the second putative N-linked glycosylation site at amino acid position 81 (81N→D) in GP5 was lost, leaving GP5 of MLV with only the conserved putative N-linked glycosylation site at amino acid position 56. Taken together, these data clearly demonstrate that the failure of MLV to infect CD3+ T lymphocytes was determined by the amino acid changes in the GP2, GP4, GP5, and M proteins but not the GP3 protein. In contrast, the reduced monocyte tropism of the MLV strain appears to be due to the amino acid changes in the GP3 protein. However, additional amino acid substitutions in the GP2, GP4, GP5, and M proteins may also have contributed to the altered monocyte tropism seen. Since the N protein encoded by ORF7 is not exposed on the virus surface, it is unlikely that this protein has contributed to the change in the cellular tropism of EAV. Therefore, it is more likely that the tropism of EAV for CD3+ T lymphocytes and CD14+ monocytes was altered due to the changes in major and minor envelope proteins of EAV.
FIG. 7.
Predicted membrane topology of minor (GP2, GP3, and GP4) and major (GP5 and M) EAV envelope proteins. Amino acid (a.a.) substitutions that occurred during extensive cell culture passage of the VB strain of EAV, resulting in the HK116 and MLV strains of the virus, are indicated. A predicted model of the disulfide-bonded structure of covalently linked minor envelope proteins GP2, GP3, and GP4 based on previous experimental studies (19, 25, 63-65) is depicted. Intramolecular (solid line) and intermolecular (dotted line) cysteine bridges are depicted. Major envelope proteins GP5 and M are covalently linked by a disulfide bond (S-S) formed between Cys-8 in the M protein and Cys-34 in the GP5 protein (54).
DISCUSSION
In contrast to attenuated strains of EAV, highly pathogenic strains are highly associated with PBMCs and cause high-titer viremia (2, 5, 35). However, until now, the interaction between EAV and PBMCs has not been fully characterized and the specific cell types infected with EAV were not identified. Therefore, the primary objective of this study was to unequivocally establish which components of the PBMC population were susceptible to EAV infection and then identify the viral proteins responsible for infection of these cells. Based on dual-color flow cytometric analysis and conventional viral titration assay, along with qrRT-PCR on infected cell culture fluids, we demonstrated that the predominant host cell type susceptible to the VB strain of EAV are CD14+ cells of the monocyte/macrophage lineage. Furthermore, under ex vivo culture conditions, these cells were permissive for complete productive replication of the VB strain. Although this is an important finding, it is perhaps not surprising in view of the fact that EAV antigens have been found in alveolar macrophages in infected horses (4, 48) and that all other members of the Arteriviridae family are monocyte/macrophage tropic (50). However, the finding that the VB strain can infect CD3+ T lymphocytes (predominantly CD4+ T cells) of some, but not all, horses is the first report of a member of this virus family being associated with T lymphocytes. Surprisingly, there was a clear difference in the horse population based on the susceptibility of their CD3+ T lymphocytes and CD14+ monocytes to in vitro VB infection. As a result, horses were categorized as group A, in which almost one-third of the monocytes, along with the CD3+ T lymphocyte (mainly CD4+ T cells) subpopulations, were susceptible to infection with the virulent EAV strain, or as group B, in which monocyte susceptibility was only half of that observed in group A horses and all CD3+ T-lymphocyte subpopulations were resistant to EAV infection. Based on this finding, we assumed that a horse's genetic background may play a significant role in determining the clinical outcome of primary infection with EAV, as reported for other species (8, 9). Efforts are under way in our laboratory to investigate if there is a genetic basis for the differences between horses in the susceptibility of their monocytes and CD3+ T lymphocytes to VB infection by analyzing possible associations with single nucleotide polymorphisms. Furthermore, an in vivo study is planned in which group A and B horses are infected with rVBS to establish the relationship between PBMC susceptibility and the severity of clinical illness or EVA, respectively.
Expression of the nsp1 and N proteins in CD3+ T lymphocytes demonstrated that the VB strain undergoes initial replication steps such as binding/entry, uncoating, and translation of nonstructural and structural proteins. However, production of progeny virus was not evident in sorted CD3+, CD4+, and CD8+ T lymphocytes from any of the horses tested based on the virus one-step growth curve results or qrRT-PCR results, despite the synthesis of the full complement of viral proteins. This indicates that either the number of progeny virions generated is below the limit of detection, a possible reflection of the relatively small numbers of T lymphocytes infected (about 5%), or there is a blockage at a later stage in viral assembly and/or release. It remains to be determined if these cells represent “dead-end” hosts or if they are fully permissive. In contrast to the VB strain, expression of MLV nsp1 or N could not be detected in CD3+ T lymphocytes from any of the horses tested, demonstrating that the amino acid substitutions that occurred during the attenuation process of the VB strain prevented entry and replication of the MLV strain in these cells. In addition, the number of CD14+ monocytes expressing the viral nsp1 or N protein following infection with the attenuated MLV strain was significantly less than that observed with the VB virus.
Data derived from this study suggested that the amino acid substitutions in major and minor envelope proteins that occurred during cell culture passage of the VB strain contributed to the altered tropism of MLV for CD3+ T lymphocytes and CD14+ monocytes. In an attempt to identify the viral proteins associated with infection of T lymphocytes and greater numbers of monocytes, we generated a panel of recombinant chimeric viruses in which the structural and nonstructural protein genes of parental rVBS were replaced with the corresponding genes of the rMLV strain. In vitro evaluation of chimeric rMLV/VBS S demonstrated conclusively that cell tropism was determined by envelope proteins of the virus. The role of the viral envelope proteins in CD3+ T-lymphocyte and macrophage tropism was further defined by using recombinant chimeric viruses in which sequences encoding the minor envelope glycoproteins (GP2, GP3, and GP4) or major envelope proteins (GP5 and M) of the MLV strain were replaced with the corresponding genes of rVBS in an rMLV backbone (designated rMLV/VBS 234 and rMLV/VBS 56, respectively). Recombinant chimeric viruses derived from these infectious cDNA clones were unable to infect T lymphocytes, and the numbers of monocytes expressing nsp1 were either lower than (rMLV/VBS 234) or equivalent to (rMLV/VBS 56) those observed when these cells were infected with parental rMLV. For the first time, evidence is presented suggesting that infection of T lymphocytes, along with the ability to infect relatively large numbers of monocytes, is dependent on cooperative interactions among five out of the six envelope proteins of EAV (GP2, GP3, GP4, GP5, and M). This, in turn, suggests that the infection process of these cell types is complex, possibly involving multiple receptor and/or receptor accessory molecules. Furthermore, the data also indicated that during extensive sequential cell culture passage, viral envelope protein genes have coevolved, resulting in a synergistic effect on cellular tropism. Interestingly, based on findings with the rVBS/HK116 S chimera, the GP3 protein is not associated with altered tropism for CD3+ T lymphocytes. Substitutions in the other two minor envelope glycoproteins (GP2 and GP4) and the two major envelope proteins (GP5 and M), however, play a critical role in changing viral tropism for these cells. The dual-color flow cytometry data clearly demonstrated that the HK116 strain was significantly different from both the VB and MLV strains. By 116 passages of the VB strain in primary HK cells, the virus lost its permissiveness for CD3+ T lymphocytes but its ability to infect CD14+ monocytes remained similar to that of the parental VB strain. Further cell culture passage of the HK116 virus in different cell lines (HK15, RK111, and ED24) to obtain the MLV strain resulted in a significant reduction of its ability to infect macrophages (27% to 5%; Fig. 6h and l, respectively). However, it is important to note that the attenuated phenotype of EAV HK116 for horses correlates with the loss of permissiveness for T lymphocytes while the ability to infect monocytes remains similar to that of the parental VB strain. This suggests that, either directly or indirectly, tropism for T lymphocytes has a significant role in the pathogenesis of VB infections in vivo. Therefore, the susceptibility of CD3+ T lymphocytes to virulent and avirulent field strains of EAV and their role in the pathogenesis of EVA warrant further in-depth investigation.
Interestingly, based on amino acid sequence comparison and the predicted membrane topology of the envelope proteins, the majority of the amino acid variations between VB and MLV, as well as HK116, are located within the predicted ectodomain of each envelope protein, except for the M protein, in which the changes are in the transmembrane and cytoplasmic domains (Fig. 7). It is known that GP5 and M exist in the virion as a heterodimer while the minor envelope proteins (GP2, GP3, and GP4) occur in particles as heterotrimeric complexes constituting a virion-exposed structure that is predicted to mediate binding of the virion to the primary receptor (65). Formation of the heterotrimeric minor envelope complex is dependent on intramolecular cysteine bonding, with GP2 and GP3 being linked via interactions with GP4 (63). The existence of these complexes, coupled with the fact that most of the amino acid substitutions appear to be nonconservative, suggests that the cell tropism of the MLV and VB strains is determined by conformational differences in structural envelope proteins that are manifested at the tertiary and quaternary levels. Therefore, the amino acid substitutions located within the various ectodomains may have direct and indirect effects on interactions with host cells, as well as associations between the viral structural proteins. Consistent with our findings, a recent published study on porcine reproductive and respiratory syndrome virus has shown that interglycoprotein interactions are critical for mediating interactions with the receptor responsible for virus entry into host cells (15). Specifically, a strong interaction between GP4 and GP5 has been demonstrated, as well as weak interactions among other minor envelope proteins, resulting in the formation of a multiprotein complex. Furthermore, in a previous study, we have also demonstrated that a change in cellular tropism and the establishment of persistent infection of HeLa cells by the VB strain of EAV were associated with amino acid substitutions in the envelope proteins (67). Reverse genetic studies further confirmed that substitutions in minor envelope proteins E and GP2 or GP3 and GP4 alone were unable to change cellular tropism and establish persistent infection of HeLa cells, but recombinant viruses with combined substitutions in the E, GP2, GP3, and GP4 proteins, as well as a single amino acid substitution in GP5, were able to alter the cellular tropism of the VB strain of EAV and favor the establishment of persistent infection in HeLa cells. Nevertheless, the situation is further complicated by the fact so little is known about the EAV receptor and whether this is restricted to a single molecule or if the virus can use multiple alternatives but potentially cell-type-specific molecules.
In summary, for the first time, we demonstrate that extensive cell culture passage of the VB strain of EAV to produce the MLV vaccine strain has altered its tropism for equine PBMCs. Evaluation of chimeric viruses demonstrated that viral tropism for PBMCs is determined by the amino acid sequences of viral envelope proteins. Specifically, the data suggest that the GP2, GP4, GP5, and M envelope proteins play a critical role in CD3+ T-lymphocyte tropism while three minor envelope proteins (GP2, GP3, and GP4), as well as the GP5 and M major envelope proteins, determine the CD14+ monocyte tropism of EAV.
Acknowledgments
This work was supported by the Frederick Van Lennep Chair endowment fund at the Maxwell H. Gluck Equine Research Center and the Kentucky Agricultural Experiment Station, College of Agriculture, University of Kentucky. Yun Young Go is supported by the Geoffrey C. Hughes Foundation graduate fellowship program.
We thank the staff at the University of Kentucky's Maine Chance Farm for their care of the horses and Jennifer Strange and Greg Bauman, Flow Cytometry Core Facility, Department of Microbiology, Immunology, and Molecular Genetics, College of Medicine, University of Kentucky, for their assistance with flow cytometry.
Footnotes
Published ahead of print on 10 March 2010.
REFERENCES
- 1.Balasuriya, U. B., J. F. Evermann, J. F. Hedges, A. J. McKeirnan, J. Q. Mitten, J. C. Beyer, W. H. McCollum, P. J. Timoney, and N. J. MacLachlan. 1998. Serologic and molecular characterization of an abortigenic strain of equine arteritis virus isolated from infective frozen semen and an aborted equine fetus. J. Am. Vet. Med. Assoc. 213:1586-1589. [PubMed] [Google Scholar]
- 2.Balasuriya, U. B., H. W. Heidner, N. L. Davis, H. M. Wagner, P. J. Hullinger, J. F. Hedges, J. C. Williams, R. E. Johnston, W. David Wilson, I. K. Liu, and N. James MacLachlan. 2002. Alphavirus replicon particles expressing the two major envelope proteins of equine arteritis virus induce high level protection against challenge with virulent virus in vaccinated horses. Vaccine 20:1609-1617. [DOI] [PubMed] [Google Scholar]
- 3.Balasuriya, U. B., C. M. Leutenegger, J. B. Topol, W. H. McCollum, P. J. Timoney, and N. J. MacLachlan. 2002. Detection of equine arteritis virus by real-time TaqMan reverse transcription-PCR assay. J. Virol. Methods 101:21-28. [DOI] [PubMed] [Google Scholar]
- 4.Balasuriya, U. B., and N. J. MacLachlan. 2004. The immune response to equine arteritis virus: potential lessons for other arteriviruses. Vet. Immunol. Immunopathol. 102:107-129. [DOI] [PubMed] [Google Scholar]
- 5.Balasuriya, U. B., E. J. Snijder, H. W. Heidner, J. Zhang, J. C. Zevenhoven-Dobbe, J. D. Boone, W. H. McCollum, P. J. Timoney, and N. J. Maclachlan. 2007. Development and characterization of an infectious cDNA clone of the virulent Bucyrus strain of equine arteritis virus. J. Gen. Virol. 88:918-924. [DOI] [PubMed] [Google Scholar]
- 6.Balasuriya, U. B., E. J. Snijder, L. C. van Dinten, H. W. Heidner, W. D. Wilson, J. F. Hedges, P. J. Hullinger, and N. J. MacLachlan. 1999. Equine arteritis virus derived from an infectious cDNA clone is attenuated and genetically stable in infected stallions. Virology 260:201-208. [DOI] [PubMed] [Google Scholar]
- 7.Blanchard-Channell, M., P. F. Moore, and J. L. Stott. 1994. Characterization of monoclonal antibodies specific for equine homologues of CD3 and CD5. Immunology 82:548-554. [PMC free article] [PubMed] [Google Scholar]
- 8.Brinton, M. A. 2001. Host factors involved in West Nile virus replication. Ann. N. Y. Acad. Sci. 951:207-219. [DOI] [PubMed] [Google Scholar]
- 9.Brinton, M. A., and A. A. Perelygin. 2003. Genetic resistance to flaviviruses. Adv. Virus Res. 60:43-85. [DOI] [PubMed] [Google Scholar]
- 10.Bryans, J. T., M. E. Crowe, E. R. Doll, and W. H. McCollum. 1957. Isolation of a filterable agent causing arteritis of horses and abortion by mares; its differentiation from the equine abortion (influenza) virus. Cornell Vet. 47:3-41. [PubMed] [Google Scholar]
- 11.Bryans, J. T., E. R. Doll, and R. E. Knappenberger. 1957. An outbreak of abortion caused by the equine arteritis virus. Cornell Vet. 47:69-75. [PubMed] [Google Scholar]
- 12.Cavanagh, D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142:629-633. [PubMed] [Google Scholar]
- 13.Claros, M. G., and G. von Heijne. 1994. TopPred II: an improved software for membrane protein structure predictions. Comput. Appl. Biosci. 10:685-686. [DOI] [PubMed] [Google Scholar]
- 14.Crawford, T. B., and J. B. Henson. 1972. Viral arteritis of horses. Adv. Exp. Med. Biol. 22:175-183. [DOI] [PubMed] [Google Scholar]
- 15.Das, P. B., P. X. Dinh, I. H. Ansari, M. de Lima, F. A. Osorio, and A. K. Pattnaik. 2010. The minor envelope glycoproteins GP2a and GP4 of porcine reproductive and respiratory syndrome virus interact with the receptor CD163. J. Virol. 84:1731-1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Del Piero, F. 2000. Equine viral arteritis. Vet. Pathol. 37:287-296. [DOI] [PubMed] [Google Scholar]
- 17.Del Piero, F., P. A. Wilkins, J. W. Lopez, A. L. Glaser, E. J. Dubovi, D. H. Schlafer, and D. H. Lein. 1997. Equine viral arteritis in newborn foals: clinical, pathological, serological, microbiological and immunohistochemical observations. Equine Vet. J. 29:178-185. [DOI] [PubMed] [Google Scholar]
- 18.de Vries, A. A., S. M. Post, M. J. Raamsman, M. C. Horzinek, and P. J. Rottier. 1995. The two major envelope proteins of equine arteritis virus associate into disulfide-linked heterodimers. J. Virol. 69:4668-4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Vries, A. A., M. J. Raamsman, H. A. van Dijk, M. C. Horzinek, and P. J. Rottier. 1995. The small envelope glycoprotein (GS) of equine arteritis virus folds into three distinct monomers and a disulfide-linked dimer. J. Virol. 69:3441-3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doll, E. R., J. T. Bryans, J. C. Wilson, and W. H. McCollum. 1968. Immunization against equine viral arteritis using modified live virus propagated in cell cultures of rabbit kidney. Cornell Vet. 48:497-524. [PubMed] [Google Scholar]
- 21.Fukunaga, Y., H. Imagawa, E. Tabuchi, and Y. Akiyama. 1981. Clinical and virological findings on experimental equine viral arteritis in horses. Bull. Equine Res. Inst. 18:110-118. [Google Scholar]
- 22.Glaser, A. L., A. A. de Vries, P. J. Rottier, M. C. Horzinek, and B. Colenbrander. 1996. Equine arteritis virus: a review of clinical features and management aspects. Vet. Q. 18:95-99. [DOI] [PubMed] [Google Scholar]
- 23.Golnik, W., Z. Michalska, and T. Michalak. 1981. Natural equine viral arteritis in foals. Schweiz. Arch. Tierheilkd. 123:523-533. [PubMed] [Google Scholar]
- 24.Harry, T. O., and W. H. McCollum. 1981. Stability of viability and immunizing potency of lyophilized, modified equine arteritis live-virus vaccine. Am. J. Vet. Res. 42:1501-1505. [PubMed] [Google Scholar]
- 25.Hedges, J. F., U. B. Balasuriya, and N. J. MacLachlan. 1999. The open reading frame 3 of equine arteritis virus encodes an immunogenic glycosylated, integral membrane protein. Virology 264:92-98. [DOI] [PubMed] [Google Scholar]
- 26.Hedges, J. F., C. D. Demaula, B. D. Moore, B. E. McLaughlin, S. I. Simon, and N. J. MacLachlan. 2001. Characterization of equine E-selectin. Immunology 103:498-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ibrahim, S., and F. Steinbach. 2007. Non-HLDA8 animal homologue section anti-leukocyte mAbs tested for reactivity with equine leukocytes. Vet. Immunol. Immunopathol. 119:81-91. [DOI] [PubMed] [Google Scholar]
- 28.Johnson, B., C. Baldwin, P. Timoney, and R. Ely. 1991. Arteritis in equine fetuses aborted due to equine viral arteritis. Vet. Pathol. 28:248-250. [DOI] [PubMed] [Google Scholar]
- 29.Jones, D. T. 2007. Improving the accuracy of transmembrane protein topology prediction using evolutionary information. Bioinformatics 23:538-544. [DOI] [PubMed] [Google Scholar]
- 30.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1994. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry 33:3038-3049. [DOI] [PubMed] [Google Scholar]
- 31.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 32.Lu, Z., A. Branscum, K. M. Shuck, J. Zhang, E. Dubovi, P. J. Timoney, and U. B. Balasuriya. 2008. Comparison of two real-time reverse transcription polymerase chain reaction assays for the detection of equine arteritis virus nucleic acid in equine semen and tissue culture fluid. J. Vet. Diagn. Invest. 20:147-155. [DOI] [PubMed] [Google Scholar]
- 33.Lunn, D. P., M. A. Holmes, D. F. Antczak, N. Agerwal, J. Baker, S. Bendali-Ahcene, M. Blanchard-Channell, K. M. Byrne, K. Cannizzo, W. Davis, M. J. Hamilton, D. Hannant, T. Kondo, J. H. Kydd, M. C. Monier, P. F. Moore, T. O'Neil, B. R. Schram, A. Sheoran, J. L. Stott, T. Sugiura, and K. E. Vagnoni. 1998. Report of the Second Equine Leucocyte Antigen Workshop, Squaw valley, California, July 1995. Vet. Immunol. Immunopathol. 62:101-143. [DOI] [PubMed] [Google Scholar]
- 34.Lunn, D. P., M. A. Holmes, and W. P. Duffus. 1991. Three monoclonal antibodies identifying antigens on all equine T lymphocytes, and two mutually exclusive T-lymphocyte subsets. Immunology 74:251-257. [PMC free article] [PubMed] [Google Scholar]
- 35.MacLachlan, N. J., U. B. Balasuriya, P. V. Rossitto, P. A. Hullinger, J. F. Patton, and W. D. Wilson. 1996. Fatal experimental equine arteritis virus infection of a pregnant mare: immunohistochemical staining of viral antigens. J. Vet. Diagn. Invest. 8:367-374. [DOI] [PubMed] [Google Scholar]
- 36.Mayall, S., E. Siedek, and A. S. Hamblin. 2001. The anti-human CD21 antibody, BU33, identifies equine B cells. J. Comp. Pathol. 124:83-87. [DOI] [PubMed] [Google Scholar]
- 37.McCollum, W. H. 1969. Development of a modified virus strain and vaccine for equine viral arteritis. J. Am. Vet. Med. Assoc. 155:318-322. [PubMed] [Google Scholar]
- 38.McCollum, W. H. 1970. Vaccination for equine viral arteritis, p.143-151. In Equine infectious diseases. II. Proceedings of the Second International Conference on Equine Infectious Diseases, Paris 1969. Karger, Basel, Switzerland.
- 39.McCollum, W. H., E. R. Doll, and J. C. Wilson. 1961. The recovery of virus from horses with experimental cases of equine arteritis using monolayer cell cultures of rabbit kidney. Am. J. Vet. Res. 23:465-469. [Google Scholar]
- 40.McCollum, W. H., E. R. Doll, J. C. Wilson, and C. B. Johnson. 1961. Propagation of equine arteritis virus in monolayer cultures of equine kidney. Am. J. Vet. Res. 22:731-735. [Google Scholar]
- 41.McCollum, W. H., E. R. Doll, J. C. Wilson, and J. Cheatham. 1962. Isolation and propagation of equine arteritis virus in monolayer cell cultures of rabbit kidney. Cornell Vet. 52:452-458. [PubMed] [Google Scholar]
- 42.McCollum, W. H., M. E. Prickett, and J. T. Bryans. 1971. Temporal distribution of equine arteritis virus in respiratory mucosa, tissues and body fluids of horses infected by inhalation. Res. Vet. Sci. 12:459-464. [PubMed] [Google Scholar]
- 43.McCollum, W. H., and P. J. Timoney. 1999. Experimental observation on the virulence of isolates of equine arteritis virus. R&W Publications (Newmarket) Limited, Newmarket, United Kingdom.
- 44.McCollum, W. H., and P. J. Timoney. 1984. The pathogenic qualities of the 1984 strain of equine arteritis virus, p. 34-84. In Grayson Foundation International Conference of Thoroughbred Breeders Organizations, Ireland 1984.
- 45.McCollum, W. H., P. J. Timoney, J. W. Lee, Jr., P. L. Habacker, U. B. R. Balasuriya, and N. J. MacLachlan. 1999. Features of an outbreak of equine viral arteritis on a breeding farm associated with abortion and fatal interstitial pneumonia in neonatal foals. R&W Publications (Newmarket) Limited, Newmarket, United Kingdom.
- 46.Molenkamp, R., H. van Tol, B. C. Rozier, Y. van der Meer, W. J. Spaan, and E. J. Snijder. 2000. The arterivirus replicase is the only viral protein required for genome replication and subgenomic mRNA transcription. J. Gen. Virol. 81:2491-2496. [DOI] [PubMed] [Google Scholar]
- 47.Moore, B. D., U. B. Balasuriya, J. F. Hedges, and N. J. MacLachlan. 2002. Growth characteristics of a highly virulent, a moderately virulent, and an avirulent strain of equine arteritis virus in primary equine endothelial cells are predictive of their virulence to horses. Virology 298:39-44. [DOI] [PubMed] [Google Scholar]
- 48.Moore, B. D., U. B. Balasuriya, J. L. Watson, C. M. Bosio, R. J. MacKay, and N. J. MacLachlan. 2003. Virulent and avirulent strains of equine arteritis virus induce different quantities of TNF-alpha and other proinflammatory cytokines in alveolar and blood-derived equine macrophages. Virology 314:662-670. [DOI] [PubMed] [Google Scholar]
- 49.Patton, J. F., U. B. Balasuriya, J. F. Hedges, T. M. Schweidler, P. J. Hullinger, and N. J. MacLachlan. 1999. Phylogenetic characterization of a highly attenuated strain of equine arteritis virus from the semen of a persistently infected standardbred stallion. Arch. Virol. 144:817-827. [DOI] [PubMed] [Google Scholar]
- 50.Plagemann, P. G., and V. Moennig. 1992. Lactate dehydrogenase-elevating virus, equine arteritis virus, and simian hemorrhagic fever virus: a new group of positive-strand RNA viruses. Adv. Virus Res. 41:99-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prickett, M. E., W. H. McCollum, and J. T. Bryans. 1972. The gross and microscopic pathology observed in horses experimentally infected with the equine arteritis virus, p. 265-272. In Equine infectious diseases. III. Proceedings of the Third International Conference on Equine Infectious Diseases, Paris 1972. Karger, Basel, Switzerland.
- 52.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 53.Senne, D. A., J. E. Pearson, and E. A. Carbrey. 1985. Equine viral arteritis: a standard procedure for the virus neutralization test and comparison of results of a proficiency test performed at five laboratories, p. 29-34. In Proceedings of the 89th Annual Meeting of the United States Animal Health Association. United States Animal Health Association, St. Joseph, MO.
- 54.Snijder, E. J., J. C. Dobbe, and W. J. Spaan. 2003. Heterodimerization of the two major envelope proteins is essential for arterivirus infectivity. J. Virol. 77:97-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Snijder, E. J., and J. J. Meulenberg. 1998. The molecular biology of arteriviruses. J. Gen. Virol. 79:961-979. [DOI] [PubMed] [Google Scholar]
- 56.Snijder, E. J., and W. J. Spaan. 2006. Arteriviruses, p. 1337-1355. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 57.Snijder, E. J., H. van Tol, K. W. Pedersen, M. J. Raamsman, and A. A. de Vries. 1999. Identification of a novel structural protein of arteriviruses. J. Virol. 73:6335-6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Timoney, P. J., and W. H. McCollum. 1993. Equine viral arteritis. Vet. Clin. North Am. Equine Pract. 9:295-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Timoney, P. J., N. W. Umphenour, and W. H. McCollum. 1988. Safety evaluation of commercial modified live equine arteritis virus vaccine for use in stallions, p. 19-27. In Equine infectious diseases. V. Proceedings of the Fifth International Conference on Equine Infectious Diseases, Lexington 1987. Karger, Basel, Switzerland.
- 60.Vaala, W. E., A. N. Hamir, E. J. Dubovi, P. J. Timoney, and B. Ruiz. 1992. Fatal, congenitally acquired infection with equine arteritis virus in a neonatal thoroughbred. Equine Vet. J. 24:155-158. [DOI] [PubMed] [Google Scholar]
- 61.van Aken, D., J. Zevenhoven-Dobbe, A. E. Gorbalenya, and E. J. Snijder. 2006. Proteolytic maturation of replicase polyprotein pp1a by the nsp4 main proteinase is essential for equine arteritis virus replication and includes internal cleavage of nsp7. J. Gen. Virol. 87:3473-3482. [DOI] [PubMed] [Google Scholar]
- 62.Wagner, H. M., U. B. Balasuriya, and N. J. MacLachlan. 2003. The serologic response of horses to equine arteritis virus as determined by competitive enzyme-linked immunosorbent assays (c-ELISAs) to structural and non-structural viral proteins. Comp. Immunol. Microbiol. Infect. Dis. 26:251-260. [DOI] [PubMed] [Google Scholar]
- 63.Wieringa, R., A. A. De Vries, S. M. Post, and P. J. Rottier. 2003. Intra- and intermolecular disulfide bonds of the GP2b glycoprotein of equine arteritis virus: relevance for virus assembly and infectivity. J. Virol. 77:12996-13004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wieringa, R., A. A. de Vries, M. J. Raamsman, and P. J. Rottier. 2002. Characterization of two new structural glycoproteins, GP(3) and GP(4), of equine arteritis virus. J. Virol. 76:10829-10840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wieringa, R., A. A. de Vries, and P. J. Rottier. 2003. Formation of disulfide-linked complexes between the three minor envelope glycoproteins (GP2b, GP3, and GP4) of equine arteritis virus. J. Virol. 77:6216-6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang, J., Y. Y. Go, N. J. MacLachlan, B. J. Meade, P. J. Timoney, and U. B. Balasuriya. 2008. Amino acid substitutions in the structural or nonstructural proteins of a vaccine strain of equine arteritis virus are associated with its attenuation. Virology 378:355-362. [DOI] [PubMed] [Google Scholar]
- 67.Zhang, J., P. J. Timoney, N. J. MacLachlan, W. H. McCollum, and U. B. Balasuriya. 2008. Persistent equine arteritis virus infection in HeLa cells. J. Virol. 82:8456-8464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ziebuhr, J., E. J. Snijder, and A. E. Gorbalenya. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81:853-879. [DOI] [PubMed] [Google Scholar]